Abstract
A microscope is an essential tool in biosciences and production quality laboratories for unveiling the secrets of microworlds. This paper describes the development of MicroHikari3D, an affordable DIY optical microscopy platform with automated sample positioning, autofocus and several illumination modalities to provide a high-quality flexible microscopy tool for labs with a short budget. This proposed optical microscope design aims to achieve high customization capabilities to allow whole 2D slide imaging and observation of 3D live specimens. The MicroHikari3D motion control system is based on the entry level 3D printer kit Tronxy X1 controlled from a server running in a Raspberry Pi 4. The server provides services to a client mobile app for video/image acquisition, processing, and a high level classification task by applying deep learning models.
1. Introduction
Microscopes are of particular importance in the advancement of biosciences and better quality production controls, as they allow us to observe the micro world at very small scales far beyond what is visible with the naked eyes, from hundreds or tens of μm—for human cells bacteria and unicellular microalgae—to nano scale—for viruses and electronics microcircuits.
Microscopy has evolved since its birth in the mid-seventeenth century with the handmade one lens microscopes created by Antonie van Leeuwenhoek [1]. The successive developments that have taken place since then have been aimed at increasing contrast in the observation of biological samples through appropriate optical and illumination systems (e.g., phase contrast microscopy, differential interference contrast (DIC), fluorescence microscopy and their variations, etc.), and improving the spatial/temporal resolution (e.g., scanning near-field optical microscopy (SNOM), stimulated emission depletion microscopy (STED), stochastic optical reconstruction microscopy (STORM), between others).
A state-of-the-art professional optical microscope has evolved into an automated image system whose acquisition and maintenance costs are beyond the reach of many research teams with tight budgets. A compromise solution for these work teams is to share or hire their infrastructure for temporary use. In this context, the availability of low-cost equipment with the required functionality it would be desirable.
The emergence of the Maker movement [2] corresponds to the impulse of the DIY philosophy for the technological development based on the availability and accessibility of open hardware and software. Since its inception, this movement has significantly impacted scientific and engineering education [3–5]. In addition, it has been able to provide low-cost equipment to diverse institutions with limited resources. This movement includes areas related to engineering such as electronics, robotics, 3D printing, and so on. These tools have somehow returned optical microscopy to its origins when scientists themselves developed their own observation instruments.
The single-board microcontroller kits such as Arduino (https://www.arduino.cc) and Raspberry Pi (https://www.raspberrypi.org/about/), smartphones, cheap digital image sensors, fused deposition modelling 3D printers, and the availability of open-source software facilitate accessibility to build flexible, low-cost microscopy platforms widely available and adapted to the needs of research teams. Two categories can be identified in the project in order to achieve a DIY low-cost digital optical microscopy system:
-
1.
Designs conceived with portability in mind. Some of them with the capability to adapt a smartphone as the image acquisition system with portability as their main advantage derived from their small size and ubiquity [6]. They are usually static optical microscopes with their observation capability limited to a single field of view (FOV) and/or quite limited movement. The most compact optical microscopes in this category employ ball lens optical systems based on Leeuwenhoek’s designs, as the Foldscope [7] and the PNNL smartphone microscope [8]. They are appropriate systems for educational purposes [9,10] and field diagnosis [11–15].
-
2.
Designs that seek to serve as a low-cost replacement for commercial systems. Therefore, they must be prepared to support the viewing of standardized slides with sample sizes that exceed a single FOV and require 2D motion (x,y) and focus (z) during observation. These developments are directed to offer flexibility, availability, and great image quality —at a cost fraction of their professional counterparts— for laboratories with limited budgets. These are specialized optical microscopes aimed at highly differentiated study areas [16–26].
The work presented in this manuscript falls into the latest mentioned category. All developments in this class must consider the design of four interconnected systems on which the final result’s performance is dependent.
-
(A)
2D positioning and focus. One of the key elements of the development of an automated microscopy system is its electromechanical positioning system ( ) and focus ( ) with the precision required for the observation of the samples. Along with these systems, the electronics that provide the logic and electrical power for the actuators must be designed (e.g., stepper motors) [18,21,24,25]. As shown in various studies, integrating the aforementioned aspects raises the system’s overall cost when compared to manual positioning [7,9–13,16,17,19,20]. The higher the needed precision of the movement, the higher the cost.
-
(B)
Optics. The optical system is responsible for the generation and, to a large extent, the quality of images. To achieve a quality comparable to that of commercial microscopes in low-cost designs, they must accept standardized objectives such as RMS thread and C-mounts.
-
(C)
Illumination. The illumination elements provide the necessary light to visualize the samples under observation. In this sense, it is very convenient to develop designs that allow using various illumination modalities to achieve contrast improvements in the final image (e.g., trans-/reflective illumination, bright/dark field, fluorescence, ultraviolet illumination, etc.). At present, low-cost systems are benefiting from the availability of high-brightness LED diodes with a wide emission spectrum that allow us to configure very flexible illumination systems.
-
(D)
Digital imaging. Digital microscopy arises as the possibility of replacing photographic films with image sensors connected to digital image processing and storage units. Among the main advantages of this process, cost reduction can be mentioned, video capture capacity, ease of storage and the possibility of applying digital image processing algorithms. The most recent CMOS sensors can reduce the size and cost of digital image capture systems, thus allowing access to an image quality like that obtained with photosensitive film photomicrography systems but at a much lower cost. Many digital microscopy systems eliminate the need for direct observation through an eyepiece by replacing it with a CMOS image sensor connected to a computer.
The integration of the mentioned systems in a functional platform requires a personalized design of couplings for its physical components. In this sense, the use of 3D printing by fused deposition modelling (FDM) is of major help in the rapid creation of system prototypes. Similarly, coordinated automation of an entire platform requires software that acts as a logical connection between all the components in the system. The use of open standards in the connection of both, physical and logical elements (e.g., RepRap, Python, etc.), is critical for the reproducibility of the designs and to the benefit of the microscopy community.
This paper presents the development of MicroHikari3D (μH3D in short), a DIY digital microscopy platform whose positioning system is based on a 3D printer kitted with an optical and a digital imaging system in place of the filament extrusion element. Section 2 describes the hardware and software systems that are used in the proposed solution. Section 3 presents the main results obtained with our first functional prototype discussing its relevance and scope. Finally, the Section 4 provides the work’s principal results and suggests areas for further research.
2. Materials and methods
This section describes in detail the design of the hardware and software that make up the MicroHikari3D microscopy platform.
2.1. Hardware
2.1.1. Electromechanical positioning system
An important limitation of static microscopy systems is the restricted field of view (FOV) of a region in the preparation. Thus, the observation of a complete slide requires motion control elements, either in the optics or of the microscope stage. The OpenFlexure Microscope [14] is a remarkably successful and popular development, albeit with limited stage movement capability ( mm) based on a well-studied bending mechanism [27] that achieves a repeatability of 1–4 μm.
Other proposals are designed with mechanical positioning systems having greater range of movement capable of performing a complete scanning of standardized slides. Among them, Incubot [25] is a significant design because, like in our proposal, it takes advantage of the positioning system offered by a low-cost entry 3D printer (∼100 USD), the Tronxy X1 3D. Moreover, this FDM printer is supplied unassembled to reduce its final retail price.
