Abstract
The process of cancer initiation and development is a dynamic and complex mechanism involving multiple genetic and non-genetic variations. With the development of high throughput techniques like next-generation sequencing, the field of cancer biology extended beyond the protein-coding genes. It brought the functional role of noncoding RNAs into cancer-associated pathways. MicroRNAs (miRNAs) are one such class of noncoding RNAs regulating different cancer development aspects, including progression and metastasis. MicroRNA-1 (miR-1) is a highly conserved miRNA with a functional role in developing skeletal muscle precursor cells and cardiomyocytes, and acts as a consistent tumor suppressor gene. In humans, two discrete genes, MIR1–1 located on 20q13.333 and MIR1–2 located on 18q11.2 loci, encode for a single mature miR-1. Dysregulation or downregulation of miR-1 has been demonstrated in multiple cancers, including lung, breast, liver, prostate, colorectal, pancreatic, medulloblastoma, and gastric cancer. A vast number of studies have shown that miR-1 affects the multiple hallmarks of cancer like proliferation, invasion and metastasis, apoptosis, angiogenesis, chemosensitization, and immune modulation. The potential therapeutic applications of miR-1 in multiple cancer pathways provide a novel platform for developing anticancer therapies. This review focuses on the different antitumorigenic and therapeutic aspects of miR-1 including how it regulates tumor development and associated immunogenic functions.
Keywords: miR-1, immunoregulation, cancer therapeutics, chemosensitivity, miRNAs
1. Introduction
MicroRNAs (miRNAs or miRs) are short non-coding RNAs consisting 21–22 nucleotides that regulate various molecular pathways and triggering the endogenous RNA-interference (RNAi) by modulating the stability or inducing mRNA degradation [1–5]. The biogenesis of miRNAs follows a systematic process; RNA-polymerase II transcribed the initial stem-loop structure or primary miRNA strand in the nucleus [6, 7]. The miRNA hairpin structure, embedded within the stem-loop primary miRNA strand, is successively processed by DROSHA and DICER (both belong to the RNase III family), liberating the mature miRNA consisting of 21–22 nucleotides [6, 7].
The mature miRNA sequence is loaded to the RNA-induced silencing complex (RISC complex) and modulates gene expression or induces degradation by binding with the 3’-untranslated region (UTR) of the target gene. The primary mode of action for miRNA usually follows a slicer-independent mechanism in which they bind with 3’-UTR of mRNA with complete or incomplete complementarity and perform gene silencing [8–11]. Because miRNAs can act through low complementarity; thus, they could have multiple targets, but the primary safety check is the restriction of imperfect base pairing. Recent studies demonstrated that miRNAs also regulate the methylation of CpG islands (present in the promoter region of different genes) and thus, regulates the epigenetic transcriptional regulations [8–11]. Most of the miRs are highly conserved among species, suggesting the importance of this highly selective pressure for their crucial roles in development, evolution, and diseased conditions [12]. Their importance was further defined through peculiar observation that mice lacking DICER (the essential enzyme for mature miRNA production) showed embryonic lethality [13]. The miRs play diverse functions in physiological and pathophysiological progressions, including metabolism [14], neurological disorders [15], diabetes [16], infectious diseases [17, 18], muscular dystrophy [19], cell-cycle progression [20], cancer development and progression [21–24], and immunity [25–27]. Interestingly, miRs represent one of the largest collections of gene regulatory molecules; therefore, they constitute an important research axis to understand different mechanisms and for the development of different therapeutic strategies.
Based on aberrant expression, miRs can categorize into two major classes: upregulated and downregulated miRs. Studies demonstrated low and high expression of miRs in normal development and various disease conditions, including viral infections, immunological disorders, and multiple types of cancers [12, 28–32]. In the case of cancer, a number of miRNAs fine-tuned the expression of oncogenes and tumor suppressors in response to a variety of extracellular signals [33–36]. The overexpression of various miRNAs has been reported in different cancers; however, the majority of miRs are found to be downregulated in multiple tumors, thus pointing towards the tumor-suppressing properties of miRs over oncogenic [4, 37, 38].
Among more than 2600 known human miRNAs (http://mirbase.org/), microRNA-1 (miR-1) is one of the highly conserved and most consistent tumor-suppressor or downregulated miRNA in various cancers [39]. In humans, two discrete genes, MIR1–1 and MIR1–2, located on genomic locus 20q13.33 and 18q11.2, respectively, encode for a single mature miR-1 having 21 nucleotides (Figure 1 &2) [40, 41]. The initial studies of miR-1 showed that miR-1–1 and miR-1–2 were expressed in cardiomyocytes and precursor cells of skeletal muscles [40]. It controls the expression of various genes including, Kruppel-like factor 4 (KLF4), heat shock protein 60 (HSP60), heart and neural crest derivatives expressed 2 (HAND2, a transcription factor that regulates the development of cardiomyocytes), stanniocalcin 2 (STC2), and transforming growth factor-beta (TGF-β) signaling [40–45]. At the same time, miR-1 genes act as a direct target of muscle differentiation regulators, including serum response factor, myoblast determination protein 1 (MyoD), and myocyte enhancer factor-2 (Mef2) [40, 46]. MiR-1 is also reported to target histone deacetylase 4 (HDAC4), which acts as a transcriptional repressor of muscle-specific gene expression and promotes myogenesis [46]. The collective outcomes of different studies suggested that miR-1 plays an important role in cardiogenesis, cell cycle regulation, cardiac conduction, myogenesis, myoblasts differentiation, myocardial infarction, chronic obstructive pulmonary disease, and heart failure [47–54]. Apart from the critical role of miR-1 in the physiological functioning of the heart, smooth, and skeletal muscles, miR-1 dysregulation has been associated with multiple cancers and immune responses [41, 55, 56]. Since the different aspects or role of miRNAs has been reported and discussed extensively [36, 51, 55, 57–62], here in the present review, we particularly discuss and summarize the tumor-associated and immunogenic roles of miR-1 in multiple cancers.
Figure 1:
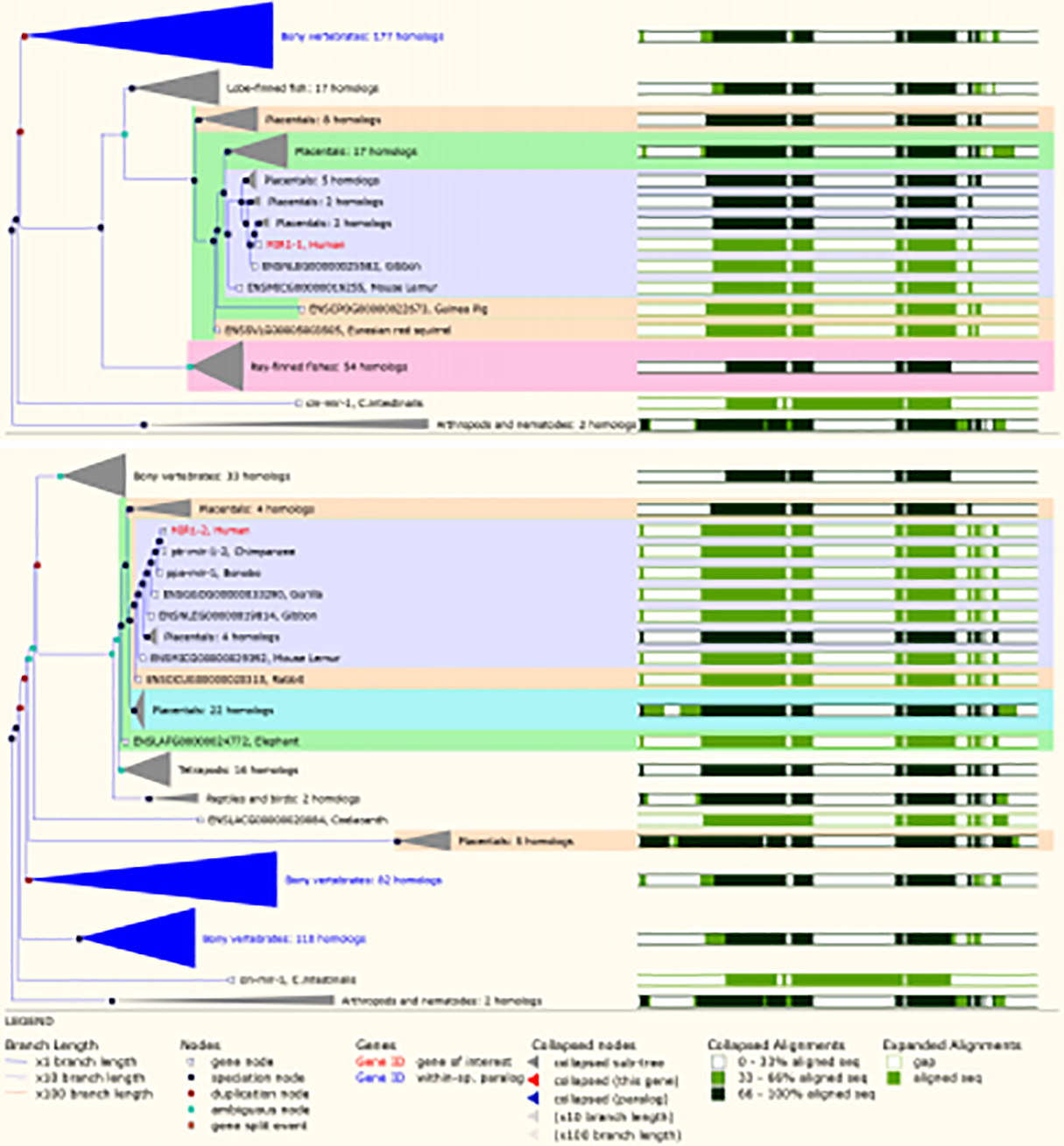
Gene tree representing the alignment of human MIR-1–1 and MIR-1–2 gene with other species. The gene tree analysis showed the conserved pattern of miR-1 across the species. Gene tree was created using Ensembl genome database (Ensemble archives103) [180].
Figure 2: Mechanism of miR-1 biogenesis and common mechanism of action.
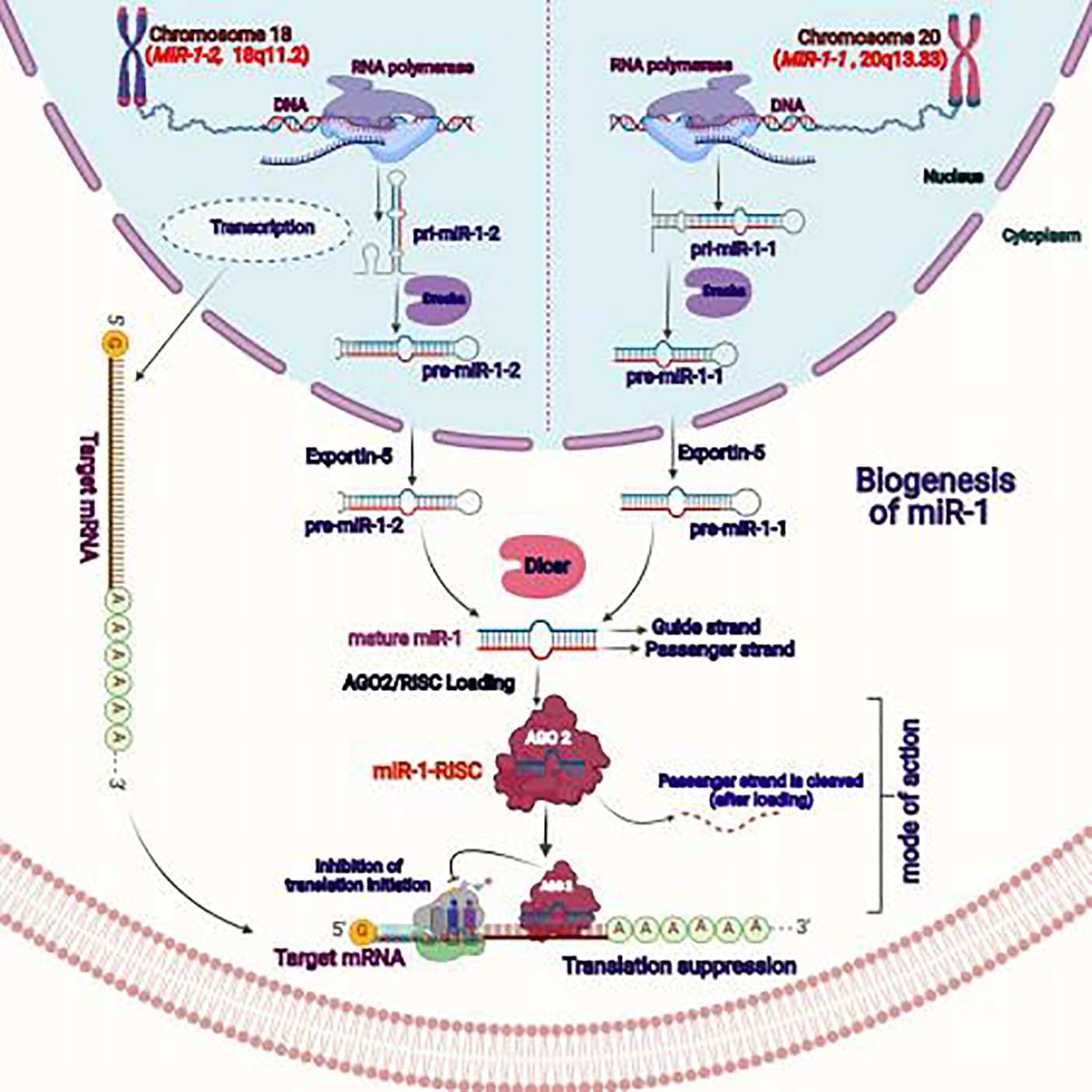
In case of humans, two discrete genes, MIR1–1 located on 20q13.333 and MIR1–2 located on 18q11.2 loci encode for a single mature miR-1. The initial stem loop or primary miR-1–1/miR-1–2 (pri-miR-1–1/pri-miR-1–2) were transcribed by RNA-polymerase II and DROSHA processed them into pre-miR-1–1 or pre-miR-1–2. The pre-miR-1 sequences then transported from the nucleus through exportin-5. Reaching out form the nucleus to cytoplasm, the pre-miR-1 sequences were processed by DICER and a single mature miR-1 was generated from both types of pre-sequences. The mature miR-1 was loaded to AGO2 that form miR-1-RISC complex for specific gene targeting or translation inhibition.
