Abstract
Bioimaging data have significant potential for reuse, but unlocking this potential requires systematic archiving of data and metadata in public databases. We propose draft metadata guidelines to begin addressing the needs of diverse communities within light and electron microscopy. We hope this publication and the proposed Recommended Metadata for Biological Images (REMBI) will stimulate discussions about their implementation and future extension.
Spectacular advances in light and electron microscopy1,2 are rapidly transforming the life sciences. For instance, scientists are now able to image molecular complexes at atomic resolution3–5, follow the fates of individual molecules in a living cell, and image the development of an organism starting from a single fertilized cell6,7. These imaging technologies are generating large amounts of complex data, the interpretation of which often requires sophisticated analyses, as in other ‘omics’ technologies. Moreover, most advanced imaging technologies are expensive, while the biological samples used in the experiments may be unique. To maximize the use of the generated data and to realize the full potential of the advances in biological imaging, these datasets need to be made available to other researchers in a timely manner, consistent with the FAIR principles—findable, accessible, interoperable and reusable8—and thus amenable to reuse.
Around the world, there are efforts to develop informatics systems for making different types of microscopy data available to the community. Sharing cryo-electron microscopy (cryo-EM) data is already quite advanced (Box 1), while sharing light microscopy data is still at an early stage. In Europe, a research infrastructure for biological and biomedical imaging called Euro-BioImaging has recently been established and is developing imaging data management and publishing solutions such as Cell-IDR and Tissue IDR9. In Japan, RIKEN launched the Systems Science of Biological Dynamics database (SSBD) in 2013, with the goal of sharing quantitative biological dynamics data including time-lapse microscopy images10. In 2016, the database expanded its remit to all bioimage data from the Japanese community. In the United States, the National Institutes of Health (NIH) has funded the establishment of the CELL Image Library11, while NIH’s BRAIN initiative is establishing specifications and resources for imaging of brain tissue (https://doryworkspace.org/, https://www.brainimagelibrary.org). In collaboration with Bioimaging North America, NIH’s 4D Nucleome project has released specifications for image-acquisition metadata12. There are also efforts that have wider geographic coverage. Global BioImaging (https://globalbioimaging.org/) has published recommendations for data formats and data repositories13, and the QUAREP-LiMi14,15 global consortium is working to establish community-driven specifications for quality assurance and testing in quantitative light microscopy.
Box 1 |. Archiving use-case: electron microscopy.
The electron microscopy (EM) field provides an example of how a well-organized (and historically relatively tightly knit) community can accomplish archiving of its raw and derived data and metadata, initially focusing on high-resolution molecular cryo-EM data and now expanding to include the vastly larger scales addressed with (and the more heterogeneous modalities of) volume EM methods.
Cryo-EM and cryo-ET have proven to be powerful tools for determining high-resolution structures of biological matter and examining the functional cellular context of macromolecular complexes. This has been possible due to technical advancements in microscope optics and detectors, sample-preparation techniques such as micropatterning grids and focused-ion-beam milling, and data-analysis pipelines including reliable automation of data-acquisition and processing workflows. Advances in the field of cryo-EM were recognized in 2017 with the Nobel Prize in Chemistry, and the method has continued to advance, now reaching truly atomic resolution3–5. In concert, cryo-ET has matured into a method that is capable of probing three-dimensional cellular context from micrometer to subnanometer scales, providing insight into biological processes such as viral infection and disease states.There is wide agreement in the cryo-EM community that detailed metadata must be recorded and deposited to public archives and that metadata standards must be reviewed over time to ensure they are fit for purpose and continue to address evolving community needs. In this sense, the cryo-EM community is setting an example for other imaging communities to follow.
