Abstract
The phenomenon of drug resistance has been a hindrance to therapeutic medicine since the late 1940s. There is a plethora of factors and mechanisms contributing to progression of drug resistance. From prokaryotes to complex cancers, drug resistance is a prevailing issue in clinical medicine. Although there are numerous factors causing and influencing the phenomenon of drug resistance, cellular transporters contribute to a noticeable majority. Efflux transporters form a huge family of proteins and are found in a vast number of species spanning from prokaryotes to complex organisms such as humans. During the last couple of decades, various approaches in analyses of biochemistry and pharmacology of transporters have led us to understand much more about drug resistance. In this review, we have discussed the structure, function, potential causes, and mechanisms of multidrug resistance in bacteria as well as cancers.
Keywords: Drug resistance, Efflux pumps, Chemotherapy, ATP-binding cassette transporters, Integrons
Introduction
Paul Ehrlich introduced us to the terms- “chemotherapy” and “the Magic bullet”, where he explained the idea of a therapy that would kill the microbe and leave the affected individual harmless. He then went on to develop a drug against Treponema pallidum (a bacterium that causes syphilis) which was the only available treatment option against bacterial infections for the next 20 years [1]. Sir Alexander Fleming discovered the antibiotic penicillin from the fungi Penicillium notatum, which changed the course of medical science. The success of this discovery was seen in World War II as it helped millions of soldiers who used antibiotics to fight against the “invisible enemy” [2]. The history of medicine catalogs numerous similar episodes that involves antibiotics. It was not until recently that we have experienced the phenomenon of drug resistance. As a fact of the matter, World Health Organization (WHO) said that drug resistance is one of the three serious challenges of public health of the twenty-first century [3].
Bacterial antibiotic resistance is a continuing and increasing concern in clinical science. Owing to a shorter lifespan, bacteria are more susceptible to genetic variation and evolution than other eukaryotic organisms. Escherichia coli has a unique identity in the microbial world as it is also an essential gut bacterium as well as a harmful pathogen. The inquilinity of E. coli with various mammals allows it to cause infections ranging from mild diarrhea to severe colitis. E. coli exhibits a wide range of resistance against drugs such as β-lactams, ampicillin, cephalosporin, quinolones, aminoglycosides, and several other antibiotics [4]. A similar resistance spectrum is shown by methicillin resistant Staphylococcus aureus (MRSA), Mycobacterium tuberculosis, Pseudomonas aeruginosa, Acinetobacter baumanii and a few other gram-negative bacteria. Multidrug resistance is mainly observed in bacteria due to a build-up of drug-resistance plasmids (R) or transposons that encode drug resistant genes, by drug efflux pumps or by both of these mechanisms [5]. Moreover, the target-protein modification that results in making the bacteria less susceptible to the drug can also aid in drug resistance. For example, the erm gene methylates the adenine at position 2058 of the 50s rRNA resulting in drug resistant bacteria [6]. Additionally, the drug resistance mechanism against naturally originating antibiotics is generally observed due to enzymatic inactivation of the drug, of which, enzymatic phosphorylation, adenylation, acetylation and hydrolysis lead to drug resistance. One of the most intriguing causes of drug resistance is the horizontal gene transfer from the same or different species of bacteria [7]. Horizontal gene transfer (HGT) is a process of transfer of a part of genetic material to a cell that is not its progeny, and it can be attained by the activity of either plasmids or transposons.
Chemotherapy has been one of the leading therapeutic regimens for cancer patients. However, clinical oncologists observe that patients develop cancer resistance against drugs that they have never been exposed to. This essentially deteriorates the effectivity of chemotherapy. In many research studies, cells in vitro are exposed to such chemotherapeutic drugs and a similar pattern of drug resistance is seen, just like in vivo. This phenomenon of resistance to multiple drugs is known as multidrug resistance (MDR), and it may be passed on to the daughter cells from the parent cancer cells. Alterations or mutations in MDR proteins [mostly adenosine triphosphate (ATP)-binding cassette (ABC) transporters] is a common mechanism for development of drug-resistance. This affects the normal cellular functions like signal transduction, uptake of extracellular materials, cellular transport and excretion, secretion of proteins or hormones, lipid transport and prevention of harmful xenobiotics, to name a few (Fig. 2). However, there is not a clear idea as to how the mechanism of MDR in vivo is responsible. Research from the past few years have concluded that P-glycoprotein (P-gp/ABCB1) is one of the major causes of MDR in cancers; although, studies about different transporters are in progress. The dynamics of P-gp are such that it can recognize multiple substrates. In this review, a few members of the ABC superfamily of transporters are described.
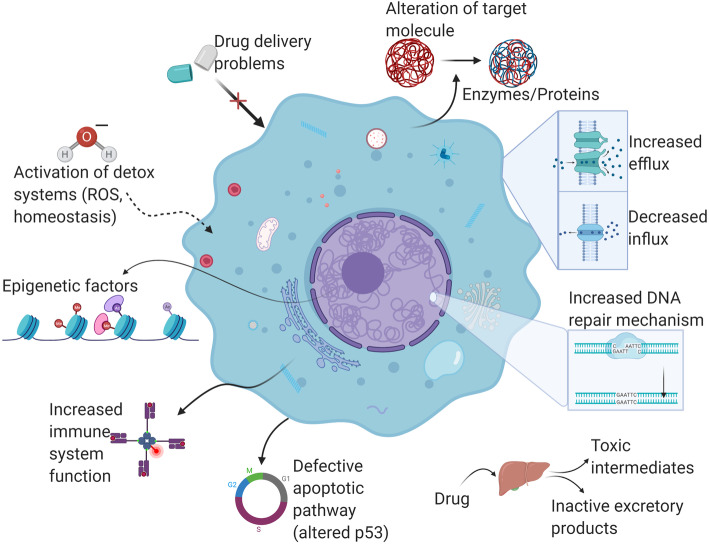
Fig. 2.
Mechanisms by which drug resistance is conferred in cancer. (In clockwise manner starting with defective apoptotic pathway) (1) the apoptotic pathway (p53 pathway) might be defective, which leads to various downstream resistance mechanisms like upregulation of Nrf2 expression, MGM2 upregulation, increased cell proliferation, etc.; (2) drug resistance is seen when there is increased or defective immune system function where hypersensitivity is observed; (3) a plethora of epigenetic factors play roles in conferring drug resistance in cancer; (4) when the detox systems (ROS, homeostasis) are activated, there is a detox of the drug from the cancer cell; (5) drug delivery problems: too low concentration or higher molecule size than required; (6) drug alteration by intrinsic enzymes or other proteins; (7) transporters of the cell membrane exhibit increased drug efflux or decreased influx; (8) even if the drug enters the cell and affects the genetic machinery, there is an increased intrinsic DNA repair mechanisms; (9) drug resistance is also seen during increased metabolism in liver
Antibacterial drug-resistance mechanisms
The bacteria develop drug resistance via multiple mechanisms. This can be either genetic or mechanistic or both. These mechanisms include acquired resistance through horizontal gene transfer or through xenobiotic transfer, protein modification of target or receptor, through drug efflux pumps or by preventing drug influx.
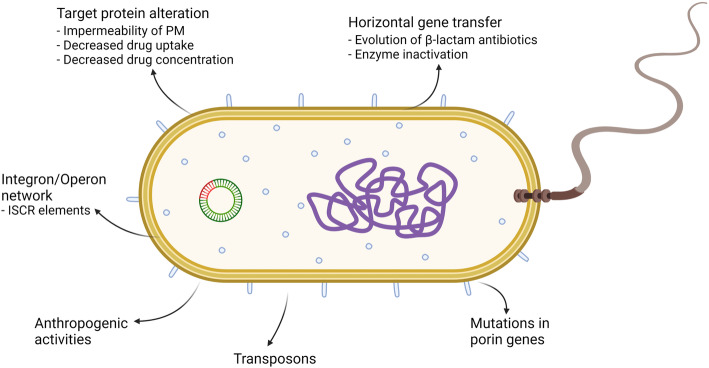
Figure 1 gives a brief representation of drug resistance mechanisms observed in bacteria. Transposons or “jumping genes” produce enzymes that facilitate the movement of genes to another locus of deoxyribonucleic acid (DNA) in either same genome or in another organism. Moreover, for drug resistance, an integral factor other than evolution is the acquisition of genes that confers antibiotic resistance, especially via HGT. It can cause large-scale changes in a bacterial genome without causing mutations and thus it is a highly effective drug-resistance mechanism [8]. Furthermore, the evolution of resistance to β-lactam antibiotics is a consequence of HGT [9]. Mutations in genes that encode for target proteins may lead to drug resistance as the drug can no longer bind to the target protein. Most instances of drug resistance arise from genes present on R plasmids. Moreover, these genes are transferred to a vulnerable bacterium in a distinct conjugation episode. At the time of discovery of R plasmids, it was found that many of these resistance genes against common antibiotics such as tetracycline, chloramphenicol, sulfonamides, and aminoglycosides were already present. However, their discovery came to light just recently as a consequence of the sequencing studies [5]. The recent discovery of integrons has also revealed a gene encoding an integrase enzyme that helps insert a resistance gene at a pre-decided site downstream of a promoter. The integron has a unique 59 bp 3′-sequence tag and the resistance gene gets marked by the tag as soon as it is integrated. After this, the gene becomes integrated into another integron, conceivably containing a completely diverse batch of resistance genes (it may contain up to eight resistance genes). It is thought that this insertion into integrons and organization into distinct operons is how resistance genes obtain high mobility. This operon will conclusively have the same transcription direction under a strong promoter which is provided by the integron framework. Tn21 is one such example of large, complex, and multiple composite transposons which consists of resistance genes against mercury, sulfonamide (sul1) and aminoglycoside (aadA1) [10–12]. Many integrons also haul enzymatic machinery to transpose the complete integron framework to other loci in the genome [13]. It was not until recently that various integrons were discovered to be associated with a downstream structure known as insertion sequences – common region(s) element (ISCR). The ISCR element mainly functions as a recruiter and a transporter to the integron framework, which results in assembly of more resistance genes [14]. Anthropogenic activities contribute a reasonable amount in conferring drug resistance to bacteria. The amount of toxic wastes, chemicals, antibiotics, metals, intermediates, and numerous xenobiotics released into the environment is immeasurable; and it brings upon ceaseless selection pressure and the maintenance of genome to populations of resistant strains in almost all the environments.
Fig. 1.
Drug resistance in bacteria. Target protein alteration. Certain modifications lead to impermeability of the cell membrane and thus decrease drug uptake. Target modification leads to a demoted drug binding. Integron Operon network. Integrons help insert a resistance gene at a pre-decided site downstream of a promoter (Example- Tn21). Anthropogenic activities. Release of toxic chemicals into the environment provides a selection and survival pressure which leads to variation and ultimately, evolution. Horizontal gene transfer. Transfer of genes from other species or from same species, but not parental cells is called HGT. Evolution of β-lactam antibiotic resistance genes is one of the results of HGT. Transposons. “Jumping genes” produces enzymes that aid in HGT. Mutation in porin genes. It can lead to decreased drug influx or increased drug efflux with the help of ion motive force, as compared to ATP hydrolysis by transporters
Mutations in porin genes can sometimes lead to drug efflux outside the cell. In addition, drug efflux pumps extrude drug molecules outside the cells. The majority of the activity of drug efflux pumps in bacteria occurs through the activity of ion motive force; however, many pumps use ATP hydrolysis as a driving factor as well. Some of the major drug efflux transporter families are major facilitator superfamily (MFS), small multidrug resistance (SMR) family, resistance/nodulation/division (RND) superfamily, multi antimicrobial extrusion (MATE) family, and ATP-binding cassette transporters (ABC) superfamily.
Bacterial efflux pumps
In the last few decades, various bacterial drug transporters were discovered and classified (Table 1). Prokaryotic efflux transporters are often referred to as half transporters. To form a functional unit, these transporters either homodimerize with themselves or heterodimerize with other transporters [48]. The normal function of a conventional MDR protein is to uptake and secrete a wide range of substrate(s) including both small molecules such as amino acids, xenobiotics, vitamins, sugars; and complex polymers such as oligopeptides, proteins, and polysaccharides. MDR proteins have an extensive uptake spectrum, allowing them to participate in a variety of cellular processes such as uptake of nutrients, cellular excretion of waste and debris, xenobiotic protection, immunity against foreign bodies, bacterial virulence, osmosis, lipid transport, and biogenesis [55–57]. The primary structure contains four domains: two nucleotide binding domains (NBDs) and two transmembrane domains (TMDs), all of which are encoded by four independent genes or by a blend of the genes or by one single gene that encodes the entire transporter [58]. Along with these four domains, there may be an extracellular substrate binding domain that transfers the substrate to the permease domain [59, 60].
Table 1.
