Abstract
The tumor suppressor protein p53 is frequently inactivated in tumors. It functions as a transcriptional activator as well as a repressor for a number of viral and cellular promoters transcribed by RNA polymerase II (Pol II) and by RNA Pol III. Moreover, it appears that p53 also suppresses RNA Pol I transcription. In this study, we examined the molecular mechanism of Pol I transcriptional inhibition by p53. We show that wild-type, but not mutant, p53 can repress Pol I transcription from a human rRNA gene promoter in cotransfection assays. Furthermore, we show that recombinant p53 inhibits rRNA transcription in a cell-free transcription system. In agreement with these results, p53-null epithelial cells display an increased Pol I transcriptional activity compared to that of epithelial cells that express p53. However, both cell lines display comparable Pol I factor protein levels. Our biochemical analysis shows that p53 prevents the interaction between SL1 and UBF. Protein-protein interaction assays indicate that p53 binds to SL1, and this interaction is mostly mediated by direct contacts with TATA-binding protein and TAFI110. Moreover, template commitment assays show that while the formation of a UBF-SL1 complex can partially relieve the inhibition of transcription, only the assembly of a UBF-SL1-Pol I initiation complex on the rDNA promoter confers substantial protection against p53 inhibition. In summary, our results suggest that p53 represses RNA Pol I transcription by directly interfering with the assembly of a productive transcriptional machinery on the rRNA promoter.
RNA polymerase I (Pol I) directs RNA synthesis from the 45S rRNA gene, a single class of genes found in multiple tandem arrayed copies in the nucleoli of eukaryotic cells. In human cells, transcription by RNA Pol I requires at least two auxiliary factors, the upstream binding factor (UBF) and the selectivity factor SL1 (27). UBF is an HMG box-containing protein that binds DNA and plays an important role in the recruitment of SL1 to the rRNA promoter (3). SL1 is a multiprotein complex consisting of TATA-binding protein (TBP) and three distinct TBP-associated factors (TAFs) (TAFI110, TAFI63, and TAFI48) (13). SL1 is required for accurate and promoter-specific Pol I transcription (4).
Regulation of Pol I transcription is extremely important because rRNA levels directly affect ribosome production and the capacity of the cell to synthesize proteins. Therefore, the expression of rRNA is highly regulated during cell growth and differentiation (22, 38). A variety of physiological or pathological stimuli, such as serum deprivation, high cell culture density, glucocorticoid treatment of lymphoid cells, heat shock, and cell differentiation can rapidly down-regulate the rate of rRNA transcription (22, 41). In addition, stimulators of cell growth and division, such as insulin and phorbol esters, or glucocorticoid treatment of nonlymphoid cells stimulate Pol I transcription (22, 41). The transcription of rRNA genes is also controlled during the cell cycle, stopping at mitosis with the concomitant disappearance of the nucleolus (47). Recent studies directed toward the identification of molecular mediators of these regulatory processes have suggested that tumor suppressor genes, such as pRb and p53, may play an important role in the regulation of Pol I transcription (6, 9, 46). These studies originated from the observation that both pRb and p53 can be localized to the nucleolus, the site of rRNA synthesis (5, 7, 14, 36, 42, 43). However, while some progress has been made on understanding the mechanisms of Pol I transcriptional repression by pRb, the role of p53 in the regulation of rRNA synthesis remains unclear.
The p53 gene encodes an important tumor suppressor that is frequently mutated in a variety of human tumors (26, 28). The inactivation of p53 results in a failure to respond properly to a variety of stress signals, leading to increased genomic instability and eventually cancer (17). Furthermore, introduction of wild-type p53 in cell culture leads to cell growth arrest or suppression of cell transformation (34, 35). Although mice that are homozygous null for p53 are developmentally competent, they are highly predisposed to tumors, indicating that p53 has an important function in protecting cells against aberrant cell growth and neoplastic transformation (16).
Functional characterization of p53 protein revealed a variety of biochemical activities, including the ability to regulate transcription (23). p53 can act as a sequence-specific transcription activator. This function requires the binding of p53 to a defined DNA element, triggering the activation of genes carrying such sequences in their promoters (25, 50). Transcriptional activation is mediated by an acidic domain at its amino terminus that binds directly to the TBP-TAF complex TFIID (33, 45). In addition, p53 is also capable of repressing promoters devoid of p53-binding sites, including c-fos, interleukin-6, PCNA, cyclin A, and bcl-2 promoters (26). Since most genes repressed by p53 are involved in promoting cell cycle progression, it has been proposed that the transcriptional repression function of p53 is critical in suppressing cell proliferation and tumor formation. This hypothesis is further supported by the observation that many tumor-derived p53 mutants have lost the ability to suppress transcription (21). The observation that the carboxy terminus of p53 is required for both TBP binding and transcriptional repression suggests that the mechanism by which p53 represses these promoters may be by TBP squelching (29, 44).
Wild-type p53 can repress not only RNA Pol II promoters but also RNA Pol III promoters (8, 10). These studies demonstrated that TFIIIB is the specific target for repression by p53. Since assembly of TFIIIB into a preinitiation complex confers substantial protection against the inhibitory effect of p53, it is likely that p53 represses RNA Pol III transcription by interfering with the preinitiation complex formation (8).
Most recently, it has been shown that expression of wild-type p53 inhibits cellular pre-rRNA synthesis (6). Point mutations in various regions of p53 that affect its transcriptional activity (175his, 270cys, and 273his) also abolished its ability to repress rRNA synthesis (6). Furthermore, the minimal rRNA promoter appears to be sufficient for p53-mediated repression of rRNA synthesis (6). However, these studies could not demonstrate a direct involvement of p53 in the repression of RNA Pol I transcription.
In this work, we have investigated in more detail the molecular mechanism of p53-mediated RNA Pol I transcriptional repression. First, we provide further evidence that the human rRNA promoter can be inhibited by overexpression of wild-type p53 but not mutant p53 (175his) in cotransfection assays. Second, we have established an in vitro cell-free transcription system that is responsive to added recombinant p53 protein. Then, we show that extracts prepared from p53−/− cells are transcriptionally more active than those prepared from p53+/+ cells. This difference in transcriptional activity is most likely due to functional inactivation of one or more Pol I factors since there is no detectable difference in the abundance of any of these transcription factors in extracts prepared from the two cell lines. In addition, we show that by binding to SL1, p53 prevents the interaction between UBF and SL1 in vitro. Finally, we show that inhibition of transcription is probably achieved by preventing the formation of a productive initiation complex at the rRNA gene (rDNA) promoter. Taken together, our data suggest that repression of Pol I transcription by p53 is not a consequence of p53-induced growth arrest, but rather due to an active repression mechanism.
MATERIALS AND METHODS
Plasmids.
Plasmids pCMV-hp53 (wild type) and pCMV-hp53 (175his) have been described previously (2). Plasmids for the expression of glutathione S-transferase (GST) fusion human p53 fragments, pGThp53N (1-160), pGThp53C1 (160-318), pGThp53C2 (318-393), and pGThp53C (160-393), have been described previously (24). Plasmids for the expression of GST fusion wild type and point mutant forms of p53 protein were constructed by inserting the p53 coding region from pCMV-hp53 (wild type) and pCMV-hp53 (175his) into pGEX-2T vector and pGEX-3X vector, respectively. Plasmids prHu3 and prHu3CAT have been described previously (52).
Transfections.
