Abstract
Cardiomyopathies such as Dilated Cardiomyopathy (DCM) and Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC) are common in large breed dogs and carry an overall poor prognosis. Research shows that these diseases have strong breed predilections, and selective breeding has historically been recommended to reduce the disease prevalence in affected breeds. Treatment of these diseases is typically palliative and aimed at slowing disease progression and managing clinical signs of heart failure as they develop. The discovery of specific genetic mutations underlying cardiomyopathies, such as the striatin mutation in Boxer ARVC and the pyruvate dehydrogenase kinase 4 (PDK4) and titin mutations in Doberman Pinschers, has strengthened our ability to screen and selectively breed individuals in an attempt to produce unaffected offspring. The discovery of these disease-linked mutations has also opened avenues for the development of gene therapies, including gene transfer and genome editing approaches. This review discusses the known genetics of cardiomyopathies in dogs, reviews existing gene therapy strategies and the status of their development in canines, and discusses ongoing challenges in the clinical translation of these technologies for treating heart disease. While challenges remain in utilizing these emerging technologies, the exponential growth of the gene therapy field holds great promise for future clinical applications.
Keywords: Dilated Cardiomyopathy, Arrhythmogenic Right Ventricular Cardiomyopathy, Gene therapy, Gene delivery, Genetic testing, Gene editing, Clustered Regularly Interspaced Short Palindromic Repeats
Introduction
Cardiomyopathies comprise an important group of cardiac disorders in humans and animals alike. Cardiomyopathies are categorized as Dilated Cardiomyopathy (DCM), Hypertrophic Cardiomyopathy, Restrictive Cardiomyopathy, or Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC) [1]. Hypertrophic cardiomyopathy is the most common form identified in humans [2] and felines [3], with prevalence approaching up to 0.4% in humans [1,4,5] and 10–15% in felines [3,6]. In canines, cardiomyopathies show a strong breed predisposition, with up to 58% of Doberman Pinschers affected by DCM [7,8] and up to ~25% of Boxers affected by ARVC [9].
Current treatments for cardiomyopathies in dogs aim to prolong the subclinical phase and treat symptoms of congestive heart failure. Drugs such as pimobendan, a phosphodiesterase 3 inhibitor, and furosemide, a loop diuretic, are two effective treatments currently available for dogs showing clinical signs, though other drugs such as antiarrhythmics, angiotensin converting enzyme inhibitors, peripheral vasodilators, and other diuretics are also frequently utilized. While medications provide improvement in quality of life and extension of the occult phase of DCM [10], the efficacy of preclinical treatment of ARVC is less clear [11,12]. Although the progression of cardiomyopathies can be delayed, no treatment currently exists which halts or reverses the disease. Prognosis for animals with congestive heart failure secondary to cardiomyopathy is generally poor, with median survival times as low as 19 weeks in dogs with DCM [13].
Due to the poor prognosis and limitations of available treatments, increased attention has been directed toward identifying novel therapies for cardiomyopathy. Multiple genetic mutations associated with the development of cardiomyopathy have been identified in humans [14,15] and animals [16,17,18,19,20]. As in humans, genetic disorders in dogs are amenable to gene therapy approaches which can potentially halt or even reverse the effects of cardiomyopathy [21,22,23,24,25,26]. This review covers the genetics of clinically significant cardiomyopathies in dogs, provides an overview of gene therapy techniques, and discusses challenges facing the clinical development of gene therapies for canine heart disease.
Known Genetics of Canine Cardiomyopathies
Increased focus throughout the last two decades on sample populations consisting largely of single breeds affected by a specific cardiomyopathy has facilitated the identification of mutations that appear to be breed-specific. The following section will discuss these genetic mutations and modes of inheritance within specific breeds of dogs (summarized in Table 1).
Table 1:
Summary of known breed-specific genetic mutations linked to cardiomyopathies in canines.
| Name | Breed affected | Disease | Gene involved | Mutation type | Biological result | Mode of inheritance | Penetrance |
|---|---|---|---|---|---|---|---|
| DCM1 | Doberman pinscher | Dilated Cardiomyopathy | Phosphodiesterase kinase 4 (PDK 4) | 16 base pair deletion | Altered cardiomyocyte metabolism with preferential glucose oxidation | Autosomal dominant | 68% |
| DCM2 | Doberman pinscher | Dilated Cardiomyopathy | Titin | Single base pair (missense) change from C to T | Incompletely understood; hypothesized changes to secondary structure resulting in titin unfolding and degeneration | Autosomal dominant | 47% |
| Striatin | Boxer | Arrhythmogenic Right Ventricular Cardiomyopathy | Striatin | 8 base pair deletion | Altered electrical conduction and structural integrity between myocytes | Autosomal dominant | 72% |
Canine Dilated Cardiomyopathy
Dilated cardiomyopathy is a disease of the cardiac muscle characterized by progressive systolic dysfunction of the heart resulting in eccentric hypertrophy (dilation) of the ventricles, predominantly the left ventricle. Individuals affected with this disease may also present with ventricular or supraventricular arrhythmias. DCM is the second most common cardiac disease in dogs, behind degenerative valvular disease [8]. Prevalence in the general canine population is estimated at 0.5% [27]. An increased prevalence is seen within certain large breeds including the Great Dane, Doberman Pinscher, Portuguese Water Dog, Irish Wolfhound, Newfoundland, Boxer, Welsh Springer Spaniels, and Cocker Spaniel, among others. Of these, the Doberman Pinscher is the breed most predisposed to DCM, with reported prevalence of up to 58% [7]. Although this breed was the initial focus of inherited canine DCM studies, recent work has identified genetic alterations in additional breeds [28,29,30].
Two separate genetic mutations are linked to DCM in Doberman Pinschers. The first is a 16-base pair deletion located at the donor splice site (5’ end) of an intron in the phosphodiesterase kinase 4 (PDK4) gene [18]. Inheritance of the PDK4 mutation is autosomal dominant, and based on a single study, prevalence may be up to 60% with 68% penetrance [18]. Phosphodiesterase kinase 4 is an important regulatory protein in cardiomyocyte energy metabolism. In the normal, healthy heart, fatty acids are the preferred energy source, and PDK4 allows for preferential oxidation of fatty acids by inhibiting glucose oxidation. The identified PDK4 mutation, named DCM1, results in decreased expression of the PDK4 protein and an energy-deficient state in cardiomyocytes due to the lower energy efficiency of glycolysis compared to fatty acid oxidation and a lifetime of reduced metabolic flexibility in the heart. Phosphodiesterase kinase 4 deficient fibroblast cells from affected Doberman Pinschers have reduced metabolic compensation capability during periods of glucose starvation in vitro [31,32].
The second genetic mutation linked to DCM in Doberman Pinschers is a single base pair change from C to T within the titin gene of affected dogs, resulting in a change in a highly conserved amino acid from glycine to arginine [19]. The mode of inheritance of the titin mutation is autosomal dominant, with a penetrance of 47% [19]. Titin is the largest protein in the body, and it contributes to both passive stiffness and active contraction of the heart muscle through unfolding and refolding of its numerous domains in response to tension. The missense mutation identified in Doberman Pinschers, named DCM2, was associated with decreased active tension and Z disc streaming [19]. It is hypothesized that changes to the secondary structure of the protein results in greater ease of unfolding and degradation; however, the pathophysiology of DCM in relation to the titin mutation is incompletely understood.
In a recent clinical study DCM2 was identified in more than 50% of the affected dogs, and 20% of affected dogs had both DCM1 and DCM2 [33]. Presence of either the DCM1 or DCM2 mutation places an individual at a higher risk of developing disease, but it is thought that the presence of both mutations heightens risk. No estimation of this combined risk exists at the time of this review. Additionally, the two mutations not only involve different genes but also occur via different mechanisms (i.e. metabolic dysfunction and contractility defects).
