Abstract
Objective:
The transcription factor Krüppel-like factor 14 (KLF14) has been associated with type 2 diabetes and high-density lipoprotein-cholesterol (HDL-C) through genome-wide association studies. The mechanistic underpinnings of KLF14’s control of metabolic processes remain largely unknown. We studied the physiological roles of KLF14 in a knockout (KO) mouse model.
Methods:
Male whole body Klf14 KO mice were fed a chow or high fat diet (HFD) and diet induced phenotypes were analyzed. Additionally, tissue-specific expression of Klf14 was determined using RT-PCR, RNA sequencing, immunoblotting and whole mount lacZ staining. Finally, the consequences of KLF14 loss-of-function were studied using RNA sequencing in tissues with relatively high Klf14 expression levels.
Results:
KLF14 loss-of-function did not affect HFD-induced weight gain or insulin resistance. Fasting plasma concentrations of glucose, insulin, cholesterol, HDL-C and ApoA-I were also comparable between Klf14+/+ and Klf14−/− mice on chow and HFD. We found that in mice expression of Klf14 was the highest in the anterior pituitary (adenohypophysis), lower but detectable in white adipose tissue and undetectable in liver. Loss of KLF14 function impacted on the pituitary transcriptome with extracellular matrix organization as the primary affected pathway and a predicted link to glucocorticoid receptor signaling.
Conclusions:
Whole body loss of KLF14 function in male mice does not result in metabolic abnormalities as assessed under chow and HFD conditions. Mostly likely there is redundancy for the role of KLF14 in the mouse and a diverging function in humans.
Keywords: Krüppel-like factor, adenohypophysis, genome-wide association studies, mouse model
1. Introduction
Krüppel-like factors (KLFs) are transcription factors characterized by three highly conserved C2H2 zinc fingers that play roles in many different biological processes ranging from pluripotency to metabolism [1, 2]. KLF14 is a relatively recently discovered member of this family. It is encoded by an intronless gene and has monoallelic maternal expression in all studied tissues in both human and mouse [3]. The physiological roles KLF14 remain largely unknown.
Potential clues into KLF14’s function came from genome-wide association studies (GWAS). Several studies associated susceptibility to type 2 diabetes with markers (rs4731702 and rs972283) close to KLF14 [4, 5]. These variants only increased risk when carried on the maternal chromosome [4] and correlated with lower expression of KLF14 in adipose tissue [4, 5]. The same variants associate with high-density lipoprotein-cholesterol (HDL-C) and triglycerides (TGs) [6–8], although the association with TGs is female-specific [8]. Follow-up studies revealed that the KLF14 locus is an adipose trans-expression quantitative trait locus (eQTL) hotspot and that the expression of several of these trans-mediated genes was significantly associated with metabolic traits [9, 10]. The association of variation near KLF14 with several metabolic traits has been replicated in several non-European populations such as African Americans [11, 12]. Some mechanistic insight was obtained by using intermediate quantitative glycemic traits and revealed that the KLF14 risk allele was characterized by primary effects on insulin sensitivity [13].
Only a few studies have addressed the mechanistic underpinnings of KLF14’s control on metabolic processes. Overexpression of KLF14 in Hepatoma carcinoma Hepa1–6 cells was shown to increase glucose uptake by stimulating the PI3K/Akt pathway [14]. KLF14 also increased expression of sphingosine kinase 1 in human and mouse endothelial cells [15]. Sphingosine kinase 1 is responsible for the generation of the signaling lipid sphingosine-1-phosphate, which is an important component of HDL and plays a role in cardiovascular protection [16]. Others have highlighted a role for KLF14 in macrophage inflammatory response impacting on atherosclerosis development [17]. KLF14 was also reported to induce ApoA-I expression in human and mouse hepatocytes offering a mechanistic explanation for the observed association with HDL-C [18]. Perhexiline was identified as a potential KLF14 activator that can increase HDL-C in several mouse models [18]. These mechanisms involve a multitude of pathways and tissues and have not been replicated so far. Here we investigated the metabolic consequences of loss of KLF14 function in mice.
2. Materials and methods
2.1. Mice
Sperm of Klf14+/− mice (C57BL/6N-Klf14tm1.1(KOMP)Vlcg) was obtained from the KOMP repository (University of California, Davis). In the Velocigene KOMP project [19], a targeted deletion was created via homologous recombination using a ZEN-UB1 cassette containing a lacZ reporter and a loxP flanked selection marker (hUbCpro-neo-p(A)). The targeted allele in these mice was designated tm1. Mice that contained the tm1 allele were bred to Cre-expressing mice resulting in the removal of the beta-actin promoter and the Neomycin gene that it activated. The resulting mice carried the tm1.1 allele and were used for this study (Fig. 1).
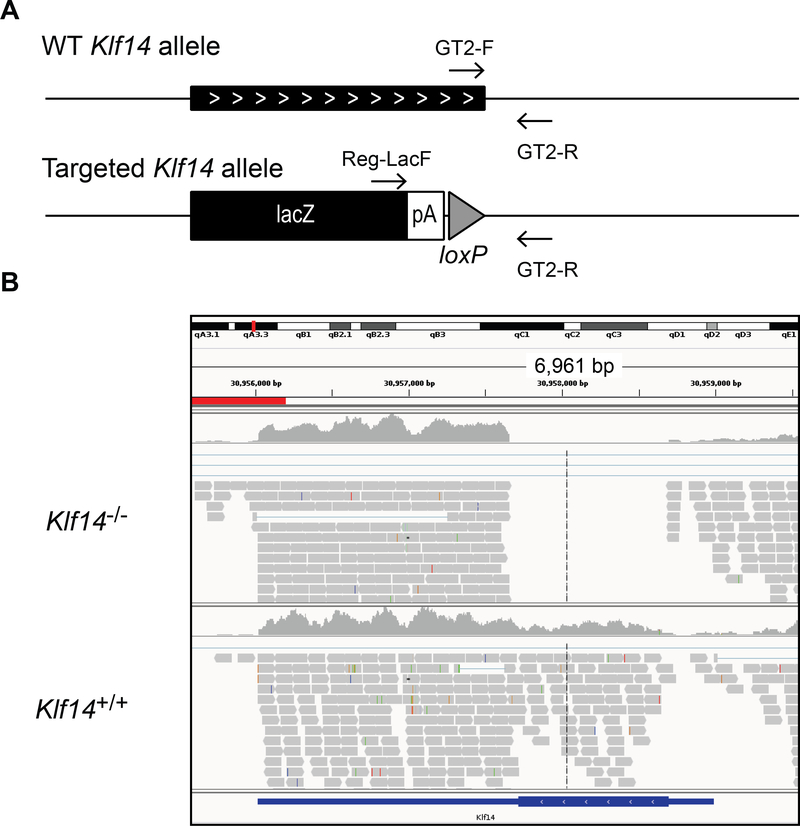
Figure 1.