For the transfection assays, HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. Cells were seeded at 7.5 × 105 per 100-mm-diameter plate 1 day prior to transfection. Cells were then transfected by the calcium phosphate method with the indicated amounts of plasmid DNA. Filler DNA (pcDNA3 or pCEP4) was used to normalize the amount of DNA used per transfection. Cells were harvested 55 to 60 h after transfection, and total RNA was isolated using the guanidinium-thiocyanate method. Thirty micrograms of total RNA was then used in a standard primer extension assay with a chloramphenicol acetyltransferase (CAT)-specific primer. The cDNA product was resolved on an 8% polyacrylamide-urea gel and subjected to autoradiography. Quantitation were performed using a PhosphorImager (Molecular Dynamics).
Cell culture.
H1299 cells were maintained in high-glucose DMEM supplemented with 10% FBS. H1299-tsp53 cells were maintained in DMEM supplemented with 10% FBS and G418 (400 μg/ml). For maintenance, both H1299 and H1299-tsp53 cells were grown at 39°C in a humidified chamber with 5% CO2. HeLa cells used for transfection were maintained in DMEM supplemented with 10% FBS. The mouse embryonic fibroblast (MEF) cells derived from normal mice (p53+/+) and p53 knockout mice (p53−/−) were maintained in DMEM supplemented with 10% FBS. Sf9 insect cells were maintained in Hink's TNM-FH medium supplemented with 10% heat-inactivated FBS.
Protein extracts and fractions.
Nuclear extracts from HeLa cells, H1299 cells, and H1299-tsp53 cells were prepared as described by Dignam et al. (15). Whole-cell extracts (WCE) from HeLa cells, H1299 cells, H1299-tsp53 cells, MEF(p53+/+) cells, and MEF(p53−/−) cells were prepared as described by Manley et al. (30). The GST-p53 proteins and their expression in bacteria and purification with glutathione-Sepharose beads have been described previously (44).
Purification of GST fusion proteins.
GST-p53 and GST-p53 mutant proteins were expressed in Escherichia coli cells by induction with IPTG (isopropyl-β-d-thiogalactopyranoside) (0.1 mM). Cell lysates in phosphate-buffered saline containing 10% glycerol and 0.5% NP-40 were incubated with glutathione affinity resin (Pharmacia) for 1 h at 4°C. The beads were then washed five times with TM10–0.3 M KCl plus 0.5% NP-40 followed by two times with elution buffer (100 mM Tris [pH 8.0], 50 mM NaCl, 0.1% NP-40). GST fusion proteins were eluted from the beads by using 20 mM reduced glutathione dissolved in elution buffer. Eluted proteins were dialyzed against TM10 buffer (50 mM Tris [pH 7.9], 100 mM KCl, 12.5 mM MgCl, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) containing 0.1 M KCl. The amount of eluted protein was estimated by Coomassie blue or silver staining of sodium dodecyl sulfate (SDS)-polyacrylamide gels.
Purification of RNA Pol I, UBF, and SL1.
HeLa cell nuclear extracts were loaded onto a heparin-agarose column, and RNA Pol I, UBF, and SL1 were eluted with a 0.1 to 1.0 M KCl gradient in TM10 buffer. Fractions containing SL1 activity, as determined by in vitro transcription assays, were pooled and dialyzed against TM10–0.2 M KCl. The SL1 pool was then loaded onto an SP-Sepharose column (Pharmacia) preequilibrated in TM10–0.2 M KCl, and after extensive washes with TM10–0.2 M KCl, SL1 was eluted with TM10–0.8 M KCl and dialyzed against TM10–0.1 M KCl. RNA Pol I used in the reconstituted transcription reaction mixture was prepared as followed: fractions eluted at 250 mM KCl from the heparin-agarose column were pooled and dialyzed against TM (50 mM Tris-HCl [pH 7.9], 12.5 mM MgCl2, 1 mM EDTA, 20% glycerol) buffer containing 0.1 M KCl and fractionated on a Sepharose 300 (Pharmacia) gel filtration column. Active fractions were then loaded onto a Q-Sepharose column (Poros) equilibrated against TM containing 0.1 M KCl. Proteins were eluted with a salt gradient of 0.1 to 0.7 M KCl. The active fractions were pooled, dialyzed to 0.125 M KCl, aliquoted, and stored at −80°C. This RNA Pol I preparation contained no detectable UBF and SL1 activity. For the experiments described in Fig. 7 (panel B), RNA Pol I was purified by heparin-agarose and DEAE-Sepharose column chromatography (eluted with a linear salt gradient of 0.08 to 0.4 M KCl) and dialyzed to 0.125 M KCl.
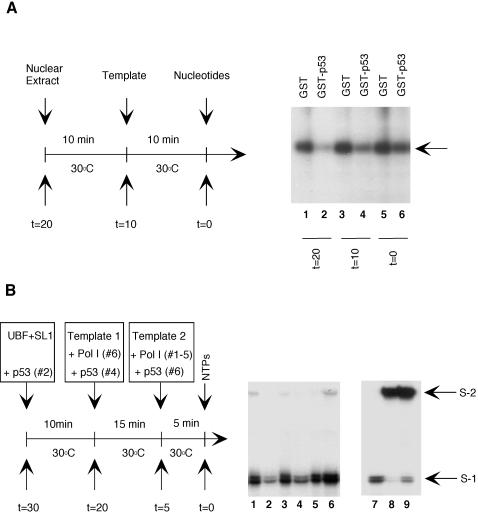
FIG. 7.
The assembly of a stable Pol I initiation complex on the rDNA promoter confers protection against repression by p53. (A). HeLa nuclear extracts were incubated for 10 min at 30°C before the addition of prHu3 (5 ng/reaction mixture); after a further 10 min at 30°C, nucleotides were added and transcription was allowed to proceed. Transcription reactions were carried out in the presence of either GST (5 pmol, lanes 1, 3, and 5) or GST-p53 (5 pmol, lanes 2, 4, and 6), which was added at the indicated time. Newly synthesized RNA transcript was detected by S1 nuclease protection assay. (B) Template commitment assays. Purified UBF, SL1, Pol I, GST-p53, 10 ng of wild-type template (template 1), and 50 ng of pseudo-wild-type template (template 2) were added to the appropriate reaction mixtures at the times indicated. Transcription reactions were started by the addition of the 4 nucleotides and performed at 30°C for 30 min. S1 and S2 represent protected fragments resulting from the transcription of the wild-type (template 1) and pseudo-wild-type (template 2) templates, respectively. In lanes 7 and 8, standard transcription reactions were carried out without GST-p53 and in the presence of either 10 ng of wild-type template (lane 7) or 50 ng of pseudo-wild-type template (lane 8). In lane 9, both templates (10 ng of wild type and 50 ng of pseudo-wild type) were added simultaneously to the transcription reaction mixture containing UBF, SL1, Pol I, and nucleotides.
UBF eluted from the heparin-agarose column was further purified by chromatography on Q-Sepharose using a linear salt gradient (0.1 to 1.0 M KCl). Fractions containing UBF were pooled and dialyzed against TM10–0.1 M KCl.
In vitro transcription assay.
