Abstract
The import motor for preproteins that are targeted into the mitochondrial matrix consists of the matrix heat shock protein Hsp70 (mtHsp70) and the translocase subunit Tim44 of the inner membrane. mtHsp70 interacts with Tim44 in an ATP-dependent reaction cycle, binds to preproteins in transit, and drives their translocation into the matrix. While different functional mechanisms are discussed for the mtHsp70-Tim44 machinery, little is known about the actual mode of interaction of both proteins. Here, we have addressed which of the three Hsp70 regions, the ATPase domain, the peptide binding domain, or the carboxy-terminal segment, are required for the interaction with Tim44. By two independent means, a two-hybrid system and coprecipitation of mtHsp70 constructs imported into mitochondria, we show that the ATPase domain interacts with Tim44, although with a reduced efficiency compared to the full-length mtHsp70. The interaction of the ATPase domain with Tim44 is ATP sensitive. The peptide binding domain and carboxy-terminal segment are unable to bind to Tim44 in the absence of the ATPase domain, but both regions enhance the interaction with Tim44 in the presence of the ATPase domain. We conclude that the ATPase domain of mtHsp70 is essential for and directly interacts with Tim44, clearly separating the mtHsp70-Tim44 interaction from the mtHsp70-substrate interaction.
Most proteins of the mitochondrial matrix are encoded by nuclear genes and are synthesized on cytosolic polysomes (29, 42, 44, 52, 62). The preproteins typically carry amino-terminal targeting sequences (presequences) that direct the proteins to the translocase of the outer mitochondrial membrane (designated TOM), consisting of receptors and a general import pore. The preproteins traverse the import pore in an (at least partially) unfolded state. The presequences then contact the translocase of the inner membrane (TIM), and the preproteins are translocated through the inner membrane channel that is apparently formed by Tim23 and Tim17 (TIM23 complex). Two driving forces for preprotein translocation are known. (i) The membrane potential ΔΨ is required for transport of the presequences across the inner membrane. ΔΨ (negative inside) is thought to exert an electrophoretic effect on the positively charged presequences (35) and/or to modulate the activity of Tim23 (5). (ii) The heat shock protein Hsp70 of the mitochondrial matrix (mtHsp70) forms an ATP-dependent import motor in cooperation with Tim44 of the inner membrane (27, 41, 60). Tim44 is a peripheral subunit of the TIM23 complex and mainly exposed to the matrix space (6, 7, 64).
The role of mtHsp70, termed Ssc1 in the yeast Saccharomyces cerevisiae, in driving the import of preproteins into the mitochondrial matrix is currently discussed in different models. A first model predicts that mtHsp70 traps unfolded preprotein segments emerging in the matrix and prevents their backsliding (known as the trapping, Brownian ratchet, or hand-over-hand model) (4, 41, 54, 55). A second model implies an active force generation by mtHsp70 such that the polypeptide chain is pulled into mitochondria; it is thought that a part of mtHsp70 is anchored to the inner membrane via Tim44 while another part binds the polypeptide chain and pulls it in by a nucleotide-induced conformational change (the pulling or motor model) (18, 36, 45, 63). The observed complex formation between mtHsp70 and Tim44 has also been incorporated into the trapping model such that the prebinding of mtHsp70 to the inner membrane increases the local concentration of mtHsp70 at the import site. When a preprotein segment arrives, the mtHsp70 is then transferred from Tim44 to the unfolded polypeptide chain, followed by a further mtHsp70 that was prebound to a second Tim44 (hand-over-hand model) (4, 41). Recent results with temperature-sensitive mutants of the essential Ssc1 revealed that a single mechanism is not enough to explain the role of mtHsp70 in protein import, indicating that both mechanisms, trapping and pulling, contribute to the translocation of preproteins (60).
While the detailed discussion of the functional mode of mtHsp70 action is ongoing (4, 24, 28, 29, 41, 46, 60), very little is known about the structural prerequisites of interaction between mtHsp70 and Tim44. Like other members of the Hsp70 family, mtHsp70 is composed of three portions: an amino-terminal ATPase domain (A) of 42 kDa, the peptide binding domain (P) of 20 kDa, and a carboxy-terminal segment (C) of 6 kDa that shows the highest variability between different Hsp70s (20). It is unknown which part of mtHsp70 is actually binding to Tim44. Information about this interaction will have interesting implications on the mode of action of mtHsp70-Tim44. In the trapping model of Bauer et al. (4), the peptide binding domain of mtHsp70 is in contact with Tim44, rendering a simultaneous contact of mtHsp70 with both preprotein and Tim44 unlikely. The pulling model, however, would predict that another portion of mtHsp70 should bind to Tim44 such that conformational changes could be converted into a pulling force on the polypeptide chain (kept by the peptide binding domain) while mtHsp70 is still in contact with Tim44.
For this report, we divided mtHsp70 into distinct portions and analyzed their interaction with Tim44 under two experimental conditions, in the two-hybrid system and in organello. Both systems revealed that the ATPase domain of mtHsp70 is the primary site of interaction with Tim44, excluding that the binding to Tim44 resembles a substrate-type interaction. Additionally, the peptide binding domain and the carboxy-terminal segment enhance the stability of binding, raising interesting comparisons to the mode of interaction of Hsp70 with other partner proteins.
MATERIALS AND METHODS
S. cerevisiae strains.
The S. cerevisiae strains used in this study are as follows: for the two hybrid-system, the reporter strain Y190 (MATa ade2-101 his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-901 tyr1-501 canR gal4Δ gal80Δ chyS URA3::GAL-lacZ LYS2::GAL-HIS3) (56); for isolation of mitochondria and protein import studies, strain PK82 (wild type; MATα his4-713 lys2 ura3-52 Δtrp1 leu2-3,112) and strain PK81 [ssc1-2; MATα ade2-101 lys2 ura3-52 Δtrp1 leu2-3,112 ssc1-2(LEU2)] (15).
Two-hybrid assay.
The two-hybrid assay was based on the method of Fields and Song (13). The tested genes were fused to the trans-activating domain of GAL4 in the vector pPC86 and to the DNA-binding domain (DB) of GAL4 in the vector pPC97 (9). The cotransformation of two-hybrid vectors into strain Y190 was performed according to Gietz and Sugino (17). The transformed yeast cells were plated onto complete synthetic medium containing 2% glucose without tryptophan (selection for pPC86), leucine (selection for pPC97), and histidine but containing 20 mM of 3-aminotriazole (selection for two-hybrid interaction of constructs). β-galactosidase filter assays were performed according to Rehling et al. (50).