2. Expression and role of miR-1 in different cancers
Several studies showed the downregulation or decreased expression of miR-1 in various human cancers, including liver cancer, lung cancer (LC), breast cancer (BC), colon cancer, medulloblastoma/glioblastoma, colorectal cancer (CRC), pancreatic cancer (PC), prostate cancer (PCa), and other cancer types associated with gastrointestinal or elementary canal and rhabdomyosarcoma [4, 41, 63–67]. These studies also demonstrated correlation between tumor types, response to treatment, and miR-1 expression. We have discussed the tumor-suppressive and immune-modulatory role of miR-1 in association with different cancers in coming sections.
2.1. Lung cancer
LC is a heterogeneous disease mainly includes two subtypes, non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) and remains one of the major leading cause of death globally [68, 69]. The outcomes of multiple studies from different LC models (cell line and mouse models) show that the downregulation of miR-1 is associated with LC [70–73]. Low expression of miR-1 was noticed in vinyl carbamate induced LC mice model compared to normal mice, and the expression was resumed when treated with the chemopreventive agent (indole-3-carbinol) [74]. In the case of NSCLC cell lines (A549), miR-1 directly targets phosphoinositide-3-kinase catalytic subunit alpha (PIK3CA) and inhibited the activation of downstream targets of the PI3K/AKT pathway (Figure 3) [75]. Overexpression of miR-1 in A549 cells showed decreased cell proliferation, migration, and invasion [75]. Another study showed that nearly 70% of NSCLC samples have low miR-1 and high PIK3CA expression, and low miR-1 patients have higher chances of lymph node metastasis and recurrence compared to moderate miR-1 and PIK3CA expression [76]. In addition, miR-1 overexpression inhibits LC growth via reducing metabolic mediators. Singh et al. have demonstrated that miR-1 plays a key role in LC metabolic reprogramming through regulating nuclear factor erythroid-2–related factor 2 (NRF2) [77].
Figure 3: Schematic illustrations for miR-1 modulated cancer associated signaling pathways and other targets.
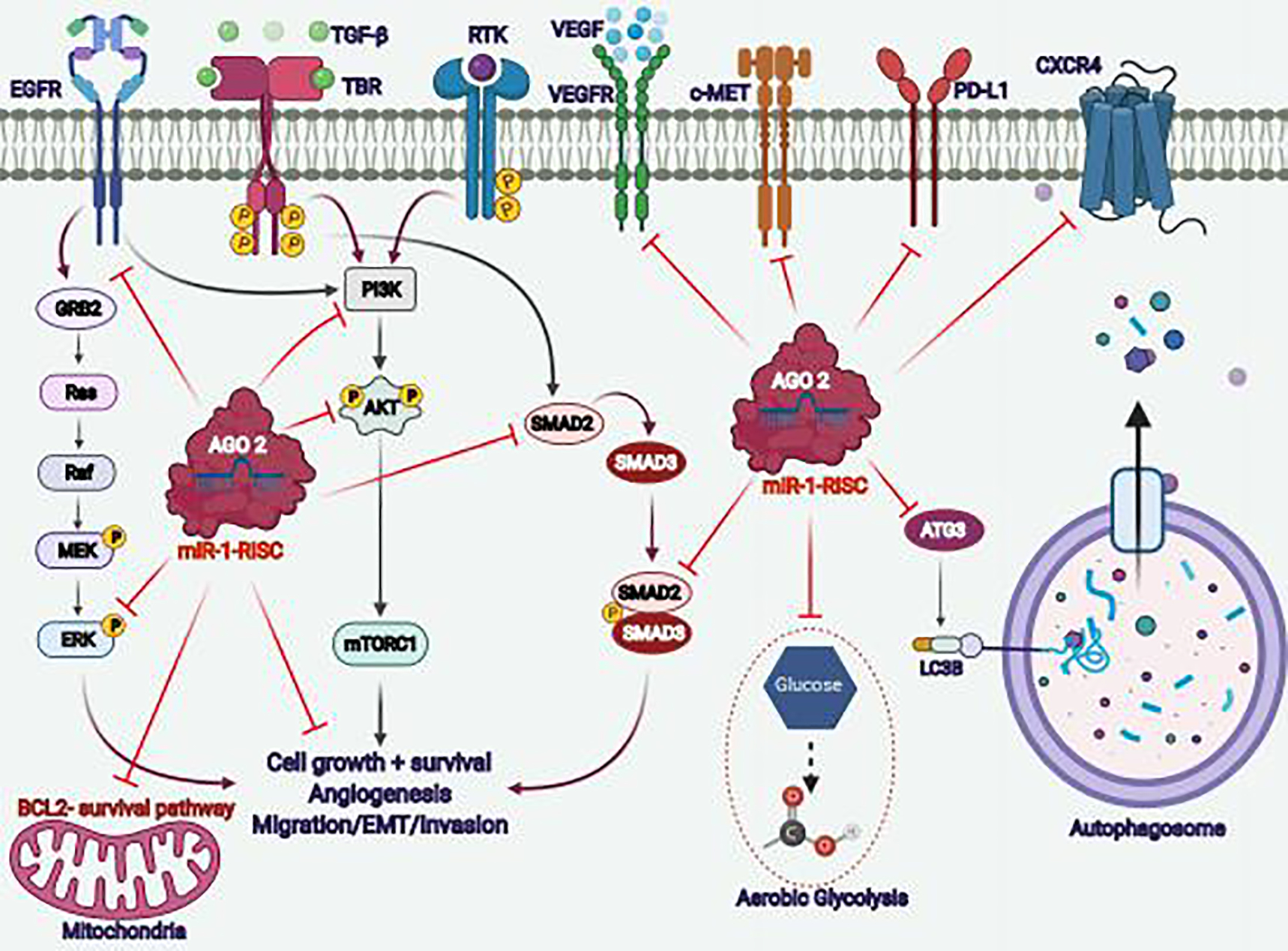
MiR-1 downregulation is implicated in the various oncogenic signaling pathways, and overexpression of miR-1 or miR-1-RISC complex decreases/inhibited the activation of transcription factors or other protein molecules in multiple pathways associated with cancer cell proliferation, cell-growth, survival, angiogenesis, metastasis, metabolism (aerobic glycolysis), and autophagy mediated networks.
Recently, Liu et al. showed that miR-1 downregulation increased the expression of FAM83A (family with sequence similarity 83, member A) in A549 and H1355 LC cells [70]. The 3-prime UTR of FAM83A has binding sites for miR-1 [70]. The miR-1 mediated downregulation of FAM83A inhibited the epidermal growth factor receptor (EGFR), mitogen-activated protein kinase (MAPK), and choline kinase alpha (CHKA) signaling. Further, the inhibition of FAM83A decreases the LC cell growth, migration, and invasion [70]. In NSCLC microarray dataset analysis, miR-1 was reported as one of the most differentially expressed miRNAs that play an essential role in disease progression [71]. Interestingly, a recent report also showed the correlation of miR-1 with cigarette smoking and malignant transformation [78]. The continuous exposure of human normal bronchial epithelial cells (BEAS-2B) to cigarette smoke to transformed into malignant cells is associated with differential expression of miR-1. The miRNA sequencing data of LC and correlation of differentially expressed genes with smoke history identified miR-1 as a negatively regulated miRNA associated with epithelial to mesenchymal transformation (EMT). Overall, this study identifies miR-1 as a predictive biomarker for smoke-induced LC [78]. Computational studies also suggest that the downregulation of miR-1 is associated with lung squamous cell carcinoma (LUSC) [79]. This study showed the association of miR-1 with p53, cell-cycle regulation, and serine/threonine metabolism associated signaling pathways [79]. Long noncoding RNAs (lncRNAs) generally contain complementary sequences for various miRNAs, thus interacting with miRNAs through different types of interactions [80, 81]. The overexpression of the RNA component of mitochondrial RNA-processing endoribonuclease (RMRP) lncRNA has been associated with multiple cancers, including LC [82, 83]. Wang et al. reported that high expression of RMRP and low miR-1 is associated with advanced-stage NSCLC and poor overall survival [73]. Loss of RMRP or induction of miR-1 in LC cell lines reduced cell growth and motility with G0/G1 cell cycle arrest. It was also reported that miR-1 targets annexin-A2 (ANXA2), a highly overexpressed protein in NSCLC [73]. Knockdown of RMRP shows high expression of miR-1 and reduced levels of ANXA2; suggesting RMRP-miR-1 axis modulates LC growth through ANXA2, and thus provides a novel therapeutic target for LC therapies [73].
Drug resistance or resistance to chemotherapy is a significant problem associated with different LC treatments. Various miRNAs demonstrated their potential role in therapy resistance [84–86]. The prominent role of miR-1 towards the chemosensitivity of LC has also been reported by Hua et al. [87]. The miRNA expression analysis of cisplatin resistance NSCLC tissues showed a downregulation of miR-1 compared to sensitive patients, and the miR-1 expression is negatively associated with light chain 3 beta (LC3B, an autophagy marker) [87]. The resistant NSCLC cell lines showed low expression of p62 with an increased LC3B-II/LC3B-I ratio, and miR-1 overexpression decreased the LC3B-II/LC3B-I ratio. This study also identifies autophagy-related 3 (ATG3) as a target of miR-1. ATG3 was found to be upregulated in resistant NSCLC cell lines and positively correlated with LC3B expression and negatively with miR-1 expression (Figure 3) [87]. ATG3 is a ubiquitin-like enzyme working as an important component of the ubiquitination process that leads to the degradation and recycling process of cytoplasmic modules, phosphatidylethanolamine conjugation and autophagy regulation [88–90]. Interestingly, the 3’-UTR of ATG3 consists of miR-1 binding sites, and miR-1 regulates its expression. The upregulation of ATG3 eliminates the miR-1 oriented inhibition of autophagy in cisplatin-resistant NSCLC cells (A549 & H1299) [87]. This study validated the role of miR-1 in chemoresistance of NSCLC cell lines and tumor tissues and showed that miR-1 upregulation induces chemosensitization and apoptosis in resistant cell lines through LC3B/ATG3-mediated autophagy inhibition (Figure 3) [87]. Thus, providing the potential role of the miR-1-ATG3 axis in relieving LC chemoresistance.
Nearly half of the LC patients harbor “driver mutations” in EGFR, and thus EGFR-tyrosine kinase inhibitors (TKI) are recommended as the first choice of therapy, but due to the acquirement of mutations and other factors the patients develop therapy resistance [91–93]. In such patients, immunotherapy becomes one of the most imperative and leading therapeutic approaches that give promising outcomes [94, 95]. However, the lymphocyte infiltration and the expression of programmed death ligand-1 (PD-L1) rationalized immunotherapy efficacy prediction. The immune microenvironment of LC tumors still under investigation to identify novel predictors for immunotherapy. A recent study evaluated the correlation between miRNAs, lymphocyte infiltration, and EGRF-TKI (before and after acquiring resistance) in LC tumor tissues and cell line models [96]. The PCR-array analysis of LC tumor samples and cell lines identifies miR-1 as the most variable miR. This study showed the role of miR-1 in the modulation of tumor immune microenvironment (TIME) [96]. It inhibited the infiltration of CD8+ T-cells and the production of C-C motif chemokine ligands (CCL5 and CCL10). Therefore, miR-1 plays a vital role in EGFR-TKI resistance and acts as a useful clinical predictor for the efficacy of immunotherapy [96]. Most of the studies (as discussed in previous sections) advocated the tumor suppressor activities of miR-1, however Kawana et al. suggests the additive role of miR-1 in EGFR-TKI resistance, thus adding a new paradigm to the future studies of miR-1, particularly in association with drug resistance and immunotherapy [96]. In addition to TIME, miR-1 plays a significant role in reprogramming normal fibroblasts to LC-associated fibroblasts through targeting CCL2 and VEGFA [97]. Overall, miR-1 studies in LC demonstrated that it acts as an essential regulator for pathogenesis, drug resistance, metastasis, and recurrence predictor, working as a potential drug candidate and providing a suitable therapeutic approach for the treatment of LC.
2.2. Breast cancer
Breast cancer (BC) is one of the primary and leading causes of death in females [69]. BC patients showed high heterogeneity, and thus despite the availability of some therapeutic options for particular BC subtypes, limited therapeutic options are available for other subtypes like triple-negative phenotypes [98, 99]. Apart from protein-based therapeutic targets, miRNAs also play a major role in BC pathogenesis [100, 101]. In the case of triple-negative BC (TNBC) patients, the miR-1 expression is highly suppressed in TNBC cells relative to normal cells (MB-157) and cells derived from luminal subtypes [102]. The overexpression of miR-1 suppressed the cell migration, invasion, activation of ERK and MEK, and induced apoptosis (Figure 3). The induction of miR-1 also enhances the chemosensitivity of BC cells [102].
Mitochondrial retrogradation and nuclear crosstalk are associated with the development and pathogenesis of BC; thus, mitochondria provide different promising drug targets for BC [103–105]. The gene enrichment analysis using different TCGA tissue samples and microarray data generated a transcription factor-miRNA hub network that showed the upregulation of MYC-associated zinc finger proteins and downregulation of miR-1 is associated with poor overall survival and prognosis in BC [65]. This hub network regulates mitochondrial functioning and acts as a diagnostic biomarker and a therapeutic target [65].
The cardiotoxicity induced by anthracycline therapy in BC patients is a severe and frequent undesirable effect [106, 107]. The cardiac biomarkers and imaging are the main parameters to study the toxicity [106, 108]. Different RNA molecules, including miRNAs and small noncoding RNA, act as the biomarkers for cardiovascular diseases and toxicity, and miR-1 is a prominent one [109, 110]. Pereira et al. recently compiled the implication of miR-1 to access the cardiotoxicity of anthracycline treatment in BC patients [111]. They studied and compared the miRNA expression in the patient samples showing anthracycline-induced cardiotoxicity and no cardiotoxicity. The downregulation of miR-1 and some other miRNAs has been reported in epirubicin-induced cardiotoxicity and myocardial infarction compared to doxorubicin-induced cardiotoxicity [111]. It means that miR-1 plays a role in chemotherapy-associated cardiotoxicity of BC patients.
