Abstract
p13suc1 (Cks) proteins have been implicated in the regulation of cyclin-dependent kinase (CDK) activity. However, the mechanism by which Cks influences the function of cyclin-CDK complexes has remained elusive. We show here that Cks1 is required for the protein kinase activity of budding yeast G1 cyclin-CDK complexes. Cln2 and Cdc28 subunits coexpressed in baculovirus-infected insect cells fail to exhibit protein kinase activity towards multiple substrates in the absence of Cks1. Cks1 can both stabilize Cln2-Cdc28 complexes and activate intact complexes in vitro, suggesting that it plays multiple roles in the biogenesis of active G1 cyclin-CDK complexes. In contrast, Cdc28 forms stable, active complexes with the B-type cyclins Clb4 and Clb5 regardless of whether Cks1 is present. The levels of Cln2-Cdc28 and Cln3-Cdc28 protein kinase activity are severely reduced in cks1-38 cell extracts. Moreover, phosphorylation of G1 cyclins, which depends on Cdc28 activity, is reduced in cks1-38 cells. The role of Cks1 in promoting G1 cyclin-CDK protein kinase activity both in vitro and in vivo provides a simple molecular rationale for the essential role of CKS1 in progression through G1 phase in budding yeast.
Cyclin-dependent kinases (CDKs) trigger a variety of events in the division cycle of eukaryotic cells (28). Multiple regulatory mechanisms conspire to ensure that CDKs are switched on at the right time and in the right place as cells proceed through the cell division program. At least four distinct posttranslational mechanisms positively regulate CDK function, including binding of cyclins, binding of p13suc1 (Cks) proteins, phosphorylation of a threonine within the T-loop of CDKs by the CDK-activating kinase (CAK), and dephosphorylation of threonine and tyrosine residues within the ATP-binding site by Cdc25 (27). Of these mechanisms, the least well understood is how binding of Cks proteins promote the function of CDKs.
The gene encoding p13suc1 was originally identified as a multicopy suppressor of cdc2ts mutations in Schizosaccharomyces pombe (15) and was subsequently isolated as a multicopy suppressor (named CKS1) of cdc28ts mutations in Saccharomyces cerevisiae (14). Characterization of budding yeast cks1 thermosensitive mutants revealed that Cks1 function is required at two points in the cell cycle: prior to start in G1 and at some point in mitosis (42). As was observed in p13suc1-depleted fission yeast (26), G2-arrested cks1ts mutants accumulate high levels of CDK activity.
The role of Cks proteins in progression through mitosis has been the subject of numerous recent studies. Xenopus extracts depleted of the Cks1 homolog p9 fail to enter mitosis due to accumulation of inhibitory phosphate on the Y15 residue of p34cdc2 (29), perhaps because p9 promotes CDK-dependent phosphorylation of the Y15 kinase Wee1 and the Y15 phosphatase Cdc25 (31). p9-depleted cycling extracts programmed with a p34cdc2 mutant that cannot be phosphorylated on Y15 no longer arrest at the G2/M transition but instead fail to initiate cyclin B destruction and consequently remain arrested in mitosis (29). These results suggest that p9 also promotes activation of cyclin B proteolysis by the anaphase-promoting complex–cyclosome (APC/C)–26S proteasome pathways (22). p9 is thought to activate cyclin B proteolysis by directly facilitating the phosphorylation of components of the APC/C, including Cdc27, by mitotic CDK (30, 36). However, there is some controversy on this point, since budding yeast Cks1 is dispensable for cyclin B ubiquitination but appears instead to promote degradation of ubiquitinated cyclin B by the 26S proteasome (19). Despite these recent advances in our understanding of how Cks proteins promote mitosis, it remains unclear why Cks1 is required for progression through Start.
Structural insights into the function of Cks proteins have emerged from X-ray crystallography of human Cks1-Cdk2 complexes (2). The binding of Cks1 does not cause significant conformational changes in either the active site or the cyclin-binding interface of Cdk2. A comparison of the Cks1-Cdk2 and cyclin A-Cdk2 (17) structures reveals that Cks1 and cyclin A bind to opposite sides of Cdk2 flanking the active site (2). The structure of the ternary complex suggests that Cks1 may help position substrates for phosphorylation by Cdk2.
In this report, we provide evidence that Cks1 is required both in vitro and in vivo for the protein kinase activity of G1 cyclin-CDK complexes from budding yeast. These findings suggest a simple explanation for why Cks1 activity is required for budding yeast cells to traverse Start.
MATERIALS AND METHODS
Preparation of cell lysates from baculovirus-infected cells.
Sf9 or Hi-5 cells were grown in suspension in supplemented Grace's insect media. For infection, 107 cells were plated per 10-cm-diameter dish and overlayed with working stocks of viruses at a multiplicity of infection of ∼10. After incubation for 40 to 44 h, cells were harvested in a clinical centrifuge at 4°C, washed once in 1× phosphate-buffered saline, and resuspended in ice-cold lysis buffer (10 mM HEPES-KOH [pH 7.4], 150 mM NaCl, 0.2% Triton X-100, 1 mM β-mercaptoethanol, 5 μg of pepstatin and 5 μg of leupeptin/ml, 2 mM phenylmethylsulfonyl fluoride (PMSF), 2 mM benzamidine, 50 mM β-glycerophosphate, 50 mM NaF, and 0.2 mM sodium orthovanadate). After a 10-min incubation on ice, lysates were sonicated three times for 10-s intervals at a setting of 4 (Branson sonifier 450) and then clarified by centrifugation at 4°C (35,000 rpm for 10 min; Sorvall RP100AT4 rotor). Supernatant fractions (typically 4 to 10 mg of protein/ml) were frozen in liquid N2 and stored at −80°C.
Activation of Cln2-Cdc28HA, immunoprecipitations, protein kinase assays, and immunoblots.
Clarified lysates from baculovirus-infected Sf9 cells were incubated in a reaction mixture consisting of activation buffer (1/10 volume of 40 mM magnesium acetate, 5 mM PMSF, 50 mM benzamidine, 10 mM dithiothreitol (DTT), 50 μg of pepstatin and 50 μg of leupeptin/ml), with or without yeast extract, Cks1, and ATP-regenerating system (1/10 volume of 500-μg/ml creatine phosphokinase, 10 mM ATP, 20 mM HEPES [pH 7.2], 10 mM magnesium acetate, 300 mM creatine phosphate). Activation mixtures were incubated for 15 min at 24°C and then chilled on ice.
