Abstract
Much of fluorescence-based microscopy involves detection of if an object is present or absent (i.e., binary detection). The imaging depth of three-dimensionally resolved imaging, such as multiphoton imaging, is fundamentally limited by out-of-focus background fluorescence, which when compared to the in-focus fluorescence makes detecting objects in the presence of noise difficult. Here, we use detection theory to present a statistical framework and metric to quantify the quality of an image when binary detection is of interest. Our treatment does not require acquired or reference images, and thus allows for a theoretical comparison of different imaging modalities and systems.
1. Introduction
Fluorescence based confocal and multiphoton microscopy are powerful tools for biological research and have provided valuable insights into many biological questions. In particular, the use of multiphoton imaging has allowed for deep access into biological tissues and is of utmost importance for biological applications where high spatial resolution imaging deep within intact tissue is required [1–3]. Deep multiphoton imaging has allowed for the visualization and detection of many types of structures [1,2]. Confocal and multiphoton imaging have also been used for dynamic imaging and cell tracking [4,5]. Nearly all confocal and multiphoton imaging experiments used to study biology first rely on the binary detection of an object. For example, in blood vessel imaging, one is tasked with identifying the blood vessels, in recoding neuronal activity one must first find the neurons, and in cell tracking one is tasked with detecting cells in each image frame.
In any such imaging experiment the microscopist must first decide which imaging approach is best. For example, a decision is made about whether to use one-photon (1P, e.g., confocal), two-photon (2P) or three-photon (3P) excitation. This decision is usually made based on the depth of where the imaging needs to take place, because different methods will provide different image quality, relative to each other, at different depths. All three-dimensionally resolved fluorescence microscopy techniques (confocal or multiphoton imaging) are fundamentally limited by out-of-focus background fluorescence, which reduces contrast, degrading quality and inhibiting detection [6–9]. A simple approach to compare the imaging performance in the past is to use the signal-to-background ratio (SBR), which is the ratio of in-focus fluorescence (i.e., signal, S) to out-of-focus fluorescence (i.e., background, B) [6–9]. In multiphoton imaging, the SBR decreases monotonically as a function of imaging depth, and eventually at a certain depth the SBR becomes too small for practical detection of objects [6,7].
One may then be tempted to consider that the SBR is an acceptable metric of image quality. Indeed, in the past a depth limit for 2P and 3P image quality has been arbitrarily defined as the depth at which SBR is reduced to unity [6,7]. However, SBR alone does not consider the noise statistics at the detector, which are described with Poisson statistics (i.e., shot noise) for a typical microscope with high gain photodetectors (e.g., photomultiplier tubes). To make this argument concrete, consider two cases both with SBR = 1: in case one and , and in case two and . In case one, a pixel containing no object is measured as counts (mean ± standard deviation) and a pixel containing an object is measured as counts. In case two these counts are and . When one is then tasked with deciding if a measurement contains an object or not, case two allows for better detection of the object when shot noise is considered (i.e., as S increases more faithful measurements of and B are obtained). Therefore, SBR alone cannot be used as a metric for image quality.
Several studies have considered how to evaluate images which can be grouped into reference or non-reference methods. Reference methods need a ground truth image, which is generally not available to the microscopist before setting up the microscope and imaging the sample [10,11]. Non-reference methods do not need a ground truth, however they vary widely from simple measures such as a signal-to-noise ratio (SNR), contrast-to-noise ratio, brightness, etc. [8,11–13], to heuristic combinations of parameters about the image such as resolution, blur, brightness etc. [14], to more complicated mathematical models [10,11,15–17], and ranking methods [10]. Generally, these methods suffer because they (1) need an image to be taken which hinders the microscopist to make predictions about which technique to use before setting up the microscope and imaging the sample, (2) do not relate to the statistics of binary detection, (3) were not developed with microscopy in mind, or (4) any combination of the three reasons listed before. Additionally, most of these techniques do not explicitly show the relationship between image quality and S and B. Two notable exceptions are an SNR used by Sandison et al. in evaluating confocal fluorescence imaging (defined as ) [8,9] and the originally introduced in [18] (and modified in [19] to include SBR) for evaluating Ca2+ spike detection in the presence of shot noise. Sandison’s SNR is a good candidate since it shows the relationship between S and B but is not grounded in binary detection theory. The modified is grounded in detection theory but applies to the problem of detection of Ca2+ spikes with the assumption that the fluorescence level detected without a spike is much greater than the maximum additional amount of fluorescence generated with a spike. Such an assumption is usually reasonable for in vivo detection of Ca2+ spikes with many indicators but cannot be generalized to confocal or multiphoton images for binary detection. Thus, there is a basic, but important, gap in the literature when it comes to quantitative assessment of image quality for binary detection in terms of S and B.
Here we present a statistical framework and metric to compare the quality of images in terms of S and B. We emphasize that this statistical framework and metric applies to binary detection. Our theory relies on making decisions about whether each pixel contains an object or not, which is similar to the case of binary optical telecommunications when one decides if each received bit is a 0 or 1 [20,21]. The utility of our theory is that it allows microscopists to make decisions about which technique is best to use without requiring a reference image or even taking an image. As practical examples, we show that our treatment can be used to compare the performance of 2P and 3P imaging as a function of the imaging depth, excitation wavelength and staining density. Our statistical metric also gives insight to why defining the depth limit of multiphoton microscopy as the depth where SBR = 1 is reasonable, even though it was chosen arbitrarily in the past.
2. Statistical theory of binary detection
We first present a statistical theory to quantify the relative image quality, for binary detection, in terms of S and B. We consider a model where we look to make a decision at every pixel (in isolation) as to whether or not that pixel contains an object. The merit of this pixel wise strategy will be discussed in section 4.
In our binary detection model, we assume that every pixel contains the same level of background fluorescence regardless of if an object is present or not and pixels which contain an object all give off the same level of signal fluorescence in addition to background. This is justified at least in deep 2P and 3P imaging where most of the background is generated away from the focus [6,7]. Additionally, we note that B can include dark counts from the detector. Under these assumptions, on average, any pixel without an object will register B photon counts at the detector, and counts if the pixel contains an object. We note that B and are the mean values of Poisson distributions assuming photon shot-noise limited performance of the imaging system.