Cryo-EM volumes (maps and tomograms) are commonly deposited in the Electron Microscopy Data Bank (EMDB; established in 2002)25, and any fitted atomic models in the Protein Data Bank (PDB; established in 1971)26. The Electron Microscopy Public Image Archive (EMPIAR) was established23 at EMBL-EBI in 2013 and has been the public resource for raw cryo-EM images that underpin the structures in EMDB. EMPIAR provides easy access to state-of-the-art raw data to facilitate methods development and validation, which will ultimately lead to better methods, better structures and a better understanding of biological questions. The EMPIAR metadata schema (http://ftp.ebi.ac.uk/pub/databases/emtest/empiar/schema) therefore defines the de facto standard for that community. It has evolved over time on the basis of feedback from depositors and workshops with community experts. Initially, EMPIAR accepted only raw datasets belonging to maps and tomograms in EMDB, which contains extensive metadata about the experiment (for example, specimen preparation, microscopy, image processing and validation). Therefore, the EMPIAR data model was designed to be lightweight and capture only information directly pertaining to the image sets (for example, number of images, image width and height). However, with the growing use of EMPIAR by the cryo-EM community, there have been increasing calls to expand its data model to incorporate more information—for example, about processing workflows and particle-picking files.
EMPIAR has slowly expanded its remit to include many more imaging modalities for which metadata are not captured in any other archive. Hence, it is consulting with the relevant communities to expand the EMPIAR data model to capture essential information about the experiment. This applies particularly to the volume EM community, which has begun to more routinely deposit its data to EMPIAR. Volume EM is a collective term for techniques that are used to acquire serial electron images through sample volumes (with a volume thickness typically in excess of 250 nm) of resin-embedded, heavy-metal-stained cells and tissues. This work is ongoing and will be further informed by the recommended metadata guidelines presented here.
Experience from other omics domains has taught us that to make data reusable, some standardization is necessary, and in particular, in reporting the metadata we need to give information describing the experiments and the samples—for instance, what instrument was used to generate the images and how the samples were prepared. To achieve this, ‘appropriate minimal’ or recommended information guidelines or standards have been adopted by various life-science communities. One of the first such initiatives was MIAME (Minimum Information About a Microarray Experiment), which was published16 in 2001 and has had a major impact on how functional genomics data are collected and reported via public repositories, and on the reusability of these data17,18. As the biological imaging field is maturing, the bioimaging community is now recognizing that it faces similar challenges. In fact, the metadata challenge in the bioimaging domain has been discussed in the European Light Microscopy Initiative (ELMI) community (https://elmi.embl.org/) since 2001, and an attempt to address it was undertaken by the OME Consortium19. In the domain of medical imaging, the challenge is partially addressed by the Digital Information and Communications in Medicine (DICOM) standard20. Nevertheless, it was reported recently that metadata on imaging methods are vastly under-reported in biomedical research21. One might argue that microscopy experiments are too complex and heterogeneous to be amenable to a standardized description. Twenty years ago, the same was often said about microarray data, but the best practices for collecting and representing metadata for various biomedical domains have evolved considerably since then. Arguably, the biological imaging field is ready for some initial data standardization and would benefit from it.
A workshop held in Hinxton, UK, in 2017 unanimously supported the establishment of a public bioimage archive to store data associated with peer-reviewed publications or systematic imaging projects22. The workshop recommended the adoption of initially flexible data standards, which could be gradually tightened as different imaging communities reach consensus. In July 2019 the BioImage Archive (https://www.ebi.ac.uk/bioimage-archive) was established at the European Bioinformatics Institute, part of the European Molecular Biology Laboratory (EMBL-EBI), and it provides the community with the means to share different types of imaging data. The BioImage Archive is a deposition database for all microscopy images associated with peer-reviewed scientific publications for which a more specialized resource is not available. It is part of a larger and developing bioimaging ‘ecosystem’ that also includes more specialized and structured image resources, such as EMPIAR for electron and X-ray microscopy images23, Cell-IDR9 for curated images of cells and Tissue-IDR for curated images of biological tissues. The BioImage Archive is built on a high-performance, high-volume data-storage system that can be used as a platform by other existing or emerging biological imaging resources.