B-cell Lymphoma 2 (BCL2), an antiapoptotic protein, plays a regulatory role in different subtypes of BC and acts as a prognostic marker [112]. In the case of TNBC, BCL2 expression works as a poor prognostic factor [113]. BCL2 plays an important role in BC cell survival and apoptotic evasion, and therapeutic targeting of BCL2 improves the efficacy of endocrine therapy and decreases the tamoxifen-induced hyperplasia of the endometrium [114]. Thus, BCL2 targeting or using BCL2 specific small molecule inhibitors provides an attractive approach in BC therapies [114, 115]. Recently, Peng et al. showed that the miR-1 is downregulated in BC cells and patients samples compared to adjacent normal control [116]. Overexpression of miR-1 decreases the BC cell proliferation, invasion and induces apoptosis in vitro and in vivo through BCL2 downregulation (Figure 3). MiR-1 overexpression inhibited the tumor growth of BC cell xenografts in nude mice models. Another exciting outcome of this study suggested that miR-1 sensitizes the BC cells towards chemotherapy (cisplatin and paclitaxel) [116]. Two independent studies reported the negative correlation between lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and miR-1 in BC cells and patient samples [117, 118]. The outcomes of these studies showed that MALAT1 promotes BC development, growth, metastasis, and EMT through miR-1 [117, 118]. Overexpression of miR-1 has been shown to decrease the expression of slug and MALAT1 with a corresponding decrease in tumor growth. Thus, these studies established miR-1 as a major tumor suppressor in BC, and targeted delivery of miR-1 may be a potential therapeutic approach for BC.
2.3. Colorectal cancer
Colorectal cancer (CRC) is the second leading cause of common cancer death in males and females in the United States [69]. Different molecules have been identified that promotes CRC development and progression, including transforming growth factor-β (TGF-β), vascular endothelial growth factor (VEGF), EGFR, B-Raf proto-oncogene serine/threonine kinase (BRAF), Kirsten rat sarcoma viral oncogene (KRAS), nicotinamide phosphoribosyltransferase (NAMPT), and SRY-box transcription factor 12 (SOX12) [119–121]. Apart from the known protein-coding genes, various miRNAs, including miR-1, also modulate CRC growth and development [122–125]. A recent study established the role of miR-1 in NAMPT and TGF-β signaling pathway that regulates the proliferation and growth of CRC cells [64]. NAMPT is a proinflammatory cytokine, and its overexpression is associated with the development of multiple cancers [126, 127]. Lv et al. reported that the 3-prime UTR of NAMPT has a putative binding site for miR-1, and TGF-β1 regulates the expression of miR-1 [64]. Overexpression of NAMPT leads to the increased expression of the TGF-β signaling pathway (Smad2-Smad3) and is associated with poor overall survival of CRC patients (Figure 3). The overexpression of miR-1 downregulated NAMPT and decreased the growth of CRC cells [64].
The in silico analysis of the CRC microarray dataset showed the downregulation of miR-1 in more than 90% of CRC cases [128]. Gene ontology analysis of CRC patients datasets showed positive results for transcription factor binding and RNA polymerase II-mediated regulations, and the hub gene analysis suggests miR-1 downregulation in the studied data sets of CRC [128]. Another interesting report has been shown that miR-1 inhibited the growth of CRC through VEGF downregulation [129]. The study results demonstrated that the miR-1 expression was downregulated in CRC tumor tissues and cell lines compared to normal tissue samples, and miR-1 expression is negatively associated with lymph node metastasis [129]. Overexpression of miR-1 inhibited the CRC cell migration and invasion, and miR-1 inhibited the expression and functional role of VEGF (Figure 3) [129]. This study reports VEGF as a direct target of miR-1 (as 3-prime UTR of VEGF consists of the miR-1 binding site) and provides the rationale of VEGF targeting through miR-1 for CRC therapies [129].
Two independent reports established the inhibitory role of miR-1 in aerobic glycolysis of CRC cells [130, 131]. Xu et al. using gain- or loss-of-function studies, established the role of miR-1 in glycolytic regulations of CRC [130]. Upregulation of miR-1 inhibited the aerobic glycolysis (Warburg effect) and lactate production in CRC cells through Smad3 and hypoxia-inducible factor 1 subunit alpha (HIF1α). MiR-1 targets HIF1α and decreases the expression of hexokinase 2 (HK2) and monocarboxylate transporter 4 (MCT4). Activated Smad3 interacts with HIF1α, however, miR-1 overexpression did not allow this interaction. It was demonstrated that miR-1 acts via Smad3/HIF1α axis in CRC and overexpression of miR-1 decreases the in vivo CRC growth through aerobic glycolysis modulation [130]. Similarly, Taniguchi et al. showed that downregulation of miR-1 induces the Warburg effect in CRC through pyruvate kinase in the muscle (PKM) and polypyrimidine tract-binding protein 1 (PTBP1) [131]. The overexpression of miR-1 induces class switching of PKM isoform from PKM2 to PKM1 by decreasing the expression of PTBP1 and reduces tumor growth and autophagy [131]. Overall, these studies showed that the downregulation of miR-1 in CRC has a diagnostic utilization, and overexpression of miR-1 is a potential therapeutic strategy for CRC patients.
2.4. Gastric cancer
Gastric cancer (GC) is also a leading cause of cancer-related deaths [69]. Due to the late diagnosis and high metastasis, few therapies are available for the treatment of GC. Multiple studies suggested that aberrant expression of miRNAs contributes towards the tumorigenesis of GC [132, 133]. Among various miRNA, the miR-1 expression has been found downregulated in GC [41, 134]. It was reported that low expression of miR-1 is associated with poor overall survival compared to higher expression in GC patients [134]. MiR-1 overexpression decreased the expression of VEGF and endothelin 1 in GC cells and inhibited cell proliferation and migration [134]. Another recent report showed that miR-1 inhibited the expression of VEGF, sorcin, and MMP-7 and decreases the invasion of GC cells [135]. Therefore, miR-1 regulates the angiogenic factor of GC and acts as a tumor suppressor.
The miR-1 expression also plays a role in developing drug resistance in GC cells [136]. It was shown that miR-1 is highly downregulated in resistant GC cells, and sorcin is upregulated [136]. In resistant GC cells, overexpression of miR-1 sensitizes the cells towards ADM/cisplatin and increases the apoptosis through sorcin targeting [136]. Stanniocalcin 2 (STC2) and MET proto-oncogene (MET) is the other target of miR-1 that regulates GC growth [45, 137]. Overexpression of miR-1 downregulated the expression of MET and STC2, decreases the GC cell migration and correlated with the tumor regression [45, 137]. Additionally, a recent report suggested the role of miR-1 in the metabolism of GC cells [66]. It has been shown that the expression of LINC00242 and glucose-6-phosphate dehydrogenase (G6PD) is very high in GC tissues and cell lines. Silencing of G6PD or LINC00242 inhibited the aerobic glycolysis of GC cells and upregulated the expression of miR-1. LINC00242 regulates the miR-1/G6PD axis in GC cells and provides a novel therapy target [66].
2.5. Liver cancer
Liver cancer or hepatocellular carcinoma (HCC) is one of the major cancers of the gastrointestinal tract. The tumor-suppressive role of miR-1 was demonstrated for the first time in liver cancer [138]. Epigenetic modifications like hypermethylation of CpG islands were also reported to downregulate the expression of miR-1 in HCC cell lines [138]. DNA methyltransferase 1 (DNMT1) regulated the methylation pattern of MIR-1 gene and showed reduced expression of miR-1 in HCC compared to matching control. Overexpression of miR-1 decreased the cell growth and clonogenic properties of HCC cells. It was shown that miR-1 performs antitumorigenic activity through FOXP1 and hepatocyte growth factor receptor (cMET) inhibition, and these molecules possess miR-1 binding sites in their 3′-UTR [138]. These observations encourage the combination of epigenetic drugs with miR-1 to treat HCC and other cancers where miR-1 expression is regulated through epigenetic pathways. Added to the antitumorigenic potential of miR-1 in HCC, one more recent study reported the downregulation of miR-1 in HCC cells compared to normal cells and suggested SOX9 as a target of miR-1 [139]. Overexpression of miR-1 inhibited HCC cell growth and decreased in vivo tumor growth through SOX9 inhibition [139].
Sorafenib (a multi tyrosine kinase inhibitor) is used as standard therapy for HCC, but its application was limited by therapy resistance [140]. It was reported that PD-L1 plays a vital role in drug resistance and immune evasion of various tumors [141, 142]. A recent study by Li et al. demonstrates the overexpression of PD-L1 in sorafenib-resistant HCC cells and depletion of PD-L1 suppressed the clonogenic potential, invasion, and drug resistance in vitro and in vivo studies [143]. Mechanistic studies showed a negative correlation between PD-L1 and miR-1 expression in sorafenib-resistant HCC cells. PD-L1 was identified as a direct target of miR-1 and showed that nuclear factor erythroid 2-related factor 2 (NRF2) expression is increased in resistance cell lines that downregulates miR-1 expression. Thus, NRF2/miR-1/PD-L1 axis contributes to the sorafenib resistance in HCC, and overexpression of miR-1 induces tumor inhibitory activities through PD-L1 downregulation [143]. This study suggests the implication of miR-1 in PD-L1 regulation and provided a novel therapeutic axis to deal with drug resistance, including immunotherapy.
Interestingly, apart from human beings, a recent report suggested the implications of miR-1 expression in canine HCC [144]. Canine HCC is a commonly diagnosed primary hepatic tumor in dogs [144, 145]. The outcomes of the study reported by Lai et al. showed that downregulation of miR-1 is associated with canine HCC, and identifies cMET as a target of miR-1 [144]. The comparison of high versus intermediate proliferating canine HCC cells showed higher cMET expression both at gene and protein level and was found directly associated with miR-1 expression [144]. Low miR-1 and high cMET conditions are associated with canine HCC cell proliferation and provided a rationale for the future exploration of the miR-1/cMET axis for therapy development.
2.6. Other cancers
Apart from the above-discussed cancers and targets, miR-1 also plays a tumor suppressive role in other cancers, including pancreatic, medulloblastoma, and prostate cancer with multiple targets as summarized in (Table 1) [146–148]. The downregulation of miR-1 also contributes to the development of pancreatic ductal adenocarcinoma (PDAC) [148]. The microarray analysis of PDAC tissues, in situ hybridization, and miRNA profiling of patient serum, suggested that miR-1 is downregulated in PDAC. The low miR-1 and high miR-214 expression are associated with poor survival of PDAC patients [148]. The outcomes of the study suggested that miR-1 plays an important role in carcinogenesis and, along with miR-214, stood as a prognosis biomarker for PDAC. In PCa, a significant downregulation of miR-1 was reported in recurrent tissues compared to non-recurrent ones and acts as a predictive marker for PCa recurrence [149]. High expression of MALAT1 and low expression of miR-1 were noticed in PCa cell lines and tumor tissues [149]. MALAT1 works like a sponge for miR-1, and knockdown of MALAT1 induces apoptosis in androgen negative PCa cells. Mechanistic studies showed that miR-1 targets KRAS in PCa cells, and MALAT1 increases KRAS expression by competing with miR-1 [149]. Upregulation of miR-1 in androgen negative PC cells inhibited the cell proliferation, migration, and induced apoptosis through KRAS targeting. Thus, the MALAT1/miR-1 axis acts as a potential therapeutic target for treating recurrent or castration-resistant PCa [149]. EGFR signaling modulates bone metastasis of multiple cancers, including PCa [147, 150]. Translocation of EGFR into nucleus downregulates miR-1 expression that promotes PCa bone metastasis. Additionally, miR-1 expression is negatively correlated with the expression of EGFR and twist family BHLH transcription factor 1 (TWIST1) in PCa tumor tissues [147]. The findings of this study demonstrated that EGFR works as a repressor for miR-1 transcription, whereas it activates TWIST1 that promotes PCa bone metastasis and restricts the antitumorigenic activity of miR-1 [147].
Table 1:
Cancer associated targets of miR-1 and related functional role.
| Name of target | Function | Type of Cancer | Reference |
|---|---|---|---|
|
| |||
| ATG3 | -chemosensitization -regulates autophagy -induces apoptosis |
NSCLC | [87] |
| CDC42 | -migration & invasion | BC | [118] |
| PD-1/PD-L1 | -immune suppression -drug resistance |
HCC, LC | [96, 143] |
| CCL2 | -suppressing the proliferation & invasion | Bladder cancer | [152] |
| VEGF | -fibroblast reprogramming -Th2 mediated inflammation -reduces tumor growth -decreases cell migration & invasion |
LC, CRC, GC | [97, 129, 134, 135, 171] |
| Bcl-2 | -decreases cell survival -chemosensitization -inhibited tumor growth -radioresistance |
BC, CRC | [116, 181, 182] |
| E-Cadherin | -metastasis -radioresistance |
BC, CRC, Cervical cancer | [116, 181, 183] |
| Slug | -cell growth & development -metastasis -EMT |
BC, LC | [117, 184] |
| EVI-1 | -inhibit cell proliferation -promote apoptosis -decreases EMT -reduced tumor growth |
BC | [185] |
| CXCL12 | -cell invasion -metastasis |
Thyroid carcinogenesis, BC, LC | [186, 187] |
| CXCR4 | -regulate SDF-1 biogenesis in cancer-associated fibroblasts -metastasis |
BC, LC | [187, 188] |
| EGFR | -angiogenesis -inhibit proliferation and invasion of tumor cells. |
HNSCC, Glioblastoma, Ovarian cancer, PCa | [153, 189, 190] |
| c-MET | -suppress growth & proliferation -decreases MCL1 |
Ovarian cancer, Osteosarcoma, HNSCC, PCa | [153, 191–194] |
| G6PD | -suppress cancer development and progression | Cervical cancer | [195] |
| CDK4 | -decreases tumor growth and metastasis -G1 cell-cycle arrest. |
GC, BC, clear cell renal cell carcinoma | [196, 197] |
| ANXA2 | -decreases cell growth and migration -reduces angiogenesis and invasion -G0/G1 cell cycle arrest -target stem cell features |
NSCLC, glioblastoma | [73, 198] |
| AKT | -inflammation -cell growth |
CRC, PCa | [194, 199] |
| PI3K | -inflammation -cell growth |
CRC | [199] |
| HDAC6, HNF4α | -decreases mucin expression -regulate bile reflexes and metaplasia |
Gastro-intestinal cancer/metaplasia | [200] |
The microarray analysis and integrated miRNA expression studies showed the downregulation of miR-1 in esophageal squamous cell carcinoma (ESCC) [151]. In situ hybridization and miRNA-mRNA network studies in ESCC cells and tumor tissues showed the downregulation of miR-1 is associated with different pathological parameters like lymph node metastasis, migration, invasion, and poor overall survival. Overexpression of miR-1 facilitates apoptosis and inhibits the invasion of ESCC cells through downregulation of fibronectin 1 (FN1), suggesting the implication of the miR-1/FN1 axis for therapy development [151]. Bladder cancer cells and tumor tissues showed low miR-1 and high C-C motif chemokine ligand 2 compared to normal tissues (CCL2) [152]. Induction of miR-1 reduced the cell proliferation and induced apoptosis through downregulation of CCL2. Since miR-1 has a binding site in 3’UTR of CCL2, suggesting that the anticancer activity of miR-1 in bladder cancer is associated with CCL2 targeting [152].
