Abstract
Circulating tumor cells (CTCs) are tumor cells that have sloughed off the primary tumor and extravasate into and circulate in the blood. Understanding of the metastatic cascade of CTCs has tremendous potential for the identification of targets against cancer metastasis. Detecting these very rare CTCs among the massive blood cells is challenging. However, emerging technologies for CTCs detection have profoundly contributed to deepening investigation into the biology of CTCs and have facilitated their clinical application. Current technologies for the detection of CTCs are summarized herein, together with their advantages and disadvantages. The detection of CTCs is usually dependent on molecular markers, with the epithelial cell adhesion molecule being the most widely used, although molecular markers vary between different types of cancer. Properties associated with epithelial-to-mesenchymal transition and stemness have been identified in CTCs, indicating their increased metastatic capacity. Only a small proportion of CTCs can survive and eventually initiate metastases, suggesting that an interaction and modulation between CTCs and the hostile blood microenvironment is essential for CTC metastasis. Single-cell sequencing of CTCs has been extensively investigated, and has enabled researchers to reveal the genome and transcriptome of CTCs. Herein, we also review the clinical applications of CTCs, especially for monitoring response to cancer treatment and in evaluating prognosis. Hence, CTCs have and will continue to contribute to providing significant insights into metastatic processes and will open new avenues for useful clinical applications.
Subject terms: Tumour biomarkers, Tumour biomarkers
Introduction
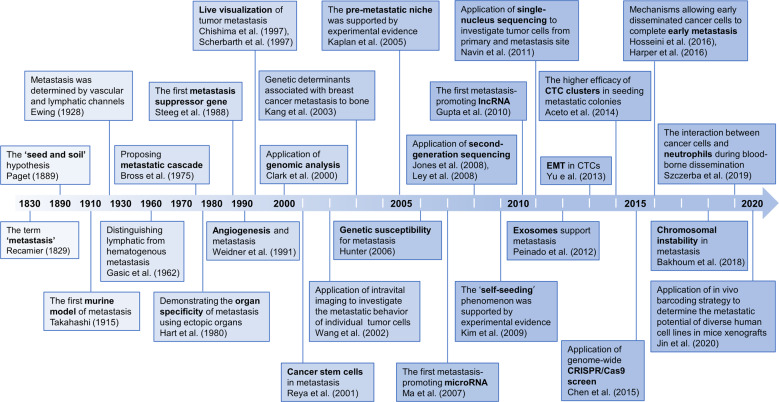
Metastasis is the most lethal feature of cancer1. Despite significant developments in cancer diagnosis and treatment over the past centuries, metastasis remains a major obstacle to improving clinical outcomes of cancer patients2. Nevertheless, we have witnessed significant progress over the past two hundred years in revealing fundamental concepts underlying the development of metastasis and in creating new technologies to facilitate cancer metastasis research. Fig 1 highlights the key discoveries and milestones in the study of cancer metastasis. The ‘seed and soil’ hypothesis by Steven Paget3 in the 1830s vividly clarified the progress of cancer metastasis. With the advances of science and technologies, particularly since the 2000s, a plethora of new technologies have been advanced, such as high-throughput sequencing4,5, transgenic mouse models6, CRISPER/Cas9 editing tools7, and single-cell sequencing8. With these powerful technologies, the biological phenomena underlying metastasis, such as epithelial-mesenchymal transition (EMT) of tumor cells9, the role of exosomes in supporting metastasis10, circulating tumor cells (CTCs) and CTC clusters in seeding metastatic colonies11, and complex interactions between tumor cells and microenvironment12, have gradually been unmasked, along with the discovery of numerous metastasis-related driver genes. The ‘black box’ of metastasis is gradually being unveiled, and effective metastasis-targeting agents are believed to be on the horizon in the near future.
Fig. 1.
The historical milestones of cancer metastasis research
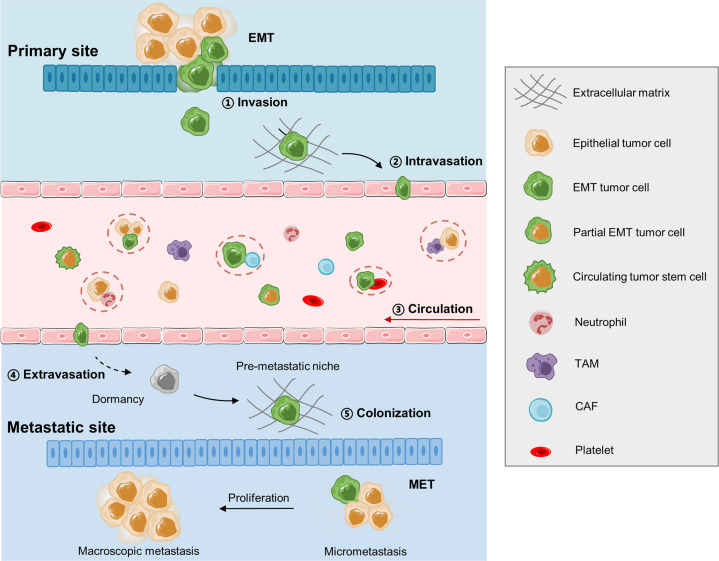
Cancer metastasis is a complex multistep process involving cancer cell invasion in the primary site, intravasation into circulation, survival in the circulation, extravasation from the circulation, and attachment to and colonization of the metastatic site (Fig. 2). CTCs are defined as tumor cells that have been sloughed from the primary tumor and are swept away by the circulatory or lymphatic systems. To date, most CTC research has focused on CTCs in the blood circulation. CTCs were first described in 1869 by Ashworth who observed “some cells” in the blood of a metastatic cancer patient with an appearance similar to tumor cells in the primary tumors13. CTCs have been assumed to be the substrate of metastasis. Although CTCs originate from the primary tumor, they are distinct from primary tumor cells14, with EMT transition properties that help them break free from the primary tumor and facilitate intravasation into the bloodstream, dissemination in clusters of CTCs to increase metastatic potential, and exhibit stemness features that enhance their ability to initiate metastasis (Fig. 2). However, most CTCs perish in the circulation, and only limited CTCs survive and infiltrate distant organs. Interactions between CTCs and the blood environment (Fig. 2), including how CTCs escape immune surveillance in the blood, have been widely implicated in the metastatic mechanisms of CTCs. It has taken more than a century for researchers to recognize the critical role of CTCs in cancer metastasis, due to the unique technical challenges required to isolate these very rare CTCs from the massive pool of circulating blood cells15. However, in the past two decades, emerging technologies for CTC isolation have allowed research on the biology of CTCs and have facilitated the clinical applications of CTCs in cancer screening, treatment response monitoring, and prognosis evaluation.
Fig. 2.
Multistep process of cancer metastasis. The complex metastatic process includes tumor cell invasion in the primary site, intravasation into circulation, survival in the circulation as CTCs and interaction with blood cells, extravasation from the circulation, attachment to and colonization of the metastatic site. EMT: epithelial to mesenchymal transition, CAF: cancer-associated fibroblast, TAM: tumor-associated macrophage
In this review, the biology of CTCs, as well as their interaction with the blood microenvironment, is fully reviewed. In addition, the growing number of highly sophisticated CTC enrichment and isolation technologies will be summarized. Finally, we also discuss the tremendous potential of CTCs in clinical applications.
The biology of CTCs
Molecular characterization of CTCs
Experimental evidence has supported the notion that tumor cells can spread even during the early stages of tumor evolution16,17. The molecular characterization of CTCs will add knowledge to the underlying mechanism of metastatic processes, contributing to early diagnosis and prevention of metastasis.
Molecular markers of CTCs
A panel of molecular markers has been used to detect CTCs in various cancers. CTC-associated markers used for different cancers are summarized in Table 118,19. As most cancers are of epithelial origin, the most common marker used for CTCs is EpCAM, a “universal” epithelial marker of cancers20. EpCAM expression varies among different cancer types21, and EpCAM-based CTC detection technologies are widely applied for cancers that strongly express EpCAM, such as breast and prostate cancer. Many studies have shown that CTCs in breast and prostate cancer are EpCAM-positive, and have validated their prognostic value in either early or metastatic stage cases22,23. Other epithelial-derived cancer types, such as pancreatic24, colorectal25, and hepatocellular cancers26, also have a considerable detection rate of EpCAM-positive CTCs. Similarly, the presence of these EpCAM-positive CTCs predicts early distant metastasis and poorer survival of patients25,27,28. However, using EpCAM as a CTC marker has limitations. It cannot be used in tumors that are EpCAM-negative or with low expression, such as neurogenic cancers. CTCs can undergo EMT, and epithelial markers, including EpCAM, are down-regulated during EMT, which affects the detection rate of EpCAM-positive CTCs. Although there are doubts as to whether EpCAM-based technologies are appropriate to detect all CTCs, numerous studies have illustrated the potential value of EpCAM-positive CTCs in clinical applications29. To some extent, EpCAM-positive CTCs are a substantial subgroup of all CTCs, thus EpCAM-positive CTCs still could be a reliable biomarker if the cancer prognosis and therapeutic efficacy is relevant to EpCAM-positive CTCs.
Table 1.
Molecular markers used to identify CTCs
| Cancer types | Epithelial markers | Mesenchymal markers | Specific markers |
|---|---|---|---|
| Breast cancer | EpCAM/CK8,18,19234,237,265–280 | Vimentin280–283 | HER237–46 |
| CK 5/7/8/18/199 | Twist253,282,284 | ER39,48–50 | |
| E-Cadherin9,280,281 | Fibronectin9,280 | AR285 | |
| N-Cadherin9,280,286 | MRP48 | ||
| SERPINE1/PAI19 | |||
| β-catenin281 | |||
| Prostate cancer | EpCAM/CK8,18,19287,288 | Vimentin102,289–291 | PSMA51–53 |
| Twist290,291 | PSA239 | ||
| EGFR51 | |||
| ARV7256,292–294 | |||
| PIM1295 | |||
| AR v567es294 | |||
| Kidney cancer | EpCAM240 | - | CD147296 |
| Bladder cancer | EpCAM/CK8,18,19297–300 | - | - |
| Colorectal cancer | EpCAM/CK8,18,19301,302 | Vimentin252,303–305 | PI3K α306 |
| Twist252,303,305 | CEA307–309 | ||
| SNAI1303,305 | PRL3252 | ||
| AKT2303,305,306 | |||
| LOXL3310 | |||
| Plastin3311 | |||
| Non-small-cell lung cancer | CK7/8/18/19312,313 | Vimentin109,313,314 | Folate receptor54–56 |
| EpCAM/CK8,18,19109,313–319 | Twist313 | Telomerase activity320 | |
| N-Cadherin314 | |||
| AXL313 | |||
| Small-cell lung cancer | EpCAM/CK8,18,1979,320–326 | Vimentin242,327 | DLL3242 |
| Pancreatic cancer | EpCAM/CK8,18,19328–332 | Vimentin257,333–335 | - |
| Twist334 | |||
| KLF8335 | |||
| Hepatocellular carcinoma | EpCAM/CK8,18,19336–340 | Vimentin336,341–343 | GPC3344,345 |
| EpCAM,CK19346 | Twist336,341–343,347 | ASGPR344 | |
| EpCAM348 | - | - | |
| Gastric cancer | EpCAM/CK244,349–351 | Vimentin352 | XAF1353 |
| CK19/CK20354 | N-Cadherin355 | MT1-MMP356 | |
| Survivin57 | |||
| HER247 | |||
| Esophageal cancer | EpCAM/CK8,18,19357 | - | - |
| Cervical cancer | EpCAM/CK8,18,19358 | - | - |
| Melanoma | - | MART-162,63 | |
| HMW-MAA59,61,65,359 | |||
| CD14661,65,359 | |||
| MAGE A362,63,66 | |||
| GalNAc-T63 | |||
| MAGE A1-6360 | |||
| hTERT360 | |||
| MLANA66 | |||
| B4GALNT166 | |||
| PAX362,66 | |||
| DCT66 |
CTC markers mainly includes the epithelial markers, the mesenchymal markers, and the cancer specific CTC markers
ARV7 androgen-receptor splice variant 7, ASGPR asialoglycoprotein receptor, CEA carcinoembryonic antigen, EGFR epidermal growth factor receptor, GPC3 glypican 3, MAGE A1-6 melanoma antigen-encoding gene family member A1-family) member A6, MRP multidrug-resistance-related proteins, PI3K α phosphatidylinositol 3-kinase α, PIM1 proviral integration site for the Moloney murine leukemia virus-1PRL3 phosphatase of regenerating liver-3, PSA prostate specific antigen, PSMA prostate-specific membrane antigen, SERPINE1/PAI1 serpin peptidase inhibitor, clade E, XAF1 XIAP-associated factor 1
Due to the EMT activity of some epithelial cancer cells, detecting only EpCAM-positive CTCs probably underestimates the actual total CTC population and misses important biological information of EpCAM-negative CTCs. In some cancer types, such as non-small-cell lung cancer (NSCLC) patients, it was even found that the quantity of EpCAM-negative CTCs was significantly larger than EpCAM-positive CTCs30. Nevertheless, the poor isolation of CTCs by EpCAM-based technologies can be rescued by using both epithelial and mesenchymal cancer markers, as well as by marker-independent detection methods. For example, in breast cancer, the use of fluorescent-magnetic nanoparticles consisting of a dual-antibody interface targeting both EpCAM and N-cadherin has contributed to high-efficiency isolation and rapid identification of CTCs31,32. In biliary tract cancer, a single-cell assay for detecting CTCs allowed identification of both epithelial CTCs and non-conventional CTCs which lacked epithelial and leukocyte markers, and therefore led to an increase of CTC positivity rate33. The EMT program of cancer cells exhibits molecular alterations, including decreased expression of epithelial markers (E-cadherin, ZO-1, claudins, and occludins) and increased expression of mesenchymal markers (vimentin, N-cadherin, fibroblast-specific protein1, and fibronectin)34. EMT is executed by EMT-related transcription factors, mainly belonging to the SNAIL, TWIST, and ZEB families34. All these EMT-related molecules can theoretically be used for EMT-CTCs targeting methods. However, many EMT-related molecules are cytoplasmic or nuclear proteins, precluding their usage in the currently available membrane molecular-based technologies of CTC detection. Proteins such as E-cadherin, vimentin, and twist were most often used in the past35(Table 1), probably because of their accessibility of detection in traditional CTC detection technologies including flow cytometry sorting, immunostaining, and fluorescence in situ hybridization (FISH) staining. However, the emergence of single-cell CTC sequencing technologies36 will make it possible to unmask the EMT status of CTCs more comprehensively, and can cover all the EMT-related molecular alternations at the RNA level.
Other biomarkers, such as human epidermal growth factor receptor-2 (HER2)37–47, estrogen receptor39,48–50, prostate-specific membrane antigen51–53, folate receptor54–56, and survivin57, have been described as CTCs markers in different cancers, with different clinical significance. These cancer-specific CTC markers are listed in Table 1. Most of these cancer-specific CTC markers are in accordance with the specific molecular markers of the primary tumor. However, there is discordance in the expression of specific markers between the primary tumor and CTCs. For example, the rates of discordance of HER2 gene amplification between CTCs and primary breast tumor were around 15%58, suggesting a clonal selection of CTCs or clonal acquisition, probably due to genetic instability. It should be mentioned that for melanoma, a skin cancer that begins in melanocytes, the detection technologies of CTCs are based on several melanoma cell adhesion molecules, such as HMW-MAA59–61, MART-162–64, CD14661,65, and MAGE A362,63,66, which are very specific molecular markers for melanoma.
The variety of CTCs markers indicates the heterogeneity of CTCs among different cancer types. Even in one patient, CTCs are spatio-temporally heterogenous, which may be the result of a spatially different microenvironment in the blood and temporal changes in therapy response. Thus, it is difficult to define the entire CTC population using the very limited molecular markers currently available. In addition, CTC markers should not be constant among different stages of cancer and treatment periods.
Genome analysis of CTCs
Genomic instability contributes to tumor evolution and the emergence of resistant tumor subclones. Monitoring tumor genomic instability, especially in terms of tumor resistance and metastases, greatly contributes to the evaluation of treatment response and precision medicine. The evaluation of CTCs assessment using noninvasive liquid biopsy is accessible for serial sampling to detect the genomic instability of the tumor.
Determining the status of EGFR and KRAS mutations is crucial for guiding treatment in NSCLC patients receiving EGFR tyrosine kinase inhibitors and colorectal cancer patients treated with anti-EGFR therapy respectively. The concordance of mutations between CTCs and matched primary or metastatic tumor tissue has attracted much attention. Using a microfluidic technique to capture CTC, Maheswaran et al. found that only two of 31 patients with mutations were overlooked from their detection assay67. They identified the EGFR activating mutation in CTCs in 92% of metastatic patients with NSCLC and detected the drug-resistant mutation T790M in CTCs of 33% of patients who responded to tyrosine kinase inhibitor therapy and in 64% of patients who exhibited clinical progression67. For the analysis of the KRAS gene mutation, the mutational concordance rate between CTCs and matched primary tumors ranged from 37% to 90% in colorectal cancer cases68–71. This difference in the concordance rate may be due to the different CTC selection protocols used in these studies. KRAS mutations are also common in pancreatic ductal adenocarcinoma (PDAC), present in 90% of PDAC cases. However, Kulemann et al. found that the discordance rate of KRAS mutation in CTCs and corresponding PDAC tumors was 42%72. Studies of gene mutation analysis in CTCs were also conducted in many cancers such as prostate cancer73,74, breast cancer75,76, hepatocellular carcinomas77, and a mutational discordance between CTCs and corresponding tumors were often found. The discordance rate was probably attributed to the different detection efficiency of CTC mutations, or the heterogeneity between CTCs and primary tumor cells. Genomic assessment of tumor tissue and CTCs can be complementary. So, a combination of mutational testing of CTCs and tumor specimens would guide treatment more precisely.
Determining copy number alternations (CNA) of CTCs helps analyze and track cancer profiles as tumors evolve. In lung cancer, Ni et al.78 found that CTCs exhibit reproducible CNAs patterns, similar to those of metastatic tumors, and different patients shared similar CNAs patterns. In small-cell lung cancer, a CNA-based classifier for CTCs correctly assigned 83.3% of patients as chemorefractory or chemosensitive79. In breast cancer, the assessment of CNAs in archived CTCs is feasible. Paoletti et al.80 found that the CNAs of CTCs and paired metastatic tumor tissue in breast cancer patients were highly concordant, although CTCs and matched tumor tissue harbored several discordant copy number alterations, suggesting that CTCs were the subclone cells of tumor tissues. In triple-negative breast cancer, CTCs with chromosome 10 and 21q CNAs are predictive of clinical progression, and their network analysis presented connected modules including HER/phosphatidylinositol-4,5-bisphosphate 3-kinase/RAS/JAK signaling81. In prostate cancer, Lambro et al.82 revealed that CNAs of CTCs were interpatient and intercell heterogeneous, and could be missed in bulk biopsy analyses. In metastatic castration-resistant prostate cancer, whole genomic copy number analysis of CTCs showed that common genomic gains in CTCs involved genes such as androgen receptor (AR), mesenchymal-to-epithelial transition (MET), ERG, and cyclin-dependent kinase 12, while common genomic losses were observed in genes such as phosphatase and tensin homolog (PTEN), RAF1, and GATA283. Similarly, Malihi et al. also observed that CNAs in genes including PTEN, RB1, TP53, and AR closely associated with genomic instability and survival in aggressive variant prostate cancer84.
Other genome analyses have also been conducted in CTCs. FISH testing was adopted in CTCs to detect biomarkers for treatment sensitivity, such as ALK FISH testing in CTCs of NSCLC patients85 and HER2 FISH testing in CTCs of breast cancer patients86,87. Recently, based on the technique of single-cell resolution DNA methylation analysis, the DNA methylome of single CTCs and CTC clusters was revealed for breast cancer patients, and indicated that the CTC cluster hypomethylation profile obtained was associated with a poor prognosis and that treatment with Na+/K+-ATPase inhibitors to dissociate CTC clusters could revert the methylation profile of CTC clusters and suppress metastasis88.
