Abstract
Objectives: Aminopeptidase N (APN) is an enzyme highly expressed in metastatic cancers and could be used in targeted cancer therapy. Our previous work showed the successful construction of CNGRC–carboxypeptidase G2 (CPG2) and CNGRC–CPG2–CNGRC fusion proteins. Our conjugates and prodrugs were effective in targeting high APN-expressing cancer cells. In the present study, we aim to produce long-acting fusion proteins to overcome 2 of the main drawbacks of antibody-directed enzyme prodrug therapy. Methods: N-terminal and N-, C-terminal fusion CPG2, CNGRC–CPG2, and CNGRC–CPG2–CNGRC, respectively, were PEGylated using polyethylene glycol (PEG) maleimide (40K). We examined the effect of PEGylation on the therapeutic efficacy of the new products. The resulting PEGylated fusion proteins were tested for their stability, ex vivo immunotoxicity, binding capacity to their target on high HT1080, and low A549 APN-expressing cells. The catalytic activity of the resulting PEGylated fusion CPG2 proteins was investigated. Pro-drug “ZD2767P” cytotoxic effect in association with PEG CPG2–CNGRC fusion proteins on cancer cells was studied. Results: Our work demonstrated that the properties of the PEGylated single-fused proteins were significantly improved over that of un-PEGylated fused CPG2, and its kinetic activity and APN-binding affinity were not negatively affected by the PEGylation. Significantly, The PEGylated single-fused CPG2 had lower immunogenicity than the un-PEGylated CPG2. Our results, however, were different in the case of the PEGylated double-fused CPG2. Although its stability in human serum under physiological conditions was not significantly affected, the kinetic activity and its binding affinity to their cellular marker (APN) were substantially reduced. When the study was performed with high and low APN-expressing cancer cell lines, using the prodrug ZD2767p, the PEGylated fusion CPG2 demonstrated cancer cell killing effects. Conclusion: We have successfully produced PEGylated-CNGRC–CPG2, which is bioactive and with lower immunogenicity in ligand-directed enzyme prodrug therapy for cancer treatment.
Keywords: PEGylated CPG2 conjugates, ligand-directed enzyme prodrug therapy, targeted cancer therapy, aminopeptidase N, immunogenicity, long-acting drugs
Introduction
The use of anticancer agents has faced the drawbacks of poor solubility, short in vivo half-life, and low specificity to tumor cells resulting in low-therapeutic efficacy and serious side effects. 1 PEGylation (conjugation of protein and/or drug with polyethylene glycol “PEG” polymer) is a well-established approach that has been increasingly widely used to improve pharmacokinetics and therapeutic properties of peptides and drugs used in cancer therapy.2–4 The process of PEGylation is about covalent attachment of the PEG moiety to the protein/drug of interest. The immense size of the polymer adds to the molecular mass of the PEGylated protein, and it enhances the hydrophilic nature of the polymer. Moreover, due to the flexible PEG chain around the protein, it wraps around the molecule masking sites or epitopes recognized by the immune cells, receptors or proteolytic enzymes, thus increasing its half-life and lowering the immunotoxicity. 5
Several therapeutic PEGylated proteins have recently been employed clinically such as those for interferon-α, granulocyte colony-stimulating factor, urate oxidase and a monoclonal antibody against tumor necrosis factor-α.6–9 We have previously succeeded in conjugating a PEG polymer to the therapeutic enzyme glucarpidase (carboxypeptidase G2 [CPG2]) known to detoxify methotrexate (MTX) in an overdose and to activate the benzoic mustard inactive prodrugs to highly cytotoxic drugs, and we have generated a PEGylated biobetter glucarpidase variant (CPG2). 10 The PEGylated CPG2 produced exhibited significantly improved properties compared with those of the non-PEGylated one, and found to enhance its kinetic activity, and in vitro serum stability in addition to being able to lower ex vivo immunotoxicity.
Targeted cancer therapy (ie, the anticancer drug directed to bind specifically to the tumor tissue) provided an effective tool to overcome the low-drug specificity, thus improving anticancer efficacy. 11 Cancer metastasis is one of the major causes of death from cancer, and recent therapies have been tailored to target certain markers involved in the cancer cell metastasis.
Aminopeptidase N (APN) is one of the enzymes known to be highly involved in processes of tumor cell invasion and neovascularisation. 12 A small peptide (asparagine–glycine–arginine) (NGR) recognized by phage display libraries that specifically bind to APN receptor has been identified. 13 Several studies have exploited such cancer marker binding peptides to design a tumor targeted drug and/or enzyme and to overcome the low specificity of anticancer therapy.14–17
Our recent study presented a developed fusion protein using the APN-binding peptide (CNGRC) conjugated to the therapeutic enzyme CPG2, 18 where the resulting constructs; N-terminal single and N-, C-termini double fusion proteins found to provide new therapeutic fusion proteins that can be used in the ligand-directed enzyme prodrug therapy (LDEPT). The fusion proteins produced showed notably enhanced enzyme kinetics of CPG2 and protein stability, and lowered ex vivo immunotoxicity compared to those of the nonfused CPG2 (wild type (WT)). Moreover, the fusion proteins preserved the APN-binding affinity in cancer cells, and in combination with the prodrug (ZD2767P), higher cytotoxic effect resulted with high APN-expressing cells pretreated with the fusion proteins.
In the current study, we combined the 2 approaches and applied PEGylation to the previously generated fusion proteins (N-terminal fusion, CNGRC–CPG2 and will designated as X-CPG2″ and N-, C-terminal fusion, CNGRC–CPG2–CNGRC and will be referred to as “X-CPG2-X”) producing PEGylated fusion proteins (PEGylated single fusion “X-CPG2-PEG”and Pegylated double fusion “X-PEG-CPG2-X” fusion proteins). We have assessed the effectiveness of PEGylated fusion proteins as potential therapeutic compounds for use in combination of a prodrug for targeted cancer therapy. We have evaluated their ex vivo stability and immunotoxicity, explored the effect of PEGylation on the fusion protein structure and the resulting effects on binding affinity of PEGylated fusion proteins to their targeted marker (APN), kinetic activity and prodrug cytotoxic effects along with PEGylated fusion proteins.
