Abstract
Direct in situ fluorescent enzyme-linked immunosorbent assay (ELISA) is rarely investigated and reported. Herein, a direct in situ high-performance HRP-labeled fluorescent immunoassay platform was constructed. The platform was developed based on a rapid in situ fluorogenic reaction between Polyethyleneimine (PEI) and p-Phenylenediamine (PPD) analogues to generate fluorescent copolymer nanoparticles (FCNPs). The formation mechanism of FCNPs was found to be the oxidation of •OH radicals, which was further proved by nitrogen protection and scavenger of •OH radicals. Meantime, the fluorescence wavelength of FCNPs could be adjusted from 471 to 512 nm by introducing various substitution groups into the PPD structure. Using cardiac troponin I (cTnI) and SARS-CoV-2 nucleocapsid protein (N-protein) as the model antigens, the proposed fluorescent ELISA exhibited a wide dynamic range of 5–180 ng/mL and a low limit of detection (LOD) of 0.19 ng/mL for cTnI, and dynamic range of 0–120 ng/mL and a LOD of 0.33 ng/mL for SARS-CoV-2 N protein, respectively. Noteworthy, the proposed method was successful applied to evaluate the cTnI and SARS-CoV-2 N protein levels in serum with satisfied results. Therefore, the proposed platform paved ways for developing novel fluorescence-based HRP-labeled ELISA technologies and broadening biomarker related clinical diagnostics.
Keywords: Fluorescent immunoassay, Wavelength-tunable, Cardiac troponin I, SARS-CoV-2 nucleocapsid protein, Horseradish peroxidase
1. Introduction
Owing to its excellent accuracy, practicality, low cost and high-throughput, enzyme-linked immunosorbent assay (ELISA) has been extensively employed in food safety analysis, environmental monitoring and biomedical diagnosis (Jarvenpaa et al., 2012; Li et al., 2020; Cohen et al., 2020; Vargas et al., 2019; Han et al., 2019; Chen et al., 2021). Usually, ELISA uses an enzyme, typically horseradish peroxidase (HRP) or alkaline phosphatase (ALP), to convert the corresponding substrate into a product with color or fluorescence for quantification (Liu et al., 2019; Yuan et al., 2012). Unfortunately, ALP-labeled enzymes can only hydrolyze product with phosphate groups and need to be in the strict alkaline environment, which limit its application (Chen et al., 2018, 2020). Alternative enzyme, HRP, gets relatively more common application in diagnosis, biosensing due to its high specific activity, stability, small molecular weight and easy preparation of pure enzyme (Cosnier et al., 2021; Li et al., 2017; Xianyu et al., 2018). HRP has a faster catalytic rate than ALP, which make it generating a strong signal in a short time. However, sensitivity of HRP based ELISA retards its application.
Until now, there are many HRP based methods applied in bioassays, such as chemiluminescence, square-wave voltammetry, fluorometry, colorimetry and electrochemistry (Lin et al., 2012; Kergaravet et al., 2012; Chen et al., 2019; Fornera and Walde, 2010; Wang et al., 2018). Among these assays, colorimetric immunoassay is the routine method adopted in laboratories and industry/clinic for detecting target analytes. Generally, HRP catalyzes the substrates with the presence of H2O2. 3,3′,5,5′-tetramethylbenzidine (TMB) is the most typical chromogenic substrate in HRP-induced immunoassay system. Nevertheless, the traditional colorimetric immunoassay suffers from low sensitivity due to the limitation of substrate though great endeavors have been put (Chen et al., 2017; Fu et al., 2018; Ellis et al., 2009; Welch et al., 2016). For example, Shiddiky's group developed an enhanced colorimetric molecular sensor for detecting auto-antibodies by coupling HRP with gold-containing nanoporous iron oxide nanoparticles (Shiddiky et al., 2017). In contrast with colorimetric immunoassay, fluorescence immunoassay has drawn increasing attention owing to the advantages of high sensitivity, fast response, and real-time monitoring (Murfin et al., 2019; Liu et al., 2020, Liu et al., 2020, Liu et al., 2020; Zhou et al., 2020). Yang and his colleagues developed a fluorescent immunoassay for alpha-fetoprotein (AFP) based on fluorescein and p-phenylenediamine (Yuan et al., 2012). Chen groups designed a fluorescent ELISA to analyze insecticidal protein toxin utilizing polymer dots and PPD (Chen et al., 2019). However, the above-mentioned biosensors achieve target detection by the internal filtration effect (IFE), which is an indirect fluorescence immunoassay. Additionally, the fluorophores of biosensors interact with the HRP, which often disturbs fluorescence detection (Zhu et al., 2012; Yuan et al., 2012; Shiang et al., 2009). It means that HRP reactions can be self-limiting due to loss of in situ fluorescent substrate of the enzyme. Hence, it is very necessary to develop fluorescent substrates of HRP for constructing direct in situ fluorescence immunoassay.
