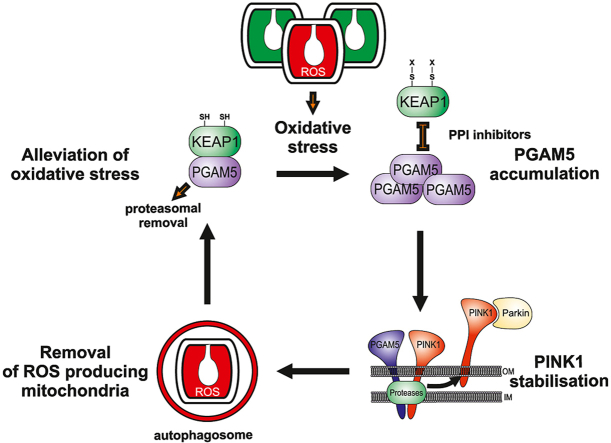
Abstract
When ROS production exceeds the cellular antioxidant capacity, the cell needs to eliminate the defective mitochondria responsible for excessive ROS production. It has been proposed that the removal of these defective mitochondria involves mitophagy, but the mechanism of this regulation remains unclear. Here, we demonstrate that moderate mitochondrial superoxide and hydrogen peroxide production oxidates KEAP1, thus breaking the interaction between this protein and PGAM5, leading to the inhibition of its proteasomal degradation. Accumulated PGAM5 interferes with the processing of the PINK1 in the mitochondria leading to the accumulation of PINK1 on the outer mitochondrial membrane. In turn, PINK1 promotes Parkin recruitment to mitochondria and sensitizes mitochondria for autophagic removal. We also demonstrate that inhibitors of the KEAP1-PGAM5 protein-protein interaction (including CPUY192018) mimic the effect of mitochondrial ROS and sensitize mitophagy machinery, suggesting that these inhibitors could be used as pharmacological regulators of mitophagy. Together, our results show that KEAP1/PGAM5 complex senses mitochondrially generated superoxide/hydrogen peroxide to induce mitophagy.
Keywords: DIV, dayin vitro; GSH-MEE, reduced glutathione ethyl ester; IMM, inner mitochondrial membrane; MAM, mitochondria-associated membranes; OMM, outer mitochondrial membrane; TMRE, tetramethyl rhodamine ethyl ester
Keywords: Oxidative stress, NRF2/KEAP1 pathway, Mitophagy, PINK1/Parkin pathway, Neurodegenerative diseases
Graphical abstract
Highlights
-
•
Increased mitochondrial ROS production disrupts the KEAP1-PGAM5 complex blocking PGAM5 processing.
-
•
Accumulating PGAM5 interferes with PINK1 processing leading to the stabilization of PINK1 on the mitochondrial membrane.
-
•
This stimulates Parkin translocation to mitochondria facilitating mitophagy and removal of ROS-producing mitochondria.
-
•
KEAP1-PGAM5 protein-protein interaction inhibitors mimic the effect of ROS and sensitize mitochondria to autophagy.
1. Introduction
While limited cellular ROS levels can be beneficial in normal physiological functions, the excessive ROS accumulation, often generated by defective mitochondria, can lead to oxidative damage of nucleic acids, proteins and lipids. Normally, excessive ROS are neutralized by scavenging systems like superoxide dismutase (SOD), catalase, glutathione peroxidase, glutathione reductase, or reduced glutathione and trigger an antioxidative response through activation of the NRF2/KEAP1 (nuclear factor erythroid 2-related factor 2/Kelch ECH associating protein 1) -dependent pathway [1,2]. Under normal conditions, NRF2 binds to KEAP1, a substrate adaptor for Cullin 3-based E3 ubiquitin ligase, thus leading to its ubiquitylation and proteasomal degradation. Under oxidative stress conditions, NRF2 dissociates from the KEAP1 and translocates into the nucleus, where it induces the expression of antioxidant enzyme genes. Such a role makes KEAP1 an attractive target for drug development against various oxidative stress-related diseases, including neurodegenerative disorders [3].
However, when mitochondrial ROS production exceeds the capacity of antioxidant defense systems, the cell needs to eliminate the defective mitochondria responsible for excessive ROS production. It has been proposed that the removal of these defective mitochondria should involve mitophagy [[4], [5], [6], [7]], but the mechanism of this regulation remains unclear.
There are several pathways by which mitophagy is induced in mammalian cells [8,9]. One of the best-characterized mitophagy pathways involves the PTEN-induced kinase 1 (PINK1) and Parkin, the products of two genes responsible for some cases of familial Parkinson's disease [[10], [11], [12]]. PINK1 is imported in mitochondria where mitochondrial protease PARL cleaves its N-terminal part, causing its release to the cytosol and subsequent degradation [[13], [14], [15], [16]]. Mitochondrial depolarization inhibits its PARL-mediated cleavage, leading to its rapid accumulation at the mitochondrial outer membrane where it recruits the E3 ubiquitin ligase Parkin to ubiquitinate mitochondrial proteins, triggering mitophagy [9,11,[17], [18], [19]]. It has been suggested that ROS may act as a trigger for PINK1/Parkin-dependent mitophagy by inducing translocation of Parkin to mitochondria [20]. However, other findings suggest that superoxide has no impact on Parkin dynamics and could promote mitophagy only following Parkin translocation to mitochondria [21]. Furthermore, one cannot exclude that some other pathways can mediate ROS-induced mitophagy. Recently it has been suggested that under oxidative stress conditions, a mitochondrial phosphatase PGAM5 induces mitophagy by dephosphorylating mitochondrial outer-membrane protein FUNDC1 [22].
Here, we demonstrate the crosstalk between PINK1-Parkin and PGAM5 pathways in the regulation of ROS-induced mitophagy. We show that oxidation of KEAP1 inhibits the proteasomal degradation of PGAM5, leading to its accumulation and interference with PINK1 processing that sensitizes mitochondria to autophagic removal. This process does not need NRF2 participation.
2. Results
2.1. Mild generation of mitochondrial superoxide and hydrogen peroxide induces Parkin-dependent mitophagy
It is generally accepted that massive ROS production induces mitochondrial depolarization, leading to activation of PINK1-mediated Parkin translocation to mitochondria and mitophagy [4]. However, how the mitophagy machinery responds to lower, more physiological mitochondrial subthreshold ROS overproduction that is not sufficient to induce the mitochondrial membrane depolarization or compromise cell viability, remains unclear.
To mimic such intrinsic ROS production, we used menadione (vitamin K3), a well-known superoxide generator in the mitochondria. Metabolism of menadione by mitochondrial NADH-dependent ubiquinone oxidoreductase (complex I) produces an unstable semi-quinone radical, and its reverse oxidation generates superoxide radical in the vicinity of the inner mitochondrial membrane, in the mitochondrial matrix. This diffusion-restricted radical is then converted to hydrogen peroxide by the mitochondrial superoxide dismutase that could then freely diffuse to mitochondrial intermembrane space and cytoplasm (Fig. S1A) [[23], [24], [25], [26], for comprehensive reviews see [27], [28], [29]]. In our model, as expected, the menadione treatment led to production of mitochondrial superoxide as well as hydrogen peroxide that was visualized both in mitochondria and cytosol (Fig. 1A, Supplementary Videos 1-3; note that in the further text, the term ROS is used to refer these species).
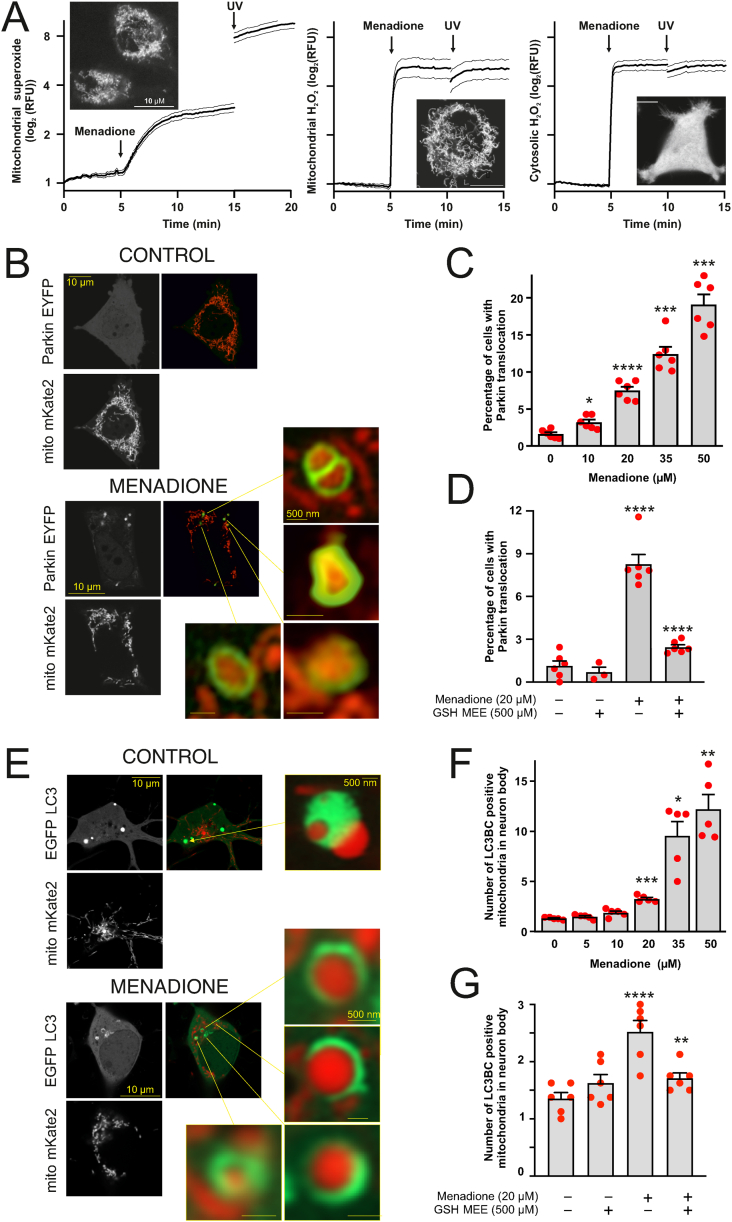
Fig. 1.
Generation of ROS in the mitochondrial inner membrane induces Parkin translocation and mitophagy, A. Menadione induces rapid production of both mitochondrial superoxide and hydrogen peroxide. PC6 cells stained with mitochondrial superoxide indicator MitoSOX™ Red (0.5 μM, left panel) or transfected with genetically encoded mitochondrial (middle panel) or cytosolic (right panel) H2O2 Hyper7 sensors were treated with 20 μM menadione and exposed for 385 nM LED Light Source for 1s at the end of the experiment to induce a massive ROS generation (positive control). Curves are depicted as mean ± SEM. Insets show subcellular localization of the indicator or sensor before the treatment. B. Menadione induces Parkin translocation. Representative superresolution Airyscan images of PC6 cells transfected with Parkin-EYFP (green) and mitochondria-targeted Kate2 (red) after treatment with DMSO or 20 μM menadione for 4 h. C. Quantification of Parkin-EYFP translocation to the mitochondria. The percentage of PC6 cells with Parkin-EYFP translocated to the mitochondria increases after menadione treatment in a concentration-dependent manner. *P < 0.05, ***P < 0.001 and ****P < 0.0001, n = 6 dishes, 20 fields per dish, Welch's ANOVA followed by Dunnett's T3 multiple comparison test. D. Co-treatment with ROS scavenger glutathione inhibits the menadione-induced Parkin translocation. The percentage of PC6 cells with Parkin-EYFP translocated to mitochondria in the presence of 20 μM menadione or/and 500 μM GSH-MEE. ****P < 0.0001, n = 3–6 dishes, 20 fields per dish, one-way ANOVA followed by Sidak's multiple comparison test. E. Menadione-induced ROS trigger mitophagy. Representative superresolution Airyscan images of cortical neurons transfected with a mix of EGFP-LC3B and GFP-LC3C (green) and mitochondria-targeted Kate2 (red) after treatment with DMSO or 20 μM menadione for 4 h. F. Quantification of the number of mitochondria co-localised with the autophagosome marker in neurons. *P < 0.05, **P < 0.01 and ***P < 0.001, n = 5 dishes, 5–10 cells per dish, one-way ANOVA followed by Dunnett's T3 multiple comparison test. G. Co-treatment with ROS scavenger glutathione inhibits the menadione-induced mitophagy in neurons. The number of mitochondrial colocalizations with EGFP-LC3B and GFP-LC3C in the presence of 20 μM menadione or/and 500 μM GSH-MEE. **P < 0.01 and ****P < 0.0001, n = 6 dishes, 8 cells per dish, one-way ANOVA followed by Sidak's multiple comparison test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Treatment of neuronal PC6 cells with relatively low menadione concentrations (10–20 μM, 4 h) produced half-maximal H2O2 amounts (Fig. S1B) without mitochondrial depolarization (Fig. S1C) or substantial loss in cell viability (Fig. S1D) and did not trigger caspase-3 dependent apoptosis (Fig. S1E). However, such a treatment induced Parkin translocation from cytosol to mitochondria (Fig. 1B and C). This translocation was indeed related to ROS generation because the cell-permeable reduced glutathione ethyl ester (GSH-MEE), scavenging the ROS (Fig. S1F), prevented the menadione-induced Parkin translocation (Fig. 1D) completely. Four-hour treatment of primary cortical neurons with menadione also induced dose-dependent mitophagy (Fig. 1E and F) that was similarly suppressed by the GSH-MEE (Fig. 1G). Similar protection was observed when 2 mM N-acetylcysteine was used (data not shown).
