Abstract
Tat stimulation of human immunodeficiency virus type 1 (HIV-1) transcription requires Tat-dependent recruitment of human positive transcription elongation factor b (P-TEFb) to the HIV-1 promoter and the formation on the trans-acting response element (TAR) RNA of a P-TEFb–Tat–TAR ternary complex. We show here that the P-TEFb heterodimer of Cdk9-cyclin T1 is intrinsically incapable of forming a stable complex with Tat and TAR due to two built-in autoinhibitory mechanisms in P-TEFb. Both mechanisms exert little effect on the P-TEFb–Tat interaction but prevent the P-TEFb–Tat complex from binding to TAR RNA. The first autoinhibition arises from the unphosphorylated state of Cdk9, which establishes a P-TEFb conformation unfavorable for TAR recognition. Autophosphorylation of Cdk9 overcomes this inhibition by inducing conformational changes in P-TEFb, thereby exposing a region in cyclin T1 for possible TAR binding. An intramolecular interaction between the N- and C-terminal regions of cyclin T1 sterically blocks the P-TEFb–TAR interaction and constitutes the second autoinhibitory mechanism. This inhibition is relieved by the binding of the C-terminal region of cyclin T1 to the transcription elongation factor Tat-SF1 and perhaps other cellular factors. Upon release from the intramolecular interaction, the C-terminal region also interacts with RNA polymerase II and is required for HIV-1 transcription, suggesting its role in bridging the P-TEFb–Tat–TAR complex and the basal elongation apparatus. These data reveal novel control mechanisms for the assembly of a multicomponent transcription elongation complex at the HIV-1 promoter.
The Tat protein encoded by the human immunodeficiency virus type 1 (HIV-1) genome strongly stimulates the synthesis of full-length HIV-1 transcripts by increasing the processivity of RNA polymerase II (Pol II). Tat recognizes the trans-acting response element (TAR) RNA stem-loop structure near the 5′ end of the nascent viral transcript and is proposed to function by recruiting a cellular cofactor(s) to Pol II in a TAR loop sequence-specific manner (for reviews, see references 8, 17, and 18).
Tat transactivation requires the C-terminal domain (CTD) of the largest subunit of Pol II (3, 29, 32, 41). Since hyperphosphorylation of the CTD correlates with processive transcriptional elongation (6) and Tat activity is sensitive to kinase inhibitors (e.g., DRB) that inhibit CTD phosphorylation (24, 25), it was proposed that Tat stimulates elongation by recruitment of a CTD kinase(s) to Pol II (reference 18 and references therein). One of the CTD kinases implicated in Tat transactivation is P-TEFb (positive transcription elongation factor b). It was originally identified as a general elongation factor in Drosophila melanogaster and later in mammalian cells (26, 27, 45). Active P-TEFb is composed of a cyclin-dependent kinase, Cdk9, and its partner cyclin T (cyclin T1 [CycT1] or the minor forms T2a and T2b [34, 37]). The in vivo assembly of the Cdk9-CycT1 heterodimer of P-TEFb requires a chaperone-dependent folding pathway involving the sequential actions of Hsp70 and a kinase-specific chaperone complex, Hsp90-Cdc37 (30). Immunodepletion of P-TEFb from HeLa nuclear extracts or inclusion of DRB in an in vitro transcription assay eliminates both basal and Tat-activated HIV-1 transcription (24, 42, 45). Supplementing the depleted extract with purified wild-type P-TEFb but not a kinase-defective mutant P-TEFb restores both activities (42). By using kinase inhibitors or introduction of dominant negative Cdk9 mutants into cells, the functional significance of P-TEFb and its kinase activity in Tat activation has also been demonstrated in vivo (13, 24).
In addition to Cdk9, the CycT1 subunit of P-TEFb also plays a critical role in Tat activation. First, CycT1 mediates the interaction of P-TEFb with Tat and the binding of recombinant CycT1 to Tat enhances the affinity and specificity of the CycT1-Tat-TAR interaction and confers dependence on TAR loop sequences (10, 37). Second, the interaction of human but not rodent P-TEFb with Tat through the cyclin box region in human CycT1 mediates a Tat-specific and species-restricted activation of HIV-1 transcription (1, 2, 9, 10). Third, overexpression of human CycT1 in nonpermissive rodent cells rescues Tat activation of HIV-1 transcription (37). This activity is attributed to a critical cysteine residue at position 261 that is present in human CycT1 but absent in the rodent homologue (1, 9, 10). This cysteine residue is located in a region termed TRM (Tat-TAR recognition motif), which is important for the interaction of human CycT1 with Tat and TAR (10). While the exact N-terminal boundary of TRM has not been determined, the C-terminal boundary of TRM is located between amino acids 250 and 262 at the C-terminal edge of the cyclin box (amino acids 1 to 272).
In addition to P-TEFb, several other cellular proteins important for Tat transactivation have also been characterized. Among these are Tat-SF1 (20, 44), the human homologue of the Saccharomyces cerevisiae transcription factor SPT5 (38), TFIIH (5, 11, 32), TFIIF (19), and a Tat-associated histone acetyltransferase (reference 17 and references therein). Tat-SF1 was biochemically identified as a Tat cofactor, and it stimulates Tat activation both in vivo and in vitro (20, 44). It was also found to have a general elongation activity under certain conditions (23, 31). Tat-SF1 interacts with Tat (44) and P-TEFb (42) and was recently found to interact with human SPT5, Pol II, and the RAP30 subunit of TFIIF as well (20). The human SPT4 and SPT5 proteins form a complex called DSIF (DRB-sensitivity-inducing factor [35]). It arrests the elongation of Pol II at sites proximal to the promoter with the help of the negative elongation factor NELF (39). It has been shown that P-TEFb positively regulates Pol II elongation by, at least in part, suppressing the activity of DSIF in a phosphorylation step that is DRB sensitive (36). While that study identified DSIF as a negative factor, other experiments have also revealed a positive role of DSIF in Tat-activated HIV-1 transcription (20, 38). Recently, Parada and Roeder (31) reported the identification of a novel RNA Pol II-containing complex that supports Tat-activated HIV-1 transcription. Interestingly, this complex contains several previously identified elongation factors, P-TEFb, Tat-SF1, and DSIF, in addition to Pol II and other unidentified polypeptides. Exactly how these elongation factors and Pol II cooperate to stimulate HIV-1 transcription is still unclear. Nevertheless, an emerging picture of the involvement of multiple factors for the control of elongation is evident, as recent studies indicated that these factors may exist in the same complex and function in combination or in a sequential manner to promote Tat stimulation of Pol II processivity.
The formation of a TAR loop-dependent P-TEFb–Tat–TAR ribonucleoprotein complex is recognized as an essential step towards the assembly of productive Pol II elongation machinery at the HIV-1 promoter. Previous analyses of the TAR loop-dependent complex used free, recombinant CycT1 (10, 37). Since the active form of P-TEFb for Tat transactivation consists of a heterodimer of Cdk9-CycT1, we wanted to understand how P-TEFb forms a stable complex with Tat and TAR. In this report, we showed that formation of the P-TEFb–Tat–TAR ternary complex requires the deactivation of two autoinhibitory mechanisms in P-TEFb. Autophosphorylation of Cdk9 overcomes the first autoinhibition by creating a favorable P-TEFb conformation and exposing the TRM region in CycT1 for efficient interaction with TAR RNA. The second autoinhibition is caused by the intramolecular interaction between the N- and C-terminal regions of CycT1, which blocks the access of TAR RNA to CycT1 TRM. Relief of this autoinhibition is provided by the interaction of the CycT1 C-terminal region with Tat-SF1. Upon release from the intramolecular interaction, the C-terminal region also interacts with Pol II and is required for efficient HIV-1 transcriptional elongation, suggesting that it may link the basal transcription elongation apparatus to the P-TEFb–Tat–TAR ternary complex. These results reveal novel control steps for the assembly at the HIV-1 promoter of a multicomponent elongation complex for Tat transactivation.
MATERIALS AND METHODS
DNA constructs and antibodies.