A follow-up workshop to discuss minimum metadata recommendations in several biological imaging fields was held in Hinxton in October 2019. Representatives from the light, electron and X-ray microscopy communities exchanged their experiences and ideas and began the process of developing the Recommended Metadata for Biological Images (REMBI) guidelines, presented here, to address the needs of these communities. A common theme in community efforts such as this is that standardized dataset annotation and deposition become more complex and time-consuming with every extra metadata element. Thus, attempts to impose requirements that are not yet sufficiently widely adopted by a given community or supported by relevant data-annotation tools may be counterproductive. However, this challenge is not dissimilar to the one the microarray community faced at the beginning of this century, and the arguments presented for and against a greater or lower level of detail in the minimum standard are similar in the two domains. In addition, the amount of information required for reuse may differ depending on the imaging technology, the scientific application and the needs of different user groups (Fig. 1). We are thus convinced that there is a need to strike the right balance between minimizing the barriers to data submission and maximizing opportunities for data reuse.
Fig. 1 |. There are at least three different categories of users of archived images, each with different needs with respect to metadata.
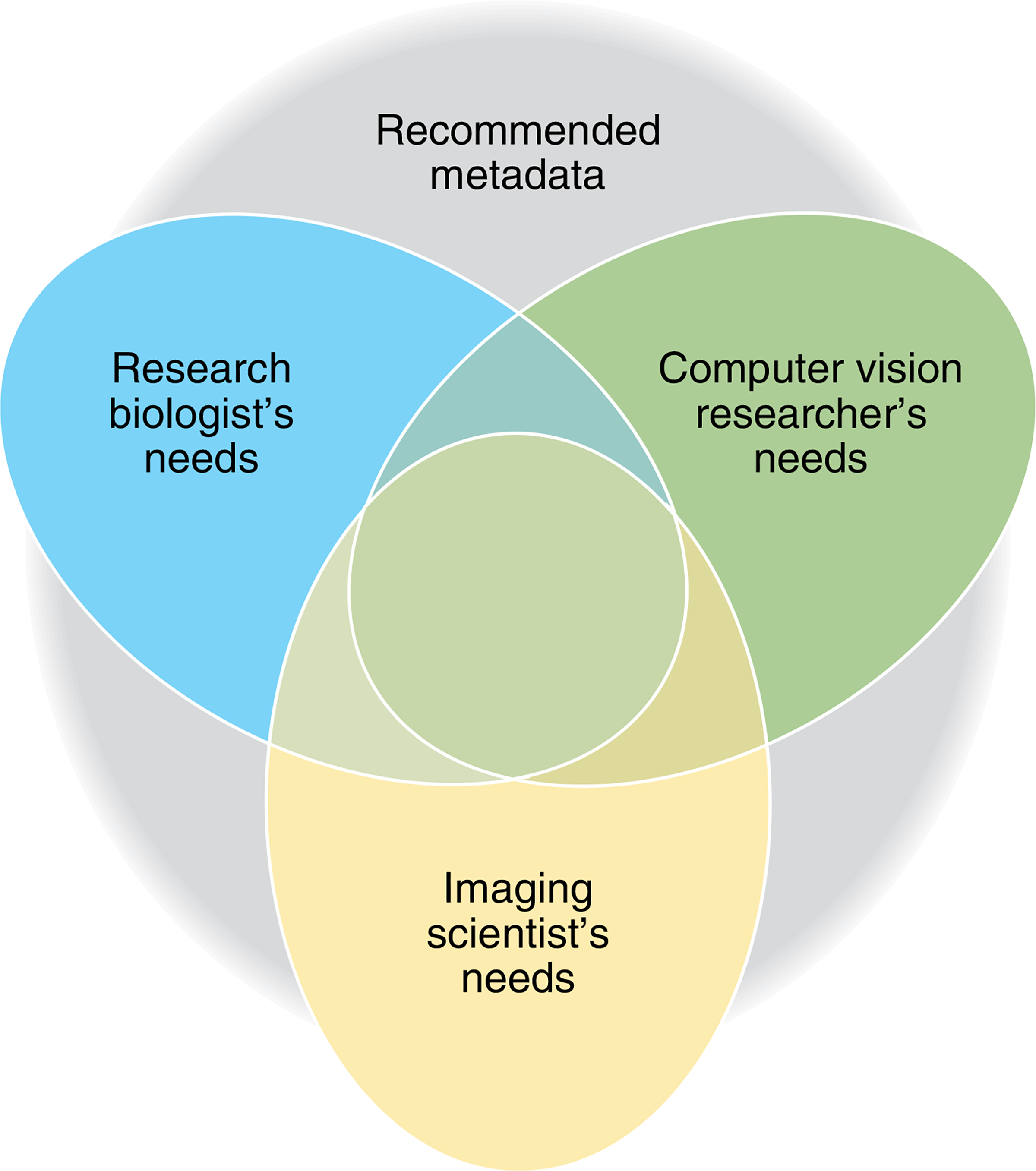
(1) Biologists and life scientists who are interested in repeating experiments, (re-)analyzing or comparing bioimage data and understanding results. For this, they need detailed information on the experimental context, such as the composition of biological samples, molecular entities, experimental interventions (for example, control vs. treatment) and how these relate to the image data. (2) Imaging scientists (microscopists and technology developers) who are interested in developing new imaging technologies. For this, they need detailed information on the image-acquisition process, such as physical properties of the image-acquisition set-up, and may benefit from some high-level information on the biological problem at hand. (3) Computer-vision researchers who develop algorithms (not limited to biological applications). Depending on the objective, they may need any of the information listed above. For example, to train a machine-learning algorithm, they would need ‘ground truth’ information such as adequately labeled images with categories (for example, control vs. treatments/phenotypes) or object outlines (segmentations).