To construct the two-hybrid fusion proteins [GAL4 activator domain (AD) or GAL4 DB fused in frame to mature-sized TIM44, SSC1(APC), SSC1(A), or SSC1(PC), the genes were first amplified from yeast genomic DNA in a PCR-based approach using VentR polymerase and the following oligonucleotides (containing the indicated restriction sites): TIM44 (1,181 bp), 5′-CACGCGGTCGACCAACCCTCGATCACCACTCC-3′ (SalI) and 5′-GGCACTAGTCAGGTGAATTGTCTAG-3′ (SpeI); SSC1(APC) (1,894 bp), 5′-GGAAGATCTACCAGTCAACCAAGGTTCAAGG-3′ (BglII) and 5′-GGCGAGCTCTTACTGCTTAGTTTCACC-3′ (SacI); SSC1(A) (1,170 bp), 5′-GGAAGATCTACCAGTCAACCAAGGTTCAAGG-3′ (BglII) and 5′-GGCCTCGAGTTAGACGTCAGTAACCTCACC-3′ (AatII); and SSC1(PC) (750 bp), 5′-GGAAGATCTACTTATTATTAGATGTTACCCC-3′ (BglII) and 5′-GGCGAGCTCTTACTGCTTAGTTTCACC-3′ (SacI). The PCR products were subcloned into SmaI-digested pGEM-4Z, and then the inserts were recovered by digestion with SalI and SpeI (TIM44) or BglII and AatII or SacI (SSC1) and ligated into SalI- and SpeI- (TIM44) or BglII- and AatII- or SacI-digested (SSC1) pPC86 or pPC97 to give the in-frame fusion genes. As Escherichia coli host, XL-1 Blue was used and transformed according to the CaCl2 method (22).
In vitro import of preproteins into isolated mitochondria.
For in vitro transcription, the precursor form pSSC1 was cloned into pGEM-4Z. First, the pSSC1 gene was amplified from yeast genomic DNA with the oligonucleotide 5′-GGTGCGGTGTATAAAAACG-3′ as upper primer and 5′-TTACTGCTTAGTTTCACCAGATTCGG-3′ as lower primer with cloned Pfu polymerase. The PCR product (2,185 bp) was purified from 0.5% agarose gel and blunt-end ligated into SmaI-digested pGEM-4Z to give the plasmid pTK201. For construction of pSSC1(PC), an inverse PCR with cloned Pfu polymerase on pTK201 was performed, using the upper primer 5′-TTATTATTAGATGTTACCCCATTG-3′ and the lower primer 5′-AACCTTGGTTGACTGCAAACGTG-3′. The PCR product was cleaned by gel extraction and blunt-end ligated to give the plasmid pTK202. Sequences of both plasmids were verified by sequencing. As templates for transcription, PCR products were used, which were obtained using the following oligonucleotides and templates in VentRpolymerase-PCR assays. The upper primer, which was used for all four constructs (5′-ATTTAGGTGACACTATAGAAGNGGGTGCGGTGTATAAAAACG-3′) carried the SP6-RNA-polymerase consensus sequence (underlined) in front of the target sequence. The following lower primers were used: pSSC1(APC) (pTK201 as template), 5′-TCACACAGGAAACAGCTATGAC-3′; pSSC1(A) (pTK201 as template), 5′-CTGCAGTCATTGGAGTGGCCTG-3′; pSSC1(AP) (pTK201 as template), 5′-GGAATTTGCTTTTTAAGCAACCAACTCCTTCAAGG-3′; pSSC1(PC) (pTK202 as template), 5′-TCACACAGGAAACAGCTATGAC-3′. The preproteins were synthesized in rabbit reticulocyte lysates in the presence of [35S]methionine and [35S]cysteine.
The S. cerevisiae strains PK81 and PK82 were grown in yeast-extract-peptone medium containing 3% glycerol (YPG medium). Mitochondria were isolated as previously described (11, 23). The import reactions were performed in bovine serum albumin (BSA)-containing buffer (250 mM sucrose, 3% [wt/vol] fatty-acid-free BSA, 80 mM KCl, 5 mM MgCl2, and 10 mM morpholinepropanesulfonic acid [MOPS]-KOH [pH 7.4]), including 2 mM ATP, 2 mM NADH, and isolated mitochondria (100 μg of mitochondrial protein/ml) at 25°C (1, 57). The import was stopped by dissipating the membrane potential (ΔΨ) by the addition of 1 μM valinomycin. Control samples (−ΔΨ) received 1 μM valinomycin before the import reaction was started.
For induction of the temperature-sensitive phenotype of ssc1-2 mitochondria, the mitochondria were reisolated by centrifugation at 16,000 × g for 10 min, resuspended in BSA-containing buffer, and incubated for 15 min at 37°C. After the samples were cooled to 4°C, they were split into halves. One half was treated with proteinase K at a final concentration of 100 μg/ml for 10 min at 0°C. The protease was inactivated by the addition of 1 mM phenylmethylsulfonyl fluoride (PMSF) to all samples and incubation for 5 min at 0°C. After two washing steps with SEM (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH [pH 7.4], 1 mM PMSF) were carried out, the pelleted mitochondria were resuspended in electrophoresis sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) and storage phosphorimaging technology (Molecular Dynamics).
Lysis of mitochondria and coimmunoprecipitation of imported proteins.
The interaction of mtHsp70 or constructs with Tim44 in organello was analyzed by a method modified after that of Voos et al. (63). Mitochondria with imported 35S-labeled proteins were pelleted (16,000 × g for 10 min) washed three times with SEM, and resuspended in lysis buffer (0.3% [vol/vol] Triton X-100 or 1% digitonin, 50 mM KCl, 20 mM Tris-HCl [pH 7.4], 3% (wt/vol) BSA, 10% (vol/vol) glycerol, 1 mM PMSF, 1× proteinase inhibitor mix [PIM; final concentration: antipain, 2 μg/ml; aprotinin, 5 μg/ml; chymotrypsin, 0.25 μg/ml; leupeptin, 1.25 μg/ml; pepstatin A, 0.5 μg/ml]), containing either 5 mM EDTA or 1 mM ATP–5 mM magnesium acetate. The lysis was performed by carefully resuspending the mixture six times, shaking it for 30 s, and incubating it for 5 min at 0°C. After a clarifying spin (16,000 × g; 5 min), the supernatants were transferred to antibodies directed against Tim44 or Ssc1 that were bound to protein A-Sepharose in lysis buffer containing BSA and 1× PIM. After being rotated end over end for 1 h at 4°C, the protein A-Sepharose beads were washed three times in lysis buffer without BSA, containing 1 mM PMSF and 1× PIM. When digitonin was used, an additional washing step in 10 mM Tris-HCl, pH 7.4, was performed to remove residual digitonin. Bound proteins were eluted by the addition of electrophoresis sample buffer, separated by SDS-PAGE, and detected by storage phosphorimaging technology. For detection of coprecipitated endogenous mtHsp70 (Ssc1), proteins that were bound to protein A-Sepharose-bound antibodies were eluted by treatment with 100 mM glycine, pH 2.5, precipitated with trichloroacetic acid, separated by SDS-PAGE, and subjected to immunodecoration.