Different hemagglutinin (HA)-tagged CycT1 cDNA fragments were generated by PCR and subcloned into the BamHI and EcoRI sites of the pcDNA3 expression vector (Invitrogen). Glutathione S-transferase (GST)–CycT1Δ1–HA was generated by subcloning the fragment containing a C-terminally truncated CycT1 (with only amino acids 1 to 333) (CycT1Δ1) into the BamHI and EcoRI sites of the pGEX-4T-1-HA expression vector, which contains the HA epitope cloned into the SalI and NotI sites of pGEX-4T-1 (Amersham Pharmacia). GST–CycT1-C was generated by subcloning the CycT1 cDNA fragment encoding a portion of the CycT1 C-terminal region (amino acids 402 to 701) into the BamHI and EcoRI sites of the pGEX-2T expression vector. C-terminally Flag-tagged Tat-SF1 cDNA was cloned into the EcoRI site of the pSV7d expression vector (Sigma). Anti-Cdk9 and anti-Pol IIa antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.), and polyclonal anti-CycT1 antibodies were generated in rabbits against the GST-CycT1-C fusion protein.
Expression and purification of P-TEFb complexes and Tat-SF1.
For P-TEFb-containing CycT1 deletion mutants, plasmids bearing the gene expressing HA-tagged, C-terminally truncated CycT1 were transiently transfected into 293T cells. P-TEFb complexes were affinity purified from the cell lysates 48 h later as described previously (42). Cdk9-Flag–HA-CycT1 complexes were generated by coexpression of HA-CycT1 and Cdk9-Flag in 293T cells followed by two sequential affinity purification steps (with anti-HA and anti-Flag antibodies) and peptide elution. A mutant P-TEFb with a kinase-defective Cdk9-HA subunit was generated and purified as described previously (42).
For making autophosphorylated P-TEFb complexes for gel shift and Tat-binding experiments, Cdk9-HA–CycT1 was first affinity purified from cell lysates with anti-HA antibody (12CA5) protein A-Sepharose beads. After extensive washes, the beads were equilibrated with kinase buffer (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol [DTT]) and the kinase reaction was initiated by the addition of ATP (0.1 mM) as described previously (42). After the reaction, ATP was removed by extensive washes and the phosphorylated P-TEFb was eluted from the beads with HA peptide. P-TEFb complexes treated with alkaline phosphatase were prepared essentially as described above, except that after the kinase reaction, 10 U of calf intestine alkaline phosphatase was incubated with the affinity beads for 5 min at 30°C before the complexes were subjected to peptide elution.
Flag-tagged Tat-SF1 was transiently expressed in 293T cells and affinity purified using anti-Flag antibody beads (Sigma) followed by Flag peptide elution as described for the purification of P-TEFb.
Tat-binding assay.
Binding of P-TEFb complexes to immobilized GST-Tat(1-48) or activation domain mutant GST-Tat(1-48, C22G) was performed as described previously (2).
TAR RNA EMSA.
32P-labeled TAR RNA was synthesized by T7 RNA polymerase from the HindIII-digested DNA templates pT7TAR (wild type) and pT7TAR(+31/+34) as described previously (42). Electrophoretic mobility shift assays (EMSA) were carried out essentially as described previously (10), and when noted in the figures, 100 μM ATP or [γ-S]ATP was also included in the incubation mixture prior to electrophoresis. To study the effect of Tat-SF1 on P-TEFb–Tat–TAR complex formation, approximately 50 ng of affinity-purified, Flag-tagged Tat-SF1 was preincubated with P-TEFb (containing ∼30 ng of Cdk9) for 10 min before being added to the gel shift reaction mixture.
Partial protease digestion of P-TEFb.
Purified P-TEFb complexes were first incubated with ATP in the kinase buffer under the same conditions described for the EMSA reaction. Trypsin diluted in D0.1 M KCl buffer (20 mM HEPES-KOH [pH 7.9], 10% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.05% NP-40, 1 mM DTT) was then added to the reaction mixture at a final concentration of 7 ng/ml, and the incubation continued for another two minutes at 30°C. HA-tagged CycT1 fragments were visualized by Western blotting with antibodies specific for HA.
Transcription assay.
Immunodepletion of P-TEFb from HeLa nuclear extracts was carried out with immobilized anti-Cdk9 and anti-CycT1 antibodies as described previously (42). In vitro transcription reactions with mixtures containing HeLa nuclear extract depleted of P-TEFb, affinity purified P-TEFb complexes, HIV-1 promoter templates, and Tat were carried out exactly as described previously (42).
Detecting CycT1-associated proteins in HeLa nuclear extract.
Approximately 0.15 mg of HeLa nuclear extracts was dialyzed against D0.1M KCl buffer and then incubated with 0.5 μg of GST or the various GST-CycT1 fusion proteins immobilized on glutathione-Sepharose beads for 3 h at 4°C. After extensive washes with D0.2M KCl buffer and then GE buffer (120 mM NaCl, 100 mM Tris-HCl [pH 8.0], 10% glycerol, 0.2 mM EDTA, 0.1% NP-40, 2 mM DTT), the bound materials were eluted with 15 mM glutathione. Eluted materials were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using antibodies specific for Tat-SF1, Pol IIa, SPT5, and RAP30.
RESULTS
Requirement of ATP for formation of a stable TAR loop-dependent P-TEFb–Tat–TAR complex.
Previously, we noticed that on a per CycT1 molecule basis, the stability of a CycT1–Tat–TAR complex as determined by EMSA was significantly higher than that of the P-TEFb–Tat–TAR complex (2, 42), implicating a negative effect of Cdk9 on formation of such a complex. To investigate the role of Cdk9 in this process, we affinity purified the Cdk9–HA-CycT1Δ1 heterodimer from 293T cells transiently expressing the HA-tagged CycT1Δ1 protein by anti-HA antibody immunoprecipitation (α-HA IP) followed by HA peptide elution (2, 42). The purified protein fraction was then tested for its ability to bind to Tat and TAR in a gel mobility shift assay (Fig. 1A). The reason for using the C-terminally truncated CycT1Δ1 protein (amino acids 1 to 333) instead of full-length CycT1 (CycT1FL) (amino acids 1 to 726) in this experiment will become clear shortly. CycT1Δ1 contains the conserved cyclin box (amino acids 1 to 272), the presence of which is sufficient for binding to Cdk9 (2, 10, 34). It also contains the TRM region important for Tat and TAR binding (10).
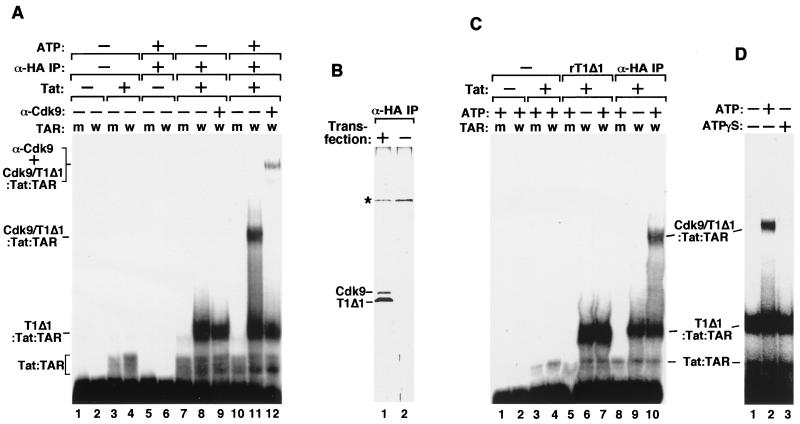
FIG. 1.
Requirement of ATP hydrolysis for formation of a stable P-TEFb–Tat–TAR ribonucleoprotein complex. (A) The Cdk9–HA-CycT1Δ1 heterodimer was affinity purified from 293T cells transiently transfected with an HA-tagged CycT1Δ1 cDNA by α-HA IP followed by HA peptide elution. The purified fraction containing the heterodimer and free HA-CycT1Δ1 was incubated with 32P-labeled wild-type TAR RNA (lanes labeled w) or the loop mutant TAR+31/+34 (lanes labeled m) in the absence (−) or presence (+) of Tat and/or ATP as indicated. The reactions were analyzed by EMSA. Lanes 9 and 12 also contained anti-Cdk9 antibodies (α-Cdk9), which caused the retardation of the Cdk9–HA-CycT1Δ1–Tat–TAR complex and also partially destabilized the complex, probably because of the polyclonal nature of the antibodies. (B) Silver-stained SDS-polyacrylamide gel showing the presence of Cdk9–HA-CycT1Δ1 and free HA-CycT1Δ1 in the α-HA IP fraction (lane 1). A nonspecific protein (∗) was revealed in a control fraction prepared in parallel from untransfected 293T cells (lane 2). (C) Free HA-CycT1Δ1 present in the α-HA IP fraction formed an ATP-independent complex with Tat and TAR in gel shift reactions. Recombinant CycT1Δ1 (rT1Δ1) was used as a reference (lanes 5 to 7) for determining the identity of the ATP-independent complex formed with the α-HA IP fraction (lanes 8 to 10). (D) Requirement of ATP hydrolysis for P-TEFb–Tat–TAR assembly. All reaction mixtures contained the α-HA IP fraction, Tat, and 32P-labeled wild-type TAR RNA. [γ-S]ATP was used in place of ATP in lane 3.