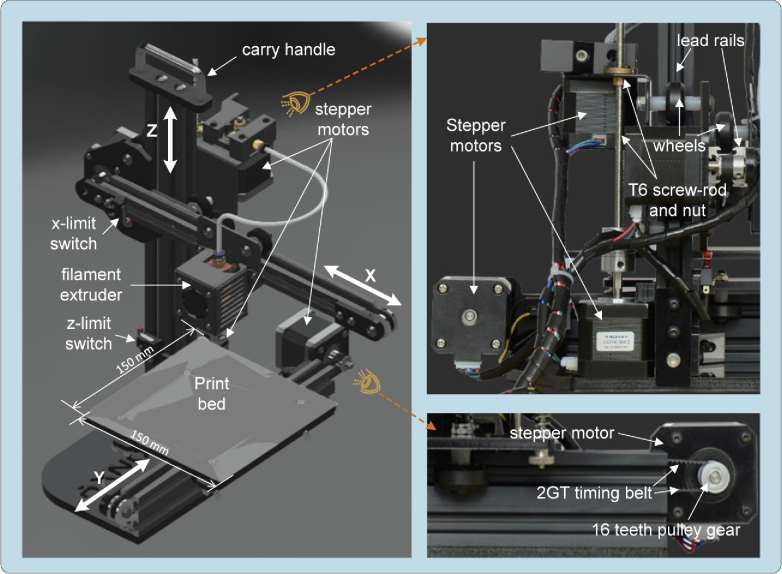
In a Tronxy X1 printer the displacement in each of the axes is achieved with a Nema 17 stepper motor (Mod. SL42STH40-1684A) used in the RepRap designs (https://reprap.org/), with a resolution per step of ). The motion transmission system on the axes is achieved with two sets of 2GT timing belts and 16 teeth pulleys (with resolution of 12 μm), whilst the axis uses a T6 screw and rod set (with resolution of 4 μm). The movement, in the three axes ( ), is guided by lead rails and high quality wheels for smooth and quiet motion with the purpose of achieving a 150 mm×150 mm×150 mm printing volume, yet sufficient for the displacements required to cover the observation of standardized size slides of mm and Petri dishes of Ø60–100 mm in spite of its reduced total volume of mm ( ). The Tronxy X1 printer kit provides a complete 3D positioning system with acceptable resolution for a competitive cost. Better resolution only can be addressed by alternative mechanical transmissions like ball bearing lineal ones but at much more cost and resigning to adopt the printer kit as mechanical support for the microscope.
The boundaries of the motion are checked with three mechanical switches (limit or stop switches) one for each axis. The limit switch in the axis can be easily adjusted to reduce the focus motion of the optical system and avoid collisions ( -stop) between the objective and the slide under observation. All the described mechanical elements are shown in Fig. 1.
Fig. 1.
Tronxy X1 3D printer mechanical elements.
An additional advantage derived from using a Tronxy X1 kit is the printers control electronics availability, which is based on the Melzi v.2.0 controller board, governed by an ATmega1284P microcontroller. To guarantee compatibility with the G-Code control language, the Repetier firmware was updated to Marlin (the most popular open-source firmware for controlling 3D printers, https://marlinfw.org/), since it allows us to deactivate the elements not used in the microscope (e.g., filament extrusion system, temperature sensor, fans extruder, and so on). The printer is powered by a 5 A/60 W (220 V∼100 V) power supply with an 12 V output.
The printer controller is connected to an external computer via an universal serial bus (USB). This computer plays a server role in charge of sending the motion orders to the printer controller and receiving the images captured by the microscope digital camera. A single board computer (SBC) based on Raspberry Pi4 (RPi4) with a 1.5 GHz BMC2711 quadcore microcontroller (50% faster than previous models) with 4 GB RAM memory was chosen to play the server role in the MicroHikari3D setup. This SBC provides physical connectivity through USB serial ports in order to connect to the illumination system and the motion controller. To facilitate remote communication, the RPi has Ethernet, WiFi (IEEE 802-11b/g/n/ac) and Bluetooth 5.0 connections. The connection to the compatible camera is made through a dedicated connection MIPI CSI-2 (ribbon cable). The RPi4 is governed by Raspberry OS, a 64 bit Debian-based Linux distribution.
2.1.2. Optical and imaging system
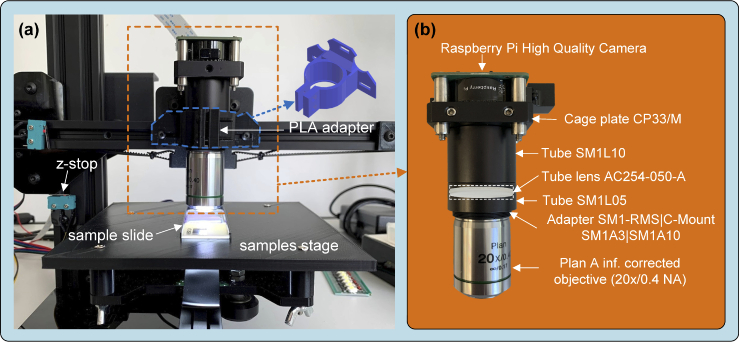
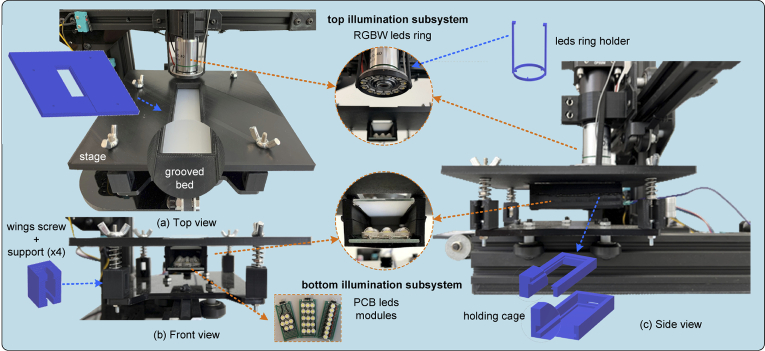
The OpenFlexure (OFM) [14,28] and Incubot [25] projects have been taken as a reference for the development of the μH3D optical system. Inverted optics were used in both works to achieve a more compact design in the first and a static superior position of the samples in the second. On the contrary, μH3D uses an upright optics arrangement located in the position originally occupied by the printer filament extruder. In our proposal, the observation surface coincides with the location of the print plate (see Fig. 2(a)) substituted by a PLA printed stage. The stage constitutes a flat surface with a convenient “grooved bed” (see the details of the current prototype in Fig. 3)) where the sample slide is laid for observation. The tilt of the stage can be levelled by adjusting the wing screws used to fix it to the carrying plate.
Fig. 2.
Optical system components in MicroHikari3D microscopy platform.
Fig. 3.
Illumination subsystems components.
A preliminary design based on OpenFlexure was made to use a 160 mm length fixed tube. This design stands out for its low cost. However, the use of 3D print-based adapters provides to the whole assembly with a certain fragility. The current design shown in Fig. 2(b) is based on commercial off-the-shelf (COTS) components that provide greater robustness to the optical system. In this design, a tube lens together with corrected to infinity objectives are used. This newer design is bulkier and increases its cost by ∼48 % with respect to the print-based version. Table 1 summarizes the elements of the μH3D optical system, along with a brief description of each one and the provider.
Table 1. Elements COTS for the optical and imaging system.
| Id. | Short description | Provider |
|---|---|---|
| RPi HQ Camera | Sony IMX477R 12.3 Mpix sensor; IR cut filter; RAW12/10/8, COMP8 output | Raspberry Pi |
| CP33/M | SM1-threaded 30 mm cage plate, 0.35" | Thorlabs Inc. |
| ER1-P4 | Cage assembly rod (x4), 1" long, Ø6 mm | Thorlabs Inc. |
| SM1L10 | SM1 lens tube, 1.0" thread depth | Thorlabs Inc. |
| SM1L05 | SM1 lens tube, 0.5" thread depth | Thorlabs Inc. |
| AC254-050-A | f=50 mm, Ø1" achromatic doublet, ARC: 400-700 nm | Thorlabs Inc. |
| SM1A3 | Adapter with external SM1 threads and internal RMS threads (for RMS objectives) | Thorlabs Inc. |
| SM1A10 (opt.) | Adapter with external SM1 threads and internal C-mount threads, 4.1 mm Spacer (for C-mount objectives) | Thorlabs Inc. |
| Objective | Plan-achromatic infinity corrected 20x/0.4 NA, RMS thread, parfocal distance 45 mm | various |
The attachment of the optical system is accomplished by means of an adapter printed in polylactide thermoplastic (PLA) fastened to the support originally occupied by the printer’s extruder as it shown in Fig. 2(a).
The image capture is carried out by the recent Raspberry Pi High Quality camera (RPi HQ) connected to the RPi4 by a MIPI CSI-2 interface. This CMOS camera is based on a 12.3 megapixel Sony IMX477 sensor with a 7.9 mm diagonal and a pixel size of . This camera is supplied with an adaptor to C-mount lenses. Table 2 shows the available modes in the RPi HQ camera and the correspondent resolutions and frames rates. All these modes can be selected from the Python programming language through the pycamera module. In our setup, the acquisition of a single image with 4056×3040 pixels last for 1.2 s.