Transcriptome analysis of CTCs
Single-cell sequencing has developed rapidly in recent years and has been applied to investigate the CTC transcriptomes. Single-cell expression profiles can distinguish CTCs from mesothelial cells and blood cells in lung adenocarcinoma, with representative markers including EpCAM for CTCs89. Single-cell sequencing-based transcriptome analysis revealed heterogeneity in the CTC subpopulation. By testing the expression of proliferation-associated genes such as the Ki-67 proliferation marker, Magbanua et al90. found that 65% of CTC in patients with metastatic breast cancer had low proliferation Ki-67 and that the 35% of patients with a high proliferation Ki-67 expression had a poor prognosis. Cheng et al.91 performed a single cell transcriptome analysis of 666 CTCs in patients with metastatic breast cancer. They determined that intra-patient CTCs were heterogeneous with regard to EMT-like and MET-like states, and CTCs were enriched for the stem-like phenotype91. Single-cell sequencing of CTCs also greatly helped in discovering driver signaling pathways that contributed to metastasis and treatment failure. RNA-Seq of single prostate CTC indicated the activation of noncanonical Wnt signaling in antiandrogen resistant patients92. Further, using mouse models, ectopic expression of Wnt5a attenuated the effects of an AR inhibitor and suppression of Wnt5a could partially restore the sensitivity in drug-resistant prostate cancer cells92. CTCs have been associated with a poor prognosis in colorectal cancer. A study of the CTC-specific transcriptome profile93 of six metastatic colorectal cancer patients characterized 410 CTC-specific genes, which were primarily related to cell movement and adhesion, such as VCL ITGB5, bone morphogenetic protein 6, transforming growth factor beta 1, and talin 1, and were related to cell death and proliferation, such as amyloid beta precursor protein, clusterin, and TIMP1.
Epithelial-to-mesenchymal transition of CTCs
EMT is the process by which epithelial tumor cells lose their intercellular adhesion and acquire mesenchymal and invasive properties. During dissemination, tumor cells detach themselves from the basement membrane through EMT activation and directly enter the circulation, serving as CTCs traveling to distant sites. When CTCs extravasate, they then undergo a reverse process termed MET and proliferate to form macro-metastases94–96. Herein, metastatic development depends on the delicate balance of the transition between these two phenotypes. The activity of EMT-MET was also proposed to play an important role in the metastatic process of CTCs97. Using mouse models, it was found that epithelial-type CTCs with a restricted mesenchymal transition had the strongest lung metastases formation capacity, whereas mesenchymal-type CTCs showed limited metastatic ability98. In breast cancer, CTCs exhibit dynamic changes in EMT composition, and mesenchymal CTCs were found to be closely associated with cancer progression9,98. A plethora of studies has shown increased EMT of CTCs rather than primary tumor cells in various cancers99–103. In a study based on the bioinformatics analysis of seven sets of gene chips, Guan et al. showed that compared with primary tumors, the main changes in CTCs involved cell adhesion, EMT, and apoptosis104. In a prospective study including 39 patients with invasive breast cancer, Tashireva et al. observed a majority of heterogeneous CTC phenotypes (22 out of 24 detectable samples) exhibiting EMT plasticity105. Interestingly, it was found that fluid shear stress can induce the EMT of CTCs via JNK signaling in breast cancer, which further confirmed the relationship between augmented EMT of CTCs and poor patient survival106.
Clinically, combining the total CTC count and the proportion of mesenchymal CTCs107 can be used to monitor therapeutic resistance and predict prognosis in cancer patients due to the significant survival differences of this criterion108. For example, since the baseline presence of total CTCs in advanced NSCLC conferred poor prognosis109, and the presence of over five EMT-CTCs indicated progressive disease110. Different numbers of total CTCs and EMT CTCs were found to play an important role in determining the prognosis of breast cancer patients. Intriguingly, it was emphasized that a better understanding of EMT-CTC subtypes and their interactions with peripheral blood mononuclear cells could help design better anti-metastatic treatments111. As CTC EMT-positive patients with neutrophil-to-lymphocyte ratios ≥3 had an 8.6 times increased risk of disease recurrence compared with CTC EMT-negative patients with lower neutrophil levels, inflammation-based scores increased the prognostic value of CTCs in primary breast cancer112. Therefore, targeting the EMT pathway may prevent tumor cell spread in early-stage patients and eradicate metastatic cells in advanced stages.
The stemness of CTCs
Many previous studies have indicated a subpopulation of aggressive CTCs with “stemness” traits in different cancers, which refer to their properties for self-renewal and tumor growth induction. In bladder cancer, studies have reported the high expression of OCT4, a crucial stemness maintenance protein113, in a subgroup of CTCs114. In breast cancer, the CD44+/CD24-/low and aldehyde dehydrogenase 1 (ALDH1) + cell phenotypes are reported to be associated with stemness. After detecting the expression of CD44, CD24, and ALDH1 of CTCs in 30 metastatic breast cancer patients, Theodoropoulos et al. found that 35.2% of 1439 CTCs were CD44+/CD24-/low, and 17.7% of 238 CTCs were ALDH1high/CD24-/low, providing evidence of CTCs stemness96. A CTC-3 cell line established from the peripheral blood cells of a breast cancer patient showed more aggressive growth than the widely used MCF-7 breast cancer cell line. Gene profiling revealed higher expression of the stemness markers in the CTC-3 cell line compared to MCF-7 cells115. In hepatocellular carcinoma (HCC), 71.4% of HCC patients were CTC positive for the cancer stem cell marker, CD44; thus, they had a significant population of CTCs with stem properties, which could contribute to tumor cell survival and dissemination116. In glioblastoma, RNA-seq analysis revealed Wnt activation-induced stemness and chemoresistance in CTCs117. In prostate cancer, the stem cell marker CD133 was observed in the majority (> 80%) of CTCs of patients with metastatic castration-resistant prostate cancer97 and a stem-like subpopulation of the C-X-C motif chemokine receptor 4+/CD133+CTCs was more prevalent in EpCAM-negative CTCs than in EpCAM-positive CTCs118.
It is crucial to understand the mechanisms regulating CTCs stemness, and interfering with the CTCs subpopulation stemness properties may more efficiently suppress cancer progression and relapse. CTCs undergo considerable levels of fluid shear flow during their dissemination, and the fluid shear flow itself may have an impact on CTCs. Using a model of breast cancer with brain metastasis, it was suggested that hemodynamic shear flow could upregulate the stemness genes of CTCs in surviving under conditions of shear flow119. EMT-like transition of CTCs by downregulating ERK and GSK3β signaling could promote the conversion of CTCs into stem-like CTCs with high sphere-forming and tumor-initiating capacity120.
CTCs and the blood microenvironment
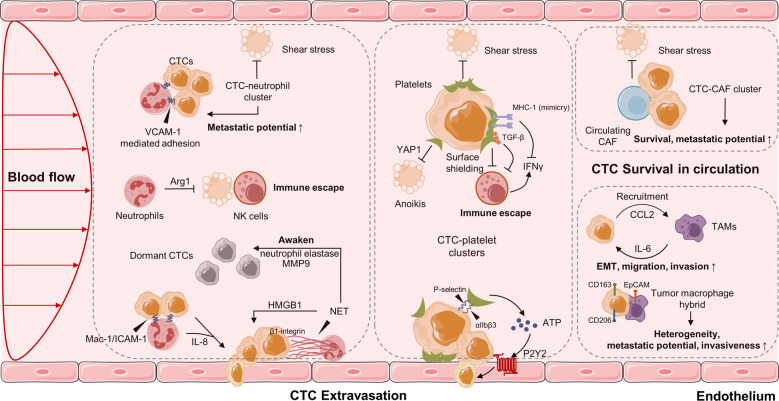
When transported in the bloodstream, a major of CTCs are constrained by detrimental shear stress, or die from anoikis, a programmed cell death mechanism due to loss of cell attachment121,122. Only a small fraction of CTCs interact tightly with platelets, neutrophils, macrophages, myeloid-derived suppressor cells (MDSCs), or cancer-associated fibroblasts (CAFs) to escape the immune system and promote their survival123,124. Recently, accumulating studies suggest that the interaction and modulation between CTCs and hostile blood microenvironment is essential for adhesion to endothelial cells, tissue invasion, and tumor metastasis (Fig. 3).
Fig. 3.
CTCs in the blood microenvironment, and their interaction with neutrophils, platelets, CAFs and TAMs. CAFs: cancer-associated fibroblasts, TAMs: tumor-associated macrophages
Interaction of CTCs with neutrophils
Neutrophils are the most abundant circulating leukocytes in humans and have recently been studied to support cancer progression125. An increased number of neutrophils in circulation is associated with poor prognosis in several types of cancers126–128. The formation of CTC-white blood cells (WBCs) clusters was previously reported within the bloodstream129. In 2019, Szczerba et al. determined that CTCs were significantly associated with neutrophils in both mouse models and breast cancer patients, exhibiting more metastatic potential with greater expression of genes that involve cell cycle progression compared to CTCs alone130. These observations are consistent with previous findings showing the proliferation role of neutrophils on tumor cells131. CTC and neutrophil binding is mediated by the cell–cell junction and possibly requires the vascular cell adhesion molecule 1130. Furthermore, neutrophils can directly adhere to CTCs through the Mac-1/ICAM-1 interaction and act as a bridge between tumor cells and the liver parenchyma, thus promoting extravasation and liver metastasis132. Thus, CTCs clusters with neutrophils anchor to the vascular endothelium for extravasation while resisting shear stress, and the process is mediated by a series of cell adhesion proteins, such as cadherin, integrin, and surface glycoprotein133–135. Intriguingly, Chen et al. found that IL-8 secreted from neutrophils was essential for neutrophil sequestration with arrested tumor cells and for the extravasation behaviors of adjacent tumor cells through the endothelial barrier136.
Neutrophils can also promote metastasis in an indirect manner. Neutrophil extracellular traps (NETs) are web-like structures formed by DNA–histone complexes and proteins released from activated neutrophils, with the ability to impact on CTC biology137. Many studies have found that NETs were able to capture CTCs in the circulation and, in doing so, promoted metastatic dissemination138,139. In vitro and in vivo experiments have shown that CTC adhesion to NET is mediated by β1-integrin expressed on both NET and cancer cells, while this effect was abrogated following DNAse I administration139. In a murine model of surgical stress, the NET formation triggered the release of high-mobility group box 1, which activated TLR9-mediated pathways in CTCs and therefore accelerated the progression of liver metastases140. In addition, NETs can also awaken dormant cancer cells and promote metastasis. Recently, Albrengues et al. elegantly demonstrated that two NET-associated proteases, neutrophil elastase and matrix metalloproteinase 9 (MMP9), concentrate at laminin, provoke its cleavage, by generating an epitope that induced awakening of dormant cancer cells by integrin activation and FAK/ERK/MLCK/YAP signaling141. In turn, tumor-expressed protein, such as protease cathepsin, has been shown to support lung metastasis of breast cancer by promoting NET formation in metastatic niches142. Coiled-coil domain containing protein 25, another protein expressed on the cancer cell membrane, could serve as a specific sensor for the DNA component NETs, which induces migration and adhesion of tumor cells143. In addition, by forming NETs, circulating neutrophils can help CTCs escape immune surveillance by suppressing the activation of peripheral leukocytes144, the function of natural killer (NK) cells145, the antitumor response of effector T cells146, and even cooperation with other immune cells (such as IL17-producing γδT cells)147. Overall, there is evidence for a pro-metastatic role of neutrophils in their interaction with CTCs, but the specific mechanism remains to be elucidated in more detail.
Interaction of CTCs with macrophages
Tumor-associated macrophages (TAMs) not only contribute to metastatic progression within the primary tumor, but also promote later stages of metastasis including dissemination and extravasation of CTCs148. Hamilton et al. tried to investigate the CTC-macrophage interactions by co-culturing peripheral blood mononuclear cells with CTC cell lines obtained from small-cell lung cancer patients. They found that CTCs were able to induce monocyte differentiation to TAMs, which secrete a host of mediators such as osteopontin, MMP9, chitinase-3-like-1, and platelet factor to promote further leukocyte recruitment, migration, and invasion149,150. In another study of colorectal cancer, the feedback loop between TAMs and cancer cells is essential for the EMT program of CTCs and intravasation into the blood stream. Mechanistically, IL6 derived from TAMs regulates invasiveness through the STAT3/miR-506-3p/FoxQ1 pathway, which in turn increases CCL2 expression of TAM-educated tumor cells to help recruit macrophages151. TAMs seem to promote CTCs acquisition of mechanical adhesiveness and endurance, thereby helping them to form protective cell clusters and confer resistance to shear stress152. The fusion of macrophages and tumor cells could be a potential mechanism of immune evasion and invasion. These macrophage-tumor cell hybrids were shown to have M2-like macrophage phenotypes (CD163) as well as epithelial markers (EpCAM)153,154, and have been isolated from the blood of patients with multiple cancers such as PDAC155, melanoma154, breast, ovarian, and colorectal cancer156. Furthermore, when transplanted into mice, they spread widely and formed lesions in distant tissues154,155. A recent study by Gast et al. revealed that fusion hybrids can increase tumor heterogeneity and metastatic behavior, further correlating with disease stage and overall survival among several cancers157. Furthermore, larger hybrid sizes were also associated with worse survival among patients with non-small cell lung cancer158. Understanding the mechanism of direct interaction and molecular fusion between CTCs and macrophages is of great significance for identifying therapeutic targets.
Interaction of CTCs with platelets
The metastasis and progression of cancer are greatly influenced by the recruitment and activation of platelets, which support the survival of CTCs as well as their seeding and outgrowth at secondary sites159,160. Platelets can bind to and form aggregates with CTCs in the blood stream, and CTCs expand the formation of aggregation by releasing prothrombotic and procoagulant microparticles or by expressing tissue factor161,162. Platelet-released mediators, such as TGF-β, have been found to accelerate EMT in CTCs and to promote invasion and metastasis163,164. Xiong et al. recently determined that the expression of the heat shock protein 47 was induced during EMT, which enhanced the cancer cell–platelet interaction through its dependent collagen secretion in breast cancer cells165. Interestingly, platelets are believed to protect CTCs against mechanical stress166 and induce resistance to anoikis, the latter being mediated by the activation of the YAP1 pathway167. Furthermore, platelets promote escape of CTCs from NK cell attack through various mechanisms including (1) platelet aggregates that produce surface shielding to defend the cytolysis effect of NK cells;168 (2) platelet-derived normal MHC-I that are transferred to the surface of the tumor cell, thus preventing the identification of NK cells;169 (3) downregulation of natural killer group 2, member D (NKG2D) in NK cells by platelet-derived TGF-β, as well as platelet-mediated shedding of NKG2D ligands, which contribute to impaired antitumor cytotoxicity;170,171 and (4) platelet-derived glucocorticoid-induced TNF-related ligand which activates GITR in NK cells and reduces their cytotoxicity172. Furthermore, NK cells and platelets can also interfere with neutrophils, T cells, and macrophages and modulate their immune function173–175. In addition to safeguard CTCs within the bloodstream, platelets are also involved in the adhesion of endothelial cells. The attachment of platelets and CTCs is mediated by platelet adhesion receptors, such as the integrin αIIbβ3 and P-selectin, thereby supporting the firm adherence of CTCs to the endothelial wall176–178. Furthermore, tumor cell-activated platelets release ATP from dense granules, which then induces the activation of the endothelial P2Y2 receptor and allows transendothelial migration of tumor cells by increasing permeability179. One study has also revealed that the interplay between integrin α6β1 on platelets and its receptor, a disintegrin, and metalloprotease 9 on CTCs is necessary for the extravasation process of cancer cells180. Platelets could also increase vascular permeability to help tumor cell extravasation. For example, a preclinical lung metastasis model showed that tumor cell-associated CD97, an G protein-coupled receptor, can initiate platelet activation thereby leading to granule secretion, including both ATP and lysophosphatidic acid release181. Similarly, the interplay between platelet-specific receptor glycoprotein VI and its ligand galectin-3 expressed on colon and breast cancer cells was revealed to promote platelet activation and ATP secretion182. Consequently, these platelet secretions favor a process of tumor metastasis by regulating vascular permeability. Recently, Xu et al. discovered that Ptx@AlbSNO can block tumor-specific platelet functions to suppress tumor EMT as well as prevent platelet adhesion around CTCs. Ptx@AlbSNO can also inhibit TGF-β secretion and enhance intratumoral immune cell infiltration to reverse the immunosuppressive TME, thereby suppressing distant metastasis183. Taken together, the close and complex crosstalk between CTCs and platelets might involve distinct molecule variants and signaling pathways and possibly represent a promising antitumor strategy, particularly attractive for the treatment of several cancers.
Interaction of CTCs with MDSCs
MDSCs are a heterogeneous subset of myeloid cells characterized by immunosuppressive properties that also promote metastatic dissemination. Under a standard protocol for isolating human MDSCs, Cassetta et al. found that polymorphonuclear (PMN)-MDSC were significantly expanded among most cancer types except melanoma compared with infection and inflammation184. CTC-MDSC clusters are thought to evade immune surveillance of the T cell response185. Indeed, a decrease in circulating MDSCs was associated with an increase in activated OX40+PD-1- T cells in patients with diffuse large B-cell lymphoma186. Furthermore, Sprouse et al. found that in vitro co-culture of CTCs derived from melanoma and breast cancer patients and PMN-MDSCs enhanced Notch activation in CTCs through direct interaction between Jagged1 (Notch1 ligands) expressed on MDSCs and the Notch1 receptor expressed on CTCs. The increased production of reactive oxygen species production of MDSCs could upregulate Notch1 receptor expression, therefore, promoting CTC proliferation187. The potential mechanisms underlying the interplay between CTCs and MDSCs remains to be determined.
Interaction of CTCs with CAFs
CAFs are one of the most abundant components in the TME and play a prominent role in tumor initiation, angiogenesis, metastasis, and drug resistance188. Mechanistically, CAFs remodel the extracellular matrix structure, which allows tumor cells to invade through the stroma and communicate with cancer cells by secreting growth factors, chemokines, and cytokines189. However, little is known about the interplay between CAFs and CTCs. Duda et al. first demonstrated that CTCs could carry CAFs from the primary tumor to the metastatic site in mouse models of lung cancer metastasis190. These host-derived CAFs directly enhance tumor cell survival and promote the formation of metastasis, while depleting CAFs from lungs significantly reduces the number of macroscopic metastasis and extends survival rate in mice. Moreover, CAFs can protect CTCs from the fluid shear forces during the dissemination process191. In a three-dimensional co-culture model, CAFs were found to induce shear resistance to prostate tumor cells through stable intercellular contact, as well as soluble factors (such as CXCL5, CCL2, and CCL7), which are associated with cell survival, invasion, and EMT. In addition to experimental models, circulating CAFs (identified by FAP and α-SMA co-expression) have been detected in the peripheral blood of patients with metastatic breast, cancer but not in patients in the early stages192, and exhibit excellent precision in metastatic diagnosis (AUC–ROC, 0.975) when isolated using a novel acoustic microstreaming platform193.
Technologies for CTCs enrichment and isolation
Over the past few years, many methods have been proposed to capture CTCs. Due to the extremely small proportion of CTCs in patients’ blood, it is still a great challenge to accurately isolate CTCs from the numerous blood cells, and especially to invent applicable methods that can efficiently detect viable CTCs for subsequent in-depth analysis. Here, we will discuss the development of CTC-related technologies over the last two decades, as they have experienced tremendous growth. We will emphasize the most innovative methods associated with nanoscale materials or novel microfluidic chips, hoping to provide a useful framework of CTC-related technologies.
Generally, there are three core strategies of CTCs technologies194, which include (1) capture and enrichment, (2) detection and identification, and (3) release. The first strategy of capture and enrichment involves a specific interaction between CTCs and materials through physical interactions or antibody–antigen interactions. The second strategy of detection, which means identifying the CTCs, refers to various methods, such as fluorescence microscopy, fluorescence spectrophotometry, flow cytometry, surface-enhanced Raman scattering, or electrical impedance. In the last strategy, released CTCs are mainly used for downstream analysis, such as genomics, transcriptomics, proteomics, and CTCs culture.
Classic CTC-related technologies based on physical properties
The physical separation enrichment method of CTCs is based on differences between CTCs and blood cells in size, density, deformability, and electrical properties. The Isolation by size of epithelial tumor cells system195 can filter blood samples through an 8-μm diameter polycarbonate TRACK-ETCH-type membrane, but it has low efficiency. An improved method, which consists of a pressure regulating system, the flexible micro spring array device196, reaches a capture efficiency of 90% with a detection of CTCs in 76% of samples. However, there are various trends of CTC counts observed from different samples, making this method not reliable for widespread use. CTCs can also be sorted using the Oncoquick system197, a density-dependent technique that allows red blood cells and WBCs to be filtered, or by Apostream198 that uses dielectric electrophoresis techniques in the microfluidic chamber to capture CTCs. These systems require large volume of blood and cannot collect the CTCs of a similar size as WBCs, which are their main limitations. Overall, the methods based on physical properties are generally inefficient, poor in purity, and lack of specificity, although the vitality is good and the cost is relatively inexpensive.