Bacterial efflux transporters
| Name/ Gene name | Other names | Organism | Family/Sub-family | Polypeptide chain length (aa) | Confers resistance to | References |
|---|---|---|---|---|---|---|
| qacA | QacA | Staphylococcus aureus | DHA2 family (MFS) | 514 | Benzalkonium chloride, Cetyl-trimethyl ammonium bromide, Ethidium bromide, Chlorhexidine, Pentamidine isethionate | [15, 16] |
| norA | – | Staphylococcus aureus | TCR family (MFS) | 388 | Fluoroquinolones | [17] |
| emrB | – | Escherichia coli | EmrA family (MFS) | 512 | 2,4-dinitrophenol, Nalidixic acid, CCCP, thiolactomycin, m-chlorophenylhydrazone | [15, 18, 19] |
| mdfA | cmlA, cmr | Escherichia coli | MdfA family (MFS) | 410 | Ethidium bromide, Tetraphenylphosphonium, Rhodamine, Daunomycin, Benzalkonium, Rifampicin, Tetracycline, Puromycin, Chloramphenicol, Erythromycin, Fluoroquinolones, extreme alkaline pH resistance | [20–25] |
| setA | yabM | Escherichia coli | SET family (MFS) | 392 | Sugar efflux, Sugar detoxification (non-metabolizable) | [26] |
| sugE | – | Escherichia coli | SMR family (DMT) | 105 | Guanidinium, Cetylpyridinium, Cetyldimethylethyl ammonium, Cetrimide cations | [27–29] |
| emrE | mvrC | Escherichia coli | SMR family (DMT) | 110 | DDAC, Ethidium, Methyl viologen, Acriflavine, Tetraphenylphosphonium, Benzalkonium, Propidium, Dequalinium, Streptomycin, Tobramycin | [30–32] |
| qacE | – | Klebsiella pneumoniae | SMR family (DMT) | 110 | Quaternary ammonium compounds | [33] |
| qacC | QacSau | Staphylococcus aureus | SMR family (DMT) | 107 | Quaternary ammonium compounds, Ethidium bromide | [33] |
| yvdS | – | Bacillus subtilis | SMR family (DMT) | 114 | [33] | |
| acrB | acrE, AcrAB-TolC | Escherichia coli | RND family | 1049 | Tetracycline, Puromycin, Chloramphenicol, Erythromycin, Rifampicin, Fusidic acid, Acriflavine, Bile salts, Cephalosporins, Crystal violet, Ethidium bromide, Fluoroquinolones, SDS, Triclosan, Triton X-100 | [34–37] |
| mmpL7 | – | Mycobacterium tuberculosis | MmpL sub-family (RND) | 920 | Phthiocerol dimycocerosate, Isoniazid | [38–40] |
| czcA | – | Ralstonia metallidurans | RND family | 1063 | CZC- Cobalt, Zinc and Cadmium resistance | [41] |
| mexB | – | Pseudomonas aeruginosa | RND family | 1046 | tetracycline, chloramphenicol, ciprofloxacin, streptonigrin, dipyridyl | [42] |
| norM | vcmA | Vibrio cholerae | MATE family | 457 | Norfloxacin, Ciprofloxacin, Ofloxacin, Daunomycin, Doxorubicin, Streptomycin, Kanamycin, Ethidium bromide, Acriflavine | [43–45] |
| yeeO | – | Escherichia coli | MATE Family | 547 | Exports peptides- pepA, pepB, pepD, pepN and flavins- FMN, FAD | [46, 47] |
| lmrA | – | Lactococcus lactis | LmrA family (ABC) | 590 | Various antibiotics | [48, 49] |
| bmrA | yvcC | Bacillus subtilis | ABC superfamily | 589 | Hoechst-33,342, Ethidium bromide, Doxorubicin | [26, 50] |
| macB | – | Escherichia coli | Macrolide Exporter family (ABC) | 648 | Macrolides | [51–53] |
| msrA | – | Staphylococcus epidermidis | ABC superfamily | 488 | Erythromycin, B-streptogramins | [26] |
| drrAB | – | Streptomyces peucetius | Drug exporter 1 family | 330 | Daunorubicin, Doxorubicin | [26, 54] |
CCCP Carbonyl cyanide m-chlorophenylhydrazine, DDAC Dodecyl dimethyl ammonium chloride, SDS Sodium dodecyl sulfate, FMN Flavin mononucleotide, FAD Flavin adenine dinucleotide
MFS superfamily consists of numerous drug efflux transporters which catalyze uniport, symport (solute:cation), antiport (solute:solute) or a combination of these processes, although, most of the members probably operate via H+ antiport. Some of the members contain 14 transmembrane segments (TMS) and others contain 12 TMS [26]. QacA and QacB fall into the category of MFS superfamily with 14 TMS and contains several acidic amino acid residues. These pumps mainly extrude biocides and dyes such as benzalkonium chloride, cetyl trimethyl ammonium bromide and ethidium bromide. QacA additionally transports out dicationic biocides such as chlorhexidine and pentamidine isethionate. The difference between these two transporters lies in the TMS10 of QacA, which aids in removal of dicationic compounds [15]. Another example of MDR transporter is EmrB of E. coli, which confers resistance against carbonyl cyanide m-chlorophenylhydrazone and antibiotics such as nalidixic acid and thiolactomycin. EmrA is a periplasmic adapter protein that connects the pump to TolC, an outer membrane channel that helps extrude the drugs directly outside the cell. EmrA and EmrB both are encoded by a chromosomal gene [15, 18]. MFS pumps containing 12 TMS includes pumps like NorA, NorB, NorC, LmrP, LmrCD, MdfA, AcrB, and many more. NorA is resistant to fluoroquinolones in Staphylococcus aureus, other cationic dyes and cationic inhibitors (Table 1) [17]. NorB and NorC are two other homologs of NorA in S. aureus that produces a similar phenotype [61]. However, all the three pumps are inhibited by reserpine [62]. Lactococcus lactis possesses LmrP, a secondary transporter of MFS family [63] that acts as a “vacuum cleaner” of the cell membrane and pumps out cationic dyes, daunomycin, tetracycline and macrolides [64]. MdfA from E. coli extrudes cationic dyes, chloramphenicol, fluoroquinolones when overexpressed in mutants lacking a constitutive RND pump called AcrB [20].
The SMR family proteins are hydrophobic in nature, with four TMSs, consist of some of the smallest transporters and are a part of drug/metabolite transporter (DMT) superfamily. These are cation-specific multidrug efflux pumps and the model example is EmrE of E. coli (about 110 amino acids in length) [30]. Moreover, the SMR family of transporters were first found in S. aureus plasmids and later, were also found in gram-negative bacteria. They transport quaternary ammonium biocides such as dodecyl dimethyl ammonium chloride (DDAC) or ethidium bromide [27, 30]. Some of the well-studied transporters include EmrE, QacC, QacE, SugE (Table 1) [27].
The RND superfamily is very well studied as transporters from this family play a vital role of drug resistance in gram-negative bacteria. A general structure for the members can be demarcated as a chain consisting of one TMS at the N-terminal linked to a comparatively hefty extracytoplasmic domain followed by six TMSs linked to another extracytoplasmic domain and finally five TMSs at the C- terminal. These transporters get linked with two other classes of proteins- TolC of E. coli, one of the outer membrane factor (OMF) family members [65]; and AcrA of E. coli, a periplasmic adapter protein of the membrane fusion protein (MFP) family [66]. This network of proteins allows them to directly transport the drugs out into the medium instead of the periplasmic space. Once transported outside, the drugs will need to tread through the outer membrane barrier which is majorly made of lipopolysaccharide, in order to enter the cell. However, this complex works harmoniously with the outer membrane and effectively prevents this entry of the drug [67]. Some of the RND family pumps exhibit a wide range of substrate specificity, for example, AcrB of E. coli can not only pump out lipophilic antibiotics but also dyes, detergents and solvents such as acriflavine, bile salts, chloramphenicol, cephalosporins, crystal violet, ethidium bromide, fluoroquinolones, sodium dodecyl sulphate, tetracyclines, triclosan, Triton X-100, etc. [34, 35]. AcrD, a homolog of AcrB can pump out aminoglycosides in E. coli. The original function of AcrB seems to be transport of bile salts as it has highest affinity to bile salts [68]. In nosocomial infections of Pseudomonas aeruginosa, increasing fluoroquinolone resistance is observed with the help of MFP family of pumps along with probably an OMF component [69]. In Mycobacterium tuberculosis, MmpL7 pump extrudes a complex, non-polar lipid- phthiocerol dimycocerosate [38].
The MATE family of transporters include NorM of Vibrio parahaemolyticus, which contains 12 transmembrane helices [43]. The members are about 450 aminoacyls long and they operate by an uncommon mechanism of drug: Na+ antiport. It extrudes fluoroquinolones and ethidium and in turn, influxes Na+ ions [26, 44].
ABC transporters have a limited role in bacterial drug resistance [70]. Both uptake and drug efflux systems have been found in this superfamily that uses energy from ATP hydrolysis, and usually both systems are localized together. The members of this superfamily contain at least two integral membrane domains (~membrane spanning domains (MSD)) and two cytoplasmic domains (~NBD), which can be found as homodimers or heterodimers. In Gram-negative bacteria, the uptake domains are found in periplasm while in Gram-positive bacteria, they are either present as lipoproteins on the outer surface of the cell membrane, or as cell-surface associated proteins [26]. It is important to note that many ABC transporters in bacteria have shown a homology to human ABC transporters. However, the uptake systems are not observed in mammalian ABC transporters. For instance, LmrA of L. lactis is homologous to a half of the P-gp protein of mammalian origin [48, 49]. Biochemical studies showed that the drug resistance mechanism is mainly a combination of drug extrusion and ATP hydrolysis [71]. Other examples of ABC transporters in bacteria include BmrA in Bacillus subtilis [72, 73] and MacB in E. coli. Interestingly, MacB is expressed with a periplasmic adapter protein MacA (in a similar fashion to AcrA of MFP family) which helps in providing resistance to macrolides when overexpressed [53].
Mammalian MDR proteins
According to the sequence homology and domain organization, there are seven ABC transporter subfamilies [74, 75] named from “A” to “G”, i.e. ABCA to ABCG, into which 49 ABC transporters have been classified [76]. The general structure of these transporters is comparable to those found in bacteria. Mammalian MDR proteins contain at least two NBDs (cytoplasmic) and TMDs (also called membrane-spanning domains – MSDs; analogous to TMS in bacteria). The cytoplasmic NBD has a similar function i.e., serving as a source of energy to produce hydrolyzed ATP, whereas the TMD is very heterogeneous in function and dynamic in nature and its functions support the binding of a drug, transport channel, dimerization/oligomerization, and trafficking [77]. This heterogeneity allows the transporter to recognize a wide range of substrates and use ATP as an energy source, irrespective of the established concentration gradient [78]. Recent updates confirm that ATP provides energy for substrate transport as well as for continuous conformation changes that allow the transporters to identify a range of substrates [79].
The cellular levels of ions, lipids, hormones, xenobiotics, and other small molecules are regulated by ABC transporters by hauling them in and out across the plasma membrane. Thus, contributing a considerable role in physiological aspects like regulating the organelles (mitochondria, lysosome, endoplasmic reticulum (ER), Golgi apparatus) [80–83]. Transporters such as P-glycoprotein (P-gp/ABCB1), multidrug resistance protein 1 (MRP1/ABCC1) and breast cancer resistance protein (BCRP/ABCG2) transports substances across the cell membrane, thereby conferring excretory and defensive physiological activities. At the blood-brain barrier, blood-testis barrier and blood-placental barrier, the entry of foreign, exogenous molecules is inhibited [84, 85]. Notably, the normal functions of ABC transporters also have a significant effect on pharmacokinetics of drugs: absorption, distribution, metabolism, excretion, and toxicity.
Genetic and mechanical aspects of MDR
What happens if something goes wrong genetically or mechanistically? A germline mutation can lead to a loss-of-function in a single ABC transporter and can be associated to diseases other than cancer like cystic fibrosis, pseudoxanthoma elasticum, Stargardt macular degeneration, Tangier disease, sitosterolaemia and harlequin ichthyosis [86]. However, these genetic anomalies can be rehabilitated by mRNA stabilization, ribosomal readthrough, correction of folding and trafficking errors, allosteric activation, modulation of protein interactions, regulation of post translational modifications, improving of protein degradation pathways and initiation of other compensatory mechanisms [87–95].
Resistance to anti-cancer drugs can be roughly described as complications in drug delivery to tumor cells or problems of impaired drug sensitivity within the cancer cells themselves as a result of genetic/epigenetic alteration. Complications in drug delivery can arise from poor absorption of oral drugs, elevated metabolism, or increased excretion. As a result, there will be lower level of drug in the blood. This means that a proper amount of drug is not reaching the tumor mass (Fig. 2) [96, 97]. Furthermore, irregular pressure gradient along with tumor vasculature was also reported to affect drug delivery [98]. Besides these factors, tumor geometry and composition of the extracellular matrix have been observed to participate in drug resistance [96, 99, 100]. Drug resistance is again observed when the cellular target of a drug is altered in conformation or geometry; or if there is increased DNA maintenance activity in tumor cells. The phenomenon of cross resistance to other structurally and mechanistically irrelevant drugs, after resistance to a single drug is attained, is categorized as MDR [101]. Drug resistance is descried by decreased drug uptake. The transporters and carrier molecules that are used for nutrient uptake are abused as “ferry” molecules by water-soluble drugs, or by the process of endocytosis. This is illustrated by drugs such as methotrexate, 5-fluorouracil (5-FU), 8-azaguanine and cisplatin [102, 103]. Activation of organized mechanisms from detoxification systems such as DNA-repair and cytochrome P450 oxidases can also lead to multidrug resistance, as seen in P-gp and CYP3A coordinated induction [104]. Besides these, other systems include superoxide dismutase, catalase, glutathione peroxidase, and antioxidants. Induction of malignancy in cancers can result in mutant or non-functional p53; however, malignancy can also be a repercussion of chemotherapy. For example, fluctuation of ceramide levels or alteration of cell-cycle machinery may activate apoptotic checkpoints, thereby allowing cancer cells to avert apoptosis (Fig. 2) [105].
One of the most important genetic aspects influencing drug response is single nucleotide variants, however, insertions, deletions, repeats, and copy number variants can also have consequences in efficacy of therapy [106–108]. These mutations and variations are observed in genes responsible for the production of drug metabolizing enzymes, drug efflux genes, drug targets, DNA maintenance machinery, apoptotic machinery, and alleles related to the immune system [109–117]. DNA maintenance machinery reinstates genomic stability to maintain homeostasis and prevent cancer development. However, mutations and variations cause loss of normal function in DNA maintenance mechanisms, which leads to carcinogenesis, accelerated tumor evolution and resistance to drugs that attack DNA such as platins, nitrogen mustards, and chloroethyl nitrosoureas. The mechanism includes the formation of a DNA adduct that interferes with active DNA replication in cancer cells [118]. The generation of replication stress in a cell drives it towards apoptosis. DNA lesions, single-strand breaks or double-strand breaks are usually repaired by various DNA damage repair pathways such as base excision repair, nucleotide excision repair, homologous recombination, non-homologous end joining, mismatch repair, trans lesion synthesis and the Fanconi anemia pathway [119–122]. Epigenetic factors also have a vital role in various cancers. DNA methylation, one of the most common epigenetic alteration, which is carried out by DNA methyltransferases (DNMTs) that covalently attach a methyl group to cytosine triphosphate of the CpG islands in the genome [123, 124]. However, inhibition of DNMTs can cause the reversal of DNA methylation and revive the expression of the silenced genes [125]. Furthermore, other important histone modifications such as acetylation, phosphorylation, ubiquitination, sumoylation and adenosine di-phosphate (ADP) ribosylation can all lead to gene silencing. Cell differentiation and cell proliferation pathways such as mitogen-activated protein kinase (MAPK), Wnt, vascular endothelial growth factor (VEGF), p53 signaling, etc. are some of the most affected pathways due to aberrant epigenetics in respective genes [126, 127]. Additionally, micro RNAs (miRNAs) are a broadly conserved class of RNAs found mostly in intronic regions that are roughly 20–25 bases long polynucleotides and do not express any protein. The main function of miRNA is to downregulate gene expression at a post-transcriptional level, but it can also activate mRNA translation on rarer occasions. The network of miRNAs is so complex that each miRNA can regulate several different mRNAs and the same mRNA can be targeted by several different miRNAs as well [128]. The importance of miRNAs lies in the expression of the MDR efflux transporters of the ABC superfamily, since miRNAs regulates the post-transcriptional activity of MDR efflux transporters in several different tumors (Fig. 2) [129]. For instance, there has been a report of downregulation of ABCB1/MDR1 encoding for P-gp by several miRNAs such as miR-331-5p and miR-27a in lymphocytic and myeloid leukemia, miR-let-7 in ovarian cancer, miR-200c and miR-195 in breast cancer, miR-30a in gastric cancer and miR-9-3p in CML, thereby reversing the phenomenon of drug resistance [130–135]. Moreover, miR-326 regulation of ABCA2 in pediatric acute lymphoblastic leukemia has been reported to have an impact on drug resistance mechanisms. Along with, miRNAs can modulate the induction of apoptosis [136].
Drug-drug interactions (DDI) are observed commonly in patients suffering from cancer, due to multiple administrations of different drugs, adjuvants, and medications to treat additional co-morbidities [137]. A classic example of pharmacokinetic DDI is the absorption of tyrosine kinase inhibitors (TKIs). Altered intra-gastric pH values and the activity of intestinal enzymes as well as drug transporters influence TKI absorption substantially. Most of the TKIs are weak bases, protonated and most soluble in the acidic setting. Consequently, an increase in pH values, such as with proton pump inhibitors, greatly decreases their solubility [138, 139]. It has been reported that about 30% of cancer patients take proton pump inhibitors (PPIs), H2-antagonists or anti-acids to relieve gastro-esophageal reflux and dyspepsia symptoms. PPIs are some of the most commonly prescribed drugs along with TKIs [140, 141]. Apart from that, there can be synergistic, antagonistic, or additive responses as well.
