Abstract
Transcriptional activators are believed to work in part by recruiting general transcription factors, such as TATA-binding protein (TBP) and the RNA polymerase II holoenzyme. Activation domains also contribute to remodeling of chromatin in vivo. To determine whether these two activities represent distinct functions of activation domains, we have examined transcriptional activation and chromatin remodeling accompanying artificial recruitment of TBP in yeast (Saccharomyces cerevisiae). We measured transcription of reporter genes with defined chromatin structure by artificial recruitment of TBP and found that a reporter gene whose TATA element was relatively accessible could be activated by artificially recruited TBP, whereas two promoters, GAL10 and CHA1, that have accessible activator binding sites, but nucleosomal TATA elements, could not. A third reporter gene containing the HIS4 promoter could be activated by GAL4-TBP only when a RAP1 binding site was present, although RAP1 alone could not activate the reporter, suggesting that RAP1 was needed to open the chromatin structure to allow activation. Consistent with this interpretation, artificially recruited TBP was unable to perturb nucleosome positioning via a nucleosomal binding site, in contrast to a true activator such as GAL4, or to perturb the TATA-containing nucleosome at the CHA1 promoter. Finally, we show that activation of the GAL10 promoter by GAL4, which requires chromatin remodeling, can occur even in swi gcn5 yeast, implying that remodeling pathways independent of GCN5, the SWI-SNF complex, and TFIID can operate during transcriptional activation in vivo.
Transcriptional activators are thought to stimulate transcription of TATA-containing promoters in part by recruiting TFIID, a multiprotein complex consisting of the TATA-binding protein (TBP) and TBP-associated factors (TAFs), to the TATA box (25, 60). Several in vitro and in vivo studies support this model. For example, transcription initiated at a mutated TATA element by induced synthesis of TBP with altered specificity was enhanced in both rate and extent in the presence of an activator that could bind upstream of the relevant promoter, consistent with activator-mediated recruitment (40). Recruitment has also been inferred from the results of “activator bypass” experiments in which artificial recruitment of TBP to promoter sites near the TATA element resulted in transcriptional activation, implying that TBP recruitment is a rate-limiting step in transcriptional activation in vivo (10, 39, 79). Most convincingly, chromatin immunoprecipitation experiments revealed TBP to be physically associated with promoters of numerous genes under activated, but not nonactivated, conditions, implying that recruitment accompanies activation (43, 46).
A potential obstacle to recruitment of TBP in vivo is posed by chromatin. Binding of TBP to nucleosomal TATA elements is greatly impeded in vitro (23, 32). Transcription is also strongly repressed by chromatin, but this repression can be largely alleviated by binding of activators (77). Genetic studies with yeast (Saccharomyces cerevisiae) support the contention that nucleosomes can also inhibit transcription in vivo, since repression of histone H4 expression results in nucleosome loss and the induction of nonactivated TATA-containing promoters (17, 29). Furthermore, the TATA elements of several yeast promoters (GAL10, PHO5, ADH2, and CHA1) are contained in nucleosomes when repressed that are disrupted upon gene activation (1, 4, 47, 51, 73), and activator-mediated chromatin remodeling has been observed even outside the context of natural promoters (53, 67). The picture that emerges from these studies is one in which activators recruit TBP (and/or perhaps other components of the general transcriptional machinery) and simultaneously alter chromatin structure to facilitate access to the promoter.
The mechanism by which activators alter chromatin structure to allow transcription factor access is a topic of great current interest. One possibility is that activators recruit any of various candidate chromatin remodeling complexes, which in turn alter local chromatin structure. This local remodeling could occur via histone acetylation following recruitment of GCN5 in the SAGA complex (72) or by alteration of histone-DNA interactions by the SWI-SNF complex, among other possibilities (14, 45, 55, 56, 65, 83). In promoters that have a nucleosome positioned in close proximity to or incorporating the TATA element, such chromatin remodeling might be required to make the TATA element accessible to TFIID.
Consistent with a role for SWI-SNF in remodeling local chromatin structure to allow access by general transcription factors, we and others have presented evidence for SWI-SNF participation in transcriptional activation at a step subsequent to activator binding (7, 27, 65, 69). Specifically, we found that a MEL1 reporter gene transcribed from the GAL10 promoter, which has a nucleosomal TATA element, showed a greater dependence on SWI-SNF for activation than a GAL4-CYC1-lacZ reporter, which has a relatively accessible TATA element (65). Additionally, activation of GAL10-MEL1 induced by artificial recruitment of the RNA polymerase II holoenzyme via a GAL4-GAL11 fusion protein was almost completely dependent on SWI-SNF (65). Since the GAL4 binding sites in the GAL10 promoter are in a nonnucleosomal region (9, 47), we suggested that SWI-SNF exerts its effect at a step subsequent to activator binding to facilitate a required remodeling of chromatin at the GAL10 promoter, possibly at the TATA element. Transcription of GAL10-MEL1 induced by classical activation domains, such as that of GAL4, was not completely SWI-SNF dependent (65). This suggests that additional SWI-SNF-independent chromatin remodeling activities could also be recruited by activators such as GAL4 to the GAL10 promoter. Although in vitro studies demonstrate that TFIID and TBP cannot bind to a TATA element positioned in a nucleosome (23, 32), there is evidence that TAFII250 (and the yeast homolog TAFII130) has histone acetyltransferase activity (50). Thus, it is conceivable that recruitment of TBP and the associated TAFs by classical activation domains allows sufficient local chromatin remodeling to allow transcriptional activation. In this paper, we have tested this possibility by examining the ability of artificially recruited TBP to activate transcription of promoters having a defined chromatin structure and to remodel chromatin.
MATERIALS AND METHODS
Plasmids.
For the experiment shown in Fig. 2A, we employed the expression vector for GAL4-yTBP of Xiao et al. (79) (a generous gift of J. Lis). For other experiments with GAL4-TBP, we constructed a GAL4-TBP expression plasmid as follows. A GAL4-GAL11 expression plasmid (65) was digested with PvuII to remove sequences encompassing the ADH1 promoter, the GAL4 DNA-binding domain (amino acids 1 to 93), and GAL11(799–1081). This fragment was subcloned into pRS412 (8) to generate pRS412GAL4-GAL11. Yeast TBP (yTBP) was amplified by PCR with the primers 5′-TTTTTAATTGAGCTCGATGAGGAACGTTTAAAG G-3′ and 5′-GAATTCGAGCTCACATAATTTCGGCATGTCATCACC-3′, and the PCR product was digested with SacI (underlined). pRS412GAL4-GAL11 was digested with SacI to remove GAL11 sequences, and the SacI-digested PCR product containing TBP sequences was ligated to generate pRS412GAL4-TBP. Clones with the correct orientation of TBP were identified by restriction analysis. Comparisons between pRS412GAL4-TBP and the construct of Xiao et al. (79), which differs in that the first 11 amino acid residues of yTBP are replaced by the first 45 residues from Drosophila melanogaster TBP, showed that the two fusion proteins behaved indistinguishably (M. P. Ryan and R. H. Morse, unpublished results).
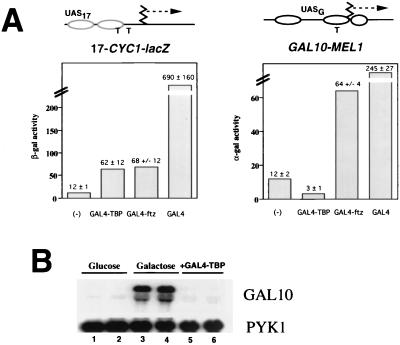
FIG. 2.