The Klf14 KO mouse. (A) Schematic representation of the WT and targeted Klf14 allele. Primers used for genotyping are indicated. (B) Integrative Genomics Viewer representation of the aligned RNA sequencing reads at the Klf14 locus in pituitary of a representative Klf14+/+ and Klf14−/− sample.
Mice were genotyped using a PCR with 3 primers (Fig. 1); Reg-LacF (5’-ACTTGCTTTAAAAAACCTCCCACA-3’), GT2-F (5’-CATGGTGGAAAGCTCTGGTT-3’) and GT2-R (5’-CGGTCTAATGACCCTGGAAA-3’). The WT allele was amplified by GT2-F and GT2-R yielding a PCR product of 186 basepairs while the tm1.1 allele was amplified by Reg-LacF and GT2-R yielding a PCR product of ~380 basepairs. In the tm1.1 allele, the complete Klf14 open reading frame is deleted. The insertion of the ZEN-Ub1 Velocigene cassette created a deletion of 1035bp between positions 30,907,660–30,908,694 of chromosome 6 (Genome Build37). The LacZ reporter is inserted just downstream of the Klf14 startcodon. The 3’-UTR of Klf14 is largely intact with the recombination event 59 basepairs downstream of the amber stopcodon. Thus the LacZ reporter can be used as an in situ reporter to determine the gene expression profile of Klf14 [20–22].
2.2. Phenotyping protocol
All animal experiments were approved by the IACUC of the Icahn School of Medicine at Mount Sinai and comply with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). We used male mice only and the entire cohort was obtained by heterozygote breeding pairs. Mice were on chow until 8–9 weeks of age. At this time, mice in the high fat diet (HFD) groups were switched to the TD.06414 diet (Envigo), which contains 60% kcal from fat and is typically used for diet-induced obesity. EchoMRI was performed to determine body composition measurements of fat, lean, free water, and total water masses at 16 weeks of age. An intraperitoneal glucose tolerance test (IPGTT) was performed at 19 weeks of age for which the mice were fasted overnight and then injected with glucose. Mice on the chow diet received 2g glucose per kg body weight (2g/kg), whereas mice on the HFD received 1g/kg glucose. Mice were euthanized at 20–21 weeks of age after overnight food withdrawal. They were first anesthetized with pentobarbital (100mg/kg IP) and then exsanguinated via the vena cava inferior. Blood was collected for the preparation of EDTA plasma and organs were snap frozen in liquid nitrogen and stored at −80°C for future analyses.
2.3. Whole mount lacZ staining
Whole mount lacZ staining to identify KLF14 expressing tissues was performed as described [21]. Mice were euthanized using CO2 asphyxiation and 30 seconds after spontaneous breathing stopped, they were perfused with ice cold 4% paraformaldehyde in PBS (pH 8.0) via the left cardiac ventricle. After fixation, tissues of interest were dissected and collected in jars with ice cold PBS (pH 8.0). After an additional 30 minutes of fixing, tissues were washed with PBS and stained overnight in lacZ staining solution (PBS pH8.0 containing 2mM MgCl2, 0.01% deoxycholic acid, 0.02% IGEPAL CA-630, 0.1% X-Gal in DMF, 5mM potassium ferrocyanide and 5mM potassium ferricyanide) at 4°C. After staining, the tissues were rinsed with PBS, postfixed overnight and subsequently cleared in 50% and then 70% glycerol in PBS.
2.4. Plasma chemistry
Plasma was used for the determination of glycerol (Sigma), TGs (Infinity, Thermo Scientific), total cholesterol (Infinity, Thermo Scientific), HDL-C (Crystal Chem) and insulin (Crystal Chem). Values for TGs were obtained after subtraction of free glycerol. Plasma ApoA-I was quantified using immunoblot. For this assay, 0.5μL of plasma was separated on a Bolt™ 4–12% Bis-Tris Plus Gel, blotted onto nitrocellulose and detected using an Apolipoprotein A1 polyclonal antibody (PA1–23059, Thermo Fisher Scientific). ApoA-I was quantified using IRDye 800CW secondary antibodies (LI-COR) and Image Studio Lite software (version 5.2, LI-COR). Equal loading was checked by Ponceau S staining. Fast Protein Liquid Chromatography (FPLC) of lipoproteins in pooled plasma samples for each cohort was performed by the Mouse Metabolic Phenotyping Center (MMPC) at Vanderbilt University School of Medicine. Corticosterone was determined using ELISA (Immuno-Biological Laboratories America; IB79175) and an EnSpire 2300 multimode plate reader (PerkinElmer).
2.5. RNA isolation, cDNA synthesis and quantitative PCR
RNA was isolated using QIAzol lysis reagent followed by purification using the RNeasy kit (Qiagen). Given the intronless nature of the Klf14 gene, a DNase digestion was carried out during the RNA purification. cDNA synthesis was performed with the SuperScript IV first strand synthesis system (Thermo Fisher Scientific) using random hexamers as primers. qRT-PCR was performed using the ABI Prism 7900HT with Bio-rad iQ SYBR Green Mastermix. Data were analyzed using the Applied Biosystems SDS software. All samples were analyzed in duplicate and expression values were normalized against Rplp0. Primers sequences are provided in Table S1.
2.6. Immunoblotting
There are currently antibodies against 3 different KLF14 epitopes commercially available. Please note that multiple vendors offer apparently identical antibodies. We used PA5–23784 (Thermo Fisher), HPA044729 (Sigma-Aldrich) and TA341450 (Origene). All these antibodies are raised against human KLF14 epitopes (amino acids 254–283, 276–310 and 2–51, respectively) and only the epitope for PA5–23784 is 100% conserved between human and mouse KLF14. We tested the reactivity of these antibodies on overexpressed mouse KLF14. Mouse KLF14 was overexpressed in HEK-293 cells through transient transfection of a plasmid encoding Myc-DDK-tagged mouse Klf14 (Origene, MR221172). Both PA5–23784 and HPA044729 were able to recognize mouse KLF14 in immunoblotting. Immunoblotting was performed as described (see 2.4).
2.7. RNA sequencing analysis
RNA samples were submitted to the Genomics Core Facility at the Icahn Institute and Department of Genetics and Genomic Sciences. cDNA libraries were prepared using the Illumina TruSeq RNA Library Preparation kit (#RS-122–2001). Samples were run on Illumina HiSeq 2500 at a read length of 100nt single end, and a depth of about 25 million per sample. Raw and processed data were returned, count files were generated by aligning the fastq files to the mouse genome mm10 (GRCm38.75) with STAR and counting overlaps with exons grouped at gene level by featureCounts [23].