The ABC superfamily of transporters
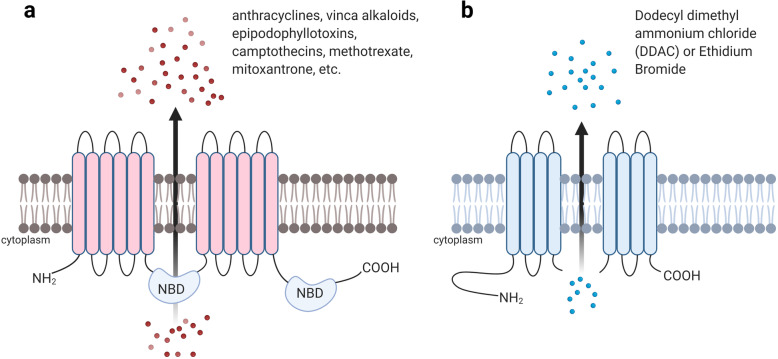
During the past years, various drug transporters pertaining to human cancers were identified and some of the latest are summarized in Table 2. ABCB1/MDR1 (also called P-gp) transporter was the first multi-drug transporter to be discovered as a surface phospho-glycoprotein that transports drugs and phospholipids across the membranes (Fig. 3). It is found on chromosome 7p21 and has 170 kDa of molecular weight [142, 143]. The structure of P-gp comprises of two homologous NBDs and two homologous TMDs with at least three sites for the binding of substrates/inhibitors. Interestingly the overexpression of P-gp alone confers drug-resistance to a huge number of neutral and cationic hydrophobic chemotherapeutic substrates including taxanes, epipodophyllotoxins, vinca alkaloids, anthracyclines, BCR-ABL TKIs, and epidermal growth factor receptor (EGFR) TKIs [101, 144–152].
Table 2.
ABC transporters in human cancers
| Name/ Gene name | Other Names | Genomic Location | Organ/tissue localization | MW (kDa) | Resistance conferred to | Modulators | References |
|---|---|---|---|---|---|---|---|
| MDR1/ ABCB1 | P-gp, GP170, CLCS, ABC20 | 7p21 | Blood-brain barrier, Bone marrow, Placenta, Gut mucosa, Liver, Kidney | 170 | Taxanes, Epipodophyllotoxins, Vinca Alkaloids, Anthracyclines, BCR-ABL TKIs, EGFR TKIs, ALK TKIs (Crizotinib, Ceritinib) | Sapitinib, Ibrutinib, RN486, Erlotinib, Lapatinib, Tariquidar, Elacridar, Zosuquidar | [101, 142–160] |
| BCRP/ ABCG2 | ABCP, MXR1, CD338, ABC15 | 4q22.1 | Placental syncytiotrophoblasts, Small intestine, Epithelial tissue of colon, Canalicular membrane in the liver, Microvessel endothelium of human brain, in the veins and blood vessels | 72 | Nucleoside analogs, Anthracyclines, Flavopiridols, Methotrexate, Methotrexate polyglutamates, E217βG, Camptothecin-derived topoisomerase I inhibitors, GSK1070916, Tivantinib, Pevonedistat, Tozasertib, Barasertib | Erlotinib, Lapatinib, Icotinib | [153, 159, 161–176] |
| PRP/ ABCB6 | DUH13, PSHK2, ABC14 | 2q35.5 | Brain, Retina, Testis, Gall bladder, Intestine | 93.8 | Paclitaxel, 5-FU, Epirubicin, Cyclophosphamide, Daunorubicin, SN-38, Vincristine | Verteporfin, Tomatine HCl, Benzethonium chloride | [177–182] |
| MRP1/ ABCC1 | GS-X, ABC29 | 16p13.1 | Placenta, BBB, Lungs, Testis, Skeletal/Cardiac muscles, Kidney, Intestine | 171.6 | Anthracyclines, Vinca alkaloids, Epipodophyllotoxins, Camptothecins, GSH conjugates, Methotrexate, Mitoxantrone, Imatinib, Arsenite, Colchicine, Flutamide, Betulin, Saquinavir, Ritonavir, Indinavir | Everolimus, GSK1904529A, Rapamycin, Tipifarnib, TAK-733, Delavirdine, Indomethacin, Verapamil | [145, 183–194] |
| MRP2/ ABCC2 | cMOAT, cMRP, ABC30 | 10q24.2 | Placenta, BBB, Lungs, Kidney, Liver, Intestine | 174.2 | LTC4, E217βG, GSH, Taxanes, Anthracyclines, Vinca alkaloids, Methotrexate, Mitoxantrone, Etoposide, Irinotecan, Cisplatin, SN-38, Saquinavir, Ritonavir, Lopinavir, Indinavir | Curcumin, Piperine, Rhinacanthin-C, Probenecid | [59, 195–202] |
| MRP3/ ABCC3 | cMOAT2, MOATD, MLP2, ABC31 | 17q21.33 | Placenta, Colon, Prostate, Kidney, Liver, Small intestine | 169.3 | Glutathione, Bile salts, LTC4, Glucuronide conjugates, DNP-SG, Vincristine | Fidaxomicin, Suramin, Lamivudine, Tenofovir | [203–208] |
| MRP4/ ABCC4 | MOAT-B, ABC32 | 13q32.1 | Blood, Pancreas, Adrenal gland, Prostatea, Kidneya, Livera | 149.5 | cAMPb, cGMPb, Loop diuretics, Cephalosporins, Topotecan, Imatinib, PMEA, 6-MP, 6-TG, Methotrexate, Plant polyphenols, Resveratrol, Quercetin, Adefovir, Ganciclovir, Tenofovir, Zidovudine | Micafungin, Rofecoxib, Indomethacin, Verapamil | [194, 209–217] |
| MRP5/ ABCC5 | SMRP, MOATC, ABC33 | 3q27.1 | Brain, Testis, Skeletal/Cardiac Muscles | 160.6 | cAMPb, cGMPb, Folates, 6-MP, 5-FU, Methotrexate, Stavudine | Zaprinast, Benzbromarone, Sulfinpyrazone, Sildenafil, Sulfinpyrazone | [216, 218–222] |
| MRP6/ ABCC6 | MLP1, MOAT-E, ABC34 | 16p13.11 | Kidney, Liver | 164.9 | Cyclopentapeptide BQ123, Etoposide, Teniposide, Doxorubicin, Daunorubicin, LTC4, n-ethylmaleimide-glutathione | Indomethacin, Benzobromarone | [194, 223–225] |
| MRP7/ ABCC10 | SIMRP7 | 6p21.1 | Brain, Lungs, Testis, Blood, Prostate, Ovary, Kidney | 161.6 | Taxanes, Vinca alkaloids, 2′,3′-dideoxycytidine, AraC, PMEA, Epothilone B, LTC4, E217βG | Nilotinib, Lapatinib, Tandutinib, PD-173074, Tariquidar | [211, 226–237] |
| MRP8/ ABCC11 | EWWD, CFTR | 16q12.1 | Placentaa, Braina, Livera | 154.3 | DHEAS, E1S, E217βG, LTC4, DNP-SG, AraC, 5-FU, Methotrexate, PMEA, 2′,3′ − dideoxycytidine, Adefovir | [194, 238–242] |
aIndicates low levels of expression
bIndicates that inhibition of MRP4 results in impaired cAMP and cGMP transport
Fig. 3.
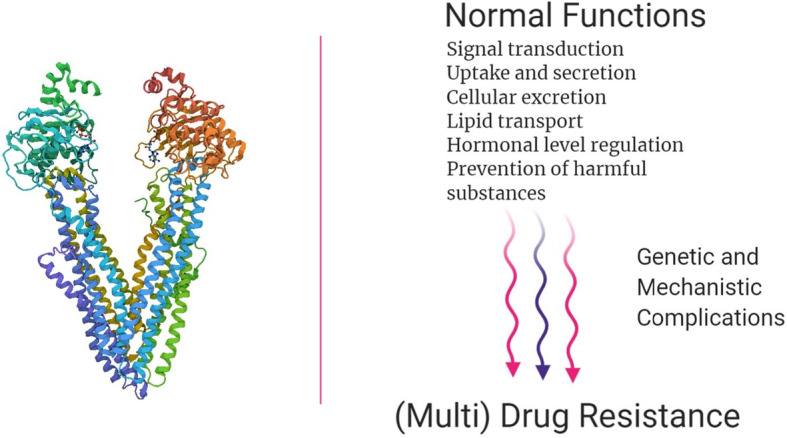
ABCB1 transporter (as a representative example of ABC superfamily of transporters). Normal functions of ABC transporters. (structure from RCSB PDB)
ABCG2/BCRP (Breast cancer resistance protein) is a 72 kDa MDR transporter with the gene located on chromosome 4q22.1, consisting of one TMD and one NBD, sometimes also called a “half transporter” [161, 162]. It gets activated upon homodimerization or oligomerization with itself or with other MDR transporters; a very similar mechanism to that of bacterial drug efflux transporters [153, 163, 243, 244]. It is one of the most widely distributed transporters and its expression is seen on the plasma membrane, highly expressed in placental syncytiotrophoblasts, the apical surface of small intestines, epithelial tissue of colon, canalicular membrane in the liver, microvessel endothelium of human brain and in the veins and blood vessels [164–167]. ABCG2 confers resistance to nucleoside analogs, anthracyclines, flavopiridols, methotrexate, organic dyes, and anionic conjugates, TKIs and camptothecin-derived topoisomerase I inhibitors [163, 168, 169].
ABCB6/PRP (P-gp related protein) is a 93.8 kDa protein with its gene located on chromosome 2q35.5. It is a mitochondrial transporter that can import heavy metals and regulate porphyrin biosynthesis and hence plays a role in cell cycle progression [177]. It is localized in mitochondria, ER and golgi apparatus. It has been reported that ABCB6 confers resistance to camptothecin and camptothecin-11 (CPT-11) in A549 lung cancer cells [178]. It also confers resistance to paclitaxel/FEC (5-FU, epirubicin, and cyclophosphamide) in breast cancer [179], daunorubicin in acute myeloid leukemia [180], and 5-FU, SN-38, and vincristine in Arsenic resistant KB-3-1 cells (KAS cells) [181].
ABCC1/MRP1 was first observed in anthracycline-resistant cell lines H69AR and HL60/Adr [183–185]. It is a 171.6 kDa protein with its gene located on chromosome 16p13.1. ABCC1 has three MSDs and two NBDs with an unusual MSD0 domain. It has a low degree of similarity with ABCB1 (15%), however, the resistance profile is quite comparable [186]. The normal function of MRP1 is xenobiotic detoxification of intermediates of phase II enzymatic reactions [245]. It confers resistance to drugs by ATP- and GSH-dependent export, however, it hydrolyzes ATP with a lower efficiency. ABCC1 has resistance against anthracyclines, vinca alkaloids, epipodophyllotoxins, camptothecins, methotrexate, and mitoxantrone [145, 187–189]. However, ABCC1 does not confer resistance to taxanes.
BCR-ABL TKIs- Breakpoint cluster regions- Abelson tyrosine kinase inhibitors; EGFR TKIs- Epidermal growth factor receptor TKIs; ALK TKIs- Anaplastic lymphoma kinase TKIs; E217βG- Estradiol 17 β-D glucuronide; 5-FU- 5-Fluorouracil; LTC4- Leukotriene C4; GSH- Glutathione; PMEA- 9-(2-phosphonyl methoxyethyl) adenine; 6-MP- 6-Mercaptopurine; 6-TG- 6-Thioguanine; cAMP- Cyclic adenosine monophosphate; cGMP- Cyclic guanosine monophosphate; DHEAS- Dehydroepiandrosterone 3-sulfate; DNP-SG- S-(2,4-dinitrophenyl) glutathione.
ABCC2/MRP2 gene is located on the chromosome 10q24.2. It encodes for MRP2 protein, which is found to be 174.2 kDa in molecular weight. It consists of two NBDs, two membrane spanning domains (MSDs), an NH2 terminal MSD0 with five transmembrane helices, and an intracellular linker segment L0. This kind of structural organization is also observed in MRP1, − 3, − 6 and − 7; while the MRP4, − 5, − 8 and − 9 do not have the MSD0 domain [185, 195, 246, 247]. MRP2 is also known as canalicular multi-specific organic anion transporter (cMOAT), named after its function of transporting amphipathic anionic conjugates of phase II conjugation reactions into the bile [245]. It is principally expressed at hepatocyte canalicular membrane, epithelial cells of the gall bladder, and in apical membranes of the proximal tubule in human kidneys [59, 196, 248]. MRP2 administers resistance against a wide spectrum of drugs like glutathione, glucuronates and sulfates [195]. It confers resistance to cisplatin as well [197].
ABCC3/MRP3 plays a crucial role in conferring multidrug resistance to cancer cells. It is a 169.3 kDa protein with gene located on chromosome 17q21.33 and it bears 58% structural similarity with MRP1 (highest similarity) with three TMDs and two NBDs [206, 207]. MRP3 is an organic anion transporter with a higher affinity for GSH conjugates and it may also play an important role in bile circulation. It provides resistance against leukotriene C4 (LTC4), S-(2,4-dinitrophenyl) glutathione (DNP-SG), glutathione sulfate and glucuronide conjugates [203]. It has been found that ABCC3 expression is correlated with doxorubicin resistance in Lung cancer patients [204, 205].
ABCC4/MRP4 is one of the shortest members of the ABC-superfamily of transporters; with only 1325 amino acids in its polypeptide sequence. The gene encoding MRP4 is located on chromosome 13q32.1 and the translated protein weighs about 149.4 kDa [209]. MRP4 has a typical structure containing two NBDs and two MSDs; and each MSD consists of six TMDs [216]. MRP4 further regulates the synthesis and efflux of prostaglandins and thereby play a role in inflammation [249]. Substrates of MRP4 include cyclic adenosine monophosphate (cAMP), cyclic guanosine monophosphate (cGMP), adefovir, ganciclovir, loop diuretics, cephalosporins, topotecan, PMEA, 6-mercaptopurine (6-MP), 6-thioguanine (6-TG), etc. [210–212, 214]. Recent updates added a few more natural products like plant polyphenols, resveratrol, and quercetin that are transported by MRP4 [213].
ABCC5/MRP5 weighs 160.6 kDa and the gene is found on the chromosome 3q27.1. This transporter is highly localized in brain, heart, skeletal muscles, and lungs [209, 220]. It is similar to MRP4, except, it is devoid of MSD0 and contains a 90 aminoacyl long hydrophilic extension in the sixth TMD [218–220]. MRP5 is an organic anionic transporter, which can transport nucleotides and nucleotide analogs like cAMP, cGMP, 6-MP, 5-FU, and antifolates like methotrexate [221].
ABCC6/MRP6 has a 41% structural similarity with MRP1 and is comprised of two NBDs and three MSDs with five, six and six TMDs, respectively. It is 164.9 kDa in molecular weight and its gene is located on chromosome 16p13.11 [223, 250]. It is mainly localized in kidney and liver and has low or miniscule levels in other tissues [223]. It functions as a calcium transporter to regulate tissue calcification. It confers resistance against cyclopentapeptide BQ123, low resistance against etoposide, teniposide, anthracyclines and cisplatin. It can also expel out glutathione (GSH) conjugates like LTC4 and n-ethylmaleimide-glutathione [224, 225].
