Abstract
We reported previously that the N-terminal D1 catalytic domain of receptor protein-tyrosine phosphatase α (RPTPα) forms a symmetrical, inhibited dimer in a crystal structure, in which a helix-turn-helix wedge element from one monomer is inserted into the catalytic cleft of the other monomer. Previous functional studies also suggested that dimerization inhibits the biological activity of a CD45 chimeric RPTP and the catalytic activity of an isolated RPTPς D1 catalytic domain. Most recently, we have also shown that enforced dimerization inhibits the biological activity of full-length RPTPα in a wedge-dependent manner. The physiological significance of such inhibition is unknown, due to a lack of understanding of how RPTPα dimerization is regulated in vivo. In this study, we show that transiently expressed cell surface RPTPα exists predominantly as homodimers, suggesting that dimerization-mediated inhibition of RPTPα biological activity is likely to be physiologically relevant. Consistent with our published and unpublished crystallographic data, we show that mutations in the wedge region of D1 catalytic domain and deletion of the entire D2 catalytic domain independently reduced but did not abolish RPTPα homodimerization, suggesting that both domains are critically involved but that neither is essential for homodimerization. Finally, we also provide evidence that both the RPTPα extracellular domain and the transmembrane domain were independently able to homodimerize. These results lead us to propose a zipper model in which inactive RPTPα dimers are stabilized by multiple, relatively weak dimerization interfaces. Dimerization in this manner would provide a potential mechanism for negative regulation of RPTPα. Such RPTPα dimers could be activated by extracellular ligands or intracellular binding proteins that induce monomerization or by intracellular signaling events that induce an open conformation of the dimer.
Protein-tyrosine phosphorylation plays a vital role in many cellular processes including growth and differentiation (18, 54). Cellular levels of tyrosine phosphorylation are maintained by a balance between protein-tyrosine kinase (PTK) and protein-tyrosine phosphatase (PTP) activity (18). At present, more than 75 PTP family members have been identified, and it has been suggested that the human genome could encode more than a hundred PTPs (54). The PTP superfamily is subdivided into three subfamilies: the dual-specificity PTPs, the intracellular PTPs, and the receptor-like PTPs (RPTPs) (49). Most RPTPs have tandem catalytic domains, with the majority of catalytic activity residing in the membrane-proximal catalytic domain (D1). While it is well established that ligand binding to receptor PTKs results in dimerization, transautophosphorylation, and kinase activation (16), how the activity of RPTPs is regulated remains poorly understood. Only a handful of RPTPs have been found to bind to other proteins via their extracellular domains (ECDs), and until recently none of these interacting proteins had been found to modulate the activity of the cognate RPTP (1, 34, 38, 40, 41, 62). However, the discovery that the secreted factor pleiotrophin interacts with and inhibits the activity of RPTPβ (also called RPTBζ) in vitro and in vivo (33) indicates that regulatory ligands for RPTPs do exist.
Based upon emerging structural and functional evidence, it has been proposed that, whereas dimerization activates receptor PTKs, dimerization may inhibit RPTPs (61). In two independent crystal forms the membrane-proximal catalytic domain (D1) of murine RPTPα exists as a symmetric dimer, in which a helix-turn-helix wedge-shaped element on each monomer inserts into the active site of the dyad-related monomer, resulting in mutual active-site occlusion (3). In principle, RPTPα dimers of this sort would lack catalytic activity, and a fraction of RPTPα elutes from gel filtration columns with a size larger than expected for a monomer, suggesting that RPTPα may indeed have the ability to dimerize or oligomerize (6). Recently, we showed that RPTPα containing a Pro137Cys mutation in the ECD dimerizes constitutively via a disulfide bond and has greatly reduced biological activity in vivo, providing the evidence that dimerization can indeed inhibit the biological activity of a full-length RPTP (20). Consistent with this, EGF-induced dimerization functionally inhibits the biological activity of an EGF receptor-CD45 chimera expressed in a T-cell line in a wedge-dependent fashion (11, 31). Moreover, RPTPδ D2 inhibits RPTPς-D1 activity in vitro (58).
Although artificial dimerization can inhibit the biological activity of some RPTPs, whether RPTPs dimerize physiologically and whether this results in functional inhibition is largely unknown. CD45, which is required for T- and B-cell receptor signaling, has been chemically cross-linked in lysates and to a lesser extent in intact cells (53), and recombinant CD45 cytoplasmic domain dimerizes in solution, emphasizing the dimerization potential of CD45 (13). Moreover, mutation of the wedge motif in CD45 in the mouse germ line leads to an immunoproliferative syndrome in vivo (R. Majeti and A. Weiss, personal communication), implying that wedge-mediated CD45 dimerization suppresses CD45 biological activity. However, even though a conserved wedge motif is present upstream of D1 in most RPTPs, not all RPTPs may form dimers in a manner similar to RPTPα and CD45. For instance, D1 of RPTPμ does not exist as a wedge-mediated dimer in the crystal structure (17), nor is the cytoplasmic region (D1+D2) of LAR present as a dimer in the crystal structure (37). Therefore, it is important to determine whether RPTPs form dimers in the cell and to understand the structural basis for dimers, if they exist.
In this study, we have used RPTPα as a model system to investigate both the efficiency and the structural determinants of homodimerization in vivo. RPTPα contains a rather short 123-amino-acid N-terminal ECD, a single transmembrane domain (TMD), two intracellular PTP domains D1 and D2 (see Fig. 1) (21, 24, 32, 45). Unlike most RPTPs, in which only D1 is catalytically active, both PTP domains in RPTPα are active, although D1 possesses substantially greater catalytic activity than does D2 (5, 29, 30, 59). RPTPα is widely expressed in mammalian tissues (41) and has been implicated in a variety of signaling pathways (2, 7, 8, 10, 19, 27, 36, 51, 56, 63, 65). For example, RPTPα has been shown to play a role in both cellular differentiation and cellular transformation by directly dephosphorylating phosphorylated Tyr527 in c-Src, leading to enhanced c-Src catalytic activity (8, 65). Recently, it was shown that RPTPα null cells (RPTPα−/− cells) derived from RPTPα knockout mice have greatly reduced c-Src PTK activity and are defective in cell adhesion and spreading, all of which are restored upon ectopic expression of RPTPα (42, 50). Moreover, the binding of the c-Src SH2 domain to the C-terminal P.Tyr789 in RPTPα results in displacement of P.Tyr527 from the SH2 domain, thus allowing RPTPα to dephosphorylate P.Tyr527 and thereby specifically activate c-Src (64). RPTPα has been found to be overexpressed in late-stage colon carcinomas (52), where c-Src is commonly found to be activated. RPTPα localization to focal adhesions requires Tyr789 at the C terminus (26), and p130Cas, which is localized to focal adhesions, has recently been shown to interact with and be a substrate for RPTPα (4). No ligand has been found for RPTPα, but RPTPα interacts with the GPI-linked protein contactin in neuronal cells to form a complex that may be linked to the intracellular Src family PTK Fyn (62). Although no regulatory ligand is known, RPTPα in vitro activity is enhanced by tetradecanoyl phorbol acetate treatment of cells, which results in protein kinase C-mediated phosphorylation of Ser180 and Ser204 (9, 55).
FIG. 1.
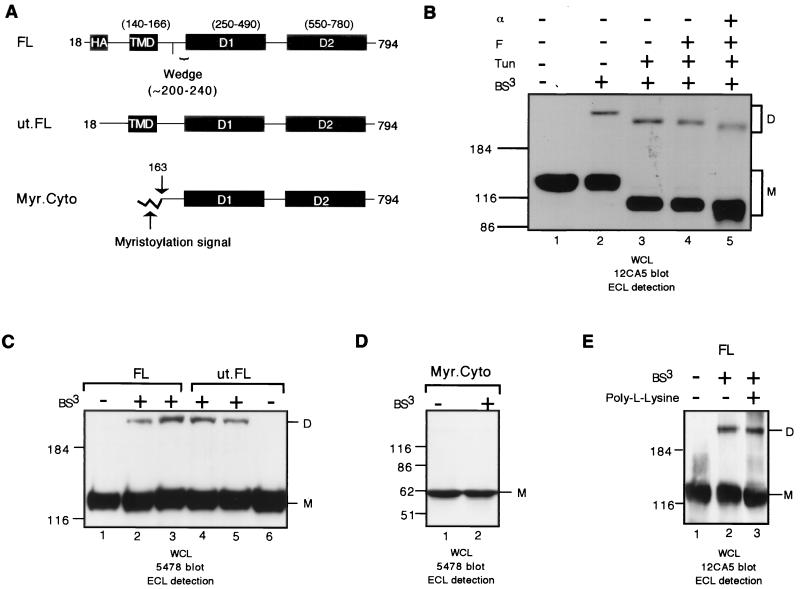
RPTPα homodimerizes on the cell surface. (A) A schematic of RPTPα constructs used in this figure. Amino acids are numbered, and boundaries of the various structural domains, including the TMD, D1, and D2, are indicated according to the original (untagged) polypeptide (45). The boundary of the wedge region is indicated according to the D1 crystal structure (3). The construct FL contains an HA tag which was inserted between amino acids 19 and 20 and is exposed by signal peptide cleavage. The construct ut.FL corresponds to the full-length untagged RPTPα. The construct Myr.Cyto contains a myristoylation signal for membrane attachment. For details of vector construction, refer to Materials and Methods. Mock cross-linking and cross-linking on intact 293 cells transiently expressing FL (B and E), ut.FL (C), and Myr.Cyto (D) is shown. (E) Cells were cultured on plates without or with poly-l-lysine coating. Shown are the results of an immunoblotting analysis with anti-HA tag MAb 12CA5 on whole-cell lysates (WCL) using ECL detection (B and E) or polyclonal antiserum 5478 (C and D). BS3: −, mock cross-linking without BS3; +, cross-linking with BS3. Tun, transfected 293 cells were exposed to tunicamycin at 200 ng/ml; F, lysate was deglycosylated with N-glycosidase F in vitro; α, lysate was deglycosylated with endo-α-N-acetylgalactosaminidase in vitro. M, monomers; D, dimers. The positions of RPTPα monomer (M) and dimer (D) are indicated at the right side of the figure. The positions of molecular-weight markers (in kilodaltons) are indicated on the left side of the figure. Similar labels are used throughout the study.
Based on surface cross-linking studies, we provide the first evidence that RPTPα homodimerizes efficiently on the cell surface via multiple domains, suggesting that dimerization-mediated negative regulation of RPTPα biological activity is likely to be physiologically relevant.
MATERIALS AND METHODS
Expression vectors, site-directed mutagenesis, and antisera.