A DCM-linked mutation termed R9H was recently identified in a pedigree of Welsh Springer Spaniels with a high incidence of left ventricular dilation, poor systolic function, arrhythmia, and early sudden cardiac death [28]. A single base pair change from G to A within the phospholamban gene, results in an amino acid change from arginine to histidine [28]. The mode of inheritance is autosomal dominant, and penetrance is suspected to be extremely high as all dogs found to carry the mutation developed the disease [28]. An identical mutation has also been identified in humans, though penetrance is reported to be much lower [34]. Phospholamban is a key inhibitor of Sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a (SERCA2a), which is responsible for calcium reuptake in cardiomyocytes [35]. The R9H mutation results in failure of inhibition of phospholamban and consequently decreased calcium reuptake by SERCA2a [28].
Evidence of familial autosomal recessive early-onset DCM in Giant Schnauzers has also been studied, and a 22-bp deletion resulting in a frameshift mutation in RNA-binding motif protein 20 was identified [36,37]. This mutation has also been identified in humans, though it is associated with much higher incidence of arrhythmias in humans [38] compared to dogs [36]. RNA-binding motif protein 20 is involved in the splicing of many important cardiac genes, and abnormal splicing of titin is considered responsible for the development of DCM with this mutation [38]. Additionally, aberrant splicing of CAM-kinase and Ryanodine receptor 2 are thought to occur with this mutation, resulting in a proarrhythmic effect due to heightened release of calcium from the sarcoplasmic reticulum and L-type calcium channel activation [38].
No other genes linked to DCM in dogs have been identified at the time of this review, but studies have identified potential loci of interest in other predisposed breeds, including the Portuguese Water Dog [39] and Irish Wolfhound [40].
Canine Arrhythmogenic Right Ventricular Cardiomyopathy
Arrhythmogenic Right Ventricular Cardiomyopathy is characterized clinically by ventricular arrhythmias and occasionally systolic dysfunction, and histologically by replacement of myocardial tissue with fibrofatty infiltrates. While the disease typically affects the right ventricle, the left ventricle may also be affected, and the high prevalence of ARVC in Boxer dogs has resulted in use of the term “Boxer Cardiomyopathy”. An 8-base pair deletion in the 3’ untranslated region of the striatin gene was identified in Boxer dogs [41] with autosomal dominant ARVC inheritance, and ~72% penetrance [41]. The deletion changes the secondary structure of the mRNA which may be linked to the reduction in striatin expression in affected Boxer dogs. Striatin is a protein localized to the intercalated discs, which contain gap junctions, responsible for facilitating electrical conduction between myocardial cells, and desmosomes, responsible for holding myocytes together during contraction. The role of the intercalated disc in intercellular impulse conduction and structural integrity may explain the conduction abnormalities and histologic changes observed when striatin is disrupted. More severe disease in homozygotes compared to heterozygotes has been observed based upon number of ventricular premature complexes during 24-ambulatory electrocardiogram (Holter monitor) [41].
Other Inherited Canine Arrhythmogenic Diseases
A genetic mutation associated with an inherited ventricular arrhythmia in Rhodesian Ridgebacks was identified in a family of dogs with history of arrhythmias and sudden death and no evidence of structural heart disease [30]. A single base pair change from G to A results in a conserved glycine to serine change in the QIL1 gene [29]. Autosomal recessive inheritance is proposed, though autosomal dominant with incomplete penetrance cannot be ruled out [29]. The QIL1 gene product is involved in the Mitochondrial Contact Site and Cristae Organizing System complex assembly, which is important for cristae stability and respiratory chain function [42]. The arrhythmias observed with this mutation may be a result of irregular respiration and diminished ATP production on action potential production and myocyte conduction [29].
Inherited ventricular arrhythmias in German Shepherd dogs with no evidence of structural heart disease may underlie sudden cardiac death [43]. Sudden cardiac death appears to occur more frequently in dogs younger than 1.5 years and typically occurs during sleep or periods of rest following exercise [43,44]. Due to these associations, abnormal development of the autonomic innervation of the heart is hypothesized to trigger early afterdepolarization and ventricular arrhythmias [44]. Although no causative mutation has been identified to date, pedigrees suggest either incomplete penetrance of a single gene defect and/or polygenic or multifactorial inheritance [43].
Genetic Testing in Dogs
The development of genetic tests has enabled informed breeding and clinical recommendations in predisposed dogs. North Carolina State University offers a range of genetic tests for canine cardiomyopathies, including the Doberman Pinscher DCM1f and DCM2g mutations and Boxer Striatinh mutation. While these tests can guide breeding and symptomatic screening, it is important to understand their limitations. None of the cardiomyopathy-linked mutations identified thus far result in 100% penetrance. While the presence of the mutation increases the risk of developing disease, other factors (exercise, diet, etc.) influence presentation [45]. Furthermore, genetic variants that remain to be discovered also contribute to the current manifestation of cardiomyopathies in dogs that are not attributed to known disease-linked mutations. Despite these limitations, genetic testing remains an important resource to guide breeding recommendations and estimate the likelihood of DCM development.
Current Gene Therapy Techniques
Gene therapy is a broad term referring to gene based therapeutic approaches for combating disease, including inherited genetic disorders [21]. Gene therapy for inherited disorders can be categorized based on approach: 1) correction of a recessive gene deficiency by delivery of a wild type cDNA (gene transfer), 2) RNA interference of mutant gene transcripts by delivery of miRNAs (gene silencing), or 3) direct editing of the genome to correct a mutation or otherwise manipulate the DNA sequence in a way that results in improved gene function (genome editing). Alternatively, the approach can be used to alter circulating or cellular protein levels for therapeutic effect (“drug effect”), to treat diseases that are not inherited or for which the causative mutation is unknown [21]. Each approach requires careful selection of a delivery vehicle and consideration for an optimal route of delivery to ensure that the material avoids degradation by the host’s cells, arrives at the desired location, and achieves adequate transduction levels to produce the desired functional improvements. Additionally, consideration must be given to potential off-target effects. Finally, duration of therapeutic expression (long-term vs. short-term) and potential toxicity or immunogenicity of either the delivery vehicle or its product are also important treatment considerations. In this section, we discuss gene therapy approaches for cardiac disease.
Gene Transfer Techniques
The first cardiac gene therapy attempts aimed to promote angiogenesis in coronary and peripheral artery disease in humans [46]. Intracardiac injections of plasmids encoding angiogenic growth factors such as VEGF-A and FGF2 were conducted to stimulate angiogenesis, followed by use of adeno-associated virus (AAV) vectors as delivery systems; however, after over 150 clinical trials, no successful promotion of angiogenesis has been achieved [46].
Similarly, attempts have been made to apply gene therapy in heart failure in humans. One main area of focus has been the modulation of ionic calcium handling in cardiomyocytes, which is essential for maintaining normal cardiac function. Sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a (SERCA2a) mediates Ca2+ uptake into the sarcoplasmic reticulum (SR) in cardiomyocytes, and the expression level and activity of this pump is reduced in failing hearts [35]. The activity of the SERCA2a pump is inhibited by a protein called phospholamban, which is deactivated by phosphorylation. One of the first trials utilizing gene therapy in heart failure involved transfer of 1 × 1013 vector genomes (vg) of AAV1-SERCA2a into the coronary arteries of failing human hearts. Unfortunately, despite successes including increase in time to clinical events, decreased frequency of cardiovascular events, and decrease in mean duration of cardiovascular hospitalizations seen in initial trials [47], this treatment failed to improve clinical outcomes, with no significant differences observed in exercise ability, quality of life, or cardiac biomarker levels between the treatment and placebo groups during the 12 months of follow-up [48]. Ultimately, analyses of cardiac samples from deceased SERCA2a/AAV1 treated patients revealed that AAV transduction was lower than that expected for a significant therapeutic effect suggesting that inefficient transduction was a factor leading to treatment failure.