We now consider how to use the measurement, y, at a particular pixel (i.e., y measured photon counts at the pixel) to determine if the pixel contains an object or not. Note that y will be distributed according to Poisson statistics, which is used to define two hypotheses: and which represent, respectively, the absence and presence of an object. Thus, given a specific measurement, y, the probability of obtaining y under is,
| (1) |
and of obtaining y under is,
| (2) |
In order to decide between the two hypotheses, one can consider a likelihood ratio of the form, , and decision rule [18,22,23],
| (3) |
where the 0 and 1 refer to selecting or respectively, and is a threshold parameter, which is set by minimizing an overall risk [22,23],
| (4) |
where is the probability of choosing when is true, is the incurred cost by choosing when is true, and is the prior probability of occurring. Note that and are commonly referred to as priors and that is the staining density in a fluorescence image, which is defined as the fraction of volume containing fluorophore. The staining density is approximately the inverse of the labeling inhomogeneity factor, , used in [6] and [7]. Provided that , is optimally chosen as [22,23],
| (5) |
Taking logarithms and doing some manipulation, Eq. (3) can be equivalently written as,
| (6) |
where,
| (7) |
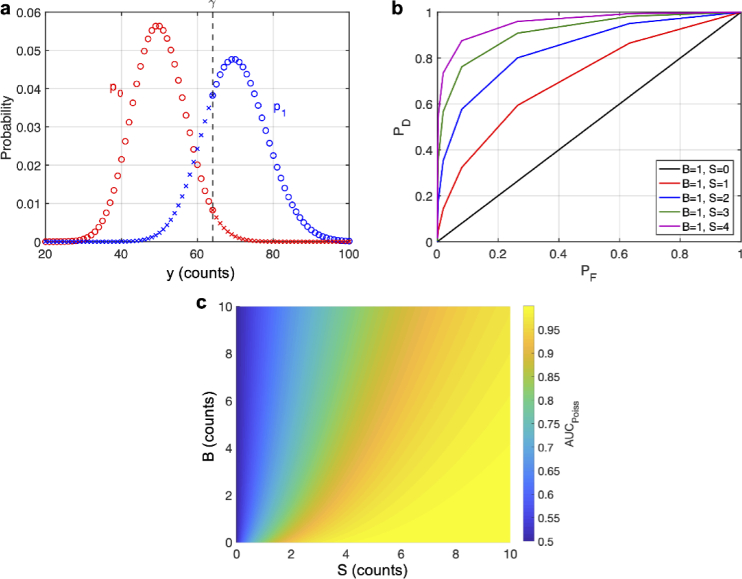
Note that can be written as an integer, as done here, since y is distributed according to a Poisson distribution and can never give a non-integer measurement. Thus can be understood as a threshold which discriminates between two Poisson distributions, and (Fig. 1(a)).
Fig. 1.
Statistical Theory. (a) Graph of the probability distributions (in red) and (in blue) with arbitrary means. A decision threshold, , is shown as a black dashed line at an arbitrary location. Blue circles show true positives and red circles show true negatives, while red x’s show false positives and blue x’s show false negatives. The points on the cutoff have both symbols to indicate that randomization may be used here. (b) ROC curves using randomization for various values of S and B. (c) Integrating under the ROC curve gives the AUC which is shown for various values of S and B. The infinite sum in Eq. (15) was computed by setting the upper limit to 100. The accuracy of this was checked by computing the sum with the upper limit set to 200, which showed little difference (see Fig. S1).
Although the priors may be well known (e.g., the mouse brain vasculature has a volume fraction of approximately 2% [2]), the choice of costs is somewhat arbitrary, and a small change can potentially give a very different true positive or detection probability, which is defined as [22–24],
| (8) |
and a very different false positive or false alarm probability, which is defined as [22–24],
| (9) |
Here is the cumulative probability distribution for . Equations (8) and (9) are graphically illustrated in Fig. 1(a). To circumvent the arbitrariness of the costs, we consider a receiver operating characteristic (ROC), which plots the best possible given a [18,23–25]. One can then integrate under the ROC curve to get the area under the curve (AUC), which gives a measure of detection fidelity, and more importantly eliminates the need for choosing an arbitrary decision threshold [18]. The larger the AUC the better the detection fidelity. The concept of ROC analysis and the AUC has been widely applied to diagnostic tests [26,27], where the ROC and AUC are typically estimated from experimental data. However, this has not been used before for assessing the quality (for binary detection) of a fluorescence image. Probabilistically speaking, AUC is the probability of an ideal observer correctly identifying an object when presented with two measurements, one which contains an object and the other which does not [18]. Additionally, the AUC can be thought of as the average when averaged over all possible . Mathematically speaking, one finds the best possible by specifying an acceptable false alarm rate, , and maximizing over all possible decision rules such that the actually achievable of the test never exceeds , i.e. [22,23],
| (10) |
Using the decision rule specified in Eq. (6), this amounts to solving [22–24],
| (11) |
which amounts to finding the smallest integer such that [24].
It is known that Eq. (10) is solved optimally when the specified ( ) and achievable ( ) false alarm probability are the same (provided it is possible to have ) [22,23], however, the decision rule specified in Eq. (6) does not allow for arbitrary and to be obtained since the resulting ROC curve is only defined at discrete points due to the Poisson distribution of the photon counts [23,24]. Therefore, one needs to construct continuous ROC curves. In other words, one needs to be able to achieve an arbitrary and . The optimum solution to Eq. (10) is known to be a randomized decision rule [22–25],
| (12) |
where q is the probability of choosing . In this case first is found, via Eq. (11), and then one will have that
| (13) |
and
| (14) |
where the value of q is selected so the achievable matches the specified [22–25]. Thus, it is now possible to choose an arbitrary and . By inspection of Eqs. (13) and (14) one sees that given the same value of , will be a linear function of and thus the randomized rule amounts to connecting the discrete and found without randomization with straight lines [24] (Fig. 1(b)). Thus, using the randomized rule, the AUC can be found by summing the areas of the trapezoids,
| (15) |
Note in Eq. (15), that , and that . Since the AUC is completely determined by S and B, i.e., , and the focus of this work is to assess the image quality (in the context of binary detection) in terms of S and B, we have plotted the AUC as a function of S and B in Fig. 1(c). Here we approximated the infinite sum by setting the upper limit to 100 and confirmed the accuracy by comparing this to the values computed with an upper limit of 200 (Fig. S1). As can be seen from Fig. 1(c), the contour for a constant AUC does not follow a straight line starting from the origin, which means the AUC is not the same for a constant SBR. Indeed, given a higher value of S and the same SBR, the AUC is also higher (Fig. 1(c)).