The expression vector pSG5 was previously described (14). All constructs used in this study were subcloned into pSG5. Construct ut.FL corresponds to the untagged wild-type full-length murine RPTPα. Construct FL corresponds to a full-length murine RPTPα with a hemagglutinin (HA) epitope inserted between amino acids 19 and 20 (Fig. 1) (7, 8). Construct FL.137C contains a Pro137Cys single-amino-acid substitution (Fig. 1) (20). Construct Myr.Cyto corresponds to a myristoylated form of the RPTPα cytoplasmic domain, containing residues 163 to 794 of RPTPα and an N-terminal myristoylation signal (residues 1 to 11 of mouse c-Src) (Fig. 1). To prepare this construct, a HindIII/BsrGI fragment (encoding residues 1 to 162 of RPTPα) of the RPTPα cDNA in the pSG.HA.RPTPα expression vector was replaced with a double-stranded oligonucleotide composed of a sense strand (5′-agctt gccgac ATG GGG AGT AGC AAG AGC AAG CCT AAG GAC CCC ct-3′; an HindIII site-compatible end is underlined; the initiation codon is italicized; the sequence coding for c-Src amino acid residues 1 to 11 is capitalized) and the antisense strand (5′-gta cag GGG GTC CTT AGG CTT GCT CTT GCT ACT CCC CAT gtc ggc a-3′; the BsrGI-compatible end is underlined; the antisense sequence of residues 1 to 11 of c-Src is capitalized).
The FL.P210L.P211L and FL.E234A constructs correspond to FL with a P210L.P211L double mutation and a E234A single mutation, respectively (see Fig. 2) (8). The construct Δ224-235 contains an internal deletion of amino acids 224 to 235, corresponding to the entire wedge sequence, and was prepared by site-directed mutagenesis using the primer 224-235 (5′-GAA GAG GAG ATT AAC CGG GCT GCA GCT TTC AAC GCT CTC CCT-3′) (see Fig. 2).
FIG. 2.
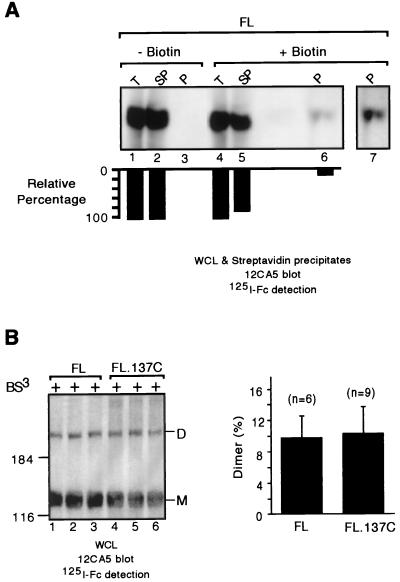
RPTPα appears to exist on the cell surface predominantly as homodimers. (A) 293 cells transiently expressing FL were surface biotinylated or not biotinylated. Cells were lysed, and biotinylated proteins were precipitated with streptavidin beads and separated by SDS-PAGE, and the biotinylated FL was detected by immunoblotting using MAb 12CA5 followed by 125I-labeled sheep anti-mouse IgG F(ab′)2. The blot was quantified using a PhosphorImager as described in Materials and Methods. Top and bottom panels are the immunoblot and quantification, respectively. The loading for each of the lanes was standardized using an equivalent amount of whole-cell lysate. Lane 7 is a longer exposure of lane 6. + or − biotin, labeled or not labeled with biotin; WCL, total whole-cell lysate; SN, whole-cell lysate supernatant after streptavidin bead precipitation; P, streptavidin precipitate. (B) BS3 cross-linking was performed on 293 cells transiently transfected with either FL or FL.137C. Whole-cell lysates of cross-linked cells were separated by SDS-PAGE and probed using MAb 12CA5 followed by 125I-labeled sheep anti-mouse IgG F(ab′)2 and then quantified using a PhosphorImager as described in Materials and Methods. Shown are the results of an immunoblotting analysis. The right panel shows quantitation of the gel in left panel. n, number of replicates. Note that FL and FL.137C are similarly localized to the cell surface (20).
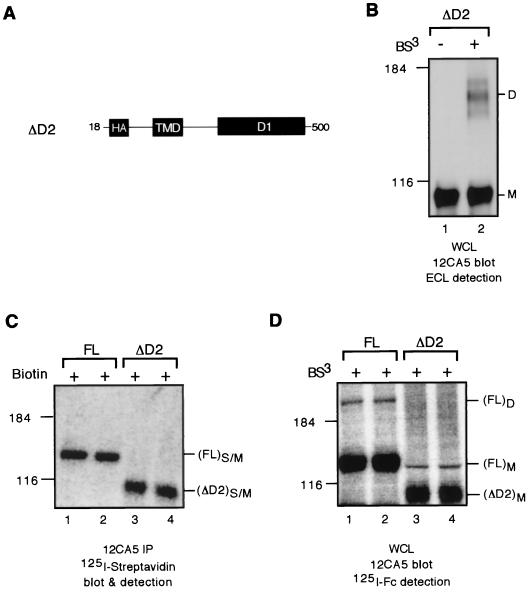
Construct ΔD2 corresponds to a truncated HA-tagged RPTPα lacking residues 501 to 794 corresponding to D2 (see Fig. 3). It was constructed by PCR amplification using as template pSG.HA.RPTPα and as primers RPTPα.1.Hind(s) (5′-tacga aagcttg ccgac ATG GAT TCC TGG TTC ATT CTT G-3′; the HindIII site and the initiation Met codon are italicized and underlined, respectively) and RPTPα.500(a).Kpn (5′-agtc ggtacc CTA CAG TTC TGT GTC CCC ATA CAG-3′; the KpnI site and the termination codon are italicized and underlined, respectively). The PCR product was digested with HindIII and KpnI and subsequently cloned into pSG5. Construct ΔCyto corresponds to a truncated HA-tagged RPTPα lacking residues 201 to 794 corresponding to almost the entire cytoplasmic domain (see Fig. 3). It was constructed similarly to the ΔD2 construct using as primers RPTPα.1.Hind(s) and RPTPα.200(a).Kpn (5′-agtc ggtacc CTA GGC CAG AAG TGG TAC ACT TTG-3′; the KpnI site and the termination codon are italicized and underlined, respectively).
FIG. 3.
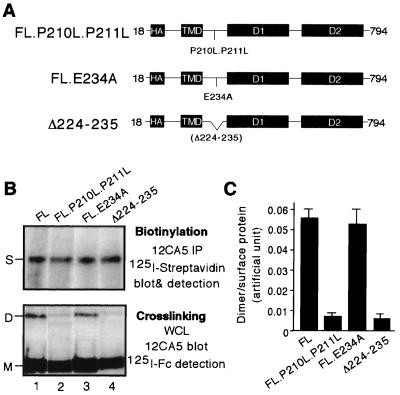
Mutations in the wedge diminish but do not abolish RPTPα oligomerization. (A) A schematic of RPTPα wedge mutant constructs, including point mutants FL.P210L.P211L and FL.E234A and deletion mutant Δ224-235. (B) For the top panel, transiently transfected 293 cells were biotinylated. Whole-cell lysates were immunoprecipitated with MAb 12CA5 to isolate the total RPTPα proteins, which were then subjected to SDS-PAGE and probed with 125I-labeled streptavidin to determine the levels of surface-expressed RPTPα protein. For the bottom panel, transiently transfected 293 cells were cross-linked with BS3. Whole-cell lysates were subjected to immunoblotting analysis with MAb 12CA5 followed by 125I-labeled sheep anti-mouse IgG F(ab′)2 to determine the levels of RPTPα dimers. The bands representing FL.P210L.P211L dimers and Δ224-235 dimers are faint but detectable by PhosphorImager analysis. Biotinylation and cross-linking were done on parallel dishes from the same transfection. All the constructs were expressed to a similar level on the cell surface. (C) Quantification of dimerization efficiency based on average of three replicates. The dimer/surface protein value is the ratio of the levels of RPTPα dimers over surface-expressed RPTPα, which were determined from the bottom and top portions of panel B, respectively, using a PhosphorImager. S, surface-expressed RPTPα (monomer).
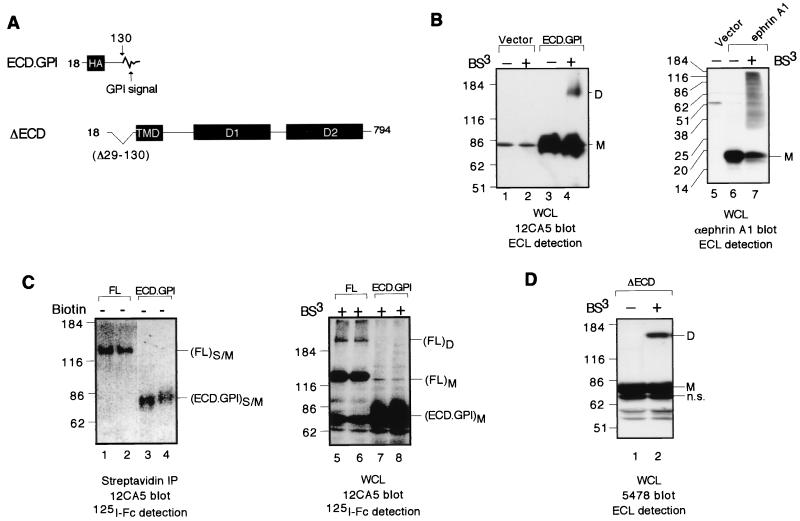
Construct ECD.GPI corresponds to an HA-tagged RPTPα ECD containing RPTPα residues 1 to 129 and a C-terminally tagged glycosylphosphatidylinositol (GPI) linkage signal sequence (see Fig. 5). It was constructed by replacing a PstI/KpnI fragment of the FL construct with a double-stranded oligonucleotide (ephrin A1.GPI.top, 5′-GGT CCA CGC CTC TTC CCA CTT GCC TGG ACT GTG CTG CTC CTT CCA CTT CTG CTG CTG CAA ACC CCG TGA G gta c-3′; ephrin A1.GPI.bot, 5′-C TCA CGG GGT TTG CAG CAG CAG AAG TGG AAG GAG CAG CAC AGT CCA GGC AAG TGG GAA GAG GCG TGG ACC tgc a-3′; the KpnI-compatible end is underlined; the PstI-compatible end is italicized; the ephrin A1 coding sequence is capitalized). Construct ΔECD corresponds to an untagged RPTPα containing the entire TMD and cytoplasmic domains but lacking residues 29 to 130 corresponding to most of the ECD (see Fig. 5). To prepare this construct, an EcoNI/PstI fragment (corresponding to residues 27 to 129) from the RPTPα cDNA in pSG.RPTPα (8) was deleted, resulting in the expression vector pSG.RPTPα.ΔECD.
FIG. 5.