A different approach focused on modulation of Ca2+ uptake through overexpression of constitutively active I-1c. I-1c is a truncated form of inhibitor 1 (I-1), a protein which inhibits protein phosphatase 1. Protein phosphatase 1 is a phosphatase which regulates several cell processes including glycogen metabolism, cell division, muscle contraction, and signal contraction [49]. Inhibition of this phosphatase by overexpression of its inhibitor I-1c results in enhanced Phospholamban phosphorylation, which ultimately results in increased activity of SERCA2a [25]. Intracoronary gene transfer of constitutively active I-1c using BNP116, a chimeric vector derived from naturally occurring AAV2 and AAV8 capsids, has shown promise in pigs [23]. High dose (3 × 1012 vg) and low-dose (1 × 1013 vg) injections of BNP116.I-1c were used, and both groups showed improved cardiac function compared to control pigs based on echocardiographic assessment two months after experimentally induced myocardial infarction.
A third approach in attempted to restore Ca2+ homeostasis through overexpression of adenylyl cyclase 6, an important cardiac second messenger which increases SERCA2a Ca2+ uptake and reduces phospholamban expression [50]. A single intra-coronary injection of an adenoviral vector Ad5 expressing Adenylyl cyclase 6 cDNA was administered to human patients with heart failure, at various doses. Echocardiographic examination was performed at four and twelve weeks following treatment with adenylyl cyclase 6. Improvement in ejection fraction was noted in the two highest dose groups at four weeks but not at twelve weeks, with nonischemic heart failure patients showing the greatest response.
Together, these studies demonstrate the potential and challenges of cardiac gene transfer. The observed difficulties in transducing target cells have led to the identification and/or engineering of new AAV vectors for more efficient and specific cell targeting and transduction.
Gene Silencing Techniques
Gene silencing involves blocking messenger ribonucleic acid (mRNA) function through inhibition of protein translation, predominantly via RNA interference and antisense oligonucleotides (ASO) [51]. RNA interference uses short, synthetic, double-stranded RNA called small interfering RNA which are specifically designed to pair with a target mRNA and cause degradation of the mRNA [53,54]. While antisense techniques work similarly, ASOs are typically single-stranded RNA molecules which complement the target mRNA and cause either degradation of the target mRNA or blocking of translation through different mechanisms such as modification of splicing and steric hindrance [54,55].
A recent example of the application of ASO technology in canine disease is canine Duchenne Muscular Dystrophy (DMD), a disease characterized by progressive degeneration of skeletal and smooth muscles, including the heart. Dystrophin is a large scaffolding protein which links the cytoskeleton with the sarcolemma of muscle tissue and helps to preserve myofibril integrity [54]. Loss of this structural function results in the muscle weakness and atrophy observed in individuals affected with DMD. Thousands of mutations within the dystrophin gene have been linked to DMD, and many of these are concentrated in the region of exons 45–50, and cause a frameshift that results in failure to express a functional dystrophin protein due to premature stop codons [26]. An ASO was designed to bind near the splice acceptor site of exon 51 and mask splice signals, resulting in skipping of exon 51 and restoration of the open reading frame with improved production of a truncated but functional dystrophin protein in canine patients [55].
Additionally, ASO technology has been used in an in vitro study of human titin-based DCM [56]. A 2-bp insertion in exon 326 of the titin gene produces a premature stop codon in one form of inherited DCM in humans, resulting in a truncated protein. Through lentiviral-mediated ASO transfection of human iPSC-derived cardiomyocytes, exon 326 skipping was achieved, with associated improvement in protein expression, sarcomere structure, and contractile performance [56].
Gene Editing Techniques
While the studies discussed above show the promise of gene transfer for heart failure, the discovery of clustered regularly interspaced short palindromic repeats (CRISPR) gene editing techniques has broadened the scope of gene therapy beyond gene delivery to include the transfer of factors able to correct genomic variants. Gene editing technologies take advantage of components of natural bacterial adaptive immune response mechanisms in order to activate, deactivate, or alter specific genes [24,59,60]. Clustered regularly interspaced short palindromic repeats and CRISPR associated protein 9 (Cas9) based approaches use short RNA sequences called guide RNAs (gRNA) in combination with the Cas9 endonuclease to identify and cut specific nucleic acid sequences [Figure 1]. A short sequence of DNA called a protospacer adjacent motif located near the target sequence is also required for Cas9 cleavage [59]. Numerous variants of Cas9 enzymes are found within different bacterial species [60], and subsequent directed evolution of Cas9 enzymes has led to the capability to target almost any DNA sequence [60,62,63].
Figure 1:
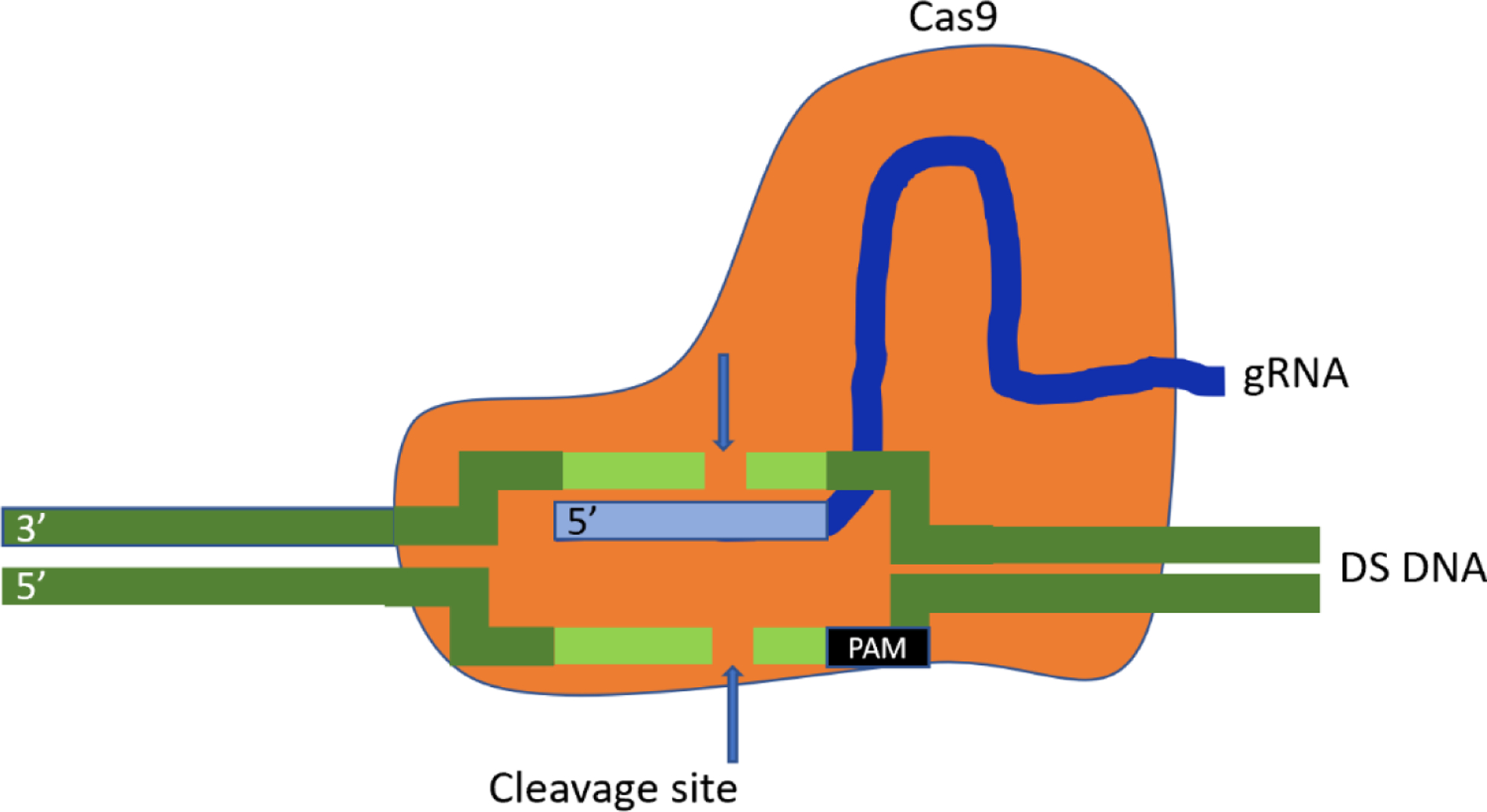
Schematic diagram of the Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR associated protein 9 (Cas9) system. Cas9 (orange) forms a complex with a guide RNA (gRNA, blue), creating a sequence-specific endonuclease. The gRNA recognizes a target sequence (light green) ending with a 3’ protospacer adjacent motif sequence, enabling double-stranded cleavage of the target DNA.
