Abstract
Increased translation of p27 mRNA correlates with withdrawal of cells from the cell cycle. This raised the possibility that antimitogenic signals might mediate their effects on p27 expression by altering complexes that formed on p27 mRNA, regulating its translation. In this report, we identify a U-rich sequence in the 5′ untranslated region (5′UTR) of p27 mRNA that is necessary for efficient translation in proliferating and nonproliferating cells. We show that a number of factors bind to the 5′UTR in vitro in a manner dependent on the U-rich element, and their availability in the cytosol is controlled in a growth- and cell cycle-dependent fashion. One of these factors is HuR, a protein previously implicated in mRNA stability, transport, and translation. Another is hnRNP C1 and C2, proteins implicated in mRNA processing and the translation of a specific subset of mRNAs expressed in differentiated cells. In lovastatin-treated MDA468 cells, the mobility of the associated hnRNP C1 and C2 proteins changed, and this correlated with increased p27 expression. Together, these data suggest that the U-rich dependent RNP complex on the 5′UTR may regulate the translation of p27 mRNA and may be a target of antimitogenic signals.
The amount of p27 is a critical determinant for the decision of cells in G1 to either withdraw from or commit to the cell cycle and enter S phase. p27 inhibits cyclin E-cdk2 (56). This kinase is both necessary and rate limiting for S-phase entry (42, 43, 50) and increases threefold as G1 cells commit to DNA replication (11, 28). Once activated in mid-G1, it triggers a positive feedback loop, both inactivating Rb (22, 26) and promoting p27 degradation (41, 55, 61), ultimately culminating in the transition to S phase.
Small changes in the amount of p27 protein can have dramatic phenotypic consequences: mice heterozygous for p27 have half the wild-type amount of protein and display intermediate growth phenotypes (27). Furthermore, carcinogen-induced tumor development is similar in p27 heterozygous mice and in animals completely lacking p27 (16). These consequences can be attributed to the role of p27 as a mediator of antimitogenic signals (7, 9, 12, 45, 59). In the absence of p27, cells exposed to signals that induce growth arrest fail to withdraw from the cell cycle in a timely fashion, undergoing more mitotic divisions until other pathways mediate their withdrawal from the cell cycle (7, 12, 59). The nature of these collaborating or redundant pathways is not always clear; however, other cdk inhibitors and the Rb-like protein p130 have been implicated in fibroblasts, at least with regard to inactivation of cyclin E-cdk2 (9).
Regardless of the potential for redundancy, the failure of p27−/− cells to respond appropriately to growth arrest signals leads to disease. In luteal cells, the lack of p27 leads to a perturbation of estradiol signaling following conception and prevents embryo implantation (59). The organization of the ear, specifically the ability to hear, also becomes compromised (8, 31), and p27-deficient animals develop tumors (10, 17, 27, 40, 45). Thus, an understanding of how the availability of p27 is controlled would impact our understanding of how tissue organization occurs and how cells communicate with each other.
p27 protein is most abundant in G1 cells and decreases precipitously as cells enter S phase, remaining low throughout the remainder of the cell cycle (35). The expression of p27 can be controlled at the levels of gene transcription (29), translation (1, 23, 35), sequestration (57), nuclear localization (58), and proteolysis (41, 44). Proteolysis of p27 is dependent on cdk2 (41, 55, 61) and possibly skp2 (6, 50, 60), which conspire to regulate ubiquitin-dependent proteolytic degradation of p27, a phenomenon that might insure irreversibility of the commitment decision, as these proteins are activated or produced just prior to or contemporaneously with the G1/S transition. A number of groups have suggested that signals promoting growth arrest may act by directly interfering with p27 proteolysis; however, the cause-and-effect relationship is not entirely clear because p27 proteolysis is dependent on proteins and activities that occur once cells are committed to S phase.
On the other hand, growth arrest is accompanied by an increase in the translation of p27 mRNA above a basal state observed in asynchronous cells. In quiescent tetradecanoyl phorbolacetate (TPA)-treated HL-60 cells, the synthesis of p27 protein is increased, correlating with an increase in the amount of p27 mRNA associated with polysomes (35). Likewise, the rate of p27 synthesis is increased in cells arrested in mid-G1 by lovastatin (23). Additionally, translation of p27 mRNA continues into S phase (and presumably G2 phase), but proteolysis of the protein prevents its accumulation (35). Thus, the translation rate of p27 mRNA can vary in a signal-dependent manner: a basal rate in growing cells and an elevated rate (induced) in growth-arrested cells.
The following observations prompted us to look at the translational regulation of p27 mRNA as a mechanism contributing to growth arrest in G1 cells. First, the steady-state amount of p27 is critical to the commitment process, and this is the sum of the synthesis and degradation rates. Second, since proteolysis is dependent on cdk2 activity and skp2, both of which appear following commitment to the cell cycle, it would seem that they could not effectively control p27 accumulation in the early G1 cell, which is deciding between proliferation and growth arrest. However, if translation could be induced in a cell cycle phase-dependent manner, one would expect that the change in synthesis rate might overcome the proteolytic barrier and p27 would accumulate. In this report, we demonstrate that p27 mRNA translation in both basal (proliferating) and induced (nonproliferating) states requires a U-rich sequence in the 5′ untranslated region (5′UTR) of p27 mRNA. This sequence promotes polysome association of the mRNA. Two proteins, designated p33 and p40/41, in cytosolic extracts from asynchronous cells could be cross-linked to the 5′UTR. These factors were enriched in nocadazole-treated cells (G2/M arrest) and lovastatin-treated cells (G1 arrest) compared to hydroxyurea-treated cells (G1/S-phase arrest). We identify p33 as HuR, which binds to the U-rich element independently of other proteins, and p40/41 as hnRNP C1/C2. We discuss the cell cycle-regulated formation of these RNPs in light of the role that translational regulation of p27 may have in the response to antimitogenic signals.
MATERIALS AND METHODS
Cell culture and drug treatments.
HeLa S3 cells were maintained in suspension culture in minimal Eagle's medium (MEM) without Ca2+ and supplemented with 10% enriched calf serum (Gemini). 293T cells were maintained in Dulbecco's modified Eagle's medium (DME) supplemented with 4.5 g of glucose per liter (DME HG), 2 mM glutamine, and 10% fetal bovine serum (FBS; Gemini). MDA468 cells were grown in a 1:1 mixture of DME HG and F12 plus nonessential amino acids supplemented with 10% FBS and 2 mM glutamine. Nocodazole (Sigma) and hydroxyurea (Sigma) were used at 2 μM for 12 h and 2 mM for 24 h, respectively, in all cell lines. Lovastatin (Merck) was used at 30 μM for 48 h. Actinomycin D (Sigma) was used at 5 μg/ml.
Oligonucleotides.
The oligonucleotides used were SSM16 (5′GCTGTCCTTAAGAGCTATGGAAGTTTTCTT3′), SSM17 (5′CATTCAGCGGCCGCACAGCTCGAATTAAGAAT3′), SSM23 (5′GCTGTCGAATTCTCCTAGAGCTCGGGCCGT3′), T7SSM23 (5′TCCTAATACGACTCACTATAGGTCCTAGAGCTCGGGCCGT3′), SSM30 (5′CATTCAGGATCCCTTTCTCCCGGGTCTGCA3′), SSM31 (5′GCTGTCGGATCCATGGAAGACGCCAAAAAC3′), SSM32 (5′CATTCAGTATGCGGCCGCTTACAATTTGGACTTTCCGCC3′), SSM40 (5′GCGGTTCCATCCTCTAGAGGAT3′), SSM45 (5′GGACTCAGATCTTCGAGAT3′), SSM46 (5′CATTCAGCTAGCCCGAACAAAACAAAGCGC3′), SSM47 (5′CATTCAGCTAGCTGCAGACCCGGGAGAAAG3′), SSM48 (5′GTATTCCGCGTACGTGATGTTCA3′), SSM51 (5′CAGCGCAAGTGGAATGCCGATGCTCAGAATCACAAACCC3′), SSM52 (5′GGGTTTGTGATTCTGAGCATCGGCATTCCACTTGCGCTG3′), SSM53 (5′GCTGTCGGATCCATGTCAAACGTGCGAGTG3′), and SSM55 (5′CATTCAGTATGCGGCCGCTCAGTGGTGGTGGTGGTG3′).
The sequence of the 42-nucleotide transcript and RNA from Curachem includes nucleotides 77 to 118, as indicated in Fig. 3.