For immunoprecipitation and histone H1 kinase assays, 3 to 10 μl of a 10-μl activation reaction mixture was diluted with 200 μl of immunoprecipitation buffer (IPB; 50 mM β-glycerophosphate [pH 7.5], 100 mM NaCl, 50 mM NaF, 5 mM EDTA, 2 mM sodium orthovanadate, 0.2% Triton X-100, 1 mM DTT, 0.5 mM PMSF, 5 μg of pepstatin and 5 μg of leupeptin/ml) and supplemented with 0.75 μl of 12CA5 antihemagglutinin (anti-HA) ascites fluid. After a 45-min incubation on ice, 20 μl of a 50% slurry of protein A-Sepharose beads was added, and tubes were incubated on a rotating wheel for 1 h at 4°C. Immune complexes were washed twice with 1 ml of IPB (minus protease inhibitors, NaF, and sodium orthovanadate). All samples were then washed twice with 1 ml of kinase assay buffer (KAB; 10 mM HEPES [pH 7.2], 10 mM MgCl2, 50 mM NaCl, 2 mM EDTA, 1 mM DTT, 0.02% Triton X-100) and supplemented with 10 μl of KAB adjusted to 150 μg of histone H1/ml, 11 μM ATP, and 1.5 μCi of [γ-32P]ATP (4,500 Ci/mmol)/μl. Following a 15-min incubation at room temperature, kinase reactions were terminated by addition of 7 μl of 3× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and heated for 3 min at 95°C. Boiled samples were subjected to SDS-PAGE, and the 32P radiolabel was quantitated by imaging dried gels with a Molecular Dynamics PhosphorImager.
Immunoblotting was performed with either anti-myc monoclonal antibody 9E10 (9) or anti-HA monoclonal antibody 12CA5 (11) ascites fluids (purchased from BabCo) used at a dilution of 1/1,000 or affinity-purified anti-Cdc28 polyclonal antibodies. Filter-bound antibodies were visualized using horseradish peroxidase-conjugated secondary antibodies and Amersham's enhanced chemiluminescence (ECL) kit. For the experiment shown in Fig. 6C, Cln33×HA immunoreactivity was quantitated using a Molecular Dynamics STORM system.
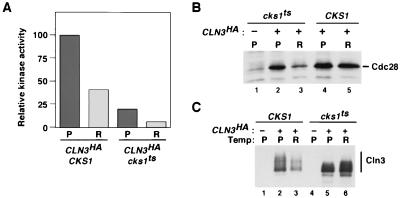
FIG. 6.
Mutant cks1-38 cells fail to assemble active Cln3-Cdc28 protein kinase complexes. (A) GAL-CLN33×HA CKS1 and GAL-CLN33×HA cks1-38 cultures grown in galactose medium at 25°C were split, and half of each culture was shifted to 38.5°C for 3 h. Total cell extract proteins (10 mg) prepared from cells maintained at 25°C (P) or shifted to 37°C (R) were immunoprecipitated with anti-HA monoclonal antibody 12CA5, and immunoprecipitates were divided in thirds and assayed for their content of Cdc28 (B) and Cln33×HA antigens (not shown) by immunoblotting and for histone H1 kinase activity (A). Kinase activity shown in panel A is normalized to Cln33×HA levels. (C) Cln33×HA present in total cell lysate of untagged controls (lanes 1 and 4), GAL-CLN33×HA CKS1 cells (lanes 2 and 3), and GAL-CLN33×HA cks1-38 cells (lanes 5 and 6) was evaluated by immunoblotting with anti-HA.
Preparation of yeast extracts.
Yeast extracts used as a source of Cln2-Cdc28 activator were prepared as described previously (5) from RJD875 (trp1 ura3 his2 ade1 bar1 cln1Δ cln2Δ cln3Δ leu2::GAL-CLN3::LEU2 cdc28-4 pep4::URA3 MATα) cells arrested in G1 by growth in yeast extract-peptone-dextrose (YPD) medium for 5.5 h at 24°C.
To examine the effect of mutant Cks1 on Cln2-Cdc28 activity, we assayed the level of Cln2-Cdc28 activity in extracts prepared from isogenic CKS1 and cks1-38 strains as follows. Yeast strains RJD1091 (BF264-15D; ade1 his2 leu2-3,112, trp1-1a cks1::LEU2 cln2::CLN23×HA::LEU2 MATa plus pSE271/cks1-38 [TRP1 CEN1 ARS cks1-ts38]) and RJD1093 (BF264-15D; ade1 his2 leu2-3,112 trp1-1a cks1::LEU2 cln2::CLN23×HA::LEU2 MATa plus pSE271/CKS1 [TRP1 CEN1 ARS CKS1]) were grown in 1 liter of YPD at 24°C to an optical density at 600 nm (OD600) of ∼0.5. Each 1-liter culture was split into two 500-ml cultures, one of which was placed at 24°C and the other at 38.5°C, for 3.5 h. Cultures were harvested by centrifugation (7 min at 7,000 rpm; Sorvall GSA rotor), and cell pellets were washed once with 50 ml of ice-cold 100 mM NaCl, frozen in liquid nitrogen, thawed, and resuspended in 3 ml of glass bead lysis buffer (GBLB; 18 mM Tris [pH 8.0], 8.8 mM MgCl2, 0.9 mM EDTA, 4.5% glycerol, 260 mM ammonium sulfate, 88 mM NaCl, 50 mM NaF, 50 mM β-glycerophosphate, 0.2 mM sodium orthovanadate, 1 mM DTT, 2 mM PMSF, 2 mM benzamidine, 5 μg of pepstatin and 5 μg of leupeptin/ml). The cell suspension was vortexed with 2 ml of glass beads (0.5 mm in diameter), and the resulting lysate was collected from the beads and centrifuged at 4°C for 10 min in a microcentrifuge, followed by 10 min at 40,000 rpm in a Sorvall microultracentrifuge (RP100AT4 rotor). Clarified lysates were used for immunoprecipitation and immunoblotting of Cln23×HA as described in the legend to Fig. 5. Cln3-Cdc28 activities in untagged (RJD539; ade1 his2 leu2 trp1 ura3 MATa), CKS1 CLN33×HA (RJD1280; cln3::LEU2::GAL1::CLN33×HA cks1::LEU2 ade1 leu2 his2 trp1 ura3 MATa pSE271-CKS1), and cks1-38 CLN33×HA (RJD1279; cln3::LEU2::GAL1::CLN33×HA cks1::LEU2 ade1 leu2 his2 trp1 ura3 MATa pSE271-cks1-38) cells were evaluated as described previously (18). RJD539, -1279, and -1280 are isogenic to each other and to BF264-15d.
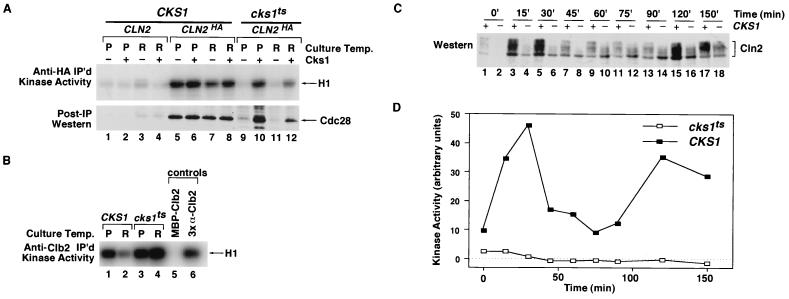
FIG. 5.
