Graphical abstract
Keywords: Hydroxychloroquine, Enantiomers, Antiviral, SARS-CoV-2, COVID-19
Abbreviations: SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; MERS, Middle East Respiratory Syndrome; COVID-19, coronavirus disease 2019; remdesivir-TP, remdesivir triphosphate complex; FDA, U.S. Food and Drug Administration; CPE, cytopathic effect; dpd, day post drug administration; r.t., room temperature
Abstract
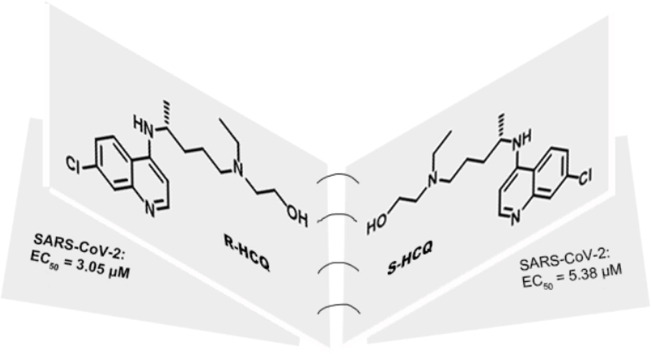
Since the end of 2019, the outbreak of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has evolved into a global pandemic. There is an urgent need for effective and low-toxic antiviral drugs to remedy Remdesivir’s limitation. Hydroxychloroquine, a broad spectrum anti-viral drug, showed inhibitory activity against SARS-CoV-2 in some studies. Thus, we adopted a drug repurposing strategy, and further investigated hydroxychloroquine. We obtained different configurations of hydroxychloroquine side chains by using chiral resolution technique, and successfully furnished R-/S-hydroxychloroquine sulfate through chemical synthesis. The R configuration of hydroxychloroquine was found to exhibit higher antiviral activity (EC50 = 3.05 μM) and lower toxicity in vivo. Therefore, R-HCQ is a promising lead compound against SARS-CoV-2. Our research provides new strategy for the subsequent research on small molecule inhibitors against SARS-CoV-2.
1. Introduction
East Respiratory Syndrome (MERS), Severe Acute Respiratory Syndrome (SARS) and Ebola virus have caused a large number of infections and deaths. Most recently, at the end of 2019, a new type of coronavirus,1 severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2, its genome sequence is highly consistent with SARS-CoV)2, suddenly appeared.3 Due to its strong contagiousness (especially for elderly people,4., 5. people with underlying diseases and immunocompromised individuals), this virus is raging widely and developed into a global epidemic. As the virus spreads, several mutated strains have been reported,6, 7., 8. which may cause easier human-to-human transmission a higher mortality rate before effective vaccines and therapeutic drugs are available. So far, there are more than 240 million people infected by coronavirus disease 2019 (COVID-19) with nearly five million recorded deaths worldwide,9 It highlights the challenges governments and medical institutions face in responding to sudden outbreaks. Therefore, there is an urgent need to develop highly effective drugs against novel coronavirus to protect people's health.
Development of highly effective new drugs for a new virus is costly and time-consuming. Although many small molecule inhibitors against different targets of SARS-CoV-2 have been discovered,10., 11. most of them have only undergone partial preclinical or clinical research,12., 13., 14., 15., 16., 17., 18. and whether they can be formulated into drugs remains to be determined. In the face of a rapidly spreading and increasingly serious epidemic, these inhibitors cannot solve the current dilemma immediately.
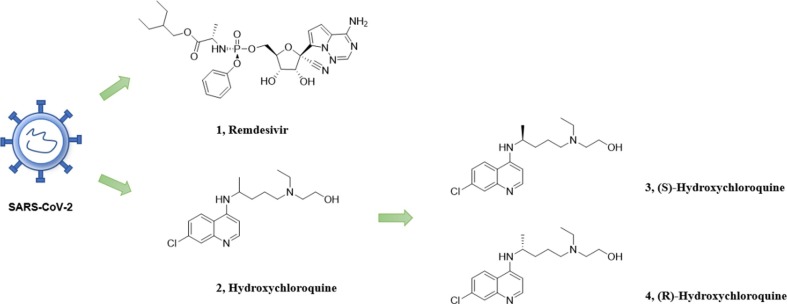
In light of the challenges, drug repurposing may be an ideal strategy. Drug repurposing has many advantages19., 20., 21. including saving time, money, and the guarantee of safety with clinical experiences. There are many successful applications of this strategy in the past, such as thalidomide, ketoconazole, finasteride and so on. The World Health Organization launched a multinational randomized trial called “the solidarity trial” in 2020 to study the efficacy of some promising molecules or drug combinations against COVID-19.22 Its subset includes Remdesivir and hydroxychloroquine (Fig. 1 ).
Fig. 1.
Representative antiviral drugs and enantiomers of hydroxychloroquine.