Downregulation of miR-1 has also been reported in the case of head and neck squamous cell carcinoma (HNSCC) tumor tissues [153]. The restoration of miR-1 expression decreases the aggressiveness of HNSCC cells. Combinatorial analysis of cancer pathways showed that miR-1 modulates ECM-receptor, FAK, and MAPK signaling pathways. Among different selected molecules, EGFR and cMET were presented as a direct target of miR-1 and their (EGFR/cMET) overexpression was reported in HNSCC tumor tissues [153]. MiR-1 overexpression decreases the expression of EGFR and cMET that further provides a preclinical rationale to target these oncogenic regulators through miR-1. HOX transcript antisense RNA (HOTAIR, a lncRNA) and Yin Yang 1 (YY1) are also involved in the tumorigenesis of medulloblastoma [146, 154]. Low expression of miR-1 and high expression of HOTAIR and YY1 were noticed in medulloblastoma cell lines and tumor tissues. It was shown that HOTAIR negatively regulates the miR-1 expression, and YY1 has a putative binding site for miR-1; thus, HOTAIR increases the expression of YY1. The knockdown of HOTAIR or overexpression of miR-1 decreased the tumor growth of medulloblastoma cell lines and reduced EMT markers (vimentin, fibronectin), and induces apoptosis [146]. Overall, the discussion of miR-1 associated cancer pathways and prominent cancer drug targets of multiple cancers strengthens our understanding of the consistent tumor suppressor nature of this miRNA. Future investigation may lead to a better understanding of miR-1 modulated targets that may augment the development of different therapeutic strategies for cancers.
3. MiR-1 and immunity
It is now well established that miRNAs play a key regulatory role in multiple physiological processes, including immune responses, and the number of miRNAs implicated in immunomodulation is continuously increasing [155, 156]. Local or innate immune response has emerged as an essential part of different aspects of cancer development, such as inflammation, metastasis, angiogenesis, modulation of TIME, and chemo-/immunotherapy response [157–159]. Tumors are not only infiltered by molecules of the innate or adaptive immune system, but the cells of both arms of the immune system constitute the tumor microenvironment [160–162]. Based on the fine-tuning of the innate and adaptive immune system, immunity can act as an anti- or pro-tumorigenic factor [162]. One such component that regulates the nature of the immune response is miRNAs, and among them, miR-1 has been known to regulate production and release of chemokines/cytokines, expression of costimulatory molecules, miRNA shuttling through exosomes, regulation of immune homeostasis, inflammation, and cancer immunotherapy response [56, 143, 163–170].
The expression status of miR-1 has been found to determine immunotherapy response as PD-L1 is a direct target of it [143]. It was demonstrated that sorafenib-resistance cancer cells have higher expression of PD-L1 and low miR-1 [143]. It means that low miR-1 is a predictive factor for immunotherapy response, and miR-1 combination therapies with standard chemotherapy or tyrosine kinase inhibitor could be novel approaches to treat such cancers. It was reported in LC that miR-1 expression determines the sensitivity of EGFR-TKI as it regulates the lymphocyte infiltration, monocyte migration, and the production of CCL-5/−10 chemokines [96]. Few studies are available that projected the role of miR-1 directly in tumor immunity, but multiple studies are available that showed that miR-1 modulates the immune-related pathways in various diseases like cardiac injuries and lung diseases [171, 172]. Interestingly similar pathways are found dysregulated in various cancers, thus putting forward the implications of miR-1 in tumor-immunity. On this line, we discussed some important immune regulation studies of miR-1.
Interestingly, miR-1 has been shown to modulate T-helper type 2 cell (Th2)-mediated inflammatory diseases like asthma [171]. VEGF is a key mediator of Th2 mediated inflammation in the lung and other diseases [173, 174]. It was shown that overexpression of VEGF in lung epithelium cells decreased the expression of miR-1, and this was recapitulated in animal models of Th2 inflammation. The external delivery of miR-1 through the intranasal route inhibited the inflammatory response to house dust, ovalbumin, and IL-13. VEGF inhibition blocks the Th2-mediated inflammatory response, and the response was restored through miR-1 inhibition. Pull-down assays of Argonaute showed that myeloproliferative leukemia protein (MPL) was a direct target of miR-1 [171]. During Th2 oriented inflammation, VEGF modulates the expression of MPL through miR-1. Knockdown of MPL leads to inhibit Th2-mediated inflammation, decreases mucin production, and expression of P-selectin [171]. Apart from these observations in Th2 animal models, similar outcomes were noticed in human primary endothelial cells (HUVECs), which suggested that miR-1 might play a crucial immunological role in different lung diseases, including LC [171]. The P-selectin is a vital adhesion molecule present on lung epithelium cells, and aberrant expression is associated with multiple malignancies, angiogenesis, and modulation of the CXCL12-CXCR4 axis [175].
P-selectin recruits T cells and eosinophils to lung epithelium and augments the inflammation through Th2 cytokines [176]. Additional studies are required to describe the functional relevance of miR-1, VEGF, MPL, and P-selectin in regulating Th2 response in inflammation-associated diseases, including cancer. A similar observation was reported recently by Korde et al. in asthma and chronic rhinosinusitis that the overexpression of miR-1 in lung endothelium decreased the trafficking of eosinophils or IL-13 oriented inflammation in murine models [172]. The miR-1 enrolled thrombopoietin receptor and P-selectin to miRNA RISC complex for degradation and decreased the expression of these genes in epithelial cells [172]. Jiang et al. recently reported the role of miR-1 in inflammation and atherosclerosis [177]. It was demonstrated that miR-1 regulates the activation of NF-kB that activates the expression of various proinflammatory cytokines and immune cells [177]. The demonstrated functional role of miR-1 and VEGF in asthmatic lung inflammation, mucin regulation, NF-kB regulation, and T-cell recruitment suggested the implications of the miR-1/VEGF axis in other diseases such as cancer, where these molecules play a crucial role in disease progression and therapy development.
4. Concluding remarks and future perspectives
Collecting outlines of multiple studies show the key developmental and tumor-suppressive roles of miR-1. The miR-1 can regulate the expression of various key regulatory targets at the post-transcriptional level that manifests the pathogenesis of various human disorders. The expression of miR-1 plays a particular role in tumor growth, therapy response, and immune response. The initial studies demonstrated the development specific role of miR-1 in skeleton muscle and cardiomyocytes, whereas recent studies suggested the prevalent role of miR-1 in inflammatory diseases, various cancers, and other pathological conditions [40, 41, 171, 172]. The functional diversity of miR-1 can be realized through its varied targets (Figure 3).
Several associated fields need detailed future investigations to continue the progress of miR-1 implications in cancer research and therapy development. One such field of miR-1 is a detailed study of tumor-associated immune functions that needs the improved implications of high-throughput RNA sequencing cross-linked with immunoprecipitation [178, 179]. In most cases, miR-1 or other miRNAs facilitate their functional activities at the protein level that make immune target identification particularly difficult through transcriptomic data. The development of better understanding and selective dispensation of miR-1 clusters in the immune system is highly desired. The promising efforts for in vivo delivery of miR-1 mimics to combat inflammation [171, 172] further opened the exciting options of miR-1 mediated therapeutic targeting of various malignancies. The therapeutic targeting of various cancers through miR-1 needs formulation optimization and development of delivery methods, that will open a window to utilize this molecule in therapy development.
Acknowledgments
We thank our colleagues for their valuable suggestions, critical reading, and useful comments on this review.Our apology to colleagues for not citing their work in this review owing to space limitations. Figures were created using BioRender.com. Lung cancer studies in our laboratory are supported by the National Institutes of Health (NIH) grants R01CA218545 and R01CA241752 to MWN. The work of SKB is supported by NIH R01CA247471, R01CA195586, and P01 CA217798. The work of RS is supported by the National Cancer Institute of the NIH under award numbers P30CA033572, U54CA209978, R01CA247471, and R01CA218545. Interpretations, opinions, and conclusions presented in this manuscript are those of the authors and does not necessarily represent the official views of the National Institutes of Health and other funding agencies.
Abbreviations
- EGFR
Epidermal growth factor receptor
- NSCLC
Non-small cell lung cancer
- SCLC
small cell lung cancer
- KRAS
Kirsten rat sarcoma viral oncogene homolog
- TK
Tyrosine kinases
- PARP
ADP-ribose polymerase
- STC2
Stanniocalcin 2
- G6PD
glucose-6-phosphate dehydrogenase
- ACT
Adoptive cell transfer
- VEGF
Vascular endothelial growth factor
- miRNAs
microRNAs
- siRNA
small interfering RNA
- RISC
RNA-induced silencing complex
- KLF4
Kruppel-like factor 4
- HSP60
heat shock protein 60
- HAND2
heart and neural crest derivatives expressed 2
- STC2
stanniocalcin 2
- TGF-β
transforming growth factor-beta
- MyoD
myoblast determination protein 1
- Mef2
myocyte enhancer factor-2
- cMET
hepatocyte growth factor receptor
- PIK3CA
phosphoinositide-3-kinase catalytic subunit alpha
- FAM83A
family with sequence similarity 83 member A
- MAPK
mitogen-activated protein kinase
- CHKA
choline kinase alpha
- FN1
Fibronectin 1
- lncRNAs
Long noncoding RNAs
- RMRP
RNA-processing endoribonuclease
- ANXA2
targets annexin-A2
- ATG3
autophagy related 3
- LC3B
light chain 3 beta
- PKM
pyruvate kinase in muscle
- PTBP1
polypyrimidine tract-binding protein 1
- NRF2
nuclear factor erythroid 2-related factor 2
- HIF1α
hypoxia inducible factor 1 subunit alpha
- HK2
hexokinase 2
- MCT4
monocarboxylate transporter 4
- SOX12
SRY-box transcription factor 12
- YY1
Yin Yang 1
- BRAF
B-Raf proto-oncogene serine/threonine kinase
- CRC
Colorectal cancer
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- CCL
C-C motif chemokine ligands
Footnotes
Conflict of interest statement
SKB is co-founder of Sanguine Diagnostics and Therapeutics, Inc. Other authors declare no competing interests.