Comparison of transcriptional activation by activators and by artificial recruitment of TBP. (A) Transcriptional activation of the two reporter genes indicated was measured (strain CY296) for at least three independent clones for each sample. Averages with standard errors are given at the top of each column. The chromatin structure determined for each reporter gene (65) is indicated; the ellipses represent positioned nucleosomes; the shaded ellipses in the schematic diagram for GAL4-CYC1-lacZ (17-CYC1-lacZ) represent nucleosomes having poorly defined positions. The T's represent TATA elements. For the GAL4 measurements, cells containing either pRS416GAL4 (for GAL4-CYC1-lacZ) or pRS424GAL4 (for GAL10-MEL1) were grown in medium containing 2% galactose. (B) GAL10 message was compared to that of a PYK1 control by Northern analysis of RNA isolated from yeast cells (strain PKY999) grown in glucose (lanes 1 and 2) or galactose (lanes 3 and 4) or for cells expressing GAL4-TBP (lanes 5 and 6).
To construct an expression plasmid for LexA-TBP, TBP was amplified by PCR with the primers 5′-GAATTCGACCTCACATAATTTCGGCATGTCATCACC-3′ and 5′-TTTTTAACCATGGCCGATGAGGAACGTTTAAAGG-3′. The PCR product was digested with NcoI (underlined) and BamHI (present in the TBP sequence) and cloned into pEG202-94ATT (24). LexA-TBP sequences were excised with BamHI and SphI. pRS412GAL4-TBP was digested with BamHI and SphI to remove GAL4-TBP sequences, and the LexA-TBP sequences were ligated to generate pRS412LexA-TBP.
The reporter plasmids pBM150SKMEL1 and 314–17Δ80lacZΔNco, containing the reporter genes GAL10-MEL1 and GAL4-CYC1-lacZ, respectively, were previously described (65), as were expression plasmids for GAL4-ER-VP16 (67), GAL4-ftz (19), LexA-GAL4 (30), LexA-SNF2 (44), and GAL4-GCN4 (82).
To construct the CHA1-LexA-MEL1 reporter gene, the yeast CHA1 promoter was amplified by PCR with primers 5′-ATAATTTCTGGATCCATTAATCGATGTGTCCTTGTTTCC-3′ and 5′-CAATCAAGAGAATTCCTGTCTCTTGTCTATCCAGCAC-3′, and the product was digested with BamHI and EcoRI (underlined) to yield a fragment encompassing from −24 to −407 in the CHA1 promoter with respect to the initiating ATG. This fragment was then used to replace the EcoRI-BamHI fragment encompassing the GAL10 promoter of pBM150SKMEL1 (65) to yield a CEN-containing, URA3-marked plasmid carrying the CHA1-MEL1 reporter gene. After the serine inducibility of this reporter gene had been verified, a LexA binding site was introduced into the HindIII site at −154 relative to the initiating ATG. The oligonucleotide 5′-AGCTGGTACTGTATGTACATACAGTACC-3′ was kinased and annealed, and the DNA was ligated to HindIII-digested pBM150CHA1MEL1, thereby destroying the HindIII site and introducing a 24-bp LexA binding site (underlined) to generate the plasmid pBM150CHA1LexAMEL1.
Cell growth, yeast transformation, and α- and β-galactosidase assays.
Yeast cells (see Table 1 for yeast strains used in this study) were grown at 30°C in complete synthetic dropout medium (Bio 101) containing 2% glucose unless otherwise indicated. Transformations were performed by a standard method (31), and α- and β-galactosidase assays were performed as described previously (65). For induction of the CHA1 promoter, cells were grown in medium including 1 mg of serine per ml for 4 to 5 h; the results did not differ when serine was included during overnight growth.
TABLE 1.
List of S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| tsTBP | MATa ura3-52 trp1 ade2-101 tbp (YCplac22-TBPts1) | 13 |
| wtTBP | MATa ura3-52 trp1-ade2-101 tbp (YC86-TBP) | 13 |
| YNN281 | MATa trp1 Δhis3-Δ200 ura3-52 lys-801a ade2-10 | 64 |
| YNN282 | MATα trp1 Δhis3-Δ200 ura3-52 lys-801a ade2-10 | 64 |
| PKY999 | MATα ade2-101 arg4-1 his3-Δ200 leu2-3,112 lys2-801 trp1-901 ura3-52 thr tyr hhf1:HIS3 hhf2:LEU2 [pUK499(HHF2)] | 35 |
| CY296 | MATa gal4Δ::LEU2 lys2-801 ade2-101 leu2-Δ1 his3-Δ200 ura3-Δ99 trp1-Δ99 | 65 |
| LYY11 | MATα gcn4-2 bas1-2 bas2-2 ura3-52 leu2-3,112 with a GAL4 site replacing the strong GCN4 site at HIS4 | 82 |
| LYY13 | Same as LYY11, but with a mutated RAP1 site at HIS4 | 82 |
| FY1312 | MATα leu2Δ1 trp1Δ63 ura3-52 lys2-173R2 snf2Δ::LEU2 | 63 |
| FY1352 | MATa leu2Δ1 his3Δ200 ura3-52 lys2-173R2 snf2Δ::LEU2 gcn5Δ::HIS3 | 63 |
| FY1353 | MATα leu2Δ1 his3Δ200 ura3-52 lys2-173R2 | 63 |
| FY1354 | MATα leu2Δ1 his3Δ200 ura3-52 lys2-173R2 gcn5Δ::HIS3 | 63 |
Plasmid TALS was introduced into yeast as previously described (64, 67). To generate yeast cells having GAL4-TBP as their only source of TBP (see Fig. 1), yeast cells expressing a temperature-sensitive TBP (tsTBP) (Table 1) were transformed with pRS412GAL4-TBP, and the cells were plated onto complete synthetic medium lacking adenine (CSM-Ade). Transformants were grown in liquid CSM-Ade for several days with dilution of an aliquot of cells into fresh medium each day, and plated onto CSM-Ade plates. Colonies were patched onto CSM-Ade and replica plated onto CSM-Ade and CSM-Ade-Trp plates to identify cells that had lost the tsTBP plasmid but retained the GAL4-TBP plasmid.
FIG. 1.
GAL4-TBP retains TBP function. (A) Yeast cells containing only wtTBP, tsTBP, or GAL4-TBP (see Materials and Methods) were incubated on rich medium for 2 days at either 30 or 37°C, as indicated. (B) Western blot against protein extracts from strains expressing wtTBP (WT), tsTBP, or GAL4-TBP, as indicated, using antibody against TBP (upper panel) or tubulin (lower panel). (C) Yeast cells containing only wtTBP, tsTBP, or GAL4-TBP (two independent clones) were grown in liquid culture (YPD medium) at 30°C for 10 h, placed in a 37°C water bath for 5 min, and then grown at 37°C, and the OD was measured at the time points shown.
Chromatin structure and topoisomer analysis.
Analysis of chromatin structure by the indirect end-label technique was done as described previously (5). TALS chromatin was probed by using a 200-bp fragment extending counterclockwise from an EcoRV site that is 782 bp clockwise from the center of the GAL4 binding site (see Fig. 5A). For analysis of the CHA1-LexA-MEL1 reporter plasmid under induced conditions, 1 mg of serine per ml was included during chromatin preparation at all stages. CHA1-LexA-MEL1 chromatin was probed with a 276-bp BamHI-SalI fragment from pBM150SKMEL1 (65). Topoisomer analysis was performed as described previously (52, 54). Indirect end-label analysis was repeated at least twice for each condition examined, and topoisomer analysis was repeated at least three times.