Miscellaneous methods.
Cell lysates were prepared by resuspending yeast cells from overnight cultures in electrophoresis sample buffer. Standard techniques were used for the manipulation of yeast and E. coli DNA and immunodecoration (2, 21).
RESULTS
The ATPase domain of mtHsp70 (or Ssc1) interacts with Tim44 in the two-hybrid system.
To assay for interactions between mtHsp70 and Tim44 in the yeast two-hybrid system (9, 10, 13), yeast Ssc1 was used as the full-length mature protein, henceforth termed Ssc1(APC), as well as split into the ATPase domain [Ssc1(A)] and the peptide binding domain plus the carboxy-terminal segment [Ssc1(PC)]. Each construct was cloned in frame to the Gal4 AD. Full-length mature Tim44 was cloned to the Gal4 DB. Physical interaction between the domains was expected to lead to β-galactosidase expression and His prototrophy. Full-length Ssc1(APC) fused to the carboxy-terminus of the AD led to the efficient growth of transformants on medium lacking histidine with Tim44 fused to the carboxy-terminus of the DB (Fig. 1A), while no growth was observed when DB-Tim44 was tested with the AD itself (Fig. 1A). The ATPase domain Ssc1(A) fused to the AD led to reduced, but significant, growth with DB-Tim44 (Fig. 1A). No growth, however, was detectable when the peptide binding domain plus the carboxy-terminal segment Ssc1(PC) fused to the AD were tested together with DB-Tim44 (Fig. 1A). Similar results were obtained with the β-galactosidase filter assay (data not shown). Moreover, an interaction between two Tim44 molecules, one fused to the DB and one to the AD, was indicated by both growth and the β-galactosidase assay (results not shown) in agreement with the recent biochemical observation of a homodimer formation of Tim44 (41). We controlled the expression of the AD-Ssc1(PC) construct by immunodecoration of total cell extracts with an antiserum directed against Hsp70s. AD-Ssc1(PC) was efficiently expressed (Fig. 1B, lane 3), as were AD-Ssc1(A), AD-Ssc1(APC), and DB-Tim44 (Fig. 1B, lanes 2, 4, and 6).
FIG. 1.
Interaction of Tim44 and the ATPase domain of mtHsp70 (Ssc1) in the two-hybrid system. (A) Two-hybrid interaction of Ssc1(APC) and Ssc1(A) with Tim44. The reporter yeast strain Y190 was transformed with the two-hybrid plasmids expressing the indicated fusion proteins. Transformants were grown for 2 days at 30°C on minimal medium lacking tryptophan and leucine. One clone of each transformation was streaked out on minimal medium lacking tryptophan, leucine, and histidine but containing 3-aminotriazole and cultured for 3 days at 30°C. (B) The two-hybrid fusion proteins are expressed in yeast. The yeast strain Y190 was transformed as described in the legend to panel in A. One clone of each transformation was cultured overnight in minimal medium lacking tryptophane and leucine. Cells of a 1-ml liquid culture were resuspended in 15 μl of electrophoresis sample buffer and separated by SDS-PAGE. The proteins were detected by immunodecoration with antibodies directed against Hsp70 or Tim44. endog., endogenous (authentic) Hsp70s and Tim44 reacting with the antibodies.
We conclude that the two-hybrid analysis suggests an interaction between Tim44 and the ATPase domain of mtHsp70, although the interaction is not as efficient as that between Tim44 and full-length mtHsp70. To assess the significance of this observation we went to a biochemical approach and analyzed the interaction between Ssc1 constructs and Tim44 in its physiological environment, i.e., in organello.
The ATPase domain of Ssc1 associates with Tim44 in organello but with lower stability than full-length Ssc1.
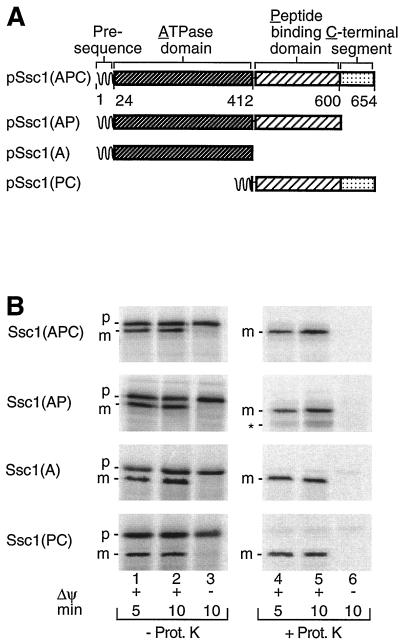
We prepared several constructs consisting of one or more domains of Ssc1 (Fig. 2A): Ssc1(AP) lacked the carboxy-terminal segment but contained the ATPase domain A and the peptide binding domain P; Ssc1(A) only contained the ATPase domain; and Ssc1(PC) lacked the ATPase domain but contained the peptide binding domain and the carboxy-terminal segment. Each construct received the entire Ssc1 presequence, to allow efficient import into isolated mitochondria (plus four amino acids of the mature part to preserve the cleavage site of the processing peptidase). The preproteins were synthesized in rabbit reticulocyte lysate in the presence of [35S]methionine and [35S]cysteine and incubated with isolated S. cerevisiae mitochondria. In the presence of a membrane potential (ΔΨ) across the inner membrane, a considerable fraction of the preproteins was imported into mitochondria as evidenced by the proteolytic processing to the mature-sized forms (Fig. 2B, lanes 1 and 2) and transport to a protease-protected location (Fig. 2B, lanes 4 and 5). The precursor forms that accumulated also in the absence of a ΔΨ (Fig. 2B, lane 3) were located on the mitochondrial surface, as they were digested by added protease (Fig. 2B, lane 6).
FIG. 2.