Transcription assays using HeLa nuclear extracts or using fractions containing RNA Pol I, SL1, and UBF activities were carried out as previously described in the presence of 100 μg of α-amanitin/ml (52). In vitro-synthesized RNAs were detected by S1 nuclease analysis using a 5′-end-labeled single-strand DNA oligonucleotide complementary to the region from −20 to +40 of the rDNA gene. For transcription assays that include GST or GST fusion p53 proteins, the purified GST fusion proteins were allowed to incubate with HeLa nuclear extracts or a mixture of RNA Pol I, SL1, and UBF activity containing fractions at 30°C for 10 min before initiating the transcription reaction by adding template DNA prHu3 and nucleotides. For p53-responsive transcription assays, 3 to 10 ng of template DNA was used in each reaction mixture. Template commitment assays were performed in the presence of wild-type promoter and pseudo-wild-type promoter templates. Pseudo-wild-type template contained a 200-bp fragment of pUC13 inserted after base +40 of the rDNA gene. 5′-end-labeled oligonucleotides were made identical to the coding strand of the rDNA promoter between −20 and +40 of the wild-type template or between −20 and +80 of the coding strand of the pseudo-wild-type template.
Nonspecific RNA polymerase assay.
Random RNA polymerization assays were performed with 20 mM Tris (pH 7.9)–3 mM MnCl2–0.66 mM dithiothreitol–20% glycerol. Each reaction mixture contained 5 μg of nicked herring sperm DNA, 10 μg of protein extract, 1 mg of bovine serum albumin/ml, 1.2 mM ATP, 1.2 mM CTP, 1.2 mM GTP, 0.005 mM UTP, and 200 μCi of [3H]UTP. For RNA Pol I activity, α-amanitin was added to a final concentration of 100 μg/ml. After incubation at 30°C for 20 min, the reaction mixture was spotted on DE81 filters that were then washed extensively with Na2HPO4. Incorporated counts were determined by counting in a liquid scintillation counter (Beckmann).
Protein-protein interactions and Western blot analysis.
Purified GST fusion p53 proteins (∼1 μg) were incubated with an aliquot of fraction SL1 (purified from HeLa extracts on heparin-agarose and S-Sepharose columns) on ice for 4 to 6 h in 50 μl of TM10 buffer containing 0.1 M KCl and 0.1% NP-40. The mixture was then centrifuged at 15,000 × g for 30 min, and the supernatant was mixed with Flag-tagged UBF (∼1 μg) immobilized on anti-Flag M2 resin (Kodak) to precipitate the SL1 complex. The resulting complex was washed extensively in TM10 buffer containing 0.1 M KCl and 0.1% NP-40, and bound proteins were eluted and precipitated essentially as previously described (51). Pull-down assays with 35S-labeled proteins were performed as follows: equal molar amounts of each GST fusion protein were allowed to bind to glutathione beads for 1 h in TM10–0.1 M KCl plus 0.1% NP-40. All binding reactions were performed at 4°C with constant mixing on a nutator. The beads were then washed four times with the binding buffer. Human UBF, TBP, and TAFs were synthesized using the TNT-coupled in vitro transcription and translation system (Promega). In vitro-translated 35S-methionine-labeled proteins were then added to the beads, and the reaction mixture was incubated overnight at 4°C. The beads were then washed five times with TM10–0.1 M KCl plus 0.1% NP-40 buffer, boiled in SDS sample buffer, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Gels were dried, and radiolabeled proteins were detected by autoradiography. Bound radiolabeled proteins were also quantified using a PhosphorImager (Molecular Dynamics). Western blot analysis was performed as previously described (52). The antibodies against human TBP, TAFI48, TAFI63, TAFI110, and UBF were produced by immunizing rabbits with full-length proteins. Antibodies against p53 were pAb122, pAb1801 (Santa Cruz), and CM5 (Novo Laboratories). Antiserum against the 180-kDa RNA Pol I subunit was a generous gift from L. Rothblum.
RESULTS
p53 represses human rRNA promoter in vivo.
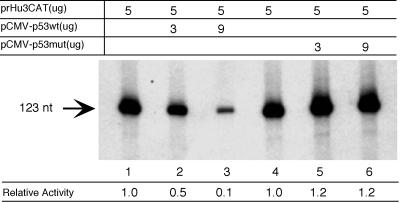
To study the inhibitory effects of p53 on the rRNA synthesis, we transfected HeLa cells with a human rRNA promoter construct (prHu3CAT) together with a plasmid expressing either the wild type (pCMV-hp53wt) or a mutant form of p53 (pCMV-hp53mut; amino acid 175 mutated to histidine). Figure 1 shows the primer extension analysis of the RNA isolated from transfected cells. Transfection of the promoter construct produced a 123-nucleotide primer extension fragment that corresponds to an RNA molecule initiated at the bona fide RNA Pol I transcription initiation site. The result of this experiment indicated that transcription from the human rRNA promoter was efficiently repressed (∼10-fold) when the expression vector encoding human p53 was cotransfected (lanes 2 and 3) compared to when the empty vector was used (lane 1 and 4). Importantly, inhibition of Pol I transcription appeared to correlate with the level of p53 expression in transfected cells. On the other hand, no repression was observed when the cells were cotransfected with the expression vector encoding a point mutant form of p53 (175His) (lanes 5 and 6). This point mutant form of p53 (Arg175 to His) is one of the hot spots for missense p53 mutations in tumors and is unable to suppress cell growth in culture. Taken together, our data suggest that transcription by RNA Pol I can be repressed by p53 in human cells and that this repression depends on a functionally active p53.
FIG. 1.
Repression of a human ribosomal RNA gene construct by wild-type p53. The human ribosomal RNA gene promoter construct prHu3CAT was cotransfected with a plasmid containing either wild-type p53 (pCMV-hp53wt, lanes 1 to 3) or mutant p53 (pCMV-hp53mut [175His], lanes 4 to 6) into HeLa cells using the calcium phosphate transfection method. The Pol I-specific transcript from prHu3CAT (123 nucleotides) was detected by primer extension using a probe specific for the reporter construct. The primer extension data were quantified using a PhosphorImager and are shown as relative activity. The amount of each plasmid (in micrograms) used in each transfection assay is indicated. Empty vector DNA (pCMV) was added to each reaction mixture to keep the total amount of plasmid in each transfection assay at 20 μg. Wild-type p53 and point mutant p53 (His175) were expressed at comparable level in HeLa cells (data not shown).
Pol I transcriptional activity but not the cellular level of Pol I factors is inhibited in p53-expressing cells.