Differential gene expression analysis was conducted with R packages limma (with voom transformation) [24] and DESeq2 [25]. Low count genes were removed in the limma analysis, genes were kept if they had at least one count per million in at least 4 samples. Cut off for differential expression was chosen at an adjusted p value (Benjamini, Hochberg) of <0.05.
2.8. Pathway enrichment analysis and transcription factor binding analysis
Pathway enrichment analysis was performed using the ClueGo v2.2.5 and CluePedia v1.2.5 plugins in Cytoscape (v. 3.4.0) with the GO, KEGG and Reactome databases (11/2016 download) [26, 27]. Cut off for pathway enrichment was chosen at an adjusted p value (Benjamini, Hochberg) of <0.05.
Iregulon v1.3 was used to create a gene regulatory network [28]. The following options Motif collection: 10K (9713 PWMs), Putative regulatory region: 10kb centered around TSS (10 species) alongside the program default settings for Recovery and TF prediction options were selected for the analysis.
2.9. Statistics
Data are displayed as the mean ± the standard deviation (SD) or standard error of the mean (SEM) as indicated in the figure legends. Differences between Klf14+/+ and Klf14−/− mice were evaluated using a two-sided t-test, one-way analysis of variance with Bonferroni’s multiple comparison test or a Kruskal-Wallis test with Dunn’s multiple comparison post-tests as indicated (GraphPad Prism 6). Significance is indicated in the figures. Deviation of expected Mendelian segregation of genotypes was calculated with a Χ2 test. Deviation of expected sex ratio of births was calculated with a binomial test.
3. Results
3.1. Characterization of Klf14 KO mice
Mice with a targeted deletion of Klf14 were created by the KOMP. We confirmed the correct targeting of Klf14 using multiple assays. Genotyping results using PCR on genomic DNA were consistent with the VelociGene KOMP design (Fig. 1A). Sequencing of genomic DNA confirmed the correct insertion of the lacZ reporter in the Klf14 locus. The Klf14+/− and Klf14−/− animals express lacZ instead of Klf14 (see 3.3 and 3.4) and alignment of RNA sequencing reads is consistent with a deletion of the complete open reading frame in Klf14−/− animals (Fig. 1B).
Klf14−/− mice were generated through heterozygote breeding pairs. Segregation of genotypes was Mendelian (59 Klf14+/+, 96 Klf14+/−, 55 Klf14−/−, Χ2 not significant) with equal male-female ratio (98 male, 112 female; sex ratio of births is 88, binomial test not significant). Male and female Klf14−/− animals were fertile and appeared healthy.
3.2. Diet-induced obesity and associated metabolic phenotypes are not affected in male Klf14 KO mice
Given KLF14’s association with metabolic variables, we performed metabolic phenotyping of Klf14+/+, Klf14+/− and Klf14−/− mice on chow and high fat diet (HFD). Feeding a HFD induced body weight gain, increased fat mass and caused glucose intolerance as measured by the IPGTT (Fig. 2A–C, Table 1). HFD feeding also increased the fasting plasma concentration of glucose, cholesterol, HDL-C and ApoA-I, but decreased TG levels. Unexpectedly, the loss of KLF14 function did not affect any of these diet-induced phenotypes (Fig. 2A–C, Table 1). Klf14+/+ and Klf14−/− mice on chow and HFD did not differ in body weight, lean and fat body mass and fasting-induced body weight loss (Fig. 2A–C, Table 1). Glucose excursion during an IPGTT was also not different between Klf14+/+ and Klf14−/− mice (Fig. 2A). Plasma clinical analytes indicative of metabolic health including concentrations of fasting glucose, insulin, TGs and cholesterol were also similar between Klf14+/+ and Klf14−/− mice (Table 1).
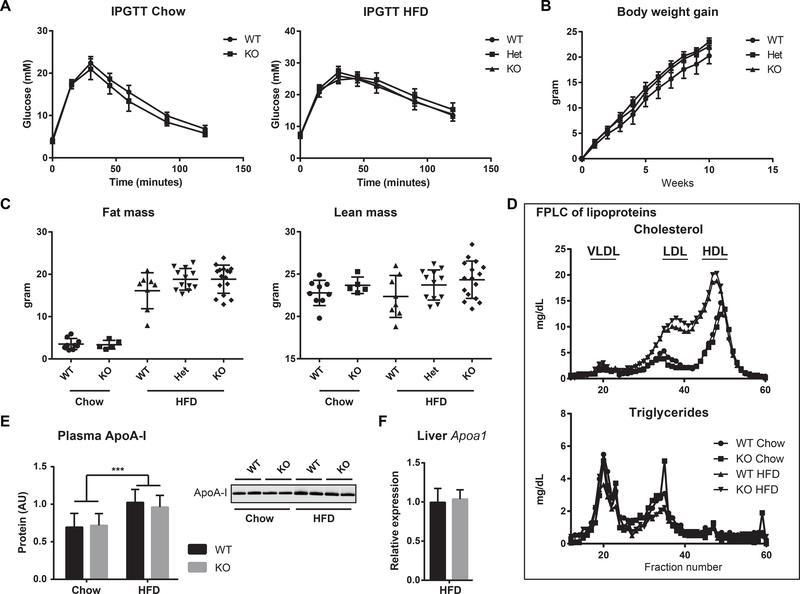
Figure 2.
Metabolic studies in the Klf14 KO mouse. (A) IPGTT in mice on chow and HFD. Error bars in the IPGTT curves indicate SEM. (B) Weight gain of Klf14+/+, Klf14+/− and Klf14−/− mice after switching to a HFD. Error bars indicate SEM. (C) Fat and lean mass as measured by EchoMRI in mice on chow and HFD. The scatter dot plots include mean and SD. (D) FPLC separation of a pooled plasma sample for each cohort. All fractions were assayed for cholesterol and triglycerides. The position of the lipoproteins is indicated in the chromatogram. (E) ApoA-I protein levels in plasma and one representative immunoblot of plasma ApoA-I protein. Error bars indicate SD, group size is 6, *** P< 0.001. (F) Expression of Apoa1 normalized to 36B4 (Rplp0) in liver of Klf14+/+ (n=4) and Klf14−/− (n=3) mice on HFD. Error bars indicate SD.
Table 1.