Mutant cks1-38 cells fail to assemble active Cln2-Cdc28 protein kinase complexes. (A) Absence of active Cln23×HA-Cdc28 complexes in cks1-38 cell extract. Total cell extract proteins (3 mg) prepared from CLN2 CKS1, CLN23×HA CKS1, and CLN23×HA cks1-38 cells grown at either the permissive (P; 24°C) or restrictive (R; 38.5°C) temperature were immunoprecipitated (IP) with anti-HA monoclonal antibody 12CA5, and immunoprecipitates were divided in half and assayed for their content of histone H1 kinase activity (top) or Cdc28 antigen (bottom). Samples in even-numbered lanes were supplemented with 3.9 μg of purified Cks1 prior to the immunoprecipitation step. Relative kinase activities in lanes 1 to 12 (top) are 1, 1, 2, 1, 25, 29, 15, 23, 2, 20, 2, and 8, respectively. (B) cks1-38 extracts contain high levels of Clb2-Cdc28 kinase activity. Total cell extract proteins (1 mg) from the experiment described for panel A were incubated with anti-Clb2 affinity-purified polyclonal antibodies, and immunoprecipitates were assayed for their content of histone H1 kinase activity. MBP-Clb2, same as sample shown in lane 1, except 20 μg of purified MBP-Clb2 was added prior to the immunoprecipitation step; 3× α-Clb2, same as sample shown in lane 1, except threefold more antibody was used. These two controls indicate that immunoprecipitation was specific and that the antibody was used at saturating levels for the assays depicted in lanes 1 to 4. The relative protein kinase activities in lanes 1 to 6 are 18, 6, 24, 36, 1, and 13, respectively. (C) Accumulation of phosphorylated Cln23×HA is strongly delayed in cks1-38 cells released from a pheromone arrest. CKS1 (odd-numbered lanes) and cks1-38 (even-numbered lanes) cultures were arrested in G1 phase with α-factor, shifted to 38.5°C for 1 h, and then released by being washed into medium lacking α-factor at 38.5°C. Lysates (100 μg) prepared from cells withdrawn at the indicated times were fractionated by SDS-PAGE and immunoblotted with anti-HA monoclonal antibody 12CA5 to detect Cln23×HA. Note that both the level and phosphorylation state of Cln2 are diminished in cks1-38 cells. (D) Cln23×HA-Cdc28 activity fails to accumulate in cks1-38 cells released from a pheromone arrest. Samples (3 mg) from the experiment shown in panel C were immunoprecipitated with 12CA5. Immunoprecipitates were assayed for their content of histone H1 kinase activity, which was evaluated by SDS-PAGE followed by phosphorimaging.
Cell extracts for the α-factor block-release experiment depicted in Fig. 5C and D were prepared as follows. Yeast strains RJD1091 and RJD1093 were grown in 500 ml of YPD at 24°C to an OD600 of ∼0.5, supplemented with 5 μg of α-factor/ml, and incubated an additional 3 h at 24°C, followed by 1.5 h at 38.5°C. G1-arrested cells were collected by filtration, washed with 500 ml of yeast extract-peptone, and resuspended in 500 ml of YPD prewarmed to 38.5°C. At 15-min intervals following release from α-factor, 45-ml aliquots of the culture were harvested by centrifugation, and cell pellets were washed with 10 ml of 100 mM NaCl and frozen in liquid nitrogen. Once aliquots at all time points were collected, cell pellets were thawed, resuspended in 1 ml of GBLB, and lysed by vortexing with 1 ml of glass beads (0.5 mm in diameter). Cell lysates were clarified by centrifugation in a Sorvall microultracentrifuge (10 min at 35,000 rpm; RP100AT4 rotor) and used for immunoprecipitation and immunoblotting of Cln23×HA as described in the legend of Fig. 5.
Recombinant proteins. (i) Cks1.
A 4-ml overnight culture of BL21(DE3) plus pLysS transformed with the plasmid pCKS1-1, which expressed Cks1 from the T7 promoter, was inoculated into 1 liter of Luria broth–100 μg of ampicillin/ml and grown at 37°C to an OD600 of 0.5. The culture was then cooled to 24°C in an ice water bath, and IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 0.3 mM to induce synthesis of Cks1. After a 4-h induction at 24°C, cells were harvested (Sorvall GSA rotor; 7,000 rpm for 7 min), washed once with 250 ml of 50 mM Tris (pH 7.6)–100 mM NaCl, and resuspended in 10 ml of 1× phosphate-buffered saline. To prepare a lysate, the cell suspension was frozen in liquid nitrogen, thawed on ice, and sonicated for 30 s at setting 5 (Branson sonifier 450). The resulting lysate was clarified by centrifugation in an IEC clinical centrifuge, and the supernatant was incubated in a boiling-water bath for 5 min and then centrifuged for 10 min at 40,000 rpm (Beckman 60Ti rotor). The supernatant fraction was supplemented with ammonium sulfate (1 g per 6.1 ml of supernatant), incubated for 30 min at 4°C, and centrifuged at 40,000 rpm for 10 min (Beckman 60Ti rotor). Precipitated proteins were redissolved in 10 ml of 50 mM Tris (pH 7.6)–100 mM NaCl–2 mM EDTA and dialyzed against three changes of 50 mM Tris (pH 7.6)–100 mM NaCl. The dialysate was chromatographed on a Sepharose CL-6B column (107 cm by 16-mm diameter), and fractions containing Cks1 were pooled and concentrated in a 15-ml Millipore centrifugal concentrator (nominal molecular mass cutoff = 10 kDa).
(ii) Cdc28HA.
A recombinant baculovirus that expresses Cdc28HA was generated by cloning a blunted, CDC28HA-containing BstBI/XbaI fragment from pSF19 (39a) into the SmaI site of pVL1392.
(iii) Cln2mycHis6.
Epitope-tagged Cln2 was generated by PCR amplification of CLN2 from a pAS101 template (provided by A. Sil) using the oligonucleotides RDO53 (GGGGGATCCCATATGGCTAGTGCTGAACC) and RDO54 (GGGCTGCAGCTATATTACTTGCGGCCGCTGGGTATTGCCCATACC). The resulting PCR fragment, which contains a unique NotI site at the 3′ end of the CLN2 coding sequence, was digested with BamHI plus PstI and subcloned into the corresponding sites of pUC119 to generate pUC119-Cln2(NotI). Complementary oligonucleotides (RDO59: GGCCTCTAGAGGAGCAGAAATTAATCAGCGAAGAGGACCTCCTCAGGCATCATCACCATCATCACG; RDO60: GGCCCGTGATGATGGTGATGATGCCTGAGGAGGTCCTCTTCGCTGATTAATTTCTGCTCCTCTAGA) encoding the bipartite mycHis6 epitope were hybridized, kinased, and ligated into the unique NotI site of pUC119-CLN2(NotI) to yield pUC119-CLN2mycHis6. Finally, the BamHI/PstI fragment of pUC119-CLN2mycHis6 was cloned into the BamHI/PstI sites of pVL1393. All PCR-amplified coding sequences were validated by DNA sequencing.
(iv) MBP-Clb2.