Separation of mtHsp70 into domains and import into isolated mitochondria. (A) Ssc1 constructs used. (B) Import into mitochondria. Rabbit reticulocyte lysates with radiolabeled mitochondrial preproteins (2% [vol/vol] of import reaction mixture) were incubated with isolated yeast wild-type mitochondria (25 μg of mitochondrial protein/100 μl of import reaction mixture) for the indicated times in the presence (+) or absence (−) of a ΔΨ. Import was stopped by the addition of 1 μM valinomycin. Half of the samples were treated with proteinase K (Prot. K). After reisolation of the mitochondria and separation by SDS-PAGE, imported proteins were detected by digital autoradiography. The asterisk indicates a fragment of Ssc1(AP) that was generated in small amounts by proteinase K upon incubation of the construct with mitochondria in the presence of a ΔΨ, apparently representing an incompletely imported membrane-spanning form of the protein; the quantitations shown in the following figures did not include this fragment but are only based on the fully imported mature forms. m, mature form; p, precursor form.
All subsequent experiments on the interaction between Ssc1 constructs and Tim44 were performed with the fully imported (i.e., protease-protected) mature-sized constructs. The experimental approach is outlined in Fig. 3A. After import of the constructs into mitochondria for 10 min in the presence of a ΔΨ, the ionophore valinomycin was added to dissipate the ΔΨ, and proteins that were not fully imported were degraded by added proteinase K. The mitochondria were reisolated, washed, and lysed in Triton X-100 in the absence or the presence of ATP, followed by coimmunoprecipitation with antibodies directed against Tim44. To assess the validity of the approach, we first assessed the association of the full-length protein Ssc1(APC) with Tim44. In the absence of ATP, a fraction of Ssc1(APC) was recovered with anti-Tim44 (Fig. 3B, top panel, lane 2), whereas no Ssc1(APC) was found in association with Tim44 in the presence of ATP (Fig. 3B, top panel, lane 3). This agrees with the nucleotide-sensitive interaction observed for the interaction of endogenous Ssc1 and Tim44 (26, 33, 48, 54, 55, 61). The constructs Ssc1(AP) and Ssc1(A) showed a reduced, yet significant, interaction with Tim44 in an ATP-sensitive manner (Fig. 3B, middle panels, lanes 2 and 3), whereas no interaction was observed between Ssc1(PC) and Tim44 (Fig. 3B, lower panel, lanes 2 and 3). We quantified the efficiency of interaction of the constructs with Tim44 in comparison to that of the full-length (wild-type) Ssc1(APC). Ssc1(AP) yielded 40 to 50% of the wild-type efficiency, while the ATPase domain alone [Ssc1(A)] gave only 10% efficiency (Fig. 3C).
FIG. 3.
ATP-sensitive interaction of the Ssc1 ATPase domain with Tim44. (A) Experimental approach. (B) ATP-sensitive coprecipitation with anti-Tim44. Rabbit reticulocyte lysate with radiolabeled mitochondrial preproteins (25% [vol/vol] of import reaction mixture) was incubated with isolated yeast mitochondria (50 μg of mitochondrial protein/100 μl of import reaction mixture) for 10 min in the presence of a ΔΨ. Import was stopped by the addition of 1 μM valinomycin, and the mitochondria were treated with proteinase K. A fraction of the import reaction mixture was taken as a control (lanes 1 and 4). The rest was split into halves and lysed in buffer containing Triton X-100 or digitonin in the absence or presence of ATP as indicated. Then, a coimmunoprecipitation with antibodies directed against Tim44 was performed. The proteins were separated by SDS-PAGE and detected by digital autoradiography. (C) Efficiency of coprecipitation. The amount of each Ssc1 construct coprecipitated with anti-Tim44 was quantified from 15 independent experiments. The coprecipitated amount of Ssc1(APC) under the Triton X-100 or digitonin condition was set to 100%, respectively. Error bars indicate the standard errors of the mean.
We wondered if the low efficiency of coprecipitation of the ATPase domain Ssc1(A) with Tim44 might be attributable to the conditions of the coprecipitation in the detergent Triton X-100. Indeed, the use of milder conditions by employing the detergent digitonin considerably enhanced the yield of coprecipitation for Ssc1(A) (Fig. 3B, third panel from top, lane 5). A quantitative analysis revealed a threefold enhancement compared to the Triton X-100 conditions (Fig. 3C). In case of the full-length Ssc1(APC), the efficiency of coprecipitation was comparable between digitonin-lysed mitochondria and Triton X-100-lysed mitochondria (Fig. 3B, top panel, lanes 2 and 5). Similarly, the efficiency of coprecipitation of Ssc1(AP) with Tim44 in digitonin yielded a coprecipitation efficiency (∼50% of full-length Ssc1) close to that in Triton X-100 (Fig. 3C). In all cases, the specificity of association was demonstrated by complete disruption in the presence of ATP (Fig. 3B, lane 6). Despite the milder conditions, we still did not observe any association between Ssc1(PC) and Tim44 (Fig. 3B, bottom panel, lanes 5 and 6; Fig. 3C).
We performed a number of control experiments to determine the efficiency and specificity of the coprecipitation approach. (i) About 2% of total imported Ssc1(APC) was found in a complex with Tim44 (Fig. 3B). This quantitative assessment agreed well with the properties of endogenous Ssc1, which is about 50-fold more abundant than Tim44 (7, 12, 41, 60). The coprecipitation of endogenous Ssc1 was analyzed by immunodecoration of anti-Tim44 precipitates. Tim44 was quantitatively precipitated (Fig. 4A, lanes 2 and 3), while ∼2% of Ssc1 was found in the precipitate in the absence of ATP (Fig. 4A, lane 2). When the mitochondria were treated with apyrase to further deplete the levels of ATP, the yield of coprecipitation of Ssc1 with anti-Tim44 was not increased (data not shown) (61). (ii) Ssc1(APC) synthesized in reticulocyte lysate was not precipitated by anti-Tim44 antibodies in the absence of mitochondria, either with or without a presequence (Fig. 4B, topmost two panels). (iii) There was a concern that the Ssc1 constructs might interact with Tim44 because of their properties as preproteins during the translocation across the mitochondrial membranes, and thus the longer constructs would have a greater chance of interaction with Tim44. This possibility seemed unlikely for several reasons: the imported Ssc1(APC) behaves like the endogenous Ssc1; the interaction of Ssc1 constructs with Tim44 is fully ATP sensitive, i.e., like mature Ssc1; the proteinase K treatment largely degrades preproteins that are still spanning the mitochondrial membranes (53); and the different lengths of the proteins do not correlate with the efficiency of interaction with Tim44 [68 kDa for Ssc1(APC), 62 kDa for Ssc1(AP), 42 kDa for Ssc1(A), and 26.5 kDa for Ssc1(PC)]. To further rule out the possibility, we selected two long preproteins, the precursor of F1-ATPase subunit β fused to β-lactamase (F1β-bla; a mature-sized protein of 87 kDa) and the precursor of Fo-ATPase subunit 9 fused to F1β (Su9-F1β; 66 kDa) (34), imported them under the conditions outlined in Fig. 3, and performed a coimmunoprecipitation with anti-Tim44. As expected, no association with Tim44 was observed, either with Triton X-100 (Fig. 4B, lower two panels) or with digitonin (Fig. 4C, lower two panels). (iv) Moreover, we wanted to exclude that the lack of association between Ssc1 constructs and Tim44 in the presence of ATP was caused by an ATP-dependent proteolytic activity in the mitochondrial extracts. We therefore determined the total amount of the Ssc1 constructs under the coprecipitation conditions by performing a precipitation with anti-Ssc1 antibodies. The precipitable amounts of the Ssc1 constructs were not diminished by the presence of ATP (Fig. 4D).