Classic CTC-related technologies based on biological properties
Biological property-based technology is another important method for CTC detection. Based on antibody–antigen interaction, CTCs are usually positively enriched using epithelial (EpCAM) and mesenchymal (vimentin) markers as well as negatively enriched by using CD45 to deplete unwanted leukocytes199. EpCAM-dependent techniques are most commonly used by researchers. The CellSearch system200, the only FDA-approved device for clinical use, employs EpCAM antibody-coated ferromagnetic beads to enrich CK + /CD45-/DAPI + CTC and remove CK-/CD45 + /DAPI + WBCs. However, CTCs strongly adhere to the surface of the equipment in antibody interaction-based methods, making them difficult to be released. This deficiency can be resolved in another EpCAM-dependent, MagsWeeper system201, which uses a magnetic rod to enrich CTCs and eliminate cells not bound to magnetic beads, allowing the release of CTCs for the following biochemical analysis. CanpatrolTM 202 is another representative of the EpCAM-dependent technique, which provides morphological, cytological, and genetic characterization of individual CTCs. In summary, techniques based EpCAM are extensively-used. However, because the CTC surface antigen has high heterogenicity, CTCs that have low expression of EpCAM may not be enriched, causing inaccurate results, while methods based on physical properties do not have this limitation. Therefore, combining the advantages of different technologies or looking for CTCs with high sensitivity and specific tumor markers have gradually won the attention of researchers.
Recently, with the development of microfluidic chips, nanomaterials, and next-generation sequencing, researchers have many advanced technologies to stimulate progress in CTC-related technologies. Importantly, researchers are trying to reach higher levels of CTC-related technologies in several key parameters: yield, purity, enrichment ratio, throughput, viability, sensitivity, specificity, release rate, accessibility for further analysis, and simplicity of equipment operation).
Recent CTC-related technologies: microfluidic-based and nanotechnology-based techniques
Besides the classic CTC-related technologies discussed above, some newer technologies, such as microfluidic-based and nanotechnology-based techniques have been developed. Microfluidic-based cell sorting approaches use “intrinsic” (e.g., fluid dynamic forces) versus “extrinsic” external forces (e.g., magnetic, electric field, acoustic, and optical forces) to separate cells, and then select target cells from a sample of heterogeneous cells through different physical and biological properties203. The CTC-chip is a silicon microfluidic platform on which the CTCs are captured on the slides of molecular marker coated posts. The CTC-chip204 can separate viable CTCs from whole blood without pre-labeling or processing of samples, resulting in increased cell activity and separation purity. A modified chip-based platform using gold nanoparticles on a herringbone chip (NP-HB CTC-Chip205) easily detaches viable CTCs and safely releases cells for further analysis by utilizing a chemical ligand-exchange reaction with gold nanoparticles on a herringbone chip. Furthermore, the monolithic CTC-iChip206 has high-efficiency WBC depletion and allows the characterization of CTCs with epithelial and mesenchymal characteristics. Although these microfluidic chips have greatly contributed to the development of the detection of CTCs (i.e., improved capture efficiency, viability, and depletion of WBCs), they are not widely applied for clinical use due to limitations, which include long set-up time, high initial cost, bulky instrumentation, and limited ability to perform single-cell molecular analysis.
In an attempt to capture CTCs in an automated manner (Table 2), Zhang et al. successfully sorted MCF-7 cells from a 5 mL volume of diluted blood within 23 m with a recovery rate of 85%207. Even more remarkable, Jia et al. developed a less costly self-driving micro-cavity array chip to achieve cell loading, lysing, isothermal amplification, and signal read-out on a single chip208. This novel chip can perform genetic analysis at the single-cell level, it has great potential in personalized therapy and efficacy monitoring. Furthermore, another automated and integrated microfluidic system proposed by Wang et al. is reported to achieve CTC capture and identification within 90 m. With the advantages of automation, stability, economy, and user-friendly operation, this system provides broad prospects for cancer screening and prognosis, especially in HCC209. Additionally, Lee et al. invented a microfluidic-based integrated system to achieve simultaneous on-chip isolation and characterization of circulating tumors utilizing differences in magnetic field gradient and immune fluorescence. Furthermore, this novel system can differentiate on-chip eight different subtypes of heterogenic CTCs, guiding the diagnosis and prognosis of breast cancer210. By combining the microfluidic technology and in situ molecular profiling techniques, the On-chip Post-processing Enabling chip platform has the ability to perform molecular analyses of single CTC from metastatic breast cancer and metastatic pancreatic cancer patients without any off-chip processes, suggesting its potential implementation of early molecular detection for cancer metastasis211. Taken together, less costly automated and integrated microfluidic systems that allow easy CTCs detection and cell analysis have great clinical value.
Table 2.
The CTC detection technologies in recent three years (from 2018 to 2021), mainly including microfluidic chip and nanotechnology-base methods
| Technology | Key features | Strengths | Weakness | Author (year) |
|---|---|---|---|---|
| Microfluidic chip | ||||
| Automated microfluidic method |
• A fully integrated microfluidic device and a set of robust fluid-driven and control units • A flow regulatory chip and two cell separation chips |
• Label-free and simple • High recovery rate (85%) • Rapid processing time (23 m) |
• A need of sample dilution to avoid cell–cell interaction • Quantitative design rules are still lacking for channels |
Zhang et al.207 |
|
Self-driving micro-cavity array chip |
• Integrate sample detection structure and vacuum driving system. • Use the “film-polydimethylsiloxane chip-film” structure and oil sealing method during amplification reaction to minimize water loss. |
• Less costly and simple • An excellent linear • Mutational gene profiling of single CTC • Achieve cell loading, lysing, isothermal amplification, and signal read-out on one chip. |
• Air diffuse across the thin polydimethylsiloxane walls under some conditions. | Jia et al.208 |
| Automated and integrated microfluidic method | • Integrate a 3D printed off-chip multisource reagent platform, a bubble retainer, and a single CTC capture microchip. |
• Achieve CTC capture and identification within 90 m. • Decrease immunostaining time and antibody consumption by 90%. • Detect CTCs from various cancers. |
• Deficiencies in patient recruitment | Wang et al.209 |
| Integrated microfluidic method | • Use magnetic field gradient and immune-fluorescence differences. |
• Isolate CTCs with an efficiency of >99%. • Differentiate eight different subtypes of heterogenic CTCs • Simultaneous on-chip isolation and discrimination of CTCs. |
• Limitation of distinguishable fluorescence wavelength light of microscope | Lee et al.210 |
| Nanotechnology-based methods | ||||
| Magnetic nanoparticles | ||||
| Tannic acid-functionalized magnetic nanoparticles | • Tannic acid interact with glycocalyx on cancer cells |
• Inhibit the nonspecific adhesion of peripheral blood mononuclear cell. • 95.1% of capture purity for MCF-7 cells |
• EpCAM-dependent | Ding et al.214 |
| Peptide-Based Magnetic Nanoparticle | • Use N-Cadherin recognition peptide-functionalized magnetic nanoparticles |
• High capture efficiency (about 85%) of mesenchymal CTCs from spiked human blood • Distinguished epithelial and mesenchymal subgroups. |
• Unmentioned | Jia et al.213 |
| CoFe2O4@Ag magnetic nanohybrids |
• Gold electrodes are modified with MXene nanosheets and an HB5 aptamer is immobilized on the MXene layers • CoFe2O4@Ag magnetic nanohybrids is bonded to the HB5. |
• A wide linear range of 102-106 cells/mL, low detection limit of 47 cells/mL • Good sensitivity and selectivity in the detection of HER2-posetive cells in blood samples. • Cost-effective and environmentally friendly |
• Ag has some aggregations. • The magnetic property of magnetic probes can affect the isolation of magnetic cells. |
Vajhadin et al.216 |
| Gold nanoparticles | ||||
| Two-dimensional nanozyme with gold nanoparticles | • Au nanoparticles-loaded two-dimensional bimetallic PdMo nanozymes assembled with an aptamer composed of a thiol-modified EpCAM |
Excellent analytical performance • A satisfactory CTCs release reaching a range of 93.7–97.4% and good cell viability |
• Long preparation time of the Au@PdMo nanozymes • Complex detection procedure |
Yang et al.224 |
| AuNP–anchored black phosphorus nanosheets |
• Use electrochemical detection • Use BP@AuNPs@aptamer as a probe combined with immunomagnetic separation |
Good analytical performance with a detection limit of 2 cell mL−1. • Good practicality in detecting MCF-7 cells |
• EpCAM-dependent | Liu et al.221 |
| Antibody-Functional Microspheres Integrated Filter Chip |
• Consist of a semicircle arc and arrays • Use interfacial zinc oxide coating with nanostructure on the surface of the microsphere to increase specific surface area |
• Separate large-scale CTC-microspheres. • High capture efficiency (>90%) for different tumor cell lines • Easy to find and isolate the CTCs |
• Unmentioned | Wang et al.225 |
With the progress of nanomaterials, nanotechnology-based methods are becoming promising tools for CTC detection at an early-stage disease and for the monitoring of cancer development, as well as in vivo imaging212. Nanomaterials have a large surface-to-volume ratio and allow CTC isolation at high specificity and CTC detection at high sensitivity by adsorbing numbers of targeting ligands to bind specific molecules on cancer cells. At present, studies have reported many types of nanomaterials (Table 2) for CTC detection, including magnetic nanoparticles213–217, gold nanoparticles218–220, and quantum dots221–223. For example, studies have shown that the utilization of tannic acid-functionalized magnetic nanoparticles214, CoFe2O4@Ag magnetic nanohybrids216, and peptide-based magnetic nanoparticle213, enhances the capture efficiency of CTCs in breast cancer patients. Among them, peptide-based magnetic nanoparticles can distinguish epithelial and mesenchymal CTC subgroups and allow analysis at the single-cell level, the detection effects of which are supported by magnetic nanoparticles and microfluidic-based integrated systems210. Regarding gold nanoparticles, there have been significant developments. For example, the cytosensor proposed by Yang et al.224 showed excellent analytical performance, with a wide linear range, satisfactory CTC release (93.7–97.4%), and good cell viability. Liu et al. reported that gold nanoparticle-modified black phosphorus nanosheets improved the stability in detecting CTC218. Furthermore, the combination of a microfluidic system and gold nanoparticles presents a wider range of applications. Wang et al. synthesize an interfacial zinc oxide coating with a nanostructure on the microsphere surface, which increases the specific surface area and thus leads to an improved capturing efficiency of CTCs225. In addition, the utilization of multicolor magnetic surface-enhanced Raman scattering nanotags and chip-based immunomagnetic separation could detect four different surface protein markers on individual tumor cells in a quantitative and simultaneous manner, thus facilitating the separation of CTC subpopulations217.
Although nanotechnology-based techniques can provide broad prospects for CTC research in various tumors in a cost-effective and simple manner, there are limitations and challenges. First, many factors (e.g., binding of nanoparticle probes, aggregation) can affect nanoparticle-based detections, leading to decreased reliability and reproducibility. Second, most nanoparticle-based assays are prepared for academic studies, and they are still unrealistic for widely clinical translation. Third, there is possible toxicity of nanoparticles.
In the era of precision medicine, CTCs analysis has great clinical value. Tools must be sharpened first if workers are to do their job well. Therefore, CTC-related technologies are the underlying foundation to the application of CTCs in precision medicine. Herein, we reviewed previous CTCs-related technologies based on physical and biological properties, and the most recent development of techniques associated with microfluidic-based and nanoparticle-based approaches. Although there are strengths and weakness between different methods, we believe that an effective combination of these techniques may benefit CTCs research in many ways, especially the in-depth analysis and possibility in clinical applications.
Clinical applications of CTCs
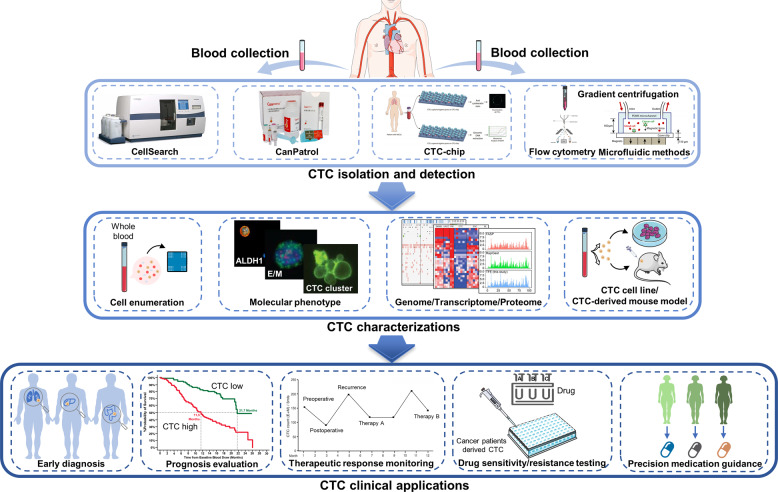
Clinically, CTCs are now used as surrogate biomarkers for many solid cancers. Numerous studies have been carried out, mainly in breast cancer, prostate cancer, lung cancer, liver cancer, pancreatic cancer, gastric cancer, and melanoma. Although the clinical guidelines have not included the clinical use of CTCs, besides the inclusion of CTCs as part of the cM0 tumor classification (i.e., no clinical of overt metastasis but the detection of tumor cells in blood), many studies have predicted the great potential of CTCs in clinical applications. In this section, we will mainly present the role of CTCs as biomarkers for diagnosis, prognostication, and therapy monitoring in different cancers (Fig. 4).
Fig. 4.
Overview of the CTC isolation and detection, characterizations, and clinical applications
Early diagnosis of cancer
As a non-invasive method, CTC detection is attractive in assisting cancer diagnosis. Studies of CTCs used for early diagnosis of cancer in the past three years are listed in Table 3. A tumor lesion already has more than 109 tumor cells by the time they are detectable in patients using current imaging procedures15, such as computed tomography, magnetic resonance imaging, and positron emission tomography. Diagnosis of cancers as early as possible, especially for fast-progressing cancers, is the best way to defeat them. Studies have determined that CTCs are correlated with tumor stage, but the clinical utility of CTCs in cancer detection or even in early cancer diagnosis is still a matter of debate. CTCs are considered a surrogate marker of metastatic activity, but whether metastatic dissemination of CTCs in patients occurs early during tumor formation is still controversial. However, in mouse models, early dissemination seeding metastasis has been found in breast226,227 and pancreatic228,229 carcinogenesis, indicating that CTC circulation is likely to be a very early event in cancer progression. In Barrière et al.’s study230, CTCs were detected in 41% of T1 stage and 47% of axillary lymph node-negative breast cancer patients, both of which are early-stage breast cancer. In the study by Thery et al.231 the CTC positivity rate was 21% and 24% in lymph node-negative and positive breast cancer, respectively. Based on this hypothesis, CTCs can be detected earlier before the primary tumor is visible on imaging studies, while the biggest challenge of the application of CTC application in early cancer diagnosis is indeed their scarcity and isolation. The limited sensitivity of CTC detection methods hinders their use as an effective biomarker in early cancer diagnostics.
Table 3.