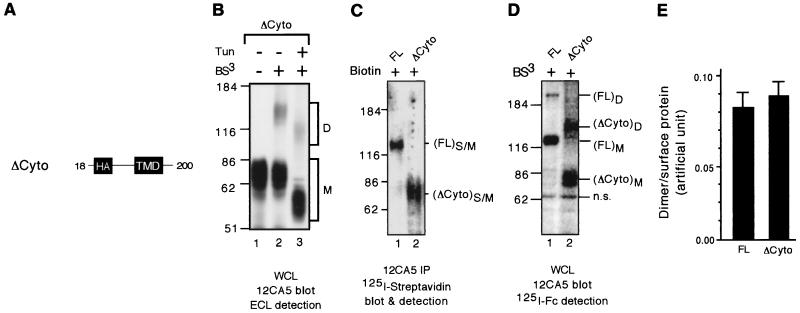
ΔCyto homodimerizes on the cell surface with high efficiency. (A) A schematic of the construct ΔCyto lacking the entire cytoplasmic domain. (B) 293 cells transiently transfected with FL or ΔCyto were treated or not treated with tunicamycin at 200 ng/ml and subsequently cross-linked with BS3. Whole-cell lysates were subjected to immunoblotting analysis with MAb 12CA5 using ECL detection. (C) Transiently transfected 293 cells were biotinylated. Whole-cell lysates were precipitated with streptavidin beads to isolate the total RPTPα proteins, which were then subjected to SDS-PAGE and probed with 125I-labeled streptavidin to determine the levels of surface-expressed RPTPα proteins. (D) Transiently transfected 293 cells were cross-linked with BS3. Whole-cell lysates were subjected to immunoblotting analysis using MAb 12CA5 followed by 125I-labeled sheep anti-mouse IgG F(ab′)2 to determine the levels of RPTPα dimers. Biotinylation (C) and cross-linking (D) were done on parallel dishes from the same transfection. Shown in panels C and D are images from PhosphorImager analysis. S/M, surface-expressed monomeric proteins. (E) Quantification of dimerization efficiency based on average of three replicates. The dimer/surface protein value is the ratio of the levels of RPTPα dimers over surface-expressed RPTPα, which were determined from panels C and D, respectively, using a PhosphorImager. n.s., nonspecific band.
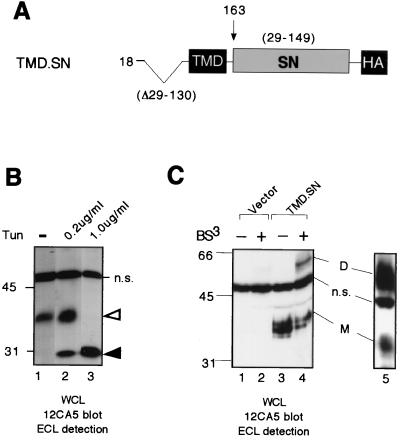
Construct TMD.SN corresponds to a fusion protein of RPTPα TMD and staphylococcal nuclease (SN) (residue 27 to the very C-terminal residue 149 of the mature protein) with a C-terminal HA tag (see Fig. 6). The construct was prepared by replacing the BsrGI/BglII fragment (encoding residues 165 to 794, the entire cytoplasmic domain) of RPTPα cDNA in the expression vector pSG.RPTPα.ΔECD with a BsrGI- and BglII-digested PCR product encoding the mature SN. The PCR product was amplified using as template pSN/GpA (28) and as 5′ primer BsrGI.SN.1(S) (5′-GCA ACT TCA ACT AAA AAA TTA CAT AAA GAA CC-3′, corresponding to the sense sequence of residues 2 to 11 of matured SN) and 3′ primer BglII.Stop.HA.SN150(A) (5′-tttt agatct TCA GGC ATA ATC TGG CAC ATC ATA AGG GTA ACC CAT ggc TGG ACC TGA ATC AGC GTT GTC TTC GCT CC; the BglII site is in lowercase and is italicized; the stop codon is capitalized; the HA tag is capitalized and underlined; the antisense sequence of residues 149 to 141 of SN is capitalized and italicized).
FIG. 6.
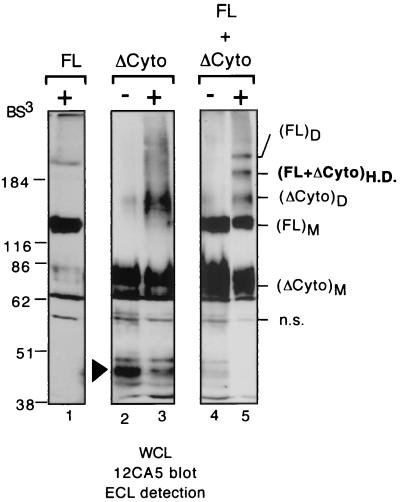
RPTPα oligomers are homodimers. Mock cross-linking and cross-linking on 293 cells transiently transfected was done with the construct FL alone (lane 1), with ΔCyto alone (lanes 2 and 3), or with both FL and ΔCyto simultaneously (lanes 4 and 5). Whole-cell lysates were subjected to immunoblotting analysis with MAb 12CA5 using ECL detection. Note that a band corresponding to either partially glycosylated or degraded ΔCyto is present in the transfections of ΔCyto alone (lanes 2 and 3) but is virtually undetectable in the cotransfections (lanes 4 and 5). (FL+ΔCyto)H.D. is the cross-linked FL-ΔCyto heterodimer.
Antiserum 5478 is a rabbit polyclonal antibody raised against a glutathione S-transferase (GST) fusion protein of the RPTPα cytoplasmic domain and purified as previously described (10). 12CA5 is a mouse monoclonal antibody (MAb) against the HA tag.
Cell culture and transient transfection.
HEK293 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum at 37°C and 10% CO2. Transient transfection of 293 cells was done using the calcium phosphate precipitation method. Briefly, cells were seeded onto 50-mm tissue culture dishes with or without poly-l-lysine coating 24 h prior to transfection at the dilution of 1 confluent 100-mm dish to 20 50-mm dishes. Poly-l-lysine does not affect cross-linking of RPTPα (Fig. 2C) but prevents cells from detaching during the many solution changes in the cross-linking procedure. At the start of transfection (0 h), 4 ml of fresh medium containing 25 μM chloroquine was added to each of the dishes. Plasmid DNA was mixed with 500 μl of 250 mM CaCl2, to which 500 μl 2× HBS (50 mM HEPES; 10 mM KCl; 280 mM NaCl; 1.5 mM Na2HPO4; 12 mM dextrose, pH 7.05) was subsequently added. The mix was immediately added to the medium, and cells were then incubated at 37°C and 5% CO2. At 10 h, the cells were washed twice with phosphate-buffered saline (PBS) and incubated in fresh medium at 37°C and 10% CO2. Chemical cross-linking or cell surface biotinylation were performed at approximately 72 h.
Cell surface chemical cross-linking.
The entire procedure was performed on intact cells at 4°C. Transiently transfected 293 cells in 50-mm dishes were washed three times with PBS and incubated with freshly prepared cross-linker solution [bis(sulfosuccinimidyl)suberate (BS3; Pierce) at 3 mg/ml in PBS (pH 7.1) without Ca2+ and Mg2+] for ∼60 min and then washed three times with PBS and incubated in PBS solution containing 50 mM Tris-HCl (pH 7.5) for 20 min to quench residual BS3. The cells were then lysed in radioimmunoprecipitation assay (RIPA) buffer (58) for 30 min. The lysates were cleared by centrifugation, and the samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After separation, the proteins on the gels were transferred to Immobilon (Millipore, Bedford, Mass.) and subjected to immunoblotting to detect the RPTPα proteins. To visualize the dimeric or monomeric RPTPα proteins, membranes were probed with affinity-purified polyclonal antisera 5478 or MAb 12CA5 and detected by enhanced chemiluminescence (ECL) as previously described (7). To quantitate the monomers and dimers, membranes were probed with MAb 12CA5 as above, washed three times with TBST (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.05% Tween 20), blocked with TBST containing 5% milk for 30 min, washed once with TBST, and blocked with TBST containing 1% bovine serum albumin (BSA) for 20 min. 125I-labeled sheep anti-mouse immunoglobulin G (IgG) F(ab′)2 fragment (NEN Life Science Products, Inc.) was then added to the blocking solution at a final concentration of 0.5 μCi/ml, and the incubation was continued for another 1.5 h. The membranes were then washed three times with TBST, dried, and analyzed using a PhosphorImager (Molecular Dynamics).
Cell surface biotinylation.
To biotinylate surface proteins, transiently transfected HEK293 cells in 50-mm dishes were washed three times with PBS, incubated for 20 min with freshly prepared biotinylation buffer (50 mM sodium phosphate; 110 mM NaCl; 0.1% NaN3, pH 8.5) containing EZ-Link-Sulfo-NHS-LC-Biotin (Pierce) at 0.4 mg/ml, and washed three times with PBS containing 0.1% NaN3. Cells were then lysed in RIPA buffer. The level of biotinylated RPTPα was determined by two different methods. In the first method, total RPTPα protein was isolated by immunoprecipitation from whole-cell lysates, and the biotinylated RPTPα was then detected by immunoblotting using 125I-labeled streptavidin (Amersham). Briefly, the lysates were incubated with MAb 12CA5 (approximately 0.5 to 1.0 μg of IgG per sample) for 1 h with gentle rotation. Protein A-Sepharose beads (25 μl of 50% bead suspension solution per sample) were subsequently added, and the incubation was continued for another 1.5 h. Beads were then washed three times with RIPA buffer and boiled in Laemmli sample buffer. The immunoprecipitates were separated by SDS-PAGE and, after separation, the proteins on the gel were transferred to Immobilon. The membranes were blocked with TBST containing 5% milk at 4°C overnight for 30 min, washed once with TBST, and then blocked with TBST containing 1% BSA for 20 min. 125I-labeled streptavidin was then added to the blocking solution. The incubation was continued for another 1.5 h. The membranes were then washed three times with TBST and analyzed by PhosphorImager analysis. In the second method, biotinylated proteins were isolated using streptavidin-agarose beads (Sigma) by the procedures described above. After SDS-PAGE, the level of biotinylated RPTPα was then detected and quantified by immunoblotting using MAb 12CA5 and secondary antiserum 125I-labeled sheep anti-mouse IgG F(ab′)2 fragment as described above. We have optimized the biotinylation experiments using different incubation times and biotin concentrations. The conditions described above represent those under which biotinylation efficiency is maximized.
Inhibition of glycosylation in vivo and deglycosylation in vitro.
To inhibit N-linked glycosylation, transfected HEK293 cells were washed three times with PBS 12 h after the initiation of transfection and then incubated in fresh medium containing either control solvent (dimethyl sulfoxide) or tunicamycin (Sigma) at the desired concentration. After incubation for another 12 h, the cells were cross-linked and/or lysed in lysis buffer (50 mM HEPES, pH 7.0; 150 mM NaCl; 1.5 mM MgCl2; 1 mM EGTA; 10% [vol/vol] glycerol; 1% Triton X-100; 1.0 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol). For in vitro deglycosylation, 5 to 30 μl of the lysates were incubated with 1 to 4 μU of either N-glycosidase F (Sigma) to remove N-linked glycosyl groups or endo-α-N-acetylgalactosaminidase (Sigma) to remove O-linked glycosyl groups.