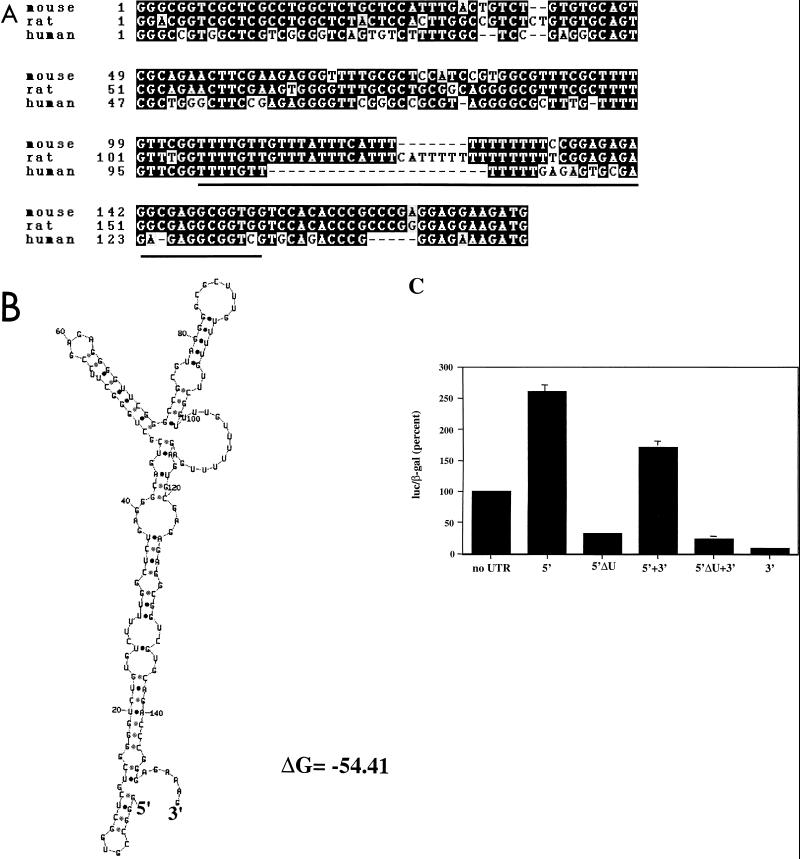
FIG. 3.
U-rich element required for the positive influence of the 5′UTR. (A) Homology between the sequences of the mouse, rat, and human p27 5′UTRs. Sequences were aligned from the ATG sequence encoding the initiating methionine at the end. Identical nucleotides in two of the three sequences are indicated in black boxes. Sequences deleted by the ΔU mutation are underlined. (B) Structural folding model of the 5′UTR. Using the mFOLD program (34), a predicted structure of the 5′UTR was made. (C) The U-rich sequence is necessary for 5′UTR function. HeLa cells were transfected as described in the legend to Fig. 1 using the constructs listed below each bar. Accumulation was normalized to that of the parental luciferase construct lacking p27 UTR sequences (set at 100%). The mean and standard deviations from three independent experiments are indicated.
The irrelevant scrambled oligonucleotide was 5′UGAUCUUGACAAUUGGCGUAAUCCAGAAGCGCAGUCAGGUUUGAAUUCAUUUGAA3′.
The 165-nucleotide gamma globin transcript used in Fig. 5C was derived from pSP65Hγ (a gift from Henry Furneaux) linearized with Sau3AI. It contains 80 nucleotides of coding sequence and 85 nucleotides of 3′UTR.
FIG. 5.
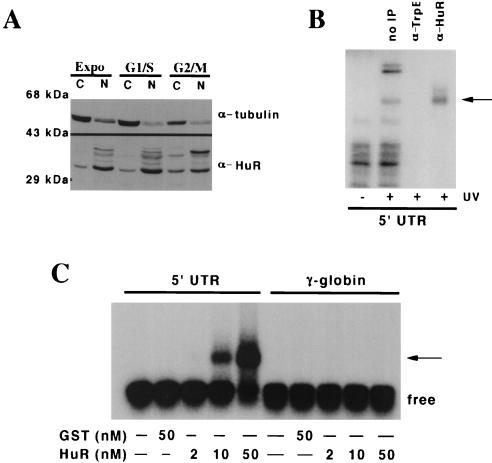
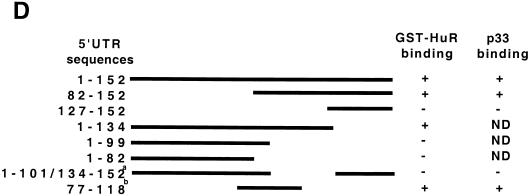
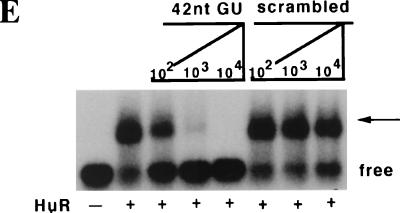
p33 is the ELAV protein HuR. (A) HuR is enriched in the cytosol of G2/M cells compared to S-phase or exponentially growing cells. Nuclear (N) and cytosolic (C) extracts were prepared from asynchronous exponentially growing 293T cells (Expo), cells treated with hydroxyurea (G1/S), and cells treated with nocadazole (G2/M), and 80 μg of each extract was resolved by SDS-PAGE, transferred to a polyvinylidene fluoride membrane, and blotted with antibodies raised against alpha-tubulin or HuR as indicated to the right of each panel. Molecular size markers are indicated on the left. The source of each extract is indicated above the panels. (B) HuR in the cytosol of G2/M cells binds to the 5′UTR. For immunoprecipitation, the cross-linking was done as described in the legend to Fig. 4C but the amounts were scaled up fivefold. One sample was not incubated with an antibody; the others were incubated with either the 19F12 monoclonal antibody to HuR or a monoclonal antibody raised against bacterial TrpE protein, as indicated above each lane. (C) Purified HuR binds to the 5′UTR of p27 mRNA. Increasing amounts of recombinant GST-HuR were purified from bacterial cultures and incubated with uniformly labeled 5′UTR RNA or a size-matched sequence of 165 nucleotides from the gamma globin mRNA. The amount of GST or GST-HuR in each binding reaction is shown below each lane. The migration of the free and bound probe (arrow) is indicated to the right of the autoradiograph. (D) The U-rich element is necessary for HuR binding. Mutants in the 5′UTR were generated by standard PCR cloning procedures and served as templates for the production transcripts to assess the binding of purified GST-HuR and p33. A schematic of the 5′ deletion series is shown, and each mutant is numbered as in Fig. 3A. The ability of a mutant transcript to bind the recombinant proteins and cross-link to p33 in G2/M extracts is indicated. Binding reactions were performed with GST-HuR or GST alone. Binding activity is scored as positive or negative. ND, not done. Under these conditions we could not detect GST binding to the transcripts. The superscript a denotes the transcript commonly used as ΔU in this work, and b denotes the sequence of the 42-nucleotide (nt) competitor used in panel E. (E) The U-rich element is sufficient to compete with HuR binding to the 5′UTR probe. As described in the legend to Fig. 4C, uniformly labeled 5′UTR transcript was incubated with 50 nM GST-HuR in the presence or absence of increasing amounts of a 42-nucleotide RNA containing the U-rich element or a scrambled oligoribonucleotide (Curachem; sequence indicated in the text). The products of the binding reaction were resolved by EMSA. The competitor used and the fold relative to the labeled transcript are indicated above each lane. The presence or absence of GST-HuR is indicated below each lane. The migration of the free probe and the bound probe (arrow) is indicated on the right of the autoradiograph.
Isolation of human p27 5′UTR.
Primers SSM23 and SSM30 were used to amplify sequences containing the 5′UTR from a genomic clone isolated from an EMBL3 SP6/T7 genomic library (Clontech). This PCR product was then cloned into the green fluorescent protein (GFP) and luciferase expression vectors as described below and sequenced using primers SSM40, SSM45, and SSM48 and GL primer 2 (Promega).
Antibodies.
Western blotting was performed as described (57). The affinity-purified anti-p27 antibody has been described (57). The 19F12 anti-HuR monoclonal antibody has been described (H. Furneaux, submitted for publication). Antibodies to cdk2 (M2; Santa Cruz Biotechnology), α-tubulin (T9026; Sigma), and β-galactosidase (Z378A; Promega) are commercially available.
Isolation of nuclear and cytoplasmic RNA and protein.
Nuclear and cytoplasmic RNA was obtained as described (53). Nuclear and cytoplasmic protein extracts were prepared as described (62).
Construction of p27ck reporters.
The cyclin binding domain mutant of human p27 PV→KK (32) was further mutated by site-directed mutagenesis using SSM51 and SSM52 with the Quickchange system (Stratagene). This generated an FDF→ADA mutation in the cdk binding domain. The p27ck− mutant was then amplified by PCR using primers SSM53 and SSM55 and directionally cloned into BamHI- and NotI-digested pSVL vectors. All clones were confirmed by direct sequencing.
Construction of luciferase reporters.