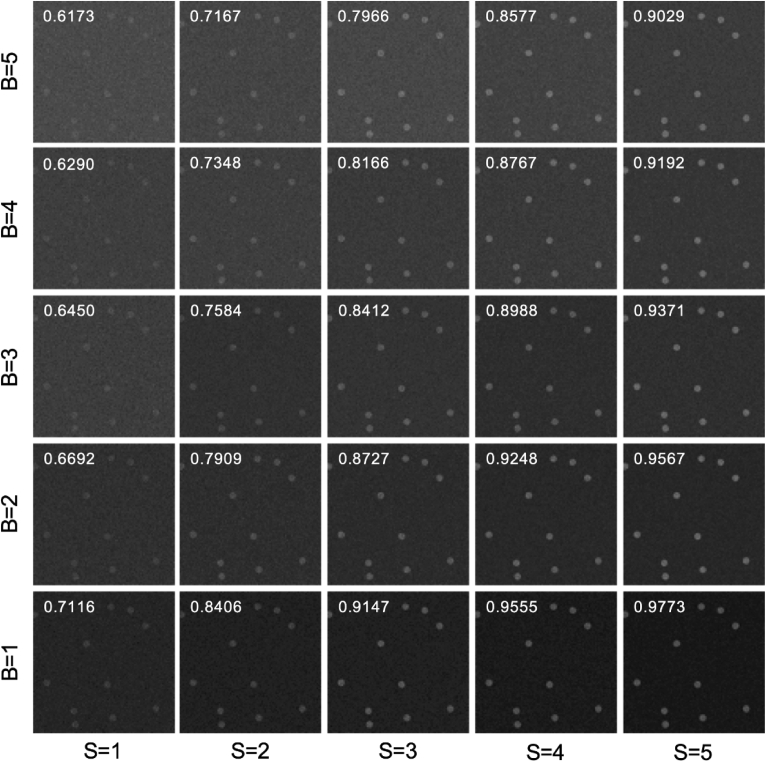
To confirm that assessing the image quality by the AUC matches reasonably with our intuition (i.e., our visual perception), we present simulated images of beads for various values of S and B as shown in Fig. 2. The images were generated by randomly choosing locations for circles of a desired size, setting pixels within these circles to contain the bead (i.e., they have an average value of ), and pixels outside to contain background (i.e., they have an average value of ). Then we randomly assigned the pixel values according to shot noise. From Fig. 2 we see that the AUC generally matches our visual perception about image quality, and images with the same SBR, but a higher S do appear to have a better quality (e.g., the quality increases along the lower left to upper right diagonal despite that all these images have SBR = 1). Again, this emphasizes the argument that SBR cannot be used as a metric of image quality.
Fig. 2.
Simulated Images. Simulated images of beads (diameter of 25 pixels on a 512 by 512 grid; staining density of 2%) are shown with Poisson noise added for various levels of S and B in units of counts/pixel. The white number on each image is the AUC computed using Eq. (15) with setting the upper limit in the infinite sum to 100. The window/level of each image is selected automatically using ImageJ.
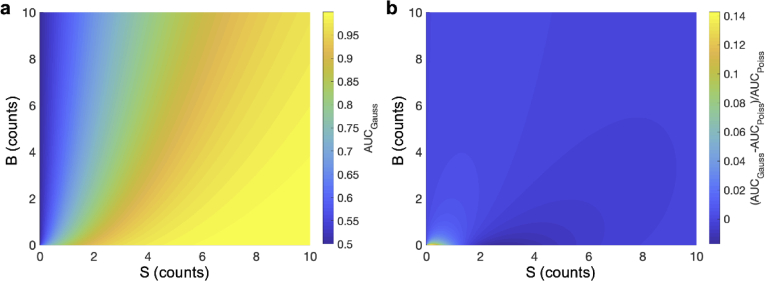
In order to better understand the dependence of AUC on S and B, we look for approximate forms to Eq. (15). To do this we approximate and as Gaussian distributions (i.e., normal distributions), which are good approximations when the counts are high, and solve for the ROC considering a single threshold, which allows the AUC to be approximated as (see Supplement 1 (1.2MB, pdf) for details),
| (16) |
where is the standard normal cumulative distribution function, and we have defined R as,
| (17) |
The AUC under the Gaussian distribution approximation ( ), and the relative error of this approximation, defined as , are shown in Fig. 3, where is the AUC calculated using Eq. (15) without the Gaussian approximation. Figure 3(b) shows that the error is minimal provided B is not small (e.g., the relative error of the approximation is approximately less than 3% when S or B are greater than 1). Note that this assumes that is non-zero, since when S is zero the AUC computed with either method is 0.5 (i.e., the smallest AUC possible), in which case practical detection is not possible. Thus, in all further discussions it will be assumed that S is non-zero.
Fig. 3.
Approximate AUC with Gaussian distributions. (a) The AUC using Gaussian distributions for the same values of S and B as used in Fig. 1(c). (b) The relative error of this approximation (defined as ) is shown for various values of S and B. Nearly all errors are small except when S and B are small, assuming is finite.
We note that since is monotonically increasing, completely characterizes the AUC in a way that captures the tradeoff between signal and background. When the photon counts are small, e.g., S and B << 1, the exact AUC (i.e., ) must be calculated using Eq. (15). In parallel to the parameter R under the Gaussian approximation we introduce the binary detection factor (BDF), defined as , as our figure of merit for image quality, which is obtained by first calculating . We note that the BDF approaches the value R when the photon counts are large (i.e., when the Gaussian approximation is valid).
3. Utility of the BDF
In this section we will demonstrate the utility of our figure of merit, BDF, by considering some practical examples. We also explore in this section how our metric can justify the arbitrary choice of SBR = 1 for defining the depth limit of multiphoton imaging.