As shown in Fig. 1A, recombinant Tat protein interacted weakly with both wild-type TAR RNA (lane 4) and the loop mutant TAR+31/+34 with four nucleotide substitutions in the apical loop (lane 3), whereas the α-HA IP fraction containing Cdk9–HA-CycT1Δ1 showed no detectable binding to either probe (Fig. 1A, lanes 5 and 6). Similar to the findings with recombinant CycT1 (10, 37), the presence of both Tat and the α-HA IP fraction in the same reaction mixture resulted in the formation of a strong, TAR loop-dependent complex with a mobility slower than that of the Tat-TAR complex (lanes 7 and 8). A new loop-dependent complex located above the center line of the gel (Fig. 1A, compare lanes 8 and 11) was produced when ATP was added to the reaction mixture. Unlike the first two complexes, this slow-migrating, ATP-dependent complex could be supershifted by anti-Cdk9 antibodies (Fig. 1A, compare lane 12 with lane 9), indicating the presence of Cdk9, most likely in the form of a Cdk9-CycT1Δ1 heterodimer, within this complex.
We also investigated the composition of the fast-migrating, ATP-independent complex derived from the α-HA IP fraction. Since this fraction was prepared from transfected cells overexpressing HA-CycT1Δ1, it probably contained both Cdk9–HA-CycT1Δ1 and free HA-CycT1Δ1. Indeed, when analyzed by SDS-PAGE and silver staining, this fraction was found to contain about twice more HA-CycT1Δ1 than Cdk9 (Fig. 1B, lane 1). In a gel mobility shift assay (Fig. 1C), the mobility of the ATP-independent complex derived from the α-HA IP fraction was identical to that of the CycT1Δ1-Tat-TAR complex formed with the recombinant CycT1Δ1 protein (Fig. 1C, compare lanes 6 and 7 with lanes 9 and 10). This finding, together with the observation that the ATP-independent complex could not be supershifted by anti-Cdk9 antibodies, suggested that this complex was very likely derived from the binding of free HA-CycT1Δ1 present in the α-HA IP fraction to Tat and TAR. Taken together, these data revealed a major difference between free CycT1 and the Cdk9-CycT1 heterodimer in that the latter requires ATP to form a high-affinity P-TEFb–Tat–TAR ternary complex.
Most of cellular P-TEFb requires ATP hydrolysis for stable interaction with Tat and TAR.
The stable association of Cdk9–HA-CycT1Δ1 with Tat and TAR requires ATP hydrolysis and not just simple ATP binding, because replacement of ATP in the binding reaction mixture with a nonhydrolyzable form of ATP, [γ-S]ATP, largely inhibited complex formation (Fig. 1D, compare lanes 2 and 3). Because P-TEFb is a Cdk-cyclin kinase complex, it is most likely a protein phosphorylation event that results in the assembly of the P-TEFb–Tat–TAR complex. To rule out the possibility that the requirement of ATP was due to the loss of phosphates on P-TEFb during cell lysis and the subsequent purification procedure, multiple phosphatase inhibitors were used in the experiment. P-TEFb thus purified still required ATP for high-affinity binding to Tat-TAR (data not shown), suggesting that most of the cellular P-TEFb may lack the key phosphorylation that is important for complex formation.
C-terminal truncation of CycT1 in addition to P-TEFb phosphorylation stabilizes the P-TEFb–Tat–TAR complex.
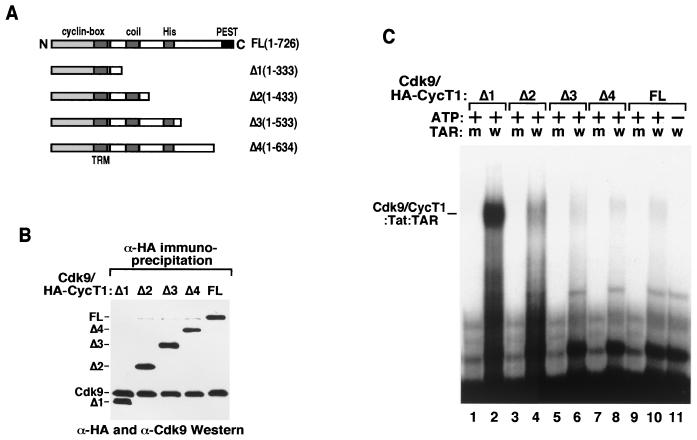
The gel mobility shift assay described above used P-TEFb complexes containing the C-terminally truncated CycT1 molecule CycT1Δ1. To examine the effect of the C-terminal region of CycT1 on ATP-dependent P-TEFb–Tat–TAR complex formation, we generated P-TEFb complexes containing HA-tagged CycT1FL (HA-CycT1FL) or a series of mutant CycT1s with C-terminal deletions (HA–CycT1Δ1 to -Δ4) (Fig. 2A). These proteins were transiently expressed in 293T cells and affinity purified by α-HA IP. Western blotting indicated that deletions of the C-terminal region of CycT1 up to position 333 did not affect its ability to interact with Cdk9 (Fig. 2B), consistent with the notion that the cyclin box of CycT1 is sufficient for Cdk9 binding.
FIG. 2.
The P-TEFb–Tat–TAR complex is stabilized by C-terminal truncation of CycT1 in addition to P-TEFb phosphorylation. (A) Diagram showing the domain structures of CycT1FL and the C-terminal truncation mutant constructs CycT1Δ1 to -Δ4. The C-terminal boundary of TRM is located between amino acids 250 and 262 at the C-terminal edge of the conserved cyclin box domain (amino acids 1 to 272). The N-terminal boundary of TRM is unclear. (B) The C-terminal truncation of CycT1 did not affect its association with Cdk9. HA-CycT1FL or HA-CycT1Δ1 to -Δ4 and their associated Cdk9 proteins were affinity purified from 293T cells transiently expressing the various HA-tagged CycT1 proteins by anti-HA IP. The levels of Cdk9 and HA-CycT1 in these preparations were analyzed by Western blotting with anti-Cdk9 and anti-HA antibodies. A faint band right below CycT1FL is a nonspecific cross-reactive protein. (C) Equal amounts of Cdk9–HA-CycT1FL and Cdk9–HA-CycT1Δ1 to -Δ4 were tested for their abilities to form ternary complexes with Tat and 32P-labeled wild-type (lanes w) or the loop mutant (lanes m) TAR RNAs in a gel mobility shift assay.
Equal amounts of each complex were next analyzed in a gel mobility shift assay. While removal of residues 334 to 726 in CycT1 (CycT1Δ1) resulted in the formation of a strong, ATP- and TAR loop-dependent complex (Cdk9-CycT1Δ1–Tat–TAR) (Fig. 2C, lanes 1 and 2), inclusion of the C-terminal residues in CycT1Δ2, CycT1Δ3, CycT1Δ4, and CycT1FL reduced the stability of the respective P-TEFb–Tat–TAR complexes (Fig. 2C, lanes 3 to 10). Compared to Cdk9-CycT1Δ1, Cdk9-CycT1FL displayed a very weak, albeit ATP- and loop-dependent, interaction with Tat and TAR (Fig. 2C, lanes 9 to 11). Similar inhibition of complex formation by the CycT1 C-terminal region was also observed with free, recombinant CycT1Δ1 to -Δ4 and CycT1FL proteins (data not shown) as well as with free CycT1 present in the α-HA IP fraction (bottom half of Fig. 2C). Thus, both the C-terminal region of CycT1 and the unphosphorylated state of P-TEFb inhibited P-TEFb–Tat–TAR complex formation.