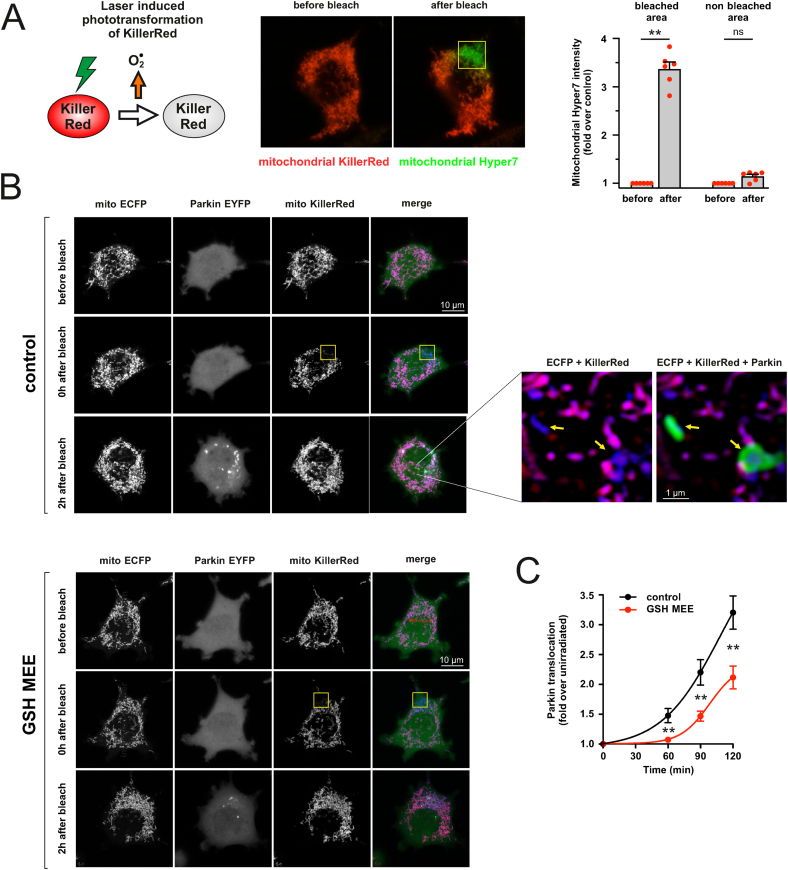
To test whether ROS generation in another mitochondrial compartment (mitochondrial matrix) gives the same effect, we used an optogenetic approach based on the genetically encoded photosensitizer, KillerRed. We targeted KillerRed to the mitochondrial matrix and irradiated few selected mitochondria with a 561 nm laser line to trigger oxidative burst in these mitochondria. As shown in Fig. 2A, the photoactivation of KillerRed led to a local ROS production as evidenced by an increase in the fluorescence of ultrafast pH-stable hydrogen peroxide sensor Hyper7 targeted to the mitochondrial matrix. Furthermore, Parkin started to accumulate onto the irradiated mitochondria as early as 60 min after irradiation producing robust Parkin translocation at 2 h (Fig. 2B). Such a translocation was strongly inhibited in the presence of GSH-MEE (Fig. 2C) and did not occur in cells expressing another mitochondria-targeted red fluorophore, Kate2 (Figs. S2A–C), suggesting that the effect of KillerRed was mainly due to ROS production. However, we were not able to monitor mitochondrial membrane potential simultaneously with KillerRed because of photobleaching.
Fig. 2.
Generation of ROS in the mitochondrial matrix induces Parkin translocation A. Photoactivation of KillerRed leads to local ROS production. Left panel: Irradiation with intense green laser line leads to KillerRed-mediated ROS production in the mitochondrial matrix. Central panel: PC6 cells were transfected with mitochondrial Hyper7 (green) and mitochondrial KillerRed (red). Sub-population of mitochondria was irradiated in a 6.7x6.7 μm frame (yellow) using a 561 nm laser line (1 iteration, using a 10% laser power, pixel time 25 μs). Images depict the cell before (left) and immediately after (right) irradiation. Only the irradiated mitochondria that lose the KillerRed signal generate ROS. Right panel: quantification of mitochondrial Hyper7 signal. **P < 0.01, n = 6, Friedman test and Dunn's multiple comparison test. B. Photoactivation of KillerRed leads to the Parkin translocation. PC6 cells were transfected with mitochondrial ECFP (blue), Parkin-EYFP (green) and mitochondrial KillerRed (red). Sub-population of mitochondria in the selected cell was irradiated in a 6.7x6.7 μm frame (yellow) using a 561 nm laser line (1 iteration, using a 10% laser power, pixel time 25 μs), and redistribution of Parkin-EYFP signal was followed over the time. Top group of images shows the cell before the irradiation (upper panel), immediately after irradiation (middle panel, note the loss of KillerRed signal in the irradiated area) and 2 h later (lower panel). Purple mitochondria in the merge panel are non-irradiated mitochondria, while the blue ones that have lost their KillerRed signal are irradiated ones generating ROS. Note the Parkin accumulations around the ROS-generating mitochondria in zoomed images (yellow arrows). Parkin translocation was visually weaker in GSH-MEE treated cells (bottom group of panels). C. Laser-induced Parkin translocation is slower and weaker in cells treated with GSH-MEE. PC6 cells were plated in separate compartments of the same dish, enabling us to visualize Parkin translocation simultaneously over 2 h under similar conditions in response to GSH-MEE (500 μM). The spatial heterogeneity of Parkin-EYFP (coefficient of variation of the intensity of individual pixels) was estimated for individual cells for each time point. **P < 0.01, n = 41–42 cells from 7 different dishes, two-way repeated-measures ANOVA and Sidak's multiple comparison test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
This set of experiments confirms that mild generation of superoxide and hydrogen peroxide in the inner mitochondrial membrane or the mitochondrial matrix, even in the absence of mitochondrial depolarization, is sufficient to induce Parkin translocation to the mitochondria and to initiate mitophagy.
2.2. ROS-induced mitophagy is mediated by KEAP1 and involves the PINK1-Parkin pathway
Theoretically, ROS could induce mitophagy by triggering specific oxidative stress response pathways. One of the main oxidative stress-induced pathways is KEAP1–NRF2 system, which protects the cell against oxidative aggression. We therefore next tested whether ROS-induced mitophagy involves KEAP1.
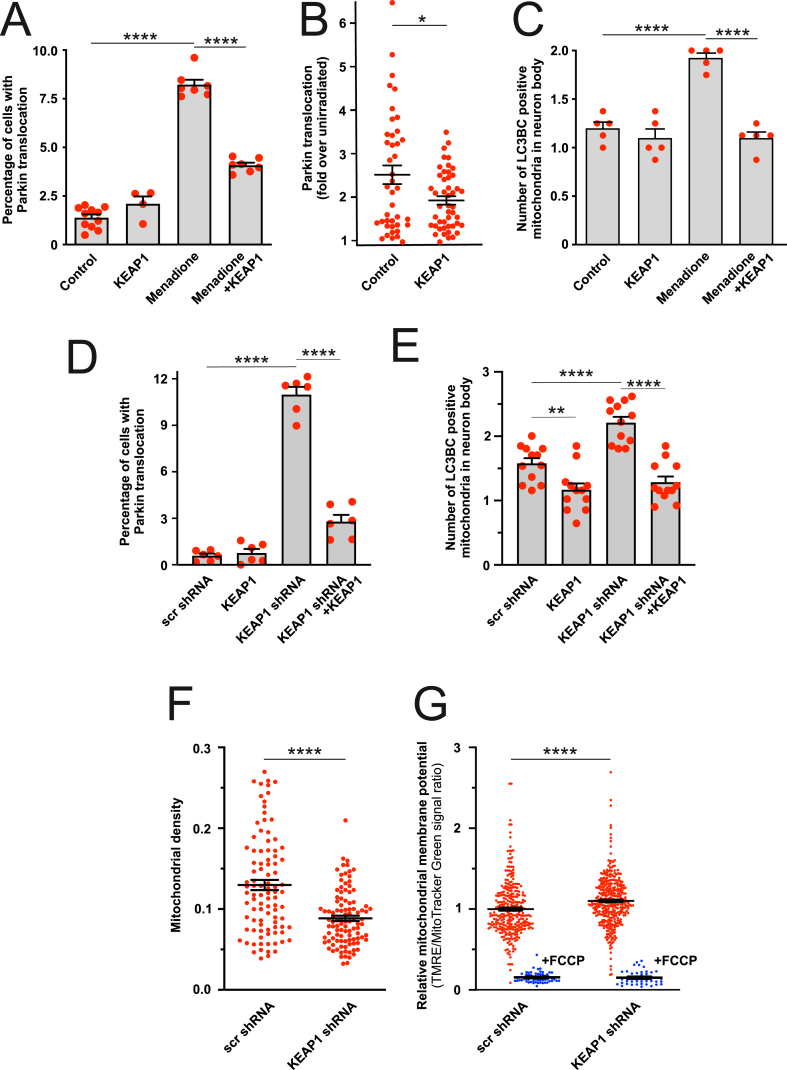
The results showed that KEAP1 overexpression dramatically decreased the menadione and KillerRed-induced Parkin translocation (Fig. 3A and B), suggesting that KEAP1 is involved in the regulation of the PINK1-Parkin pathway by ROS. In the same line of evidence, the ROS-induced mitophagy was also suppressed by KEAP1 overexpression (Fig. 3C).
Fig. 3.
ROS-induced mitophagy involves KEAP1, A-B. KEAP1 overexpression suppresses menadione- and KillerRed-induced Parkin-EYFP translocation to the mitochondria. (A) PC6 cells transfected with Parkin-EYFP and control plasmid or KEAP1 overexpressing plasmid were treated for 4 h with DMSO or menadione (20 μM). ****P < 0.0001, n = 4–12 dishes, 20 fields per dish, one-way ANOVA followed Holms-Sidak's multiple comparison test. (B) Sub-population of mitochondria in selected PC6 cells transfected with Parkin-EYFP, mitochondrial KillerRed and control or KEAP1 overexpressing plasmid were irradiated with 561 nm laser line (1 iteration, using a 10% laser power, pixel time 25 μs) and redistribution of Parkin-EYFP signal was followed for 2 h. P = 0.015, n = 41–47 cells, t-test with Welch's correction. C. KEAP1 overexpression suppresses the number of mitochondria colocalized with the autophagosome marker LC3B/LC3C in cortical neurons. Primary cortical neurons were transfected with a mix of EGFP-LC3B, GFP-LC3C, mitochondrial Kate2 and KEAP1, and then treated with DMSO or 20 μM menadione for 4 h. ****P < 0.0001, n = 5 dishes, 8 cells per dish, one-way ANOVA followed by Sidak's multiple comparison test. D. Silencing of KEAP1 expression induces Parkin-EYFP translocation to mitochondria. PC6 cells were transfected with Parkin-EYFP and scrambled shRNA or KEAP1 shRNA and shRNA insensitive human KEAP1 encoding plasmid. ****P < 0.0001, n = 6 dishes, 20 fields per dish, one-way ANOVA followed by Sidak's multiple comparison test. E. Silencing of KEAP1 expression increases mitophagy. Neurons were transfected with EGFP-LC3B, GFP-LC3C, mitochondrial Kate2, scrambled or KEAP1 shRNA and shRNA insensitive KEAP1. **P < 0.01 and ****P < 0.0001, n = 12 dishes, 5–8 cells per dish, one-way ANOVA followed by Sidak's multiple comparison test. F. KEAP1 silencing decreases mitochondrial density in axons. The neurons were transfected with neuronal marker hSyn-EGFP, mitochondrial DsRed2 and scrambled or KEAP1 shRNA and mitochondrial density was quantified at the end of the axon. ****P < 0.0001, n = 95–104 axons from 10 dishes, Mann-Whitney test. G. KEAP1 silencing does not cause mitochondrial depolarization as can be seen by relative TMRE intensity normalized to MitoTracker Green in neurons transfected with scrambled or KEAP1 shRNA (detected by mitochondrial ECFP co-transfection). Note that FCCP treatment (10 μM for 5 min) almost completely abolished TMRE fluorescence. ****P < 0.0001, n = 301–368 cells from 6 dishes, Mann-Whitney test (n = 42–58 cells from 3 dishes for FCCP treated groups). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
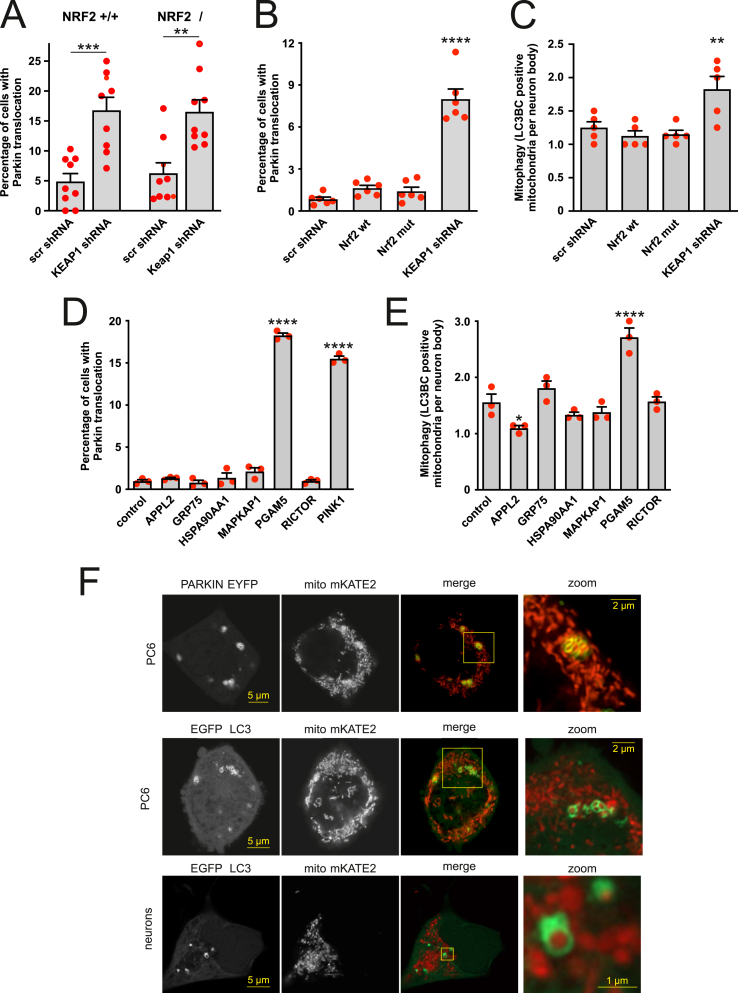
To confirm the involvement of KEAP1 in ROS-induced mitophagy, we silenced the KEAP1 expression using specific shRNAs (Fig. S3). This induced a small but significant increase in Parkin translocation from cytosol to mitochondria (Fig. 3D) and enhanced mitophagy (Fig. 3E). Significantly, normal levels of Parkin translocation and mitophagy were restored when shRNA-resistant KEAP1 was co-expressed with KEAP1 shRNA showing the specificity of the shRNA. We also observed a decrease of the mitochondrial mass (Fig. 3F) in the axons of the neurons. Furthermore, it is noteworthy that the increased mitophagy was not due to decreased mitochondrial membrane potential as the KEAP1 silencing led to slight hyperpolarization as observed earlier by others [30,31] and is probably related to higher NRF2 activity [32] (Fig. 3G).
Observed Parkin translocation in KEAP1 shRNA expressing cells required Parkin ubiquitin ligase activity because Parkin mutant T240R ligase dead mutant associated with autosomal recessive juvenile parkinsonism [33], or mutant C431 N (which can self-associate in response to mitochondrial depolarization and lacks ligase activity [34] did not translocate to the mitochondria (Fig. S4A). Besides, silencing of KEAP1 did not induce Parkin translocation (Fig. S4B) or mitophagy in PINK1 knockdown cells (Fig. S4C). Silencing of KEAP1 led to PINK1 accumulation (Figs. S4D and S4E), whereas PINK1 expression was not augmented (Fig. S4F).