Hydroxychloroquine is an early approved drug for the treatment of malaria.23 It is also used to treat diseases such as rheumatoid arthritis and lupus erythematosus.24 Studies have shown that hydroxychloroquine sulfate can inhibit the new coronavirus25., 26., 27. in vitro, and it is inferred that the activity of hydroxychloroquine is higher than chloroquine28 or Remdesivir. As a weak base, hydroxychloroquine is able to accumulate in acidic organelles, increasing the pH of endosomal/lysosomal to inhibit viral replication29 and prevents viral fusion into the cell.30 Additionally, cytokines IL-6 and IL-10 have been reported to be increased in response to SARS-CoV-2 infection.31 Both chloroquine and hydroxychloroquine have immunomodulatory effects and can suppress the immune response to avoid multiorgan failure and death caused by cytokine storm.32., 33. In a clinical trial treatment of 36 COVID-1934 patients, patients who received hydroxychloroquine therapy had a nasopharyngeal swab virus negative rate of 57.1% on the 6th day, while patients who received hydroxychloroquine in combination with azithromycin had 100% negative swab and the rate in the untreated control group was 12.5%. This shows that hydroxychloroquine, alone and combination with azithromycin are effective against SARS-CoV-2. Based on this trail with such as small number of patients, the US Food and Drug Administration approved hydroxychloroquine as a clinical sympathetic drug. In another study, chloroquine was found to be able to reduce the SARS-CoV-2 viral load and shorten the duration of viremia, with no serious adverse events observed.25 However, a comparative analysis of patients who received chloroquine, hydroxychloroquine or concomitant macrolide therapy in various regions of the world with other patients who did not receive these drugs found that it is impossible to prove that the use of chloroquine, hydroxychloroquine alone or together with macrolides is beneficial.35 On the contrary, these two drugs may lead to an increase in the frequency of ventricular arrhythmias and reduce the patient's survival.36., 37., 38. The WHO announced the failure of the “the solidarity trial”, which means that hydroxychloroquine did not achieve the desired effect in the treatment of COVID-19.39., 40.
Since the hydroxychloroquine used clinically is in racemic form, it is possible that only one of its enantiomers had antiviral activity, and other enantiomer may have toxicity, thus leading to undesirable results. In fact, in two earlier studies in rheumatoid arthritis, the pharmacokinetics of hydroxychloroquine were shown to be enantioselective, with higher blood levels of R-hydroxychloroquine than S-hydroxychloroquine in patients following the administration of racemic hydroxychloroquine,41., 42. implying a higher clearance of S-hydroxychloroquine,43., 44. which may suggest that a particular configuration of hydroxychloroquine plays an important role in the actual treatment.45 In the present study, therefore, we investigated the antiviral activity of the enantiomers of hydroxychloroquine and their toxicity in mice, trying to find an optimal enantiomer to inhibit SRAS-CoV-2 virus. Unlike other reported studies of hydroxychloroquine isomers against SARS-CoV-2,45., 46. we found for the first time that R-hydroxychloroquine had higher replication inhibitory activity against SARS-CoV-2 compared to S-hydroxychloroquine and was less toxic in mice.
2. Results
2.1. Synthesis
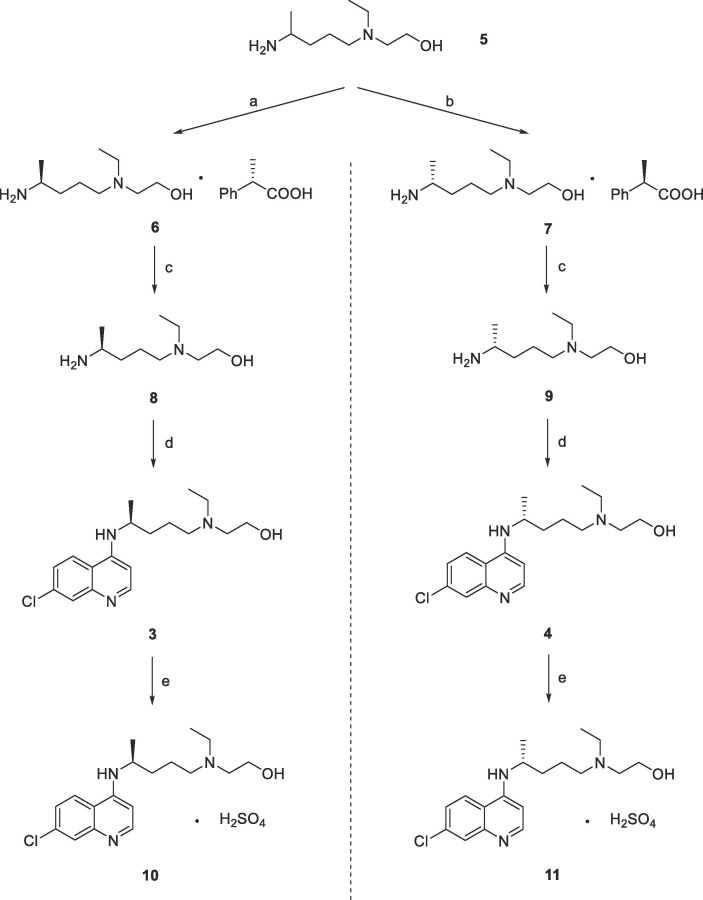
The synthetic routes of (S)-hydroxychloroquine sulfate and (R)- hydroxychloroquine sulfate were depicted in Schemes 1 . The synthesis began with the recrystallization of commercially available starting materials 5 with mandelic acid of different configurations as reported methods47. Chiral amine 8 or 9 was prepared from the mandelate 6 or 7 using sodium hydroxide to remove the corresponding mandelic acid. Subsequently, treatment of the amine with 4,7-dichloroquinoline gave compound 3 and 4, respectively, with good yields, followed by reaction with sulfuric acid furnished the final products hydroxychloroquine sulfate 10 and 11.
Scheme 1.
Synthesis of target compounds 10 and 11. Reagents and conditions: (a) S-(+)-mandelic acid, 2-propanol, recrystallization, 52%; (b) R-(−)-mandelic acid, 2-propanol, crystallization, 56%; (c) NaOH, tert-butyl methyl ether, 2 h, 63–83%; (d) 4,7-dichloroquinoline, TEA, K2CO3, 135 °C, 24 h, 62–68%; (e) H2SO4, EtOH, reflux, 67–85%.
