Abstract
Introduction
The development of a human immunodeficiency virus 1 (HIV‐1) vaccine remains a formidable challenge. An effective vaccine likely requires the induction of broadly neutralizing antibodies (bNAbs), which likely involves the use of native‐like HIV‐1 envelope (Env) trimers at some or all stages of vaccination. Development of such trimers has been very difficult, but much progress has been made in the past decade, starting with the BG505 SOSIP trimer, elucidation of its atomic structure and implementing subsequent design iterations. This progress facilitated understanding the weaknesses of the Env trimer, fuelled structure‐guided HIV‐1 vaccine design and assisted in the development of new vaccine designs. This review summarizes the relevant literature focusing on studies using structural biology to reveal and define HIV‐1 Env sites of vulnerability; to improve Env trimers, by creating more stable versions; understanding antibody responses in preclinical vaccination studies at the atomic level; understanding the glycan shield; and to improve “on‐target” antibody responses versus “off‐target” responses.
Methods
The authors conducted a narrative review of recently published articles that made a major contribution to HIV‐1 structural biology and vaccine design efforts between the years 2000 and 2021.
Discussion
The field of structural biology is evolving at an unprecedented pace, where cryo‐electron microscopy (cryo‐EM) and X‐ray crystallography provide complementary information. Resolving protein structures is necessary for defining which Env surfaces are accessible for the immune system and can be targeted by neutralizing antibodies. Recently developed techniques, such as electron microscopy‐based polyclonal epitope mapping (EMPEM) are revolutionizing the way we are analysing immune responses and shed light on the immunodominant targets on new vaccine immunogens. Such information accelerates iterative vaccine design; for example, by reducing undesirable off‐target responses, while improving immunogens to drive the more desirable on‐target responses.
Conclusions
Resolving high‐resolution structures of the HIV‐1 Env trimer was instrumental in understanding and improving recombinant HIV‐1 Env trimers that mimic the structure of viral HIV‐1 Env spikes. Newly emerging techniques in structural biology are aiding vaccine design efforts and improving immunogens. The role of structural biology in HIV‐1 vaccine design has indeed become very prominent and is unlikely to diminish any time soon.
Keywords: broadly neutralizing antibodies, reverse vaccinology 2.0, structure‐based vaccine design
1. INTRODUCTION
After more than 30 years of extensive research, an effective human immunodeficiency virus 1 (HIV‐1) vaccine is still not available. The major challenge for an HIV‐1 vaccine is the enormous viral diversity, and an effective HIV‐1 vaccine should induce an immune response that could cope with this diversity [1, 2, 3, 4]. The only relevant target for vaccine design efforts is the HIV‐1 envelope glycoprotein (Env) trimer, the protein that initiates viral entry and the only target for broadly neutralizing antibodies (bNAbs) [5, 6].
Env is a trimeric protein complex composed of three gp120 and three gp41 subunits that are held together by weak non‐covalent interactions [7, 8]. Env is synthesized as a gp160 polyprotein precursor, which is posttranslationally cleaved in its gp120 and gp41 subunits. During synthesis, the protein is decorated by N‐linked glycans that comprise approximately 50% of its total mass [9, 10, 11]. The N‐linked glycans play various essential roles in the viral life cycle, for example, in protein folding, infectivity by binding to lectin receptors on immune cells and immune evasion [12, 13, 14, 15, 16, 17].
Approximately 20% to 30% of people living with HIV (PLHIV) develop bNAbs albeit usually after several years of infection [18]. In recent years, many bNAbs have been isolated and some have remarkable neutralization breadth [19]. Passive immunization studies in rhesus macaques have shown that bNAbs can protect against simian/human immunodeficiency virus (SHIV) infection [20, 21, 22]. The protective efficacy of the VRC01 bNAb was tested in two recent randomized clinical trials [23]. The outcome of the trials was that VRC01 was not able to protect against HIV‐1 acquisition overall, but did result in a sieving of VRC01‐resistant viruses, suggesting that VRC01 was able to prevent transmission of VRC01‐sensitive viruses. The implication is that one bNAb specificity will most likely not be enough to induce broad protection, and an effective HIV‐1 vaccine will need to induce multiple neutralizing specificities.
Env protein structures are playing an increasingly important role in vaccine design efforts. Resolving protein structures at the atomic level is necessary to define the surfaces that are accessible for the immune system, to determine sites that are targeted by monoclonal antibodies (MAbs), to design immunogens that mimic the proteins found on viral surfaces and to stabilize the desirable conformations [24]. This approach follows from “reverse vaccinology” in which complete genome sequencing of a pathogen is used to select for surface‐expressed proteins to be used in a vaccine [25]. The term “reverse vaccinology” was repurposed in 2002 to describe the utilization of antibodies to select or design antigens with the appropriate binding properties [26]. A next iteration, “reverse vaccinology 2.0” involves the use of antibodies and high‐resolution structures for the design of vaccine antigens [27].
While these concepts are widely applied in the HIV‐1 vaccine design field, such structure‐based vaccine design approaches also accelerated the development of novel vaccines against other pathogens. An example includes respiratory syncytial virus (RSV), for which vaccine development was accelerated when the crystal structure of the pre‐fusion F glycoprotein was resolved, using the D25 neutralizing antibody (NAb) specific for the pre‐fusion form [28, 29]. Similar to HIV‐1 Env and other class I viral fusion proteins, the RSV F glycoprotein is very unstable and transitions very easily to its post‐fusion conformation. The apex targeting D25 MAb stabilized the F glycoprotein in its pre‐fusion conformation, resulting in the high‐resolution pre‐fusion structure of the protein. This structure allowed for the development of a recombinant pre‐fusion‐stabilized F glycoprotein [24, 28]. As structure‐guided design is now an integral part of HIV‐1 vaccine development, we review here the role of structural biology in defining the sites of vulnerability of HIV‐1 Env, and how new vaccine candidates are designed in an iterative manner by using the structural knowledge of immunogens and the antibodies they induce in (pre)clinical immunization experiments. Our focus is on native‐like Env trimers, but we will treat other approaches where relevant as well.
2. METHODS
We conducted a narrative review of the relevant literature focusing on studies that use structural biology to reveal and define HIV‐1 Env sites of vulnerability. This information was used to improve soluble Env trimers by several strategies: (a) by creating more stable versions; (b) to understand antibody responses in preclinical vaccination studies at the atomic level; (c) to understand the glycan shield; and (d) to improve “on‐target” antibody responses versus “off‐target” responses. Peer‐reviewed papers and grey literature that reported on the HIV‐1 Env structure, structural improvement of the Env trimer, Env glycosylation and clinical studies were identified between the years 2000 and 19 April 2021. We searched through grey literature and keyword searches in PubMed for the following terms: reverse vaccinology, reverse vaccinology 2.0, structure‐based vaccine design, HIV‐1 vaccine design, HIV‐1 Env structures, HIV‐1 Env structure‐based vaccine design, HIV‐1 broadly neutralizing antibodies, HIV‐1 Env glycosylation. We also identified studies in the articles’ reference lists.
3. DISCUSSION
3.1. Structure‐based definition of the Env trimer
“Reverse vaccinology” involves the recombinant generation of the NAb‐relevant surface proteins. However, creating stable mimics of the native Env trimers was not achieved until 2013 (Table 1). Since then, much progress has been made in developing faithful mimics of the HIV‐1 Env spike. A major initial step involved the design of the BG505 SOSIP.664 prototype native‐like Env trimer, based on the clade A transmitter/founder virus BG505 [30]. Several modifications were introduced to stabilize the trimer: a disulphide bond (“SOS”) between residues 501 in the gp120 subunit and 605 in the gp41 subunit; an Isoleucine‐to‐Proline (“IP”) substitution at position 559 in the first heptad repeat (HR1) of gp41 that prevents a helix from forming, thereby locking the protein in the pre‐fusion state; in improved furin cleavage site to facilitate complete precursor cleavage between gp120 and gp41; and the removal of the membrane‐proximal, transmembrane and cytoplasmic tail domains [31]. The BG505 SOSIP.664 trimer closely resembled the native viral Env in both structure and antigenicity [31, 32, 33, 34, 35]. It also allowed obtaining the first medium‐resolution structures of an Env trimer by both cryo‐electron microscopy (cryo‐EM) and X‐ray crystallography techniques. The structures revealed that the gp120 subunits have a globular conformation, which are assembled into a trimer through key interactions in the V1V2 and V3 domains. The gp41 domain forms a pedestal with the gp120 subunits docked on it, combining into a mushroom‐like shape for the functional trimer [36, 37, 38]. The high‐resolution cryo‐EM structure of the JRFL native viral Env also confirmed that the BG505 SOSIP.664 trimer closely mimics viral Env [39] (Table 1). The structure and conformational flexibility of SOSIP trimers were also studied using hydrogen‐deuterium exchange (HDX) and double electron‐electron resonance (DEER) spectroscopy, revealing that these regions are more conformationally flexible than others [40, 41].