Once a targeted double-stranded cut has been made, the DNA must be repaired, and this occurs via one of two mechanisms: non-homologous end joining or homology directed repair (HDR) [61]. Non-homologous end joining is a fast, efficient method of DNA repair in which the ends of a double-stranded break are ligated [Figure 2a]. This mechanism may occur in any phase of the cell cycle but can be error-prone and generates genetic insertion/deletions [62]. In contrast, HDR relies on a DNA template in order to enable precise repair of a break and preserve genetic integrity [63] [Figure 2b]. This process, however, is thought to be mostly limited to the S- or G2-phase of the cell cycle, thus limiting efficiency of HDR in mature non-replicating cells [64].
Figure 2:
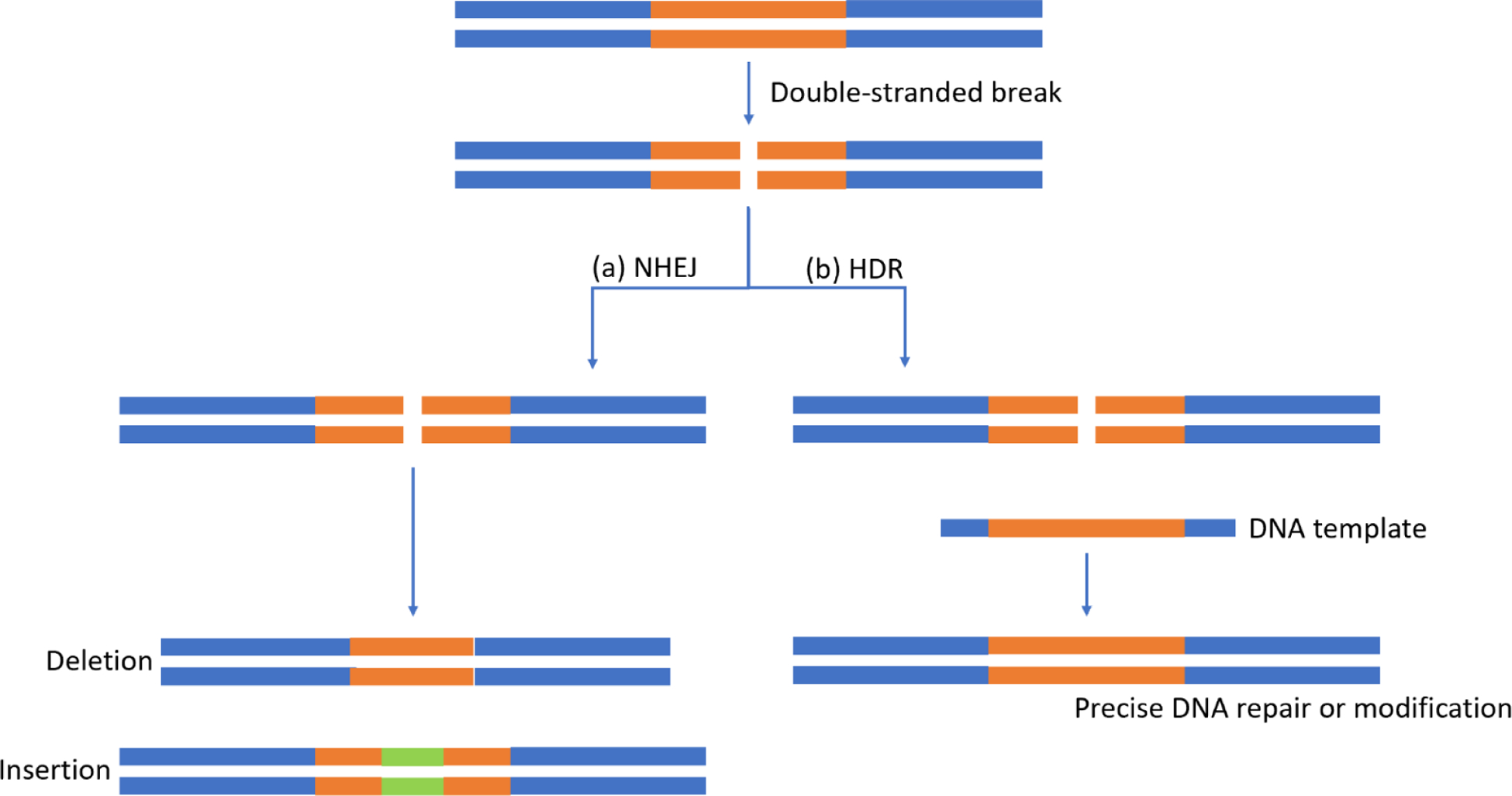
Schematic diagram of possible double-stranded break repair mechanisms. (a) Non-homologous end joining is a fast, efficient pathway which frequently results in insertions and/or deletions. (b) Homology directed repair is a slower pathway which uses a deoxyribonucleic acid (DNA) template, allowing for precise repair and possibly DNA modification.
A recent example of the application of gene editing technology in canine disease is that of DMD, mentioned earlier. Building on the discovery that ASO-mediated exon skipping can successfully restore the open reading frame, a CRISPR/Cas9 construct was designed to edit the splice acceptor site of exon 51, resulting in exon 51 skipping and production of functional dystrophin in dogs [26]. Using an AAV9 vector, 1.2 × 1013 vg was injected into one of the cranialis tibialis muscles of study dogs, resulting in restoration of approximately 60% of wildtype levels of dystrophin in the treated muscle and only 2% of wildtype levels of dystrophin in the untreated muscle 6 weeks after treatment. Systemic high-dose injection of 1 × 1014 vg using an AAV9 vector was demonstrated to restore up to 70% of wildtype levels of dystrophin expression in skeletal muscle and up to 92% in cardiac muscle 8 weeks following treatment.
These previous gene therapy studies have guided ongoing research and will help to further inform future studies. Failures in effective transduction of target cells have led to creative approaches to generate engineered serotypes with the capacity to target and transduce specific tissues types more efficiently [65]. The successful restoration of the open reading frame in DMD using either an antisense oligonucleotide or CRISPR gene editing demonstrates the repertoire of effective strategies available to present day scientists. In combination with improved technology and an increased understanding of inherited disease, the review and critical assessment of previous experiments allows us to continue to build this new branch of science and may lead to profound improvement in the care and quality of life of patients of all species.