CLB2 was amplified by PCR from a pC2408 template (provided by K. Nasmyth) using oligonucleotides RDO70 (GAACGGTCGACTCAGAATTCTTCATGCAAGGTCATTATATCATAGCC) and RDO71 (GAACGGCTAGCATATGTCGCGGCCGCTATCCAACCCAATAGAAAACAC). The resulting fragment was digested with SalI plus NheI and cloned into the XbaI/SalI sites of pRS306 (37), which had been previously modified by filling in the unique NotI site (pRS306ΔNotI). All PCR-amplified coding sequences were validated by DNA sequencing. CLB2 coding sequences were excised from this plasmid by digestion with NdeI (filled in with Klenow fragment) and SalI and inserted into the EcoRI (filled in with Klenow fragment) and SalI sites of pMAL-c1 (New England BioLabs). Maltose-binding protein (MBP)–Clb2 was expressed and purified on amylose resin by standard methods (New England BioLabs).
MBP-Sic1 was prepared as described previously (45). The Far1 fragment used for kinase assays in Fig. 3 contained the 60 N-terminal amino acids of Far1 followed by a hexahistidine tag (provided by F. Cross). Far1(1-60)His6 was expressed and purified by nitrilotriacetic acid affinity chromatography as described previously (Qiagen).
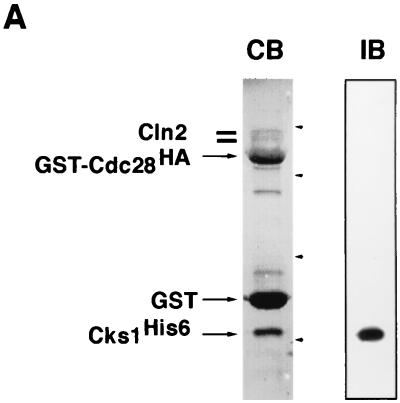
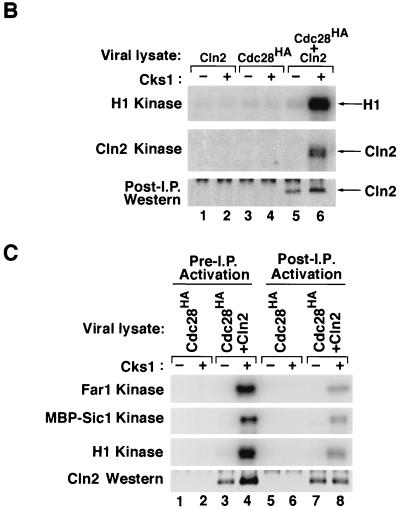
FIG. 3.
Cks1 binds to Cln2-Cdc28 and activates phosphorylation of multiple substrates. (A) Lysates from insect cells coinfected with baculovirus vectors encoding Cln2, GST-Cdc28HA, and Cks1His6 were adsorbed to glutathione resin, and specifically bound proteins were revealed by staining with Coomassie blue (CB) or immunoblotting with anti-His6 antibodies (IB). Arrowheads, migration of molecular mass markers (from top to bottom, 68, 46, 30, and 21.5 kDa). (B) Lysates (40 μg) of Sf9 cells infected with recombinant baculoviruses encoding Cln2MH6 or Cdc28HA, as indicated, were either mock treated or mixed with 390 ng of purified Cks1 for 15 min at 24°C. Reaction mixtures were immunoprecipitated (I.P.) with anti-HA monoclonal antibody 12CA5, and immunoprecipitates were divided in thirds and assayed for their quantity of histone H1 kinase (top), Cln2MH6 kinase (middle), or Cln2MH6 antigen (bottom; detected by immunoblotting with affinity-purified anti-Cln2 polyclonal antibody). (C) Lanes 1 to 4, immunoprecipitates prepared as described for panel B were directly assayed for Far1 or MBP-Sic1 or histone H1 kinase activity. Lanes 5 to 8, Cks1-free immunoprecipitates prepared from Sf9 lysates were subdivided into aliquots which were then either mock treated or supplemented with 390 ng of Cks1. After a brief incubation, kinase complexes were washed and assayed for Far1, MBP-Sic1, or histone H1 kinase activity as indicated. All samples were also evaluated by immunoblotting with an antimyc monoclonal antibody (bottom).
RESULTS
Activation of baculovirus-expressed Cln2-Cdc28 complexes requires a factor from yeast extract.
Cyclin-CDK subunits that have been expressed from recombinant baculoviruses typically assemble into active protein kinase complexes (4, 23, 25). Thus, to characterize the budding yeast Cln2-Cdc28 complex, we constructed recombinant baculoviruses to express Cln2 and Cdc28 either singly or in combination in insect Sf9 cells. The Cln2 and Cdc28 subunits were tagged at their C termini with a dual myc-hexahistidine epitope (Cln2MH6) and an HA epitope (Cdc28HA), respectively, to facilitate the recovery of Cln2-Cdc28 complexes from cell lysates. Following immunoprecipitation with the anti-HA monoclonal antibody 12CA5, immune complexes were assayed for histone H1 protein kinase activity and evaluated by SDS-PAGE (see Materials and Methods). Unexpectedly, Sf9 cells coinfected with recombinant baculoviruses encoding Cln2MH6 and Cdc28HA failed to express histone H1 protein kinase activity (Fig. 1, lane 5), even though both subunits were produced as judged by immunoblotting (data not shown).
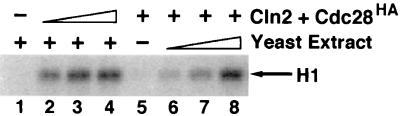
FIG. 1.
Yeast extract activates baculovirus-expressed Cln2-Cdc28 protein kinase. Lysates from Sf9 cells (5 μg) coinfected with recombinant baculoviruses encoding Cln2MH6 and Cdc28HA were incubated either alone (lane 5) or in the presence of 18, 54, and 180 μg of extract from Cln-depleted cdc28ts cells (lanes 6 to 8, respectively; relevant genotype: cdc28ts cln1Δ cln2Δ cln3Δ GAL-CLN3). In a parallel experiment, 180 μg of yeast extract was incubated alone (lane 1) or in the presence of 2.5, 5, and 7.5 μg of lysate from baculovirus-infected Sf9 cells expressing Cln2MH6 plus Cdc28HA (lanes 2 to 4, respectively). Following incubation for 15 min at 24°C in the presence of an ATP-regenerating system, all reaction mixtures were immunoprecipitated with anti-HA monoclonal antibody 12CA5, assayed for histone H1 kinase activity, fractionated on an SDS-polyacrylamide gel, and quantitated with a PhosphorImager. Relative kinase activities in lanes 1 to 8 are 1, 9, 14, 15, 2, 4, 7, and 17, respectively.
Recombinant glutathione S-transferase (GST)-Cln2 isolated from Escherichia coli can activate Cdc28HA in crude yeast extract (5). Thus, we examined whether yeast extract contains a factor that is required for the production of active Cln2-Cdc28 protein kinase. Yeast extract prepared from a cdc28ts mutant strain depleted of G1 cyclins was added to lysates prepared from Sf9 insect cells coinfected with recombinant baculoviruses encoding Cln2MH6 and Cdc28HA. Following a brief incubation at 24°C in the presence of an ATP-regenerating system, Cdc28HA and associated proteins were recovered and assayed for histone H1 kinase activity as described above. Whereas the yeast extract itself had no 12CA5-precipitable histone H1 kinase activity (Fig. 1, lane 1), it promoted the recovery of active Cln2MH6-Cdc28HA from insect cell lysates (lanes 6 to 8). No histone H1 kinase activity was recovered upon incubation of yeast extract with Sf9 lysates containing only Cdc28HA or Cln2MH6 (data not shown).