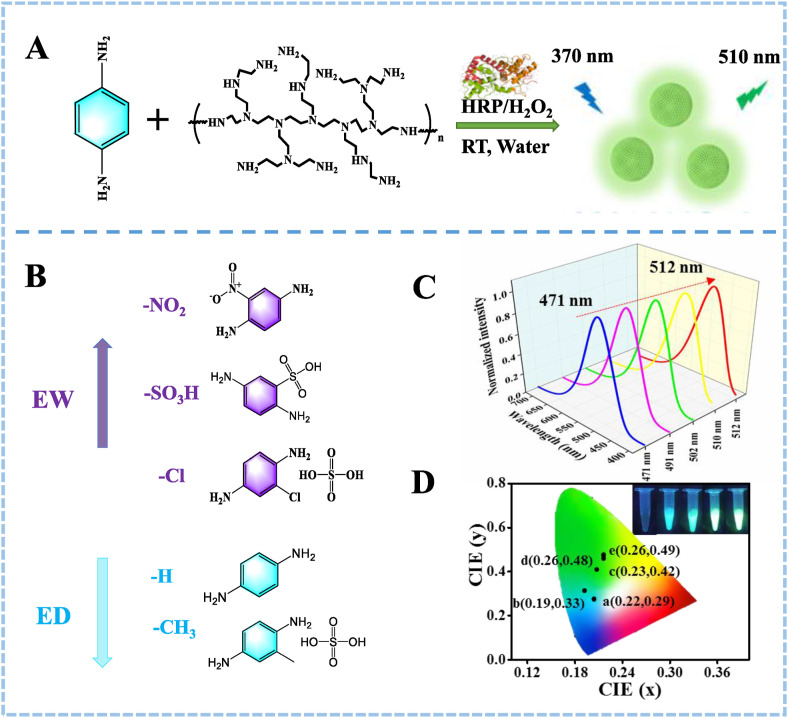
Herein, we developed a facile and direct in situ HRP-induced fluorescent immunoassay platform. The proposed fluorescent ELISA was based on the in situ fluorogenic reactions between polyethyleneimine (PEI) and p-phenylenediamine (PPD) triggered by HRP in the presence of H2O2 to rapidly generate potent green fluorescent copolymer nanoparticles (FCNPs) (Scheme 1 A). The FCNPs were formed through oxidation of •OH radicals as proved by nitrogen protection and scavenger (ascorbic acid) of •OH radicals. Meantime, the fluorescence wavelength of FCNPs could be adjusted from 471 to 512 nm by introducing various substitution groups (electron-donating groups and electron-withdrawing groups) into the PPD structure. Additionally, we speculated that the strong fluorescence original of FCNPs was also attributed to PEI with many cationic reactive primary amino groups and high charge density. The in situ fluorogenic reaction platform could not only reduce the interference of background signal, but also exhibit outstanding features such as stability, wavelength-tunability, highly fluorescence quantum yield. Noteworthy, using the cTnI and SARS-CoV-2 nucleocapsid protein (N protein) as the models, the proposed fluorescent ELISA displayed high sensitivity for detecting cTnI and SARS-CoV-2 N protein with a low limit of detection (LOD) of 0.19 ng/mL and 0.33 ng/mL, respectively. Overall, as developed fluorescent platform provide great potential for rapid and sensitive analysis of other protein biomarkers in clinical diagnostics, fundamental discovery, and other biomedical applications.
Scheme 1.
(A) Principle of HRP-triggered formation of FNCPs. (B) The chemical structure of PPD derivatives. The order of the substituent X is from neutral to strong ED or EW properties. (C) Fluorescence spectra of PPD derivatives and PEI catalyzed by HRP in the presence of H2O2. (D) Calculated CIE coordinates from the FL spectra of the different wavelength-tunable FCNPs. Inset: the corresponding photographs of wavelength-tunable FCNPs under ultraviolet light.
2. Experimental section
2.1. Reagents and instruments
All chemicals and related instruments have been listed in Supporting Information.
2.2. Preparation of FCNPs and calculation its relative quantum yield
Briefly, horseradish peroxidase (120 mU/mL) and hydrogen peroxide (500 μM) were dissolved in Tris-HCl buffer (10 mM, pH = 7.4). After incubating at 25 °C for 15 min on a shaker, Polyethyleneimine (5 mg/mL, MW = 70000) and p-phenylenediamine (60 μM) were added in it. The polymeric nanocluster was obtained by mixture at room temperature within minutes. The relative quantum yield of FCNPs was calculated by the following equation:
Where Φ is the fluorescence quantum yield, F is the integrated area of emitted fluorescence spectra, and A is the absorbance at the excitation wavelength. The subscript P and D implied product of PEI-PPD and quinine sulfate, respectively.
2.3. Analytical performance
All analysis performance processes have been listed in Supporting Information.
3. Results and discussion
3.1. Establishment and optimization of HRP sensing system
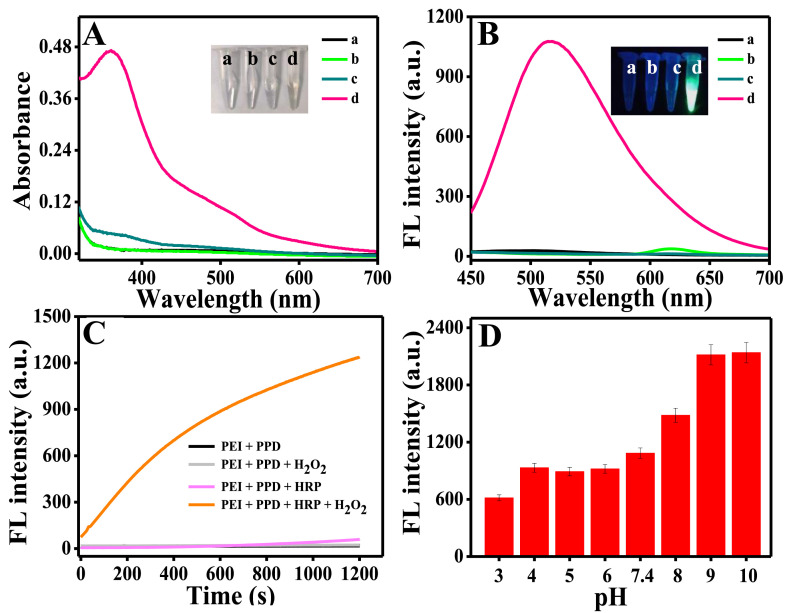
To prove the feasibility of PEI and PPD as substrates for HRP activity detection in the presence of H2O2, we measured the absorption and fluorescence spectra of the system. As showed in Fig. 1 A and B, the hybrid solution of PEI and PPD (line a), and the hybrid solution of PEI, PPD and HRP (line b) did not present absorption peak at 370 nm and fluorescence signal at 510 nm. The hybrid solution of PEI-PPD-H2O2 (line c) had an absorption peak at 370 nm but no fluorescence signal at 510 nm. While adding HRP into the mixed solution of PEI-PPD-H2O2 (line d), a sharp absorption peak at 370 nm and a strong fluorescence signal at 510 nm was observed, indicating that the HRP catalyzed H2O2 to produce hydroxyl radicals (•OH) which led to oxidative polymerization reaction between PPD and PEI. Besides, the color and fluorescence change of solutions under visible light and ultraviolet light also confirmed the corresponding process (inset image of Fig. 1A and B). Thus, an HRP sensing design is successfully constructed based on the effective fluorogenic and chromogenic reaction using PPD and PEI as the substrates (Scheme 1A).