In Vivo Gene Delivery Techniques
While many different inexpensive and efficient methods of gene delivery to cells in vitro and ex vitro exist, delivery to cells and tissues in vivo is more challenging. In vivo delivery is clinically necessary, but successful transduction can be difficult to achieve due to interactions with non-target cells and the host immune system [66]. The ideal delivery vector will be non-immunogenic with high specificity for the target cell or tissue types, and result in sufficient transduction and transgene expression levels to produce the desired therapeutic effect. Gene delivery systems are typically categorized as viral vs. nonviral. While nonviral delivery systems are typically less immunogenic, their lower ability to effectively target mature cells such as cardiomyocytes and reduced persistence over time make these systems, with the possible exception of nanoparticle delivery systems [69,70,71], less effective for cardiac gene therapy. Thus, this review will focus on commonly used viral vector systems.
Typical viruses used as vectors for gene delivery include adenovirus, adeno-associated virus, lentivirus, human immunodeficiency virus, and herpes simplex virus [70], among others. Portions of plasmids containing DNA/RNA and/or CRISPR/Cas9 constructs (gene editing materials) are packaged into the virus, which delivers these materials to the target cell for expression. Several different virus strains have been identified with different characteristics including varied expression duration (long-term vs. short-term), diverse somatic cell line targets, and different packaging capacities.
Adenovirus
Adenovirus, a medium-sized non-enveloped virus, is a widely used viral vector in gene transfer. Adenoviruses naturally cause diseases in mammals; therefore, it was not unexpected that the original adenovirus vectors evoked a strong immune response from hosts. Second- and third-generation adenoviruses containing deletions were engineered to be less immunogenic [70]. While genetic material can gain entry into the nucleus of infected cells and be transcribed using such a system, integration of the donor DNA into the host genome does not occur and therefore short-term expression is achieved.
Adeno-associated virus
Adeno-associated virus is a small non-enveloped virus in the Parvoviridae family. It is capable of infecting both dividing and non-dividing cells and has been found to have low pathogenicity and low toxicity, although repeated administration of AAV vectors triggers an immune response [66]. Adeno-associated virus has evolved to enter cells through interactions with surface molecules on target cells. Differences in sugar-binding preferences and secondary receptors confer the varying tissue selectivity that distinguishes the different viral serotypes (AAV 1–9), and as a result one variant may be chosen over another in order to preferentially transduce a particular cell type [71]. The recombinant AAVs (rAAVs) used in gene delivery studies retain only the inverted terminal repeats of the original viral genome. While rAAVs do not undergo host genome integration, they do persist for long periods of time as episomes in the nucleus of cells [72]. Due to the lack of host genome integration, repeated administration may be necessary in some gene therapy approaches where the target cell population is replaced over time, as the rAAV episomes become diluted out among daughter nuclei with cell division.
One of the greatest limitations of this viral vector is the small packaging capacity of approximately 4.8 kilobases [24,75]. This is a challenge for delivery of large genes (e.g. dystrophin or titin) and also for CRISPR gene editing approaches which require packaging of a Cas9 gene and gRNA in addition to template DNA for HDR. As an example, wildtype Cas9 derived from Streptococcus pyogenes (spCas9) is approximately 4.1 kb, leaving little room for addition of gRNA and template DNA [24]. One method of circumventing this issue is the use of dual vectors, with one AAV containing the Cas9 enzyme and another containing the gRNA and DNA template [74]. This approach, however, requires that each target cell be transduced with both AAV vectors for the desired gene editing to occur. Another method of adjusting to the size constraints of AAVs is using smaller Cas9 variants. While spCas9 is the original Cas9 enzyme found in Streptococcus pyogenes, other Cas9 enzymes with differing properties including smaller sizes have been isolated from other bacterial species, ranging from Francisella novicida (~1.6 kb) to Campylobacter jejuni (~984 bp) [61]. Yet another approach is split-Cas9 systems utilizing split-inteins, which can be thought of as protein introns. Cas9 halves connected to split-inteins may be administered separately, and trans-splicing of inteins allows for reconstitution of the complete Cas9 following transfection [75].
Retrovirus
Retroviruses are enveloped viruses containing an RNA genome. They are particularly useful for long-term expression due to integration of DNA into the host genome [76]. As random genome integration of retroviruses is problematic, safer vectors designed for targeted integration are being developed to minimize undesired host mutagenesis. There is also concern regarding the potential for pathogenicity when using this viral vector, as replication-competent cells may be produced. Another significant drawback of this type of viral vector is the inability to infect nondividing cells; however, lentivirus, a specific type of retrovirus, is capable of infecting both dividing and nondividing cells.
Lentivirus is a type of retrovirus mainly derived from human immunodeficiency virus 1 which differs from other retroviruses in its ability to translocate across the nuclear membrane and therefore transduce nonreplicating cells [77]. This ability makes lentiviruses particularly useful in gene editing of mature somatic cells such as cardiomyocytes. Safer lentiviral vectors have been engineered through removal of accessory virulence factors and splitting of the viral genome into separate plasmids, which reduce the risk of creating recombinant viruses [78]. Insertion of lentiviral vectors near oncogenes in the host genome and subsequent tumor formation remains a concern.
Although not covered in this review, multiple additional viruses have been adapted as vectors in gene therapy systems [72,76]. The continued development of vectors with improved ability to target and transduce specific cells will enable future genetic therapeutic approaches.
Challenges in Gene Therapy
While gene therapy holds great promise for the treatment of inherited and non-inherited disorders, there are several challenges in the potential clinical application. Gene transfer and gene editing both require vectors to carry enzymes and/or genetic material to target cells. As discussed in the previous section, size constraints of certain vectors are an issue particularly when attempting to transfer large genes (gene transfer) or when using larger Cas9 enzymes (gene editing) [48,72,24,76]. In the case of gene transfer, the use of minigenes (truncated but functional forms of a gene) shows promise [79]. In gene editing, packaging of the Cas9 enzyme and gRNA/DNA template into separate plasmids, use of smaller Cas9 variants, and use of split-Cas9 systems are potential options [63,24,76].
As discussed previously, some vectors that insert into the host genome also carry the risk of oncogenesis [80,82,83,84]. Avoidance of these types of vectors is not always possible, especially if viral integration into the host genome for long-term expression of a gene is desired. The alteration of existing viral vectors including deletion of accessory virulence factors in order to reduce oncogenesis and risk of recombination is another potential solution, though the risk is not eliminated [80].
Immunogenicity is a large challenge in the clinical application of gene therapy since many commonly used viral vectors naturally infect humans and animals and therefore have the potential to stimulate both innate and adaptive immune responses [85,86]. An immune response against a gene therapy vector may eliminate the vector and/or transfected cells, potentially reducing the duration and intensity of transgene protein expression [85]. Treatments must be optimized to use the lowest possible therapeutic dose to achieve adequate transduction of target cells and avoid stimulation of the host immune response, as the latter can be fatal [86,87].
Clustered regularly interspaced short palindromic repeats technology continues to improve exponentially, with the number of known Cas9 systems and the repertoire of delivery vectors quickly expanding. Progress thus far gives hope for the development of new, more effective treatments for inherited diseases. However, it is important to note the unique limitations and challenges faced by this new branch of therapeutics. First, efficiency of gene editing systems is a limiting factor. While the existence of many Cas9 variants theoretically enables targeting of almost any portion of the genome, the insertion of specific DNA sequences is limited by HDR, which is much lower in efficiency compared to the more error-prone process of non-homologous end joining. As discussed in the previous section, HDR is significantly limited in nonreplicating cells such as cardiomyocytes, reducing the efficiency of gene editing. Additionally, Cas9 enzymes have variable specificities, depending on the variant and the sequence to be targeted. Off-target endonuclease activity may result in the creation of undesired mutations and unpredictable consequences. Due to this risk, extensive in vitro testing and optimization is essential.
