Abstract
Background:
Epigenetic alterations play an important role in tumorigenesis. Hypermethylation of CpG islands within the promoter regions of tumor suppressor genes (TSGs) and histone deacetylation lead to the silencing of the genes resulting in cancer induction. The suppressor of cytokine signaling (SOCS) family is an important negative regulator of cytokine signaling and deregulation of this family has been reported in several cancers, the protein of the SOCS family inhibit the cytokine-activated Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling pathway to modulate cellular responses. Previously, we evaluated the effects of DNA demethylating agents and histone deacetylase inhibitors on hepatocellular carcinoma (HCC). The current study aimed to investigate the effect of decitabine (5-aza-2ˈ-deoxycytidine, 5-aza-CdR) in comparison to vorinostat (suberoylanilide hydroxamic acid, SAHA) on DNMT1, DNMT3a and DNMT3b, HDAC 1-3, SOCS 1, SOCS 3, JAK2, and STAT3 gene expression, cell growth inhibition, and apoptosis induction of HCC HLE and LCL-PI 11 cell lines.
Material and Methods:
The HLE and LCL-PI 11 cells were treated with 5-aza-CdR and SAHA and then the MTT assay, flow cytometry assay, and quantitative real-time RT-PCR were achieved to determine cell viability, cell apoptosis, and relative gene expression respectively.
Results:
The result indicated that both compounds inhibited cell growth, induced apoptosis, and down-regulated DNMT1, DNMT3a DNMT3b, HDAC 1-3, JAK2, and STAT3 and up-regulated HDAC 1-3, SOCS 1, and SOCS 3 genes expression significantly. The apoptotic effect of SAHA was stronger than that of 5-Aza-CdR.
Conclusion:
5-Aza-CdR and SAHA can induce cell growth inhibition and apoptosis induction through the JAK/STAT pathway.
Key Words: Decitabine, vorinostat, SOCS, JAK/STAT, HCC
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent and lethal cancers worldwide. The molecular mechanisms and pathways leading to the diseases are extremely complicated including genomic, genetic, and epigenetic changes (Pogribny et al., 2014). There are several evidences that epigenetic alterations play an important role in tumorigenesis, cancer progression, and pathogenesis. These epigenetic changes include DNA methylation and histone modifications which lead to heritable changes in gene expression without alterations in chromatin structure. Hypermethylation of CpG islands within the promoter regions of tumor suppressor genes (TSGs) leads to the silencing of the genes resulting in cancer induction. In HCC, hypermethylation of TSGs involved in cancer induction and progression has been demonstrated (Hoshida et al., 2010). DNA methyltransferases (DNMTs) are the enzymes that are overexpressed in cancer cells. DNMTs transfer a methyl group to one of the four bases that constitute the coding sequence of DNA and catalyze the methylation of CpG islands resulting in DNA methylation. Mammalian DNMTs can be divided into DNMT1, DNMT3a, DNMT3b, and DNMT3l (Turek-Plewa et al., 2005).
A significant increase in the mRNA levels of DNMT1, DNMT3a, and DNMT3b has been reported in HCCs (Oh. Et al., 2007). Two DNMT inhibitors (DNMTIs) have been approved by the US Food and Drug Administration (FDA) including decitabine (5-aza-2ˈ-deoxycytidine, 5-aza-CdR) and azacytidine (Vidaza; Celgene) (Gnyszka et al., 2013). In addition to DNMTIs, flavonoids are a class of plant secondary metabolites that have been reported to interfere in the prevention and initiation of cancer by modulating different mechanisms and pathways related to cellular differentiation, proliferation, and apoptosis (Ravishankar et al., 2013). It has been demonstrated that bioflavonoids such as quercetin, genistein, fisetin can inhibit DNMT activity (Nebbioso et al., 2012). Previously, we evaluated the effects of DNA demethylating agent 5-aza-CdR on DNMT1 gene expression and apoptosis induction and also cell viability in the HCC WCH-17 cell line [Kavoosi et al., 2019; Sanaei et al., 2020; Sanaei et al., 2019).
As mentioned above, histone modification modulates chromatin structure and affects gene transcription and expression. This modification is a reversible process and result of a balance between the opposing activities of histone deacetylases (HDACs) and histone acetyltransferases (HATs). There are 18 potential human HDACs classified into four classes, including class I (HDAC1, 2, 3, and 8), class II which divided into two subclasses: IIa (HDAC4, 5, 6, 7, and 9) and IIb (HDAC6 and 10), class III which are NAD-dependent protein deacetylases and/or ADP ribosylases and class IV which contains only HDAC11 (Li et al., 2016). By removal of acetyl groups from histones, these enzymes create a chromatin conformation that prevents the transcription of genes that encode several proteins involved in both cancer initiation and cancer progression (Glozak et al., 2007). HDAC1-3 activity increase HCC (Quint et al., 2011; Rikimaru et al., 2007). HDAC inhibitors (HDACIs) can regulate gene expression through the acetylation of histones and non-histone proteins without changing the DNA sequence. Based on chemical structure, these agents can be classified into several groups, including hydroxamic acids [vorinostat, (suberoylanilide hydroxamic acid, SAHA)], aminobenzamides (mocetinostat, entinostat), carboxylic acids (sodium butyrate and valproate), epoxyketones (trapoxins), cyclic peptides (romidepsin and apicidin), and hybrid molecules (West. et al., 2014). The suppressor of cytokine signaling (SOCS) family is an important negative regulator of cytokine signaling and deregulation of this family has been reported in several cancers, the protein of the SOCS family plays a key role in the negative regulation of cytokine signal transduction. These proteins inhibit the cytokine-activated Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling pathways to modulate cellular responses. The SOCS family consists of eight members, including SOCS-1 to SOCS-7 and cytokine-inducible SH2 protein. SOCS1 and 3 appear to have tumor suppressor activity in HCC cells (Kim et al., 2015; Puhr et al., 2009). Hypermethylation and deacetylation of the SOCS family have been shown in numerous solid cancers (Kim. Et al., 2015). Our previous finding indicated that histone deacetylase inhibitors valproic acid (VPA) and trichostatin A (TSA) can induce apoptosis in HCC (Kavoosi et al., 2018; Sanaei et al., 2018; Sanaei et al., 2017; Sanaei et al., 2019). The aim of the current study was to investigate the effect of 5-aza-CdR in comparison to SAHA on DNMT1, DNMT3a and DNMT3b, HDAC 1-3, SOCS 1, SOCS 3, JAK2, and STAT3 gene expression, cell growth inhibition, and apoptosis induction of HCC HLE and LCL-PI 11 cell lines.