Budding yeast Cdc28 can potentially be activated by CAK, which phosphorylates T169 (8, 20, 44), and by Mih1 phosphatase (35), which dephosphorylates the Y19 residue of Cdc28 that is phosphorylated by Swe1 protein kinase (1). Neither of these factors was responsible for the activation of recombinant Cln2MH6-Cdc28HA because yeast extract efficiently promoted Cln2MH6-Cdc28HA activation even in the absence of ATP and in the presence of the potent phosphatase inhibitors sodium fluoride and sodium orthovanadate (data not shown). Moreover, immunopurified Cln2MH6-Cdc28HA complexes could be activated in the absence of crude lysate (see Fig. 3C).
The yeast extract Cln2-Cdc28 activator is Cks1.
In preliminary attempts to characterize the Cln2MH6-Cdc28HA activator in yeast extract, we tested its sensitivity to heat. Surprisingly, boiled yeast extract fully retained its ability to activate baculovirus-expressed Cln2MH6-Cdc28HA (Fig. 2A). Eukaryotic cells express a small protein that binds tightly to the CDK subunit of cyclin-CDK complexes and that is known by a variety of names including p13, suc1, and Cks (3, 14, 33). Depending on the species, p13suc1 or Cks1 ranges in size from 9 to 19 kDa. Given that the S. pombe p13suc1 protein is heat stable (39), we tested whether the Cln2MH6-Cdc28HA activator might correspond to Cks1, which is the budding yeast homolog of p13suc1 (14). Untreated or boiled E. coli extract that contained Cks1 could substitute for yeast extract to activate Cln2MH6-Cdc28HA complexes in insect cell lysate (Fig. 2A), whereas control E. coli extract had no effect (not shown). Half-maximal activation of Cln2MH6-Cdc28HA complexes in crude Sf9 cell extracts (Fig. 2C) was obtained with ∼100 nM purified Cks1 (Fig. 2B). Based on these observations, we conclude that the heat-stable Cln2MH6-Cdc28HA activator present in yeast extract is Cks1.
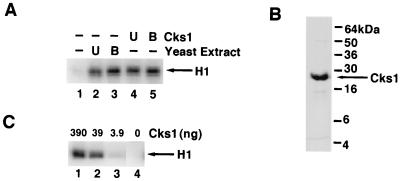
FIG. 2.
Cks1 activates Cln2-Cdc28 protein kinase. (A) Cks1 substitutes for yeast extract in the activation of Cln2MH6-Cdc28HA. Extracts prepared from Sf9 cells coinfected with Cln2MH6- and Cdc28HA-expressing baculoviruses were mixed with the indicated components, incubated at 24°C for 15 min, immunoprecipitated with anti-HA monoclonal antibody 12CA5, and assayed for histone H1 protein kinase activity. Relative kinase activities in lanes 1 to 5 are 1, 11, 14, 12, and 14, respectively. U and B, E. coli lysate (40 μg) containing Cks1 or yeast extract (100 μg) that was either untreated (U) or boiled for 5 min (B). (B) Purified Cks1. Cks1 (10 μg) purified from E. coli was fractionated on an SDS–15% polyacrylamide gel and stained with Coomassie blue. (C) Titration of Cks1. The indicated amounts of Cks1 were mixed with lysate (5 μg) prepared from Sf9 cells coinfected with recombinant baculoviruses encoding Cln2MH6 and Cdc28HA, and the mixtures were processed as described for panel A. Relative kinase activities in lanes 1 to 4 are 22, 16, 2, and 1, respectively.
Cks1 assembles with and activates the ability of Cln2-Cdc28 to phosphorylate multiple substrates.
Cks proteins are well known for binding tightly to cyclin B-CDK complexes. To test if Cks1 likewise assembles with Cln2-Cdc28, all three proteins were expressed from baculovirus vectors in insect cells. Coomassie blue staining and immunoblot analysis confirmed that Cln2-Cdc28 complexes bound to Cks1 beads (data not shown) and that both Cln2 and Cks1His6 were recovered along with GST-Cdc28HA on glutathione resin (Fig. 3A; these complexes exhibited high histone H1 kinase activity [data not shown]). These results confirm a previous report that Cdc28 and Cks1 copurify with Cln2 from yeast cells (46).
Histone H1 is a convenient reporter for CDK activity but is most likely not a physiological substrate for most CDKs. Cln2 (24), Sic1 (45), and Far1 (16) have been implicated as authentic substrates of Cln2-Cdc28 complexes. Thus, we tested whether recombinant Cln2MH6-Cdc28HA complexes were able to phosphorylate these substrates and whether phosphorylation was dependent on Cks1. Cln2MH6-Cdc28HA immunoprecipitated from Cks1-treated Sf9 lysates phosphorylated both histone H1 and the associated Cln2MH6 subunit (Fig. 3B, lane 6, top and middle). In contrast, neither substrate was phosphorylated by immunoprecipitates prepared from lysates containing only Cln2MH6 or Cdc28HA (lanes 1 to 4) or from coinfected lysates that were not supplemented with Cks1 (lane 5). Similar results were obtained with Far1 and MBP-Sic1 (Fig. 3C, lanes 1 to 4). Thus, Cks1 activates the ability of Cln2MH6-Cdc28HA to phosphorylate multiple substrates.
Cks1-dependent activation of Cdc28 is Cln specific.
Since expression of many different cyclin-CDK combinations in Sf9 cells has yielded active protein kinase complexes in the absence of added Cks proteins, we examined whether the Cks1-dependent activation of Cln2MH6-Cdc28HA was a peculiarity of the budding yeast CDK or of the G1 cyclin. Whereas coinfection of Sf9 cells with baculoviruses encoding Cln2MH6 and Cdc28HA yielded little precipitable protein kinase activity unless Cks1 was added prior to immunoprecipitation with anti-HA antibodies, Sf9 lysates containing Clb4 plus Cdc28HA or Clb5 plus Cdc28HA yielded substantial histone H1 kinase activity even in the absence of added Cks1 (Fig. 4). Besides Cln2-Cdc28 complexes, Cln1-Cdc28 complexes expressed in insect cells require Cks1 for protein kinase activity (38; J. W. Harper, personal communication).
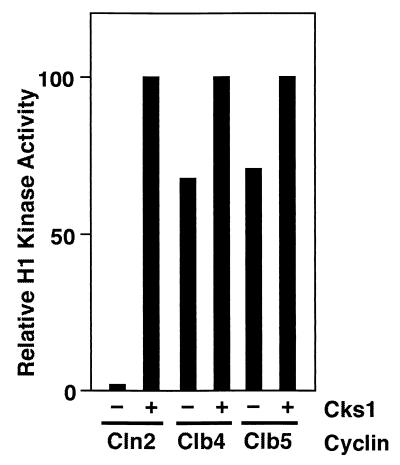
FIG. 4.