Many inherited diseases including inherited cardiomyopathies are multifactorial; that is, they are influenced by lifestyle and environment in addition to typically involving several different genes, some of which remain unidentified [16,17,18,19,20,40,41,48]. Gene editing of specific targeted mutations will likely not provide a single treatment which can be broadly applied to all affected patients with diseases which are known to be caused my many unique mutations (e.g. human hypertrophic cardiomyopathy). However, the smaller number of uniform mutations identified in dogs may make this species more amenable to genome editing, and gene delivery to address molecular abnormalities (such as calcium cycling) remains an alternative approach.
The incomplete penetrance of many mutations linked to inherited diseases also makes it difficult to predict which individuals will develop the disease phenotype and to what extent. Here the question arises as to which patients should be treated with gene therapy and when such a treatment should be performed. It is reasonable to assume in the case of inherited cardiac disorders, treatment should be encouraged prior to the onset of irreversible structural and hemodynamic changes; however, the variable length of the occult stage of some diseases makes this determination difficult.
Finally, the determination of whether a treatment was a success (i.e., the target gene was successfully edited as intended) or a failure can be difficult. This uncertainty is linked to the multifactorial nature of disease and variation in disease phenotypes among affected individuals. Successful genetic treatment of a patient may still result in clinical disease due to a separate, unidentified mutation or other environmental factors. Conversely, a treated patient may fail to develop an inherited disorder, but determination of treatment success cannot be assumed since it is difficult to predict if this patient would have ever developed clinical disease even without genetic therapy. Prospective studies evaluating treatments must be sufficiently powered to overcome these obstacles in determining success or failure, and natural history studies are essential to the design of clinical trials with appropriate outcome measures for the target patient population.
Repeated genetic screening using non-target cells (e.g., cheek swab) from a treated individual would be expected to continue showing a mutant genotype due to the theoretic cell type specificity of gene editing (i.e., only cardiomyocytes should show genetic changes after treatment). Genetic screening of the target cell type following treatment would help to confirm treatment success; however, in the case of cardiomyopathies, repeated screening is not practical antemortem, as cardiomyocytes are not routinely sampled in live animals. Another method of gauging treatment success may include selecting patients which have developed some signs of structural disease (e.g., left ventricular (LV) thickening and diastolic dysfunction in hypertrophic cardiomyopathy or left ventricular dilation and systolic dysfunction in DCM), then performing follow-up examinations following treatment to monitor disease improvement or progression.
Conclusion
The discovery of specific genetic mutations linked to certain cardiomyopathies has strengthened our ability to screen and estimate risk of cardiac disease development in certain breeds. While the availability of commercial tests for these genetic mutations is helpful, incomplete penetrance and the multifactorial nature of cardiomyopathies continues to complicate interpretation of test results. Genetic testing should be viewed as a method to assess risk of disease in a breeding animal or beloved pet rather than a method of diagnosis, and interpretation of results under the guidance of a qualified professional is recommended.
The discovery of these disease-linked mutations has also opened avenues for seeking different forms of treatment. While gene therapy holds promise, challenges such as identification of vectors with adequate packaging capacity and efficient target cell transduction must be overcome to make clinical application widely feasible. Additionally, the complexity of breed-specific mutations necessitates the investigation of different approaches as opposed to a universal treatment to optimize treatment success. While significant limitations and challenges in genetic editing have been identified, the continued exponential growth of this branch of research shows great promise for future clinical applications.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institutes of Health [grant number T32 HG008958].
Abbreviations
- AAV
adeno-associated virus
- ARVC
arrhythmogenic right ventricular cardiomyopathy
- ASO
antisense oligonucleotide
- Cas9
CRISPR associated protein 9
- CRISPR
clustered regularly interspaced short palindromic repeats
- DCM
dilated cardiomyopathy
- DMD
Duchenne muscular dystrophy
- gRNA
guide ribonucleic acid
- HDR
homology directed repair
- mRNA
messenger ribonucleic acid
- PDK4
phosphodiesterase kinase 4
- SERCA2a
sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors have no conflicts of interest to declare in the completion of this manuscript.
NCSU DCM1 Genetic Testing, North Carolina State Veterinary Hospital, Raleigh, NC, USA.
NCSU DCM2 Genetic Testing, North Carolina State Veterinary Hospital, Raleigh, NC, USA.
Boxer ARVC Genetic Testing, North Carolina State Veterinary Hospital, Raleigh, NC, USA.
References
- [1].McKenna WJ, Maron BJ, Thiene G. Classification, epidemiology, and global burden of cardiomyopathies. Circ Res 2017;121:722–30. [DOI] [PubMed] [Google Scholar]
- [2].Maron BJ, Doerer JJ, Haas TS, Tierney DM, Mueller FO. Sudden deaths in young competitive athletes: Analysis of 1866 deaths in the United States, 1980–2006. Circulation 2009;119:1085–92. [DOI] [PubMed] [Google Scholar]
- [3].Payne JR, Brodbelt DC, Luis Fuentes V. Cardiomyopathy prevalence in 780 apparently healthy cats in rehoming centres (the CatScan study). J Vet Cardiol 2015;17:S244–57. [DOI] [PubMed] [Google Scholar]
- [4].Maron BJ, McKenna WJ, Danielson GK, Kappenberger LJ, Kuhn HJ, Seidman CE, et al. American College of Cardiology/European Society of Cardiology Clinical Expert Consensus Document on Hypertrophic Cardiomyopathy: A Report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the European S. J Am Coll Cardiol 2003;42:1687–713. [DOI] [PubMed] [Google Scholar]
- [5].Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults: Echocardiographic analysis of 4111 subjects in the CARDIA study. Circulation 1995;92:785–9. [DOI] [PubMed] [Google Scholar]
- [6].Freeman LM, Rush JE, Stern JA, Huggins GS, Maron MS. Feline Hypertrophic Cardiomyopathy: a spontaneous large animal model of human HCM. Cardiol Res 2017;8:139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wess G, Schulze A, Butz V, Simak J, Killich M, Maeurer J, et al. Prevalence of dilated cardiomyopathy in Doberman Pinschers in various age groups. J Vet Intern Med 2010;24:533–8. [DOI] [PubMed] [Google Scholar]
- [8].Dutton E, López-Alvarez J. An update on canine cardiomyopathies – is it all in the genes? J Small Anim Pract 2018;59:455–64. [DOI] [PubMed] [Google Scholar]
- [9].Stern JA, Meurs KM, Spier AW, Koplitz SL, Baumwart RD. Ambulatory electrocardiographic evaluation of clinically normal adult Boxers. J Am Vet Med Assoc 2010;236:430–3. [DOI] [PubMed] [Google Scholar]