RESULTS
RPTPα oligomerizes on the cell surface with high efficiency.
We showed previously that RPTPα D1 exists as a dimer in two independent crystal forms (3). To determine whether RPTPα dimerizes on the cell surface, BS3-mediated chemical cross-linking was performed on intact 293 human embryonic kidney cells (293 cells) transiently transfected with an N-terminally HA-tagged full-length RPTPα expression vector and plated on poly-l-lysine (FL; Fig. 1A). BS3, which reacts with free NH2 groups in proteins, is not membrane permeant due to its charged nature and therefore only cross-links surface-expressed proteins. Using MAb 12CA5, which specifically recognizes the HA tag, immunoblotting analysis of whole-cell lysate of mock-cross-linked cells showed that FL RPTPα migrated as an ∼130-kDa band (Fig. 1B, lane 1), representing fully glycosylated FL protein. Immunoblotting analysis of whole-cell lysate of BS3-cross-linked cells revealed an additional band of ∼230 kDa (Fig. 1B, lanes 2), indicating that RPTPα oligomerizes on the cell surface. The apparent size of the ∼230-kDa band suggests that it contains RPTPα homodimers.
As one means of confirming that the ∼230-kDa band indeed contains exclusively RPTPα protein, we determined whether its size was reduced by either tunicamycin (an inhibitor of N-linked glycosylation) treatment in vivo, and/or by N-glycosidase F (an enzyme that removes N-linked sugars) and endo-α-N-acetylgalactosaminidase (an enzyme that removes O-linked sugars) treatment in vitro in a manner similar to the RPTPα monomer. The mature 140-kDa RPTPα protein contains both N-linked and O-linked sugars, and the unmodified RPTPα precursor is ∼90 kDa (45). When tunicamycin-treated cells were treated with BS3 and analyzed as described above, the apparent size of the ∼130-kDa monomer was reduced to ∼100 kDa (Fig. 1B, lane 3 versus lane 2), representing FL RPTPα with reduced N-linked glycosylation. We observed a parallel reduction in the size of the ∼230-kDa band upon tunicamycin treatment in vivo (lane 3 versus lane 2). Furthermore, consistent with the notion that tunicamycin inhibits N-linked glycosylation, we found that the sizes of both the lower- and the higher-molecular-weight bands were further reduced by deglycosylation in vitro using endo-α-N-acetylgalactosaminidase but not by N-glycosidase F (lane 5 versus lane 3 and lane 4 versus lane 3). Taken together, these results are consistent with the ∼230-kDa band being an RPTPα homodimer rather than a hetero-oligomer with another unknown protein (see Fig. 6 and text).
To confirm that cross-linking of the HA-tagged RPTPα is not a consequence of the N-terminal HA tag, we performed cross-linking on 293 cells transiently expressing ut.FL, a full-length RPTPα construct lacking the HA tag (Fig. 1A). Immunoblotting analysis of cross-linked 293 cells expressing either FL or ut.FL showed that antiserum 5478, which was raised against a GST fusion of the entire RPTPα cytoplasmic domain, specifically detected similar levels of the ∼230-kDa bands (Fig. 1C, lanes 1 to 3 versus lanes 4 and 5), clearly demonstrating that the homodimerization of FL is not due to the HA tag.
To confirm that BS3 cross-linking truly reflects cross-linking of RPTPα via its ECD outside the cell, we constructed an expression vector for a myristoylated form of the RPTPα cytoplasmic domain (Myr.Cyto; Fig. 1A). Myr.Cyto contains the entire RPTPα cytoplasmic domain and a N-terminal myristoylation signal corresponding to residues 1 to 11 of murine c-Src. A similar strategy has been used to produce membrane-localized forms of numerous proteins (see, for example, references 23, 39, and 43). We expected Myr.Cyto to be membrane localized but not to be cross-linked by BS3 in intact cells, since it contains no ECD. As expected, immunoblotting analysis of both mock-cross-linked and cross-linked 293 cells expressing Myr.Cyto showed that antiserum 5478 specifically detected a single band of ∼62 kDa (Fig. 1D, lane 1), corresponding to monomeric Myr.Cyto protein. However, no higher-molecular-weight forms were observed upon BS3 cross-linking (Fig. 1D, lane 1 versus lane 2), indicating that BS3 had not cross-linked the Myr.Cyto protein.
We were concerned that the poly-l-lysine coating of the culture dishes could affect cross-linking due to the high density of lysyl NH2 groups, which might compete for BS3 or result in cross-linking of RPTPα to poly-l-lysine. We therefore determined the effect of poly-l-lysine on RPTPα cross-linking efficiency. The presence of poly-l-lysine did not affect RPTPα cross-linking (Fig. 1E, compare lane 2 to lane 3), confirming that the RPTPα ECD was cross-linked in a specific fashion.
To determine the extent of RPTPα homodimerization on the cell surface, we measured in parallel the efficiency with which transiently expressed FL RPTPα was transported to the cell surface and was cross-linked by BS3 in intact 293 cells. We determined the fraction of FL that was localized on the cell surface and therefore accessible to BS3 cross-linking by surface biotinylation followed by streptavidin precipitation (Fig. 2A). By comparing the amounts of RPTPα in total lysates with the amounts that were bound to streptavidin or left in the supernatant, we found that ∼15% of the total FL protein from biotinylated transiently transfected 293 cells bound to streptavidin beads (lanes 4 to 7). The binding of the FL protein to streptavidin beads was specifically due to biotinylation, since no FL protein from control (unbiotinylated) transiently transfected 293 cells was bound to streptavidin beads (Fig. 2B, lanes 1 to 3). These results suggest that only a relatively small fraction (∼15%) of FL RPTPα molecules are localized to the cell surface in transfected 293 cells. This result is consistent with our previous finding that the majority of transiently expressed FL is localized intracellularly, as determined by immunofluorescence staining (55). When we quantified the cross-linking efficiency of transiently expressed FL, we found that ∼10% of the total transiently expressed FL protein was present as a dimer (Fig. 2B, lanes 1 to 3; Fig. 2C). Given the result that only approximately 15% of the total FL is on the cell surface, the ∼10% cross-linking efficiency indicates that the majority of RPTPα exists on the cell surface as homodimers.
To confirm that most RPTPα on the cell surface is dimeric, we compared the cross-linking efficiency of FL and FL.137C RPTPα (Fig. 2B). FL.137C is a cysteine mutant that dimerizes constitutively in cells via a disulfide bond and is localized to the cell surface with an efficiency similar to that of FL (20). The results showed that FL was cross-linked with an efficiency similar to that of FL.137C (Fig. 2B), indicating that RPTPα indeed exists predominantly on the cell surface as a homodimer, at least under our experimental conditions.
The wedge structure is important but not essential for RPTPα oligomerization.
RPTPα D1 dimers in crystals are stabilized by interactions between the active site of one monomer and the wedge of the dyad axis-related monomer. The wedge structure itself is stabilized by residues P210 and P211 at the base, and several residues at the tip, including E234, participate in protein-protein interactions (3). We showed that FL.137C dimerizes in vivo and has reduced biological activity. Furthermore, the biological activity of FL.137C can be restored by the P210L.P211L double mutation but not by other mutations, including E234A in the wedge, probably because the P210L.P211L mutation causes a more significant structural effect (20). These results suggest, both structurally and functionally, that the wedge is important for RPTPα oligomerization. Accordingly, the cross-linking efficiency of FL was compared to that of several wedge mutants, including FL.P210L.P211L, FL.E234A, and Δ224-235, a deletion that is predicted to eliminate the entire wedge structure (Fig. 3A). When the wild type (FL) and the wedge mutants were expressed to similar levels on the cell surface as determined by biotinylation (Fig. 3B, top panel), dimeric forms of the proteins were readily detectable for FL and FL.E234A but not for FL.P210L.P211L and Δ224-235 (Fig. 3B, bottom panel, lanes 1 and 3 versus lanes 2 and 4). Quantitative analysis demonstrated that both the P210L.P211L double mutation and the wedge deletion reduced oligomerization efficiency of RPTPα by approximately 80% on the cell surface (Fig. 3C). The fact that the Δ224-235 and FL.P210L.P211L proteins have reduced dimerization potential indicates that the wedge structure is important for RPTPα homodimerization. The result that the Δ224-235 and FL.P210L.P211L proteins dimerized with similarly low efficiency confirmed our previous speculation that the P210L.P211L double mutation likely disrupts the wedge structure. Finally, the finding that Δ224-235 mutant still dimerized suggests that there are other oligomerization domain(s) in addition to the wedge in RPTPα. The fact that the wedge mutations affect RPTPα cross-linking in a fashion stereochemically consistent with the crystallographic data confirmed that BS3-mediated cross-linking of RPTPα is specific.
The C-terminal catalytic domain D2 is important but not essential for RPTPα oligomerization.
To investigate the possibility that RPTPα has oligomerization domains in addition to the wedge, we tested the oligomerization potential of an RPTPα deletion mutant lacking the C-terminal D2 (ΔD2) (Fig. 4A). Immunoblotting analysis of 293 cells transiently transfected with the ΔD2 expression vector showed that MAb 12CA5 specifically recognized a band of ∼100 kDa (Fig. 4B, lane 1), a finding consistent with the predicted size of a fully glycosylated ΔD2 protein. After cross-linking, a new band of ∼160 kDa was detected (Fig. 4B, lane 2), indicating that ΔD2 oligomerizes. To assess the dimerization potential of D2, we compared the dimerization efficiency of the ΔD2 and wild-type FL proteins. When the two proteins were expressed on the cell surface to similar levels as determined by surface biotinylation (Fig. 4C, lanes 1 and 2 versus lanes 3 and 4), FL dimers but no ΔD2 dimers were apparent (Fig. 4D, lanes 1 and 2 versus lanes 3 and 4), indicating that ΔD2 has reduced cross-linking efficiency compared to FL. Taken together, these results suggest that D2 participates but is not essential for RPTPα homodimerization.
FIG. 4.
Deletion of D2 diminishes but does not abolish RPTPα oligomerization. (A) A schematic of the D2 deletion mutant construct. (B) 293 cells transiently expressing ΔD2 protein were cross-linked or not cross-linked with BS3. Shown are the results of an immunoblotting analysis with anti-HA tag MAb 12CA5 on whole-cell lysates using ECL detection. (C) Transiently transfected 293 cells were biotinylated. Whole-cell lysates were immunoprecipitated with MAb 12CA5 to isolate the total RPTPα proteins, which were then subjected to SDS-PAGE and probed with 125I-labeled streptavidin to determine the levels of surface-expressed RPTPα protein. (D) Transiently transfected 293 cells were cross-linked with BS3. Whole-cell lysates were subjected to immunoblotting analysis using MAb 12CA5 followed by 125I-labeled sheep anti-mouse IgG F(ab′)2 to determine the levels of RPTPα dimers. Biotinylation (C) and cross-linking (D) were done on parallel dishes from the same transfection. Shown in panels C and D are images obtained via PhosphorImager analysis. S/M, surface-expressed monomeric proteins.