3.1. Quantitative comparison of 2P and 3P imaging for deep tissue imaging
It is qualitatively understood that 3P imaging is advantageous for deep tissue imaging while 2P imaging is preferred in the shallow regions [2,19,28,29]. Some previous attempts to calculate the cross-over depth, defined as the depth where 3P imaging begins to outperform 2P imaging, were based on the signal strength alone, but such calculations establish the upper limit of the depth where 2P imaging is preferred, as taking the SBR into account will shorten this depth [19]. The metric developed here enables us to perform a rigorous and quantitative comparison of 2P and 3P imaging by properly considering the contributions of both S and SBR. To illustrate how this is done we perform this calculation with typical parameters for 2P and 3P imaging, with 2P and 3P cross-sections reflecting those of GCaMP6s [19].
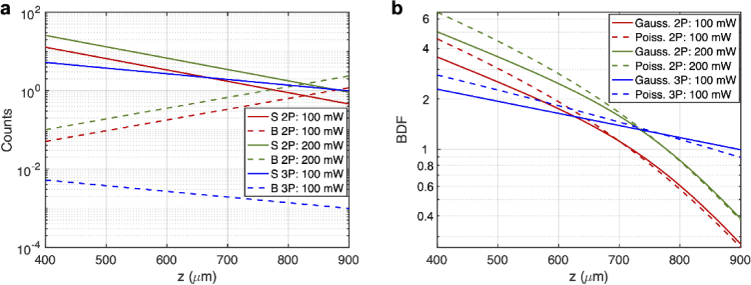
We first calculate the 2P and 3P signal as a function of imaging depth assuming a diffraction-limited focus. Because Fig. 1(c) shows that the AUC (and consequently the BDF) for the same SBR is larger with larger S, we consider the cases where S can be made maximum (likewise one can also note that for the same SBR). For biological imaging, we note that the maximum S will typically depend on the maximum average power allowed in the tissue. We thus constrain this problem to a maximum average power allowed at the surface, . In our calculations, we choose average powers at the brain surface which are at or less than the maximum power established by previous work for mouse brain imaging: mW for 1300 nm 3P imaging [19] and 200 mW for 920 nm 2P Imaging [30]. Another fundamental consideration is the maximum pulse energy at the focus, which is limited by fluorophore saturation (i.e., ground state depletion) and nonlinear damage. We choose to constrain our problem to a maximum allowable saturation level (i.e., the probability of excitation per molecule per pulse), , where is a constant related to the temporal pulse shape, is the n-photon cross-section, is the pulse full width at half maximum, and is the peak intensity at the focus [31]. In our calculations we choose (of which small changes have minimal effect on calculating the signal; see Supplement 1 (1.2MB, pdf) ). The resulting pulse energy at this excitation level is also commonly used for most 2P and 3P deep tissue imaging. Using the maximum average power at the surface and an allowable saturation level, along with knowledge of the concentration of fluorophore, C, the n-photon cross-section, , pulse full width at half maximum, , pulse shape, effective attenuation length, , excitation wavelength, , collection efficiency, , fluorophore quantum efficiency, , tissue refractive index, , numerical aperture, NA, and pixel dwell time, T, the n-photon signal, , can be completely determined (see Supplement 1 (1.2MB, pdf) for details). We chose a constant pulse energy at the focus throughout the imaging depth (i.e., the same level of saturation) to achieve the best imaging performance possible for both 2P and 3P imaging. We note that our calculation is for demonstration purposes, and we have selected the imaging parameters commonly used for 2P and 3P imaging of the mouse brain. as a function of depth is shown in Fig. 4(a).
Fig. 4.
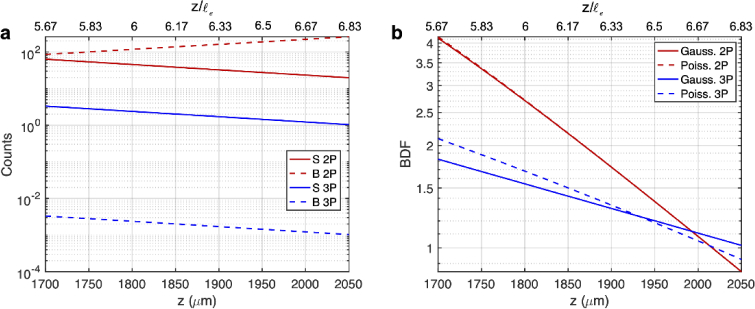
Comparison of 2P and 3P image quality when imaging the same fluorophores (e.g., GCaMP6). (a) Calculation of the S and B as a function of imaging depth for 2P and 3P imaging. S was calculated under an average power at the surface (100 mW for 3P imaging and 100 mW or 200 mW for 2P imaging) and saturation constraint (20%). Other parameters used are as follows: for 2P imaging: GM, , , nm, , , NA=1, fs, , , , ; for 3P imaging: , , , nm, , , NA=1, fs, , , , . B was subsequently calculated from S using the SBR data. SBR values above were set to to account for the finite dynamic range of a real detector. (b) S and B can then be used to calculate . Note that in the Gaussian case . The upper limit for the infinite sum in Eq. (15) was set to 2000. The accuracy of this was checked by computing the sum with the upper limit set to 3000, which showed little difference (see Fig. S2).
We then proceed to calculate the 2P and 3P background as a function of depth by using the calculated signal and the 2P and 3P SBR (i.e., ). For simplicity, we consider the bulk background (i.e., fluorescence generation in the light cone above the focus) but not the defocus background (i.e., the fluorescence generated by the side lobes of a distorted point-spread function, See Fig. 3(a) in [2]), which is important when imaging through a highly scattering layer such as the mouse corpus callosum or the intact mouse skull [32]. For the 2P bulk background we note that in [6] it states that the depth limit (defined as the depth where SBR = 1) increases approximately one for a sevenfold increase in . Assuming that , and together with the computed values in Fig. 6 of [6], the SBR can be found to be an exponential function of depth. Additionally, since the SBR could be impractically high (which is not practical due to the limited dynamic range of a real detector) at shallow depths, we forced any SBR value above to . For 3P imaging we note that the SBR is very large for the combinations of imaging depth and staining density considered in this paper [7,33], and so we set the 3P SBR also at a constant value of . The 2P and 3P SBR values used in our calculation are shown in Fig. S2. The resulting calculated values of are shown in Fig. 4(a). Here we chose to use corresponding to a staining density of 2% (which is typical for mouse brain vasculature [2]). We note that over the depth range shown in Fig. 4(a), B in 3P imaging decreases with depth. This is merely a consequence of the fact that the SBR is held constant at over this depth range (Fig. S2) while the 3P signal decreases with depth.