It is important to point out that P-TEFb complexes containing the C-terminal region of CycT1 demonstrated somewhat higher affinity for Tat than the complex (Cdk9-CycT1Δ1) without this region (2). Thus, the C-terminal region inhibited P-TEFb–Tat–TAR formation not by disrupting the interaction between P-TEFb and Tat but more likely by suppressing the interaction of the P-TEFb–Tat complex with TAR RNA.
Autophosphorylation of Cdk9 is required for P-TEFb–Tat–TAR complex formation.
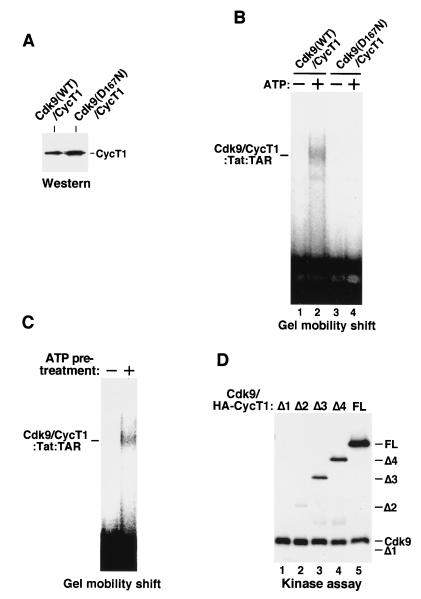
Given the fact that high-affinity P-TEFb–Tat–TAR assembly requires ATP hydrolysis, we wanted to confirm that it is the Cdk9 kinase of P-TEFb and not some other contaminating kinases or ATPases in the affinity-purified fraction which hydrolyzes ATP and stimulates P-TEFb–Tat–TAR assembly. We prepared P-TEFb heterodimers that contained either wild-type or a kinase-defective Cdk9 with a point mutation changing Asp167 to Asn (Fig. 3A). (Since the HA tag is appended to Cdk9, no free CycT1 is present in these α-HA IP fractions.) Because of the inhibitory activity of the CycT1 C-terminal region, a large amount of wild-type P-TEFb containing CycT1FL was used to detect the formation of the ternary complex (Fig. 3B, lanes 1 and 2). Compared to wild-type P-TEFb, the kinase-defective Cdk9(D167N)-CycT1 heterodimer failed to form a stable complex with Tat and TAR both in the presence and absence of ATP (Fig. 3B, lanes 3 and 4). This binding defect, in addition to the failure of this kinase-defective P-TEFb to phosphorylate the Pol II CTD, probably contributed to its severe deficiency in mediating Tat activation (42).
FIG. 3.
Autophosphorylation of Cdk9 is required for efficient P-TEFb–Tat–TAR complex formation. (A) P-TEFb complexes containing either HA-tagged wild-type Cdk9 (WT) or a kinase-defective Cdk9 (D167N) were affinity purified by α-HA IP and normalized for their CycT1 levels by Western blotting with anti-CycT1 antibodies. (B) The kinase activity of Cdk9 is required for P-TEFb–Tat–TAR formation. The two P-TEFb complexes prepared for panel A were compared in a gel mobility shift assay for forming P-TEFb–Tat–TAR complexes in the presence (+) or absence (−) of ATP. Since the HA tag was attached to Cdk9, no free CycT1 was present in these α-HA IP fractions. (C) Autophosphorylation of P-TEFb is required for P-TEFb–Tat–TAR complex formation. Purified Cdk9-HA–CycT1 immobilized on anti-HA antibody beads was incubated with ATP in a kinase reaction mixture. Upon removal of ATP, the phosphorylated complex was eluted from the beads and incubated with Tat and 32P-labeled wild-type TAR RNA in a reaction mixture without ATP. An equal amount of unphosphorylated P-TEFb was used as a control. (D) Phosphorylation of CycT1 by Cdk9 is not required for P-TEFb–Tat–TAR formation. Cdk9–HA-CycT1FL and Cdk9–HA-CycT1Δ1 to -Δ4 complexes were prepared, and their levels were normalized as described for Fig. 2B. Equal amounts of these complexes were analyzed in in vitro kinase reactions. Note that HA-CycTΔ1 was not phosphorylated by Cdk9. However, Cdk9–HA-CycT1Δ1 formed an ATP-dependent complex with Tat-TAR.
Since the Cdk9 kinase activity is important for P-TEFb–Tat–TAR assembly, we wanted to confirm that autophosphorylation of P-TEFb, but not its phosphorylation of Tat or TAR, is crucial for this process. Instead of adding ATP directly to the incubation of P-TEFb with Tat and TAR, Cdk9-HA–CycT1 purified from the stable cell line B4 (2) was immobilized on anti-HA antibody beads and subjected to a kinase reaction. After extensive washes to remove ATP, phosphorylated Cdk9-HA–CycT1 was eluted from the beads and incubated with Tat and TAR in a reaction mixture without ATP. As shown in Fig. 3C, ATP-pretreated P-TEFb associated with Tat and TAR while the unphosphorylated P-TEFb did not. Thus, autophosphorylation of P-TEFb is critical for P-TEFb–Tat–TAR formation.
When the P-TEFb heterodimer undergoes autophosphorylation in vitro, Cdk9 phosphorylates itself as well as the associated CycT1 molecule (42). To determine whether autophosphorylation of Cdk9 or its phosphorylation of CycT1 is critical for P-TEFb–Tat–TAR formation, we performed in vitro kinase reactions to compare the levels of Cdk9 and CycT1 phosphorylation among P-TEFb complexes containing HA-tagged CycT1 with different C-terminal deletions. All complexes were normalized for their Cdk9 and CycT1 levels by immunoblotting with anti-Cdk9 and anti-HA antibodies (Fig. 2B). While deletions of CycT1 C-terminal residues to position 333 did not affect the ability of Cdk9 to autophosphorylate, phosphorylation of CycT1 by Cdk9 gradually diminished as more amino acids were deleted from the C terminus of CycT1 (Fig. 3D). It is important to note that CycT1Δ1 was not phosphorylated by Cdk9 (Fig. 3D, lane 1; refer to Fig. 2B, lane 1, for the position of CycT1Δ1), probably because of a lack of phosphorylation sites. Yet the complex containing this mutant (Cdk9–HA-CycT1Δ1) can still be induced by phosphorylation to form a stable P-TEFb–Tat–TAR complex (Fig. 1), indicating that the phosphorylation of Cdk9 but not CycT1 is crucial for this process.
Autophosphorylation of Cdk9 does not affect P-TEFb–Tat binding but induces conformational changes in P-TEFb for better TAR recognition.
Autophosphorylation of Cdk9 may facilitate P-TEFb–Tat–TAR formation by increasing the binding of P-TEFb to either Tat protein or TAR RNA. To distinguish between these two possibilities, we compared unphosphorylated, in vitro autophosphorylated, and alkaline phosphatase-treated Cdk9-HA–CycT1 heterodimers (see Materials and Methods) for their abilities to interact with wild-type GST–Tat(1-48) proteins. GST-Tat(1-48, C22G), which contains a point mutation in the Tat activation domain, was used as a control to demonstrate the specificity of the interaction. Western blotting with anti-CycT1 antibodies indicated that autophosphorylation of Cdk9-HA only minimally increased the binding of Cdk9-HA–CycT1 to wild-type Tat (Fig. 4A, compare lanes 2 and 4) but that phosphatase treatment slightly decreased the binding (compare lanes 2, 4, and 5). In contrast, adding the same amount of phosphatase to a binding reaction mixture prior to the gel shift electrophoresis inhibited the formation of the Cdk9-T1Δ1–Tat–TAR complex but not the complex containing free T1Δ1 (T1Δ1-Tat-TAR) (Fig. 4B). These results indicated that autophosphorylation of Cdk9 did not significantly affect the P-TEFb–Tat interaction. Rather, it probably enhanced the binding of the P-TEFb–Tat complex to TAR RNA to facilitate P-TEFb–Tat–TAR complex formation.
FIG. 4.