The TMD and the ECD also participate in RPTPα homodimerization.
Our data indicate that both D1 and D2 participate in RPTPα homodimerization but that neither of them is essential. To determine whether the entire cytoplasmic domain is essential for RPTPα homodimerization and whether the TMD and the ECD also participate in RPTPα homodimerization, we made a construct, ΔCyto, which lacks the entire cytoplasmic domain but contains the ECD and the TMD (Fig. 5A). Immunoblotting analysis of 293 cells transiently transfected with the ΔCyto expression vector showed that MAb 12CA5 specifically recognized an ∼80-kDa band (Fig. 5B, lane 1), most likely representing fully glycosylated ΔCyto protein. The expected molecular size of fully glycosylated ΔCyto is ∼65 kDa (∼40 kDa contributed by glycosylation) instead of the observed 80 kDa. The SDS gel mobility of a protein in which carbohydrate contributes most of the mass is hard to predict, but it seems likely that the protein will run significantly slower than expected because the charge/mass ratio of the carbohydrate to which SDS does not bind is expected to be lower than that of SDS-saturated protein. After cross-linking of ΔCyto-expressing cells, a new band of ∼140 kDa was also detected (Fig. 4C, lane 2), indicating that ΔCyto oligomerized. Tunicamycin treatment in vivo to inhibit N-linked glycosylation caused a parallel reduction in the apparent size of the 80- and 140-kDa protein species (Fig. 4C, lane 3 versus lane 2), confirming that the ∼140-kDa band is a ΔCyto oligomer (we show later that the ΔCyto oligomer is a homodimer [see Fig. 6 and associated text]).
Taken together, these results demonstrate that the entire cytoplasmic domain is not essential for RPTPα oligomerization and confirm the existence of additional oligomerization domain(s) in the ECD and/or the TMD (see below). To assess the dimerization potential of the motif(s) within the ECD and/or the TMD, we compared the dimerization efficiency of ΔCyto proteins and of the wild-type FL protein. When the two proteins were expressed on the cell surface to similar levels (Fig. 5C, lane 1 versus lane 2), they were cross-linked with similar efficiency (Fig. 5D and E), suggesting that either the ECD or the TMD can homodimerize efficiently.
RPTPα oligomers are homodimers.
We concluded that the oligomers detected in the previous experiments are RPTPα homodimers. However, the apparent sizes of most of the various oligomers judged by their migration in SDS-polyacrylamide gels are somewhat less than twice the sizes of the corresponding monomers (FL = ∼130 kDa, FL oligomers = ∼230 kDa; ΔCyto = ∼80 kDa, ΔCyto oligomer = ∼140 kDa). To rule out the formal possibility that the oligomers are RPTPα heterodimers or hetero-oligomers with other proteins, the potential of FL and ΔCyto to heterodimerize was determined by a cross-linking experiment with 293 cells that had been cotransfected with both expression vectors. Immunoblotting analysis using MAb 12CA5 detected a novel band of ∼200 kDa migrating between FL oligomers and fg.ΔCyto oligomers (Fig. 6, lane 5 versus lanes 1 and 3), suggesting that it was a FL-ΔCyto heterodimer. The results show that RPTPα homodimerizes and that the oligomers observed in Fig. 1 to 5 are RPTPα homodimers. We explain the fact that the dimers migrate faster than expected as most likely being due to the nonlinear backbone structures of the cross-linked molecules.
Since there are four sites in the RPTPα ECD that can potentially react with BS3 (Lys36, Lys45, Lys49, and the α-NH2 group at the N terminus of the mature polypeptide after signal peptide cleavage), more than two molecules of RPTPα can potentially be cross-linked in one complex. However, we did not observe higher-order oligomers based on the apparent size of the bands, suggesting that RPTPα homodimerizes but does not form higher-order oligomers. However, we cannot formally exclude the possibility that only one of the sites can be efficiently cross-linked, preventing the cross-linking of more than two RPTPα molecules in the same complex. Additionally, if only a small fraction of FL indeed forms higher-order oligomers, the levels of these oligomers may be below the limit of detection.
The RPTPα ECD dimerizes weakly and is not essential for oligomerization.
To determine whether the ECD participates in RPTPα homodimerization, we determined the dimerization potential of ECD.GPI, a protein corresponding to RPTPα ECD with a C-terminally fused GPI linkage signal sequence (Fig. 7A). The GPI membrane anchor for the ECD was derived from human ephrin A1, consisting of residues 185 to 205 from the very C terminus, which contains no Lys residues that could react with BS3. Immunoblotting analysis of 293 cells transiently transfected with the ECD.GPI expression vector showed that MAb 12CA5 specifically recognized a band of ∼85 kDa (Fig. 7B, compare lane 3 to lane 1). The expected size of fully glycosylated ECD.GPI is ∼56 kDa (∼40 kDa from glycosylation). Based on the reasoning used for the ΔCyto protein, we believe that the ∼85-kDa protein represents fully glycosylated ECD.GPI protein. After cross-linking, a new band of ∼150 kDa was detected (Fig. 7B, lane 4). Since the GPI moiety itself would not be expected to dimerize, we deduce that ECD.GPI dimerizes via the ECD. Nevertheless, to exclude the possibility that the dimerization of ECD.GPI is due to the GPI moiety, the potential of ephrin A1 itself to homodimerize was determined. Immunoblotting analysis of 293 cells transiently transfected with the ephrin A1 expression vector showed that ephrin A1 antibodies specifically recognized a band of ∼20 kDa, corresponding to the full-length ephrin A1 protein (Fig. 7B, compare lanes 6 and 5). After cross-linking, a whole array of new bands ranging from ∼30 kDa to more than 200 kDa was detected (Fig. 7B, lane 7), probably representing various ephrin A1 hetero-oligomers. In contrast to RPTPα ECD.GPI, there was no apparent ephrin A1 homodimer band based on its expected molecular size. Therefore, the homodimerization of the ECD.GPI fusion protein is unlikely to be mediated by the GPI moiety per se, and we conclude that the ECD has an intrinsic ability to homodimerize. Quantitative analysis showed that, when the ECD.GPI fusion protein was expressed on the cell surface to a level similar to that of the FL (Fig. 7C, lanes 1 and 2 versus lanes 3 and 4), FL homodimers but not ECD.GPI homodimers were readily detected after cross-linking (Fig. 7C, lanes 5 and 6 versus lanes 7 and 8), suggesting that ECD by itself has a much weaker dimerization potential than the full-length FL protein.
FIG. 7.
The ECD possesses relatively weak dimerization potential and is not required for the homodimerization of the full-length RPTPα. (A) A schematic of RPTPα constructs used in this figure. (B) Mock cross-linking and cross-linking on 293 cells transiently transfected with the construct ECD.GPI (lanes 1 to 4) or ephrin A1 (lanes 5 to 7). Shown are the results of immunoblotting analysis with MAb 12CA5 of whole-cell lysates using ECL detection. (C) In the left panel, transiently transfected 293 cells were biotinylated. Streptavidin-agarose beads were used to isolate the total biotinylated surface proteins, which were then subjected to immunoblotting analysis using MAb 12CA5 followed by 125I-labeled sheep anti-mouse IgG F(ab′)2 to determine the levels of surface-expressed RPTPα protein. In the right panel, transiently transfected 293 cells were cross-linked with BS3. Whole-cell lysates were subjected to immunoblotting analysis using MAb 12CA5 followed by 125I-sheep anti-mouse IgG F(ab′)2 to determine the levels of RPTPα dimers. n.s., nonspecific band. (D) Mock cross-linking and cross-linking on 293 cells transiently transfected with the construct ΔECD. Whole-cell lysates were subjected to immunoblotting analysis with anti-RPTPα serum 5478 using ECL detection.
To establish whether the ECD is required for RPTPα homodimerization, we determined the dimerization potential of ΔECD, an RPTPα mutant lacking most of the ECD (Fig. 7A). Immunoblotting analysis using anti-RPTPα serum 5478 showed that ΔECD is expressed as an ∼83-kDa protein that can be cross-linked by BS3 (Fig. 7D), suggesting that ECD is not required for dimerization. The fact that an ΔECD dimer can be generated by cross-linking is somewhat surprising, since only the α NH2 group at the N terminus of the mature protein is left to mediate cross-linking (unless it occurs via the carbohydrate side chains, which would not be expected to react with BS3). Taken together, these results lead us to conclude that the ECD probably participates in but is not essential for RPTPα homodimerization. However, we cannot quantitatively compare the cross-linking efficiency of ΔECD to that of FL, since the two constructs have different numbers of reactive extracellular NH2 groups for cross-linking as well as biotinylation.
The RPTPα TMD efficiently dimerizes in vivo.
Some TMDs are known to be able to dimerize (e.g., glycophorin A). To determine whether the TMD also plays a role in RPTPα homodimerization, we prepared an expression vector that expresses the RPTPα TMD (residues 140 to 164) fused to SN (TMD.SN) (Fig. 8A). Mature SN, a 150 residue bacterial protein, is known to exist exclusively as a monomer and has been fused to other proteins, including the glycophorin A transmembrane domain, for dimerization studies (28, 48). Two stretches (residues 18 to 28 and 131 to 139) from the RPTPα ECD were also included in the construct. Residues 18 to 28, immediately C terminal to the signal peptide, are required for correct cleavage of the signal peptide. Residues 131 to 139, containing both polar and charged residues in the juxtamembrane region, are required for membrane localization of the fusion protein. We reasoned that these two short stretches should not obscure our analysis of the TMD, since each of the stretches by themselves is probably too short to form a functional dimerization domain. Moreover, the two stretches are unlikely to interact with each other to form a composite functional dimerization domain, since they are derived from the two extreme ends of the ECD. As for ΔECD (Fig. 7A), only the α NH2 group at the N terminus of the mature TMD.SN fusion is left to react with BS3.
FIG. 8.
The TMD of RPTPα is a potent dimerization domain. (A) A schematic of RPTPα constructs used in this figure. (B) 293 cells transiently transfected with TMD.SN were left untreated (lane 1) or were treated with tunicamycin at 200 ng/ml (lane 2) or 1,000 ng/ml (lane 3). Whole-cell lysates were subjected to immunoblotting analysis with MAb 12CA5 using ECL detection. Open arrow, most likely a N-glycosylated TMD.SN protein; closed arrow, most likely a nonglycosylated TMD.SN protein; n.s., a nonspecifically recognized band. (C) Mock cross-linking and cross-linking on intact cells by BS3 was performed on 293 cells transiently transfected with pSG5 vector alone or the TMD.SN construct. Whole-cell lysates were subjected to immunoblotting analysis with MAb 12CA5 using ECL detection.