Materials and Methods
Materials
The human hepatocellular carcinoma HLE and LCL-PI 11 cell lines were purchased from the National Cell Bank of Iran Pasteur Institute. 5-aza-CdR, SAHA, 3-(4,5-dimethyl-2-thiazyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), and Dulbecco’s modified Eagle medium (DMEM) were supplied by Sigma–Aldrich (Sigma–Aldrich, Louis, MI, USA). The Annexin V and propidium iodide (PI) apoptosis kit was purchased from Life Technologies. Dimethyl sulfoxide (DMSO) was purchased from Merck Co. (Darmstadt, Germany). Total RNA extraction kit (TRIZOL reagent) and real-time polymerase chain reaction (PCR) kits (qPCR MasterMix Plus for SYBR Green I dNTP) were obtained from Applied Biosystems Inc. (Foster, CA, USA). This work was approved by the Ethics Committee of Jahrom University of Medical science with a code number of IR.JUMS.REC.1396.154.
Cell culture
Both cell lines were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 1% nonessential amino acids, 1% antibiotics (penicillin/streptomycin), and grown in a humidified incubator at 37°C containing 5% CO2. SAHA and 5-Aza-CdR were dissolved at a concentration of 100 μM in DMSO to prepare a stock solution and all of the other test concentrations were provided by dilution of this solution. The final DMSO concentration did not exceed 0.1% and all control groups were administered 0.1% DMSO concentration.
Cell viability assay
The effect of 5-Aza-CdR and SAHA on the HLE and LCL-PI 11 cell viability was measured by MTT assay. First, the HLE and LCL-PI 11 cells were cultured with a culture medium. After enough confluency (more than 80%), 4 ×105 cells per well were transferred into 96-well plates and allowed to adhere overnight. After 24 hours of cell seeding, the cells were treated with medium containing different doses of 5-Aza-CdR (1, 2.5, 5, 10, and 20 μM) and SAHA (1, 2.5, 5, 10, and 20 μM) except control groups for different periods, an equal volume of solvent, DMSO, was added to control experiments. After exposure to the various concentrations of the compounds for 24 and 48 h, the cells were trypsinized and the viable cell population was determined using the MTT assay.
Flow cytometric analysis of apoptosis
To detection apoptotic cells, the apoptotic cells were assessed using the Annexin V-FITC/PI detection kit. In this regard, the HLE and LCL-PI 11 cells were cultured in 24-well plates at a density of 4 × 105 cells/well and incubated for 24h before treatment with a medium containing SAHA and 5-Aza-CdR. After 24h, the cells treated with 5-Aza-CdR and SAHA, based on IC50 values indicated in table 1, for 24 and 48h. After treatment times, all the adherent cells were harvested with trypsin- EDTA washed with cold PBS, and resuspended in Binding buffer (1×). Annexin-V-(FITC) and PI were used for staining according to the protocol. Finally, the apoptotic cells were counted by the FACScanTM flow cytometer (Becton Dickinson, Heidelberg, Germany).
Table 1.
IC50 Values
| Drug | Duration/h | IC50 value/ μM | LogIC50 | R squared |
|---|---|---|---|---|
| 5-Aza-CdR | 24 | 2.62 | 0.4193 | 0.9765 |
| 5-Aza-CdR | 48 | 1.4 | 0.1464 | 0.9478 |
| SAHA | 24 | 4.19 | 0.6228 | 0.989 |
| SAHA | 48 | 3.37 | 0.5278 | 0.9064 |
Determination of gene expression by quantitative real-time RT-PCR
To determine DNMT1, DNMT3a, DNMT3b, HDAC 1-3, SOCS 1, SOCS 3, JAK2, and STAT3 genes expression in HLE and LCL-PI 11 cells, the cells were treated with 5-Aza-CdR and SAHA, based on IC50 values indicated in table 1, for 24 and 48 h. After exposure to the compounds, the total RNA of the control and treated cells was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol and then pretreated with RNase-free DNase (Qiagen) to remove the genomic DNA prior to cDNA synthesis. The RNA concentration was determined using a Biophotometer (Eppendorf, Hamburg, Germany). Total RNA (100 ng) was reverse transcribed to cDNA using the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Real-time RT-PCR was performed with the Maxima SYBR Green ROX qPCR Master Mix Kit (Fermentas) as described previously (Sanaei et al., 2018). The primer sequences were obtained from our previous and other published articles (Sanaei et al., 2018; Nemati et al., 2018, Sutherland et al., 2004; Zhou et al., 2014; Wang et al., 2017) which their sequences are shown in Table 2. GAPDH was used as an endogenous control.
Table 2.