P-TEFb phosphorylation does not affect P-TEFb–Tat binding but induces conformational changes in P-TEFb for better TAR recognition. (A) Equal amounts of in vitro-phosphorylated P-TEFb (Cdk9-HA–CycT1; lane 4), unphosphorylated P-TEFb (lanes 1 to 3), and alkaline phosphatase (AP; 10 U)-treated P-TEFb (lane 5) complexes were incubated with wild-type (WT) GST–Tat(1-48) proteins bound to glutathione-Sepharose beads. After washes, the amount of P-TEFb bound to Tat was examined by Western blotting with anti-CycT1 antibodies (α-CycT1). The GST–Tat(1-48, C22G) mutant protein, which contains a point mutation in the Tat activation domain, was used as a control (lane 3). (B) Alkaline phosphatase treatment inhibited the formation of the Cdk9–T1Δ1-Tat-TAR complex but not the complex containing free T1Δ1. The binding and gel shift conditions are the same as described for lane 11 of Fig. 1A, except that 10 U of AP was included in the binding reaction mixture in lane 3 prior to electrophoresis. (C) P-TEFb phosphorylation resulted in changes in trypsin sensitivity in the CycT1 C-terminal region. Purified Cdk9-Flag–HA-CycT1 complexes were first subjected to in vitro kinase reactions in the presence or absence of ATP, and then trypsin was added to the reaction mixtures. The cleaved products were detected by Western blotting with an anti-HA monoclonal antibody (α-HA) which recognizes the HA tag at the N terminus of CycT1. The position of a cross-reactive protein doublet is indicated by an ∗.
To explain the positive effect of Cdk9 autophosphorylation on the binding of the P-TEFb–Tat complex to TAR RNA, we asked whether phosphorylation induces a conformational change in P-TEFb which may be favorable for TAR recognition. Partial proteolysis was carried out on phosphorylated and unphosphorylated P-TEFb consisting of HA-CycT1 and Cdk9-Flag to look for phosphorylation-induced changes in trypsin-sensitive sites. Cleaved fragments of HA-CycT1 were visualized by Western blotting with anti-HA antibody. The patterns of CycT1 cleavage shown in Fig. 4C differed between the two forms of P-TEFb. In general, more cleaved fragments in the range of 37 to 55 kDa and fewer uncleaved HA-CycT1 fragments were observed with unphosphorylated P-TEFb than with the phosphorylated form. However, a novel CycT1 fragment of ∼48 kDa was observed only in phosphorylated P-TEFb. Since all the cleaved CycT1 fragments contained an HA tag at their N termini, their sizes allowed the mapping of the major trypsin-sensitive sites to a region C terminal to the cyclin box. We were, however, unable to detect trypsin-cleaved fragments of Cdk9-Flag by anti-Flag antibody Western blotting under this condition (data not shown), probably because it was protected by the associated CycT1. Nevertheless, the observed alteration of trypsin sensitivity in the C-terminal region of CycT1 suggested a phosphorylation-induced conformational change in CycT1 and/or Cdk9, which probably exposes the TAR-binding surface in CycT1 for possible P-TEFb–TAR interaction.
The N- and C-terminal regions of CycT1 interact in an intramolecular manner to inhibit P-TEFb–Tat–TAR complex formation.
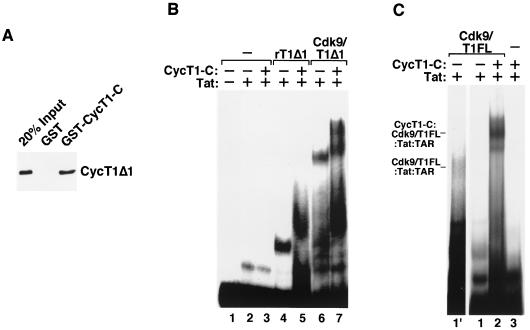
The data presented above suggested that P-TEFb needs to overcome two separate barriers in order to bind TAR RNA in conjunction with Tat. The unphosphorylated state of Cdk9 constitutes the first barrier, which can be overcome through Cdk9 autophosphorylation. The induced conformational change in P-TEFb probably facilitates P-TEFb–TAR binding. The second barrier resides in the C-terminal region of CycT1, which interfered with the binding of the P-TEFb–Tat complex to TAR RNA (Fig. 2C). To explore the mechanism for this inhibition, we asked whether the C-terminal region of CycT1 can fold back to interact with the CycT1 N-terminal region, thereby masking the TAR-binding surface. First, a GST pull-down assay was performed to test the interaction of CycT1Δ1, which contains the N-terminal cyclin box and its immediate flanking region (Fig. 2A), with GST–CycT1-C, which contains a CycT1 C-terminal fragment (amino acids 402 to 701). As shown in Fig. 5A, CycT1Δ1 was able to interact with GST-CycT1-C but not with GST alone, indicating a possible interaction between the N- and C-terminal regions of CycT1.
FIG. 5.
The CycT1 C-terminal region interacts with the N-terminal region to inhibit P-TEFb–Tat–TAR complex formation. (A) HA-tagged recombinant CycT1Δ1 proteins were incubated with equal amounts of GST or GST–CycT1-C (CycT1 C-terminal fragment, amino acids 402 to 701) bound to glutathione-Sepharose beads. After washes, bound CycT1Δ1 was detected by Western blotting with the anti-HA monoclonal antibody 12CA5. Twenty percent of the CycT1Δ1 used in the binding reaction mixture was shown as input. (B) Binding of CycT1-C to recombinant CycT1Δ1 (rT1Δ1) and the Cdk9–CycT1Δ1 complex (Cdk9/T1Δ1) in trans did not impede their interactions with Tat and TAR. Recombinant CycT1Δ1 (∼200 ng) or Cdk9-CycT1Δ1 (∼300 ng) was incubated in the presence or absence of CycT1-C (∼400 ng) for 10 min, followed by the addition of Tat (100 ng) and 32P-labeled TAR RNA. The reaction products were analyzed by gel mobility shift assay. (C) Binding of CycT1-C to Cdk9-CycT1FL disrupted the intramolecular interaction in CycT1 and stabilized the P-TEFb–Tat–TAR complex. Reaction mixtures containing CycT1-C (∼400 ng), Cdk9-CycT1FL (∼300 ng), Tat (100 ng), TAR, and ATP were analyzed as described for panel B. Lane 1′ is an eight-times-longer exposure of lane 1 and shows the position of Cdk9-CycT1FL–Tat–TAR in the gel.
Assuming that the hypothetical intramolecular interaction in CycT1 indeed inhibits the P-TEFb–TAR interaction, two possible mechanisms may account for this inhibition. The C-terminal region may directly contact and block the TAR-binding surface located in the N-terminal region, or it may contact a different N-terminal domain, but the intramolecular interaction would create steric hindrance to prevent the P-TEFb–TAR interaction. To distinguish between these two possibilities, recombinant CycT1-C was added to gel shift reaction mixtures to test its effect on the binding of free CycT1Δ1 or the Cdk9-CycT1Δ1 heterodimer to TAR RNA. Adding CycT1-C in trans did not inhibit TAR binding but rather resulted in a further retardation in the mobilities of the two complexes (Fig. 5B, lanes 4 to 7). In a control reaction, CycT1-C alone did not affect the Tat-TAR interaction (Fig. 5B, lane 3). The lack of inhibition by CycT1-C suggested that the binding surface between the CycT1 N and C termini does not overlap with the TAR recognition surface. Furthermore, when the N- and C-terminal regions were physically separated, their interaction in trans no longer produced the steric hindrance observed in the intact CycT1, and hence no inhibition was observed.
To further test the hypothesis that the intramolecular interaction produces a steric hindrance that causes the autoinhibition of CycT1-TAR binding, we attempted to disrupt this interaction by challenging full-length P-TEFb (Cdk9-CycT1FL) with excess amounts of recombinant CycT1-C. Compared with the weak P-TEFb–Tat–TAR complex formed in the absence of CycT1-C (Fig. 5C, lane 1′, an eight-times-longer exposure of lane 1), binding of CycT1-C to Cdk9-CycT1FL resulted in a dramatic enhancement of the P-TEFb–TAR interaction (compare lanes 1 and 2). Thus, the autoinhibition of TAR binding can be relieved by the disruption of the intramolecular interaction in CycT1 with CycT1-C. These results are consistent with the notions that the C-terminal region of CycT1 negatively regulates P-TEFb–Tat–TAR complex formation through interacting with the N-terminal region and that this intramolecular interaction sterically blocks the binding of the P-TEFb–Tat complex to TAR.
The C-terminal region of CycT1 interacts with Tat-SF1 and Pol II in HeLa nuclear extracts.