The reporters were generated by subcloning the simian virus 40 (SV40) promoter (nucleotides 22 to 239) from the pGL-2 promoter (Promega) directly into the XhoI and HindIII sites of pEGFP-1 (Clontech) to create pSVG. The p27 5′UTR was amplified from a human genomic clone by PCR using primers SSM23 and SSM30. The p27 3′UTR was amplified from pBΔ5′ p27 with primers SSM16 and SSM17. These products were directionally cloned into pSVG. GFP sequences in the pSVG series were replaced with a luciferase PCR product using primers SSM31 and SSM32 and the pGL2 promoter vector. This created the pSVL series of constructs. To generate clones with an SV40 polyadenylation signal, an XhoI-NotI-digested pEGFP-1 vector was ligated to and XhoI-AflII-digested pSVL insert.
Deletion of the U-rich element of the 5′UTR was performed by PCR using p5′SVL and primers SSM45, SSM46, SSM47, and SSM48, which replaced the U-rich element with an NheI site.
Transfection assay.
For the transfection of HeLa cells, we combined 8 × 106 cells with 10 μg of pSVL construct, 5 μg of pCMV-β, and 5 μg of pCDNA3 (Invitrogen) in a final volume of 800 μl in a 0.4-cm cuvette. Cells were electroporated at 0.28 kV and 960 μF using a Bio-Rad electroporator and then plated for 24 h before harvesting. 293T and MDA468 cells were transfected using CaPO4 (Gibco-BRL) according to the manufacturer's instructions. 293T cells were harvested 24 h after the CaPO4 precipitate was washed off, and MDA468 were used 48 h afterwards.
RNase protection assay.
To generate a luciferase probe for the RNase protection assay, we subcloned a BamHI-NotI PCR product from pGL-2 promoter into pBluescript II (Clontech). This clone (pB-LUC) was digested with PacI and transcribed with T7 RNA polymerase (Gibco-BRL), resulting in a 375-nucleotide antisense probe. For β-galactosidase, we subcloned an AccI-NdeI (blunt) fragment of pCMV-β (Clontech) to AccI- and SmaI-digested pBluescript II. This clone (pB-βAcc-Nde) was digested with AccI and transcribed with T7 RNA polymerase, resulting in a 249-nucleotide antisense transcript. The sizes of the protected luciferase and β-galactosidase transcripts detected in RNA from transfected cells were 325 and 193 nucleotides, respectively. RNase protection was performed as described (51).
Endogenous c-myc transcripts were detected using mouse pTri c-myc/exon 3 (Ambion). The human and mouse exon 3 sequences diverge at two nucleotides and thus yielded three protected fragments.
Polysome gradients.
Continuous sucrose gradients (10 ml, 15 to 40%) were prepared and run as described previously (35), except for the following modifications. Each 1-ml fraction was collected directly into 120 μl of 8.3% sodium dodecyl sulfate (SDS) and 83 mM EDTA. Then 200 μg of proteinase K was added, and samples were incubated at 37°C for 15 min and extracted with phenol-chloroform.
EMSA and UV cross-linking assay.
For the electrophoretic mobility shift assay (EMSA), 5′UTR RNA transcripts were prepared by PCR amplification of the appropriate pSVL template using T7SSM23 and SSM30 and transcribed using T7 RNA polymerase. Radiolabeled probes and glutathione S-transferase (GST)-HuR were prepared as described (24). The specific activity of each probe was 1 × 104 to 5 × 104 cpm/pmol.
Binding reactions using extracts as a source of protein were performed in a 20-μl volume containing 20 fmol of transcript, 15 μg of protein extract, 50 μg of tRNA, 50 mM Tris (pH 7.0), and 5 μg of bovine serum albumin. Reactions were performed essentially as described (24).
For UV cross-linking experiments, following the incubation the binding reaction was irradiated at 1,200 μJ/cm2 in a Stratalinker (Stratagene). RNase A (Sigma) and RNase T1 (Calbiotech) were then added to 500 μg/ml and 250 U/ml, respectively, and incubated at 30°C for 15 min prior to SDS-polyacrylamide gel electrophoresis (PAGE).
Purification of hnRNP C1/C2.
HeLa nuclear extract (60 mg) was loaded onto a 5-ml HiTrap Q-Sepharose column and eluted with a 0.25 to 0.5 M linear KCl gradient. p40/41 activity eluted after the major protein peak at 0.4 M KCl. Pooled fractions from the Q column were loaded on a 1-ml methyl-Sepharose column and eluted with a linear gradient of 1 to 0 M (NH4)2SO4. p40/41 activity eluted at 0.45 M (NH4)2SO4. These fractions were pooled, loaded on a 1-ml HiTrap SP-Sepharose column, and eluted with a 0.05 to 1 M KCl gradient; the peak activity eluted at 0.45 M KCl. The Coomassie-stained p40 and p41 bands were excised for analysis by mass spectrometry fingerprinting (13).
RESULTS
A U-rich element in the 5′UTR of p27 mRNA facilitates polysome association.
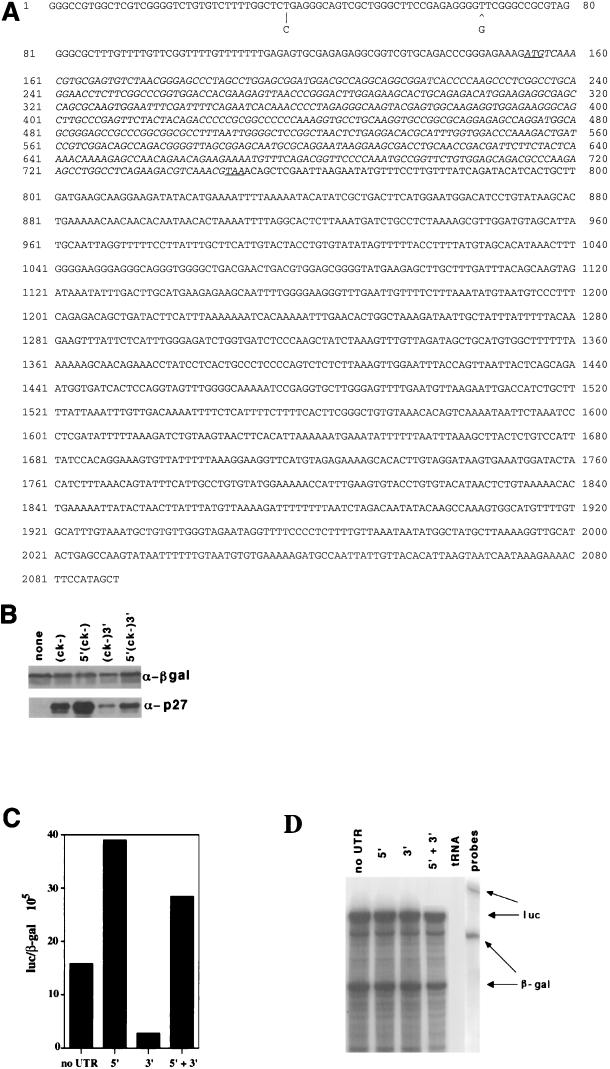
To better understand the translational regulation of p27 and the role it plays during the cell cycle and growth arrest, we needed to first isolate the 5′ and 3′ UTRs of human p27. We obtained the 3′UTR sequence by defining a contig in the GenBank dbest database and sequenced overlapping clones on both strands (Fig. 1A). We obtained 5′UTR sequence by sequencing a genomic clone encoding the p27 locus (unpublished data). This sequence largely agreed with that published by Minami and colleagues (36); the only exceptions (indicated in Fig. 1A) are a T-to-C change and the absence of a G nucleotide at positions 37 and 66/67, respectively. These differences could be due to polymorphisms or sequencing errors. We reasoned that cis-acting regulatory sequences might be in either the 5′UTR, the 3′UTR, or the open reading frame (ORF) or any combination of these elements.
FIG. 1.
5′UTR of p27 mRNA contains sequences that enhance expression of heterologous reporters. (A) Sequence of the coding stand of p27 mRNA. 3′UTR sequence was obtained by identifying and sequencing expressed sequence tags with homology to the p27 ORF (AA206936, R18972, and AA180937). To determine the 5′UTR sequence, we combined analysis of 5′RACE using HL-60 mRNA as the template and sequences obtained from a human genomic clone. Nucleotide differences between the 5′UTR sequence and that reported by Minami and colleagues (36) are indicated below the sequence. The initiation and termination codons are underlined. (B) The 5′UTR is required to overcome suppressive effects of the 3′UTR on p27 accumulation. 293T cells were transfected with 10 μg of an expression vector encoding a noncyclin/cdk binding form of p27 (p27ck−), with different combinations of p27 UTRs as defined above each lane. Then 5 μg of a plasmid expressing the β-galactosidase gene (β-gal) from the cytomegalovirus promoter was cotransfected. Transfection conditions and protein extraction are described in the text. Following extraction, lysates were normalized for β-galactosidase activity, and equivalent units were resolved by SDS-PAGE and Western blotted with anti-β-galactosidase antibodies (βgal) or anti-p27 antibodies as indicated to the right. (C) The effect of the UTRs is independent of sequences within the ORF. Experimentally similar to panel B, but luciferase replaced the p27ck− ORF in the expression vector and we used HeLa cells. The UTRs used are shown below each bar, and the accumulation of luciferase normalized for β-galactosidase is shown on the y axis. This experiment was repeated greater than five times with similar results, and a representative experiment is shown. (D) The presence of the p27 UTR sequences did not affect the yield of mRNA. The data in this panel are matched to the transfection shown in panel C. We determined the amount of luciferase (luc) and β-galactosidase (β-gal) mRNA by RNase protection assay in 10 μg of cytosolic HeLa mRNA. The protected luciferase (325 nucleotides) and β-galactosidase (193 nucleotides) transcripts are indicated with arrows. The mobilities of the unprotected probes are shown in the rightmost lane. We used 10 μg of tRNA as a negative control.