Cks1 is not required for protein kinase activity of Clb-Cdc28HA complexes. Sf9 cells were coinfected with a recombinant baculovirus encoding Cdc28HA plus an additional virus encoding either Cln2MH6, Clb4, or Clb5, as indicated. Lysates of infected cells were either mock treated or supplemented with 390 ng of purified Cks1, incubated at 24°C for 10 min, and immunoprecipitated with anti-HA monoclonal antibody 12CA5. Immunoprecipitates were assayed for their content of histone H1 kinase activity, which was assessed by SDS-PAGE followed by phosphorimaging. In this experiment, the relative maximal H1 kinase activities obtained were 1.0 for Clb4-Cdc28, 0.69 for Cln2-Cdc28, and 0.42 for Clb5-Cdc28.
Cks1 stabilizes Cln2-Cdc28 complexes and can activate preexisting complexes.
To determine how Cks1 influences the activity of Cln2MH6-Cdc28HA, we first examined whether Cks1 influences the assembly of the complex. Immunoblotting revealed that greater amounts of Cln2MH6 consistently coimmunoprecipitated with Cdc28HA from insect cell lysates that were supplemented with Cks1 (Fig. 3B, bottom, lane 5 versus lane 6; Fig. 3C, bottom, lane 3 versus lane 4). Although Cks1 enhanced the stability of the Cln2MH6-Cdc28HA complex, there was substantial Cln2MH6 antigen in Cdc28HA precipitates prepared in the absence of Cks1, even though these precipitates had little or no protein kinase activity (Fig. 3B and C). The addition of Cks1 to Cln2MH6-Cdc28HA immunoprecipitates prepared in the absence of Cks1 led to the appearance of Cln2MH6-dependent protein kinase activity towards multiple substrates (Fig. 3C, lanes 5 to 8; note that although the level of protein kinase activity is higher in lane 4 than in lane 8, there is significantly less Cln2MH6 in the latter samples). Thus, activation of Cln2MH6-Cdc28HA by Cks1 can proceed in the absence of insect cell lysate proteins and ATP. Taken together, these data suggest that Cks1 has two functions: it can promote the assembly of Cln2MH6 with Cdc28HA and it can activate preassembled Cln2MH6-Cdc28HA complexes.
Cks1 is required for Cln2-Cdc28 activity in vivo.
Temperature-sensitive mutant cks1-38 cells arrest cell division in both G1 and M phases at the nonpermissive temperature (42). To determine whether Cks1 function is required for the appearance of active Cln2-Cdc28 complexes in vivo, we examined the level of histone H1 kinase activity associated with epitope-tagged Cln2 (Cln23×HA) in asynchronous and G1-synchronized populations of CKS1 and cks1-38 cells (Fig. 5). Both histone H1 kinase activity and Cdc28 antigen were recovered in anti-HA immunoprecipitates prepared from asynchronous populations of CKS1 CLN2HA cells (Fig. 5A, lanes 5 and 7) but not from CKS1 cells that did not express tagged Cln23×HA (lanes 1 and 3). In contrast, little or no histone H1 kinase activity and Cdc28 antigen were detected in anti-HA precipitates prepared from asynchronous cultures of cks1-38 cells maintained at the permissive temperature (24°C; lane 9) or shifted to the restrictive temperature (38.5°C; lane 11). Immunoblot analysis confirmed that these cks1-38 cultures contained normal levels of Cdc28 and Cln23×HA antigens (data not shown). Moreover, recovery of both Cln23×HA-associated protein kinase activity and Cdc28 antigen was restored by the addition of purified Cks1 to the cks1-38 lysate prior to the immunoprecipitation step (Fig. 5A, lanes 10 and 12), indicating that the Cdc28 and Cln23×HA subunits present in cks1-38 cell extracts were competent to form an active kinase complex.
Defective assembly of active Cln2-Cdc28 complexes was also observed in synchronous G1 phase cultures of cks1-38 cells (Fig. 5C and D), suggesting that the results observed in Fig. 5A are not an artifact due to arrest of cks1-38 cells in G2 phase. Cln23×HA expressed in synchronous cultures of G1 phase cks1-38 cells at 38.5°C migrated upon SDS-PAGE largely as a hypophosphorylated species relative to the migration seen in CKS1 cells (Fig. 5C). Given that Cln2 assembled into active complexes is hyperphosphorylated by Cdc28 (24), this observation provides further evidence that Cln2-Cdc28 protein kinase activity was dramatically reduced in intact cks1-38 cells.
Although Cln2-Cdc28 protein kinase activity was severely reduced in cks1-38 cells, precipitation of the same extracts used above with anti-Clb2 antibodies revealed that these cells contained elevated levels of Clb-associated histone H1 kinase (Fig. 5B, lane 2 versus lane 4). The elevated level of Clb2-Cdc28 activity seen in cks1-38 cells is consistent with the observation that Cks1 is required for Clb2 turnover (19).
Cks1 is required for Cln3-Cdc28 activity in vivo.
Cks1 is important for activation of recombinant Cln1-Cdc28 and Cln2-Cdc28 complexes, and cks1ts cells fail to transit from G1 to S phase. Given that either Cln1, Cln2, or Cln3 is able to sustain progression through G1 phase in budding yeast cells (34), we reasoned that Cks1 might also be important for Cln3-Cdc28 activity. To test this, we examined the formation of active Cln3-Cdc28 complexes in cks1-38 strains. GAL-CLN33×HA CKS1 and GAL-CLN33×HA cks1-38 cells were grown in galactose medium at 25°C and one-half of each culture was shifted to 38.5°C for 3 h. Cln33×HA-Cdc28 activity and Cln33×HA plus Cdc28 antigens were evaluated by protein kinase assay and immunoblotting of anti-HA immunoprecipitates. Cln33×HA-associated histone H1 kinase activity was substantially diminished in cks1-38 cells grown at either the permissive or restrictive temperature (Fig. 6A). As was observed for Cln2, little Cdc28 was recovered in Cln3HA precipitates prepared from cks1-38 cells incubated at the nonpermissive temperature (Fig. 6B). Interestingly, Cdc28 was recovered in association with Cln3 from cks1-38 cells grown at the permissive temperature (Fig. 6B), even though these complexes had little protein kinase activity (Fig. 6A). Taken together with the data shown in Fig. 3C and 5A, this observation suggests that Cks1 stimulates both the assembly of Cln proteins with Cdc28 and the activity of preformed Cln-Cdc28 complexes.
The accumulation of Cln3 is negatively regulated by its Cdc28-dependent phosphorylation (48). Consistent with the reduced Cln3-associated Cdc28 protein kinase activity detected in vitro, Cln33×HA accumulated to high levels, but primarily as hypophosphorylated forms, in cks1-38 cells (Fig. 6C).
DISCUSSION
Cks1 activates budding yeast G1 cyclin-CDK complexes.
