Abstract
Purpose
Wallerian degeneration (WD) is an antegrade degenerative process distal to peripheral nerve injury. Numerous genes are differentially regulated in response to the process. However, the underlying mechanism is unclear, especially the early response. We aimed at investigating the effects of sciatic nerve injury on WD via CLDN 14/15 interactions in vivo and in vitro.
Methods
Using the methods of molecular biology and bioinformatics analysis, we investigated the molecular mechanism by which claudin 14/15 participate in WD. Our previous study showed that claudins 14 and 15 trigger the early signal flow and pathway in damaged sciatic nerves. Here, we report the effects of the interaction between claudin 14 and claudin 15 on nerve degeneration and regeneration during early WD.
Results
It was found that claudin 14/15 were upregulated in the sciatic nerve in WD. Claudin 14/15 promoted Schwann cell proliferation, migration and anti-apoptosis in vitro. PKCα, NT3, NF2, and bFGF were significantly upregulated in transfected Schwann cells. Moreover, the expression levels of the β-catenin, p-AKT/AKT, p-c-jun/c-jun, and p-ERK/ERK signaling pathways were also significantly altered.
Conclusion
Claudin 14/15 affect Schwann cell proliferation, migration, and anti-apoptosis via the β-catenin, p-AKT/AKT, p-c-jun/c-jun, and p-ERK/ERK pathways in vitro and in vivo. The results of this study may help elucidate the molecular mechanisms of the tight junction signaling pathway underlying peripheral nerve degeneration.
Keywords: Nerve regeneration, Schwann cells, Sciatic nerve, Tight junctions, Wallerian degeneration, Claudin 14/15
Introduction
Wallerian degeneration (WD) is a complex phenomenon that occurs distal to peripheral nerve injury.1,2 Structural changes develop at the distal end of the damaged nerve and lead to complete nerve disintegration.3, 4, 5 Changes in the expression levels of numerous genes and proteins happen during degeneration and create conditions conducive to nerve repair and regeneration.6 This process depends mainly on the Schwann cells (SCs) in the peripheral nervous system (PNS). SCs are glial cells that form part of the axon sheath surrounded by peripheral nerves.7,8 Moreover, macrophages are associated with the inflammatory responses induced by nerve damage. However, the specific response mechanism has not been fully elucidated.9
The cell junction complex comprises gap junction, hemides, adherens, and tight junction (TJ) proteins.10 TJ consists of the closed proteins claudins (CLDNs), the occlusion protein occludin, junction adhesion molecules (JAMs), three intact membrane proteins, and closed small loop proteins (ZO-1, ZO-2, and ZO-3). The cytosolic protein composition11 increases the mechanical strength, barrier function, and cell polarity of intercellular connections. CLDNs are the main components of TJ, constitute its skeleton, and maintain its various functions. Claudin 14 (CLDN 14) is affected by calcium ions. Foods with high calcium levels promote CLDN 14 mRNA and upregulate CLDN 14.12, 13, 14, 15 CLDN 15 has cation-selective permeability. CLDNs affect extracellular loop interactions often restricted to tight junction structures. The loss of function and structure often found contribute to cell apoptosis. CLDNs may also interact with the extracellular components to alter cell behavior. The expression changes of CLDNs may induce apoptotic response and cells migration.12, 13, 14, 15, 16 However, the roles of CLDN 14 and CLDN 15 in nerve degeneration and regeneration are seldom reported.
Previously, we analyzed a rat distal sciatic nerve transection model by DNA and protein chips. A large number of genes were either up regulated or down regulated in the early WD degeneration after sciatic nerve injury. We have reported some key factors such as CLDN 14, CLDN 15, ITG, Acvr1c, Birc3, Bid and CCL etc., which regulated gene expression in injured rat sciatic nerves during WD. We have explored the effect of CLDN 14 and CLDN 15 which regulate the early WD following rat sciatic nerve injury in vivo and in vitro.16,17 Nevertheless, the molecular mechanisms of CLDN 14 and CLDN 15 interactions in early WD are poorly understood. We hypothesized that CLDN 14/15 may participate in degeneration and regeneration after nerve injury. In the present study, then, we investigated the effects of sciatic nerve injury on WD via CLDN 14/15 interactions in vivo and in vitro.
Methods
Animal models for WD
Male Sprague-Dawley rats were acquired from the Experimental Animal Center of Nantong University, Nantong, Jiangsu, China. They each weighed about 220 g and were randomly divided into six groups with six animals per group. They were then anesthetized and their sciatic nerves were incised. The rats were sacrificed on day 4, 7, 14, and 21 after treatment. One group of rats was treated immediately after sciatic nerve surgery.6, 7, 8 The expression levels of CLDN 14/15 in the injured sciatic nerves were measured by RT-PCR at 0 h, 0.5 h, 4 days, 7 days, 14 days, and 28 days after surgery. The 0-h rats underwent a sham operation. The experiments were repeated three times and the means were calculated.
SC culture and transfection
One-day-old rats were selected for sciatic nerve SC culture.17,18 The SCs were grown in 10% (w/v) fetal bovine serum (FBS) and 100 IU mL-l penicillin and 100 μg mL-l streptomycin (Sigma-Aldrich Corp., St Louis, MO, USA). The cells were treated with anti-Thy1.1 antibody to remove any fibroblasts. Immunofluorescence with the specific SC marker S100 confirmed that the cell culture was 98% SC. The SCs were then transfected with CLDN 14 and CLDN 15 siRNAs (Ribobio, Guangzhou, China. The transfection efficiency was about 50%–60% in purified SCs), and with pEGFP-c1-CLDN 14 and pEGFP-c1-CLDN 15 recombinant plasmids (The transfection efficiency was about 180%–220%). The siRNA primers are presented in Table 1. CLDN 14/15-siRNA were mixed and injected into the sciatic nerve injuries (The transfection efficiency was about 30%–40%). pEGFP-c1-CLDN 14/15 recombinant plasmids were mixed and injected into the sciatic nerve injuries (The transfection efficiency was about 120%–160%).
Table 1.