To investigate which of these elements controlled translation, we subcloned the entire p27 cDNA or versions lacking either the 5′ or 3′ UTR into a vector containing the SV40 early promoter and polyadenylation signal. To prevent expression of p27 from altering the distribution of cells in the cell cycle but maintaining the presumptive secondary structure of the full-length mRNA, we mutated 10 nucleotides encoding four amino acids required for cyclin-cdk binding and inhibition (p27ck−) (61). Transfection of 293T cells with these constructs did not lead to an alteration in cell cycle distribution (data not shown). Cells were cotransfected with a β-galactosidase expression vector to allow for normalization of p27 expression, and we examined the accumulation of p27 by immunoblotting (Fig. 1B). Expression of protein from a construct containing both the 5′ and 3′ UTRs was nearly equivalent to that observed in the control construct without any additional UTR sequences. From this point, we use the phrase no UTR to represent the basal construct which contains the UTR sequences provided by the expression vector but not additional p27 mRNA sequences. However, expression of p27ck− was greatly diminished when the 3′UTR was present alone, and the 5′UTR alone could promote expression. Thus, the 5′UTR can suppress or overcome the negative effect of the 3′UTR on accumulation of p27ck−. This suggests that there is a positive regulatory element in the 5′UTR. To determine if the UTR-dependent effects required sequences in the ORF, we replaced the p27 coding sequence with luciferase and repeated the analysis. Similar UTR-dependent changes in luciferase expression were detected when normalized for β-galactosidase. The 5′UTR was able to increase luciferase expression 2- to 3-fold, whereas the 3′UTR inhibited luciferase expression more than 10-fold. When both UTRs were present on the luciferase reporter, the 5′UTR was able to suppress many of the negative effects of the 3′UTR (Fig. 1C). The yield of each transcript as measured by RNase protection was equivalent in all samples, suggesting that the activity measurements do not reflect differences in steady-state mRNA levels (Fig. 1C). This eliminated the possibility that the sequences within the p27 ORF contributed to the UTR-dependent effects.
The steady-state amount of protein can be affected by a number of factors: the rate of mRNA synthesis and export from the nucleus, the stability of the mRNA in the cytosol, and the incorporation of the mRNA into polysomes. To determine how the 5′UTR sequences affected the accumulation and utilization of luciferase mRNA, we examined the amount of mRNA in the cytosol, the half-life of cytosolic mRNA, and the association of the mRNA in polysomes using the 5′-plus-3′ UTR construct and the 3′UTR construct alone (Fig. 2). When normalized for the amount of cotransfected β-galactosidase mRNA, the amount of mRNA in the cytosol of transfected cells was equivalent for the two constructs (Fig. 2A), suggesting that the accumulation of mRNA in the cytosol was not affected in a UTR-dependent manner. We also did not observe any significant UTR-dependent changes in the half-life of these messages in the presence of actinomycin D. Endogenous c-myc mRNA had a half-life of approximately 40 min in the same samples, confirming that the drug treatment was effective (Fig. 2B). Actinomycin D incubations as long as 12 h were also performed and revealed no UTR-specific differences in mRNA half-life (data not shown). Thus, the accumulation of luciferase as controlled by the 5′ and 3′ UTRs was likely due to translational affects.
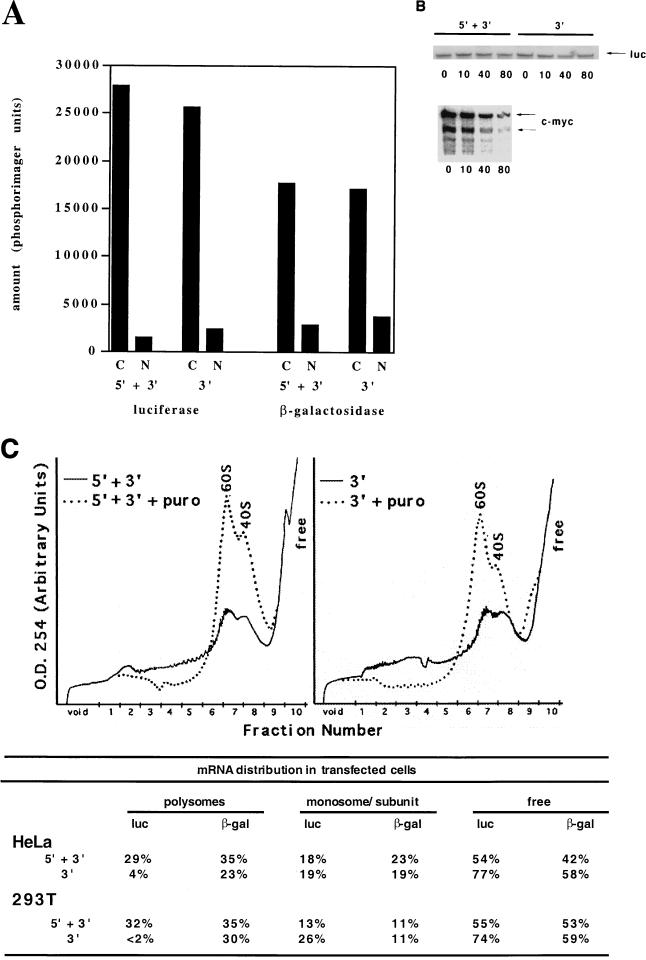
FIG. 2.
5′UTR of p27 mRNA contains a sequence that enhanced translation of the mRNAs that encode it. (A) RNA localization. Luciferase expression vectors flanked by the 5′ and 3′ UTRs or the 3′UTR alone were cotransfected into HeLa cells with a β-galactosidase expression vector as described in the legend to Fig. 1. Cytosolic (C) and nuclear (N) RNAs were isolated as described in the text, and the amount of luciferase and β-galactosidase mRNA in 10 μg was determined by the RNase protection assay. Below each sample we indicate the source of the RNA and the probe used: cytosol (C) or nuclear (N), with the 5′ and 3′ UTRs (5′ + 3′) or 3′UTR alone (3′). Quantitation was done on a phosphorimager using MacBas software. Representative data are shown; the experiment was repeated twice with two different extraction conditions. (B) RNA half-life. HeLa cells were transfected as in panel A and treated with actinomycin D (5 μg/ml) for the times indicated (minutes) above each lane prior to extraction of cytosolic mRNA. The UTR-containing construct used is indicated above each set of samples. As a control for the actinomycin treatment, a similar analysis of the endogenous c-myc mRNA level in cells transfected with the 5′ and 3′ expression constructs is shown. (C) Polysome association. As in panel A, 293T or HeLa cells were cotransfected with the luciferase expression vector flanked by either the 5′ and 3′ UTRs or the 3′UTR alone with another plasmid expressing β-galactosidase. Following this, the cells were divided into two populations, with one receiving puromycin (150 μg/ml) for an additional 30 min. Cytosolic RNA was prepared as described (35) and fractionated on continuous sucrose gradients. Fractions (1 ml) were collected (numbered below each recording), and their absorbance at 254 nm was determined during collection. Mock-treated samples are indicated by the solid line and puromycin (puro)-treated samples by the dotted line. The migration of free mRNA, 40S, and 60S are indicated. Half of each fraction was subsequently used as a source of RNA for the RNase protection assay as described in the legend to Fig. 1. Each panel contains the data from a single experiment using either the 5′ and 3′ UTRs or the 3′UTR alone. In the table we indicate the amount of mRNA in polysomal (puromycin sensitive), monosome or subunit associated, and free RNA as a percent of the total. Two representative transfections in two different cell lines are shown.
To further establish this point, we fractionated cytosolic extracts from transfected cells on continuous sucrose gradients and monitored absorbance at 254 nm to follow RNP complexes (Fig. 2C). To specifically identify the region of the gradient containing polysomes, we treated parallel cultures with puromycin and fractionated the RNA. Puromycin causes mRNA to accumulate in the monosome and subunit fractions of the gradient (Fig. 2C). Simply comparing the amount of puromycin-sensitive material in each fraction, it was clear that the luciferase mRNAs containing both the 5′ and 3′ UTRs were enriched in the heavier regions of the gradient compared to the luciferase mRNAs containing the 3′UTR alone. The distribution of cotransfected β-galactosidase mRNA, however, was not significantly different between these two populations of cells (Fig. 2C). This firmly established that the 5′UTR contained sequences that facilitated polysome association in cycling cells.