Clinical applications of CTCs in recent three years (from 2019 to 2021)
| Cancer type | Author & year | CTCs utility | Detection methods | CTC marker | Patients | Main findings | Clinical Trial No. |
|---|---|---|---|---|---|---|---|
| Breast cancer | Paoletti et al.232 | prognostic value | CellSearch | EpCAM, CK | 549 mBC | In mBC patients with 1st line chemotherapy, CTC counts was associated with mortality. | NCT00382018 |
| Mego et al.361 | prognostic value | RT-qPCR | TWIST, SNAIL1, SLUG, ZEB1 | 427 stage I-III BC | CTC EMT was detected in 18% of patients, which was associated with worse DFS. | NA | |
| Magbanua et al.233 | prognostic value | CellSearch | EpCAM, CK | 742 untreated BC | CTC positivity associated with reduced DRFS. | NA | |
| Strati et al.253 | prognostic value | density-based isolation, RT-qPCR | EpCAM, TWIST1, CD24/CD44,ALDH1 | 100 early BC | Detection of TWIST1 overexpression and stem-cell transcripts in EpCAM+ CTCs provides prognostic information. | NA | |
| Wang et al.255 | therapeutic monitoring | CellSearch | EpCAM, CK | 160 early BC | CTC counts were lower in surgical group than trastuzumab group, but the OS rate in surgical group was higher. | NA | |
| Radovich et al.279 | prognostic value | microfluidic device | EpCAM | 123 early triple-negative BC | Positive CTC and circulating tumor DNA after neoadjuvant chemotherapy associated with inferior DFS and OS. | NCT02101385 | |
| Zhang et al.362 | recurrence monitoring | Cyttel detection | CD133, CEP8 | 135 early Luminal A BC | There were no differences in DFS and OS between CTC monitoring and routine re-examination monitoring group. | NA | |
| Magbanua et al.234 | prognostic value, therapeutic monitoring | CellSearch | EpCAM, CK | 294 ER + mBC | CTCs ≥5 was detected in 31% of the patients. Letrozole with bevacizumab gain better OS than without bevacizumab in patients with CTC ≥ 5. | NCT00601900 | |
| Wang et al.46 | prognostic value, guiding therapy | CellSearch | EpCAM, CK | 105 HER2- advanced BC | HER2 + CTCs ≥ 2 associated with shorter survival and higher risk for disease progression (HR 2.16). Those received anti-HER2 targeted therapies had improved PFS. | NA | |
| Stefanovic et al.363 | no prognostic value | CellSearch | EpCAM, CK | 261 mBC | CTCs had no prognostic value in different receptor change pattern subgroups. | NA | |
| Papadaki et al.254 | prognostic value | density-based isolation | CK, CD47, PD-L1 | 198 (100 early BC, 98 mBC) | CTCs expressing CD47 and PD-L1 have independent poor prognostic implications in mBC. | NA | |
| Jin et al.364 | diagnostic value | CytoSorter | EpCAM, CK | 130 BC | CTC detection rates in BC patients at Tis and T1-4 stages were 50%, 81.67%, 91.07%, 100%, and 100%, respectively. | NA | |
| Silveira et al.235 | prognostic value | CellSearch | EpCAM, CK | 198 HER2- mBC | CTC count ≥1 and ≥5 was detected in 37% and 22% of the patients at 4 weeks of treatment, respectively. CTCs levels at four weeks had a significant prognostic impact on PFS and OS. | NCT01745757 | |
| Paoletti et al.236 | prognostic value | CellSearch | EpCAM, CK | 121 ER + /HER2- mBC | CTCs ≥5 at baseline was detected in 36% of patients. Elevated CTC at 1 month was associated with worse PFS. | NCT01701050 | |
| Magbanua et al.237 | prognostic value | CellSearch | EpCAM, CK | 469 mBC | Intermediate or high CTC trajectory pattern was associated with poor prognosis. | NCT00785291 | |
| Shliakhtunou et al.365 | guiding therapy | magnetic isolation, RT-qPCR | EpCAM, Survivin, HER2-neu | 228 BC | CTC-oriented personalized adjuvant chemotherapy (turn to taxanes or add gemcitabine) can 100% eradicate CTCs, and increase 5-year DFS by 7.4% and OS by 11.6% | NA | |
| Prostate cancer | Sieuwert et al.366 | prognostic value | CellSearch, RT-qPCR | EpCAM, CK, AR-Vs | 118 mCRPC | CTC-adjusted detection of AR-V1 after two cycles of cabazitaxel was an independent prognostic factor for OS. | NA |
| Kruijff et al.238 | prognostic value | CellSearch | EpCAM, CK | 120 mCRPC | CTC count was independently associated with PFS and OS in mCRPC patients with cabazitaxel treatment. | NA | |
| Graf et al.256 | therapeutic monitoring | Streck tubes | CK, AR-V7 | 193 mCRPC | Patients with detectable nuclear-localized AR-V7 in CTCs had superior survival with taxanes over ARSIs. | NA | |
| Cie´slikowski et al.239 | diagnostic value | CellSearch, EPISPOT, GILUPI CellCollector | EpCAM, CK, PSA, FGF2 | 104 PC | High CTC counts related to high-risk prostate cancer patients with occult metastases at the time of diagnosis. | NA | |
| Schonhoft et al.367 | prognostic value | Epic Sciences | CK, AR | 294 mCRPC | Chromosomal instability in CTCs was associated with poor OS in patients treated with AR signaling inhibitors and taxanes. | NA | |
| Armstrong et al.368 | prognostic value | AdnaTest, Epic Sciences | CK, AR, AR-V7 | 118 mCRPC | AR-V7 in CTCs was independently associated with shorter PFS and OS with abiraterone or enzalutamide. | NA | |
| Xu et al.369 | diagnostic value | Parsortix | CK, vimentin | 155 PC | Combining the PSA, CTCs and the 12-gene panel, the AUC of clinically significant prostate cancer prediction was 0.927. | NA | |
| Sperger et al.370 | prognostic value | VERSA | EpCAM | 147 mCRPC | A transcriptional profile detectable in CTCs can serve as an independent prognostic marker in mCRPC. | NCT01942837, NCT01942837 | |
| Renal cell carcinoma | Zhang et al.371 | no prognostic value | CanPatrol-ITMCTCs | EpCAM, CK, Beclin vimentin, TWIST, | 199 RCC | No differences in the OS and DFS of RCC between the different numbers of CTCs and Beclin1 expression. | NA |
| Basso et al.240 | prognostic value | CellSearch | EpCAM, CK | 195 RCC | Patients with ≥ 3 CTCs had a shorter OS. | NA | |
| Non-small-cell lung cancer | Chemic et al.241 | prognostic value | CellSearch | EpCAM,CK | 100 NSCLC | Pulmonary venous-CTCs were detected in 48% of 100 patients, serving as early predictors of NSCLC recurrence after surgery. | NA |
| Li et al.372 | prognostic value | CytoploRare Kit, FR ligand-TaqMan | CD45, CD14 | 347 NSCLC | The median follow-up time was 38 months. Preoperative CTC concentration was an independent prognostic factor. | NA | |
| Small-cell lung cancer | Messaritakis et al.242 | prognostic value | CellSearch, Ficoll-Hypaque | Notch 1–4 receptors, CK, CD45, vimentin | 108 SCLC | The detection of DLL3 + /CD45- CTCs at baseline and progression was related to decreased PFS and OS, respectively. | NA |
| Wang et al.373 | prognostic value | EpCAM-independent | EpCAM, CD45, DAPI | 138 SCLC | The high number of CTC predicted adverse prognosis. | NA | |
| Hepatocellular Carcinoma | Ha et al.374 | prognostic value | Tapered slit filter | CK | 105 HCC | Postoperative CTCs was detected in 23.8% of HCC patients and it may serve as an independent predictor of recurrence. | NA |
| Chen et al.375 | no prognostic value | Canpatrol | EpCAM, CK, vimentin, TWIST | 256 HCC | CTC count and EMT status were not correlated with clinical stages or predictive of HCC recurrence. | NA | |
| Cheng et al.376 | diagnostic value | CanPatrol | EpCAM, CK, vimentin, TWIST | 113 HCC | Mesenchymal CTCs were increased in late-stage HCC patients. The cut-off value CTCs ≥ 1 was for the diagnosis of HCC. | NA | |
| Sun et al.243 | prognostic value | CellSearch | EpCAM, CK | 197 HCC | CTC count ≥3 was associated with higher risk of postoperative extrahepatic metastases. | NA | |
| Lei et al.377 | prognostic value | CanPatrol | EpCAM, CK, vimentin, TWIST, Nanog | 160 HCC | The numbers of EpCAM mRNA+ CTCs and Nanog mRNA+ CTCs were correlated with postoperative HCC recurrence, with Nanog > 6.7 (HR = 2.33) being the most crucial marker. | NA | |
| Pancreatic cancer | Wei et al.257 | diagnostic value, treatment monitoring | CytoQuest, microfluidic chip | vimentin, EpCAM, CK | 100 PDAC | Vimentin+ CTCs were detected in 76% of patients with PDAC. Preoperatively higher counts was correlated with shortened RFS. | NA |
| Zhao et al.334 | prognostic value | CanPatrol | EpCAM, vimentin, TWIST | 107 PDAC | CTCs were detected in 78.5% of PDAC patients. Patients with ≥ 6 total CTCs had significantly decreased OS and PFS. | NA | |
| Hugenschmidt et al.331 | guiding therapy, prognostic value | CellSearch | EpCAM | 209 patients | CTC-positive (≥1 CTC/7.5 mL) preoperatively showed a detrimental outcome despite successful tumor resections. | NCT01919151 | |
| Gastric cancer | Szczepanik et al.378 | prognostic value | flow cytometry | CD45, CD44, CK | 228 GC | CK + /CD44 + cells were significantly more common among patients with distant metastases. | NA |
| Miki et al.379 | prognostic value | Ficoll | EpCAM, CEA, CK | 150 GC | The number of EpCAM − /CEA + cells was higher in patients with stage II–III and IV than in patients with stage I. A lower number of these cells indicated a higher 3-year RFS. | NA | |
| Kuroda et al.380 | prognostic value | Ficoll | EpCAM, FGFR2, CD45 | 100 GC | FGFR2 + CTCs (≥5 cells/10 mL blood) showed poorer RFS. | NA | |
| Nevisi et al.244 | prognostic value | CellSearch | EpCAM, HER2 | 105 GC | HER2-expression on CTCs was an independent prognostic factor for both overall and progression-free survival. | NA | |
| Colorectal cancer | Wang et al.381 | prognostic value | magnetic isolation | chromosome enumeration probe | 130 CRC with stage II-III | Postoperative CTCs were significantly correlated with poor RFS. | NA |
| Bidard et al.382 | prognostic value | CellSearch | EpCAM, CK | 153 CRC with liver metastasis | Baseline CTCs ≥3 was detected in 19% of the patients. CTC ≥ 3 at baseline and 4 weeks after therapy showed shorter OS. | NCT01442935 | |
| Wang et al.383 | prognostic value | Cyttel | Chromosomes 8 and 17 H1 | 121 CRC | CTCs were detected in 58.7% of CRC patients. Advanced CRC patients with CTC-positive had worse PFS and OS. | NA | |
| Messaritaki et al.384 | prognostic value | Density gradient isolation, RT-qPCR | CEACAM5, EpCAM | 198 advanced CRC | CEACAM5 was a dynamic adverse prognostic CTC biomarker in patients with metastatic CRC. | NA | |
| Su et al.252 | prognostic value | CellSearch | EpCAM, CK, vimentin, twist, PRL-3 | 156 CRC | CTCs were detected in 100% of CRC patients. The count of mesenchymal and PRL-3+ CTCs ≥ 12 was significantly associated with recurrence and shorter DFS. | NA | |
| Sastre et al.245 | prognostic value | CellSearch | EpCAM, CK | 1202 metastatic CRC | Baseline CTCs ≥3 was detected in 41% of the patients. | NCT01640405, NCT01640444 | |
| Pan et al.385 | prognostic value | magnetic isolation | CK19 | 149 CRC | CTC counts were associated with TNM stages. The change escalated more rapidly in the CTC-positive group. | NA |
AR androgen receptor, BC breast cancer, CK cytokeratin, CTC circulating tumor cell, CRC colorectal cancer, DAPI 4’,6-diamidino-2-phenylindole, DFS disease free survival, DRFS distant recurrence-free survival, EMT epithelial-mesenchymal transition, GC gastric cancer, HCC hepatocellular carcinoma, HER2 human epidermal growth factor receptor-2, mBC metastatic breast cancer, mCRPC metastatic castration-resistant prostate cancer, NA not applicable, NSCLC non-small-cell lung cancer, OS overall survival, PC prostate cancer, PDAC pancreatic ductal adenocarcinoma, PD-L1 programmed cell death ligand-1, PFS progression free survival, qRT-PCR quantitative real time polymerase chain reaction, RCC renal cell carcinoma, RFS recurrence free survival
Evaluation of the cancer prognosis
The prognostic value of CTCs has been extensively studied. CellSearch is the only FDA-approved system for CTC detection used clinically. Based on CellSearch system46,232–245, CTCs represent an independent prognostic factor. Studies evaluating other CTC detection systems such as CanPatol, CTC-chip obtained the similar results. CTC enumeration is the main target of investigation, with a cut-off value of ≥5 for positivity, which usually indicates a worse prognosis. It is generally considered that increased CTC counts are correlated with higher likelihood of metastasis and cancer aggressiveness. In a meta-analysis pooling 2239 breast cancer patients including 21 studies246, the CTC count before neoadjuvant chemotherapy had a detrimental and decremental impact on patient survival, and patients with one, two, three to four, and five or more CTCs displayed a HR of death (95% CI) of 1.09 (0.65–1.69), 2.63 (1.42–4.54), 3.83 (2.08–6.66), and 6.25 (4.34–9.09), respectively. Furthermore, elevated baseline CTCs levels were associated with inferior survival, the presence of CTC clusters often predicted poor prognosis247–249, and increasing CTC counts or failure to clear CTCs during treatment was also a prognostic factor for worse survival234,250,251. Many studies have found that the molecular phenotypes of CTCs have strong prognostic value. EMT and stemness are the main molecular phenotypes of CTCs studied clinically. CTCs with expression of mesenchymal252 or stemness253-related markers associated with inferior survival. The expression of other molecular markers, such as HER246, CD47254, PD-L1254 also have prognostic implications. Most studies investigate the prognostic value of CTCs at a single timepoint, while intriguingly, some studies have taken CTC dynamics into account. Magbanua et al.237 developed a novel latent mixture model to stratify groups with similar CTC trajectory patterns during the treatment course, and they found that analysis of serial CTCs can further stratify the patients of poor prognosis into distinct prognostic subgroups. The dynamic changes of CTCs may act as a surrogate prognosis biomarker over the long course of cancer progression. Given the rapid advancements in the accessibility and strengthening of sequencing technologies at the single-cell level, we may expect that in the future, genomic/transcriptional profiles of CTCs may serve as an outstanding prognostic marker, representing biological information that is more comprehensive and more closely related to prognosis. Studies of CTCs for predicting prognosis in recent three years are summarized in Table 3.
Monitoring of the therapeutic response
In many clinical trials, CTCs have been used as useful biomarkers for monitoring cancer treatment responses234,255–257, either combined with imaging examinations, serum biomarkers or alone. Researchers prefer to involve CTCs in the evaluation of therapeutic efficiency, in view of the higher sensitivity of CTCs than imaging examination in some cases9. As a non-invasive method, the detection of CTCs may also contribute to avoiding frequent radiation exposure from imaging studies during the evaluation of treatment response. Most studies found a decrease or clearance in CTC counts was associated with a good therapeutic response, while the increase of CTC counts signified the opposite258,259. The Response Evaluation Criteria in Solid Tumors (RECIST) guidelines are the most often used standard for evaluating therapeutic response in solid tumors. However, in some studies, changes in CTC following therapy were not correlated with RECIST responses in cancer patients260. Indeed, CTCs assessment has not been included in the RECIST guidelines. Some CTCs measurement technologies have been recently developed to achieve genotyping for CTCs, which can also detect crucial gene mutations, such as ER39,49, HER239,49, EGFR261, KRAS262, and TP53263, thus helping clinicians in treatment personalization and resistance options at the time of tumor progression. Studies using CTCs to monitor treatment response in recent three years are summarized in Table 3.
The great potential for CTCs in the clinical application of cancer diagnostics has emerged, although clinically, its use as a surrogate biomarker for cancer screening, treatment monitoring, and prognosis predicting is still limited. Once metastasis occurs, repeat biopsies of metastatic lesions are usually difficult to obtain, and different metastases are heterogenous even in the same patients. CTCs testing using peripheral blood samples is convenient, and may be more representative of the traits of metastatic cells, which are derived from different metastatic lesions in patients. Nevertheless, there is still a lack of guidelines for the clinical use of CTCs, such as a standardized CTCs detecting assay for different cancers, a combination diagnostic scheme with other clinical examinations, and indications for the appropriate timepoints for blood sampling.
Discussion
Studies investigating CTCs have the great potential to reveal the fundamental processes of metastases, including the mechanisms involved in extravasation of CTCs from the primary tumor, how CTCs interact with blood cells to survive in the circulatory microenvironment, and how CTCs intravasate into the distant metastatic site to initiate new lesions. Significant molecular traits of CTCs can greatly contribute to identify targets for anti-metastatic therapies. Only a small proportion of CTCs can finally generate metastases, thus studies focusing on these strongly metastatic CTCs may provide deeper insights into CTCs-related therapeutic targets.
Various CTCs detecting technologies have emerged, however, the sensitivity and specificity of these technologies still need to be further improved. Epithelial marker-based CTC detection technology, such as the CellSearch system has opened a new era for CTC analysis and clinical applications, but their drawbacks are rapidly being acknowledged and appreciated by researchers. EMT is a crucial trait of metastatic cancer cells, indicating insufficient capture efficiency of epithelial marker-based CTC detection technology. However, mesenchymal marker-based detection technologies may also be contaminated by non-CTCs, such as tumor-associated fibroblasts and endothelial clusters, which induce the risk of false positivity. Nevertheless, it is intriguing that recent studies have reported that those non-cancerous tumor-derived cells presented in cancer patients are also important surrogate biomarkers for cancer patients264. Cancer-type-specific molecular markers for CTCs are likely another option, as CTCs of different cancer types possess different molecular markers. However, the sensitivity and specificity of known cancer-type-specific CTC markers are not satisfactory. Physical-property-based CTC detection technologies also have the problem of contamination by non-CTCs, especially for those with similar physical properties as CTCs. Microfluidic-based and nanotechnology-based CTC detection technologies have become popular in recent years, while the efficiency of these technologies still needs further large-scale clinical validation. High cell detection efficiency and contamination removal capability are the two key strengths of a successful CTC detection technology, while substantial technical optimization of CTC detection is urgently needed to achieve these requirements.
Comprehensive characterization of CTCs is lacking. The limited amount of genomic DNA, RNA, and protein content of CTCs is a bottleneck for exploring their genome, transcriptome, epigenome, and proteome properties. Nonetheless, the emerging genome and transcriptome studies of CTCs have recently profited from the fast-evolving technology of single-cell sequencing, while the proteome studies of CTCs are still elusive due to the very limited technologies for proteome exploring at a single-cell level. However, the study of the CTC proteome is imminent, not only because it can provide a picture of the biological characterization of CTCs, but also because it can help to discover CTC-specific membrane proteins which may help optimize CTC detection.
As for the solid tumor microenvironment, the blood microenvironment around CTCs also plays a significant role in tumor survival and invasion capacity. However, knowledge of the underlying mechanisms behind the survival of CTCs is still limited, as it is a complex process that involves not only shear forces and fluid mechanics but also soluble factors and tumor-associated extracellular vesicles, which are not detailed here. Furthermore, it remains to be confirmed whether CTC clusters are more suitable for interacting with other blood components or adapting to shear forces than single migratory CTCs. If combined with specific biomarkers for strategically detecting CTCs and the interaction of CTCs with associated peripheral blood cells, we could improve the clinical practicability and monitoring power of CTCs by obtaining more comprehensive information on tumor burden and immune status of patients.
Although CTCs have shown initial promise in clinical applications, many challenges must still be overcome before CTC analysis can be widely applied in the clinic. Today, the clinical application of CTCs mainly depends on the analysis of CTC cell enumeration and molecular phenotypes. A more comprehensive characterization of CTCs based on their genome, transcriptome, and proteome with high-throughput sequencing will further benefit clinical application, but also add to the complexity and difficulty of data analysis. CTCs will be a crucial component of “Precision medicine” in the future, as the phenotypic, genotypic, and functional characterization can provide an opportunity to study drug susceptibility that is related to metastasis. The genome and transcriptome analysis of CTCs can unveil potential drug targets. Viable CTCs for drug sensitivity/resistance testing over the therapy course can guide precision medication. However, the culture of CTCs is very challenging: (1) limited methods are available to isolate viable CTCs, which also yield low numbers of CTCs and (2) a favorable circulatory microenvironment for CTC survival is difficult to mimic. Very limited CTC-derived cell lines from cancer patients have been established. Optimization of CTC culture conditions will be needed.
Furthermore, CTCs, circulating tumor DNA (ctDNA), and exosomes are all present in liquid biopsy samples. An exploration of the advantages and disadvantages of each substrate present in the liquid biopsies, and how to better incorporate them into clinical application is needed to achieve more precise diagnoses. Among liquid biopsy methods, CTCs have tremendous advantages, as isolated CTCs can be viable, which can optimize CTC-derived explants or three-dimensional organoid cultures for functional testing or for drug-screening assays. The study of CTCs is attractive, and CTC detection may likely become an essential component of cancer management in the future. As the picture becomes clearer, we are fully confident about the promising potentials of CTCs.
Funding
This study was supported by the funding of the National Natural Science Foundation of China (Grant No. 82172344, 81702866, Jiaojiao Zhou), Natural Science Foundation of Zhejiang Province (Grant No. LY21H160039, Jiaojiao Zhou), Fundamental Research Funds for the Central Universities (Grant No. 2021FZJD009, Jiaojiao Zhou) and the National Natural Science Foundation of China (Grant No. 82072900, Yiding Chen).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Danfeng Lin, Lesang Shen, Meng Luo, and Kun Zhang
Contributor Information
Yiding Chen, Email: ydchen@zju.edu.cn.
Jiaojiao Zhou, Email: zhoujj@zju.edu.cn.