Table 2. Frame size and rate for RPi HQ camera depending on selected mode (mode 0 for automatic selection).
| Mode | Size ( ) | Aspect Ratio | Frame rates (fps) | FOV | Binning |
|---|---|---|---|---|---|
| 1 | 2028×1080 | 169:90 | 0.1-50 | partial | 2×2 binned |
| 2 | 2028×1520 | 4:3 | 0.1-50 | full | 2×2 binned |
| 3 | 4056×3040 | 4:3 | 0.005-10 | full | none |
| 4 | 1332×990 | 74:55 | 50.1-120 | partial | 2×2 binned |
2.1.3. Illumination system
The two illumination subsystems implemented in μH3D are displayed in the Fig. 3).
Top subsystem for reflective illumination. It consists of a NeoPixel WS2812B 12 RGBW leds ring attached to the optical system. This ring has a four channel input, one for each colour (red, green, blue and white) coded with 8 bits per channel. The ring is held close to the objective by a PLA printed holder. This led ring allows us to illuminate the sample to a convenient wave length for specific microscopy modalities.
Bottom subsystem for transmissive illumination. This subsystem is inside a convenient holding cage back-attached (epoxied) to the microscope stage (see the side view in Fig. 3). The lower part of the holding cage houses one of the customized PCB LED modules ( cm) to obtain various illumination modalities (three of those LED modules can be seen in the image in Fig. 3). In the upper part of the holding cage, a filter could be installed (e.g., diffuser). The holding cage has been designed so that both, a LED module and a filter can be easily installed employing grooves (see details in Fig. 3) with the purpose of guiding their location in place.
The logic control of both illumination subsystems is carried out by an Arduino UNO board equipped with the ATmega328P microcontroller (https://store.arduino.cc/products/arduino-uno-rev3/). This controller generates the appropriate signals for the leds ring of the upper subsystem. In the case of the lower subsystem, a logical signal from the Arduino UNO activates the illumination by means of a relay driven by a logical signal through a transistor. To supply the necessary power consumed by the illumination system (∼5 W), the main power source of the Tronxy X1 printer is used, since the required consumption is compensated with the power released for the filament extruder that is replaced by the optical system. To facilitate the supply of the required power to both illumination subsystems (top/bottom), two DC/DC converter XL4015 modules (a quite popular DC/DC buck converter for electronic projects with microcontrollers) are used for the working voltage of each subsystem.
2.1.4. Performance analysis
To validate the usefulness of μH3D in the daily tasks of an optical microscope, it is essential to quantify its performance in terms of image distortion and optical resolution. In our proposal, the distortion in the acquired image derives from both the optical subsystem properties (i.e., lens, tube lenses, camera, etc.), and from the tilt between the plane of the stage and the optical axis. This tilt results in images that are partially out of focus since the focus plane is not perpendicular to the axis of the optical system.
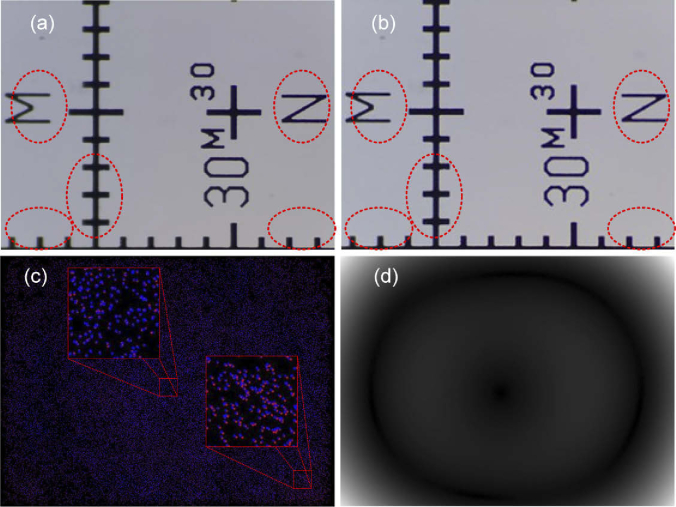
To correct the tilt mentioned above, a test image corresponding to a pattern such as the one shown in image 4(a) was used. Once the optimal focus position is reached, the test image reveals the areas blurred by tilt deviation (see red ovals in Fig. 4(a)). At this moment it is possible to manually adjust the levelling by tightening the stage’s wings screws until the image is focused on the entire FOV (see image in Fig. 4(b)). The small residual focus error obtained after applying this calibration method will be corrected by applying the focus stacking method described in Section 3.1 (see Fig. 8).
Fig. 4.
Distortion analysis: (a) Test image to correct tilt distortion pointed with red ovals, (b) Image after tilt correction, (c) Correspondent points obtained by "Distortion Correction" plugin, and (d) Distortion magnitude computed with "Distortion Correction" plugin.
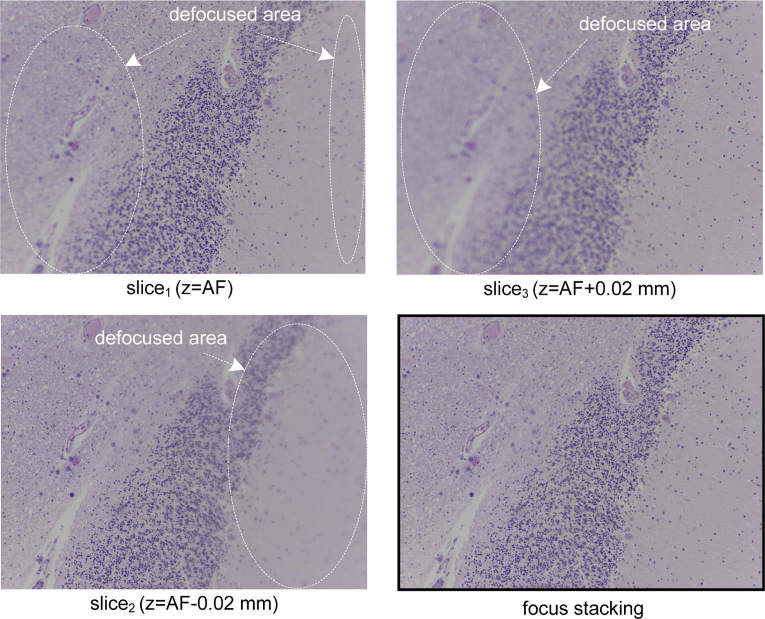
Fig. 8.
Focus stack with three images and result of the focus stacking algorithm.
Once the distortion produced by the tilt of the stage was corrected, the quantitative analysis of the distortion was carried out. To accomplish this study, the plugin "Distortion Correction" [29] for Fiji (ImageJ) was used. This plugin allows us to quantify the optical distortion from a tile of size 3×3 overlapped images. The overlapping between consecutive images should be 50% both horizontally and vertically. The distortion magnitude is obtained by estimating spatial displacements by computation based on SHIFT (i.e., scale-invariant feature transform) to detect corresponding features in the images. Moreover, the plugin provides the correction of such distortion, although in our case we are only interested in evaluating the distortion. The image in Fig. 4(c) shows the deviation between corresponding points projected on a common coordinate system. The degree of overlapping of the blue dots over the red ones provides a measure of distortion. Figure 4(d) shows the magnitude of distortion with brighter pixels for larger distortion in the test image corresponding with greater separation between points in Fig. 4(c).
The value obtained for the distortion indicates that our system does not present distortions with a marked direction that suggests the lack of tilt errors in the stage.
To analyse the optical resolution of the system, an image of the USAF test (i.e., 1951 USAF resolution test chart) acquired with μH3D has been used. This analysis has been carried out using the plugin ASI_MTF [30] for ImageJ. This plugin provides both the calculation of the MTF (Modulation Transfer Function) and the sigma value for the PSF (Point Spread Function) in the image regions selected for each of the selected USAF test groups.