Table 1.
Key developments in structure‐guided HIV‐1 vaccine designs in the past decade
| Year | Development | Reference |
|---|---|---|
| 2013 | First structure of a native‐like trimer in complex with PG9 | Julien et al., 2013 [44] |
| 2013 | Development of the BG505 SOSIP.664 trimer | Sanders et al., 2013 [31] |
| 2013 | First cryo‐EM structure of a native‐like Env trimer | Lyumkis et al., 2013 [36] |
| 2013 | First crystal structure of a native‐like Env trimer | Julien et al., 2013 [37] |
| 2014 | First structure of the complete pre‐fusion conformation gp41 | Pancera et al., 2014 [38] |
| 2014 | First dynamics of SOSIP trimers using hydrogen‐deuterium exchange analysis | Guttman et al., 2014 [40] |
| 2016 | First cryo‐EM structure of a native HIV‐1 viral envelope | Lee et al., 2016 [39] |
| 2016 | Development of the eOD‐GT8 germline‐targeting inmmunogen | Jardine et al., 2016 [118] |
| 2017 | Development of the germline‐targeting BG505 SOSIP GT1 trimer | Medina‐Ramirez et al., 2017 [125] |
| 2018 | Evaluation of site‐specific glycosylation on virion‐derived Envs | Struwe et al., 2018; Cao et al., 2018 [95, 96] |
| 2018 | Analysis of conformational dynamics native‐like Env trimers using DEER spectroscopy | Stadtmueller et al., 2018 [41] |
| 2018 | First in‐human phase I clinical trial started with the eOD‐GT8 60mer vaccine candidate | Clinicaltrials.gov [136] |
| 2018 | First in‐human phase I clinical trial started with a native‐like Env trimer | Clinicaltrials.gov [137] |
| 2018 | Development of electron microscopy‐based polyclonal epitope wrapping (EMPEM) | Bianchi et al., 2018 [112] |
| 2020 | First in‐human phase I clinical trial started with a germline‐targeting native‐like Env trimer | C1inicaltrials.gov [124] |
3.2. Structure‐based definition of targets of vulnerability on the Env trimer
A plethora of bNAbs have been isolated from PLHIV that provide templates for vaccine design according to the approaches defined by “reverse vaccinology 2.0”. While structures of bNAbs in complex with gp120 were solved before and generated important structural information on some bNAb epitopes, the availability of the SOSIP trimer allowed many more bNAb epitopes to be determined structurally including epitopes requiring quaternary structures such as those located at the trimer apex and the gp120/gp41 interface [42, 43]. While at very low resolution, the first structure to be described was that of the bNAb PG9 in complex with the BG505 SOSIP.664 trimer [44]. The development of the BG505 SOSIP.664 trimer also helped in revealing many sites of vulnerability on Env [45, 46]. These bNAbs target eight well‐defined clusters comprising of (1) the V1V2 quaternary structure‐dependent epitope cluster, involving the N156 and N160 glycans and electropositive residues in the C‐strand of the V2 [47]. Inducing neutralizing breadth to this epitope can be achieved by a limited amount of mutations, making this an attractive vaccine target [48, 49]. The bNAbs targeting this site possess a very long CDRH3, which is required to navigate through the glycan shield. (2) The N332 centred supersite of vulnerability, which can be subdivided into a V3‐glycan epitope cluster and an outer domain (OD) cluster, but both involve the N332 glycan [46]. This site is being explored in immunogen design efforts discussed below. (3) The CD4 binding site (CD4bs) is targeted by the most potent bNAbs discovered and a main feature is that bNAbs targeting this site need to accommodate the N276 glycan [50]. Many bNAbs to this site, those belonging to the VRC01‐class, are derived from VH1‐2 and have a light chain with a short five amino acid CDRL3. (4) The highly glycosylated “silent” face on Env is a relatively newly discovered epitope that comprises of the glycans N262, N295 and N448. Only a few bNAbs against this site have been isolated, but some of them show remarkable breadth, making the silent face an additional target for vaccine design efforts [51, 52]. (5) The gp120‐gp41 interface comprises of protein domains and glycans of both the gp120 and gp41 subunits and recognize the pre‐fusion closed state of Env [43]. (6) The Fusion peptide epitope cluster comprises of domains in gp120, gp41 and the N‐terminal part of the fusion peptide (FP) [43]. This epitope cluster overlaps with the previous one. (7) The gp41‐domain is targeted by the 3BC176 and 3BC315 bNAbs and these bNAbs neutralize the virus by increasing the decay of the Env trimer [53]. (8) Finally, the membrane proximal external region (MPER) is a highly conserved, hydrophobic region in gp41, and is targeted by bNAbs that are very broad, but have moderate potency [19]. While the above classification of epitopes is useful, in fact, the Env trimer surface appears to form one contiguous target for bNAbs. When multiple bNAbs are plotted on the trimer surface virtually no uncovered surface remains (Figure 1) [46]. Furthermore, while the extensive glycosylation on Env was long thought to create an immunologically “silent face” and act as a “glycan shield” to protect the underlying Env protein domains against NAbs [6, 54], the definition of bNAb epitopes reveals that the glycan shield itself is, paradoxically, a target for bNAbs. As some of these clusters are capable of inducing very potent bNAbs, they are of particular interest for vaccine design efforts, and we further discuss below sites that are being explored as potential vaccine targets.
Figure 1.
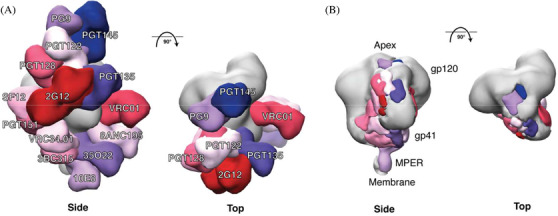
bNAb epitopes mapped onto the three‐dimensional structure of the BG505 SOSIP.664 trimer. (a) Side and top views of the bNAbs labelled in different colours that are modelled onto an EM density map of the BG505 SOSIP.664 trimer (coloured in grey). The figure includes bNAbs recognizing eight well‐defined sites of vulnerability: PG9 and PGT145 (V2apex), PGT122 and PGT128 (N332‐glycan); PGT135 and 2G12 (OD‐glycan) both involve the N332 glycan; VRC01 (CD4bs); SF12 (silent face); PGT151, 8ANC195 and 35O22 (gp120‐gp41 interface); VRC34.01 (fusion peptide); 3BC315 (gp41); and 10E8 (MPER). Only one copy of each epitope per trimer is shown for clarity. Thus, the model does not indicate the stoichiometry of bNAb binding, only the location of the epitope. (b) Side and top views of the bNAb footprints displayed in (a). This figure is an updated version of fig. 1 from de Taeye et al., 2016 [53] (we thank Gabe Ozorowski for preparing it).
3.3. Structure‐based immunogen improvement
While class I viral fusion proteins are intrinsically unstable, a number of tricks can be applied to stabilize them in their pre‐fusion conformation. We briefly review a few important ones that are now routinely used, but there are other possibilities (Figure 2). For a more extensive review, see Torrents de la Peña et al., 2018 [55].
Figure 2.
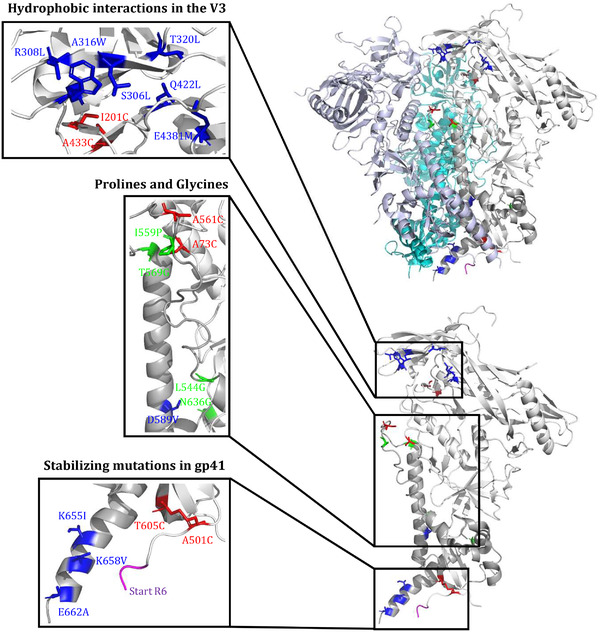
Amino acid substitutions that are routinely used to stabilize soluble native‐like trimers. The amino acid substitutions were modelled on the BG505 SOSIP.664 trimer (see text for details). The gp120 subunit is coloured in white and the gp41 subunit in light gray. In gp120: blue ‐ S306L, R308L, A316W, T320L, E381M and Q422L hydrophobic residues; red ‐ I201C‐A433C and A73C‐A561C disulphide bonds; green ‐ I559P, L544G, T569G and N636G stabilizing mutations; magenta ‐ adjacent residues of the R6 furin cleavage site or flexible linkers. In gp41: red ‐ A501C‐T605C disulphide bond; blue ‐ D589V, K655I, K658V and E662A hydrophobic stabilizing mutations. A more extensive list of stabilizing mutations was reviewed previously [55].