Fig. 1.
(A) Absorption and (B) fluorescence emission spectra of PEI + PPD (a), mixture of HRP + PEI + PPD (b), mixture of H2O2 + PEI + PPD (c) and the mixture of HRP + H2O2 + PEI + PPD (d), respectively. The final concentrations of PEI, PPD, HRP and H2O2 in the reaction solutions are 5 mg/mL, 60 μM, 120 mU/mL and 500 μM, respectively. (C) Time-fluorescence curve of PEI-PPD in Tris-HCl buffer system with H2O2 (gray), HRP (pink) and HRP-H2O2 (orange). (D) Fluorescence intensity of FNCPs change at different pH (3–10). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The catalytic activity of HRP toward PEI and PPD in the presence of H2O2 was tested in the Tris-HCl buffer (pH 7.4) at 25 °C. After incubation of the solution containing HRP and H2O2, the mixed solution of PEI and PPD exhibited vigorous fluorescence at 510 nm. The increase gradually levelled off in 15–20 min, indicating oxidation induced polymerization of PEI-PPD to form FCNPs in the presence of H2O2. By contrast, in the presence of only HRP or H2O2, the solution showed no apparent fluorescence changes, implying that the direct oxidation of PEI and PPD by HRP or H2O2 alone was kinetically sluggish (Fig. 1C). The result confirmed that •OH played a vital role in oxidative polymerization of PEI and PPD.
To obtain better sensing performance, we optimized the pH and incubation time of reaction. As shown in Fig. 1D and Fig. S1, with the increase of pH and incubation time, the fluorescence intensities at 510 nm of the copolymer dot also increased. The pH and the incubation time were chosen 7.4 and 15 min, respectively. Then, we also optimized the concentration of PEI, PPD and H2O2. As displayed in Fig. S2, Fig. S3 and Fig. S4, as the concentration of PEI, PPD and H2O2 increased, the value of F1/F0 increased rapidly. 5 mg/mL of PEI, 60 μM of PPD and 500 μM of H2O2 were selected in the subsequent experiments.
As the emission spectral of reaction could be modulated by introducing various substitutions, we administrated electron donating groups (EDGs) and electron withdrawing groups (EWGs) to achieve wavelength tunability of FCNPs. The structures of substituent were showed in Scheme 1B, as named in bold type. As indicated in Table S1, FCNPs-1 exhibited the emission maximum at 510 nm. By introducing chlorine into the system, the resulting FCNPs-2 showed a blue-shifted emission maximum of 502 nm. However, by replacing chlorine moiety with sulfonic acid or nitro, leading to a blue-shift emission maximum of 491 and 471 nm of FCNPs-3 and FCNPs-4, respectively. The blue-shift of 11 and 31 nm could be attributed to the stronger electron withdrawing of the sulfonic acid group and nitro than the chlorine group. Moreover, upon adding a methyl group FCNPs-5 showed to a bathochromic shift (peak at 512 nm). This could be attributed to the donor capacity of the methyl group. Therefore, the introduction of different substituents in the PPD structure realized the tunability of fluorescence emission wavelength of FCNPs from 471 to 512 nm (Scheme 1C). Thus, it is feasible to build a wavelength-tunable sensing platform which is attributed to the degree of conjugation caused by different electron donating groups and electron withdrawing groups on the benzene ring (Lei et al., 2019; Feng et al., 2013). In addition, in view of the FCNPs of different wavelengths, we calculated the transparent color perception of the sensing platform through CIE, which further confirmed the construction of the wavelength tunable sensing platform (Scheme 1D).