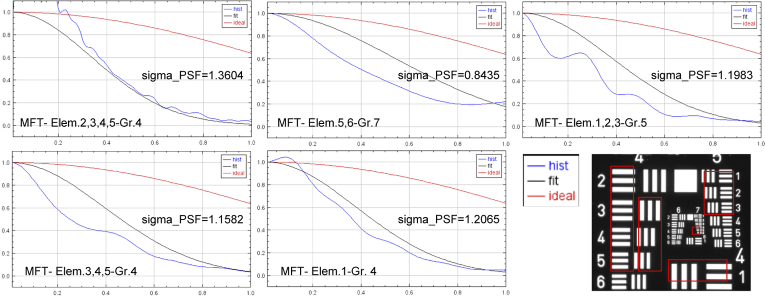
Figure 5 shows the MTF graphs obtained for the selected elements (marked in red) in the USAF test image acquired for the resolution analysis. The graphs in Fig. 5 show the MTF values obtained with respect to the spatial frequency components. All these graphs also display a legend with the sigma values for the PSF computed for each selected group.
Fig. 5.
MTF graphs and sigma PSF values for each selected group (marked in red) in USAF test image (bottom-right).
It can be concluded that with μH3D it is possible to observe in sufficient detail up to element 6 of group 7 in the USAF test image, which corresponds to an optical resolution of 228 lp/mm equivalent to 2.9 μm/line in our system. This value is consistent with the empirical observation in images of sample specimens acquired with μH3D (see details in Fig. 7(a)).
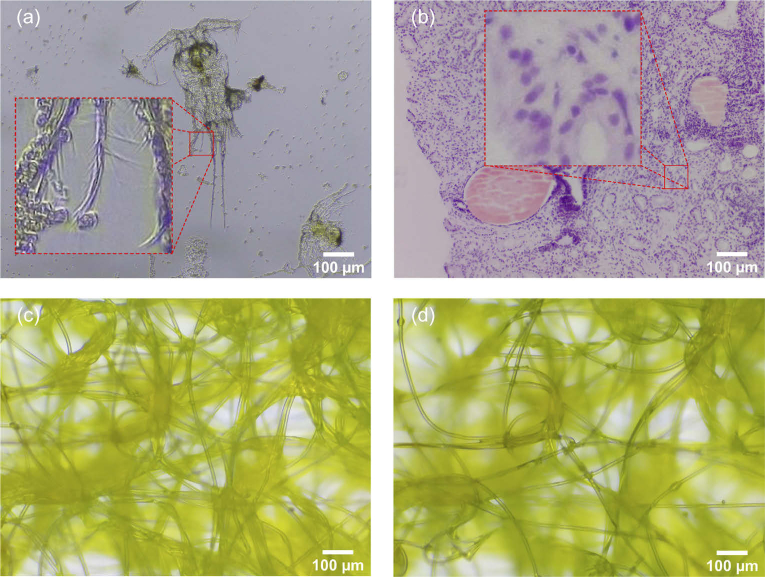
Fig. 7.
RGB raw images captured by μH3D: (a) copepod with bight field transmission illumination and a square section of side 75 μm zoomed in; (b) stained tissue biopsy with trans- and reflective illumination and a square section of side 75 μm; (c) synthetic fibre with transmission bright field illumination; and (d) same fibre with trans- and reflective illumination. These images where captured at size pixels with an achromatic objective 20x, NA 0.4, ∞/0.17.
2.2. Software
To provide the platform with modularity, a client-server architecture has been used (see Fig. 6) in which the resources and services provided by the platform are accessible through a representational state transfer application programming interface (API REST) [31,32]. This interface uses GET, POST, PUT and DELETE requests to create, update, and delete resources. The status between server and client is carried out by exchanging files in HTML, XLM and JSON formats. Design using API REST defines a set of restrictions about how services are published, and how requests are made to them. This API is independent of the programming language used for their implementation. Thus, the programming of any client can be decoupled of the operating system and the hardware platform it uses.
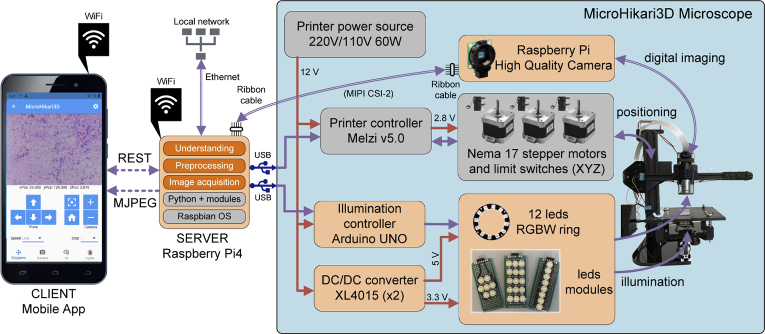
Fig. 6.
MicroHikari3D architecture and functional components.
In μH3D, the JSON format is used to exchange the state between the server and the client. JSON is an open standard independent of the programming language for both, the client and the server. The values that can be retrieved and modified on the platform are the following:
Camera parameters. Resolution of live and snapshot image capture modes, ISO, exposure compensation, saturation, sharpening filter, and zoom.
Motion parameters. Speed in mm/s, absolute displacement from reference position and relative motion from previous position.
Illumination parameters. Selective switching on and off for the top and bottom illumination subsystems, and selection of emission colour of the top illumination subsystem (RGBW).
In turn, the server provides an MJPEG video stream from the RPi HQ camera that allows remote viewing of the images captured by the microscope at a maximum resolution of pixels with a 4:3 full frame aspect ratio and 10 fps as maximum frame rate (see Table 2).
In the implementation of the architecture, the server is implemented in the SBC RPi4. This choice is determined by its affordable cost and connection compatibility with the CMOS RPi HQ camera. The server is programmed using Python and a set of lightweight open-source modules that provide the functionality required on the server side. Among the most important utilized modules are:
Flask and Flask-RESTful. Flask is a minimalist framework for web application programming. Along with Flask-RESTful, it provides HTTP server functionality and REST API programming.
Picamera. Access interface to RPi HQ camera through which the acquisition of digital images from the microscope is possible.
OpenCV. Interface to computer vision library and image processing with a multitude of algorithms that constitute the state of the art in the field and applicable to digital microscopy images.
TensorFlow Lite. Lightweight framework aimed at developing machine learning algorithms (e.g., image classification) on resource-limited hardware.
The services provided by the MicroHikari3D server fall into one of the three categories into which digital microscopy tasks can be classified:
-
a)
Acquisition. Tasks aimed at acquiring digital images such as autofocus, focus stacking, whole slide scanning, etc.
-
b)
Preprocessing. Image enhancement tasks such as noise reduction (e.g., by Gaussian filters), background correction (e.g.; by background division), contrast enhancement (e.g., by histogram equalization), etc.
-
c)
Understanding. Tasks that interpret the information present in the image such as recognition, identification, classification, tracking, etc.
3. Results and discussion
After the full assembly of all the components, we get a functional microscopy platform for a fraction of the cost of a professional one. Table 3 resumes the cost of each constituent system reaching a total cost of approximately 540 USD (∼500 €). It should be noticed that this cost is strongly linked to the quality of the chosen optics, hence a premium objective can easily reach the same cost of the entire system displayed —the considered cost for the 20x objective in Table 3 was about 70 $ (in USD)—.
Table 3. Approximate costs of MicroHikari3D platform.
| Short description | Cost (USD) |
|---|---|
| Tronxy X1 3D printer | 120 |
| Controllers and electronics | 96 |
| Illumination system | 24 |
| Optical system | 244 |
| CMOS Camera | 56 |
|
| |
| Total | 540 |
Figure 7 displays two samples captured with μH3D. These raw RGB images were captured by the 20× objective used in the preliminary design of the platform.
The following sections explain the results and algorithms implemented in the server side corresponding to the acquisition step (autofocus, focus stacking and whole slide scanning) and the understanding step of the image (classification by deep learning models).
3.1. Autofocus and focus stacking
In an automated digital microscopy platform, the autofocus (AF) is aimed at obtaining a focused image. There are numerous studies that analyse and compare autofocus algorithms applied to digital microscopy [33–37]. The algorithm implemented in the μH3D server is based on the Laplacian variance of the image [33]. This has been chosen for the compromise it offers between the quality of the result and its simplicity. It is based on the intuitive idea that focused images have fewer levels of grey variability (i.e., sharper edges) and therefore more high frequency components than unfocused images.