CLDN 14/15 small interfering (si) RNA primers.
| Gene | Sequence |
|---|---|
| si-CLDN 14 | F: 5′-CGAAUGAUGUGGUGCAGAAU-3′ |
| R: 5′-UUCUGCACCACAUCAUUCGU-3′ | |
| si-CLDN 15 | F: 5′-GGAACGUCAUCACCACUAACA-3′ |
| R: 5′-UUAGUGGUGAUGACGUUCCCA-3′ | |
| NC | F: 5′-UUCUCCGAACGUGUCACGUTT-3′ |
| R: 5′-ACGUGACACGUUCGGAGAATT-3′ |
siRNA: small interfering RNA, F: forward, R: reverse, NC: negative control.
RNA isolation and qRT-PCR
RNA was extracted from the modeled distal sciatic nerves at 0 h, 0.5 h, 4 days, 7 days, 14 days, and 28 days and the in vitro SCs were subjected to qRT-PCR. The qRT-PCR primers used are presented in Table 2. The relative expression levels of each mRNA were calculated by the 2−ΔΔCt method. The data were statistically analyzed and differences were considered significant at p < 0.05.
Table 2.
qRT-PCR primers.
| Gene | Sequence |
|---|---|
| CLDN 14 | F:5′-AGACCACCTTCGCGGTGTT-3′ |
| CLDN 15 | R:5′-CGCTTGGCAGGGTGTTTGGTCATA-3′ |
| F:5′-CGGGCAGAAGCAATCAGAC-3′ | |
| R:5′-AAGACTGAGGAGGGAGAAGGTT-3′ | |
| Bcl2 | F:5′-GCAGAGATGTCCAGTCAGC-3′ |
| R:5′-CCCACCGAACTCAAAGAAGG-3″ | |
| Bax | F:5′-TGCAGAGGATGATTGCTGAC-3′ |
| R:5′-GATCAGCTCGGGCACTTTAG-3′ | |
| bFGF | F:5′-CCCGCACCCTATCCCTTCACAGC-3′ |
| R:5′-CACAACGACCAGCCTTCCACCCAAA-3′ | |
| NT3 | F:5′-GACAAGTCCTCAGCCATTGACATTC-3′ |
| R:5′-CTGGCTTCTTTACACCTCGTTTCAT-3′ | |
| Nf2 | F:5′-CTGGGATTGGGTTCATGGGTGGAT-3′ |
| R:5′-AGGAAGCCCGAGAAGCAGAGCG-3′ | |
| PKCα | F:5′-GAACACATGATGGACGGGGTCACGAC-3′ |
| R:5′-CGCTTGGCAGGGTGTTTGGTCATA-3′ | |
| GAPDH | F:5′-TGGAGTCTACTGGCGTCTT-3′ |
| R:5′-TGTCATATTTCTCGTGGTTCA-3′ |
F: forward, R: reverse, bcl2: B-cell lymphoma 2, Bax: Bcl-2-associated X protein, bFGF: basic fibroblast growth factor, NT3: neurotrophin-3, Nf2: neurofibromin 2, PKCα: protein kinase C α GAPDH: glyceraldehyde 3-phosphate dehydrogenase.
Western blot analysis
The distal protein extracts from the modeled sciatic nerves at 0 h, 0.5 h, 4 days, 7 days, 14 days, and 28 days and the extract proteins of the SCs cultured in vitro were measured by western blot. The classical signaling pathway proteins AKT, p-AKT, ERK, p-ERK, c-Jun, and p-c-Jun were used to assess the effects of CLDN 14/15 on WD.
Cell migration assay
The migration of cultured SCs was evaluated in Transwell cell culture chambers (Costar, Cambridge, MA, USA). The SCs were transferred to the upper chamber of each Transwell and complete medium was injected into each of the lower chambers. At a predetermined time point, the unmigrated cells were removed with cotton swabs. Methanol was added to fix the migrated cells and they were stained by dropwise addition of crystal violet solution. The cells were observed under a microscope (Leica Microsystems, Wetzlar, Germany) and counted.
Cell proliferation assay
The proliferation of cultured SCs was determined with a Cell-Light EdU DNA Cell Proliferation Assay Kit (Ribobio, Guangzhou, China). Transfected SCs were resuspended in complete medium, counted, and transferred to poly-L-lysine-coated 96-well plates. The SCs were fixed with formaldehyde in phosphate-buffered saline (PBS), observed under a microscope (Leica Microsystems, Wetzlar, Germany) and subjected to proliferation analysis.
Flow cytometry
Cultured SCs were analyzed for apoptosis with a Flow Collect Annexin Red Kit (Millipore, Bedford, MA, USA). The cells were digested with 0.125% (w/v) trypsin for 20 s, centrifuged, and resuspended in assay buffer. Annexin V solution was added to each sample. The suspensions were then incubated and resuspended. Then 7-AAD was added to the suspensions and they were incubated in the dark. The cells were then examined for apoptosis by flow cytometry (BD Biosciences, San Jose, CA, USA).
Immunofluorescence
The cultured SCs were fixed, washed with Tris-buffered saline (TBS), permeabilized with 0.1% (w/v) Triton X-100, blocked with 10% (v/v) goat serum, and diluted with primary and secondary antibodies. The SCs were imaged with a Zeiss bright field fluorescence microscope (Carl Zeiss AG, Oberkochen, Germany) and Improvision image analysis software (Improvision, Coventry, UK).
Statistical analysis
Data were processed by statistical analysis using SPSS 15.0 for Windows (SPSS, IL, USA). Group differences were analyzed using one-way analyses of variance as appropriate. All data are expressed as mean ± standard deviation. A p value less than 0.05 is considered statistically significant.
Results
CLDN 14 and CLDN 15 expression in injured sciatic nerves and SCs
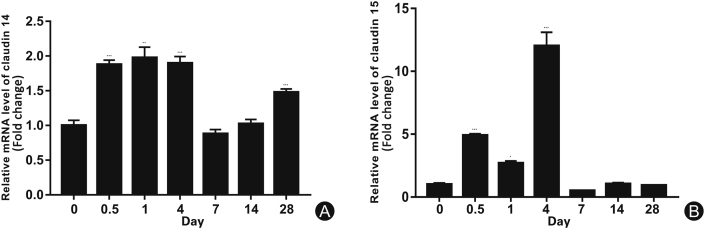
The expression levels of CLDN 14 (Fig. 1A) and CLDN 15 (Fig. 1B) at various time points in the distal part of the sciatic nerve were evaluated by RT-PCR. At the early stage of WD, the protein expression levels of CLDN 14 and CLDN 15 at the distal end of the sciatic nerve initially increased then decreased. Data were processed by ANOVA and Scheffé’s post hoc tests (p < 0.05).