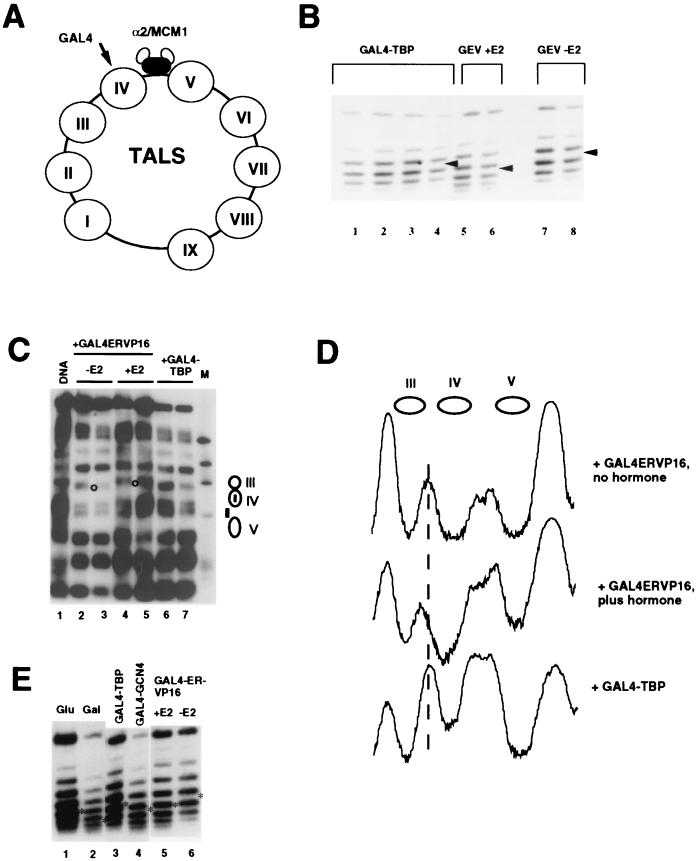
FIG. 5.
Inability of GAL4-TBP to remodel TALS chromatin. (A) Schematic diagram of the TALS episome. (B) Topology of TALS from α cells. DNA was isolated from separate clones of yeast cells (strain PKY999) harboring TALS and expressing GAL4-TBP or GAL4-ER-VP16 (GEV) in the presence or absence of 100 nM β-estradiol, as indicated. Topoisomers were resolved on a 1.5% agarose gel containing 30 μg of chloroquine diphosphate per ml and visualized by hybridization with a TALS-specific probe after blotting to a nylon membrane. The uppermost band in each lane corresponds to nicked circular plasmid, and the bands below correspond to individual topoisomers, with more rapidly migrating bands being more positively supercoiled. The Gaussian centers of the distributions are indicated by the arrowheads; lanes 1 to 4 have the same center, as do lanes 5 and 6 and lanes 7 and 8. (C) Indirect end-label analysis of TALS chromatin. MNase cleavage sites in naked DNA (lane 1) and chromatin (lanes 2 to 7) were determined relative to an EcoRV site 782 bp clockwise from the center of the GAL4 binding site. Chromatin was isolated from YNN282 cells expressing GAL4-ER-VP16 in the presence (+E2) or absence (−E2) of 100 nM β-estradiol or in the presence of GAL4-TBP, as indicated, and digested with 20 (lanes 2, 4, and 6) or 50 (lanes 3, 5, and 7) U of MNase per ml. Naked DNA was digested with 20 U of MNase per ml. The marker lane (M) contains ΦX DNA digested with HaeIII. The positions of nucleosomes III to V are indicated by ellipses to the right, as are the GAL4 binding site in nucleosome IV and the α2-MCM1 operator (small rectangles). The open circles between lanes 2 and 3 and 4 and 5 indicate cleavage sites differing in the presence and absence of hormone when GAL4-ER-VP16 is expressed. (D) Densitometric scans of regions from lanes 2, 4, and 7 in panel C. (E) Topology of TALS from a cells. DNA was prepared from YNN281 cells, fractionated and visualized as in panel B, in cells expressing the indicated GAL4 fusion proteins or in cells grown in glucose or galactose without exogenously expressed GAL4 derivatives (lanes 1 and 2). The asterisks mark the centers of the Gaussian distributions of topoisomers. Because the lanes are not shown in the same order in which they were electrophoresed on the gel, some unevenness is present in the migration of the upper, nicked circular plasmid bands; topoisomers are aligned as they were on the gel.
Northern analysis.
RNA was extracted from yeast (66), and 10 μg was run on agarose-formaldehyde gels (71). Gels were blotted onto nylon membranes in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), UV cross-linked, and hybridized (11) with probes labeled by random priming. Blots were stripped by boiling membranes in a mixture of 0.015 M NaCl, 0.1× SSC, and 1% sodium dodecyl sulfate (SDS) prior to hybridization with another probe. Northern blots were quantitated by using scanned images on a Molecular Dynamics PhosphorImager.
Western analysis.
Western analysis was performed with whole-cell extracts. Two optical density at 600 nm (OD600) units of cells (A600 = 0.5 to 2) was added to 2 ml of 50 mM Tris (pH 7.5)–10 mM NaN3 on ice, spun down, resuspended in 30 ml of ESB (2% SDS, 80 mM Tris [pH 6.8], 10% glycerol, 1.5% dithiothreitol, 0.1 mg of bromphenol blue per ml) with protease inhibitors (leupeptin and pepstatin A at 1 μg/ml and phenylmethyl sulfonyl fluoride at 1 mM) and quickly transferred to microcentrifuge tubes for a 3-min incubation at 100°C. Glass beads were added to reach the meniscus, and the samples were vortexed at top speed for 2 min. An additional 70 μl of ESB was added, and the samples were heated to 100°C for 1 min. Following standard SDS-polyacrylamide gel electrophoresis the proteins were electroblotted to Millipore polyvinylidene difluoride membrane. The blots were treated with 5% blocking solution from Amersham's enhanced chemiluminescence ECL kit or 5% powdered milk in TBS-T (20 mM Tris [pH 7.6], 137 mM NaCl, 0.1% Tween 20) and incubated for about 1 h with primary antibody directed against the N terminus of yTBP (a generous gift of Steve Buratowski). Following washing, the blots were incubated for about 1 h with the secondary antibody, horseradish peroxidase-linked antirabbit immunoglobulin, and then developed as per the manufacturer's instructions. After decay of the chemiluminescence, the blots were reprobed with monoclonal antitubulin antibodies (a gift from M. Joan Curcio), followed by peroxidase-linked antimouse immunoglobulin and again developed according to the manufacturer's instructions.
RESULTS
Retention of TBP function in a GAL4-TBP fusion.
To test whether TFIID recruitment aids in transcriptional activation by perturbing chromatin structure, a GAL4-TBP fusion protein, consisting of the GAL4 DNA-binding domain and full-length yTBP, was used in conjunction with reporters for transcription and/or chromatin structure perturbation containing GAL4 sites. To confirm that the TBP moiety of GAL4-TBP was functional, a GAL4-TBP expression vector was introduced into yeast expressing a tsTBP from a TRP1-marked plasmid (kindly provided by K. Struhl) (13) and lacking wild-type TBP (wtTBP). Growth of the resulting transformants in medium containing tryptophan resulted in loss of the plasmid expressing the tsTBP, demonstrating that GAL4-TBP can fulfill TBP functions needed for growth (Fig. 1A). Western blotting of extracts from cells depending on GAL4-TBP for growth, using antibody against yTBP, demonstrated loss of wtTBP and the presence of a new protein containing the TBP epitope migrating at the predicted size for the GAL4-TBP fusion protein (Fig. 1B). Yeast cells expressing GAL4-TBP grew as well as those expressing wtTBP at 30 and 37°C (the nonpermissive temperature for tsTBP) (Fig. 1C), indicating that fusing TBP to the GAL4 DNA-binding domain does not appreciably impair TBP function. Since TFIID is probably required for transcription at most polymerase II promoters in yeast (41), this further implies that TAFs are normally associated with GAL4-TBP, as has been found for native yTBP (62).
Transcriptional activation by GAL4-TBP.