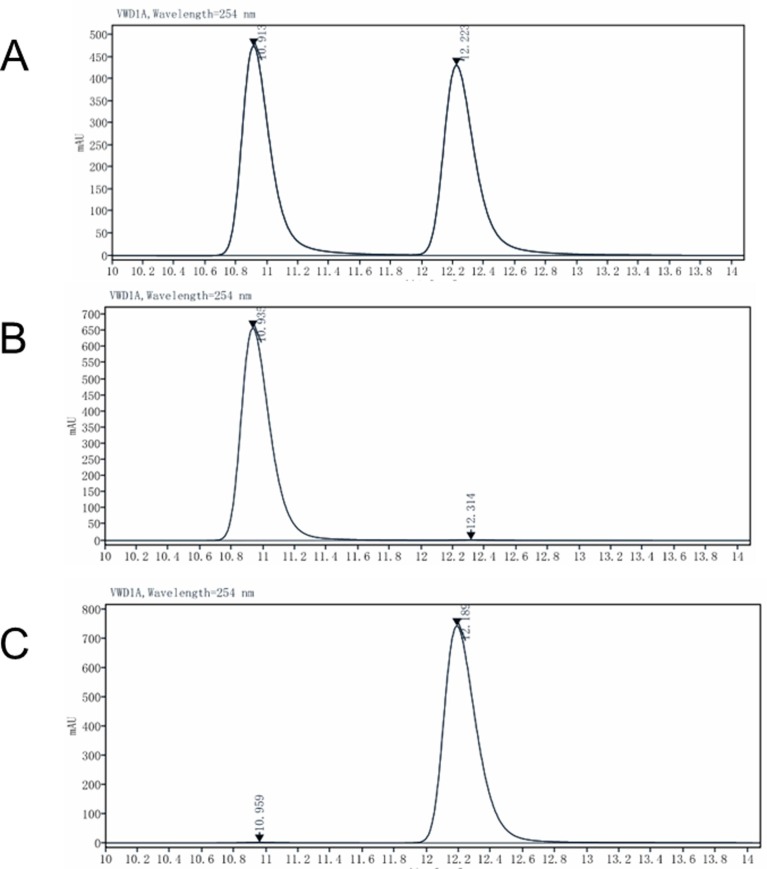
In order to test the purity and enantiomeric excess values of the asymmetric synthesis of hydroxychloroquine, we performed a new chiral HPLC analysis method, and the results are shown in Fig. 2 and Fig. S1 in Supporting information, SI. The purity of hydroxychloroquine exceeded 95%, and the enantiomeric excess values of hydroxychloroquine were all higher than 99%. The results confirm that we had indeed obtained a pair of enantiomers with high purity.
Fig. 2.
Chromatograms for racemic hydroxychloroquine and its enantiomers. A: racemic hydroxychloroquine (Rac-HCQ); B: (R)- hydroxychloroquine (R-HCQ); C: (S)- hydroxychloroquine (S-HCQ).
2.2. Preliminary biochemical evaluation of hydroxychloroquine and its Enantiomers
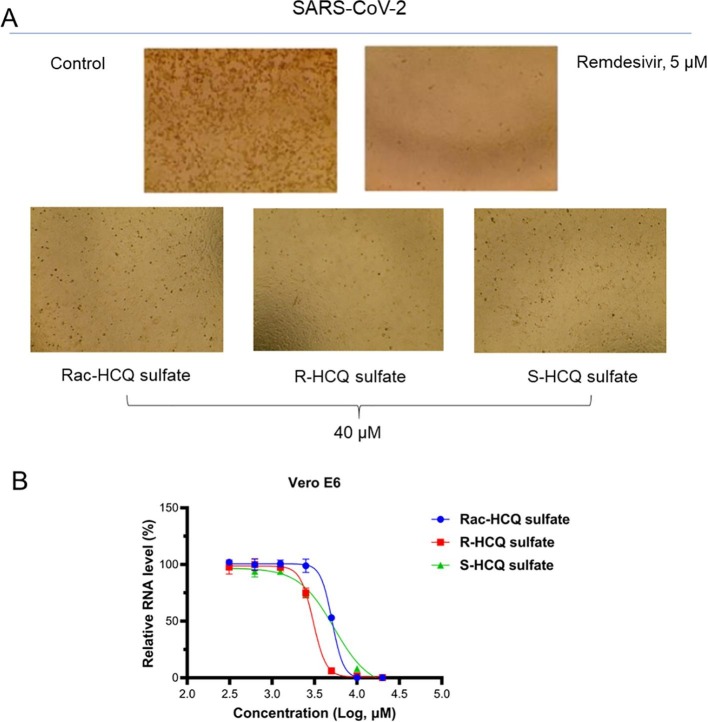
In order to evaluate the inhibitory effect of hydroxychloroquine and its enantiomers on the cytopathic effect (CPE) caused by the new coronavirus infection, we conducted an observational assay on the correlation between drug treatment and CPE. As shown in Fig. 3 A, obvious cytopathic changes occurred after Vero E6 cells were infected with SARS-CoV-2, while Remdesivir treatment completely inhibited the occurrence of lesions. The drug treatment group (racemic hydroxychloroquine sulfate, R-Hydroxychloroquine sulfate and S-hydroxychloroquine sulfate) also showed complete inhibition of cytopathic changes caused by the virus at 40 μM. These results indicated that the enantiomers of hydroxychloroquine had inhibitory activity against SARS-CoV-2.
Fig. 3.
Preliminary biochemical evaluation of hydroxychloroquine and its enantiomers. A: CPE observation trial of Rac-HCQ sulfate, R-HCQ sulfate and S-HCQ sulfate using Remdesivir as a positive control; B: viral RNA level detection trial.