ABCC10/MRP7 gene is found on chromosome 6p21.1 and is a 161.6 kDa protein. It consists of three MSDs and two NBDs [226, 251, 252]. It is found to be expressed in tissues like skin, testes, spleen, stomach, colon, kidney, heart, and brain (sparse proportions) [226]. Although, its expression in the pancreas, liver, placenta, and spleen is much higher [227]. MRP7 is a lipophilic anion transporter with functions in transporting GSH conjugates and glucuronides, and tissue detoxification. Substrates of MRP7 include glucuronides (17-β-D glucuronide) and the glutathione conjugate of LTC4 [228]. HEK293 cells with overexpression of ABCC10 confers resistance to taxanes, vinca alkaloids, 2′,3′-dideoxycytidine, 9-(2-phosphonyl methoxyethyl) adenine (PMEA), and epothilone B [211, 226, 228–235, 253].
ABCC11/MRP8 is another transporter similar to MRP4 and MRP5, holding two MSDs, two NBDs and 12 TMDs. It weighs 154.3 kDa and the gene is located on chromosome 16q12.1. It is broadly apportioned throughout the body with high levels in breast, brain, liver, placenta, and testes [238]. It is found that MRP8 is an important part of central and peripheral nervous system as it transports dehydroepiandrosterone 3-sulfate (DHEAS), a neuromodulatory steroid [239]. MRP8 is associated with cellular transport of DHEAS, E1S, estradiol 17 β-D glucuronide (E217βG), some nucleotide analogs, lipophilic anions like LTC4 and DNP − SG, AraC, 5-FU, methotrexate, PMEA and 2′,3′ − dideoxycytidine [240–242].
In addition to ABC transporters, there are extracellular vesicles (EVs) present in the cells that can carry out drug efflux. EVs are about 30–1000 nm sized particles enclosed by a phospholipid bilayer, which cannot replicate [254, 255]. EVs were conventionally addressed as exosomes, microparticles, apoptotic bodies, microvesicles, etc. depending on their biogenesis, size, and content [256]. Interestingly, ABC transporters like P-gp, MRP1 and BCRP are associated with EVs [254, 257]. It is highly probable that the direction of these transporters might be reversed in some but not all EVs, such that there is an influx of drugs into the EVs. Influx of drugs into the EVs will lead to a decreased drug concentration inside the cell, resulting in drug resistance [257–259].
Discussion
This review provides a brief overview and updates on the biochemistry and pharmacology of drug resistance in bacteria and cancer cells. Extensive research of over four decades has now culminated in the identification of thousands of ABC ATPases [260] and it is quite indisputable that all the three phyla of life share the ABC superfamily genes in some variation [55–57, 59, 261–268]. For instance, LmrA and EmrE found in bacterium Lactococcus lactis are structurally and functionally comparable to P-gp in humans (Fig. 4). The structure of MsbA of bacterial origin is also structurally comparable to inward facing conformation of mammalian P-gp [49, 269]. MDR in bacteria constitutes a huge, mutual reservoir of resistance determinants to most families of antimicrobial agents across numerous higher species, including humans. Modifications in cell membrane and its associated proteins lead to many causes of MDR in bacteria. Although, recent advances in imaging and organizational studies of MDR proteins have led us to better understand the molecular mechanisms of multidrug transport. The contribution of man-made activities like release of toxic chemicals into the ecosystem has built up an evolutionary survival pressure on bacteria. Both bacteria and cancer have similar ways to circumvent the phenomenon of cell death: a simpler way for bacteria is to swim away from the cytotoxic environment, which corresponds to metastasis in tumors. Another course of action is the formation of biofilms, where the bacterial colony creates a survivable surrounding, which corresponds to formation of altered tumor microenvironments including a vasculature. Although, these approaches of drug resistance are quite reversible in nature. Furthermore, while comparing the causalities of intrinsic or acquired resistance in bacteria and cancers (comparing Figs. 1 and 2), it is found that the basic idea behind the various mechanisms is supported by the DNA repair mechanisms. For instance, homologous recombination, DNA mismatch repairs, cell cycle regulation, etc. are very much similar mechanisms of MDR in both bacteria and cancer. These causalities lead to heritable resistances in both parties along with pre-existing genetic variations within the population and the origination of de novo mutations. The intrinsic or acquired resistance to drugs are more of a permanent nature as compared to previous reversible approaches [270]. Lambert et al. [270] discusses about stress-induced mutagenesis, which is usually advantageous to the whole population. The survival pressure exerted on bacteria promotes adaptive mutations. These adaptive mutations generally originate from point mutations, and in addition, external oxidative stress recurrently affects the accuracy of DNA transcription. Subsequently, it will lead to the production of mutant protein with a non-permanent alteration to the DNA template; also known as transcriptional mutagenesis [271]. However, in cancer, the occurrence is often, and it is referred to genetic instability. Conversely, it is safe to assume that genetic instability is an organized tactic to speed up adaptation which will finally be beneficial to malignancies.
Fig. 4.
Comparative representation P-gp and EmrE. a ABCB1 (P-gp) transporter, one of the most frequently overexpressed transporters in human cancers. ABCB1 confers resistance to anthracyclines, vinca alkaloids, epipodophyllotoxins, camptothecins, methotrexate, mitoxantrone, etc. b EmrE transporter of Escherichia coli, belonging to SMR family of drug efflux transporters. EmrE confers resistance to dodecyl dimethyl ammonium chloride (DDAC) or Ethidium bromide
Genetic heterogeneity of cancer cells in a single population is acquired by exposure to various chemotherapeutic drugs. Thus, in any cell population of a tumor mass already exposed to chemotherapy, there may be multiple mechanisms of MDR observed, which is termed as multifactorial MDR [101]. Additionally, the normal functions of the ABC transporters discussed above have a consequential effect on the pharmacokinetics of drugs through their absorption, distribution, metabolism, excretion, and toxicity. Inhibition of these transporters produces changes in the pharmacokinetics, toxicities, DDIs, and other complexities [272]. DDIs are preeminent in patients who have been prescribed drugs with a narrow therapeutic index and inherent toxicity, bringing about poor patient compliance and therapeutic decline. Since the activity of ABC transporters in MDR is not readily identified, it is necessary to develop a reliable discovery system. A reliable discovery system would help researchers determine the optimal combination of drug(s) and inhibitor(s), which in turn would aid clinicians in using the ABC transporters as clinical targets and cancer biomarkers.
Acknowledgements
The authors are thankful to the Assistantship received from the College of Pharmacy and Health Sciences, St. John’s University, New York.
Code availability
Not applicable.
Abbreviations
- 5-FU
5-Fluorouracil
- 6-MP
6-Mercaptopurine
- 6-TG
6-Thioguanine
- ABC
ATP-binding cassette transporters
- ADP
Adenosine di-phosphate
- ALK
Anaplastic lymphoma kinase
- ATP
Adenosine tri-phosphate
- BCRP
Breast cancer resistance protein
- cAMP
Cyclic adenosine mono-phosphate
- cGMP
Cyclic guanosine mono-phosphate
- DDAC
Dodecyl dimethyl ammonium chloride
- DDI
Drug-drug interactions
- DHEAS
Dehydroepiandrosterone 3-sulfate
- DNA
Deoxyribonucleic acid
- DNMT
DNA methyltransferases
- DNP-SG
S-(2,4-dinitrophenyl) glutathione
- E2-17βG
Estradiol 17 β-D glucuronide
- EGFR
Epidermal growth factor receptor
- ER
Endoplasmic reticulum
- EVs
Extracellular vesicles
- HGT
Horizontal gene transfer
- ISCR
Insertion sequences-common regions
- LTC4
Leukotriene C4
- MAPK
Mitogen-activated protein kinase
- MATE
Multi-antimicrobial extrusion
- MDR
Multidrug resistance
- MFP
Membrane fusion protein
- MFS
Major facilitator superfamily
- miRNA
Micro RNAs
- MRSA
Methicillin-resistant Staphylococcus aureus
- MSD
Membrane spanning domain
- NBD
Nucleotide binding domain
- OMF
Outer membrane factor
- P-gp
P-glycoprotein
- PMEA
9-(2-Phosphonyl methoxyethyl) adenine
- PPI
Proton pump inhibitors
- RND
Resistance/nodulation/division
- rRNA
Ribosomal ribonucleic acid
- SMR
Small multidrug resistance
- TKI
Tyrosine kinase inhibitors
- TMD
Transmembrane domain
- TMS
Transmembrane segments
- VEGF
Vascular endothelial growth factor
- WHO
World Health Organization
Authors’ contributions
All authors contributed substantially to this work. HP wrote and prepared the original draft. Z-XW, YC, and LB wrote, reviewed, and edited the manuscript. Z-SC supervised the study. All authors approved the submitted version.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there is no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gaynes R. The discovery of penicillin—new insights after more than 75 years of clinical use. Emerg Infect Dis. 2017;23(5):849. doi: 10.3201/eid2305.161556. [DOI] [Google Scholar]
- 2.Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Rev Infect Dis. 1980;2(1):129–139. doi: 10.1093/clinids/2.1.129. [DOI] [PubMed] [Google Scholar]
- 3.Shankar PR, Balasubramanium R. Antimicrobial resistance: global report on surveillance 2014. Australas Med J (Online) 2014;7:237. [Google Scholar]
- 4.Poirel L, Madec JY, Lupo A, Schink AK, Kieffer N, Nordmann P, et al. Antimicrobial resistance in Escherichia coli. Microbiol Spectr. 2018;6(4):289–316. doi: 10.1128/microbiolspec.ARBA-0026-2017. [DOI] [PubMed] [Google Scholar]
- 5.Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78(1):119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39(3):577–585. doi: 10.1128/aac.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spratt BG. Resistance to antibiotics mediated by target alterations. Science. 1994;264(5157):388–393. doi: 10.1126/science.8153626. [DOI] [PubMed] [Google Scholar]
- 8.Daubin V, Szollosi GJ. Horizontal gene transfer and the history of life. Cold Spring Harb Perspect Biol. 2016;8(4):a018036. doi: 10.1101/cshperspect.a018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63(3):507–522. doi: 10.1128/MMBR.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall RM, Stokes H. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica. 1993;90(2–3):115–132. doi: 10.1007/BF01435034. [DOI] [PubMed] [Google Scholar]
- 12.Rowe-Magnus DA, Mazel D. The role of integrons in antibiotic resistance gene capture. Int J Med Microbiol. 2002;292(2):115–125. doi: 10.1078/1438-4221-00197. [DOI] [PubMed] [Google Scholar]
- 13.Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4(8):608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 14.Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev. 2006;70(2):296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60(4):575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357(9264):1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 17.Neyfakh AA, Borsch C, Kaatz G. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob Agents Chemother. 1993;37(1):128–129. doi: 10.1128/aac.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomovskaya O, Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci U S A. 1992;89(19):8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borges-Walmsley MI, Beauchamp J, Kelly SM, Jumel K, Candlish D, Harding SE, et al. Identification of oligomerization and drug-binding domains of the membrane fusion protein EmrA. J Biol Chem. 2003;278(15):12903–12912. doi: 10.1074/jbc.M209457200. [DOI] [PubMed] [Google Scholar]
- 20.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol. 2001;183(20):5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179(7):2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohn C, Bouloc P. The Escherichia coli cmlA gene encodes the multidrug efflux pump Cmr/MdfA and is responsible for isopropyl-beta-D-thiogalactopyranoside exclusion and spectinomycin sensitivity. J Bacteriol. 1998;180(22):6072–6075. doi: 10.1128/JB.180.22.6072-6075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Evidence for chloramphenicol/H+ antiport in Cmr (MdfA) system of Escherichia coli and properties of the antiporter. J Biochem. 1998;124(1):187–193. doi: 10.1093/oxfordjournals.jbchem.a022078. [DOI] [PubMed] [Google Scholar]
- 24.Lewinson O, Adler J, Poelarends GJ, Mazurkiewicz P, Driessen AJ, Bibi E. The Escherichia coli multidrug transporter MdfA catalyzes both electrogenic and electroneutral transport reactions. Proc Natl Acad Sci U S A. 2003;100(4):1667–1672. doi: 10.1073/pnas.0435544100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewinson O, Padan E, Bibi E. Alkalitolerance: a biological function for a multidrug transporter in pH homeostasis. Proc Natl Acad Sci U S A. 2004;101(39):14073–14078. doi: 10.1073/pnas.0405375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saier MH, Jr, Paulsen IT. Phylogeny of multidrug transporters. Semin Cell Dev Biol. 2001;12(3):205–213. doi: 10.1006/scdb.2000.0246. [DOI] [PubMed] [Google Scholar]
- 27.Paulsen IT, Skurray RA, Tam R, Saier MH, Jr, Turner RJ, Weiner JH, et al. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol Microbiol. 1996;19(6):1167–1175. doi: 10.1111/j.1365-2958.1996.tb02462.x. [DOI] [PubMed] [Google Scholar]
- 28.Chung YJ, Saier MH., Jr Overexpression of the Escherichia coli sugE gene confers resistance to a narrow range of quaternary ammonium compounds. J Bacteriol. 2002;184(9):2543–2545. doi: 10.1128/jb.184.9.2543-2545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kermani AA, Macdonald CB, Gundepudi R, Stockbridge RB. Guanidinium export is the primal function of SMR family transporters. Proc Natl Acad Sci U S A. 2018;115(12):3060–3065. doi: 10.1073/pnas.1719187115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuldiner S, Granot D, Mordoch SS, Ninio S, Rotem D, Soskin M, et al. Small is mighty: EmrE, a multidrug transporter as an experimental paradigm. Physiology. 2001;16(3):130–134. doi: 10.1152/physiologyonline.2001.16.3.130. [DOI] [PubMed] [Google Scholar]
- 31.Ninio S, Rotem D, Schuldiner S. Functional analysis of novel multidrug transporters from human pathogens. J Biol Chem. 2001;276(51):48250–48256. doi: 10.1074/jbc.M108231200. [DOI] [PubMed] [Google Scholar]
- 32.Nasie I, Steiner-Mordoch S, Schuldiner S. New substrates on the block: clinically relevant resistances for EmrE and homologues. J Bacteriol. 2012;194(24):6766–6770. doi: 10.1128/JB.01318-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jack DL, Yang NM, Saier MH., Jr The drug/metabolite transporter superfamily. Eur J Biochem. 2001;268(13):3620–3639. doi: 10.1046/j.1432-1327.2001.02265.x. [DOI] [PubMed] [Google Scholar]
- 34.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64(2):159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 35.Poole K, Srikumar R. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr Top Med Chem. 2001;1(1):59–71. doi: 10.2174/1568026013395605. [DOI] [PubMed] [Google Scholar]
- 36.Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc Natl Acad Sci U S A. 2012;109(41):16696–16701. doi: 10.1073/pnas.1210093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5(1):e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402(6757):79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 39.Camacho LR, Constant P, Raynaud C, Laneelle MA, Triccas JA, Gicquel B, et al. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem. 2001;276(23):19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- 40.Domenech P, Reed MB, Barry CE., 3rd Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect Immun. 2005;73(6):3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldberg M, Pribyl T, Juhnke S, Nies DH. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J Biol Chem. 1999;274(37):26065–26070. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 42.Guan L, Ehrmann M, Yoneyama H, Nakae T. Membrane topology of the xenobiotic-exporting subunit, MexB, of the MexA,B-OprM extrusion pump in Pseudomonas aeruginosa. J Biol Chem. 1999;274(15):10517–10522. doi: 10.1074/jbc.274.15.10517. [DOI] [PubMed] [Google Scholar]
- 43.Morita Y, Kataoka A, Shiota S, Mizushima T, Tsuchiya T. NorM of Vibrio parahaemolyticus is an Na+−driven multidrug efflux pump. J Bacteriol. 2000;182(23):6694–6697. doi: 10.1128/jb.182.23.6694-6697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamae S, Ueda O, Yoshimura F, Hwang J, Tanaka Y, Nikaido H. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob Agents Chemother. 2001;45(12):3341–3346. doi: 10.1128/AAC.45.12.3341-3346.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huda MN, Morita Y, Kuroda T, Mizushima T, Tsuchiya T. Na+−driven multidrug efflux pump VcmA from Vibrio cholerae non-O1, a non-halophilic bacterium. FEMS Microbiol Lett. 2001;203(2):235–239. doi: 10.1111/j.1574-6968.2001.tb10847.x. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi M, Tabata K, Yagasaki M, Yonetani Y. Effect of multidrug-efflux transporter genes on dipeptide resistance and overproduction in Escherichia coli. FEMS Microbiol Lett. 2010;304(1):12–19. doi: 10.1111/j.1574-6968.2009.01879.x. [DOI] [PubMed] [Google Scholar]
- 47.McAnulty MJ, Wood TK. YeeO from Escherichia coli exports flavins. Bioengineered. 2014;5(6):386–392. doi: 10.4161/21655979.2014.969173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lubelski J, Konings WN, Driessen AJ. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol Mol Biol Rev. 2007;71(3):463–476. doi: 10.1128/MMBR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Veen HW, Callaghan R, Soceneantu L, Sardini A, Konings WN, Higgins CF. A bacterial antibiotic-resistance gene that complements the human multidrug-resistance P-glycoprotein gene. Nature. 1998;391(6664):291–295. doi: 10.1038/34669. [DOI] [PubMed] [Google Scholar]
- 50.Orelle C, Gubellini F, Durand A, Marco S, Levy D, Gros P, et al. Conformational change induced by ATP binding in the multidrug ATP-binding cassette transporter BmrA. Biochemistry. 2008;47(8):2404–2412. doi: 10.1021/bi702303s. [DOI] [PubMed] [Google Scholar]
- 51.Lin HT, Bavro VN, Barrera NP, Frankish HM, Velamakanni S, van Veen HW, et al. MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA. J Biol Chem. 2009;284(2):1145–1154. doi: 10.1074/jbc.M806964200.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu S, Zgurskaya HI. MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity. J Bacteriol. 2013;195(21):4865–4872. doi: 10.1128/JB.00756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol. 2001;183(19):5639–5644. doi: 10.1128/JB.183.19.5639-5644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guilfoile PG, Hutchinson CR. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc Natl Acad Sci U S A. 1991;88(19):8553–8557. doi: 10.1073/pnas.88.19.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 56.Higgins CF. ABC transporters: physiology, structure and mechanism--an overview. Res Microbiol. 2001;152(3–4):205–210. doi: 10.1016/s0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- 57.Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol Life Sci. 2004;61(6):682–699. doi: 10.1007/s00018-003-3336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higgins CF, Hiles ID, Salmond GP, Gill DR, Downie JA, Evans IJ, et al. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986;323(6087):448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- 59.Holland IB, Cole SP, Kuchler K, Higgins CF. ABC proteins: from bacteria to man. San Diego: Elsevier; 2003.