Fig. 1.
Expression of CLDN 14 and CLDN 15 at various time points in the distal part of rat sciatic nerve injury. (A) Real-time CLDN 14 gene expression at various distal points after sciatic nerve transection; (B) Real-time CLDN 15 gene expression at various distal points after sciatic nerve transection. ∗p < 0.05.
CLDN 14/15 expression levels influenced SC proliferation
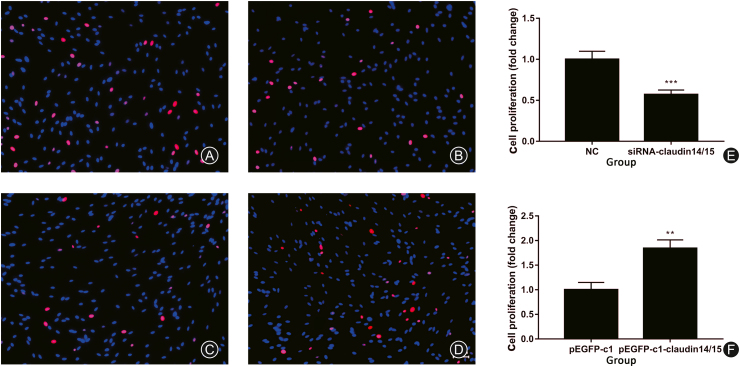
We measured the expression levels of the CLDN 14/15 siRNAs and overexpression of the CLDN 14/15 plasmids after they were transfected into SCs. CLDN 14 and CLDN 15 were downregulated. An EdU assay on SC proliferation was also performed. It revealed that after inhibition of CLDN 14/15 expression, SC proliferation in both the experimental and control groups (Fig. 2A and B) significantly decreased (Fig. 2E). Thereafter, CLDN 14/15 were upregulated and proportions of proliferating SCs in the experimental and control groups (Fig. 2C and D) significantly increased (Fig. 2F). Therefore, CLDN 14/15 upregulation may promote SC proliferation in vitro.
Fig. 2.
EdU proliferation assay after SCs transfection. (A) NC control group for CLDN 14 and CLDN 15 siRNA transfection; (B) CLDN 14 and CLDN 15 siRNA target group; (C) pEGFP-c1 empty plasmid control group; (D) pEGFP-c1-CLDN 14/15 plasmid. The experimental group was expressed. Blue indicates SC nuclei and red indicates the number of proliferating cells according to EdU. (E, F) Graphs of the cell proliferation rates according to EdU.
∗∗p < 0.01; ∗p < 0.05. Bar = 50 μm.
CLDN 14/15 expression affected SC migration
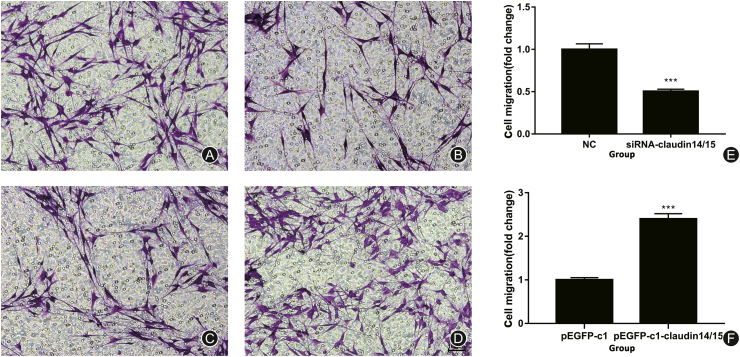
The CLDN 14/15 siRNA and overexpressing recombinant plasmids were transfected into SCs and migration was analyzed by a Transwell assay. CLDN 14/15 were significantly downregulated after their expression was inhibited (Fig. 3A and B). After CLDN 14/15 were upregulated, the experimental and control groups were compared. The proportions of cellular migration (Fig. 3C and D) had significantly increased (Fig. 3F). Therefore, CLDN 14/15 promotes SC migration.
Fig. 3.
Transwell cell migration assay after SC transfection. (A) NC control group for siRNA interference in CLDN 14 and CLDN 15 cells; (B) Experimental group of CLDN 14 and CLDN 15 siRNA interference; (C) pEGFP-c1 empty vector control group; (D) pEGFP-c1-CLDN 14/15 plasmid. The experimental group was expressed. Purple indicates cells migrating through the chamber. (E, F) Results of the Transwell cell mobility assay.
∗∗∗p < 0.001. Bar = 100 μm.
