Abstract
The Pharmacogene Variation Consortium (PharmVar) catalogues star (*) allele nomenclature for the polymorphic human CYP2C9 gene. Genetic variation within the CYP2C9 gene locus impacts the metabolism or bioactivation of many clinically important drugs including NSAIDs, phenytoin, anti-diabetic agents and angiotensin receptor blocker. Variable CYP2C9 activity is of particular importance regarding efficacy and safety of warfarin and siponimod as indicated in their package inserts. This GeneFocus provides a comprehensive overview and summary of CYP2C9 and describes how haplotype information catalogued by PharmVar is utilized by the Pharmacogenomics Knowledgebase (PharmGKB) and the Clinical Pharmacogenetics Implementation Consortium (CPIC).
CYP2C9 Brief History
CYP2C9 is the most abundantly expressed human CYP2C isoform in the liver (1, 2). Data from the 1970s suggested that polymorphic expression affects metabolism of tolbutamide (3) but was not related to CYP2D6 (4). A combination of protein purification and cDNA cloning approaches eventually identified CYP2C9 as the enzyme responsible for tolbutamide hydroxylation in humans (5, 6). A role for this enzyme in phenytoin hydroxylation was also demonstrated (7). Initially it was thought that the CYP2C9 gene product was also responsible for mephenytoin metabolism, but this was refuted in 1991 after the discovery of CYP2C19 (8). Subsequent studies on S-warfarin and diclofenac hydroxylation demonstrated that CYP2C9 was the main enzyme responsible for these reactions (9, 10). After that, many additional CYP2C9 substrates spanning a variety of drug classes, have been identified and are further discussed below.
The existence of CYP2C9 variants became evident when comparing cDNA sequences that initially pointed to the existence of two common nonsynonymous variants (c.430C>T, p.R144C, rs1799853 and c.1075A>C, p.I359L, rs1057910), which are now defining variants of the haplotypes described as CYP2C9*2 and CYP2C9*3, respectively (11). In the early studies of these variants, decreased metabolism of tolbutamide and S-warfarin in vivo and in vitro correlated with the presence of CYP2C9*2 and *3 alleles (12, 13). Further sequencing during the early 1990s resulted in the discovery of additional variants, including the decreased function CYP2C9*5 allele (c.1080C>G, p.D360E, rs28371686) (14) and the nonfunctional CYP2C9*6 allele (c.818delA, L273frameshift, rs9332131) (15) among others (16).
Another milestone in the history of CYP2C9 is the US Food and Drug Administration (FDA) revision of labeling for two drugs, warfarin with a boxed warning added based in part on CYP2C9 pharmacogenetics and siponimod (MAYZENT®) with testing required due to a contraindication for patients with a CYP2C9*3/*3 genotype. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for various CYP2C9 gene-drug pairs including warfarin, phenytoin and nonsteroidal anti-inflammatory drugs (NSAIDs) have been published (17–19). Guidelines have also been published by the Royal Dutch Pharmacogenetics Working Group (DPWG) and the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) information for those can be accessed through PharmGKB (20).
Status of Nomenclature before PharmVar
CYP2C9 star allele nomenclature was maintained by the Human Cytochrome P450 Allele Nomenclature Database since 2000 until its transition to PharmVar in 2017 (21). The pharmacogenetics community accepted this as the central resource for cataloguing CYP variation and it was utilized by knowledge resources (e.g., PharmGKB), the pharmacogenetics testing and implementation communities, including clinical genetic testing laboratories and the CPIC, as well as to inform research.
A total of 60 CYP2C9 haplotypes, CYP2C9*1 through *60, were submitted to this database before it was transitioned to the Pharmacogene Variation (PharmVar) Consortium in 2017 (22). Although only exons were required to be sequenced for submission, sequence variants in the upstream gene region or in flanking introns, may also have been submitted and included in haplotype definitions. These regions were, however, not consistently interrogated and captured by the database. Initially, so-called ‘suballeles’, e.g., CYP2C9*1A-D, were catalogued, but eventually no longer considered for independent naming. PharmVar has reviewed all transitioned star alleles and updated or revised their definitions as necessary to conform to PharmVar standards (23).
In this review, star (*) allele sequence variants are shown according to their relative position in the CYP2C9 NM_000771.4 reference transcript sequence with the ‘A’ of the ATG translation start codon being +1; corresponding protein coordinates are also provided. For example, the CYP2C9*2 allele-defining variant (rs1799853) is referred to as c.430C>T (p.R144C) indicating that this variant causes an arginine to cysteine change at amino acid position 144.
Resources cited throughout this review are summarized in Table 1.
Table 1.
Online CYP2C9 Resources - Links to Sites and Online Resources Referenced Throughout the Review
| Sources and References | References |
|---|---|
| PharmVar | |
CYP2C9 Gene Page
|
(94) |
| Standards | (23) |
| Allele Designation and Evidence Level Criteria Document | (81) |
| CYP2C9 Gene Expert Panel Roster | (82) |
| P450 Nomenclature Site – Archive | (95) |
| PharmGKB | |
| CYP2C9 Gene Page | (20) |
Gene-Specific Information Tables for CYP2C9
|
(36) |
| CYP2C9 Drug Label Annotations | (24) |
| PGx Publication Tips | (86) |
| CPIC | |
| Guidelines Standard Operating Procedure (SOP) for Assigning Allele Function Standardized terms for clinical pharmacogenetic test results Gene/Drug Pairs
|
(96) (80) (66) (27) |
| FDA | |
| FDA Pharmacogenomic Biomarkers in Drug Labeling | (25, 40) |
| FDA Table of Pharmacogenetic Associations | (26) |
| Other Resources | |
| Drug Interactions Flockhart Table™ | (40) |
| GTR:Genetic Testing Registry | (76) |
| Human Genome Variation Society (HGVS) Nomenclature | (97) |
| NCBI Reference Sequences Database | (83) |
| Locus Reference Genomic (LRG) Project | (98) |
| Genetic Testing Reference Materials Coordination Program (GeT-RM) | (87) |
Clinical Relevance
The CYP2C9 enzyme is part of the CYP450 superfamily contributing to the metabolism of many clinically used drugs, including warfarin, phenytoin, multiple NSAIDs (e.g. celecoxib, flurbiprofen, lornoxicam, ibuprofen, piroxicam, tenoxicam, and meloxicam), losartan, irbesartan, sulfonylureas (e.g. tolbutamide and gliclazide) and siponimod. Additional information on drugs metabolized by CYP2C9 can be found in the PharmGKB drug label annotations (24), the FDA Table of Pharmacogenomic Biomarkers in Drug Labeling (25), the FDA Table of Pharmacogenetic Associations (26), and the CPIC drug-gene pairs (27). CYP2C9 polymorphisms have significant consequences for narrow therapeutic index drugs, including warfarin and phenytoin (17, 19). In contrast, while CYP2C9 variation influences the oral clearance of losartan (28) it is not clear whether this translates into clinically significant effects.