To examine the effect of these three compounds on the removal of viral RNA, we further tested the level of viral RNA in the cell supernatant. Consistent with the above results, all three drugs showed effective inhibition on the replication of the virus (Fig. 3B). Among them, R-HCQ exhibited strongest inhibitory activity (EC50 = 3.05 μM), while the inhibitory levels of hydroxychloroquine in racemate (EC50 = 5.09 μM) and S configuration (EC50 = 5.38 μM) were comparable. These data implied that R-HCQ might be the most active one of the enantiomers of hydroxychloroquine against SARS-CoV-2.
2.3. In vivo acute toxicity evaluation
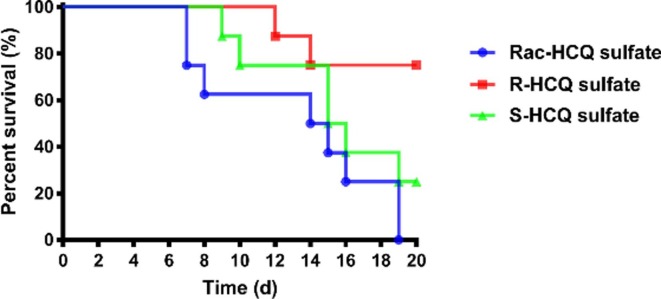
Having obtained in vitro antiviral activities of these three compounds, we further evaluated their toxicity in vivo in a double-dose acute toxicity assay. When administered orally at a dose of 230 mg/kg with the compounds, the mice treated with Rac-HCQ, R-HCQ and S-HCQ behaved differently. As described in Fig. 4 , the mice of the racemate group died at the 7th day post drug administration (dpd), and the mice of the S configuration group appeared malaise at 9 dpd, while the mice of the R configuration group performed well until the 12 dpd. More mice died after 15 dpd in the racemate group and S configuration group, and relatively stable number still maintained in R configuration group. Hence, the compound R-HCQ could be better tolerated in mice when compared to Rac-HCQ and S-HCQ.
Fig. 4.
The survival curve of mice in acute toxicity assay.
3. Discussion
The outbreak of SARS-CoV-2 has evolved into an emergent global pandemic. Although Remdesivir has been approved by the FDA as the first drug for the treatment of COVID-19, its effectiveness is still limited and the treatment was accompanied by many side effects, such as gastrointestinal symptoms and heart and lung failure. More importantly, the clinical application of Remdesivir did not reduce the mortality caused by COVID-19. Therefore, it is still urgent to find effective and low-toxic antiviral drugs that can control the infection.
We adopted the idea of drug repurposing and focus on hydroxychloroquine. The data showed for the application of the racemate hydroxychloroquine as an anti-COVID-19 drug for clinical research is, at best, mixed. We speculated that the reason for the inconsistent clinical effects of hydroxychloroquine may be due to the lower activity or higher toxicity of one of its enantiomers, which affects the antiviral effect of the racemate. Therefore, we obtained hydroxychloroquine with different configuration of side chains by using the technique of chiral resolution, and successfully furnished R- and S-hydroxychloroquine sulfate through chemical synthesis. The pair of enantiomers with an enantiomeric excess greater than 99% were obtained and we further tested the inhibitory activity of racemic hydroxychloroquine, R configuration hydroxychloroquine, and S configuration hydroxychloroquine against SARS-CoV-2 virus in Vero E6 cells. In vitro CPE observation and viral RNA level detection assays showed that both racemic hydroxychloroquine and optically pure hydroxychloroquine have inhibitory but limited activity on SARS-CoV-2. When compared to Rac-HCQ, R-HCQ showed more efficient antiviral activity, while the activity of S-HCQ was comparable to that of the Rac-HCQ. We further conducted the acute toxicity studies in vivo, the toxicity of R-HCQ was lower than that of racemate and S-HCQ. We will conduct long-term toxicity assay on the two enantiomers to complete the toxicity studies, and test the antiviral activity of R-HCQ in vivo.
4. Conclusion
Through chiral resolution and chemical synthesis, we successfully synthesized optically pure hydroxychloroquine. Among them, R configuration hydroxychloroquine exhibited higher antiviral activity (EC50 = 3.05 μM) than S configuration and racemic hydroxychloroquine. Acute toxicity tests in vivo showed that S-hydroxychloroquine had higher toxicity with racemates, while R configuration has lower toxicity. This is the first report that R-hydroxychloroquine has better in vivo toxicity than S-hydroxychloroquine. Based on the content above, the synthesis and biological functions of R-HCQ may be an important inspiration in promoting this field. Taken together, R-HCQ might be a promising lead compound for further investigation of antiviral drugs against SARS-CoV-2.
5. Materials and methods
5.1. Chemistry general methods
Reagents and solvents from commercial sources were used without further purification. The progress of all reactions was monitored by TLC using EtOAc/n-hexane or DCM/MeOH as solvent system, and spots were visualized by irradiation with UV light (254 nm) or staining with phosphomolybdic acid. Flash chromatography was performed using silica gel (300–400 mesh). 1H NMR and 13C NMR spectra were recorded on a Bruker Avance ARX-300 or a Bruker Avance ARX-400. Chemical shifts are reported in ppm, and multiplicity of signals are denoted as: s = singlet, d = doublet, t = triplet and m = multiplet. The low resolution ESIMS was recorded on an Agilent 1200 HPLC-MSD mass spectrometer and the high resolution on an Applied Biosystems Q-STAR Elite ESI-LC-MS/MS mass spectrometer. Anhydrous toluene and tetrahydrofuran (THF) were freshly distilled from sodium with benzophenone as the indicator. All other solvents were reagent grade. All moisture sensitive reactions were carried out in flame dried flask under argon atmosphere. The purity of the final compounds was determined by Agilent 1260 series HPLC system using the following conditions: chiralpak IH30CE-WB024 column (DAICEL, 0.46 cm I.D. × 25 cm × 3 μm) with the solvent system (elution conditions: mobile phase A consisting of n-hexane/diethylamine with a ratio of 100/0.1; mobile phase B consisting of isopropanol/methanol/diethylamine with a ratio of 75/25/0.1), with monitoring 254 nm. A flow rate of 0.8 mL/min was used and the column temperature is 35 °C. The retention time was reported as tR (min). The purity of final compounds is >95%.