3.3.1. Trimer stabilization using disulphide bonds
The initial step of the SOSIP design involved the introduction of a well‐placed disulphide bond between gp120 and gp41 described over two decades ago [32]. The structures of the BG505 SOSIP.664 trimer sparked the designs of more stable versions of the Env trimer by adding additional disulphide bonds. Such hyperstabilization of native‐like Env trimers can reduce their flexibility and increase their stability, which could be beneficial for promoting interactions of B‐cell receptors (BCRs) with bNAb epitopes. Several studies increased the stability of the Env trimer by introducing disulphide bonds [56, 57]. The DS‐Env trimer, a variant of the BG505 SOSIP.664 trimer, is conformationally fixed by a disulphide bond at positions 201 and 433 that prevents conformational changes associated with CD4 binding, resulting in reduced binding of non‐NAbs targeting the V3 and CD4bs [57]. Torrent de la Pena et al., created a hyperstable BG505 SOSIP trimer by introducing several new disulphide bonds, including one between gp120 and gp41 from different protomers covalently linking them, leading to improved stability and antigenicity [58]. A similar approach was used to stabilize the glycoprotein complex (GPC) of Lassa virus (LASV), and involved a disulphide bond to covalently link the GP1 and GP2 subunits [59].
3.3.2. Trimer stabilization using prolines
The second critical component of the original SOSIP design involves a proline substitution in HR1 that prevents conformational changes down the fusion pathway, thereby stabilizing the Env trimers in the pre‐fusion state. Proline substitutions at other positions, as well as glycine substitutions, can have similar stabilizing effects [34, 60, 61]. This method proved to be a universal method for stabilizing class I fusion proteins. Its next use was for the stabilization for RSV F proteins, but applications to LASV GPC [59], Ebola virus GP, and MERS‐CoV and SARS‐CoV Spike (S) proteins swiftly followed [62, 63, 64, 65]. The latter configuration was adopted in SARS‐CoV‐2 vaccine design, and the Pfizer/BioNTech, Moderna, Curevac, Janssen/J&J, and Novavax COVID‐19 vaccines all include the proline stabilization method [66, 67].
3.3.3. Trimer stabilization using hydrophobic residues
While the BG505 SOSIP.664 trimer is a good mimic of the native Env spike, it can still fluctuate between a closed and more open state(s). This flexibility can lead to the exposure of the V3 [68, 69, 70]. Several strategies to further stabilize the trimer involved further reducing flexibility and silencing of the immunodominant V3. Using structure‐guided design, the A316W mutation was introduced to strengthen the hydrophobic interactions at the trimer apex, reducing the propensity of the V3 to “flip out” of its position underneath the V1V2 domain [71]. The introduction of this tryptophan significantly reduced the V3‐directed responses in rabbits and mice [71]. By using structure‐based design and mammalian display, other hydrophobic residues were introduced between the V3 and V1V2‐domains to reduce spontaneous exposure of the V3 by introducing leucine residues at positions 306 and 308 to create hydrophobic interactions with the tryptophan at position 316, by the “so‐called” MD39 mutations, in the trimer core (A204I, T320L, E381M, Q422L), or in gp41 (D589V, K655I, K658V, E662A) [72, 73, 74, 75, 76].
3.3.4. Trimer stabilization by removing the furin cleavage site
The SOSIP platform includes an improved furin cleavage site to allow for complete precursor cleavage, which requires coexpression of the furin protease. This configuration was also applied successfully to LASV GPC. The BG505 SOSIP structures allowed for the design of single chain (SC) or native flexibly linked trimer (NFL) platforms, in which the furin cleavage site is replaced by a flexible linker allowing for the production of native‐like trimers independent of the presence of furin [60, 77, 78, 79]. These flexible linkers have a specific length to allow for the natural association of the Env subunits, and these platforms often contain some or all of the SOSIP modifications to assist in stabilization. Replacement of the cleavage site in combination with proline stabilization has also aided the development of SARS‐CoV‐2 vaccines [66].
3.4. Broadening the panel of native‐like trimers
The enormous sequence diversity of HIV‐1 is a key challenge in designing an effective vaccine. The vaccine should induce bNAb responses that can cope with this diversity [1, 2, 3, 4]. For MAbs to be effective against highly mutable pathogens, they have to undergo a mutation process called somatic hypermutation (SHM), to produce high‐affinity MAbs that can tolerate this sequence diversity. Developing immunization strategies that drive this process will be key for an effective HIV‐1 vaccine. In silico studies revealed that cocktail and sequential immunization regiments could be good strategies to drive the SHM process [80, 81, 82]. Therefore, increasing the number of native‐like trimers based on different genotypes would be a good starting point. The high‐resolution structures of the BG505 SOSIP.664 trimer helped in the designs of more stable versions of this protein and helped in the identification of residues involved in its inherent stability [31, 36, 37, 38, 58, 71, 79]. The identification of these residues made it possible to successful apply specific stabilizing mutations to other Env sequences. In recent years, many native‐like trimers, based on different platforms, have been developed including ones based on a clade A sequence (92UG037.8), clade B sequences (B41, AMC008, AMC009, AMC011, JRFL), clade C sequences (DU422, ZM197M, CZA97.012, 16055), recombinant B/C sequence (LT5.J4b12C) and consensus sequences [56, 57, 60, 69, 71, 73, 74, 75, 76, 77, 78, 79, 83, 84, 85, 86, 87, 88, 89]. The generation of native‐like trimers from any desired Env sequence was taken a step further by Rutten et al., 2018, who devised a universal approach in order to obtain diverse high‐quality pre‐fusion closed Env trimers [76]. A structure‐based screening approach was used to develop a “repair and stabilize” (RnS) method, by removing rare amino acids and optimizing regions critical in the folding process. This approach was applicable to diverse HIV‐1 Env trimers in order to improve expression and stabilization [76]. The procedure was improved by the development of a pipeline using an automated process called ADROITrimer. This process combined the DS‐SOSIP design with RnS to create a large number of pre‐fusion‐stabilized Env trimers [90].
3.5. Structure‐based interpretation of the glycan shield
Concurrent with the developments in the Env structural biology field, new mass spectrometry (MS) glycan analysis platforms facilitated the dissection of glycans with unprecedented detail. These techniques, when applied to native‐like recombinant trimers, allowed for a detailed characterization of the Env glycan shield, including the composition of each glycan, the level of occupancy at potential N‐linked glycosylation site (PNGS) and how this affects binding of bNAbs [91].
HIV‐1 Env has a particular dense glycan shield and this density imposes steric constrains on early glycan trimming enzymes, leading to the large abundance of underprocessed oligomannose‐type glycans [91]. These underprocessed glycans form two specific “mannose patches” on Env: (a) the intrinsic mannose patch (IMP), which is highly conserved among all HIV‐1 clades; and (b) the trimer‐associated mannose patch (TAMP), which only forms on native‐like Env trimers [92]. Glycan density thus influences glycan processing and epitope presentation. This was corroborated when PNGS surrounding the 2G12 epitope were removed, thereby increasing the processing of these glycans, which impacted 2G12 binding. A similar effect was observed for the N160 glycan, which showed increased processing after the removal of the N156 or N197 sites, affecting the binding of apex targeting bNAbs [93]. These examples underpin the importance of understanding the role of glycan networks and glycan composition on conserving bNAb epitopes for HIV‐1 Env immunogen design. The glycan density was shown to be important for the induction of neutralizing breadth, as during natural HIV‐1 infection viruses that have an intact glycan shield are more prone to induce bNAb responses compared to the ones that have holes in the glycan shield [94]. The BG505 sequence lacks the conserved N241 and N289 PNGS and in the BG505 infant, neutralization breadth was first observed after the introduction of the N241 glycan showing the importance of filling glycan holes [94].
At least equally important from an immunogen design perspective was the quantitative measurement of PGNS occupancy on both viral Envs and recombinant SOSIP trimers, showing that several sites on recombinant trimers are underoccupied [95, 96]. Underoccupancy creates artificial glycan holes that are immunogenic, but induce NAb responses that are unable to neutralize the corresponding virus on which the respective sites are removed [97, 98]. The importance of artificial glycan holes will be discussed below.