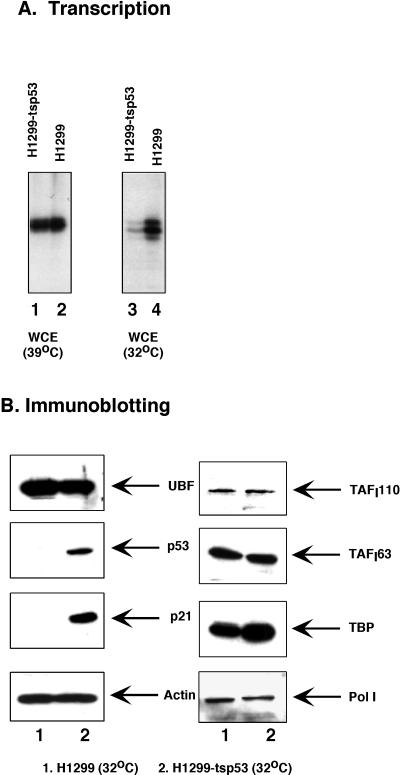
Studies by other investigators and us have revealed that the synthesis of RNA from an rRNA promoter can be repressed by the expression of wild-type p53 protein (this work and reference 6). The decrease in Pol I transcriptional activity caused by expression of p53 could be explained as either a direct effect on the Pol I transcriptional apparatus or an indirect effect attributable to a decrease in the abundance of any of the Pol I factors. To address these questions and investigate this process further, we took advantage of a pair of established human cell lines, the H1299 cells and the H1299-tsp53 cells. The H1299 cell line contains a homozygous deletion of the p53 gene, whereas the H1299-tsp53 cell line carries a temperature-sensitive form of p53 (20). tsp53 is in the wild-type conformation at permissive temperature (32°C) and is in the mutant form at elevated temperature (39°C). The two cell lines were cultured for 24 to 48 h at the permissive and nonpermissive temperatures, respectively, and subsequently, WCE were made from these two cell lines. The Pol I transcriptional activity of these extracts was then assessed in an in vitro transcription assay using the same amount of total proteins. As shown in Fig. 2A, extracts prepared from H1299 and H1299-tsp53 cells grown at the nonpermissive temperature have comparable Pol I transcriptional activity (lanes 1 and 2, respectively). On the other hand, when cells were grown at the permissive temperature, Pol I transcriptional activity of H1299 cells is about three- to fourfold higher than that of H1299-tsp53 cells (lanes 3 and 4, respectively). Since the only difference between H1299-tsp53 cells grown at 32 and 39°C is the conformational change of p53 protein, our results demonstrated a correlation between a decreased level of Pol I transcriptional activity and the presence of a functional p53. We were also able to observe an elevated level of Pol I transcriptional activity (∼two- to threefold) using extracts prepared from primary MEF cells from p53 knockout mice (p53−/−) compared to that of control cells from normal mice (p53+/+) (data not shown). These results indicate that, even under physiological concentration, p53 has an inhibitory effect on Pol I transcriptional activity.
FIG. 2.
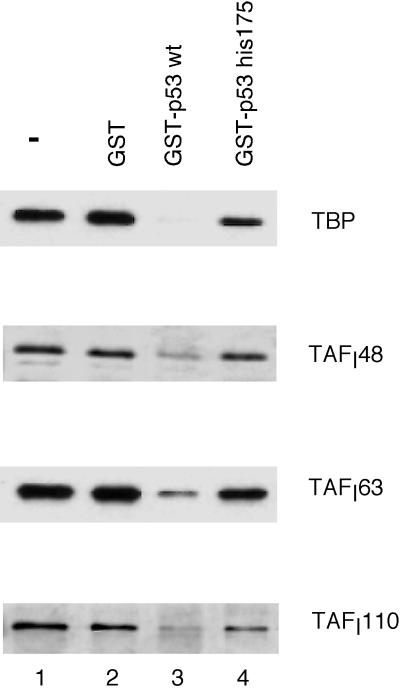
Effect of p53 expression on Pol I transcriptional activity and on the abundance of Pol I factors. (A) RNA Pol I transcriptional activity is elevated in p53−/− cells. WCE were prepared from H1299 cells (p53−/−, homozygous deletion) and H1299-tsp53 cells (from H1299, stably expressing a temperature-sensitive p53 mutant [135val]) grown at either 39 or 32°C. In vitro transcription assays were carried out using 10 μg of each WCE and 100 ng of prHu3. (B) The abundance of Pol I factors is not affected by the expression of wild-type p53. WCE were prepared from H1299 cells (lane 1) and H1299-tsp53 cells (lane 2) grown at 32°C for 48 h. An equal amount of total protein was separated by SDS-PAGE and immunoblotted using antibodies against UBF, p53, p21, TAFI110, TAFI63, TAFI48, TBP, and the 180-kDa subunit of RNA Pol I, as indicated.
To investigate whether a decrease in the abundance in Pol I, UBF, or SL1 could account for the reduced Pol I transcriptional activity in p53-expressing cells, we determined the amounts of these factors present in the extracts by Western blot analysis. Equal amounts of WCE prepared from H1299 and H1299-tsp53 cells grown at 32°C were separated by SDS-PAGE, and subsequent immunoblotting was carried out using antibodies against each specific factor (Fig. 2B). As expected, expression of p53 and the cyclin-dependent kinase inhibitor p21 can be observed in H1299-tsp53 cells (lane 2) but not in H1299 cells (lane 1). However, no detectable difference in the abundance of UBF and two subunits of SL1 (TAFI110, and TAFI63) was observed between these two cell lines. We noticed a slight increase in cellular TBP level in H1299-tsp53 cells. This increase in TBP can be consistently seen in the presence of a functional p53. However, it is very unlikely that such an increase in cellular TBP level will result in a fourfold decrease in Pol I transcriptional activity in p53-expressing cells. To estimate the cellular level of RNA Pol I in these two cell lines, we determined the abundance of the 180-kDa subunit of RNA Pol I. Western blot analysis revealed a minor decrease of this Pol I subunit in H1299-tsp53 cells. However, when promoter-independent RNA Pol I activity was tested with nicked calf thymus DNA in the presence of α-amanitin, we did not observe any difference in the extracts prepared from the two cell lines (data not shown). Taken together, our results indicate that the expression of a functional p53 has little or no effect on the cellular level of UBF, SL1, and RNA Pol I. Therefore, we can rule out the possibility that p53 represses Pol I transcription indirectly by down-regulating the expression of Pol I factors.
p53 represses human RNA Pol I transcription in vitro.
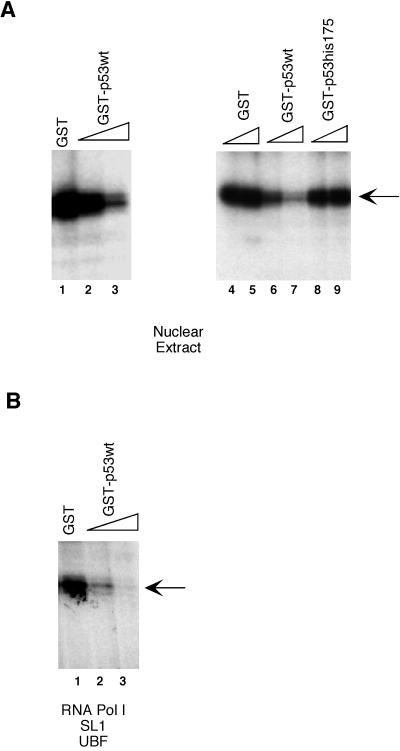
To establish that repression of Pol I was the result of a direct effect of p53, we then proceeded to test the effect of p53 on RNA Pol I transcription in an in vitro-reconstituted transcription system. Full-length wild-type p53 and mutant p53 (175His) were expressed as GST fusion forms in bacteria and purified using glutathione-Sepharose resin. When purified recombinant GST-p53 was added to an in vitro cell-free transcription system containing HeLa nuclear extracts, transcription from the human rRNA promoter decreased dramatically (Fig. 3A, lanes 2 and 3 and lanes 6 and 7), whereas an equal amount of GST alone had little or no effect (Fig. 3, lanes 1, 4, and 5). Importantly, the decrease in Pol I transcriptional activity appeared to correlate with the amount of p53 added. Quantitation of the transcription assay data indicated that p53 represses Pol I transcription by approximately eightfold. Similar results were also obtained using recombinant p53 purified from baculovirus-infected Sf9 cells (data not shown). To provide further support for a direct inhibition by p53, a point mutant form of p53 (175His) that is inactive in transient-cotransfection assays was then tested in the p53-responsive system. As shown in Fig. 3A, the Pol I transcription was unaffected by the addition of the mutant form of p53 (lanes 8 and 9). Thus, our data indicate that p53 acts directly on the Pol I transcription machinery to inhibit rRNA synthesis.