Real-Time PCR Primers Sequences Used in the Current Study
| Primer name | Primer sequences (5’ to 3’) | Product length | Reference |
|---|---|---|---|
| DNMT1 Forward | GAG GAA GCT GCT AAG GAC TAG TTC | 206 | 23 |
| DNMT1 Reverse | ACT CCA CAA TTT GAT CAC TAA ATC | 23 | |
| DNMT3a Forward | GGA GGC TGA GAA GAA AGC CAA GGT | 370 | 23 |
| DNMT3a Reverse | TTT GCC GTC TCC GAA CCA CAT GAC | 23 | |
| DNMT3b Forward | TAC ACA GAC GTG TCC AAC ATG GGC | 195 | 23 |
| DNMT3b Reverse | GGA TGC CTT CAG GAA TCA CAC CTC | 23 | |
| HDAC 1 Forward | AAC TGG GGA CCT ACGG | 230 | 24 |
| HDAC 1 Reverse | ACT TGG CGT GTC CTT | 24 | |
| HDAC 2 Forward | GTT GCT CGA TGT TGG AC | 212 | 24 |
| HDAC 2 Reverse | CCA GGT GCA TGA GGTA | 24 | |
| HDAC 3 Forward | TAGGGATGAGATACAGACAAGG | 103 | 25 |
| HDAC 3 Reverse | GAAGCAGGGAAGAAATAAGG | 25 | |
| SOCS-1 Forward | AGAC CCCTTCTCACCTCTTG | 251 | 26 |
| SOCS-1 Reverse | CTGCACAGCA GAAAATAAAGC | 26 | |
| SOCS-3 Forward | TCCCCCCAGAAGAGCCTATTAC | 107 | 26 |
| SOCS-3 Reverse | TCCG ACAGAGATGCTGAAGAGTG | 26 | |
| JAK2 Forward | AGCCTATCGGCATGGAATATCT | 163 | 27 |
| JAK2 Reverse | TAACACTGCCATCCCAAGACA | 27 | |
| STAT3 Forward | ACCAGCAGTATAGCCGCTTC | 124 | 27 |
| STAT3 Reverse | GCCACAATCCGGGCAATCT | 27 | |
| GAPDH Forward | AGCCACATCGCTCAGACAC | 66 | 28 |
| GAPDH Reverse | GCCCAATACGACCAAATCC | 28 |
Statistical analysis
The database was set up with the SPSS 16.0 software package (SPSS Inc., Chicago, Illinois, USA) and Graph Pad Prism 8.0 for data analysis. Results are expressed as mean ±standard deviation (SD) for n=3 independent experiments. Statistical comparisons between groups were performed with ANOVA (oneway ANOVA) and the Turkey test. A significant difference was considered as P < 0.05.
Results
In vitro effects of 5-Aza-CdR and SAHA on the HLE and LCL-PI 11 cells growth
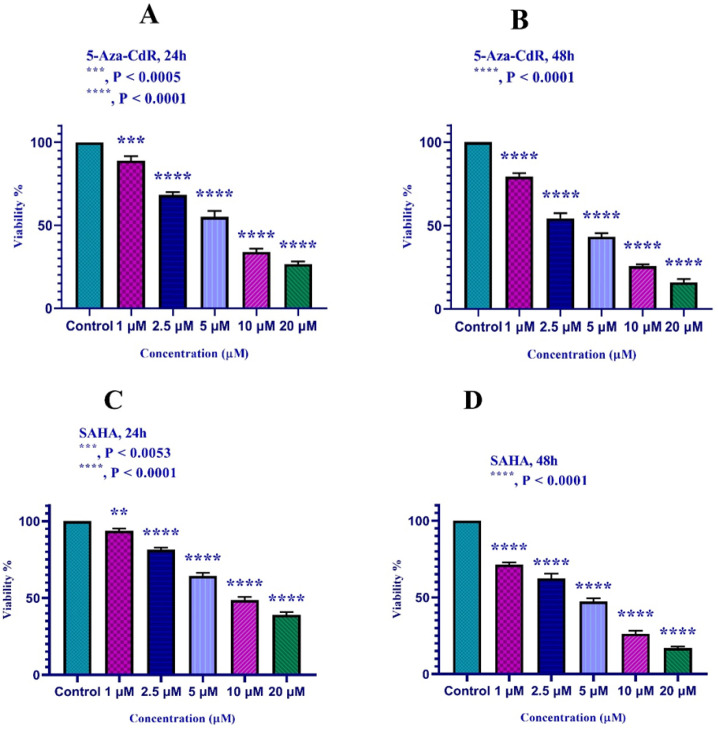
The HLE and LCL-PI 11 cells were treated with different doses of 5-Aza-CdR and SAHA vs control groups for 24 and 48h. As mentioned above, the inhibitory effects of the compounds were evaluated by MTT assay. The results indicated that both compounds can inhibit HLE and LCL-PI 11 cell growth significantly versus control groups. As shown in figure 1, 5-Aza-CdR and SAHA significantly inhibited cell growth with all mentioned doses (p < 0.001). IC50 values for 5-Aza-CdR and SAHA are indicated in Table 1.
Figure 1.