Methods
Materials
Cell lines used were purchased from American Type Culture Collection, including MDA-MB468 (human breast adenocarcinoma, HTB-132), MDA-MB231 (human breast adenocarcinoma, HTB-26), A549 (human lung carcinoma, CCL-158), HT-1080 (human fibrosarcoma, CCL-121), HepG2 (human hepatocellular carcinoma, HB-8065), FaDu (human pharyngeal squamous cell carcinoma, HTB-43) cell lines. For fusion protein PEGylation the reagent used was Y-shape PEG Maleimide MW 40 000 (JenKem Technology), dialysis of protein was done using Spectra/Por 7 Dialysis Tubing (10 kDa and 50 kDa MWCO) (Fischer Scientific). The kits used for the immunogenicity assay Pierce™ High Capacity Endotoxin Removal Resin Pierce™ and LAL Chromogenic Endotoxin Quantitation purchased from thermo Fisher Scientific, and Cell Counting Kit – 8 purchased from sigma. The antibodies used were: the primary antibodies (Mouse 6x-His Tag Monoclonal Antibody [HIS.H8] from Invitrogen, Rabbit polyclonal anti Xen-CPG2 antibody from GE Healthcare and Rabbit anti-PEG antibody [PEG-B-47] [ab51257] from ABCAM) and HRP-labeled secondary antibodies (goat anti-Mouse immunoglobulin G (IgG) H&L [ab205719] and goat anti-Rabbit IgG H&L [ab6721] from ABCAM). The prodrug used (ZD2767P) supplied as solid crystalline powder by the Inovapharm Ltd and 2 mM stock solution was prepared by dissolving 1 mg in 1 mL sterile DMSO. Protein purification was carried using nickel affinity chromatography, Ni-NTA resin (Sigma), as a DNA marker Quick-Load® Purple1kbDNA Ladder (NEB) was used, for protein marker See Blue Plus2 Prestained ladder (198-10 kDa) (Thermo Fischer Scientific) was used.
PEGylation and Purification of the CNGRC–CPG2 Fusion Proteins
Purified fusion proteins (x–CPG2 and x–CPG2–x) in addition to the WT CPG2 protein, were dialyzed in 1 × PBS pH6 and 9mg/mL of the protein was mixed with 3-fold molar excess of Y-shape PEG Maleimide MW 40 000 (Y-MAL-40 K) (JenKem Technology) which targets cysteine (-SH) moiety in the protein, with immediate rapid vigorous mixing by vortex and incubated in shaking incubator at 30 °C, 200 RPM for 10 h. The PEGylated protein mixture was then subjected to Ni–NTA purification to remove the excess nonreactive PEGylation reagent and dialyzed in 1 × PBS followed with further purification using AKTA fast protein liquid chromatography (AKTA FPLC) purification system and gel filtration. Finally, the pure form of the PEGylated CPG2 conjugate was isolated and quantified. The resulting proteins were visualized on 10% Sodium dodecyl-sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels by staining with Coomassie brilliant blue.
Circular Dichroism Spectroscopy of the PEGylated CPG2 Fusion Proteins:
Prior to circular dichroism (CD) scanning purified PEGylated CPG2 fusion proteins were dialyzed in Distilled water at least 3 times each for 18 h, then any aggregates or precipitated proteins were removed by centrifugation at maximum speed (12 000 rpm) 4 °C/30 min. Protein absorbances obtained from the ANanoDrop2000 spectrophotometer was used to calculate protein concentrations(using the extension coefficient of each protein that were taken as ε = 23 380 M−1 cm−1 for WT CPG2, 23 505 M−1 cm−1 for X-CPG2 and 23 630 for X-CPG2-X measured using a Nanodrop 2000) to adjust required concentration for CD measurement. The CD scanning of the PEGylated fusion proteins was carried from 180 to 260 nm in the far ultraviolet (UV) spectral region using Chirascan™ Plus CD Spectrometer (Applied Photophysics). Prepared samples (about 70 µL of 10 µM) concentration were placed in a SUPRASIL Quartz cuvette demountable rectangular “.2 mm light-path length” (Hellma®). The applied CD parameters were as previously mentioned, 10 band width 1 nm, scan time/point of .5 s, and for each sample 4 scans were taken, which were averaged and smoothed using the CD analysis software. The produced spectra were subtracted from an averaged CD spectrum of the used blank (Milli Q water) baseline. The resulting normalized averaged readings along with the molecular weights and number of amino acid residues (PEG-WT as 392 AA and 41 761.48 Da, for PEG X-CPG2 403 AA and 42 668.53 Da and for X-PEGCPG2-X 410 AA and 43 316.25 Da.) were further analyzed by CD data deconvolution analysis using the CDNN (version 2.1) software tool in the spectral range of (180-260 nm) and 0.02 cm light-path length of the cuvette.
Enzymatic Activity and Plasma Stability of PEGylated CPG2 Fusion Proteins
As previously described, CPG2 catalytic activity was assessed employing MTX. 19 Briefly, purified PEGylated CPG2 fusion protein (50 µg/mL) was added to a mixture of MTX with its buffer (0.1 M Tris-HCl pH 7.3 containing 0.2 mM ZnSO4 and 5 µL of MTX “0.45 mM”) and incubated at 37 οC. The decrease in absorbance was measured every 10 min using NANODROP 1000 spectrophotometer (Thermo Scientific). Different MTX concentrations were used to determine the catalytic activity of the PEGylated CPG2 fusion proteins. The purified PEGylated proteins (2.12 µg/mL) were incubated with different MTX concentrations (0.03 up to 0.42 mM), and the rate of MTX hydrolysis was measured for every 2 min using Infinite M200 PRO NanoQuant Plate Reader (TECAN) at 320 nm. To determine Km, Kcat, and Vmax values of each protein Michaelis–Menten equation was employed using Graph Pad PRISM 6 software. A unit of CPG2 represents the amount of enzyme (mg) required for hydrolysis of 1 mM of MTX t 37 °C. The molar extinction coefficient for MTX (8300) used to calculate enzyme activity/mL. To investigate plasma stability of the PEGylated CPG2 fusion proteins, the purified proteins (0.1 µL/µg) were incubated with the separated plasma from normal healthy blood donors at 37 °C for 14 days. The catalytic activity of CPG2 was tested every 48 h by taking a sample from the incubated protein–plasma mixture and carry the previously mentioned MTX assay.