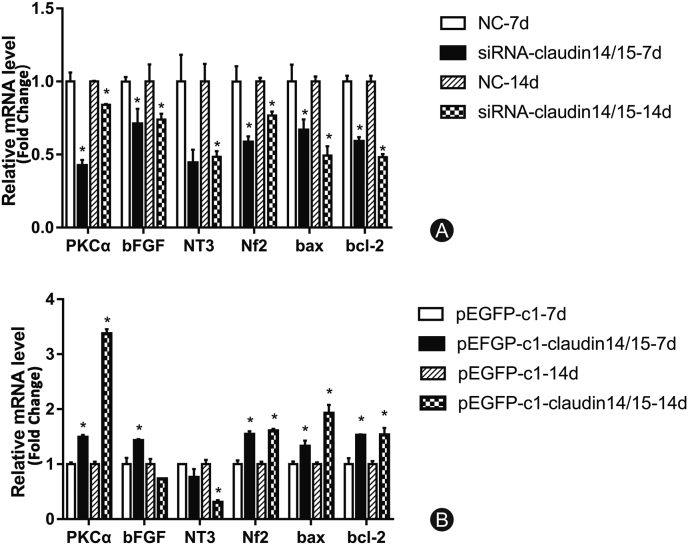
Changes in the expression of related genes after SC transfection
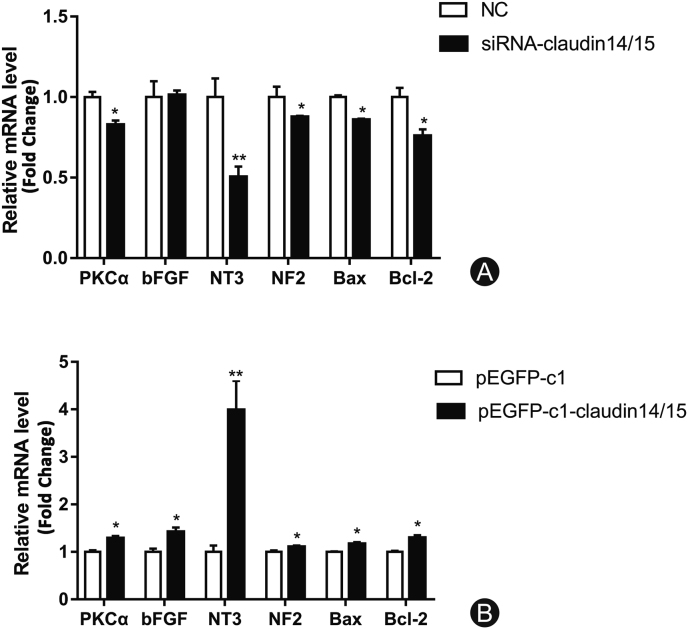
RT-PCR was used to detect changes in the expression of certain neurotrophic factors after CLDN 14/15 interference and overexpression. There were significant differences in the expression of factors regulating SC migration, proliferation, and apoptosis after CLDN 14/15 interference and overexpression in SCs. The mRNA levels of PKC-α, NT3, NF2, Bax, and Bcl-2 changed significantly after CLDN 14/15 downregulation (Fig. 4A). After subsequent CLDN 14/15 upregulation, PKC-α and bFGF were detected. The mRNA levels of NT3, NF2, Bax, and Bcl-2 changed significantly (Fig. 4B). Thus, CLDN 14/15 may affect expression of the genes regulating SC function.
Fig. 4.
RT-PCR analysis of related gene expression after interference and overexpression of CLDN 14- and CLDN 15-transfected SCs. (A) Differential expression patterns of certain factors after CLDN 14 and CLDN 15 interference according to RT-PCR. NC is the negative control group and siRNA-CLDN 14/15 is the experimental group. (B) Differential expression of the related factors after CLDN 14 and CLDN 15 overexpression. pEGFP-c1 is the empty control group and pEGFP-c1-CLDN 14/15 is the overexpressing experimental group.
∗p < 0.05; ∗∗p < 0.01.
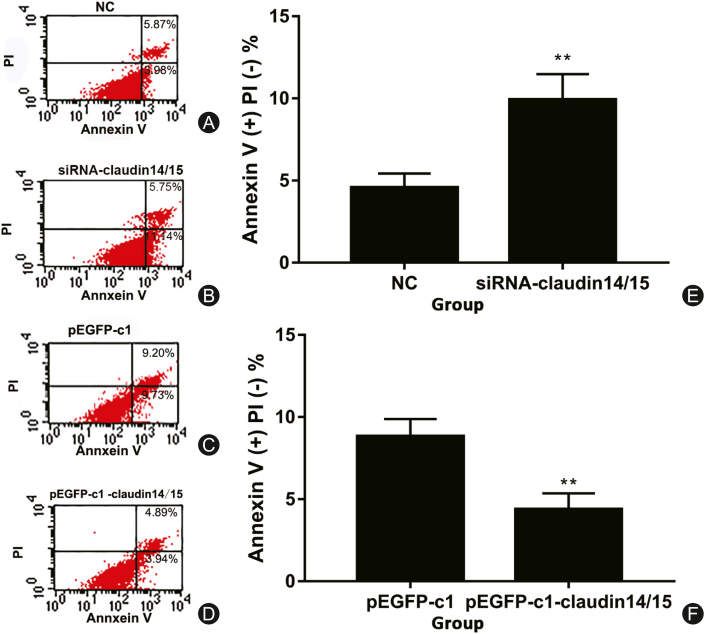
Effects of CLDN 14/15 on SC apoptosis
The recombinant plasmids bearing siRNA and overexpressing CLDN 14/15 were transfected into SCs. CLDN 14/15 were downregulated and SC apoptosis was evaluated. After siRNA interfered with CLDN 14/15 expression, SC apoptosis was detected by flow cytometry. The apoptosis rate was higher than that of the control group (Fig. 5A, B, and 5E). In contrast, the apoptosis rate of SCs transfected with recombinant plasmids overexpressing CLDN 14/15 was significantly reduced (Fig. 5C, D, and 5F). Therefore, CLDN 14/15 may have anti-apoptotic effects on SCs.
Fig. 5.
Detection of apoptosis by flow cytometry with Annexin V after transfection. (A) Control group of cells transfected with CLDN 14 and CLDN 15 siRNA-NC; (B) Interference assay group of CLDN 14 and CLDN 15 siRNA-CLDN 14/15; (C) pEGFP-c1 empty vector control group; (D) pEGFP-c1-CLDN 14/15 overexpressing plasmid experimental group. (A–D) Apoptosis ratio analyses; (E, F) Apoptosis rates after cell transfection.
∗∗p < 0.01.