References
- 1.Sethi N, Kang Y. Unravelling the complexity of metastasis-molecular understanding and targeted therapies. Nat. Rev. Cancer. 2011;11:735–748. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8:98–101. [PubMed] [Google Scholar]
- 4.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. USA. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosseini H, et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540:552–558. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aceto N, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashworth TR. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Australas. Med. J. 1869;14:146–149. [Google Scholar]
- 14.Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene. 2016;35:1216–1224. doi: 10.1038/onc.2015.192. [DOI] [PubMed] [Google Scholar]
- 15.Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat. Rev. Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 16.Harper KL, et al. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature. 2016;540:588–592. doi: 10.1038/nature20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosseini H, et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540:552–558. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lianidou ES, Markou A. Circulating tumor cells in breast cancer: detection systems, molecular characterization, and future challenges. Clin. Chem. 2011;57:1242–1255. doi: 10.1373/clinchem.2011.165068. [DOI] [PubMed] [Google Scholar]
- 19.Thanh Huong P, et al. Emerging role of circulating tumor cells in gastric cancer. Cancers. 2020;12:695–716. doi: 10.3390/cancers12030695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gires O, Pan M, Schinke H, Canis M, Baeuerle PA. Expression and function of epithelial cell adhesion molecule EpCAM: where are we after 40 years? Cancer Metastasis Rev. 2020;39:969–987. doi: 10.1007/s10555-020-09898-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 22.Criscitiello C, Sotiriou C, Ignatiadis M. Circulating tumor cells and emerging blood biomarkers in breast cancer. Curr. Opin. Oncol. 2010;22:552–558. doi: 10.1097/CCO.0b013e32833de186. [DOI] [PubMed] [Google Scholar]
- 23.Gorin MA, et al. Circulating tumour cells as biomarkers of prostate, bladder, and kidney cancer. Nat. Rev. Urol. 2017;14:90–97. doi: 10.1038/nrurol.2016.224. [DOI] [PubMed] [Google Scholar]
- 24.Varillas JI, et al. Microfluidic isolation of circulating tumor cells and cancer stem-like cells from patients with pancreatic ductal adenocarcinoma. Theranostics. 2019;9:1417–1425. doi: 10.7150/thno.28745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcuello M, et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Asp. Med. 2019;69:107–122. doi: 10.1016/j.mam.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Xia W, et al. In vivo coinstantaneous identification of hepatocellular carcinoma circulating tumor cells by dual-targeting magnetic-fluorescent nanobeads. Nano Lett. 2021;21:634–641. doi: 10.1021/acs.nanolett.0c04180. [DOI] [PubMed] [Google Scholar]
- 27.Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol. cancer. 2019;18:114. doi: 10.1186/s12943-019-1043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gall TMH, Belete S, Khanderia E, Frampton AE, Jiao LR. Circulating tumor cells and cell-free DNA in pancreatic ductal adenocarcinoma. Am. J. Pathol. 2019;189:71–81. doi: 10.1016/j.ajpath.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Eslami SZ, Cortes-Hernandez LE, Alix-Panabieres C. Epithelial cell adhesion molecule: an anchor to isolate clinically relevant circulating tumor cells. Cells. 2020;9:1836–1852. doi: 10.3390/cells9081836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, et al. Label-free isolation and mRNA detection of circulating tumor cells from patients with metastatic lung cancer for disease diagnosis and monitoring therapeutic efficacy. Anal. Chem. 2015;87:11893–11900. doi: 10.1021/acs.analchem.5b03484. [DOI] [PubMed] [Google Scholar]
- 31.Wang ZL, et al. High-efficiency isolation and rapid identification of heterogeneous circulating tumor cells (CTCS) using dual-antibody-modified fluorescent-magnetic nanoparticles. ACS Appl. Mater. Interface. 2019;11:39586–39593. doi: 10.1021/acsami.9b14051. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, et al. Antifouling hydrogel-coated magnetic nanoparticles for selective isolation and recovery of circulating tumor cells. J. Mater. Chem. B. 2021;9:677–682. doi: 10.1039/d0tb02380a. [DOI] [PubMed] [Google Scholar]
- 33.Reduzzi C, et al. A novel circulating tumor cell subpopulation for treatment monitoring and molecular characterization in biliary tract cancer. Int J. Cancer. 2020;146:3495–3503. doi: 10.1002/ijc.32822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu. Rev. Pathol. 2018;13:395–412. doi: 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
- 35.Okabe T, et al. Mesenchymal characteristics and predictive biomarkers on circulating tumor cells for therapeutic strategy. Cancers. 2020;12:3588–3610. doi: 10.3390/cancers12123588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, et al. Using single-cell sequencing technology to detect circulating tumor cells in solid tumors. Mol. Cancer. 2021;20:104. doi: 10.1186/s12943-021-01392-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wulfing P, et al. HER2-positive circulating tumor cells indicate poor clinical outcome in stage I to III breast cancer patients. Clin. Cancer Res. 2006;12:1715–1720. doi: 10.1158/1078-0432.CCR-05-2087. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi N, et al. Prognostic value of HER2-positive circulating tumor cells in patients with metastatic breast cancer. Int J. Clin. Oncol. 2012;17:96–104. doi: 10.1007/s10147-011-0260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beije N, et al. Prognostic impact of HER2 and ER status of circulating tumor cells in metastatic breast cancer patients with a HER2-negative primary tumor. Neoplasia. 2016;18:647–653. doi: 10.1016/j.neo.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang CH, Chang CJ, Yeh KY, Chang PH, Huang JS. The prognostic value of HER2-positive circulating tumorcells in breast cancer patients: a systematic review and meta-analysis. Clin. Breast Cancer. 2017;17:341–349. doi: 10.1016/j.clbc.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Jaeger BAS, et al. The HER2 phenotype of circulating tumor cells in HER2-positive early breast cancer: a translationalresearch project of a prospective randomized phase III trial. PLoS One. 2017;12:e0173593. doi: 10.1371/journal.pone.0173593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brouwer A, et al. HER-2 status of circulating tumor cells in a metastatic breast cancer cohort: a comparative study oncharacterization techniques. PLoS One. 2019;14:e0220906. doi: 10.1371/journal.pone.0220906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen W, et al. Detection of HER2-positive circulating tumor cells using the liquidbiopsy system in breast cancer. Clin. Breast Cancer. 2019;19:e239–e246. doi: 10.1016/j.clbc.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Jacot W, et al. Actionability of HER2-amplified circulating tumor cells in HER2-negative metastatic breast cancer: the CirCeT-DM1 trial. Breast Cancer Res. 2019;21:121. doi: 10.1186/s13058-019-1215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nanou A, Zeune LL, Bidard FC, Pierga JY, Terstappen L. HER2 expression on tumor-derived extracellularvesicles and circulating tumor cells in metastatic breast cancer. Breast Cancer Res. 2020;22:86. doi: 10.1186/s13058-020-01323-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C, et al. Prognostic value of HER2 status on circulating tumor cells in advanced-stage breast cancer patients withHER2-negative tumors. Breast Cancer Res. Treat. 2020;181:679–689. doi: 10.1007/s10549-020-05662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishima Y, et al. Detection of HER2 amplification in circulating tumor cells of HER2-negative gastric cancer patients. Target Oncol. 2017;12:341–351. doi: 10.1007/s11523-017-0493-6. [DOI] [PubMed] [Google Scholar]
- 48.Gradilone A, et al. Circulating tumor cells (CTCs) in metastatic breast cancer (MBC): prognosis, drug resistance andphenotypic characterization. Ann. Oncol. 2011;22:86–92. doi: 10.1093/annonc/mdq323. [DOI] [PubMed] [Google Scholar]
- 49.Somlo G, et al. Multiple biomarker expression on circulating tumor cells in comparison to tumor tissues from primary andmetastatic sites in patients with locally advanced/inflammatory, and stage IV breast cancer, using a novel detection technology. Breast Cancer Res. Treat. 2011;128:155–163. doi: 10.1007/s10549-011-1508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forsare C, et al. Evolution of estrogen receptor status from primary tumors to metastasis and serially collected circulating tumor cells. Int. J. Mol. Sci. 2020;21:2885–2897. doi: 10.3390/ijms21082885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Todenhofer T, et al. Preliminary experience on the use of the adnatest (R) system for detection of circulating tumor cells inprostate cancer patients. Anticancer Res. 2012;32:3507–3513. [PubMed] [Google Scholar]
- 52.Friedlander TW, et al. Detection and characterization of invasive circulating tumor cells derived from men with metastaticcastration-resistant prostate cancer. Int. J. Cancer. 2014;134:2284–2293. doi: 10.1002/ijc.28561. [DOI] [PubMed] [Google Scholar]
- 53.Yin C, et al. Molecular profiling of pooled circulating tumor cells from prostate cancer patients using a dual-antibodyfunctionalizedmicrofluidic device. Anal. Chem. 2018;90:3744–3751. doi: 10.1021/acs.analchem.7b03536. [DOI] [PubMed] [Google Scholar]
- 54.Chen L, et al. Combined use of EpCAM and FRalpha enables the high-efficiency capture of circulating tumor cells in nonsmallcell lung cancer. Sci. Rep. 2018;8:1188. doi: 10.1038/s41598-018-19391-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, et al. Folate receptor-positive circulating tumor cells as a predictive biomarker for the efficacy of first-linepemetrexed-based chemotherapy in patients with non-squamous non-small cell lung cancer. Ann. Transl. Med. 2020;8:631. doi: 10.21037/atm-19-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei S, et al. Effect of vein-first vs artery-first surgical technique on circulating tumor cells and survival in patients with nonsmallcell lung cancer: a randomized clinical trial and registry-based propensity score matching analysis. JAMA Surg. 2019;154:e190972. doi: 10.1001/jamasurg.2019.0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao W, et al. Using detection of survivin-expressing circulating tumor cells in peripheral blood to predict tumor recurrencefollowing curative resection of gastric cancer. J. Surg. Oncol. 2011;103:110–115. doi: 10.1002/jso.21777. [DOI] [PubMed] [Google Scholar]
- 58.Krishnamurthy S, et al. Discordance in HER2 gene amplification in circulating and disseminated tumor cells in patients withoperable breast cancer. Cancer Med. 2013;2:226–233. doi: 10.1002/cam4.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bidard FC, et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastaticuveal melanoma. Int. J. Cancer. 2014;134:1207–1213. doi: 10.1002/ijc.28436. [DOI] [PubMed] [Google Scholar]
- 60.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lucci A, et al. Circulating tumor cells and early relapse in node-positive melanoma. Clin. Cancer Res. 2020;26:1886–1895. doi: 10.1158/1078-0432.CCR-19-2670. [DOI] [PubMed] [Google Scholar]
- 62.Hoshimoto S, et al. Assessment of prognostic circulating tumor cells in a phase III trial of adjuvant immunotherapy aftercomplete resection of stage IV melanoma. Ann. Surg. 2012;255:357–362. doi: 10.1097/SLA.0b013e3182380f56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoshimoto S, et al. Association between circulating tumor cells and prognosis in patients with stage III melanoma withsentinel lymph node metastasis in a phase III international multicenter trial. J. Clin. Oncol. 2012;30:3819–3826. doi: 10.1200/JCO.2011.40.0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiniwa Y, et al. Usefulness of monitoring circulating tumor cells as a therapeutic biomarker in melanoma with BRAFmutation. BMC Cancer. 2021;21:287. doi: 10.1186/s12885-021-08016-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hall CS, et al. Circulating tumor cells in stage IV melanoma patients. J. Am. Coll. Surg. 2018;227:116–124. doi: 10.1016/j.jamcollsurg.2018.04.026. [DOI] [PubMed] [Google Scholar]
- 66.Lin SY, et al. Prospective molecular profiling of circulating tumor cells from patients with melanoma receivingcombinatorial immunotherapy. Clin. Chem. 2020;66:169–177. doi: 10.1373/clinchem.2019.307140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fabbri F, et al. Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRASmutation status in pure CTCs. Cancer Lett. 2013;335:225–231. doi: 10.1016/j.canlet.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 69.Buim ME, et al. Detection of KRAS mutations in circulating tumor cells from patients with metastatic colorectal cancer. Cancer Biol. Ther. 2015;16:1289–1295. doi: 10.1080/15384047.2015.1070991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalikaki A, et al. KRAS genotypic changes of circulating tumor cells during treatment of patients with metastatic colorectalcancer. PloS One. 2014;9:e104902. doi: 10.1371/journal.pone.0104902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kondo Y, et al. KRAS mutation analysis of single circulating tumor cells from patients with metastatic colorectal cancer. BMC Cancer. 2017;17:311. doi: 10.1186/s12885-017-3305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kulemann B, et al. Pancreatic cancer: circulating tumor cells and primary tumors show heterogeneous KRAS mutations. Sci. Rep. 2017;7:4510. doi: 10.1038/s41598-017-04601-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lohr JG, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Faugeroux V, et al. An accessible and unique insight into metastasis mutational content through whole-exome sequencing ofcirculating tumor cells in metastatic prostate cancer. Eur. Urol. Oncol. 2020;3:498–508. doi: 10.1016/j.euo.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 75.Deng G, et al. Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. BMC Cancer. 2014;14:456–467. doi: 10.1186/1471-2407-14-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fernandez V, et al. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breastcancer patients. Breast Cancer Res. 2014;16:445. doi: 10.1186/s13058-014-0445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kelley RK, et al. Circulating tumor cells in hepatocellular carcinoma: a pilot study of detection, enumeration, and nextgenerationsequencing in cases and controls. BMC Cancer. 2015;15:206. doi: 10.1186/s12885-015-1195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ni X, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc. Natl Acad. Sci. USA. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carter L, et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients withchemosensitive and chemorefractory small-cell lung cancer. Nat. Med. 2017;23:114–119. doi: 10.1038/nm.4239. [DOI] [PubMed] [Google Scholar]
- 80.Paoletti C, et al. Comprehensive mutation and copy number profiling in archived circulating breast cancer tumor cellsdocuments heterogeneous resistance mechanisms. Cancer Res. 2018;78:1110–1122. doi: 10.1158/0008-5472.CAN-17-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ortolan E, et al. Blood-based genomics of triple-negative breast cancer progression in patients treated with neoadjuvantchemotherapy. ESMO Open. 2021;6:100086. doi: 10.1016/j.esmoop.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lambros MB, et al. Single-cell analyses of prostate cancer liquid biopsies acquired by apheresis. Clin. Cancer Res.: . J. Am. Assoc. Cancer Res. 2018;24:5635–5644. doi: 10.1158/1078-0432.CCR-18-0862. [DOI] [PubMed] [Google Scholar]
- 83.Gupta S, et al. Whole genomic copy number alterations in circulating tumor cells from men with abiraterone orenzalutamide-resistant metastatic castration-resistant prostate cancer. Clin. Cancer Res.: . J. Am. Assoc. Cancer Res. 2017;23:1346–1357. doi: 10.1158/1078-0432.CCR-16-1211. [DOI] [PubMed] [Google Scholar]
- 84.Malihi PD, et al. Single-cell circulating tumor cell analysis reveals genomic instability as a distinctive feature of aggressiveprostate cancer. Clin. Cancer Res.: . J. Am. Assoc. Cancer Res. 2020;26:4143–4153. doi: 10.1158/1078-0432.CCR-19-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pailler E, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-celllung cancer. J. Clin. Oncol. 2013;31:2273–2281. doi: 10.1200/JCO.2012.44.5932. [DOI] [PubMed] [Google Scholar]
- 86.Mayer JA, et al. FISH-based determination of HER2 status in circulating tumor cells isolated with the microfluidic CEEplatform. Cancer Genet. 2011;204:589–595. doi: 10.1016/j.cancergen.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 87.Frithiof H, Aaltonen K, Ryden L. A FISH-based method for assessment of HER-2 amplification status in breast cancercirculating tumor cells following CellSearch isolation. OncoTargets Ther. 2016;9:7095–7103. doi: 10.2147/OTT.S118502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gkountela S, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176:98–112.e114. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dong Y, Wang Z, Shi Q. Liquid biopsy based single-cell transcriptome profiling characterizes heterogeneity ofdisseminated tumor cells from lung adenocarcinoma. Proteomics. 2020;20:e1900224. doi: 10.1002/pmic.201900224. [DOI] [PubMed] [Google Scholar]
- 90.Magbanua MJM, et al. Expanded genomic profiling of circulating tumor cells in metastatic breast cancer patients to assessbiomarker status and biology over time (CALGB 40502 and CALGB 40503, Alliance) Clin. Cancer Res.: . J. Am. Assoc. Cancer Res. 2018;24:1486–1499. doi: 10.1158/1078-0432.CCR-17-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng Y-H, et al. Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells. Nat. Commun. 2019;10:2163–2173. doi: 10.1038/s41467-019-10122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miyamoto D, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barbazan J, et al. Molecular characterization of circulating tumor cells in human metastatic colorectal cancer. PloS One. 2012;7:e40476. doi: 10.1371/journal.pone.0040476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kallergi G, et al. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastaticbreast cancer patients. Breast Cancer Res. 2011;13:R59. doi: 10.1186/bcr2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Balasubramanian P, et al. Multiparameter analysis, including EMT markers, on negatively enriched blood samples frompatients with squamous cell carcinoma of the head and neck. PLoS One. 2012;7:e42048. doi: 10.1371/journal.pone.0042048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Theodoropoulos PA, et al. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients withbreast cancer. Cancer Lett. 2010;288:99–106. doi: 10.1016/j.canlet.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 97.Armstrong AJ, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelialand mesenchymal markers. Mol. Cancer Res. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu X, et al. Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source ofmetastasis. Sci. Adv. 2019;5:eaav4275. doi: 10.1126/sciadv.aav4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thiery JP, Lim CT. Tumor dissemination: an EMT affair. Cancer Cell. 2013;23:272–273. doi: 10.1016/j.ccr.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 100.Pecot CV, et al. A novel platform for detection of CK+ and CK- CTCs. Cancer Discov. 2011;1:580–586. doi: 10.1158/2159-8290.CD-11-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun YF, et al. Circulating tumor cells from different vascular sites exhibit spatial heterogeneity in epithelial andmesenchymal composition and distinct clinical significance in hepatocellular carcinoma. Clin. Cancer Res.: . J. Am. Assoc. Cancer Res. 2018;24:547–559. doi: 10.1158/1078-0432.CCR-17-1063. [DOI] [PubMed] [Google Scholar]
- 102.Xu L, et al. The novel association of circulating tumor cells and circulating megakaryocytes with prostate cancer prognosis. Clin. Cancer Res. 2017;23:5112–5122. doi: 10.1158/1078-0432.CCR-16-3081. [DOI] [PubMed] [Google Scholar]
- 103.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications forcancer. Nat. Rev. Mol. Cell Bio. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 104.Guan Y, et al. Identification of key genes and functions of circulating tumor cells in multiple cancers through bioinformaticanalysis. BMC Med. Genom. 2020;13:140. doi: 10.1186/s12920-020-00795-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tashireva LA, et al. Heterogeneous manifestations of epithelial-mesenchymal plasticity of circulating tumor cells in breast cancer patients. Int. J. Mol. Sci. 2021;22:2504–2521. doi: 10.3390/ijms22052504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xin Y, Li K, Yang M, Tan Y. Fluid shear stress induces EMT of circulating tumor cells via jnk signaling in favor of their survival during hematogenous dissemination. Int. J. Mol. Sci. 2020;21:8115–8130. doi: 10.3390/ijms21218115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Horimoto Y, et al. Analysis of circulating tumour cell and the epithelial mesenchymal transition (EMT) status duringeribulin-based treatment in 22 patients with metastatic breast cancer: a pilot study. J. Transl. Med. 2018;16:287. doi: 10.1186/s12967-018-1663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guan XW, et al. The prognostic and therapeutic implications of circulating tumor cell phenotype detection based on epithelial-mesenchymal transition markers in the first-line chemotherapy of HER2-negative metastatic breast cancer. CancerCommun. 2019;39:1–10. doi: 10.1186/s40880-018-0346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lindsay CR, et al. A prospective examination of circulating tumor cell profiles in non-small-cell lung cancer molecularsubgroups. Ann. Oncol. 2017;28:1523–1531. doi: 10.1093/annonc/mdx156. [DOI] [PubMed] [Google Scholar]