The Env glycans and the glycan shield are conformationally dynamic, much more so than the underlying Env protein domains. A recent study described an approach that combined cryo‐EM, computational modelling and MS to visualize the glycan shield structure and dynamics [99]. This approach was able to detect subtle changes in PNGS occupancy, glycan compositions and glycan dynamics that can impact the structure of the glycan shield and epitope accessibility [99]. Further in silico studies using molecular dynamics simulations attempted to predict glycan movement for HIV‐1 and SARS‐CoV‐2 [100, 101]. A better understanding and visualization of the dynamics of the glycan shield helps to understand how bNAbs cope with the glycan shield and how we need to consider this in vaccine design efforts.
3.6. Structure‐based analysis of Ab responses against native‐like trimers
3.6.1. Mapping Ab responses by traditional serology
The extensive studies on the prototype BG505 SOSIP.664 trimer in rabbit and rhesus macaque immunization studies underlined how the glycan shield ‐ and its holes ‐ shape the NAb response [70]. Serum neutralization experiments with mutant viruses revealed several NAb specificities, but with limited breadth. A major proportion of the autologous Tier‐2 NAb responses was directed to the large glycan hole around positions 241 and 289, which is specific to the BG505 strain [70, 97]. The serum neutralization experiments also identified neutralizing epitopes residing in the V1 and C3/V5 regions [102, 103, 104]. The BG505 SOSIP.664 trimer also induced strong Tier‐1 neutralizing responses targeting a linear epitope in the V3 of gp120. The V3 is not exposed on most primary viruses, and these specificities are therefore unable to neutralize such viruses [70]. Finally, the removal of the transmembrane domain and cytoplasmic tail creates a neo‐epitope at the trimer base, which is immunodominant [105]. Animal vaccination studies using NFL trimers yielded very similar results as BG505 SOSIP trimers, with strain‐specific responses to the N241/N289 glycan hole dominating [106, 107].
3.6.2. Mapping Ab responses by structural analysis of MAbs from immunized animals
Traditional serology studies are sometimes difficult to interpret because the mutations might have allosteric effects. Furthermore, they do not reveal approach angles, atomic contacts and other structural features. MAbs have been isolated from several animal species immunized with the BG505 trimer [97, 102, 108, 109, 110, 111]. EM analysis of these MAbs in complex with the BG505 trimer revealed that many of the strain‐specific NAbs indeed targeted the N241/N289 glycan hole created by the absence of these glycans [97, 102]. Other studies identified sites leading to narrow‐neutralization breadth, in the C3/V4, C3/V5 and the V1 regions [102, 108, 109, 110, 111]. While these specificities are unlikely to be amendable to developing neutralizing breadth, occasionally MAbs are found that are. An example is a BG505 trimer‐induced MAb that targets the gp120/gp41 interface with significant overlap with the epitope of the VRC34 bNAb [98].
3.6.3. Mapping Ab responses by EMPEM
MAb isolation is a laborious process and it always remains unclear how representative a given MAb is for the overall serological response. In addition, it is difficult to appreciate the totality of the polyclonal response based on a limited set of MAbs. Electron microscopy‐based polyclonal epitope mapping (EMPEM) is a recently developed technique that allows for a visual snapshot of the specificities present in serum samples [111, 112] (Table 1). The values of the EMPEM technique in delineating serum responses was revealed as it uncovered previously unidentified epitopes (described below), but also confirmed the known N241/N289 glycan hole and base targeting specificities [112]. The newly discovered antibody specificities included ones that target the cleft of the trimer (COT) between two protomers, and one around the N241/N289 glycan hole with no neutralization activity [112]. In macaques, similar responses were observed, but also additional epitopes were identified targeting the fusion peptide, V1/V3 and an epitope in the gp120/gp41 interface [111]. The base specificities are not new, but EMPEM analysis revealed that the base responses were dominating very early after the first prime [112]. The visualization of the different specificities present in one serum is a major asset to the toolkit to analyse antibody serum responses. When performed in sequential samples, it also allows visualizing the evolving antibody specificities over time, in response to different immunizations.
3.7. Structure‐based modification of the glycan shield
3.7.1. Filling holes in the glycan shield
While glycans play important roles in bNAb responses, holes in the glycan shield caused by the absence of PNGS that are present in most primary virus isolates appear to dominate the antibody response after vaccination with Env trimer immunogens. These glycan holes induce NAb responses of narrow breadth [70, 97, 113]. Filling glycan holes could therefore be a strategy to focus the immune response to more desirable targets. This strategy has been successfully applied to BG505, where the absence of the highly conserved PNGS at positions 241 and 289 in the BG505 sequence creates a large immunodominant hole that induces the majority of the strain‐specific NAbs (Figure 3) [70, 97]. As the NAb responses to this hole are “dead‐end” specificities that cannot be broadened, filling this hole may be a good strategy, and it can easily be achieved by restoring the missing PNGS [102, 104, 113].
Figure 3.
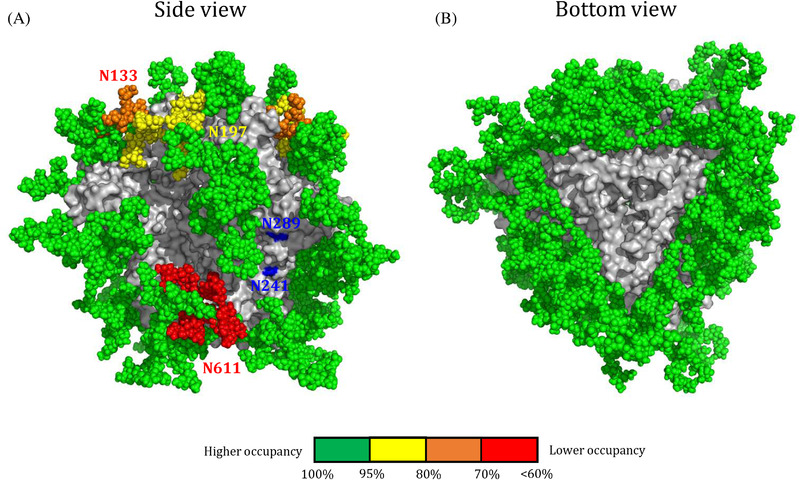
Sources for glycan holes found on stabilized soluble native‐like trimers. (a) Side and bottom view of the glycosylated BG505 SOSIP.664 trimer. The glycans were modelled on the crystal structure of BG505 SOSIP.664 (PDB: 5CEZ) using the Glyprot tool from Glycosciences. The missing glycans at positions 241 and 289 on gp120 are indicated in blue. Colour coding reflects the occupancy of each PNGS, similar as described in [114]. Green, full occupancy (>95%); yellow, 80% to 95%; orange, 70% to 80%; red, >60% occupancy. Lower occupancy of a PNGS results in an artificial glycan hole that can be immunogenic. (b) Bottom view of the glycosylated BG505 SOSIP.664 trimer. The removal of the trimer from the membrane creates a large immunodominant hole.
3.7.2. Filling artificial holes in the glycan shield
Glycan holes can also exist by underoccupancy of PNGS, creating artificial glycan holes that can distract the immune system from more desirable targets. Several PNGS are underoccupied on BG505 SOSIP trimers, while they are fully occupied on the parental virus (Figure 3). The fact that holes created by PNGS underoccupancy are immunogenic was revealed by isolation of MAbs from BG505 SOSIP.664 immunized rhesus macaques that target the N611 site. These MAbs can only neutralize when the N611 glycan is removed, indicating that the site is fully occupied on the virus [70, 97, 98]. High‐resolution structures of Env in complex with the N611 targeting non‐NAb RM20E1 revealed that the N611 glycan is indeed underoccupied to an extend of approximately 60% [98, 114]. A sequon engineering strategy increased the occupancy of several PNGS on the BG505 SOSIP trimer, as well on several other Env isolates, thereby eliminating artificial glycan holes [114]. The strategy exploited observations on model proteins from the mid‐1990s that N‐X‐T sequons have a two to three‐fold higher propensity to become occupied compared to N‐X‐S sequons [115]. The replacement of N‐X‐S sequons by N‐X‐T sequons enhanced PNGS occupancy overall, although some site‐specific adjustments needed to be made [114]. Increasing PNGS occupancy should reduce off‐target responses and steer the immune response to more desirable targets.
3.7.3. Covering the trimer base using glycans
MAb isolation and in particular EMPEM analyses revealed that the trimer base is highly immunodominant after immunization with soluble Env trimers. The truncation of the Env trimer from the membrane, in effect creates a large glycan hole that is highly immunogenic but only induces non‐NAbs because it is absent from virus‐associated Env (Figure 3). As these responses might also be distracting from more desirable NAb responses, covering this large hole might therefore be necessary. This might not be very straightforward. Kulp et al. (2017) addressed this by redesigning a BG505 trimer by swapping the orientation of gp120 and gp41 subunits and placing a linker between the two subunits [73, 75]. This linker allowed for the introduction of PNGS to shield the glycan base, resulting in reduced binding of base‐targeting non‐NAbs [75]. Additional efforts to close this hole on Env‐based immunogens are underway and include the presentation of Env on nanoparticles [116, 117]. In a similar approach, glycans were used to mask the “backside” of the eOD‐GT8 immunogen (see below: CD4 binding site‐focused approaches) resulting in the MUT16 immunogen [118, 119].