The most common clinical application of CYP2C9 genotype information described to date is its use, together with VKORC1 and possibly CYP4F2, to guide warfarin dosing (17). Individuals with one or two decreased or no function alleles have decreased metabolism of the more potent S-enantiomer of warfarin and increased risk of bleeding with usual warfarin doses (i.e., 5 mg per day) and thus, a lower warfarin dose is required to achieve therapeutic anticoagulation (17, 29). Three multi-site clinical trials have examined the efficacy of CYP2C9 plus VKORC1 genotype-guided warfarin of which two trials demonstrated favorable effects of a genotype-guided approach on the outcome of improved anticoagulation control (30) or reduction in risk for bleeding, thromboembolism, death or supratherapeutic anticoagulation following total joint arthroplasty (31). The third trial, COAG, showed no improvement in anticoagulation control with a genotype-guided approach (32). However, in contrast to the other trials that were conducted predominantly in European ancestry populations, nearly 30% of participants in the COAG trial were of African ancestry, in whom genotype-guided dosing led to worse anticoagulation control compared to a non-genotype guided approach. All three trials limited genotyping to the CYP2C9*2 and *3 alleles, which are the most common decreased or no function alleles in European ancestry patients but occur less often in those with African ancestry. The CYP2C9*5, *6, *8 (c.449G>A, p.R150H, rs7900194) and *11 (c.1003C>T, p.R335W, rs28371685) alleles are predominantly found in people with African ancestry, and data show that not accounting for these alleles leads to significant over-estimation of warfarin dose requirements in African Americans (33). Failure to account for these alleles in the COAG trial may have contributed to the trial’s negative outcomes. CPIC guidelines were revised in 2017 to state that genotype should only be used to guide warfarin dosing in African ancestral patients if the CYP2C9*5, *6, *8, and *11 alleles are tested (17). The Association for Molecular Pathology (AMP) includes these variants, in addition to the CYP2C9*2 and *3 alleles, on their Tier 1 allele recommendations (34). CYP2C9 testing options are discussed in detail elsewhere (35). Additional CYP2C9 alleles may be common in other populations worldwide (36).
CYP2C9 decreased and no function alleles similarly lead to increased exposure to other CYP2C9 substrates, which may increase the risk for serious adverse effects, including neurotoxicity with phenytoin, gastrointestinal bleeding (37) and adverse cardiovascular effects with NSAIDs, and bradycardia with siponimod (38). Siponimod is contraindicated for poor metabolizers (PM) with the CYP2C9*3/*3 genotype, who are expected to have little to no enzyme activity. Thus, genotyping is required prior to siponimod initiation (39). Lower phenytoin maintenance doses are recommended for patients with genotypes associated with significant reductions in enzyme activity (19). For NSAIDs, the consequences of decreased CYP2C9-mediated metabolism are expected to be greatest for drugs with a long elimination half-life (e.g., piroxicam, tenoxicam, melocoxicam) and less significant for NSAIDs with shorter half-lives (e.g., celecoxib and ibuprofen). This is reflected by CPIC guidelines recommending that in PMs (e.g., CYP2C9*2/*3 or *3/*3 genotypes), piroxicam, tenoxicam and melocoxicam be avoided, whereas celecoxib and ibuprofen may be used but should be started at lower than usual doses (18).
Other Factors that can Influence CYP2C9 activity
CYP2C9 is inhibited and induced by a wide range of drugs. Sulfamethoxazole, fluconazole, voriconazole, metronidazole, and amiodarone are examples of CYP2C9 inhibitors (40). Amiodarone represents a risk for anticoagulated patients since it is often prescribed concomitantly with warfarin. The drug-drug interaction between amiodarone and warfarin is well documented (41) and is associated with a 6–65% decrease in warfarin dose requirement (42). On the other hand, known inducers of CYP2C9 are rifampicin, clotrimazole, nifedipine, hyperforin (an active component of the herbal drug St John’s wort), phenobarbital, phenytoin, carbamazepine, dicloxacillin, flucloxacillin and tamoxifen (43–47). Although several mechanisms are known to be involved in CYP2C9 upregulation, the most prominent is by nuclear receptor binding to cis-regulatory elements in the promoter region (1, 48), such as the pregnane X receptor (PXR), constitutive androstane receptor (CAR), and the glucocorticoid, estrogen and vitamin D (VDR) receptors. Finally, cytochrome b5 has been shown to modulate catalytic activity of CYP2C9 (49), and it has been suggested that variation in its gene (CYB5A) might contribute to variable CYP2C9-mediated drug metabolism.
Age-related and sex differences
Sex-related differences in CYP2C9 expression have been investigated to a limited extent and to date there is no evidence supporting meaningful differences between males and females (50). Data obtained in liver microsomes with tolbutamide as substrate (51, 52) and from phenotyping studies with losartan and tolbutamide as substrates (53, 54) did not reveal differences in activity between males and females, with the exception of one study that found slower metabolism of losartan in females compared with males (53). It has also been reported that females taking oral contraceptives show impairment of losartan metabolism (55), although this observation was not replicated using tolbutamide as substrate (56). However, considering that estrogens upregulate CYP2C9 expression via the estrogen receptor (57), studies on sex differences should consider this possible confounder.
Age-related differences in CYP2C9 expression have been reported. CYP2C9 metabolic activity is low in fetal livers and increases linearly immediately from birth up to 20 years (58, 59); the greatest increase in activity is observed in the first two years of life (60). Findings for the effect of age on CYP2C9 contrast with CYP2C19 where levels appear to be higher in children than in adults (61). Also, there are pediatric-specific recommendations for warfarin treatment based on CYP2C9 and VKORC1 genotype, but these only apply to patients of European ancestry and are limited to the CYP2C9*2 and *3 alleles (17).
CYP2C9 and the Pharmacogenomics Knowledge Base (PharmGKB)
The PharmGKB collects, curates and disseminates knowledge about the impact of human genetic variation on drug response (62). The PharmGKB CYP2C9 gene page allows structured access to gene-specific pharmacogenomic knowledge (20). Information is presented in sections including prescribing information, drug label annotations, clinical annotations, variant annotations, and curated pathways. Prescribing information encompasses: 1) annotations of clinical guidelines from sources such as CPIC, DPWG, CPNDS, 2) a list of drug labels that include prescribing information for a CYP2C9 genotype or phenotype with respective excerpts and links to the label annotation (see below), and 3) “Rx study annotations” that provide genotype-based drug dosing reported in individual journal articles. Seven CPIC guideline annotations (one providing no dosing recommendations), nine DPWG guideline annotations (3 with recommendations and 6 providing no drug-specific recommendations) and 46 “Rx study annotations” are available as of March 2021 for CYP2C9 with overlapping CYP2C9-drug pairs.
PharmGKB extracts pharmacogenomic-relevant information from agency-approved drug labels and applies a “pharmacogenetics (PGx) level” category, based on the level of action implied in each label (e.g., Testing required, Actionable PGx, Testing recommended or Informative PGx). On the CYP2C9 page, annotations can be accessed (as of March 2021) for 15 US FDA approved labels, 3 European Medicines Agency (EMA) approved labels, 1 Pharmaceuticals and Medical Devices Agency, Japan (PMDA) approved label, 2 Health Canada (HCSC) approved labels, and 8 Swiss Agency of Therapeutic Products (Swissmedic) approved labels. Currently, PharmGKB contains 68 CYP2C9 related clinical annotations, which are evidence-rated genotype level summaries for specific variant/allele-drug combinations based on curated literature (variant annotations). Pharmacokinetic pathways depicting CYP2C9 in drug metabolism are available for 36 drugs, although the significance for CYP2C9 involvement varies by drug. PharmGKB and CPIC work collaboratively to develop gene-specific resources that accompany each CPIC guideline, including allele definition mapping, allele functionality, allele frequency, and diplotype to phenotype mapping files in a standardized format. Gene-specific information tables for CYP2C9 are available from PharmGKB (36). In addition, the Pharmacogenomics Clinical Annotation Tool (PharmCAT) is currently under development to facilitate the interpretation and reporting of pharmacogenomic-based dosing recommendations, including those for CYP2C9 (63).