To examine transcriptional activation by GAL4-TBP, we employed two plasmid-based reporter genes whose chromatin structure we had previously characterized (65). One of these reporter genes, GAL10-MEL1, contains the MEL1 coding sequence fused to the GAL10 promoter. As in the endogenous GAL10 gene, the GAL4 binding sites are nonnucleosomal and the TATA element is positioned within a nucleosome (9, 47, 65). The second reporter gene, GAL4-CYC1-lacZ, contains a single GAL4 binding site upstream of the CYC1 promoter and the lacZ reporter gene. In contrast to GAL10-MEL1, the TATA elements in the CYC1 promoter are relatively accessible (65). We reasoned that if TFIID plays an important role in chromatin remodeling, GAL4-TBP would be expected to activate transcription regardless of the chromatin structure at the TATA element. If, on the other hand, transcriptional activators provide chromatin remodeling capabilities that TFIID cannot fulfill, then the two reporter genes might respond differently to activation by GAL4-TBP.
Activation of GAL10-MEL1 and GAL4-CYC1-lacZ was assessed by measuring α-galactosidase (the MEL1 gene product) and β-galactosidase activity colorimetrically (Fig. 2A). GAL4-TBP weakly activated GAL4-CYC1-lacZ. Although β-galactosidase activity was low compared to that observed with GAL4, GAL4-TBP induced transcription of GAL4-CYC1-lacZ at levels similar to those of the weak activator GAL4-ftz (Fig. 2A) and a “mini-GAL4” comprising amino acids 1 to 100 plus 840 to 857 of GAL4 (78; Ryan and Morse, unpublished). In contrast to GAL4-CYC1-lacZ, GAL10-MEL1 was not activated by GAL4-TBP, although both the weak activators GAL4-ftz (Fig. 2A) and GAL4(1–100 + 840–857) (Ryan and Morse, unpublished) showed significant activation. These results were obtained by using the GAL4-TBP expression vector of Xiao et al. (79) in strain CY296; essentially identical results were obtained in the yeast strain PKY999 (35) by using the expression vector for GAL4-TBP we constructed ourselves, which was used in Fig. 1 (Ryan and Morse, unpublished). GAL4-TBP also failed to support growth of gal4− yeast (strain CY296) in galactose (Ryan and Morse, unpublished), and Northern analysis confirmed that the endogenous GAL10 gene was not induced in cells expressing GAL4-TBP (Fig. 2B) (essentially identical results were obtained with strain CY296 harboring expression vectors for GAL4 or GAL4-TBP). In contrast, both GAL4 and GAL4-ftz allowed gal4− yeast to grow in galactose when the proteins were expressed from plasmids (Ryan and Morse, unpublished). The differential ability of GAL4-TBP to activate these two promoters cannot easily be explained by distance effects, because the nearest of the four GAL4 binding sites in the GAL10 promoter is only 105 bp from the TATA element, while the GAL4 binding site in GAL4-CYC1-lacZ is 120 bp from the nearest CYC1 TATA element.
These results demonstrate that activation of transcription by TBP recruitment is promoter dependent and suggest that the ability to achieve such activation may depend on the chromatin structure of the promoter. More specifically, the data suggest that if the TATA element is accessible, at least some transcriptional activity can be induced via artificial recruitment of TBP, whereas a nucleosomal TATA element may prevent such activation. To test this idea further, we turned to the yeast CHA1 promoter. The CHA1 gene is activated in the presence of serine or threonine and has been reported to have a nucleosomal TATA element when not activated and an accessible upstream activation site that is constitutively bound by the activator CHA4 (51). We constructed a CHA1-MEL1 reporter gene by fusing a 384-bp fragment from the CHA1 promoter (−407 to −24 with respect to the initiating ATG codon) to the MEL1 coding sequence on a URA3-marked, CEN-containing plasmid. After determining that the fusion gene retained the serine inducibility of the native CHA1 gene, we inserted a LexA binding site at the HindIII site that lies just upstream of the TATA-containing nucleosome in the native CHA1 promoter. We then transformed the resulting plasmid into yeast cells alone or with expression vectors for LexA-TBP, LexAGAL4, or LexASNF2.
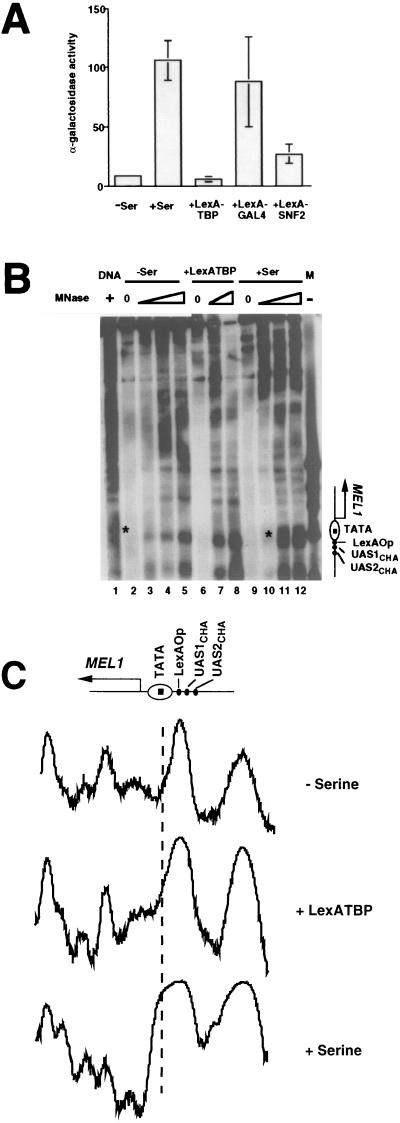
Measurement of α-galactosidase activity in the resulting transformants showed that the CHA1-LexA-MEL1 reporter gene showed little transcriptional activity in its uninduced state and substantial activity in the presence of 1 mg of serine per ml (Fig. 3A). In accord with prediction, expression of LexATBP gave no activation of the reporter gene over its uninduced level. Control experiments showed that LexATBP expression allowed significant activation of a reporter gene having four LexA sites upstream of a CYC1-lacZ reporter gene (Ryan and Morse, unpublished) and that CHA1-LexA-MEL1 is strongly activated by LexAGAL4 and less strongly activated by LexASNF2 (Fig. 3A). The chromatin structure of the CHA1 promoter was examined by digesting chromatin and naked DNA with various concentrations of micrococcal nuclease (MNase); MNase cleavage sites were mapped relative to a SalI site 5′ of the CHA1 promoter. Previous examination of the chromatin structure of the native CHA1 promoter showed changes in cleavage by MNase and DNase I upon induction, especially near the TATA element, suggestive of disruption of a positioned nucleosome, although naked DNA controls were absent (51). Our data are consistent with such an assignment, because an MNase cleavage site near the TATA element present in naked DNA is protected in the uninduced reporter gene (Fig. 3B, lanes 1 to 5; see the band marked by an asterisk in lane 1). This site is cleaved when the reporter gene is induced by serine (Fig. 3B, lanes 11 and 12, asterisk), but not in the presence of LexATBP (lanes 7 and 8). Densitometric scans of lanes 4, 7, and 11 are shown in Fig. 3C. In addition to the cleavage near the TATA element that is specific to the induced (with serine) state, other differences in relative intensity can be seen between the samples with and without serine. The cleavage pattern seen in the presence of LexATBP most resembles that of the uninduced state. Consistent with previous work (51), the site at which the LexA operator was inserted maps to the edge of the putative TATA-containing nucleosome, a region very accessible to MNase. Thus, although the LexA site should be accessible to LexATBP, LexATBP is unable to perturb the nucleosome containing the CHA1 TATA element and transcription remains uninduced.
FIG. 3.