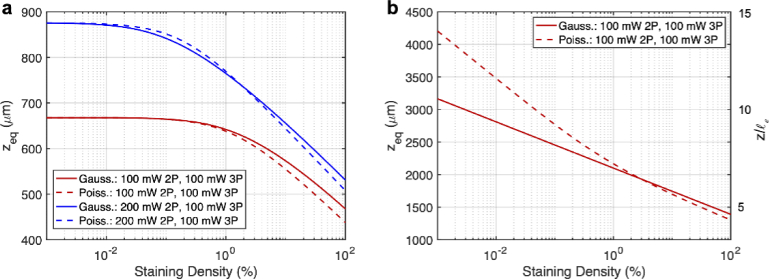
Fig. 6.
Calculations of . is calculated as a function of the staining density using the parameters in Fig. 4, and Fig. 5 for panels (a) and (b), respectively. Calculations with the Poisson BDF and the Gaussin approximation are both shown. For the Poisson case the upper limit for the infinite sum in Eq. (15) was set to 2000, and the accuracy of this was checked by computing the sum with the upper limit set to 3000, which showed little difference (see Fig. S4).
We used the calculated values of S and B to calculate the BDF using both the Poisson and Gaussian statistics. Since S and B are both determined by the imaging depth, we plot the BDF as a function of depth in Fig. 4(b). From our calculations we see that, if the average power at the surface is the same for 2P and 3P imaging (i.e., = 100 mW), 2P imaging is better at shallower depths due to the higher signal, and 3P imaging outperforms 2P imaging after about 620 µm, which corresponds to about at the 2P imaging wavelength.
We then repeated these calculations for 2P imaging using a higher average surface power of 200 mW (which may be of more practical interest), while keeping the 3P imaging average surface power at 100 mW (Fig. 4). We see that the new 2P BDF is about times larger, as predicted by theory, and the new cross-over depth at which 3P imaging outperforms 2P imaging is around 730 µm, which corresponds to about at the 2P imaging wavelength. These results are consistent with previous experimental investigations which show that 2P imaging generally works better at depths shorter than about 700 µm in the mouse brain [2,19,28,29,34]. Figure 4(b) also shows that the Gaussian approximation (i.e., using R instead of BDF) predicts similar cross-over depths (Fig. 4(b)).
The above calculation uses two different excitation wavelengths for 2P (920 nm) and 3P (1300 nm) imaging. This is practical for comparing 2P and 3P imaging of the same fluorophore (e.g., GFP, GCaMP6, fluorescein, etc.), and the longer excitation wavelength and stronger excitation confinement of 3P excitation both contribute to the advantages of 3P imaging when imaging deep. Our metric can also delineate the two advantages of 3P imaging (i.e., longer excitation wavelength and stronger excitation confinement) by comparing 2P and 3P imaging with the same excitation wavelength, e.g., imaging near IR dyes with 2P excitation and green dyes with 3P excitation at around 1300 nm. As an example, we repeat our calculations, except assume the same excitation wavelength of 1300 nm for both 2P and 3P imaging and the same of 300 µm (Fig. 5). Additionally, since we expect the depth at which 3P imaging outperforms 2P imaging to be larger we change the pixel dwell time to 50 µs, which is typical for deep tissue imaging.
Fig. 5.
Comparison of image quality for 2P imaging of near IR fluorophores (e.g., long wavelength Alexa dyes) and 3P imaging of green fluorophores (e.g., GFP). (a) Calculation of the S and B as a function of depth for 2P and 3P. S was calculated using an average power at the surface (100 mW) and saturation constraint (20%). Other parameters used are as follows: 2P imaging: GM, , , nm, , , NA=1, fs, , , , ; 3P imaging: , , , nm, , , NA=1, fs, , , , . B was subsequently calculated from S using SBR data. SBR values above were set to to account for the finite dynamic range of a real detector. (b) S and B can then be used to calculate . Note that in the Gaussian case The upper limit for the infinite sum in Eq. (15) was set to 2000. The accuracy of this was checked by computing the sum with the upper limit set to 3000, which showed little difference (see Fig. S3).
From Fig. 5(b), we see that 3P imaging outperforms 2P imaging after about 2000 µm, which corresponds to about . We note that the cross-over depth is deeper (even in unit of ) than the previous calculations in Fig. 3, simply because the 2P excitation also benefits from the long excitation wavelength and has less attenuation in this case. Figure 5 shows the intrinsic advantage of 3P microscopy for deep tissue imaging due to its stronger excitation confinement.
A recent paper [33] has also shown that it is possible to perform mixed 2P and 3P excitation of the same dye, providing the additional possibility for multicolor 3P imaging. Our theory can also be straightforwardly extended to more complicated cases such as mixed 2P and 3P excitation.
3.2. Deep imaging 3P advantage dependance on staining density
It is well known the excitation confinement advantage of 3P imaging (as compared to 2P imaging) is more pronounced with densely labeled samples, and thus 3P is generally better than 2P for deep imaging in densely labeled samples. We also note that our calculations in section 3.1 are a function of the staining density (since ), and thus the 3P advantage for deep imaging as a function of staining density can be quantified. In order to do so, we repeat our calculations form section 3.1, except we vary the staining density (e.g. ) and solve for the depth at which the BDF is the same for 2P and 3P imaging, which we denote as (Fig. 6).
From Fig. 6(a) one sees that below a certain value of staining density (about 0.1% for the 100 mW 2P and 100 mW 3P case, and about 0.01% for the 200 mW 2P case and 100 mW 3P case), does not change much. This can be understood because eventually the staining density becomes so small, that very little background is generated. In these cases, one finds that the confinement advantage of 3P excitation no longer matters and only the wavelength advantage plays out. This can be confirmed by noticing that the maximum value of in Fig. 6(a) matches the depth where the 2P and 3P signal cross in Fig. 5(a). In this sense one can consider a ‘densely labeled sample’ to mean a sample where it is necessary to consider the background generation when comparing the performance of 2P and 3P imaging in terms of .