3.2. Characterization of FCNPs
The FCNPs were synthesized in the presence of HRP and H2O2. The optical properties of FCNPs were characterized by absorption and fluorescence spectra. As seen from Fig. 2 A, the obtained FCNPs displayed an absorption peaks at 370 nm and the light yellow mixture solution display intensively green fluorescence under 365 nm (inset of Fig. 2A). The maximum excitation and emission wavelength of FCNPs were located at 370 nm and around 510 nm, and the relative fluorescence quantum yield (QY) was about 12.4% with quinine sulfate (QY = 54%) as a reference. The TEM and AFM confirmed morphology of FCNPs. The FCNPs had a diameter of 3 nm (Fig. 2B) and a height of about 2.5 nm (Fig. 2D, E and 2F), indicating that the FCNPs were approximate elliptical structure. In addition, the high resolution TEM (HRTEM) image displayed lattice fringes, which was close to the facet of graphite. It showed that the nanoparticles separated well from each other. The diffraction rings were indexed to the planes of the FCNPs crystal structure (Fig. 2C). The FCNPs were further characterized by FTIR to evaluate the groups. As shown in Fig. 2G, the signals at 1598 cm−1 and 1501 cm−1 were derived from the stretching vibration of C N double bonds. The absorption peaks at 1189 cm−1and 1285 cm−1 were assigned to the stretching vibrations of C–N bonds, which implying that Michael addition reaction might occur between aromatic ring and amine group. Besides, the FT-IR measurement of the stretching vibration mode showed that the fluorescent FCNPs also had other functional groups, such as N–H (3307 cm−1 and 3038 cm−1) and C–H (2704 cm−1). Furthermore, time scanning further confirmed the stability of FCNPs after reaction completely terminated (Fig. S5).
Fig. 2.
(A) Absorption (black), fluorescence excitation (blue), and emission (red) spectra of the FNCPs. Inset photos display the FNCPs under sunlight (left) and 365 nm ultraviolet light (right), respectively. (B) TEM image of the FNCPs. The scale bar is 50 nm. Inset is the size distribution histogram. (C) HRTEM image of FNCPs. (D) AFM image of FNCPs. (E) Corresponding height image and (F) Height distribution. (G) FTIR of FNCPs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Mechanism investigation of FCNPs formation
PEI and PPD mixture incubated for 1 day at room temperature generated strong green fluorescence under UV light (Figs. S6A and S6B). We speculated that FCNPs were formed via oxidation polymerization of PEI and PPD. To confirm that, we conducted nitrogen protection. As shown in Fig. 3 B, the mixture solution of PEI and PPD displayed extremely low fluorescence with nitrogen protection compared with the regular group (with oxygen), suggesting the indispensability of the oxygen for this process. Furthermore, we introduced ascorbic acid, the scavenger of •OH radicals (Song et al., 2016) into the reaction. As showed in Fig. S7A, fluorescence of FCNPs gradually decrease with increasing of ascorbic acid, suggesting that the •OH was effectively inhibited by ascorbic acid. The above results indicated that •OH radicals were the main active substances which triggered formation of FCNPs was the oxidation polymerization of PEI and PPD.
Fig. 3.
(A) Scheme of the preparation of FCNPs based on PPD and PEI. (B) Fluorescence emission spectra of FNCPs with N2 protection (blue line) and without N2 protection (green line). (C) Fluorescence emission spectra of the mixture solution about PEI + HRP + H2O2 with OPD (a), MPD (b) and PPD (c). All above substance concentrations are 60 μM. (D) Fluorescence emission spectra of the mixture solution of PPD + HRP + H2O2 with different molecular weights of PEI (1 M), 0 (a), 600 (b), 1800 (c), 10000 (d) and 70000 (e). (E) Fluorescence emission spectra of the mixture solution of PPD + HRP + H2O2 in the presence of PEI (a), ETA (b), EDA (c), PEA (d) and NaOH (e). All substance concentrations are 5 mg/mL. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The reaction process of FCNPs was further investigated by introducing common reductants as the terminating agent. Under the similar experimental conditions, the fluorescence intensity of the sensing system was suppressed after adding reductants. These results illustrated that ascorbic acid and glutathione had a good capacity as a terminating agent and could make fluorescence intensity of the system leveled off for a long time (Fig. S7B).
To further study the effect of different precursors on the formation of FCNPs, we administered substrates with similar structure to PPD and PEI. First, we examined the impact of isomers of phenylenediamine, including OPD, MPD and PPD on the formation of FCNPs. Under the same reaction conditions, MPD and OPD didn't lead to similar fluorogenic (Fig. 3D). The amino group was an electron-donating group when located in ortho and counter position. It attached to the benzene ring with an unshared electron pair led to p-π hyper conjugation on the benzene ring and increased the density of the electron cloud on the aromatic ring (Maity et al., 2020; Rahemi et al., 2020). Thus, this could help to explain why MPD and PEI did not produce fluorescence. As for the gap between PPD and OPD, it might be due to the higher steric hindrance of the ortho-substituents, which led to a decrease in conjugation (Roberto et al., 2018). When PPD interacted with PEI, FCNPs was formed due to p-π hyper conjugation and lower steric hindrance. Moreover, several tests were conducted to illustrate the generation process of the FCNPs. The PPD was incubated with HRP and H2O2 for 15 min before adding PEI, the mixed solution of PPD displayed an intense absorption at around 500 nm. After reaction with PEI, the system showed an absorption peak around 370 nm, which was consistent with that of the FCNPs (Fig. S8A). Besides, in the presence of HRP and H2O2, the mixed solutions of PPD did not generate fluorescence. But after adding PEI, a strong fluorescence emission peak near 510 nm was present, same as that of FCNPs (Fig. S8B). Summarily, the results revealed that PPD firstly formed an intermediate, and then interacted with PEI to form FCNPs. There were other studies addressing oxidation of PPD with absorption peaks at 500 nm in the presence of HRP and H2O2, which was consistent with our results. In addition, as mentioned before, the FT-IR spectrum of FCNPs showed a sharp peak at 1501 cm−1 and 1598 cm−1 (C N stretching vibration), which further confirmed the formation of the Bandrowski's base intermediate. Thus, the results demonstrate that during the formation of FCNPs, PPD first undergoes oxidative polymerization to form a Bandrowski's base intermediate, and then reacts with PEI to form the final product (Zhang et al., 2017).