Immunoblotting analysis of 293 cells transiently transfected with the TMD.SN expression vector showed that MAb 12CA5 specifically recognized a band of ∼35 kDa in mock-cross-linked cells (Fig. 8B, cf. lanes 3 and 1). Since a potential N-glycosylation site containing Asn21 is present in the construct, we reasoned that the ∼35-kDa protein most likely represents singly N-glycosylated SN.TMD protein. Consistent with such a notion, tunicamycin treatment in vivo to inhibit N-linked glycosylation reduced the apparent size from ∼35 to ∼28 kDa (Fig. 8B, lanes 2 and 3 versus lane 1), which likely represents an unglycosylated protein. After cross-linking, a new band of ∼50 kDa was also detected (Fig. 8B, cf. lanes 4 and 2), indicating that the TMD.SN homodimerizes. The intensity of the ∼50-kDa dimer band ranged from approximately 30% (Fig. 8B, lane 4) to 80% (Fig. 8B, lane 5) of the total TMD.SN protein. Considering that probably not all the TMD.SN protein is localized to the cell surface and accessible to BS3 cross-linking, these results suggest that nearly all of the TMD.SN protein on the cell surface exist as homodimers. Taken together, our results suggest that the TMD is potentially a potent dimerization domain. As for ΔECD, however, we cannot quantitatively compare the cross-linking efficiency of TMD.SN to that of FL because the two constructs have different numbers of reactive extracellular NH2 groups for cross-linking as well as biotinylation.
DISCUSSION
Several studies have strongly suggested that dimerization inhibits the biological activity of certain RPTPs (11, 20, 31, 58). In this study, we showed that homodimers of RPTPα can readily be observed after BS3-mediated cross-linking on the surface of intact transiently transfected HEK293 cells. Several lines of evidence suggest that the cross-linking is due to an intrinsic propensity of RPTPα to homodimerize: first, the cross-linking of RPTPα did not result in heterodimerization with other proteins and was unaffected by the presence of poly-l-lysine, a polymer with many reactive primary NH2 groups, suggesting that RPTPα cross-linking does not occur in a promiscuous fashion (Fig. 1D); second, wedge mutations reduced the cross-linking of RPTPα in a fashion stereochemically consistent with our previous crystallographic data (Fig. 3), demonstrating that the cross-linking has strict structural requirements; third, deletion of D2 reduces RPTPα homodimerization (Fig. 4), which is consistent with our observation that D2 forms a dimer in crystal structure (A. M. Bilwes, J. den Hertog, T. Hunter, and J. P. Noel, unpublished data); and, finally, in contrast to the cross-linking of ephrin A1-overexpressing cells which resulted in the formation of a whole array of ephrin A1 oligomers (Fig. 7B), cross-linking of cells expressing the many different RPTPα constructs consistently resulted in the exclusive formation of homodimers. Furthermore, by performing cross-linking and biotinylation experiments in parallel and by comparing the cross-linking efficiency between wild-type RPTPα (FL) and disulfide-bond stabilized RPTPα homodimer (FL.137C), we conclude that the majority of cell surface RPTPα is homodimerized in transiently transfected 293 cells. In conjunction with previous observations that CD45 also dimerizes (13, 53), these results support the notion that dimerization-mediated negative regulation of PTP activity is physiologically relevant for RPTPα as well as for other RPTPs.
We demonstrated that RPTPα homodimerization can be mediated by multiple domains, including the ECD, the TMD, D2, and the wedge structure immediately N-terminal to D1. The finding that wedge mutations reduced cross-linking confirms the previous crystallographic data on RPTPα (3) as well as functional studies on RPTPα (20) and CD45 (31). The finding that deletion of D2 significantly reduced RPTPα homodimerization is also consistent with the existence of a D2 crystal dimer. The current study is the first to show that the ECD and the TMD of an RPTP also dimerize. The ECDs of many of the RPTPs are large and contain well-characterized structural moieties such as immunoglobulin-like domains. The ECD of RPTPα, however, is short and lacks any obvious structural motifs. It is therefore somewhat unexpected and intriguing that the RPTPα ECD dimerizes. The finding that RPTPα TMD homodimerizes is reminiscent of the previous reports on some other transmembrane proteins. Dimerization of glycophorin A via its TMD, for example, has been extensively investigated. In fact, many of the dimerization determinants within glycophorin A TMD were mapped using SN fusion proteins, a strategy we adopted in the current study (12, 28, 35). Dimerization via the TMD has also been implicated in the activation of the ErbB2/Neu receptor PTK (60), and FGFR3 (44, 47).
The fact that wedge mutants and D2 deletion mutant had much-reduced dimerization efficiency compared to the full-length RPTPα demonstrates that both the wedge and D2 are important although not essential for RPTPα homodimerization. However, although we showed that both the ECD and the TMD by themselves can homodimerize, we have been unable to assess the role of these two domains in the context of the native receptor by chemical cross-linking. We cannot compare the cross-linking efficiency of the ECD deletion construct (ΔECD) and the TMD fusion construct (TMD.SN) to that of the wild-type RPTPα (FL) due to the difference in the number of extracellular lysine residues they have available for cross-linking and biotinylation. We have attempted to assess the role of the TMD by inserting single alanine residues into several positions in the TMD of both the full-length RPTPα and TMD.SN fusion construct, but none of these single insertions reduced the level of cross-linked homodimers significantly. Other experimental approaches, such as fluorescence resonance energy transfer (FRET), will be needed to study RPTPα proteins with a modified ECD and TMD. In this connection, using RPTPα chimeras in which the ECD, TMD, and D1 of RPTPα are fused to two different GFP derivatives, we have recently shown that RPTPα dimerization can be detected in living cells by FRET analysis, and, by analyzing a panel of deletion mutants, found that the TMD was required and sufficient for dimerization of these chimeras (L. G. Tertoolen, J. C. Blanchetot, G. Jiang, J. Overvoorde, T. W. J. Gadella, T. Hunter, and J. den Hertog, submitted for publication).
So far, we have not determined the exact contribution of each of the individual dimerization domains toward the stable homodimerization of RPTPα. One of the difficulties in reaching a clear conclusion lies in our observation that a structural domain in isolation may behave somewhat differently than in the context of the full-length receptor. For instance, truncation mutants lacking the entire cytoplasmic domain (ΔCyto, Fig. 5) dimerized with high efficiency, suggesting that the cytoplasmic domain is not essential for dimerization. On the other hand, both wedge mutations and the D2 deletion significantly reduced RPTPα dimerization potential (Fig. 3 and 4), suggesting otherwise. We believe that it is reasonable to assume that results based on the mutated full-length receptor forms are more relevant than those based on truncated receptors or isolated domains. Accordingly, it appears that the wedge structure and D2 may have a relatively larger contribution than the ECD and/or TMD toward the stability of the dimer of the full-length receptor. Although this appears to be inconsistent with the fact that the ΔCyto truncation mutant homodimerizes as efficiently as the wild-type protein, the lack of bulky cytoplasmic domains may allow the TMDs to be aligned more closely in the truncated protein dimer than in full-length RPTPα, therefore exaggerating the TMD-TMD interaction.
The fact that cross-linking of transiently expressed full-length RPTPα was observed in the absence of added ligand(s) suggests that RPTPα homodimerizes in a ligand-independent fashion. However, given that several widely expressed surface proteins and extracellular matrix components have been found to bind to and potentially act as ligands for RPTPs (1, 34, 38, 40, 41, 62), one cannot exclude the possibility that homodimerization of full-length RPTPα is actually mediated by an unidentified ligand present in the tissue culture system. The fact that the RPTPα ECD deletion mutant still dimerized (Fig. 7D) unequivocally demonstrates that the efficient homodimerization of the truncated RPTPα can occur in a ligand-independent fashion. However, since some truncated receptor PTKs lacking all or part of the ECD undergo ligand-independent dimerization, whereas the full-length receptors do not (15, 22, 57), the fact that the RPTPα ECD deletion mutant dimerized does not necessarily imply that the dimerization of full-length RPTPα is also ligand independent. So far, we have been unable to cross-link endogenous RPTPα in embryonic fibroblasts or retrovirally transduced RPTPα in RPTPα−/− embryonic fibroblasts derived from RPTPα−/− mice (50). One possible explanation is that cross-linked dimers exist but are below the limit of detection, since we only detected a fairly faint band of the monomeric RPTPα protein in these cells. Another reason may be that these cells express a secreted ligand that promotes its dissociation or an intracellular protein that binds RPTPα and prevents dimerization that is absent or present at lower levels in 293 cells. Indeed, it may be necessary to overexpress RPTPα to override such dissociation factors and thereby detect RPTPα dimerization. The reported association of RPTPα with contactin via its ECD is an example of an interaction that might prevent RPTPα dimerization (62). Likewise, proteins interacting with the intracellular domain of RPTPα, in a manner similar to the interaction of LIP1 and the catenin-cadherin complex with intracellular domain of LAR (25, 46), could reduce the extent of RPTPα dimerization.
