Abstract
Background
HLA‐G is a non‐classical class I gene of the human Major Histocompatibility encoding molecules with immune‐modulatory properties. Expression of HLA‐G is being largely studied in pathological conditions, such as tumors, viral infections, inflammation, and autoimmune diseases, grafted tissues, among others. HLA‐G +3142C/G (rs1063320: dbSNP database) polymorphism is located in 3′ UTR of HAL‐G and plays a key role in determining the magnitude of gene and protein expression. The detection of HLA‐G +3142C/G polymorphism in the most published report is done through polymerase chain reaction followed by enzymatic digestion. Therefore, it is so interesting to develop a rapid and sensitive assay to genotype HLA‐G +3142C/G polymorphism. High‐resolution melt analysis (HRM) is a technology that is based on the analysis of the melting profile of PCR products through gradual temperature increase. The aim of this work is to apply high‐resolution melt method for genotyping the HLA‐G +3142C/G polymorphism.
Methods
DNA from 118 individuals was extracted from whole blood with QIAamp® DNA blood mini kit (Qiagen, Germany). Primer couple was designed using Primer 3 online tools so as to have only one SNP in the target sequence for high HRM efficiency. Positive Controls were identified using DNA sequencing and used as reference when assigning genotypes for trial samples.
Results
We were able to recognize the three genotypes with similar accuracy than DNA sequencing using high resolution melting method. Hardy‐Weinberg equilibrium test shows that our population is in equilibrium for the studied SNP. Genotypes frequencies of +3142C/G polymorphism in Tunisian general population are 0.475 for heterozygote G/C, 0.186 for homozygote G/G and 0.339 for homozygote C/C.
Conclusion
HRM is a cost‐effective method suitable for SNP genotyping.
HLAG SNPs molecular identification and QPCR‐HRM analysis.

1. INTRODUCTION
Single nucleotide polymorphisms (SNP) are the most common genetic variation occurring in the human genome. These variations are mainly studied to characterize the genetic component of complex diseases and elucidate their molecular bases. In these last years, huge effort had been devoted to perform genomic‐wide association study (GWAS) in order to identify genetic susceptibility to multifactorial diseases (Hirschhorn & Daly, 2005; McCarthy et al., 2008; Xue et al., 2018). Currently, two major platforms are being used for large‐scale SNP genotyping: Illumina (San Diego, CA) and Affymetrix (Santa Clara, CA) (Bush & Moore,). The validation of GWAS‐based associations could be done by replicating the association test for SNP of interest using other techniques. This implies the need for middle throughput platform. In addition, GWAS‐based associated SNPs are usually replicated in a specific group, to evaluate their ethnic‐specific effect.
In parallel, candidate gene approach is an alternative to GWAS to select relevant polymorphisms and investigate their association to pathological conditions. This approach is based on the knowledge of the physiopathology of the disease and the suspicion of defined genes. Thus, several SNP genotyping techniques had been developed or applied to analyze these single variations, such as DNA sequencing, polymerase chain reaction (PCR)‐restriction fragments length polymorphism (RFLP), probe‐based real‐time PCR, or mass spectrometry. However, there is a continuous need to develop accurate, rapid, and cost‐effective technologies for SNP analysis.
High‐resolution melting (HRM) is a relatively new method introduced in 2003 (Wittwer et al., 2003). This technique consists of subjecting PCR products to gradual temperature increase and recording in parallel the level of fluorescence. As the temperature rises, double‐stranded PCR product melts and dye is released from DNA. Once released, the dye becomes non‐fluorescent. The decrease of fluorescence level corresponds to the melting curve. DNA sequences with different base composition results in different melting curves. The gradual melting: 0.1–0.3℃/s and the robustness of the optical system of HRM capable‐thermocycler enable discrimination of samples down to single base‐pair changes. HRM has proven to be a sensitive assay for mutation discovery and SNP genotyping (Cho et al., 2008; Liew et al., 2004; Smith et al., 2008).