To further verify the mechanism, substances with similar structure to PPD with PEI were investigated. As shown in Fig. S9A, after incubation with HRP and H2O2, addition PEI into the mixed solution contain 2,5-Diaminobenzene-1,4-diol dihydrochloride or 2,3,5,6-Tetramethyl-1,4-phenylene- diamine could hardly generate a fluorescence signals. We speculated that this was because the reaction sites on the benzene ring structure of DMDDC or TMPDA in the mixed solution were replaced by carboxyl and methyl groups, and no further oxidation could occur. In other words, PEI reacts with oxidized intermediates to form FCNPs, instead of directly reacting with PPD to produce fluorescence (Scheme 1).
Next, the role of PEI in the reactants was carefully studied. PEI, as a water-soluble polymer, is a colorless or light yellow viscous liquid and its molecular weight will change with the degree of polymerization. We investigated the effect of molecular weight of PEI (MW = 600, 1800, 10000, 70000) on the reaction system. The FL intensity of FCNPs gradually enhanced as the molecular weights increasing (Fig. 3D), implying that long-chain PEI precursors were more liable to achieve folding and warping. To eliminate the possibility of alkali-induced fluorophores formation, we introduced NaOH and other bases to replace PEI in the system (Fig. 3E). The results showed that only PEI and EDA could react with PPD in the presence of HRP and H2O2 to form a blue and green fluorescent copolymer, respectively. PEI, as a polymer, had more cationic reactive primary amino groups and higher charge density than EDA. So we speculated that the formation of the green fluorescent copolymer was associated with the structure of PEI itself.
3.4. Quantitation analysis of HRP activity
After optimization, we use the assay to analyze HRP activity. The relationship between HRP concentration and FL intensity at 510 nm was investigated in detail. As presented in Fig. 4 A, when HRP increased from 0 to 150 mU/mL, FL intensity of the system at around 510 nm augmented gradually, and it displayed a two-stage good linear relationship (Fig. 4B). The linear fitting equation was to be F1/F0 = 1.72009 + 5.2548CHRP (R1 2 = 0.95896) and F1/F0 = 53.78294 + 0.24271CHRP (R2 2 = 0.97629). The limit of detection was 0.13 mU/mL, which was superior to other reported ones (Table S2). Furthermore, when the HRP concentration was 1 mU/mL, obvious fluorescent color changes could be easily read under ultraviolet light (Fig. 4C). The selectivity of our proposed assay was investigated by comparing with other common biomolecules, such as ALP, ACP, Ppase, BSA, Pepsin, Cyt-c, Tyrosinase, Trypsin and GOx. All the tests were performed in 1.5 mL centrifuge tubes. As shown in Fig. 4D, those biomolecules, except HRP, with concentration of 120 mU/mL did not lead to noticeable fluorescence change. These results indicated that our detection system exhibited the excellent selectivity and selectivity toward HRP.
Fig. 4.
(A) Fluorescence emission spectra of FNCPs toward various concentrations of HRP. (B) Linear range of F1/F0 against concentrations of HRP from 0 to 150 mU/mL. F1 and F0 was the fluorescence intensity of the FNCPs in the presence and absence of HRP, respectively. (C) Photographs under ultraviolet light. (D) Selectivity investigation of the sensing system for HRP activity. The concentrations of HRP and other proteins are both 120 mU/mL.
3.5. Fluorescent immunoassay for cTnI and SARS-CoV-2 N protein
Inspired by comprehensive application of HRP in ELISA, we initiated to explore the application of HRP activity based fluorescence turn-on sensing system in the HRP-labeled immunoassay target antigen. The cTnI, the biomarker of myocardial infarction (AMI), was adopted as our target antigen (Kergaravet et al., 2012; Yuan et al., 2012; Anabel et al., 2021; Carlo et al., 2021). The mouse anti-cTnI monoclonal antibody, goat anti-cTnI antibody and HRP-conjugated rabbit anti-goat IgG were employed as the capture antibody, primary antibody and secondary antibody, respectively. Fig. 5 A vividly displayed the strategy of HRP-labeled immunoassay for the detection of cTnI. The specific antibody was pre-immobilized on a 96-well plate to capture the target cTnI. Subsequently, through the specific recognition of the antigen and antibody, the goat anti-cTnI antibody and HRP secondary antibody labels were fixed on the plate. After that H2O2, followed by PEI and PPD, were added into the plate. As expected, the fluorescence intensity gradually increased with the increase of HRP concentration (Fig. 5B), implying gradual binding of HRP labeled on the antibody. The inset graph displayed the linear relationship between the fluorescence intensity and cTnI concentrations from 5 to 180 ng/mL. The fitted linear equation could be expressed as F1/F0 = 1.7885 + 0.0589CcTnI, R2 = 0.99362 (Fig. 5C). The detection limit was 0.19 ng/mL, as calculated from 3σ/S, which was superior to those previously reported assay for cTnI detection (Table S3) (Liu et al., 2016; Wu et al., 2010, 2017; Guo et al., 2019; Song et al., 2011; Dorraj et al., 2015; Zuo et al., 2016; Tsaloglou et al., 2014; Periyakaruppan et al., 2013). Comprehensive consideration of performance, cost, and detection time, our method still showed obvious overall advantages compared with other fluorescence-based methods (Liu et al., 2020, Liu et al., 2020, Liu et al., 2020; Jarvenpaa et al., 2012; Zhao et al., 2019; Liu et al., 2020, Liu et al., 2020, Liu et al., 2020) (Table S4). To validate the sensing ability and specificity of our sensor toward target antigen cTnI, nonspecific proteins, including alpha fetoprotein, lysozyme, pepsin and trypsin were administrated, and some co-existing reducing species, such as AA, GSH, Cys, as well as metal ions (Fe3+, Cu2+) had been added the system for target sensing. None of those substances induced obvious increase of fluorescence signal (Fig. 5D and Fig. S11), suggesting excellent selectivity of our assay for detecting cTnI.