FIG. 3.
Repression of RNA Pol I transcription in vitro by recombinant p53. (A) HeLa nuclear extracts were preincubated for 20 min at 30°C with either GST (lanes 1, 4, and 5, 2, 2, and 6 pmol, respectively, as judged by silver-stained gels), an increasing amount of GST-p53 (lanes 2 and 3 and lanes 6 and 7, 2 and 6 pmol, respectively), or an increasing amount of GST-p53(his175) (lanes 8 and 9, 2 and 6 pmol, respectively) before the addition of transcription template prHu3 (5 ng/reaction mixture) and nucleotides to initiate transcription. GST, GST-p53, and GST-p53(his175) were expressed in E. coli and were affinity purified using glutathione-Sepharose beads. (B) Transcription assays were carried out using column-purified fractions (see Materials and Methods) containing RNA Pol I, UBF, and SL1 in place of HeLa nuclear extracts as described in panel A. Newly synthesized RNA transcript was detected by S1 nuclease protection assay as described in Materials and Methods. Arrows indicate transcript.
To further investigate whether a minimal set of Pol I factors is sufficient to support the p53-mediated transcriptional repression, HeLa nuclear extracts were replaced with a mixture of RNA Pol I, UBF, and SL1 fractions purified from HeLa cells in the following in vitro transcription assays. As shown in Fig. 3B, addition of purified recombinant GST-p53 (lanes 2 and 3) resulted in a dramatic decrease in the level of rRNA synthesis. As seen in the previous experiment, the repression was proportional to the amount of recombinant p53 added. Taken together, our results suggest that purified RNA Pol I, UBF, and SL1 are sufficient to support the p53-mediated Pol I transcriptional repression.
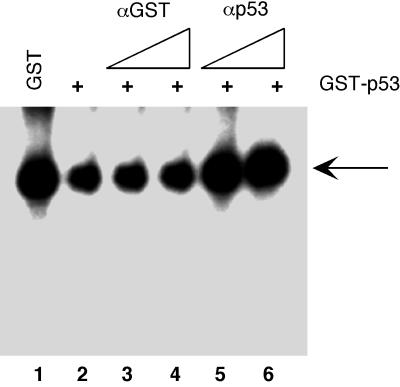
To confirm that the p53 portion of the fusion protein is indeed the functional domain repressing Pol I transcription, we tested the effect of purified GST-p53 on in vitro rRNA synthesis following preincubation with antibodies against either GST or p53. As shown in Fig. 4, the ability of GST-p53 to repress Pol I transcription is dramatically reduced following preincubation with a purified polyclonal antibody raised against p53 (CM5; Novo Laboratories; lanes 5 and 6). Importantly, the level of transcription after the addition of p53 antibodies is comparable to that from the reaction in which GST alone was added to the transcription reaction mixture (lane 1). The neutralizing effect appeared to be specific, since preincubating GST-p53 with a polyclonal antibody against the GST portion (lanes 3 and 4) has little or no effect on the ability of GST-p53 to repress Pol I transcription in vitro (lane 2). These observations rule out the possibility that the inhibitory effect of GST-p53 is due to the contaminant and demonstrate that Pol I transcription is directly repressed by p53.
FIG. 4.
Effect of neutralizing antibodies on the Pol I transcriptional repression by GST-p53. HeLa nuclear extracts were preincubated for 20 min at 30°C with 3 pmol of either GST (lane 1) or GST-p53 (lanes 2 to 6) before the addition of transcription template prHu3 (5 ng/reaction mixture) and nucleotides to initiate transcription. Prior to use, the GST-p53 was incubated for 2 h on ice with antibodies against GST (lanes 3 and 4, 0.3 and 0.6 μg, respectively) or with antibodies against p53 (lanes 5 and 6, 0.3 and 0.6 μg, respectively). Both GST and GST-p53 were expressed in E. coli and were affinity purified using glutathione-Sepharose beads. Newly synthesized RNA transcript was detected by S1 nuclease protection assay.
p53 inhibits the interaction between UBF and SL1.
One important step in stimulation of Pol I transcription is the formation of a stable complex between UBF and SL1 at the ribosomal DNA promoter. To assess the possible effect of p53 on the recruitment of SL1, we performed the following protein-protein interaction assays. GST fusion proteins were first expressed and purified using glutathione-Sepharose resin as described in Materials and Methods. The purified proteins were then incubated with an SL1 fraction purified from HeLa cells. After the incubation, the mixture was added to Flag-tagged UBF that was immobilized on anti-Flag M2 affinity resin. Flag-tagged UBF was then immunoprecipitated using Flag-antibody resin, and the immunoprecipitation products were separated by SDS-PAGE. SL1 was then detected by Western blot analysis using antibodies against each of the individual SL1 subunits (TBP, TAFI48, TAFI63, and TAFI110). As shown in Fig. 5, preincubation of SL1 with GST-p53 resulted in a dramatic decrease in the formation of a UBF-SL1 complex, as indicated by a decrease of the four SL1 subunits in the immunoprecipitation products (lane 3). In the control experiment in which SL1 was preincubated with GST protein alone (lane 2), the formation of a UBF-SL1 complex was unaffected compared to when no protein (lane 1) was used in the preincubation with SL1. Thus, our data indicate that p53 can directly interfere with the formation of a preinitiation complex between UBF and SL1. To further assess the functional significance of this interference on UBF-SL1 complex formation by p53 in the Pol I transcriptional repression, we asked whether the p53 (175His) mutant had any effect in this protein-protein interaction assay. Unlike wild-type p53, the point mutant form of p53 (GST-p53his175) has no effect on the UBF-SL1 interaction (lane 4). The inability of this point mutant form of p53 to interfere with UBF-SL1 complex formation correlates with its inability to repress Pol I transcription both in vivo and in vitro.
FIG. 5.
Effects of wild-type and mutant (His175) p53 proteins on UBF-SL1 interaction in vitro. (A) GST (lane 2), GST fusion p53 protein (lanes 3), and GST fusion mutant p53 (His175) (lane 4) were expressed in E. coli and were affinity purified using glutathione-Sepharose beads. An aliquot of the partially purified SL1 fraction (purified by heparin-agarose and S-Sepharose column chromatography; approximately 0.6 μg) was first incubated with each purified GST fusion protein (approximately 2 μg) for 4 to 6 h before being immunoprecipitated with Flag-tagged UBF (∼ 1 μg) immobilized on anti-Flag M2 beads. Flag-tagged UBF was expressed in Sf9 cells using a recombinant baculovirus. The immunoprecipitation products were separated by SDS-PAGE and the SL1 complex was detected by Western blotting using antibodies against each individual subunit (TBP, TAFI48, TAFI63, and TAFI110), as indicated. Lane 1, SL1 input.
P53 interacts with SL1.