Altogether, these data allow suggesting that ROS-dependent mitophagy involves KEAP1 and depends on PINK1-Parkin.
2.3. KEAP1-dependent mitophagy does not involve NRF2
It has been shown [35,36] that inactivation of KEAP1 leads to the expression of pro-mitophagy genes via activation of NRF2. However, the silencing of the KEAP1 in NRF2-deficient mouse embryonic fibroblasts also induced Parkin translocation (Fig. 4A), suggesting another pathway controlling mitophagy. Moreover, overexpression of wt or KEAP1-insensitive constitutively active NRF2 (ETGE>GAGA) did not induce Parkin translocation or mitophagy (Fig. 4B and C).
Fig. 4.
KEAP1-dependent mitophagy does not involve NRF2 but another KEAP1 substrate PGAM5. A. Silencing of KEAP1 induces Parkin-EYFP translocation both in wt and NRF2-deficient mouse embryonic fibroblasts (MEFs). MEF-s were transfected with Parkin-EYFP and scrambled or KEAP1 shRNA encoding plasmid. **P < 0.01 and ***P < 0.001, n = 9 dishes, 20–35 fields per dish, one-way ANOVA followed by Sidak's multiple comparison test. B. NRF2 does not induce Parkin-EYFP translocation. PC6 cells were transfected with Parkin-EYFP, wt or constitutively active NRF2, or with KEAP1 shRNA. ****P < 0.0001 compared with the scrambled shRNA-expressing group, n = 6 dishes, 20 fields per dish, one-way ANOVA followed by Dunnett's multiple comparison test. C. NRF2 does not induce mitophagy. Primary cortical neurons were transfected with a mix of EGFP-LC3B and GFP-LC3C, mitochondrial Kate2 and wt or constitutively active NRF2, or with scrambled shRNA or KEAP1 shRNA. **P < 0.01 compared with the scrambled shRNA-expressing group, n = 5 dishes, 8 fields per dish, one-way ANOVA followed by Dunnett's multiple comparison test. D. Overexpression of PGAM5 induces Parkin-EYFP translocation. PC6 cells were transfected with Parkin-EYFP and plasmids of interest. ****P < 0.0001 compared with the control group, n = 3 dishes, 20 fields per dish, one-way ANOVA followed by Dunnett's multiple comparison test. E. Overexpression of PGAM5 induces mitophagy. Neurons were transfected with EGFP-LC3B, GFP-LC3C, mitochondrial Kate2 and plasmids of interest. *P < 0.05 and ****P < 0.0001 when compared with control group, n = 3 dishes, 7 fields per dish, one-way ANOVA followed by Dunnett's multiple comparison test. F. Representative superresolution Airyscan images of PC6 cells transfected with PGAM5, Parkin-EYFP and mitochondrial Kate 2 (upper panels) or PC6 cells (middle panels) or neurons (lower panels) transfected with PGAM5, EGFP-LC3B, GFP-LC3C and mitochondrial Kate2.
In this context, one should consider that NRF2 is not the only substrate for the KEAP1-Cullin3-RBX1 E3-ubiquitin ligase complex. Many other proteins are known to possess the KEAP1 degron E(T/S)GE but whose substrate statuses are not known. We therefore hypothesized that there could exist another protein(s) degraded by KEAP1-CUL3-RBX1 E3-ubiquitin ligase that mediate(s) the activation of mitophagy. It is unlikely that the KEAP1-CUL3-RBX1 E3-ubiquitin ligase complex could degrade PINK1 itself as the latter does not have the E(T/S)GE motif.
To identify possible candidates, we performed a search among PINK1 interacting/stabilizing proteins for ETGE motifs using interaction database BioGRID and respective proteomic datasets [37]. This search revealed a group of proteins including APPL2, GRP75, HSP90AA1, MAPKAP1, PGAM5 and RICTOR. We next overexpressed these proteins to test which of them could induce Parkin translocation to mitochondria and subsequent mitophagy. The results depicted in Fig. 4D and E show that among these proteins, only PGAM5 was able to induce Parkin translocation to mitochondria and mitophagy.
The majority of the PC6 cells overexpressing PGAM5 had a fragmented mitochondrial network, and some cells also had mitochondrial aggregates or clumps (Fig. 4F). These clumps were often surrounded by Parkin or LC3. Mitochondrial fragmentation and clumps were also visible in PGAM5 overexpressing neurons leading to massive toxicity 2–3 days after transfection.
2.4. KEAP1 interacts with full-length PGAM5 and controls its degradation in the cytosol
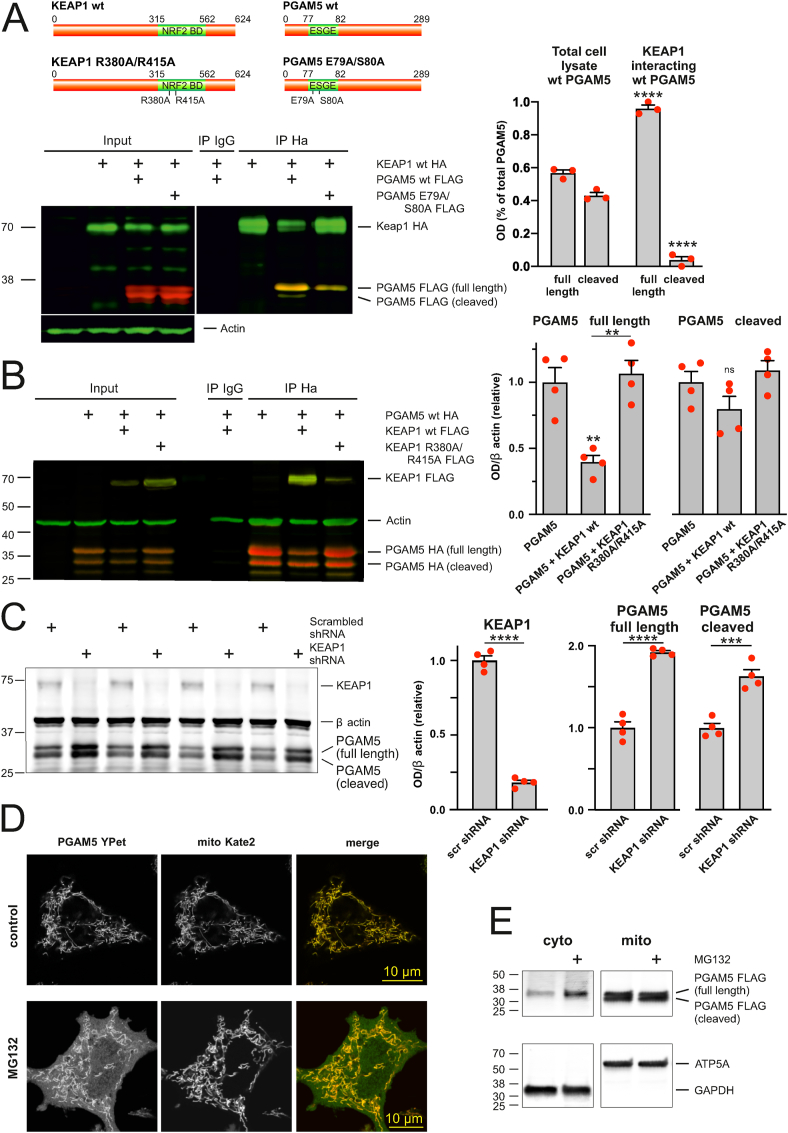
PGAM5 is found in two forms, a full-length mitochondrial outer membrane protein and a short form cleaved by IMM-resident proteases that lacks the mitochondrial membrane targeting sequence [[38], [39], [40]]. Our first task was therefore to find out which form of PGAM5 interacts with KEAP1. We immunoprecipitated KEAP1 and identified co-immunoprecipitating PGAM5 forms. Fig. 5A shows that while both the full-length and cleaved forms of PGAM5 were present in the total cell lysate, only the full-length form was co-immunoprecipitating with KEAP1. Note also that the mutated PGAM5 E79A S80A where the glutamate and serine residues of the ESGE motif in PGAM5 were replaced with alanine interacts with KEAP1 less efficiently (Fig. 5A).
Fig. 5.
KEAP1 controls the degradation of full-length PGAM5. A. KEAP1 interacts only with full-length PGAM5. The left panel shows that full-length PGAM5-FLAG, but not cleaved PGAM5, co-immunoprecipitates with KEAP1-HA in HEK cells. Note also that PGAM5 E79A/S80A co-immunoprecipitates with KEAP1 less efficiently than PGAM5-WT-FLAG. The right panel shows quantification of full length and cleaved PGAM5-WT from total cell lysate and KEAP1 interacting fraction. ****P < 0.0001 compared with respective total cell lysate group, n = 3 independent IP-s, one-way ANOVA followed by Sidak's multiple comparison test. B. KEAP1 overexpression induces degradation of PGAM5. The left panel shows the representative Western blot image of PGAM5 expression in the presence of KEAP1-FLAG or KEAP1 R380A/R415A-FLAG in HEK cells. Note also that KEAP1-FLAG but not KEAP1 R380A/R415A-FLAG is co-immunoprecipitating with PGAM5-HA. The right panel shows quantification of full length and cleaved PGAM5 from total cell lysate. **P < 0.01, ns: not significant, n = 4 independent experiments, one-way ANOVA followed by Sidak's multiple comparison test. C. KEAP1 silencing increases the level of endogenous PGAM5. Representative Western blot image (left) and analysis of PGAM5 expression (right) in PC6 cells expressing scrambled or KEAP1 shRNA. ***P < 0.001 and ****P < 0.0001, n = 4, t-test. D. PGAM5 is accumulating in the cytosol after proteasome inhibition. Representative superresolution Airyscan images showing PC6 cells expressing PGAM5-YPet and mitochondrially targeted Kate2 treated with DMSO or 25 μM MG132 for 5 h. E. Proteasome inhibition leads to cytosolic accumulation of full-length PGAM5. Representative Western blot image of PGAM5-Flag levels in cytosolic and mitochondrial fractions of PC6 cells treated DMSO or 25 μM MG132 for 5 h. ATP5A and GAPDH as markers of mitochondria and cytosol, respectively, demonstrate the purity of fractions.
It has been previously shown that PGAM5 is ubiquitinated by the KEAP1-dependent ubiquitin ligase complex [41,43], but it has been suggested that it may not be efficiently targeted for proteasomal degradation after ubiquitination [42]. Therefore, we tested whether or not the PGAM5 degradation is under the control of KEAP1. Quantification data on Fig. 5B shows that the overexpression of wt KEAP1 led to degradation of the full-length PGAM5 form while the cleaved form remained relatively stable. There was no PGAM5 degradation when we expressed KEAP1 mutant having R380A/R415A substitutions in Kelch domain, and this mutant was not co-immunoprecipitating with PGAM5 (Fig. 5B). These data are also supported by the reverse experiment where silencing the expression of endogenous KEAP1 led to the PGAM5 accumulation (Fig. 5C).
These experiments show that KEAP1 is involved in the degradation of PGAM5 and that only the full-length form of PGAM5 can interact with KEAP1.
Our next question was where exactly the KEAP1-dependent proteasomal degradation of PGAM5 occurs. Theoretically, this could happen before PGAM5 is entering into the mitochondria (similar to NRF2 degradation before it could enter into the nucleus) or later, where it is already in the mitochondria and released into the OMM. The latter possibility is supported by the earlier findings suggesting the PGAM5 tethers a ternary complex containing KEAP1 and NRF2 to mitochondria [42].
We treated the PC6 cells expressing fluorescent PGAM5-YPet with proteasome inhibitor MG132 and followed the fate of the PGAM5 in cytosol and mitochondria. The superresolution microscopy images demonstrate that while under control conditions, the PGAM5-YPet was localized mainly in the mitochondria (although in some cells where expression levels were very high, it formed the filamentous aggregates as has been shown earlier [43]), it started to accumulate in the cytosol when its proteasomal degradation was blocked (Fig. 5D). Moreover, further Western blot analysis demonstrates that it is specifically the full-length form of PGAM5 accumulating in the cytosol in response to proteasomal inhibition (Fig. 5E).
Together, these findings suggest that KEAP1 controls the degradation of soluble full-length cytosolic PGAM5 before it is inserted into the outer mitochondrial membrane.
2.5. PGAM5 mediates ROS/KEAP1 related mitophagy
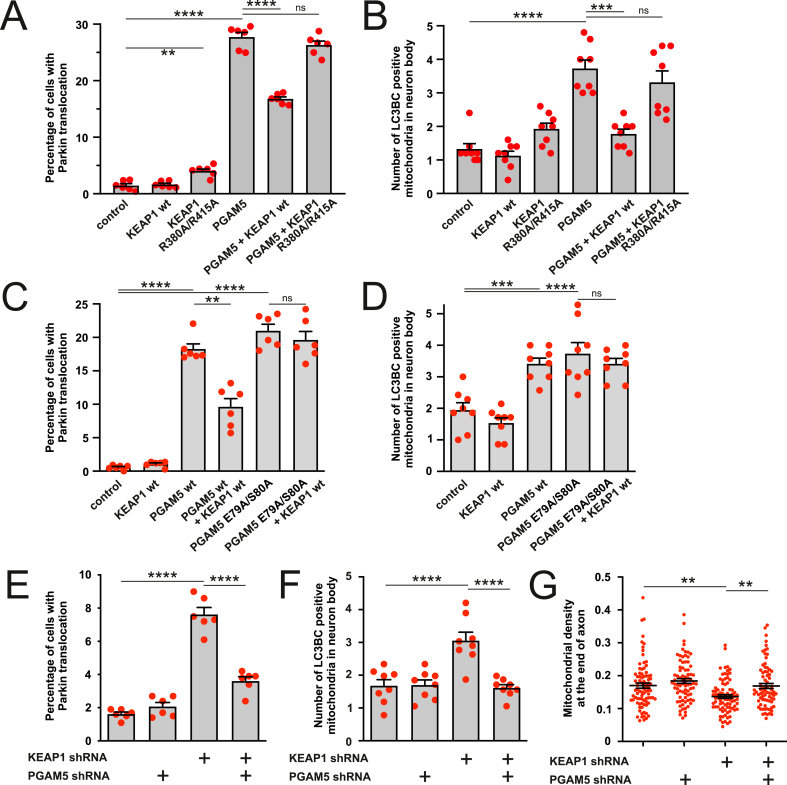
The previous set of experiments demonstrated that PGAM5 accumulates after KEAP1 downregulation, but these experiments left an open question of whether it is the PGAM5 accumulation that thrives the KEAP1 mediated mitophagy. Fig. 6 shows that overexpressed KEAP1 effectively suppressed the PGAM5-induced Parkin translocation to the mitochondria (Fig. 6A) as well as mitophagy (Fig. 6B), while KEAP1 R380A/R415A had no such effects. On the other hand, in contrast to wt PGAM5, PGAM5 E79A/S80A-induced Parkin translocation and mitophagy were insensitive to KEAP1 co-expression (Fig. 6C and D). Furthermore, silencing of KEAP1 could not induce Parkin translocation in cells where PGAM5 was knocked down (Fig. 6E and Fig. S5A). KEAP1 silencing was also not able to induce mitophagy (Fig. 6F and Fig. S5B) or affect the mitochondrial density (which depends on the mitophagy; Fig. 6G and Fig. S5C) in the absence of PGAM5. Thus, KEAP1 needs PGAM5 to control mitophagy.