The Laplacian operator is calculated by convoluting the kernel over the image (with being the width and height in pixels of the image ). Before the convolution, the original RGB image is converted into a grey scale image. The mathematical operation can be described as:
| (1) |
with being:
| (2) |
After applying the operator to a image, a new array is obtained. A value of the variance is computed for this array using the next equation:
| (3) |
with being the average of the Laplacian.
Based on the focus value (i.e., the Laplacian variance) it is necessary to define a strategy to reach the optimal focal position after a 2D stage motion. In μH3D a simple strategy in two phases is implemented: coarse and fine focus. In both phases a maximum for the Laplacian variance is sought in successive images acquired at different focus positions ( ). Coarse and fine focus differ in the step amplitude taken between two consecutive positions. The final image in focus is the one with the largest Laplacian variance on it. The fine focus takes about 4.54 s to be completed, while the complete autofocus process with the coarse and fine steps lasts for about 22.28 s. It should be noticed that these times are quite dependent on the time required to compute the Laplacian on the images.
Fine focus is especially important when a small blur is achieved between successively acquired images. That is the case in sequential scanning of a whole slide with little defocus between successive FOVs and when multiple focal planes are preset in the same FOV due to 3D structures in the sample, uneven stage, and the very limited depth of field (DOF) for the optical system. In those cases, it is desirable to apply a focus stacking or extended depth of field strategy (extDOF) to obtain an image with the relevant regions of interest in focus. Another very efficient autofocusing strategy proposed in the context of the Openflexure utilizes the MPEG stream on the basis that sharper images require more functions for encoding the stream and threfore it requires more storage space [14].
In the server of μH3D it is possible to activate an extDOF strategy based on focus stacking with three (±1 slices from AF), five (±2 AF) or seven (±3 AF) focus planes. Successive slices are acquired with the finest resolution from the reference position at best in focus plane (i.e., AF-0.02 mm. and AF+0.02 mm). Several works had been carried out to improve autofocus capabilities in microscopy platforms [33–36,38,39]. Among them, the algorithm implemented in μH3D for extDOF [40] —by multifocus fusion— is derived from the work of Forster et al. [39]. This algorithm is based on the complex discrete Daubechies wavelet transform (CDWT) value for each image in the focus stack and combining the coefficients of the CDWT at each scale appropriately. The result after the inverse discrete wavelet transform of the proper combination of coefficients produces an extended DOF in focus image. Figure 8 shows the result obtained by the extDOF algorithm for a three slice focus stack. The focal stacking of a three slice focus stack lasts for 33.5 s and it increases linearly with the number of slices in the focus stack (i.e., 50.5 s for 5 slices and 66.7 s for 7 slices).
3.2. Whole slide scanning
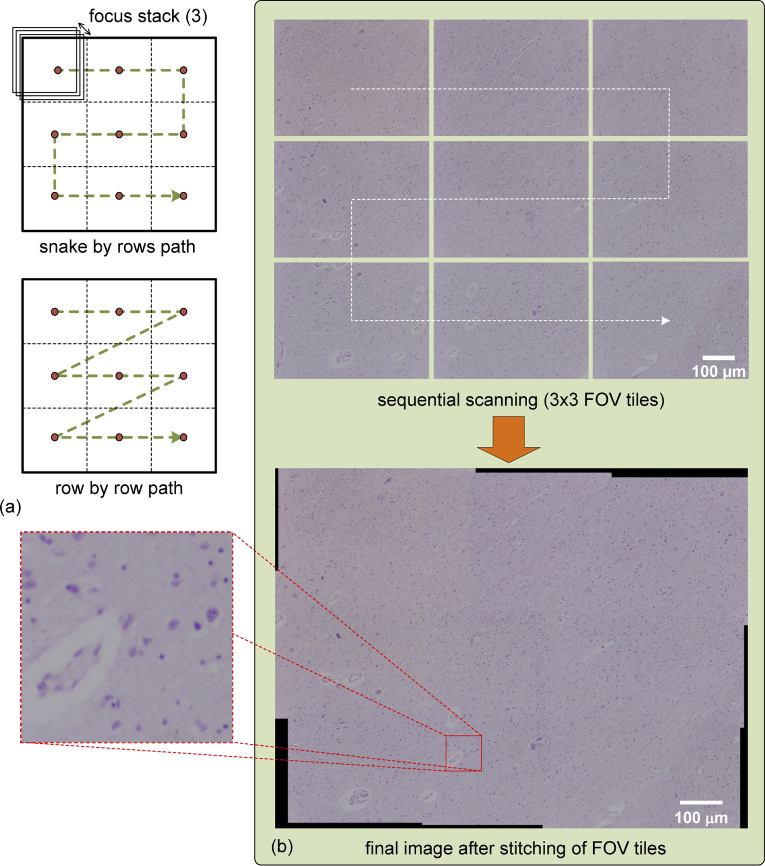
Very often in microscopy studies it is impossible to cover the whole slide of a sample in a single FOV. In these cases it is imperative to look over the complete slide taking several captures by sequential scanning to get a set of tiles that can be stitched together in order to get a single whole image or panorama [41,42]. The method implemented in μH3D is based on the Fourier Shift Theorem for computing all possible translations between pairs of images, achieving the best overlap in terms of cross-correlation [42,43]. There are two strategies to deal with the sequential scanning of the slide depending on the path followed to reach consecutive points for image acquisition (see two possible strategies in the Fig. 9(a)). In the first strategy, the whole slide is divided into an array of FOVs covered by a snake by rows path. In the second one, a row by row path is followed with the acquisition of images towards the same direction. The snake by rows path is the shortest path, thus requires less time to be completed (∼4% for a 3×3 FOV scanning in μH3D). Its main disadvantage arises from mechanical backlash, because motion in different directions during acquisition makes it difficult to compensate mechanical backlash in timing belts and pulley gears.
Fig. 9.
(a) Two strategies of whole slide scanning implemented in μH3D and (b) the result of tiles stitching to obtain a whole slide image with an small square section of side 75 μm zoomed in to appreciate the final image quality.
The size of the scanned surface depends on the objective magnification, hence with more magnification, less area is covered by the same array size of FOVs tiles. Once the array size has been set, a careful calculation should be carried out to obtain the motion of the stage to reach the desired point where each tile will be acquired, remembering that close tiles have to overlap at least 30% over each other. Moreover, the acquisition in each position could use focus stacking with the purpose of getting better quality on each tile at the cost of more processing time. Once all the tiles have been acquired, stitching of the tiles is carried out (see Fig. 9(b)) for the result with a tile set).
Table 4 displays the total processing time (acquisition + stitching) for eight experiments in which several parameters are changed: the size of the FOV tiles array (3×3 and 5×5), the working frequency of the processor (1.5 and 2.0 GHz) and the size of the final image after stitching.
Table 4. Total processing time for automatic scanning depending on FOV array size, processing frequency and image size ( with application of extended DOF by focus stacking).
| Id. | FOVs | Freq. (GHz) | Size (w ×h) | Time (s) |
|---|---|---|---|---|
| 1* | 3×3 | 1.5 | 9670×7137 | 843 |
| 2* | 3×3 | 2.0 | 9682×7136 | 721 |
| 3 | 5×5 | 1.5 | 6203×4646 | 795 |
| 4 | 5×5 | 2.0 | 6203×4646 | 584 |
| 5 | 3×3 | 1.5 | 9782×7134 | 704 |
| 6 | 3×3 | 2.0 | 9692×7135 | 582 |
| 7 | 3×3 | 1.5 | 3886×2885 | 233 |
| 8 | 3×3 | 2.0 | 3955×2885 | 192 |
The comparison between experiments in Table 4 allows us to conclude that focus stacking during tiles acquisition increases the computation time but not as much as when the size of FOVs array rises. In order to alleviate heavy computation loads in the μH3D server, a processing pipeline has been implemented. With this pipeline processing strategy, it is possible to compute the focus stacking in a FOV while the next one is being acquired. After the FOV array has been acquired, the stitching process starts. The stitching represents a high percentage over the total slide scanning time, for 3×3 FOV arrays we get 165.5 s, and this value rises up to 567.8 s for 5×5 arrays (with individual FOV sizes of 4045×3040 pixels and a 30% of overlapping).