Availability of data and materials
Not applicable, all information in this review can be found in the reference list.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sun K, Lai EC, Adult-specific functions of animal microRNAs, Nat Rev Genet 14(8) (2013) 535–48. 10.1038/nrg3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kloosterman WP, Plasterk RH, The diverse functions of microRNAs in animal development and disease, Dev Cell 11(4) (2006) 441–50. 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- [3].Ambros V, The functions of animal microRNAs, Nature 431(7006) (2004) 350–5. 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- [4].Annese T, Tamma R, De Giorgis M, Ribatti D, microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis, Front Oncol 10 (2020) 581007. 10.3389/fonc.2020.581007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Khan AW, Nuclear functions of microRNAs relevant to the cardiovascular system, Transl Res 230 (2021) 151–163. 10.1016/j.trsl.2020.11.004 [DOI] [PubMed] [Google Scholar]
- [6].Kim YK, Kim B, Kim VN, Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis, Proc Natl Acad Sci U S A 113(13) (2016) E1881–9. 10.1073/pnas.1602532113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN, The nuclear RNase III Drosha initiates microRNA processing, Nature 425(6956) (2003) 415–9. 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- [8].Arif KMT, Elliott EK, Haupt LM, Griffiths LR, Regulatory Mechanisms of Epigenetic miRNA Relationships in Human Cancer and Potential as Therapeutic Targets, Cancers (Basel) 12(10) (2020). 10.3390/cancers12102922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Liang G, Weisenberger DJ, DNA methylation aberrancies as a guide for surveillance and treatment of human cancers, Epigenetics 12(6) (2017) 416–432. 10.1080/15592294.2017.1311434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gerthoffer W, Epigenetic Targets for Oligonucleotide Therapies of Pulmonary Arterial Hypertension, Int J Mol Sci 21(23) (2020). 10.3390/ijms21239222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Farooqi AA, Fayyaz S, Poltronieri P, Calin G, Mallardo M, Epigenetic deregulation in cancer: Enzyme players and non-coding RNAs, Semin Cancer Biol (2020). 10.1016/j.semcancer.2020.07.013 [DOI] [PubMed] [Google Scholar]
- [12].DeVeale B, Swindlehurst-Chan J, Blelloch R, The roles of microRNAs in mouse development, Nat Rev Genet (2021). 10.1038/s41576-020-00309-5 [DOI] [PubMed] [Google Scholar]
- [13].Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ, Dicer is essential for mouse development, Nat Genet 35(3) (2003) 215–7. 10.1038/ng1253 [DOI] [PubMed] [Google Scholar]
- [14].Heyn GS, Correa LH, Magalhaes KG, The Impact of Adipose Tissue-Derived miRNAs in Metabolic Syndrome, Obesity, and Cancer, Front Endocrinol (Lausanne) 11 (2020) 563816. 10.3389/fendo.2020.563816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Szeto RA, Tran T, Truong J, Negraes PD, Trujillo CA, RNA processing in neurological tissue: development, aging and disease, Semin Cell Dev Biol (2020). 10.1016/j.semcdb.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Eliasson L, Esguerra JLS, MicroRNA Networks in Pancreatic Islet Cells: Normal Function and Type 2 Diabetes, Diabetes 69(5) (2020) 804–812. 10.2337/dbi19-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cao W, Stricker E, Hotz-Wagenblatt A, Heit-Mondrzyk A, Pougialis G, Hugo A, Kuzmak J, Materniak-Kornas M, Lochelt M, Functional Analyses of Bovine Foamy Virus-Encoded miRNAs Reveal the Importance of a Defined miRNA for Virus Replication and Host-Virus Interaction, Viruses 12(11) (2020). 10.3390/v12111250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bochnakian A, Zhen A, Zisoulis DG, Idica A, KewalRamani VN, Neel N, Daugaard I, Hamdorf M, Kitchen S, Lee K, Pedersen IM, Interferon-Inducible MicroRNA miR-128 Modulates HIV-1 Replication by Targeting TNPO3 mRNA, J Virol 93(20) (2019). 10.1128/JVI.00364-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].van Westering TLE, Lomonosova Y, Coenen-Stass AML, Betts CA, Bhomra A, Hulsker M, Clark LE, McClorey G, Aartsma-Rus A, van Putten M, Wood MJA, Roberts TC, Uniform sarcolemmal dystrophin expression is required to prevent extracellular microRNA release and improve dystrophic pathology, J Cachexia Sarcopenia Muscle 11(2) (2020) 578–593. 10.1002/jcsm.12506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bueno MJ, Malumbres M, MicroRNAs and the cell cycle, Biochim Biophys Acta 1812(5) (2011) 592–601. 10.1016/j.bbadis.2011.02.002 [DOI] [PubMed] [Google Scholar]
- [21].Bandi N, Vassella E, miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner, Mol Cancer 10 (2011) 55. 10.1186/1476-4598-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ortiz-Quintero B, Extracellular MicroRNAs as Intercellular Mediators and Noninvasive Biomarkers of Cancer, Cancers (Basel) 12(11) (2020). 10.3390/cancers12113455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Svoronos AA, Engelman DM, Slack FJ, OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer, Cancer Res 76(13) (2016) 3666–70. 10.1158/0008-5472.CAN-16-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Abolghasemi M, Tehrani SS, Yousefi T, Karimian A, Mahmoodpoor A, Ghamari A, Jadidi-Niaragh F, Yousefi M, Kafil HS, Bastami M, Edalati M, Eyvazi S, Naghizadeh M, Targhazeh N, Yousefi B, Safa A, Majidinia M, Rameshknia V, MicroRNAs in breast cancer: Roles, functions, and mechanism of actions, J Cell Physiol 235(6) (2020) 5008–5029. 10.1002/jcp.29396 [DOI] [PubMed] [Google Scholar]
- [25].Eniafe J, Jiang S, MicroRNA-99 family in cancer and immunity, Wiley Interdiscip Rev RNA (2020) e1635. 10.1002/wrna.1635 [DOI] [PubMed] [Google Scholar]
- [26].Zhang X, Chapat C, Wang P, Choi JH, Li Q, Luo J, Wiebe S, Kim SH, Robichaud N, Karam IF, Dai D, Hackett AP, Lin R, Alain T, Yang L, Jafarnejad SM, Sonenberg N, microRNA-induced translational control of antiviral immunity by the cap-binding protein 4EHP, Mol Cell (2021). 10.1016/j.molcel.2021.01.030 [DOI] [PubMed] [Google Scholar]
- [27].Vierbuchen T, Fitzgerald KA, Long non-coding RNAs in antiviral immunity, Semin Cell Dev Biol 111 (2021) 126–134. 10.1016/j.semcdb.2020.06.009 [DOI] [PubMed] [Google Scholar]
- [28].Mehta A, Baltimore D, MicroRNAs as regulatory elements in immune system logic, Nat Rev Immunol 16(5) (2016) 279–94. 10.1038/nri.2016.40 [DOI] [PubMed] [Google Scholar]
- [29].Chen Y, Zhao Y, Jin W, Li Y, Zhang Y, Ma X, Sun G, Han R, Tian Y, Li H, Kang X, Li G, MicroRNAs and their regulatory networks in Chinese Gushi chicken abdominal adipose tissue during postnatal late development, BMC Genomics 20(1) (2019) 778. 10.1186/s12864-019-6094-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bhowmick SS, Bhattacharjee D, Rato L, Integrated analysis of the miRNA-mRNA next-generation sequencing data for finding their associations in different cancer types, Comput Biol Chem 84 (2020) 107152. 10.1016/j.compbiolchem.2019.107152 [DOI] [PubMed] [Google Scholar]
- [31].Wyler E, Mosbauer K, Franke V, Diag A, Gottula LT, Arsie R, Klironomos F, Koppstein D, Honzke K, Ayoub S, Buccitelli C, Hoffmann K, Richter A, Legnini I, Ivanov A, Mari T, Del Giudice S, Papies J, Praktiknjo S, Meyer TF, Muller MA, Niemeyer D, Hocke A, Selbach M, Akalin A, Rajewsky N, Drosten C, Landthaler M, Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy, iScience 24(3) (2021) 102151. 10.1016/j.isci.2021.102151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ricafrente A, Nguyen H, Tran N, Donnelly S, An Evaluation of the Fasciola hepatica miRnome Predicts a Targeted Regulation of Mammalian Innate Immune Responses, Front Immunol 11 (2020) 608686. 10.3389/fimmu.2020.608686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pagotto S, Colorito ML, Nicotra A, Apuzzo T, Tinari N, Protasi F, Stassi G, Visone R, Di Franco S, Veronese A, A perspective analysis: microRNAs, glucose metabolism, and drug resistance in colon cancer stem cells, Cancer Gene Ther (2021). 10.1038/s41417-021-00298-5 [DOI] [PubMed] [Google Scholar]
- [34].Esquela-Kerscher A, Slack FJ, Oncomirs - microRNAs with a role in cancer, Nat Rev Cancer 6(4) (2006) 259–69. 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- [35].Kai K, Dittmar RL, Sen S, Secretory microRNAs as biomarkers of cancer, Semin Cell Dev Biol 78 (2018) 22–36. 10.1016/j.semcdb.2017.12.011 [DOI] [PubMed] [Google Scholar]
- [36].Huang H, Weng H, Chen J, m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer, Cancer Cell 37(3) (2020) 270–288. 10.1016/j.ccell.2020.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Garzon R, Marcucci G, Croce CM, Targeting microRNAs in cancer: rationale, strategies and challenges, Nat Rev Drug Discov 9(10) (2010) 775–89. 10.1038/nrd3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Toden S, Zumwalt TJ, Goel A, Non-coding RNAs and potential therapeutic targeting in cancer, Biochim Biophys Acta Rev Cancer 1875(1) (2021) 188491. 10.1016/j.bbcan.2020.188491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kozomara A, Birgaoanu M, Griffiths-Jones S, miRBase: from microRNA sequences to function, Nucleic Acids Res 47(D1) (2019) D155–D162. 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhao Y, Samal E, Srivastava D, Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis, Nature 436(7048) (2005) 214–20. 10.1038/nature03817 [DOI] [PubMed] [Google Scholar]
- [41].Safa A, Bahroudi Z, Shoorei H, Majidpoor J, Abak A, Taheri M, Ghafouri-Fard S, miR-1: A comprehensive review of its role in normal development and diverse disorders, Biomed Pharmacother 132 (2020) 110903. 10.1016/j.biopha.2020.110903 [DOI] [PubMed] [Google Scholar]
- [42].Shan ZX, Lin QX, Deng CY, Zhu JN, Mai LP, Liu JL, Fu YH, Liu XY, Li YX, Zhang YY, Lin SG, Yu XY, miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes, FEBS Lett 584(16) (2010) 3592–600. 10.1016/j.febslet.2010.07.027 [DOI] [PubMed] [Google Scholar]
- [43].Xie C, Huang H, Sun X, Guo Y, Hamblin M, Ritchie RP, Garcia-Barrio MT, Zhang J, Chen YE, MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4, Stem Cells Dev 20(2) (2011) 205–10. 10.1089/scd.2010.0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Connolly M, Garfield BE, Crosby A, Morrell NW, Wort SJ, Kemp PR, miR-1–5p targets TGF-betaR1 and is suppressed in the hypertrophying hearts of rats with pulmonary arterial hypertension, PLoS One 15(2) (2020) e0229409. 10.1371/journal.pone.0229409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ke J, Zhang BH, Li YY, Zhong M, Ma W, Xue H, Wen YD, Cai YD, MiR-1–3p suppresses cell proliferation and invasion and targets STC2 in gastric cancer, Eur Rev Med Pharmacol Sci 23(20) (2019) 8870–8877. 10.26355/eurrev_201910_19282 [DOI] [PubMed] [Google Scholar]
- [46].Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ, The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation, Nat Genet 38(2) (2006) 228–33. 10.1038/ng1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mishima Y, Stahlhut C, Giraldez AJ, miR-1–2 gets to the heart of the matter, Cell 129(2) (2007) 247–9. 10.1016/j.cell.2007.04.008 [DOI] [PubMed] [Google Scholar]
- [48].Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D, Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2, Cell 129(2) (2007) 303–17. 10.1016/j.cell.2007.03.030 [DOI] [PubMed] [Google Scholar]
- [49].Gagan J, Dey BK, Layer R, Yan Z, Dutta A, Notch3 and Mef2c proteins are mutually antagonistic via Mkp1 protein and miR-1/206 microRNAs in differentiating myoblasts, J Biol Chem 287(48) (2012) 40360–70. 10.1074/jbc.M112.378414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shrivastava A, Haase T, Zeller T, Schulte C, Biomarkers for Heart Failure Prognosis: Proteins, Genetic Scores and Non-coding RNAs, Front Cardiovasc Med 7 (2020) 601364. 10.3389/fcvm.2020.601364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Canas JA, Rodrigo-Munoz JM, Sastre B, Gil-Martinez M, Redondo N, Del Pozo V, MicroRNAs as Potential Regulators of Immune Response Networks in Asthma and Chronic Obstructive Pulmonary Disease, Front Immunol 11 (2020) 608666. 10.3389/fimmu.2020.608666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lewis A, Riddoch-Contreras J, Natanek SA, Donaldson A, Man WD, Moxham J, Hopkinson NS, Polkey MI, Kemp PR, Downregulation of the serum response factor/miR-1 axis in the quadriceps of patients with COPD, Thorax 67(1) (2012) 26–34. 10.1136/thoraxjnl-2011-200309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Moreira-Costa L, Barros AS, Lourenco AP, Leite-Moreira AF, Nogueira-Ferreira R, Thongboonkerd V, Vitorino R, Exosome-Derived Mediators as Potential Biomarkers for Cardiovascular Diseases: A Network Approach, Proteomes 9(1) (2021). 10.3390/proteomes9010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shirazi-Tehrani E, Firouzabadi N, Tamaddon G, Bahramali E, Vafadar A, Carvedilol Alters Circulating MiR-1 and MiR-214 in Heart Failure, Pharmgenomics Pers Med 13 (2020) 375–383. 10.2147/PGPM.S263740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Han C, Yu Z, Duan Z, Kan Q, Role of microRNA-1 in human cancer and its therapeutic potentials, Biomed Res Int 2014 (2014) 428371. 10.1155/2014/428371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Xu H, Jiang Y, Xu X, Su X, Liu Y, Ma Y, Zhao Y, Shen Z, Huang B, Cao X, Inducible degradation of lncRNA Sros1 promotes IFN-gamma-mediated activation of innate immune responses by stabilizing Stat1 mRNA, Nat Immunol 20(12) (2019) 1621–1630. 10.1038/s41590-019-0542-7 [DOI] [PubMed] [Google Scholar]