Fig. 5.
(A) Schematic representation of fluorescent immunoassay. (B) Fluorescence emission spectra and (C) F1/F0 at 510 nm as a function of cTnI activities (0–200 ng/mL). Inset: the linear relationship from 5 to 180 ng/mL. (D) Selectivity investigation of the developed fluorescent immunoasay against cTnI or other control enzymes/proteins (200 ng/mL). (E) Fluorescence emission spectra and (F) F1/F0 value change as a function of SARS-CoV-2 N protein (0–120 ng/mL).
In order to further verify the versatility of the proposed platform, we used the SARS-CoV-2 N protein replace the cTnI as the model antigen. The mouse anti-N protein antibody, rabbit anti-N protein antibody and HRP-conjugated goat anti-rabbit IgG were employed as the capture antibody, primary antibody and secondary antibody, respectively. As shown in Fig. 5E, the fluorescence spectra of the system were recorded at different concentration of N protein from 0 to 120 ng/mL. An up-to-3.5-fold fluorescence intensity enhancement was observed. A linear relationship between F1/F0 and the N protein concentration was achieved (Fig. 5F). The fitted linear data were expressed as F1/F0 = 1.0396 + 0.1181CN-protein (R1 2 = 0.9785) ranging from 0 to 10 ng/mL and F1/F0 = 2.0264 + 0.0104CN-protein (R2 2 = 0.9724) ranging from 10 to 120 ng/mL, where F was the fluorescence intensity at 510 nm of system. The detection limit of our fluorescence assay was 0.33 ng/mL and the sensitivity was satisfactory.
3.6. Detection of cTnI in clinical samples and SARS-CoV-2 N protein in serum
Encouraged by the above results, we further applied the sensor to the determination of cTnI in patient samples. cTnI was a highly specific principle biomarker for acute myocardial infarction (AMI) (Amodio et al., 2007) having clinical cutoff level of about 0.06–1.5 ng/mL (Christenson and Azzazy, 2009) and levels greater than 2 ng/mL indicated an increased risk for serious heart disease. As shown in Fig. S10, the result of our developed fluorescent ELISA exhibited a well linear relationship when the concentration of cTnI was in the range from 0 to 100 ng/mL, implying its potential for detecting cTnI in patient samples. Subsequently, the cTnI levels in two control and four patients samples were measured by utilizing our HRP based ELISA detection system and TMB-based commercial standard ELISA kit (Table S5). Noteworthy, the cTnI concentration determined with our system was in good agreement with the results obtained by commercial TMB-based standard ELISA kit. Additionally, we listed the cost of our methods and commercial kit tests (Table S6, Table S7 and Table S8). Compared with the traditional ELISA for testing a cTnI sample ($4.58, 20 min), the proposed sensor was cheaper ($1.42) and faster (17 min). These results suggested that as developed HRP based fluorescent ELISA detection system had reliable cTnI assessing capability in clinical samples, and it held great potential for sensitive detection of other disease biomarkers.
SARS-CoV could cause epidemics that threaten the life of the world, such as the current Coronavirus Disease 2019. Detection of SARS-CoV-2 N protein is of great value for disease diagnosis. For SARS-CoV-2 N protein sensing, diluted human serum samples (1%) was utilized. As shown in Table S9, diluted human serum samples with different concentration of N protein (0–120 ng/mL) were applied to the system. The results were consistent with the spiked N protein and exhibit an excellent recovery in the range of 96.7–106.0%, indicating that the assay could be successfully applied in N protein monitoring the real biological sample. It suggested that our system was a universal fluorescent immunoassay platform and held great potential for sensitive detection of other disease biomarkers.
4. Conclusions
In summary, we constructed a high-performance direct in situ fluorescent immunoassay platform based on a rapid fluorogenic reaction between PEI and PPD analogues triggered by HRP. This fluorescent immunoassay system displayed a few outstanding features: (i) A series of facile and effective substrates of HRP-H2O2 system are developed; (ii) The obtained FCNPs by in situ fluorogenic reaction between PEI and PPD analogues showed strong fluorescence and stability. (iii) The fluorescence wavelength of FCNPs could be adjusted from 471 to 512 nm by introducing various substitution groups. (iv) Using cTnI and SARS-CoV-2 N protein as the model antigen, the proposed fluorescent ELISA exhibited a wide dynamic range of 5–180 ng/mL and 0–120 ng/mL for cTnI and N protein detection with a low limit of detection (LOD) of 0.19 ng/mL and 0.33 ng/mL, respectively. (v) The proposed method achieved cTnI detection in clinical samples with well consistency of traditional methods, and for SARS-CoV-2 N protein detection it also gets satisfied results. (vi) Compared with the traditional ELISA for testing a cTnI sample ($4.58, 20 min), the proposed sensor is cheaper ($1.42) and faster (17 min). This was the first attempt to use new substrates (PEI and PPD) of HRP to design a high-performance universal fluorescent immunoassay platform. It paved ways for developing new types of fluorescence-based HRP-labeled ELISA strategy, which could be used in the detection of pathogens, such as SARS-CoV-2.