CYP2C9 and the Clinical Pharmacogenetics Implementation Consortium (CPIC)
The CPIC develops structured, evidence-based, clinical practice guidelines for drugs affected by pharmacogenetic variation (64, 65). Several CYP2C9 gene-drug pairs have been prioritized through consideration of multiple factors such as the available body of literature, severity of clinical consequences, availability of alternative therapies and whether a prescribing change (drug choice or dose) is warranted. To date, three CPIC guidelines have been published that include CYP2C9: warfarin (17), phenytoin/fosphenytoin (19), and NSAIDs (18). Each guideline has multiple components, including CYP2C9 phenotype-specific therapeutic recommendations, a systematic evidence review, and implementation resources to support the translation of the guideline into electronic health records (EHRs) with example clinical decision support (CDS) text.
Genotype to Phenotype Translation
An individual has two CYP2C9 haplotypes, one on each chromosome, which constitutes a diplotype. For example, a CYP2C9*2/*3 diplotype assignment indicates that one chromosome (or allele) has a single nucleotide variation (SNV) defining the CYP2C9*2 haplotype and the second chromosome (or allele) has a SNV defining the CYP2C9*3 haplotype.
For CYP2C9 phenotypic classification, individuals are categorized into the following CPIC-recommended phenotype categories: poor (PM), intermediate (IM), and normal (NM) (formerly EM, extensive) metabolizers (66). To facilitate CYP2C9 genotype to phenotype translation the Activity Score (AS) system first developed for the CYP2D6 gene (67) has been adopted. Briefly, a value of 0, 0.5, or 1 is assigned to each allele reflecting no function, decreased function or normal function, respectively. The sum of the values assigned to each allele is a diplotype AS. For example, CYP2C9*2/*3 has values of 0.5 (CYP2C9*2, decreased function allele) plus 0 (CYP2C9*3, no function allele) giving rise to an AS of 0.5. Individuals with activity scores of 0 and 0.5 are categorized as PMs, AS of 1 and 1.5 as IMs, and AS of 2 as NMs. By utilizing the AS system, different prescribing guidance can be provided within a phenotype category to tailor specific allele function combinations. For example, in the CPIC guideline for CYP2C9 and nonsteroidal anti-inflammatory drugs (18), the recommendation differs between an AS of 1 (a combination of two decreased function alleles or one no function allele with a normal function allele) and an AS of 1.5 (a combination of a normal function allele with a decreased function) within the CYP2C9 IM category. The Diplotype-Phenotype-Table provided by the PharmGKB and CPIC serves as a reference for calculating the AS of each genotype (36).
Need for standardized genetic variation definitions and reporting of functional/clinical impact
In order to guide drug therapy, it is imperative to understand how CYP2C9 allelic variation can impact CYP2C9 function. Although many alleles have been observed in phenotypic PMs and their causative genetic variations described (e.g., CYP2C9*2, *3, and *4, etc.), some alleles have unknown or uncertain function. In vitro characterization of allelic variants often produces results that are inconsistent among test systems that may be attributed to differences in the substrates used and between the experimental variables (see CYP2C9 functionality table for a detailed summary (36)). Although in silico prediction tools are improving, in vivo validation is still the gold standard. Therefore, for any given allele (except for those shown to completely abolish activity) caution should be taken when extrapolating functional data from one drug or substrate to another. Ideally, one would be able to assess the in vivo function of each individual CYP2C9 haplotype with each individual CYP2C9 substrate, which would refine the phenotype predicting capacity of CYP2C9 genetic testing. In addition, co-medications (drug–drug interactions) may not affect all CYP2C9 variants equally, and there is still limited or no information regarding genetic variability for many minority populations.
For many drug metabolizing enzymes, the combination of sequence variations that define haplotypes is critical to precisely predict enzyme function. A notable example is CYP2C9*3. Its defining variant (c.1075A>C, p.I359L, rs1057910) is also present in CYP2C9*18, which additionally harbors a missense variant (c.1190A>C, p.D397A, rs72558193).
As previously reported in the CYP2D6 GeneFocus (68), clinical pharmacogenetic programs have successfully been implemented over the past years, but numerous challenges remain to accelerate adoption (69). Standardization is a key area that continues to represent an opportunity for all pharmacogenetic stakeholders to improve upon, including for laboratory processes, test ordering, result reporting, and data representation. This is in alignment with recent reports, which emphasized that clinically actionable pharmacogenetic information must be accurately represented in electronic health records by using a harmonized system for genotype and phenotype information (70, 71). Although many pharmacogenetic laboratories utilize star nomenclature as recommended by PharmVar and CPIC, inter-laboratory differences in testing approaches and reporting remain. Clinical testing for CYP2C9 can be performed on a variety of platforms using different methodologies and genotyping data can be reported in different ways, such as chromosomal or genomic position on reference sequence (RefSeq), amino acid change, dbSNP rsID, and/or using star (*) allele nomenclature (21, 72).
A study performed by the Genetic Testing Reference Material Program (GeT-RM) concluded that many pharmacogenetic variants interrogated were not consistent across commercial and laboratory platforms (73). Similar findings were reported by Moyer et al., when they surveyed laboratories offering pharmacogenetic services for CYP2D6 and CYP2C19 genotyping (74). To address this, the AMP and College of American Pathologists (CAP) have recently published recommendations for alleles to be included in clinical CYP2C9 testing (34). In brief, the recommendations are based on allele function, allele frequencies across populations and ethnicities, and the availability of reference materials. The AMP working group recommended Tier 1 alleles (minimum panel of variant alleles), which includes CYP2C9*2, *3, *5, *6, *8, and *11, and Tier 2 (extended panel of variant alleles) that includes CYP2C9*12 (c.1465C>T, p.P489S, rs9332239), *13 (c.269T>C, p.L90P, rs72558187) and *15 (c.485C>A, p.S162X, rs72558190). The utilization of star allele nomenclature as provided by PharmVar (75) will not only ensure that each star allele represents a unique and fully defined haplotype but also minimize “mis-interpretation” of a genotype result and its clinical implication(s).
The two end-user groups benefiting the most from standardized allele designations are clinicians and patients. Standardized terms and language will help clinicians to convey and explain results and patients to understand what the results mean for them. Consistent nomenclature is critical for the integration of pharmacogenetics into EHR, as well as for the establishment of clinical decision support algorithms and the design of clinical support tools such as interruptive alerts (70, 71). For example, drug/allele combinations for alerts require systematic annotations within the EHR, using standardized nomenclature and terms. Finally, the analysis of pharmacogenetic clinical correlations will benefit from harmonized nomenclature of the gene variants, as well as support consistent test interpretation by providers across healthcare systems.
The CYP2C9 Gene Locus
The CYP2C9 gene is a member of the CYP2C family, has 9 exons and is translated into 490 amino acids. The gene is located on chromosome 10q23.33 spanning a region of approximately 390 kb. In addition to CYP2C9 the locus also harbors CYP2C8, CYP2C18 and CYP2C19 (Figure 1). Genotyping assays need to employ CYP2C9-specific regions for primer design (e.g., using intronic sequences) to avoid amplification from any of the other genes in the locus, in particular CYP2C19. Many commercial laboratories offer CYP2C9 testing (76) using a variety of test platforms.