- [57].Hydbring P, Wang Y, Fassl A, Li X, Matia V, Otto T, Choi YJ, Sweeney KE, Suski JM, Yin H, Bogorad RL, Goel S, Yuzugullu H, Kauffman KJ, Yang J, Jin C, Li Y, Floris D, Swanson R, Ng K, Sicinska E, Anders L, Zhao JJ, Polyak K, Anderson DG, Li C, Sicinski P, Cell-Cycle-Targeting MicroRNAs as Therapeutic Tools against Refractory Cancers, Cancer Cell 31(4) (2017) 576–590 e8. 10.1016/j.ccell.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jiang Q, Isquith J, Zipeto MA, Diep RH, Pham J, Delos Santos N, Reynoso E, Chau J, Leu H, Lazzari E, Melese E, Ma W, Fang R, Minden M, Morris S, Ren B, Pineda G, Holm F, Jamieson C, Hyper-Editing of Cell-Cycle Regulatory and Tumor Suppressor RNA Promotes Malignant Progenitor Propagation, Cancer Cell 35(1) (2019) 81–94 e7. 10.1016/j.ccell.2018.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ma B, Li H, Qiao J, Meng T, Yu R, Immune-related miRNA signature identifies prognosis and immune landscape in head and neck squamous cell carcinomas, Biosci Rep 40(11) (2020). 10.1042/BSR20201820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Singh PK, Brand RE, Mehla K, MicroRNAs in pancreatic cancer metabolism, Nat Rev Gastroenterol Hepatol 9(6) (2012) 334–44. 10.1038/nrgastro.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chen D, Yang X, Liu M, Zhang Z, Xing E, Roles of miRNA dysregulation in the pathogenesis of multiple myeloma, Cancer Gene Ther (2021). 10.1038/s41417-020-00291-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Di Leva G, Garofalo M, Croce CM, MicroRNAs in cancer, Annu Rev Pathol 9 (2014) 287–314. 10.1146/annurev-pathol-012513-104715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hazini A, Dieringer B, Pryshliak M, Knoch KP, Heimann L, Tolksdorf B, Pappritz K, El-Shafeey M, Solimena M, Beling A, Kurreck J, Klingel K, Fechner H, miR-375- and miR-1-Regulated Coxsackievirus B3 Has No Pancreas and Heart Toxicity But Strong Antitumor Efficiency in Colorectal Carcinomas, Hum Gene Ther 32(3–4) (2021) 216–230. 10.1089/hum.2020.228 [DOI] [PubMed] [Google Scholar]
- [64].Lv X, Zhang J, Zhang J, Guan W, Ren W, Liu Y, Xu G, A Negative Feedback Loop Between NAMPT and TGF-beta Signaling Pathway in Colorectal Cancer Cells, Onco Targets Ther 14 (2021) 187–198. 10.2147/OTT.S282367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yan LR, Wang A, Lv Z, Yuan Y, Xu Q, Mitochondria-related core genes and TF-miRNA-hub mrDEGs network in breast cancer, Biosci Rep 41(1) (2021). 10.1042/BSR20203481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Deng P, Li K, Gu F, Zhang T, Zhao W, Sun M, Hou B, LINC00242/miR-1–3p/G6PD axis regulates Warburg effect and affects gastric cancer proliferation and apoptosis, Mol Med 27(1) (2021) 9. 10.1186/s10020-020-00259-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C, The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation, J Clin Invest 119(8) (2009) 2366–78. 10.1172/JCI38075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Khan P, Siddiqui JA, Maurya SK, Lakshmanan I, Jain M, Ganti AK, Salgia R, Batra SK, Nasser MW, Epigenetic landscape of small cell lung cancer: small image of a giant recalcitrant disease, Semin Cancer Biol (2020). 10.1016/j.semcancer.2020.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2020, CA Cancer J Clin 70(1) (2020) 7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- [70].Liu PJ, Chen YH, Tsai KW, Yeah HY, Yeh CY, Tu YT, Yang CY, Involvement of MicroRNA-1-FAM83A Axis Dysfunction in the Growth and Motility of Lung Cancer Cells, Int J Mol Sci 21(22) (2020). 10.3390/ijms21228833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jin X, Guan Y, Zhang Z, Wang H, Microarray data analysis on gene and miRNA expression to identify biomarkers in non-small cell lung cancer, BMC Cancer 20(1) (2020) 329. 10.1186/s12885-020-06829-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sheervalilou R, Lotfi H, Shirvaliloo M, Sharifi A, Nazemiyeh M, Zarghami N, Circulating MiR-10b, MiR-1 and MiR-30a Expression Profiles in Lung Cancer: Possible Correlation with Clinico-pathologic Characteristics and Lung Cancer Detection, Int J Mol Cell Med 8(2) (2019) 118–129. 10.22088/IJMCM.BUMS.8.2.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wang Y, Luo X, Liu Y, Han G, Sun D, Long noncoding RNA RMRP promotes proliferation and invasion via targeting miR-1–3p in non-small-cell lung cancer, J Cell Biochem 120(9) (2019) 15170–15181. 10.1002/jcb.28779 [DOI] [PubMed] [Google Scholar]
- [74].Melkamu T, Zhang X, Tan J, Zeng Y, Kassie F, Alteration of microRNA expression in vinyl carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol, Carcinogenesis 31(2) (2010) 252–8. 10.1093/carcin/bgp208 [DOI] [PubMed] [Google Scholar]
- [75].Yu QQ, Wu H, Huang X, Shen H, Shu YQ, Zhang B, Xiang CC, Yu SM, Guo RH, Chen L, MiR-1 targets PIK3CA and inhibits tumorigenic properties of A549 cells, Biomed Pharmacother 68(2) (2014) 155–61. 10.1016/j.biopha.2014.01.005 [DOI] [PubMed] [Google Scholar]
- [76].Zhao Q, Zhang B, Shao Y, Chen L, Wang X, Zhang Z, Shu Y, Guo R, Correlation between the expression levels of miR-1 and PIK3CA in non-small-cell lung cancer and their relationship with clinical characteristics and prognosis, Future Oncol 10(1) (2014) 49–57. 10.2217/fon.13.242 [DOI] [PubMed] [Google Scholar]
- [77].Singh A, Happel C, Manna SK, Acquaah-Mensah G, Carrerero J, Kumar S, Nasipuri P, Krausz KW, Wakabayashi N, Dewi R, Boros LG, Gonzalez FJ, Gabrielson E, Wong KK, Girnun G, Biswal S, Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis, J Clin Invest 123(7) (2013) 2921–34. 10.1172/JCI66353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wang J, Yu XF, Ouyang N, Zhao S, Yao H, Guan X, Tong J, Chen T, Li JX, MicroRNA and mRNA Interaction Network Regulates the Malignant Transformation of Human Bronchial Epithelial Cells Induced by Cigarette Smoke, Front Oncol 9 (2019) 1029. 10.3389/fonc.2019.01029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Li X, Qin M, Huang J, Ma J, Hu X, Clinical significance of miRNA1 and its potential target gene network in lung squamous cell carcinoma, Mol Med Rep 19(6) (2019) 5063–5078. 10.3892/mmr.2019.10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yang S, Wang Y, Lin Y, Shao D, He K, Huang L, LncMirNet: Predicting LncRNA-miRNA Interaction Based on Deep Learning of Ribonucleic Acid Sequences, Molecules 25(19) (2020). 10.3390/molecules25194372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wang X, Yin H, Zhang L, Zheng D, Yang Y, Zhang J, Jiang H, Ling X, Xin Y, Liang H, Fang C, Ma J, Zhu J, The construction and analysis of the aberrant lncRNA-miRNA-mRNA network in non-small cell lung cancer, J Thorac Dis 11(5) (2019) 1772–1778. 10.21037/jtd.2019.05.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Yang M, Ke H, Zhou W, LncRNA RMRP Promotes Cell Proliferation and Invasion Through miR-613/NFAT5 Axis in Non-Small Cell Lung Cancer, Onco Targets Ther 13 (2020) 8941–8950. 10.2147/OTT.S255126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Leng Q, Lin Y, Zhan M, Jiang F, An integromic signature for lung cancer early detection, Oncotarget 9(37) (2018) 24684–24692. 10.18632/oncotarget.25227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA, MicroRNAome genome: a treasure for cancer diagnosis and therapy, CA Cancer J Clin 64(5) (2014) 311–36. 10.3322/caac.21244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Si W, Shen J, Zheng H, Fan W, The role and mechanisms of action of microRNAs in cancer drug resistance, Clin Epigenetics 11(1) (2019) 25. 10.1186/s13148-018-0587-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Fadejeva I, Olschewski H, Hrzenjak A, MicroRNAs as regulators of cisplatin-resistance in non-small cell lung carcinomas, Oncotarget 8(70) (2017) 115754–115773. 10.18632/oncotarget.22975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hua L, Zhu G, Wei J, MicroRNA-1 overexpression increases chemosensitivity of non-small cell lung cancer cells by inhibiting autophagy related 3-mediated autophagy, Cell Biol Int 42(9) (2018) 1240–1249. 10.1002/cbin.10995 [DOI] [PubMed] [Google Scholar]
- [88].Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E, Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p, J Biol Chem 277(16) (2002) 13739–44. 10.1074/jbc.M200385200 [DOI] [PubMed] [Google Scholar]
- [89].Gaudet P, Livstone MS, Lewis SE, Thomas PD, Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium, Brief Bioinform 12(5) (2011) 449–62. 10.1093/bib/bbr042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Yamada Y, Suzuki NN, Hanada T, Ichimura Y, Kumeta H, Fujioka Y, Ohsumi Y, Inagaki F, The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation, J Biol Chem 282(11) (2007) 8036–43. 10.1074/jbc.M611473200 [DOI] [PubMed] [Google Scholar]
- [91].Oxnard GR, Binder A, Janne PA, New targetable oncogenes in non-small-cell lung cancer, J Clin Oncol 31(8) (2013) 1097–104. 10.1200/JCO.2012.42.9829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cagle PT, Raparia K, Portier BP, Emerging Biomarkers in Personalized Therapy of Lung Cancer, Adv Exp Med Biol 890 (2016) 25–36. 10.1007/978-3-319-24932-2_2 [DOI] [PubMed] [Google Scholar]
- [93].Akamatsu H, Ninomiya K, Kenmotsu H, Morise M, Daga H, Goto Y, Kozuki T, Miura S, Sasaki T, Tamiya A, Teraoka S, Tsubata Y, Yoshioka H, Hattori Y, Imamura CK, Katsuya Y, Matsui R, Minegishi Y, Mizugaki H, Nosaki K, Okuma Y, Sakamoto S, Sone T, Tanaka K, Umemura S, Yamanaka T, Amano S, Hasegawa K, Morita S, Nakajima K, Maemondo M, Seto T, Yamamoto N, The Japanese Lung Cancer Society Guideline for non-small cell lung cancer, stage IV, Int J Clin Oncol 24(7) (2019) 731–770. 10.1007/s10147-019-01431-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Anagnostou VK, Brahmer JR, Cancer immunotherapy: a future paradigm shift in the treatment of non–small cell lung cancer, Clinical Cancer Research 21(5) (2015) 976–984. [DOI] [PubMed] [Google Scholar]
- [95].Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D, Ready N, Gainor J, Aren Frontera O, Havel L, Steins M, Garassino MC, Aerts JG, Domine M, Paz-Ares L, Reck M, Baudelet C, Harbison CT, Lestini B, Spigel DR, Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer, N Engl J Med 373(2) (2015) 123–35. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kawana S, Saito R, Miki Y, Kimura Y, Abe J, Sato I, Endo M, Sugawara S, Sasano H, Suppression of tumor immune microenvironment via microRNA-1 after epidermal growth factor receptor-tyrosine kinase inhibitor resistance acquirement in lung adenocarcinoma, Cancer Med 10(2) (2021) 718–727. 10.1002/cam4.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Shen H, Yu X, Yang F, Zhang Z, Shen J, Sun J, Choksi S, Jitkaew S, Shu Y, Reprogramming of Normal Fibroblasts into Cancer-Associated Fibroblasts by miRNAs-Mediated CCL2/VEGFA Signaling, PLoS Genet 12(8) (2016) e1006244. 10.1371/journal.pgen.1006244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Gong Y, Liu YR, Ji P, Hu X, Shao ZM, Impact of molecular subtypes on metastatic breast cancer patients: a SEER population-based study, Sci Rep 7 (2017) 45411. 10.1038/srep45411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hoeferlin LA, Chalfant CE, Park MA, Challenges in the treatment of triple negative and HER2-overexpressing breast cancer, Journal of surgery and science 1(1) (2013) 3. [PMC free article] [PubMed] [Google Scholar]
- [100].Kanchan RK, Siddiqui JA, Mahapatra S, Batra SK, Nasser MW, microRNAs Orchestrate Pathophysiology of Breast Cancer Brain Metastasis: Advances in Therapy, Mol Cancer 19(1) (2020) 29. 10.1186/s12943-020-1140-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Jafri MA, Al-Qahtani MH, Shay JW, Role of miRNAs in human cancer metastasis: Implications for therapeutic intervention, Semin Cancer Biol 44 (2017) 117–131. 10.1016/j.semcancer.2017.02.004 [DOI] [PubMed] [Google Scholar]
- [102].Yang L, Cai N, Zhao L, MicroRNA-1 regulates the growth and chemosensitivity of breast cancer cells by targeting MEK/ERK pathway, J BUON 25(5) (2020) 2215–2220. [PubMed] [Google Scholar]
- [103].Vyas S, Zaganjor E, Haigis MC, Mitochondria and Cancer, Cell 166(3) (2016) 555–566. 10.1016/j.cell.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Ma Y, Bai RK, Trieu R, Wong LJ, Mitochondrial dysfunction in human breast cancer cells and their transmitochondrial cybrids, Biochim Biophys Acta 1797(1) (2010) 29–37. 10.1016/j.bbabio.2009.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Seyfried TN, Arismendi-Morillo G, Mukherjee P, Chinopoulos C, On the Origin of ATP Synthesis in Cancer, iScience 23(11) (2020) 101761. 10.1016/j.isci.2020.101761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].López-Sendón J, Álvarez-Ortega C, Zamora Auñon P, Buño Soto A, Lyon AR, Farmakis D, Cardinale D, Canales Albendea M, Feliu Batlle J, Rodríguez Rodríguez I, Classification, prevalence, and outcomes of anticancer therapy-induced cardiotoxicity: the CARDIOTOX registry, European heart journal 41(18) (2020) 1720–1729. [DOI] [PubMed] [Google Scholar]