Guidelines must take into account that microscopy technology development is highly dynamic, that there are many existing file formats (with new formats appearing regularly), and that datasets are becoming larger and more complex. Recognizing the enormous heterogeneity of biological imaging methods and the wide range of scales (from subnanometer to centimeter scale), the workshop established three working groups to address metadata recommendations for different subdomains: (1) the Electron Cryo-Microscopy and Cryo-Tomography working group; (2) the Volume EM and Correlative Imaging working group; and (3) the Light Microscopy working group, which covered cell-, tissue- and organism-level imaging. While these types of imaging each require specific types of metadata, they are all applied to study biological systems, and therefore commonalities are to be expected. The working groups converged on a common high-level structure of the recommended metadata guidelines (Fig. 2 and Supplementary Information).
Fig. 2 |. Different categories of metadata that are covered by REMBI.
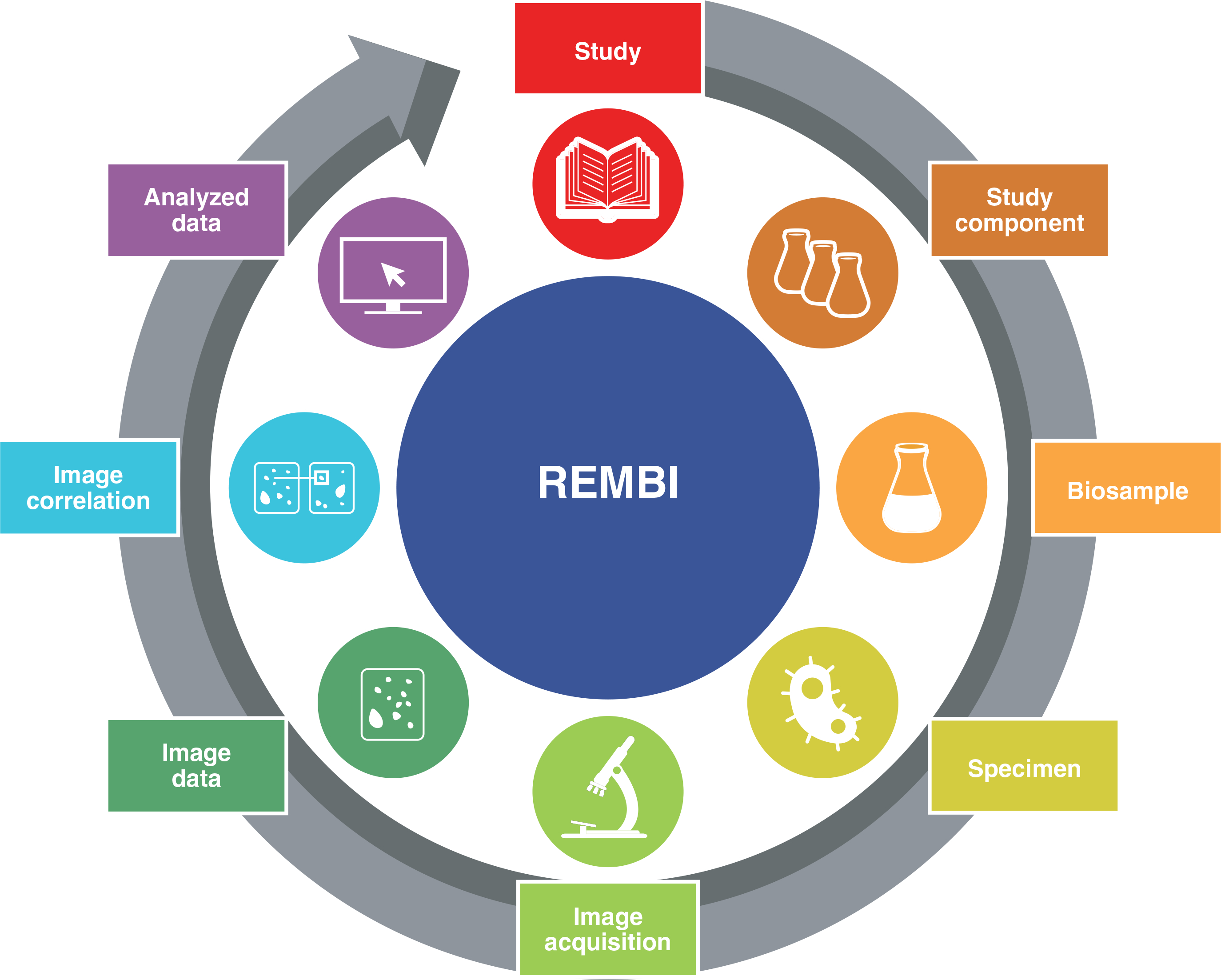
The “study” module describes the top-level metadata elements, in alignment with existing generic standards such as Dublin Core, DataCite Metadata, and schema.org. For example, in a correlative study comprising serial block-face scanning electron microscopy (SBF-SEM) and confocal images, one of the study components would contain all information on the EM image stack, the other study component would correspond to the confocal stack, and a transformation description would allow an overlay of the two types of image. Data that retain spatial fidelity to underlying images (for example, label maps, volume renderings) are described in the “image data” module, whereas “analyzed data” (for example, volumetric analyses, image segment features, counts) contains image-derived measurements, typically presented in tabular form. For more details, see the Supplementary Information.