T-Cell Proliferation Assay for Immunotoxicity
T-cell proliferation assay was used as an indicator for the immunotoxicity of PEGylated CPG2 fusion proteins. The purified protein samples were first tested for the level of endotoxin using Pierce LAL Chromogenic Endotoxin Quantitation Kit. The samples with high endotoxin level were cleaned up using Pierce™ High Capacity Endotoxin Removal Resin until get the level of endotoxin ˂0.1 EU/mL. Peripheral blood mononuclear cells (PBMCs) from normal blood samples were isolated (1 × 10 6 cells/mL in 96 well plate) and incubated with 10 µg/mL purified PEGylated CPG2 fusion proteins (endotoxin level ˂ 0.1 EU/mL) in X-VIVO™ 15 media 48 h at 37 °C in CO2 incubator. Negative (PBS only) and positive (LPS) controls were also subjected to similar conditions. CCK-8 kit was used to determine T-cell proliferation rate. As indicated by the manufacturer (Sigma), the CCK-8 working solution (10 µL) was added to each well, followed by incubation at 37 °C for 4 h. The resulting color intensity was measured using Infinite M200 PRO NanoQuant Plate Reader (TECAN).
The Binding Capacity of PEGylated CPG2 Fusion Proteins to Cancer Cell Lines Expressing Different Levels of APN
As in our previous study, 18 we used 2 cell lines as models for high (HT1080) and low (A549) APN-expressing cell lines. In a 96 well plate 25 000 cells/well were seeded, and the following day the cells were fixed with 4% paraformaldehyde. After washing with 1 × phosphate buffered saline tween (PBST), 5% bovine serum albumin was added used for blocking (1 h) at room temperature. Next, the cells were properly washed at least 3 times with 1 × PBST, prior to adding varying concentrations of purified PEGylated CPG2 fusion proteins (0.063, 0.127, 0.192, and 0.255 µM) in addition to the vehicle PBS “0 µM” (negative control). The plate was incubated for 1 hour at room temperature, then washed twice with 1 × PBST followed by incubation with anti-His6-HRP monoclonal antibody (1:1000). After washing with 1 × PBST twice, the substrate for HRP, TMB was added (100 µL/well) and the developing color was stopped after 10 to 15 min with 2 N H2SO4. Infinite M200 PRO NanoQuant Plate Reader (TECAN) was used to measure the resulting color absorbance at 450 nm. The resulting absorbance readings were employed to calculate the dissociation constant (Kd) applying nonlinear regression analysis using GraphPad PRISM.
In Vitro MTX and/or ZD2767P Prodrug Cytotoxic Efficacy on Cells Pretreated With PEGylated CPG2 Fusion Proteins
The 2 groups of cells: high (HT1080, MDA-MB468 and HepG2) and low (A549, MDA-MB231 and FaDu) APN-expressing cells lines were plated in 96 well plate (20 000 cells/well). Next day, prior to treatment of cells with MTX or the prodrug, cells were incubated for 2 hours with 80 µg/mL of the purified PEGylated CPG2 fusion proteins followed by washing with 1 × PBS to eliminate any unbound PEGylated CPG2 fusion proteins. Then the controls were either treated with the PEGylated CPG2c fusion protein only “negative” or the MTX alone “positive”, or the test groups treated either with 5 nM of MTX or the prodrug ZD2767P (0.1 µM). The plate was incubated at 37 °C for 1 to 2 h, then washed and replaced with their complete media and incubated at 37 °C and 5% CO2 for 2 days. To test the cell viability after 2 days, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a tetrazole (MTT) assay was used. Briefly, working MTT solution (5 mg/mL) was added to each well, mixed, and incubated for 4 h. Next, the cells were washed with PBS before solubilizing the formed crystals in MTT solvent, and the resulting color was measured at 570 nm.
Data Analysis
All treatments were performed in triplicate, and the statistical significance was determined using Student's t-test (2-tailed) where the results considered significant at P ≤ .05. Dissociation constant Kd was obtained using GraphPad PRISM 5 software and the nonlinear regression analysis.
Results
Preparation and Identification of the PEGylated CPG2 Fusion Proteins
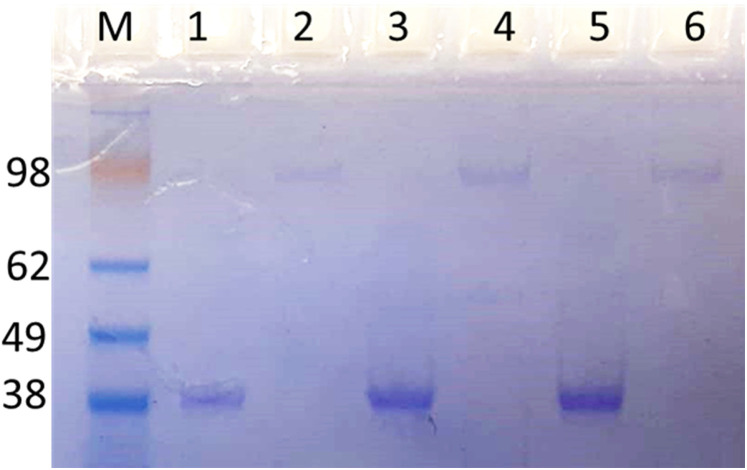
Successful reaction of PEG with the fusion proteins was demonstrated by an increase in the molecular weight of the fusion proteins indicated in the band shift (>100 KDa) by Coomassie blue stained SDS-PAGE. Figure 1 shows the purified Pegylated products after separation from the unconjugated PEG with AKTA FPLC purification system and gel filtration. The approximate molecular weight of the CPG2 is 40 kDa and the PEG is the same molecular weight and the ratio of PEG to CPG2 s about 1:1.