Figure 1. Overview of the gene locus and allelic variation.
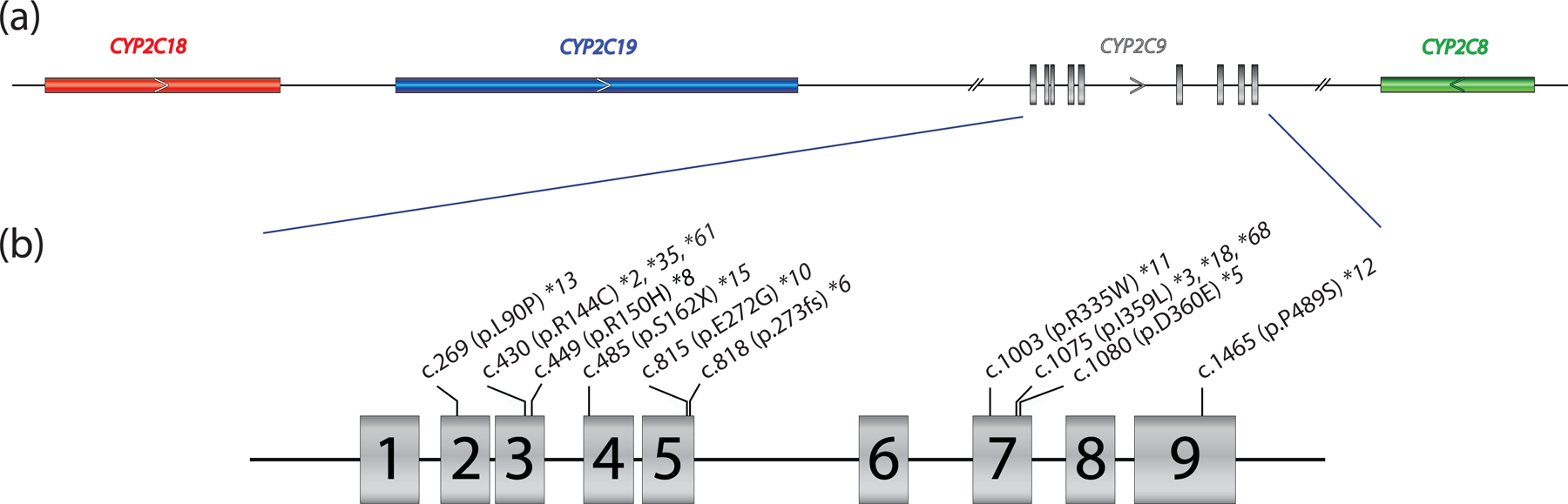
Panel (a) provides a graphical overview of the CYP2C gene locus containing CYP2C18, CYP2C19, CYP2C9, and CYP2C8. The latter is encoded on the reverse strand (arrow) while the other three genes are encoded on the forward strand. CYP2C9 is composed of nine exons, covering 51.5 kb (GRCh37, 96,698,415 – 96,749,147; GRCh38, 94,938,658 – 94,990,091). Panel (b) shows the core sequence variations defining CYP2C9*2,*3, *5, *6, *8, *10, *11, *12, *13 and *15 representing Tier 1 and Tier 2 alleles per AMP allele recommendation guidelines (34). p.R144C is also present on CYP2C9*35 and *61 and is thus not unique to CYP2C9*2. Likewise, p.I359L is also part of several haplotypes, i.e., CYP2C9*3, *18, and *68.
Although no copy number variation (CNV) alleles have been defined to date, CYP2C9 can, in rare cases, be part of large CNV events affecting the CYP2C locus as described (77) and summarized by the CYP2C19 GeneFocus (61). CNVs may implicate the entire CYP2C9 gene, present as partial gene deletions or duplications, and often additional genes flanking the CYP2C gene locus are also involved. CYP2C9 CNVs present at low frequency in the population (0.009%). CNVs affecting CYP2C9 are not routinely tested and are most often detected as incidental findings by array platforms such as array comparative genomic hybridization (aCGH), performed for unrelated diagnostic purposes.
CYP2C9 allele, genotype, and phenotype frequencies across populations
The CYP2C9 frequency table available at PharmGKB (36) summarizes population-based allele frequencies reported in the literature. Studies were considered for inclusion if 1) the ethnicity of the population was clearly indicated, 2) either allele frequencies or genotype frequencies were reported, 3) the methodology, by which the genes were genotyped was indicated, and 4) the study represented an original publication. The ethnicities/locations reported in the articles were mapped into seven geographically defined groups (American, Central/South Asian, East Asian, European, Near Eastern, Oceanian, and Sub-Saharan African) and two admixed groups (African American/Afro-Caribbean and Latino), using the biogeographical grouping system developed by PharmGKB (78). The CYP2C9 frequency table is periodically updated and contains multiple tabs summarizing ‘allele frequencies by biogeographical group’, ‘diplotype frequencies by biogeographical group’, ‘phenotype frequency’, and ‘references’; the latter describes allele frequencies for each publication included in the listing, which also allows the user to customize allele frequencies as needed. There are, however, limitations regarding the accuracy of allele frequencies as follows: 1) frequencies are based on published allele data (limited or unavailable for some populations and many alleles), 2) most studies test for a limited number of allelic variants that may lead to an underestimation of certain alleles. For example, c.430C>T (p.R144C) is often defaulted to a CYP2C9*2 assignment, although this SNV is also present on CYP2C9*35 and CYP2C9*61 (Figure 2). Likewise, if no SNVs are found, CYP2C9*1 is assigned, which inflates the frequency of this allele. Therefore, all calculations based on allele frequencies are estimates at best and should be used with caution.
Figure 2. Allele assignment by ‘default’, a commonly used strategy.