CRediT authorship contribution statement
Jinhua Liu: Methodology, Writing – original draft, overall supervise, writing the final manuscript. Guotong Ruan: Started the work, synthesis, properties studies, initial manuscript, Writing – original draft. Wenlin Ma: Supplemented the experimental data, modified manuscript. Yujie Sun: Properties studies and supplemented the experimental data. Haidong Yu: Writing – review & editing, gave some suggestions. Zhihui Xu: Provided serum samples, Writing – review & editing. Changmin Yu: Writing – review & editing, gave some suggestions. Hai Li: Provided AFM characterization. Cheng-wu Zhang: Initial manuscript writing, Writing – review & editing, writing the final manuscript. Lin Li: Initial manuscript writing, Writing – review & editing, Writing – original draft.
Declaration of competing interest
There are no conflicts of interest.
Acknowledgements
The authors acknowledge the National Science Foundation of China (Grant nos. 22174065) and financial support from the National Key R&D Program of China (2020YFA0709900).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113823.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Anabel V., Itziar E., Ana M.P.C., Beatriz M., Concepcion P., Reynaldo V. Biosens. Bioelectron. 2021;183:113203. [Google Scholar]
- Cohen L., Cui N.W., Cai Y.M., Garden P.M., Li X., Weitz D.A., Walt D.R. ACS Nano. 2020;14:9491–9501. doi: 10.1021/acsnano.0c02378. [DOI] [PubMed] [Google Scholar]
- Chen C.X., Zhao J.H., Lu Y.Z., Sun J., Yang X.R. Anal. Chem. 2018;90:3505–3511. doi: 10.1021/acs.analchem.7b05325. [DOI] [PubMed] [Google Scholar]
- Chen L., Li Y., Miao L.Y., Pang X.L., Li T., Qian Y.J., Li He. Biosens. Bioelectron. 2021;188:113308. doi: 10.1016/j.bios.2021.113308. [DOI] [PubMed] [Google Scholar]
- Chen C.X., Zhao D., Wang B., Ni P.J., Jiang Y.Y., Zhang C.H., Yang F., Lu Y.Z., Sun J. Anal. Chem. 2020;92:4639–4646. doi: 10.1021/acs.analchem.0c00224. [DOI] [PubMed] [Google Scholar]
- Chen X., Sun L.H., Li Z.L., Jiang H., Ma J.M. Microchimica Acta. 2019;186:731–737. doi: 10.1007/s00604-019-3831-9. [DOI] [PubMed] [Google Scholar]
- Chen X., Chen L., Choo J. Anal. Chem. 2017;89:124–137. doi: 10.1021/acs.analchem.6b02251. [DOI] [PubMed] [Google Scholar]
- Christenson R.H., Azzazy H.M. Clin. Biochem. 2009;42:150–157. doi: 10.1016/j.clinbiochem.2008.09.105. [DOI] [PubMed] [Google Scholar]
- Dorraj G.S., Rassaee M.J., Latifi A.M., Pishgoo B., Tavallaei M. J. Biotechnol. 2015;208:80–86. doi: 10.1016/j.jbiotec.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Ellis W.C., Tran C.T., Denardo M.A., Fischer A., Ryabov A.D., Collins T.J. J. Am. Chem. Soc. 2009;131:18052–18053. doi: 10.1021/ja9086837. [DOI] [PubMed] [Google Scholar]
- Fu G.L., Sanjay S.T., Wan Z., Brekken R.A., Kirken R.A., Li X.J. Anal. Chem. 2018;90:5930–5937. doi: 10.1021/acs.analchem.8b00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J.J., Huang H., Zhou D.L., Cai L.Y., Tu Q.Q., Wang A.J. J. Mater. Chem. C. 2013;1:4720–4725. [Google Scholar]
- Fornera S., Walde P. Anal. Biochem. 2010;407:293–295. doi: 10.1016/j.ab.2010.07.034. [DOI] [PubMed] [Google Scholar]
- Guo X.Y., Zong L.J., Jiao Y.C., Han Y.F., Zhang X.P., Xu J., Li L., Zhang C.W., Liu Z.P., Ju Q., Liu J.H., Xu Z.H., Yu H.D., Huang W. Anal. Chem. 2019;91:9300–9307. doi: 10.1021/acs.analchem.9b02557. [DOI] [PubMed] [Google Scholar]
- Jarvenpaa M.L., Kuningas K., Niemi I., Hedberg P., Ristiniemi N., Pettersson K., Lovgren T. Clin. Chim. Acta. 2012;414:70–75. doi: 10.1016/j.cca.2012.08.027. [DOI] [PubMed] [Google Scholar]
- Kergaravet S.V., Pividori M.I., Hernandez S.R. Talanta. 2012;88:468–476. doi: 10.1016/j.talanta.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Li D.Y., Cui Y.L., Morisseau C., Gee S.J., Bever C.S., Liu X.J., Wu J., Hammock B.D., Ying Y.B. Anal. Chem. 2017;89:6248–6256. doi: 10.1021/acs.analchem.7b01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.Y., Morisseau C., McReynolds C.B., Duflot T., Bellien J.M., Nagra R.M., Taha A.Y., Hammock B.D. Anal. Chem. 2020;92:7334–7342. doi: 10.1021/acs.analchem.0c01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Zhao J., Wang S., Lu S., Sun J., Yang X. Sensor. Actuator. B Chem. 2020;306:127583. [Google Scholar]