- [107].Abdel-Qadir H, Austin PC, Lee DS, Amir E, Tu JV, Thavendiranathan P, Fung K, Anderson GM, A population-based study of cardiovascular mortality following early-stage breast cancer, JAMA cardiology 2(1) (2017) 88–93. [DOI] [PubMed] [Google Scholar]
- [108].Alexandre J, Cautela J, Ederhy S, Damaj GL, Salem JE, Barlesi F, Farnault L, Charbonnier A, Mirabel M, Champiat S, Cardiovascular toxicity related to cancer treatment: a pragmatic approach to the American and European cardio- oncology guidelines, Journal of the American Heart Association 9(18) (2020) e018403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gioffre S, Chiesa M, Cardinale DM, Ricci V, Vavassori C, Cipolla CM, Masson S, Sandri MT, Salvatici M, Ciceri F, Latini R, Staszewsky LI, Pompilio G, Colombo GI, D’Alessandra Y, Circulating MicroRNAs as Potential Predictors of Anthracycline-Induced Troponin Elevation in Breast Cancer Patients: Diverging Effects of Doxorubicin and Epirubicin, J Clin Med 9(5) (2020). 10.3390/jcm9051418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Pinchi E, Frati P, Aromatario M, Cipolloni L, Fabbri M, La Russa R, Maiese A, Neri M, Santurro A, Scopetti M, miR- 1, miR- 499 and miR- 208 are sensitive markers to diagnose sudden death due to early acute myocardial infarction, Journal of cellular and molecular medicine 23(9) (2019) 6005–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Pereira JD, Tosatti JAG, Simoes R, Luizon MR, Gomes KB, Alves MT, microRNAs associated to anthracycline-induced cardiotoxicity in women with breast cancer: A systematic review and pathway analysis, Biomed Pharmacother 131 (2020) 110709. 10.1016/j.biopha.2020.110709 [DOI] [PubMed] [Google Scholar]
- [112].Eom YH, Kim HS, Lee A, Song BJ, Chae BJ, BCL2 as a Subtype-Specific Prognostic Marker for Breast Cancer, J Breast Cancer 19(3) (2016) 252–260. 10.4048/jbc.2016.19.3.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Honma N, Horii R, Ito Y, Saji S, Younes M, Iwase T, Akiyama F, Differences in clinical importance of Bcl-2 in breast cancer according to hormone receptors status or adjuvant endocrine therapy, BMC Cancer 15 (2015) 698. 10.1186/s12885-015-1686-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Vaillant F, Merino D, Lee L, Breslin K, Pal B, Ritchie ME, Smyth GK, Christie M, Phillipson LJ, Burns CJ, Mann GB, Visvader JE, Lindeman GJ, Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer, Cancer Cell 24(1) (2013) 120–9. 10.1016/j.ccr.2013.06.002 [DOI] [PubMed] [Google Scholar]
- [115].Oakes SR, Vaillant F, Lim E, Lee L, Breslin K, Feleppa F, Deb S, Ritchie ME, Takano E, Ward T, Fox SB, Generali D, Smyth GK, Strasser A, Huang DC, Visvader JE, Lindeman GJ, Sensitization of BCL-2-expressing breast tumors to chemotherapy by the BH3 mimetic ABT-737, Proc Natl Acad Sci U S A 109(8) (2012) 2766–71. 10.1073/pnas.1104778108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Peng J, Yuan C, Wu Z, Wang Y, Yin W, Lin Y, Zhou L, Lu J, Upregulation of microRNA1 inhibits proliferation and metastasis of breast cancer, Mol Med Rep 22(1) (2020) 454–464. 10.3892/mmr.2020.11111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Jin C, Yan B, Lu Q, Lin Y, Ma L, Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development, Tumour Biol 37(6) (2016) 7383–94. 10.1007/s13277-015-4605-6 [DOI] [PubMed] [Google Scholar]
- [118].Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, Zhang Y, Xing Y, Xi T, MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42, Biochem Biophys Res Commun 472(1) (2016) 262–9. 10.1016/j.bbrc.2016.02.102 [DOI] [PubMed] [Google Scholar]
- [119].Martini G, Troiani T, Cardone C, Vitiello P, Sforza V, Ciardiello D, Napolitano S, Della Corte CM, Morgillo F, Raucci A, Cuomo A, Selvaggi F, Ciardiello F, Martinelli E, Present and future of metastatic colorectal cancer treatment: A review of new candidate targets, World J Gastroenterol 23(26) (2017) 4675–4688. 10.3748/wjg.v23.i26.4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Li J, Zheng Y, Li X, Dong X, Chen W, Guan Z, Zhang C, UCHL3 promotes proliferation of colorectal cancer cells by regulating SOX12 via AKT/mTOR signaling pathway, Am J Transl Res 12(10) (2020) 6445–6454. [PMC free article] [PubMed] [Google Scholar]
- [121].Du F, Chen J, Liu H, Cai Y, Cao T, Han W, Yi X, Qian M, Tian D, Nie Y, Wu K, Fan D, Xia L, SOX12 promotes colorectal cancer cell proliferation and metastasis by regulating asparagine synthesis, Cell Death Dis 10(3) (2019) 239. 10.1038/s41419-019-1481-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Urh K, Zlajpah M, Zidar N, Bostjancic E, Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis, Biomedicines 9(2) (2021). 10.3390/biomedicines9020179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Dias F, Almeida C, Teixeira AL, Morais M, Medeiros R, LAT1 and ASCT2 Related microRNAs as Potential New Therapeutic Agents against Colorectal Cancer Progression, Biomedicines 9(2) (2021). 10.3390/biomedicines9020195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Huang J, Liu H, Zhao Y, Luo T, Liu J, Liu J, Pan X, Tang W, MicroRNAs Expression Patterns Predict Tumor Mutational Burden in Colorectal Cancer, Front Oncol 10 (2020) 550986. 10.3389/fonc.2020.550986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Xu B, Shen X, Yang Z, Zhao T, Liu B, Gao S, Shi Y, Bao C, Plasma miR-1, but not Extracellular Vesicle miR-1, Functions as a Potential Biomarker for Colorectal Cancer Diagnosis, Clin Lab 67(1) (2021). 10.7754/Clin.Lab.2020.200344 [DOI] [PubMed] [Google Scholar]
- [126].Gholinejad Z, Kheiripour N, Nourbakhsh M, Ilbeigi D, Behroozfar K, Hesari Z, Golestani A, Shabani M, Einollahi N, Extracellular NAMPT/Visfatin induces proliferation through ERK1/2 and AKT and inhibits apoptosis in breast cancer cells, Peptides 92 (2017) 9–15. 10.1016/j.peptides.2017.04.007 [DOI] [PubMed] [Google Scholar]
- [127].Guo Q, Han N, Shi L, Yang L, Zhang X, Zhou Y, Yu S, Zhang M, NAMPT: A potential prognostic and therapeutic biomarker in patients with glioblastoma, Oncol Rep 42(3) (2019) 963–972. 10.3892/or.2019.7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Wang JY, Huang JC, Chen G, Wei DM, Expression level and potential target pathways of miR-1–3p in colorectal carcinoma based on 645 cases from 9 microarray datasets, Mol Med Rep 17(4) (2018) 5013–5020. 10.3892/mmr.2018.8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Zhu D, Sun Y, Zhang D, Dong M, Jiang G, Zhang X, Zhou J, miR1 inhibits the progression of colon cancer by regulating the expression of vascular endothelial growth factor, Oncol Rep 40(2) (2018) 589–598. 10.3892/or.2018.6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Xu W, Zhang Z, Zou K, Cheng Y, Yang M, Chen H, Wang H, Zhao J, Chen P, He L, Chen X, Geng L, Gong S, MiR-1 suppresses tumor cell proliferation in colorectal cancer by inhibition of Smad3-mediated tumor glycolysis, Cell Death Dis 8(5) (2017) e2761. 10.1038/cddis.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Taniguchi K, Sakai M, Sugito N, Kumazaki M, Shinohara H, Yamada N, Nakayama T, Ueda H, Nakagawa Y, Ito Y, Futamura M, Uno B, Otsuki Y, Yoshida K, Uchiyama K, Akao Y, PTBP1-associated microRNA-1 and −133b suppress the Warburg effect in colorectal tumors, Oncotarget 7(14) (2016) 18940–52. 10.18632/oncotarget.8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Peng Y, Croce CM, The role of MicroRNAs in human cancer, Signal Transduct Target Ther 1 (2016) 15004. 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Hwang J, Min BH, Jang J, Kang SY, Bae H, Jang SS, Kim JI, Kim KM , MicroRNA Expression Profiles in Gastric Carcinogenesis, Sci Rep 8(1) (2018) 14393. 10.1038/s41598-018-32782-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Xie M, Dart DA, Guo T, Xing XF, Cheng XJ, Du H, Jiang WG, Wen XZ, Ji JF, MicroRNA-1 acts as a tumor suppressor microRNA by inhibiting angiogenesis-related growth factors in human gastric cancer, Gastric Cancer 21(1) (2018) 41–54. 10.1007/s10120-017-0721-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Lin XQ, Wu W, Chen X, Chen RP, Wu F, Chen ZF, Huang EJ, Chen C, mir-1 inhibits migration of gastric cancer cells, Front Biosci (Landmark Ed) 25 (2020) 452–462. [DOI] [PubMed] [Google Scholar]
- [136].Deng LM, Tan T, Zhang TY, Xiao XF, Gu H, miR1 reverses multidrug resistance in gastric cancer cells via downregulation of sorcin through promoting the accumulation of intracellular drugs and apoptosis of cells, Int J Oncol 55(2) (2019) 451–461. 10.3892/ijo.2019.4831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Han C, Zhou Y, An Q, Li F, Li D, Zhang X, Yu Z, Zheng L, Duan Z, Kan Q, MicroRNA-1 (miR-1) inhibits gastric cancer cell proliferation and migration by targeting MET, Tumour Biol 36(9) (2015) 6715–23. 10.1007/s13277-015-3358-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, Ghoshal K, Jacob ST, Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis, Cancer Res 68(13) (2008) 5049–58. 10.1158/0008-5472.CAN-07-6655 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [139].Zhang H, Zhang Z, Gao L, Qiao Z, Yu M, Yu B, Yang T, miR-1–3p suppresses proliferation of hepatocellular carcinoma through targeting SOX9, Onco Targets Ther 12 (2019) 2149–2157. 10.2147/OTT.S197326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Niu L, Liu L, Yang S, Ren J, Lai PBS, Chen GG, New insights into sorafenib resistance in hepatocellular carcinoma: Responsible mechanisms and promising strategies, Biochim Biophys Acta Rev Cancer 1868(2) (2017) 564–570. 10.1016/j.bbcan.2017.10.002 [DOI] [PubMed] [Google Scholar]
- [141].Tsai TF, Lin JF, Lin YC, Chou KY, Chen HE, Ho CY, Chen PC, Hwang TI, Cisplatin contributes to programmed death-ligand 1 expression in bladder cancer through ERK1/2-AP-1 signaling pathway, Biosci Rep 39(9) (2019). 10.1042/BSR20190362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Wu L, Cai S, Deng Y, Zhang Z, Zhou X, Su Y, Xu D, PD-1/PD-L1 enhanced cisplatin resistance in gastric cancer through PI3K/AKT mediated P-gp expression, Int Immunopharmacol 94 (2021) 107443. 10.1016/j.intimp.2021.107443 [DOI] [PubMed] [Google Scholar]
- [143].Li D, Sun FF, Wang D, Wang T, Peng JJ, Feng JQ, Li H, Wang C, Zhou DJ, Luo H, Fu ZQ, Zhang T, Programmed Death Ligand-1 (PD-L1) Regulated by NRF-2/MicroRNA-1 Regulatory Axis Enhances Drug Resistance and Promotes Tumorigenic Properties in Sorafenib-Resistant Hepatoma Cells, Oncol Res 28(5) (2020) 467–481. 10.3727/096504020X15925659763817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Lai YC, Ushio N, Rahman MM, Katanoda Y, Ogihara K, Naya Y, Moriyama A, Iwanaga T, Saitoh Y, Sogawa T, Sunaga T, Momoi Y, Izumi H, Miyoshi N, Endo Y, Fujiki M, Kawaguchi H, Miura N, Aberrant expression of microRNAs and the miR-1/MET pathway in canine hepatocellular carcinoma, Vet Comp Oncol 16(2) (2018) 288–296. 10.1111/vco.12379 [DOI] [PubMed] [Google Scholar]
- [145].Liptak JM, Dernell WS, Monnet E, Powers BE, Bachand AM, Kenney JG, Withrow SJ, Massive hepatocellular carcinoma in dogs: 48 cases (1992–2002), J Am Vet Med Assoc 225(8) (2004) 1225–30. 10.2460/javma.2004.225.1225 [DOI] [PubMed] [Google Scholar]
- [146].Zhang J, Li N, Fu J, Zhou W, Long noncoding RNA HOTAIR promotes medulloblastoma growth, migration and invasion by sponging miR-1/miR-206 and targeting YY1, Biomed Pharmacother 124 (2020) 109887. 10.1016/j.biopha.2020.109887 [DOI] [PubMed] [Google Scholar]
- [147].Chang YS, Chen WY, Yin JJ, Sheppard-Tillman H, Huang J, Liu YN, EGF Receptor Promotes Prostate Cancer Bone Metastasis by Downregulating miR-1 and Activating TWIST1, Cancer Res 75(15) (2015) 3077–86. 10.1158/0008-5472.CAN-14-3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Cheng Q, Han LH, Zhao HJ, Li H, Li JB, Abnormal alterations of miR-1 and miR-214 are associated with clinicopathological features and prognosis of patients with PDAC, Oncol Lett 14(4) (2017) 4605–4612. 10.3892/ol.2017.6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Wei W, Leng J, Shao H, Wang W, MiR-1, a Potential Predictive Biomarker for Recurrence in Prostate Cancer After Radical Prostatectomy, Am J Med Sci 353(4) (2017) 315–319. 10.1016/j.amjms.2017.01.006 [DOI] [PubMed] [Google Scholar]
- [150].Xiong A, Li C, Xu J, Yang X, Nie W, Zhong H, Chu T, Zhang W, Zhong R, Pan F, Shen Y, Lou Y, Zhang B, Han B, Zhang X, Solid subtype predicts early bone metastases in sensitive EGFR-mutated lung adenocarcinoma patients after surgery, Lung Cancer 154 (2021) 124–130. 10.1016/j.lungcan.2021.02.029 [DOI] [PubMed] [Google Scholar]
- [151].Wei Q, Li X, Yu W, Zhao K, Qin G, Chen H, Gu Y, Ding F, Zhu Z, Fu X, Sun M, microRNA-messenger RNA regulatory network of esophageal squamous cell carcinoma and the identification of miR-1 as a biomarker of patient survival, J Cell Biochem 120(8) (2019) 12259–12272. 10.1002/jcb.28166 [DOI] [PubMed] [Google Scholar]
- [152].Wang W, Shen F, Wang C, Lu W, Wei J, Shang A, Wang C, MiR-1–3p inhibits the proliferation and invasion of bladder cancer cells by suppressing CCL2 expression, Tumour Biol 39(6) (2017) 1010428317698383. 10.1177/1010428317698383 [DOI] [PubMed] [Google Scholar]