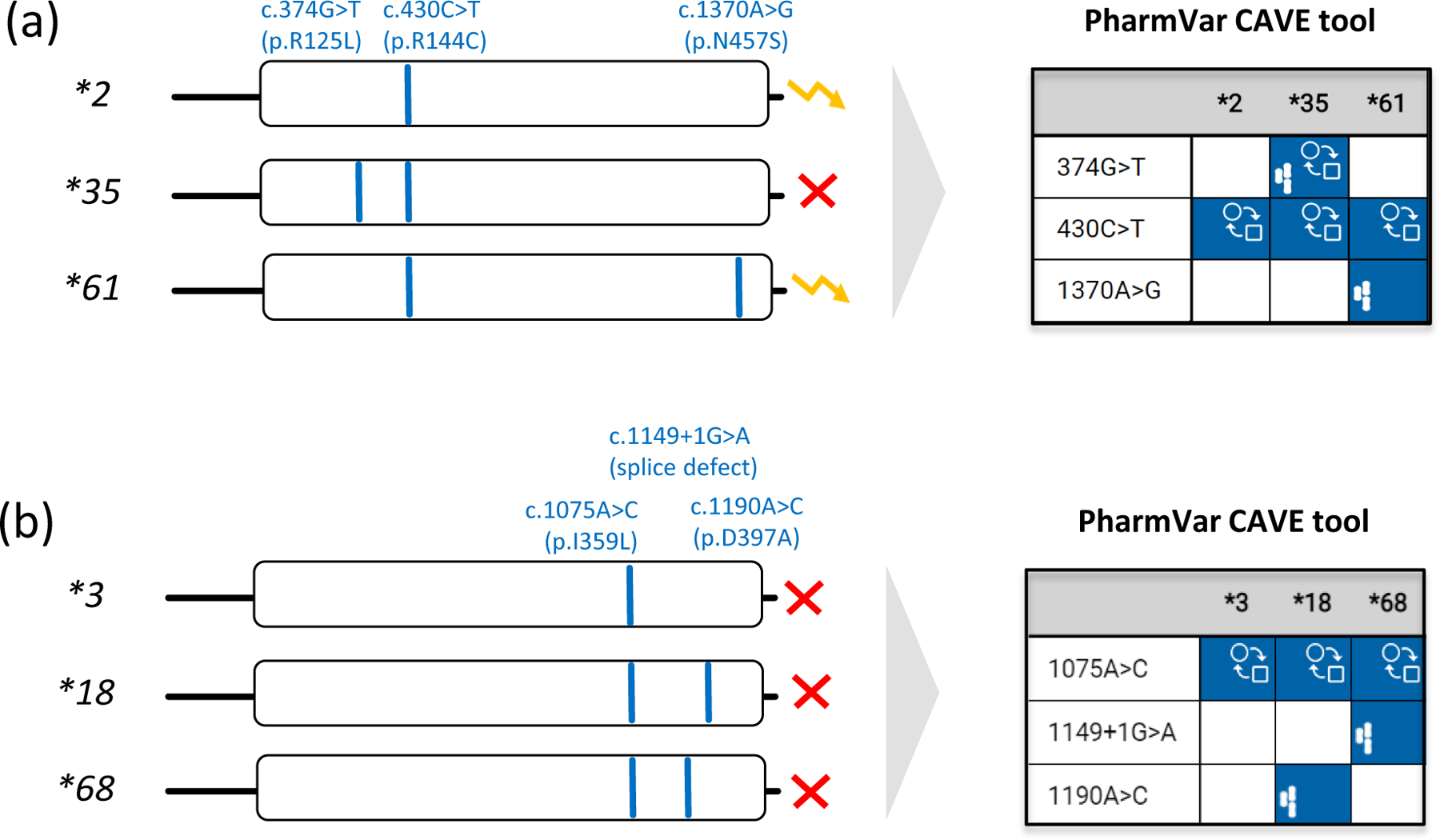
Pharmacogenetic test platforms typically comprise the more commonly observed SNVs and do not test for all star (*) alleles catalogued by PharmVar. Consequently, some alleles may not be identified or receive an assignment by ‘default’. Therefore, it is important to understand which SNVs are tested and how alleles were called and translated into phenotype. Panel (a) shows that the relatively common SNV at c.430C>T (p.R144C) is not only a core SNV of CYP2C9*2, but also *35 and *61. Unless c.374G>T and c.1370A>G are also tested, a CYP2C9*35 or *61 allele will be called as a *2 potentially leading to an incorrect phenotype assignment. Since CYP2C9*2 and *61 are both classified as decreased function by CPIC, defaulting CYP2C9*61 as *2 will not impact the patient’s phenotype call. Panel (b) demonstrates that c.1075A>C (p.I359L) is also present in CYP2C9*18 and the recently discovered *68 alleles. c.1075A>C on its own causes severely decreased function and thus, CYP2C9*3 is classified by CPIC as ‘no function’ allele. CYP2C9*68 has an additional SNV interfering with splicing predicted to be deleterious/probably damaging (93). It remains unknown though whether p.D379A on CYP2C9*18 also impacts function. The yellow ‘zigzag’ arrow and the red X denote decreased and no function, respectively.
Comparative
Allele
ViewEr (CAVE) outputs are shown for both examples to demonstrate the utility of this tool. The graph visualizes which core alleles have the tested SNVs and may be not be identified by limited testing and default allele assignments. Blue boxes indicate the presence of core SNVs. The function ( ) symbol indicates that a core SNV alters function and the PharmVar (
) symbol indicates that a core SNV alters function and the PharmVar ( ) symbol highlights that a core SNV is unique to a star allele. SNV positions refer to the transcript (NM_000771.4).
) symbol highlights that a core SNV is unique to a star allele. SNV positions refer to the transcript (NM_000771.4).
There is considerable variation among the estimated frequencies for individual alleles across and within the biogeographical groups. The decreased function CYP2C9*2 allele has been found at higher frequencies in European (13%), Central/South Asian (11%), Near Eastern (13%), and Latino (8%) populations, but is less frequent in other populations (≤3%). Likewise, CYP2C9*3, a no function allele, is most frequent in Central/South Asian (11%), European (7.6%), and Near Eastern (8.3%), compared to other populations (≤4%). Other allelic variants impacting activity including CYP2C9*5 (≤1.2%), *6 (≤0.9%), *8 (≤7.6%) and *11 (≤2.6%) are most often, but not exclusively, observed in individuals with African ancestry.
Frequency information from the literature is scarce for CYP2C9 alleles across populations, especially for CYP2C9*12 and higher, and limited to CYP2C9*2 and *3 in Oceanians. Absence of a reported allele frequency does not necessarily indicate absence of an allele in that population and does not rule out that someone in that population might have the allele. It is important to keep in mind that the allele frequencies for each biogeographical group are averages of aggregated allele frequencies from multiple publications, each reporting on smaller, more specific study populations. The allele frequencies within each biogeographical group can range widely depending on the specific study population. For example, CYP2C9*2 frequencies reportedly range from 0.5% (Ecuadorian Mestizos) to 19.6% (Brazilian admixed population), with both studies contributing to the allele frequency of the Latino biogeographical group (8%). Similarly, CYP2C9*3 frequencies ranged from 1% in (Malay) to 15% (Vietnamese), both contributing to the allele frequency in the East Asian biogeographical group (4%). Therefore, reported allele frequencies for each biogeographical population is an estimate that cannot be applied to individual patients. Detailed allele frequency information for individual studies and across biogeographical groups can be accessed through the PharmGKB/CPIC Frequency table (36).
Considering CYP2C9*1 through *71, hundreds of allele combinations are possible, and thus, the number of possible genotypes in a population or patient cohort can be quite large. The actual number of combinations that occur in a specific biogeographical population may be significantly less, however, depending on the number of alleles and their frequencies. Phenotype frequencies calculated from the averaged allele frequencies (i.e. not observed phenotype frequencies in populations) across populations are provided in the ‘Phenotype frequency’ tab in the PharmGKB/CPIC CYP2C9 Frequency Table (36). We stress, however, to view all calculated phenotype group frequencies (including those shown in the CYP2C9 gene-specific reference table) with caution due to the limitations regarding the accuracy of allele frequencies, as well as the method used to translate genotype into phenotype and inconsistencies in the classification of ‘population’, ‘ethnicity’, or ‘race’ (79).
CYP2C9 Allele function
PharmVar displays allele clinical function as determined by CPIC, using their respective terms (increased, normal, decreased, no, uncertain or unknown function) (66). Alleles that have not been assigned a function by CPIC are shown as ‘N/A’ (function not assigned). The filter option on the PharmVar CYP2C9 gene page allows the user to sort alleles by functional status.
In 2019, CPIC drafted a standardized protocol that describes in more detail the criteria used for assigning clinical function to alleles that are part of CPIC guidelines to harmonize the process across guidelines (80). CYP2C9 was the first gene for which the developed protocol was applied. A group of CYP2C9 experts discussed literature evidence for each CYP2C9 allele and agreed upon consensus function assignments. It should be noted that CPIC’s primary focus is to assign allele function based on clinical actionability, not solely on molecular or biochemical function. Alleles with no clinical function may have some residual activity which is exemplified by CYP2C9*3. This allele is assigned no function under ‘allele clinical functional status’ but decreased function under ‘allele functional status’ given that its biochemical function is so minimal that, clinically, it is deemed a no function allele.