Fig. 6.
PGAM5 mediates the KEAP1-dependent mitophagy. A. KEAP1 but not KEAP1 R380A/R415A suppresses PGAM5-induced Parkin translocation. PC6 cells were transfected with Parkin-EYFP, wt KEAP1 or KEAP1 R380A/R415A and PGAM5. **P < 0.01 and ****P < 0.0001, n = 6 dishes, 25 fields per dish, Ordinary one-way ANOVA followed by Holm-Sidak's multiple comparison test. B. KEAP1 but not KEAP1 R380A/R415A suppresses PGAM5-induced mitophagy. Primary cortical neurons were transfected with a mix of EGFP-LC3B and GFP-LC3C, mitochondrially targeted Kate2, wt KEAP1 or KEAP1 R380A/R415A and PGAM5. ***P < 0.001 and ****P < 0.0001, n = 8 dishes, 5 fields per dish, Welch's ANOVA followed by Dunnett's T3 multiple comparison test. C. PGAM5 E79A/S80A-induced Parkin translocation is insensitive to KEAP1. PC6 cells were transfected with Parkin-EYFP, wt PGAM5 or PGAM5 E79A/S80A and wt KEAP1. **P < 0.01 and ****P < 0.0001, n = 6 dishes, 20 fields per dish, Welch's ANOVA followed by Dunnett's T3 multiple comparison test. D. PGAM5 E79A/S80A-induced mitophagy is insensitive to KEAP1. Primary cortical neurons were transfected with a mix of EGFP-LC3B and GFP-LC3C, mitochondrial Kate2, wt PGAM5 or PGAM5 E79A/S80A and wt KEAP1. ***P < 0.001 and ****P < 0.0001, n = 8 dishes, 7 fields per dish, Welch's ANOVA followed by Dunnett's T3 multiple comparison test. E. KEAP1 silencing does not induce Parkin translocation in the absence of PGAM5. PC6 cells were transfected with Parkin-EYFP and scrambled shRNA, KEAP1 shRNA, or/and PGAM5 shRNA expressing plasmids. ****P < 0.0001, n = 6 dishes, 20 fields per dish, one-way ANOVA followed Holms-Sidak's multiple comparison test. Interaction between the treatment P < 0.0001, Two-way ANOVA. F. KEAP1 silencing does not induce mitophagy in the absence of PGAM5. Primary cortical neurons were transfected with a mix of EGFP-LC3B and GFP-LC3C, mitochondrial Kate2 and scrambled shRNA, KEAP1 shRNA or/and PGAM5 shRNA expressing plasmids. ****P < 0.0001, n = 8 dishes, 7 cells per dish, one-way ANOVA followed by Sidak's multiple comparison test. Interaction between the treatment P = 0.0003, Two-way ANOVA. G. KEAP1 silencing does not lead to mitochondrial loss in the absence of PGAM5. The neurons were transfected with neuronal marker hSyn-EGFP, mitochondrial DsRed2 and KEAP1 shRNA or/and PGAM5 shRNA and mitochondrial density was quantified at the end of the axon. **P < 0.01, n = 76–85 axons from 8 dishes, Kruskal-Wallis test followed by Dunn's multiple comparisons test.
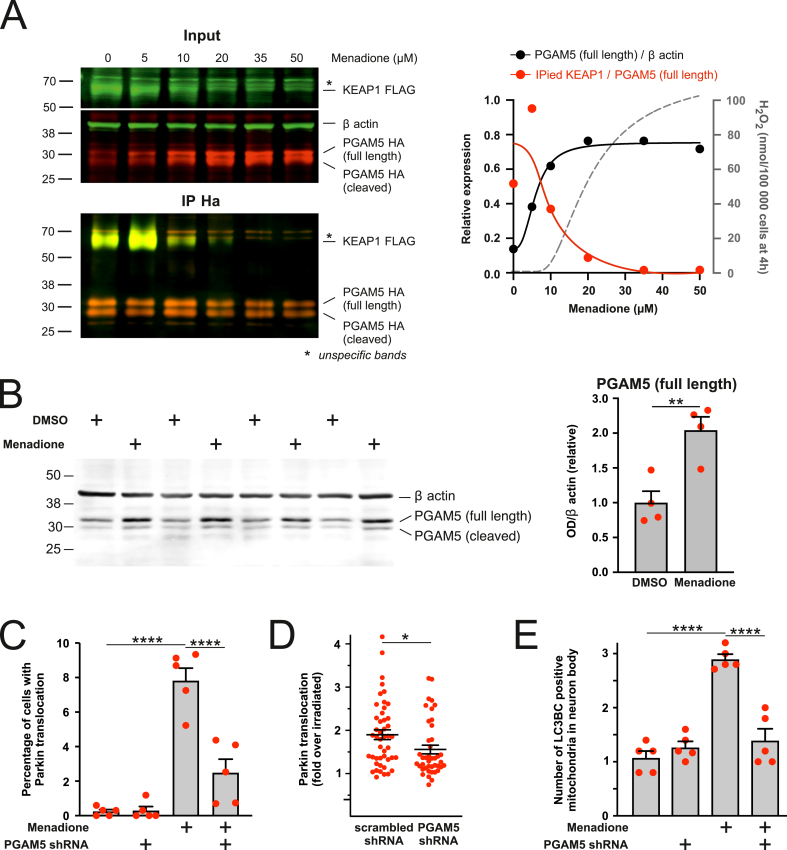
We further examined whether the inactivation of KEAP1 by mitochondria-produced ROS would lead to a dissociation of PGAM5 from KEAP1 and PGAM5 accumulation and whether this would be sufficient to induce mitophagy. Indeed, as demonstrated in Fig. 7A, menadione broke the interaction between the overexpressed KEAP1 and PGAM5 starting from 10 μM concentration leading to a parallel increase in PGAM5. Moreover, the levels of endogenous PGAM5 were significantly increased in cells treated with relatively low menadione concentration (20 μM for 4 h) (Fig. 7B). PGAM5 silencing inhibited the Parkin translocation in menadione-treated (Fig. 7C and Fig. S5D) or laser-irradiated cells (Fig. 7D) and menadione-induced mitophagy (Fig. 7E).
Fig. 7.
PGAM5 mediates the ROS-induced KEAP1-dependent mitophagy, A. KEAP1 interaction with PGAM5 is disrupted by menadione-induced ROS. Left panel - original blots of PGAM5-FLAG co-immunoprecipitates with KEAP1-HA in HEK cells treated with different concentrations of menadione for 4 h. Right panel - quantification of KEAP1 in immunoprecipitated fraction as well as the amount of full-length PGAM5-WT in total cell lysate. The grey curve depicts the amount of H2O2 produced by the cells at these menadione concentrations (note that these data are derived from the standard curve depicted in Fig. S1B). n = 1. B. Menadione induces accumulation of full-length PGAM5. Representative Western blot image and analysis of endogenous full-length PGAM5 expression in PC6 cells treated with DMSO or 20 μM menadione for 4 h. **P < 0.01, n = 4, t-test. C. PGAM5 silencing inhibits menadione-induced Parkin translocation. PC6 cells expressing Parkin-EYFP and scrambled or PGAM5 shRNA were treated with DMSO or 20 μM menadione for 4 h. ****P < 0.0001, n = 5 dishes, 20 fields per dish, one-way ANOVA followed Holms-Sidak's multiple comparison test. Interaction between the treatments P = 0.0002, Two-way ANOVA. D. PGAM5 silencing inhibits laser irradiation-induced Parkin translocation. PC6 cells were transfected with mitochondrial ECFP, Parkin-EYFP, mitochondrial KillerRed and scrambled or PGAM5 shRNA encoding plasmid. Sub-population of mitochondria in the selected cell was irradiated using a 561 nm laser, and redistribution of Parkin-EYFP signal was visualized 2 h later. *P = 0.018, n = 42–46 cells from 8 individual dishes, Mann Whitney test. E. PGAM5 silencing inhibits menadione-induced mitophagy. Primary cortical neurons expressing EGFP-LC3B, GFP-LC3C, Kate2 targeted to the mitochondria and scrambled or PGAM5 shRNAs were treated with DMSO or 20 μM menadione for 4 h. ****P < 0.0001, n = 5 dishes, 5 cells per dish, one-way ANOVA followed by Sidak's multiple comparison test. Interaction between the treatments P < 0.0001, Two-way ANOVA.
Altogether, these experiments demonstrate that a moderate mitochondrial ROS production inactivating KEAP1 is sufficient to inhibit PGAM5 degradation and induce mitophagy.
2.6. PGAM5-induced Parkin translocation does not involve the phosphatase activity of PGAM5 and could be related to the proteolytic processing of PINK1
We next investigated the mechanism of the PGAM5-mediated mitophagy. It has been earlier demonstrated that PGAM5 could promote mitophagy by dephosphorylating FUNDC1 or dynamin-related protein 1 (DRP1) at the mitochondrial surface [[44], [45], [46], [47]]. However, silencing of FUNDC1 or DRP1 did not abolish PGAM5-induced Parkin translocation (Figs. S6A and B), discarding the involvement of these PGAM5 substrates. Interestingly, DRP1 shRNA, but not FUNDC1 shRNA, blocked PGAM5-induced mitophagy (Figs. S6C and D). This allows us to suggest that although DRP1 is not required at the early stages of mitophagy, it is still required at later stages of mitophagy when mitochondria should undergo fission to be eliminated by autophagosomes.
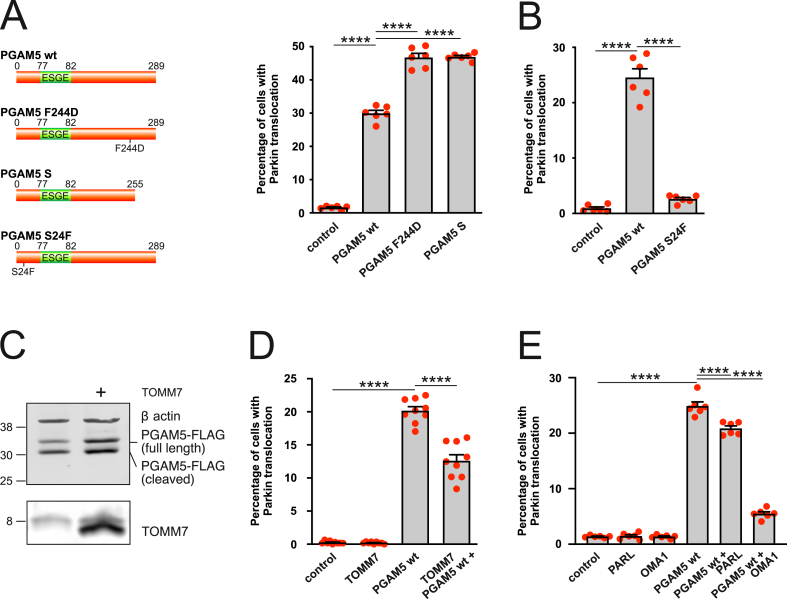
These findings led us to suggest that PGAM5 phosphatase activity might not necessarily be needed for PGAM5-induced mitophagy. We therefore introduced the F244D mutation to its dimer interface to prevent the oligomerization and abrogate phosphatase activity [43,48]. Fig. 8A demonstrates that phosphatase dead PGAM5 mutant induced Parkin translocation even more strongly than wt PGAM5. We observed similar effects when we overexpressed short isoform of PGAM5 (isoform 2, UniProt #Q96HS1-2) that had reduced ability to form dimers and lacked detectable phosphatase activity [49].
Fig. 8.
PGAM5 interferes with PINK1 processing independently of its phosphatase activity. A. PGAM5 mutant and isoform lacking phosphatase activity induce Parkin translocation. PC6 cells were transfected with Parkin-EYFP and PGAM5 wt, PGAM5 F244D or short PGAM5 isoform (isoform 2, UniProt #Q96HS1-2). ****P < 0.0001, n = 6 dishes, 20 fields per dish, Welch's ANOVA followed by Dunnett's T3 multiple comparisons test. B. PGAM5 mutant lacking PARL/OMA1 cleaving site does not induce Parkin translocation. PC6 cells were transfected with Parkin-EYFP and PGAM5 or PGAM5 S24F. ****P < 0.0001, n = 6 dishes, 20 fields per dish, Welch's ANOVA followed by Dunnett's T3 multiple comparisons test. C. TOMM7 overexpression increases the fraction of full-length PGAM5 in HEK293 cells expressing PGAM5-flag and TOMM7-myc. D. TOMM7 overexpression protects partially against PGAM5 induced Parkin translocation. PC6 cells were transfected with Parkin-EYFP, PGAM5 wt or/and TOMM7. ****P < 0.0001, n = 9 dishes, 20 fields per dish, Welch's ANOVA followed by Dunnett's T3 multiple comparisons test. E. OMA1 and PARL protect partially against PGAM5 induced Parkin translocation. PC6 cells were transfected with Parkin-EYFP, PGAM5 wt and PARL or OMA1. ****P < 0.0001, n = 6 dishes, 20 fields per dish, One-way ANOVA followed by Sidak's multiple comparisons test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We next suggested that PGAM5 might affect the processing of PINK1 in the mitochondria by competing for cleavage by IMM-resident proteases [50]. Both PGAM5 and PINK1 are cleaved by IMM-resident proteases PARL and OMA1 (an IMM-resident zinc metalloprotease), and they could compete for cleavage by IMM-resident proteases [50]. We therefore introduced S24F mutation into PGAM5, shown to abolish the cleaving site for PARL [39] and presumably also for OMA1 [51]. Notably, this mutant was no longer able to induce Parkin translocation (Fig. 8B), suggesting that PGAM5 requires the intact PARL/OMA1 cleavage site to activate the PINK1/Parkin pathway.