Next, we wanted to identify sequences in the 5′UTR that were responsible for this positive effect and determine if they interact with proteins in a cell cycle phase-specific manner. Sequence comparisons of the 5′UTR from mouse, human, and rat (Fig. 3A) showed the presence of a highly conserved U-rich element. Other conserved regions, notably those between this element and the initiating methionine, were noted as well. Inspection of the secondary structure of the 5′UTR (Fig. 3B) revealed that the U-rich element could form into a loop surrounded by a stable stem structure. Similar stem-loop structures have been implicated in the regulation of translation initiation in ferritin and other mRNAs (37, 38, 52). To test the significance of the U-rich loop, we deleted the nucleotides that comprise the loop and a number of additional nucleotides at the 3′ end (underlined in Fig. 3A). The removal of the 3′ stem nucleotides was done to ensure complete disruption of the stem-loop structure. In constructs either containing or lacking the 3′UTR, deletion of the 5′UTR U-rich element reduced luciferase expression, even below that observed in the parental vector (Fig. 3C). In all cases, the amount of mRNA and the half-life of the mRNA were not significantly affected (data not shown). This indicated that the U-rich element was required for effective translation of this message, and in its absence either the sequence of the 5′UTR or the structure formed by it was capable of inhibiting translation. From these studies we concluded that the 5′UTR contained a U-rich element that participated in the incorporation of p27 mRNA into polysomes in cycling cells.
5′UTR binds complexes in a cell cycle phase-dependent manner.
Having defined a role for the 5′UTR in translation of p27 mRNA and noting that the U-rich element participated, we next wanted to determine if we could identify proteins that would interact with the 5′UTR in a U-rich element-dependent manner. We used an RNA EMSA to determine whether protein complexes could form on the 5′UTR. We made cytosolic extract from asynchronous cultures of 293T cells and incubated them with a uniformly labeled 5′UTR transcript in the presence of a 100,000-fold molar excess of nonspecific tRNA. A slower-migrating complex was detected in asynchronous extracts (Fig. 4A). Under the conditions used, approximately 50% of the RNA remains free (when binding is maximal, i.e., with the G2-M extract), as determined by EMSA (Fig. 4A). As expected, increasing the amount of protein increased the amount of RNA bound, but the specificity of interaction was compromised at higher ratios of protein to transcript (data not shown).
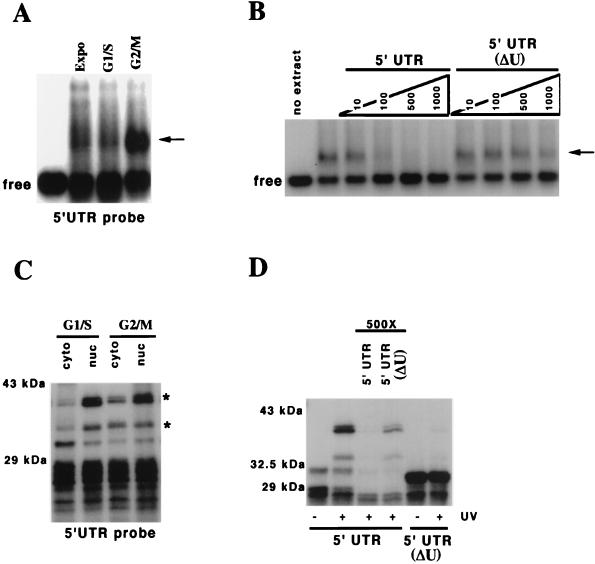
FIG. 4.
RNP complexes form on the 5′UTR in a cell cycle phase-specific manner dependent on the U-rich element. (A) An RNP complex forms on the 5′UTR of p27 mRNA. Cytosolic extract was prepared from asynchronously exponentially growing 293T cells (Expo), cells treated with 2 mM hydroxyurea (G1/S), and cells treated with 2 μM nocadazole (G2/M), and 15 μg of extract was combined with 20 fmol of uniformly labeled 5′UTR transcript, and free and bound probe were separated by electrophoresis through native 1% agarose gels before autoradiography. The cytosolic G2/M extract contained the greatest amount of binding activity. (B) Effective competition for the 5′UTR binding species required the U-rich element. Similar to panel A, but during the incubation of the G2/M cytosolic extract with probe, we added either the 5′UTR or a 5′UTR lacking the U-rich element (ΔU) as an unlabeled competitor. The molar excess of competitor to transcript is indicated above each lane. The first lane on the left shows the migration of probe without any extract added. The second lane shows the probe with G2/M cytosolic extract and no competitor. (C) p33 and p40/41 can be UV cross-linked to the 5′UTR and are enriched in cytosolic extract obtained from G2/M-phase cells. Following the treatments in panel A, nuclei and cytosol were separated, and extracts prepared for binding to the uniformly labeled 5′UTR transcript. Subsequently, the reaction products were cross-linked by UV as described in the text and RNA degraded, and the resulting products were resolved by SDS-PAGE and detected by autoradiography. Only the products marked with asterisks were specifically cross-linked by UV. The other bands were extract dependent but not UV dependent (for example, see Fig. 4D and 5B). It is clear that both nuclear and cytosolic proteins can be cross-linked; however, the amount of the proteins is increased in the extract prepared from G2/M-enriched cells. (D) p33 and p40/41 binding require the U-rich element. As in panel C, the G2/M extract was incubated with uniformly labeled probe; however, the middle two lanes included a 500-fold molar excess of either unlabeled competitor 5′UTR or 5′UTR lacking the U-rich element (ΔU). Furthermore, using a uniformly labeled 5′UTR lacking the U-rich element, we were unable to detect any specific binding proteins (last two lanes on the right). Probes are indicated below the autoradiograph. UV treatment is indicated below each lane. Competitors are shown above the appropriate lanes. Molecular size markers are shown on the left.
We next asked if this binding activity was enriched at particular stages of the cell cycle. To accomplish this, we prepared cytosolic extracts from hydroxyurea-treated or nocadazole-treated cells. These treatments lead to accumulation of cells in G1/S phase and in late G2/M phase, respectively. Incubation of these extracts with the 5′UTR transcript and resolution of bound and free RNA by agarose gel electrophoresis demonstrated that a complex was enriched in the extract derived from nocadazole-treated cells (Fig. 4A). This complex could be competed by a 100-fold molar excess of unlabeled 5′UTR RNA but not by a 1,000-fold molar excess of a similar competitor lacking the U-rich element (Fig. 4B).
To determine the molecular weight of the protein(s) directly contacting the 5′UTR, we performed UV cross-linking experiments. We prepared nuclear and cytosolic extract from the hydroxyurea- and nocadazole-treated cells, subjected the binding reactions to UV, and digested the RNA before resolution of the labeled proteins on SDS-PAGE. Three cross-linked species, a doublet migrating at 40/41 kDa and a 33-kDa protein, were enriched in the cytosolic extract prepared from late G2/M cells compared to S-phase cells. Similar proteins were at least equally abundant or more abundant in the nuclear extracts (Fig. 4C). In extracts from S-phase cells, p40/41 and p33 were largely nuclear, and there was very little cytosolic protein. In cytosolic extracts from asynchronous cells, all three species could be detected (data not shown). Importantly, the cross-linking of these proteins to the 5′UTR was dependent on the U-rich element (Fig. 4D). These interactions could be competed with by a 500-fold molar excess of unlabeled competitor containing the U-rich element but not by the unlabeled transcript lacking this element (Fig. 4D). Consequently, from these data we concluded that a 5′UTR U-rich element-dependent binding activity could be detected in the cytosol of G2/M cells. It was not clear from these studies if these proteins bound to the same or different RNAs.
p33 protein is HuR.
We next wanted to identify the proteins that interacted with the U-rich element. Members of the ELAV family of RNA binding proteins were candidates for a number of reasons. The size of the cross-linked species was similar to the reported 36-kDa size of HuR. There is evidence that the localization of HuR changes during the cell cycle, becoming more cytosolic in G2 cells (Fig. 5A) (3), and that HuR can shuttle between the nucleus and cytosol (14, 15, 46). In addition, HuR has been implicated in regulating mRNA stability and transport (18, 25, 30, 33, 39, 46) as well as translation (2).
To determine if HuR was binding to the 5′UTR, we attempted to immunoprecipitate proteins cross-linked to the 5′UTR in late G2/M cytosolic extract. Using a monoclonal antibody raised against HuR (Furneaux, submitted), we were able to immunoprecipitate the p33 species, which was not precipitated with an unrelated isotype-matched antibody raised against TrpE (Fig. 5B).