3.3. Classification by deep learning models
After whole slide scanning, higher level tasks could be carried out with the purpose of understanding the information present in the image. One of the most time demanding tasks in pathology deals with screening for early cancer diagnosis, thus automatic classification of tissue images could significantly alleviate the work load of pathologists. Automatic image classification has been a subject of study for decades [44], being addressed by diverse machine learning algorithms. However, the more recent tendency is to apply Deep Neural Networks (i.e.; deep learning) because its promising results and the unnecessary handcrafted selection of images features.
TensorFlow (TF, https://www.tensorflow.org/lite) is the Google open source framework to develop and deploy deep learning models. TensorFlow Lite (TFLite) is the tool to deploy inference in limited computing devices (i.e.; mobile, embedded and IoT devices), based on models built with TF. Thus, any deep learning model developed with TF can be deployed in an RPi for inference with TFLite after converting the model to the proper tflite format. Four well known classification models had been trained for inference in μH3D using TFLite: 1. MobileNetV2 [45,46], 2. EfficientNet0 Lite [47], 3. ResNet50 [48], and 4. InceptionV3 [49].
These models had been trained on the AIDPATH dataset (http://aidpath.eu/) [50] employing images from breast and kidney biopsies. Table 5 shows the composition of the dataset with the images for each class, and the percentages devoted to training (80%), validation (10%) and testing (10%).
Table 5. Composition of AIDPATH dataset [50].
| Class | Training | Validation | Test |
|---|---|---|---|
| Malignant | 2676 | 334 | 334 |
| Benign | 2464 | 308 | 308 |
| Background | 2080 | 260 | 260 |
| Total | 7220 | 902 | 902 |
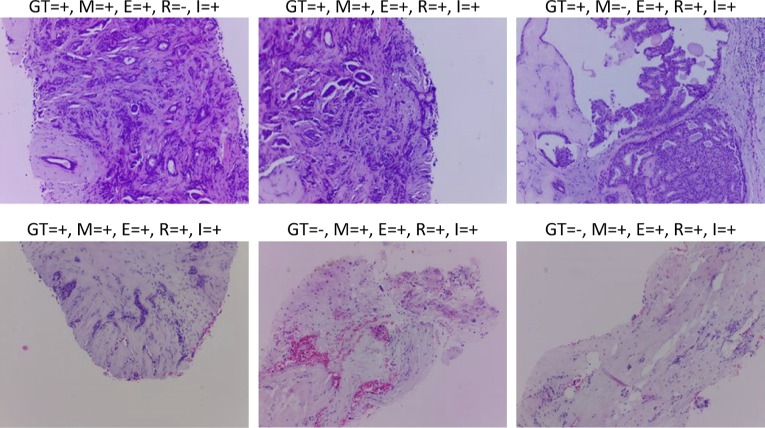
The training of the four models has lasted 25 epochs with a learning rate of 0.001 and a batch size of 32. The final model should be converted to TFLite format and deployed in the RPi being served to the μH3D client. The available models in the server could be consulted from the client by a GET request to recover a JSON file with the info about the deployed models. The inference is carried out on the RPi server after the client requires the classification of the current images by the selected model from the four available ones (i.e., Mobilenet V2, EfficienteNet0 Lite, ResNet50, and InceptionV3.). Figure 10 shows six samples from a classification test round of 36 real breast tissue samples with Hematoxylin-Eosin staining acquired with μH3D. The samples were previously labelled by a collaborator pathologist.
Fig. 10.
Breast tissue samples with Hematoxylin-Eosin staining acquired with μH3D and classified by the four inference models in the server upon client request. Over each sample a legend displays the correspondent values of ground truth (GT) and classification results (i.e., +/- for positive/negative to cancer) for each model: (M)obilenet V2, (E)fficienteNet0 Lite, (R)esNet50, and (I)nceptionV3.
Table 6 summarizes the classification performance obtained by each model after training and testing with AIDPATH dataset, accuracy on 36 samples taken with μH3D, and the inference time in the RPi. As shown in the table the accuracy is quite similar, although the fastest inference is carried out by the MobileNet V2 model. The inference results obtained in 36 samples acquired by μH3D show consistency with the values obtained in the training AIDPATH dataset. The inference process can even be accelerated by using tensor processing units (TPU) available for RPi through a USB Coral accelerator (https://coral.ai/products/accelerator).
Table 6. Accuracy of classification models during training on AIDPATH dataset and results of inference on a round of 36 real samples images acquired by μH3D.
| Model | Acc. training (%) | Acc. samples (%) | Inference time (ms) |
|---|---|---|---|
| MobileNet V2 | 91.9 | 80.56 | 128 |
| EfficientNet0 Lite | 92.1 | 94.44 | 159 |
| ResNet50 | 92.5 | 83.33 | 900 |
| InceptionV3 | 92.2 | 94.44 | 1090 |
The result obtained by the EfficientNet0 Lite is remarkable considering the fact that it achieves the highest accuracy on the training dataset, and on the classification round of real samples. Moreover, it obtains the second-lowest inference time of the available models.
3.4. Client mobile app
The μH3D’s client-server architecture for remote control of the microscope platform allows for a variety of client types. For that purpose, a first preliminary mobile Android client has been developed with Flutter (https://flutter.dev/), the Google UI toolkit to build multiplatform applications using the programming language Dart.
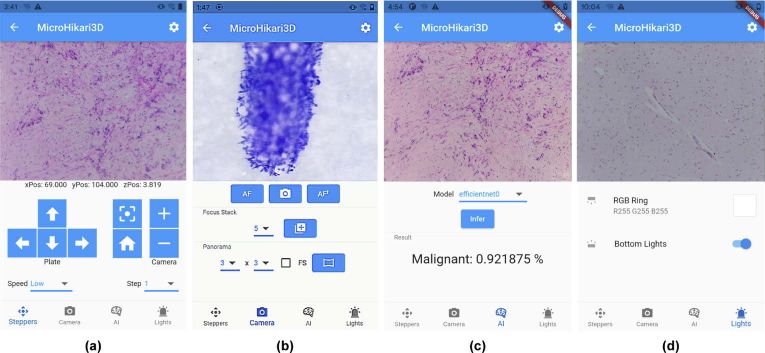
The mobile client screen is divided in two parts. The top middle screen shows the live preview image acquired by the microscope camera, and the bottom middle screen is occupied —upon request— by a control panel among four possible ones (see Fig. 11):
-
(a)
Motors. It allows us to control the stepper motors for the microscope stage 2D motion ( ) and the focus ( ), setting the speed and motion step. Moreover, this panel provides convenient functions for homing and recentering the stage.
-
(b)
Camera. It provides controls to several capture modes from the camera live preview: snapshot, focus stacking (with a setting for the number of planes), and whole slide scanning or panorama (by setting the scanning mode and FOV array size).
-
(c)
AI (Artificial Intelligence). It enables the automatic classification of the image shown (live preview) on the screen with the AI model selected among the four models available.
-
(d)
Illumination. It provides the controls to activate the illumination modules of the microscope and to set the specific RBG colour for the ring of leds from the top illumination subsystem by a convenient colour picker.
Fig. 11.
The live preview from the microscope and the four main panels in the mobile μH3D Android client: (a) Motors; (b) Camera; (c) AI; and (d) Illumination.
In addition to the control panels described previously, the client app has a setting section for the imaging system where it is possible to adjust e.g. resolution (for snapshot and video modes), ISO, sharpness, saturation, and other parameters.
After describing the results achieved with our microscopy platform, a comparison with similar platforms has been carried out and summarized in Table 7. This table shows that μH3D outperforms OpenFlexure [14] and Incubot 3D [25] in almost every reviewed characteristic. OpenFlexure stands out for its illumination modalities and overall cost, however its compactness restricts the application of complex algorithms such as whole slide scanning. Some other works as OpenWSI [22] and Octopi [23] offer truly relevant performance features, however they are significantly more expensive than our proposal. In the future we plan to extend the number of lighting modalities supported by the μH3D.