The expert consensus for allele functions is included in the supporting material for CYP2C9-based CPIC guidelines and can be accessed on PharmGKB (36). The table includes per allele the activity value, allele functional status, allele clinical functional status (displayed on PharmVar), and references that were reviewed during the assignment process.
PharmVar Nomenclature and CYP2C9 allele designation
PharmVar stores and displays allelic data consistently across genes, relying on public standards and data sources wherever possible. The standardized nomenclature follows criteria developed by gene experts. The “Allele Designation and Evidence Level Criteria” document describes the nomenclature system and provides examples (81). For example, a new star (*) number is only issued if a haplotype contains a sequence variant that: 1) results in an amino acid change, e.g., CYP2C9*2 harbors an arginine to cysteine change (c.430C>T, p.R144C); 2) causes a frameshift, e.g., CYP2C9*6 harbors a frameshift at position p.L273 (c.818delA); 3) affects splicing; or 4) changes expression levels causing decreased or increased function. Importantly, new haplotypes that contain previously characterized variants that obliterate gene function are catalogued under the original star (*) allele number as a suballele. For example, any haplotype having a novel variant in addition to the CYP2C9*15 defining c.485C>A (p.S162X) variant will be designated as a *15 suballele, and considered to have no function, regardless of the functional status of the novel variant.
The PharmVar CYP2C9 gene expert panel
International experts representing research, clinical testing, and implementation interests were recruited from PharmVar members to serve on the CYP2C9 expert panel. The panel also includes PharmGKB/CPIC representation to ensure that the nomenclature is consistent with CPIC guidelines and to facilitate dissemination to a greater audience through PharmGKB as well as other databases, such as ClinGen. The composition of the panel can be found on the PharmVar website (82). Table 2 summarizes the alleles reviewed and accepted by the panel to date.
Table 2.
Novel allele(s) and confirmed suballele(s)
| Core Allele Designation | Novel alleles/suballeles |
|---|---|
| *1 | *1.005 – *1.013 |
| *2 | *2.004 |
| *8 | *8.002 – *8.005 |
| *61 | *61.001, *61.002 |
| *62 – *71 | *62.001 – *71.001 |
Submissions for known alleles. Their original haplotype was confirmed, and/or evidence level raised from ‘Limited’ (Lim) or ‘Moderate’ (Mod) to ‘Definitive’ (Def):
*2.001, *3.002, *5.001, *8.001, *9.001, *11.001, *61.002
The PharmVar CYP2C9 gene page
The PharmVar CYP2C9 gene details all allelic variants defined by PharmVar. Sequence variations are mapped to the latest genomic and cDNA reference sequences (RefSeqs), issued by the NCBI Reference Sequences database (83) and the GRCh37 (NC_000010.10) and GRCh38 (NC_000010.11) genome builds. For CYP2C9, the current genomic and transcript RefSeqs are NG_008385.2 and NM_000771.4, respectively. CYP2C9 was transitioned from the original nomenclature site to PharmVar on Sept 26, 2017 (original content is available through the ‘Archive’ link on the PharmVar homepage).
A Locus Reference Genomic (LRG) record has been issued from the LRG Project, a NCBI (RefSeq) and EMBL-EBI (Ensembl/GENCODE) initiative (https://www.lrg-sequence.org/). LRGs were created to serve as reference standards that ‘never change’ or version after their release. The CYP2C9 LRG (LRG_1195) matches 100% with the NG_008385.2 RefSeq and was used by PharmVar instead of NG_008385.2. However, considering the ongoing Matched Annotation from NCBI and EMBL-EBI (MANE) project (https://www.ncbi.nlm.nih.gov/refseq/MANE/), genomic RefSeqs, derived from MANE Select transcripts, are viewed as the new gold standard. Thus, PharmVar will return to using the genomic RefSeq NG_008385.2.
On the PharmVar CYP2C9 gene page, the user can easily cross-reference genomic and cDNA position(s) by choosing the respective reference sequence or genome build of interest; there is also the option of two count modes, i.e., counting from the first nucleotide in the reference sequence or the ATG translation start codon being +1. Variant annotations are now also provided according to HGVS along the more traditional PharmVar display format (84). Figure 3 provides an excerpt of the page illustrating CYP2C9*2 and *8. Additional details and an example are provided in the gene’s Read Me document available through at the PharmVar CYP2C9 gene page.
Figure 3. Overview of core allele and suballele categorization.
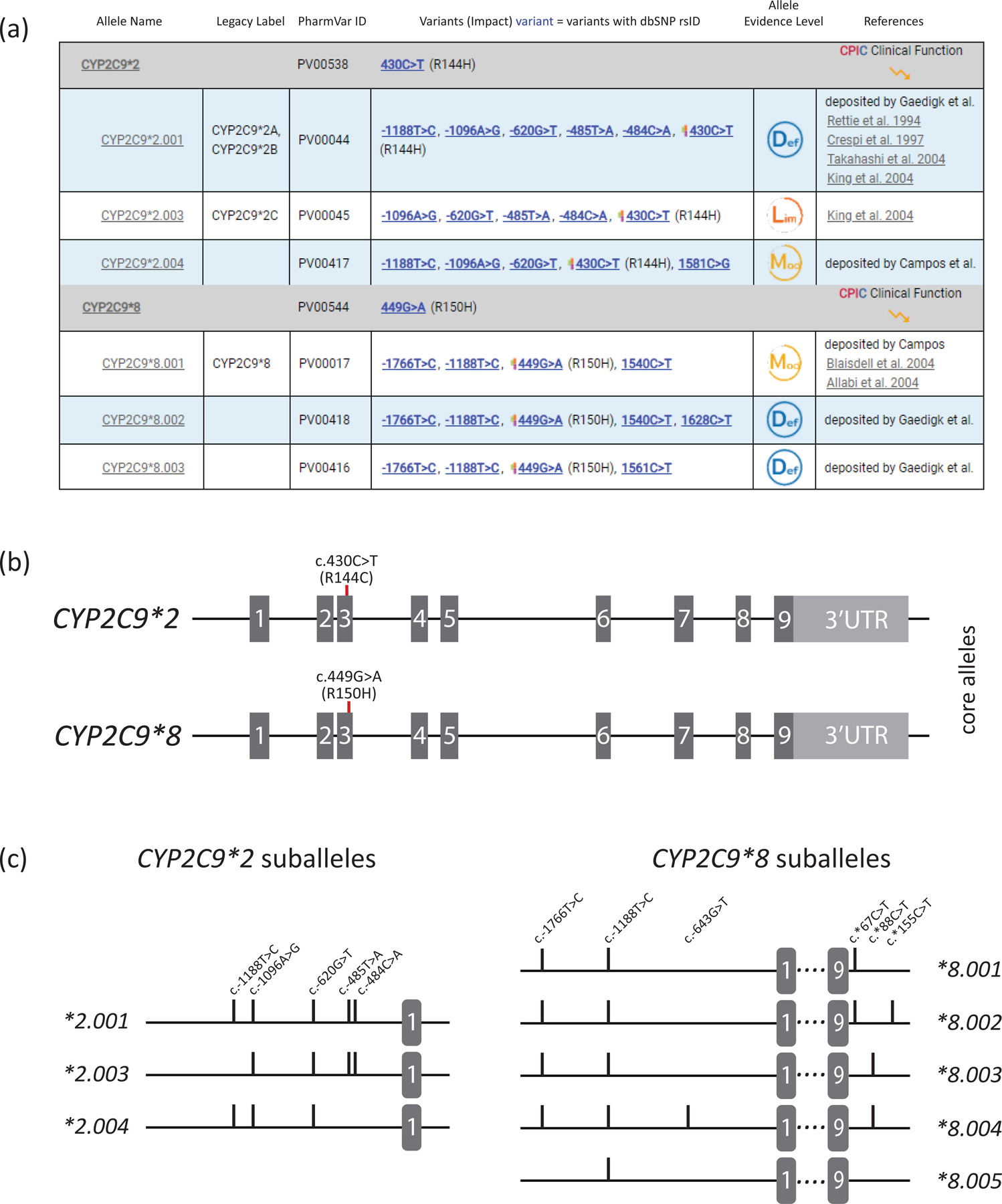
Panel (a) shows the CYP2C9*2 and *8 core allele definitions (gray bar) with NM_000771.4 as the reference sequence. Core SNVs, PharmVar ID (PVID), and evidence level is shown for each allele. All currently defined CYP2C9*2 suballeles and selected CYP2C9*8 suballeles are displayed underneath the core allele bar. Legacy allele designations are cross-referenced (e.g., *2.001 corresponds to *2A and *2B which have been merged). Panel (b) is a graphical representation of the CYP2C9*2 and *8 core alleles. Each is characterized by a single core SNV is highlighted in red (CYP2C9*2 by c.430C>T, p.R144C and CYP2C9*8 by c.449G>A, R150H). While c.449G>A is unique to CYP2C9*8, c.430C>T is not only found in the CYP2C9*2 haplotype (see Figure 2 for more details). Gray boxes represent the nine exons (scale is approximated); 3’UTR denotes the 3’ untranslated region. Panel (c) shows the CYP2C9*2 and *8 suballeles defined to date. As shown, suballeles of CYP2C9*2 only differ in their upstream region (graph only shows this portion of the gene), while those for CYP2C9*8 also vary in their 3’UTR regions (graph showing respective regions). ‘Lim’, ‘Mod’ and ‘Def’ symbols denote ‘Limited’, ‘Moderate’ and ‘Definitive’ haplotype evidence levels.