Analyzed variables in WT and Klf14 KO mice on chow and high fat diet.
| KW | Chow diet |
post | High fat diet |
post | ||||
|---|---|---|---|---|---|---|---|---|
| Sig. | Klf14 +/+ | Klf14 −/− | Sig. | Klf14 +/+ | Klf14 +/− | Klf14 −/− | Sig. | |
|
| ||||||||
| Body weight (g) | **** | 27.7 ± 1.9 | 28.6 ± 1.4 | # | 40.8 ± 6.8 | 45.0 ± 3.5 | 45.4 ± 3.1 | # |
| Lean mass (g) | # | 22.8 ± 1.5 | 23.7 ± 1.0 | 22.4 ± 2.5 | 23.7 ± 1.8 | 24.3 ± 2.2 | ||
| Fat mass (g) | **** | 3.5 ± 1.3 | 3.4 ± 1.0 | # | 16.1 ± 4.3 | 18.8 ± 2.5 | 18.8 ± 3.3 | # |
| Fasting-induced BW loss (g) | # | 3.1 ± 0.4 | 2.9 ± 0.7 | 2.3 ± 0.5 | 2.7 ± 0.6 | 2.7 ± 0.5 | ||
| AUC BW gain (g * week) | # | ND | ND | 111 ± 37 | 130 ± 27 | 125 ± 25 | ||
| Glucose (mmol/L) | ** | 5.0 ± 0.9 | 4.9 ± 0.5 | # | 7.6 ± 1.7 | 6.9 ± 1.9 | 6.7 ± 1.6 | # |
| Insulin (ng/mL) | # | 1.3 ± 0.8 | 1.3 ± 0.5 | 1.2 ± 0.3 | 1.1 ± 0.3 | 0.9 ± 0.2 | ||
| Triglycerides (mmol/L) | *** | 0.48 ± 0.14 | 0.42 ± 0.26 | # | 0.28 ± 0.06 | 0.22 ± 0.06 | 0.25 ± 0.11 | # |
| Glycerol (mmol/L) | # | 0.36 ± 0.08 | 0.31 ± 0.08 | 0.34 ± 0.05 | 0.35 ± 0.05 | 0.37 ± 0.07 | ||
| Cholesterol (mmol/L) | **** | 1.6 ± 0.2 | 1.5 ± 0.2 | # | 3.2 ± 0.8 | 3.2 ± 0.9 | 3.4 ± 0.7 | # |
| HDL (mmol/L) | **** | 1.3 ± 0.2 | 1.2 ± 0.1 | # | 2.1 ± 0.3 | 2.1 ± 0.5 | 2.1 ± 0.4 | # |
| ApoA-I (relative) | ** | 0.7 ± 0.2 | 0.7 ± 0.2 | # | 1.0 ± 0.2 | ND | 1.0 ± 0.2 | # |
| Corticosterone (ng/mL) | # | 179 ± 72 | 189 ± 74 | 229 ± 44 | ND | 221 ± 93 | ||
| AUC IPGTT (dose 2g/kg; mmol/L * minute) | # | 1132 ± 355 | 1039 ± 372 | ND | ND | ND | ||
| AUC IPGTT (dose 1g/kg; mmol/L * minute) | # | ND | ND | 1510 ± 449 | 1673 ± 587 | 1572 ± 364 | ||
| Heart weight (mg) | # | 126 ± 17 | 124 ± 12 | 111 ± 10 | 121 ± 17 | 124 ± 17 | ||
| Liver weight (g) | ** | 1.12 ± 0.12 | 1.16 ± 0.22 | # | 1.47 ± 0.61 | 1.70 ± 0.51 | 1.66 ± 0.44 | # |
Values are the average ± SD. The values are sampled the following cohorts: Chow diet, 11 Klf14+/+ and 6 Klf14−/−; High fat diet, 8 Klf14+/+, 11 Klf14+/− and 15 Klf14−/−. Statistical analysis of the data was performed using a nonparametric Kruskal-Wallis (KW) test with Dunn’s multiple comparison post tests for WT versus KO. AUC stands for area under the baseline corrected curve. Significance (Sig.) is indicated as follows
= not significant
< 0.05
< 0.01
< 0.001 and
< 0.0001. ND denotes not done.
The variation in body weight gain and fat mass was considerably larger in the Klf14+/+ mice when compared to the Klf14+/− and Klf14−/− mice (Fig. 2B, C), but this could be explained by 2 low weight gainers in the Klf14+/+ group. High phenotypic variation with low and high weight gainers in diet-induced obesity is typically observed in male C57BL/6 mice [29, 30]. The difference between the groups is smaller if we remove the 2 low weight gainers in the Klf14+/+ group. Based on these observations, we conclude that loss of KLF14 function in male C57BL/6 mice does not affect diet-induced obesity phenotypes.
Liver-specific deletion of Klf14 was recently shown to decrease HDL-C concentration. This effect was explained by the ability of KLF14 to modulate expression of ApoA-I [18]. However, in contrast to these reported findings, we did not observe changes in plasma lipoprotein profile, cholesterol, HDL-C and ApoA-I in Klf14−/− mice on chow or on HFD (Table 1, Fig. 2D, E). Consistently, expression of Apoa1 in liver was not affected in Klf14−/− mice on HFD (Fig. 2F). Moreover, we were unable to reliably detect Klf14 expression in liver (see 3.3).
3.3. Klf14 is highly expressed in pituitary and testis and undetectable in liver
Previous work has shown that Klf14 expression was higher in embryonic and extra-embryonic tissues when compared to expression in adult tissues and was undetectable in the adult liver [3]. The publicly available GeneAtlas data (BioGPS) and EST profiles (UniGene) further suggest that Klf14 is highly expressed in mouse pituitary. Expression levels of Klf14 are much lower in all other surveyed tissues. This observation is confirmed by other studies that profiled brain region-specific expression profiles and showed higher expression of Klf14 in the pituitary compared to most other brain regions [31, 32]. To further study the expression profile of Klf14 in the adult mouse, we surveyed cDNA of 12 different tissues by PCR. Klf14 expression was readily detectable in pituitary and several white adipose tissue (WAT) depots and was virtually undetectable in most other tissues including the liver and brown adipose tissue (BAT; Fig. 3A). In order to more accurately quantify and compare Klf14 expression between different tissues, we performed RNA sequence analysis on RNA extracted from pituitary, inguinal WAT, gastrocnemius muscle, heart and liver tissues. While most sequencing reads mapping to Klf14 were identified in pituitary, inguinal WAT also had significant Klf14 expression (Fig. 3B). On the other hand, very low read counts were found in muscle and heart, and no Klf14 mapped reads were identified in liver from two separate mouse cohorts (Fig. 3B). Unfortunately, we were unable to detect KLF14 protein in pituitary and liver (Fig. S1). KLF14 expression results could therefore not be further corroborated at the protein level.