- 60.Young J, Holland IB. ABC transporters: bacterial exporters-revisited five years on. Biochim Biophys Acta. 1999;1461(2):177–200. doi: 10.1016/s0005-2736(99)00158-3. [DOI] [PubMed] [Google Scholar]
- 61.Truong-Bolduc QC, Strahilevitz J, Hooper DC. NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50(3):1104–1107. doi: 10.1128/AAC.50.3.1104-1107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeMarco CE, Cushing LA, Frempong-Manso E, Seo SM, Jaravaza TA, Kaatz GW. Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51(9):3235–3239. doi: 10.1128/AAC.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolhuis H, Poelarends G, van Veen HW, Poolman B, Driessen AJ, Konings WN. The Lactococcal lmrP gene encodes a proton motive force-dependent drug transporter. J Biol Chem. 1995;270(44):26092–26098. doi: 10.1074/jbc.270.44.26092. [DOI] [PubMed] [Google Scholar]
- 64.Mazurkiewicz P, Driessen AJ, Konings WN. Energetics of wild-type and mutant multidrug resistance secondary transporter LmrP of Lactococcus lactis. Biochim Biophys Acta. 2004;1658(3):252–261. doi: 10.1016/j.bbabio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 65.Paulsen IT, Park JH, Choi PS, Saier MH., Jr A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol Lett. 1997;156(1):1–8. doi: 10.1111/j.1574-6968.1997.tb12697.x. [DOI] [PubMed] [Google Scholar]
- 66.Dinh T, Paulsen IT, Saier M. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol. 1994;176(13):3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vaara M. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother. 1993;37(11):2255–2260. doi: 10.1128/aac.37.11.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zgurskaya HI, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci U S A. 1999;96(13):7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56(1):20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 70.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72(2):317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Veen HW, Higgins CF, Konings WN. Multidrug transport by ATP binding cassette transporters: a proposed two-cylinder engine mechanism. Res Microbiol. 2001;152(3–4):365–374. doi: 10.1016/s0923-2508(01)01208-6. [DOI] [PubMed] [Google Scholar]
- 72.Chami M, Steinfels E, Orelle C, Jault JM, Di Pietro A, Rigaud JL, et al. Three-dimensional structure by cryo-electron microscopy of YvcC, an homodimeric ATP-binding cassette transporter from Bacillus subtilis. J Mol Biol. 2002;315(5):1075–1085. doi: 10.1006/jmbi.2001.5309. [DOI] [PubMed] [Google Scholar]
- 73.Steinfels E, Orelle C, Fantino JR, Dalmas O, Rigaud JL, Denizot F, et al. Characterization of YvcC (BmrA), a multidrug ABC transporter constitutively expressed in Bacillus subtilis. Biochemistry. 2004;43(23):7491–7502. doi: 10.1021/bi0362018. [DOI] [PubMed] [Google Scholar]
- 74.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 75.Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22(47):7468–7485. doi: 10.1038/sj.onc.1206948. [DOI] [PubMed] [Google Scholar]
- 76.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11(7):1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 77.Yin JY, Huang Q, Yang Y, Zhang JT, Zhong MZ, Zhou HH, et al. Characterization and analyses of multidrug resistance-associated protein 1 (MRP1/ABCC1) polymorphisms in Chinese population. Pharmacogenet Genomics. 2009;19(3):206–216. doi: 10.1097/FPC.0b013e328323f680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res. 2001;42(7):1007–1017. doi: 10.1016/S0022-2275(20)31588-1. [DOI] [PubMed] [Google Scholar]
- 79.Esser L, Zhou F, Pluchino KM, Shiloach J, Ma J, Tang WK, et al. Structures of the multidrug transporter P-glycoprotein reveal asymmetric ATP binding and the mechanism of polyspecificity. J Biol Chem. 2017;292(2):446–461. doi: 10.1074/jbc.M116.755884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Burke MA, Ardehali H. Mitochondrial ATP-binding cassette proteins. Transl Res. 2007;150(2):73–80. doi: 10.1016/j.trsl.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 81.Chapuy B, Koch R, Radunski U, Corsham S, Cheong N, Inagaki N, et al. Intracellular ABC transporter A3 confers multidrug resistance in leukemia cells by lysosomal drug sequestration. Leukemia. 2008;22(8):1576–1586. doi: 10.1038/leu.2008.103. [DOI] [PubMed] [Google Scholar]
- 82.Kashiwayama Y, Seki M, Yasui A, Murasaki Y, Morita M, Yamashita Y, et al. 70-kDa peroxisomal membrane protein related protein (P70R/ABCD4) localizes to endoplasmic reticulum not peroxisomes, and NH2-terminal hydrophobic property determines the subcellular localization of ABC subfamily D proteins. Exp Cell Res. 2009;315(2):190–205. doi: 10.1016/j.yexcr.2008.10.031. [DOI] [PubMed] [Google Scholar]
- 83.Tsuchida M, Emi Y, Kida Y, Sakaguchi M. Human ABC transporter isoform B6 (ABCB6) localizes primarily in the Golgi apparatus. Biochem Biophys Res Commun. 2008;369(2):369–375. doi: 10.1016/j.bbrc.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 84.de Lange EC. Potential role of ABC transporters as a detoxification system at the blood-CSF barrier. Adv Drug Deliv Rev. 2004;56(12):1793–1809. doi: 10.1016/j.addr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 85.Kannan P, John C, Zoghbi SS, Halldin C, Gottesman MM, Innis RB, et al. Imaging the function of P-glycoprotein with radiotracers: pharmacokinetics and in vivo applications. Clin Pharmacol Ther. 2009;86(4):368–377. doi: 10.1038/clpt.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tarling EJ, de Aguiar Vallim TQ, Edwards PA. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol Metab. 2013;24(7):342–350. doi: 10.1016/j.tem.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447(7140):87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 88.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426(6968):905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 89.Robert R, Carlile GW, Pavel C, Liu N, Anjos SM, Liao J, et al. Structural analog of sildenafil identified as a novel corrector of the F508del-CFTR trafficking defect. Mol Pharmacol. 2008;73(2):478–489. doi: 10.1124/mol.107.040725. [DOI] [PubMed] [Google Scholar]
- 90.Basseville A, Tamaki A, Ierano C, Trostel S, Ward Y, Robey RW, et al. Histone deacetylase inhibitors influence chemotherapy transport by modulating expression and trafficking of a common polymorphic variant of the ABCG2 efflux transporter. Cancer Res. 2012;72(14):3642–3651. doi: 10.1158/0008-5472.CAN-11-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ma T, Vetrivel L, Yang H, Pedemonte N, Zegarra-Moran O, Galietta LJ, et al. High-affinity activators of cystic fibrosis transmembrane conductance regulator (CFTR) chloride conductance identified by high-throughput screening. J Biol Chem. 2002;277(40):37235–37241. doi: 10.1074/jbc.M205932200. [DOI] [PubMed] [Google Scholar]
- 92.Hillebrand M, Verrier SE, Ohlenbusch A, Schafer A, Soling HD, Wouters FS, et al. Live cell FRET microscopy: homo- and heterodimerization of two human peroxisomal ABC transporters, the adrenoleukodystrophy protein (ALDP, ABCD1) and PMP70 (ABCD3) J Biol Chem. 2007;282(37):26997–27005. doi: 10.1074/jbc.M702122200. [DOI] [PubMed] [Google Scholar]
- 93.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5(7):596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 94.Grove DE, Rosser MF, Ren HY, Naren AP, Cyr DM. Mechanisms for rescue of correctable folding defects in CFTRDelta F508. Mol Biol Cell. 2009;20(18):4059–4069. doi: 10.1091/mbc.E08-09-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Genin EC, Gondcaille C, Trompier D, Savary S. Induction of the adrenoleukodystrophy-related gene (ABCD2) by thyromimetics. J Steroid Biochem Mol Biol. 2009;116(1–2):37–43. doi: 10.1016/j.jsbmb.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 96.Pluen A, Boucher Y, Ramanujan S, McKee TD, Gohongi T, di Tomaso E, et al. Role of tumor-host interactions in interstitial diffusion of macromolecules: cranial vs. subcutaneous tumors. Proc Natl Acad Sci U S A. 2001;98(8):4628–4633. doi: 10.1073/pnas.081626898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jain RK. Delivery of molecular and cellular medicine to solid tumors. Adv Drug Deliv Rev. 2001;46(1–3):149–168. doi: 10.1016/s0169-409x(00)00131-9. [DOI] [PubMed] [Google Scholar]
- 98.Jain RK. Normalizing tumor vasculature with anti-angiogenic therapy: a new paradigm for combination therapy. Nat Med. 2001;7(9):987–989. doi: 10.1038/nm0901-987. [DOI] [PubMed] [Google Scholar]
- 99.Green SK, Frankel A, Kerbel RS. Adhesion-dependent multicellular drug resistance. Anticancer Drug Des. 1999;14(2):153–168. [PubMed] [Google Scholar]
- 100.Durand RE, Olive PL. Resistance of tumor cells to chemo- and radiotherapy modulated by the three-dimensional architecture of solid tumors and spheroids. Methods Cell Biol. 2001;64:211–233. doi: 10.1016/s0091-679x(01)64015-9. [DOI] [PubMed] [Google Scholar]
- 101.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 102.Shen DW, Goldenberg S, Pastan I, Gottesman MM. Decreased accumulation of [14C] carboplatin in human cisplatin-resistant cells results from reduced energy-dependent uptake. J Cell Physiol. 2000;183(1):108–116. doi: 10.1002/(SICI)1097-4652(200004)183:1<108::AID-JCP13>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 103.Shen D, Pastan I, Gottesman MM. Cross-resistance to methotrexate and metals in human cisplatin-resistant cell lines results from a pleiotropic defect in accumulation of these compounds associated with reduced plasma membrane binding proteins. Cancer Res. 1998;58(2):268–275. [PubMed] [Google Scholar]
- 104.Schuetz EG, Beck WT, Schuetz JD. Modulators and substrates of P-glycoprotein and cytochrome P4503A coordinately up-regulate these proteins in human colon carcinoma cells. Mol Pharmacol. 1996;49(2):311–318. [PubMed] [Google Scholar]
- 105.Liu YY, Han TY, Giuliano AE, Cabot MC. Ceramide glycosylation potentiates cellular multidrug resistance. FASEB J. 2001;15(3):719–730. doi: 10.1096/fj.00-0223com. [DOI] [PubMed] [Google Scholar]
- 106.Deenen MJ, Cats A, Beijnen JH, Schellens JH. Part 1: background, methodology, and clinical adoption of pharmacogenetics. Oncologist. 2011;16(6):811–819. doi: 10.1634/theoncologist.2010-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Olivera G, Sendra L, Herrero MJ, Berlanga P, Gargallo P, Yáñez Y, et al. Pharmacogenetics implementation in the clinics: information and guidelines for germline variants. Cancer Drug Resist. 2019;2(1):53–68. doi: 10.20517/cdr.2018.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Turajlic S, Sottoriva A, Graham T, Swanton C. Resolving genetic heterogeneity in cancer. Nat Rev Genet. 2019;20(7):404–416. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 109.Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359(6375):582–587. doi: 10.1126/science.aao4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Daigo S, Takahashi Y, Fujieda M, Ariyoshi N, Yamazaki H, Koizumi W, et al. A novel mutant allele of the CYP2A6 gene (CYP2A6*11 ) found in a cancer patient who showed poor metabolic phenotype towards tegafur. Pharmacogenetics. 2002;12(4):299–306. doi: 10.1097/00008571-200206000-00005. [DOI] [PubMed] [Google Scholar]
- 111.Kim DH, Sriharsha L, Xu W, Kamel-Reid S, Liu X, Siminovitch K, et al. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res. 2009;15(14):4750–4758. doi: 10.1158/1078-0432.CCR-09-0145. [DOI] [PubMed] [Google Scholar]
- 112.Kjersem JB, Skovlund E, Ikdahl T, Guren T, Kersten C, Dalsgaard AM, et al. FCGR2A and FCGR3A polymorphisms and clinical outcome in metastatic colorectal cancer patients treated with first-line 5-fluorouracil/folinic acid and oxaliplatin +/− cetuximab. BMC Cancer. 2014;14:340. doi: 10.1186/1471-2407-14-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li J, Cusatis G, Brahmer J, Sparreboom A, Robey RW, Bates SE, et al. Association of variant ABCG2 and the pharmacokinetics of epidermal growth factor receptor tyrosine kinase inhibitors in cancer patients. Cancer Biol Ther. 2007;6(3):432–438. doi: 10.4161/cbt.6.3.3763. [DOI] [PubMed] [Google Scholar]
- 114.Marin JJ, Briz O, Monte MJ, Blazquez AG, Macias RI. Genetic variants in genes involved in mechanisms of chemoresistance to anticancer drugs. Curr Cancer Drug Targets. 2012;12(4):402–438. doi: 10.2174/156800912800190875. [DOI] [PubMed] [Google Scholar]
- 115.Mathijssen RH, Marsh S, Karlsson MO, Xie R, Baker SD, Verweij J, et al. Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin Cancer Res. 2003;9(9):3246–3253. [PubMed] [Google Scholar]
- 116.Mukerjee G, Huston A, Kabakchiev B, Piquette-Miller M, van Schaik R, Dorfman R. User considerations in assessing pharmacogenomic tests and their clinical support tools. NPJ Genom Med. 2018;3:26. doi: 10.1038/s41525-018-0065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rodriguez-Antona C, Gomez A, Karlgren M, Sim SC, Ingelman-Sundberg M. Molecular genetics and epigenetics of the cytochrome P450 gene family and its relevance for cancer risk and treatment. Hum Genet. 2010;127(1):1–17. doi: 10.1007/s00439-009-0748-0. [DOI] [PubMed] [Google Scholar]