Figure 1.
Identification of PEGylated CPG2 proteins. The resulting PEG–CPG2 fusion proteins (WT “PEG-CPG2”, single “PEG X-CPG2” and double “PEG X-CPG2-X” fusion proteins) were separated on SDS-PAGE and stained with Coomassie brilliant blue. Un-PEGylated proteins (WT, X-CPG2, and X-CPG2-X) in lanes 1, 3, and 5, and PEGylated proteins are shown in lanes 2, 4, and 6, respectively. Abbreviations: PEG, polyethylene glycol; CPG2, carboxypeptidase G2.
Circular Dichroism Spectral Analysis of the PEGylated Fusion Proteins
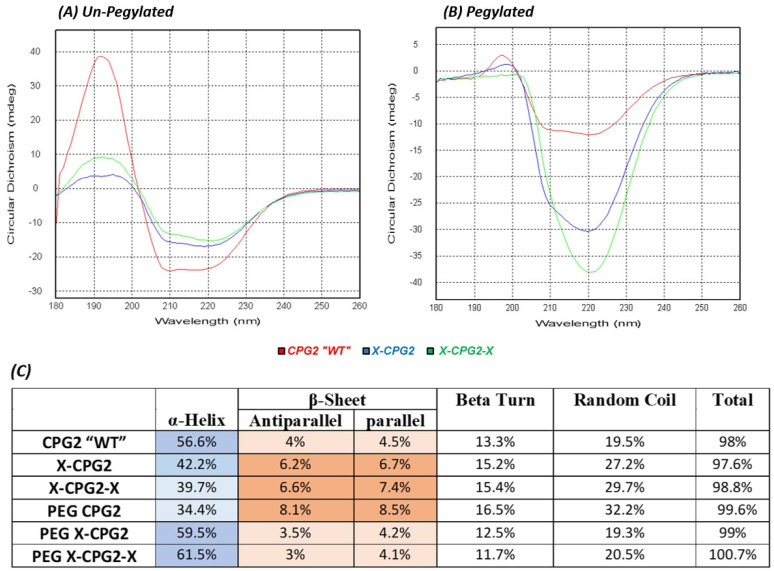
The secondary structure of the PEGylated fusion proteins was examined by CD spectroscopy. As the results in Figure 2 show the far UV spectra combined for the un-PEGylated proteins (WT, X-CPG2, and X-CPG2-X), CD spectrum showed negative peaks (−15 mdeg) at 208 and 220 nm) for the un-PEGylated fusion proteins.
Figure 2.
Circular dichroism spectra of CPG2 fusion proteins before and after PEGylation. (A) The combined far UV spectra of CPG2 “WT”, single “X-CPG2” and double “X-CPG2-X” fusion proteins. (B) The combined far UV spectra of PEG CPG2, PEG X-CPG2, and PEG X-CPG2-X fusion proteins. (C) Table presents the results of CDNN deconvulation analysis of the PEGylated and non-PEGylated CPG2 fusion proteins CD spectra, showing their secondary structure composition. Darker shades indicate an increase in the percentage composition. Abbreviations: UV, ultraviolet; PEG, polyethylene glycol; CPG2, carboxypeptidase G2; CD, circular dichroism.
Differential Effect of PEGylation on Enzymatic Activity and Stability of the CPG2 Fusion Proteins
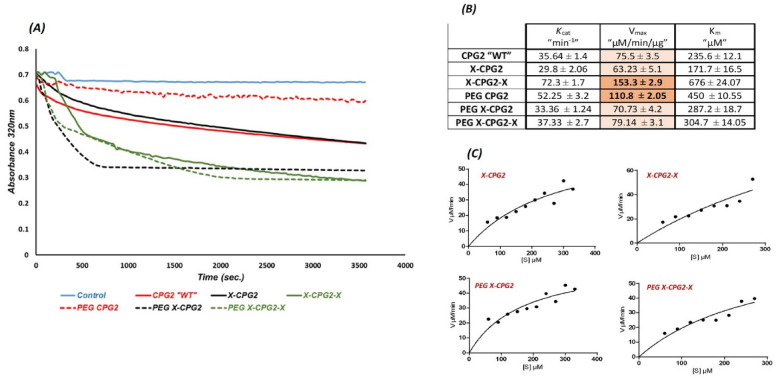
To investigate the catalytic activity of the PEGylated CPG2 fusion proteins they were incubated with different concentrations of MTX and the change in color was measured (Figure 3[A]). Figure 3(B) shows the resulting kinetic parameters following calculation with Graph pad PRISM 6 software, maximal initial velocity (Vmax), Michaels–Menten constant (Km) and turnover number (Kcat). Compared with the kinetic parameters of the non-PEGylated CPG2 fusion proteins, PEGylation of CPG2 fusion proteins resulted in a notable decrease of Vmax in case of double fused CPG2 (PEG X-CPG2-X), whereas the enzyme activity was not significantly affected for the PEGylated single fused protein (PEG X-CPG2).
Figure 3.
Enzyme kinetics of the PEGylated and non-PEGylated CPG2 fusion proteins. (A) The catalytic activity of CPG2 presented in the graph of MTX hydrolysis rate by CPG2. (B) The enzyme kinetics parameters (Km, Vmax, and Kcat) of the PEGylated and non-PEGylated CPG2 fusion proteins, Graph pad PRISM 6 software was used to calculate the parameters. (C) Michaelis–Menten plots of the CPG2 (in fusion proteins) enzymatic action on MTX hydrolysis. Abbreviations: MTX, methotrexate; PEG, polyethylene glycol; CPG2, carboxypeptidase G2.