FIG. 4.
The coprecipitation of Ssc1 constructs with anti-Tim44 is specific. (A) Efficiency of coprecipitation of endogenous Ssc1 with anti-Tim44. Isolated yeast mitochondria were lysed with Triton X-100 and subjected to coprecipitation with antibodies directed against Tim44. Precipitated Ssc1 and Tim44 were identified by immunodecoration. A total of 5% of the nonprecipitated material is shown in sample 1. (B) Neither Ssc1 in reticulocyte lysate nor control preproteins in mitochondria are coprecipitated with anti-Tim44. Topmost two panels: the precursor form (p) and the mature-sized form (m) of Ssc1(APC) were synthesized and radiolabeled in reticulocyte lysates and subjected to coprecipitation with anti-Tim44 in the presence of Triton X-100. Bottom three panels: the precursors of Ssc1(APC), F1β-bla and Su9-F1β, were imported into mitochondria. The mitochondria were lysed with Triton X-100-containing buffer and subjected to coprecipitation with anti-Tim44 as described in the legend to Fig. 3B. A total of 2% of the material subjected to immunoprecipitation (reticulocyte lysate or mitochondria, respectively) is shown as a control (sample 1). (C) Control preproteins are not coprecipitated with anti-Tim44 under digitonin conditions. The experiment was performed as described for the bottom three images of panel B except that the mitochondria were lysed with digitonin instead of Triton X-100. (D) The Ssc1 constructs are stable upon lysis in the presence of ATP. Rabbit reticulocyte lysates containing the radiolabeled Ssc1 constructs were incubated with isolated mitochondria as described in the legend to Fig. 3B. After lysis in Triton X-100-containing buffer in the absence or presence of ATP, a precipitation with antibodies directed against Ssc1 was performed. The amount of precipitated proteins was determined by digital autoradiography, and the ratio of the signals in lane 2 to those in lane 1 is shown.
We conclude that the ATPase domain of Ssc1 alone is able to specifically interact with Tim44 in an ATP-sensitive manner, although with a reduced stability. The additional presence of the peptide binding domain and, further, the carboxy-terminal segment enhance the stability and efficiency of interaction. However, no interaction with Tim44 is observed when the ATPase domain is lacking [Ssc1(PC) construct].
The binding of imported Ssc1 constructs to Tim44 is not impaired in ssc1-2 mitochondria.
Like other Hsp70s (32), Ssc1 can self-associate to homooligomers under low-ATP conditions (dimer, trimer, and tetramer) (3). Although under the high-ATP conditions in the mitochondrial matrix, mtHsp70 should be preferentially present as a monomer, it could not be ruled out a priori that the ATPase domain does not bind to Tim44 directly but forms an oligomer with endogenous full-length Ssc1 that would mediate the interaction with Tim44. To investigate this possibility, we used mitochondria from the temperature-sensitive yeast mutant strain ssc1-2, which carries a point mutation in the SSC1 gene (30). When the mutant mitochondria are incubated at a nonpermissive temperature, the Ssc1-2 protein is strongly impaired in binding to Tim44 (54, 60, 63).
The Ssc1 constructs were efficiently imported into the ssc1-2 mitochondria (Fig. 5A, lanes 1 and 4). The association with Tim44 was determined after lysis of the mitochondria with Triton X-100 (Fig. 5A, lanes 2 and 3) or digitonin (Fig. 5A, lanes 5 and 6). We observed a nucleotide-sensitive pattern of interaction that was comparable to that observed with wild-type mitochondria, i.e., Ssc1(AP) and Ssc1(A) were found in association with Tim44 (Fig. 5A, middle panels), whereas Ssc1(PC) did not interact (Fig. 5A, bottom panel). To exclude that long preproteins in transit were kept in complex with Tim44 in ssc1-2 mitochondria, the control experiment with F1β-bla and Su9-F1β was performed and did not reveal any coprecipitation with anti-Tim44 (Fig. 5B).
FIG. 5.
The association of Ssc1 constructs with Tim44 is not impaired in ssc1-2 mutant mitochondria. (A) Coprecipitation of Ssc1 constructs with anti-Tim44. The experiment was performed essentially as described in the legend to Fig. 3B with the following modifications. ssc1-2 mitochondria were used. The mutant phenotype was induced by incubation of the mitochondria for 15 min at 37°C as described in Materials and Methods. Triton X-100 or digitonin was used for lysis of the mitochondria as indicated. No difference in the association of Ssc1 constructs was observed when wild-type mitochondria were treated at 37°C or not (results not shown). (B) Control proteins are not coprecipitated with anti-Tim44 from ssc1-2 mitochondria. The experiment was performed as described in panel A except that the preproteins F1β-bla and Su9-F1β were included. (C) Association of Ssc1 constructs with Tim44 in wild-type and ssc1-2 mitochondria. The amounts of Ssc1 constructs coprecipitated with anti-Tim44 were determined from wild-type and ssc1-2 mitochondria, under both Triton X-100 and digitonin conditions (means of 15 independent experiments for each condition are shown, as described in the legends to Fig. 3 and 5A). Columns 1 to 3 and 5 to 7 show the ratios of coprecipitation of imported constructs between ssc1-2 and wild-type mitochondria. Columns 4 and 8 show the ratios of coprecipitation with anti-Tim44 for endogenous Ssc1-2 versus wild-type Ssc1 (determined by immunodecoration as described in Materials and Methods).