- 118.Bellon SF, Coleman JH, Lippard SJ. DNA unwinding produced by site-specific intrastrand cross-links of the antitumor drug cis-diamminedichloroplatinum (II) Biochemistry. 1991;30(32):8026–8035. doi: 10.1021/bi00246a021. [DOI] [PubMed] [Google Scholar]
- 119.Smelick GS, Heffron TP, Chu L, Dean B, West DA, Duvall SL, et al. Prevalence of acid-reducing agents (ARA) in cancer populations and ARA drug-drug interaction potential for molecular targeted agents in clinical development. Mol Pharm. 2013;10(11):4055–4062. doi: 10.1021/mp400403s. [DOI] [PubMed] [Google Scholar]
- 120.Tian H, Gao Z, Li H, Zhang B, Wang G, Zhang Q, et al. DNA damage response--a double-edged sword in cancer prevention and cancer therapy. Cancer Lett. 2015;358(1):8–16. doi: 10.1016/j.canlet.2014.12.038. [DOI] [PubMed] [Google Scholar]
- 121.Yamanaka K, Chatterjee N, Hemann MT, Walker GC. Inhibition of mutagenic translesion synthesis: a possible strategy for improving chemotherapy? PLoS Genet. 2017;13(8):e1006842. doi: 10.1371/journal.pgen.1006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 124.Zhang C, Ge S, Wang J, Jing X, Li H, Mei S, et al. Epigenomic profiling of DNA methylation for hepatocellular carcinoma diagnosis and prognosis prediction. J Gastroenterol Hepatol. 2019;34(10):1869–1877. doi: 10.1111/jgh.14694. [DOI] [PubMed] [Google Scholar]
- 125.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19(5):698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 126.Calvisi DF, Pascale RM, Feo F. Dissection of signal transduction pathways as a tool for the development of targeted therapies of hepatocellular carcinoma. Rev Recent Clin Trials. 2007;2(3):217–236. doi: 10.2174/157488707781662715. [DOI] [PubMed] [Google Scholar]
- 127.Yuan YH, Wang HY, Lai Y, Zhong W, Liang WL, Yan FD, et al. Epigenetic inactivation of HOXD10 is associated with human colon cancer via inhibiting the RHOC/AKT/MAPK signaling pathway. Cell Commun Signal. 2019;17(1):9. doi: 10.1186/s12964-018-0316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20(1):21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Assaraf YG, Brozovic A, Goncalves AC, Jurkovicova D, Line A, Machuqueiro M, et al. The multi-factorial nature of clinical multidrug resistance in cancer. Drug Resist Updat. 2019;46:100645. doi: 10.1016/j.drup.2019.100645. [DOI] [PubMed] [Google Scholar]
- 130.Feng DD, Zhang H, Zhang P, Zheng YS, Zhang XJ, Han BW, et al. Down-regulated miR-331-5p and miR-27a are associated with chemotherapy resistance and relapse in leukaemia. J Cell Mol Med. 2011;15(10):2164–2175. doi: 10.1111/j.1582-4934.2010.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boyerinas B, Park SM, Murmann AE, Gwin K, Montag AG, Zillhardt M, et al. Let-7 modulates acquired resistance of ovarian cancer to Taxanes via IMP-1-mediated stabilization of multidrug resistance 1. Int J Cancer. 2012;130(8):1787–1797. doi: 10.1002/ijc.26190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen J, Tian W, Cai H, He H, Deng Y. Down-regulation of microRNA-200c is associated with drug resistance in human breast cancer. Med Oncol. 2012;29(4):2527–2534. doi: 10.1007/s12032-011-0117-4. [DOI] [PubMed] [Google Scholar]
- 133.Yang G, Wu D, Zhu J, Jiang O, Shi Q, Tian J, et al. Upregulation of miR-195 increases the sensitivity of breast cancer cells to Adriamycin treatment through inhibition of Raf-1. Oncol Rep. 2013;30(2):877–889. doi: 10.3892/or.2013.2532. [DOI] [PubMed] [Google Scholar]
- 134.Li C, Zou J, Zheng G, Chu J. MiR-30a decreases multidrug resistance (MDR) of gastric cancer cells. Med Sci Monit. 2016. 10.12659/MSM.898415. [DOI] [PubMed]
- 135.Li Y, Zhao L, Li N, Miao Y, Zhou H, Jia L. [Corrigendum] miR-9 regulates the multidrug resistance of chronic myelogenous leukemia by targeting ABCB1. Oncol Rep. 2019;41(5):3148. doi: 10.3892/or.2019.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ghodousi ES, Rahgozar S. MicroRNA-326 and microRNA-200c: two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J Cell Biochem. 2018;119(7):6024–6032. doi: 10.1002/jcb.26800. [DOI] [PubMed] [Google Scholar]
- 137.Riechelmann RP, Tannock IF, Wang L, Saad ED, Taback NA, Krzyzanowska MK. Potential drug interactions and duplicate prescriptions among cancer patients. J Natl Cancer Inst. 2007;99(8):592–600. doi: 10.1093/jnci/djk130. [DOI] [PubMed] [Google Scholar]
- 138.Herbrink M, Nuijen B, Schellens JH, Beijnen JH. Variability in bioavailability of small molecular tyrosine kinase inhibitors. Cancer Treat Rev. 2015;41(5):412–422. doi: 10.1016/j.ctrv.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 139.Mathijssen RH, Sparreboom A, Verweij J. Determining the optimal dose in the development of anticancer agents. Nat Rev Clin Oncol. 2014;11(5):272–281. doi: 10.1038/nrclinonc.2014.40. [DOI] [PubMed] [Google Scholar]
- 140.Sharma M, Holmes HM, Mehta HB, Chen H, Aparasu RR, Shih YT, et al. The concomitant use of tyrosine kinase inhibitors and proton pump inhibitors: prevalence, predictors, and impact on survival and discontinuation of therapy in older adults with cancer. Cancer. 2019;125(7):1155–1162. doi: 10.1002/cncr.31917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ghosh S. Cisplatin: the first metal based anticancer drug. Bioorg Chem. 2019;88:102925. doi: 10.1016/j.bioorg.2019.102925. [DOI] [PubMed] [Google Scholar]
- 142.Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta. 1976;455(1):152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 143.Ueda K, Cornwell MM, Gottesman MM, Pastan I, Roninson IB, Ling V, et al. The mdr1 gene, responsible for multidrug-resistance, codes for P-glycoprotein. Biochem Biophys Res Commun. 1986;141(3):956–962. doi: 10.1016/s0006-291x(86)80136-x. [DOI] [PubMed] [Google Scholar]
- 144.Schinkel AH, Jonker JW. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev. 2003;55(1):3–29. doi: 10.1016/s0169-409x(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 145.Sodani K, Patel A, Kathawala RJ, Chen ZS. Multidrug resistance associated proteins in multidrug resistance. Chin J Cancer. 2012;31(2):58–72. doi: 10.5732/cjc.011.10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tiwari AK, Sodani K, Dai CL, Ashby CR, Jr, Chen ZS. Revisiting the ABCs of multidrug resistance in cancer chemotherapy. Curr Pharm Biotechnol. 2011;12(4):570–594. doi: 10.2174/138920111795164048. [DOI] [PubMed] [Google Scholar]
- 147.Findling-Kagan S, Sivan H, Ostrovsky O, Nagler A, Galski H. Establishment and characterization of new cellular lymphoma model expressing transgenic human MDR1. Leuk Res. 2005;29(4):407–414. doi: 10.1016/j.leukres.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 148.Liu Yin JA, Wheatley K, Rees JK, Burnett AK, Party UMALW. Comparison of ‘sequential’ versus ‘standard’ chemotherapy as re-induction treatment, with or without cyclosporine, in refractory/relapsed acute myeloid leukaemia (AML): results of the UK Medical Research Council AML-R trial. Br J Haematol. 2001;113(3):713–726. doi: 10.1046/j.1365-2141.2001.02785.x. [DOI] [PubMed] [Google Scholar]
- 149.Sauna ZE, Smith MM, Muller M, Kerr KM, Ambudkar SV. The mechanism of action of multidrug-resistance-linked P-glycoprotein. J Bioenerg Biomembr. 2001;33(6):481–491. doi: 10.1023/a:1012875105006. [DOI] [PubMed] [Google Scholar]
- 150.Peng XX, Tiwari AK, Wu HC, Chen ZS. Overexpression of P-glycoprotein induces acquired resistance to imatinib in chronic myelogenous leukemia cells. Chin J Cancer. 2012;31(2):110–118. doi: 10.5732/cjc.011.10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mahon F-X, Hayette S, Lagarde V, Belloc F, Turcq B, Nicolini F, et al. Evidence that resistance to nilotinib may be due to BCR-ABL, Pgp, or Src kinase overexpression. Cancer Res. 2008;68(23):9809–9816. doi: 10.1158/0008-5472.CAN-08-1008. [DOI] [PubMed] [Google Scholar]
- 152.Marchetti S, de Vries NA, Buckle T, Bolijn MJ, van Eijndhoven MA, Beijnen JH, et al. Effect of the ATP-binding cassette drug transporters ABCB1, ABCG2, and ABCC2 on erlotinib hydrochloride (Tarceva) disposition in in vitro and in vivo pharmacokinetic studies employing Bcrp1−/−/Mdr1a/1b−/−(triple-knockout) and wild-type mice. Mol Cancer Ther. 2008;7(8):2280–2287. doi: 10.1158/1535-7163.MCT-07-2250. [DOI] [PubMed] [Google Scholar]
- 153.Shi Z, Peng XX, Kim IW, Shukla S, Si QS, Robey RW, et al. Erlotinib (Tarceva, OSI-774) antagonizes ATP-binding cassette subfamily B member 1 and ATP-binding cassette subfamily G member 2-mediated drug resistance. Cancer Res. 2007;67(22):11012–11020. doi: 10.1158/0008-5472.CAN-07-2686. [DOI] [PubMed] [Google Scholar]
- 154.Katayama R, Sakashita T, Yanagitani N, Ninomiya H, Horiike A, Friboulet L, et al. P-glycoprotein mediates Ceritinib resistance in anaplastic lymphoma kinase-rearranged non-small cell lung cancer. EBioMedicine. 2016;3:54–66. doi: 10.1016/j.ebiom.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schinkel AH. P-Glycoprotein, a gatekeeper in the blood-brain barrier. Adv Drug Deliv Rev. 1999;36(2–3):179–194. doi: 10.1016/s0169-409x(98)00085-4. [DOI] [PubMed] [Google Scholar]
- 156.Sparreboom A, van Asperen J, Mayer U, Schinkel AH, Smit JW, Meijer DK, et al. Limited oral bioavailability and active epithelial excretion of paclitaxel (Taxol) caused by P-glycoprotein in the intestine. Proc Natl Acad Sci U S A. 1997;94(5):2031–2035. doi: 10.1073/pnas.94.5.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gao HL, Gupta P, Cui Q, Ashar YV, Wu ZX, Zeng L, et al. Sapitinib reverses anticancer drug resistance in colon cancer cells overexpressing the ABCB1 transporter. Front Oncol. 2020;10:574861. doi: 10.3389/fonc.2020.574861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dong XD, Zhang M, Ma X, Wang JQ, Lei ZN, Teng QX, et al. Bruton's tyrosine kinase (BTK) inhibitor RN486 overcomes ABCB1-mediated multidrug resistance in cancer cells. Front Cell Dev Biol. 2020;8:865. doi: 10.3389/fcell.2020.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dai CL, Tiwari AK, Wu CP, Su XD, Wang SR, Liu DG, et al. Lapatinib (Tykerb, GW572016) reverses multidrug resistance in cancer cells by inhibiting the activity of ATP-binding cassette subfamily B member 1 and G member 2. Cancer Res. 2008;68(19):7905–7914. doi: 10.1158/0008-5472.CAN-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Palmeira A, Sousa E, Vasconcelos MH, Pinto MM. Three decades of P-gp inhibitors: skimming through several generations and scaffolds. Curr Med Chem. 2012;19(13):1946–2025. doi: 10.2174/092986712800167392. [DOI] [PubMed] [Google Scholar]
- 161.Matsuo H, Takada T, Ichida K, Nakamura T, Nakayama A, Takada Y, et al. Identification of ABCG2 dysfunction as a major factor contributing to gout. Nucleosides Nucleotides Nucleic Acids. 2011;30(12):1098–1104. doi: 10.1080/15257770.2011.627902. [DOI] [PubMed] [Google Scholar]
- 162.Zhang H, Patel A, Ma SL, Li XJ, Zhang YK, Yang PQ, et al. In vitro, in vivo and ex vivo characterization of ibrutinib: a potent inhibitor of the efflux function of the transporter MRP1. Br J Pharmacol. 2014;171(24):5845–5857. doi: 10.1111/bph.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mao Q, Unadkat JD. Role of the breast cancer resistance protein (ABCG2) in drug transport. AAPS J. 2005;7(1):E118–E133. doi: 10.1208/aapsj070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cooray HC, Blackmore CG, Maskell L, Barrand MA. Localisation of breast cancer resistance protein in microvessel endothelium of human brain. Neuroreport. 2002;13(16):2059–2063. doi: 10.1097/00001756-200211150-00014. [DOI] [PubMed] [Google Scholar]
- 165.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci U S A. 1998;95(26):15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Maliepaard M, Scheffer GL, Faneyte IF, van Gastelen MA, Pijnenborg AC, Schinkel AH, et al. Subcellular localization and distribution of the breast cancer resistance protein transporter in normal human tissues. Cancer Res. 2001;61(8):3458–3464. [PubMed] [Google Scholar]
- 167.Rocchi E, Khodjakov A, Volk EL, Yang C-H, Litman T, Bates SE, et al. The product of the ABC half-transporter gene ABCG2 (BCRP/MXR/ABCP) is expressed in the plasma membrane. Biochem Biophys Res Commun. 2000;271(1):42–46. doi: 10.1006/bbrc.2000.2590. [DOI] [PubMed] [Google Scholar]
- 168.Sun YL, Kathawala RJ, Singh S, Zheng K, Talele TT, Jiang WQ, et al. Zafirlukast antagonizes ATP-binding cassette subfamily G member 2-mediated multidrug resistance. Anti-Cancer Drugs. 2012;23(8):865–873. doi: 10.1097/CAD.0b013e328354a196. [DOI] [PubMed] [Google Scholar]
- 169.Wang DS, Patel A, Sim HM, Zhang YK, Wang YJ, Kathawala RJ, et al. ARRY-334543 reverses multidrug resistance by antagonizing the activity of ATP-binding cassette subfamily G member 2. J Cell Biochem. 2014;115(8):1381–1391. doi: 10.1002/jcb.24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wu ZX, Mai Q, Yang Y, Wang JQ, Ma H, Zeng L, et al. Overexpression of human ATP-binding cassette transporter ABCG2 contributes to reducing the cytotoxicity of GSK1070916 in cancer cells. Biomed Pharmacother. 2021;136:111223. doi: 10.1016/j.biopha.2021.111223. [DOI] [PubMed] [Google Scholar]