In vitro Effects of 5-Aza-CdR and SAHA on HLE (part A and B) and LCL-PI 11 (part C and D) cells viability. Values are means of three experiments in triplicate. Asterisks (*) demonstrate significant differences between treated and untreated control groups. Results are expressed as mean ±standard deviation (SD) for n=3 independent experiments. Part A: ***, P < 0.0005; ****, P < 0.0001; Part B: ****, P < 0.0001; Part C: ***, P < 0.0053, ****, P < 0.0001; Part D: ****, P < 0.0001
Effects of 5-Aza-CdR and SAHA on the HLE and LCL-PI 11 cells apoptosis
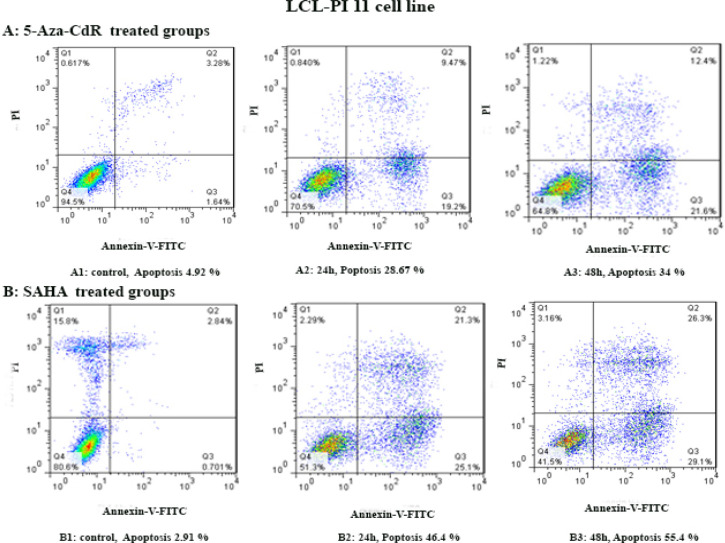
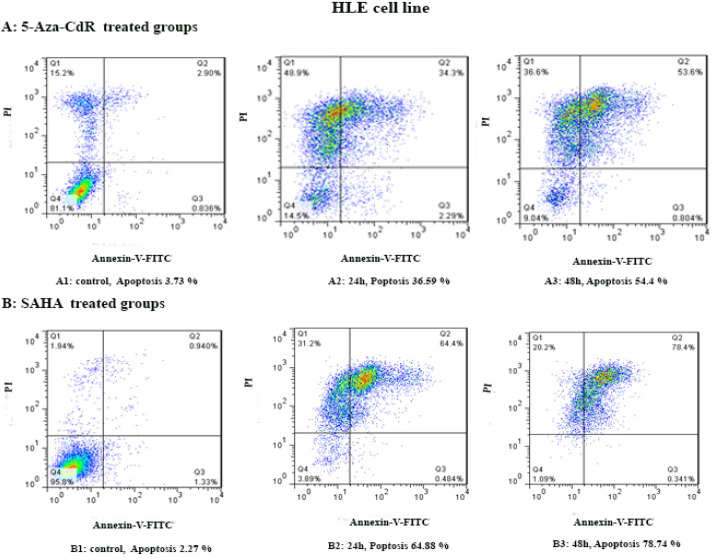
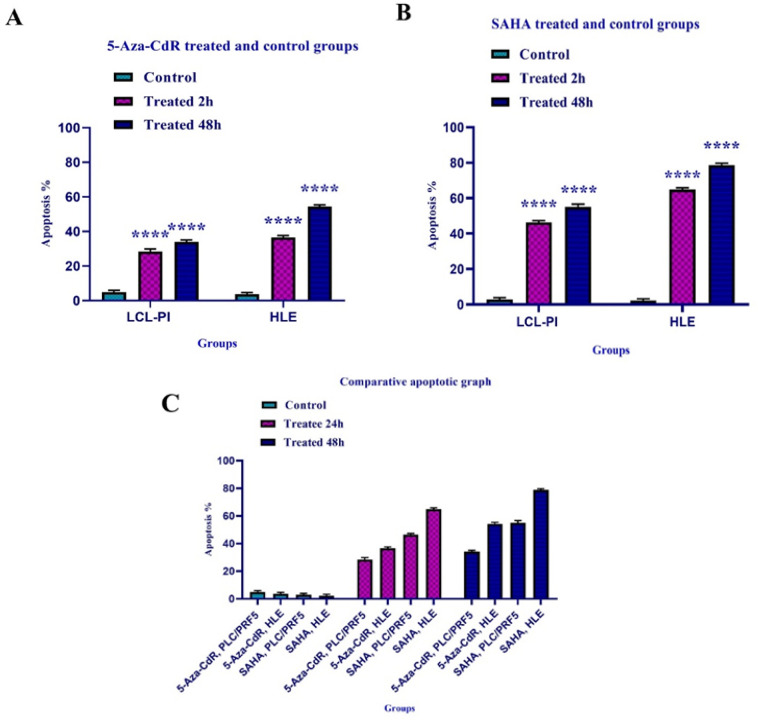
By flow cytometry assay, the apoptotic cells were determined. In this regard, the cells were treated with 5-Aza-CdR and SAHA for 24 and 48 h except for the control groups. Both compounds induced apoptosis significantly as indicated in Figures 2 and 3. The percentage of apoptotic cells is indicated in table 3. Relative analysis indicated that SAHA induced apoptosis more significantly than 5-Aza-CdR (P<0.0001), Figure 4. Maximal apoptosis was seen in the group which received SAHA for 48h.
Figure 2.
The Apoptotic Effect of 5-Aza-CdR and SAHA on LCL-PI 11 Cell Line. Results were obtained from three independent experiments. Quadrant (Q) 2 and 3, late and primary apoptosis respectively, were calculated in this graph
Figure 3.
The Apoptotic Effect of 5-Aza-CdR and SAHA on the HLE Cell Line. Results were obtained from three independent experiments. Quadrant (Q) 2 and 3, late and primary apoptosis respectively, were calculated in this graph
Table 3.