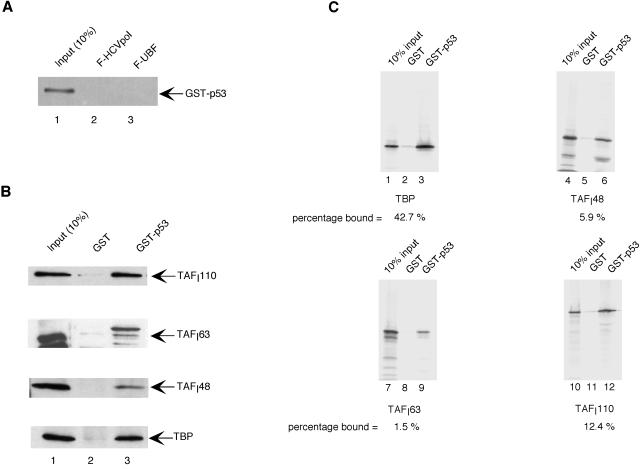
The inability of SL1 to bind to UBF in the presence of wild-type p53 could be explained if p53, by binding to either UBF or SL1, prevents the protein-protein interactions between these two factors. To address whether p53 binds to UBF, the same molar amount of GST-p53 used in the previous assay was incubated with immobilized Flag-tagged UBF, and the bound material was then resolved by SDS-PAGE and subjected to Western blot analysis using p53 antibodies. As shown in Fig. 6A, there was no detectable interaction between p53 and UBF (lane 3). On the other hand, when we performed in vitro GST pull-down studies with GST-p53 and SL1, we observed a significant interaction between GST-p53 and SL1, as determined by the presence of TBP and TAFIs in the pull-down fraction (Fig. 6B). These results suggest that p53 may prevent the interaction between SL1 and UBF by binding directly to SL1. To identify the SL1 subunits involved in the interaction, we tested GST-p53 and GST alone for the ability to bind in vitro-translated 35S-methionine-labeled TBP, TAFI48, TAFI63, and TAFI110. The results of this experiment show that p53 binds well to TBP (Fig. 6C, lanes 1 to 3) and with lower affinity to TAFI110 (lanes 10 to 12), while there is little or no significant interaction (below 10% of input) between p53 and TAFI48 (lanes 4 to 6) or TAFI63 (lanes 7 to 9).
FIG. 6.
p53 binds to SL1. (A) Flag-tagged UBF (lane 3) and HCV polymerase (HCVpol) (lane 2) were immobilized on anti-Flag M2 beads and incubated with purified GST-p53 (2 μg). After extensive washing, bound material was detected by Western blot analysis using p53 antibodies (pAB122). Lane 1 shows 10% of the input material used in reactions 2 and 3. (B) An aliquot of the partially purified SL1 fraction (purified by heparin-agarose and S-Sepharose column chromatography; approximately 0.6 μg) was incubated for 4 to 6 h with either GST (lane 2; ∼2 μg) or GST-p53 (lane 3; ∼2 μg), which were immobilized on glutathione resin. After extensive washing, bound proteins were resolved on an SDS-polyacrylamide gel and SL1 was detected by Western blot analysis using antibodies against TAFI110, TAFI63, TAFI48, and TBP. (C) In vitro-translated 35S-methionine-labeled TBP (lanes 1 to 3), TAFI48 (lanes 4 to 6), TAFI63 (lanes 7 to 9), and TAFI110 (lanes 10 to 12) were added to either GST (lanes 2, 5, 8, and 11) or GST-p53 (lanes 3, 6, 9, and 12) immobilized on glutathione beads, and reaction mixtures were incubated overnight at 4°C. After extensive washes, bound proteins were resolved by SDS-PAGE and visualized by autoradiography. 35S-methionine-labeled proteins were quantitated by PhosphorImager analysis. Lanes 1, 4, 7, and 10 show 10% of the proteins used in the pull-down assays.
Assembly of the initiation complex on the rDNA promoter confers protection against p53.
The ability of p53 to block the interaction between UBF and SL1 suggests that p53 might represses Pol I transcription by preventing the formation of a stable transcription initiation complex. To test this hypothesis, we first investigated whether the assembly of a Pol I initiation complex can prevent the repressive effect of p53. GST alone or GST-p53 was added to an aliquot of HeLa cell nuclear extracts either 10 min prior to the addition of the transcription template prHu3 (Fig. 7A, lanes 1 and 2), simultaneously with the addition of the template (lanes 3 and 4), or 10 min after the addition of the template (lanes 5 and 6). Nucleotides were then added to initiate transcription. Since the UBF-SL1 complex can be assembled on the rDNA promoter rapidly at 30°C, our assay allows us to detect the ability of p53 to repress a preformed initiation complex. Relative to GST alone, GST-p53 was found to inhibit Pol I transcription by about fivefold when added prior to the addition of template and about 2.5-fold when added simultaneously with the template. On the other hand, when GST-p53 was added 10 min after the template, Pol I transcriptional activity was minimally affected (1.4-fold).
To further investigate whether the association of UBF with SL1 at the ribosomal DNA promoter was sufficient for preventing the inhibitory effect of p53, we performed template commitment assays using purified UBF, SL1, and RNA Pol I. As observed in the previous experiments, p53 inhibited Pol I transcription when incubated with UBF and SL1 before the addition of the DNA template (Fig. 7, lane 3). Interestingly, the inhibitory function was partly retained even when p53 was added to the reaction mixture in the presence of a preformed UBF-SL1 complex (lane 4). Quantitation of the transcription assays using a phosphorimager indicated that Pol I transcription was inhibited approximately threefold in lane 2 and 1.5-fold in lane 4, compared to lanes 1, 3, and 5. On the other hand, when an UBF-SL1-Pol I complex was preassembled on the rDNA promoter, p53 was unable to repress transcription (lane 6). The lower degree of p53 inhibition in this assays is probably due to an overall decrease in the activity of factors such as SL1, which is particularly sensitive to incubation at temperatures above 4°C (W. Zhai and L. Comai, unpublished observation).
DISCUSSION
A high level of rRNA transcription is necessary for cell growth and proliferation. When quiescent cells are stimulated by mitogenic factors, the production of rRNA increases, resulting in the assembly of more ribosomes and increased protein synthesis. Conversely, a 50% reduction in translation is sufficient to cause proliferating cells to withdraw from the cell cycle. Clearly, the production of ribosomes and protein synthesis correlate closely with the growth rate of the cell. The rapid growth of tumor cells requires elevated rates of biosynthesis, and a higher level of rRNA transcription is frequently observed in these cells. It is therefore conceivable that p53, as a tumor suppressor protein, has the ability to place a physiological constraint upon Pol I transcription. The first evidence suggesting that p53 may be directly involved in regulating Pol I transcription is provided by immunohistochemistry studies showing that p53 can be localized to the nucleolus under certain physiological conditions (7, 36). Recent studies by Budde and Grummt have also indicated that p53 induction correlates with a down-regulation of Pol I transcription; however, they concluded that this effect might be a consequence of growth arrest induced by p53 (6).
In this study, we have investigated the molecular mechanism of p53-mediated Pol I transcriptional repression. An important step in this analysis is the establishment of an in vitro cell-free transcription system that can faithfully reproduce the in vivo inhibitory effect of p53 on Pol I transcription. These assays indicate that RNA Pol I, UBF, and SL1 are sufficient to reconstitute the repression of transcription by p53 in vitro. In addition, transcription experiments with a series of p53 deletion mutants indicate that the carboxy-terminal region of p53 is necessary, but not sufficient, to repress Pol I transcription (unpublished results). These results suggest that efficient repression of Pol I activity requires the cooperative action of the amino- and carboxy-terminal domains of p53. This model fits well with the observation that growth suppression by p53 requires both its amino-terminal and carboxy-terminal domains (39). In addition, it provides further support to the concept that the repression of rRNA gene transcription is an important step in growth suppression mediated by p53.