The processing of PINK1, and perhaps PGAM5, also involves TOMM7 (a small accessory protein of the TOM complex) mediating the lateral release of PINK1 to the OMM [52]. It has been demonstrated that TOMM7 competes with IMM-resident proteases for PINK1 [16]. We speculated that TOMM7 might also be involved in the processing of PGAM5. We overexpressed TOMM7 to saturate the IMM-resident proteases to check if this upregulation prevents the cleavage of PGAM5 and subsequently suppresses the Parkin translocation. Fig. 8 demonstrates that despite increased levels of full-length PGAM5 (Fig. 8C) when co-expressed with TOMM7, the PGAM5-induced Parkin translocation remained weak (Fig. 8D). Thus, it seems that the accumulation of full-length PGAM5 alone via escaping from protease cleavage in mitochondrial intermembrane space is not sufficient to activate PINK1/Parkin pathway. This hypothesis is indirectly supported by a finding that accelerated cleavage of PGAM5 by overexpression of IMM-resident proteases suppressed the PGAM5-induced Parkin translocation (Fig. 8E).
Altogether these data suggest that the pro-mitophagic effect of PGAM5 does not require its phosphatase activity and is likely to be related to the inhibition of proteolytic degradation of PINK1. This is indirectly supported by our results demonstrating that PGAM5 overexpression led to the accumulation of PINK1 (Fig. S7A) and specifically the full-length, uncleaved PINK1 (Fig. S7B). Moreover, Parkin translocation induced by PINK1 overexpression is only slightly suppressed in the absence of PGAM5 (Fig. S7C).
2.7. KEAP1–PGAM5 protein-protein interaction inhibitors activate mitophagy
Impaired mitophagy and uncontrolled accumulation of dysfunctional organelles are common denominators of a diverse range of pathologies. We therefore asked whether the pharmacological activation of the KEAP1-PGAM5-PINK1 pathway by KEAP1 inhibitors might be used to promote the efficient removal of damaged mitochondria and restore the energetic status of the cell. Several KEAP1 inhibitors of various chemical classes are currently at various stages of clinical development, and we decided to test whether these compounds could be used to activate the KEAP1-mediated mitophagy.
However, all tested electrophilic KEAP1 inhibitors that covalently modify cysteine residues in KEAP1, such as dimethyl fumarate (DMF), omaveloxone (RTA408) and RA839, depolarized mitochondria (Fig. S8A) after 24 h treatment. Although all of them induced a very strong Parkin translocation at a similar concentration range (Fig. S8B), it was impossible to differentiate between a specific KEAP1-mediated and depolarization-induced mitophagy.
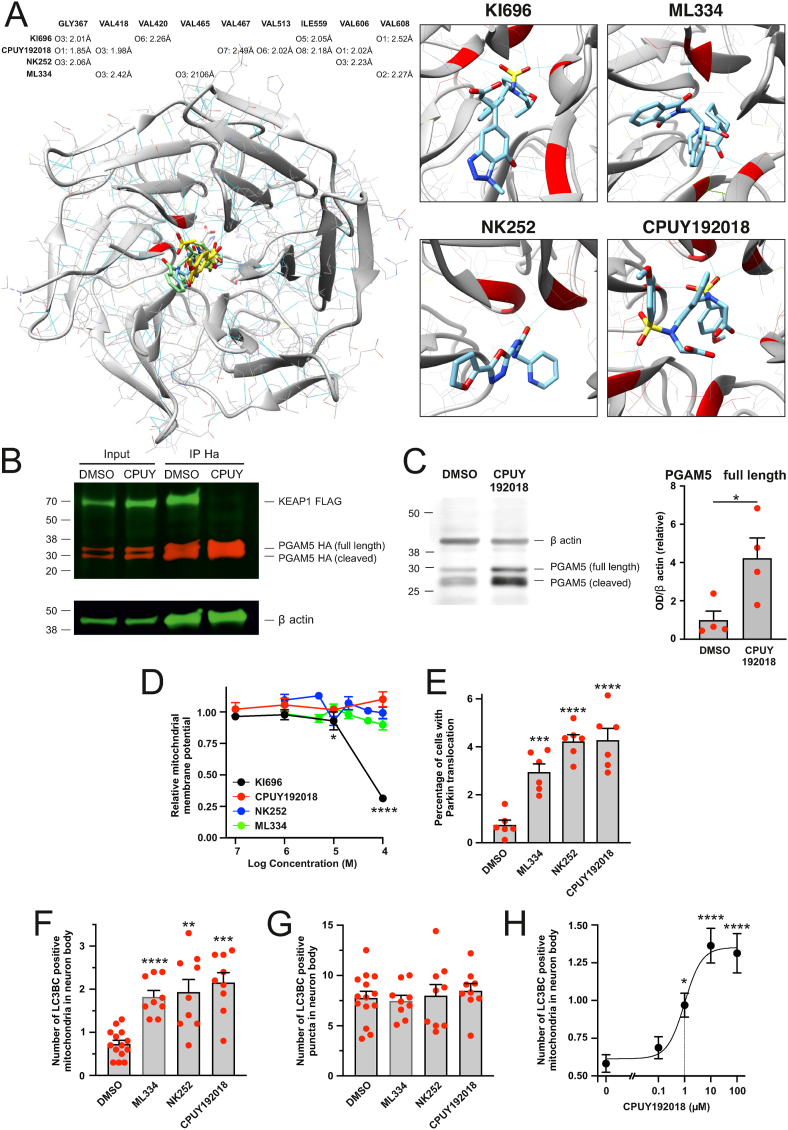
We therefore decided to continue with more specific non-electrophilic inhibitors that disrupt the protein-protein interaction between KEAP1 and its partners like NRF2. We selected four structurally different compounds, NK252, ML334, CPUY192018 and KI696, known to activate NRF2, and performed a molecular docking using a web program SwissDock [53,54] that is based on the docking software EADock DSS. Energetically most favorable interactions between a KEAP1 and these small molecular ligands are summarised in Fig. 9A. EADock DSS software predicted stable hydrogen bond interactions for all these compounds with different glycine, valine or isoleucine residues in KEAP1 through either carbonyl or sulfonyl group while CPUY192018 formed more hydrogen bonds with KEAP1 than others (Fig. 9A). Most of these residues lie in the NRF2 interaction pocket of KEAP1 close to the amino acids that make electrostatic contacts with the glutamate residues within the ET(S)GE motif [55], suggesting that these inhibitors should also block the interaction between the KEAP1 and PGAM5 that utilizes the ESGE motif for binding.
Fig. 9.
Inhibitors of KEAP1–PGAM5 protein-protein interaction enhance mitophagy. A. Molecular docking simulation showing energetically most favorable interactions between a KEAP1 and non-electrophilic KEAP1 inhibitors. The table is showing hydrogen-bond pairings in KEAP1-inhibitor complexes. The length of the hydrogen bond is given in angstroms. B. Treatment with CPUY192018 disrupts KEAP1-PGAM5 interaction. PC6 cells were co-transfected with KEAP1-FLAG and PGAM5-HA and treated for 24 h with DMSO or 100 μM CPUY192018. KEAP1-FLAG was co-immunoprecipitating with PGAM5-HA in DMSO but not in CPUY192018 treated cells. C. Treatment with CPUY192018 increases the level of endogenous PGAM5. PC6 cells were treated with 100 μM CPUY192018 for 24 h. The left panel shows a representative Western blot and the right panel quantitative analysis. *P < 0.05, n = 4 samples, t-test. D. Effect of non-electrophilic KEAP1 inhibitors on mitochondrial membrane potential. PC6 cells were treated with KEAP1 inhibitors for 24 h, after which the cells were stained with ratiometric mitochondrial membrane potential sensor JC10. *P < 0.05 and ****P < 0.0001 when compared with DMSO treated group, n = 7–12 wells per data point, One-way ANOVA followed by Dunn's multiple comparisons test. E. Non-electrophilic KEAP1 inhibitors induce Parkin-EYFP translocation to mitochondria. PC6 cells expressing Parkin-EYFP were treated with DMSO or 100 μM of KEAP1 inhibitors for 24 h ***P < 0.001 and ****P < 0.0001 when compared with DMSO treated group, n = 6 dishes per group, 20 fields per dish, One-way ANOVA followed by Dunnett's multiple comparisons test. F and G. Non-electrophilic KEAP1 inhibitors enhance mitophagy but not general autophagy. Neurons expressing mitochondrial marker Kate2 and autophagosome markers EGFP-LC3B and GFP-LC3C were treated with DMSO or 100 μM of KEAP1 inhibitors for 24 h. The number of LC3 positive puncta colocalizing with mitochondria (F) as well as the total number of LC3 positive puncta (G) was counted by a blinded observer. **P < 0.01, ***P < 0.001 and ****P < 0.0001 when compared with DMSO treated group, n = 9–14 dishes (10 cells per dish), Welch's ANOVA followed by Dunnett's T3 multiple comparisons test. H. Treatment of primary cortical neurons with CPUY192018 (24 h) induces a concentration-dependent increase in mitochondrial colocalization with autophagosome markers EGFP-LC3B/GFP-LC3C. *P < 0.05 and ****P < 0.0001, when compared with DMSO, treated group, n = 16–18 dishes from 4 independent experiments (10 cells per dish), Kruskal-Wallis test followed by Dunn's multiple comparisons test, the dotted line shows the EC50.
To test that, we treated the cells with CPUY192018, immunoprecipitated PGAM5, and visualized the amount of co-immunoprecipitating KEAP1. As demonstrated in Fig. 9B, CPUY192018 blocked the interaction between the KEAP1 and PGAM5 completely. Moreover, it led to a significant, 4-fold increase in the level of the endogenous PGAM5, also suggesting the blockage of proteasomal degradation (Fig. 9C).
Moreover, 3 out of 4 of these inhibitors, NK252, ML334 and CPUY192018 (but not KI696), did not affect the mitochondrial membrane potential (Fig. 9D); nevertheless, they were able to induce a slight but statistically significant increase in Parkin translocation (Fig. 9E). All these inhibitors also induced considerable mitophagy without affecting global autophagy (Fig. 9F and G). Relevantly, CPUY192018, known to have Kd around 39.8 nM in isothermal titration calorimetry (ITC) assay [56], was able to induce mitophagy already at submicromolar concentrations (EC50 0.998 μM, 95% Cl 0.23–4.23 μM; Fig. 9H). It should also be mentioned that 100 μM CPUY192018 did not affect axonal development in neuronal culture, suggesting to be relatively nontoxic under these conditions (average axonal length was 3398 ± 158 and 3425 ± 180 μm in DMSO and CPUY192018 treated groups, respectively (n = 156 and 167 axons from 11 to 12 dishes)).
Thus, we can demonstrate that compounds inhibiting KEAP1-PGAM5 interaction could serve as a mitophagy enhancer and could accelerate the removal of damaged mitochondria.
3. Discussion
Depending on the level, localization and circumstances, reactive oxygen species could initiate diverse cellular responses like triggering signaling pathways involved in cell protection or initiating coordinated activation of mitochondrial fission and autophagy to optimize clearance of abnormal mitochondria to protect spreading the damage to the neighboring mitochondria (for review see Ref. [57]). Too low or too high ROS levels may have deleterious consequences. When low, ROS cannot provide proper cellular functioning by regulating a significant number of biochemical reactions. Being too high, they impair various cell components, including mitochondria, which are, in turn, important sources of ROS. These ROS-damaged and ROS-producing mitochondria should be eliminated so that feedback mechanisms linking elevated mitochondrial ROS generation to the destruction of low-quality mitochondria producing ROS are essential for cell viability. Impairment of such mechanisms associated with insufficient mitophagy could contribute to the pathogenesis of various pathologies such as neurodegenerative diseases like Alzheimer's disease and Parkinson's disease [11,12,[58], [59], [60]].
In this paper, we discover a novel mechanism of mitophagy regulation by mitochondrial ROS generation. We demonstrate that moderate mitochondrial ROS production, either by menadione generating superoxide radicals in the vicinity of the inner mitochondrial membrane or superoxide optogenetically produced in the mitochondrial matrix, triggers small but significant Parkin translocation from cytosol to mitochondria. Our data shows that this ROS-induced Parkin translocation and enhanced mitophagy involves KEAP1 but not its primary substrate, NRF2. Instead, we have discovered that the levels of another KEAP1 binding partner, PGAM5, are effectively controlled by mitochondrial ROS and that even moderate oxidative stress could lead to PGAM5 accumulation.
We further demonstrated that PGAM5 accumulation increases the level of PINK1. We could not clearly establish the precise molecular mechanism of how PGAM5 accumulation is leading to PINK1 stabilization.
Nevertheless, we demonstrate that this mechanism does not involve FUNDC1 or DRP1, and it is independent of the dimerization and phosphatase activity of PGAM5. Since PGAM5 and PINK1 are cleaved in mitochondrial intermembrane space by the same proteases, PARL and OMA1 [13,16,39,51], PGAM5 might interfere with PINK1 at the level of its processing. Our data partially support this hypothesis because PGAM5 with mutated PARL (and OMA1?) cleavage site is less efficient in induction of Parkin translocation. However, as this construct is still cleaved, although less extensively [our data, 40], we cannot exclude the involvement of other proteases.
Our data also show that the role of PGAM5 in mitophagy is considerably more complex than previously thought. PGAM5 is involved in the dephosphorylation of FUNDC1 (an LC3 interacting protein) that helps to recruit autophagic isolation membrane to mitochondria under hypoxic conditions [[44], [46],61,62]. PGAM5 also promotes mitochondrial fission, which could contribute to mitophagy [63]. PGAM5 dephosphorylates Drp1 at Ser-637, triggering its GTPase activity leading to mitochondrial fragmentation [[64], [65]]. It has been shown that PGAM5 could also migrate to mitochondria-associated membranes and dephosphorylates Drp1with the assistance of the MAM protein Stx17 [45]. Thus, the role of PGAM5 in the mitochondrial fission regulation seems to be based mainly on Drp1 and future work is needed to investigate whether it could modulate functions of other proteins involved in fission (Fis1, Mff). PGAM5 has been earlier shown to protect PINK1 from degradation and to be indispensable for PINK1/Parkin-dependent mitophagy [38,45]. Moreover, genetic deficiency of PGAM5 has been shown to cause Parkinson's-like movement disorder in mice [38]. Here we demonstrate that PGAM5 being required for PINK1/Parkin dependent mitophagy is a mediator between oxidative stress and the mitophagy machinery.