This technique is suitable to analyze particular SNP that have consequential effect on protein structure or/and function and gene expression. The HLA‐G gene (OMIM accession number: 142871) is characterized with a particular DNA variation: NC_000006.12: g.29830972C>G, reference SNP ID number: rs1063320, commonly called HLA‐G +3142C/G. This variation is located in the 3’ untranslated region and associated with differential HLA‐G expression. In silico analysis and functional study demonstrated that the presence of guanine in the position +3142 of HLA‐G gene increases the affinity of three microRNAs (miR‐148a, miR‐148b, and miR‐152) to the HLA‐G mRNA with consequent downregulation of HLA‐G expression (Tan et al., 2007). The modulatory effect of this polymorphism has led to several study investigating its association with different pathological conditions (Ben Fredj et al., 2016; Guberina et al., 2017; Poomarimuthu et al., 2017; Veit et al., 2014; Zidi et al., 2016).
HLA‐G +3142C/G polymorphism is mainly identified through polymerase chain reaction followed by PCR products digestion with BaeG1 enzyme (Agnihotri et al., 2017; Consiglio et al., 2011; Cordero et al., 2009; Hashemi et al., 2017). However, PCR‐RFLP is a time‐consuming technique and requires post‐PCR manipulation. Therefore, it is so interesting to develop more rapid and sensitive assay to genotype the HLAG +3142C/G polymorphism.
The aim of his work is to apply, for the first time, the HRM method for genotyping the SNP: HLA‐G +3142C/G.
2. METHODS
2.1. Study samples and ethical compliance
Ethics committee of Pasteur Institute of Tunis approved this study. All subjects enrolled gave their agreement to participate with written consent. This study was carried out in 118 individuals recruited from the external consultant service of Pasteur Institute of Tunis. Genomic DNA was extracted from peripheral blood mononuclear cells using a QIAamp® DNA Blood mini kit (Qiagen, Germany) according to the manufacturer's protocol. DNA was quantified using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE), and concentration adjusted to 30ng/µL. Only DNA samples with consistent DO260/280 ratio (between 1.8 and 2) were selected for this work.
2.2. Primer design
We first extracted HLA‐G gene (GenBank reference: NC_000006.12) sequence with all reported polymorphisms from Ensembl genome browser ( http://www.ensembl.org/index.html). Then we designed primers using primer3 online tool (http://primer3.ut.ee/). Two pairs of primers met our predefined criteria: a PCR product size not exceeding 150bp and containing a single DNA variation: rs1063320 (Table1).
TABLE 1.
Sequences of the primers meeting the defined selection criteria for HLA‐G+3142C/G SNP identification
| SNP ID | Primers sequence | Amplicon size | |
|---|---|---|---|
| rs1063320 |
F5′‐ACAGAAGTAAGTTATAGCTCAGTG−3′ R5′‐CTTCCCCAATCACCTTTCCT−3′ |
— | 95 bp* |
|
HamF5′‐TCAGATTCTAATTTTAATACAGAAGTA−3′ Ham R5′‐CCTCTTCCTCATGCTGAACT−3′ |
Used primers | 139 bp |
Abbreviation: bp, base pair.
The first pair of primers generate a 95bp PCR product. The polymorphism of interest is located at +1bp downstream of the forward primer (5′→3′). The second pair of primers generate a 139bp PCR product. In this case, the polymorphism of interest is located at +16 downstream of the forward primer (5′→3′). Primers sequences are listed in Table 1. The second primers were retained because of the central amplicon location of the polymorphism rs1063320.
2.3. Experimental determination of optimal annealing temperature
Gradient PCR of 58–65℃ was performed in order to determine the optimal annealing temperature. Optimal results were obtained at 60℃ and 61℃ (figure 3, supplementary information).