The Percentage of Apoptotic Cells Treated with 5-Aza-CdR and SAHA for 24 and 48 h
| Drug | Cell line | Duration (h) | Apoptosis (%) | P-value |
|---|---|---|---|---|
| 5-Aza-CdR | LCL-PI 11 | 24 | 28.67 | 0.001 |
| 5-Aza-CdR | LCL-PI 11 | 48 | 34 | 0.001 |
| 5-Aza-CdR | HLE | 24 | 36.59 | 0.001 |
| 5-Aza-CdR | HLE | 48 | 54.4 | 0.001 |
| SAHA | LCL-PI 11 | 24 | 46.4 | 0.001 |
| SAHA | LCL-PI 11 | 48 | 55.4 | 0.001 |
| SAHA | HLE | 24 | 64.88 | 0.001 |
| SAHA | HLE | 48 | 78.74 | 0.001 |
Figure 4.
The Apoptotic Effect of 5-Aza-CdR (part A) and SAHA (part B) on HLE and LCL-PI 11 Cells versus Control Groups. The effect of 5-Aza-CdR in comparison to SAHA on HLE and LCL-PI 11 cells has been shown in part C. Results were obtained from three independent experiments and were expressed as mean ± standard error of the mean. The results of the statistical analysis indicate significant differences between treated and untreated cells. ****, P<0.0001
Effects of 5-Aza-CdR and SAHA on gene expression
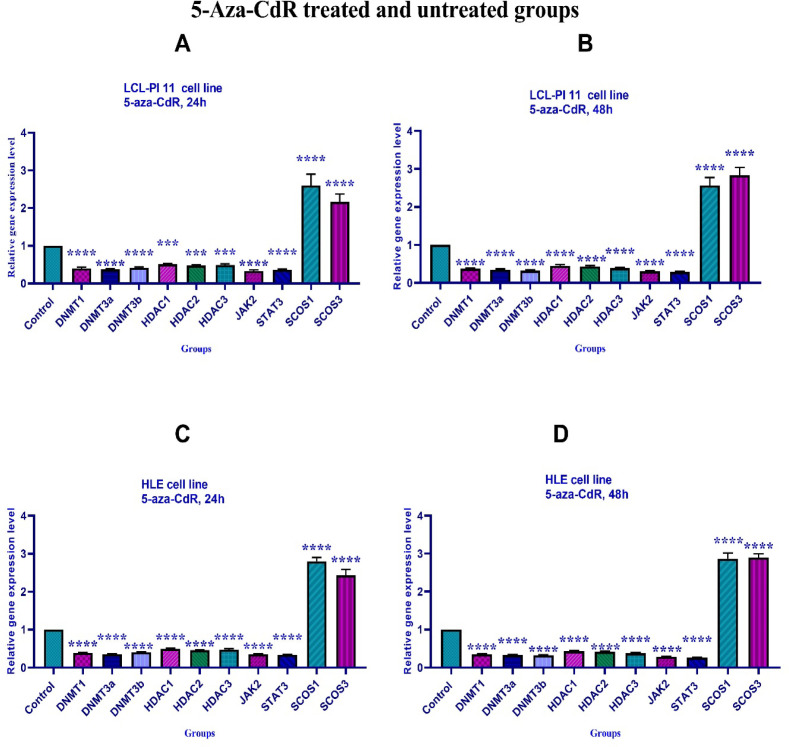
To determine the effect of 5-Aza-CdR and SAHA on DNMT1, DNMT3a, DNMT3b, HDAC 1-3, SOCS 1, SOCS 3, JAK2, and STAT3 genes expression, real-time RT-PCR was carried out. The results indicated that both compounds significantly down-regulated DNMT1, DNMT3a, DNMT3b, HDAC 1-3, JAK2, and STAT3 and up-regulated SOCS 1, and SOCS 3 gene expression at different periods (24 and 48 h), Figures 5 and 6.
Figure 5.
The Relative Expression Level of DNMT1, DNMT3a, DNMT3b, HDAC 1-3, SOCS 1, SOCS 3, JAK2, and STAT3 Genes in the HLE and LCL-PI 11 Cell Lines Treated with 5-Aza-CdR. A significant difference was seen between treated and untreated control groups. Asterisks (*) indicate significant differences between the treated and untreated control groups. Results are expressed as mean±standard deviation (SD) for n=3 independent experiments. Part A: ***, P<0.0008, ****, P<0.0001; Part B: ****, P<0.0001; Part C: ****, P<0.0001; Part D: ****, P<0.0001
Figure 6.
The Relative Expression Level of DNMT1, DNMT3a, DNMT3b, HDAC 1-3, SOCS 1, SOCS 3, JAK2, and STAT3 Genes in the HLE and LCL-PI 11 Cell Lines Treated with SAHA. A significant difference was seen between treated and untreated control groups. Asterisks (*) indicate significant differences between the treated and untreated control groups. Results are expressed as mean±standard deviation (SD) for n=3 independent experiments. Part A: ****, P<0.0001; Part B: ****, P<0.0001; Part C: ****, P<0.0001; Part D: ****, P<0.0001
Discussion
Acetylation of lysine is regulated by the opposing activity of two groups of enzymes including HDACs and HATs. The HATs catalyze the transfer of an acetyl group to the ε-amino group of lysine thereby neutralize the lysine’s positive charge by which weak the interactions between histones and DNA and facilitates transcription. There are two major groups of HATs including type-A and Type-B (Bannister et al., 2011). A well-known characteristic of cancer is the deregulation of posttranslational histone modifications in particular histone acetylation. Several studies have demonstrated that a histone deacetylation is a common event in human cancer (Ropero et al., 2007). In addition to histone deacetylation, aberrant expression of DNMTs leads to DNA methylation which is closely associated with tumorigenesis (Jin et al., 2013). Several compounds such as butyric acid, SAHA, and TSA can inhibit HDAC activity thereby inhibit the cell growth of transformed cells and induce apoptosis (Marks et al., 2000). Previously, we reported the effect of zebularine on class I HDACs (HDACs 1, 2, 3) and class II HDACs (HDACs 4, 5, 6) gene expression in colon cancer LS 180 and LS 174T cell lines (Sanaei et al., 2020; Sanaei et al., 2020).