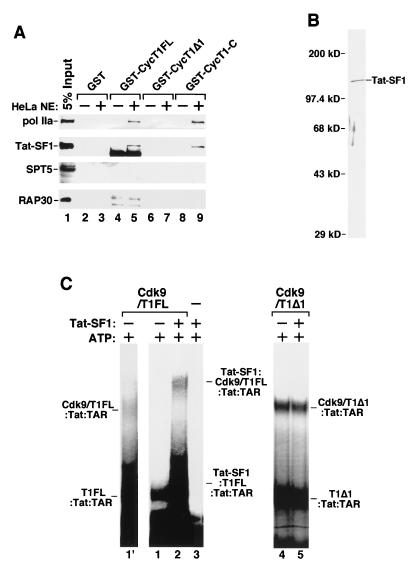
The above results indicated that the key to relieving the second autoinhibition of P-TEFb–Tat–TAR complex formation is to disrupt the intramolecular interaction in CycT1. Other than by artificially disrupting this interaction by causing competition with recombinant CycT1-C or removing the C-terminal region, we investigated the possibility that the CycT1-associated proteins may relieve the autoinhibition by interacting with and stabilizing the C-terminal inhibitory region of CycT1. First, we examined whether the C-terminal region of CycT1 may interact with components of a recently characterized Pol II elongation complex (31). Equal amounts of immobilized GST, GST-CycT1FL, GST-CycT1Δ1, and GST–CycT1-C proteins were incubated with HeLa nuclear extracts, and their abilities to bind to several known elongation factors and Pol II were analyzed by immunoblotting. Small fractions of RNA Pol IIa and Tat-SF1 in the nuclear extract were found to interact with CycT1FL and its C-terminal region (Fig. 6A, lanes 5 and 9) but not with the N-terminal region (CycT1Δ1; lane 7) or GST alone (lane 3). Unlike Pol IIa and Tat-SF1, little SPT5 (a subunit of DSIF) or RAP30 (a subunit of TFIIF) in the nuclear extract was found to associate with CycT1 under this condition (Fig. 6A), although a weak interaction between P-TEFb and DSIF was detected in a purified system (data not shown).
FIG. 6.
Tat-SF1 interacts with the C-terminal region of CycT1 and represses the autoinhibitory activity of this region. (A) The C-terminal region of CycT1 interacted with Tat-SF1 and RNA Pol IIa in HeLa nuclear extracts (NE). HeLa nuclear extracts were incubated with equal amounts of GST, GST-CycT1FL, GST-CycT1Δ1, and GST-CycT1-C proteins bound to glutathione-Sepharose beads. After washes, the bound proteins were analyzed by Western blotting with antibodies specific for Tat-SF1, Pol IIa, RAP30, and SPT5. Five percent of the nuclear extracts used in the binding reaction mixture were shown as input. The nonspecific bands shown in lanes 4 and 5 in anti-Tat-SF1 and anti-RAP30 antibody panels are cross-reactive bacterial proteins. (B) Silver-stained SDS-polyacrylamide gel showing Flag-tagged Tat-SF1 affinity purified from transfected 293T cells. (C) Binding of Tat-SF1 to the C-terminal region of CycT1 enhanced the P-TEFb–TAR interaction. Equal amounts of Cdk9–HA-CycT1FL (Cdk9/T1FL; lanes 1′, 1, and 2) and Cdk9–HA-CycT1Δ1 (Cdk9/T1Δ1; lanes 4 and 5) were incubated with Tat, 32P-labeled TAR, and ATP in the presence or absence of purified Tat-SF1. Reaction products were analyzed by gel mobility shift assay. Lane 1′ is an eight-times-longer exposure of lane 1 and shows the position of Cdk9–T1FL-Tat-TAR.
Binding of Tat-SF1 to the CycT1 C-terminal region stabilizes the P-TEFb–Tat–TAR complex.
Since the CycT1 C-terminal region interacts with Pol IIa and Tat-SF1, we investigated whether the association with Tat-SF1 would stabilize the binding of the P-TEFb–Tat complex to TAR. We affinity purified Flag-tagged Tat-SF1 protein from transiently transfected 293T cells, and the purified protein was analyzed by SDS-PAGE and silver staining (Fig. 6B). When tested in a gel mobility shift assay, Tat-SF1 did not recognize TAR RNA by itself (data not shown) or in the presence of Tat (Fig. 6C, lane 3), in spite of having two putative RNA recognition motifs (44). Because of the strong autoinhibition caused by the intramolecular interaction in CycT1, wild-type P-TEFb (Cdk9-CycT1FL) produced a very weak, albeit ATP-dependent (data not shown), P-TEFb–Tat–TAR complex visible only after a prolonged exposure of the autoradiogram (Fig. 6C, lane 1′ is an eight-times-longer exposure of lane 1). Importantly, preincubation of Flag-tagged Tat-SF1 with P-TEFb supershifted the P-TEFb–Tat–TAR complex and significantly enhanced the assembly of a multiprotein complex that most likely contained Tat-SF1, Cdk9-CycT1FL, Tat, and TAR (Fig. 6C, lane 2). The involvement of Tat-SF1 in forming this supershifted complex was also revealed by the observation that inclusion of anti-Flag or anti-Tat-SF1 antibody in the binding reaction mixture inhibited the complex formation (data not shown). This stimulatory effect of Tat-SF1 depended on the presence of ATP in the reaction mixture, suggesting that Tat-SF1 cannot overcome the first autoinhibitory step in P-TEFb, which is relieved only through Cdk9 autophosphorylation. In the same reaction as that shown in lane 2 of Fig. 6C, Tat-SF1 also interacted with and increased the binding to TAR RNA by free CycT1 (CycT1FL) present in the α-HA–CycT1 IP fraction. As predicted, the cooperative TAR binding promoted by Tat-SF1 requires the C-terminal region of CycT1, as Cdk9-CycT1Δ1 interacted strongly with Tat and TAR independently of Tat-SF1 (Fig. 6C, lanes 4 and 5). These results demonstrated an important role of Tat-SF1 in overcoming the intramolecular inhibition in CycT1 by binding to the CycT1 C-terminal region. In contrast to Tat-SF1, inclusion of purified calf thymus RNA Pol II in a binding reaction mixture did not lead to an enhanced binding of P-TEFb to Tat-TAR (data not shown).
We noticed that the stimulation of complex formation by Tat-SF1 did not reach the level attained by the deletion of the CycT1 C-terminal region (Fig. 6C, compare lane 2 with lanes 4 and 5). Assuming that the affinity of Cdk9-CycT1FL for Tat-TAR during transcription would reach the same level attained by Cdk9-CycT1Δ1, it is possible that other cellular factors may need to work together with Tat-SF1 to fully stabilize the P-TEFb–Tat–TAR complex.
The C-terminal region of CycT1 is required for efficient HIV-1 transcriptional elongation.
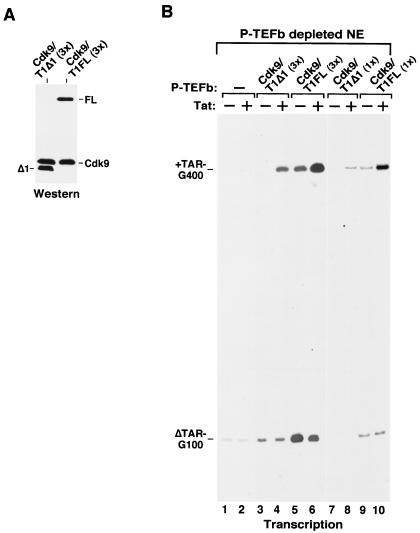
The binding of the CycT1 C-terminal region to Pol II and Tat-SF1 and the requirement of this region for the assembly of a multiprotein complex at TAR RNA suggested that this region may be important for basal and Tat-activated HIV-1 transcription. We analyzed the abilities of Cdk9-CycT1FL and Cdk9-CycT1Δ1 to mediate Tat activation in HeLa nuclear extracts immunodepleted of the endogenous P-TEFb. Immunoblotting analysis indicated only a trace amount of P-TEFb left in the depleted extract (data not shown), which was probably responsible for the very low level of HIV-1 transcription observed in this extract (Fig. 7B, lanes 1 and 2). Supplementing the depleted extract with two concentrations (1× and 3×) of wild-type P-TEFb (Cdk9-CycT1FL) allowed Tat to specifically increase the level of transcripts elongating beyond 1,000 nucleotides from an HIV-1 promoter containing the wild-type TAR element (pHIV+TAR-G400 [43]) but not from an internal control promoter with a mutant TAR (pHIVΔTAR-G100) (Fig. 7B, lanes 5, 6, 9, and 10). The reduced fold of Tat activation as a result of more P-TEFb (3×) being added to the reaction mixture (2.8-fold in lanes 5 and 6 versus 6.1-fold in lanes 9 and 10 after normalization to internal controls) was probably due to an efficient elongation mediated by high levels of P-TEFb, which partially bypassed the requirement for Tat.