2.4. Identification of positive controls
The high‐resolution melting method requires prior identification of positive controls to assign to each sample the corresponding genotype. Then, we sequenced 20 samples randomly chosen to detect the three possible genotypes (HLA‐G+3142GG, HLA‐G+3142CC, and HLA‐G+3142CG). Sequencing data served also to validate the accuracy of HRM genotyping results.
HLA‐G target sequence was firstly amplified. PCR was performed under cycling conditions of 95℃ for 5 min, 95℃ for 40 s, 60℃ for 1 min, 72℃ for 35 s and for 35 cycles with a final extension of 10 min at 72℃. After purification, 5 μL of bigdye terminator cycle sequencing kit v.1.1 (Applied Biosystems), 1 μL of reverse an forward primer (10 µM), and 5 μL purified PCR product were mixed. Dideoxynucleotides (ddntp) incorporation was carried under the following thermal program: 25 temperature cycles (96℃ for 10 s, 50℃ for 5 s, and 60℃ for 4 min). Sequencing reaction was set up in ABI PRISM 3100 Genetic Analyzer (Applied Boisystems).
2.5. Real‐time polymerase chain reaction‐High resolution melting
Real‐time PCR and HRM were performed in 96 well‐plates using LightCycler® 480 real‐time PCR system (Roche, Germany). The 15 µl PCR reaction mixture contained 7.5 µL of master mix resolight (Roche, Germany), 300 nM (final concentration) of forward and reverse primer (Table 1), and 30 ng of genomic DNA. The PCR‐HRM program consists of an initial denaturation–activation step at 95℃ for 8 min, 45‐cycle program: denaturation at 95℃ for 10 s, annealing at 60℃ for 30s (touchdown from 64℃ to 60℃ at the ramp of 0.5℃ per second) and extension at 72℃ for 15 s) followed by a high resolution melting phase: from 70℃ to 90℃, with a ramp of 0.2℃ per second. Positive controls and no‐template negative controls were included in each run.
2.6. Hardy–Weinberg equilibrium test
Deviation from Hardy–Weinberg (HW) Equilibrium was tested for the HLA‐G +3142C/G polymorphism using the online tool: http://www.oege.org/software/hwe‐mr‐calc.shtml.
3. RESULTS
3.1. HLA‐G gene amplification
The primers designed for the amplification of HLA‐G target sequence are reported in Table 1. Real‐time amplification curves are summarized in S1 (supplementary information).
We performed melting peaks analysis and electrophoretic migration to check the specificity of the PCR product. The mean Cycle threshold value is 24. A single peak is present at approximately 81.4℃ (S2, supplementary information) corresponding to a band of 139 bp in gel agarose's (S3, supplementary information). The designed primers allowed a specific amplification of HLA‐G target sequence.
3.2. High‐resolution melting analysis
HLA‐G +3142C/G genotypes were assigned from HRM curves analyzed with the light cycler gene scanning software (Roche), using sequenced samples as reference.
Firstly, raw melting curve data were adjusted by putting bars in pre‐melting melting zone and post‐melting zone in order to restrict and focus the analysis to the active melting zone as shown in S3, supplementary information.
Then melting curves normalization was performed (Figure 1). This curve analyzing option allows the elimination of fluorescence variance between samples, and discrimination is based only on amplicon melting behavior. At this stage, heterozygous ‘G/C’ are easily distinguishable from homozygous by their atypical shape due to heteroduplexs formation (mismatches at SNP site: C:C and G:G in this case).
FIGURE 1.