- 171.Wu ZX, Yang Y, Teng QX, Wang JQ, Lei ZN, Wang JQ, et al. Tivantinib, a c-Met inhibitor in clinical trials, is susceptible to ABCG2-mediated drug resistance. Cancers (Basel). 2020;12(1). 10.3390/cancers12010186. [DOI] [PMC free article] [PubMed]
- 172.Wei LY, Wu ZX, Yang Y, Zhao M, Ma XY, Li JS, et al. Overexpression of ABCG2 confers resistance to pevonedistat, an NAE inhibitor. Exp Cell Res. 2020;388(2):111858. doi: 10.1016/j.yexcr.2020.111858. [DOI] [PubMed] [Google Scholar]
- 173.Kathawala RJ, Espitia CM, Jones TM, Islam S, Gupta P, Zhang YK, et al. ABCG2 overexpression contributes to pevonedistat resistance. Cancers (Basel). 2020;12(2). 10.3390/cancers12020429. [DOI] [PMC free article] [PubMed]
- 174.Michaelis M, Selt F, Rothweiler F, Wiese M, Cinatl J., Jr ABCG2 impairs the activity of the aurora kinase inhibitor tozasertib but not of alisertib. BMC Res Notes. 2015;8:484. doi: 10.1186/s13104-015-1405-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Marchetti S, Pluim D, van Eijndhoven M, van Tellingen O, Mazzanti R, Beijnen JH, et al. Effect of the drug transporters ABCG2, Abcg2, ABCB1 and ABCC2 on the disposition, brain accumulation and myelotoxicity of the aurora kinase B inhibitor barasertib and its more active form barasertib-hydroxy-QPA. Investig New Drugs. 2013;31(5):1125–1135. doi: 10.1007/s10637-013-9923-1. [DOI] [PubMed] [Google Scholar]
- 176.Chen ZS, Robey RW, Belinsky MG, Shchaveleva I, Ren XQ, Sugimoto Y, et al. Transport of methotrexate, methotrexate polyglutamates, and 17beta-estradiol 17-(beta-D-glucuronide) by ABCG2: effects of acquired mutations at R482 on methotrexate transport. Cancer Res. 2003;63(14):4048–4054. [PubMed] [Google Scholar]
- 177.Polireddy K, Chavan H, Abdulkarim BA, Krishnamurthy P. Functional significance of the ATP-binding cassette transporter B6 in hepatocellular carcinoma. Mol Oncol. 2011;5(5):410–425. doi: 10.1016/j.molonc.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yasui K, Mihara S, Zhao C, Okamoto H, Saito-Ohara F, Tomida A, et al. Alteration in copy numbers of genes as a mechanism for acquired drug resistance. Cancer Res. 2004;64(4):1403–1410. doi: 10.1158/0008-5472.can-3263-2. [DOI] [PubMed] [Google Scholar]
- 179.Park S, Shimizu C, Shimoyama T, Takeda M, Ando M, Kohno T, et al. Gene expression profiling of ATP-binding cassette (ABC) transporters as a predictor of the pathologic response to neoadjuvant chemotherapy in breast cancer patients. Breast Cancer Res Treat. 2006;99(1):9–17. doi: 10.1007/s10549-006-9175-2. [DOI] [PubMed] [Google Scholar]
- 180.Varatharajan S, Abraham A, Karathedath S, Ganesan S, Lakshmi KM, Arthur N, et al. ATP-binding casette transporter expression in acute myeloid leukemia: association with in vitro cytotoxicity and prognostic markers. Pharmacogenomics. 2017;18(3):235–244. doi: 10.2217/pgs-2016-0150. [DOI] [PubMed] [Google Scholar]
- 181.Minami K, Kamijo Y, Nishizawa Y, Tabata S, Horikuchi F, Yamamoto M, et al. Expression of ABCB6 is related to resistance to 5-FU, SN-38 and vincristine. Anticancer Res. 2014;34(9):4767–4773. [PubMed] [Google Scholar]
- 182.Polireddy K, Khan MM, Chavan H, Young S, Ma X, Waller A, et al. A novel flow cytometric HTS assay reveals functional modulators of ATP binding cassette transporter ABCB6. PLoS One. 2012;7(7):e40005. doi: 10.1371/journal.pone.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bakos E, Evers R, Szakacs G, Tusnady GE, Welker E, Szabo K, et al. Functional multidrug resistance protein (MRP1) lacking the N-terminal transmembrane domain. J Biol Chem. 1998;273(48):32167–32175. doi: 10.1074/jbc.273.48.32167. [DOI] [PubMed] [Google Scholar]
- 184.Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258(5088):1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 185.Kruh GD, Belinsky MG. The MRP family of drug efflux pumps. Oncogene. 2003;22(47):7537–7552. doi: 10.1038/sj.onc.1206953. [DOI] [PubMed] [Google Scholar]
- 186.Leschziner G, Zabaneh D, Pirmohamed M, Owen A, Rogers J, Coffey AJ, et al. Exon sequencing and high resolution haplotype analysis of ABC transporter genes implicated in drug resistance. Pharmacogenet Genomics. 2006;16(6):439–450. doi: 10.1097/01.fpc.0000197467.21964.67. [DOI] [PubMed] [Google Scholar]
- 187.Anreddy N, Gupta P, Kathawala RJ, Patel A, Wurpel JN, Chen Z-S. Tyrosine kinase inhibitors as reversal agents for ABC transporter mediated drug resistance. Molecules. 2014;19(9):13848–13877. doi: 10.3390/molecules190913848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Assaraf YG, Rothem L, Hooijberg JH, Stark M, Ifergan I, Kathmann I, et al. Loss of multidrug resistance protein 1 expression and folate efflux activity results in a highly concentrative folate transport in human leukemia cells. J Biol Chem. 2003;278(9):6680–6686. doi: 10.1074/jbc.M209186200. [DOI] [PubMed] [Google Scholar]
- 189.Wang Y-J, Zhang Y-K, Kathawala RJ, Chen Z-S. Repositioning of tyrosine kinase inhibitors as antagonists of ATP-binding cassette transporters in anticancer drug resistance. Cancers. 2014;6(4):1925–1952. doi: 10.3390/cancers6041925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chen XY, Yang Y, Wang JQ, Wu ZX, Li J, Chen ZS. Overexpression of ABCC1 confers drug resistance to betulin. Front Oncol. 2021;11:640656. doi: 10.3389/fonc.2021.640656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Peterson BG, Tan KW, Osa-Andrews B, Iram SH. High-content screening of clinically tested anticancer drugs identifies novel inhibitors of human MRP1 (ABCC1) Pharmacol Res. 2017;119:313–326. doi: 10.1016/j.phrs.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 192.Gupta P, Xie M, Narayanan S, Wang YJ, Wang XQ, Yuan T, et al. GSK1904529A, a potent IGF-IR inhibitor, reverses MRP1-mediated multidrug resistance. J Cell Biochem. 2017;118(10):3260–3267. doi: 10.1002/jcb.25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Williams GC, Liu A, Knipp G, Sinko PJ. Direct evidence that saquinavir is transported by multidrug resistance-associated protein (MRP1) and canalicular multispecific organic anion transporter (MRP2) Antimicrob Agents Chemother. 2002;46(11):3456–3462. doi: 10.1128/aac.46.11.3456-3462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yang AK, Zhou ZW, Wei MQ, Liu JP, Zhou SF. Modulators of multidrug resistance associated proteins in the management of anticancer and antimicrobial drug resistance and the treatment of inflammatory diseases. Curr Top Med Chem. 2010;10(17):1732–1756. doi: 10.2174/156802610792928040. [DOI] [PubMed] [Google Scholar]
- 195.Jedlitschky G, Hoffmann U, Kroemer HK. Structure and function of the MRP2 (ABCC2) protein and its role in drug disposition. Expert Opin Drug Metab Toxicol. 2006;2(3):351–366. doi: 10.1517/17425255.2.3.351. [DOI] [PubMed] [Google Scholar]
- 196.Schaub TP, Kartenbeck J, Konig J, Spring H, Dorsam J, Staehler G, et al. Expression of the MRP2 gene-encoded conjugate export pump in human kidney proximal tubules and in renal cell carcinoma. J Am Soc Nephrol. 1999;10(6):1159–1169. doi: 10.1681/ASN.V1061159. [DOI] [PubMed] [Google Scholar]
- 197.Kawabe T, Chen ZS, Wada M, Uchiumi T, Ono M, Akiyama S, et al. Enhanced transport of anticancer agents and leukotriene C4 by the human canalicular multispecific organic anion transporter (cMOAT/MRP2) FEBS Lett. 1999;456(2):327–331. doi: 10.1016/s0014-5793(99)00979-5. [DOI] [PubMed] [Google Scholar]
- 198.Wu L, Chen Y, Liu H, Zhan Z, Liang Z, Zhang T, et al. Emodin-induced hepatotoxicity was exacerbated by probenecid through inhibiting UGTs and MRP2. Toxicol Appl Pharmacol. 2018;359:91–101. doi: 10.1016/j.taap.2018.09.029. [DOI] [PubMed] [Google Scholar]
- 199.Bi X, Yuan Z, Qu B, Zhou H, Liu Z, Xie Y. Piperine enhances the bioavailability of silybin via inhibition of efflux transporters BCRP and MRP2. Phytomedicine. 2019;54:98–108. doi: 10.1016/j.phymed.2018.09.217. [DOI] [PubMed] [Google Scholar]
- 200.Chaisit T, Siripong P, Jianmongkol S. Rhinacanthin-C enhances doxorubicin cytotoxicity via inhibiting the functions of P-glycoprotein and MRP2 in breast cancer cells. Eur J Pharmacol. 2017;795:50–57. doi: 10.1016/j.ejphar.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 201.Ge S, Yin T, Xu B, Gao S, Hu M. Curcumin affects phase II disposition of resveratrol through inhibiting efflux transporters MRP2 and BCRP. Pharm Res. 2016;33(3):590–602. doi: 10.1007/s11095-015-1812-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Agarwal S, Pal D, Mitra AK. Both P-gp and MRP2 mediate transport of Lopinavir, a protease inhibitor. Int J Pharm. 2007;339(1–2):139–147. doi: 10.1016/j.ijpharm.2007.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang H, Fu LW. Multidrug resistance-associated proteins and their roles in multidrug resistance. Yao Xue Xue Bao. 2011;46(5):479–486. [PubMed] [Google Scholar]
- 204.Young LC, Campling BG, Cole SP, Deeley RG, Gerlach JH. Multidrug resistance proteins MRP3, MRP1, and MRP2 in lung cancer: correlation of protein levels with drug response and messenger RNA levels. Clin Cancer Res. 2001;7(6):1798–1804. [PubMed] [Google Scholar]
- 205.Zhao Y, Lu H, Yan A, Yang Y, Meng Q, Sun L, et al. ABCC3 as a marker for multidrug resistance in non-small cell lung cancer. Sci Rep. 2013;3:3120. doi: 10.1038/srep03120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Konig J, Rost D, Cui Y, Keppler D. Characterization of the human multidrug resistance protein isoform MRP3 localized to the basolateral hepatocyte membrane. Hepatology. 1999;29(4):1156–1163. doi: 10.1002/hep.510290404. [DOI] [PubMed] [Google Scholar]
- 207.Kool M, van der Linden M, de Haas M, Scheffer GL, de Vree JM, Smith AJ, et al. MRP3, an organic anion transporter able to transport anti-cancer drugs. Proc Natl Acad Sci U S A. 1999;96(12):6914–6919. doi: 10.1073/pnas.96.12.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ali I, Welch MA, Lu Y, Swaan PW, Brouwer KLR. Identification of novel MRP3 inhibitors based on computational models and validation using an in vitro membrane vesicle assay. Eur J Pharm Sci. 2017;103:52–59. doi: 10.1016/j.ejps.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kool M, de Haas M, Scheffer GL, Scheper RJ, van Eijk MJ, Juijn JA, et al. Analysis of expression of cMOAT (MRP2), MRP3, MRP4, and MRP5, homologues of the multidrug resistance-associated protein gene (MRP1), in human cancer cell lines. Cancer Res. 1997;57(16):3537–3547. [PubMed] [Google Scholar]
- 210.Zelcer N, Reid G, Wielinga P, Kuil A, van der Heijden I, Schuetz JD, et al. Steroid and bile acid conjugates are substrates of human multidrug-resistance protein (MRP) 4 (ATP-binding cassette C4) Biochem J. 2003;371(Pt 2):361–367. doi: 10.1042/BJ20021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hopper-Borge E, Chen ZS, Shchaveleva I, Belinsky MG, Kruh GD. Analysis of the drug resistance profile of multidrug resistance protein 7 (ABCC10): resistance to docetaxel. Cancer Res. 2004;64(14):4927–4930. doi: 10.1158/0008-5472.CAN-03-3111. [DOI] [PubMed] [Google Scholar]
- 212.Rius M, Nies AT, Hummel-Eisenbeiss J, Jedlitschky G, Keppler D. Cotransport of reduced glutathione with bile salts by MRP4 (ABCC4) localized to the basolateral hepatocyte membrane. Hepatology. 2003;38(2):374–384. doi: 10.1053/jhep.2003.50331. [DOI] [PubMed] [Google Scholar]
- 213.Wu CP, Calcagno AM, Hladky SB, Ambudkar SV, Barrand MA. Modulatory effects of plant phenols on human multidrug-resistance proteins 1, 4 and 5 (ABCC1, 4 and 5) FEBS J. 2005;272(18):4725–4740. doi: 10.1111/j.1742-4658.2005.04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chen ZS, Lee K, Kruh GD. Transport of cyclic nucleotides and estradiol 17-beta-D-glucuronide by multidrug resistance protein 4. Resistance to 6-mercaptopurine and 6-thioguanine. J Biol Chem. 2001;276(36):33747–33754. doi: 10.1074/jbc.M104833200. [DOI] [PubMed] [Google Scholar]
- 215.Lempers VJ, van den Heuvel JJ, Russel FG, Aarnoutse RE, Burger DM, Bruggemann RJ, et al. Inhibitory potential of antifungal drugs on ATP-binding cassette transporters P-glycoprotein, MRP1 to MRP5, BCRP, and BSEP. Antimicrob Agents Chemother. 2016;60(6):3372–3379. doi: 10.1128/AAC.02931-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Borst P, de Wolf C, van de Wetering K. Multidrug resistance-associated proteins 3, 4, and 5. Pflugers Arch. 2007;453(5):661–673. doi: 10.1007/s00424-006-0054-9. [DOI] [PubMed] [Google Scholar]
- 217.Imaoka T, Kusuhara H, Adachi M, Schuetz JD, Takeuchi K, Sugiyama Y. Functional involvement of multidrug resistance-associated protein 4 (MRP4/ABCC4) in the renal elimination of the antiviral drugs adefovir and tenofovir. Mol Pharmacol. 2007;71(2):619–627. doi: 10.1124/mol.106.028233. [DOI] [PubMed] [Google Scholar]