We directly compared the coprecipitation efficiencies of Ssc1 constructs with Tim44 from ssc1-2 mitochondria and wild-type mitochondria by determining the ratio of ssc1-2 to wild-type efficiencies in Triton X-100 and digitonin (Fig. 5C). Additionally, we analyzed the association of endogenous Ssc1 of ssc1-2 and wild-type mitochondria with Tim44 by immunodecoration of anti-Tim44 immunoprecipitates. Fig. 5C demonstrates that the mutant protein Ssc1-2 was strongly impaired in interaction with Tim44 under both Triton X-100 and digitonin conditions. In contrast, the imported Ssc1 constructs interacted with Tim44 of ssc1-2 mitochondria with a much higher yield in a manner similar to that in wild-type mitochondria [and somewhat higher for Ssc1(AP) and Ssc1(A) in Triton X-100 (Fig. 5C). Therefore, the association of imported Ssc1 constructs with Tim44 in ssc1-2 mitochondria does not correlate with the properties of the endogenous Ssc1-2, excluding that the interaction of Ssc1 constructs with Tim44 is indirectly mediated by dimerization with endogenous Ssc1.
DISCUSSION
Tim44 plays a central and essential role in the action of mtHsp70 in driving the import of preproteins into the matrix. The characterization of the mode of interaction between Tim44 and mtHsp70 is thus an important step towards an understanding of the mitochondrial import motor. Here, we report the first analysis of how the distinct domains of mtHsp70 or Ssc1 contribute to the interaction with Tim44. Of the three portions of mtHsp70, the ATPase domain, the peptide binding domain, and the carboxy-terminal segment, only the ATPase domain is able to bind to Tim44 by itself. In the absence of ATP, the ATPase domain remains associated with Tim44, whereas the addition of ATP causes a dissociation, similar to the interaction between Tim44 and full-length mtHsp70. However, the stability of interaction between the ATPase domain and Tim44 is significantly reduced compared to the wild-type situation. In the detergent Triton X-100, the ATPase domain achieves only about 10% of the binding yield that the full-length mtHsp70 does. With a milder detergent, digitonin, the interaction is about threefold more stable. The presence of the peptide binding domain markedly stabilizes the mtHsp70-Tim44 interaction, while the full yield of association is only obtained when the carboxy-terminal segment is present too.
The interactions between mtHsp70 domains and Tim44 were analyzed under the physiological conditions, i.e., in a mitochondrial context. With the combined evidence of the following results, we exclude the possibility that alternative or indirect explanations are conceivable. (i) The association of the ATPase domain alone, but not the peptide binding domain (plus carboxy-terminal segment) alone, with Tim44 is demonstrated by two independent assays, after import into mitochondria and in the two-hybrid system. (ii) The yield and ATP sensitivity of interaction with Tim44 are comparable for imported and endogenous mtHsp70. The mtHsp70 constructs are not simply coprecipitated with Tim44 as preproteins in transit according to their length, since even longer control preproteins are not found in complex with Tim44 under both conditions (Triton X-100 and digitonin). (iii) The ATP-dependent lack of association is not explained by an ATP-dependent breakdown of the imported mtHsp70 constructs, since they are stable independently of the level of ATP. (iv) The interaction of the ATPase domain with Tim44 is not indirectly mediated by an oligomerization with endogenous mtHsp70. By using ssc1-2 mitochondria containing a mutant mtHsp70 that is strongly impaired in binding to Tim44, we can directly demonstrate that the interaction of the mtHsp70 domains with Tim44 is independent of the properties of the endogenous mtHsp70.
The observations made here give some interesting hints in the discussion about the role of mtHsp70 in the mitochondrial import motor. In the trapping model, a simple single-site binding of mtHsp70 to Tim44 would be sufficient to increase the local concentration at the protein import site. In particular, it has been suggested in this model that mtHsp70 is bound to Tim44 via the peptide binding domain, and thus a pulling action would be difficult to explain (4). In contrast, the primary interaction of Tim44 with the ATPase domain of mtHsp70 observed here proves that the binding of Tim44 is clearly different from a substrate-like interaction but is well compatible with the proposed formation of a ternary complex of mtHsp70 with Tim44 and preprotein substrate, an essential prerequisite of the pulling model (18, 26, 36, 45, 60, 63). This view is supported by the observation that mtHsp70 carrying an iodinated substrate peptide could associate with Tim44 (26). Future studies will have to address if the substantial stabilization of the mtHsp70-Tim44 interaction by the peptide binding domain and the carboxy-terminal segment plays a role in permitting conformational changes of mtHsp70 that is still bound to Tim44.
Moro et al. (41) proposed that Tim44 forms a dimer and recruits two molecules of mtHsp70. A dimerization of Tim44 is supported by our two-hybrid analysis. The binding of mtHsp70 constructs to Tim44 does not involve an oligomerization with preexisting mtHsp70 molecules. Moreover, the interaction of radiochemical, i.e., tiny, amounts of imported mtHsp70 with Tim44 occurs with the same efficiency as that of large amounts of preexisting mtHsp70. Since the amount of in vitro-imported mtHsp70 molecules is several orders of magnitude lower than the amount of endogenous mtHsp70 (43), it is thus highly unlikely that a homooligomerization of the imported mtHsp70 molecules is a prerequisite for binding to Tim44. We conclude that individual mtHsp70 molecules in a monomeric form bind to Tim44.
Besides Tim44, several other partner proteins of 70-kDa heat shock proteins have been described, including Hip, BAG-1, and Hop in the eukaryotic cytosol-nucleus, GrpE (Mge1) in the prokaryotic-mitochondrial system, and DnaJ (Hsp40) in both the eukaryotic and prokaryotic systems (8, 14, 25, 39, 59). The cofactors Hip and BAG-1 bind to the ATPase domain of the heat shock cognate protein Hsc70 of the mammalian cytosol in a competitive manner, while the peptide binding domain and the carboxy-terminal segment of Hsc70 do not seem to be involved in binding. The cofactor Hop binds to the carboxy-terminal domain of Hsc70 and acts as an Hsc70-Hsp90-organizing component of chaperone complexes in mammals and in yeast. Thus, the characteristics of interaction between Hsc70 and these cofactors are distinct from the Hsp70-Tim44 interaction. Moreover, the interaction of the mitochondrial GrpE, Mge1, with mtHsp70 is distinct from the mtHsp70-Tim44 interaction. This was demonstrated by the finding of a ternary complex between mtHsp70, Tim44, and Mge1 (27, 37, 55, 61), i.e., that Tim44 and Mge1 bind to distinct sites of mtHsp70. However, some similarity in the mode of interaction with Hsp70 can be seen between Tim44 and DnaJ. Tim44 is not a DnaJ homolog, since it does not contain a J domain, the ∼70-amino-acid residue domain conserved between all J proteins (31, 40, 49), but Tim44 is considered to perform a partially analogous function. A short segment of 18 amino acid residues of Tim44 shares a weak similarity with a segment of DnaJ, in particular when primary and secondary structure analyses are combined. This J-related segment of Tim44 is essential for its function and modulates the interaction with mtHsp70 (38, 47, 48). Interestingly, the corresponding segment of DnaJ has recently been shown to represent a binding site to DnaK (19). Additionally, the ATPase domain of DnaK has been reported to represent an important site for interaction with DnaJ, although an involvement of the peptide binding domain was also proposed (16, 58). These results resemble the situation found with Tim44 and mtHsp70, i.e., primary interaction via the ATPase domain but enhancement and/or stabilization by the additional mtHsp70 domains. Since no involvement of a true mitochondrial DnaJ in protein import has been observed (51, 65), Tim44 may take over DnaJ-like functions at the protein import site of the mitochondrial inner membrane.