- 110.Satelli A, et al. Epithelial-mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin. Cancer Res. 2015;21:899–906. doi: 10.1158/1078-0432.CCR-14-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brechbuhl HM, et al. Analysis of circulating breast cancer cell heterogeneity and interactions with peripheral bloodmononuclear cells. Mol. Carcinog. 2020;59:1129–1139. doi: 10.1002/mc.23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Miklikova S, et al. Inflammation-based scores increase the prognostic value of circulating tumor cells in primary breast cancer. Cancers. 2020;12:1134–1148. doi: 10.3390/cancers12051134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu Y, et al. SUMOylation represses Nanog expression via modulating transcription factors Oct4 and Sox2. PloS One. 2012;7:e39606. doi: 10.1371/journal.pone.0039606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang R, et al. Co-expression of stem cell and epithelial mesenchymal transition markers in circulating tumor cells ofbladder cancer patients. OncoTargets Ther. 2020;13:10739–10748. doi: 10.2147/OTT.S259240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao P, et al. Establishment and characterization of a CTC cell line from peripheral blood of breast cancer patient. J. Cancer. 2019;10:6095–6104. doi: 10.7150/jca.33157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wan S, et al. New labyrinth microfluidic device detects circulating tumor cells expressing cancer stem cell marker andcirculating tumor microemboli in hepatocellular carcinoma. Sci. Rep. 2019;9:18575. doi: 10.1038/s41598-019-54960-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu T, et al. Circulating glioma cells exhibit stem cell-like properties. Cancer Res. 2018;78:6632–6642. doi: 10.1158/0008-5472.CAN-18-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yin J, et al. Circulating tumor cells enriched by the depletion of leukocytes with bi-antibodies in non-small cell lung cancer:potential clinical application. PloS One. 2015;10:e0137076. doi: 10.1371/journal.pone.0137076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jin J, Tang K, Xin Y, Zhang T, Tan Y. Hemodynamic shear flow regulates biophysical characteristics and functionsof circulating breast tumor cells reminiscent of brain metastasis. Soft Matter. 2018;14:9528–9533. doi: 10.1039/c8sm01781f. [DOI] [PubMed] [Google Scholar]
- 120.Choi H, et al. Hydrodynamic shear stress promotes epithelial-mesenchymal transition by downregulating ERK and GSK3?activities. Breast Cancer Res. 2019;21:6. doi: 10.1186/s13058-018-1071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Follain G, et al. Fluids and their mechanics in tumour transit: shaping metastasis. Nat. Rev. Cancer. 2020;20:107–124. doi: 10.1038/s41568-019-0221-x. [DOI] [PubMed] [Google Scholar]
- 122.Strilic B, Offermanns S. Intravascular survival and extravasation of tumor cells. Cancer cell. 2017;32:282–293. doi: 10.1016/j.ccell.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 123.Rejniak KA. Circulating tumor cells: when a solid tumor meets a fluid microenvironment. Adv. Exp. Med. Biol. 2016;936:93–106. doi: 10.1007/978-3-319-42023-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Garrido-Navas C, et al. Cooperative and escaping mechanisms between circulating tumor cells and blood constituents. Cells. 2019;8:1382–1391. doi: 10.3390/cells8111382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shaul ME, Fridlender ZG. Tumour-associated neutrophils in patients with cancer. Nat. Rev. Clin. Oncol. 2019;16:601–620. doi: 10.1038/s41571-019-0222-4. [DOI] [PubMed] [Google Scholar]
- 126.Aliustaoglu M, et al. The effect of peripheral blood values on prognosis of patients with locally advanced gastric cancerbefore treatment. Med. Oncol. 2010;27:1060–1065. doi: 10.1007/s12032-009-9335-4. [DOI] [PubMed] [Google Scholar]
- 127.Hu S, et al. The preoperative peripheral blood monocyte count is associated with liver metastasis and overall survival incolorectal cancer patients. PloS One. 2016;11:e0157486. doi: 10.1371/journal.pone.0157486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang Y, et al. Circulating neutrophils predict poor survival for HCC and promote HCC progression through p53 andSTAT3 signaling pathway. J. Cancer. 2020;11:3736–3744. doi: 10.7150/jca.42953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stott SL, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl Acad. Sci. USA. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Szczerba BM, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566:553–557. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- 131.Hattar K, et al. Interactions between neutrophils and non-small cell lung cancer cells: enhancement of tumor proliferationand inflammatory mediator synthesis. Cancer Immunol., Immunother. 2014;63:1297–1306. doi: 10.1007/s00262-014-1606-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Spicer JD, et al. Neutrophils promote liver metastasis via Mac-1-mediated interactions with circulating tumor cells. Cancer Res. 2012;72:3919–3927. doi: 10.1158/0008-5472.CAN-11-2393. [DOI] [PubMed] [Google Scholar]
- 133.Strell C, Lang K, Niggemann B, Zaenker KS, Entschladen F. Surface molecules regulating rolling and adhesion toendothelium of neutrophil granulocytes and MDA-MB-468 breast carcinoma cells and their interaction. Cell. Mol. life Sci. 2007;64:3306–3316. doi: 10.1007/s00018-007-7402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huh SJ, Liang S, Sharma A, Dong C, Robertson GP. Transiently entrapped circulating tumor cells interact withneutrophils to facilitate lung metastasis development. Cancer Res. 2010;70:6071–6082. doi: 10.1158/0008-5472.CAN-09-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rowson-Hodel AR, et al. Membrane Mucin Muc4 promotes blood cell association with tumor cells and mediates efficientmetastasis in a mouse model of breast cancer. Oncogene. 2018;37:197–207. doi: 10.1038/onc.2017.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen MB, et al. Inflamed neutrophils sequestered at entrapped tumor cells via chemotactic confinement promote tumorcell extravasation. Proc. Natl Acad. Sci. USA. 2018;115:7022–7027. doi: 10.1073/pnas.1715932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Masucci MT, Minopoli M, Del Vecchio S, Carriero MV. The emerging role of neutrophil extracellular traps (NETs)in tumor progression and metastasis. Front. Immunol. 2020;11:1749. doi: 10.3389/fimmu.2020.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cools-Lartigue J, et al. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. Investig. 2013;123:3446–3458. doi: 10.1172/JCI67484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Najmeh S, et al. Neutrophil extracellular traps sequester circulating tumor cells via ?1-integrin mediated interactions. Int. J. Cancer. 2017;140:2321–2330. doi: 10.1002/ijc.30635. [DOI] [PubMed] [Google Scholar]
- 140.Tohme S, et al. Neutrophil extracellular traps promote the development and progression of liver metastases after surgicalstress. Cancer Res. 2016;76:1367–1380. doi: 10.1158/0008-5472.CAN-15-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Albrengues J, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science. 2018;361:1353–1367. doi: 10.1126/science.aao4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xiao Y, et al. Cathepsin C promotes breast cancer lung metastasis by modulating neutrophil infiltration and neutrophilextracellular trap formation. Cancer Cell. 2021;39:423–437.e427. doi: 10.1016/j.ccell.2020.12.012. [DOI] [PubMed] [Google Scholar]
- 143.Yang L, et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature. 2020;583:133–138. doi: 10.1038/s41586-020-2394-6. [DOI] [PubMed] [Google Scholar]
- 144.Zhang J, et al. Circulating tumor-associated neutrophils (cTAN) contribute to circulating tumor cell survival by suppressingperipheral leukocyte activation. Tumour Biol.: J. Int. Soc. Oncodev. Biol. Med. 2016;37:5397–5404. doi: 10.1007/s13277-015-4349-3. [DOI] [PubMed] [Google Scholar]
- 145.Spiegel A, et al. Neutrophils suppress intraluminal nk cell-mediated tumor cell clearance and enhance extravasation ofdisseminated carcinoma cells. Cancer Discov. 2016;6:630–649. doi: 10.1158/2159-8290.CD-15-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kumagai Y, et al. Surgical stress increases circulating low-density neutrophils which may promote tumor recurrence. J. Surg. Res. 2020;246:52–61. doi: 10.1016/j.jss.2019.08.022. [DOI] [PubMed] [Google Scholar]
- 147.Coffelt SB, et al. IL-17-producing ?? T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 2015;522:345–348. doi: 10.1038/nature14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Doak GR, Schwertfeger KL, Wood DK. Distant relations: macrophage functions in the metastatic niche. TrendsCancer. 2018;4:445–459. doi: 10.1016/j.trecan.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hamilton G, Rath B, Klameth L, Hochmair MJ. Small cell lung cancer: recruitment of macrophages by circulatingtumor cells. Oncoimmunology. 2016;5:e1093277. doi: 10.1080/2162402X.2015.1093277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hamilton G, Rath B. Circulating tumor cell interactions with macrophages: implications for biology and treatment. Transl. Lung Cancer Res. 2017;6:418–430. doi: 10.21037/tlcr.2017.07.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wei C, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulatingtumor cell-mediated colorectal cancer metastasis. Mol. Cancer. 2019;18:64. doi: 10.1186/s12943-019-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Osmulski PA, et al. Contacts with macrophages promote an aggressive nanomechanical phenotype of circulating tumorcells in prostate cancer. Cancer Res. 2021;81:4110–4123. doi: 10.1158/0008-5472.CAN-20-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shabo I, et al. Macrophage traits in cancer cells are induced by macrophage-cancer cell fusion and cannot be explained bycellular interaction. BMC Cancer. 2015;15:922. doi: 10.1186/s12885-015-1935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Clawson GA, et al. Macrophage-tumor cell fusions from peripheral blood of melanoma patients. PloS One. 2015;10:e0134320. doi: 10.1371/journal.pone.0134320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Clawson GA, et al. “Stealth dissemination” of macrophage-tumor cell fusions cultured from blood of patients withpancreatic ductal adenocarcinoma. PloS One. 2017;12:e0184451. doi: 10.1371/journal.pone.0184451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Manjunath Y, et al. Tumor-cell-macrophage fusion cells as liquid biomarkers and tumor enhancers in cancer. Int. J. Mol. Sci. 2020;21:1872. doi: 10.3390/ijms21051872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gast CE, et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage andsurvival. Sci. Adv. 2018;4:eaat7828. doi: 10.1126/sciadv.aat7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Manjunath Y, et al. Circulating giant tumor-macrophage fusion cells are independent prognosticators in patients withNSCLC. J. Thorac. Oncol.: . Publ. Int. Assoc. Study Lung Cancer. 2020;15:1460–1471. doi: 10.1016/j.jtho.2020.04.034. [DOI] [PubMed] [Google Scholar]
- 159.Schlesinger M. Role of platelets and platelet receptors in cancer metastasis. J. Hematol. Oncol. 2018;11:125. doi: 10.1186/s13045-018-0669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gaertner F, Massberg S. Patrolling the vascular borders: platelets in immunity to infection and cancer. Nat. Rev. Immunol. 2019;19:747–760. doi: 10.1038/s41577-019-0202-z. [DOI] [PubMed] [Google Scholar]
- 161.Thomas GM, et al. Tissue factor expressed by circulating cancer cell-derived microparticles drastically increases theincidence of deep vein thrombosis in mice. J. thrombosis Haemost. 2015;13:1310–1319. doi: 10.1111/jth.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stark K, et al. Distinct pathogenesis of pancreatic cancer microvesicle-associated venous thrombosis identifies newantithrombotic targets in vivo. Arteriosclerosis, thrombosis, Vasc. Biol. 2018;38:772–786. doi: 10.1161/ATVBAHA.117.310262. [DOI] [PubMed] [Google Scholar]
- 163.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelialmesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20:576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Guo Y, Cui W, Pei Y, Xu D. Platelets promote invasion and induce epithelial to mesenchymal transition in ovariancancer cells by TGF-? signaling pathway. Gynecol. Oncol. 2019;153:639–650. doi: 10.1016/j.ygyno.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 165.Xiong G, et al. Hsp47 promotes cancer metastasis by enhancing collagen-dependent cancer cell-platelet interaction. Proc. Natl Acad. Sci. USA. 2020;117:3748–3758. doi: 10.1073/pnas.1911951117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Egan K, Cooke N, Kenny D. Living in shear: platelets protect cancer cells from shear induced damage. Clin. Exp. Metastasis. 2014;31:697–704. doi: 10.1007/s10585-014-9660-7. [DOI] [PubMed] [Google Scholar]
- 167.Haemmerle M, et al. Platelets reduce anoikis and promote metastasis by activating YAP1 signaling. Nat. Commun. 2017;8:310. doi: 10.1038/s41467-017-00411-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nieswandt B, Hafner M, Echtenacher B, Männel DN. Lysis of tumor cells by natural killer cells in mice is impededby platelets. Cancer Res. 1999;59:1295–1300. [PubMed] [Google Scholar]
- 169.Placke T, et al. Platelet-derived MHC class I confers a pseudonormal phenotype to cancer cells that subverts the antitumorreactivity of natural killer immune cells. Cancer Res. 2012;72:440–448. doi: 10.1158/0008-5472.CAN-11-1872. [DOI] [PubMed] [Google Scholar]
- 170.Kopp HG, Placke T, Salih HR. Platelet-derived transforming growth factor-beta down-regulates NKG2D therebyinhibiting natural killer cell antitumor reactivity. Cancer Res. 2009;69:7775–7783. doi: 10.1158/0008-5472.CAN-09-2123. [DOI] [PubMed] [Google Scholar]
- 171.Maurer S, et al. Platelet-mediated shedding of NKG2D ligands impairs NK cell immune-surveillance of tumor cells. Oncoimmunology. 2018;7:e1364827. doi: 10.1080/2162402X.2017.1364827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Placke T, Salih HR, Kopp HG. GITR ligand provided by thrombopoietic cells inhibits NK cell antitumor activity. J. Immunol. 2012;189:154–160. doi: 10.4049/jimmunol.1103194. [DOI] [PubMed] [Google Scholar]
- 173.Ren J, et al. Platelet TLR4-ERK5 axis facilitates net-mediated capturing of circulating tumor cells and distant metastasisafter surgical stress. Cancer Res. 2021;81:2373–2385. doi: 10.1158/0008-5472.CAN-20-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rachidi S, et al. Platelets subvert T cell immunity against cancer via GARP-TGFβ axis. Sci. Immunol. 2017;2:7911–7937. doi: 10.1126/sciimmunol.aai7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xiang B, et al. Platelets protect from septic shock by inhibiting macrophage-dependent inflammation via thecyclooxygenase 1 signalling pathway. Nat. Commun. 2013;4:2657. doi: 10.1038/ncomms3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bendas G, Borsig L. Cancer cell adhesion and metastasis: selectins, integrins, and the inhibitory potential of heparins.Int. J. Cell Biol. 2012;2012:676731. doi: 10.1155/2012/676731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lonsdorf AS, et al. Engagement of αIIbβ3 (GPIIb/IIIa) with ανβ3 integrin mediates interaction of melanoma cells with platelets: a connection to hematogenous metastasis. J. Biol. Chem. 2012;287:2168–2178. doi: 10.1074/jbc.M111.269811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Coupland LA, Chong BH, Parish CR. Platelets and P-selectin control tumor cell metastasis in an organ-specificmanner and independently of NK cells. Cancer Res. 2012;72:4662–4671. doi: 10.1158/0008-5472.CAN-11-4010. [DOI] [PubMed] [Google Scholar]
- 179.Schumacher D, Strilic B, Sivaraj KK, Wettschureck N, Offermanns S. Platelet-derived nucleotides promote tumorcelltransendothelial migration and metastasis via P2Y2 receptor. Cancer Cell. 2013;24:130–137. doi: 10.1016/j.ccr.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 180.Mammadova-Bach E, et al. Platelet integrin ?6?1 controls lung metastasis through direct binding to cancer cell-derivedADAM9. JCI Insight. 2016;1:e88245. doi: 10.1172/jci.insight.88245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ward Y, et al. Platelets promote metastasis via binding tumor CD97 leading to bidirectional signaling that coordinatestransendothelial migration. Cell Rep. 2018;23:808–822. doi: 10.1016/j.celrep.2018.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mammadova-Bach E, et al. Platelet glycoprotein VI promotes metastasis through interaction with cancer cell-derivedgalectin-3. Blood. 2020;135:1146–1160. doi: 10.1182/blood.2019002649. [DOI] [PubMed] [Google Scholar]
- 183.Xu Y, et al. Blockade of platelets using tumor-specific NO-releasing nanoparticles prevents tumor metastasis and reversestumor immunosuppression. ACS Nano. 2020;14:9780–9795. doi: 10.1021/acsnano.0c01687. [DOI] [PubMed] [Google Scholar]
- 184.Cassetta L, et al. Differential expansion of circulating human MDSC subsets in patients with cancer, infection and inflammation. J. Immunother. Cancer. 2020;8:1223–1235. doi: 10.1136/jitc-2020-001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu Q, Liao Q, Zhao Y. Myeloid-derived suppressor cells (MDSC) facilitate distant metastasis of malignancies byshielding circulating tumor cells (CTC) from immune surveillance. Med. Hypotheses. 2016;87:34–39. doi: 10.1016/j.mehy.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 186.Jiménez-Cortegana C, et al. Circulating myeloid-derived suppressor cells and regulatory T cells as immunological biomarkers in refractory/relapsed diffuse large B-cell lymphoma: translational results from the R2-GDP-GOTEL trial. J. Immunother. Cancer. 2021;9:2323–2334. doi: 10.1136/jitc-2020-002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sprouse ML, et al. PMN-MDSCs enhance CTC metastatic properties through reciprocal interactions via ROS/Notch/nodal signaling. Int. J. Mol. Sci. 2019;20:1916–1935. doi: 10.3390/ijms20081916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019;18:99–115. doi: 10.1038/s41573-018-0004-1. [DOI] [PubMed] [Google Scholar]
- 189.Gaggioli C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading andfollowing cells. Nat. Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 190.Duda DG, et al. Malignant cells facilitate lung metastasis by bringing their own soil. Proc. Natl Acad. Sci. USA. 2010;107:21677–21682. doi: 10.1073/pnas.1016234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ortiz-Otero N, et al. Cancer associated fibroblasts confer shear resistance to circulating tumor cells during prostate cancermetastatic progression. Oncotarget. 2020;11:1037–1050. doi: 10.18632/oncotarget.27510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ao Z, et al. Identification of cancer-associated fibroblasts in circulating blood from patients with metastatic breast cancer. Cancer Res. 2015;75:4681–4687. doi: 10.1158/0008-5472.CAN-15-1633. [DOI] [PubMed] [Google Scholar]
- 193.Jiang, R. et al. Rapid isolation of circulating cancer associated fibroblasts by acoustic microstreaming for assessing metastatic propensity of breast cancer patients. Lab. Chip 969-981 (2020). [DOI] [PubMed]
- 194.Shen Z, Wu A, Chen X. Current detection technologies for circulating tumor cells. Chem. Soc. Rev. 2017;46:2038–2056. doi: 10.1039/c6cs00803h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vona G, et al. Isolation by size of epithelial tumor cells-a new method for the immunomorphological and molecularcharacterization of circulating tumor cells. Am. J. Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Harouaka RA, et al. Flexible micro spring array device for high-throughput enrichment of viable circulating tumor cells. Clin. Chem. 2014;60:323–333. doi: 10.1373/clinchem.2013.206805. [DOI] [PubMed] [Google Scholar]
- 197.Rosenberg R, et al. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumorcells in blood. Cytometry. 2002;49:150–158. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- 198.Gupta V, et al. ApoStream(), a new dielectrophoretic device for antibody independent isolation and recovery of viablecancer cells from blood. Biomicrofluidics. 2012;6:24133. doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sajay BN, et al. Microfluidic platform for negative enrichment of circulating tumor cells. Biomed. Microdevices. 2014;16:537–548. doi: 10.1007/s10544-014-9856-2. [DOI] [PubMed] [Google Scholar]
- 200.Allard WJ, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects orpatients with nonmalignant diseases. Clin. Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 201.Talasaz AH, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood usinga magnetic sweeper device. Proc. Natl Acad. Sci. USA. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wu S, et al. Enrichment and enumeration of circulating tumor cells by efficient depletion of leukocyte fractions. Clin. Chem. Lab. Med. 2015;53:337. doi: 10.1515/cclm-2015-5000. [DOI] [PubMed] [Google Scholar]
- 203.Nasiri R, et al. Microfluidic-based approaches in targeted cell/particle separation based on physical properties: fundamentalsand applications. Small. 2020;16:e2000171. doi: 10.1002/smll.202000171. [DOI] [PubMed] [Google Scholar]
- 204.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Park MH, et al. Enhanced isolation and release of circulating tumor cells using nanoparticle binding and ligand exchangein a microfluidic chip. J. Am. Chem. Soc. 2017;139:2741–2749. doi: 10.1021/jacs.6b12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fachin F, et al. Monolithic chip for high-throughput blood cell depletion to sort rare circulating tumor cells. Sci. Rep. 2017;7:10936–10946. doi: 10.1038/s41598-017-11119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang X, Zhu Z, Xiang N, Long F, Ni Z. Automated microfluidic instrument for label-free and high-throughput cellseparation. Anal. Chem. 2018;90:4212–4220. doi: 10.1021/acs.analchem.8b00539. [DOI] [PubMed] [Google Scholar]