- 218.Haimeur A, Conseil G, Deeley RG, Cole SP. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Curr Drug Metab. 2004;5(1):21–53. doi: 10.2174/1389200043489199. [DOI] [PubMed] [Google Scholar]
- 219.Belinsky MG, Bain LJ, Balsara BB, Testa JR, Kruh GD. Characterization of MOAT-C and MOAT-D, new members of the MRP/cMOAT subfamily of transporter proteins. J Natl Cancer Inst. 1998;90(22):1735–1741. doi: 10.1093/jnci/90.22.1735. [DOI] [PubMed] [Google Scholar]
- 220.McAleer MA, Breen MA, White NL, Matthews N. pABC11 (also known as MOAT-C and MRP5), a member of the ABC family of proteins, has anion transporter activity but does not confer multidrug resistance when overexpressed in human embryonic kidney 293 cells. J Biol Chem. 1999;274(33):23541–23548. doi: 10.1074/jbc.274.33.23541. [DOI] [PubMed] [Google Scholar]
- 221.Ritter CA, Jedlitschky G, Meyer zu Schwabedissen H, Grube M, Kock K, Kroemer HK. Cellular export of drugs and signaling molecules by the ATP-binding cassette transporters MRP4 (ABCC4) and MRP5 (ABCC5) Drug Metab Rev. 2005;37(1):253–278. doi: 10.1081/dmr-200047984. [DOI] [PubMed] [Google Scholar]
- 222.Reid G, Wielinga P, Zelcer N, De Haas M, Van Deemter L, Wijnholds J, et al. Characterization of the transport of nucleoside analog drugs by the human multidrug resistance proteins MRP4 and MRP5. Mol Pharmacol. 2003;63(5):1094–1103. doi: 10.1124/mol.63.5.1094. [DOI] [PubMed] [Google Scholar]
- 223.Kool M, van der Linden M, de Haas M, Baas F, Borst P. Expression of human MRP6, a homologue of the multidrug resistance protein gene MRP1, in tissues and cancer cells. Cancer Res. 1999;59(1):175–182. [PubMed] [Google Scholar]
- 224.Madon J, Hagenbuch B, Landmann L, Meier PJ, Stieger B. Transport function and hepatocellular localization of mrp6 in rat liver. Mol Pharmacol. 2000;57(3):634–641. doi: 10.1124/mol.57.3.634. [DOI] [PubMed] [Google Scholar]
- 225.Belinsky MG, Chen ZS, Shchaveleva I, Zeng H, Kruh GD. Characterization of the drug resistance and transport properties of multidrug resistance protein 6 (MRP6, ABCC6) Cancer Res. 2002;62(21):6172–6177. [PubMed] [Google Scholar]
- 226.Hopper E, Belinsky MG, Zeng H, Tosolini A, Testa JR, Kruh GD. Analysis of the structure and expression pattern of MRP7 (ABCC10), a new member of the MRP subfamily. Cancer Lett. 2001;162(2):181–191. doi: 10.1016/s0304-3835(00)00646-7. [DOI] [PubMed] [Google Scholar]
- 227.Takayanagi S, Kataoka T, Ohara O, Oishi M, Kuo MT, Ishikawa T. Human ATP-binding cassette transporter ABCC10: expression profile and p53-dependent upregulation. J Exp Ther Oncol. 2004;4(3):239–246. [PubMed] [Google Scholar]
- 228.Chen ZS, Hopper-Borge E, Belinsky MG, Shchaveleva I, Kotova E, Kruh GD. Characterization of the transport properties of human multidrug resistance protein 7 (MRP7, ABCC10) Mol Pharmacol. 2003;63(2):351–358. doi: 10.1124/mol.63.2.351. [DOI] [PubMed] [Google Scholar]
- 229.Bessho Y, Oguri T, Ozasa H, Uemura T, Sakamoto H, Miyazaki M, et al. ABCC10/MRP7 is associated with vinorelbine resistance in non-small cell lung cancer. Oncol Rep. 2009;21(1):263–268. doi: 10.3892/or_00000217. [DOI] [PubMed] [Google Scholar]
- 230.Hopper-Borge E, Xu X, Shen T, Shi Z, Chen Z-S, Kruh GD. Human multidrug resistance protein 7 (ABCC10) is a resistance factor for nucleoside analogues and epothilone B. Cancer Res. 2009;69(1):178–184. doi: 10.1158/0008-5472.CAN-08-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hopper-Borge EA, Churchill T, Paulose C, Nicolas E, Jacobs JD, Ngo O, et al. Contribution of Abcc10 (Mrp7) to in vivo paclitaxel resistance as assessed in Abcc10−/− mice. Cancer Res. 2011;71(10):3649–3657. doi: 10.1158/0008-5472.CAN-10-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hu S, Niu H, Inaba H, Orwick S, Rose C, Panetta JC, et al. Activity of the multikinase inhibitor sorafenib in combination with cytarabine in acute myeloid leukemia. J Natl Cancer Inst. 2011;103(11):893–905. doi: 10.1093/jnci/djr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Malofeeva EV, Domanitskaya N, Gudima M, Hopper-Borge EA. Modulation of the ATPase and transport activities of broad-acting multidrug resistance factor ABCC10 (MRP7) Cancer Res. 2012;72(24):6457–6467. doi: 10.1158/0008-5472.CAN-12-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Oguri T, Ozasa H, Uemura T, Bessho Y, Miyazaki M, Maeno K, et al. MRP7/ABCC10 expression is a predictive biomarker for the resistance to paclitaxel in non-small cell lung cancer. Mol Cancer Ther. 2008;7(5):1150–1155. doi: 10.1158/1535-7163.MCT-07-2088. [DOI] [PubMed] [Google Scholar]
- 235.Rudin D, Li L, Niu N, Kalari KR, Gilbert JA, Ames MM, et al. Gemcitabine cytotoxicity: interaction of efflux and deamination. J Drug Metab Toxicol. 2011;2(107):1–10. doi: 10.4172/2157-7609.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Anreddy N, Patel A, Sodani K, Kathawala RJ, Chen EP, Wurpel JN, et al. PD173074, a selective FGFR inhibitor, reverses MRP7 (ABCC10)-mediated MDR. Acta Pharm Sin B. 2014;4(3):202–207. doi: 10.1016/j.apsb.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Tiwari AK, Sodani K, Dai CL, Abuznait AH, Singh S, Xiao ZJ, et al. Nilotinib potentiates anticancer drug sensitivity in murine ABCB1-, ABCG2-, and ABCC10-multidrug resistance xenograft models. Cancer Lett. 2013;328(2):307–317. doi: 10.1016/j.canlet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bera TK, Lee S, Salvatore G, Lee B, Pastan I. MRP8, a new member of ABC transporter superfamily, identified by EST database mining and gene prediction program, is highly expressed in breast cancer. Mol Med. 2001;7(8):509–516. doi: 10.1007/BF03401856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Bortfeld M, Rius M, Konig J, Herold-Mende C, Nies AT, Keppler D. Human multidrug resistance protein 8 (MRP8/ABCC11), an apical efflux pump for steroid sulfates, is an axonal protein of the CNS and peripheral nervous system. Neuroscience. 2006;137(4):1247–1257. doi: 10.1016/j.neuroscience.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 240.Chen ZS, Guo Y, Belinsky MG, Kotova E, Kruh GD. Transport of bile acids, sulfated steroids, estradiol 17-beta-D-glucuronide, and leukotriene C4 by human multidrug resistance protein 8 (ABCC11) Mol Pharmacol. 2005;67(2):545–557. doi: 10.1124/mol.104.007138. [DOI] [PubMed] [Google Scholar]
- 241.Honorat M, Terreux R, Falson P, Di Pietro A, Dumontet C, Payen L. Localization of putative binding sites for cyclic guanosine monophosphate and the anti-cancer drug 5-fluoro-2′-deoxyuridine-5′-monophosphate on ABCC11 in silico models. BMC Struct Biol. 2013;13:7. doi: 10.1186/1472-6807-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Guo Y, Kotova E, Chen ZS, Lee K, Hopper-Borge E, Belinsky MG, et al. MRP8, ATP-binding cassette C11 (ABCC11), is a cyclic nucleotide efflux pump and a resistance factor for fluoropyrimidines 2′,3′-dideoxycytidine and 9′-(2′-phosphonylmethoxyethyl)adenine. J Biol Chem. 2003;278(32):29509–29514. doi: 10.1074/jbc.M304059200. [DOI] [PubMed] [Google Scholar]
- 243.Goler-Baron V, Assaraf YG. Structure and function of ABCG2-rich extracellular vesicles mediating multidrug resistance. PLoS One. 2011;6(1):e16007. doi: 10.1371/journal.pone.0016007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Robey RW, Polgar O, Deeken J, To KW, Bates SE. ABCG2: determining its relevance in clinical drug resistance. Cancer Metastasis Rev. 2007;26(1):39–57. doi: 10.1007/s10555-007-9042-6. [DOI] [PubMed] [Google Scholar]
- 245.Kruh GD, Zeng H, Rea PA, Liu G, Chen ZS, Lee K, et al. MRP subfamily transporters and resistance to anticancer agents. J Bioenerg Biomembr. 2001;33(6):493–501. doi: 10.1023/a:1012827221844. [DOI] [PubMed] [Google Scholar]
- 246.Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst. 2000;92(16):1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- 247.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 248.Rost D, Konig J, Weiss G, Klar E, Stremmel W, Keppler D. Expression and localization of the multidrug resistance proteins MRP2 and MRP3 in human gallbladder epithelia. Gastroenterology. 2001;121(5):1203–1208. doi: 10.1053/gast.2001.28648. [DOI] [PubMed] [Google Scholar]
- 249.Kochel TJ, Reader JC, Ma X, Kundu N, Fulton AM. Multiple drug resistance-associated protein (MRP4) exports prostaglandin E2 (PGE2) and contributes to metastasis in basal/triple negative breast cancer. Oncotarget. 2017;8(4):6540–6554. doi: 10.18632/oncotarget.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Cai J, Daoud R, Alqawi O, Georges E, Pelletier J, Gros P. Nucleotide binding and nucleotide hydrolysis properties of the ABC transporter MRP6 (ABCC6) Biochemistry. 2002;41(25):8058–8067. doi: 10.1021/bi012082p. [DOI] [PubMed] [Google Scholar]
- 251.Kruh GD, Guo Y, Hopper-Borge E, Belinsky MG, Chen ZS. ABCC10, ABCC11, and ABCC12. Pflugers Arch. 2007;453(5):675–684. doi: 10.1007/s00424-006-0114-1. [DOI] [PubMed] [Google Scholar]
- 252.Deng W, Dai CL, Chen JJ, Kathawala RJ, Sun YL, Chen HF, et al. Tandutinib (MLN518) reverses multidrug resistance by inhibiting the efflux activity of the multidrug resistance protein 7 (ABCC10) Oncol Rep. 2013;29(6):2479–2485. doi: 10.3892/or.2013.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sun YL, Chen JJ, Kumar P, Chen K, Sodani K, Patel A, et al. Reversal of MRP7 (ABCC10)-mediated multidrug resistance by tariquidar. PLoS One. 2013;8(2):e55576. doi: 10.1371/journal.pone.0055576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Namee NM, O'Driscoll L. Extracellular vesicles and anti-cancer drug resistance. Biochim Biophys Acta Rev Cancer. 2018;1870(2):123–136. doi: 10.1016/j.bbcan.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 255.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 257.Sousa D, Lima RT, Vasconcelos MH. Intercellular transfer of cancer drug resistance traits by extracellular vesicles. Trends Mol Med. 2015;21(10):595–608. doi: 10.1016/j.molmed.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 258.Samuel P, Fabbri M, Carter DRF. Mechanisms of drug resistance in cancer: the role of extracellular vesicles. Proteomics. 2017;17(23–24). 10.1002/pmic.201600375. [DOI] [PubMed]
- 259.Gong J, Luk F, Jaiswal R, George AM, Grau GE, Bebawy M. Microparticle drug sequestration provides a parallel pathway in the acquisition of cancer drug resistance. Eur J Pharmacol. 2013;721(1–3):116–125. doi: 10.1016/j.ejphar.2013.09.044. [DOI] [PubMed] [Google Scholar]
- 260.Bouige P, Laurent D, Piloyan L, Dassa E. Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr Protein Pept Sci. 2002;3(5):541–559. doi: 10.2174/1389203023380486. [DOI] [PubMed] [Google Scholar]
- 261.Dano K. Active outward transport of daunomycin in resistant Ehrlich ascites tumor cells. Biochim Biophys Acta. 1973;323(3):466–483. doi: 10.1016/0005-2736(73)90191-0. [DOI] [PubMed] [Google Scholar]
- 262.Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol. 2004;11(10):918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 263.Locher KP. Structure and mechanism of ABC transporters. Curr Opin Struct Biol. 2004;14(4):426–431. doi: 10.1016/j.sbi.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 264.van der Does C, Tampe R. How do ABC transporters drive transport? Biol Chem. 2004;385(10):927–933. doi: 10.1515/BC.2004.121. [DOI] [PubMed] [Google Scholar]
- 265.Schmitt L, Tampe R. Structure and mechanism of ABC transporters. Curr Opin Struct Biol. 2002;12(6):754–760. doi: 10.1016/s0959-440x(02)00399-8. [DOI] [PubMed] [Google Scholar]
- 266.Lage H. ABC-transporters: implications on drug resistance from microorganisms to human cancers. Int J Antimicrob Agents. 2003;22(3):188–199. doi: 10.1016/s0924-8579(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 267.Kerr ID. Structure and association of ATP-binding cassette transporter nucleotide-binding domains. Biochim Biophys Acta. 2002;1561(1):47–64. doi: 10.1016/s0304-4157(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 268.Jones PM, George AM. Subunit interactions in ABC transporters: towards a functional architecture. FEMS Microbiol Lett. 1999;179(2):187–202. doi: 10.1111/j.1574-6968.1999.tb08727.x. [DOI] [PubMed] [Google Scholar]
- 269.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Acad Sci U S A. 2007;104(48):19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lambert G, Estevez-Salmeron L, Oh S, Liao D, Emerson BM, Tlsty TD, et al. An analogy between the evolution of drug resistance in bacterial communities and malignant tissues. Nat Rev Cancer. 2011;11(5):375–382. doi: 10.1038/nrc3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Viswanathan A, You HJ, Doetsch PW. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284(5411):159–162. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- 272.Choi YH, Yu AM. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr Pharm Des. 2014;20(5):793–807. doi: 10.2174/138161282005140214165212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.