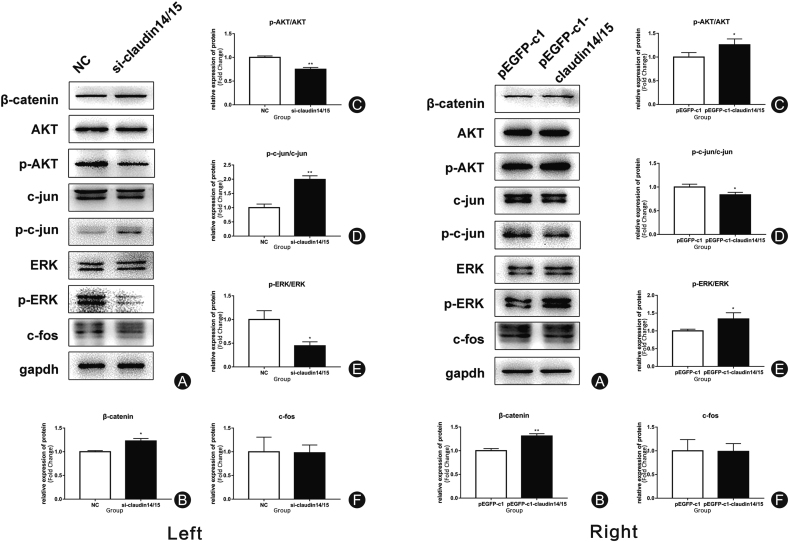
Differential expression of related proteins in SCs
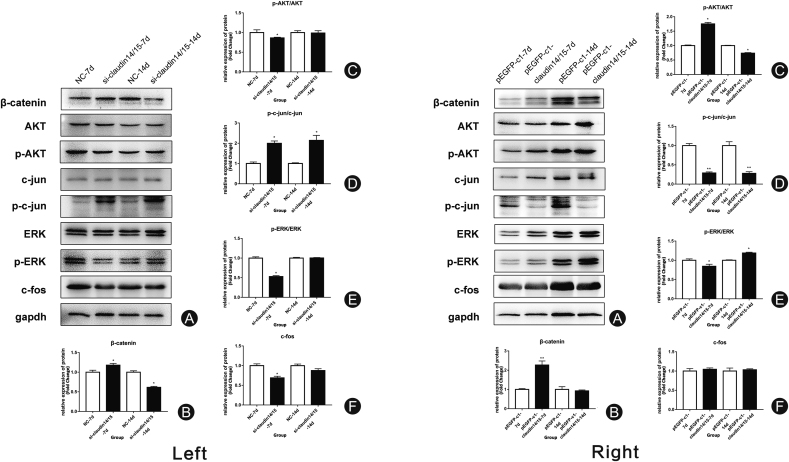
After detecting the differences in expression of the related genes, we used western blot to identify the changes in expression of the signaling pathway proteins involved in SC migration, proliferation, and apoptosis (Fig. 6A). The expression levels of β-catenin, p-AKT/AKT, pc-jun/c-jun, and p-ERK/ERK had significantly changed after interference (Fig. 6B–E). There were no significant differences between treatments in terms of c-fos expression (Fig. 6F). The expression levels of the related proteins after overexpression were evaluated by western blot. The expression levels of β-catenin, p-AKT/AKT, pc-jun/c-jun, and p-ERK/ERK had significantly changed after overexpression (Fig. 6B–E). There were no significant differences between treatments in terms of c-fos expression (Fig. 6F).
Fig. 6.
Changes in the expression of related proteins after CLDN 14/15 siRNA transfection into interfering SC cells. Left: (A) Band expression map of related proteins after CLDN 14 and CLDN 15 siRNA interference; (B–F) Statistical analyses of β-catenin, p-AKT/AKT, c-jun/p-c-jun, p-ERK/ERK, and c-fos protein. NC is a negative control group for transfected cells. The siRNA-CLDN 14/15 is a cell interference test group transfected with CLDN 14 and CLDN 15. ∗∗p < 0.01; ∗p < 0.05. Right: Relative changes in the expression of related proteins after transfection of CLDN 14/15-overexpressing plasmids. (A) Western blot of related protein expression after transfection of CLDN 14- and CLDN 15-overexpressing plasmids; (B–F): Statistical analyses of β-catenin, p-AKT/AKT, c-jun/p-c-jun, p-ERK/ERK, and c-fos proteins. The pEGFP-c1 is a negative control group transfected with an empty plasmid. The pEGFP-c1-CLDN 14/15 is an overexpressing experimental group.
∗∗p < 0.01; ∗p < 0.05.
Changes in expression of related genes after SC transfection
The preceding experiments demonstrated that CLDN 14/15 may interact with SCs. Moreover, cytokines and pathway proteins interact with CLDN 14/15 to influence and regulate SC function. Here, qRT PCR was used to measure changes in the expression of certain neurotrophic factors after 7 days and 14 days of CLDN 14/15 interference and overexpression. There were significant differences in the expression levels of the factors controlling SC migration, proliferation, and apoptosis after CLDN 14/15 interference and overexpression (Fig. 7). The mRNA levels of NT3, Nf2, PKC-α and bFGF significantly changed at 7 days and 14 days after CLDN 14/15 interference and overexpression. For this reason, CLDN 14/15 may influence the expression of several genes governing SC function.
Fig. 7.
RT-PCR analysis of related gene interference and overexpression in CLDN 14- and CLDN 15-transfected SCs at 7 days and 14 days. (A) RT-PCR assay of differential expression of certain factors after CLDN 14 and CLDN 15 interference at 7 days and 14 days. NC is a negative control group. The siRNA-CLDN 14/15 is an experimental group. (B) CLDN 14 and CLDN 15 overexpression. Differences in the expression levels of post-correlation factors between day 7 and 14. The pEGFP-c1 is the empty control group. The pEGFP-c1-CLDN 14/15 is the overexpressing experimental group.
∗p < 0.05.