3.7.4. Introducing new holes in the glycan shield
Filling the N241/N289 glycan hole on BG505 SOSIP trimers reduces the responses directed to this specific site. Opening holes elsewhere on the trimer, by knocking‐out a PNGS can redirect the immune response to these newly formed holes, providing important proof of concept that this strategy might be valuable [104]. Such strategies are being explored to focus antibody responses to the CD4bs by removing glycans that restrict access to this bNAb epitope cluster [120, 121, 122]. In anecdotal cases, this has led to the elicitation of cross‐neutralizing antibody responses directed to the CD4bs [120, 123, 124]. Removing glycans might be particularly relevant for germline‐targeting immunogen approaches, that is, approaches based on targeting and activating bNAb precursor B cells with specific genetic signatures [118, 119, 125]. One component of germline‐targeting design often involves enhancing accessibility of the site of interest, while another component involves the structure‐based design of modifications to enhance the interaction with desirable naïve B cells.
3.8. Role of native‐like trimer immunogens in driving bNAb responses
In the above sections, we described the development and fine‐tuning of native‐like trimers, what type of antibody specificities they induce and how they can be further modified. However, they are unlikely to induce bNAb responses by themselves, but they form a platform for further, epitope‐specific, immune focusing and/or they provide the final “polishing” steps to finish off a sequential vaccination regimen to mature antibody lineages to become broadly neutralizing (see also Williams et al. [126]). We review three prominent epitope‐focused approaches that involve native‐like trimers at some stage in the regimen. The structures of bNAbs in complex with the HIV‐1 Env trimer and the delineation of the sites of vulnerability on Env are the basis of immunogen designs that focuses on specific epitopes. Epitope‐focused vaccines can be “germline‐agnostic” embracing antibodies derived from different genetic germline signatures, or they can target specific germline classes [43].
3.8.1. Fusion peptide‐focused vaccine approach
While it has long been thought that the FP was hidden inside the interior of the pre‐fusion Env trimers and was poorly immunogenic, the highly conserved FP became a prominent target following the discovery of the FP targeting bNAbs VRC34 and ACS202 [127, 128, 129]. Xu et al. investigated the ability of a simple FP peptide to prime FP‐directed Ab responses, choosing the eight N‐terminal residues of the FP as these where shown to be contacted by VRC34 [127, 129]. Structure‐based design was used to engineer FP‐containing immunogens that comprised epitope scaffolds and native‐like Env trimers. The FP scaffolds were created by conjugating the eight N‐terminal residues, as well as the N88 glycan that contributes to the VRC34 and ACS202 epitopes to keyhole limpet haemocyanin (KLH) as a carrier [129]. An immunization scheme evaluated in nonhuman primates with five FP‐KLH primes followed by three trimer boosts, led to neutralizing breadth in a few animals, showing that FP epitope‐focused vaccine design merits further study [129].
3.8.2. N332 supersite‐focused approaches
The relatively conserved V3‐glycan “supersite” is also an interesting target, as very potent bNAbs are induced against this epitope. Furthermore, the majority of PLHIV produce glycan‐dependent bNAbs to this epitope [130]. The PGT121‐class bNAbs, targeting the N332 supersite in the V3, belong to the most potent and well‐characterized bNAbs described today. Steichen et al. designed a gl‐targeting gp120 molecule that specifically binds PGT121 gl‐reverted MAbs, by using mammalian display‐directed evolution [73]. These mutations were then transferred to a stabilized BG505 trimer, resulting in a trimer termed 11MUTB. Multivalent display of these trimers on liposomes enhanced the activation of B cells that express gl‐PGT121 as their BCR, compared to the soluble trimers, and elicited MAbs with neutralization breadth in a KI‐mouse model [73, 131]. The RC1 trimer, based on 11MUTB, lacks the N133 and N137 glycans but also the N156 glycan as it was hypothesized that this would further improve binding of PGT121‐class precursors [132]. In a further iteration, termed RC1‐4fill, PNGS at positions 230, 241, 289 and 344 were introduced to fill glycan holes (see above) and focus the immune response further to the N332 supersite in the V3. The designs were able to activate and expand B cells in mice, rabbits and macaques that resemble the human V3‐glycan “supersite” targeting germline‐MAbs [132].
3.8.3. CD4 binding site‐focused approaches
The CD4bs is an interesting site for epitope‐focused vaccine design because some of the most potent bNAbs target this functionally conserved site. The VRC01‐class of CD4bs bNAbs have received much attention in this regard because VRC01‐class bNAbs have been identified in multiple PLHIV, and they have distinct genetic signatures that include a VH1‐2‐derived heavy chain and a light chain with a short five amino acid CDRL3. Designing immunogens that could activate gl‐VRC01‐class B‐cell precursors in vivo might be key in developing an effective vaccine that can induce bNAbs targeting the CD4bs. Native‐like trimers, such as SOSIP trimers, do not readily induce CD4bs‐directed MAbs very easily nor do they interact with gl‐VRC01‐class MAbs. Reasons are steric constraints because of the location of the CD4bs in the cleft between two protomers, steric hindrance by the N276 glycan and specific atomic contacts with the gl‐MAbs [133, 134]. The development of the VRC01 lineage was most likely initiated by viruses that lacked the N276 glycan [135]. Several immunogens have been designed to activate precursor VRC01‐class B cells. These immunogens include an engineered outer domain (eOD‐GT8) and the BG505 SOSIP GT1 trimer [118, 125, 136]. An updated version of this trimer (BG505 SOSIP GT1.1) entered a phase I clinical trial, as did eOD‐GT8 that showed very encouraging results [124, 137, 138]. In addition to germline targeting approaches for the CD4bs, other germline agnostic approaches involved the deletion of glycans to approach accessibility of the CD4bs [120, 121, 122].
4. CONCLUSIONS
The development of the native‐like BG505 SOSIP.664 trimer and its first high‐resolution structure fueled the field of Env structure‐based vaccine design. The Env trimer structures allowed for the identification of modifications to improve the stability, antigenicity and yield of native‐like trimer vaccine candidates, but also allowed for the development of native‐like trimers based on different isolates and clades. Native‐like SOSIP trimers have now entered phase I clinical trials[124, 139, 140, 141, 142, 143, 144] (Table 1; Kim et al. [145]), and the outcome of these trials will inform whether native‐like Env vaccine candidates will induce autologous Tier‐2 NAb responses in humans. High‐resolution structures of bNAbs and their germline counterparts in complex with native‐like HIV‐1 Env trimers aided in the identification of their epitopes. New emerging techniques, such as EMPEM, allowing for an increasing understanding of what immunogens induce, will further guide iterative vaccine design efforts with increasing speed. This information will help in reducing undesirable off‐target responses and improve driving on‐target responses towards the development of bNAbs. Finally, rationally designed nanoparticle platforms, not reviewed here, facilitate the multivalent presentation of Env trimers, thereby increasing B‐cell activation and enhancing the magnitude of the antibody response.
COMPETING INTERESTS
The International AIDS Vaccine Initiative (IAVI) has previously filed a patent relating to the BG505 SOSIP.664 trimer: U.S. provisional application 61/772,739, entitled “HIV‐1 Envelope Glycoprotein”, with R.W.S. among the co‐inventors.
AUTHORS’ CONTRIBUTIONS
RD: Writing, original draft preparation and figure preparation. RWS: Writing, review and editing.
FUNDING
Work by the authors in this area is supported by the U.S. National Institutes of Health under grant P0 AI110657; by the Bill and Melinda Gates Foundation through the Collaboration for AIDS Vaccine Discovery (CAVD), grants OPP1132237 and INV‐002022; by the European Union's Horizon 2020 research and innovation programme under grant agreement No. 681137; and by a Vici grant from the Netherlands Organization for Scientific Research (NWO).
ACKNOWLEDGEMENTS
None.