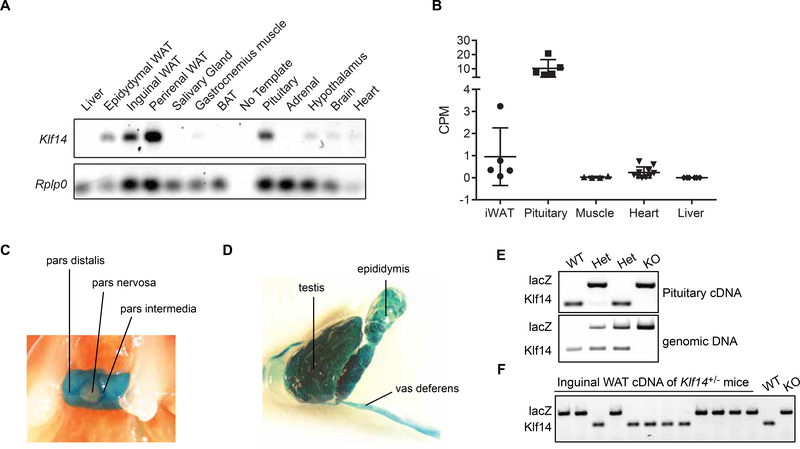
Figure 3.
Expression analysis of Klf14 in the mouse. (A) Semi-quantitative analysis of Klf14 expression across 12 mouse tissues. (B) Count per million mapped Klf14 reads derived from RNA sequencing data from 5 different tissues. The scatter dot plots include mean and SD. (C) Whole mount LacZ staining of pituitary of a male Klf14+/− mouse. (D) Whole mount LacZ staining of testis, epididymis and vas deferens of a Klf14−/− mouse. (E) Allele-specific expression of Klf14 in mouse pituitary. The Klf14 and lacZ expressing alleles were amplified from pituitary cDNA and genomic DNA from the same animal. (F) Allele-specific expression of Klf14 in inguinal WAT of Klf14+/− mice.
Given the central role of the pituitary in neuroendocrine processes, we further studied the expression of Klf14 in this tissue using whole mount lacZ staining. The pituitary is composed of two separate parts; the anterior pituitary or adenohypophysis (pars distalis and pars intermedia) and the posterior pituitary or neurohypophysis (pars nervosa). In order to visualize expression of Klf14 in situ, we used the lacZ reporter that is expressed in Klf14+/− and Klf14−/− animals under control of the Klf14 promoter. Pituitary displayed strong lacZ staining in the pars distalis and intermedia, indicating that the adenohypophysis is the major Klf14 expressing tissue (Fig. 3C). Testis, epididymis and vas deferens were also positive for lacZ, which could indicate that Klf14 is expressed in (developing) spermatozoa (Fig. 3D). Other tissues with lower levels of lacZ staining include gallbladder, muscle and several brain regions including hypothalamus and striatum (not shown). We observed no lacZ staining in liver, WAT and BAT. Overall our whole mount lacZ staining is consistent with pituitary being a primary Klf14 expressing tissue in mouse.
Theoretically Klf14+/− animals should express one WT Klf14 allele and one KO (lacZ) allele. However, we noted that some Klf14+/− animals were negative for lacZ staining. This is consistent with the reported imprinting at the Klf14 locus in mouse oocytes [3]. To demonstrate the allele-specific expression at the Klf14 locus in pituitary, we amplified the WT and KO allele on cDNA generated from adult Klf14+/+, Klf14+/− and Klf14−/− animals. As expected, Klf14+/+ and Klf14−/− animals expressed the Klf14 or lacZ allele respectively, while the Klf14+/− animals expressed either the Klf14 allele or the LacZ allele (Fig. 3E). Amplification on genomic DNA showed the presence of both alleles in these animals (Fig. 3E) indicating that Klf14 is imprinted in adult mouse pituitary. Given the eQTL for KLF14 in humans, we amplified the WT and KO allele on cDNA isolated from inguinal WAT of Klf14+/− animals on HFD and found that the Klf14+/− animals expressed lacZ or Klf14 demonstrating that Klf14 is also imprinted in adult adipose tissue (Fig. 3F).
3.4. Survey of tissue expression of Klf14
We surveyed frozen section lacZ staining of tissues of this Klf14 KO mouse model, which are publicly available at the International Mouse Phenotyping Consortium website [20–22]. Pituitary was positive in pars distalis and pars intermedia confirming the results of the whole mount lacZ staining. Smooth muscle cell layers were lacZ positive in vasculature (e.g. in BAT and quadriceps muscle) and in the walls of the digestive system, urinary bladder and seminal vesicles. There was also abundant lacZ staining in the male reproductive system with the smooth muscle cell layers, the apical cytoplasm of the epithelium in epididymis and the ciliated epithelium in the vas deferens being positive. The more mature layers of the seminiferous tubules in testis were also lacZ positive consistent with expression in (developing) spermatozoa. In addition, lacZ staining was also observed in the cartilage and chondrocytes of the trachea.
3.5. RNA sequencing reveals a role for KLF14 in pituitary function
In order to shed light on the physiological functions of KLF14 in mice, we performed RNA sequencing analysis of RNA isolated from inguinal WAT and pituitary of Klf14+/+ and Klf14−/− animals on chow. Interestingly, we found no differentially expressed genes (DEGs) in the inguinal WAT samples when Klf14+/+ mice were compared to Klf14−/− mice using DESeq2 or limma methods. Mouse genotyping errors and other sample process mishandling were excluded as the cause of the lack of DEGs as evidenced by Integrative Genomics Viewer analysis of Klf14 reads and appropriate sample clustering in principal component analysis (data not shown). In contrast, we found 240 DEGs in pituitary of the same mice by DESeq2 (153 up and 87 down), of which 118 were detected also by limma methodology (Fig. 4A and Table S2). Expression of Klf14 itself was most significantly altered with an almost ~2-fold upregulation suggesting the existence of a positive feed-back mechanism regulating pituitary Klf14 expression levels. Q-PCR analysis of selected DEGs; Cldn7, Rcn1, Megf10 and Vwde, validated the results obtained by RNA sequencing analysis (Fig. 4B).
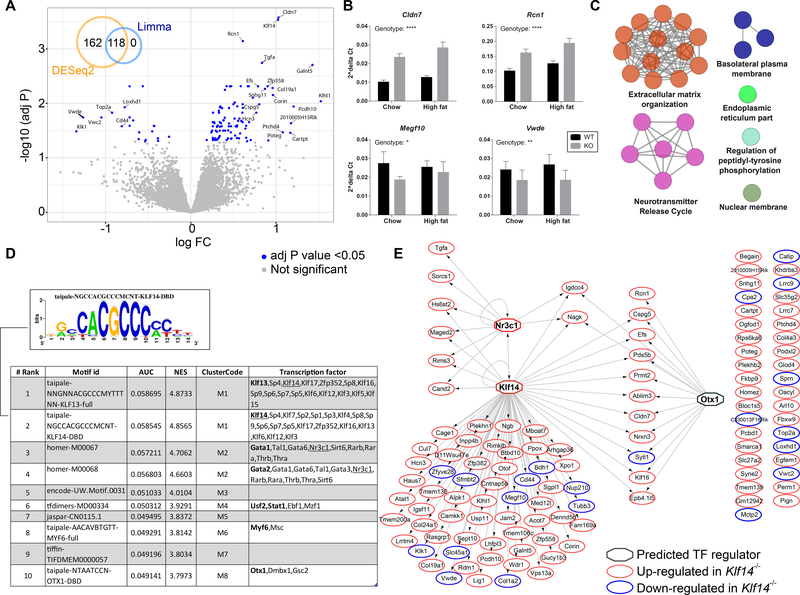
Figure 4.