In summary, the ATPase domain of mtHsp70 is the primary site of interaction with Tim44, while the peptide binding domain and the C-terminal segment play a stabilizing role. The interaction of mtHsp70 with Tim44 is fundamentally different from the interaction with preprotein substrates and is distinct from the interaction of most cofactors with Hsp70 or Hsc70 proteins. However, the mtHsp70-Tim44 interaction shows some similarity to the Hsp70-DnaJ interaction, supporting the view that Tim44 has been developed instead of a DnaJ at the mitochondrial preprotein import site.
ACKNOWLEDGMENTS
We thank Elizabeth Craig for the ssc1-2 strain and discussion; Jan Brix, Falk Martin, Alessio Merlin, and Martin Moczko for experimental advice; and Nicole Zufall for expert technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 388, the Forschungsschwerpunktprogramm des Landes Baden-Württemberg, and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Alconada A, Gärtner F, Hönlinger A, Kübrich M, Pfanner N. Mitochondrial receptor complex from Neurospora crassa and Saccharomyces cerevisiae. Methods Enzymol. 1995;260:263–286. doi: 10.1016/0076-6879(95)60144-9. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidmann J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 3.Azem A, Oppliger W, Lustig A, Jenö P, Feifel B, Schatz G, Horst M. The mitochondrial hsp70 chaperone system. Effect of adenine nucleotides, peptide substrate, and mGrpE on the oligomeric state of mhsp70. J Biol Chem. 1997;272:20901–20906. doi: 10.1074/jbc.272.33.20901. [DOI] [PubMed] [Google Scholar]
- 4.Bauer M F, Hofmann S, Neupert W, Brunner M. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 2000;10:25–31. doi: 10.1016/s0962-8924(99)01684-0. [DOI] [PubMed] [Google Scholar]
- 5.Bauer M F, Sirrenberg C, Neupert W, Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. doi: 10.1016/s0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- 6.Berthold J, Bauer M F, Schneider H-C, Klaus C, Dietmeier K, Neupert W, Brunner M. The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ATP driving system. Cell. 1995;81:1085–1093. doi: 10.1016/s0092-8674(05)80013-3. [DOI] [PubMed] [Google Scholar]
- 7.Blom J, Dekker P J T, Meijer M. Functional and physical interactions of components of the yeast mitochondrial inner-membrane import machinery (MIM) Eur J Biochem. 1995;232:309–314. doi: 10.1111/j.1432-1033.1995.tb20813.x. [DOI] [PubMed] [Google Scholar]
- 8.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 9.Chevray P M, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien C T, Bartel P L, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daum G, Böhni P C, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 12.Dekker P J T, Martin F, Maarse A C, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 14.Frydman J, Höhfeld J. Chaperones get in touch: the Hip-Hop connection. Trends Biochem Sci. 1997;22:87–92. doi: 10.1016/s0968-0004(97)01005-0. [DOI] [PubMed] [Google Scholar]
- 15.Gambill B D, Voos W, Kang P J, Miao B, Langer T, Craig E A, Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gässler C S, Buchberger A, Laufen T, Mayer M P, Schröder H, Valencia A, Bukau B. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 18.Glick B S. Can Hsp70 proteins act as force-generating motors? Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- 19.Greene M K, Maskos K, Landry S J. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta R S, Golding G B. Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol. 1993;37:573–582. doi: 10.1007/BF00182743. [DOI] [PubMed] [Google Scholar]
- 21.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [PubMed] [Google Scholar]
- 22.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 23.Hartl F-U, Ostermann J, Guiard B, Neupert W. Successive translocation into and out of the mitochondrial matrix: targeting of proteins to the intermembrane space by a bipartite signal peptide. Cell. 1987;51:1027–1037. doi: 10.1016/0092-8674(87)90589-7. [DOI] [PubMed] [Google Scholar]
- 24.Hebert D N. Protein unfolding: mitochondria offer a helping hand. Nat Struct Biol. 1999;6:1084–1085. doi: 10.1038/70006. [DOI] [PubMed] [Google Scholar]
- 25.Höhfeld J. Regulation of the heat shock conjugate Hsc70 in the mammalian cell: the characterization of the anti-apoptotic protein BAG-1 provides novel insights. Biol Chem. 1998;379:269–274. [PubMed] [Google Scholar]
- 26.Horst M, Oppliger W, Feifel B, Schatz G, Glick B S. The mitochondrial protein import motor: dissociation of mitochondrial hsp70 from its membrane anchor requires ATP binding rather than ATP hydrolysis. Protein Sci. 1996;5:759–767. doi: 10.1002/pro.5560050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horst M, Oppliger W, Rospert S, Schönfeld H J, Schatz G, Azem A. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 1997;16:1842–1849. doi: 10.1093/emboj/16.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang S, Ratliff K S, Schwartz M P, Spenner J M, Matouschek A. Mitochondria unfold precursor proteins by unraveling them from their N-termini. Nat Struct Biol. 1999;6:1132–1138. doi: 10.1038/70073. [DOI] [PubMed] [Google Scholar]
- 29.Jensen R E, Johnson A E. Protein translocation: is Hsp70 pulling my chain? Curr Biol. 1999;9:R779–R782. doi: 10.1016/S0960-9822(00)80012-3. [DOI] [PubMed] [Google Scholar]
- 30.Kang P J, Ostermann J, Shilling J, Neupert W, Craig E A, Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 31.Kelley W L. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 32.Kim D, Lee Y, Corry P. Constitutive HSP70: oligomerization and its dependence on ATP binding. J Cell Physiol. 1992;153:353–361. doi: 10.1002/jcp.1041530215. [DOI] [PubMed] [Google Scholar]
- 33.Kronidou N G, Oppliger W, Bolliger L, Hannavy K, Glick B S, Schatz G, Horst M. Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc Natl Acad Sci USA. 1994;91:12818–12822. doi: 10.1073/pnas.91.26.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahlke K, Pfanner N, Martin J, Horwich A L, Hartl F-U, Neupert W. Sorting pathways of mitochondrial inner membrane proteins. Eur J Biochem. 1990;192:551–555. doi: 10.1111/j.1432-1033.1990.tb19260.x. [DOI] [PubMed] [Google Scholar]