Table 7. Comparison of low cost automated optical microscopes.
| Description | OpenWSI [22] | OpenFlexure [14] | Incubot 3D [25] | H3D [ours] |
|---|---|---|---|---|
| range (mm) | 300x180 | 12x12 | 130x130 | 130x130 |
| range (mm) | 45 | 4 | 130 | 130 |
| Objectives 160 mm | × | ✓ | × | ✓ |
| Objectives | ✓ | ✓ | ✓ | ✓ |
| Objectives C-mount | × | × | × | ✓ |
| Illumination modalities | trans- | trans- or epi- | reflect- | trans- or/and reflect- |
| Camera | DFK 33UX183 | Pi Cam2|webcam | Pi Cam2 | RPi HQ Cam |
| Video streaming | ✓ | ✓ | × | ✓ |
| Focus stacking | ✓ | × | × | ✓ |
| Whole slide scanning | ✓ | × | × | ✓ |
| AI classifier | × | × | × | ✓ |
| Total cost (USD) | ∼2 500 | ∼230 | ∼1 250 | ∼540 |
4. Conclusion
As a conclusion after the results described in the previous sections, μH3D constitutes a novel proposal for an automated microscope DIY platform that takes advantage of the mechanical control support provided by an entry-level FDM printer kit —based on RepRap project— quite affordable, with motion resolution of 12 μm ( ) and 4 μm ( ). The optical and illumination systems are built with available commercial off-the-shelf elements providing flexibility to adopt several microscopy modalities. The assemblage of the whole systems is achieved by customized adapters and holders printed in PLA by an auxiliary 3D printer. The main control of μH3D is achieved by a server-client architecture implemented in the server side by a RPi4 whilst the client side is implemented by a mobile app with the live preview of the digital image acquired by the RPi HQ CMOS camera attached to the optical system.
The software deployed with μH3D provides valuable characteristics —unavailable on many more costly counterpart platforms— such as:
Autofocus (AF) and extended depth of field by focus stacking.
Whole slide scanning by automated sequential tile capturing and stitching.
Intelligent classification of images by inference with deep learning models.
As a result of MicroHikari3D’s success, efforts are being made to improve it and explore new possibilities, as follows.
New light microscopy modalities such as dark field and fluorescence. These could extend the application of the proposed microscopy system and it could be used in laboratories with scarcity of resources.
Development of a client desktop application. This software could provide support to more processing demanding algorithms and μH3D integration with pre-existent open source software tools (e.g., ImageJ).
Execution optimization to reduce the time for whole slide scanning. Being the more time-consuming task, its optimization would strongly impact in the workload of related processes.
Miscellaneous functionality. Several new features could be explored such as: assisted levelling system for the stage, bracketing for stack exposition fusion, real time tracking of live specimens, time-lapse acquisition, etc.
Acknowledgments
This work was supported in part by Junta de Comunidades de Castilla-La Mancha under project HIPERDEEP (Ref. SBPLY/19/180501/000273).
Funding
Junta de Comunidades de Castilla-La Mancha10.13039/501100011698 (SBPLY/19/180501/000273).
Disclosures
The authors declare no conflicts of interest.
Data availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request. The software for the server and the client, and more miscellaneous material related to the project MicroHikary3D has been made available in three public GitHub repositories [51–53].
References
- 1.Wollman A. J. M., Nudd R., Hedlund E. G., Leake M. C., “From animaculum to single molecules: 300 years of the light microscope,” Open Biol. 5(4), 150019 (2015). 10.1098/rsob.150019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatch M., The Maker Movement Manifesto: Rules for Innovation in the New World of Crafters, Hackers, and Tinkerers (McGraw-Hill Education, 2014). [Google Scholar]
- 3.Zhang C., Anzalone N. C., Faria R. P., Pearce J. M., “Open-source 3d-printable optics equipment,” PLoS One 8(3), e59840 (2013). 10.1371/journal.pone.0059840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chagas A. M., “Haves and have nots must find a better way: the case for open scientific hardware,” PLoS Biol. 16(9), e3000014 (2018). 10.1371/journal.pbio.3000014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katunin P., Cadby A., Nikolaev A., “An open-source experimental framework for automation of high-throughput cell biology experiments,” bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 6.Kornilova A., Kirilenko I., Iarosh D., Kutuev V., Strutovsky M., “Smart mobile microscopy: towards fully-automated digitization,” arXiv arXiv:2105.11179 (2021).
- 7.Cybulski J. S., Clements J., Prakash M., “Foldscope: origami-based paper microscope,” PLoS One 9(6), e98781 (2014). 10.1371/journal.pone.0098781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.“PNNL smartphone microscope,” (2015). Accessed 25 Sep 2021, https://www.pnnl.gov/available-technologies/pnnl-smartphone-microscope.
- 9.Anselmi F., Grier Z., Soddu M. F., Kenyatta N., Odame S. A., Sanders J. I., Wright L. P., “A low-cost do-it-yourself microscope kit for hands-on science education,” in Optics Education and Outreach V , Gregory G. G., ed. (SPIE, 2018). [Google Scholar]
- 10.Vos B. E., Blesa E. B., Betz T., “Designing a high-resolution, LEGO-based microscope for an educational setting,”The Biophysicist (2021). DOI: 10.35459/tbp.2021.000191 [DOI]
- 11.Dong S., Guo K., Nanda P., Shiradkar R., Zheng G., “FPscope: a field-portable high-resolution microscope using a cellphone lens,” Biomed. Opt. Express 5(10), 3305 (2014). 10.1364/BOE.5.003305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu W., Pirovano G., O’Neal P. K., Gong C., Kulkarni N., Nguyen C. D., Brand C., Reiner T., Kang D., “Smartphone epifluorescence microscopy for cellular imaging of fresh tissue in low-resource settings,” Biomed. Opt. Express 11(1), 89 (2020). 10.1364/BOE.11.000089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aidukas T., Eckert R., Harvey A. R., Waller L., Konda P. C., “Low-cost, sub-micron resolution, wide-field computational microscopy using opensource hardware,” Sci. Rep. 9(1), 7457 (2019). 10.1038/s41598-019-43845-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins J. T., Knapper J., Stirling J., Mduda J., Mkindi C., Mayagaya V., Mwakajinga G. A., Nyakyi P. T., Sanga V. L., Carbery D., White L., Dale S., Lim Z. J., Baumberg J. J., Cicuta P., McDermott S., Vodenicharski B., Bowman R., “Robotic microscopy for everyone: the OpenFlexure microscope,” Biomed. Opt. Express 11(5), 2447–2460 (2020). 10.1364/BOE.385729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wincott M., Jefferson A., Dobbie I. M., Booth M. J., Davis I., Parton R. M., “Democratising microscopi: a 3D printed automated XYZT fluorescence imaging system for teaching, outreach and fieldwork,” Wellcome Open Res. 6, 63 (2021). 10.12688/wellcomeopenres.16536.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltran-Parrazal L., Morgado-Valle C., Serrano R. E., Manzo J., Vergara J. L., “Design and construction of a modular low-cost epifluorescence upright microscope for neuron visualized recording and fluorescence detection,” J. Neurosci. Methods 225, 57–64 (2014). 10.1016/j.jneumeth.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 17.Vera R. H., Schwan E., Fatsis-Kavalopoulos N., Kreuger J., “A modular and affordable time-lapse imaging and incubation system based on 3d-printed parts, a smartphone, and off-the-shelf electronics,” PLoS One 11(12), e0167583 (2016). 10.1371/journal.pone.0167583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneidereit D., Kraus L., Meier J. C., Friedrich O., Gilbert D. F., “Step-by-step guide to building an inexpensive 3D printed motorized positioning stage for automated high-content screening microscopy,” Biosens. Bioelectron. 92, 472–481 (2017). 10.1016/j.bios.2016.10.078 [DOI] [PubMed] [Google Scholar]
- 19.Chagas A. M., Prieto-Godino L. L., Arrenberg A. B., Baden T., “The €100 lab: a 3D-printable open-source platform for fluorescence microscopy, optogenetics, and accurate temperature control during behaviour of zebrafish, drosophila, and caenorhabditis elegans,” PLoS Biol. 15(7), e2002702 (2017). 10.1371/journal.pbio.2002702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diederich B., Richter R., Carlstedt S., Uwurukundo X., Wang H., Mosig A., Heintzmann R., “UC2 – a 3D-printed general-purpose optical toolbox for microscopic imaging,” in Imaging and Applied Optics 2019 (COSI, IS, MATH, pcAOP) , (OSA, 2019). [Google Scholar]
- 21.Gürkan G., Gürkan K., “Incu-Stream 1.0: An open-hardware live-cell imaging system based on inverted bright-field microscopy and automated mechanical scanning for real-time and long-term imaging of microplates in incubator,” IEEE Access 7, 58764–58779 (2019). 10.1109/ACCESS.2019.2914958 [DOI] [Google Scholar]
- 22.Guo C., Bian Z., Jiang S., Murphy M., Zhu J., Wang R., Song P., Shao X., Zhang Y., Zheng G., “OpenWSI: a low-cost, high-throughput whole slide imaging system via single-frame autofocusing and open-source hardware,” Opt. Lett. 45(1), 260 (2020). 10.1364/OL.45.000260 [DOI] [Google Scholar]
- 23.Li H., Soto-Montoya H., Voisin M., Valenzuela L. F., Prakash M., “Octopi: open configurable high-throughput imaging platform for infectious disease diagnosis in the field,” bioRxiv (2019).