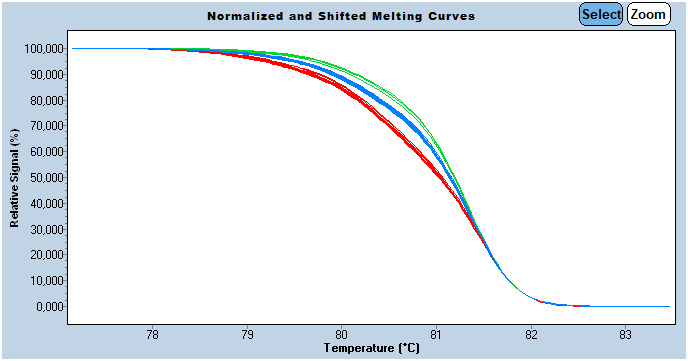
Normalized melt curve of HLA‐G target sequence; Normalization allows the elimination of fluorescence variance between samples, and discrimination is based only on amplicon melting behavior. Sensitivity was set to 0.35
Finally, small differences, especially between homozygous are best visualized using difference curves option. In this plot, sample curves are subtracted from a single reference. In our work, the C/C genotype was selected as reference (Figure 2).
FIGURE 2.
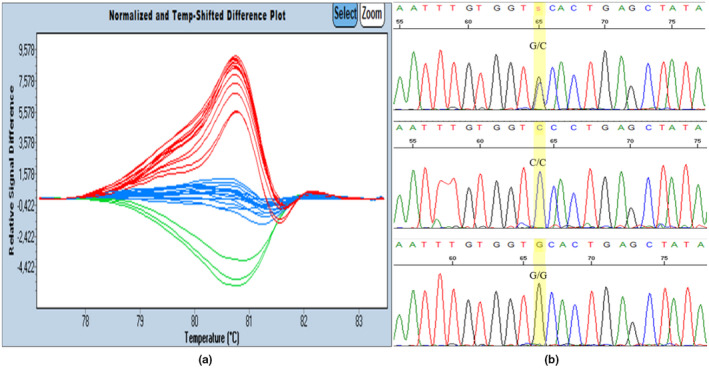
High resolution melting Difference plots and nucleotide sequencing chromatograms showing the HLA‐G +3142C/G polymorphism. (a) Normalized difference curves of HLA‐G target sequence. Blue curves: C/C samples, red curves: C/G samples, green curves: G/G samples. (b) Nucleotide sequencing chromatograms showing the HLAG +3142C/G polymorphism
High resolution melting analysis allowed the detection of three clusters. Each cluster or group of curves correspond to a specific genotype. Positive controls were included in each PCR experiment and allowed automatic attribution of the corresponding genotype to each cluster.
3.3. Genotype verification
Among the 118 samples analyzed with HRM, 20 DNA samples, randomly chosen, were subjected to DNA sequencing. Genotyping result of the two techniques showed 100% concordance validating the reliability of HRM results.
3.4. Distribution of HLA‐G +3142C/G polymorphism in our Tunisian sample
The HLA‐G +3142C/G genotype distribution was compared with Hardy–Weinberg (HW) expectations using chi‐squared test. Hardy‐Weinberg test indicates that our sample is in HW equilibrium for the +3142C/G polymorphism (X 2 = 0.09, p = 0.75) (Table 2). Genotype and alleles frequencies were determined for the studied SNP. The frequencies of C and G allele in our sample are respectively 0.577 and 0.423. The most widespread genotype is heterozygous C/G with a frequency of 0.475. Homozygous genotype frequencies C/C and G/G are respectively 0.339 and 0.186 (Table 2).
TABLE 2.
Genotype and allele frequencies for the rs1063320 SNP.
| SNP | N | Freq* | HWE | ||
|---|---|---|---|---|---|
| X2 | p | ||||
| rs1063320 | Genotypes | 0.09 | 0.75 | ||
| C/C | 40 | 0.339 | |||
| G/G | 22 | 0.186 | |||
| C/G | 56 | 0.475 | |||
| Alleles | |||||
| C | 136 | 0.577 | |||
| G | 100 | 0.423 | |||
Abbreviation: Freq, frequencies; X2 , Chi‐squared test; HWE, Hardy Weinberg equilibrium; N, Number; p, p value.