Each allele is listed in sequential order on the CYP2C9 gene page and cross-references with its legacy name (if existing), variants (including core SNVs; see Core Allele section below), evidence level, and clinical function as assigned by CPIC. A ‘Compare View’ allows the viewer to toggle between the standard allele table and the Comparative Allele ViewEr (CAVE) tool. The CYP2C9 gene page also includes ‘Read Me’ and ‘Change Log,’ documents, as well as links to other websites with CYP2C9 information including a link to PharmGKB’s gene information.
CYP2C9 haplotype evidence levels
PharmVar designates the “Haplotype Evidence Level” for each of the star alleles reported on the CYP2C9 gene page. Evidence levels are displayed as symbols indicating ‘Def’ (definitive), ‘Mod’ (moderate) or ‘Lim’ (limited) levels of support for a given haplotype reflecting the level of certainty that a haplotype exists in its reported form evidence (note that evidence levels in support of allele function can be found on the PharmGKB (36)). This three-tiered system represents a modified ClinVar classification system; more detailed information is provided in the ‘Allele Designation and Evidence Criteria Level’ document (81). This type of information (e.g., whether an allele was sequenced across the gene, how haplotype was determined) was not always systematically captured prior to PharmVar. For existing haplotype definitions, a literature review was conducted in order to assign evidence levels. Many alleles are currently labeled as ‘Lim’ because their definitions do not include any upstream region or do not extend 2 kb upstream, which is required by PharmVar allele designation requirements. This was the case for many allelic variants, including CYP2C9*1.002 and *1.003, as well as *4, *6, *21-*24 and *31-*60. Other alleles, such as CYP2C9*7, *10, *15-*20, *61 were labeled as ‘Mod’ despite being fully sequenced because the phase of the variants was computationally inferred and has not been validated. The value of evidence levels is centered on providing users with as much information on haplotype reliability as possible and enabling users to quickly parse haplotypes based on robust, high evidence as required for ‘Def’, versus other haplotypes with ‘Lim’ or ‘Mod’ evidence levels. PharmVar solicits submissions for all alleles labeled ‘Lim’ and ‘Mod’ to ultimately raise their evidence levels to ‘Def’. Moreover, PharmVar also encourages encore submissions for alleles with single citations and shown as ‘Def’ to further corroborate a haplotype definition.
See the CYP2C9 gene page (20) for current star allele definitions and their assigned evidence levels, including suballeles. Selected citations supporting respective haplotype definitions can also be found here.
PharmVar IDs
Each characterized haplotype receives a PharmVar ID (PVID). The PVID is a unique numeric identifier analogous to dbSNP rsIDs. Star allele names are driven by functional grouping, i.e., they are not guaranteed to be permanent and can be subject to change. Additional changes may be necessary in the future as more information becomes available. If an allele’s star designation is updated to a new star number, the PVID of the haplotype remains constant and does not change (no example for CYP2C9). In contrast, if a haplotype definition changes (e.g., through the addition or removal of variants) a new PVID will be assigned. Original PVIDs and their haplotype definitions can be tracked in the database via the PVID Lookup function. The CYP2C9 experts decided to not include intronic variants of unknown functional consequence as part of the required region to interrogate for a CYP2C9 haplotype, therefore intronic variants were removed from CYP2C9*26, *27, and *29 allele definitions and *26.001, *27.001, *29.001 received new PVIDs.
Core allele definitions
For already defined alleles, there is a growing number of suballeles that share one or more ‘core’ defining sequence variant(s). Although suballele information can be valuable for e.g., design of test platforms (sequence or genotype-based) and the interpretation of genotyping test results, there is no need to distinguish suballeles for phenotype prediction because all alleles under a star number are presumed to be functionally equivalent. Thus, even if a test is capable of distinguishing suballeles, from a functional standpoint, these can be simply reported using core allele definitions (e.g., CYP2C9*1, *3, *8, etc.).
A core allele is defined only by sequence variations that cause an amino acid change or impact function by changing expression levels or interfere with splicing and are present in all suballeles within an allele group (85). With this rule-based system, suballeles are collapsed into a single ‘core’ definition representing all suballeles categorized under a star (*) number. For example, CYP2C9*2 suballeles share the c.430C>T (p.R144C) SNV that fulfills this rule. Thus, this SNV constitutes the CYP2C9*2 core allele definition. Of importance, a sequence variant found in a core allele definition is not necessarily unique to that haplotype as illustrated in Figure 3.
One challenge with core allele definitions is that a definition may change over time as new information becomes available. However, this scenario is less likely for CYP2C9 given that most core alleles are defined by a single variant.
The core alleles are the basis of the CYP2C9 allele definition table used in CPIC guidelines and by PharmGKB (Table 1). The CYP2C9 core allele definitions are also utilized for clinical annotations in PharmGKB.