RNA sequencing analysis of pituitary in the Klf14 KO mouse. (A) Volcano plot displaying the log2 fold change versus -log10 of the adj P value for all genes in the comparison of Klf14+/+ versus Klf14−/− in the pituitary of chow fed animals. The inset Venn diagram shows the overlap of genes detected as differentially expressed using either DESeq2 or limma packages. (B) qPCR validation of selected DEGs. Gene expression was calculated using the ΔCt method using Rplp0 as the reference gene. All 4 genes were significantly altered in a two-way ANOVA. The significance for the variation in genotype is indicated in the figure; **** P < 0.0001, ** P < 0.01, and * P < 0.05. (C) A network displaying pathways significantly enriched for the 118 DEGs in pituitary. The color of the node represents the pathway term grouping. All nodes are significantly enriched at an adj P value <0.05. (D) A table summarizing the results of the Iregulon analysis, which lists the top transcription binding motifs and their associated transcription factors found enriched in the cis-regulatory regions of the 118 DEGs. The underlined TFs are the more likely candidates based on the input data. (E) A network representing the predicted targets (as oval nodes) of the top potential transcriptional regulators (as octagonal nodes). The color of the oval nodes indicates whether that gene was up- or down-regulated in the pituitary of Klf14−/− mice when compared to Klf14+/+ mice.
Canonical pathway enrichment analysis of the 118 DEGs in pituitary revealed significant enrichment in pathways associated with extracellular matrix organization due to several collagen encoding genes, and neurotransmitter release cycle amongst other pathways (Fig. 4C and Table S3). Given that KLF14 is a transcription factor, we assessed whether the DEGs were enriched in KLF14 response elements, which would imply that they are direct KLF14 targets. We used the iRegulon plugin within cytoscape in order to identify ‘regulons’ using motif discovery in a set of co-regulated genes [28]. A regulon is defined as consisting of a transcription factor and its predicted direct transcriptional targets, which within their cis-regulatory control elements contain common transcription factor binding sites. The top transcription factor binding sites found enriched in the 118 DEGs was for elements related to KLF13 and KLF14 (Fig. 4D). Given the high degree of conservation of the C2H2 zinc fingers, it is not surprising to find multiple candidate KLF family members as potential binding transcription factors. Our results suggest that the genes with a KLF13/14 motif are potentially direct KLF14 targets in pituitary (Fig. 4E). The human orthologs of two DEGs (Maged2 and Slc35g2) were also identified as having trans eQTLs at the KLF14 locus [10], but this is not a significant overlap in a hypergeometric test. Of significance was the observation that Nr3c1, the glucocorticoid nuclear hormone receptor was not only significantly upregulated in the Klf14−/− mice, but also predicted to have both a KLF14 as well as an NR3C1 binding element (Fig. 4E). This suggests that KLF14 may impact the glucocorticoid signaling axis, but analysis of plasma corticosterone levels did not reveal differences between Klf14+/+ and Klf14−/− animals (Table 1).
4. Discussion
We investigated the physiological functions of KLF14 by studying a whole body KO mouse model and focused on the metabolic role of this transcription factor by investigating insulin resistance and HDL-C in mice on chow and HFD. Our results indicate that KLF14 does not play a role in the development of diet-induced insulin resistance in male C57BL/6 mice. Moreover, our results do not support a major role of hepatic KLF14 in the regulation of ApoA-I and HDL-C levels, as was previously reported by others [18]. In fact, we found that expression of Klf14 in mouse liver is virtually undetectable and that pituitary was the tissue with highest Klf14 expression. RNA sequencing analysis of pituitary samples from Klf14+/+ and Klf14−/− mice revealed that the loss of KLF14 function impacted on the pituitary transcriptome with extracellular matrix organization as the primary affected pathway. Analysis of transcription factor binding sites revealed a link to glucocorticoid receptor signaling.
Two reports describing the mechanism underlying the regulation of metabolic processes by KLF14 have focused on liver as the primary organ involved. Yang et al showed that Klf14 mRNA and KLF14 protein expression is decreased in liver, adipose and muscle of C57BL/6 mice on HFD and db/db mice when compared to control animals [14]. In subsequent experiments the authors use ectopic KLF14 expression in the mouse hepatoma cell line Hepa1–6 and show that KLF14 stimulates insulin signaling via the classical PI3K/Akt pathway [14]. Consistent with these observations, Guo et al also showed that Klf14 mRNA and KLF14 protein expression was significantly decreased in liver of C57BL/6 mice in response to a HFD and described similar results in ob/ob mice as well [18]. They showed that overexpression of human KLF14 in liver of mice that were fed a HFD, increased HDL-C, cholesterol efflux and ApoA-I expression. Conversely, they showed that liver-specific deletion of Klf14 (floxed Klf14 gene and Alb-Cre) decreased HDL-C and ApoA-I expression. The authors offered a mechanistic explanation by demonstrating that the ApoA-I promoter contained one KLF14-binding site. Moreover, perhexiline was identified as a KLF14 activator that raises HDL-C, ApoA-I and cholesterol efflux in vivo and reduced atherosclerosis development in Apoe−/− mice [18]. Our results do not support a role for KLF14 in hepatic ApoA-I production. The reason for this discrepancy is currently unclear, but may be caused by the use of different transgenic mouse models. Guo et al use a model with a floxed Klf14 gene and Alb-Cre to delete Klf14 in the liver. Expression of the Alb-Cre transgene is progressive with age, but can be detected in late fetal and neonatal stages of hepatocyte development [33]. The KOMP Klf14 mouse model used in this study is a constitutive whole body KO, and its metabolic phenotype could therefore be compromised through early compensatory mechanisms. Regardless, we found no evidence for expression of Klf14 in adult mouse liver as we were unable to amplify Klf14 cDNA and did not find Klf14 mapped reads in our liver RNA sequencing data. Surprisingly, both Yang et al [14] and Guo et al [18] report the use of male C57BL/6 mice similar to our studies. Moreover, the lack of expression of Klf14 in liver in our RNA sequencing experiments does not appear to be mouse strain or institution specific because we analyzed two different mouse cohorts generated in different animal facilities: a second generation progeny of a cross between C57BL/6 and 129/P (B6129PF2) and second, a more advanced C57BL/6N backcross (4 generations [34]). Indeed, our data on the absence of Klf14 expression in liver are consistent with previously published work [3] and publicly available data sources. We would also like to stress that assessing mRNA expression of Klf14 by PCR requires DNase treatment of isolated RNA given the intronless nature of Klf14. Our well validated whole body Klf14 KO mouse model did not display decreased ApoA-I mRNA nor did the animals have changes in plasma lipoprotein profile, cholesterol, HDL-C and ApoA-I, which can be viewed as consistent with the absence of expression of Klf14 in the liver.