REFERENCES
- 1. Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, et al. Diversity considerations in HIV‐1 vaccine selection. Science. 2002;296:2354–60. [DOI] [PubMed] [Google Scholar]
- 2. Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV‐1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–23. [DOI] [PubMed] [Google Scholar]
- 3. Spira S, Wainberg MA, Loemba H, Turner D, Brenner BG. Impact of clade diversity on HIV‐1 virulence, antiretroviral drug sensitivity and drug resistance. J Antimicrob Chemother. 2003;51:229–40. [DOI] [PubMed] [Google Scholar]
- 4. Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV‐1 subtype diversity. N Engl J Med. 2008;358:1590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gelderblom HR, Hausmann EH, Ozel M, Pauli G, Koch MA. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–6. [DOI] [PubMed] [Google Scholar]
- 6. Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV‐1. Nature. 2003;422:307–12. [DOI] [PubMed] [Google Scholar]
- 7. McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, Reyes GR, et al. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. [DOI] [PubMed] [Google Scholar]
- 8. Wyatt R, Sodroski J. The HIV‐1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–8. [DOI] [PubMed] [Google Scholar]
- 9. Earl PL, Moss B, Doms RW. Folding, interaction with GRP78‐BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991;65:2047–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doms RW, Lamb RA, Rose JK, Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993;193:545–62. [DOI] [PubMed] [Google Scholar]
- 11. Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–82. [PubMed] [Google Scholar]
- 12. Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, et al. DC‐SIGN, a dendritic cell‐specific HIV‐1‐binding protein that enhances trans‐infection of T cells. Cell. 2000;100:587–97. [DOI] [PubMed] [Google Scholar]
- 13. Montefiori DC, Robinson WE, Mitchell WM. Role of protein N‐glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1988;85:9248–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walker BD, Kowalski M, Goh WC, Kozarsky K, Krieger M, Rosen C, et al. Inhibition of human immunodeficiency virus syncytium formation and virus replication by castanospermine. Proc Natl Acad Sci U S A. 1987;84:8120–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reitter JN, Means RE, Desrosiers RC. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–84. [DOI] [PubMed] [Google Scholar]
- 16. Li Y, Luo L, Rasool N, Kang CY. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J Virol. 1993;67:584–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Binley JM, Ban Y‐EA, Crooks ET, Eggink D, Osawa K, Schief WR, et al. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J Virol. 2010;84:5637–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Gils MJ, Euler Z, Schweighardt B, Wrin T, Schuitemaker H. Prevalence of cross‐reactive HIV‐1‐neutralizing activity in HIV‐1‐infected patients with rapid or slow disease progression. AIDS. 2009;23:2405–14. [DOI] [PubMed] [Google Scholar]
- 19. Sok D, Burton DR. Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol. 2018;19:1179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, et al. Broadly neutralizing human anti‐HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moldt B, Rakasz EG, Schultz N, Chan‐Hui P‐Y, Swiderek K, Weisgrau KL, et al. Highly potent HIV‐specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 2012;109:18921–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shingai M, Donau OK, Plishka RJ, Buckler‐White A, Mascola JR, Nabel GJ, et al. Passive transfer of modest titers of potent and broadly neutralizing anti‐HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211:2061–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corey L, Gilbert PB, Juraska M, Montefiori DC, Morris L, Karuna ST, et al. Two randomized trials of neutralizing antibodies to prevent HIV‐1 acquisition. N Engl J Med. 2021;384:1003–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graham BS, Gilman MSA, McLellan JS. Structure‐based vaccine antigen design. Annu Rev Med. 2019;70:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. 2000;3:445–50. [DOI] [PubMed] [Google Scholar]
- 26. Burton DR. Antibodies, viruses and vaccines. Nat Rev Immunol. 2002;2:706–13. [DOI] [PubMed] [Google Scholar]
- 27. Rappuoli R, Bottomley MJ, D'Oro U, Finco O, De Gregorio E. Reverse vaccinology 2.0: human immunology instructs vaccine antigen design. J Exp Med. 2016;213:469–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion‐specific neutralizing antibody. Science. 2013;340:1113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CMM, Lukens M V, et al. Generation of stable monoclonal antibody‐producing B cell receptor‐positive human memory B cells by genetic programming. Nat Med. 2010;16:123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffenberg S, Powell R, Carpov A, Wagner D, Wilson A, Kosakovsky PS, et al. Identification of an HIV‐1 clade A envelope that exhibits broad antigenicity and neutralization sensitivity and elicits antibodies targeting three distinct epitopes. J Virol. 2013;87:5372–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sanders RW, Derking R, Cupo A, Julien J‐P, Yasmeen A, de Val N, et al. A next‐generation cleaved, soluble HIV‐1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non‐neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion‐associated structure. J Virol. 2000;74:627–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanders RW, Schiffner L, Master A, Kajumo F, Guo Y, Dragic T, et al. Variable‐loop‐deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J Virol. 2000;74:5091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Binley JM, Sanders RW, Master A, Cayanan CS, Wiley CL, Schiffner L, et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2002;76:2606–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lyumkis D, Julien J‐P, de Val N, Cupo A, Potter CS, Klasse P‐J, et al. Cryo‐EM structure of a fully glycosylated soluble cleaved HIV‐1 envelope trimer. Science. 2013;342:1484–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Julien J‐P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, et al. Crystal structure of a soluble cleaved HIV‐1 envelope trimer. Science. 2013;342:1477–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, et al. Structure and immune recognition of trimeric pre‐fusion HIV‐1 Env. Nature. 2014;514:455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee JH, Ozorowski G, Ward AB. Cryo‐EM structure of a native, fully glycosylated, cleaved HIV‐1 envelope trimer. Science. 2016;351:1043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guttman M, Garcia NK, Cupo A, Matsui T, Julien J‐P, Sanders RW, et al. CD4‐induced activation in a soluble HIV‐1 Env trimer. Structure. 2014;22:974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stadtmueller BM, Bridges MD, Dam K‐M, Lerch MT, Huey‐Tubman KE, Hubbell WL, et al. DEER spectroscopy measurements reveal multiple conformations of HIV‐1 SOSIP envelopes that show similarities with envelopes on native virions. Immunity. 2018;49:235–46.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burton DR. Advancing an HIV vaccine; advancing vaccinology. Nat Rev Immunol. 2019;19:77–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kwong PD, Mascola JR. HIV‐1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity. 2018;48:855–71. [DOI] [PubMed] [Google Scholar]
- 44. Julien J, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, et al. Asymmetric recognition of the HIV‐1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A. 2013;110:4351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burton DR, Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol. 2016;34:635–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, et al. Comprehensive antigenic map of a cleaved soluble HIV‐1 envelope trimer. PLoS Pathog. 2015;11:e1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Doria‐Rose NA, Georgiev I, O'Dell S, Chuang G‐Y, Staupe RP, McLellan JS, et al. A short segment of the HIV‐1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J Virol. 2012;86:8319–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee JH, Andrabi R, Su C‐Y, Yasmeen A, Julien J‐P, Kong L, et al. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic β‐hairpin structure. Immunity. 2017;46:690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roark RS, Li H, Williams WB, Chug H, Mason RD, Gorman J, et al. Recapitulation of HIV‐1 Env‐antibody coevolution in macaques leading to neutralization breadth. Science. 2021;371:eabd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Medina‐Ramírez M, Sanders RW, Sattentau QJ. Stabilized HIV‐1 envelope glycoprotein trimers for vaccine use. Curr Opin HIV AIDS. 2017;12:241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou T, Zheng A, Baxa U, Chuang G‐Y, Georgiev IS, Kong R, et al. A neutralizing antibody recognizing primarily N‐linked glycan targets the silent face of the HIV envelope. Immunity. 2018;48:500–13.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schoofs T, Barnes CO, Suh‐Toma N, Golijanin J, Schommers P, Gruell H, et al. Broad and potent neutralizing antibodies recognize the silent face of the HIV envelope. Immunity. 2019;50:1513–29.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de Taeye SW, Moore JP, Sanders RW. HIV‐1 envelope trimer design and immunization strategies to induce broadly neutralizing antibodies. Trends Immunol. 2016;37:221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–11. [DOI] [PubMed] [Google Scholar]