- 24.Salido J., Sánchez C., Ruiz-Santaquiteria J., Cristóbal G., Blanco S., Bueno G., “A low-cost automated digital microscopy platform for automatic identification of diatoms,” Appl. Sci. 10(17), 6033 (2020). 10.3390/app10176033 [DOI] [Google Scholar]
- 25.Merces G. O., Kennedy C., Lenoci B., Reynaud E. G., Burke N., Pickering M., “The incubot: a 3D printer-based microscope for long-term live cell imaging within a tissue culture incubator,” HardwareX 9, e00189 (2021). 10.1016/j.ohx.2021.e00189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Krishnamurthy D., Li E., Vyas P., Akireddy N., Chai C., Prakash M., “Squid: simplifying quantitative imaging platform development and deployment,” bioRxiv (2020).
- 27.Sharkey J. P., Foo D. C. W., Kabla A., Baumberg J. J., Bowman R. W., “A one-piece 3D printed flexure translation stage for open-source microscopy,” Rev. Sci. Instrum. 87(2), 025104 (2016). 10.1063/1.4941068 [DOI] [PubMed] [Google Scholar]
- 28.Stirling J., Sanga V. L., Nyakyi P. T., Mwakajinga G. A., Collins J. T., Bumke K., Knapper J., Meng Q., McDermott S., Bowman R., “The OpenFlexure project. the technical challenges of co-developing a microscope in the UK and Tanzania,” in 2020 IEEE Global Humanitarian Technology Conference (GHTC) (IEEE, 2020). [Google Scholar]
- 29.Kaynig V., Fischer B., Müller E., Buhmann J. M., “Fully automatic stitching and distortion correction of transmission electron microscope images,” J. Struct. Biol. 171(2), 163–173 (2010). 10.1016/j.jsb.2010.04.012 [DOI] [PubMed] [Google Scholar]
- 30.Maddox E., “Plugin ASI_MTF,” Github, 2020, https://github.com/emx77/ASI_MTF.
- 31.Masse M., REST API Design Rulebook (O’Reilly Media, Inc, 2011). [Google Scholar]
- 32.Avraham S. B., “That is REST–a simple explanation for beginners,” Medium (2017), accessed 25 Sep. 2021. Shortened URL: https://t.ly/lxBa
- 33.Pech-Pacheco J., Cristobal G., Chamorro-Martinez J., Fernandez-Valdivia J., “Diatom autofocusing in brightfield microscopy: a comparative study,” in Proceedings 15th International Conference on Pattern Recognition. ICPR-2000 (IEEE Comput. Soc, 2000). [Google Scholar]
- 34.Sun Y., Duthaler S., Nelson B. J., “Autofocusing in computer microscopy: selecting the optimal focus algorithm,” Microsc. Res. Tech. 65(3), 139–149 (2004). 10.1002/jemt.20118 [DOI] [PubMed] [Google Scholar]
- 35.Yazdanfar S., Kenny K. B., Tasimi K., Corwin A. D., Dixon E. L., Filkins R. J., “Simple and robust image-based autofocusing for digital microscopy,” Opt. Express 16(12), 8670–8677 (2008). 10.1364/OE.16.008670 [DOI] [PubMed] [Google Scholar]
- 36.Redondo R., Bueno G., Valdiviezo J. C., Nava R., Cristóbal G., Déniz O., García-Rojo M., Salido J., del Milagro Fernández M., Vidal J., Escalante-Ramírez B., “Autofocus evaluation for brightfield microscopy pathology,” J. Biomed. Opt. 17(3), 036008 (2012). 10.1117/1.JBO.17.3.036008 [DOI] [PubMed] [Google Scholar]
- 37.Pertuz S., Puig D., Garcia M. A., “Analysis of focus measure operators for shape-from-focus,” Pattern Recognit. 46(5), 1415–1432 (2013). 10.1016/j.patcog.2012.11.011 [DOI] [Google Scholar]
- 38.Yeo T., Ong S., Jayasooriah, Sinniah R., “Autofocusing for tissue microscopy,” Image Vis. Comput. 11(10), 629–639 (1993). 10.1016/0262-8856(93)90059-P [DOI] [Google Scholar]
- 39.Forster B., Ville D. V. D., Berent J., Sage D., Unser M., “Complex wavelets for extended depth-of-field: A new method for the fusion of multichannel microscopy images,” Microsc. Res. Tech. 65(1-2), 33–42 (2004). 10.1002/jemt.20092 [DOI] [PubMed] [Google Scholar]
- 40.Aimonen P., “Fast and easy focus stacking,” Github, 2020, https://github.com/PetteriAimonen/focus-stack.
- 41.Brown M., Lowe D. G., “Automatic panoramic image stitching using invariant features,” Int. J. Comput. Vis. 74(1), 59–73 (2007). 10.1007/s11263-006-0002-3 [DOI] [Google Scholar]
- 42.Preibisch S., Saalfeld S., Tomancak P., “Globally optimal stitching of tiled 3D microscopic image acquisitions,” Bioinformatics 25(11), 1463–1465 (2009). 10.1093/bioinformatics/btp184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mcmaster J., “xystitch. Microscope image stitching,” Github, 2020, https://github.com/JohnDMcMaster/xystitch.
- 44.Lorente Ò., Riera I., Rana A., “Image classification with classic and deep learning techniques,” arXiv arXiv:2105.04895 (2021).
- 45.Howard A. G., Zhu M., Chen B., Kalenichenko D., Wang W., Weyand T., Andreetto M., Adam H., “MobileNets: Efficient convolutional neural networks for mobile vision applications,” arXiv arXiv:1704.04861 (2017).
- 46.Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.-C., “MobileNetV2: Inverted residuals and linear bottlenecks,” The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2018 (2018), pp. 4510–4520. [Google Scholar]
- 47.Tan M., Le Q. V., “EfficientNet: rethinking model scaling for convolutional neural networks,” International Conference on Machine Learning (2019). [Google Scholar]
- 48.He K., Zhang X., Ren S., Sun J., “Deep residual learning for image recognition,” arXiv arXiv:1512.03385 (2015).
- 49.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z., “Rethinking the inception architecture for computer vision,” arXiv arXiv:1512.00567 (2015).
- 50.Bueno G., Fernández-Carrobles M. M., Deniz O., García-Rojo M., “New trends of emerging technologies in digital pathology,” Pathobiology 83(2-3), 61–69 (2016). 10.1159/000443482 [DOI] [PubMed] [Google Scholar]
- 51.Salido J., “MicroHikari3D: an automated DIY digital microscopy platform with deep learning capabilities: software,” Github, 2021, https://github.com/UCLM-VISILAB/uH3D-server. [DOI] [PMC free article] [PubMed]
- 52.Salido J., “MicroHikari3D: an automated DIY digital microscopy platform with deep learning capabilities: software,” Github, 2021, https://github.com/UCLM-VISILAB/uH3D-client. [DOI] [PMC free article] [PubMed]
- 53.Salido J., “MicroHikari3D: an automated DIY digital microscopy platform with deep learning capabilities: software,” Github, 2021, https://github.com/UCLM-VISILAB/uH3D-misc. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request. The software for the server and the client, and more miscellaneous material related to the project MicroHikary3D has been made available in three public GitHub repositories [51–53].