The purpose of the proposed guidelines is to provide a framework for discussing different aspects of useful sharing of imaging data with the goal of reaching community-wide consensus on the level of detail that is optimal. The workshop participants agreed that it is important to distinguish recommended metadata requirements from particular data models: the former concern the semantic requirements of what annotation is needed to understand and reuse image data whereas the latter concern the syntactic representation of these metadata elements by computer software. There is also a third layer, specifying the implementation of a data model in a deposition system for a particular archive, along with the user interface of such a system.
In the field of cryo-EM, a tradition of detailed data annotation and deposition to a public repository is well established (Box 1). Standardizing metadata for light microscopy is challenging, as it covers a wide range of imaging modalities spanning several temporal and spatial scales, including single-molecule localization microscopy, wide-field or confocal microscopy, optical projection tomography, and light-sheet microscopy. The plethora of experimental set-ups (for example, high-content screening, light-sheet microscopy and digital pathology), file formats and compression methods, and the increasing complexity of datasets, are all complicating factors. Acknowledging that this subdomain produces datasets to address an extremely wide range of research questions, the working group concluded that it is currently difficult to expand the recommended metadata required for archival deposition beyond the basic information needed to open a dataset and access the pixel data such that visualization or reanalysis is possible. While such an approach does not immediately ensure full experimental reproducibility or provide a biological understanding of the sample, imaging conditions or other contextual information, it can serve as a starting point. The standard will of course evolve and be subject to refinement by the community as standardization progresses in the field. We hope that this publication will accelerate this process by facilitating discussions in the community, eventually producing a consensus view on metadata that allows experimental reproducibility and is fully consistent with the FAIR principles.