We next asked if the binding of HuR to the p27 5′UTR was dependent on other factors in the G2/M extract, i.e., p40/41. Purified recombinant HuR was incubated with labeled transcript and assayed for complex formation. GST-HuR bound to the p27 5′UTR and not a control γ-globin transcript, whereas GST did not (Fig. 5C). Using deletion analysis and binding assays, we identified a 42-nucleotide region overlapping the U-rich element which was required for HuR binding (Fig. 5D, and unpublished data). An RNA encoding this 42-nucleotide region effectively competed for HuR binding, whereas an irrelevant oligoribonucleotide of similar base composition did not (Fig. 5E). This indicated that HuR alone could bind to the U-rich sequences in the 5′UTR of p27. Together these data indicate that the 33-kDa species associated with the p27 mRNA in a G2/M-phase-specific manner is HuR. Furthermore, HuR binding to the 5′UTR site is not dependent on other factors present in the cell extract. To our knowledge, this is the first time a binding site for a mammalian ELAV protein has been defined in the 5′UTR of an mRNA.
p40/41 is hnRNP C1/C2.
To identify the p40/41 binding activity, we used ion-exchange chromatography and the UV cross-linking assay described above to purify these proteins. After determining the linear range of the assay, we defined 1 unit as 200 counts of cross-linking activity on a phosphorimager screen.
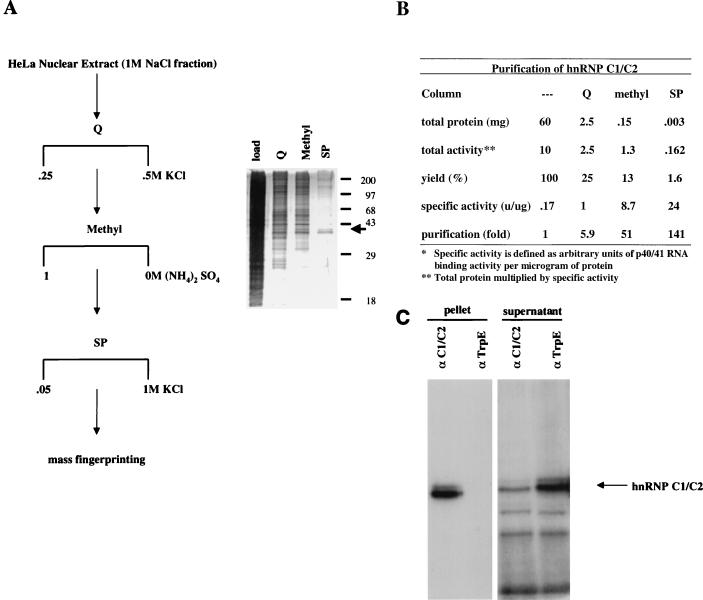
We were able to detect p40/41 binding activity in the cytoplasm of nocadozole-treated cells, but using EMSA and cross-linking we found that a 1 M NaCl extraction of HeLa nuclei had the highest specific activity (data not shown). Therefore, we used this as the starting material for purification. Preliminary binding studies suggested that p40/41 could be absorbed to and eluted from Q, methyl, and SP matrices; thus, we used these three columns to purify p40/41 binding activity from 60 mg of HeLa nuclear extract (Fig. 6A). This purification scheme resulted in a 141-fold purification of p40/41 (Fig. 6B). The SP fractions were run on SDS-PAGE gels and stained with silver (Fig. 6A) and Coomassie. A doublet migrating at 40 kDa cofractionated with p40/41 5′UTR binding activity. The Coomassie-stained p40 and p41 bands were excised for analysis by mass spectrometry fingerprinting (13).
FIG. 6.
p40/41 is hnRNP C1/C2. (A and B) Purification of the p40/41 binding activity. The purification procedure and silver-stained gel monitoring purification are shown. 5′UTR binding activity was measured by UV cross-linking of eluted fractions as described in the legend to Fig. 4C. To obtain sequence from the p40/41 species, the final fraction of the SP column was resolved by preparative SDS-PAGE; the region was excised and subjected to tryptic digestion, and the products were analyzed by mass spectroscopy. The purification table is shown. Sizes are shown in kilodaltons. (C) hnRNP C1/C2 binds to the 5′UTR. Cross-linking and immunoprecipitation were done as described in the legend to Fig. 5B using the monoclonal antibody to C1/C2 (4F4) in place of HuR. The products in the precipitate and the supernatant were resolved by SDS-PAGE. Following immunoprecipitation with 4F4, p40/41 is enriched in the immunoprecipitate and appropriately diminished in the supernatant. Its migration is indicated to the right of the autoradiogram. The pellet and supernatant from the immunoprecipitation are indicated above each lane.
Peptide mass fingerprinting of p40 and p41 positively identified them as hnRNP C1 and C2, a pair of previously described RNA binding proteins. hnRNP C1 and C2 are alternatively spliced products of the same gene and differ by 13 amino acids near the middle of the protein (5). These two proteins migrate as a doublet on SDS-PAGE gels, with C2 migrating more slowly than C1 (47). Monoclonal antibodies raised against hnRNP C1/C2 (47) are able to immunoprecipitate the p27 5′UTR p40/41 binding activity from HeLa nuclear extract, whereas a monoclonal against bacterial TrpE cannot (Fig. 6C). We conclude that the p40/41 binding activity consists of the hnRNP C1/C2 proteins.
hnRNP C1 and C2 are nuclear proteins proposed to be involved in mRNA processing (48). They bind to U-rich sequences on mRNA with high affinity (21). Recent work has suggested that there may be a cytoplasmic role for these proteins in the cytoplasm of differentiated cells bound to translationally induced platelet-derived growth factor (PDGF) mRNA (54). It is therefore possible that the C proteins could regulate translation when they are in the cytoplasm.
Translation and formation of RNP complexes in growth-arrested cells.
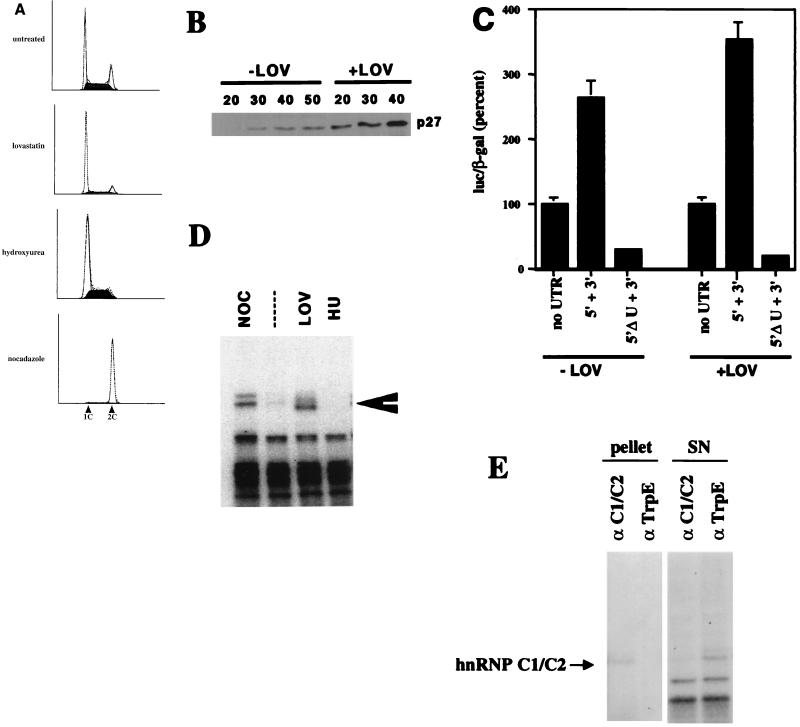
Having established above that RNP complexes containing HuR and/or hnRNP C1/C2 would form on the 5′UTR, we next wanted to determine if these proteins were cytosolic and accessible to the p27 mRNA in cells where translation of p27 mRNA is enhanced. We and others had previously shown that the rate of p27 synthesis was increased in growth-arrested cells compared to asynchronous populations or cells in S phase (1, 23, 35). Using MDA468 cells, we observed lovastatin-induced accumulation of G1 cells and a reduction in S-phase cells, correlating with accumulation of p27 (Fig. 7A and B). this increase could be due to both suppression of proteolysis (49) and increased translation (23). To focus on the translational control mechanism, we first examined the expression of luciferase from the UTR-dependent reporters. Lovastatin enhanced expression from the 5′UTR in a manner dependent on the U-rich element, indicating that this element was required for expression in growth-arrested cells (Fig. 7C).
FIG. 7.