Figure 6(b) does not exhibit the same behavior as Fig. 6(a), because here the 2P signal will always be higher than the 3P signal since the same excitation wavelength is used. In this case the notion of a ‘densely labeled sample’ is less clear because only the excitation confinement advantage of 3P comes into play, and thus the background always needs to be considered in determining despite how small it may actually be. Figure 6(b) also shows that when the staining density becomes very small that BDF and R are no longer predicting a similar . This can be understood since the recorded signal counts become low and so the Gaussian approximation is no longer valid. If the integration time was made longer, then the predicted by BDF would be closer to that predicted by R. We have varied the maximum SBR value between 200 and 5000 and compared the results with Fig. 6 (maximum SBR of 1000), we found negligible difference in .
3.3. Justification of SBR=1 for defining the depth limit of multiphoton imaging
Our statistical metric also gives insight to why defining the depth limit of multiphoton microscopy as the depth where SBR = 1 is reasonable, even though it was chosen arbitrarily in the past [6,7]. For simplicity we will use our metric under the Gaussian approximation (i.e., using ). We consider two extremes in SBR, where the SBR is much less and much greater than unity and consider how R (i.e., the image quality) behaves when all other parameters are held constant. In this case we will have that
| (18) |
for n-photon excitation, where K is a proportionality constant. Additionally, we assume that the SBR behaves as an exponential function with depth as shown by previous work [6,7]. That is,
| (19) |
where A and are constants. When , then from Eq. (17),
| (20) |
When , on the other hand,
| (21) |
In both cases, decreases exponentially as a functional of imaging depth. However, when , a much faster decrease occurs in R as compared to the case when . Figure 7 shows R against z on a semi-log plot. For the slope is proportional to , and when the slope is proportional to (Fig. 7). Such a slope change in the exponential function is identical to the situation of imaging two different layers of tissues where the deeper layer is much more scattering than the superficial layer.
Fig. 7.
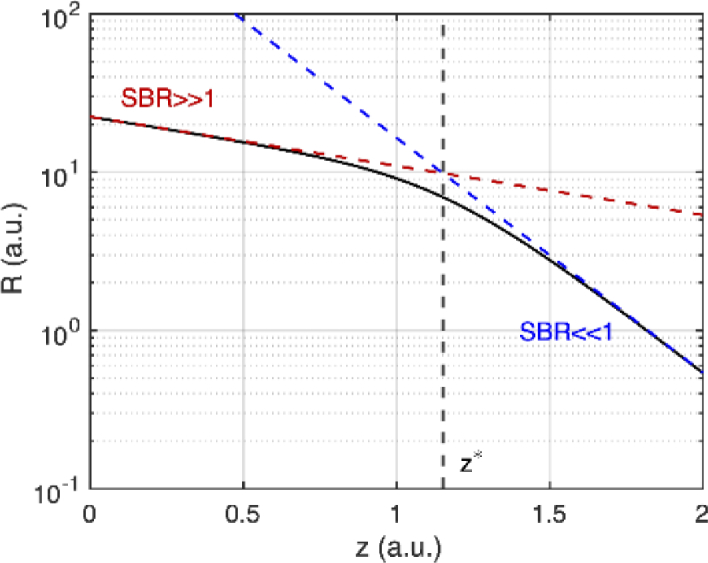
R as a function of imaging depth on a semi-log plot. Red and blue dashed lines show the approximations made when SBR>>1 and SBR<<1. The dashed black line shows the approximate depth where the slope changes.
To get an idea of the depth z where this slope change happens, we equate Eqs. (20) and (21). Denoting this depth as , one finds that , and that . This suggests that when the SBR falls below 2, R will decrease at a much faster rate (Fig. 7), as if encountering a much more scattering layer of tissue. Thus, one could consider SBR=2 as a reasonable criterion for the depth limit. We note that this is close to the arbitrarily selected SBR = 1 criterion.
4. Discussion
Much of the literature and our understanding of fluorescence imaging have delt with how imaging parameters, such as NA, pixel dwell time, cross-section, etc., affect the signal strength, resolution, background generation, etc. [1,2,6,31]. However, knowledge of how signal and background photon counts influence the quality of an image, in terms of the binary detection of objects, has remained largely unexplored from a statistical point of view. The metric , thus fills this gap and enables a quantitative treatment of this issue. Furthermore, as explained in sections 3.1 and 3.2, this metric can be used to evaluate different imaging modalities and instruments, and quantify the dependence of when different modalities outperform each other as a function of experimentally defined parameters such as imaging depth, excitation wavelength, and staining density. Additionally, our metric requires only knowledge of S and B, and so one does not need a reference image, or even to produce an image. This means that one can evaluate and design new imaging systems for biological applications, without the need to first build the instrument and then use it to produce an image.
In section 3.1 we only considered how 2P and 3P imaging compare, but one could also consider confocal imaging too. Given that the confocal point-spread-function is nearly the same as the 2P point-spread-function (neglecting the Stokes shift in the fluorescence emission), one would expect that, using the same excitation wavelength, confocal imaging should always be better than 2P imaging since the one-photon excited signal is stronger. Indeed, long wavelength one-photon confocal imaging can achieve greater than 1 mm imaging depth [35,36]. However, this does not consider the important differences in out-of-focus photobleaching and photodamage, and the significant loss of fluorescence signal in confocal detection. While our metric provides a quantitative assessment of the image quality (i.e., binary detection) in terms of S and B, other factors must also be considered for the best imaging approach, particularly when comparing between linear and nonlinear imaging modalities where the excitation confinement is fundamentally different.
Interestingly, our metric R is similar to the SNR used by Sandison et al. [8,9], although their SNR was not derived based on a binary detection problem. Indeed, R is nearly the same as Sandison’s SNR with the one difference being the factor of 2 in front of the B in our metric. This factor of 2 can be intuitively understood as follows: to measure S one would need two measurements, one with the object ( ) and one without ( ), and then subtract B from . Simple uncertainty propagation (assuming Poisson statistics) will show that the result would be (signal ± standard deviation), suggesting that our metric represents the effective SNR that could actually be measured.
Our result also validates the arbitrary choice of SBR = 1 for defining the depth limit when one considers the decrease in R versus imaging depth with all else held the same. From our analysis we found that a SBR below 2 marks the cutoff for when R will decrease more rapidly with depth. Although not exactly SBR = 1, this result is close to the arbitrarily set limit. Interestingly, a similar argument can be carried out for Ca2+ spike estimation with the modified to include SBR [19], in which case one finds that a SBR below unity marks the cutoff, matching the arbitrary choice. Thus, the SBR limit, as defined by the rate of decrease in detection ability as a function of depth, will depend on the specific detection problem at hand.