Another interesting question is the association between mitophagy and apoptosis, as these processes are both affected by reactive oxygen species. ROS promotes cell survival at lower amounts, whereas higher amounts activate death processes such as apoptosis. Similarly, the role of PGAM5 in apoptosis seems to be associated with the degree of stress. Under mild stress conditions, PGAM5 phosphorylates Bcl-xL facilitating the sequestration of proapoptotic proteins such as Bak and Bax, thereby indirectly inhibiting apoptosis [22]. Conversely, PGAM5 recruits Drp1 and Bax to mitochondria under severe stress, increasing mitochondrial fission and mitochondrial outer membrane permeabilization, leading to mitochondria-triggered apoptosis [64]. In our experiments, the menadione concentrations inducing mitophagy did not provoke apoptosis but instead slightly inhibited it. One may speculate that increased mitophagy helps to remove damaged ROS-producing mitochondria (which could serve as a source of apoptotic mediators), thus contributing to an antiapoptotic activity. This is an intriguing question that should be studied further.
Current findings also shed more light on the complexity of KEAP1 dependent pathways. It is not only the KEAP1-NRF2 pathway, which activates cytoprotective responses to stresses caused by ROS, but also the KEAP1-PGAM5 pathway. It has been thought that the role of the KEAP1 interaction with PGAM5 is to relocate the KEAP1 from the cytosol to the outer mitochondrial membrane [42,66]. Although PGAM5 has been shown to be ubiquitinated by the KEAP1-dependent ubiquitin ligase complex [41,42], it has been suggested that it may not be efficiently targeted for proteasome-dependent degradation after ubiquitination [42]. Here we show that PGAM5 behaves similarly to the “canonical” substrate of KEAP1, NRF2. PGAM5 is partially degraded under basal conditions allowing only a fraction of synthesized PGAM5 to enter the mitochondria while at oxidative stress, the PGAM5 degradation is inhibited so that this protein is able to migrate to mitochondria to facilitate mitophagy.
Inhibition of the protein-protein interaction between the transcription factor NRF2 and its repressor KEAP1 has emerged as a promising strategy to target oxidative stress in diseases [3,67,68], but what has usually not been taken into account that all these inhibitors bind to NRF2 interaction pocket and also break interactions between KEAP1 and its other substrates. Indeed, our data demonstrate that these inhibitors block the KEAP1-PGAM5 protein-protein interaction leading to an accumulation of PGAM5, which induces mild mitophagy. However, one cannot fully exclude also a partial involvement of NRF2 of in this process.
Interestingly, we also noticed that all electrophilic KEAP1 inhibitors and one of the protein-protein interaction (PPI) inhibitors led to abrupt dissipation of mitochondrial membrane potential. This suggests that potential direct mitochondrial toxicity should always be taken into account when studying the KEAP1 inhibitors. There is currently only one KEAP1 inhibitor, DMF, in clinical use to treat psoriasis and remitting-relapsing multiple sclerosis, but there are many more in phase I-III clinical studies [3]. Notably, recent clinical studies describing the patients with dimethyl fumarate-associated renal Fanconi syndrome identified mitochondrial toxicity in proximal tubules [69].
In summary, the data obtained allow us to propose a mechanism by which elevated mitochondrial ROS production reduces mitochondrial mass via increased mitophagy and, in such a way, negatively regulates the source of ROS. This mechanism does not need mitochondrial depolarization and could be operating under conditions of a relatively moderate ROS increase. Oxidants react with cysteine sensors within KEAP1, causing a conformational change and the inability of KEAP1 to target its substrates for degradation. Among these targets, we have identified PGAM5 as a central element, which is not any more degraded and is starting to accumulate in the cytosol under oxidative stress conditions. Such an accumulation induces stabilization of PINK1, probably by affecting its intramitochondrial processing, leading finally to Parkin translocation to mitochondria triggering mitophagy. In such a way, enhanced ROS level ensures negative feedback eliminating mitochondria, the main source of ROS. Knowledge of these mechanisms might have important medical implications because reduced elimination of damaged and/or non-functional mitochondria is a hallmark of several neurodegenerative diseases like Alzheimer's and Parkinson's disease. Under these conditions, pharmacological stimulation of the KEAP1/PGAM5 pathway could permit to activate weakened mitophagy. Further studies are needed to identify whether these mitophagy activators could bolster mitochondrial health and ameliorate cell viability in neurodegenerative pathologies.
4. Material and methods
4.1. Reagents and resources
| REAGENT or RESOURCE |
SOURCE |
IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Ms anti-FLAG M2 | SIGMA Aldrich | Cat# F1804 |
| Rb anti-FLAG | SIGMA Aldrich | Cat# F7425 |
| Rb anti-Rabbit | Abcam | Cat#a b9110 |
| Novus | Cat# NPB2-21581 | |
| Ms anti-Myc | Invitrogen | Cat# 46-0603 |
| Rb anti-Myc | Abcam | Cat# ab9106 |
| Novus | Cat# NB600-336 | |
| Ms anti-β-Actin | SIGMA Aldrich | Cat# A2228 |
| Rb anti-KEAP1 | Abcam | Cat# ab139729 |
| Rb anti-Pink1 | Novus | Cat# BC100-94 |
| Rb anti-VDAC1 | Abcam | Cat# ab15895 |
| Ms anti-ATG5a | Abcam | Cat# Ab14748 |
| Ms anti-GADPH | Abcam | Cat# Ab8245 |
| Rb anti-GFP | Abcam | Cat# ab290 |
| Rb anti-PGAM5 | Atlas Antibodies | Cat# HPA036978 |
| Secondary antibodies: | ||
| Goat anti-Mouse IR Dye 680LT | LICOR | Cat# 926-68020 |
| Goat anti-Mouse IRDye 800CW | LICOR | Cat# 926-32210 |
| Goat anti-Rabbit IRDye 800CW | LICOR | Cat# 926-32211 |
| Goat anti-Rabbit IR Dye 680LT | LICOR | Cat# 926-68021 |
| Chemicals, Peptides and Recombinant Proteins | ||
| Menadione sodium bisulfite | Sigma-Aldrich | Cat# M2518 |
| Hydrogen peroxide solution 30% | Sigma-Aldrich | Cat# H1009 |
| FCCP | Tocris | Cat# 0453 |
| GSH-MEE | Sigma-Aldrich | Cat# G1404-25 MG |
| MG132 | Tocris | Cat# 1748 |
| DMF | Tocris | Cat# 4512 |
| ML334 | Tocris | Cat# 5625 |
| RTA408 | CAYMAN Chemical Company | Cat# 17854 |
| RA839 | Tocris | Cat# 5707 |
| NK252 | Tocris | Cat# 5293 |
| CPUY192018 | AOBIOUS, INC | Cat# AOB 9974 |
| KI696 | MedChem Express | Cat# HY-101140 |
| TMRE | Sigma-Aldrich | Cat# 87917 |
| MitoTracker™ Green FM | Thermo Fischer Scientific | Cat# M7514 |
| JC10 | Enzo Life Sciences | Cat# ENZ-52305 |
| Collagen, type IV | Sigma-Aldrich | Cat# C5533 |
| Poly-l-lysine hydrobromide | Sigma-Aldrich | Cat# P6282 |
| RPMI Medium (IX) | Gibco, ThermoFisher | Cat# A1049101 |
| Scientific | ||
| DMEM, high glucose, GlutaMAX™ Supplement, pyruvate | Gibco, ThermoFisher | Cat# 31966021 |
| Scientific | ||
| Fetal Bovine Serum | Gibco, ThermoFisher | Cat# 10270106 |
| Scientific | ||
| Horse serum | Gibco, ThermoFisher | Cat# 26050088 |
| Scientific | ||
| B27™ | Gibco, ThermoFisher | Cat# 17504044 |
| Scientific | ||
| B27™ Supplement minus antioxidants | Gibco, ThermoFisher | Cat# 10889038 |
| Scientific | ||
| RIPA Lysis and Extraction Buffer | Thermo Scientific | Cat# 89900 |
| Intercept® Blocking buffer | Li-COR | Cat# 927-70001 |
| IP lysis buffer (20 mM HEPES, 100 mMNaCl, 5 mM EDTA, 1% Triton X-100, 10% glycerol, pH 7.4) | Prepared in the lab | |
| Qproteome Mitochondria Isolation Kit | Qiagen | Cat# 37612 |
| RNAeasy mini kit | Qiagen | Cat# 74104 |
| SuperScript III RT Kit | Invitrogen | Cat# 18080-044 |
| FastSYBR Green Master Mix | Fermentas | Cat# K0229 |
| Life Sciences | ||
| RnaseZAP | Ambion | Cat# AM9780, |
| Cat# AM9782 | ||
| DC™ Protein Assay Reagent A, B, C | BIO-RAD | Cat# 500-0113 |
| Cat# 500-0114 | ||
| Cat# 500-0115 | ||
| Critical Commercial Assays | ||
| ROS-Glo™ H2O2 assay | Promega | Cat# G8820 |
| CytoTox-Glo™ Assay | Promega | Cat# G9290 |
| Caspase-Glo® 3/7 Assay System | Promega | Cat# G8090 |
| MitoSOX™ Red | Thermo Fischer Scientific | Cat# M36008 |
| Experimental Models: Cell Lines | ||
| PC6-3 | Gift from Dan Lindholm | RRID: CVCL_7101 |
| HEK293 | ATCC | Cat# CRL-1573 |
| HELA | ATCC | Cat# CRM-CCL-2 |
| NRF2 deficient mouse embryonic fibroblasts | Gift from Henrik Luuk | |
| Oligonucleotides | ||
| rPGAM5-F-AGATGAACTGGCTTCCAGGC | TAG Copenhagen A/S | |
| rPGAM5-R-CCTGTTCCCGACCTAATGGG | ||
| rPINK1-F-ACCTGGAGAAGGCCAAACAC | TAG Copenhagen A/S | |
| rPINK1-R-CTGGAGGATCCTGCCGAGATA | ||
| rCycA-F-GAGCACTGGGGAGAAAGGAT | TAG Copenhagen A/S | |
| rCycA-R-CTTGCCATCCAGCCACTCAG | ||
| rKEAP1-F-TCTTCAACCTGTCACACTGC | TAG Copenhagen A/S | |
| rKEAP1-r-ATCTCACACTTCTGCAGCTG | ||
| Recombinant DNA | ||
| KEAP1 shRNA | SABiosciences | Cat# KR47610 N |
| AAGGTCATGGAGAGGCTAATT (no 2) | ||
| GGACTACCTGGTGCAGATCTT (no 3) | ||
| PGAM5 shRNA | OriGene | Cat# TR702898 (TI711593) |
| ATTGAGGCTGCCTTCAGGAACTACATCCA | ||
| PINK1 shRNA | SABiosciences | Cat# KR55105 N |
| CTGCAATGCCGCTGTGTATGA | ||
| DRP1 shRNA | SABiosciences | Cat# KR45077 N |
| CGAGCAGGGTGTTCTGAACTT | ||
| FUNDC1 shRNA | Origene | Cat# TR702819 |
| Mito-CFP | Clontech. | Cat# 632432 |
| DsRed2-Mito | Clontech. | Cat# 632421 |
| mKate2-mito | Evrogen | Cat# FP187 |
| KillerRed-dMito | Evrogen | Cat# FP964 |
| MLS-HyPer7 | Addgene | Cat# 136470 |
| hSyn-EGFP | Addgene | Cat# 50465 |
| EGFP-LC3B | Addgene | Cat# 24920 |
| Cat# 11546 | ||
| Cat# 24920 | ||
| GFP-LC3C | MRC reagents | Cat# DU40779 |
| Parkin-EYFP and Parkin-EYFP C431 N | Gift from Richard Youle | |
| Parkin-EYFP T340R | generated from Parkin-EYFP by Mutagenex Inc. | |
| PINK1-EYFP | Addgene | Cat# 101874 |
| PINK1 | Gift from Emma Deas | |
| HSP90AA1 | SinoBiological | Cat# HG11445-UT |
| Myc-RICTOR | Addgene | Cat#11367 |
| MAPKAP1-Myc | Addgene | Cat#12576 |
| APPL2 | Harvard PlasmID Database | CloneID: HsCD00335054 |
| GRP75 | Gift from Gyorgy Szabadkai | |
| Flag-NRF2 WT and Flag-NRF2 ETGE to GAGA | Generated in our lab | |
| FLAG-KEAP1 and FLAG-KEAP1 R380A-R415A | Generated in our lab | |
| HA2-KEAP1 | Addgene | Cat# 21556 |
| PGAM5-Myc-DDK | Origene | Cat# RC229840 |
| PGAM5-Myc, PGAM5-Myc E79A/S80A, PGAM5-Myc F244D, PGAM5-Myc S24F | Generated from the previous plasmid by EUROFINS | |
| PGAM5-HA | MRC reagents #DU46867 | |
| PGAM5-YPet | Synthesized by Thermo Fischer | |
| PGAM5-Myc-DDK transcript variant 3 (isoform 2, UniProt #Q96HS1-2) | OriGene | Cat# RC201678 |
| TOMM7- Myc-DDK | Origene | Cat# RC210040 |
| PARL-FLAG | Addgene | Cat# 13639 |
| OMA1-FLAG | Addgene | Cat# 61719 |
| HyPer7 | Addgene | Cat# 136466 |
| MLS-HyPer7 | Addgene | Cat# 136470 |
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors, Vinay Choubey (vinay.choubey@ut.ee) and Allen Kaasik (allen.kaasik@ut.ee).
4.2. Cell cultures
Primary rat cortical neuron cultures were prepared from <1 day old neonatal Wistar rats as described earlier [69]. Neurons were plated in BME supplemented with 10% FBS, 2 mM GlutaMAX™-I and 100 μg/ml gentamicin on 35 mm glass bottom dishes (MatTek, MA, USA) precoated with poly‐l‐lysine (106 cells per dish in 2 ml of cell suspension). After incubating for 3 h, the medium was changed to Neurobasal™ A medium containing B‐27™ or B‐27™ plus supplement, 2 mM GlutaMAX™‐I and 100 μg/ml gentamicin. In experiments involving menadione, an antioxidant-free B-27™ supplement was used.
PC6 cells were grown in RPMI‐1640 medium supplemented with 10% horse serum and 5% FBS on collagen IV‐coated 35 mm glass bottom dishes or PEI-coated plastic dishes (60 and 100 mm). HEK293 and HeLa cells were cultured in DMEM containing GlutaMAX™‐I supplemented with 10% FBS on poly‐l‐lysine coated 100‐mm plastic dishes.
Mouse embryonic fibroblasts were cultured in DMEM containing 2 mM GlutaMAX™‐I, supplemented with 10% FBS and a 1% solution of penicillin and streptomycin.
4.3. Transfection
Primary cortical neurons were transfected at DIV2-3. For the transfection of cells growing on glass bottom dishes, the conditioned medium was replaced with 100 μl Opti-MEM I medium containing 2% Lipofectamine® 2000 and 1–2 μg total DNA with an equal amount of each different plasmid. The dishes were incubated with transfection mixture for 2–3 h following that fresh neurobasal medium (Phenol red-free, supplemented with B27 and Glutamax-I) was added.