5.2. Experimental procedures
(S)-2-((4-aminopentyl)(ethyl)amino)ethan-1-ol (S)- mandelate (6). A solution of 2-((4-aminopentyl)(ethyl)amino)ethan-1-ol (8.0 g, 45.9 mmol) in 2-propanol (100 mL) was added to a solution of S-(+)-mandelic acid (3.5 g, 23.0 mmol) in 2-propanol (20 mL). The mixture was stirred overnight at room temperature. Filtration gave white crystals which were recrystallized twice more from 2-propanol (100 mL and 80 mL respectively) to afford 6 (7.7 g, 52%) as white crystals.
(R)-2-((4-aminopentyl)(ethyl)amino)ethan-1-ol (R)- mandelate (7). Compound 7 was prepared from 5 and R-mandelic acid in a similar manner as described for 6. White crystals. (8.1 g, 56%).
(S)-2-((4-aminopentyl)(ethyl)amino)ethan-1-ol (8). To a solution of (S)-2-((4-aminopentyl)(ethyl)amino)ethan-1-ol (S)- mandelate (7 g, 21.6 mmol) in tert-butyl methyl ether (50 mL) was cooled to 0 °C. This clear solution was dropwise treated with 1 M NaOH aq. to adjust the pH of the mixture to 12. Upon complete addition, the reaction was warmed to r.t. over 2 h. The aqueous layer was extracted sextic with tert-butyl methyl ether (60 mL), and the organic layer washed with brine and dried over anhydrous Na2SO4, filtered and concentrated under reduced pressure to give the title compound (3.3 g, 83%) as colorless oil, which was used in the next step without further purification. 1H NMR (300 MHz, Chloroform-d): δ 3.52 (t, J = 5.4 Hz, 2H), 2.88 (h, J = 6.3 Hz, 1H), 2.60–2.49 (m, 4H), 2.44 (t, J = 7.3 Hz, 2H), 1.54–1.39 (m, 2H), 1.34–1.24 (m, 2H), 1.10–0.95 (m, 6H). 13C NMR (75 MHz, Chloroform-d): δ 77.27, 58.33, 54.93, 53.28, 47.22, 46.78, 37.77, 24.17, 24.04, 11.80. HRMS (ESI+): m/z calculated for C9H23N2O (M + 1)+ 175.1805 found 175.1808.
(R)-2-((4-aminopentyl)(ethyl)amino)ethan-1-ol (9). Compound 9 was prepared from 7 in a similar manner as described for 8. Colorless oil (2.6 g, 80%). 1H NMR (400 MHz, Chloroform-d): δ 3.52 (t, J = 5.4 Hz, 2H), 2.87 (h, J = 6.3 Hz, 1H), 2.60–2.50 (m, 4H), 2.44 (t, J = 7.3 Hz, 2H), 1.52–1.38 (m, 2H), 1.34–1.25 (m, 2H), 1.05 (d, J = 6.3 Hz, 3H), 1.00 (t, J = 7.1 Hz, 3H). 13C NMR (75 MHz, Chloroform-d): δ 77.26, 58.34, 54.91, 53.27, 47.21, 46.86, 37.82, 24.20, 24.11, 11.83. HRMS (ESI+): m/z calculated for C9H23N2O (M + 1)+ 175.1805 found 175.1808.
(S)-2-((4-((7-chloroquinolin-4-yl) amino) pentyl) (ethyl) amino) ethan-1-ol (3). The mixture of (S)-2-((4-aminopentyl) (ethyl)amino)ethan-1-ol (1.21 g, 7.0 mmol), 4,7-dichloroquinoline (1.1 g, 5.8 mmol), triethylamine (0.4 mL, 7.0 mmol), and potassium carbonate (400 mg, 2.9 mmol) were heat to 135 °C for overnight without any solvent. After reaction was complete, indicated by TLC, the resulting mixture was cooled down to room temperature. The mixture was extracted with DCM (3 × 100 mL) and the combined organic layers were dried over anhydrous Na2SO4, filtered and concentrated under reduced pressure. The resulting residue was passed through a silica gel column chromatography (0–5% methanol in DCM) to provide the title compound (1.3 g, yield 68%) as colorless oil. 1H NMR (300 MHz, Chloroform-d): δ 8.50 (d, J = 5.4 Hz, 1H), 7.94 (d, J = 2.2 Hz, 1H), 7.71 (d, J = 9.0 Hz, 1H), 7.35 (dd, J = 8.9, 2.2 Hz, 1H), 6.40 (d, J = 5.5 Hz, 1H), 4.96 (d, J = 7.7 Hz, 1H), 3.79–3.63 (m, 1H), 3.56 (t, J = 5.4 Hz, 2H), 2.61–2.54 (m, 4H), 2.50 (q, J = 4.4, 2.5 Hz, 2H), 1.75–1.52 (m, 4H), 1.32 (d, J = 6.3 Hz, 3H), 1.02 (t, J = 7.1 Hz, 3H). 13C NMR (75 MHz, Chloroform-d): δ 151.87, 149.20, 149.03, 134.88, 128.68, 125.20, 121.13, 117.20, 99.13, 58.29, 54.80, 52.98, 48.33, 47.48, 34.28, 23.94, 20.41, 11.63. [α]20 D= +95.8 (c = 1, EtOH). HRMS (ESI+): m/z calculated for C18H27ClN3O (M + 1)+ 336.1837 found 336.1841.