It is important to use a metric that is appropriate for the specific detection problem at hand. For example, for a problem involving Ca2+ spike detection, one should use as this metric was developed specifically for Ca2+ imaging. For binary detection of an object, on the other hand, one should use the BDF developed here. The BDF or (or other metrics) should not be applied universally, and specific metrics should be used for specific problems which they are designed for. We also note that not only will the depth limit determined by the SBR change with a different metric (such as or BDF), but also the cross-over depth will change.
The fact that one universal metric, in terms of S and B, may not adequately describe the detection quality of any detection problem can of course have implications in scenarios where more than one detection problem needs to be applied. For example, in Ca2+ imaging one first needs to perform a binary detection problem to detect the neurons, and then once the neurons are identified, needs to be able to detect spikes in time. This two-step process is most obvious in schemes such as adaptive excitation [37]. Because of the multiple metrics in this problem, an image which may be acceptable in one sub-problem may not be acceptable in the other, and the microscopist should take careful consideration of all sub-problems when evaluating systems in these applications.
We also note that in many applications, and published images, many frames of the same field of view are taken and averaged. Our theory can be extended to this case with some slight modification. In many cases most images are viewed digitally meaning that averaging a certain number of frames and summing the same number of frames look no different when displayed. However, the latter case means that each pixel will behave with poison statistics, and our theory can be used. We note there will now be an effective signal, and effective background, , where N is the number of frames added. Thus, we would have , and so R would still provide a comparison provided that N is held constant.
The careful reader will note that we considered a pixel wise metric, that is to say a decision is made at each pixel, in isolation from its neighbors, as to whether or not it contains an object. We note that this mirrors a similar approach to binary optical communication where a decision is made for every bit [20,21]. In imaging, the detection of object pixels from background pixels using this approach is the same as hard thresholding an image (potentially with randomization). While this is a somewhat simplistic view, it is sufficient for a relative comparison between images. A better image should allow for more pixels to be correctly classified, and since we assume that the only thing that changes between different imaging modalities is S and/or B, the comparison of how many pixels are correctly classified provides a relative comparison of image quality. We note that such a relative comparison is adequate for making decisions about which imaging instruments (e.g., 2P or 3P imaging), dyes, and parameters to use.
Although the AUC is the most common metric for comparing ROC curves, it gives a global metric [26,27]. When not all values of may be acceptable, one may want to consider instead a partial AUC where only the area under the ROC curve in the acceptable range of is considered [26,27]. We note that such analysis could be carried out in a similar manner as presented here.
One could consider a more complicated case for detecting a group of pixels containing an object. For example, for a bead of a pre-defined size one would have a circle of pixels that contain an object. We note that this detection problem could also be combined with an estimation problem (e.g., determining where the bead is centered in the image). We did not explore these problems here since it is not needed in our pixel-wise approach and could be considered in future work. We also note that this work did not consider multi-level detections, which could also be examined in the future.
Acknowledgments
The authors acknowledge fruitful discussions with Alejandro Simon, Yi Wang, and Farhan Rana.
Funding
National Science Foundation10.13039/100000001 (DBI-1707312).
Disclosures
The authors declare no conflicts of interest.
Data Availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
Supplemental document
See Supplement 1 (1.2MB, pdf) for supporting content.
References
- 1.Helmchen F., Denk W., “Deep tissue two-photon microscopy,” Nat. Methods 2(12), 932–940 (2005). 10.1038/nmeth818 [DOI] [PubMed] [Google Scholar]
- 2.Wang T., Xu C., “Three-photon neuronal imaging in deep mouse brain,” Optica 7(8), 947–960 (2020). 10.1364/OPTICA.395825 [DOI] [Google Scholar]
- 3.Stutzmann G. E., Parker I., “Dynamic multiphoton imaging: a live view from cells to systems,” Physiology 20(1), 15–21 (2005). 10.1152/physiol.00028.2004 [DOI] [PubMed] [Google Scholar]
- 4.Germain R. N., Miller M. J., Dustin M. L., Nussenzweig M. C., “Dynamic imaging of the immune system: progress, pitfalls and promise,” Nat Rev Immunol 6(7), 497–507 (2006). 10.1038/nri1884 [DOI] [PubMed] [Google Scholar]
- 5.Wang J., Hossain M., Thanabalasuriar A., Gunzer M., Meininger C., Kubes P., “Visualizing the function and fate of neutrophils in sterile injury and repair,” Science 358(6359), 111–116 (2017). 10.1126/science.aam9690 [DOI] [PubMed] [Google Scholar]
- 6.Theer P., Denk W., “On the fundamental imaging-depth limit in two-photon microscopy,” J. Opt. Soc. Am. A 23(12), 3139–3149 (2006). 10.1364/JOSAA.23.003139 [DOI] [PubMed] [Google Scholar]
- 7.Akbari N., Xu C., “Theoretical and experimental investigation of the depth limit of three-photon microscopy,” Proc. SPIE 11648, 116481A (2021). 10.1117/12.2582771 [DOI] [Google Scholar]
- 8.Sandison D. R., Webb W. W., “Background rejection and signal-to-noise optimization in confocal and alternative fluorescence microscopes,” Appl. Opt. 33(4), 603–615 (1994). 10.1364/AO.33.000603 [DOI] [PubMed] [Google Scholar]
- 9.Sandison D. R., Piston D. W., Webb W. W., “Background rejection and optimization of signal-to-noise in confocal microscopy,” in Three-Dimensional Confocal Microscopy: Volume Investigation of Biological Specimens , Stevens J. K., Mills L. R., Trogadis J. E., eds. (Academic Press Inc., 1994). [Google Scholar]