- [153].Koshizuka K, Hanazawa T, Fukumoto I, Kikkawa N, Matsushita R, Mataki H, Mizuno K, Okamoto Y, Seki N, Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma, J Hum Genet 62(1) (2017) 113–121. 10.1038/jhg.2016.47 [DOI] [PubMed] [Google Scholar]
- [154].Zhao L, Chen T, Tang X, Li S, Liang R, Wang Y, Medulloblastoma malignant biological behaviors are associated with HOTAIR/miR-483–3p/CDK4 axis, Ann Transl Med 8(14) (2020) 886. 10.21037/atm-20-5006 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [155].Momen-Heravi F, Bala S, miRNA regulation of innate immunity, J Leukoc Biol (2018). 10.1002/JLB.3MIR1117-459R [DOI] [PubMed] [Google Scholar]
- [156].Self-Fordham JB, Naqvi AR, Uttamani JR, Kulkarni V, Nares S, MicroRNA: Dynamic Regulators of Macrophage Polarization and Plasticity, Front Immunol 8 (2017) 1062. 10.3389/fimmu.2017.01062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Kitamura T, Qian BZ, Pollard JW, Immune cell promotion of metastasis, Nat Rev Immunol 15(2) (2015) 73–86. 10.1038/nri3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Chen DS, Mellman I, Oncology meets immunology: the cancer-immunity cycle, immunity 39(1) (2013) 1–10. [DOI] [PubMed] [Google Scholar]
- [159].Yi M, Xu L, Jiao Y, Luo S, Li A, Wu K, The role of cancer-derived microRNAs in cancer immune escape, J Hematol Oncol 13(1) (2020) 25. 10.1186/s13045-020-00848-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Paladini L, Fabris L, Bottai G, Raschioni C, Calin GA, Santarpia L, Targeting microRNAs as key modulators of tumor immune response, J Exp Clin Cancer Res 35 (2016) 103. 10.1186/s13046-016-0375-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Rossi JF, Lu ZY, Massart C, Levon K, Dynamic Immune/Inflammation Precision Medicine: The Good and the Bad Inflammation in Infection and Cancer, Front Immunol 12 (2021) 595722. 10.3389/fimmu.2021.595722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Grivennikov SI, Greten FR, Karin M, Immunity, inflammation, and cancer, Cell 140(6) (2010) 883–99. 10.1016/j.cell.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Ewelina K, Eljaszewicz A, Kazimierczyk R, Tynecka M, Zembko P, Tarasiuk E, Kaminski K, Sobkowicz B, Moniuszko M, Tycinska A, Altered microRNA dynamics in acute coronary syndrome, Postepy Kardiol Interwencyjnej 16(3) (2020) 287–293. 10.5114/aic.2020.99263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Sondergaard HB, Airas L, Christensen JR, Nielsen BR, Bornsen L, Oturai A, Sellebjerg F, Pregnancy-Induced Changes in microRNA Expression in Multiple Sclerosis, Front Immunol 11 (2020) 552101. 10.3389/fimmu.2020.552101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Crouser ED, Julian MW, Bicer S, Ghai V, Kim TK, Maier LA, Gillespie M, Hamzeh NY, Wang K, Circulating exosomal microRNA expression patterns distinguish cardiac sarcoidosis from myocardial ischemia, PLoS One 16(1) (2021) e0246083. 10.1371/journal.pone.0246083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Xiu MX, Liu ZT, Tang J, Screening and identification of key regulatory connections and immune cell infiltration characteristics for lung transplant rejection using mucosal biopsies, Int Immunopharmacol 87 (2020) 106827. 10.1016/j.intimp.2020.106827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Sarma A, Phukan H, Halder N, Madanan MG, An in-silico approach to study the possible interactions of miRNA between human and SARS-CoV2, Comput Biol Chem 88 (2020) 107352. 10.1016/j.compbiolchem.2020.107352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Zhang Y, Wang D, Shen D, Luo Y, Che YQ, Identification of exosomal miRNAs associated with the anthracycline-induced liver injury in postoperative breast cancer patients by small RNA sequencing, PeerJ 8 (2020) e9021. 10.7717/peerj.9021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Zuo H, Weng K, Luo M, Yang L, Weng S, He J, Xu X, A MicroRNA-1-Mediated Inhibition of the NF-kappaB Pathway by the JAK-STAT Pathway in the Invertebrate Litopenaeus vannamei, J Immunol 204(11) (2020) 2918–2930. 10.4049/jimmunol.2000071 [DOI] [PubMed] [Google Scholar]
- [170].Qi X, Li Z, Li H, Wang T, Zhang Y, Wang J, MicroRNA-1 Negatively Regulates Peripheral NK Cell Function via Tumor Necrosis Factor-Like Weak Inducer of Apoptosis (TWEAK) Signaling Pathways During PPRV Infection, Front Immunol 10 (2019) 3066. 10.3389/fimmu.2019.03066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Takyar S, Vasavada H, Zhang JG, Ahangari F, Niu N, Liu Q, Lee CG, Cohn L, Elias JA, VEGF controls lung Th2 inflammation via the miR-1-Mpl (myeloproliferative leukemia virus oncogene)-P-selectin axis, J Exp Med 210(10) (2013) 1993–2010. 10.1084/jem.20121200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Korde A, Ahangari F, Haslip M, Zhang X, Liu Q, Cohn L, Gomez JL, Chupp G, Pober JS, Gonzalez A, Takyar SS, An endothelial microRNA-1-regulated network controls eosinophil trafficking in asthma and chronic rhinosinusitis, J Allergy Clin Immunol 145(2) (2020) 550–562. 10.1016/j.jaci.2019.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Kim HY, DeKruyff RH, Umetsu DT, The many paths to asthma: phenotype shaped by innate and adaptive immunity, Nat Immunol 11(7) (2010) 577–84. 10.1038/ni.1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Lee CG, Link H, Baluk P, Homer RJ, Chapoval S, Bhandari V, Kang MJ, Cohn L, Kim YK, McDonald DM, Elias JA, Vascular endothelial growth factor (VEGF) induces remodeling and enhances TH2-mediated sensitization and inflammation in the lung, Nat Med 10(10) (2004) 1095–103. 10.1038/nm1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Ma SN, Mao ZX, Wu Y, Liang MX, Wang DD, Chen X, Chang PA, Zhang W, Tang JH, The anti-cancer properties of heparin and its derivatives: a review and prospect, Cell Adh Migr 14(1) (2020) 118–128. 10.1080/19336918.2020.1767489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Banerjee ER, Triple selectin knockout (ELP−/−) mice fail to develop OVA-induced acute asthma phenotype, J Inflamm (Lond) 8 (2011) 19. 10.1186/1476-9255-8-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Jiang F, Chen Q, Wang W, Ling Y, Yan Y, Xia P, Hepatocyte-derived extracellular vesicles promote endothelial inflammation and atherogenesis via microRNA-1, J Hepatol 72(1) (2020) 156–166. 10.1016/j.jhep.2019.09.014 [DOI] [PubMed] [Google Scholar]
- [178].Zhao J, Luo R, Xu X, Zou Y, Zhang Q, Pan W, High-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation (HITS-CLIP) reveals Argonaute-associated microRNAs and targets in Schistosoma japonicum, Parasit Vectors 8 (2015) 589. 10.1186/s13071-015-1203-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Seok H, Ham J, Jang ES, Chi SW, MicroRNA Target Recognition: Insights from Transcriptome-Wide Non-Canonical Interactions, Mol Cells 39(5) (2016) 375–81. 10.14348/molcells.2016.0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, Billis K, Boddu S, Marugan JC, Cummins C, Davidson C, Dodiya K, Fatima R, Gall A, Giron CG, Gil L, Grego T, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, Kay M, Lavidas I, Le T, Lemos D, Martinez JG, Maurel T, McDowall M, McMahon A, Mohanan S, Moore B, Nuhn M, Oheh DN, Parker A, Parton A, Patricio M, Sakthivel MP, Abdul Salam AI, Schmitt BM, Schuilenburg H, Sheppard D, Sycheva M, Szuba M, Taylor K, Thormann A, Threadgold G, Vullo A, Walts B, Winterbottom A, Zadissa A, Chakiachvili M, Flint B, Frankish A, Hunt SE, Kostadima IIG,M, Langridge N, Loveland JE, Martin FJ, Morales J, Mudge JM, Muffato M, Perry E, Ruffier M, Trevanion SJ, Cunningham F, Howe KL, Zerbino DR, Flicek P, Ensembl 2020, Nucleic Acids Res 48(D1) (2020) D682–D688. 10.1093/nar/gkz966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Wu Y, Pu N, Su W, Yang X, Xing C, Downregulation of miR-1 in colorectal cancer promotes radioresistance and aggressive phenotypes, J Cancer 11(16) (2020) 4832–4840. 10.7150/jca.44753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Tang Y, Zheng J, Sun Y, Wu Z, Liu Z, Huang G, MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2, Int Heart J 50(3) (2009) 377–87. 10.1536/ihj.50.377 [DOI] [PubMed] [Google Scholar]
- [183].Cheng Y, Yang M, Peng J, Correlation the between the regulation of miRNA-1 in c-Met-induced EMT and cervical cancer progression, Oncol Lett 17(3) (2019) 3341–3349. 10.3892/ol.2019.9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Tominaga E, Yuasa K, Shimazaki S, Hijikata T, MicroRNA-1 targets Slug and endows lung cancer A549 cells with epithelial and anti-tumorigenic properties, Exp Cell Res 319(3) (2013) 77–88. 10.1016/j.yexcr.2012.10.015 [DOI] [PubMed] [Google Scholar]
- [185].Wu L, Wang T, He D, Li X, Jiang Y, miR-1 inhibits the proliferation of breast cancer stem cells by targeting EVI-1, Onco Targets Ther 11 (2018) 8773–8781. 10.2147/OTT.S188836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Li B, Wang Z, Wu H, Xue M, Lin P, Wang S, Lin N, Huang X, Pan W, Liu M, Yan X, Qu H, Sun L, Li H, Wu Y, Teng W, Wang Z, Zhou X, Chen H, Poznansky MC, Ye Z, Epigenetic Regulation of CXCL12 Plays a Critical Role in Mediating Tumor Progression and the Immune Response In Osteosarcoma, Cancer Res 78(14) (2018) 3938–3953. 10.1158/0008-5472.CAN-17-3801 [DOI] [PubMed] [Google Scholar]
- [187].Leone V, D’Angelo D, Rubio I, de Freitas PM, Federico A, Colamaio M, Pallante P, Medeiros-Neto G, Fusco A, MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1alpha, J Clin Endocrinol Metab 96(9) (2011) E1388–98. 10.1210/jc.2011-0345 [DOI] [PubMed] [Google Scholar]
- [188].Li J, Guan J, Long X, Wang Y, Xiang X, mir-1-mediated paracrine effect of cancer-associated fibroblasts on lung cancer cell proliferation and chemoresistance, Oncol Rep 35(6) (2016) 3523–31. 10.3892/or.2016.4714 [DOI] [PubMed] [Google Scholar]
- [189].Wang X, Huang X, Yang Z, Gallego-Perez D, Ma J, Zhao X, Xie J, Nakano I, Lee LJ, Targeted delivery of tumor suppressor microRNA-1 by transferrin-conjugated lipopolyplex nanoparticles to patient-derived glioblastoma stem cells, Curr Pharm Biotechnol 15(9) (2014) 839–46. 10.2174/1389201015666141031105234 [DOI] [PubMed] [Google Scholar]
- [190].Chang J, Xu W, Du X, Hou J, MALAT1 silencing suppresses prostate cancer progression by upregulating miR-1 and downregulating KRAS, Onco Targets Ther 11 (2018) 3461–3473. 10.2147/OTT.S164131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Qu W, Chen X, Wang J, Lv J, Yan D, MicroRNA-1 inhibits ovarian cancer cell proliferation and migration through c-Met pathway, Clin Chim Acta 473 (2017) 237–244. 10.1016/j.cca.2017.07.008 [DOI] [PubMed] [Google Scholar]
- [192].Zhu K, Wang W, Green tea polyphenol EGCG suppresses osteosarcoma cell growth through upregulating miR-1, Tumour Biol 37(4) (2016) 4373–82. 10.1007/s13277-015-4187-3 [DOI] [PubMed] [Google Scholar]
- [193].Leonardo L, Laura P, Serena BM, miR-1 and miR-133b expression in canine osteosarcoma, Res Vet Sci 117 (2018) 133–137. 10.1016/j.rvsc.2017.12.002 [DOI] [PubMed] [Google Scholar]
- [194].Gao S, Zhao Z, Wu R, Wu L, Tian X, Zhang Z, MiR-1 inhibits prostate cancer PC3 cells proliferation through the Akt/mTOR signaling pathway by binding to c-Met, Biomed Pharmacother 109 (2019) 1406–1410. 10.1016/j.biopha.2018.10.098 [DOI] [PubMed] [Google Scholar]
- [195].Hu T, Chang YF, Xiao Z, Mao R, Tong J, Chen B, Liu GC, Hong Y, Chen HL, Kong SY, Huang YM, Xiyang YB, Jin H, miR-1 inhibits progression of high-risk papillomavirus-associated human cervical cancer by targeting G6PD, Oncotarget 7(52) (2016) 86103–86116. 10.18632/oncotarget.13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Liu C, Zhang S, Wang Q, Zhang X, Tumor suppressor miR-1 inhibits tumor growth and metastasis by simultaneously targeting multiple genes, Oncotarget 8(26) (2017) 42043–42060. 10.18632/oncotarget.14927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Xiao H, Zeng J, Li H, Chen K, Yu G, Hu J, Tang K, Zhou H, Huang Q, Li A, Li Y, Ye Z, Wang J, Xu H, MiR-1 downregulation correlates with poor survival in clear cell renal cell carcinoma where it interferes with cell cycle regulation and metastasis, Oncotarget 6(15) (2015) 13201–15. 10.18632/oncotarget.3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Bronisz A, Wang Y, Nowicki MO, Peruzzi P, Ansari K, Ogawa D, Balaj L, De Rienzo G, Mineo M, Nakano I, Ostrowski MC, Hochberg F, Weissleder R, Lawler SE, Chiocca EA, Godlewski J, Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1, Cancer Res 74(3) (2014) 738–750. 10.1158/0008-5472.CAN-13-2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Josse C, Bouznad N, Geurts P, Irrthum A, Huynh-Thu VA, Servais L, Hego A, Delvenne P, Bours V, Oury C, Identification of a microRNA landscape targeting the PI3K/Akt signaling pathway in inflammation-induced colorectal carcinogenesis, Am J Physiol Gastrointest Liver Physiol 306(3) (2014) G229–43. 10.1152/ajpgi.00484.2012 [DOI] [PubMed] [Google Scholar]
- [200].Wang N, Chen M, Ni Z, Li T, Zeng J, Lu G, Wang J, Zhang J, Wu S, Shi Y, HDAC6/HNF4alpha loop mediated by miR-1 promotes bile acids-induced gastric intestinal metaplasia, Gastric Cancer 24(1) (2021) 103–116. 10.1007/s10120-020-01108-x [DOI] [PMC free article] [PubMed] [Google Scholar]