(R)-2-((4-((7-chloroquinolin-4-yl) amino) pentyl) (ethyl) amino) ethan-1-ol (4). Compound 4 was prepared from 9 in a similar manner as described for 3. Colorless oil (1.6 g, yield 62%). 1H NMR (300 MHz, Chloroform-d): δ 8.51 (d, J = 5.4 Hz, 1H), 7.94 (d, J = 2.2 Hz, 1H), 7.71 (d, J = 9.0 Hz, 1H), 7.35 (dd, J = 9.0, 2.2 Hz, 1H), 6.40 (d, J = 5.5 Hz, 1H), 4.96 (d, J = 7.7 Hz, 1H), 3.69 (dt, J = 13.5, 6.6 Hz, 1H), 3.56 (t, J = 5.4 Hz, 2H), 2.63–2.55 (m, 4H), 2.54–2.49 (m, 2H), 1.77–1.53 (m, 4H), 1.32 (d, J = 6.4 Hz, 3H), 1.02 (t, J = 7.1 Hz, 3H). 13C NMR (75 MHz, Chloroform-d): δ 151.58, 149.23, 148.90, 135.02, 128.39, 125.27, 121.36, 117.17, 99.07, 77.47, 77.05, 76.62, 58.14, 54.94, 53.05, 48.39, 47.64, 34.16, 23.74, 20.41, 11.40. [α]20 D = −86.5 (c = 1, EtOH). HRMS (ESI+): m/z calculated for C18H27ClN3O (M + 1)+ 336.1837 found 336.1840.
(S)-2-((4-((7-chloroquinolin-4-yl) amino) pentyl) (ethyl) amino) ethan-1-ol sulfate (10). A solution of (S)-2-((4-((7-chloroquinolin-4-yl) amino) pentyl) (ethyl) amino) ethan-1-ol (500 mg, 1.5 mmol) in EtOH (5 mL) was placed under an atmosphere of Ar and cooled to 0 °C. This clear solution was dropwise treated with a solution of 1 M sulfuric acid in EtOH (1.5 mL, 1.5 mmol). Upon complete addition, the reaction was warmed to r.t. for 2 h, then reflux for 6 h with the formation of a white precipitate. The solid was collected by filtration, washed with hexane, and dried to give 550 mg (85%) for the title compound as a white power. 1H NMR (300 MHz, Deuterium Oxide): δ 8.22 (d, J = 7.2 Hz, 1H), 8.11 (d, J = 9.0 Hz, 1H), 7.70 (d, J = 2.0 Hz, 1H), 7.51 (d, J = 9.0 Hz, 1H), 6.79 (d, J = 7.3 Hz, 1H), 4.14–4.02 (m, 1H), 3.82 (t, J = 5.2 Hz, 2H), 3.22 (q, J = 5.2, 4.7 Hz, 6H), 1.79 (s, 4H), 1.36 (d, J = 6.4 Hz, 3H), 1.25–1.17 (m, 3H). 13C NMR (75 MHz, Deuterium Oxide): δ 155.28, 142.08, 139.11, 137.99, 127.13, 124.00, 118.91, 115.07, 98.46, 55.20, 53.67, 51.92, 49.40, 48.18, 31.80, 19.91, 18.69, 7.80. [α]20 D= +81.3 (c = 1, H2O).
(R)-2-((4-((7-chloroquinolin-4-yl) amino) pentyl) (ethyl) amino) ethan-1-ol sulfate (11). Compound 11 was prepared from 4 in a similar manner as described for 10. White solid (232 mg, yield 67%). 1H NMR (300 MHz, Deuterium Oxide): δ 8.12 (d, J = 7.2 Hz, 1H), 7.98 (d, J = 9.1 Hz, 1H), 7.56 (d, J = 2.0 Hz, 1H), 7.38 (dd, J = 9.1, 2.1 Hz, 1H), 6.69 (d, J = 7.4 Hz, 1H), 4.05–3.93 (m, 1H), 3.74 (q, J = 5.4 Hz, 2H), 3.13 (t, J = 10.8 Hz, 6H), 1.71 (t, J = 6.5 Hz, 4H), 1.27 (d, J = 6.5 Hz, 3H), 1.13 (td, J = 7.3, 1.6 Hz, 3H). 13C NMR (75 MHz, Deuterium Oxide): δ 155.19, 142.04, 139.05, 137.91, 127.07, 123.96, 118.84, 114.99, 98.46, 55.17, 53.65, 51.90, 49.39, 48.14, 31.76, 19.92, 18.66, 7.84. [α]20 D = −89.3 (c = 1, H2O).
5.3. Cell culture
African green monkey kidney Vero E6 cells were obtained from Shanghai Cell Bank, Chinese Academy of Sciences. Cells were cultured at 37 °C with 5% CO2 in Dulbecco’s modified Eagle medium (DMEM) (Gibco, Carlsbad, USA) containing 2 mmol/L l-glutamine, 50 U/mL penicillin, 100 mg/mL streptomycin, and 10% (v/v) fetal bovine serum (Gibco, Carlsbad, USA). Vero E6 cells after SARS-CoV-2 infection were maintained in DMEM containing 2 mmol/L l-glutamine, 50 U/mL penicillin, 100 mg/mL streptomycin, and 2% (v/v) fetal bovine serum.
5.4. Cell Counting Kit-8 (CCK-8) assay
The Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) was used to evaluate cell viability and cytotoxicity according to the manufacturer’s instructions. Briefly, Vero E6 cells were dispensed into 96-well plate (1.0 × 104 cells/well), cultured in medium supplemented with different concentrations of the compounds for 48 h. After removal of the medium, the cells were incubated with fresh serum-free medium containing 10% CCK-8 for 1 h at 37 °C and then the absorbances at 450 nm were measured using a microplate reader (Bio-Rad, Hercules, USA).