- [10].Summerfield NJ, Boswood A, O’Grady MR, Gordon SG, Dukes-Mcewan J, Oyama MA, et al. Efficacy of pimobendan in the prevention of congestive heart failure or sudden death in Doberman Pinschers with preclinical dilated cardiomyopathy (the PROTECT study). J Vet Intern Med 2012;26:1337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Meurs KM, Stern JA, Reina-Doreste Y, Spier AW, Koplitz SL, Baumwart RD. Natural History of Arrhythmogenic Right Ventricular Cardiomyopathy in the Boxer Dog: A Prospective Study. J Vet Intern Med 2014;28:1214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Caro-Vadillo A, García-Guasch L, Carretón E, Montoya-Alonso J, Manubens J. Arrhythmogenic right ventricular cardiomyopathy in boxer dogs: a retrospective study of survival. Vet Rec 2013;172:268. [DOI] [PubMed] [Google Scholar]
- [13].Martin MWS, Stafford Johnson MJ, Strehlau G, King JN. Canine dilated cardiomyopathy: A retrospective study of prognostic findings in 367 clinical cases. J Small Anim Pract 2010;51:428–36. [DOI] [PubMed] [Google Scholar]
- [14].Ashrafian H, Watkins H. Reviews of Translational Medicine and Genomics in Cardiovascular Disease: New Disease Taxonomy and Therapeutic Implications. Cardiomyopathies: Therapeutics Based on Molecular Phenotype. J Am Coll Cardiol 2007;49:1251–64. [DOI] [PubMed] [Google Scholar]
- [15].Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: A review for genetics professionals. Genet Med 2010;12:655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Meurs KM, Sanchez X, David RM, Bowles NE, Towbin JA, Reiser PJ, et al. A cardiac myosin binding protein C mutation in the Maine Coon cat with familial hypertrophic cardiomyopathy. Hum Mol Genet 2005;14:3587–93. [DOI] [PubMed] [Google Scholar]
- [17].Meurs KM, Norgard MM, Ederer MM, Hendrix KP, Kittleson MD. A substitution mutation in the myosin binding protein C gene in ragdoll hypertrophic cardiomyopathy. Genomics 2007;90:261–4. [DOI] [PubMed] [Google Scholar]
- [18].Meurs KM, Lahmers S, Keene BW, White SN, Oyama MA, Mauceli E, et al. A splice site mutation in a gene encoding for PDK4, a mitochondrial protein, is associated with the development of dilated cardiomyopathy in the Doberman pinscher. Hum Genet 2012;131:1319–25. [DOI] [PubMed] [Google Scholar]
- [19].Meurs KM, Friedenberg SG, Kolb J, Saripalli C, Tonino P, Woodruff K, et al. A missense variant in the titin gene in Doberman pinscher dogs with familial dilated cardiomyopathy and sudden cardiac death. Hum Genet 2019;138:515–24. [DOI] [PubMed] [Google Scholar]
- [20].Meurs KM, Stern JA, Sisson DD, Kittleson MD, Cunningham SM, Ames MK, et al. Association of dilated cardiomyopathy with the striatin mutation genotype in boxer dogs. J Vet Intern Med 2013;27:1437–40. [DOI] [PubMed] [Google Scholar]
- [21].Sleeper M, Bish LT, Haskins M, Ponder KP, Sweeney HL. Status of therapeutic gene transfer to treat cardiovascular disease in dogs and cats. J Vet Cardiol 2011;13:131–40. [DOI] [PubMed] [Google Scholar]
- [22].Schwab DM, Tilemann L, Bauer R, Heckmann M, Jungmann A, Wagner M, et al. AAV-9 mediated phosphatase-1 inhibitor-1 overexpression improves cardiac contractility in unchallenged mice but is deleterious in pressure-overload. Gene Ther 2018;25:13–9. [DOI] [PubMed] [Google Scholar]
- [23].Ishikawa K, Fish KM, Tilemann L, Rapti K, Aguero J, Santos-Gallego CG, et al. Cardiac I-1c overexpression with reengineered AAV improves cardiac function in swine ischemic heart failure. Mol Ther 2014;22:2038–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lau CH, Suh Y. In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease. F1000Research 2017;6:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fish KM, Ladage D, Kawase Y, Karakikes I, Jeong D, Ly H, et al. AAV9.I-1c delivered via direct coronary infusion in a porcine model of heart failure improves contractility and mitigates adverse remodeling. Circ Hear Fail 2013;6:310–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science (80-) 2018;362:86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sisson D, Thomas W, Keene B. Primary myocardial disease in the dog. In: Ettinger SJ, Feldman EC, editors. Textb. Vet. Intern. Med 5th ed., Philadelphia: Saunders; 2000, p. 874–95. [Google Scholar]
- [28].Yost O, Friedenberg SG, Jesty SA, Olby NJ, Meurs KM. The R9H phospholamban mutation is associated with highly penetrant dilated cardiomyopathy and sudden death in a spontaneous canine model. Gene 2019;697:118–22. [DOI] [PubMed] [Google Scholar]
- [29].Meurs KM, Friedenberg SG, Olby NJ, Condit J, Weidman J, Rosenthal S, et al. A QIL1 variant associated with ventricular arrhythmias and sudden cardiac death in the juvenile rhodesian ridgeback dog. Genes (Basel) 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Meurs KM, Weidman JA, Rosenthal SL, Lahmers KK, Friedenberg SG. Ventricular arrhythmias in Rhodesian Ridgebacks with a family history of sudden death and results of a pedigree analysis for potential inheritance patterns. J Am Vet Med Assoc 2016;248:1135–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bolfer L, Estrada AH, Larkin C, Conlon TJ, Lourenco F, Taggart K, et al. Functional consequences of PDK4 deficiency in Doberman Pinscher fibroblasts. Sci Rep 2020;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Taggart K, Estrada A, Thompson P, Lourenco F, Kirmani S, Suzuki-Hatano S, et al. PDK4 deficiency induces intrinsic apoptosis in response to starvation in fibroblasts from Doberman Pinschers with dilated cardiomyopathy. Biores Open Access 2017;6:182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Meurs KM, Stern JA, Adin D, Keene BW, DeFrancesco TC, Tou SP. Assessment of PDK4 and TTN gene variants in 48 Doberman Pinschers with dilated cardiomyopathy. J Am Vet Med Assoc 2020;257:1041–4. [DOI] [PubMed] [Google Scholar]
- [34].Young HS, Ceholski DK, Trieber CA. Deception in simplicity: Hereditary phospholamban mutations in dilated cardiomyopathy. Biochem Cell Biol 2015;93:1–7. [DOI] [PubMed] [Google Scholar]
- [35].Park WJ, Oh JG. SERCA2a: A prime target for modulation of cardiac contractility during heart failure. BMB Rep 2013;46:237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Harmon MW, Leach SB, Lamb KE. Dilated cardiomyopathy in standard schnauzers: Retrospective study of 15 cases. J Am Anim Hosp Assoc 2017;53:38–44. [DOI] [PubMed] [Google Scholar]
- [37].Gilliam DH. Molecular Genetic Studies of Canine Inherited Diseases Including SAMS, Neuronal Ceroid Lipofuscinosis and Dilated Cardiomyopathy. University of Missouri, 2016. [Google Scholar]
- [38].Van Den Hoogenhof MMG, Beqqali A, Amin AS, Van Der Made I, Aufiero S, Khan MAF, et al. RBM20 mutations induce an arrhythmogenic dilated cardiomyopathy related to disturbed calcium handling. Circulation 2018;138:1330–42. [DOI] [PubMed] [Google Scholar]
- [39].Werner P, Raducha MG, Prociuk U, Sleeper M, Henthorn PS. A novel locus for Dilated Cardiomyopathy maps to canine chromosome 8. Genomics 2008;91:517–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Philipp U, Vollmar A, Häggström J, Thomas A, Distl O. Multiple loci are associated with Dilated Cardiomyopathy in Irish Wolfhounds. PLoS One 2012;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Meurs KM, Mauceli E, Lahmers S, Acland GM, White SN, Lindblad-Toh K. Genome-wide association identifies a deletion in the 3’ untranslated region of Striatin in a canine model of arrhythmogenic right ventricular cardiomyopathy: Identification of Striatin deletion in canine ARVC. Hum Genet 2010;128:315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Guarani V, McNeill EM, Paulo JA, Huttlin EL, Fröhlich F, Gygi SP, et al. QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. Elife 2015;4:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Moise NS, Meyers-Wallen V, Flahive WJ, Valentine BA, Scarlett JM, Brown CA, et al. Inherited ventricular arrhythmias and sudden death in German shepherd dogs. J Am Coll Cardiol 1994;24:233–43. [DOI] [PubMed] [Google Scholar]