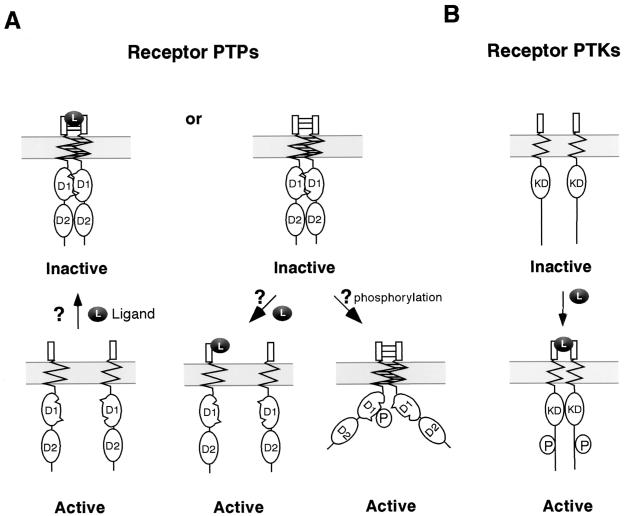
In conjunction with our previous observation that dimerization inhibits RPTPα biological activity (20), the current results lead us to propose that, in its inactive state, RPTPα exists as homodimers. Furthermore, based on the observation that multiple domains appears to mediate RPTPα homodimerization, we propose a zipper model in which RPTPα homodimers are stabilized by weak interactions between multiple dimer interfaces (Fig. 9A). In addition to the types of symmetric interactions depicted, it remains to be seen whether asymmetric interactions, such as a D1-D2 interaction, may also play a role in RPTPα intermolecular interactions, as suggested by studies of RPTPδ and RPTPς (58). The inactive dimeric state of RPTPα could in principle be induced by ligand binding (Fig. 9A, left). If this were true, we note that it is the opposite of how ligands regulate receptor PTK activity, where ligand-induced dimerization leads to activation (16) (Fig. 9B). Alternatively, based on the zipper model where multiple domains are involved in receptor homodimerization, it is possible that RPTPα is constitutively dimeric and inactive (Fig. 9A, right top). In this case, RPTPα may be activated by ligand(s) that stabilize the monomeric state of the receptor, thus preventing dimerization, or by an intracellular signaling event(s), such as phosphorylation, that induces an open conformation of the intracellular domains (Fig. 9A, right). In fact, we showed that both the EGF-bound EGFR-CD45 chimeric receptor (31) and disulfide-bonded full-length RPTPα dimers (20) with wedge mutation(s) are biologically active even though they are dimerized via their ECDs, supporting the notion that RPTP dimers can be enzymatically active. Considering that wedge mutations reduce dimerization efficiency as much as 80% (Fig. 3), it is possible that active dimers are unstable and may act as a transition state between inactive dimers and active monomers. The notion that RPTP dimerization may be regulated by intracellular signaling events is particularly attractive for receptors such as RPTPα, which has a short ECD without any recognizable protein-protein interaction domains. In fact, we showed previously that RPTPα is activated by phosphorylation of Ser180 and Ser204, which lie in the juxtamembrane domain close to the wedge (9, 55). We are currently testing whether such phosphorylation can decrease RPTPα homodimerization.
FIG. 9.
A model for the regulation of RPTPs via dimerization. (A) Hypothetical model for regulation of RPTPs via dimerization. In the inactive state, RPTPs are dimerized via wedge-active site interaction in D1, interaction via the TMD, and interaction via the ECD. In the active state, the receptors are either monomers or dimers in which dimerization via D1 no longer occurs due to phosphorylation. Ligand binding can either destabilize or stabilize dimers. (B) Classical model of activation of receptor PTKs. Ligand binding leads to receptor dimerization, transautophosphorylation, and subsequent kinase activation. KD, kinase domain; L, ligand; P, phosphorylation.
In summary, we provide evidence that RPTPα has the potential to homodimerize efficiently in vivo via multiple interacting domains, including the wedge structure, the TMD, and the ECD. The results presented here are consistent with our observation that dimerization decreases catalytic activity via a wedge region interaction (20), as we originally proposed (3). The results therefore suggest that dimerization-mediated downregulation of RPTPα biological activity is most likely physiologically relevant. Given that dimerization has been shown to inhibit several different RPTPs and that the wedge element is conserved, we believe that there will be other examples of RPTPs that are regulated in a similar manner through dimerization. However, this does not mean that all RPTPs will be regulated in this fashion. For instance, D1 of RPTPμ does not form a wedge-mediated dimer in the crystal structure (17), even though the helix-turn-helix that forms the wedge is present, and instead forms a different type of dimer. Additionally, the cytoplasmic portion (D1+D2) of LAR did not form a dimer in the recently reported crystal structure (37). Thus, it will obviously be important to determine which RPTPs can dimerize and whether this inhibits their activity and to investigate exactly how RPTP dimerization is regulated. In this connection, it will be very interesting to determine whether pleiotrophin, which binds to RPTPβ and inhibits its activity (33), induces RPTPβ dimerization.
ACKNOWLEDGMENTS
We thank Nigel Carter for providing us with the ephrin A1 construct and antibodies and Gunnar von Heijne, Ismael Mingarr, and Mark Lemmon for providing the pSN/GpA plasmid. We are very grateful to Walter Eckhart, Joel Leverson, Claudio Joazeiro, and Peter Blume-Jensen for insightful discussions and critical review of the manuscript.
This work was supported by USPHS grants CA14195 and CA39780 from the National Cancer Institute. G.J. was the recipient of a postdoctoral fellowship from the American Cancer Society. T.H. is a Frank and Else Schilling American Cancer Society Research Professor.
REFERENCES
- 1.Barnea G, Grumet M, Milev P, Silvennoinen O, Levy J B, Sap J, Schlessinger J. Receptor tyrosine phosphatase β is expressed in the form of proteoglycan and binds to the extracellular matrix protein tenascin. J Biol Chem. 1994;269:14349–14352. [PubMed] [Google Scholar]
- 2.Bhandari V, Lim K L, Pallen C J. Physical and functional interactions between receptor-like protein-tyrosine phosphatase α and p59fyn. J Biol Chem. 1998;273:8691–8698. doi: 10.1074/jbc.273.15.8691. [DOI] [PubMed] [Google Scholar]
- 3.Bilwes A M, den Hertog J, Hunter T, Noel J P. Structural basis for inhibition of receptor protein-tyrosine phosphatase α by dimerization. Nature. 1996;382:555–559. doi: 10.1038/382555a0. [DOI] [PubMed] [Google Scholar]
- 4.Buist, A., C. Blanchetot, L. G. Tertoolen, and J. den Hertog. Identification of p130Cas as an in vivo substrate of receptor protein-tyrosine phosphatase α. J. Biol. Chem., in press. [DOI] [PubMed]
- 5.Buist A, Zhang Y L, Keng Y F, Wu L, Zhang Z Y, den Hertog J. Restoration of potent protein-tyrosine phosphatase activity into the membrane-distal domain of receptor protein-tyrosine phosphatase α. Biochemistry. 1999;38:914–922. doi: 10.1021/bi981936b. [DOI] [PubMed] [Google Scholar]
- 6.Daum G, Regenass S, Sap J, Schlessinger J, Fischer E H. Multiple forms of the human tyrosine phosphatase RPTPα. Isozymes and differences in glycosylation. J Biol Chem. 1994;269:10524–10528. [PubMed] [Google Scholar]
- 7.den Hertog J, Hunter T. Tight association of GRB2 with receptor protein-tyrosine phosphatase α is mediated by the SH2 and C-terminal SH3 domains. EMBO J. 1996;15:3016–3027. [PMC free article] [PubMed] [Google Scholar]
- 8.den Hertog J, Pals C E, Peppelenbosch M P, Tertoolen L G, de Laat S W, Kruijer W. Receptor protein tyrosine phosphatase α activates pp60c-src and is involved in neuronal differentiation. EMBO J. 1993;12:3789–3798. doi: 10.1002/j.1460-2075.1993.tb06057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Hertog J, Sap J, Pals C E, Schlessinger J, Kruijer W. Stimulation of receptor protein-tyrosine phosphatase α activity and phosphorylation by phorbol ester. Cell Growth Differ. 1995;6:303–307. [PubMed] [Google Scholar]
- 10.den Hertog J, Tracy S, Hunter T. Phosphorylation of receptor protein-tyrosine phosphatase α on Tyr789, a binding site for the SH3-SH2-SH3 adaptor protein GRB-2 in vivo. EMBO J. 1994;13:3020–3032. doi: 10.1002/j.1460-2075.1994.tb06601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai D M, Sap J, Schlessinger J, Weiss A. Ligand-mediated negative regulation of a chimeric transmembrane receptor tyrosine phosphatase. Cell. 1993;73:541–554. doi: 10.1016/0092-8674(93)90141-c. [DOI] [PubMed] [Google Scholar]
- 12.Engelman D M, Adair B D, Brunger A, Flanagan J M, Hunt J F, Lemmon M A, Treutlein H, Zhang J. Dimerization of glycophorin A transmembrane helices: mutagenesis and modeling. Soc Gen Physiol Ser. 1993;48:11–21. [PubMed] [Google Scholar]
- 13.Felberg J, Johnson P. Characterization of recombinant CD45 cytoplasmic domain proteins. Evidence for intramolecular and intermolecular interactions. J Biol Chem. 1998;273:17839–17845. doi: 10.1074/jbc.273.28.17839. [DOI] [PubMed] [Google Scholar]
- 14.Green S, Issemann I, Sheer E. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haley J D, Hsuan J J, Waterfield M D. Analysis of mammalian fibroblast transformation by normal and mutated human EGF receptors. Oncogene. 1989;4:273–283. [PubMed] [Google Scholar]
- 16.Heldin C H. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann K M, Tonks N K, Barford D. The crystal structure of domain 1 of receptor protein-tyrosine phosphatase μ. J Biol Chem. 1997;272:27505–27508. doi: 10.1074/jbc.272.44.27505. [DOI] [PubMed] [Google Scholar]
- 18.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 19.Jacob K K, Sap J, Stanley F M. Receptor-like protein-tyrosine phosphatase α specifically inhibits insulin-increased prolactin gene expression. J Biol Chem. 1998;273:4800–4809. doi: 10.1074/jbc.273.8.4800. [DOI] [PubMed] [Google Scholar]
- 20.Jiang G, den Hertog J, Su J, Noel J, Sap J, Hunter T. Dimerization inhibits the activity of receptor-like protein-tyrosine phosphatase α. Nature. 1999;401:606–610. doi: 10.1038/44170. [DOI] [PubMed] [Google Scholar]
- 21.Jirik F R, Janzen N M, Melhado I G, Harder K W. Cloning and chromosomal assignment of a widely expressed human receptor-like protein-tyrosine phosphatase. FEBS Lett. 1990;273:239–242. doi: 10.1016/0014-5793(90)81094-5. [DOI] [PubMed] [Google Scholar]
- 22.Khazaie K, Dull T J, Graf T, Schlessinger J, Ullrich A, Beug H, Vennstrom B. Truncation of the human EGF receptor leads to differential transforming potentials in primary avian fibroblasts and erythroblasts. EMBO J. 1988;7:3061–3071. doi: 10.1002/j.1460-2075.1988.tb03171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohn A D, Barthel A, Kovacina K S, Boge A, Wallach B, Summers S A, Birnbaum M J, Scott P H, Lawrence J C, Jr, Roth R A. Construction and characterization of a conditionally active version of the serine/threonine kinase Akt. J Biol Chem. 1998;273:11937–11943. doi: 10.1074/jbc.273.19.11937. [DOI] [PubMed] [Google Scholar]