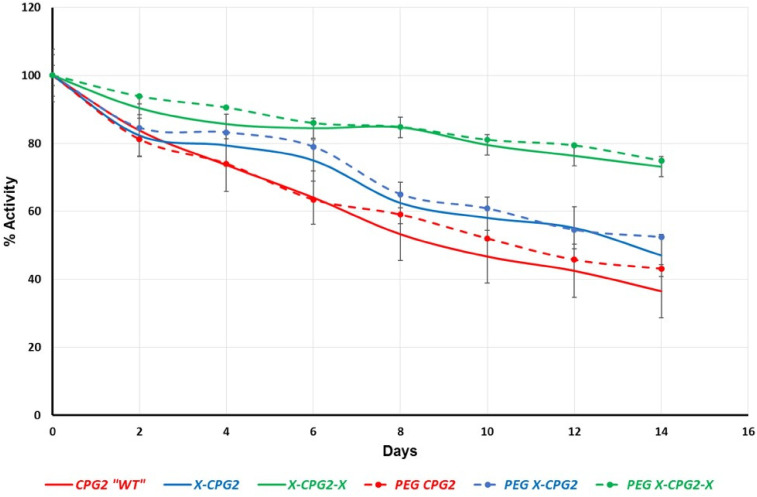
In vitro serum stability of the produced PEGylated CPG2 fusion proteins’ catalytic activity was examined at physiologic conditions. In agreement with our previous results, 10 there was a trend of improvement in the enzyme stability of PEG-WT compared with non-PEGylated WT-CPG2 (Figure 4). Interestingly, both single and double CPG2 fusion proteins (PEG X-CPG2 and PEG X-CPG2-X) stability was not significantly affected following PEGylation.
Figure 4.
PEGylated CPG2 fusion proteins stability in human serum. Serum samples from healthy donors were incubated at 37 °C with 0.1 µL/µg of the purified PEGylated and non-PEGylated CPG2 fusion proteins. The enzymatic (MTX hydrolytic) activity of CPG2 in each sample was tested every 48 h, and the percentage of remaining catalytic activity was plotted following normalization to a 100% activity at 0 time point. Abbreviations: MTX, methotrexate; PEG, polyethylene glycol; CPG2, carboxypeptidase G2.
Ex Vivo Immunotoxicity of the PEGylated CPG2 Fusion Proteins
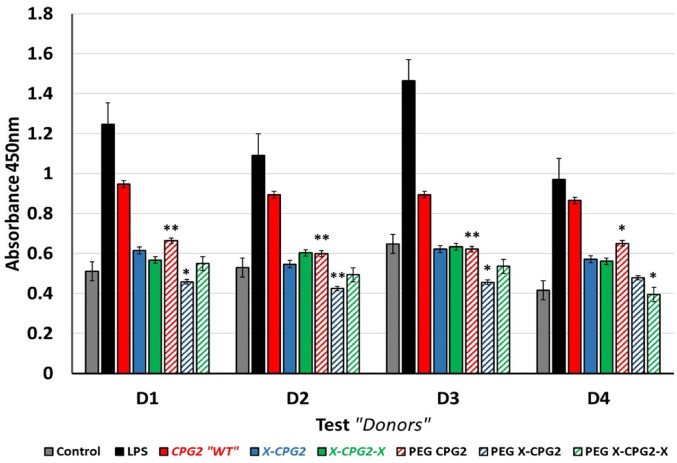
In order to investigate the potential relative risk of PEGylated CPG2 fusion protein immunotoxicity, T-cell proliferation assay was employed. 20 Endotoxin-free purified PEGylated CPG2 fusion proteins were incubated with PBMCs from normal healthy donors, and the response of T-cell proliferation was assessed after 48 h. As positive and negative controls, the PBMCs were incubated with LPS or the vehicle (PBS) respectively. As shown in Figure 5, LPS exhibited a significant induction of T-cell proliferation 2-fold higher than the proliferation level of negative control (PBS). Moreover, PEGylated single CPG2 fusion proteins presented a remarkable reduction in the T-cell proliferation compared with the non-PEGylated ones.
Figure 5.
Ex vivo T-cell proliferation assay. Study of the immunotoxicity of PEGylated and non-PEGylated CPG2 fusion proteins. PBMCs from normal healthy donors were co-incubated with the CPG2 fusion proteins “10 µg/mL”, in addition to the vehicle (PBS) as negative control and LPS as positive control. After 48 h cells incubated with the resulting CCK-8 solution for 4 h, and the resulting absorbance at 450 nm plotted for each donor (D). Abbreviation: LPS: lipolysaccharide; MTX, methotrexate; PEG, polyethylene glycol; CPG2, carboxypeptidase G2.
Diverse Cellular Binding Effect of the PEGylated CPG2 Fusion Protein in Vitro
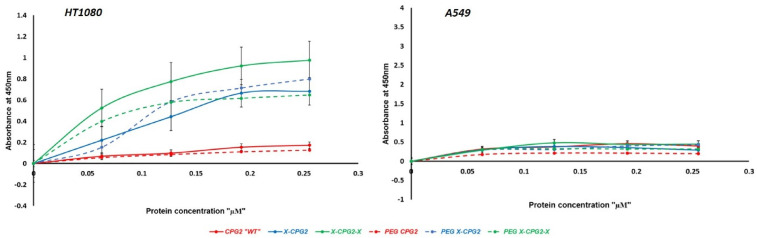
Binding activity of the produced PEGylated CPG2 fusion proteins to APN was assessed using differentially APN expressing cell lines. All PEGylated CPG2 fusion proteins displayed selective binding to the high APN expressing cell line (Figure 6) as in the case of non-PEGylated ones. However, the results showed that PEGylation slightly increased the binding affinity of the single fusion protein (PEG X-CPG2), whereas in case of the PEGylated double fused CPG2 protein the binding was remarkably lowered. Moreover, compared with the non-PEGylated CPG2 fusion proteins, there was no significant difference in the binding affinity of PEGylated single fusion protein. However, the PEGylated double fusion protein (PEG X-CPG2-X) showed a notable lowered binding affinity (Kd 0.657 ± 0.26 µM) compared with the non-PEGylated double fusion protein (Kd 0.404 ± 0.11 µM).
Figure 6.