Inability of artificially recruited TBP to activate transcription or remodel chromatin at the CHA1 promoter. (A) α-Galactosidase activity was measured in YNN282 cells from a CHA1-LexA-MEL1 reporter gene, carried on a CEN-containing plasmid, under uninduced (−Ser) or induced (+Ser) conditions or in the presence of the indicated LexA fusion proteins. At least three independent determinations were made for each activity; the standard error was too small to be visible for the uninduced sample. (B) Indirect end-label analysis of the CHA1-LexA-MEL1 reporter gene. MNase cleavage sites in naked DNA (lane 1) and chromatin (lanes 2 to 12) were determined relative to a SalI site 623 bp 5′ of the CHA1 TATA element. The locations of some salient features of the modified CHA1 promoter are indicated to the right, and the asterisks next to lanes 1 and 11 indicate a cleavage site present in naked DNA and under induced conditions only. Chromatin was isolated from YNN282 cells under uninduced (−Ser) or induced (+Ser) conditions or in the presence of LexATBP, as indicated, and digested by using 5 (lanes 3, 7, and 10), 20 (lanes 4, 8, and 11), or 50 (lanes 5 and 12) U of MNase per ml. Naked DNA was digested with 4 U of MNase per ml. Controls lacking MNase are indicated, and the marker lane (M) contains ΦX DNA digested with HaeIII. (C) Densitometric traces from lanes 4, 7, and 11 of panel B. The dashed line indicates the approximate location of the TATA element, which is depicted schematically, along with other genetic landmarks, at the top.
To test further the effect of chromatin structure on the ability of artificially recruited TBP to activate transcription, we took advantage of recent findings from our laboratory regarding activation of the HIS4 promoter (82). Activation of this promoter by GCN4 requires a RAP1 binding site (15), although RAP1 cannot activate HIS4 by itself; rather, RAP1 appears to function at HIS4 to open the chromatin to allow access by GCN4 (15, 82). Chromatin opening by RAP1 is also needed for HIS4 activation by GAL4 at a weak binding site, but not at a strong binding site (82). Since GAL4 has a fairly strong ability to access nucleosomal binding sites and to remodel chromatin (5, 53, 67, 68, 81) whereas GAL4-TBP does not (Fig. 3B and see below), we asked whether activation by GAL4-TBP of a modified HIS4 promoter containing a strong GAL4 binding site would require help from RAP1.
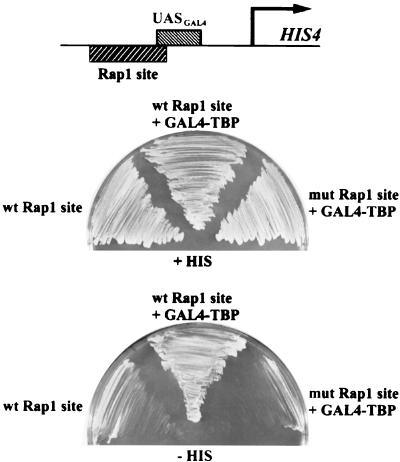
Using a modified HIS4 promoter having a GAL4 site replacing the GCN4 binding site, we found that RAP1 alone could not support robust growth on plates containing synthetic medium lacking histidine (CSM-His), in agreement with previous results (Fig. 4) (82). Expression of GAL4-TBP in yeast having a mutated RAP1 site in the modified HIS4 promoter also did not support growth (Fig. 4), in contrast to the strong activator GAL4, which can support growth on CSM-His media in the same cells (82). Thus, TBP is unable to provide some function that is provided by a true activation domain, such as that of GAL4, and that is required for transcriptional activation at the modified HIS4 promoter lacking a RAP1 binding site. In contrast, when GAL4-TBP was expressed in cells having the wild-type RAP1 site at the modified HIS4 promoter, growth on CSM-His was restored (Fig. 4), indicating that artificial recruitment of TBP to the promoter is sufficient to activate HIS4 transcription when RAP1 can also bind the promoter and strongly suggesting that the function provided by a true activation domain, but not by TBP, is chromatin opening. These data therefore suggest that two functions, chromatin opening and recruitment of the general transcriptional machinery, are required to achieve transcriptional activation. Similarly, RAP1 is required for transcription of HIS4 independent of BAS1, BAS2, and GCN4 in sit mutants, and sit1 and sit2 encode mutant alleles of the two largest subunits of RNA polymerase II (15). Thus, chromatin opening by RAP1 is likely also necessary for activator-independent transcription of HIS4 by these mutant alleles of RNA polymerase II. The finding that chromatin opening and recruitment of the general transcription machinery can be separated in the contrived circumstance of the experiment just described is consistent with the possibility that they may occur by distinct avenues even when both functions are provided by a single classical activation domain (20).
FIG. 4.
Activation of a modified HIS4 gene by GAL4-TBP requires a RAP1 binding site. (Top) structure of the modified HIS4 gene in which the major GCN4 binding site is replaced by a strong GAL4 binding site. Yeast cells containing the modified HIS4 gene with either a wild-type (wt) or mutant (mut) RAP1 binding site (strains LYY11 and LYY13; see Table 1) were grown for 2 days at 30°C on plates containing or lacking histidine, in the presence or absence of GAL4-TBP, as indicated.
Failure of GAL4-TBP to remodel chromatin.
As a further test of the ability of GAL4-TBP to remodel chromatin, we used the TALS minichromosome, which has been useful for assessing activator-dependent perturbations in chromatin (Fig. 5A). TALS contains a single GAL4 binding site derived from the GAL3 promoter near an α2-MCM1 operator (64, 67). When TALS is introduced into yeast α cells, the episome is packaged into nucleosomes which are strongly positioned by the α2-MCM1 complex (64). The GAL4 binding site is about 40 bp from the edge of a nucleosome adjacent to the α2-MCM1 operator, and this nucleosome is strongly perturbed in the presence of GAL4 and GAL4-ER-VP16 plus β-estradiol, but not by nonactivating derivatives of GAL4 (67, 68). One criterion for assessing this perturbation is a change in the topology of TALS DNA. Because each nucleosome introduces one negative supercoil into closed circular DNA, perturbations in chromatin structure that lead to the loss of one nucleosome result in the loss of one negative supercoil and a change in plasmid topology.
To examine whether GAL4-TBP can remodel chromatin, TALS topology was examined in yeast cells containing either GAL4-TBP or the chimeric activator GAL4-ER-VP16, whose ability to activate transcription in yeast is estrogen dependent. DNA was rapidly isolated from yeast to preserve the in vivo topology and electrophoresed on chloroquine-containing gels to resolve individual topoisomers, and TALS DNA was visualized by Southern blotting and hybridization (Fig. 5B). Individual bands represent distinct topoisomers of the plasmid, with the fastest-migrating band being the most positively supercoiled molecule under the conditions used; the band at the top of each lane corresponds to nicked circular molecules. These populations of topoisomers are present as Gaussian distributions whose centers can be precisely assessed (52, 54), and changes in topology caused by perturbation of chromatin structure are evident on the gel by a shift in the center of distribution of the topoisomers. As observed previously, the TALS minichromosome loses an average of 0.7 negative supercoil in the presence of GAL4-ER-VP16 and 100 nM β-estradiol, consistent with loss of one nucleosome from most of the plasmid molecules (5, 65, 67, 68) (Fig. 5B). In contrast, in cells expressing GAL4-TBP, TALS shows slightly more negative supercoiling than in the presence of GAL4-ER-VP16 without hormone (Fig. 5B). Since GAL4-ER-VP16 causes a loss of 0.2 negative supercoil per TALS molecule in the absence of hormone (65, 67, 68), we conclude that TALS topology is unaffected by GAL4-TBP. MNase cleavage in TALS chromatin is also unaffected by GAL4-TBP (Fig. 5C). Whereas a characteristic shift in the MNase cleavage site between nucleosomes IV and III (Fig. 5C, see lanes 2 and 3 and 4 and 5; see also scans in panel D) is seen upon addition of β-estradiol in the presence of GAL4-ER-VP16, no such changes are seen in the presence of GAL4-TBP (lanes 6 and 7).