As agreement on recommended metadata is emerging, data-deposition tools that facilitate collection of these metadata (including submission tools for the BioImage Archive) will be developed, testing these standards in practice. For instance, the SSBD repository currently uses its own metadata template (comprising 11 required input fields), but the templates will be revised as an accepted standard emerges. Intelligent software strategies, such as autofilling common fields and automatic ‘data harvesting’ of information from log files, should be used to lower the barriers for data upload and to increase the quality of the captured information. Development and adoption of metrics to assess the completeness and correctness of uploads may encourage better deposition practices, resulting in wider use of and greater trust in the shared data in the community. Implementing recommended criteria in a way that encourages submission of additional structured metadata in the archive submission systems will facilitate dataset annotation beyond the required minimum, as better documented datasets will benefit from enhanced reusability and gain broader visibility. On the basis of the experience gained and the community feedback regarding the practicalities of data submissions and reuse, the standards will need to be kept up to date and to evolve with the science, technology and practices of bioimaging. However, the ultimate test of this effort will be the extent to which biological imaging data deposited in relevant archives will be reused (Fig. 3). The lessons from microarray data show that the earliest mode of data reuse may be related to testing new data-analysis tools, rather than providing biological insights, which happened later18.
Fig. 3 |. Imaging data are already being reused.
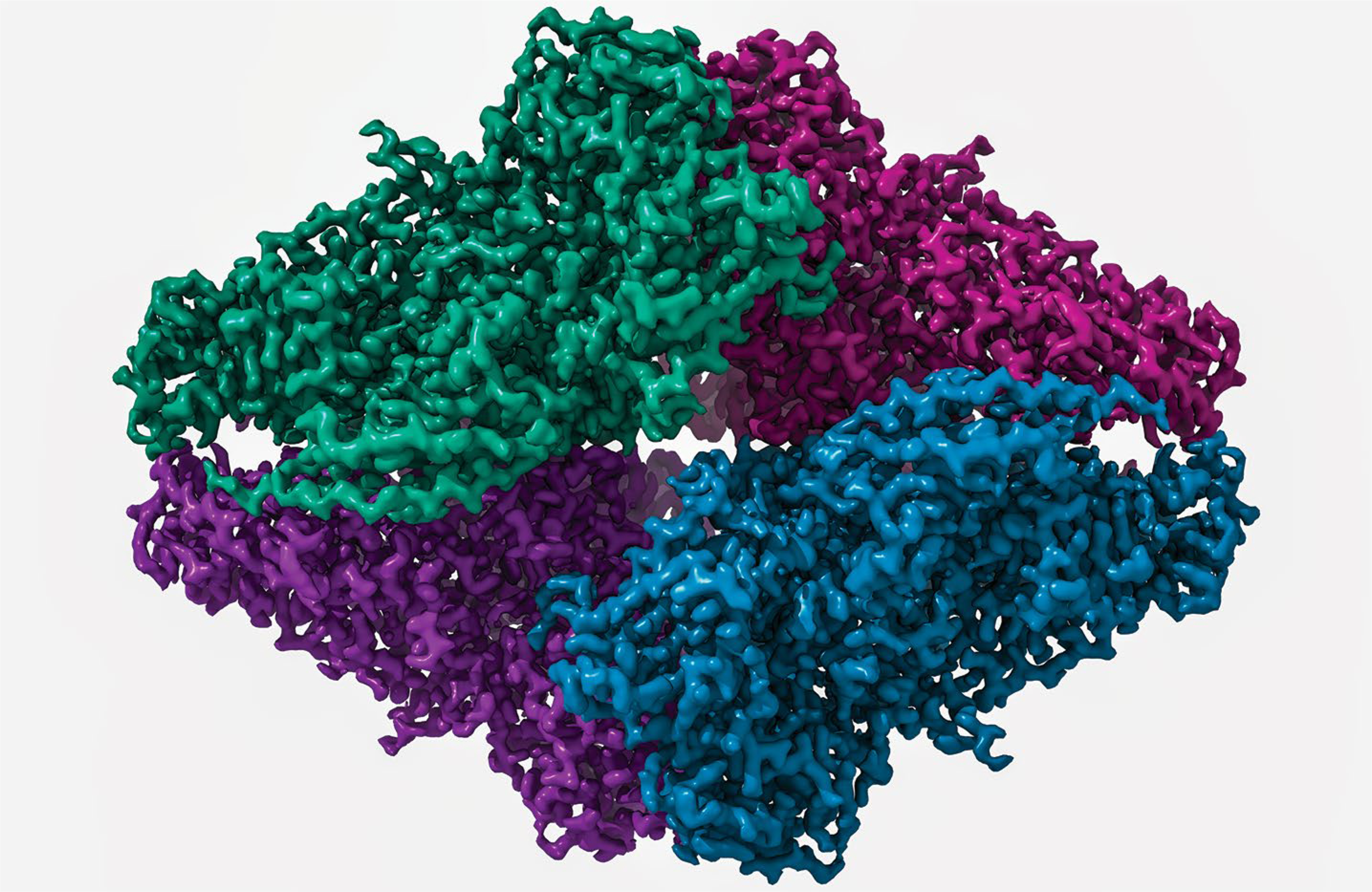
An example of a widely reused dataset is EMPIAR entry EMPIAR-10061 (https://empiar.org/10061), which contains the raw cryo-EM data (12.4 TB in size) underpinning what was a breakthrough structure and at the time the highest resolution cryo-EM structure available, the 2.2 Å resolution structure of β-galactosidase24. Several groups have reprocessed the data