Lovastatin treatment induces changes in the accumulation of p27 and the migration of hnRNP C1/C2. (A) Flow cytometry. MDA468 cells were treated with either lovastatin, hydroxyurea, or nocadazole as described in the text, and the cell cycle position was determined by propidium iodide staining and flow cytometry as described (57). Below each DNA histogram we have labeled the G1 and G2 peaks as 1C and 2C, respectively. Each treatment is indicated to the left of the DNA content profile. (B) Lovastatin treatment induces accumulation of p27 protein. Following treatment of MDA468 cells with either lovastatin (+LOV) or solvent controls (−LOV), protein extracts were prepared and increasing amounts were resolved by SDS-PAGE and transferred to polyvinylidene fluoride, and the amount of p27 determined by immunoblot with enhanced chemiluminescence as a detection reagent. The amount of extract loaded is indicated above each lane (in micrograms). (C) Lovastatin treatment increases translation of the templates containing the 5′ and 3′ UTRs of p27 mRNA in a U-rich element-dependent manner. At 24 h following cotransfection of MDA468 cells with 10 μg of luciferase reporter constructs flanked by the indicated UTRs and 5 μg of CMV-β-galactosidase, the cells were exposed to 30 μM lovastatin or solvent vehicle (+LOV and −LOV, respectively), and activity was measured 48 h later as described in the legend to Fig. 3C. To account for any affect of lovastatin on general cell metabolism, all the data were normalized to the expression of the constructs containing only the vector UTRs. (D) A p40/41 binding activity can be detected in the cytosol of lovastatin-treated cells. Cytosolic extract was prepared from the treated and control cells in panel A, and following a binding reaction to a uniformly labeled 5′UTR transcript, associated proteins were detected by UV cross-linking as described in the legend to Fig. 4C. (E) The p40/41 binding protein in lovastatin-treated cells is hnRNP C1/C2. After scaling up the binding reactions fivefold, following cross-linking and RNA digestion, immunoprecipitation was done as described in the legend to Fig. 5B, using the monoclonal antibody to either C1/C2 (4F4) or TrpE, as indicated above each lane. The products in the precipitate and in the supernatant were resolved by SDS-PAGE, and p40/41 was enriched in the immunoprecipitate and appropriately diminished in the supernatant. Its migration is indicated to the right of the autoradiogram. The pellet and supernatant (SN) from the precipitation are indicated above each lane.
Lovastatin treatment did not affect the steady-state levels of mRNA (data not shown). A faster-migrating form of p40/41 could be cross-linked to the 5′UTR in the extract from lovastatin-treated cells (Fig. 7D). We were able to immunoprecipitate this complex with antibodies specific to hnRNP C1/C2 (Fig. 7E), suggesting that this mobility shift is due to modification of hnRNP C1/C2. We suspect that the differences in the mobility of hnRNP C1/C2 in nocadazole- and lovastatin-treated cells could be due to phosphorylation (47). On longer exposures, HuR was also detected (data not shown).
DISCUSSION
Since its first description by three independent groups in late 1996 and early 1997, remarkably little attention has been paid to the translational regulation of p27. This is surprising since this mode of regulation corresponds best with the role of p27 in growth arrest. In lovastatin-treated cells (23), confluence-arrested BALB/c 3T3 cells (1), and TPA-treated HL-60 cells (35), the accumulation of p27 was attributed to changes in the rate of synthesis, not to changes in the rate of degradation. From studies of PDGF-induced cell cycle reentry of BALB/c 3T3 cells, it appeared that translation was inhibited as cells acquired competence to enter the cell cycle, but that the further reduction in p27 as cells entered S phase was due to proteolysis. This was consistent with the findings in HL-60 cells: there was an increase in the amount of p27 mRNA associated with polysomes in G0 cells compared to G1 cells. The rate of p27 proteolysis was the same in G0 and G1 cells but increased as cells entered S phase.
Three major conclusions can be drawn from this work. First, the U-rich element located in the 5′UTR of p27 mRNA is necessary in both cycling and noncycling cells for efficient translation. Second, this element interacts with HuR and hnRNP C1/C2 in a cell cycle phase-specific manner. Third, an induction of modified hnRNP C1/C2 complexes on the 5′UTR correlates with an induction of p27 translation.
The U-rich element in the 5′UTR of p27 provides a high-affinity binding site for HuR (Kd, ∼10 nM; data not shown). This is the first report of a mammalian ELAV protein binding to the 5′UTR of an mRNA; however, similar interactions have been observed in lower metazoans. In Drosophila melanogaster, sex-lethal (sxl), another ELAV family member, binds to the 5′UTR of the male-specific lethal (msl2) transcript in female flies (4). In this system, sxl negatively regulates translation of the msl2 transcript (4, 19). In mammalian cells, HuR has been implicated in regulating mRNA stability through 3′UTR AU-rich sequences (46). Antic and colleagues have also shown that Hel-N1, a neuron-specific ELAV family member, regulates translation of neurofilament M mRNA, presumably through a 3′UTR binding site (2). These data together suggest that cytoplasmic HuR may regulate translation of specific mRNAs. Attempts to modulate HuR expression and correlate this to p27 expression have not been successful. However, we have never been able to completely eliminate the expression of HuR in cells (unpublished data). This indicates that HuR might not be a limiting component of the complex. An attractive possibility is that HuR binding might facilitate changes in the UTR structure that may allow recruitment of other proteins.
We have also identified hnRNP C1/C2 as p27 5′UTR binding proteins. These proteins are abundant nuclear factors that are thought to be involved in mRNA processing (21). The C proteins have also been implicated in translational regulation of c-sis mRNA, which contains an internal ribosome entry site that is induced in differentiated cells (54). Thus, these proteins may have signal-dependent cytoplasmic roles that modulate translation of specific mRNAs. We detect C-protein binding activity in G2/M-arrested cells and a modified form of this activity in cells that have induced p27 translation. The binding of these factors to the 5′UTR is dependent on the U-rich element, providing a strong link between binding and translation. However, until we develop a cell-free cell cycle phase-specific translation extract, not dependent on rabbit reticulocyte lysate, we are unable to directly examine the role of hnRNP C1/C2 or HuR in the translation of p27 mRNA.
Furthermore, it should be noted that the proteins that we observed cross-linked to the 5′UTR are likely to be only a subset of proteins in a larger RNP complex, most of which may not even contact the RNA in a manner allowing radiolabel transfer. The exact phase of the cell cycle during which these binding activities become cytoplasmic is unclear. We could detect these activities in asynchronous cell extracts as well as in extracts from G2/M cells isolated by centrifugal elutriation (data not shown). However, the magnitude of the 5′UTR binding activities in these extracts was reduced compared to that of nocodazole-treated cells. This suggests that there may be a very specific window of time in which these interactions could occur. We propose that these 5′UTR binding activities may persist, perhaps due to nuclear membrane breakdown, from G2/M through early G1. This would be an ideal time frame for p27 mRNA to be receptive to signals that induce its translation.
Finally, with the exception of ferritin mRNA (52), there is very little information on how UTR binding proteins can affect induced translation. Further studies on the p27 mRNA will provide a counterpoint to that experimental system. In contrast, there is extensive knowledge of proteins involved in the basal translation machinery and how these interact to regulate translation. Our previous work and the results presented here identified cell cycle-regulated changes in p27 translation, mapped sequences involved in this to the 5′UTR, and begun to identify some of the proteins which interact with these sequences. This should provide the foundation for developing appropriate in vitro systems that will allow further mechanistic analysis of induced translation and the roles of these proteins in that process and, importantly, as a function of cell cycle status.
ACKNOWLEDGMENTS
We thank James Roberts (FHCRC, Seattle, Wash.), Ken Marians (MSKCC), and Gino Vairo (AMRAD, Australia) and members of the laboratory for discussions during completion of these studies and comments on the manuscript. We thank Henry Furneaux (MSKCC) and Myriam Gorospe (National Institute of Aging, NIH) for sharing unpublished data and results with HuR reduction experiments. We thank Serafin Pinol-Roma (Mt. Sinai, New York, N.Y.), Stacy Blain (MSKCC), and Merck for their generosity with the 4F4 hnRNP C1/C2 antibody, the p27ck− construct, and lovastatin, respectively. We thank Paul Tempst (MSKCC) and the protein sequencing facility for mass fingerprinting p40/41.
This work was supported by funds from the National Institutes of Health (GM52597, A.K.) and the National Cancer Institute (Cancer Center grant CA08748 to Memorial Sloan-Kettering Cancer Center). A.V. is supported by an FPI fellowship of the Spanish Ministry for Education and Culture. A.K. is a Pew Scholar in Biomedical Sciences, an Irma T. Hirschl Scholar, and the incumbent of the Frederick R. Adler Chair for Junior Faculty at Memorial Sloan-Kettering Cancer Center.