In vitro binding strength of PEGylated and non-PEGylated CPG2 fusion proteins to the APN-expressing cancer cell lines. Low “A549” and high “HT1080” APN expressing cancer cell lines were cultured in 96-well plate with either PBS (as negative control) or increasing concentrations of the purified PEGylated and non-PEGylated CPG2 fusion proteins (0.063, 0.127, 0.192, and 0.255 µM). After 1 hour, cells were washed twice and ELISA assay using HRP-tagged anti-His6 antibody was utilized to explore the binding capacities of the PEGylated fusion proteins. Abbreviations: APN, Aminopeptidase N; PEG, polyethylene glycol; HRP, horseradish peroxidase; ELISA, enzyme-linked immunosorbent assay
Varied Cytotoxicity of MTX and the Prodrug ZD2767P on Cancer Cell Lines pretreated With PEGylated CPG2 Fusion Proteins
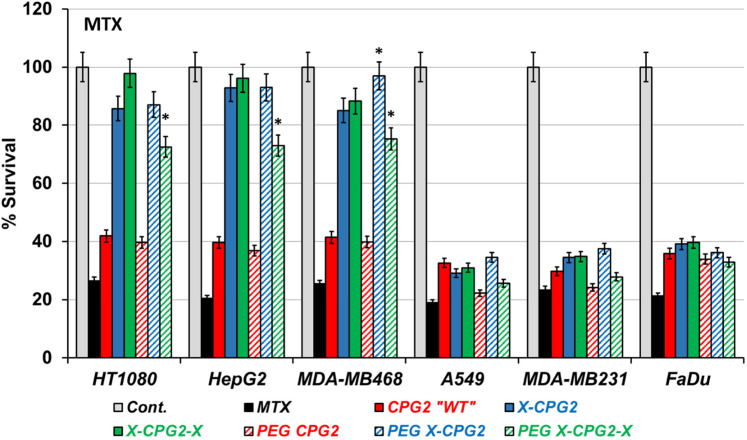
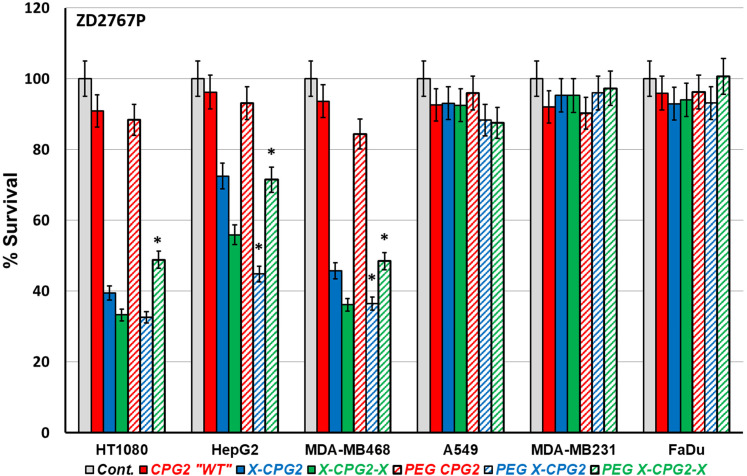
High and low APN-expressing cancer cell lines pretreated with the produced PEGylated CPG2 fusion proteins were employed to investigate the cytotoxic effect of MTX and the prodrug ZD2767P. Figure 7 shows the percentage survival calculated from the MTT assay results, with the cells treated with MTX, generally cells pretreated with PEGylated CPG2 fusion proteins exhibited up to 2-fold higher cell survival in the high APN-expressing cell lines compared with the low ones. There was a notable increase in the cell survival in case of cells treated with PEGylated CPG2 single fusion protein compared with the non-PEGylated ones; however, a significant decrease in cell survival resulted upon treatment with PEGylated CPG2 double fusion protein. Following treatment with pro-drug ZD2767P (Figure 8) a remarkable decrease in cell survival (high cell death) noticed with PEGylated single fusion protein, but PEGylation of double fusion protein (PEG X-CPG2-X) increased cell survival (low cell death) with the prodrug.
Figure 7.
The effect of MTX on cell viability following treatment with PEGylated and non-PEGylated fusion proteins. High “HT1080, HepG2 and MDA-MB468” and low “A549, MDA-MB231 and FaDu” APN-expressing cancer cell lines were incubated with the purified PEGylated fusion proteins, followed by treatment with MTX along with PBS “ negative control” and MTX only “positive control for 48 h. The MTT assay was employed to obtain the percentage of cell viability. The resulting survival results are normalized to the negative control (taken as 100% survival). Abbreviations: MTX, methotrexate; APN, aminopeptidase N; PEG, polyethylene glycol.
Figure 8.
The cytotoxic effect of the prodrug ZD2767P in combination with PEGylated and non-PEGylated fusion proteins. Cancer cell lines expressing high and low APN levels were treated with the prodrug ZD2767P following incubation with the purified PEGylated fusion proteins. Cells were treated with PBS as negative control, and then the cell viability level was assessed using MTT assay. The resulting survival results are normalized to the negative control as 100% survival. Abbreviations: APN, aminopeptidase N; PEG, polyethylene glycol.
Discussion
As a membrane protein highly expressed in invasive tumor cells, 12 APN is an ideal cancer marker for targeting tumor tissues. We have previously succeeded in engineering and generating a fusion protein composed of the therapeutic enzyme CPG2 and a peptide moiety (CNGRC) known to specifically bind APN. The single and double CPG2–CNGRC fusion proteins produced showed “biobetter” features in binding specificity, stability and in vitro cytotoxicity. 18 Several PEGylation strategies have been established to improve the therapeutic potency of proteins/drugs through enhancing their stability and lowering immunotoxicity. 21 To enhance the therapeutic efficacy and to reduce the protein immunogenicity of the produced fusion proteins here we conjugated the previously generated CNGRC–CPG2 fusion proteins with PEG polymer and studied the effect of PEGylation on the anti-cancer targeting effectiveness.