PC6 and MEFs cells on the glass-bottomed dishes (coated with Collagen IV) were transfected as described above, but transfection was completed with the addition of RPMI (supplemented with 10% HS and 5% FBS) or DMEM medium (supplemented with 10% FBS) without antibiotics or antimycotic.
For biochemical analyses, HEK, PC6, or HeLa cells were transfected in either 60-mm or 100 mm plastic dishes coated with poly l-lysine or polyethyleneimine (PEI, for PC6 cells). The medium was replaced with 2 ml or 3 ml of Opti-MEM I medium containing 2% Lipofectamine® 2000 and 20–25 μg of total DNA with an equal amount of each plasmid of interest. After an incubation of 3–4 h with transfection mixture, a fresh medium was added. For experiments with shRNA-expressing plasmids containing a neomycin resistance gene (shRNA efficiency testing), the PC6 cell medium was supplemented with 200 μg/ml G418 for 6–7 days. In all cases when the specific shRNAs were transfected, the same amount of scrambled shRNA was included in the control group(s). In case of overexpression, either empty vector, pcDNA3-VN, EGFP/EYFP, firefly or Renilla luciferase was used as compensation.
4.4. Parkin translocation
PC6 cells and MEFs transfected with EYFP-Parkin and plasmids of interest were visualized 2–3 days later using an Olympus IX70 inverted microscope (Tokyo, Japan) equipped with a WLSM PlanApo 40x/0.90 water immersion objective and Olympus DP70 CCD camera (Tokyo, Japan). In menadione experiments, the antioxidant containing RPMI was replaced with antioxidant and serum-free DMEM and treated with menadione or DMSO for 4 h. For each dish, at least 20 fields per dish were randomly captured for further analysis of the percentage of cells showing EYFP-Parkin translocation.
4.5. Optogenetic ROS generation and Parkin translocation
The PC6 cells were plated in collagen IV- and poly-l-lysine-coated 35 mm glass-bottom dishes subdivided into four individual compartments (Cellvis, Mountain View, CA, cat. No. D35C4-20-1.5-N) to allow simultaneous analysis of various treatments and diverse transfections. Cells were transfected one day after plating with Parkin-EYFP, mitochondria-targeted KillerRed and plasmids of interest, and visualized 3–4 days later. Sub-population of mitochondria was then irradiated with a 561 nm laser line of LSM 780 confocal microscope (using Plan-Apochromat 63x/1.4 oil immersion objective and ROI size of 6.73 x 6.73 μm) to induce localized mitochondrial ROS burst. The subcellular changes in the localization of Parkin-EYFP were visualized using a 514 nm laser line 60, 90 and 120 min later.
To quantify the strength of Parkin translocation from cytosol to mitochondria, we then quantified the spatial heterogeneity (coefficient of variation of the intensity of individual pixels) of the Parkin-EYFP signal from individual cells for each time point using ImageJ. Spatial heterogeneity was relatively low (around 0.2) when Parkin-EYFP was homogeneously cytosolic and reached close to 1 in cells where most cytosolic Parkin-EYFP translocated to mitochondria.
4.6. Mitophagy
Primary cortical neurons were transfected at DIV 2–3 with EGFP-LC3B, GFP-LC3C, mitochondrial Kate2 and plasmids of interest. Co-localization of LC3 dots with mitochondrial Kate2 in neuronal somas was visualized 2–3 days later on LSM 510 (LCI Plan-Neofluar 63x/1.3 water immersion objective). In menadione experiments, the neurons were grown in Neurobasal™ A medium containing antioxidant-free B‐27® supplement (menadione was not inducing mitophagy in neurons grown in antioxidant containing B‐27® supplemented media) and treated with menadione 4 h before visualization. For each dish, approximately 5–10 cells per dish per condition were randomly captured, and images were further analyzed by a blinded observer.
4.7. Mitochondrial membrane potential
For mitochondrial membrane potential measurements in neurons, the neurons growing in glassbottom dishes were transfected with mitochondrial ECFP and scrambled or KEAP1 shRNA at DIV 2–3. 3–4 days post-transfection or post-incubation with inhibitors, the cells were incubated with 5 nM Tetramethyl rhodamine ethyl ester (TMRE) and 100 nM MitoTracker™ Green FM (Thermo Fischer Scientific, cat. no. M7514) for 30 min at 37 °C in the dark. Cells were then washed with Krebs and imaged using a laser scanning confocal microscope LSM 510 Duo (Carl Zeiss Microscopy GmbH, Göttingen, Germany) equipped with an LCI Plan-Neofluar 63 × /1.3 water immersion objective. ECFP, MitoTracker Green™ and TMRE signals were acquired sequentially using the 405, 488 and 561 laser lines, respectively. Cells were also treated with 10 μM FCCP for 5 min to give maximal dissipation of the Δѱm. TMRE and MitoTracker Green signal from non-transfected or transfected individual neurons was measured using ImageJ. The data is presented as TMRE/MitoTracker signal ratio (to exclude the effect of different mitochondrial density) and normalized further to non-transfected cells in the same image (to minimize the variation of the staining intensity between the dishes).
For mitochondrial membrane potential measurement in PC6 cells, the PC6 cells were grown overnight on a Nunc™ MicroWell™ 96-Well Optical-Bottom Plates (Thermo Fischer Scientific) and then were treated with compounds of interest. After that, the old media was removed and fresh media containing JC10 dye (Enzo Life Sciences) was added to each well, and the plate was incubated for 30 min at 37 °C.
Both JC10 Fluorescence was measured using the Flex station II (Molecular Devices) using absorption/emission of 485/538 nm for a monomeric green form of JC10 and 544/590 nm, for aggregate red form. The results are presented as a red to green emission ratio that should only depend on the membrane potential and no other factors like shape or density.
4.8. Mitochondrial density
For mitochondrial density measurements, the neurons were transfected at DIV3-5 with neuronal marker hSyn-EGFP, mitochondrial DsRed2 and plasmids of interest. At DIV9–10, the segments of the axons close to the end of the axon from randomly selected neurons were visualized using an Olympus IX70 inverted microscope (WLSM PlanApo 40x/0.90 water immersion objective). Mitochondrial length measurements were performed as described previously [70].
4.9. ROS production
To measure ROS induced by menadione, we used ROS-Glo™ H2O2 assay (Promega, G8820). PC6 cells were grown overnight on an opaque white 96 well plate. On the next day, the cells were treated with menadione (different concentrations) alone or in combination with cell-permeable reduced glutathione ethyl ester (GSH-MEE) and subjected to ROS-Glo™ H2O2 assay (Promega). Briefly, the H2O2 substrate solution were added, bringing the final volume to 100 μl per well. The plate was incubated at 37 °C in a 5% CO2 incubator. After 4 h incubation, 100 μl of the ROS-Glo detection solution was added to each well and further incubated the plate for 30 min at room temperature. The luminescence was recorded using the Glomax Multi Detection System (Promega).
4.10. Dead cell protease activity
The CytoTox-Glo™ Assay (Promega, G9290) was used to measure the dead-cell protease activity. Briefly, PC6 cells were seeded in an opaque white 96 well plate and treated with menadione the same way mentioned above. 50 μl of CytoTox-Glo™ reagent was added to target wells of the plate and mixed briefly by orbital shaking. The plate was incubated for 15 min at room temperature, and afterward, the luminescence was measured.
4.11. shRNA efficiency testing
PC6 cells were seeded in 35 mm plastic dishes (Thermo Scientific) precoated with collagen type IV. The next day, cells were transfected with plasmids containing shRNAs targeted against rat KEAP1, PGAM5, PINK1, or non-targeted scrambled shRNA and GFP to monitor transfection efficiency. At 24 h post-transfection, cells were treated every day with G418 disulfate salt (200 μg/ml) (Sigma) for selection. After 6 days of transfection, cells were first imaged and then harvested and subjected for expression analysis. Briefly, total RNA was extracted using RNAeasy mini kit (Qiagen), and the first-strand synthesis was performed using 5 μg of total RNA with Maxima First Stand cDNA Synthesis Kit (Thermo). cDNAs were subjected to qPCR using specific primers for CYC (as housekeeping gene), KEAP1, PGAM5 and PINK1 on QuantStudio 12K Flex from Applied Biosystems by Life technologies. Acquired data were analyzed by the delta-delta Ct method and normalized to transfection efficiency estimated separately.
4.12. Mitochondria isolation
PC6 cells were seeded in 100-mm plastic dishes (Thermo Scientific) precoated with collagen type IV. The next day, cells were transfected with PGAM5-WT-Flag alone or plasmids containing shRNA against rat KEAP1 or non-targeting ± KEAP1-flag. Cells were treated with DMSO or FCCP or 25 μM MG132 for 4 h followed by the harvesting of the cells for mitochondrial isolation. Mitochondrial-enriched fractions were prepared using Qproteome mitochondrial isolation kit (Qiagen) following the manufacturer's instructions. Mitochondrial-enriched fractions were solubilized in RIPA buffer (Thermo) containing Complete protease inhibitor cocktail (Roche). Protein content in cytosolic and mitochondrial fractions was evaluated by DC protein assay method (Bio-Rad).
4.13. Co-immunoprecipitation
For immunoprecipitation, the transfected or non-transfected HEK293 or PC6 cells were collected (in some experiments post menadione or CPUY192018 treatment) in their culture media and pelleted at 500 g for 5 min at 4 °C. Cells were washed in PBS, pelleted at 500 g for 5 min at 4 °C, and then lysed in ice-cold IP lysis buffer containing 20 mM HEPES, 100 mM NaCl, 5 mM EDTA, 1% Triton X-100, 10% glycerol pH 7.4 and protease inhibitor cocktail (Roche). Cell lysates were passed through a 26G needle 10 times before incubating on ice for 30 min with shaking at 350 rpm. The insoluble fraction was removed by centrifuging at 30 000 g for 10 min at 4 °C. The protein concentrations in the lysates were estimated using DC protein assay method (Bio-Rad) according to the manufacturer's protocol. Lysates were incubated overnight on a rotary shaker at 4 °C in Pierce spin columns with 5 μg of rabbit anti-Flag, rabbit anti-HA, or Rabbit IgG antibodies. The following day 100 μl protein G-sepharose 4B beads (50% suspension in IP lysis buffer) were added and further incubated for 5 h on a rotary shaker at 4 °C. Beads were then washed 5 times with PBS, and immunoprecipitates were eluted twice in 2x Lane marker reducing sample buffer (Pierce) in PBS at 99 °C for 10 min.
4.14. Western blotting
RIPA-solubilized mitochondrial fractions, RIPA-solubilized total cell lysates from inhibitor-treated or shRNA transfected cells, cytosolic fractions, inputs, and elutes from immunoprecipitation experiments were analyzed using Western blotting (WB). Equivalent amounts of proteins were resolved on 10% or 12% or 4–20% gradients (Bio-Rad) polyacrylamide gels by SDS-PAGE. Resolved proteins were transferred to Immobilon® PSQ membrane (ISEQ00010, Millipore) in 0.1 M Tris-base, 0.192 M glycine and 10% (w⁄w) methanol using an electrophoretic transfer system with cold-block. The membranes were blocked using Odyssey blocking buffer (LI-COR Bioscience, 927–40000) at room temperature for 1 h. After blocking, the membranes were incubated sequentially overnight with different primary antibodies based on the requirement of the experiment. Antibody dilutions used were rabbit (Rb) anti-PGAM5 1:1000, mouse (Ms) anti-Actin 1:4000, Rb anti-KEAP1 1:1000, Ms anti-ATG5A 1:1000, Rb anti-VDAC1 1:2000, Rb anti-flag 1:1000, Ms anti-flag 1:1000, Rb anti-PINK 1:500, Rb anti-GFP 1:5000 and Goat anti-HA 1:1000. Incubations were followed by 3x washing in Tris Buffered Saline containing 0.1% Tween-20 followed by incubation with the appropriate secondary antibody (1:5000) either goat anti-rabbit IRDye 800CW or 680LT or goat anti-mouse IRDye 800CW or 680LT or donkey anti-goat IRDye 680LT (all from LI-COR Biosciences) for 1 h at room temperature. Immuno-reactive bands were detected by the Odyssey Infrared Imaging System.
4.15. Nuclear NRF2
PC6 cells were transfected with KEAP1 shRNA or scrambled shRNA and NRF2-GFP plasmids. After 72 h of transfection, cells were checked for NRF2-GFP expression under the fluorescence microscope (Olympus IX70). For each experimental condition, 10 images per dish were randomly captured. Cells were also treated with proteasome inhibitor MG132 to induce maximal nuclear translocation of NRF2. Cells demonstrating predominantly cytosolic or nuclear NRF2-GFP expression were counted using ImageJ, and the results are presented as the percentage of cells showing predominantly nuclear NRF2-GFP signal.
4.16. Statistical analysis
Data are presented as the mean ± SEM or Tukey boxplot. The D'Agostino-Pearson omnibus test was used to test the normality of distribution. To test the equality of variances, we used the F test for two conditions or the Brown and Forsythe test for more than two conditions. Student t-tests, Mann–Whitney U-tests, one-way ANOVA and repeated-measures ANOVA followed by Sidak post hoc test, Welch's ANOVA followed by Games–Howell test, or Kruskal–Wallis tests followed by the Dunn test were used to compare differences between experimental samples and control groups. Two-way ANOVAs were used to analyze interactions between two factors. P-values of <0.05 were considered statistically significant.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
☐The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Acknowledgments
We thank Dr. R. Youle, E. Deas and G. Szabadkai for providing the plasmids used in this study and Dr. H. Luuk and D. Lindholm for cell lines. We thank Ulla Peterson for her preparation of primary neurons.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102186.
Contributor Information
Vinay Choubey, Email: Vinay.Choubey@ut.ee.
Allen Kaasik, Email: Allen.Kaasik@ut.ee.
Funding
This work was supported by grants from the Estonian Research Council (PRG400), the European Regional Development Fund (Project No. 2014-2020.4.01.15-0012) and the European Union's Horizon 2020 research and innovation program under grant agreement 692202. A.K. was supported by Chan Zuckerberg Initiative and I.I. by EMBO Installation grant 2524 and Estonian Research Council grant PUT124.