to even higher resolution and published and deposited the resulting EM maps. The dataset has been used by several developers of cryo-EM processing software to improve and test their algorithms, and it was used in the development of two deep-learning methods for automated particle picking, and to demonstrate cloud-based data processing. Details and literature references can be found at https://empiar.org/reuse.
The recommended imaging metadata standard described here will be adopted by the BioImage Archive, EMPIAR, Cell-IDR and Tissue-IDR. We hope that other existing and future archives will also adopt REMBI and engage with us to help shape the future development of the standard, in the spirit of the worldwide drive toward FAIR data sharing. To facilitate this, we encourage interested parties to contact us at rembi@ebi.ac.uk. We encourage scientific journals to support the deposition of bioimaging data in such FAIR resources, and funders to make data deposition a condition of grant funding. We also hope that instrument manufacturers and software developers, as well as large facilities and centers, will increasingly support recording of the recommended metadata automatically (in agreed formats), thereby minimizing the burden on the data submitters and minimizing data entry errors. Finally, we call on all scientists who use imaging methods in their published work to consider depositing their data and the associated rich metadata in the appropriate archives.
The current version of REMBI, including examples from the fields covered by the three working groups, is available as Supplementary Information, as well as from http://bit.ly/rembi_v1.
Supplementary Material
Acknowledgements
The workshop was hosted and funded by EMBL-EBI. We are grateful to J. Christiaens, R. Sherry and C. Karikides for logistical and administrative support. We thank the workshop participants C. Lore, O. Selchow, S. Tille and K. Wadel for their valuable contributions to the discussion. Figures 1 and 2 were created by S. Phillips and Fig. 3 by O. Salih. Finally, we would like to thank A. Reed for help with the preparation of the manuscript. Travel was funded by the individual participants.