Protein structure and function are interrelated. PEGylation induces various forms of conformational changes when PEG is conjugated with the protein;, such structural changes have proven to affect protein stability, binding and catalytic activity. 22 CD spectroscopy is one of the reliable methods for rapid evaluation of the protein secondary structure. 23 The results of CD spectra (Figure 2) show that PEGylation of the CPG2 fusion proteins has introduced significant changes to the protein structures. It is verified that the site of PEG polymer-binding with therapeutic proteins and the induced conformational changes reported having some effect on the active site function resulting in activity changes. 24 These conformational changes might explain the lower activity of the PEGylated double-fused CPG2 (Figure 4), also as shown in our previous studies. 10
Several challenges have been facing the use of therapeutic proteins such as fast degradation by serum protease and their immunogenicity. 21 Two different studies found that conjugation of CPG2 either with the fusion peptide (CNGRC) or PEG polymer formed fusion proteins and PEGylated CPG2, respectively with higher serum stability and lower ex vivo immunotoxicity compared with the nonfused or non-PEGylated CPG2.10,18 In this study, we produced new PEGylated fused CPG2 proteins which tested for their stability and immunotoxicity. Our data showed that the serum stability of the PEGylated CPG2 fusion proteins was not changed in relation to the non-PEGylated ones (Figure 4). More significantly, however, the PEGylated single fused CPG2 exhibit lower level of immunotoxicity compared to the non-PEGylated one (Figure 5).
For assessment of the immunotoxicity of therapeutic proteins/peptides, it is essential to evaluate their preclinical safety using appropriate in vivo (animal) system. However, still a possible risk is there to develop immunotoxic effect in human subject even if it was found to be safe in animal trials.25,26 This provides a good reason to test such new compounds safety before moving to clinical trials. Consequently, ex vivo assays using human PBMCs employed to investigate species-specific immunotoxicity of the new therapeutic compounds. We utilized the ex vivo assay using hPBMCs to evaluate our produced PEGylated fusion proteins immunotoxicity, but still, further assessment is required to identify their immunogenicity.
The binding of the CPG2 fusion proteins to their targeted marker on tumor cells is crucial in the process of directed cytotoxic effect. Since PEGylation is known to induce changes in the structure of the conjugated proteins, 22 the binding affinity to APN might also be affected. As shown in Figure 6 with PEGylated single fusion there was a notable increase in the binding affinity, whereas significantly reduced with double fusion protein (PEG X-CPG2-X) PEGylation. However, binding capacity needs to be further described using fluorescence-based a quantitative ligand-binding assay or use flow cytometry to analyze ligand binding to proteins. The conjugation of PEG polymer with protein through covalent linkage provides steric shielding. Although this shielding offers protection of the protein (enzyme) against proteolytic enzymes, it might cause some hindrance on reaching the receptor or the active site.27,28
Furthermore, the effect of PEGylation depends on the degree of PEGylation and molecular weight of PEG used. Several studies confirmed the differential effect of PEG with low and high molecular weight (5-40 kDa) on the function and residual activity of the protein/enzyme.29,30 Hence, PEGylation with lower molecular weight PEG found to reduce PEGylation degree allowing higher protein activity due to the effect of both steric hindrance and molecular. Whereas higher degree of PEGylation found to favor the stability of the produced protein. 27 Moreover, providing more sites for the PEG group to bind on the protein results in higher degree of PEGylation, thus PEGylation of the double fusion protein (X-CPG2-X) provided more sites for the PEG to bind compared with the single fusion protein (X-CPG2) resulting in higher PEGylation degree of the double fusion protein. This induced structural changes in case of the double fusion CPG2 which might lead to the reduction of the protein binding to the APN receptor, in addition to the its effect on CPG2 enzymatic activity and stability.
We followed 2 approaches to investigate the application of our new PEGylated fusion proteins in cancer targeted therapy, LDEPT, one using the cytotoxic drug, MTX and the other using the prodrug (ZD2767P). In case of the prodrug ZD2767P, it shows, as expected, an increase in cell death (high cytotoxic effect) in combination with the PEGylated single fusion protein (PEG X-CPG2). However, with the PEGylated double fusion protein (PEG X-CPG2-X) the cytotoxic effect was reduced compared with that of the non-PEGylated one (Figure 7). This could be explained by the reduced binding capability of the double PEGylated fusion CPG2. In the case of MTX, since MTX is known to be degraded by CPG2 and become nonactive (nontoxic), the finding would be the opposite of that for the prodrug. Our data showed that in the case of the PEGylated single fused CPG2, there was higher cell survival on account of the ability of CPG2 to degrade MTX and with less toxic effects (Figure 8), supporting the higher efficacy of PEGylated single fusion protein comparing to the non-PEGylated one.
The use of Protein PEGylation in biomedical research has solved many problems in the process of drug manufacturing such as higher solubility, reduce antigenicity and immunogenicity of the target protein, increase the half life time of the circulation drugs and many other advantages. The protein PEGylation however have many limitations and disadvantages. One of the main limitations is the production of the anti-PEG by the patient and the hypersensitivity towards PEG. In this work we showed that PEGylation of CPG2 produced products with lower immunogenicity than the un-pegylated fused CPG2. This is on one hand partially overcome one of the main obstacles in the antibody-directed enzyme prodrug therapy but on the other hand the anti-PEG and the sensitivity forwards PEG could be an obstacle for the extensive use of this drug for cancer targeted therapy.
The current and future research works need to explore new protocols to solve these PEGylation associated problems.
Conclusion
To overcome the immunogenicity, one of the main pitfalls of directed enzyme prodrug therapy, we have successfully produced 2 new long-acting peptide-fused CPG2 for LDEPT, PEG–CNGRC–CPG2 and PEG. The first is PEGylated N-terminal CCPG2 and PEGylated N-, C-terminal fused CPG2. Both PEGylated products showed lower immunogenicity than the free fused CPG2. The PEGylated double-fused CPG2, however, has lower APN binding and less bioactivity that the free fused CPG2. Our work paves the way for in vivo studies and perhaps clinical investigations.
Acknowledgments
The authors thank Fatma Rashidi for carrying out some technical work including the CD spectral.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: These studies were supported by the Qatar National Research Fund (grant number: NPRP6-065-3-012).