5.5. In vitro CPE observation and viral RNA level detection trials
A clinical isolate of SARS-CoV-2 (GenBank: MT121215.1), was propagated in Vero E6 cells and the viral titer was determined as 50% tissue culture infectious dose (TCID50) per milliliter (mL) by CPE quantification. All the infection experiments were performed in the biosafety level-3 (BSL-3) laboratory of Fudan University.
Before virus infection, the drugs were diluted to the corresponding concentration by maintenance medium (DMEM with 2% FBS). Each well was added 100 μL of medium containing the corresponding concentration of the drugs and placed in a CO2 incubator for continuous cultivation for 1 h at 37 °C. After pretreatment of cells and drugs, each well was added two times the final concentration of diluted drug 60 μL. At the same time, cell control (CC) and virus control (VC) were set up with 120 μL and 60 μL of maintenance medium, respectively. Subsequently, in addition to the cell control, the virus (diluted to 100 TCID50/50 μL) was dropped vertically onto the 96-well plate to make the volume of the virus-drug mixture 120 μL (Note: After co-incubating 60 μL of virus dilution and 60 μL of drug dilution, use 100 μL of virus-drug mixture to infect the cells so that the final amount of virus infected cells is 100 TCID50/well). After mixing the added virus-antibody on a shaker and removed the supernatant (100 μL) of the culture plate seeded with cells, the residue was added 100 μL/well of the virus-drug mixture. Cells were placed in a 37 °C CO2 incubator for 48 h and observed the cytopathic changes with an inverted microscope to determine the antiviral ability of the drugs. Viral RNA level detection follows the steps below: Initially, cells were placed in a 37 °C CO2 incubator for continuous culture for 1 h, and the supernatant medium was discarded. After washed twice with PBS, 100 μL of maintenance medium was added to each well, and placed in a 37 °C CO2 incubator for continuous culture for 48 h. Subsequently, 100 μL of the supernatant was added to 300 μL of TRIzol LS (Invitrogen, Carlsbad, USA) to extract viral RNA for RT-qPCR detection following the manufacturer’s instructions. RT-qPCR was performed by using Verso 1-step RT-qPCR Kit (Thermo Fisher, Waltham, USA) on CFX96™ Real-Time PCR System (Bio-Rad, Hercules, CA). The PCR primers targeting the N gene (nt608-706) of SARS-CoV-2 were: 5′-GGGGAACTTCTCCTGCTAGAAT-3′/5′-CAGACATTTTGCTCTCAAGCTG-3′ (forward/reverse).
5.6. In vivo acute toxicity assay
All the procedures for animal handling, care, and the treatment in this study were performed according to the guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of China Pharmaceutical University following the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). The compounds Rac-HCQ sulfate, R-HCQ sulfate and S-HCQ sulfate were dissolved in physiological saline. Sixteen male and 16 female mice (25 g) were divided into four groups (n = 8): male and female control groups and male and female test groups. All mice were fasted overnight and then administered intragastrically with the vehicle or 230 mg/ kg of test compounds twice a day. The death, daily behavior, and body weight of the mice were monitored during the subsequent 20 days.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the National Natural Science Foundation (81773559), and the Double First-Class University Project (CPU2018GY03) (S.J.) This work was also supported by Major science and technology project for the prevention and treatment of major infectious diseases (2018ZX10301208), and National base cultivation project (20DZ2210404).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bmc.2021.116523.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ceccarelli M., Berretta M., Venanzi Rullo E., Nunnari G., Cacopardo B. Differences and similarities between Severe Acute Respiratory Syndrome (SARS)-CoronaVirus (CoV) and SARS-CoV-2. Would a rose by another name smell as sweet? Eur Rev Med Pharmacol Sci. 2020;24:2781–2783. doi: 10.26355/eurrev_202003_20551. [DOI] [PubMed] [Google Scholar]
- 3.Perrella A., Carannante N., Berretta M., et al. Novel Coronavirus 2019 (Sars-CoV2): a global emergency that needs new approaches? Eur Rev Med Pharmacol Sci. 2020;24:2162–2164. doi: 10.26355/eurrev_202002_20396. [DOI] [PubMed] [Google Scholar]
- 4.Zhou F., Yu T., Du R., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nord J.E., Shah P.K., Rinaldi R.Z., Weisman M.H. Hydroxychloroquine cardiotoxicity in systemic lupus erythematosus: a report of 2 cases and review of the literature. Semin Arthritis Rheu. 2004;33:336–351. doi: 10.1016/j.semarthrit.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Korber B., Fischer W.M., Gnanakaran S., et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wrapp D., Wang N., Corbett K.S., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom; 2020. https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-rapid-increase-sars-cov-2-variant-united-kingdom#no-link [accessed 2021-01-11].
- 9.WHO Coronavirus Disease (COVID-19) Dashboard; 2021. https://covid19.who.int/ [accessed 2021-01-11].