- 10.Koho S., Fazeli E., Eriksson J. E., Hänninen P. E., “Image quality ranking method for microscopy,” Sci. Rep. 6(1), 28962 (2016). 10.1038/srep28962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanciu S. G., Ávila F. J., Hristu R., Bueno J. M., “A study on image quality in polarization-resolved second harmonic generation microscopy,” Sci. Rep. 7(1), 15476 (2017). 10.1038/s41598-017-15257-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanniyappan U., Wang B., Yang C., Litorja M., Suresh N., Wang Q., Chen Y., Pfefer T. J., “Performance test methods for near-infrared fluorescence imaging Pejman Ghassemi,” Med. Phys. 47(8), 3389–3401 (2020). 10.1002/mp.14189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bray M.-A., Fraser A. N., Hasaka T. P., Carpenter A. E., “Workflow and metrics for image quality control in large-scale high-content screens:,” J Biomol Screen 17(2), 266–274 (2012). 10.1177/1087057111420292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanciu S. G., Stanciu G. A., Coltuc D., “Automated compensation of light attenuation in confocal microscopy by exact histogram specification,” Microsc. Res. Tech. 73(3), 165–175 (2010). 10.1002/jemt.20767 [DOI] [PubMed] [Google Scholar]
- 15.Dima A. A., Elliott J. T., Filliben J. J., Halter M., Peskin A., Bernal J., Kociolek M., Brady M. C., Tang H. C., Plant A. L., “Comparison of segmentation algorithms for fluorescence microscopy images of cells,” Cytometry 79(7), 545–559 (2011). 10.1002/cyto.a.21079 [DOI] [PubMed] [Google Scholar]
- 16.Nill N. B., Bouzas B., “Objective image quality measure derived from digital image power spectra,” Opt. Eng. 31(4), 813–825 (1992). 10.1117/12.56114 [DOI] [Google Scholar]
- 17.Ferzli R., Karam L. J., “A no-reference objective image sharpness metric based on the notion of just noticeable blur (JNB),” IEEE Trans. on Image Process. 18(4), 717–728 (2009). 10.1109/TIP.2008.2011760 [DOI] [PubMed] [Google Scholar]
- 18.Wilt B. A., Fitzgerald J. E., Schnitzer M. J., “Photon shot noise limits on optical detection of neuronal spikes and estimation of spike timing,” Biophysical Journal 104(1), 51–62 (2013). 10.1016/j.bpj.2012.07.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang T., Wu C., Ouzounov D. G., Gu W., Xia F., Kim M., Yang X., Warden M. R., Xu C., “Quantitative analysis of 1300-nm three-photon calcium imaging in the mouse brain,” eLife 9, e53205 (2020). 10.7554/eLife.53205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agrawal G. P., Fiber-Optic Communication Systems (John Wiley & Sons, 2002), Chap. 4. [Google Scholar]
- 21.Keiser G., Optical Fiber Communications (McGraw Hill, 2000), Chap. 7. [Google Scholar]
- 22.Poor H. V., An Introduction to Signal Detection and Estimation , Second (Springer-Verlag Berlin Heidelberg, 1994, Chap. 2. [Google Scholar]
- 23.Levy B. C., Principles of Signal Detection and Paramater Estimtion (Springer Science and Business Media LLC, 2008), Chap. 2. [Google Scholar]
- 24.Johnson S. E., “Target detection with randomized thresholds for lidar applications,” Appl. Opt. 51(18), 4139–4150 (2012). 10.1364/AO.51.004139 [DOI] [PubMed] [Google Scholar]
- 25.Scharf L. L., Statistical Signal Processing: Detcetion, Estimation, and Time Series Analysis (Addison-Wesley, 1991), Chap. 4. [Google Scholar]
- 26.Hajian-Tilaki K., “Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation,” Casp. J. Intern. Med. 4(2), 627–635 (2013). [PMC free article] [PubMed] [Google Scholar]
- 27.Lasko T. A., Bhagwat J. G., Zou K. H., Ohno-Machado L., “The use of receiver operating characteristic curves in biomedical informatics,” Journal of Biomedical Informatics 38(5), 404–415 (2005). 10.1016/j.jbi.2005.02.008 [DOI] [PubMed] [Google Scholar]
- 28.Takasaki K., Abbasi-Asl R., Waters J., “Superficial bound of the depth limit of two-photon imaging in mouse brain,” eNeuro 7(1), ENEURO.0255-19.2019 (2020). 10.1523/ENEURO.0255-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouzounov D. G., Wang T., Wang M., Feng D. D., Horton N. G., Cruz-Hernandez J. C., Cheng Y.-T., Reimer J., Tolias A. S., Nishimura N., Xu C., “In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain,” Nat. Methods 14(4), 388–390 (2017). 10.1038/nmeth.4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Podgorski K., Ranganathan G., “Brain heating induced by near-infrared lasers during multiphoton microscopy,” J. Neurophysiol. 116(3), 1012–1023 (2016). 10.1152/jn.00275.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu C., Webb W. W., “Multiphoton excitation of molecular fluorophores and nonlinear laser microscopy,” in Topics in Fluorescence Spectroscopy , Lakowicz J. R., ed. (Plenum Press, 2002). [Google Scholar]
- 32.Wang T., Ouzounov D. G., Wu C., Horton N. G., Zhang B., Wu C.-H., Zhang Y., Schnitzer M. J., Xu C., “Three-photon imaging of mouse brain structure and function through the intact skull,” Nat. Methods 15(10), 789–792 (2018). 10.1038/s41592-018-0115-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hontani Y., Xia F., Xu C., “Multicolor three-photon fluorescence imaging with single-wavelength excitation deep in mouse brain,” Sci. Adv. 7(12), eabf3531 (2021). 10.1126/sciadv.abf3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yildirim M., Sugihara H., So P. T. C., Sur M., “Functional imaging of visual cortical layers and subplate in awake mice with optimized three-photon microscopy,” Nat. Commun. 10(1), 177 (2019). 10.1038/s41467-018-08179-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia F., Wu C., Sinefeld D., Li B., Qin Y., Xu C., “In vivo label-free confocal imaging of the deep mouse brain with long-wavelength illumination,” Biomed. Opt. Express 9(12), 6545–6555 (2018). 10.1364/BOE.9.006545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia F., Gevers M., Fognini A., Mok A. T., Li B., Akabri N., Zadeh I. E., Qin-Dregely J., Xu C., “Short-wave infrared confocal fluorescence imaging of deep mouse brain with a superconducting nanowire single-photon detector,” ACS Photonics 8(9), 2800–2810 (2021). 10.1021/acsphotonics.1c01018 [DOI] [Google Scholar]
- 37.Li B., Wu C., Wang M., Charan K., Xu C., “An adaptive excitation source for high-speed multiphoton microscopy,” Nat. Methods 17(2), 163–166 (2020). 10.1038/s41592-019-0663-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.