DNA methyltransferase inhibitors such as 5-aza-CR and vidaza have recently been approved as an anticancer drugs. The 5-aza-CR is the most widely used DNMTs inhibitor characterized 25 years ago, this compound is a cytidine analog that inhibits DNA methyltransferases (Stresemann et al., 2006). In the present study, we report that 5-Aza-CdR and SAHA significantly down-regulated DNMT1, DNMT3a, DNMT3b and HDAC 1-3, JAK2, and STAT3 and up-regulated SOCS 1, and SOCS 3 gene expression at different periods (24 and 48 h) in HLE and LCL-PI 11 cell lines. In line with our report, the inhibitory and apoptotic effect of SAHA has been reported on HCC HepG2 (Wang et al., 2013), cervix cancer, colon, and rectum carcinoma (Anantharaju et al., 2017). As we reported, SAHA could induce apoptosis by down-regulation of DNMTs. Other studies have been reported that SAHA induces the expression of apoptosis signal-regulating kinase 1 (ASK1), which induces apoptosis via death receptor and intracellular apoptotic pathways. Furthermore, this compound activates the expression of pro-apoptotic proteins, including Bak and Bax, and inhibits the expression of anti-apoptosis proteins, including Bcl-xL and Bcl-2 resulting in apoptotic induction in tumor cells. Another pathway is the induction of transcription of CDK inhibitor p21/wafl leads to the reduction of cell proliferation and induction of apoptosis (Tan et al., 2006). SAHA can arrest tumor cells in the G0/G1 phase and induce apoptosis which may be related to the increased expression of p21/waf1. It can also induce apoptosis by influencing the expression of p27 and p53 (Hirata et al., 2011; Grimes et al., 2011). Several studies have demonstrated that SAHA can activate caspase-3 to promote apoptosis by activation of TRAIL-DISC in HCC (Liu et al., 2018).
Apoptosis induction of HCC cells by DNMTs down-regulation was one of the results of our work. Inconsistent with our finding, it has been shown that 5-aza-CdR reactivates TSGs through down-regulation of DNMT1, DNMT3a, and 3b resulting in rapid demethylation (Ghoshal et al., 2008). Similarly, it has been indicated that 5-Aza-CdR inhibits the activity of DNMT3a and DNMT3b in gastric cancer cell lines SGC7901 and BGC823 thereby up-regulate tumor suppressor gene RASSF1A. The effect of 5-Aza-CdR co-treatment with sodium butyrate on DNMT3a and DNMT3b inhibition is significantly higher than that of 5-AzaCdR alone (Shen et al., 2008). In mouse hippocampus-derived neuronal HT22 cell line, 5-Aza-CdR down-regulates the expression of mRNA and protein DNMT1 and 3a (but no DNMT 3b) (Yang et al., 2017). In myelodysplastic syndrome (MDS), 5-Aza-CdR can re-activate the expression of the p15INK4B gene in MUTZ-1 cells by demethylation of the p15INK4B gene through inhibition of DNMT3a gene expression which is a mechanism of 5-Aza-CdR in the treatments of MDS (Tong et al., 2004). It has been shown that SAHA inhibits cell growth and induces apoptosis in pancreatic cancer by inhibiting HDAC1, 3, and 4 (Kumagai et al., 2007). In uterine cancer, it has been reported that SAHA efficiently suppresses MES-SA uterine sarcoma cell growth by down-regulation of HDACs class I (HDAC2 and 3) and class II (HDAC7).
Our findings indicated that SAHA down-regulated HDAC 1-3 significantly. Similarly, our previous work demonstrated that histone deacetylase inhibitor trichostatin A can down-regulate class I HDACs (HDACs 1, 2, 3) and class II HDACs (HDACs 4, 5, 6) gene expression in colon cancer LS 180 and LS 174T cell lines (Sanaei et al., 2020; Sanaei et al., 2020). In line with our report, it has been shown that SAHA can down-regulate HDACs class I (HDAC2 and 3) and class II (HDAC7) resulting in p21WAF1 upregulation and apoptosis induction in uterine cancer (Hrzenjak et al., 2010).
Similar to our result, other researchers have indicated that histone deacetylase inhibitor TSA suppresses the growth of colorectal cancer cells, and induces apoptosis through SOCS1 and SOCS3 upregulation and JAK2/STAT3 signaling inhibition (Xiong et al., 2012). Furthermore, in vitro studies have shown that 5-Aza and TSA can reactivate the SOCS-3 gene in acute myeloid leukemia (AML) cells (Johan et al., 2015). Inconsistently, our previous work indicated that valproic acid and zebularine can up-regulate SOCS-1 and SOCS-3 gene expression leads to cell growth inhibition in the colon carcinoma SW48 cell line (Sanaei et al., 2020). In summary 5-Aza-CdR and SAHA can induce apoptosis through the JAK/STAT pathway. We didn’t evaluate the protein assessment of the mentioned genes. Therefore, the evaluation of these proteins is recommended.
In conclusion, 5-Aza-CdR and SAHA can epigenetically induce apoptosis through up-regulation of SOCS 1 and SOCS 3 gene expression and JAK2/STAT3 signaling inhibition in HLE and LCL-PI 11 cell lines.