4. DISCUSSION
HLA‐G +3142C/G polymorphism is a class three SNP. This class exhibits low Tm difference between homozygous and could be resolved with HRM analysis only in case of nearest‐neighbor asymmetry around the base change (Liew et al., 2004). HLA‐G +3142C/G polymorphism has nearest‐neighbor asymmetry: it is surrounded by G base and A base (not complementary): 5′G‐(C/G)‐A3′. In this work, we successfully developed an HRM assay to genotype HLA‐G polymorphism: rs1063320. We were able to recognize the three genotypes of the studied SNP. Genotyping result of the twenty samples subjected to DNA sequencing showed 100% concordance with HRM Genotyping results.
According to the literature review, there are two PCR‐based protocols commonly used to identify this polymorphism: PCR‐RFLP and hydrolysis probe‐based real‐time PCR. This is the first application of HRM to identify HLA‐G polymorphism: rs1063320.
The accuracy of genotyping HLA‐G +3142C/G variant by RFLP has created a debate. Indeed bortolotti and collaborators evaluated hydrolysis probe‐based real‐time PCR genotyping assay for rs1063320 SNP. This work has reported that 19% of the individuals genotyped as heterozygous by PCR‐RFLP were identified as homozygous for the G allele (GG) by hydrolysis probe‐based real‐time PCR and sequencing methods. The authors explained this discrepancy by high rates of partial digestion in the PCR‐RFLP assay and, therefore, suggested that the PCR‐RFLP technique was unreliable (Bortolotti et al., 2012). In response, Zambra and collaborators who originally described the PCR‐RFLP protocol, conducted a re‐evaluation of their polymerase chain reaction‐restriction fragment length polymorphism genotyping method. They compared PCR‐RFLP results with DNA sequencing and concluded that RFLP assay is effective and reliable in genotyping the HLA‐G +3142 C/G polymorphism (Zambra & Response to Bortolotti et al.,).
We used DNA sequencing to identify positive controls and validate HRM genotyping results. Genotype frequencies of HLA‐G +3142C/G polymorphism in our Tunisian sample are 0.475 for heterozygote G/C, 0.339 for homozygote C/C and 0.186 for homozygote G/G (G allele: 0.423, C allele: 0.577).
Based on the released data of 1000 genome project phase 3, the SNP rs1063320 has heterogeneous distribution among the world population. The minor allele is C in African, South American and Asian populations. While the minor allele in European populations is G. Allelic distribution in subpopulations of each population mentioned above are almost equal except TSI (Tuscany in Italy) which is the only European subpopulation among the six European subpopulation (CEU, FIN, GBR, IBS and TSI) characterized with the minor allele C (Table 3). The European population has the closest genotypic distribution of HLA‐G +3142C/G polymorphism (0.222 GG, 0.333 CC, and 0.444 GC) compared to our Tunisian sample (0.186 GG, 0.339 CC, and 0.475 GC).
TABLE 3.
Genotype frequencies in Europe (CEU), Japan (JPT), Columbia (CLM), Yoruba (YRI), populations, and the Tunisian sample
| SNP | Population | Genotype | ||
|---|---|---|---|---|
| rs1063320 | C/C | G/C | G/G | |
| CEU* | 0.333 | 0.444 | 0.222 | |
| JPT* | 0.058 | 0.385 | 0.558 | |
| CLM* | 0.213 | 0.479 | 0.309 | |
| YRI* | 0.120 | 0.463 | 0.417 | |
| BEB* | 0.105 | 0.407 | 0.488 | |
| Our sample | 0.339 | 0.475 | 0.186 | |
Data extracted from 1000 genome project phase 3.
HRM method is a cost‐effective technology able to differentiate primary mutations and identify novel mutations (Cui et al., 2013). The key steps for efficient SNP genotyping with HRM are: the use of saturating dye (eva green, LCG green, Syto 9, etc.), primer design avoiding, as much as possible, hairpin and primer‐dimer formation, targeting an amplicon not exceeding 150 bp, and standardization of DNA samples concentration prior to real‐time PCR (10 ng/µl to 50 ng/µl). Furthermore, for optimal efficiency, target sequence must contain a single variation and all analyzed DNA samples should be extracted using the same protocol. This is to minimize as much as possible differences in salt concentration and ensure sample to sample uniformity.