- Liu G., Zhao J., Yan M., Zhu S., Dou W., Sun J., Yang X. Sci. China Chem. 2020;63:554–560. [Google Scholar]
- Liu X., Wang Y., Chen P., McCadden A., Palaniappan A., Zhang J., Liedberg B. ACS Sens. 2016;1:1416–1422. [Google Scholar]
- Liu Y., Pan M., Wang W., Jiang Q., Wang F., Pang D.W., Liu X. Anal. Chem. 2019;91:2086–2092. doi: 10.1021/acs.analchem.8b04517. [DOI] [PubMed] [Google Scholar]
- Liu S.Y., Yan A.M., Guo Z.W., Fang Y.Y., Dong Q.J., Li R.R., Ni S.S., Sun Y., Yang W.C., Yang G.F. ACS Nano. 2020;14:4244–4254. doi: 10.1021/acsnano.9b09493. [DOI] [PubMed] [Google Scholar]
- Lei Z.H., Sun C.X., Pei P., Wang S.F., Li D.D., Zhang X., Zhang F. Angew. Chem. Int. Ed. 2019;58:8166–8171. doi: 10.1002/anie.201904182. [DOI] [PubMed] [Google Scholar]
- Maity S., Brain D., Chakraborty S., Kolay S., Patra A. ACS Sustain. Chem. Eng. 2020;8:18335–18344. [Google Scholar]
- Murfin L.C., Weber M., Park S.J., Kim W.T., Lopez-Alled C.M., McMullin C.L., Fabienne P.C., Kohn K.K., Wenk J., Bull S.T., Yoo J.Y., Kim H.M., James T.D., Lewis S.E. J. Am. Chem. Soc. 2019;141:19389–19396. doi: 10.1021/jacs.9b09813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periyakaruppan A., Gandhiraman R.P., Meyyappan M., Koehne J.E. Anal. Chem. 2013;85:3858–3863. doi: 10.1021/ac302801z. [DOI] [PubMed] [Google Scholar]
- Rahemi V., Trashin S., Hafideddine Z., Van Doorslaer S., Meynen V., Gorton L., Wael K.D. Anal. Chem. 2020;92:3643–3649. doi: 10.1021/acs.analchem.9b04617. [DOI] [PubMed] [Google Scholar]
- Roberto M.H., Beatriz J.S., Alberto E. Anal. Chem. 2018;90:9830–9837. [Google Scholar]
- Shiang Y.C., Huang C.C., Chang H.T. Chem. Commun. 2009;23:3437–3439. doi: 10.1039/b901916b. [DOI] [PubMed] [Google Scholar]
- Song B., Choi J.L., Zhu Y.T., Geng Z.S., Zhang L., Lin Z.Y., Tuan C.C., Moon K.S., Wong C.P. Chem. Mater. 2016;28:9110–9121. [Google Scholar]
- Song S.Y., Han Y.D., Kim K., Yang S.S., Yoon H.C. Biosens. Bioelectron. 2011;26:3818–3824. doi: 10.1016/j.bios.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Tsaloglou M.N., Jacobs A., Morgan H. Anal. Bioanal. Chem. 2014;406:5967–5976. doi: 10.1007/s00216-014-7997-z. [DOI] [PubMed] [Google Scholar]
- Vargas E., Teymourian H., Tehrani F., Eksin E., Sanchez-Tirado E., Warren P., Erdem A., Dassau E., Wang J. Angew. Chem. Int. Ed. 2019;58:6376–6379. doi: 10.1002/anie.201902664. [DOI] [PubMed] [Google Scholar]
- Wang Q., Xue R., Guo H., Wei Y.L., Yang W. J. Electroanal. Chem. 2018;817:184–194. [Google Scholar]
- Welch N.G., Scoble J.A., Easton C.D., Williams C.C., Bradford B.J., Mamedova L.K., Pigram P.J., Muir B.W. Anal. Chem. 2016;88:10102–10110. doi: 10.1021/acs.analchem.6b02531. [DOI] [PubMed] [Google Scholar]
- Wu Q., Sun Y., Zhang D., Li S., Zhang Y., Ma P., Yu Y., Wang X. Biosens. Bioelectron. 2017;96:288–293. doi: 10.1016/j.bios.2017.05.023. [DOI] [PubMed] [Google Scholar]
- Wu J., Cropek D.M., West A.C., Banta S. Anal. Chem. 2010;82:8235–8243. doi: 10.1021/ac101657h. [DOI] [PubMed] [Google Scholar]
- Xianyu Y.L., Wu J., Chen Y.P., Zheng W.S., Xie M.X., Jiang X.Y. Angew. Chem. Int. Ed. 2018;57:7503–7507. doi: 10.1002/anie.201801815. [DOI] [PubMed] [Google Scholar]
- Yuan L., Lin W.Y., Xie Y.N., Chen B., Zhu S.S. J. Am. Chem. Soc. 2012;134:1305–1315. doi: 10.1021/ja2100577. [DOI] [PubMed] [Google Scholar]
- Zhou W., Liu M.M., Liu Z.C., Yang T. J. Am. Chem. Soc. 2020;142:7532–7541. doi: 10.1021/jacs.0c00771. [DOI] [PubMed] [Google Scholar]
- Zhu A., Qu Q., Shao X., Kong B., Tian Y. Angew. Chem. Int. Ed. 2012;51:7185–7189. doi: 10.1002/anie.201109089. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Schmid Y.R.F., Luginbuhl S., Wang Q., Dittrich P.S., Walde P. Anal. Chem. 2017;89:5484–5493. doi: 10.1021/acs.analchem.7b00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wang S., Lu S., Liu G., Sun J., Yang X. Anal. Chem. 2019;91:7828–7834. doi: 10.1021/acs.analchem.9b01553. [DOI] [PubMed] [Google Scholar]
- Zuo J., Zhao X., Ju X., Qiu S., Hu W., Fan T., Zhang J. Electroanalysis. 2016;28:2044–2049. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.