ORCID iDs: Layla Al-mansoori https:orcid.org/0000-0003-4028-1954
Sayed K. Goda https://orcid.org/0000-0002-3450-9257
References
- 1.Carter PJ. Introduction to current and future protein therapeutics: a protein engineering perspective. Exp Cell Res. 2011;317(9):1261-1269. [DOI] [PubMed] [Google Scholar]
- 2.Dozier JK, Distefano MD. Site-Specific PEGylation of therapeutic proteins. Int J Mol Sci. 2015;16(10):25831-25864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turecek PL, Bossard MJ, Schoetens F, Ivens IA. PEGylation of biopharmaceuticals: a review of chemistry and nonclinical safety information of approved drugs. J Pharm Sci. 2016;105(2):460-475. [DOI] [PubMed] [Google Scholar]
- 4.Mishra P, Nayak B, Dey RK. PEGylation in anti-cancer therapy: an overview. Asian J Pharm Sci. 2016;11(3):337-348. [Google Scholar]
- 5.Reichert C, Borchard G. Noncovalent PEGylation, An innovative subchapter in the field of protein modification. J Pharm Sci. 2016;105(2):386-390. [DOI] [PubMed] [Google Scholar]
- 6.Nyborg AC, Ward C, Zacco A, et al. A therapeutic uricase with reduced immunogenicity risk and improved development properties. PLoS One. 2016;11(12):e0167935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gowin K, Jain T, Kosiorek H, et al. Pegylated interferon alpha - 2a is clinically effective and tolerable in myeloproliferative neoplasm patients treated off clinical trial. Leuk Res. 2017;54:73-77. [DOI] [PubMed] [Google Scholar]
- 8.Hong J, Lee B, Kang K, et al. Characterisation of the site-specific monoPEGylated rhG-CSF analogue pegteograstim. Biologicals. 2018;51:54-61. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Zhan P, De Clercq E, Lou H, Liu X. Current drug research on PEGylation with small molecular agents. Prog Polym Sci. 2013;38(3):421-444. [Google Scholar]
- 10.AlQahtani AD, Al-Mansoori L, Bashraheel SS, et al. Production of “biobetter” glucarpidase variants to improve drug detoxification and antibody directed enzyme prodrug therapy for cancer treatment. Eur J Pharm Sci. 2019;127:79-91. [DOI] [PubMed] [Google Scholar]
- 11.Sharma SK, Bagshawe KD. Translating antibody directed enzyme prodrug therapy (ADEPT) and prospects for combination. Expert Opin Biol Ther. 2017;17(1):1-13. [DOI] [PubMed] [Google Scholar]
- 12.Wickstrom M, Larsson R, Nygren P, Gullbo J. Aminopeptidase N (CD13) as a target for cancer chemotherapy. Cancer Sci. 2011;102(3):501-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasqualini R, Koivunen E, Kain R, et al. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000;60(3):722-727. [PMC free article] [PubMed] [Google Scholar]
- 14.Li JJ, Chang SF, Liau II, et al. Targeted antitumor prodrug therapy using CNGRC-yCD fusion protein in combination with 5-fluorocytosine. J Biomed Sci. 2016;23:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillen KP, Kurkjian C, Harrison RG. Targeted enzyme prodrug therapy for metastatic prostate cancer - a comparative study of L-methioninase, purine nucleoside phosphorylase, and cytosine deaminase. J Biomed Sci. 2014;21:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J, Zhang D, Ying X, et al. A novel NGR-conjugated peptide targets DNA damage responses for radiosensitization. Curr Cancer Drug Targets. 2015;15(6):533-541. [DOI] [PubMed] [Google Scholar]
- 17.Di Matteo P, Mangia P, Tiziano E, et al. Anti-metastatic activity of the tumor vascular targeting agent NGR-TNF. Clin Exp Metastasis. 2015;32(3):289-300. [DOI] [PubMed] [Google Scholar]
- 18.Al-mansoori L, Bashraheel SS, Qahtani ADA, O’Connor CD, Elsinga P, Goda SK. In vitro studies on CNGRC-CPG2 fusion proteins for ligand-directed enzyme prodrug therapy for targeted cancer therapy. Oncotarget. 2020;11:619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCullough JL, Chabner BA, Bertino JR. Purification and properties of carboxypeptidase G 1. J Biol Chem. 1971;246(23):7207-7213. [PubMed] [Google Scholar]
- 20.Fu K, Cheng Q, Liu Z, et al. Immunotoxicity assessment of rice-derived recombinant human serum albumin using human peripheral blood mononuclear cells. PLoS One. 2014;9(8):e104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jevsevar S, Kunstelj M, Porekar VG. PEGylation of therapeutic proteins. Biotechnol J. 2010;5(1):113-128. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence PB, Price JL. How PEGylation influences protein conformational stability. Curr Opin Chem Biol. 2016;34:88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2006;1(6):2876-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veronese FM, Pasut G. PEGylation, successful approach to drug delivery. Drug Discov Today. 2005;10(21):1451-1458. [DOI] [PubMed] [Google Scholar]
- 25.Frantz S. Pharma faces major challenges after a year of failures and heated battles. Nat Rev Drug Discov. 2007;6(1):5-7. [DOI] [PubMed] [Google Scholar]
- 26.Suntharalingam G, Perry MR, Ward S, et al. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N Engl J Med. 2006;355(10):1018-1028. [DOI] [PubMed] [Google Scholar]
- 27.Qi F, Hu C, Yu W, Hu T. Conjugation with eight-Arm PEG markedly improves the In vitro activity and prolongs the blood circulation of staphylokinase. Bioconjugate Chem. 2018;29(2):451-458. [DOI] [PubMed] [Google Scholar]
- 28.Mu Q, Hu T, Yu J. Molecular insight into the steric shielding effect of PEG on the conjugated staphylokinase: biochemical characterization and molecular dynamics simulation. PLoS One. 2013;8(7):e68559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgenstern J, Baumann P, Brunner C, Hubbuch J. Effect of PEG molecular weight and PEGylation degree on the physical stability of PEGylated lysozyme. Int J Pharm. 2017;519(1-2):408-417. [DOI] [PubMed] [Google Scholar]
- 30.Wan X, Zhang J, Yu W, Shen L, Ji S, Hu T. Effect of protein immunogenicity and PEG size and branching on the anti-PEG immune response to PEGylated proteins. Process Biochem. 2017;52:183-191. [Google Scholar]