- 55. Torrents de la Peña A, Sanders RW. Stabilizing HIV‐1 envelope glycoprotein trimers to induce neutralizing antibodies. Retrovirology. 2018;15:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Torrents de la Peña A, de Taeye SW, Sliepen K, LaBranche CC, Burger JA, Schermer EE, et al. Immunogenicity in rabbits of HIV‐1 SOSIP trimers from clades A, B, and C, given individually, sequentially, or in combination. J Virol. 2018;92:e01957‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Do Kwon Y, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, et al. Crystal structure, conformational fixation and entry‐related interactions of mature ligand‐free HIV‐1 Env. Nat Struct Mol Biol. 2015;22:522–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Torrents de la Peña A, Julien JP, de Taeye SW, Garces F, Guttman M, Ozorowski G, et al. Improving the immunogenicity of native‐like HIV‐1 envelope trimers by hyperstabilization. Cell Rep. 2017;20:1805–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hastie KM, Zandonatti MA, Kleinfelter LM, Heinrich ML, Rowland MM, Chandran K, et al. Structural basis for antibody‐mediated neutralization of Lassa virus. Science. 2017;356:923–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guenaga J, Garces F, de Val N, Stanfield RL, Dubrovskaya V, Higgins B, et al. Glycine substitution at helix‐to‐coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity. 2017;46:792–803.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qiao H, Pelletier SL, Hoffman L, Hacker J, Armstrong RT, White JM. Specific single or double proline substitutions in the “spring‐loaded” coiled‐coil region of the influenza hemagglutinin impair or abolish membrane fusion activity. J Cell Biol. 1998;141:1335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rutten L, Gilman MSA, Blokland S, Juraszek J, McLellan JS, Langedijk JPM. Structure‐based design of prefusion‐stabilized filovirus glycoprotein trimers. Cell Rep. 2020;30:4540–50.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hsieh C‐L, Goldsmith JA, Schaub JM, DiVenere AM, Kuo H‐C, Javanmardi K, et al. Structure‐based design of prefusion‐stabilized SARS‐CoV‐2 spikes. Science. 2020;369:1501–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, et al. Immunogenicity and structures of a rationally designed prefusion MERS‐CoV spike antigen. Proc Natl Acad Sci U S A. 2017;114:E7348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Walls AC, Xiong X, Park Y‐J, Tortorici MA, Snijder J, Quispe J, et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–39.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sanders RW, Moore JP. Virus vaccines: proteins prefer prolines. Cell Host Microbe. 2021;29:327–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Krammer F. SARS‐CoV‐2 vaccines in development. Nature. 2020;586:516–27. [DOI] [PubMed] [Google Scholar]
- 68. Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, et al. Conformational dynamics of single HIV‐1 envelope trimers on the surface of native virions. Science. 2014;346:759–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, et al. A native‐like SOSIP.664 trimer based on a HIV‐1 subtype B Env gene. J Virol. 2015;89:3380–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, et al. HIV‐1 neutralizing antibodies induced by native‐like envelope trimers. Science. 2015;349:aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. de Taeye SW, Ozorowski G, Torrents de la Peña A, Guttman M, Julien J‐P, van den Kerkhof TLGM, et al. Immunogenicity of stabilized HIV‐1 envelope trimers with reduced exposure of non‐neutralizing epitopes. Cell. 2015;163:1702–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. De Taeye SW, De La Peñ AT, Vecchione A, Scutigliani E, Sliepen K, Burger JA, et al. Stabilization of the gp120 V3 loop through hydrophobic interactions reduces the immunodominant V3‐directed non‐neutralizing response to HIV‐1 envelope trimers. J Biol Chem. 2018;293:1688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, et al. HIV vaccine design to target germline precursors of glycan‐dependent broadly neutralizing antibodies. Immunity. 2016;45:483–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chuang G‐Y, Geng H, Pancera M, Xu K, Cheng C, Acharya P, et al. Structure‐based design of a soluble prefusion‐closed HIV‐1 env trimer with reduced CD4 affinity and improved immunogenicity. J Virol. 2017;91:e02268‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, et al. Structure‐based design of native‐like HIV‐1 envelope trimers to silence non‐neutralizing epitopes and eliminate CD4 binding. Nat Commun. 2017;8:1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rutten L, Lai Y‐T, Blokland S, Truan D, Bisschop IJM, Strokappe NM, et al. A universal approach to optimize the folding and stability of prefusion‐closed HIV‐1 envelope trimers. Cell Rep. 2018;23(2):584–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, et al. Single‐chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV‐1 Env. J Virol. 2015;89:5318–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, et al. Cleavage‐independent HIV‐1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, et al. Structure‐guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J Virol. 2015;90:2806–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chaudhury S, Reifman J, Wallqvist A. Simulation of B cell affinity maturation explains enhanced antibody cross‐reactivity induced by the polyvalent malaria vaccine AMA1. J Immunol. 2014;193:2073–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang S, Mata‐Fink J, Kriegsman B, Hanson M, Irvine DJ, Eisen HN, et al. Manipulating the selection forces during affinity maturation to generate cross‐reactive HIV antibodies. Cell. 2015;160:785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shaffer JS, Moore PL, Kardar M, Chakraborty AK. Optimal immunization cocktails can promote induction of broadly neutralizing Abs against highly mutable pathogens. Proc Natl Acad Sci U S A. 2016;113:E7039–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ringe RP, Yasmeen A, Ozorowski G, Go EP, Pritchard LK, Guttman M, et al. Influences on the design and purification of soluble, recombinant native‐like HIV‐1 envelope glycoprotein trimers. J Virol. 2015;89:12189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Julien J‐P, Lee JH, Ozorowski G, Hua Y, Torrents de la Peña A, de Taeye SW, et al. Design and structure of two HIV‐1 clade C SOSIP.664 trimers that increase the arsenal of native‐like Env immunogens. Proc Natl Acad Sci U S A. 2015;112:11947–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, et al. Well‐ordered trimeric HIV‐1 subtype B and C soluble spike mimetics generated by negative selection display native‐like properties. PLoS Pathog. 2015;11:e1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kumar R, Ozorowski G, Kumar V, Holden LG, Shrivastava T, Patil S, et al. Characterization of a stable HIV‐1 B/C recombinant, soluble, and trimeric envelope glycoprotein (Env) highly resistant to CD4‐induced conformational changes. J Biol Chem. 2017;292:15849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sliepen K, Han BW, Bontjer I, Mooij P, Garces F, Behrens AJ, et al. Structure and immunogenicity of a stabilized HIV‐1 envelope trimer based on a group‐M consensus sequence. Nat Commun. 2019;10:2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Schorcht A, van den Kerkhof TLGM, Cottrell CA, Allen JD, Torres JL, Behrens A‐J, et al. Neutralizing antibody responses induced by HIV‐1 envelope glycoprotein SOSIP trimers derived from elite neutralizers. J Virol. 2020;94:e01214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Aldon Y, McKay PF, Allen J, Ozorowski G, Felfödiné Lévai R, Tolazzi M, et al. Rational design of DNA‐expressed stabilized native‐like HIV‐1 envelope trimers. Cell Rep. 2018;24:3324–38.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rawi R, Rutten L, Lai Y‐T, Olia AS, Blokland S, Juraszek J, et al. Automated design by structure‐based stabilization and consensus repair to achieve prefusion‐closed envelope trimers in a wide variety of HIV strains. Cell Rep. 2020;33:108432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Behrens A‐J, Struwe WB, Crispin M. Glycosylation profiling to evaluate glycoprotein immunogens against HIV‐1. Expert Rev Proteomics. 2017;14:881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Behrens A‐J, Crispin M. Structural principles controlling HIV envelope glycosylation. Curr Opin Struct Biol. 2017;44:125–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Seabright GE, Cottrell CA, van Gils MJ, D'addabbo A, Harvey DJ, Behrens A‐J, et al. Networks of HIV‐1 envelope glycans maintain antibody epitopes in the face of glycan additions and deletions. Structure. 2020;28:897–909.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wagh K, Kreider EF, Li Y, Barbian HJ, Learn GH, Giorgi E, et al. Completeness of HIV‐1 envelope glycan shield at transmission determines neutralization breadth. Cell Rep. 2018;25:893–908.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cao L, Pauthner M, Andrabi R, Rantalainen K, Berndsen Z, Diedrich JK, et al. Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat Commun. 2018;9:3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Struwe WB, Chertova E, Allen JD, Seabright GE, Watanabe Y, Harvey DJ, et al. Site‐specific glycosylation of virion‐derived HIV‐1 Env is mimicked by a soluble trimeric immunogen. Cell Rep. 2018;24:1958–66.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, et al. Holes in the glycan shield of the native HIV envelope are a target of trimer‐elicited neutralizing antibodies. Cell Rep. 2016;16:2327–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cottrell CA, van Schooten J, Bowman CA, Yuan M, Oyen D, Shin M, et al. Mapping the immunogenic landscape of near‐native HIV‐1 envelope trimers in non‐human primates. PLoS Pathog. 