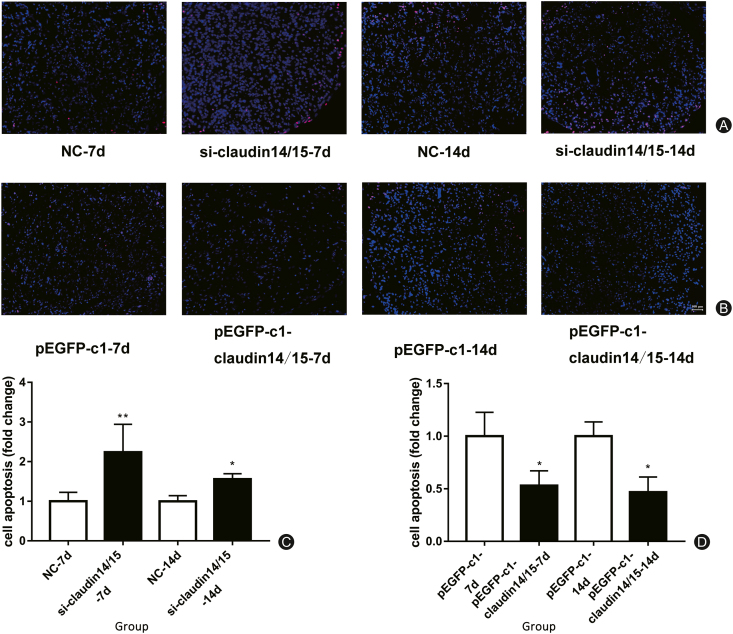
Detection of apoptosis in TUNEL tissue
We cultured and purified SCs in vitro, transfected them with CLDN 14/15, and monitored their apoptosis by flow cytometry with Annexin V-FITC. It was noted that CLDN 14/15 inhibited SC apoptosis. We then induced WD in a rat sciatic nerve transection model. We connected the proximal and distal nerves at the damaged end with a silicone tube. After 7 days and 14 days subculture, tissue samples were frozen. After they were sectioned, TUNEL apoptosis was detected in them. The nuclei of apoptotic cells stained red while those of healthy cells stained blue. After in vivo transfection with CLDN 14/15 interference, apoptosis significantly increased. In contrast, tissues transfected with overexpressing CLDN 14/15 presented with significantly reduced apoptosis (Fig. 8).
Fig. 8.
TUNEL apoptosis detection. (A) Apoptosis staining at 7 d and 14 d after siRNA transfection in vivo; (B) Apoptosis staining at 7 d and 14 d after transfection; (C) Statistical analysis of A chart; (D) Statistical analysis of B chart.
∗∗p < 0.01; ∗∗∗p < 0.001.
Differential expression of related proteins after CLDN 14/15 siRNA transfectionin vivo
After matrigel treatment, RVG and CLDN 14/15-siRNA were mixed and injected into the sciatic nerve injuries. They were transfected 7 days and 14 days later and the efficiencies of CLDN 14/15 interference on the protein expression level were analyzed by western blot. The CLDN 14/15 interference efficiencies statistically differed at 7 days and 14 days. Western blot detected substantial changes in the signaling pathway protein levels at the distal end of the sciatic nerve injuries. β-catenin was upregulated after 7 days interference and downregulated after 14 days interference. The p-AKT/AKT, p-ERK/ERK, and c-fos pathway proteins were downregulated 7 days after interference but their expression levels had not significantly changed after 14 days interference. The p-c-jun/c-jun was significantly upregulated at 7 days and 14 days after interference (Fig. 9).
Fig. 9.
Changes in the expression of related proteins at 7 days and 14 days after transfection with CLDN 14 and CLDN 15 siRNA in vivo. Left: (A) Western blot band diagram showing expression of related proteins at 7 days and 14 days after CLDN 14 and CLDN 15 siRNA transfection; (B–F) Statistical analyses of β-catenin, p-AKT/AKT, c-jun/p-c-jun, p-ERK/ERK, and c-fos proteins. NC represents the negative control group with CLDN 14 and CLDN 15 siRNA interference. The siRNA-CLDN 14/15 indicates the experimental group with CLDN 14 and CLDN 15 siRNA interference. ∗p < 0.05. Right: Changes in the expression of related proteins after CLDN 14 and CLDN 15 transfection for 7 days and 14 days. (A) Western blot band diagram showing the expression of related proteins at 7 days and 14 days after CLDN 14 and CLDN 15 overexpression in vivo; (B–F) Statistical analysis of β-catenin, p-AKT/AKT, c-jun/p-c-jun, p-ERK/ERK, and c-fos proteins. The pEGFP-c1 is the negative control empty plasmid group. The pEGFP-c1-CLDN 14/15 is the overexpressing experimental group.
∗p < 0.05; ∗∗p < 0.01.
Differential expression of related proteins after CLDN 14/15 plasmid transfectionin vivo
Western blot was used to evaluate CLDN 14/15 protein expression levels in response to plasmid transfection for 7 days and 14 days. The signaling pathway protein expression levels were measured by western blotting. The results showed that the expression of β-catenin increased significantly after 7 days of overexpression but did not change after 14 days. The p-AKT/AKT was upregulated after 7 days overexpression. The pc-jun/c-jun was significantly downregulated after 7 days and 14 days overexpression. The p-ERK/ERK was significantly downregulated after 7 days overexpression and significantly upregulated after 14 days expression. There was no significant difference in c-fos expression level between 7 days and 14 days overexpression (Fig. 9).
Discussion
WD consists of a series of signal responses and physiological changes in the PNS in response to sciatic nerve injury. This process changes proximal nerve fibers and neuronal cell bodies and degenerate distal nerve fibers.14 Immunohistochemistry (IHC) and other assays disclosed that nerve regeneration comprised gradual nerve fiber extension in the distal direction several hours after nerve injury. Therefore, early WD activation may be significant in nerve degeneration and regeneration.15, 16, 17, 18, 19 Using bioinformatics technology, we found that CLDN 14 and CLDN 15 are vital to the signal network regulating sciatic nerve injury regeneration.
CLDNs comprise a small family of transmembrane proteins integral to tight junctions. They form a cell compartment barrier that controls molecule flow in the epithelial cell gap.20, 21, 22, 23, 24 CLDNs all have the same membrane organization but different tissue distribution patterns.25 Tight junction proteins are expressed in the inner ear and have various distributions and localizations there.26 CLDN 14 is expressed in the Corti hair cells and supporting cells and is barrier limiting potassium and other cations.22,27 It is critical for tight junction formation and highly expressed in the kidney and liver as well.28,29 CLDN 14 may also participate in calcium reabsorption.30 CLDN 15 has selective cation permeability. CLDN 15 knockout mice have the unique giant intestine phenotype. Their gut is longer and wider than that of wild type mice.31,32
Previously, we used a rat sciatic nerve transection model to study the effects of CLDN 14 and CLDN 15 signals on SC function in early WD. CLDN 14 promotes SC proliferation and migration and regulates their function via the c-Jun pathway. CLDN 15 inhibits SC proliferation, promotes their apoptosis, and also regulates their function via the c-Jun pathway. Earlier studies reported interactions between CLDN 14 and CLDN 16. CLDN 14 may be physiologically bound to the CLDN complex and play a negative regulatory role in it.21, 22, 23, 24
Therefore, the aims of this experiment were to determine whether: (1) CLDN 14 collaborates with CLDN 15, (2) this interaction is involved in the signal-regulating network regenerated after sciatic nerve injury, and (3) the latter affects the biological functions of SCs which play important roles in peripheral nerve regeneration.