- 24.Krueger N X, Streuli M, Saito H. Structural diversity and evolution of human receptor-like protein tyrosine phosphatases. EMBO J. 1990;9:3241–3252. doi: 10.1002/j.1460-2075.1990.tb07523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kypta R M, Su H, Reichardt L F. Association between a transmembrane protein tyrosine phosphatase and the cadherin-catenin complex. J Cell Biol. 1996;134:1519–1529. doi: 10.1083/jcb.134.6.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lammers R, Lerch M M, Ullrich A. The carboxyl-terminal tyrosine residue of protein-tyrosine phosphatase α mediates association with focal adhesion plaques. J Biol Chem. 2000;275:3391–3396. doi: 10.1074/jbc.275.5.3391. [DOI] [PubMed] [Google Scholar]
- 27.Lammers R, Moller N P, Ullrich A. The transmembrane protein tyrosine phosphatase α dephosphorylates the insulin receptor in intact cells. FEBS Lett. 1997;404:37–40. doi: 10.1016/s0014-5793(97)00080-x. [DOI] [PubMed] [Google Scholar]
- 28.Lemmon M A, Flanagan J M, Hunt J F, Adair B D, Bormann B J, Dempsey C E, Engelman D M. Glycophorin A dimerization is driven by specific interactions between transmembrane alpha-helices. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 29.Lim K L, Kolatkar P R, Ng K P, Ng C H, Pallen C J. Interconversion of the kinetic identities of the tandem catalytic domains of receptor-like protein-tyrosine phosphatase PTPα by two point mutations is synergistic and substrate-dependent. J Biol Chem. 1998;273:28986–28993. doi: 10.1074/jbc.273.44.28986. [DOI] [PubMed] [Google Scholar]
- 30.Lim K L, Lai D S, Kalousek M B, Wang Y, Pallen C J. Kinetic analysis of two closely related receptor-like protein-tyrosine-phosphatases, PTPα and PTPɛ. Eur J Biochem. 1997;245:693–700. doi: 10.1111/j.1432-1033.1997.00693.x. [DOI] [PubMed] [Google Scholar]
- 31.Majeti R, Bilwes A M, Noel J P, Hunter T, Weiss A. Dimerization-induced inhibition of receptor protein tyrosine phosphatase function through an inhibitory wedge. Science. 1998;279:88–91. doi: 10.1126/science.279.5347.88. [DOI] [PubMed] [Google Scholar]
- 32.Matthews R J, Cahir E D, Thomas M L. Identification of an additional member of the protein-tyrosine phosphatase family: evidence for alternative splicing in the tyrosine phosphatase domain. Proc Natl Acad Sci USA. 1990;87:4444–4448. doi: 10.1073/pnas.87.12.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng K, Rodriguez-Pena A, Dimitrov T, Chen W, Yamin M, Noda M, Deuel T F. Pleiotrophin signals increased tyrosine phosphorylation of b-catenin through inactivation of the intrinsic catalytic activity of the receptor type protein tyrosine phosphatase β/ζ. Proc Natl Acad Sci USA. 2000;97:2603–2608. doi: 10.1073/pnas.020487997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milev P, Friedlander D R, Sakurai T, Karthikeyan L, Flad M, Margolis R K, Grumet M, Margolis R U. Interactions of the chondroitin sulfate proteoglycan phosphacan, the extracellular domain of a receptor-type protein tyrosine phosphatase, with neurons, glia, and neural cell adhesion molecules. J Cell Biol. 1994;127:1703–1715. doi: 10.1083/jcb.127.6.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mingarro I, Whitley P, Lemmon M A, von Heijne G. Ala-insertion scanning mutagenesis of the glycophorin A transmembrane helix: a rapid way to map helix-helix interactions in integral membrane proteins. Protein Sci. 1996;5:1339–1341. doi: 10.1002/pro.5560050712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moller N P, Moller K B, Lammers R, Kharitonenkov A, Hoppe E, Wiberg F C, Sures I, Ullrich A. Selective down-regulation of the insulin receptor signal by protein-tyrosine phosphatases α and ɛ. J Biol Chem. 1995;270:23126–23131. doi: 10.1074/jbc.270.39.23126. [DOI] [PubMed] [Google Scholar]
- 37.Nam H J, Poy F, Krueger N X, Saito H, Frederick C A. Crystal structure of the tandem phosphatase domains of RPTP LAR. Cell. 1999;97:449–457. doi: 10.1016/s0092-8674(00)80755-2. [DOI] [PubMed] [Google Scholar]
- 38.O'Grady P, Thai T C, Saito H. The laminin-nidogen complex is a ligand for a specific splice isoform of the transmembrane protein tyrosine phosphatase LAR. J Cell Biol. 1998;141:1675–1684. doi: 10.1083/jcb.141.7.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panesar M, Papillon J, McTavish A J, Cybulsky A V. Activation of phospholipase A2 by complement C5b-9 in glomerular epithelial cells. J Immunol. 1997;159:3584–3594. [PubMed] [Google Scholar]
- 40.Peles E, Nativ M, Campbell P L, Sakurai T, Martinez R, Lev S, Clary D O, Schilling J, Barnea G, Plowman G D, Schlessinger J. The carbonic anhydrase domain of receptor tyrosine phosphatase β is a functional ligand for the axonal cell recognition molecule contactin. Cell. 1995;82:251–260. doi: 10.1016/0092-8674(95)90312-7. [DOI] [PubMed] [Google Scholar]
- 41.Peles E, Schlessinger J, Grumet M. Multi-ligand interactions with receptor-like protein tyrosine phosphatase β: implications for intercellular signaling. Trends Biochem Sci. 1998;23:121–124. doi: 10.1016/s0968-0004(98)01195-5. [DOI] [PubMed] [Google Scholar]
- 42.Ponniah S, Wang D Z, Lim K L, Pallen C J. Targeted disruption of the tyrosine phosphatase PTPα leads to constitutive downregulation of the kinases Src and Fyn. Curr Biol. 1999;9:535–538. doi: 10.1016/s0960-9822(99)80238-3. [DOI] [PubMed] [Google Scholar]
- 43.Ramarao M K, Cohen J B. Mechanism of nicotinic acetylcholine receptor cluster formation by rapsyn. Proc Natl Acad Sci USA. 1998;95:4007–4012. doi: 10.1073/pnas.95.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet J M, Maroteaux P, Le Merrer M, Munnich A. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- 45.Sap J, D'Eustachio P, Givol D, Schlessinger J. Cloning and expression of a widely expressed receptor tyrosine phosphatase. Proc Natl Acad Sci USA. 1990;87:6112–6116. doi: 10.1073/pnas.87.16.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serra-Pages C, Kedersha N L, Fazikas L, Medley Q, Debant A, Streuli M. The LAR transmembrane protein tyrosine phosphatase and a coiled-coil LAR-interacting protein co-localize at focal adhesions. EMBO J. 1995;14:2827–3288. doi: 10.1002/j.1460-2075.1995.tb07282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiang R, Thompson L M, Zhu Y Z, Church D M, Fielder T J, Bocian M, Winokur S T, Wasmuth J J. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78:335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- 48.Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983;22:181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- 49.Stone R L, Dixon J E. Protein-tyrosine phosphatases. J Biol Chem. 1994;269:31323–31326. [PubMed] [Google Scholar]
- 50.Su J, Muranjan M, Sap J. Receptor protein tyrosine phosphatase α activates Src-family kinases and controls integrin-mediated responses in fibroblasts. Curr Biol. 1999;9:505–511. doi: 10.1016/s0960-9822(99)80234-6. [DOI] [PubMed] [Google Scholar]
- 51.Su J, Yang L T, Sap J. Association between receptor protein-tyrosine phosphatase RPTPα and the Grb2 adaptor. Dual Src homology (SH) 2/SH3 domain requirement and functional consequences. J Biol Chem. 1996;271:28086–28096. doi: 10.1074/jbc.271.45.28086. [DOI] [PubMed] [Google Scholar]
- 52.Tabiti K, Smith D R, Goh H S, Pallen C J. Increased mRNA expression of the receptor-like protein tyrosine phosphatase α in late stage colon carcinomas. Cancer Lett. 1995;93:239–248. doi: 10.1016/0304-3835(95)03816-f. [DOI] [PubMed] [Google Scholar]
- 53.Takeda A, Wu J J, Maizel A L. Evidence for monomeric and dimeric forms of CD45 associated with a 30-kDa phosphorylated protein. J Biol Chem. 1992;267:16651–16659. [PubMed] [Google Scholar]
- 54.Tonks N K, Neel B G. From form to function: signaling by protein tyrosine phosphatases. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- 55.Tracy S, van der Geer P, Hunter T. The receptor-like protein-tyrosine phosphatase, RPTPα, is phosphorylated by protein kinase C on two serines close to the inner face of the plasma membrane. J Biol Chem. 1995;270:10587–10594. doi: 10.1074/jbc.270.18.10587. [DOI] [PubMed] [Google Scholar]
- 56.Tsai W, Morielli A D, Cachero T G, Peralta E G. Receptor protein tyrosine phosphatase alpha participates in the m1 muscarinic acetylcholine receptor-dependent regulation of Kv1.2 channel activity. EMBO J. 1999;18:109–118. doi: 10.1093/emboj/18.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uren A, Yu J C, Karcaaltincaba M, Pierce J H, Heidaran M A. Oncogenic activation of the αPDGFR defines a domain that negatively regulates receptor dimerization. Oncogene. 1997;14:157–162. doi: 10.1038/sj.onc.1200810. [DOI] [PubMed] [Google Scholar]
- 58.Wallace M J, Fladd C, Batt J, Rotin D. The second catalytic domain of protein tyrosine phosphatase δ (PTPδ) binds to and inhibits the first catalytic domain of PTPs. Mol Cell Biol. 1998;18:2608–2616. doi: 10.1128/mcb.18.5.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Pallen C J. The receptor-like protein tyrosine phosphatase HPTPα has two active catalytic domains with distinct substrate specificities. EMBO J. 1991;10:3231–3237. doi: 10.1002/j.1460-2075.1991.tb04886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiner D B, Liu J, Cohen J A, Williams W V, Greene M I. A point mutation in the neu oncogene mimics ligand induction of receptor aggregation. Nature. 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 61.Weiss A, Schlessinger J. Switching signals on or off by receptor dimerization. Cell. 1998;94:277–280. doi: 10.1016/s0092-8674(00)81469-5. [DOI] [PubMed] [Google Scholar]
- 62.Zeng L, D'Alessandrini L, Kalousek M B, Vaughan L, Pallen C J. Protein tyrosine phosphatase α (PTPα) and contactin form a novel neuronal receptor complex linked to the intracellular tyrosine kinase fyn. J Cell Biol. 1999;147:707–714. doi: 10.1083/jcb.147.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng X M, Pallen C J. Expression of receptor-like protein tyrosine phosphatase α in rat embryo fibroblasts activates mitogen-activated protein kinase and c-Jun. J Biol Chem. 1994;269:23302–23309. [PubMed] [Google Scholar]
- 64.Zheng X M, Resnick R J, Shalloway D. A phosphotyrosine displacement mechanism for activation of Src by PTPα. EMBO J. 2000;19:964–978. doi: 10.1093/emboj/19.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng X M, Wang Y, Pallen C J. Cell transformation and activation of pp60c-src by overexpression of a protein tyrosine phosphatase. Nature. 1992;359:336–339. doi: 10.1038/359336a0. [DOI] [PubMed] [Google Scholar]