Work on IDR by J.R.S., F.W., A.B. and U.S. is supported by the Wellcome Trust (212962/Z/18/Z) and BBSRC (BB/R015384/1). W.C. was supported by NIH R01GM079429. L.C. was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001999), the UK Medical Research Council (FC001999) and the Wellcome Trust (FC001999). M.C.D. would like to acknowledge the Wellcome Trust (212980/Z/18/Z) for funding. J.E. was supported by a grant from the European Commission H2020-ES-RI-INFRAEOSC “EOSC-Life” (Grant Agreement 824087). D.G. was supported by NIH 8U01DA047733–05 and NSF 1917206. Work on EMPIAR by A.P., A.I., C. Catavitello and G.J.K. is supported by UKRI-MRC (MR/P019544/1), the Wellcome Trust (221371/Z/20/Z) and EMBL-EBI. G.G.M. is supported by grant PTDCBII-BTI323752017-FCT and is part of the national Portuguese infrastructure PPBI, supported by PPBI-POCI-01–0145-FEDER-022122 from Fundação para a Ciência e Tecnologia / FEDER. T.M. and H.P. were supported in part by NIH Common Fund Award 5UM1HG006370–10. K.N. was supported by federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. C.S.-D.-C. was supported by the Chan Zuckerberg Initiative (Imaging Scientist award no. 2019–198155) and by NIH grant U01CA200059. V.U., M.B., E.B. and J. McEntyre are supported by EMBL internal funding. C. Cawthorne was supported by the Fonds Wetenschappelijk Onderzoek (FWO 1001719N). P.P.-G. was supported by CROCOVAL (ANR-18-CE45–0015) and is part of the national infrastructure “France BioImaging” supported by the ANR PIA1 (ANR-10-INBS-04). S.O. was supported by the Core Research for Evolutionary Science and Technology (CREST) grant no. JPMJCR1511, Japan Science and Technology Agency (JST). M.P. is supported by BBSRC Bioimaging UK community network grant (BB/S018689/1). P.Z. is supported by the Wellcome Trust (206422/Z/17/Z) and BBSRC (BB/S003339/1).
Footnotes
Competing interests
J.R.S. is a founder of and holds equity in Glencoe Software Inc., which builds commercial image data solutions. E.G. is an employee and shareholder of Johnson & Johnson.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41592–021-01166–8.
Peer review information Nature Methods thanks Ben Giepmans, Maryann Martone and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
References
- 1.Schermelleh L et al. Nat. Cell Biol. 21, 72–84 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Fernandez-Leiro R & Scheres SH Nature 537, 339–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K, Pintilie GD, Li S, Schmid MF & Chiu W Cell Res. 30, 1136–1139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakane T et al. Nature 587, 152–156 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yip KM, Fischer N, Paknia E, Chari A & Stark H Nature 587, 157–161 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Huisken J, Swoger J, Del Bene F, Wittbrodt J & Stelzer EH Science 305, 1007–1009 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Duncan LH et al. J. Vis. Exp. 10.3791/59533 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkinson MD et al. Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams E et al. Nat. Methods 14, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tohsato Y, Ho KH, Kyoda K & Onami S Bioinformatics 32, 3471–3479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orloff DN, Iwasa JH, Martone ME, Ellisman MH & Kane CM Nucleic Acids Res. 41, D1241–D1250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer M et al. Preprint at bioRxiv 10.1101/2021.04.25.441198 (2021). [DOI] [Google Scholar]
- 13.Swedlow JR et al. Nat. Methods 10.1038/s41592-021-01113-7 (2021). [DOI] [Google Scholar]
- 14.Nelson G et al. Preprint at https://arxiv.org/abs/2101.09153 (2021). [Google Scholar]
- 15.Boehm U et al. Nat. Methods 10.1038/s41592-021-01162-y (2021). [DOI] [Google Scholar]
- 16.Brazma A et al. Nat. Genet. 29, 365–371 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Ioannidis JP et al. Nat. Genet. 41, 149–155 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Rung J & Brazma A Nat. Rev. Genet. 14, 89–99 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Linkert M et al. J. Cell Biol. 189, 777–782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mildenberger P, Eichelberg M & Martin E Eur. Radiol. 12, 920–927 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Marques G, Pengo T & Sanders MA eLife 9, e55133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellenberg J et al. Nat. Methods 15, 849–854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iudin A, Korir PK, Salavert-Torres J, Kleywegt GJ & Patwardhan A Nat. Methods 13, 387–388 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Bartesaghi A et al. Science 348, 1147–1151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawson CL et al. Nucleic Acids Res. 44, D396–D403 (2016). (D1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.wwPDB Consortium. Nucleic Acids Res. 47, D520–D528 (2019). (D1). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