Although the above experiments provided a strong indication that GAL4-TBP is ineffective at remodeling chromatin, it seemed possible that the α2-MCM1 complex, which helps to dictate nucleosome positioning on the TALS plasmid in yeast α cells, might play a part in preventing remodeling via TBP. The α2-MCM1 repressor inhibits transcription in yeast, probably by both chromatin-mediated and chromatin-independent mechanisms (12, 74), and the latter could conceivably involve blocking TBP function. We therefore also examined the plasmid topology of TALS in yeast a cells. Although nucleosome positioning is weaker in TALS in a than in α cells, it is still nonrandom (38; Ryan and Morse, unpublished), and changes in topology slightly larger than those seen in α cells are seen in the presence of GAL4 or GAL4-ER-VP16 plus hormone (Fig. 5E). When GAL4-TBP was introduced into yeast a cells harboring TALS, no change in topology was observed (Fig. 5E). We conclude that GAL4-TBP has little if any ability to remodel chromatin via nucleosomal GAL4 binding sites.
Transcriptional activation at the GAL10 promoter in gcn5 swi yeast cells.
Previously, we found that activation of the GAL10-MEL1 reporter by GAL4-GAL11 was almost completely abolished in swi1Δ yeast cells, implying that chromatin remodeling is required for activation of this reporter gene. Activation of GAL10-MEL1 by GAL4 is reduced substantially, but not completely, in swi1Δ yeast, suggesting that GAL4 can induce remodeling of GAL10-MEL1 by a pathway independent of SWI-SNF. The data presented so far suggest that recruitment of TFIID is not sufficient to allow remodeling and transcriptional activation of GAL10-MEL1, and so some other chromatin remodeling activity must be recruited. One obvious possibility is the SAGA complex. This complex contains ADA2, which can interact with the GAL4 activation domain in vitro (49), and GCN5, which can acetylate the histone H3 and H4 amino termini, possibly altering chromatin structure in a way that facilitates transcription factor access (26, 42). This idea is buttressed by studies showing that recruitment of the SAGA complex by the VP16 and GCN4 activation domains can allow transcriptional activation of nucleosomal templates in vitro (72).
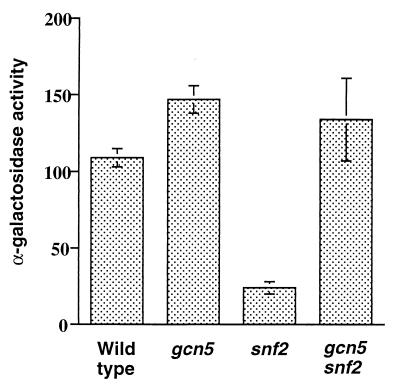
We examined activation of the GAL10-MEL1 reporter gene by GAL4 in gcn5Δ yeast grown in galactose medium and found a reproducible slight increase in transcriptional activity (Fig. 6). However, these cells retain the SWI-SNF complex, which could be redundant with GCN5 at this promoter. We therefore examined GAL10-MEL1 activation by GAL4 in snf2 gcn5 cells (Fig. 6). Transcriptional activation by GAL4 was significantly decreased in snf2 cells, consistent with our results obtained with swi1Δ yeast cells. Surprisingly, transcription was increased again, approximately to wild-type levels, in the double mutant. We conclude that the hypothesized remodeling activity recruited by GAL4 in swi− yeast cells is not the SAGA complex (or the ADA-GCN complex).
FIG. 6.
Transcriptional activity of GAL10-MEL1 in wild-type, gcn5, snf2, and gcn5 snf2 yeast cells (strains FY1314, FY1352, FY1353, and FY1354) (Table 1). α-Galactosidase activity was measured from strains harboring the indicated gene disruptions after growth in raffinose-plus-galactose medium. Each value was derived from at least three independent determinations, and standard errors are indicated.
DISCUSSION
Transcription in eukaryotes is often initiated by activator proteins that bind to upstream sites in promoters, and a strong case, built on an impressive wealth of evidence, has been made that these activators use protein-protein interactions to recruit components of the transcriptional apparatus (6, 18, 21, 43, 46, 60). However, discussions of the recruitment model have not generally explicitly considered how this mechanism allows repressive effects of chromatin to be overcome. Two basic possibilities can be imagined: either recruitment of general transcription factors somehow suffices to overcome repression by chromatin, or proteins distinct from the GTFs are also recruited that alter chromatin structure to allow transcription.
Transcriptional activators can perturb nearby chromatin from both nucleosomal and nonnucleosomal binding sites, and this perturbation is not seen with nonactivating derivatives (67, 70). We have tested whether TBP can mimic an activation domain in this regard by examining whether GAL4-TBP and LexA-TBP fusion proteins can remodel chromatin via nucleosomal or nonnucleosomal GAL4 or LexA binding sites and whether such fusion proteins can activate promoters that require chromatin remodeling for activated transcription. We find that a GAL4-TBP fusion protein is unable to remodel chromatin via a nucleosomal GAL4 binding site in the TALS episome, in contrast to GAL4-based activators (Fig. 5). We also find that LexATBP is unable to perturb a TATA-containing nucleosome in a modified CHA1 promoter that contains a LexA binding site adjacent to this nucleosome (Fig. 3). Furthermore, both the CHA1 promoter and the GAL10 promoter, which also has a nucleosomal TATA element, are refractory to activation by artificially recruited TBP, whereas a CYC1-lacZ reporter gene having a relatively accessible TATA element can be activated by a GAL4-TBP fusion protein (Fig. 2 and 3). Finally, the HIS4 promoter can be activated by artificial recruitment of TBP only when a RAP1 site is present to allow chromatin opening by RAP1 (which does not activate HIS4 by itself) (Fig. 4).
These results suggest that in the absence of a true activator, recruitment of TBP does not allow chromatin remodeling that is needed for high levels of transcription. This remodeling may be required to allow the initial (artificial) recruitment of TBP, as is likely at the modified HIS4 promoter, or to allow access of the TBP moiety to the TATA element, as is likely at the GAL10 and modified CHA1 promoters. Although we have not directly demonstrated binding of GAL4-TBP and LexA-TBP, respectively, to these promoters, it seems likely that this occurs, given that (i) the binding sites are accessible to nucleases (9, 47, 51, 65) (Fig. 3); (ii) GAL4 can bind to the GAL1-10 promoter under nonactivating conditions (i.e., in glucose medium) (16); (iii) both LexA-TBP and GAL4-TBP can activate some promoters via LexA or GAL4 binding sites (Fig. 2 and data not shown); and (iv) GAL4-TBP can activate the modified HIS4 promoter when a RAP1 site is present, indicating that it is able to bind to a region of open chromatin (82) (Fig. 4). However, whether the failure of artificially recruited TBP is caused by the inability of the fusion protein to bind to the cognate LexA or GAL4 binding site or the inability of TBP to access the TATA element after the initial binding is largely irrelevant to the interpretations of the results. In either case, the results imply that if an activator were imagined that could recruit TBP but could not remodel chromatin, the activator would fail to activate transcription, either because of its failure to bind to its own binding site or its inability to open chromatin to allow TBP binding to the TATA element following its recruitment. GAL4-TBP is likely to be associated with TAFs to form GAL4-TFIID in the cell, based on its ability to complement tbp− yeast cells and on the finding that a majority of TBP is associated with TAFs in yeast (62). Thus, our findings suggest that in spite of the histone acetyltransferase activity of yeast TAFII130 (50), TFIID is considerably poorer at remodeling chromatin than are true activators.
Previous studies have also reported variable transcriptional activation by artificially recruited TBP at different promoters in yeast, but chromatin structure has not generally been examined (10, 22, 36, 39, 79). Similarly, experiments done in mammalian cells have been performed with transiently transfected DNA, which is not incorporated into organized chromatin in the same fashion as genomic DNA (2, 48, 57, 80). Interestingly, however, sufficient activation by artificially recruited TBP is seen at the HIS3 locus to allow growth in the presence of 3-aminotriazole (which requires HIS3 activation) (10, 36, 39), and this promoter contains a poly(dA-dT) sequence that creates an open chromatin structure (34).