Both Yang et al [14] and Guo et al [18] show KLF14 expression in liver at the protein level, which is remarkable given the absence of Klf14 mRNA in this tissue. The rabbit polyclonal antibody employed by Guo et al [18] (and possibly also by Yang et al [14]) is raised against the C-terminal region of human KLF14 (PA5–23784; amino acids 254–283) and is 100% conserved between human and mouse KLF14. By using the PA5–23784 and HPA044729 antibodies, we were able to detect overexpressed mouse KLF14. However, with both antibodies we were unable to detect a specific signal in whole pituitary extracts, nuclear pituitary extract and whole liver extracts (Fig. S1). Combined our data indicate that KLF14 expression in relatively low in pituitary and likely absent in liver.
Similar to mouse, human KLF14 is expressed at higher levels in fetal tissue and placenta than in adult tissues and is absent in liver and lymphoblasts [3]. GTEx data confirm that overall expression of KLF14 is low, but some expression is noted in testis, adrenal gland, spleen, visceral and subcutaneous adipose and breast (RPKM 1.6 and lower) [35]. Liver is the lowest KLF14 expressing organ of all the 53 tissues in the GTEx dataset. Importantly GTEx identified cis eQTLs for the KLF14 expressing tissues testis and subcutaneous adipose [35]. This supports the earlier GWAS studies that linked type 2 diabetes risk to a locus near KLF14 that also harbors a cis eQTL in subcutaneous adipose [4, 5, 9, 10].
We failed to find significant consequences of KLF14 loss of function in mice on insulin resistance and adipose tissue gene expression. Male Klf14−/− animals on a C57BL/6 background had similar IPGTT, fasting insulin and fasting glucose when compared to Klf14+/+ under chow and HFD conditions. Moreover, RNA sequencing analysis of inguinal WAT did not reveal differential expression between male Klf14+/+ and Klf14−/− animals on a chow diet. There are several potential explanations for these observations. First, the metabolic phenotype could have been compensated for through developmental or other processes. Second, the effect of Klf14 deletion on insulin resistance may be very small and thus our study could have been underpowered to detect such an effect. However, the numbers of animals that we used in our studies are not atypical for diet induced obesity studies in KO mouse models. We can therefore argue that any effect missed under these conditions is probably not biologically relevant. Alternatively, the regulation of adipose tissue gene expression by KLF14 may be more specific for humans and therefore not observed in mice. Indeed, there is some evidence that KLF14 has been undergoing human-specific accelerated evolution [3]. Moreover, KLF14 in humans does not appear to be highly expressed in pituitary (GTEx), indicating divergent roles between both species. Lastly, the function of KLF14 may be sex-specific and therefore not readily detectable in male mice. Indeed a conference abstract by Small et al suggests that the molecular cis and trans regulatory effects are present in both sexes, but the cellular and whole body effects (type 2 diabetes, lipids, hip circumference) are female-specific [36].
Despite the lack of effect of Klf14 deletion in adipose tissue, we did observe a significant effect on gene expression in the pituitary. Given the expression profile of Klf14 in mouse pituitary, we anticipate that this is driven by changes in gene expression in the adenohypophysis. The DEGs were particularly enriched in KLF14 transcriptional binding motifs independently providing validity of the KO mouse model. We observed a previously uncharacterized link between KLF14 and glucocorticoid signaling through regulation of Nr3c1 expression. This observation extends the known links between KLF4, KLF13 and KLF15 with glucocorticoid signaling [37–39]. Glucocorticoid signaling plays an important role in stress responses propagated by the HPA axis. We did not observe any signs of major HPA axis dysfunction such as changes in plasma corticosterone, glucose or insulin resistance. We speculate that perturbation of the HPA axis in Klf14−/− mice should be studied upon acute restraint stress. As the HPA axis is critical for metabolic control, our finding in mice may be relevant to explaining the GWAS links found in human adipose.
In summary, whole body loss of KLF14 function in the mouse did not result in metabolic abnormalities as assessed under chow or HFD conditions. Most likely there is redundancy for the role of KLF14 in the mouse and a divergent role for KLF14 in humans.
Supplementary Material
Acknowledgements
The authors thank Andrew Shin and Henry Ruiz of the Mount Sinai Diabetes, Obesity and Metabolism Institute for performing echoMRI analyses, Cindy Else and Ying Dai of the Comparative Pathology Laboratory for help with the whole mount lacZ staining, Carla Harris of the Vanderbilt Mouse Metabolic Phenotyping Center for performing the lipoprotein analyses, and Robin Sgueglia of the Biomarker Analytic Research Core at Albert Einstein College of Medicine for performing plasma corticosterone measurements. We acknowledge the help of the shared resource facilities at the Icahn School of Medicine at Mount Sinai (Colony Management, Real Time Polymerase Chain Reaction (qPCR), Mouse Genetics and Gene Targeting, Genomics Core and the Comparative Pathology Laboratory). This study was supported by grants to the Vanderbilt Mouse Metabolic Phenotyping Center (U24DK59637), the Diabetes Research Center (P60DK020541) and the ICTR Einstein-Montefiore (CTSA UL1TR001073).