Because p53 has the ability to repress a variety of Pol II promoters, we could not rule out the possibility that a down-regulation of the synthesis of mRNAs encoded by genes encoding factors required for rRNA synthesis may contribute to the repression of Pol I transcription. However, using Western blot analysis, we showed that there isn't any significant change in the abundance of UBF, SL1, or RNA Pol I to account for the inhibition of Pol I transcription.
An important finding of our analysis is that wild-type p53 prevents the formation of the UBF-SL1 complex. Moreover, a cancer-associated p53 point mutant (175His) does not interfere with UBF-SL1 complex formation nor does it repress Pol I transcription. These results provide further evidence that inhibition of the protein-protein interaction between UBF and SL1 by p53 plays an important role in repression of Pol I transcription. Arg175 is a structurally very important p53 mutation because of its role in the stabilization of the L2 and L3 loops (11). This mutant retains its ability to bind to DNA (53); however, several lines of evidence suggest that it may not fold properly (11).
In addition to blocking the interaction between UBF and SL1, our data suggest that p53 can be still partially effective in Pol I inhibition in the absence of a stable transcription initiation complex at the rDNA promoter. Template commitment assays show that only the preassembly of a UBF-SL1-Pol I complex on the rDNA promoter confers full protection against the repression by p53. Therefore, it is possible that in the absence of the polymerase, p53 can destabilize a preformed UBF-SL1 complex at the rDNA promoter, such that only the subsequent recruitment of the polymerase provides a p53-resistant, transcriptionally competent, multiprotein complex. Preliminary in vitro protein binding studies addressing whether p53 could disrupt a preformed UBF-SL1 complex did not yield results consistent enough to allow a clear interpretation. Therefore, a more detailed analysis is necessary to implicate this or other mechanisms.
In eukaryotic cells, transcriptions by RNA Pol I, Pol II, and Pol III share many common themes. Transcription regulators, such as simian virus 40 large T antigen and pRb, have been shown to regulate transcription by all three polymerases and, apparently, in similar fashions. In the Pol II system, p53 can inhibit the transcription from a variety of TATA-containing promoters. It has been proposed that the carboxy-terminal domain of p53, by interacting with TBP, prevents the basal transcription factors from binding to the TATA element and thereby represses transcription from these promoters (32, 40). In other instances, Pol II transcriptional repression by p53 may be mediated by its interaction with factors other than TBP, such as the TAFs (45) and the CCAAT-binding factor (1, 49). In the Pol III system, p53 acts as a general repressor for all promoters. Repression of Pol III transcription by p53 has been proposed to occur through its interaction with TFIIIB, the TBP-BRF complex (8, 10). The binding of p53 to TFIIIB results in a complete inactivation of this factor that can be rescued by the addition of more TFIIIB to the transcription reaction (8). However, it is unclear whether p53 interferes with the ability of TFIIIB to interact with TFIIIC and/or Pol III or inactivates TFIIIB by some other mechanism. The mechanism of Pol I transcriptional repression by p53, as shown in this study, is quite similar in several aspects to that of the Pol II and Pol III systems. First, we demonstrate that repression of Pol I transcription by p53 is primarily a direct effect. Addition of p53 to either HeLa cell nuclear extracts or to a purified system containing RNA Pol I, UBF, and SL1 resulted in a dramatic decrease in Pol I transcriptional activity. Second, p53 represses Pol I transcription initiation by interfering with the formation of a stable initiation complex. Upon incubation with p53, SL1 is no longer able to form a complex with UBF. In analogy to the Pol II and Pol III systems, we also show that p53 targets the TBP-TAFI complex SL1. We have determined that p53 binds to SL1 in vitro, and in vitro interaction studies indicate that at least one subunit of the SL1 complex, TBP, can strongly interact with p53. SL1 is far less abundant than UBF and the RNA Pol I in the cell, and it is likely the limiting factor in Pol I transcription (22, 41). p53 is also a relatively low-abundance cellular protein. Therefore, its is reasonable to speculate that negative regulation of Pol I transcription by molecules such as p53 may be more readily attained by targeting the SL1 complex. Unfortunately, we were not able to coimmunoprecipitate p53 and SL1 from cell extracts, possibly suggesting that this protein-protein interaction is not very stable. We also found that both the wild type and the mutant p53 (175His) can interact with SL1 in vitro (unpublished results). Because the in vitro binding assay also indicate that each SL1 subunit binds to some degree to p53 compared to the GST control, we cannot completely rule out the possibility that the interaction observed in vitro is not the result of a specific affinity, but casually due to ionic or hydrophobic interactions.
Results from a number of studies have indicated that p53 plays a role in the control of protein synthesis. It has been shown that p53 is capable of repressing the translation of certain mRNAs (18, 37). It has also been reported that a subpopulation of cytoplasmic p53 is covalently linked to a single specific RNA—the 5.8S rRNA (19). In addition, p53 has been found in a complex with the ribosomal L5 protein and mdm2, suggesting that p53 may control ribosome biogenesis and/or translational regulation in the cell (31). Further support to this concept had been provided by the demonstration that p53 can control the synthesis of 5S rRNA, a Pol III-transcribed gene (8, 10). Since the 5S rDNA and the large rDNA genes transcribed by Pol I are used stoichiometrically in the assembly of ribosomes, the cell must use a mechanism to coordinate the transcription of these genes. Thus, p53, which controls cell growth, may have to put a restraint on the expression of the rDNA genes transcribed by Pol I as well as the 5S rDNA gene transcribed by Pol III. Such a role of a master regulator may be true not only for p53 but also for the retinoblastoma gene product, pRb. pRb is a tumor suppressor protein whose deregulation leads to uncontrolled cell cycle progression, elevated rates of biosynthesis, and eventually, tumor formation. As recently demonstrated by Cavanaugh et al., pRb participates in the repression of rRNA synthesis in differentiated U937 cells (9). Furthermore, pRb has also been shown to inhibit transcription by Pol III (12, 48). Although there is only limited sequence homology between pRb and p53, the two tumor suppressors share many functional features, such as to repress cell growth, and it has been proposed that they play a direct role in the inhibition of transcription by RNA Pol I and Pol III. More importantly, as seen for pRb, the expression of p53 can be detected both in the nucleus and in the nucleolus in nondividing and relatively stable cells (7, 43). Therefore, modulation of Pol I and Pol III transcription by p53 and pRb may be part of a similar strategy adopted by these tumor suppressor proteins for the control of cell growth.
ACKNOWLEDGMENTS
We are grateful to Thomas Shenk for pGThp53N, pGThp53C1, pGThp53C2, and pGThp53C constructs, Axel Schonthal for mouse embryonic fibroblast cells, Nancy Rabb-Traub and Arnold Berk for H1299 and H1299-tsp53 cells, and Lawrence Rothblum for antiserum against the large subunit of RNA Pol I. The technical assistance of Tiffany Bui is acknowledged.
This work was funded by a grant to L.C. (R01 GM53949) from the National Institutes of Health.