FIG. 7.
The C-terminal region of CycT1 is required for efficient HIV-1 transcriptional elongation. (A) P-TEFb complexes containing either HA-tagged CycT1Δ1 or CycT1FL were normalized for their Cdk9 and CycT1 levels by Western blotting with anti-Cdk9 and anti-HA antibodies. (B) Equal amounts of the two P-TEFb complexes were added to transcription reaction mixtures containing P-TEFb-depleted HeLa nuclear extracts (NE) as well as the DNA templates pHIV+TAR-G400 and pHIVΔTAR-G100 in the presence (+) or absence (−) of Tat protein. The amount of P-TEFb analyzed in lanes 3 to 6 was three times (3×) higher than that in lanes 7 to 10. +TAR-G400 and ΔTAR-G100 are RNase T1-resistant RNA fragments transcribed from the two G-less DNA cassettes (400 and 100 bp) inserted, respectively, into pHIV+TAR-G400 and pHIVΔTAR-G100 at a position ≈1 kb downstream of the HIV-1 promoter region.
Compared with the same amount of wild-type P-TEFb, the ability of Cdk9-CycT1Δ1 to mediate basal, TAR-independent HIV-1 transcription decreased by about 14-fold (Fig. 7B, compare the ΔTAR-G100 signals between lane 3 and lane 5). Meanwhile, the Tat-specific and TAR-dependent transcription mediated by Cdk9-CycT1Δ1 decreased by up to sixfold (compare the +TAR-G400 signals between lane 8 and lane 10; the decrease was threefold between lanes 4 and 6, which contained 3× P-TEFb). Because of the more severe reduction in basal activity, the fold of Tat activation supported by Cdk9-CycT1Δ1 was slightly better than that supported by wild-type P-TEFb.
These results indicated that the C-terminal region of CycT1 is crucial for basal HIV-1 transcription, as this domain may mediate the interaction of P-TEFb with Tat-SF1, Pol II, and perhaps other components of the elongation apparatus. The lack of stable interaction between Cdk9-CycT1Δ1 and the elongation apparatus may result in inefficient phosphorylation of the Pol II CTD and hence a marked reduction in basal elongation. These results are consistent with the previous findings that truncation of the C-terminal region of Drosophila CycT1 reduced the basal activity to about 10% (33, 34).
Our data also indicated that the CycT1 C-terminal region is important for Tat-specific and TAR-dependent HIV-1 transcription, although it has a less pronounced effect than in basal elongation. Unlike in the Tat-independent elongation process, it is possible that the presence of Tat can at least recruit Cdk9-CycT1Δ1 to the HIV-1 promoter (Fig. 2C), and the existence of alternative weak interactions between the Cdk9-CycT1Δ1–Tat–TAR complex and the general elongation apparatus independent of the CycT1 C-terminal region may be responsible for the observed weak Tat-specific transcription mediated by Cdk9-CycT1Δ1. Previous reports of the associations of Tat with the Pol II holoenzyme (4, 31) and Tat-SF1 (44) may provide such alternative interactions.
It is important to note that adding an amount of Cdk9-CycT1Δ1 about three times larger than that of endogenous P-TEFb in HeLa nuclear extract into P-TEFb-depleted reaction mixtures (lanes 3 and 4 of Fig. 7B) appeared to further strengthen these alternative interactions, leading to a diminished requirement of the CycT1 C-terminal region for efficient Tat-specific transcription (compare lanes 4 and 6). In agreement with this observation, overexpression of C-terminally truncated human CycT1 in rodent cells has been shown to support Tat-dependent HIV-1 transcription with full to about half the capacity of full-length CycT1 (10, 16).
DISCUSSION
The formation of a TAR loop-dependent P-TEFb–Tat–TAR complex is essential for Tat-specific and TAR-dependent stimulation of HIV-1 transcription. The data presented here provide a mechanistic view of how a high-affinity P-TEFb–Tat–TAR complex is assembled. Interestingly, assembly of this complex is a regulated process involving the relief of two autoinhibitory mechanisms in P-TEFb. Most of the P-TEFb heterodimers isolated from human cells appear to be intrinsically inactive in forming stable P-TEFb–Tat–TAR complexes. P-TEFb undergoes conformational changes in at least two controlled steps and requires the help from another elongation factor(s) in order to form a high-affinity complex on TAR RNA.
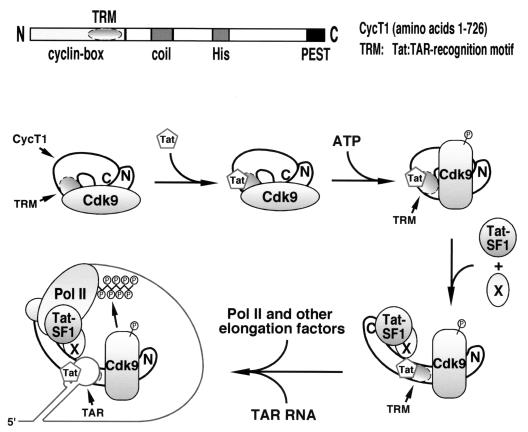
Previous alanine-scanning mutagenesis of the CycT1 TRM identified residues that are critical for the interaction with Tat and others that are required specifically for binding of the Tat-CycT1 complex to TAR RNA (10). Thus, the CycT1 TRM makes independent contacts with Tat and TAR. As depicted in the diagram shown in Fig. 8, the data presented here are consistent with the model that the TRM subdomain required for P-TEFb–TAR interaction is blocked, either directly or indirectly, by both unphosphorylated Cdk9 and the C-terminal region of CycT1, which folds back to interact with the N-terminal region. In contrast, the part of TRM specific for Tat binding appears to be accessible irrespective of the phosphorylation state of Cdk9 (Fig. 4A) and the intramolecular interaction in CycT1 (2). Relief of the first autoinhibition requires Cdk9 autophosphorylation, which alters the conformation in CycT1 and/or Cdk9 and unmasks the critical TRM subdomain for possible interaction with TAR RNA. It is interesting that a mutant P-TEFb with a kinase-defective Cdk9 subunit was incapable of forming a stable complex with Tat and TAR (Fig. 3C), suggesting that the failure of this mutant elongation factor to mediate Tat activation is not simply due to its defective CTD kinase activity (42) but rather to its inability to bind TAR RNA and to be recruited to the HIV promoter at an earlier stage.
FIG. 8.
Model for the assembly of the P-TEFb–Tat–TAR complex through relief of two built-in autoinhibitory mechanisms in P-TEFb. See the text for details. It is important to point out that in this diagram the order of events depicted between the first and last steps is purely hypothetical.
Autophosphorylation of Cdk9 only partially exposes the subdomain of TRM for TAR recognition (Fig. 8). The efficient interaction between P-TEFb and TAR RNA also requires suppression of the autoinhibitory activity of the CycT1 C-terminal region. The intramolecular interaction of the CycT1 C-terminal region with the N-terminal half probably creates steric hindrance that blocks the access to TRM by TAR RNA (Fig. 8). To disrupt this intramolecular interaction and relieve the autoinhibition, transcription elongation factor Tat-SF1 interacts with the C-terminal region and markedly enhances the binding of the Tat-SF1–P-TEFb–Tat complex to TAR RNA (Fig. 6C). However, the effect of Tat-SF1 did not approach the level attained by the deletion of the CycT1 C-terminal domain. One possibility is that another cellular factor(s) (Fig. 8) works together with Tat-SF1 to fully stabilize the complex. Indeed, in addition to Tat-SF1, the CycT1 C-terminal region was also shown to interact with Pol IIa and perhaps other components of the Pol II elongation apparatus. Upon its release from the intramolecular interaction, this region may function as a bridge linking the P-TEFb–Tat–TAR ternary complex with the basal elongation apparatus. In support of this model, when a physiological amount of P-TEFb was analyzed in transcription reactions, the C-terminal region of CycT1 was found to be required for both basal and Tat-activated HIV-1 transcription (Fig. 7), although other minor interactions between the P-TEFb–Tat–TAR complex and the basal elongation apparatus may also contribute to the assembly of a highly processive, multicomponent Pol II elongation machinery (Fig. 8). Tat-SF1 was biochemically identified as a Tat-specific cellular cofactor (44). Our results provide a plausible explanation for the Tat-specific elongation activity of this factor. Since Tat-SF1 binds to Tat and contains two RNA recognition motifs (44), future experiments are necessary to determine whether these properties of Tat-SF1 contribute to cooperative TAR recognition by Tat-SF1, P-TEFb, and Tat.