Author Contribution Statement
All authors generated the ideas and contributed to the writing of the manuscript, acquisition of data, analysis, interpretation of data, critical revision of the manuscript for important intellectual content, statistical analysis, and technical or material support. All authors approved the final revision.
Acknowledgments
Thanks are given to our group members for performing the MTT assay, real-time PCR, and flow cytometry. This work was supported by the adjutancy of research of Jahrom medical University-Iran.This article was supported by the adjutancy of research of Jahrom medical University-Iran.
Conflict of interest
The authors report no conflict of interest.
References
- Anantharaju PG, Reddy DB, Padukudru MA, et al. Induction of colon and cervical cancer cell death by cinnamic acid derivatives is mediated through the inhibition of Histone Deacetylases (HDAC) PLoS One. 2017;12:e0186208. doi: 10.1371/journal.pone.0186208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Datta J, Majumder S, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–41. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Glozak M, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- Gnyszka A, Jastrzebski Z, Flis S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013;33:2989–96. [PubMed] [Google Scholar]
- Grimes MJ, Picchi MA, Mitchell HD, et al. Combination therapy with vidaza and entinostat suppresses tumor growth and reprograms the epigenome in an orthotopic lung cancer model. Cancer Res. 2011;71:454–62. doi: 10.1158/0008-5472.CAN-10-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, Nakajima K, et al. Wnt antagonist DKK1 acts as a tumor suppressor gene that induces apoptosis and inhibits proliferation in human renal cell carcinoma. Int J Cancer. 2011;128:1793–803. doi: 10.1002/ijc.25507. [DOI] [PubMed] [Google Scholar]
- Hoshida Y, Toffanin S, Lachenmayer A, et al. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis. 2010;30:35–51. doi: 10.1055/s-0030-1247131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrzenjak A, Moinfar F, Kremser M-L, et al. Histone deacetylase inhibitor vorinostat suppresses the growth of uterine sarcomas in vitro and in vivo. Mol Cancer. 2010;9:49–54. doi: 10.1186/1476-4598-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Laban M, Leung CH, et al. Inhibition of histone deacetylase 2 increases apoptosis and p21 Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differentiation. 2005;12:395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- Johan MF, Jusoh SAM. AB119 Induction of suppressor of cytokine signaling-3 in FLT3-ITD positive MV4-11 acute myeloid leukemia cells in response to 5-Azacytidine and Trichostatin A. Ann Transl Med. 2015;3:78–85. [Google Scholar]
- Kavoosi F, Sanaei M. Comparative analysis of the effects of Valproic Acid and Tamoxifen on proliferation, and apoptosis of human hepatocellular carcinoma WCH 17 CellLin. Iran J Ped Hematol Oncol. 2018;8:12–20. [Google Scholar]
- Kavoosi F, Sanaei M. Effects of 5-aza-2ˈ-deoxycytidine and Valproic Acid on Epigeneticmodifying DNMT1 Gene Expression, Apoptosis Induction and Cell Viability in Hepatocellular Carcinoma WCH-17 cell line. Iran J Ped Hematol Oncol. 2019;9:83. [Google Scholar]
- Kim M-H, Kim M-S, Kim W, et al. Suppressor of cytokine signaling (SOCS) genes are silenced by DNA hypermethylation and histone deacetylation and regulate response to radiotherapy in cervical cancer cells. PLoS One. 2015;10:e0123133. doi: 10.1371/journal.pone.0123133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T, Wakimoto N, Yin D, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (Vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int J Cancer. 2007;121:656–65. doi: 10.1002/ijc.22558. [DOI] [PubMed] [Google Scholar]
- Li Y, Seto E. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect Med. 2016;6:026831. doi: 10.1101/cshperspect.a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K-Y, Wang L-T, Hsu S-H. Modification of epigenetic histone acetylation in hepatocellular carcinoma. Cancers (Basel) 2018;10:8–14. doi: 10.3390/cancers10010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92:1210–6. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- Nebbioso A, Carafa V, Benedetti R, Altucci L. Trials with ‘epigenetic’drugs: an update. Mol Oncol. 2012;6:657–82. doi: 10.1016/j.molonc.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemati M, Ajami N, Estiar MA, et al. Deregulated expression of HDAC3 in colorectal cancer and its clinical significance. Adv Clin Exp Med. 2018;27:305–11. doi: 10.17219/acem/66207. [DOI] [PubMed] [Google Scholar]
- Oh B-K, Kim H, Park H-J, et al. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int J Mol Med. 2007;20:65–73. [PubMed] [Google Scholar]
- Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014;342:223–30. doi: 10.1016/j.canlet.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhr M, Santer FR, Neuwirt H, et al. Down-regulation of suppressor of cytokine signaling-3 causes prostate cancer cell death through activation of the extrinsic and intrinsic apoptosis pathways. Cancer Res. 2009;69:7375–84. doi: 10.1158/0008-5472.CAN-09-0806. [DOI] [PubMed] [Google Scholar]
- Quint K, Agaimy A, Di Fazio P, et al. Clinical significance of histone deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of survival in HCC. Virchows Archiv. 2011;459:129–39. doi: 10.1007/s00428-011-1103-0. [DOI] [PubMed] [Google Scholar]