Abbreviations
- BAT
brown adipose tissue
- DEG
differentially expressed genes
- FPLC
Fast Protein Liquid Chromatography
- GWAS
genome-wide association studies
- HDL-C
high-density lipoprotein-cholesterol
- HFD
high fat diet
- IPGTT
intraperitoneal glucose tolerance test
- KLF
Krüppel-like factors
- KO
knockout
- eQTL
expression quantitative trait locus
- TG
triglyceride
- WAT
white adipose tissue
References
- [1].Turner J, Crossley M, Mammalian Kruppel-like transcription factors: more than just a pretty finger, Trends Biochem Sci, 24 (1999) 236–240. [DOI] [PubMed] [Google Scholar]
- [2].Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S, Kruppel-like transcription factors: a functional family, Int J Biochem Cell Biol, 40 (2008) 1996–2001. [DOI] [PubMed] [Google Scholar]
- [3].Parker-Katiraee L, Carson AR, Yamada T, Arnaud P, Feil R, Abu-Amero SN, et al. , Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution, PLoS Genet, 3 (2007) e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P, Besenbacher S, et al. , Parental origin of sequence variants associated with complex diseases, Nature, 462 (2009) 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, et al. , Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis, Nat Genet, 42 (2010) 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, Clarke R, et al. , Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis, PLoS Genet, 5 (2009) e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Global Lipids Genetics C, Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, et al. , Discovery and refinement of loci associated with lipid levels, Nat Genet, 45 (2013) 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. , Biological, clinical and population relevance of 95 loci for blood lipids, Nature, 466 (2010) 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, Kong A, et al. , Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes, Nat Genet, 43 (2011) 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Civelek M, Wu Y, Pan C, Raulerson CK, Ko A, He A, et al. , Genetic Regulation of Adipose Gene Expression and Cardio-Metabolic Traits, Am J Hum Genet, 100 (2017) 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen G, Bentley A, Adeyemo A, Shriner D, Zhou J, Doumatey A, et al. , Genome-wide association study identifies novel loci association with fasting insulin and insulin resistance in African Americans, Hum Mol Genet, 21 (2012) 4530–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ng MC, Saxena R, Li J, Palmer ND, Dimitrov L, Xu J, et al. , Transferability and fine mapping of type 2 diabetes loci in African Americans: the Candidate Gene Association Resource Plus Study, Diabetes, 62 (2013) 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dimas AS, Lagou V, Barker A, Knowles JW, Magi R, Hivert MF, et al. , Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity, Diabetes, 63 (2014) 2158–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang M, Ren Y, Lin Z, Tang C, Jia Y, Lai Y, et al. , Kruppel-like factor 14 increases insulin sensitivity through activation of PI3K/Akt signal pathway, Cell Signal, 27 (2015) 2201–2208. [DOI] [PubMed] [Google Scholar]
- [15].de Assuncao TM, Lomberk G, Cao S, Yaqoob U, Mathison A, Simonetto DA, et al. , New role for Kruppel-like factor 14 as a transcriptional activator involved in the generation of signaling lipids, J Biol Chem, 289 (2014) 15798–15809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sattler K, Levkau B, Sphingosine-1-phosphate as a mediator of high-density lipoprotein effects in cardiovascular protection, Cardiovasc Res, 82 (2009) 201–211. [DOI] [PubMed] [Google Scholar]
- [17].Wei X, Yang R, Wang C, Jian X, Li L, Liu H, et al. , A novel role for the Kruppel-like factor 14 on macrophage inflammatory response and atherosclerosis development, Cardiovasc Pathol, 27 (2017) 1–8. [DOI] [PubMed] [Google Scholar]
- [18].Guo Y, Fan Y, Zhang J, Lomberk GA, Zhou Z, Sun L, et al. , Perhexiline activates KLF14 and reduces atherosclerosis by modulating ApoA-I production, J Clin Invest, 125 (2015) 3819–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, et al. , A conditional knockout resource for the genome-wide study of mouse gene function, Nature, 474 (2011) 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].West DB, Pasumarthi RK, Baridon B, Djan E, Trainor A, Griffey SM, et al. , A lacZ reporter gene expression atlas for 313 adult KOMP mutant mouse lines, Genome Res, 25 (2015) 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tuck E, Estabel J, Oellrich A, Maguire AK, Adissu HA, Souter L, et al. , A gene expression resource generated by genome-wide lacZ profiling in the mouse, Dis Model Mech, 8 (2015) 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Koscielny G, Yaikhom G, Iyer V, Meehan TF, Morgan H, Atienza-Herrero J, et al. , The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data, Nucleic Acids Res, 42 (2014) D802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. , STAR: ultrafast universal RNA-seq aligner, Bioinformatics, 29 (2013) 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Smyth GK, limma: Linear Models for Microarray Data, in: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S (Eds.) Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Springer New York, New York, NY, 2005, pp. 397–420. [Google Scholar]
- [25].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol, 15 (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. , ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks, Bioinformatics, 25 (2009) 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bindea G, Galon J, Mlecnik B, CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data, Bioinformatics, 29 (2013) 661–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, et al. , iRegulon: from a gene list to a gene regulatory network using large motif and track collections, PLoS Comput Biol, 10 (2014) e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Koza RA, Nikonova L, Hogan J, Rim JS, Mendoza T, Faulk C, et al. , Changes in gene expression foreshadow diet-induced obesity in genetically identical mice, PLoS Genet, 2 (2006) e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Anunciado-Koza RP, Manuel J, Koza RA, Molecular correlates of fat mass expansion in C57BL/6J mice after short-term exposure to dietary fat, Ann N Y Acad Sci, 1363 (2016) 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kasukawa T, Masumoto KH, Nikaido I, Nagano M, Uno KD, Tsujino K, et al. , Quantitative expression profile of distinct functional regions in the adult mouse brain, PLoS One, 6 (2011) e23228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hovatta I, Zapala MA, Broide RS, Schadt EE, Libiger O, Schork NJ, et al. , DNA variation and brain region-specific expression profiles exhibit different relationships between inbred mouse strains: implications for eQTL mapping studies, Genome Biol, 8 (2007) R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Weisend CM, Kundert JA, Suvorova ES, Prigge JR, Schmidt EE, Cre activity in fetal albCre mouse hepatocytes: Utility for developmental studies, Genesis, 47 (2009) 789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Diekman EF, van Weeghel M, Wanders RJ, Visser G, Houten SM, Food withdrawal lowers energy expenditure and induces inactivity in long-chain fatty acid oxidation-deficient mouse models, FASEB J, 28 (2014) 2891–2900. [DOI] [PubMed] [Google Scholar]
- [35].GTEx-Consortium, Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans, Science, 348 (2015) 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Small K, Todorcevic M, Civelek M, El-Sayed Moustafa J, Mahajan A, Horikoshi M, et al. , Adipose- and maternal- specific regulatory variants at KLF14 influence Type 2 Diabetes risk in women via a female-specific effect on adipocyte physiology and body composition, in: ASHG 2015 Annual Meeting, Baltimore, MD, 2015. [Google Scholar]
- [37].Shimizu N, Yoshikawa N, Ito N, Maruyama T, Suzuki Y, Takeda S, et al. , Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle, Cell Metab, 13 (2011) 170–182. [DOI] [PubMed] [Google Scholar]
- [38].Cruz-Topete D, He B, Xu X, Cidlowski JA, Kruppel-like Factor 13 Is a Major Mediator of Glucocorticoid Receptor Signaling in Cardiomyocytes and Protects These Cells from DNA Damage and Death, J Biol Chem, 291 (2016) 19374–19386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sevilla LM, Latorre V, Carceller E, Boix J, Vodak D, Mills IG, Perez P, Glucocorticoid receptor and Klf4 co-regulate anti-inflammatory genes in keratinocytes, Mol Cell Endocrinol, 412 (2015) 281–289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.