- 35.Martin J, Mahlke K, Pfanner N. Role of an energized inner membrane in mitochondrial protein import. ΔΨ drives the movement of presequences. J Biol Chem. 1991;266:18051–18057. [PubMed] [Google Scholar]
- 36.Matouschek A, Azem A, Ratliff K, Glick B S, Schmid K, Schatz G. Active unfolding of precursor proteins during mitochondrial protein import. EMBO J. 1997;16:6727–6736. doi: 10.1093/emboj/16.22.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merlin A, von Ahsen O, Craig E A, Dietmeier K, Pfanner N. A mutant form of mitochondrial GrpE suppresses the sorting defect caused by an alteration in the presequence of cytochrome b2. J Mol Biol. 1997;273:1–6. doi: 10.1006/jmbi.1997.1300. [DOI] [PubMed] [Google Scholar]
- 38.Merlin A, Voos W, Maarse A C, Meijer M, Pfanner N, Rassow J. The J-related segment of Tim44 is essential for cell viability: a mutant Tim44 remains in the mitochondrial import site, but inefficiently recruits mtHsp70 and impairs protein translocation. J Cell Biol. 1999;145:961–972. doi: 10.1083/jcb.145.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minami Y, Höhfeld J, Ohtsuka K, Hartl F-U. Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J Biol Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- 40.Misselwitz B, Staeck O, Rapoport T A. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 41.Moro F, Sirrenberg C, Schneider H-C, Neupert W, Brunner M. The TIM17 · 23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J. 1999;18:3667–3675. doi: 10.1093/emboj/18.13.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 43.Palmisano A, Zara V, Hönlinger A, Vozza A, Dekker P J T, Pfanner N, Palmieri F. Targeting and assembly of the oxoglutarate carrier: general principles for biogenesis of carrier proteins of the mitochondrial inner membrane. Biochem J. 1998;333:151–158. doi: 10.1042/bj3330151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfanner N, Craig E A, Hönlinger A. Mitochondrial preprotein translocase. Annu Rev Cell Dev Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- 45.Pfanner N, Meijer M. Protein sorting: pulling in the proteins. Curr Biol. 1995;5:132–135. doi: 10.1016/s0960-9822(95)00033-9. [DOI] [PubMed] [Google Scholar]
- 46.Pilon M, Schekman R. Protein translocation: how Hsp70 pulls it off. Cell. 1999;97:679–682. doi: 10.1016/s0092-8674(00)80780-1. [DOI] [PubMed] [Google Scholar]
- 47.Rassow J, Dekker P J T, van Wilpe S, Meijer M, Soll J. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol. 1999;286:105–120. doi: 10.1006/jmbi.1998.2455. [DOI] [PubMed] [Google Scholar]
- 48.Rassow J, Maarse A C, Krainer E, Kübrich M, Müller H, Meijer M, Craig E A, Pfanner N. Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J Cell Biol. 1994;127:1547–1556. doi: 10.1083/jcb.127.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rassow J, Voos W, Pfanner N. Partner proteins determine multiple functions of Hsp70. Trends Cell Biol. 1995;5:207–212. doi: 10.1016/s0962-8924(00)89001-7. [DOI] [PubMed] [Google Scholar]
- 50.Rehling P, Marzioch M, Niesen F, Wittke E, Veenhuis M, Kunau W H. The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J. 1996;15:2901–2913. [PMC free article] [PubMed] [Google Scholar]
- 51.Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell. 1994;77:249–259. doi: 10.1016/0092-8674(94)90317-4. [DOI] [PubMed] [Google Scholar]
- 52.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 53.Schleyer M, Neupert W. Transport of proteins into mitochondria: translocational intermediates spanning contact sites between outer and inner membranes. Cell. 1985;43:339–350. doi: 10.1016/0092-8674(85)90039-x. [DOI] [PubMed] [Google Scholar]
- 54.Schneider H-C, Berthold J, Bauer M F, Dietmeier K, Guiard B, Brunner M, Neupert W. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature. 1994;371:768–774. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- 55.Schneider H-C, Westermann B, Neupert W, Brunner M. The nucleotide exchange factor MGE exerts a key function in the ATP-dependent cycle of mt-Hsp70-Tim44 interaction driving mitochondrial protein import. EMBO J. 1996;15:5796–5803. [PMC free article] [PubMed] [Google Scholar]
- 56.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 57.Söllner T, Rassow J, Pfanner N. Analysis of mitochondrial protein import using translocation intermediates and specific antibodies. Methods Cell Biol. 1991;34:345–358. doi: 10.1016/s0091-679x(08)61689-1. [DOI] [PubMed] [Google Scholar]
- 58.Suh W C, Burkholder W F, Lu C Z, Zhao X, Gottesman M E, Gross C A. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci USA. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takayama S, Bimston D N, Matsuzawa S, Freeman B C, Aime-Sempe C, Xie Z, Morimoto R I, Reed J C. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voisine C, Craig E A, Zufall N, von Ahsen O, Pfanner N, Voos W. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell. 1999;97:565–574. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- 61.von Ahsen O, Voos W, Henninger H, Pfanner N. The mitochondrial protein import machinery. Role of ATP in dissociation of the Hsp70 · Mim44 complex. J Biol Chem. 1995;270:29848–29853. doi: 10.1074/jbc.270.50.29848. [DOI] [PubMed] [Google Scholar]
- 62.Voos W, Martin H, Krimmer T, Pfanner N. Mechanisms of protein translocation into mitochondria. Biochim Biophys Acta. 1999;1422:235–254. doi: 10.1016/s0304-4157(99)00007-6. [DOI] [PubMed] [Google Scholar]
- 63.Voos W, von Ahsen O, Müller H, Guiard B, Rassow J, Pfanner N. Differential requirement for the mitochondrial Hsp70-Tim44 complex in unfolding and translocation of preproteins. EMBO J. 1996;15:2668–2677. [PMC free article] [PubMed] [Google Scholar]
- 64.Weiss C, Oppliger W, Vergères G, Demel R, Jenö P, Horst M, de Kruijff B, Schatz G, Azem A. Domain structure and lipid interaction of recombinant yeast Tim44. Proc Natl Acad Sci USA. 1999;96:8890–8894. doi: 10.1073/pnas.96.16.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Westermann B, Neupert W. Mdj2p, a novel DnaJ homolog in the mitochondrial inner membrane of the yeast Saccharomyces cerevisiae. J Mol Biol. 1997;272:477–483. doi: 10.1006/jmbi.1997.1267. [DOI] [PubMed] [Google Scholar]