- 10.Cannalire R., Cerchia C., Beccari A.R., Di Leva F.S., Summa V. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. J Med Chem. 2020 doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman R.L., Kania R.S., Brothers M.A., et al. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J Med Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Namchuk M.N. Early returns on small molecule therapeutics for SARS-CoV-2. ACS Infect Dis. 2021;7:1298–1302. doi: 10.1021/acsinfecdis.0c00874. [DOI] [PubMed] [Google Scholar]
- 13.Owen D.R., Allerton C.M., Anderson A.S., et al. An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021 doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 14.Riva L., Yuan S., Yin X., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy J., Goren A., Cadegiani F.A., et al. Proxalutamide reduces the rate of hospitalization for COVID-19 male outpatients: a randomized double-blinded placebo-controlled trial. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.668698. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Cadegiani F.A., McCoy J., Gustavo Wambier C., et al. Proxalutamide significantly accelerates viral clearance and reduces time to clinical remission in patients with mild to moderate COVID-19: results from a randomized, double-blinded, placebo-controlled trial. Cureus. 2021;13:e13492. doi: 10.7759/cureus.13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khoo S.H., Fitzgerald R., Fletcher T., et al. Optimal dose and safety of molnupiravir in patients with early SARS-CoV-2: a Phase I, open-label, dose-escalating, randomized controlled study. J Antimicrob Chemother. 2021 doi: 10.1093/jac/dkab318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Do T.N.D., Donckers K., Vangeel L., et al. A robust SARS-CoV-2 replication model in primary human epithelial cells at the air liquid interface to assess antiviral agents. Antiviral Res. 2021;192:105122. doi: 10.1016/j.antiviral.2021.105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercorelli B., Palù G., Loregian A. Drug repurposing for viral infectious diseases: how far are we? Trends Microbiol. 2018;26:865–876. doi: 10.1016/j.tim.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pushpakom S., Iorio F., Eyers P.A., et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 21.Rosa S.G.V., Santos W.C. Clinical trials on drug repositioning for COVID-19 treatment. Revista panamericana de salud publica = Pan Am J Publ Health. 2020;44 doi: 10.26633/RPSP.2020.40. e40-e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kupferschmidt K., Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020;367:1412–1413. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- 23.Parhizgar A.R., Tahghighi A. Introducing new antimalarial analogues of chloroquine and amodiaquine: a narrative review. Iran J Med Sci. 2017;42:115–128. [PMC free article] [PubMed] [Google Scholar]
- 24.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases. Lancet Infect Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. BioScience Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 26.Xueting Yao F.Y., Zhang M., Cui C., et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J., Cao R., Xu M., et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discovery. 2020;6 doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao X., Ye F., Zhang M., et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L.F., Lin Y.S., Huang N.C., et al. Hydroxychloroquine-inhibited dengue virus is associated with host defense machinery. J Interferon Cytokine Res. 2015;35:143–156. doi: 10.1089/jir.2014.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mauthe M., Orhon I., Rocchi C., et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrezenmeier E., Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 33.Zhao M. Cytokine storm and immunomodulatory therapy in COVID-19: role of chloroquine and anti-IL-6 monoclonal antibodies. Int J Antimicrob Ag. 2020;55 doi: 10.1016/j.ijantimicag.2020.105982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gautret P., Lagier J.C., Parola P., et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Pathak D.S.K., Salunke D.A.A., Thivari D.P. No benefit of hydroxychloroquine in COVID-19: results of systematic review and meta-analysis of randomized controlled trials. Diab Metab Synd: Clin Res Rev. 2020;14:1673. doi: 10.1016/j.dsx.2020.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giudicessi J.R., Noseworthy P.A., Friedman P.A., Ackerman M.J. Urgent guidance for navigating and circumventing the QTc-prolonging and torsadogenic potential of possible pharmacotherapies for coronavirus disease 19 (COVID-19) Mayo Clin Proc. 2020;95:1213–1221. doi: 10.1016/j.mayocp.2020.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang W., Cao Z., Han M., et al. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. Bmj-British Med J. 2020;369 doi: 10.1136/bmj.m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borba M.G.S., Val F.F.A., Sampaio V.S., et al. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection a randomized clinical trial. Jama Netw Open. 2020;3:e208857. doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PubMed] [Google Scholar]
- 39.“Solidarity” clinical trial for COVID-19 treatments; 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments [accessed 2021-01-11].
- 40.Maisonnasse P., Guedj J., Contreras V., et al. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature. 2020;585:584–587. doi: 10.1038/s41586-020-2558-4. [DOI] [PubMed] [Google Scholar]
- 41.Tett S.E., McLachlan A.J., Cutler D.J., Day R.O. Pharmacokinetics and pharmacodynamics of hydroxychloroquine enantiomers in patients with rheumatoid arthritis receiving multiple doses of racemate. Chirality. 1994;6:355–359. doi: 10.1002/chir.530060420. [DOI] [PubMed] [Google Scholar]
- 42.McLachlan A.J., Tett S.E., Cutler D.J., Day R.O. Disposition of the enantiomers of hydroxychloroquine in patients with rheumatoid arthritis following multiple doses of the racemate. Br J Clin Pharmacol. 1993;36:78–81. doi: 10.1111/j.1365-2125.1993.tb05897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brocks D.R., Skeith K.J., Johnston C. Hematologic disposition of hydroxychloroquine enantiomers. J Clin Pharmacol. 1994;34:1088–1097. doi: 10.1002/j.1552-4604.1994.tb01986.x. [DOI] [PubMed] [Google Scholar]
- 44.Ducharme H.F.J., Ducharme M.P., Khalil S.K., Wainer I.W. Enantioselective disposition of hydroxychloroquine after a single oral dose of the racemate to healthy subjects. Br J Clin Pharmacol. 1995;40:127–133. doi: 10.1111/j.1365-2125.1995.tb05768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D'Acquarica I., Agranat I. Chiral switches of chloroquine and hydroxychloroquine: potential drugs to treat COVID-19. Drug Discov Today. 2020;25:1121–1123. doi: 10.1016/j.drudis.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li G., Sun J., Huang Y.-Y., et al. Enantiomers of chloroquine and hydroxychloroquine exhibit different activities against SARS-CoV-2 in vitro, evidencing S-hydroxychloroquine as a potentially superior drug for COVID-19. bioRxiv. 2020 [Google Scholar]
- 47.Blaney P.M., Byard S.J., Carr G., et al. A practical synthesis of the enantiomers of hydroxychloroquine. Tetrahedron Asymmetry. 1994;5:1815–1822. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.