We provide evidence that Cks1 is required for the protein kinase activity of Cln2-Cdc28 and Cln3-Cdc28 protein kinase complexes. Cks1 potently activates the ability of immunoisolated Cln2-Cdc28 complexes to phosphorylate multiple substrates but has only a modest effect on the activity of Clb-Cdc28 complexes. Immunoprecipitation experiments suggest that Cks1 promotes the assembly of active Cln2-Cdc28 complexes in vivo and in vitro. However, intact Cln2-Cdc28 complexes recovered from insect cells in the absence of yeast Cks1 are nonetheless activated by added Cks1, suggesting that Cks1 may promote Cln2-Cdc28 protein kinase activity by an additional mechanism besides complex formation.
A previous analysis of the phenotype of cks1-38 mutants failed to reveal the defect in G1 cyclin-Cdc28 protein kinase activity reported here (42). This discrepancy is probably due to the fact that, in the prior work, Cdc28 protein kinase activity was monitored using an antibody directed against Cdc28. G1 cyclins account for only a small fraction of total Cdc28 protein kinase activity (47), and Cks1 is not required for the protein kinase activity of the more abundant Clb-Cdc28 complexes (Fig. 4 and 5B; the small contribution of Cks1 to Clb-Cdc28 protein kinase activity is probably obscured by the accumulation of Clb2 in cks1-38 cells). Thus, since anti-Cdc28 antibodies are expected to retrieve both Cln-Cdc28 and Clb-Cdc28 complexes, it is not surprising that Tang and Reed (42) failed to detect the effect reported here.
Mechanism of Cln2-Cdc28 activation by Cks1.
How does Cks1 promote Cln2-Cdc28 protein kinase activity? First, Cks1 appears to enhance the interaction between Cln2 and Cdc28. It is difficult to envision how Cks1 stabilizes the interaction of Cln2 with Cdc28 since Cks1 and cyclin A bind to opposite sides of Cdk2 and since the binding of Cks1 to the C-terminal lobe of Cdk2 has little effect on the conformation of the cyclin-binding interface (2). However, the binding of cyclin B to p34cdc2 enhances the binding of human Cks2 (7), suggesting that cyclin and Cks proteins may interact cooperatively with CDKs. Another possibility is that the long C-terminal tail following the cyclin box of Cln2 may form stabilizing contacts with Cks1 bound to the carboxy-terminal lobe of Cdc28.
Although Cks1 strongly stabilizes the interaction between Cln2 and Cdc28 in yeast extract, it has a far less dramatic effect on the assembly of stable Cln2-Cdc28 complexes in insect cell extract. We do not understand the reason for this discrepancy. Perhaps the far greater concentration of Cln2 and Cdc28 subunits expressed in insect cells favors complex assembly even if Cks1 is absent. Alternatively, insect cells may contain a factor (insect Cks1-like protein?) that stabilizes Cln2-Cdc28 association but that is insufficient to sustain protein kinase activity. Although it is unclear how the binding of Cks1 activates preformed Cln2-Cdc28 complexes isolated from insect cells, it seems unlikely that this effect is brought about through conformational changes propagated from the Cks1 binding site to the active site of Cdc28, because binding of Cks1 does not evoke substantial rearrangements within the catalytic site of Cdk2 (2) and because Cks1 has little effect on the activity of Clb-Cdc28 complexes.
We suggest four alternative hypotheses to explain how Cks1 selectively activates Cln2-Cdc28 complexes. First, Cln2 may exert both positive and negative effects on Cdc28. Positive regulation would presumably be accomplished in the same manner as is observed for cyclin A: binding of Cln2 rearranges key residues in the active site of Cdc28, favoring a geometry that permits transfer of the γ phosphate of ATP to bound substrate (17). In contrast, other domains within Cln2 (e.g., the C-terminal tail) might act negatively on Cdc28 unless displaced via binding of Cks1. A second possibility is that Cln proteins may bind Cdc28 more weakly than do Clb proteins and that Cks1 may be required to stabilize the Cln-Cdc28 complex. Third, Cks1 may provide substrate interaction sites that are normally provided by the cyclin in Clb-Cdc28 complexes (21) but that are otherwise absent from Cln-Cdc28 complexes. Last, Cks1 may promote Cln2-Cdc28 activity by acting directly on Cln2. Although it seems likely that the target of action of Cks1 is the α5 helix-L14 loop within the C-terminal lobe of Cdc28, this last possibility is suggested by the observation that a Cdc28 mutant (Cdc28-1N) that binds Cks1 poorly (2) nonetheless proceeds through Start and arrests at G2/M instead (32, 41).
Cdc37 has also been reported to promote interactions between Cln2 and Cdc28 (13). Unlike cdc37-1 mutants, cks1-38 cells contain normal levels of Cdc28 and accumulate high levels of Clb-Cdc28 activity. Cdc37 is thought to promote the assembly of cyclin-CDK complexes by promoting conformational maturation of the CDK subunit (10, 40). Thus, Cdc37 and Cks1 probably promote Cln2-Cdc28 activity by different means.
Is Cks1 required for the activity of other cyclin-CDK complexes?
To date, all cyclin-CDK complexes that have been expressed in insect cells (including cyclin A-Cdc2, cyclin B-Cdc2, cyclin D-Cdk4, and cyclin E-Cdk2) are active in the absence of any added proteins (4, 23, 25). Thus, the role of Cks1 in Cln2-Cdc28 activation may be unique to this particular combination. However, because mammalian cyclin-Cdk complexes may recruit endogenous Cks1-like proteins in insect cells, it is possible that mammalian G1 cyclins also require Cks proteins to activate their cognate CDKs. It is unclear whether insect cell Cks1-like proteins bind to Cln2-Cdc28, but if they do they are not sufficient to sustain protein kinase activity in vitro.
There is precedent for assembly factors contributing to the formation of active cyclin-CDK complexes in animal cells. Besides the Cdc37 example noted above, Mat1 is required for the assembly of cyclin H-Cdk7 (CAK) complexes (6, 12, 43). Further work will be required to test the generality of the observations we report here and to deduce the exact mechanism by which Cks1 promotes the activation of Cln2-Cdc28 complexes.
ACKNOWLEDGMENTS
We acknowledge M. Weinreich and B. Stillman for providing recombinant baculoviruses encoding Clb4 and Clb5. We also thank M. Olson for MBP-Clb2 and anti-Clb2 serum, R. Booher for Cks1 plasmids, S. Reed for cks1-38, F. Cross for the Far1 fragment and Cln33×HA strains, Wade Harper for baculoviruses encoding Cks1 and GST-Cdc28HA, members of W. Dunphy's laboratory for advice on baculovirus expression, and W. Dunphy, R. Feldman, G. Turner, R. Verma, and D. Patra for discussions and comments on the manuscript.
R.J.D. is a Searle and Markey Scholar, and this work was supported by the Searle Program/The Chicago Community Trust, The Lucille P. Markey Charitable Trust, and the National Institutes of Health (NIH RO1 GM52466).