- 208.Jia Z, et al. Single-cell genetic analysis of lung tumor cells based on self-driving micro-cavity array chip. Talanta. 2021;226:122172. doi: 10.1016/j.talanta.2021.122172. [DOI] [PubMed] [Google Scholar]
- 209.Wang J, et al. A fully automated and integrated microfluidic system for efficient ctc detection and its application inhepatocellular carcinoma screening and prognosis. ACS Appl. Mater. Interfaces. 2021;13:30174–30186. doi: 10.1021/acsami.1c06337. [DOI] [PubMed] [Google Scholar]
- 210.Lee J, Kwak B. Simultaneous on-chip isolation and characterization of circulating tumor cell sub-populations. Biosens. Bioelectron. 2020;168:112564. doi: 10.1016/j.bios.2020.112564. [DOI] [PubMed] [Google Scholar]
- 211.Lee AC, et al. OPENchip: an on-chip in situ molecular profiling platform for gene expression analysis and oncogenicmutation detection in single circulating tumour cells. Lab Chip. 2020;20:912–922. doi: 10.1039/c9lc01248f. [DOI] [PubMed] [Google Scholar]
- 212.Zhang Y, Li M, Gao X, Chen Y, Liu T. Nanotechnology in cancer diagnosis: progress, challenges and opportunities. J. Hematol. Oncol. 2019;12:137. doi: 10.1186/s13045-019-0833-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jia F, et al. Novel peptide-based magnetic nanoparticle for mesenchymal circulating tumor cells detection. Anal. Chem. 2021;93:5670–5675. doi: 10.1021/acs.analchem.1c00577. [DOI] [PubMed] [Google Scholar]
- 214.Ding P, et al. Tannic acid (TA)-functionalized magnetic nanoparticles for EpCAM-independent circulating tumor cell(CTC) isolation from patients with different cancers. ACS Appl. Mater. Interfaces. 2021;13:3694–3700. doi: 10.1021/acsami.0c20916. [DOI] [PubMed] [Google Scholar]
- 215.Li F, et al. Nondestructive capture, release, and detection of circulating tumor cells with cystamine-mediated folic aciddecorated magnetic nanospheres. J. Mater. Chem. B. 2020;8:9971–9979. doi: 10.1039/d0tb01091j. [DOI] [PubMed] [Google Scholar]
- 216.Vajhadin F, et al. MXene-based cytosensor for the detection of HER2-positive cancer cells using CoFe2O4@Ag magneticnanohybrids conjugated to the HB5 aptamer. Biosens. Bioelectron. 2021;195:113626. doi: 10.1016/j.bios.2021.113626. [DOI] [PubMed] [Google Scholar]
- 217.Wilson RE, et al. Immunomagnetic capture and multiplexed surface marker detection of circulating tumor cells withmagnetic multicolor surface-enhanced Raman scattering nanotags. ACS Appl. Mater. Interfaces. 2020;12:47220–47232. doi: 10.1021/acsami.0c12395. [DOI] [PubMed] [Google Scholar]
- 218.Liu S, Luo J, Jiang X, Li X, Yang M. Gold nanoparticle-modified black phosphorus nanosheets with improvedstability for detection of circulating tumor cells. Mikrochim. Acta. 2020;187:397. doi: 10.1007/s00604-020-04367-8. [DOI] [PubMed] [Google Scholar]
- 219.Dou B, Xu L, Jiang B, Yuan R, Xiang Y. Aptamer-functionalized and gold nanoparticle array-decorated magneticgraphene nanosheets enable multiplexed and sensitive electrochemical detection of rare circulating tumor cells in whole blood. Anal. Chem. 2019;91:10792–10799. doi: 10.1021/acs.analchem.9b02403. [DOI] [PubMed] [Google Scholar]
- 220.Sheng W, Chen T, Tan W, Fan ZH. Multivalent DNA nanospheres for enhanced capture of cancer cells inmicrofluidic devices. ACS Nano. 2013;7:7067–7076. doi: 10.1021/nn4023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhang Y, et al. Gold nanorods-mediated efficient synergistic immunotherapy for detection and inhibition of postoperativetumor recurrence. Acta Pharm. Sin. B. 2021;11:1978–1992. doi: 10.1016/j.apsb.2021.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhang J, et al. Sensitive signal amplifying a diagnostic biochip based on a biomimetic periodic nanostructure for detectingcancer exosomes. ACS Appl. Mater. Interfaces. 2020;12:33473–33482. doi: 10.1021/acsami.0c06785. [DOI] [PubMed] [Google Scholar]
- 223.Cui H, et al. Rapid and efficient isolation and detection of circulating tumor cells based on ZnS:Mn(2+) quantum dots andmagnetic nanocomposites. Talanta. 2019;202:230–236. doi: 10.1016/j.talanta.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 224.Yang W, et al. Reversible capturing and voltammetric determination of circulating tumor cells using two-dimensionalnanozyme based on PdMo decorated with gold nanoparticles and aptamer. Mikrochim. Acta. 2021;188:319. doi: 10.1007/s00604-021-04927-6. [DOI] [PubMed] [Google Scholar]
- 225.Su Y, et al. Antibody-functional microsphere-integrated filter chip with inertial microflow for size-immune-capturing anddigital detection of circulating tumor cells. ACS Appl Mater. Interfaces. 2019;11:29569–29578. doi: 10.1021/acsami.9b09655. [DOI] [PubMed] [Google Scholar]
- 226.Husemann Y, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 227.Hosseini H, et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540:552–558. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Rhim AD, et al. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology. 2014;146:647–651. doi: 10.1053/j.gastro.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Yamaguchi J, et al. Premalignant pancreatic cells seed stealth metastasis in distant organs in mice. Oncogene. 2021;40:2273–2284. doi: 10.1038/s41388-021-01706-8. [DOI] [PubMed] [Google Scholar]
- 230.Barrière G, Riouallon A, Renaudie J, Tartary M, Rigaud M. Mesenchymal and stemness circulating tumor cells in early breast cancer diagnosis. BMC Cancer. 2012;12:114–120. doi: 10.1186/1471-2407-12-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Thery L, et al. Circulating tumor cells in early breast. Cancer JNCI Cancer Spectr. 2019;3:pkz026. doi: 10.1093/jncics/pkz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Paoletti C, et al. Circulating tumor cell clusters in patients with metastatic breast cancer: a SWOG S0500 translationalmedicine study. Clin. Cancer Res. 2019;25:6089–6097. doi: 10.1158/1078-0432.CCR-19-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Magbanua MJM, et al. Synchronous detection of circulating tumor cells in blood and disseminated tumor cells in bonemarrow predicts adverse outcome in early breast cancer. Clin. Cancer Res. 2019;25:5388–5397. doi: 10.1158/1078-0432.CCR-18-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Magbanua MJM, et al. Clinical significance of circulating tumor cells in hormone receptor-positive metastatic breastcancer patients who received letrozole with or without Bevacizumab. Clin. Cancer Res. 2020;26:4911–4920. doi: 10.1158/1078-0432.CCR-20-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Bortolini Silveira A, et al. Multimodal liquid biopsy for early monitoring and outcome prediction of chemotherapy inmetastatic breast cancer. NPJ Breast Cancer. 2021;7:115. doi: 10.1038/s41523-021-00319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Paoletti C, et al. Circulating tumor cell number and endocrine therapy index in ER positive metastatic breast cancerpatients. NPJ Breast Cancer. 2021;7:77. doi: 10.1038/s41523-021-00281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Magbanua MJM, et al. Serial analysis of circulating tumor cells in metastatic breast cancer receiving first-linechemotherapy. J. Natl Cancer Inst. 2021;113:443–452. doi: 10.1093/jnci/djaa113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.de Kruijff IE, et al. Circulating tumor cell enumeration and characterization in metastatic castration-resistant prostate cancer patients treated with cabazitaxel. Cancers. 2019;11:1212–1225. doi: 10.3390/cancers11081212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cieslikowski WA, et al. Circulating tumor cells as a marker of disseminated disease in patients with newly diagnosed high-risk prostate cancer. Cancers. 2020;12:160–175. doi: 10.3390/cancers12010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Basso U, et al. Prognostic role of circulating tumor cells in metastatic renal cell carcinoma: a large, multicenter, prospectivetrial. Oncologist. 2021;26:740–750. doi: 10.1002/onco.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Chemi F, et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat. Med. 2019;25:1534–1539. doi: 10.1038/s41591-019-0593-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Messaritakis I, et al. Characterization of DLL3-positive circulating tumor cells (CTCs) in patients with small cell lungcancer (SCLC) and evaluation of their clinical relevance during front-line treatment. Lung Cancer. 2019;135:33–39. doi: 10.1016/j.lungcan.2019.06.025. [DOI] [PubMed] [Google Scholar]
- 243.Sun YF, et al. Postoperative circulating tumor cells: an early predictor of extrahepatic metastases in patients withhepatocellular carcinoma undergoing curative surgical resection. Cancer Cytopathol. 2020;128:733–745. doi: 10.1002/cncy.22304. [DOI] [PubMed] [Google Scholar]
- 244.Matsushita D, et al. Clinical significance of circulating tumor cells in the response to trastuzumab for HER2-negativemetastatic gastric cancer. Cancer Chemother. Pharm. 2021;87:789–797. doi: 10.1007/s00280-021-04251-z. [DOI] [PubMed] [Google Scholar]
- 245.Sastre J, et al. Association between baseline circulating tumor cells, molecular tumor profiling, and clinical characteristics ina large cohort of chemo-naive metastatic colorectal cancer patients prospectively collected. Clin. Colorectal Cancer. 2020;19:e110–e116. doi: 10.1016/j.clcc.2020.02.014. [DOI] [PubMed] [Google Scholar]
- 246.Bidard FC, et al. Circulating tumor cells in breast cancer patients treated by neoadjuvant chemotherapy: a meta-analysis. J. Natl Cancer Inst. 2018;110:560–567. doi: 10.1093/jnci/djy018. [DOI] [PubMed] [Google Scholar]
- 247.Murlidhar V, et al. Poor prognosis indicated by venous circulating tumor cell clusters in early-stage lung cancers. CancerRes. 2017;77:5194–5206. doi: 10.1158/0008-5472.CAN-16-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jansson S, Bendahl PO, Larsson AM, Aaltonen KE, Ryden L. Prognostic impact of circulating tumor cellapoptosis and clusters in serial blood samples from patients with metastatic breast cancer in a prospective observational cohort. BMC Cancer. 2016;16:433. doi: 10.1186/s12885-016-2406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wang C, et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. BreastCancer Res. Treat. 2017;161:83–94. doi: 10.1007/s10549-016-4026-2. [DOI] [PubMed] [Google Scholar]
- 250.Wang C, et al. Improved prognostic stratification using circulating tumor cell clusters in patients with metastatic castration-resistant prostate cancer. Cancers. 2021;13:268–280. doi: 10.3390/cancers13020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lozano R, et al. Value of early circulating tumor cells dynamics to estimate docetaxel benefit in metastatic castration-resistant prostate cancer (mCRPC) patients. Cancers. 2021;13:2334–2344. doi: 10.3390/cancers13102334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Su P, et al. Mesenchymal and phosphatase of regenerating liver-3 status in circulating tumor cells may serve as a crucialprognostic marker for assessing relapse or metastasis in postoperative patients with colorectal cancer. Clin. Transl. Gastroenterol. 2020;11:e00265. doi: 10.14309/ctg.0000000000000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Strati A, Nikolaou M, Georgoulias V, Lianidou ES. Prognostic significance of TWIST1, CD24, CD44, and ALDH1 transcript quantification in EpCAM-positive circulating tumor cells from early stage breast cancer patients. Cells. 2019;8:652–667. doi: 10.3390/cells8070652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Papadaki MA, et al. Clinical relevance of immune checkpoints on circulating tumor cells in breast cancer. Cancers. 2020;12:376–396. doi: 10.3390/cancers12020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wang XQ, Liu B, Li BY, Wang T, Chen DQ. Effect of CTCs and INHBA level on the effect and prognosis ofdifferent treatment methods for patients with early breast cancer. Eur. Rev. Med. Pharm. Sci. 2020;24:12735–12740. doi: 10.26355/eurrev_202012_24172. [DOI] [PubMed] [Google Scholar]
- 256.Graf RP, et al. Clinical utility of the nuclear-localized AR-V7 biomarker in circulating tumor cells in improving physiciantreatment choice in castration-resistant prostate cancer. Eur. Urol. 2020;77:170–177. doi: 10.1016/j.eururo.2019.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wei T, et al. Vimentin-positive circulating tumor cells as a biomarker for diagnosis and treatment monitoring in patientswith pancreatic cancer. Cancer Lett. 2019;452:237–243. doi: 10.1016/j.canlet.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 258.Economos C, Morrissey C, Vessella RL. Circulating tumor cells as a marker of response: implications for determiningtreatment efficacy and evaluating new agents. Curr. Opin. Urol. 2012;22:190–196. doi: 10.1097/MOU.0b013e3283519b58. [DOI] [PubMed] [Google Scholar]
- 259.Li Y, Wu S, Bai F. Molecular characterization of circulating tumor cells-from bench to bedside. Semin. Cell Dev. Biol. 2018;75:88–97. doi: 10.1016/j.semcdb.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 260.Massard C, et al. RECIST response and variation of circulating tumour cells in phase 1 trials: a prospective multicentricstudy. Eur. J. Cancer. 2017;83:185–193. doi: 10.1016/j.ejca.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 261.Ntzifa A, Kotsakis A, Georgoulias V, Lianidou E. Detection of EGFR mutations in plasma cfDNA and paired CTCs of NSCLC patients before and after osimertinib therapy using crystal digital PCR. Cancers. 2021;13:2376–2394. doi: 10.3390/cancers13112736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Pan X, Zhang X. Utility of circulating tumor cells and DNA in the management of advanced colorectal cancer. FutureOncol. 2020;16:1289–1299. doi: 10.2217/fon-2020-0073. [DOI] [PubMed] [Google Scholar]
- 263.Garrido-Navas MC, et al. The polemic diagnostic role of tp53 mutations in liquid biopsies from breast, colon and lung cancers. Cancers. 2020;12:3343–3359. doi: 10.3390/cancers12113343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Mong J, Tan MH. Size-based enrichment technologies for non-cancerous tumor-derived cells in blood. TrendsBiotechnol. 2018;36:511–522. doi: 10.1016/j.tibtech.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 265.Cristofanilli M, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 266.Liu MC, et al. Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. J. Clin. Oncol. 2009;27:5153–5159. doi: 10.1200/JCO.2008.20.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Pestrin M, et al. Final results of a multicenter phase II clinical trial evaluating the activity of single-agent lapatinib inpatients with HER2-negative metastatic breast cancer and HER2-positive circulating tumor cells. A proof-of-concept study. BreastCancer Res. Treat. 2012;134:283–289. doi: 10.1007/s10549-012-2045-1. [DOI] [PubMed] [Google Scholar]
- 268.Pierga, J. Y. et al. High independent prognostic and predictive value of circulating tumor cells compared with serum tumormarkers in a large prospective trial in first-line chemotherapy for metastatic breast cancer patients. Ann. Oncol. 23, 618-624 [DOI] [PubMed]
- 269.Giordano A, et al. Circulating tumor cells in immunohistochemical subtypes of metastatic breast cancer: lack of predictionin HER2-positive disease treated with targeted therapy. Ann. Oncol. 2012;23:1144–1150. doi: 10.1093/annonc/mdr434. [DOI] [PubMed] [Google Scholar]
- 270.Jiang ZF, et al. Circulating tumor cells predict progression-free and overall survival in Chinese patients with metastaticbreast cancer, HER2-positive or triple-negative (CBCSG004): a multicenter, double-blind, prospective trial. Ann. Oncol. 2013;24:2766–2772. doi: 10.1093/annonc/mdt246. [DOI] [PubMed] [Google Scholar]
- 271.Pierga JY, et al. Circulating tumor cells and brain metastasis outcome in patients with HER2-positive breast cancer: theLANDSCAPE trial. Ann. Oncol. 2013;24:2999–3004. doi: 10.1093/annonc/mdt348. [DOI] [PubMed] [Google Scholar]
- 272.Smerage JB, et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J. Clin. Oncol. 2014;32:3483–3489. doi: 10.1200/JCO.2014.56.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Hall C, et al. Circulating tumor cells after neoadjuvant chemotherapy in stage I-III triple-negative breast cancer. Ann. Surg. Oncol. 2015;22:S552–S558. doi: 10.1245/s10434-015-4600-6. [DOI] [PubMed] [Google Scholar]
- 274.Hall CS, et al. Prognostic value of circulating tumor cells identified before surgical resection in nonmetastatic breastcancer patients. J. Am. Coll. Surg. 2016;223:20–29. doi: 10.1016/j.jamcollsurg.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Riethdorf S, et al. Prognostic impact of circulating tumor cells for breast cancer patients treated in the neoadjuvant“geparquattro” trial. Clin. Cancer Res. 2017;23:5384–5393. doi: 10.1158/1078-0432.CCR-17-0255. [DOI] [PubMed] [Google Scholar]
- 276.Sparano J, et al. Association of circulating tumor cells with late recurrence of estrogen receptor-positive breast cancer: asecondary analysis of a randomized clinical trial. JAMA Oncol. 2018;4:1700–1706. doi: 10.1001/jamaoncol.2018.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Rossi G, et al. Cell-free DNA and circulating tumor cells: comprehensive liquid biopsy analysis in advanced breast cancer. Clin. Cancer Res. 2018;24:560–568. doi: 10.1158/1078-0432.CCR-17-2092. [DOI] [PubMed] [Google Scholar]
- 278.Trapp E, et al. Presence of circulating tumor cells in high-risk early breast cancer during follow-up and prognosis. J. NatlCancer Inst. 2019;111:380–387. doi: 10.1093/jnci/djy152. [DOI] [PubMed] [Google Scholar]
- 279.Radovich M, et al. Association of circulating tumor DNA and circulating tumor cells after neoadjuvant chemotherapy withdisease recurrence in patients with triple-negative breast cancer: preplanned secondary analysis of the BRE12-158 randomizedclinical trial. JAMA Oncol. 2020;6:1410–1415. doi: 10.1001/jamaoncol.2020.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhou J, et al. Epithelial-mesenchymal transition status of circulating tumor cells in breast cancer and its clinical relevance. Cancer Biol. Med. 2020;17:169–180. doi: 10.20892/j.issn.2095-3941.2019.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Le Du,F, et al. EpCAM-independent isolation of circulating tumor cells with epithelial-to-mesenchymal transition andcancer stem cell phenotypes using ApoStream(R) in patients with breast cancer treated with primary systemic therapy. PLoS One. 2020;15:e0229903. doi: 10.1371/journal.pone.0229903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhang SR, et al. Mesenchymal phenotype of circulating tumor cells is associated with distant metastasis in breast cancerpatients. Cancer Manag. Res. 2017;9:691–700. doi: 10.2147/CMAR.S149801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Bulfoni M, et al. In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-tomesenchymaltransition is associated with a poor prognosis. Breast Cancer Res. 2016;18:30. doi: 10.1186/s13058-016-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Papadaki MA, et al. Circulating tumor cells with stemness and epithelial-to-mesenchymal transition features arechemoresistant and predictive of poor outcome in metastatic breast Cancer. Mol. Cancer Ther. 2019;18:437–447. doi: 10.1158/1535-7163.MCT-18-0584. [DOI] [PubMed] [Google Scholar]
- 285.de Kruijff IE, et al. Androgen receptor expression in circulating tumor cells of patients with metastatic breast cancer. Int. J. Cancer. 2019;145:1083–1089. doi: 10.1002/ijc.32209. [DOI] [PubMed] [Google Scholar]
- 286.Bock C, et al. Distinct expression of cytokeratin, N-cadherin and CD133 in circulating tumor cells of metastatic breastcancer patients. Future Oncol. 2014;10:1751–1765. doi: 10.2217/fon.14.58. [DOI] [PubMed] [Google Scholar]
- 287.Goodman OB, et al. Circulating tumor cells in patients with castration-resistant prostate cancer baseline values andcorrelation with prognostic factors. Cancer Epidemiol. Biomark. Prev. 2009;18:1904–1913. doi: 10.1158/1055-9965.EPI-08-1173. [DOI] [PubMed] [Google Scholar]
- 288.Goldkorn A, et al. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: a phase iii trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2014;32:1136–113. doi: 10.1200/JCO.2013.51.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Satelli A, et al. Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as aprognostic marker in cancer patients. Sci. Rep. 2016;6:28910. doi: 10.1038/srep28910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Chen J, et al. Metabolic reprogramming-based characterization of circulating tumor cells in prostate cancer. J. Exp. Clin. Cancer Res. 2018;37:127. doi: 10.1186/s13046-018-0789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Liu H, Ding J, Wu Y, Wu D, Qi J. Prospective study of the clinical impact of epithelial and mesenchymal circulatingtumor cells in localized prostate cancer. Cancer Manag. Res. 2020;12:4549–4560. doi: 10.2147/CMAR.S253997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Antonarakis ES, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Okegawa T, et al. AR-V7 in circulating tumor cells cluster as a predictive biomarker of abiraterone acetate andenzalutamide treatment in castration-resistant prostate cancer patients. Prostate. 2018;78:576–582. doi: 10.1002/pros.23501. [DOI] [PubMed] [Google Scholar]
- 294.Tagawa ST, et al. Expression of AR-V7 and ARv(567es) in circulating tumor cells correlates with outcomes to taxanetherapy in men with metastatic prostate cancer treated in TAXYNERGY. Clin. Cancer Res. 2019;25:1880–1888. doi: 10.1158/1078-0432.CCR-18-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Markou A, et al. PIM-1 Is Overexpressed at a high frequency in circulating tumor cells from metastatic castration-resistant prostate cancer patients. Cancers. 2020;12:1188–1201. doi: 10.3390/cancers12051188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Liu S, et al. Combined cell surface carbonic anhydrase 9 and CD147 antigens enable high-efficiency capture of circulatingtumor cells in clear cell renal cell carcinoma patients. Oncotarget. 2016;7:59877–59891. doi: 10.18632/oncotarget.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Naoe M, et al. Detection of circulating urothelial cancer cells in the blood using the CellSearch System. Cancer. 2007;109:1439–1445. doi: 10.1002/cncr.22543. [DOI] [PubMed] [Google Scholar]