SCs are glial cells in the PNS that form myelin sheaths on nerve fibers. WD is critically dependent on SCs and plays an important role in peripheral nerve regeneration.33 In the present study, then, we investigated CLDN 14 and CLDN 15 in vitro and selected SCs as our research object. We created an in vivo model to verify sciatic nerve transection in rats. We obtained the distal sciatic nerve by qRT-PCR. Distal tissue CLDN 14 and CLDN 15 expression initially increased then decreased. Thus, CLDN 14 and CLDN 15 regulate the repair of the sciatic nerve in the early stage of its injury. We determined whether the interaction between CLDN 14 and CLDN 15 influences the nerve collapse process by affecting SC function. We purified, knocked down, and overexpressed CLDN 14 and CLDN 15 transfected into SCs in vitro. The CLDN 14/15 combination promoted SC proliferation. Thus, it had a positive effect on nerve degeneration. Flow cytometry detected post-transfection apoptosis. The CLDN 14/15 combination also made SCs anti-apoptotic. Moreover, the SCs formed cell cords during peripheral nerve collapse. We established that CLDN 14/15 promoted SC migration. After SC transfection, PKC-α, NT3, NF2, and bFGF expression was significantly altered. These genes encode neurotrophic factors that play important roles in nerve repair and regeneration.34, 35, 36, 37 The protein expression levels of the β-catenin, p-AKT/AKT, pc-jun/c-jun, and p-ERK/ERK pathways also markedly changed. These proteins are critical for cell proliferation, migration,38, 39, 40, 41 and overall function.
We established that the interactions between CLDN 14 and CLDN 15 substantially influence early degeneration and regeneration after rat sciatic nerve injury and we elucidated the underlying mechanisms of this process. The results of this study provide basic techniques that may eventually be applied towards nerve repair and regeneration.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 31971277 and 31950410551), the Scientific Research Foundation for Returned Scholars of the Ministry of Education of China, a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (No. KYCX 19-2050).
Ethical statement
All animal tests were conducted according to the Key Laboratory of Neuroregeneration of Jiangsu, the Ministry of Education Guidelines for the Care and Use of Laboratory Animals, and the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. The Institutional Animal Care and Use Committee of Nantong University approved all protocols.
Declaration of competing interest
The authors declare that they have no competing interest.
Acknowledgments
The authors thank Ian Haigler for his editorial assistance.
Footnotes
Peer review under responsibility of Chinese Medical Association.
References
- 1.Choi E.J., Choi Y.M., Jang E.J., et al. Neural ablation and regeneration in pain practice. Korean J Pain. 2016;29:3–11. doi: 10.3344/kjp.2016.29.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nectow A.R., Marra K.G., Kaplan D.L. Biomaterials for the development of peripheral nerve guidance conduits. Tissue Eng B Rev. 2012;18:40–50. doi: 10.1089/ten.TEB.2011.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vis C., Mol M., Kleiboer A., et al. Improving implementation of mental health for mood disorders in routine practice: systematic review of barriers and facilitating factors. JMIR Ment Health. 2018;16:e20. doi: 10.2196/mental.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knoepfel T., Furet P., Mah R., et al. 2-Formylpyridyl ureas as highly selective reversible-covalent inhibitors of fibroblast growth factor receptor 4. ACS Med Chem Lett. 2018;9:215–220. doi: 10.1021/acsmedchemlett.7b00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abioye A.I., Park S., Ripp K., et al. Anemia of inflammation during human pregnancy does not affect newborn iron endowment. J Nutr. 2018;148:427–436. doi: 10.1093/jn/nxx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubey N., Letourneau P.C., Tranquillo R.T. Guided neurite elongation and schwann cell invasion into magnetically aligned collagen in simulated peripheral nerve regeneration. Exp Neuol. 1999;158:338–350. doi: 10.1006/exnr.1999.7095. [DOI] [PubMed] [Google Scholar]
- 7.Ali A., Pisipati S., Tewari A. Words of wisdom: Re: autonomic nerve development contributes to prostate cancer progression. European Urology, Eur Urol. 2014;65:665–666. doi: 10.1016/j.eururo.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 8.Jessen K.R., Mirsky R. The repair Schwann cell and its function in regenerating nerves. J Physiol. 2016;594:3521–3531. doi: 10.1113/JP270874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cámaralemarroy C.R., Fernándezgarza N.E., Nancy E.F. Molecular inflammatory mediators in peripheral nerve degeneration and regeneration. Neuroimmunomodulation. 2010;17:314–324. doi: 10.1159/000292020. [DOI] [PubMed] [Google Scholar]
- 10.Liron E.N., Tamar B.Y. Functions of claudin tight junction proteins and their complex interactions in various physiological systems. Int Rev Cell Mol Biol. 2010;279:1–32. doi: 10.1016/S1937-6448(10)79001-8. [DOI] [PubMed] [Google Scholar]
- 11.Chiba H., Makoto O., Masaki M., et al. Transmembrane proteins of tight junctions. Biochim Biophys Acta. 2008;1778:588–600. doi: 10.1016/j.bbamem.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Peng S., Rao V.S., Adelman R.A., et al. Claudin-19 and the barrier properties of the human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2011;52:1392–1403. doi: 10.1167/iovs.10-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyamoto T., Morita K., Takemoto D., et al. Tight junctions in Schwann cells of peripheral myelinated axons: a lesson from claudin-19–deficient mice. J Cell Biol. 2005;169:527–538. doi: 10.1083/jcb.200501154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kagan J.C., Su T., Horng T., et al. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navarro X., Vivó M., ValerocabréA Neural plasticity after peripheral nerve injury and regeneration. Prog Neurobiol. 