The immunoregulatory function of HLA‐G and its implication in several pathological conditions varying from beneficial effect in inflammatory disease to unwanted tolerance in solid tumors has led to an increasing focus on polymorphisms determining the magnitude of HLA‐G gene and protein expression such as the HLA‐G +3142C/G single base change (Downs‐Kelly et al., 2007; Rizzo et al., 2012; Rouas‐Freiss et al., 2014).
5. CONCLUSION
Our work provides a new option: HRM assay to identify the highly studied polymorphism HLA‐G +3142C/G as an alternative to hydrolysis probe‐based real‐time PCR and RFLP assay. HRM is a fast, powerful and cost‐effective technology suitable for SNP genotyping especially in the middle throughput platform.
CONFLICT OF INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTION
CHELBI, H devised and supervised the project, and drafted the manuscript. BEN SALAH, H. and JELASSI, R. were involved in carrying out the experiment. Ben Salah, H performed the experimental data and statistical analysis. Ben SALAH, H; ZIDI, I; BEN AMOR, A, BIZID, S; AMMI, R; GUIZANI, L; BOURATBINE, A and AOUN, K aided in interpreting the results and worked the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The present study was approved by the Ethics Committee of Pasteur Institute of Tunis and written informed consent was obtained from all participants.
Supporting information
Supplementary Material
ACKNOWLEDGMENT
We are grateful to the individuals involved in this study for providing their general information and blood samples.
Ben Salah, H. , Jelassi, R. , Zidi, I. , Ben Amor, A. , Bizid, S. , Ammi, R. , Guizani, L. , Bouratbine, A. , Aoun, K. , & Chelbi, H. (2021). Rapid high‐resolution melting method to identify human leukocyte antigen‐G (HLA‐G) 3′ untranslated region polymorphism +3142C/G (rs1063320). Molecular Genetics & Genomic Medicine, 9, e1817. 10.1002/mgg3.1817
Funding information
This study was supported by the ministère de l'enseignement supérieur et de la recherche scientifique, Tunisia, and by Internal Collaborative Project PCI 06 Institut Pasteur de Tunis, Tunisia.
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- Agnihotri, V. et al (2017). Promising link of HLA‐G polymorphism, tobacco consumption and risk of Head and Neck Squamous Cell Carcinoma (HNSCC) in North Indian population. Human Immunology, 78(2), 172–178. 10.1016/j.humimm.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Ben Fredj, N. et al (2016). The association between functional HLA‐G 14bp insertion/deletion and +3142 C>G polymorphisms and susceptibility to multiple sclerosis. Immunology Letters, 180, 24–30. [DOI] [PubMed] [Google Scholar]
- Bortolotti, D. et al (2012). An accurate and reliable real time SNP genotyping assay for the HLA‐G +3142 bp C>G polymorphism. Tissue Antigens, 80(3), 259–262. [DOI] [PubMed] [Google Scholar]
- Bush, W. S. , & Moore, J. H. (2012). Chapter 11: Genome‐wide association studies. PLoS Computational Biology, 8(12), e1002822. 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, M. H. et al (2008). High‐resolution melting curve analysis of genomic and whole‐genome amplified DNA. Clinical Chemistry, 54(12), 2055–2058. 10.1373/clinchem.2008.109744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consiglio, C. R. et al (2011). Association of the HLA‐G gene +3142C>G polymorphism with systemic lupus erythematosus. Tissue Antigens, 77(6), 540–545. [DOI] [PubMed] [Google Scholar]
- Cordero, E. A. et al (2009). HLA‐G polymorphism influences the susceptibility to HCV infection in sickle cell disease patients. Tissue Antigens, 74(4), 308–313. 10.1111/j.1399-0039.2009.01331.x. [DOI] [PubMed] [Google Scholar]