Author contributions
VC, DS, VV and AK conceived and designed the project. AZ, VC, DS, RG, II, KT and AK designed experiments. AZ, VC, RG, MK, AV, DS, NG, ML, KT performed and analyzed experiments. II provided reagents. AZ, VC, RG, DS, VV and AK wrote the manuscript. All authors reviewed and edited the manuscript.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Kasai S., Shimizu S., Tatara Y., et al. Regulation of Nrf2 by mitochondrial reactive oxygen species in physiology and pathology. Biomolecules. 2020 Apr;10(2) doi: 10.3390/biom10020320. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deshmukh P., Unni S., Krishnappa G., et al. The Keap1–Nrf2 pathway: promising therapeutic target to counteract ROS-mediated damage in cancers and neurodegenerative diseases. Biophys Rev. 2017 Apr;9(1):41–56. doi: 10.1007/s12551-016-0244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuadrado A., Rojo A.I., Wells G., et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019 Apr;18(4):295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Nartiss Y., Steipe B., et al. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy. 2012 Oct;8(10):1462–1476. doi: 10.4161/auto.21211. [DOI] [PubMed] [Google Scholar]
- 5.Schofield J.H., Schafer Z.T. Mitochondrial reactive oxygen species and mitophagy: a complex and nuanced relationship. Antioxidants Redox Signal. 2021 Mar 1;34(7):517–530. doi: 10.1089/ars.2020.8058. [DOI] [PubMed] [Google Scholar]
- 6.Kurihara Y., Kanki T., Aoki Y., et al. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 2012 Jan 27;287(5):3265–3272. doi: 10.1074/jbc.M111.280156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank M., Duvezin-Caubet S., Koob S., et al. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim. Biophys. Acta Mol. Cell Res. 2012 Dec;1823(12):2297–2310. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Yoo S.M., Jung Y.K. A molecular approach to mitophagy and mitochondrial dynamics. Mol. Cell. 2018 Jan 31;41(1):18–26. doi: 10.14348/molcells.2018.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickles S., Vigié P., Youle R.J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 2018 Feb 19;28(4):R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palikaras K., Lionaki E., Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 2018 Sep;20(9):1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 11.Pickrell A.M., Youle R.J. The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015 Jan 21;85(2):257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu C.T. Multiple pathways for mitophagy: a neurodegenerative conundrum for Parkinson's disease. Neurosci. Lett. 2019 Apr 1;697:66–71. doi: 10.1016/j.neulet.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deas E., Plun-Favreau H., Gandhi S., et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 2011 Mar 1;20(5):867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekine S. PINK1 import regulation at a crossroad of mitochondrial fate: the molecular mechanisms of PINK1 import. J. Biochem. 2020 Mar 1;167(3):217–224. doi: 10.1093/jb/mvz069. [DOI] [PubMed] [Google Scholar]
- 15.Sekine S., Youle R.J. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol. 2018 Jan 10;16(1):2. doi: 10.1186/s12915-017-0470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekine S., Wang C., Sideris D.P., et al. Reciprocal roles of Tom7 and OMA1 during mitochondrial import and activation of PINK1. Mol. Cell. 2019 Mar 7;73(5):1028–1043. doi: 10.1016/j.molcel.2019.01.002. e5. [DOI] [PubMed] [Google Scholar]
- 17.Narendra D., Tanaka A., Suen D.F., et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008 Dec 1;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kane L.A., Lazarou M., Fogel A.I., et al. PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014 Apr 28;205(2):143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazarou M., Sliter D.A., Kane L.A., et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015 Aug 20;524(7565):309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao B., Goh J.Y., Xiao L., et al. Reactive oxygen species trigger Parkin/PINK1 pathway–dependent mitophagy by inducing mitochondrial recruitment of Parkin. J. Biol. Chem. 2017 Oct 6;292(40):16697–16708. doi: 10.1074/jbc.M117.787739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao B., Deng X., Lim G.G.Y., et al. Superoxide drives progression of Parkin/PINK1-dependent mitophagy following translocation of Parkin to mitochondria. Cell Death Dis. 2017 Oct 12;8(10) doi: 10.1038/cddis.2017.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma K., Zhang Z., Chang R., et al. Dynamic PGAM5 multimers dephosphorylate BCL-xL or FUNDC1 to regulate mitochondrial and cellular fate. Cell Death Differ. 2020 Mar;27(3):1036–1051. doi: 10.1038/s41418-019-0396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyanagi T., Yamazaki I. One-electron-transfer reactions in biochemical systems. V. Difference in the mechanism of quinone reduction by the NADH dehydrogenase and the NAD(P)H dehydrogenase (DT-diaphorase) Biochim. Biophys. Acta. 1970;216:282–294. doi: 10.1016/0005-2728(70)90220-3. [DOI] [PubMed] [Google Scholar]
- 24.Criddle D.N., Gillies S., Baumgartner-Wilson H.K., Jaffar M., Chinje E.C., Passmore S., Chvanov M., Barrow S., Gerasimenko O.V., Tepikin A.V., Sutton R., Petersen O.H. Menadione-induced reactive oxygen species generation via redox cycling promotes apoptosis of murine pancreatic acinar cells. J. Biol. Chem. 2006;281:40485–40492. doi: 10.1074/jbc.M607704200. [DOI] [PubMed] [Google Scholar]
- 25.Fukui M., Choi H.J., Zhu B.T. Rapid generation of mitochondrial superoxide induces mitochondrion-dependent but caspase-independent cell death in hippocampal neuronal cells that morphologically resembles necroptosis. Toxicol. Appl. Pharmacol. 2012;262:156–166. doi: 10.1016/j.taap.2012.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goffart S., Tikkanen P., Michell C., Wilson T., Pohjoismäki J.L.O. The type and source of reactive oxygen species influences the outcome of oxidative stress in cultured cells. Cells. 2021;10:1075. doi: 10.3390/cells10051075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brand M.D. Riding the tiger – physiological and pathological effects of superoxide and hydrogen peroxide generated in the mitochondrial matrix. Crit. Rev. Biochem. Mol. Biol. 2020 Nov;55:592–661. doi: 10.1080/10409238.2020.1828258. [DOI] [PubMed] [Google Scholar]
- 28.Brand M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 2016 Nov;100:14–31. doi: 10.1016/j.freeradbiomed.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Palma F.R., He C., Danes J.M., et al. Mitochondrial superoxide dismutase: what the established, the intriguing, and the novel reveal about a key cellular redox switch. Antioxidants Redox Signal. 2020 Apr;32(10):701–714. doi: 10.1089/ars.2019.7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmström K.M., Baird L., Zhang Y., et al. Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biol. Open. 2013 Jun 20;2(8):761–770. doi: 10.1242/bio.20134853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovac S., Angelova P.R., Holmström K.M., et al. Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim. Biophys. Acta. 2015 Apr;1850(4):794–801. doi: 10.1016/j.bbagen.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinkova-Kostova A.T., Abramov A.Y. The emerging role of Nrf2 in mitochondrial function. Free Radic. Biol. Med. 2015 Nov;88(Pt B):179–188. doi: 10.1016/j.freeradbiomed.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hattori N., Matsumine H., Asakawa S., et al. Point mutations (Thr240Arg and Ala311Stop) in the parkin gene. Biochem. Biophys. Res. Commun. 1998 Aug 28;249(3):754–758. doi: 10.1006/bbrc.1998.9134. [DOI] [PubMed] [Google Scholar]
- 34.Lazarou M., Narendra D.P., Jin S.M., et al. PINK1 drives parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J. Cell Biol. 2013 Jan 21;200(2):163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murata H., Takamatsu H., Liu S., et al. NRF2 Regulates PINK1 Expression under oxidative stress conditions. PLoS One. 2015 Nov 10;10(11) doi: 10.1371/journal.pone.0142438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.East D.A., Fagiani F., Crosby J., et al. PMI: a ΔΨm independent pharmacological regulator of mitophagy. Chem. Biol. 2014 Nov 20;21(11):1585–1596. doi: 10.1016/j.chembiol.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein C., Rakovic A., Grünewald A., et al. PINK1-interacting proteins: proteomic analysis of overexpressed PINK1. Parkinsons. Dis. 2011 Mar 16;2011 doi: 10.4061/2011/153979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu W., Karuppagounder S.S., Springer D.A., et al. Genetic deficiency of the mitochondrial protein PGAM5 causes a Parkinsons-like movement disorder. Nat. Commun. 2014 Sep 15;5:4930. doi: 10.1038/ncomms5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekine S., Kanamaru Y., Koike M., et al. Rhomboid protease PARL mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5. J. Biol. Chem. 2012 Oct 5;287(41):34635–34645. doi: 10.1074/jbc.M112.357509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng M., Lin N., Dong D., et al. PGAM5: a crucial role in mitochondrial dynamics and programmed cell death [Internet] Eur. J. Cell Biol. 2021 Jan;100(1) doi: 10.1016/j.ejcb.2020.151144. [DOI] [PubMed] [Google Scholar]
- 41.Lo S.C., Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J. Biol. Chem. 2006 Dec 8;281(49):37893–37903. doi: 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- 42.Lo S.C., Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp. Cell Res. 2008 May 1;314(8):1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruiz K., Thaker T.M., Agnew C., et al. Functional role of PGAM5 multimeric assemblies and their polymerization into filaments. Nat. Commun. 2019 Jan 31;10(1):531. doi: 10.1038/s41467-019-08393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen G., Han Z., Feng D., et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell. 2014 May 8;54(3):362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 45.Sugo M., Kimura H., Arasaki K., et al. Syntaxin 17 regulates the localization and function of PGAM5 in mitochondrial division and mitophagy. EMBO J. 2018 Nov 2;37(21) doi: 10.15252/embj.201798899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu W., Lin C., Wu K., Jiang L., et al. FUNDC 1 regulates mitochondrial dynamics at the ER –mitochondrial contact site under hypoxic conditions. EMBO J. 2016 Jul 1;35(13):1368–1384. doi: 10.15252/embj.201593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen M., Chen Z., Wang Y., et al. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy. 2016;12(4):689–702. doi: 10.1080/15548627.2016.1151580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holze C., Michaudel C., MacKowiak C., et al. Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway article. Nat. Immunol. 2018 Feb;19(2):130–140. doi: 10.1038/s41590-017-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lenhausen A.M., Wilkinson A.S., Lewis E.M., et al. Apoptosis inducing factor binding protein PGAM5 triggers mitophagic cell death that is inhibited by the ubiquitin ligase activity of X-linked inhibitor of apoptosis. Biochemistry. 2016 Jun 14;55(23):3285–3302. doi: 10.1021/acs.biochem.6b00306. [DOI] [PubMed] [Google Scholar]
- 50.Lysyk L., Brassard R., Arutyunova E., et al. Insights into the catalytic properties of the mitochondrial rhomboid protease PARL. J. Biol. Chem. 2021 Feb 5;296 doi: 10.1016/j.jbc.2021.100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wai T., Saita S., Nolte H., et al. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i ‐AAA protease YME1L. EMBO Rep. 2016 Dec;17(12):1844–1856. doi: 10.15252/embr.201642698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasson S.A., Kane L.A., Yamano K., et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013 Dec 12;504(7479):291–295. doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grosdidier A., Zoete V., Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011 Jul;39:W270–W277. doi: 10.1093/nar/gkr366. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grosdidier A., Zoete V., Michielin O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011 Jul 30;32(10):2149–2159. doi: 10.1002/jcc.21797. [DOI] [PubMed] [Google Scholar]
- 55.Canning P., Sorrell F.J., Bullock A.N. Structural basis of Keap1 interactions with Nrf2 [internet] Free Radic. Biol. Med. 2015 Nov;88(Pt B):101–107. doi: 10.1016/j.freeradbiomed.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu M.C., Ji J.A., Jiang Y.L., et al. An inhibitor of the Keap1-Nrf2 protein-protein interaction protects NCM460 colonic cells and alleviates experimental colitis. Sci. Rep. 2016 May 24;6 doi: 10.1038/srep26585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014 Jul;94(3):909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fivenson E.M., Lautrup S., Sun N., et al. Mitophagy in neurodegeneration and aging. Neurochem. Int. 2017 Oct;109:202–209. doi: 10.1016/j.neuint.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kerr J.S., Adriaanse B.A., Greig N.H., et al. Mitophagy and Alzheimer's disease: cellular and molecular mechanisms. Trends Neurosci. 2017 Mar;40(3):151–166. doi: 10.1016/j.tins.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai Q., Jeong Y.Y. Mitophagy in Alzheimer's disease and other Age-related neurodegenerative diseases. Cells. 2020 Jan 8;9(1):150. doi: 10.3390/cells9010150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu L., Feng D., Chen G., et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2012 Jan 22;14(2):177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 62.Chen Z., Siraj S., Liu L., et al. MARCH5-FUNDC1 axis fine-tunes hypoxia-induced mitophagy. Autophagy. 2017 Jul 3;13(7):1244–1245. doi: 10.1080/15548627.2017.1310789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng M., Lin N., Dong D., et al. PGAM5: a crucial role in mitochondrial dynamics and programmed cell death. Eur. J. Cell Biol. 2021 Jan;100(1):151144. doi: 10.1016/j.ejcb.2020.151144. [DOI] [PubMed] [Google Scholar]
- 64.Xu W., Jing L., Wang Q., et al. Bax-PGAM5L-Drp1 complex is required for intrinsic apoptosis execution. Oncotarget. 2015 Oct 6;6(30):30017–30034. doi: 10.18632/oncotarget.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Z., Jiang H., Chen S., et al. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012 January 20;148(1–2):228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 66.O'Mealey G.B., Plafker K.S., Berry W.L., et al. A PGAM5-KEAP1-Nrf2 complex is required for stress-induced mitochondrial retrograde trafficking. J. Cell Sci. 2017 Oct 15;130(20):3467–3480. doi: 10.1242/jcs.203216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abed D.A., Goldstein M., Albanyan H., et al. Discovery of direct inhibitors of Keap1-Nrf2 protein-protein interaction as potential therapeutic and preventive agents. Acta Pharm. Sin. B. 2015 Jul;5(4):285–299. doi: 10.1016/j.apsb.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pallesen J.S., Tran K.T., Bach A. Non-covalent small-molecule kelch-like ECH-associated protein 1-nuclear factor erythroid 2-related factor 2 (Keap1-Nrf2) inhibitors and their potential for targeting central nervous system diseases. J. Med. Chem. 2018 Sep 27;61(18):8088–8103. doi: 10.1021/acs.jmedchem.8b00358. [DOI] [PubMed] [Google Scholar]
- 69.Wan E.R., Siew K., Heptinstall L., et al. Fumaric acid ester–induced renal Fanconi syndrome: evidence of mitochondrial toxicity. Clin. Kidney J. 2021 Sept;14(9):2085–2089. doi: 10.1093/ckj/sfaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cagalinec M., Safiulina D., Liiv M., et al. Principles of the mitochondrial fusion and fission cycle in neurons. J. Cell Sci. 2013 May 15;126(Pt 10):2187–2197. doi: 10.1242/jcs.118844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.