- 298.Gallagher DJ, et al. Detection of circulating tumor cells in patients with urothelial cancer. Ann. Oncol. 2009;20:305–308. doi: 10.1093/annonc/mdn627. [DOI] [PubMed] [Google Scholar]
- 299.Rink M, et al. Detection of circulating tumour cells in peripheral blood of patients with advanced non-metastatic bladdercancer. BJU Int. 2011;107:1668–1675. doi: 10.1111/j.1464-410X.2010.09562.x. [DOI] [PubMed] [Google Scholar]
- 300.Gazzaniga P, et al. Prognostic value of circulating tumor cells in nonmuscle invasive bladder cancer: a CellSearch analysis. Ann. Oncol. 2012;23:2352–2356. doi: 10.1093/annonc/mdr619. [DOI] [PubMed] [Google Scholar]
- 301.Cohen SJ, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival inpatients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 302.Cohen SJ, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol. 2009;20:1223–1229. doi: 10.1093/annonc/mdn786. [DOI] [PubMed] [Google Scholar]
- 303.Wang W, et al. Mesenchymal marker and LGR5 expression levels in circulating tumor cells correlate with colorectal cancerprognosis. Cell Oncol. 2018;41:495–504. doi: 10.1007/s13402-018-0386-4. [DOI] [PubMed] [Google Scholar]
- 304.Wu F, et al. Associations between the epithelial-mesenchymal transition phenotypes of circulating tumor cells and theclinicopathological features of patients with colorectal cancer. Dis. Markers. 2017;2017:9474532. doi: 10.1155/2017/9474532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Zhao R, et al. Expression and clinical relevance of epithelial and mesenchymal markers in circulating tumor cells fromcolorectal cancer. Oncotarget. 2017;8:9293–9302. doi: 10.18632/oncotarget.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Ning Y, et al. Clinical relevance of EMT and stem-like gene expression in circulating tumor cells of metastatic colorectalcancer patients. Pharmacogenom. J. 2018;18:29–34. doi: 10.1038/tpj.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Bao H, et al. High expression of carcinoembryonic antigen and telomerase reverse transcriptase in circulating tumor cells isassociated with poor clinical response to the immune checkpoint inhibitor nivolumab. Oncol. Lett. 2018;15:3061–3067. doi: 10.3892/ol.2017.7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Shen C, Hu L, Xia L, Li Y. Quantitative real-time RT-PCR detection for survivin, CK20 and CEA in peripheral bloodof colorectal cancer patients. Jpn. J. Clin. Oncol. 2008;38:770–776. doi: 10.1093/jjco/hyn105. [DOI] [PubMed] [Google Scholar]
- 309.Shimada R, Iinuma H, Akahane T, Horiuchi A, Watanabe T. Prognostic significance of CTCs and CSCs of tumordrainage vein blood in Dukes' stage B and C colorectal cancer patients. Oncol. Rep. 2012;27:947–953. doi: 10.3892/or.2012.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Barbazan J, et al. A multimarker panel for circulating tumor cells detection predicts patient AQ5 outcome and therapyresponse in metastatic colorectal cancer. Int. J. Cancer. 2014;135:2633–2643. doi: 10.1002/ijc.28910. [DOI] [PubMed] [Google Scholar]
- 311.Yokobori T, et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transitionand is associated with colorectal cancer prognosis. Cancer Res. 2013;73:2059–2069. doi: 10.1158/0008-5472.CAN-12-0326. [DOI] [PubMed] [Google Scholar]
- 312.Bian J, Yan K, Liu N, Xu X. Correlations between circulating tumor cell phenotyping and 18F-fluorodeoxyglucosepositron emission tomography uptake in non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 2020;146:2621–2630. doi: 10.1007/s00432-020-03244-4. [DOI] [PubMed] [Google Scholar]
- 313.Dong J, et al. Detection of circulating tumor cell molecular subtype in pulmonary vein predicting prognosis of stage I-IIInon-small cell lung cancer patients. Front Oncol. 2019;9:1139. doi: 10.3389/fonc.2019.01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Manjunath Y, et al. PD-L1 Expression with epithelial mesenchymal transition of circulating tumor cells is associated with poor survival in curatively resected non-small cell lung cancer. Cancers. 2019;11:806–816. doi: 10.3390/cancers11060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Krebs MG, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lungcancer. J. Clin. Oncol. 2011;29:1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 316.Punnoose EA, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer:association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin. Cancer Res. 2012;18:2391–2401. doi: 10.1158/1078-0432.CCR-11-3148. [DOI] [PubMed] [Google Scholar]
- 317.Liang N, et al. Efficient isolation and quantification of circulating tumor cells in non-small cell lung cancer patients usingpeptide-functionalized magnetic nanoparticles. J. Thorac. Dis. 2020;12:4262–4273. doi: 10.21037/jtd-20-1026A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Tamminga M, et al. Circulating tumor cells in advanced non-small cell lung cancer patients are associated with worse tumorresponse to checkpoint inhibitors. J. Immunother. Cancer. 2019;7:173. doi: 10.1186/s40425-019-0649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tamminga M, et al. Analysis of released circulating tumor cells during surgery for non-small cell lung cancer. Clin. CancerRes. 2020;26,:1656–1666. doi: 10.1158/1078-0432.CCR-19-2541. [DOI] [PubMed] [Google Scholar]
- 320.Frick MA, et al. Circulating tumor cells are associated with recurrent disease in patients with early-stage non-small celllung cancer treated with stereotactic body radiotherapy. Clin. Cancer Res. 2020;26:2372–2380. doi: 10.1158/1078-0432.CCR-19-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Hiltermann TJN, et al. Circulating tumor cells in small-cell lung cancer: a predictive and prognostic factor. Ann. Oncol. 2012;23:2937–2942. doi: 10.1093/annonc/mds138. [DOI] [PubMed] [Google Scholar]
- 322.Hou JM, et al. Evaluation of circulating tumor cells and serological cell death biomarkers in small cell lung cancer patientsundergoing chemotherapy. Am. J. Pathol. 2009;175:808–816. doi: 10.2353/ajpath.2009.090078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Hou JM, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumormicroemboli in patients with small-cell lung cancer. J. Clin. Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 324.Huang CH, et al. A multicenter pilot study examining the role of circulating tumor cells as a blood-based tumor marker inpatients with extensive small-cell lung cancer. Front Oncol. 2014;4:271. doi: 10.3389/fonc.2014.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Naito T, et al. Prognostic impact of circulating tumor cells in patients with small cell lung cancer. J. Thorac. Oncol. 2012;7:512–519. doi: 10.1097/JTO.0b013e31823f125d. [DOI] [PubMed] [Google Scholar]
- 326.Normanno N, et al. Prognostic value of circulating tumor cells' reduction in patients with extensive small-cell lung cancer. Lung Cancer. 2014;85:314–319. doi: 10.1016/j.lungcan.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 327.Messaritakis I, et al. Phenotypic characterization of circulating tumor cells in the peripheral blood of patients with small celllung cancer. PLoS One. 2017;12:e0181211. doi: 10.1371/journal.pone.0181211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Bidard FC, et al. Circulating tumor cells in locally advanced pancreatic adenocarcinoma: the ancillary CirCe 07 study tothe LAP 07 trial. Ann. Oncol. 2013;24:2057–2061. doi: 10.1093/annonc/mdt176. [DOI] [PubMed] [Google Scholar]
- 329.Court CM, et al. Circulating tumor cells predict occult metastatic disease and prognosis in pancreatic cancer. Ann. Surg. Oncol. 2018;25:1000–1008. doi: 10.1245/s10434-017-6290-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Gall TM, et al. Reduced dissemination of circulating tumor cells with no-touch isolation surgical technique in patients withpancreatic cancer. JAMA Surg. 2014;149:482–485. doi: 10.1001/jamasurg.2013.3643. [DOI] [PubMed] [Google Scholar]
- 331.Hugenschmidt H, et al. Circulating tumor cells are an independent predictor of shorter survival in patients undergoingresection for pancreatic and periampullary adenocarcinoma. Ann. Surg. 2020;271:549–558. doi: 10.1097/SLA.0000000000003035. [DOI] [PubMed] [Google Scholar]
- 332.Okubo K, et al. Clinical impact of circulating tumor cells and therapy response in pancreatic cancer. Eur. J. Surg. Oncol. 2017;43:1050–1055. doi: 10.1016/j.ejso.2017.01.241. [DOI] [PubMed] [Google Scholar]
- 333.Dong X, et al. Spatial heterogeneity in epithelial to mesenchymal transition properties of circulating tumor cells associatedwith distant recurrence in pancreatic cancer patients. Ann. Transl. Med. 2020;8:676. doi: 10.21037/atm-20-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Zhao XH, et al. Molecular detection of epithelial-mesenchymal transition markers in circulating tumor cells frompancreatic cancer patients: Potential role in clinical practice. World J. Gastroenterol. 2019;25:138–150. doi: 10.3748/wjg.v25.i1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Zhu P, et al. Circulating tumor cells expressing Kruppel-like factor 8 and vimentin as predictors of poor prognosis inpancreatic cancer patients. Cancer Control. 2021;28:10732748211027163. doi: 10.1177/10732748211027163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ou H, et al. Circulating tumor cell phenotype indicates poor survival and recurrence after surgery for hepatocellularcarcinoma. Dig. Dis. Sci. 2018;63:2373–2380. doi: 10.1007/s10620-018-5124-2. [DOI] [PubMed] [Google Scholar]
- 337.Wang PX, et al. Circulating tumor cells are an indicator for the administration of adjuvant transarterial chemoembolizationin hepatocellular carcinoma: a single-center, retrospective, propensity-matched study. Clin. Transl. Med. 2020;10:e137. doi: 10.1002/ctm2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Yu JJ, et al. Effect of surgical liver resection on circulating tumor cells in patients with hepatocellular carcinoma. BMCCancer. 2018;18:835. doi: 10.1186/s12885-018-4744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Zhou J, et al. Preoperative circulating tumor cells to predict microvascular invasion and dynamical detection indicate theprognosis of hepatocellular carcinoma. BMC Cancer. 2020;20:1047. doi: 10.1186/s12885-020-07488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Zhou KQ, et al. Effect of surgical margin on recurrence based on preoperative circulating tumor cell status inhepatocellular carcinoma. EBioMedicine. 2020;62:103107. doi: 10.1016/j.ebiom.2020.103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Liu YK, et al. An improved strategy to detect the epithelial-mesenchymal transition process in circulating tumor cells inhepatocellular carcinoma patients. Hepatol. Int. 2016;10:640–646. doi: 10.1007/s12072-016-9732-7. [DOI] [PubMed] [Google Scholar]
- 342.Qi LN, et al. Circulating tumor cells undergoing EMT provide a metric for diagnosis and prognosis of patients withhepatocellular carcinoma. Cancer Res. 2018;78:4731–4744. doi: 10.1158/0008-5472.CAN-17-2459. [DOI] [PubMed] [Google Scholar]
- 343.Wang Z, et al. Correlation between postoperative early recurrence of hepatocellular carcinoma and mesenchymal circulatingtumor cells in peripheral blood. J. Gastrointest. Surg. 2018;22:633–639. doi: 10.1007/s11605-017-3619-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Court CM, et al. A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellularcarcinoma. Liver Transpl. 2018;24:946–960. doi: 10.1002/lt.25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Yi B, et al. The clinical significance of CTC enrichment by GPC3-IML and its genetic analysis in hepatocellular carcinoma. J. Nanobiotechnol. 2021;19:74. doi: 10.1186/s12951-021-00818-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Sun YF, et al. Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq inhepatocellular carcinoma. Nat. Commun. 2021;12:4091. doi: 10.1038/s41467-021-24386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Yin LC, et al. Twist expression in circulating hepatocellular carcinoma cells predicts metastasis and prognoses. Biomed. Res. Int. 2018;2018:3789613. doi: 10.1155/2018/3789613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Sun YF, et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis ofhepatocellular carcinoma after curative resection. Hepatology. 2013;57:1458–1468. doi: 10.1002/hep.26151. [DOI] [PubMed] [Google Scholar]
- 349.Lee SJ, et al. Circulating tumor cells are predictive of poor response to chemotherapy in metastatic gastric cancer. Int. J. Biol. Markers. 2015;30:e382–e386. doi: 10.5301/jbm.5000151. [DOI] [PubMed] [Google Scholar]
- 350.Uenosono Y, et al. Clinical significance of circulating tumor cells in peripheral blood from patients with gastric cancer. Cancer. 2013;119:3984–3991. doi: 10.1002/cncr.28309. [DOI] [PubMed] [Google Scholar]
- 351.Zhang Q, et al. A prospective study on the changes and clinical significance of pre-operative and post-operative circulatingtumor cells in resectable gastric cancer. J. Transl. Med. 2018;16:171. doi: 10.1186/s12967-018-1544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Liu M, et al. Prognostic significance of PD-L1 expression on cell-surface vimentin-positive circulating tumor cells in gastriccancer patients. Mol. Oncol. 2020;14:865–881. doi: 10.1002/1878-0261.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Hatakeyama K, et al. A novel splice variant of XIAP-associated factor 1 (XAF1) is expressed in peripheral bloodcontaining gastric cancer-derived circulating tumor cells. Gastr. Cancer. 2015;18:751–761. doi: 10.1007/s10120-014-0426-3. [DOI] [PubMed] [Google Scholar]
- 354.Vaiopoulos AG, et al. Detection of circulating tumor cells in colorectal and gastric cancer using a multiplex PCR assay. Anticancer Res. 2014;34:3083–3092. [PubMed] [Google Scholar]
- 355.Ishiguro Y, et al. Prognostic significance of circulating tumor cells with mesenchymal phenotypes in patients with gastriccancer: a prospective study. Ann. Surg. Oncol. 2021;28:1178–1186. doi: 10.1245/s10434-020-08827-6. [DOI] [PubMed] [Google Scholar]
- 356.Mimori K, et al. A large-scale study of MT1-MMP as a marker for isolated tumor cells in peripheral blood and bonemarrow in gastric cancer cases. Ann. Surg. Oncol. 2008;15:2934–2942. doi: 10.1245/s10434-008-9916-z. [DOI] [PubMed] [Google Scholar]
- 357.Li Y, et al. Prognostic value of circulating tumor cells detected with the CellSearch system in esophageal cancer patients: asystematic review and meta-analysis. BMC Cancer. 2020;20:581. doi: 10.1186/s12885-020-07059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Tewari KS, et al. Circulating tumor cells in advanced cervical cancer: NRG Oncology-Gynecologic Oncology Group study240 ( NCT 00803062) Mol. Cancer Ther. 2020;19:2363–2370. doi: 10.1158/1535-7163.MCT-20-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Anand K, et al. Pilot study of circulating tumor cells in early-stage and metastatic uveal melanoma. Cancers. 2019;11:856–863. doi: 10.3390/cancers11060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Kim DD, Yang CS, Chae HD, Kwak SG, Jeon CH. Melanoma antigen-encoding gene family member A1-6and hTERT in the detection of circulating tumor cells following CD45(-) depletion and RNA extraction. Oncol. Lett. 2017;14:837–843. doi: 10.3892/ol.2017.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Mego M, et al. Circulating tumor cells with epithelial-to-mesenchymal transition phenotypes associated with inferioroutcomes in primary breast cancer. Anticancer Res. 2019;39:1829–1837. doi: 10.21873/anticanres.13290. [DOI] [PubMed] [Google Scholar]
- 362.Zhang Y, et al. Utility of circulating tumor cells for detection of early-stage luminal a breast cancer. Am. J. Med Sci. 2020;360:543–551. doi: 10.1016/j.amjms.2020.01.020. [DOI] [PubMed] [Google Scholar]
- 363.Stefanovic S, et al. The lack of evidence for an association between cancer biomarker conversion patterns and CTC-status in patients with metastatic breast cancer. Int. J. Mol. Sci. 2020;21:2161–2171. doi: 10.3390/ijms21062161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Jin L, et al. Evaluation of the diagnostic value of circulating tumor cells with CytoSorter((R)) CTC capture system inpatients with breast cancer. Cancer Med. 2020;9:1638–1647. doi: 10.1002/cam4.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Shliakhtunou YA. CTCs-oriented adjuvant personalized cytostatic therapy non-metastatic breast cancer patients:continuous non-randomized prospective study and prospective randomized controlled study. Breast Cancer Res. Treat. 2021;186:439–451. doi: 10.1007/s10549-020-06036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Sieuwerts AM, et al. AR splice variants in circulating tumor cells of patients with castration-resistant prostate cancer:relation with outcome to cabazitaxel. Mol. Oncol. 2019;13:1795–1807. doi: 10.1002/1878-0261.12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Schonhoft JD, et al. Morphology-predicted large-scale transition number in circulating tumor cells identifies achromosomal instability biomarker associated with poor outcome in castration-resistant prostate cancer. Cancer Res. 2020;80:4892–4903. doi: 10.1158/0008-5472.CAN-20-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Armstrong AJ, et al. Prospective multicenter study of circulating tumor cell AR-V7 and taxane versus hormonal treatment outcomes in metastatic castration-resistant prostate cancer. JCO Precis. Oncol. 2020;4:1285–1301. doi: 10.1200/PO.20.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Xu L, et al. Noninvasive detection of clinically significant prostate cancer using circulating tumor cells. J. Urol. 2020;203:73–82. doi: 10.1097/JU.0000000000000475. [DOI] [PubMed] [Google Scholar]
- 370.Sperger JM, et al. Prospective evaluation of clinical outcomes using a multiplex liquid biopsy targeting diverse resistancemechanisms in metastatic prostate cancer. J. Clin. Oncol. 2021;39:2926–2937. doi: 10.1200/JCO.21.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Zhang P, et al. The significance of detection of circulating tumor cells and BECLIN1 in peripheral blood of patients withrenal cell carcinoma. Crit. Rev. Eukaryot. Gene Expr. 2020;30:483–492. doi: 10.1615/CritRevEukaryotGeneExpr.2020036246. [DOI] [PubMed] [Google Scholar]
- 372.Li Z, et al. Circulating tumor cells can predict the prognosis of patients with non-small cell lung cancer after resection: aretrospective study. Transl. Lung Cancer Res. 2021;10:995–1006. doi: 10.21037/tlcr-21-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Wang PP, et al. Circulating tumor cells as a new predictive and prognostic factor in patients with small cell lung cancer. J. Cancer. 2020;11:2113–2122. doi: 10.7150/jca.35308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Ha Y, et al. Circulating tumor cells are associated with poor outcomes in early-stage hepatocellular carcinoma: aprospective study. Hepatol. Int. 2019;13:726–735. doi: 10.1007/s12072-019-09994-9. [DOI] [PubMed] [Google Scholar]
- 375.Chen Y, et al. Circulating tumor cells undergoing EMT are poorly correlated with clinical stages or predictive of recurrencein hepatocellular carcinoma. Sci. Rep. 2019;9:7084. doi: 10.1038/s41598-019-43572-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Cheng Y, et al. Diagnostic value of different phenotype circulating tumor cells in hepatocellular carcinoma. J. Gastrointest. Surg. 2019;23:2354–2361. doi: 10.1007/s11605-018-04067-y. [DOI] [PubMed] [Google Scholar]
- 377.Lei Y, et al. Association of preoperative NANOG-positive circulating tumor cell levels with recurrence of hepatocellularcarcinoma. Front. Oncol. 2021;11:601668. doi: 10.3389/fonc.2021.601668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Szczepanik A, et al. CD44(+) cytokeratin-positive tumor cells in blood and bone marrow are associated with poor prognosisof patients with gastric cancer. Gastr. Cancer. 2019;22:264–272. doi: 10.1007/s10120-018-0858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Miki Y, et al. Circulating CEA-positive and EpCAM-negative tumor cells might be a predictive biomarker for recurrence inpatients with gastric cancer. Cancer Med. 2021;10:521–528. doi: 10.1002/cam4.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Kuroda K, et al. Circulating tumor cells with FGFR2 expression might be useful to identify patients with existing FGFR2-overexpressing tumor. Cancer Sci. 2020;111:4500–4509. doi: 10.1111/cas.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Wang D, et al. Prognostic models based on postoperative circulating tumor cells can predict poor tumor recurrence-freesurvival in patients with stage II-III colorectal cancer. J. Cancer. 2019;10:4552–4563. doi: 10.7150/jca.30512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Bidard FC, et al. Circulating tumor cells and circulating tumor DNA detection in potentially resectable metastatic colorectal cancer: a prospective ancillary study to the unicancer Prodige-14 Trial. Cells. 2019;8:516–528. doi: 10.3390/cells8060516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Wang L, et al. Circulating tumor cells as an independent prognostic factor in advanced colorectal cancer: a retrospectivestudy in 121 patients. Int. J. Colorectal Dis. 2019;34:589–597. doi: 10.1007/s00384-018-03223-9. [DOI] [PubMed] [Google Scholar]
- 384.Messaritakis I, et al. Evaluation of the role of circulating tumor cells and microsatellite instability status in predicting outcome of advanced CRC patients. J. Pers. Med. 2020;10:235–347. doi: 10.3390/jpm10040235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Pan RJ, et al. Detection and clinical value of circulating tumor cells as an assisted prognostic marker in colorectal cancerpatients. Cancer Manag. Res. 2021;13:4567–4578. doi: 10.2147/CMAR.S300554. [DOI] [PMC free article] [PubMed] [Google Scholar]