Although we favor the idea that a function provided by classical activation domains that is missing in artificially recruited TBP and that is needed for efficient activation is chromatin remodeling, other explanations for the failure of GAL4-TBP (or LexA-TBP) to activate specific promoters should be considered. First, it is possible that some functions of TBP, such as interactions with particular TAFs or other proteins, are compromised in TBP fusions. We have effectively ruled out this possibility by demonstrating that yeast cells carrying GAL4-TBP as the only source of TBP are phenotypically essentially wild type. Similarly, LexA-TBP was previously shown to complement tbpΔ yeast (36). Second, it is possible that TBP is subject to negative regulation which is normally overcome by classical activation domains (28). We attempted to address this possibility by measuring GAL10-MEL1 transcription in the presence and absence of GAL4-TBP in yeast harboring the bur6-1 and mot1-301 alleles, which encode mutations in the yeast homologs of NC2-α and MOT1, respectively (59). NC2-α is a subunit of the mammalian protein complex Dr1-NC2, which inhibits interactions between TBP and TFIIA and/or TFIIB in vitro (33, 37), and MOT1 displaces TBP from TATA elements in the presence of ATP (3). Neither mutation significantly increased GAL10-MEL1 transcription in the presence of GAL4-TBP (Ryan and Morse, unpublished). However, it is possible that these regulators function redundantly or that other regulators such as the Ccr4-NOT complex are involved (28). This explanation would demand that these negative regulators somehow differentially regulate TBP at different promoters and would also require that RAP1 be capable of overcoming this negative regulation at the modified HIS4 promoter. This seems a less straightforward explanation than considerations of chromatin structure, but it cannot yet be formally ruled out.
A third possible explanation for differential activity of artificially recruited TBP (or other components of the general transcription machinery), as discussed by Gaudreau et al. (22), is that nonclassical activators such as GAL4-TBP may suffer steric constraints that classical activators can overcome by interacting with multiple targets within the general transcriptional machinery, thus affording them greater flexibility. This may well account in part for the generally low transcription levels induced by artificially recruited TBP and also for part of its differential activity at different promoters (10, 22, 36, 39, 79). However, it does not readily explain the ability of RAP1 to synergize with GAL4-TBP at the modified HIS4 promoter of Fig. 4, nor does it account for the inverse correlation observed between the requirement for chromatin remodeling and the ability of artificially recruited TBP to function at the promoters examined here. Another explanation (not mutually exclusive with the preceding) for differential activity of nonclassical activators at different promoters considered by Gaudreau et al. (22) is that some promoters may require recruitment of different components of the transcriptional machinery than others. This is entirely in accord with the interpretation offered here, if the transcription machinery is allowed to include chromatin-modifying activities, as is in fact considered in the discussion of Gaudreau et al. (22).
If an inability of artificially recruited TBP to remodel chromatin is responsible for its transcriptional inactivity at the GAL10 promoter and its low activity at most other promoters examined, it seems possible that altering chromatin structure in vivo by artificial means, in conjunction with TBP recruitment, might overcome the need for a classical activation domain. However, when we examined GAL10-MEL1 expression in a yeast strain carrying the hhf2-13 mutation in histone H4, we did not see any increase in stimulation by GAL4-TBP (Ryan and Morse, unpublished). Thus, although this sin mutation alleviates the requirement for SWI-SNF for transcriptional activation of an HO-lacZ reporter gene (58, 75) and increases accessibility of nucleosomal DNA to MNase and Escherichia coli Dam methyltransferase in yeast (75), it evidently does not sufficiently perturb the local chromatin structure of the GAL10 promoter to allow activation by GAL4-TBP. We also employed an altered GAL10-MEL1 reporter having a LexA binding site between the GAL4 binding sites and the TATA element to ask whether artificial depletion of histone H4 would allow activation by LexATBP. Yeast cells in which histone H4 gene expression is under galactose control (29) and a matched control strain were grown in galactose medium and then switched to glucose medium, and expression of the MEL1 reporter was assessed by Northern analysis. We saw no increased expression upon H4 depletion by LexATBP (Ryan and Morse, unpublished). Further work will be required to understand the extent of chromatin remodeling that is needed for promoter activation, although our results using a modified HIS4 promoter indicate that perturbation of chromatin by a single protein—in this case RAP1—can suffice in at least some instances.
Our results suggest that a normal function of activators in addition to recruitment of the general transcriptional machinery is to remodel chromatin locally to allow access by general transcriptional factors such as TBP. This could occur through recruitment of chromatin-modifying complexes such as the SWI-SNF complex, or complexes capable of modifying histones, such as the GCN5-containing SAGA complex (14, 26, 45, 55, 56, 65, 72, 83). The strong and general capability of artificially recruited GAL11 to activate transcription and to remodel the PHO5 promoter (18, 21, 22) suggests that such chromatin-modifying activity can be recruited along with the holoenzyme (76). Indeed, we have shown that activation of GAL10-MEL1 by GAL4-GAL11 is almost completely abolished in swi1Δ yeast (65). This complete dependence on SWI-SNF for activation of GAL10-MEL1 by GAL4-GAL11 implicates chromatin remodeling as a requisite step in activation of this reporter gene. Since GAL4 can still achieve modest levels of transcriptional activation of GAL10-MEL1 in swi1Δ yeast, this suggests that GAL4 is able to recruit some additional chromatin remodeling activity. Since one obvious candidate was GCN5, we examined activation of GAL10-MEL1 by GAL4 in snf2 gcn5 yeast cells. Surprisingly, transcription was substantially increased relative to that of the snf2 single mutant. This is consistent with results of Recht and Osley (61) and in contrast to those of Roberts and Winston (63); the reasons for the latter discrepancy are unclear and may be strain dependent. Regardless, these results suggest that chromatin remodeling proteins other than GCN5 or the SWI-SNF complex may participate in transcriptional activation in yeast. Interestingly, activation of the CHA1 promoter with concomitant chromatin remodeling is not affected in gcn5 or swi− yeast (51); however, double mutants have not been tested for effects on CHA1 activation.
In conclusion, our results, together with a large body of earlier work, indicate that two essential functions provided by classical activation domains are recruitment of the general transcription machinery and of chromatin remodeling activity and that these functions may be separable in some instances (20). Although this is not a revolutionary idea, it has not been strongly emphasized in discussions of activation by recruitment. The experiments presented here underscore that although recruitment of the general transcription machinery may be necessary for activated levels of transcription, it is not sufficient, and may even be completely insufficient, depending on the promoter and its chromatin structure.
ACKNOWLEDGMENTS
We are grateful to Steve Buratowski, M. Joan Curcio, Michael Grunstein, Steve Hanes, Brehon Laurent, John Lis, Gregory Prelich, Kevin Struhl, and Fred Winston for generously providing antibodies, yeast strains, and plasmids. We gratefully acknowledge the Wadsworth Center Molecular Genetics Core Facility for oligonucleotide synthesis and DNA sequencing.
This work was supported by grants from the NIH to R.H.M. (R01 GM51993) and M.P.R. (F32 GM18356).