REFERENCES
- 1.Agoff S N, Hou J, Linzer D I, Wu B. Regulation of the human hsp70 promoter by p53. Science. 1993;259:84–87. doi: 10.1126/science.8418500. [DOI] [PubMed] [Google Scholar]
- 2.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 3.Bell S P, Learned R M, Jantzen H M, Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P, Pikaard C S, Reeder R H, Tjian R. Molecular mechanisms governing species-specific transcription of ribosomal RNA. Cell. 1989;59:489–497. doi: 10.1016/0092-8674(89)90032-9. [DOI] [PubMed] [Google Scholar]
- 5.Benninghoff J, Kartarius S, Teleb Z, Selter H, Unteregger G, Zwergel T, Wullich B, Montenarh M. Two different forms of p53 localized differently within cells of urogenital tumours. Cancer Lett. 1999;144:55–64. doi: 10.1016/s0304-3835(99)00187-1. [DOI] [PubMed] [Google Scholar]
- 6.Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- 7.Bukovsky A, Caudle M R, Keenan J A, Wimalasena J, Foster J S, Upadhyaya N B, van Meter S E. Expression of cell cycle regulatory proteins (p53, pRb) in the human female genital tract. J Assist Reprod Genet. 1995;12:123–131. doi: 10.1007/BF02211381. [DOI] [PubMed] [Google Scholar]
- 8.Cairns C A, White R J. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 1998;17:3112–3123. doi: 10.1093/emboj/17.11.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 10.Chesnokov I, Chu W M, Botchan M R, Schmid C W. p53 inhibits RNA polymerase III-directed transcription in a promoter-dependent manner. Mol Cell Biol. 1996;16:7084–7088. doi: 10.1128/mcb.16.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Crystal structure of a p53 tumor supppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 12.Chu W M, Wang Z, Roeder R G, Schmid C W. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J Biol Chem. 1997;272:14755–14761. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 13.Comai L, Tanese N, Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 14.Dang C V, Lee W M. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J Biol Chem. 1989;264:18019–18023. [PubMed] [Google Scholar]
- 15.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 16.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 17.El-Deiry W S. p21/p53, cellular growth control and genomic integrity. Curr Top Microbiol Immunol. 1998;227:121–137. doi: 10.1007/978-3-642-71941-7_6. [DOI] [PubMed] [Google Scholar]
- 18.Ewen M E, Oliver C J, Sluss H K, Miller S J, Peeper D S. p53-dependent repression of CDK4 translation in TGF-beta-induced G1 cell-cycle arrest. Genes Dev. 1995;9:204–217. doi: 10.1101/gad.9.2.204. [DOI] [PubMed] [Google Scholar]
- 19.Fontoura B M, Sorokina E A, David E, Carroll R B. p53 is covalently linked to 5.8S rRNA. Mol Cell Biol. 1992;12:5145–5151. doi: 10.1128/mcb.12.11.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fries K L, Miller W E, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginsberg D, Mechta F, Yaniv M, Oren M. Wild-type p53 can down-modulate the activity of various promoters. Proc Natl Acad Sci USA. 1991;88:9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog Nucleic Acid Res Mol Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- 23.Haffner R, Oren M. Biochemical properties and biological effects of p53. Curr Opin Genet Dev. 1995;5:84–90. doi: 10.1016/s0959-437x(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 24.Horikoshi N, Usheva A, Chen J, Levine A J, Weinmann R, Shenk T. Two domains of p53 interact with the TATA-binding protein, and the adenovirus 13S E1A protein disrupts the association, relieving p53-mediated transcriptional repression. Mol Cell Biol. 1995;15:227–234. doi: 10.1128/mcb.15.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kern S E, Kinzler K W, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991;252:1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- 26.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 27.Learned R M, Learned T K, Haltiner M M, Tjian R T. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell. 1986;45:847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- 28.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 29.Mack D H, Vartikar J, Pipas J M, Laimins L A. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nature. 1993;363:281–283. doi: 10.1038/363281a0. [DOI] [PubMed] [Google Scholar]
- 30.Manley J L, Fire A, Samuels M, Sharp P A. In vitro transcription: whole-cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 31.Marechal V, Elenbaas B, Piette J, Nicolas J C, Levine A J. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin D W, Munoz R M, Subler M A, Deb S. p53 binds to the TATA-binding protein-TATA complex. J Biol Chem. 1993;268:13062–13067. [PubMed] [Google Scholar]
- 33.Martin D W, Subler M A, Munoz R M, Brown D R, Deb S P, Deb S. p53 and SV40 T antigen bind to the same region overlapping the conserved domain of the TATA-binding protein. Biochem Biophys Res Commun. 1993;195:428–434. doi: 10.1006/bbrc.1993.2061. [DOI] [PubMed] [Google Scholar]
- 34.Mercer W E, Amin M, Sauve G J, Appella E, Ullrich S J, Romano J W. Wild type human p53 is antiproliferative in SV40-transformed hamster cells. Oncogene. 1990;5:973–980. [PubMed] [Google Scholar]
- 35.Michalovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 36.Mogaki M, Hirota M, Chaney W G, Pour P M. Comparison of p53 protein expression and cellular localization in human and hamster pancreatic cancer cell lines. Carcinogenesis. 1993;14:2589–2594. doi: 10.1093/carcin/14.12.2589. [DOI] [PubMed] [Google Scholar]
- 37.Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 1995;14:4442–4449. doi: 10.1002/j.1460-2075.1995.tb00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paule M R. Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. New York, N.Y: Springer-Verlag Inc.; 1998. [Google Scholar]
- 39.Pietenpol J A, Tokino T, Thiagalingam S, el-Deiry W S, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragimov N, Krauskopf A, Navot N, Rotter V, Oren M, Aloni Y. Wild-type but not mutant p53 can repress transcription initiation in vitro by interfering with the binding of basal transcription factors to the TATA motif. Oncogene. 1993;8:1183–1193. [PubMed] [Google Scholar]
- 41.Redeer R. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol. 1999;62:293–327. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- 42.Rogalsky V, Todorov G, Moran D. Translocation of retinoblastoma protein associated with tumor cell growth inhibition. Biochem Biophys Res Commun. 1993;192:1139–1146. doi: 10.1006/bbrc.1993.1535. [DOI] [PubMed] [Google Scholar]
- 43.Rubbi C, Milner J. Non-activated p53 co-localizes with sites of trasncription within both the nucleoplasm and the nucleolus. Oncogene. 2000;19:85–96. doi: 10.1038/sj.onc.1203378. [DOI] [PubMed] [Google Scholar]
- 44.Seto E, Usheva A, Zambetti G P, Momand J, Horikoshi N, Weinmann R, Levine A J, Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci USA. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thut C J, Chen J L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 46.Voit R, Schafer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weisenberger D, Scheer U. A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis. J Cell Biol. 1995;129:561–575. doi: 10.1083/jcb.129.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White R J, Trouche D, Martin K, Jackson S P, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 49.Yun J, Chae H D, Choy H E, Chung J, Yoo H S, Han M H, Shin D Y. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J Biol Chem. 1999;274:29677–29682. doi: 10.1074/jbc.274.42.29677. [DOI] [PubMed] [Google Scholar]
- 50.Zambetti G P, Bargonetti J, Walker K, Prives C, Levine A J. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- 51.Zhai W, Comai L. A kinase activity associated with simian virus 40 large T antigen phosphorylates upstream binding factor (UBF) and promotes formation of a stable initiation complex between UBF and SL1. Mol Cell Biol. 1999;19:2791–2802. doi: 10.1128/mcb.19.4.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhai W, Tuan J A, Comai L. SV40 large T antigen binds to the TBP-TAF(I) complex SL1 and coactivates ribosomal RNA transcription. Genes Dev. 1997;11:1605–1617. doi: 10.1101/gad.11.12.1605. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W, Funk W D, Wright W E, Shay J W, Deisseroth A B. Novel DNA binding of p53 mutants and their role in transcriptional activation. Oncogene. 1993;8:2555–2559. [PubMed] [Google Scholar]