- Cui, G. et al (2013). Applications of the method of high resolution melting analysis for diagnosis of Leber's disease and the three primary mutation spectrum of LHON in the Han Chinese population. Gene, 512(1), 108–112. 10.1016/j.gene.2012.09.110. [DOI] [PubMed] [Google Scholar]
- Downs‐Kelly, E. , Schade, A. E. , & Hansel, D. E. (2007). The role of HLA‐G in gastrointestinal inflammatory disease and malignancy. Seminars in Cancer Biology, 17(6), 451–458. 10.1016/j.semcancer.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Guberina, H. et al (2017). Recipient HLA‐G +3142 CC genotype and concentrations of soluble HLA‐G impact on occurrence of CMV Infection after living‐donor kidney transplantation. International Journal of Molecular Sciences, 18(11), 2338. 10.3390/ijms18112338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi, M. et al (2017). Evaluation of HLA‐G 14‐bp ins/del and +3142G>C polymorphisms with susceptibility to recurrent spontaneous abortion. Taiwanese Journal of Obstetrics & Gynecology, 56(3), 276–280. [DOI] [PubMed] [Google Scholar]
- Hirschhorn, J. N. , & Daly, M. J. (2005). Genome‐wide association studies for common diseases and complex traits. Nature Reviews Genetics, 6(2), 95–108. 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Liew, M. et al (2004). Genotyping of single‐nucleotide polymorphisms by high‐resolution melting of small amplicons. Clinical Chemistry, 50(7), 1156–1164. 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- McCarthy, M. I. et al (2008). Genome‐wide association studies for complex traits: consensus, uncertainty and challenges. Nature Reviews Genetics, 9(5), 356–369. 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Poomarimuthu, M. et al (2017). HLA‐G 3'UTR gene polymorphisms and rheumatic heart disease: a familial study among South Indian population. Pediatric Rheumatology, 15(1), 10. 10.1186/s12969-017-0140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo, R. et al (2012). New insights into HLA‐G and inflammatory diseases. Inflammation & Allergy: Drug Targets, 11(6), 448–463. [DOI] [PubMed] [Google Scholar]
- Rouas‐Freiss, N. et al (2014). The dual role of HLA‐G in cancer. Journal of Immunology Research, 2014, 359748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. D. et al (2008). Detection of epidermal growth factor receptor gene mutations in cytology specimens from patients with non‐small cell lung cancer utilising high‐resolution melting amplicon analysis. Journal of Clinical Pathology, 61(4), 487–493. 10.1136/jcp.2007.051425. [DOI] [PubMed] [Google Scholar]
- Tan, Z. et al (2007). Allele‐specific targeting of microRNAs to HLA‐G and risk of asthma. American Journal of Human Genetics, 81(4), 829–834. 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit, T. D. et al (2014). HLA‐G +3142 polymorphism as a susceptibility marker in two rheumatoid arthritis populations in Brazil. Tissue Antigens, 83(4), 260–266. 10.1111/tan.12311. [DOI] [PubMed] [Google Scholar]
- Wittwer, C. T. et al (2003). High‐resolution genotyping by amplicon melting analysis using LCGreen. Clinical Chemistry, 49(6 Pt 1), 853–860. 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- Xue, A. et al (2018). Genome‐wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nature Communications, 9(1), 2941. 10.1038/s41467-018-04951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambra, F. m b , Chies, J. a b , Alho, C. s , & Veit, T. d (2013). Response to Bortolotti et al. 2012‐a re‐evaluation of our polymerase chain reaction‐restriction fragment length polymorphism genotyping method. Tissue Antigens, 82(4), 286‐7. [DOI] [PubMed] [Google Scholar]
- Zidi, I. et al (2016). Association of HLA‐G +3142 C>G polymorphism and breast cancer in Tunisian population. Immunologic Research, 64(4), 961–968. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