REFERENCES
- 1.Booher R N, Deshaies R J, Kirschner M. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourne Y, Watson M H, Hickey M J, Holmes W, Rocque W, Reed S I, Tainer J A. Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell. 1996;84:863–874. doi: 10.1016/s0092-8674(00)81065-x. [DOI] [PubMed] [Google Scholar]
- 3.Brizuela L, Draetta G, Beach D. p13suc1 acts in the fission yeast cell division cycle as a component of the p34cdc2 protein kinase. Eur Mol Biol Org J. 1987;6:3507–3514. doi: 10.1002/j.1460-2075.1987.tb02676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai D, Gu Y, Morgan D O. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshaies R J, Kirschner M. Reconstitution of p34CDC28 activity with G1 and mitotic cyclins. Proc Natl Acad Sci USA. 1995;92:1182–1186. doi: 10.1073/pnas.92.4.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devault A, Martinez A-M, Fesquet D, Labbe J-C, Morin N, Tassan J-P, Nigg E, Cavadore J, Doree M. MAT1 (menage a trois), a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 1995;14:5027–5036. doi: 10.1002/j.1460-2075.1995.tb00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egan E A, Solomon M J. Cyclin-stimulated binding of Cks proteins to cyclin-dependent kinases. Mol Cell Biol. 1998;18:3659–3667. doi: 10.1128/mcb.18.7.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinoza F H, Farrell A, Erdjument-Bromage H, Tempst P, Morgan D O. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science. 1996;273:1714–1717. doi: 10.1126/science.273.5282.1714. [DOI] [PubMed] [Google Scholar]
- 9.Evan G J, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cellular Biology. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell A, Morgan D. Cdc37 promotes the stability of protein kinases Cdc28 and Cak1. Mol Cell Biol. 2000;20:749–754. doi: 10.1128/mcb.20.3.749-754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Field J, Nikawa J I, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 13.Gerber M R, Deshaies R J, Herskowitz I, Morgan D O. Cdc37 is required for association of the protein kinase Cdc28 with G1 and mitotic cyclins. Proc Natl Acad Sci USA. 1995;92:4651–4655. doi: 10.1073/pnas.92.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadwiger J A, Wittenberg C, Mendenhall M D, Reed S I. The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol Cell Biol. 1989;9:2034–2041. doi: 10.1128/mcb.9.5.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayles J, Beach D, Durkacz B, Nurse P. The fission yeast cell cycle control gene cdc2+: isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol Gen Genet. 1986;202:291–293. doi: 10.1007/BF00331653. [DOI] [PubMed] [Google Scholar]
- 16.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R J, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 18.Jeoung D-I, Oehlen L, Cross F. Cln3-associated kinase activity in Saccharomyces cerevisiae is regulated by the mating factor pathway. Mol Cell Biol. 1998;18:433–441. doi: 10.1128/mcb.18.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser P, Moncollin V, Clarke D J, Watson M H, Bertolaet B L, Reed S I, Bailly E. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 1999;13:1190–1202. doi: 10.1101/gad.13.9.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaldis P, Sutton A, Solomon M J. The CDK-activating kinase (CAK) from budding yeast. Cell. 1996;86:553–564. doi: 10.1016/s0092-8674(00)80129-4. [DOI] [PubMed] [Google Scholar]
- 21.Kellogg D R, Kikuchi A, Fuji-Nakata T, Turck C W, Murray A W. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King R W, Deshaies R J, Peter J M, Kirschner M. How proteolysis controls cell division. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 23.Koff A, Giordano A, Desai D, Yamashita K, Harper J W, Elledge S, Nishimoto T, Morgan D O, Franza B R, Roberts J M. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 24.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 25.Matsushime H, Ewen M E, Strom D K, Kato J Y, Hanks S K, Roussel M F, Sherr C J. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- 26.Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- 27.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 28.Murray A, Hunt T. The cell cycle, an introduction. Oxford, United Kingdom: Oxford University Press; 1993. [Google Scholar]
- 29.Patra D, Dunphy W G. Xe-p9, a Xenopus Suc1/Cks homolog, has multiple essential roles in cell cycle control. Genes Dev. 1996;10:1503–1515. doi: 10.1101/gad.10.12.1503. [DOI] [PubMed] [Google Scholar]
- 30.Patra D, Dunphy W G. Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase-promoting complex at mitosis. Genes Dev. 1998;12:2549–2559. doi: 10.1101/gad.12.16.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patra D, Wang S X, Kumagai A, Dunphy W G. The Xenopus Suc1/Cks protein promotes the phosphorylation of G(2)/M regulators. J Biol Chem. 1999;274:36839–36842. doi: 10.1074/jbc.274.52.36839. [DOI] [PubMed] [Google Scholar]
- 32.Piggott J R, Rai R, Carter B L A. A bifunctional gene involved in two phases of the yeast cell cycle. Nature. 1982;298:391–394. doi: 10.1038/298391a0. [DOI] [PubMed] [Google Scholar]
- 33.Richardson H E, Stueland C S, Thomas J, Russell P, Reed S I. Human cDNAs encoding homologs of the small p34Cdc28/Cdc2-associated protein of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Genes Dev. 1990;4:1332–1344. doi: 10.1101/gad.4.8.1332. [DOI] [PubMed] [Google Scholar]
- 34.Richardson H E, Wittenberg C, Cross F, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 35.Russell P, Moreno S, Reed S I. Conservation of mitotic controls in fission and budding yeasts. Cell. 1989;57:295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- 36.Shteinberg M, Hershko A. Role of Suc1 in the activation of the cyclosome by protein kinase Cdk1/cyclin B. Biochem Biophys Res Commun. 1999;257:12–18. doi: 10.1006/bbrc.1999.0409. [DOI] [PubMed] [Google Scholar]
- 37.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skowyra D, Craig K, Tyers M, Elledge S, Harper J. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 39.Solomon M J, Glotzer M, Lee T, Philippe M, Kirschner M W. Cyclin activation of p34cdc2. Cell. 1990;63:1013–1024. doi: 10.1016/0092-8674(90)90504-8. [DOI] [PubMed] [Google Scholar]
- 39a.Sorger P, Murray A W. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc2. Nature. 1992;355:365–368. doi: 10.1038/355365a0. [DOI] [PubMed] [Google Scholar]
- 40.Stepanova L, Leng X H, Parker S B, Harper J W. Mammalian p50(CDC37) is a protein kinase-targeting subunit of HSP90 that binds and stabilizes CDK4. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- 41.Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher A B, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 42.Tang Y, Reed S I. The Cdk-associated protein Cks1 functions both in G1 and G2 in Saccharomyces cerevisiae. Genes Dev. 1993;7:822–832. doi: 10.1101/gad.7.5.822. [DOI] [PubMed] [Google Scholar]
- 43.Tassan J-P, Jaquenoud M, Fry A M, Frutiger S, Hughes G, Nigg E A. In vitro assembly of a functional human cdk7/cyclin H complex requires MAT1, a novel 36 kD RING finger protein. Eur Mol Biol Org J. 1995;14:5608–5617. doi: 10.1002/j.1460-2075.1995.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thuret J Y, Valay J G, Faye G, Mann C. CIV1 (CAK in vivo), a novel CDK-activating kinase. Cell. 1996;86:565–576. doi: 10.1016/s0092-8674(00)80130-0. [DOI] [PubMed] [Google Scholar]
- 45.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 46.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 47.Wittenberg C, Sugimoto K, Reed S I. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34cdc2 kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- 48.Yaglom J, Linskens M H K, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of CLN3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]