- Ravishankar D, Rajora AK, Greco F, Osborn HM. Flavonoids as prospective compounds for anti-cancer therapy. Int J Biochem Cell Biol. 2013;45:2821–31. doi: 10.1016/j.biocel.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Rikimaru T, Taketomi A, Yamashita Y-i, et al. Clinical significance of histone deacetylase 1 expression in patients with hepatocellular carcinoma. Oncology. 2007;72:69–74. doi: 10.1159/000111106. [DOI] [PubMed] [Google Scholar]
- Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Zhuang L, Jiang X, et al. Apoptosis signal-regulating kinase 1 is a direct target of E2F1 and contributes to histone deacetylase inhibitor-induced apoptosis through positive feedback regulation of E2F1 apoptotic activity. J Biol Chem. 2006;281:10508–15. doi: 10.1074/jbc.M512719200. [DOI] [PubMed] [Google Scholar]
- Tong H, Lin M. Effect of 5-aza-2’-deoxycytidine on cell of high-risk patients with myelodysplastic syndrome in vitro. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2004;12:467–71. [PubMed] [Google Scholar]
- Turek-Plewa J, Jagodzinski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631. [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Salehi H. Genistein and trichostatin a induction of estrogen receptor alpha gene expression, apoptosis and cell growth inhibition in hepatocellular carcinoma HepG 2 cells. Asian Pac J Cancer Prev. 2017;18:3445. doi: 10.22034/APJCP.2017.18.12.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F. Effect of curcumin and trichostatin a on the expression of DNA methyltransfrase 1 in hepatocellular carcinoma cell line Hepa 1-6. Iran J Ped Hematol Oncol. 2018;8:193. [Google Scholar]
- Sanaei M, Kavoosi F, Roustazadeh A, Golestan F. Effect of genistein in comparison with trichostatin A on reactivation of DNMTs genes in hepatocellular carcinoma. J ClinTransl Hepatol. 2018;6:1–6. doi: 10.14218/JCTH.2018.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F. Effects of 5-aza-2ˈ-deoxycytidine and valproic acid on epigenetic-modifying DNMT1 gene expression, apoptosis induction and cell viability in hepatocellular carcinoma WCH-17 cell line. Iran J Ped Hematol Oncol. 2019;9:83–90. [Google Scholar]
- Sanaei M, Kavoosi F, Mohammadi M, Khanezad M. Effect of 5-aza-2′-deoxycytidine on p16INK4a, p14ARF, p15INK4b genes expression, cell viability, and apoptosis in PLC/PRF5 and MIA Paca-2 cell lines. Iran J Ped Hematol Oncol. 2019;9:219–28. [Google Scholar]
- Sanaei M, Kavoosi F. Investigation of the effect of zebularine in comparison to and in combination with trichostatin A on p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2, DNA methyltransferases and histone deacetylases in colon cancer LS 180 cell line. Asian Pac J Cancer Prev. 2020;21:1819–28. doi: 10.31557/APJCP.2020.21.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F. Effect of zebularine in comparison to and in combination with trichostatin A on CIP/KIP family (p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2), DNMTs (DNMT1, DNMT3a, and DNMT3b), class I HDACs (HDACs 1, 2, 3) and class II HDACs (HDACs 4, 5, 6) gene expression, cell growth inhibition and apoptosis induction in colon cancer LS 174T cell line. Asian Pac J Cancer Prev. 2020;21:2131–9. doi: 10.31557/APJCP.2020.21.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Esmi Z. The effect of 5-Aza-2′-deoxycytidine in combination to and in comparison with vorinostat on DNA methyltransferases, histone deacetylase 1, glutathione S-transferase 1 and suppressor of cytokine signaling 1 genes expression, cell growth inhibition and apoptotic induction in hepatocellular LCL-PI 11 cell line. Int J Hematol Oncol Stem Cell Res. 2020;14:45. [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Behjoo H. Effect of zebularine on SOCS-1 and SOCS-3 gene expression in colon carcinoma SW 48 cell line. Exp Oncol. 2020;42:183–7. doi: 10.32471/exp-oncology.2312-8852.vol-42-no-3.15113. [DOI] [PubMed] [Google Scholar]
- Shen W-J, Dai D-Q, Teng Y, Liu H-B. Regulation of demethylation and re-expression of RASSF1A gene in gastric cancer cell lines by combined treatment of 5-Aza-CdR and NaB. World J Gastroenterol. 2008;14:595. doi: 10.3748/wjg.14.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- Sutherland KD, Lindeman GJ, Choong DY, et al. Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene. 2004;23:7726. doi: 10.1038/sj.onc.1207787. [DOI] [PubMed] [Google Scholar]
- Wang M, Liu J, Liu Q, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21:4613–22. [PubMed] [Google Scholar]
- Wang Y-C, Yang X, Xing L-H, Kong W-Z. Effects of SAHA on proliferation and apoptosis of hepatocellular carcinoma cells and hepatitis B virus replication. World J Gastroenterol. 2013;19:5159. doi: 10.3748/wjg.v19.i31.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–9. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Du W, Zhang YJ, et al. Trichostatin A, a histone deacetylase inhibitor, suppresses JAK2/STAT3 signaling via inducing the promoter-associated histone acetylation of SOCS1 and SOCS3 in human colorectal cancer cells. Mol Carcinogenesis. 2012;51:174–84. doi: 10.1002/mc.20777. [DOI] [PubMed] [Google Scholar]
- Yang J, Tian X, Yang J, et al. 5-Aza-2′-deoxycytidine, a DNA methylation inhibitor, induces cytotoxicity, cell cycle dynamics and alters expression of DNA methyltransferase 1 and 3A in mouse hippocampus-derived neuronal HT22 cells. J Toxicol Environ Health. 2017;80:1222–9. doi: 10.1080/15287394.2017.1367143. [DOI] [PubMed] [Google Scholar]
- Zhou B, Chen H, Wei D, et al. A novel miR-219-SMC4-JAK2/Stat3 regulatory pathway in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2014;33:55. doi: 10.1186/1756-9966-33-55. [DOI] [PMC free article] [PubMed] [Google Scholar]