- [44].Moïse NS. Inherited arrhythmias in the dog: potential experimental models of cardiac disease. Cardiovasc Res 1999;44:37–46. [DOI] [PubMed] [Google Scholar]
- [45].Kousi M, Katsanis N. Genetic modifiers and oligogenic inheritance. Cold Spring Harb Perspect Med 2015;5:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cannatà A, Ali H, Sinagra G, Giacca M. Gene Therapy for the heart: lessons learned and future perspectives. Circ Res 2020;126:1394–414. [DOI] [PubMed] [Google Scholar]
- [47].Jessup M, Greenberg B, Mancini D, Cappola T, Pauly DF, Jaski B, et al. Calcium upregulation by percutaneous administration of gene therapy in cardiac disease (CUPID): A phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation 2011;124:304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Greenberg B, Butler J, Felker GM, Ponikowski P, Voors AA, Desai AS, et al. Calcium upregulation by percutaneous administration of gene therapy in patients with cardiac disease (CUPID 2): A randomised, multinational, double-blind, placebo-controlled, phase 2b trial. Lancet 2016;387:1178–86. [DOI] [PubMed] [Google Scholar]
- [49].Huang KX, Paudel HK. Ser67-phosphorylated inhibitor 1 is a potent protein phosphatase 1 inhibitor. Proc Natl Acad Sci U S A 2000;97:5824–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hammond HK, Penny WF, Traverse JH, Henry TD, Watkins MW, Yancy CW, et al. Intracoronary gene transfer of adenylyl cyclase 6 in patients with heart failure. JAMA Cardiol 2016;1:163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Choi J Gene silencing. HOPES Stanford Univ 2012. [Google Scholar]
- [52].Chery J RNA therapeutics: RNAi and antisense mechanisms and clinical applications. Postdoc J 2016;4:35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Liou S Antisense gene therapy. HOPES Stanford Univ 2010. [Google Scholar]
- [54].Gao Q, McNally E. The dystrophin complex: structure, function and implications for therapy. Compr Physiol 2015;5:1223–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Echevarría L, Aupy P, Goyenvalle A. Exon-skipping advances for Duchenne muscular dystrophy. Hum Mol Genet 2018;27:R163–72. [DOI] [PubMed] [Google Scholar]
- [56].Gramlich M, Pane LS, Zhou Q, Chen Z, Murgia M, Schötterl S, et al. Antisense-mediated exon skipping: a therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol Med 2015;7:562–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rodríguez-Rodríguez DR, Ramírez-Solís R, Garza-Elizondo MA, Garza-Rodríguez MDL, Barrera-Saldaña HA. Genome editing: A perspective on the application of CRISPR/Cas9 to study human diseases (Review). Int J Mol Med 2019;43:1559–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 2020;38:824–44. [DOI] [PubMed] [Google Scholar]
- [59].Gleditzsch D, Pausch P, Müller-Esparza H, Özcan A, Guo X, Bange G, et al. PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol 2019;16:504–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Murovec J, Pirc Ž, Yang B. New variants of CRISPR RNA-guided genome editing enzymes. Plant Biotechnol J 2017;15:917–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cebrian-Serrano A, Davies B. CRISPR-Cas orthologues and variants: optimizing the repertoire, specificity and delivery of genome engineering tools. Mamm Genome 2017;28:247–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Davis AJ, Chen DJ. DNA double strand break repair via non-homologous endjoining. Transl Cancer Res 2013;2:130–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sansbury BM, Hewes AM, Kmiec EB. Understanding the diversity of genetic outcomes from CRISPR-Cas generated homology-directed repair. Commun Biol 2019;2:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D, et al. Methodologies for improving HDR efficiency. Front Genet 2019;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Grimm D, Zolotukhin S. E Pluribus Unum: 50 Years of Research, Millions of Viruses, and One Goal-Tailored Acceleration of AAV Evolution. Mol Ther 2015;23:1819–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther 2010;17:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yan C, Quan X-J, Feng Y-M. Nanomedicine for Gene Delivery for the Treatment of Cardiovascular Diseases. Curr Gene Ther 2019;19:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Patra JK, Das G, Fraceto LF, Campos EVR, Rodriguez-Torres MDP, AcostaTorres LS, et al. Nano based drug delivery systems: Recent developments and future prospects. J Nanobiotechnology 2018;16:1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Riley MK, Vermerris W. Recent advances in nanomaterials for gene delivery—A review. Nanomaterials 2017;7:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lundstrom K Viral vectors in gene therapy. Diseases 2018;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Naso MF, Tomkowicz B, Perry WL, Strohl WR. Adeno-Associated Virus (AAV) as a vector for gene therapy. BioDrugs 2017;31:317–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Schnepp BC, Clark KR, Klemanski DL, Pacak CA, Johnson PR. Genetic Fate of Recombinant Adeno-Associated Virus Vector Genomes in Muscle. J Virol 2003;77:3495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Colella P, Ronzitti G, Mingozzi F. Emerging issues in AAV-mediated in vivo gene therapy. Mol Ther Methods Clin Dev 2018;8:87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Xu CL, Ruan MZC, Mahajan VB, Tsang SH. Viral delivery systems for CRISPR. Viruses 2019;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Truong DJJ, Kühner K, Kühn R, Werfel S, Engelhardt S, Wurst W, et al. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res 2015;43:6450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kurian K, Watson C, Wyllie A. Retroviral vectors. J Clin Pathol 2000;53:173–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Merten OW, Hebben M, Bovolenta C. Production of lentiviral vectors. Mol Ther - Methods Clin Dev 2016;3:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Milone MC, O’Doherty U. Clinical use of lentiviral vectors. Leukemia 2018;32:1529–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chamberlain JS. Gene therapy of muscular dystrophy. Hum Mol Genet 2002;11:2355–62. [DOI] [PubMed] [Google Scholar]
- [80].Modlich U, Baum C. Preventing and exploiting the oncogenic potential of integrating gene vectors. J Clin Invest 2009;119:755–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Von Kalle C, Deichmann A, Schmidt M. Vector integration and tumorigenesis. Hum Gene Ther 2014;25:475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zhou S, Fatima S, Ma Z, Wang YD, Lu T, Janke LJ, et al. Evaluating the safety of retroviral vectors based on insertional oncogene activation and blocked differentiation in cultured thymocytes. Mol Ther 2016;24:1090–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Shirley JL, de Jong YP, Terhorst C, Herzog RW. Immune responses to viral gene therapy vectors. Mol Ther 2020;28:709–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Mingozzi F, High KA. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood 2013;122:23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bessis N, GarciaCozar FJ, Boissier MC. Immune responses to gene therapy vectors: Influence on vector function and effector mechanisms. Gene Ther 2004;11:S10–7. [DOI] [PubMed] [Google Scholar]
- [86].Raper SE, Magosin S, Simoes H, Speicher L, Hughes J, Tazelaar J, et al. A pilot study of in vivo liver-directed gene transfer with an adenoviral vector in partial ornithine transcarbamylase deficiency. Hum Gene Ther 2002;13:163–75. [DOI] [PubMed] [Google Scholar]
- [87].Lehrman S Virus treatment questioned after gene therapy death. Nature 1999;401:517–8. [DOI] [PubMed] [Google Scholar]