2007;82:163–201. doi: 10.1016/j.pneurobio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Gong L.L., Zhu Y., Xu X., et al. The effects of claudin 14 during early Wallerian degeneration after sciatic nerve injury. Neural Regen Res. 2014;9:2151–2158. doi: 10.4103/1673-5374.147946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J.N., Zhang Z., Wu G.Z., et al. Claudin-15 overexpression inhibits proliferation and promotes apoptosis of Schwann cells in vitro. Neural Regen Res. 2020;15:169–177. doi: 10.4103/1673-5374.264463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raivich G., Bohatschek M., Costa D., et al. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Chattopadhyay S., Shubayev V.I. MMP-9 controls Schwann cell proliferation and phenotypic remodeling via IGF-1 and ErbB receptor-mediated activation of MEK/ERK pathway. Glia. 2010;57:1316–1325. doi: 10.1002/glia.20851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu C.H., Xiao K., Luan Z.S., et al. Early weaning increases intestinal permeability, alters expression of cytokine and tight junction proteins, and activates mitogen-activated protein kinases in pigs. J Anim Sci. 2013;91:1094–1101. doi: 10.2527/jas.2012-5796. [DOI] [PubMed] [Google Scholar]
- 21.Lu Z., Ding L., Hong H., et al. Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp Cell Res. 2011;317:1935–1946. doi: 10.1016/j.yexcr.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benyosef T., Belyantseva I.A., Saunders T.L., et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum Mol Genet. 2003;12:2049–2061. doi: 10.1093/hmg/ddg210. [DOI] [PubMed] [Google Scholar]
- 23.Arthur-Farraj P.J., Latouche M., Wilton D.K., et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 2012;75:633–647. doi: 10.1016/j.neuron.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y.H., Bao Y., Peng W., et al. Claudin-3 gene silencing with siRNA suppresses ovarian tumor growth and metastasis. Proc Natl Acad Sci USA. 2009;106:426–430. doi: 10.1073/pnas.0813348106. https://doi:.org/10.1073/pnas.0813348106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y., Yao Jun, Wei Q., et al. Genetic analysis of CLDN14 in the Chinese population affected with non-syndromic hearing loss. Int J Pediatr Otorhinolaryngol. 2018;105:6–11. doi: 10.1016/j.ijporl.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Kitajiri S.I., Furuse M., Morita K., et al. Expression patterns of claudins, tight junction adhesion molecules, in the inner ear. Hear Res. 2004;187:25–34. doi: 10.1016/s0378-5955(03)00338-1. [DOI] [PubMed] [Google Scholar]
- 27.Morita K., Fujimoto K., Tsukitaet S., et al. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. https://doi: 10.1073/pnas.96.2.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markov A.G., Aschenbach J.R., Amasheh S. Claudin clusters as determinants of epithelial barrier function. IUBMB Life. 2015;67:29–35. doi: 10.1002/iub.1347. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki H., Tani K., Tamura A., et al. Model for the architecture of claudin-based paracellular ion channels through tight junctions. J Mol Biol. 2015;427:291–297. doi: 10.1016/j.jmb.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 30.Thorleifsson G., Holm H., Edvardsson V., et al. Sequence variants in the CLDN14 gene associate with kidney stones and bone mineral density. Nat Genet. 2009;41:926–930. doi: 10.1038/ng.404. [DOI] [PubMed] [Google Scholar]
- 31.Kari J.A., Farouq M., Alshaya H., et al. Familial hypomagnesemia with hypercalciuria and nephrocalcinosis. Pediatr Nephrol. 2003;18:506–510. doi: 10.1007/s00467-003-1139-8. [DOI] [PubMed] [Google Scholar]
- 32.Ishizaki T., Chiba H., Kojima T., et al. Cyclic AMP induces phosphorylation of claudin-5 immunoprecipitates and expression of claudin-5 gene in blood-brain-barrier endothelial cells via protein kinase A-dependent and -independent pathways. Exp Cell Res. 2003;290:275–288. doi: 10.1016/s0014-4827(03)00354-9. [DOI] [PubMed] [Google Scholar]
- 33.Feoktistova M., Peter G., Kellert B., et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L.J., Liu L.P., Gu X.L., et al. Implantation of adipose-derived stem cells cures the optic nerve injury on rats through inhibiting the expression of inflammation factors in the TLR4 signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:1196–1202. doi: 10.26355/eurrev_201803_14458. [DOI] [PubMed] [Google Scholar]
- 35.Aisiku O., Dowal L., Scarlata S. Protein kinase C phosphorylation of PLCβ1 regulates its cellular localization. Arch Biochem Biophys. 2011;509:186–190. doi: 10.1016/j.abb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J., Liu Y., Soh J.W., et al. Antiapoptotic effects of vasopressin in the neuronal cell line H32 involve protein kinase Calpha and beta. J Neurochem. 2009;110:1310–1320. doi: 10.1111/j.1471-4159.2009.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keefe K.M., Sheikh I.S., Smith G.M. Targeting neurotrophins to specific populations of neurons: NGF, BDNF, and NT-3 and their relevance for treatment of spinal cord injury. Int J Mol Sci. 2017;548:1–17. doi: 10.3390/ijms18030548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu P., Han Z., Couvillon A.D., et al. Critical role of endogenous Akt/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J Biol Chem. 2004;279:49420–49429. doi: 10.1074/jbc.M407700200. [DOI] [PubMed] [Google Scholar]
- 39.Sanna M.D., Galeotti N. The HDAC1/c-JUN complex is essential in the promotion of nerve injury-induced neuropathic pain through JNK signaling. Eur J Pharmacol. 2018;825:99–106. doi: 10.1016/j.ejphar.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 40.Yu H., Zhu L., Li C., et al. ERK1/2 and AKT are vital factors in regulation of the migration of rat Schwann cells. J Vet Med Sci. 2015;77:427–432. doi: 10.1292/jvms.14-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H.F., Wang J., Tony S.S. The phosphatidylinositol 3-kinase/Akt and c-Jun N-terminal kinase signaling in cancer: alliance or contradiction? Int J Oncol. 2015;47:429–436. doi: 10.3892/ijo.2015.3052. [DOI] [PubMed] [Google Scholar]