Autoinhibition, involving intramolecular interactions that negatively regulate the function of otherwise autonomous modules, is observed in many transcription factors (for a review, see reference 14). For instance, SNAPc, a core promoter-binding factor required for transcription of RNA Pol II and Pol III snRNA promoters, was shown to have an autoinhibitory C-terminal region that represses the binding of SNAPc to DNA (28). Interestingly, like the derepression effect mediated by the interaction of Tat-SF1 with P-TEFb, repression of SNAPc DNA binding can be relieved by the interaction of SNAPc with the Oct-1 POU domain that promotes cooperative binding. Other examples of transcription factors with built-in negative control of DNA binding include Escherichia coli ς70 (7) and the largest subunit of TFIID from both Drosophila and yeast (21, 22). The similarities between P-TEFb and these transcription factors suggest the existence of a common autoinhibition and derepression mechanism to ensure specific and stable interactions of these transcription factors with either RNA recognition sequences or promoter DNA.
We noticed that most of the Cdk9-CycT1 isolated from stably transfected human 293 cells required an in vitro autophosphorylation step to form stable P-TEFb–Tat–TAR complexes, implying the lack of a key Cdk9 phosphorylation that is important for complex formation. It is not clear what effect this may have on the general and Tat-specific elongation activity of P-TEFb in vivo. Nevertheless, our observations raise an interesting possibility that the regulation of Cdk9 phosphorylation may provide a novel control step for Tat activity and HIV-1 transcription in infected cells. Future experiments to map and mutate the phosphorylated residue(s) in Cdk9 and to test the activity of the mutant construct(s) in HIV-1 transcription may allow us to test this possibility.
Although our purified in vitro system permits autophosphorylation of only Cdk9, Cdk9 can theoretically be phosphorylated and activated by other kinases in vivo. During the activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines, both of which are relevant for HIV infection, a dramatic increase in HIV-1 gene expression (12) as well as P-TEFb kinase activity has been observed (12, 15, 40). The induction of P-TEFb activity has been attributed to an increase in Cdk9 and CycT1 levels in activated peripheral blood lymphocytes and an increase of CycT1 in differentiated monocytes (12, 40). However, the pharmacological reagents (phorbol esters, ionomycin, and phytohemagglutinin) used to treat the cells are known to activate a spectrum of protein kinases, one of which may in turn phosphorylate Cdk9, stabilize the P-TEFb–Tat–TAR complex, and increase the Tat-specific activity of P-TEFb. Besides cellular kinases that may modulate P-TEFb activity, specific phosphatases may also be regulated to maintain appropriate levels of phosphorylated P-TEFb in vivo in response to environmental stimuli and perhaps also during the cell cycle. Since induction of P-TEFb activity in activated T cells and differentiated macrophages may contribute directly to high levels of HIV-1 transcription and the escape from latency of transcriptionally silent proviruses (15), it is important to investigate whether induction of P-TEFb phosphorylation may contribute directly to these processes.
ACKNOWLEDGMENTS
We thank K. Luo and S. Stroschein for valuable comments on the manuscript and K. Henning for technical support. We also thank members of the Zhou and Luo laboratories for fruitful discussions.
This work was supported by grants from the National Institutes of Health (AI-41757), the University of California Universitywide AIDS Research Program (R97-B-113), and the U.S. Army Breast Cancer Research Program (DAMD17-96-1-6137) to Q.Z.
REFERENCES
- 1.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen D, Fong Y, Zhou Q. Specific interaction of Tat with the human but not rodent P-TEFb complex mediates the species-specific Tat activation of HIV-1 transcription. Proc Natl Acad Sci USA. 1999;96:2728–2733. doi: 10.1073/pnas.96.6.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun R F, Jeang K T. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus I and HIV-1. J Biol Chem. 1996;271:27888–27894. doi: 10.1074/jbc.271.44.27888. [DOI] [PubMed] [Google Scholar]
- 4.Cujec T P, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin B M. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 7.Dombroski A J, Walter W A, Gross C A. Amino-terminal amino acids modulate sigma-factor DNA-binding activity. Genes Dev. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- 8.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 9.Fujinaga K, Taube R, Wimmer J, Cujec T P, Peterlin B M. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci USA. 1999;96:1285–1290. doi: 10.1073/pnas.96.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Martinez L F, Mavankal G, Neveu J M, Lane W S, Ivanov D, Gaynor R B. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garriga J, Peng J, Parreno M, Price D H, Henderson E E, Grana X. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene. 1998;17:3093–3102. doi: 10.1038/sj.onc.1202548. [DOI] [PubMed] [Google Scholar]
- 13.Gold M O, Yang X, Herrmann C H, Rice A P. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J Virol. 1998;72:4448–4453. doi: 10.1128/jvi.72.5.4448-4453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves B J, Cowley D O, Goetz T L, Petersen J M, Jonsen M D, Gillespie M E. Autoinhibition as a transcriptional regulatory mechanism. Cold Spring Harbor Symp Quant Biol. 1998;63:621–629. doi: 10.1101/sqb.1998.63.621. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann C H, Carroll R G, Wei P, Jones K A, Rice A P. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J Virol. 1998;72:9881–9888. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov D, Kwak Y T, Nee E, Guo J, Garcia-Martinez L F, Gaynor R B. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for tat-activation. J Mol Biol. 1999;288:41–56. doi: 10.1006/jmbi.1999.2663. [DOI] [PubMed] [Google Scholar]
- 17.Jeang K T, Xiao H, Rich E A. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 18.Jones K A. Taking a new TAK on tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 19.Kato H, Sumimoto H, Pognonec P, Chen C H, Rosen C A, Roeder R G. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 1992;6:655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- 20.Kim J B, Yamaguchi Y, Wada T, Handa H, Sharp P A. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol Cell Biol. 1999;19:5960–5968. doi: 10.1128/mcb.19.9.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kokubo T, Swanson M J, Nishikawa J I, Hinnebusch A G, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kokubo T, Yamashita S, Horikoshi M, Roeder R G, Nakatani Y. Interaction between the N-terminal domain of the 230-kDa subunit and the TATA box-binding subunit of TFIID negatively regulates TATA-box binding. Proc Natl Acad Sci USA. 1994;91:3520–3524. doi: 10.1073/pnas.91.9.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X Y, Green M R. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes Dev. 1998;12:2992–2996. doi: 10.1101/gad.12.19.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mancebo H S, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marciniak R A, Sharp P A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall N F, Peng J, Xie Z, Price D H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 27.Marshall N F, Price D H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 28.Mittal V, Ma B, Hernandez N. SNAP(c): a core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes Dev. 1999;13:1807–1821. doi: 10.1101/gad.13.14.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamoto H, Sheline C T, Corden J L, Jones K A, Peterlin B M. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:11575–11579. doi: 10.1073/pnas.93.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Keeffe B, Fong Y, Chen D, Zhou S, Zhou Q. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated Tat stimulation of HIV-1 transcription. J Biol Chem. 2000;275:279–287. doi: 10.1074/jbc.275.1.279. [DOI] [PubMed] [Google Scholar]
- 31.Parada C A, Roeder R G. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 1999;18:3688–3701. doi: 10.1093/emboj/18.13.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parada C A, Roeder R G. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 33.Peng J, Marshall N F, Price D H. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J Biol Chem. 1998;273:13855–13860. doi: 10.1074/jbc.273.22.13855. [DOI] [PubMed] [Google Scholar]
- 34.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 38.Wu-Baer F, Lane W S, Gaynor R B. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J Mol Biol. 1998;277:179–197. doi: 10.1006/jmbi.1997.1601. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 40.Yang X, Gold M O, Tang D N, Lewis D E, Aguilar-Cordova E, Rice A P, Herrmann C H. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc Natl Acad Sci USA. 1997;94:12331–12336. doi: 10.1073/pnas.94.23.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X, Herrmann C H, Rice A P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Q, Sharp P A. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Q, Sharp P A. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]