REFERENCES
- 1.Agrawal D, Hauser P, McPherson F, Dong F, Garcia A, Pledger W J. Repression of p27kip1 synthesis by platelet-derived growth factor in BALB/c 3T3 cells. Mol Cell Biol. 1996;16:4327–4336. doi: 10.1128/mcb.16.8.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antic D, Lu N, Keene J D. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atasoy U, Watson J, Patel D, Keene J D. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- 4.Bashaw G J, Baker B S. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell. 1997;89:789–798. doi: 10.1016/s0092-8674(00)80262-7. [DOI] [PubMed] [Google Scholar]
- 5.Burd C G, Swanson M S, Gorlach M, Dreyfuss G. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc Natl Acad Sci USA. 1989;86:9788–9792. doi: 10.1073/pnas.86.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrano A C, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 7.Casaccia-Bonnefil P, Tikoo R, Kiyokawa H, Friedrich V, Jr, Chao M V, Koff A. Oligodendrocyte precursor differentiation is perturbed in the absence of the cyclin-dependent kinase inhibitor p27Kip1. Genes Dev. 1997;11:2335–2346. doi: 10.1101/gad.11.18.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen P, Segil N. p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development. 1999;126:1581–1590. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- 9.Coats S, Whyte P, Fero M L, Lacy S, Chung G, Randel E, Firpo E, Roberts J M. A new pathway for mitogen-dependent cdk2 regulation uncovered in p27(Kip1)-deficient cells. Curr Biol. 1999;9:163–173. doi: 10.1016/s0960-9822(99)80086-4. [DOI] [PubMed] [Google Scholar]
- 10.Cordon-Cardo C, Koff A, Drobnjak M, Capodieci P, Osman I, Millard S S, Gaudin P B, Fazzari M, Zhang Z F, Massague J, Scher H I. Distinct altered patterns of p27KIP1 gene expression in benign prostatic hyperplasia and prostatic carcinoma. J Natl Cancer Inst. 1998;90:1284–1291. doi: 10.1093/jnci/90.17.1284. [DOI] [PubMed] [Google Scholar]
- 11.Dulic V, Lees E, Reed S I. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 12.Durand B, Fero M L, Roberts J M, Raff M C. p27Kip1 alters the response of cells to mitogen and is part of a cell-intrinsic timer that arrests the cell cycle and initiates differentiation. Curr Biol. 1998;8:431–440. doi: 10.1016/s0960-9822(98)70177-0. [DOI] [PubMed] [Google Scholar]
- 13.Erdjument-Bromage H L M, Grewal A, Annan R S, McNulty D E, Carr S A, Tempst P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- 14.Fan X C, Steitz J A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fero M L, Randel E, Gurley K E, Roberts J M, Kemp C J. The murine gene p27Kip1 is haplo-insufficient for tumour suppression. Nature. 1998;396:177–180. doi: 10.1038/24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fero M L, Rivkin M, Tasch M, Porter P, Carow C E, Firpo E, Polyak K, Tsai L H, Broudy V, Perlmutter R M, Kaushansky K, Roberts J M. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 18.Ford L P, Watson J, Keene J D, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gebauer F, Merendino L, Hentze M W, Valcarcel J. The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA. 1998;4:142–150. [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh P M, Moyer M L, Mott G E, Kreisberg J I. Effect of cyclin E overexpression on lovastatin-induced G1 arrest and RhoA inactivation in NIH3T3 cells. J Cell Biochem. 1999;74:532–543. doi: 10.1002/(sici)1097-4644(19990915)74:4<532::aid-jcb3>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Gorlach M, Burd C G, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 22.Hatakeyama M, Weinberg R A. The role of RB in cell cycle control. Prog Cell Cycle Res. 1995;1:9–19. doi: 10.1007/978-1-4615-1809-9_2. [DOI] [PubMed] [Google Scholar]
- 23.Hengst L, Reed S I. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- 24.Joseph B, Orlian M, Furneaux H. p21(waf1) mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J Biol Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- 25.Keene J D. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly B L, Wolfe K G, Roberts J M. Identification of a substrate-targeting domain in cyclin E necessary for phosphorylation of the retinoblastoma protein. Proc Natl Acad Sci USA. 1998;95:2535–2540. doi: 10.1073/pnas.95.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiyokawa H K R, Manova-Todorova K O, Soares V C, Hoffman E, Ono M, Khanam D, Hayday A C, Frohman L A, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27Kip1. Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 28.Koff A, Giordano A, Desai D, Yamashita K, Harper J W, Elledge S, Nishimoto T, Morgan D O, Franza B R, Roberts J M. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 29.Kolluri S K, Weiss C, Koff A, Gottlicher M. p27(Kip1) induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev. 1999;13:1742–1753. doi: 10.1101/gad.13.13.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy N S, Chung S, Furneaux H, Levy A P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 31.Lowenheim H, Furness D N, Kil J, Zinn C, Gultig K, Fero M L, Frost D, Gummer A W, Roberts J M, Rubel E W, Hackney C M, Zenner H P. Gene disruption of p27(Kip1) allows cell proliferation in the postnatal and adult organ of corti. Proc Natl Acad Sci USA. 1999;96:4084–4088. doi: 10.1073/pnas.96.7.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y, Hurwitz J, Massague J. Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature. 1995;375:159–161. doi: 10.1038/375159a0. [DOI] [PubMed] [Google Scholar]
- 33.Ma W J, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 34.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 35.Millard S S, Yan J S, Nguyen H, Pagano M, Kiyokawa H, Koff A. Enhanced ribosomal association of p27(Kip1) mRNA is a mechanism contributing to accumulation during growth arrest. J Biol Chem. 1997;272:7093–7098. doi: 10.1074/jbc.272.11.7093. [DOI] [PubMed] [Google Scholar]
- 36.Minami S, Ohtani-Fujita N, Igata E, Tamaki T, Sakai T. Molecular cloning and characterization of the human p27Kip1 gene promoter. FEBS Lett. 1997;411:1–6. doi: 10.1016/s0014-5793(97)00660-1. [DOI] [PubMed] [Google Scholar]
- 37.Mizokami A, Chang C. Induction of translation by the 5′-untranslated region of human androgen receptor mRNA. J Biol Chem. 1994;269:25655–25659. [PubMed] [Google Scholar]
- 38.Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 1995;14:4442–4449. doi: 10.1002/j.1460-2075.1995.tb00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myer V E, Fan X C, Steitz J A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama K I N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh D Y, Nakayama K I. Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen H, Gitig D M, Koff A. Cell-free degradation of p27kip1, a G1 cyclin-dependent kinase inhibitor, is dependent on CDK2 activity and the proteasome. Mol Cell Biol. 1999;19:1190–1201. doi: 10.1128/mcb.19.2.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohtsubo M, Roberts J M. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- 43.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Del Sal G, Chau V, Yew P R, Draetta G F, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 45.Park M S, Rosai J, Nguyen H T, Capodieci P, Cordon-Cardo C, Koff A. p27 and Rb are on overlapping pathways suppressing tumorigenesis in mice. Proc Natl Acad Sci USA. 1999;96:6382–6387. doi: 10.1073/pnas.96.11.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng S S, Chen C Y, Xu N, Shyu A B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinol-Roma S, Dreyfuss G. Cell cycle-regulated phosphorylation of the pre-mRNA-binding (heterogeneous nuclear ribonucleoprotein) C proteins. Mol Cell Biol. 1993;13:5762–5770. doi: 10.1128/mcb.13.9.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Portman D S, Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13:213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao S, Porter D C, Chen X, Herliczek T, Lowe M, Keyomarsi K. Lovastatin-mediated G1 arrest is through inhibition of the proteasome, independent of hydroxymethyl glutaryl-CoA reductase. Proc Natl Acad Sci USA. 1999;96:7797–7802. doi: 10.1073/pnas.96.14.7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Resnitzky D, Gossen M, Bujard H, Reed S I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richon V M, Venta-Perez G. Changes in E2F DNA-binding activity during induced erythroid differentiation. Cell Growth Differ. 1996;7:31–42. [PubMed] [Google Scholar]
- 52.Rouault T A, Klausner R D, Harford J B. Translational control monograph. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. Translational control of ferritin; pp. 335–362. [Google Scholar]
- 53.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci USA. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sella O, Gerlitz G, Le S Y, Elroy-Stein O. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol Cell Biol. 1999;19:5429–5440. doi: 10.1128/mcb.19.8.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 56.Sherr C J, Roberts J M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 57.Soos T J, Kiyokawa H, Yan J S, Rubin M S, Giordano A, DeBlasio A, Bottega S, Wong B, Mendelsohn J, Koff A. Formation of p27-CDK complexes during the human mitotic cell cycle. Cell Growth Differ. 1996;7:135–146. [PubMed] [Google Scholar]
- 58.Soucek T, Holzl G, Bernaschek G, Hengstschlager M. A role of the tuberous sclerosis gene-2 product during neuronal differentiation. Oncogene. 1998;16:2197–2204. doi: 10.1038/sj.onc.1201743. [DOI] [PubMed] [Google Scholar]
- 59.Tong W, Kiyokawa H, Soos T J, Park M S, Soares V C, Manova K, Pollard J W, Koff A. The absence of p27Kip1, an inhibitor of G1 cyclin-dependent kinases, uncouples differentiation and growth arrest during the granulosa->luteal transition. Cell Growth Differ. 1998;9:787–794. [PubMed] [Google Scholar]
- 60.Tsvetkov L M, Yeh K H, Lee S J, Sun H, Zhang H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- 61.Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W, Furneaux H, Cheng H, Caldwell M C, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]