REFERENCES
- 1.Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 3.Auble D T, Hansen K E, Meuller C G F, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 4.Axelrod J D, Reagan M S, Majors J. GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev. 1993;7:857–869. doi: 10.1101/gad.7.5.857. [DOI] [PubMed] [Google Scholar]
- 5.Balasubramanian B, Morse R H. Binding of Gal4p and Bicoid to nucleosomal sites in yeast in the absence of replication. Mol Cell Biol. 1999;19:2977–2985. doi: 10.1128/mcb.19.4.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barberis A J, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 7.Biggar S R, Crabtree G R. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Cavalli G, Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 11.Church G, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper J P, Roth S Y, Simpson R T. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- 13.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 14.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 15.Devlin C, Tice-Baldwin K, Shore D, Arndt K T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991;11:3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudley A M, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durrin L K, Mann R K, Grunstein M. Nucleosome loss activates CUP1 and HIS3 promoters to fully induced levels in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1621–1629. doi: 10.1128/mcb.12.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 19.Fitzpatrick V D, Ingles C J. The Drosophila fushi trazu polypeptide is a DNA-binding transcriptional activator in yeast cells. Nature. 1989;337:666–668. doi: 10.1038/337666a0. [DOI] [PubMed] [Google Scholar]
- 20.Fryer C J, Archer T K. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 21.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Hörz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 22.Gaudreau L, Keaveney M, Nevado J, Zaman Z, Bryant G O, Struhl K, Ptashne M. Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences. Proc Natl Acad Sci USA. 1999;96:2668–2673. doi: 10.1073/pnas.96.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godde J S, Nakatani Y, Wolffe A P. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP-TFIIA association with nucleosomal DNA. Nucleic Acids Res. 1995;23:4557–4564. doi: 10.1093/nar/23.22.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golemis E A, Serebriiskii I, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 3, Suppl. 39. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 25.Goodrich J A, Tjian R. TBP-TAF complexes: selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 26.Grant P A, Duggan L, Coté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 27.Gregory P D, Schmid A, Zavari M, Münsterkötter M, Hörz W. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 1999;18:6407–6414. doi: 10.1093/emboj/18.22.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 30.Hanes S D, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 31.Hill J, Ian K A, Donald G, Griffiths D E. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 33.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 34.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayne P S, Kim U-J, Han M, Mullen J R, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 36.Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 37.Kim T K, Zhao Y, Hui G, Bernstein R, Roeder R G. TATA-binding protein residues implicated in a functional interplay between negative cofactor 2 (Dr1) and general factors TFIIA and TFIIB. J Biol Chem. 1995;270:10976–10981. doi: 10.1074/jbc.270.18.10976. [DOI] [PubMed] [Google Scholar]
- 38.Kladde M P, Simpson R T. Positioned nucleosomes inhibit Dam methylation in vivo. Proc Natl Acad Sci USA. 1994;91:1361–1365. doi: 10.1073/pnas.91.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 40.Klein C, Struhl K. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science. 1994;266:280–282. doi: 10.1126/science.7939664. [DOI] [PubMed] [Google Scholar]
- 41.Komarnitsky P B, Michel B, Buratowski S. TFIID-specific TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev. 1999;13:2484–2489. doi: 10.1101/gad.13.19.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo M-H, Brownell J, Sobel R E, Ranalli T A, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 43.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 44.Laurent B C, Treitel M A, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee C-H, Murphy M R, Lee J-S, Chung J H. Targeting a SWI/SNF-related chromatin remodeling complex to the β-globin promoter in erythroid cells. Proc Natl Acad Sci USA. 1999;96:12311–12315. doi: 10.1073/pnas.96.22.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X-Y, Virbasius A, Zhu X, Green M R. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 47.Lohr D, Lopez J. GAL4/GAL80-dependent nucleosome disruption/deposition on the upstream regions of the yeast GAL1-10 and GAL80 genes. J Biol Chem. 1995;270:27671–27678. doi: 10.1074/jbc.270.46.27671. [DOI] [PubMed] [Google Scholar]
- 48.Majello B, Napolitano G, DeLuca P, Lania L. Recruitment of human TBP selectively activates RNA polymerase II TATA-dependent promoters. J Biol Chem. 1998;273:16509–16516. doi: 10.1074/jbc.273.26.16509. [DOI] [PubMed] [Google Scholar]
- 49.Melcher K, Johnston S A. GAL4 interacts with TATA-binding protein and coactivators. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizzen C A, Yang X-J, Brownell J E, Bannister A J, Owen-Hughes T, Workman J L, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 51.Moreira J M A, Holmberg S. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morse R H. Topoisomer heterogeneity of plasmid chromatin in living cells. J Mol Biol. 1991;222:133–137. doi: 10.1016/0022-2836(91)90198-f. [DOI] [PubMed] [Google Scholar]
- 53.Morse R H. Nucleosome disruption by transcription factor binding in yeast. Science. 1993;262:1563–1566. doi: 10.1126/science.8248805. [DOI] [PubMed] [Google Scholar]
- 54.Morse R H. Analysis of DNA topology in yeast chromatin. Methods Mol Biol. 1999;119:379–393. doi: 10.1385/1-59259-681-9:379. [DOI] [PubMed] [Google Scholar]
- 55.Natarajan K, Jackson B M, Zhou H, Winston F, Hinnebusch A G. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- 56.Neely K E, Hassan A H, Wallberg A E, Steger D J, Cairns B R, Wright A P H, Workman J L. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription of nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 57.Nevado J, Gadreau L, Adam M, Ptashne M. Transcriptional activation by artificial recruitment in mammalian cells. Proc Natl Acad Sci USA. 1999;96:2674–2677. doi: 10.1073/pnas.96.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pérez-Martin J, Johnson A D. The C-terminal domain of Sin1 interacts with the SWI-SNF complex in yeast. Mol Cell Biol. 1998;18:4157–4164. doi: 10.1128/mcb.18.7.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 61.Recht J, Osley M A. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 1999;18:229–240. doi: 10.1093/emboj/18.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Yeast TAFIIs in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 63.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/Mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roth S Y, Dean A, Simpson R T. Yeast α2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990;10:2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ryan M P, Jones R, Morse R H. SWI-SNF complex participation in transcriptional activation at a step subsequent to activator binding. Mol Cell Biol. 1998;18:1774–1782. doi: 10.1128/mcb.18.4.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stafford G A, Morse R H. Chromatin remodeling by transcriptional activation domains in a yeast episome. J Biol Chem. 1997;272:11526–11534. doi: 10.1074/jbc.272.17.11526. [DOI] [PubMed] [Google Scholar]
- 68.Stafford G A, Morse R H. Mutations in the AF-2/hormone binding domain of the chimeric activator GAL4.ER.VP16 inhibit hormone-dependent transcriptional activation and chromatin remodeling in yeast. J Biol Chem. 1998;273:34240–34246. doi: 10.1074/jbc.273.51.34240. [DOI] [PubMed] [Google Scholar]
- 69.Sudarsanam P, Cao Y, Wu L, Laurent B C, Winston F. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 1999;18:3101–3106. doi: 10.1093/emboj/18.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Svaren J, Schmitz J, Hörz W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsang S S, Yin X, Guzzo-Arkuran C, Jones V S, Davison A J. Loss of resolution in gel electrophoresis of RNA: a problem associated with the presence of formaldehyde gradients. BioTechniques. 1993;14:380–381. [PubMed] [Google Scholar]
- 72.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 73.Verdone L, Camilloni G, Di Mauro E, Caserta M. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol Cell Biol. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wahi M, Komachi K, Johnson A D. Gene regulation by the yeast Ssn6-Tup1 corepressor. Cold Spring Harbor Symp Quant Biol. 1999;63:447–457. doi: 10.1101/sqb.1998.63.447. [DOI] [PubMed] [Google Scholar]
- 75.Wechsler M A, Kladde M P, Alfieri J A, Peterson C L. Effects of Sin− versions of histone H4 on yeast chromatin structure and function. EMBO J. 1997;16:2086–2095. doi: 10.1093/emboj/16.8.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF transcriptional activators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 77.Workman J L, Taylor I C, Kingston R E. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991;64:533–544. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]
- 78.Wu Y, Reece R J, Ptashne M. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 79.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao H, Lis J T, Jeang K-T. Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol Cell Biol. 1997;17:6898–6905. doi: 10.1128/mcb.17.12.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu M, Simpson R T, Kladde M P. Gal4p-mediated chromatin remodeling depends on binding site position in nucleosomes but does not require DNA replication. Mol Cell Biol. 1998;18:1201–1212. doi: 10.1128/mcb.18.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu L, Morse R H. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5279–5288. doi: 10.1128/mcb.19.8.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yudovsky N, Logie C, Hahn S, Peterson C L. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]