2020;16:e1008753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Berndsen ZT, Chakraborty S, Wang X, Cottrell CA, Torres JL, Diedrich JK, et al. Visualization of the HIV‐1 Env glycan shield across scales. Proc Natl Acad Sci U S A. 2020;117:28014–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Casalino L, Gaieb Z, Goldsmith JA, Hjorth CK, Dommer AC, Harbison AM, et al. Beyond shielding: the roles of glycans in the SARS‐CoV‐2 spike protein. ACS Cent Sci. 2020;6:1722–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wei Q, Hargett AA, Knoppova B, Duverger A, Rawi R, Shen C‐H, et al. Glycan positioning impacts HIV‐1 Env glycan‐shield density, function, and recognition by antibodies. iScience. 2020;23:101711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Klasse PJ, Ketas TJ, Cottrell CA, Ozorowski G, Debnath G, Camara D, et al. Epitopes for neutralizing antibodies induced by HIV‐1 envelope glycoprotein BG505 SOSIP trimers in rabbits and macaques. PLoS Pathog. 2018;14:e1006913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Charles TP, Burton SL, Arunachalam PS, Cottrell CA, Sewall LM, Bollimpelli VS, et al. The C3/465 glycan hole cluster in BG505 HIV‐1 envelope is the major neutralizing target involved in preventing mucosal SHIV infection. PLoS Pathog. 2021;17:e1009257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ringe RP, Pugach P, Cottrell CA, LaBranche CC, Seabright GE, Ketas TJ, et al. Closing and opening holes in the glycan shield of HIV‐1 envelope glycoprotein SOSIP trimers can redirect the neutralizing antibody response to the newly unmasked epitopes. J Virol. 2019;93:e01656‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, et al. Murine antibody responses to cleaved soluble HIV‐1 envelope trimers are highly restricted in specificity. J Virol. 2015;89:10383–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bale S, Martiné A, Wilson R, Behrens A‐J, Le Fourn V, de Val N, et al. Cleavage‐independent HIV‐1 trimers from CHO cell lines elicit robust autologous tier 2 neutralizing antibodies. Front Immunol. 2018;9:1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Martinez‐Murillo P, Tran K, Guenaga J, Lindgren G, Àdori M, Feng Y, et al. Particulate array of well‐ordered HIV clade C env trimers elicits neutralizing antibodies that display a unique V2 cap approach. Immunity. 2017;46:804–17.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Pauthner M, Havenar‐Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy A V, et al. Elicitation of robust Tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity. 2017;46:1073–88.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA, et al. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell. 2019;177:1153–71.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lei L, Yang YR, Tran K, Wang Y, Chiang C‐I, Ozorowski G, et al. The HIV‐1 envelope glycoprotein C3/V4 region defines a prevalent neutralization epitope following immunization. Cell Rep. 2019;27:586–98.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nogal B, Bianchi M, Cottrell CA, Kirchdoerfer RN, Sewall LM, Turner HL, et al. Mapping polyclonal antibody responses in non‐human primates vaccinated with HIV Env trimer subunit vaccines. Cell Rep. 2020;30:3755–65.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bianchi M, Turner HL, Nogal B, Cottrell CA, Oyen D, Pauthner M, et al. Electron‐microscopy‐based epitope mapping defines specificities of polyclonal antibodies elicited during HIV‐1 BG505 envelope trimer immunization. Immunity. 2018;49:288–300.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Yang YR, McCoy LE, van Gils MJ, Andrabi R, Turner HL, Yuan M, et al. Autologous antibody responses to an HIV envelope glycan hole are not easily broadened in rabbits. J Virol. 2020;94:e01861‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Derking R, Allen JD, Cottrell CA, Sliepen K, Seabright GE, Lee W‐H, et al. Enhancing glycan occupancy of soluble HIV‐1 envelope trimers to mimic the native viral spike. Cell Rep. 2021;35:108933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kasturi L, Eshleman JR, Wunner WH, Shakin‐Eshleman SH. The hydroxy amino acid in an Asn‐X‐Ser/Thr sequon can influence N‐linked core glycosylation efficiency and the level of expression of a cell surface glycoprotein. J Biol Chem. 1995;270:14756–61. [DOI] [PubMed] [Google Scholar]
- 116. Brinkkemper M, Sliepen K. Nanoparticle vaccines for inducing HIV‐1 neutralizing antibodies. Vaccines. 2019;7:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Brouwer PJM, Sanders RW. Presentation of HIV‐1 envelope glycoprotein trimers on diverse nanoparticle platforms. Curr Opin HIV AIDS. 2019;14:302–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jardine JG, Kulp DW, Havenar‐Daughton C, Sarkar A, Briney B, Sok D, et al. HIV‐1 broadly neutralizing antibody precursor B cells revealed by germline‐targeting immunogen. Science. 2016;351:1458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Duan H, Chen X, Boyington JC, Cheng C, Zhang Y, Jafari AJ, et al. Glycan masking focuses immune responses to the HIV‐1 CD4‐binding site and enhances elicitation of VRC01‐class precursor antibodies. Immunity. 2018;49:301–11.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Dubrovskaya V, Guenaga J, de Val N, Wilson R, Feng Y, Movsesyan A, et al. Targeted N‐glycan deletion at the receptor‐binding site retains HIV Env NFL trimer integrity and accelerates the elicited antibody response. PLoS Pathog. 2017;13:e1006614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. LaBranche CC, McGuire AT, Gray MD, Behrens S, Kwong PD, Chen X, et al. HIV‐1 envelope glycan modifications that permit neutralization by germline‐reverted VRC01‐class broadly neutralizing antibodies. PLoS Pathog. 2018;14:e1007431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhou T, Doria‐Rose NA, Cheng C, Stewart‐Jones GBE, Chuang G‐Y, Chambers M, et al. Quantification of the impact of the HIV‐1‐glycan shield on antibody elicitation. Cell Rep. 2017;19:719–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Dubrovskaya V, Tran K, Ozorowski G, Guenaga J, Wilson R, Bale S, et al. Vaccination with glycan‐modified HIV NFL envelope trimer‐liposomes elicits broadly neutralizing antibodies to multiple sites of vulnerability. Immunity. 2019;51:915–29.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. ClinicalTrials.gov . Identifier: NCT04224701 Available from: https://clinicaltrials.gov/ct2/show/NCT04224701
- 125. Medina‐Ramírez M, Garces F, Escolano A, Skog P, de Taeye SW, Del Moral‐Sanchez I, et al. Design and crystal structure of a native‐like HIV‐1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J Exp Med. 2017;214:2573–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Williams WB, Wiehe K, Saunders KO, Haynes BF. Strategies for induction of HIV‐1 envelope‐reactive broadly neutralizing antibodies. J Int AIDS Soc. 2021;24(S7):47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, et al. Fusion peptide of HIV‐1 as a site of vulnerability to neutralizing antibody. Science. 2016;352:828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. van Gils MJ, van den Kerkhof TLGM, Ozorowski G, Cottrell CA, Sok D, Pauthner M, et al. An HIV‐1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat Microbiol. 2016;2:16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Xu K, Acharya P, Kong R, Cheng C, Chuang G‐Y, Liu K, et al. Epitope‐based vaccine design yields fusion peptide‐directed antibodies that neutralize diverse strains of HIV‐1. Nat Med. 2018;24:857–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Landais E, Huang X, Havenar‐Daughton C, Murrell B, Price MA, Wickramasinghe L, et al. Broadly neutralizing antibody responses in a large longitudinal sub‐saharan HIV primary infection cohort. PLoS Pathog. 2016;12:e1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Escolano A, Steichen JM, Dosenovic P, Kulp DW, Golijanin J, Sok D, et al. Sequential immunization elicits broadly neutralizing Anti‐HIV‐1 antibodies in Ig knockin mice. Cell. 2016;166:1445–58.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Escolano A, Gristick HB, Abernathy ME, Merkenschlager J, Gautam R, Oliveira TY, et al. Immunization expands B cells specific to HIV‐1 V3 glycan in mice and macaques. Nature. 2019;570:468–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. van Schooten J, van Gils MJ. HIV‐1 immunogens and strategies to drive antibody responses towards neutralization breadth. Retrovirology. 2018;15:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kepler TB, Wiehe K. Genetic and structural analyses of affinity maturation in the humoral response to HIV‐1. Immunol Rev. 2017;275:129–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lynch RM, Wong P, Tran L, O'Dell S, Nason MC, Li Y, et al. HIV‐1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J Virol. 2015;89:4201–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Jardine J, Julien J‐P, Menis S, Ota T, Kalyuzhniy O, McGuire A, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. ClinicalTrials.gov . Identifier: NCT03547245. Available from: https://www.clinicaltrials.gov/ct2/show/NCT03547245 access 7 July 2021
- 138. IAVI press release . https://www.iavi.org. Accessed October 2021
- 139. ClinicalTrials.gov . Identifier: NCT03699241. Available from: https://clinicaltrials.gov/ct2/show/NCT03699241 access 7 July 2021
- 140. ClinicalTrials.gov . Identifier: NCT03783130. Available from: https://clinicaltrials.gov/ct2/show/NCT03783130 access 7 July 2021
- 141. ClinicalTrials.gov . NCT04177355. Available from: https://clinicaltrials.gov/ct2/show/NCT04177355 access 7 July 2021
- 142. ClinicalTrials.gov . Identifier: NCT03816137. Available from: https://clinicaltrials.gov/ct2/show/NCT03816137 access 7 July 2021
- 143. ClinicalTrials.gov . Identifier: NCT03961438. Available from: https://clinicaltrials.gov/ct2/show/NCT03961438 access 7 July 2021
- 144. ClinicalTrials.gov . Identifier: NCT04046978. Available from: https://clinicaltrials.gov/ct2/show/NCT04046978 access 7 July 2021
- 145. Kim J, Vasan S, Kim JH, Ake JA. Current approaches to HIV vaccine development: a narrative review. J Int AIDS Soc. 2021;24(S7):22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]