The PharmVar Comparative Allele ViewEr
The Comparative Allele ViewEr (CAVE) tool was developed by PharmVar to easily compare core alleles (85). This tool can be accessed using the “Compare View” button on the CYP2C9 gene page. Figure 2 not only exemplifies the utility of this tool on two sets of alleles, CYP2C9*2, *35 and *61 and CYP2C9*3, *18 and *68, but also illustrates the pitfalls of allele identification if laboratories assign alleles based on few markers. In this display mode it is easy to see which core SNV(s) are shared among the selected haplotypes, whether they alter function and/or are unique to a haplotype. CYP2C9*2, *35 and *61 harbor c.430C>T (p.R144C) while CYP2C9*3, *18 and *68 have c.1075A>C (p.259L) in common. Additional unique variants are found on CYP2C9*35 (c.374G>T, p.R125L, rs72558189) and *61 (c.1370A>G, p.N457S, rs202201137) and CYP2C9*18 (c.1190A>C, p.D397A) and *68 (c.1149+1G>A, splice defect, rs542577750) which distinguishes these from CYP2C9*2 and *3, respectively.
Reporting genotype and translation into phenotype
PharmVar and PharmGKB have also collaboratively developed templates to facilitate more consistent and transparent reporting of genotype details and how genotype is translated into phenotype to be used by the community to include more detailed information as part of the submission of research findings to publishers. This information can be provided as supplemental materials of a publication to facilitate access to important data for subsequent curation. The first template file (Supplementary materials 1) collects information, including methods or platforms used for genotyping and which SNVs are interrogated; the template also provides a standardized set-up for reporting genotype results for individual subjects, as well as allele frequencies. The second template file (Supplementary materials 2) facilitates the reporting of how genotype is translated into phenotype, as well as genotype frequencies. Although it is recommended by CPIC, as well as other groups, to use their standardized translation method, not every investigator or laboratory adopts this method. Too often, papers reference previous work stating that ‘genotyping was performed as previously described’ or indicate that ‘CYP2C9 phenotype was correlated with the metabolism of a drug’ without specifying which SNVs or alleles were genotyped or how phenotype was assigned. The lack of such information can make it extremely difficult, if not impossible, for curators to compare findings or extract information for CPIC guideline development. Colleagues are therefore strongly encouraged to utilize the provided templates, or revised versions thereof, for publication of these types of information. These tables are available through the PharmVar CYP2C9 gene page under ‘More Documents’ and at PharmGKB under ‘PGx Publication Tips’ (86).
CYP2C9 reference materials
The Genetic Testing Reference Material (GeT-RM) Coordination program is a combined effort among the Centers for Disease Control, Coriell Institute for Medical Research, and members of the pharmacogenetic testing community (87). Considering the growing use of pharmacogenetic testing, established sets of well-characterized reference materials are needed for assay development, validation, quality control, and proficiency testing. To address the increasing need for reference materials, a set of 137 genomic DNA samples were characterized for 28 pharmacogenes, including CYP2C9 and “consensus” genotypes established (73). Although the most common variants were assayed, many rare alleles were not identified among the samples tested; additional samples to complement the existing materials for CYP2C9 have been identified (88). Testing and research laboratories can acquire these materials from the Coriell Institute (Camden, NJ, USA), as they are publicly available. Information for testing materials is currently not included on the PharmVar CYP2C9 gene page.
Curation efforts
Gene region mapped/required for allele definition:
CYP2C9 allele definitions include variants within the coding region, upstream region up to c.-1911 and downstream region. Intronic variants are generally not considered for allele definitions unless they affect enzyme activity. New submissions will be required to sequence the upstream region at least to −1950 bp (including c.-1911T>C) and the downstream region to 250 bp. Currently available allele definitions have some limitations however, including limited sequencing data for up- and downstream sequences and haplotypes that were computationally inferred but have not yet been independently confirmed.
Corrections, revisions, new alleles, and other updates:
Extensive curation efforts were part of the content transfer from the P450 nomenclature webpage into the PharmVar database to standardize the annotations to the above-mentioned conventions. A summary is provided in Table 3. The following sections describe general and specific efforts undertaken.
Table 3.
Summary of edits and changes during the transitioned into the PharmVar database and notable changes made thereafter
| Reason |
Change | SNVs and Affected Alleles |
|---|---|---|
| Standardization | Intronic SNVs were removed | *26, *27, *29 |
| Comments removed | *1B−D, *2A−C, *3A+B, *11A+B, *15, *24, *26, *27, *29 | |
| Revised alleles after removing SNVs that are outside of the region used to define haplotypes | *2B, *11B, *27 | |
| Merged alleles after removing SNVs that are outside the region used to define haplotypes |
*1.002 and *1.003; *2.001 and *2.002 |
|
| Retired after its only defining SNV was removed due to being outside the region used to define haplotype |
*1D
|
|
| Revised | *3A, *3B, *8 | |
| Other | Correction | *26 (c.-1565C>T was erroneously listed as 1565C>T) |
During the transition process into the PharmVar database, comments and footnotes were removed and errors corrected. References in support of allele definitions have been updated and those solely describing function removed. References for function are provided in the PharmGKB/CPIC CYP2C9 Allele Functionality table (36). Descriptors such as “Existence of the CYP2C9*2 polymorphism 430C>T on the same allele cannot be excluded” or “linkage with c.-1188T>C cannot be excluded” have been removed and thus, c.430C>T (p.R144C) and c.-1188T>C (rs4918758) are currently not part of the allele definition for CYP2C9*24 and CYP2C9*15, respectively.
Both c.-1766T>C (rs9332094) and c.-1188T>C were included into the CYP2C9*8.001 (former CYP2C9*8) haplotype definition since those variants were part of the original publication by Blaisdell et al (16) but were omitted when these alleles were first defined. The presence of c.-1766T>C and c.-1188T>C has been substantiated by new submissions to PharmVar not only confirming the updated CYP2C9*8.001 allele definition but also adding two novel suballeles, CYP2C9*8.002 and CYP2C9*8.003. Similarly, c.1425A>T (p.G475G, rs1057911) was added to the CYP2C9*3.001 and CYP2C9*3.002 allele definitions. A review by the gene expert panel of the original literature revealed that this SNV had inadvertently been omitted from the haplotype definitions; the presence of c.1425A>T (p.G475G) was also corroborated by new submissions to PharmVar. Changes and revisions made are detailed in the ‘Change Log’ document on the CYP2C9 gene page (20) and are summarized in Table 3.
As of April 2021, the CYP2C9 expert panel has designated 11 novel alleles (*61 through *71) and 17 new suballeles. In addition, seven alleles that were based on partial information are now fulfilling PharmVar allele definition requirements and their evidence level was raised to ‘Def’ (Table 2). There are still numerous allele definitions with ‘Lim’ and ‘Mod’ evidence levels for which PharmVar seeks submissions. The ‘Change Log’ document also tracks submissions and indicates the star alleles that have been updated.
Of the novel alleles, we would like to highlight CYP2C9*71. This allele has two core variants in cis (i.e. on the same allele), c.815A>G (p.E272G, rs9332130) and c.1465C>T (p.P489S, rs9332239), which are the core SNVs of CYP2C9*10 and *12, respectively. As shown in Figure 4b, long distance phasing using 10X Genomics Linked-Read technology revealed, however, that in this case, both SNVs are in cis and thus represents a novel haplotype. This finding raises concerns regarding the accuracy of the original CYP2C9*10 and *12 allele definitions (16). Interestingly, c.815A>G and c.1465C>T were initially discovered in the same individual but presumed to be in trans, which resulted in the definition of two separate star alleles each characterized by a single SNV. Neither CYP2C9*10 nor *12 have been independently confirmed to date.
Figure 4. Characterization of novel allelic variants.
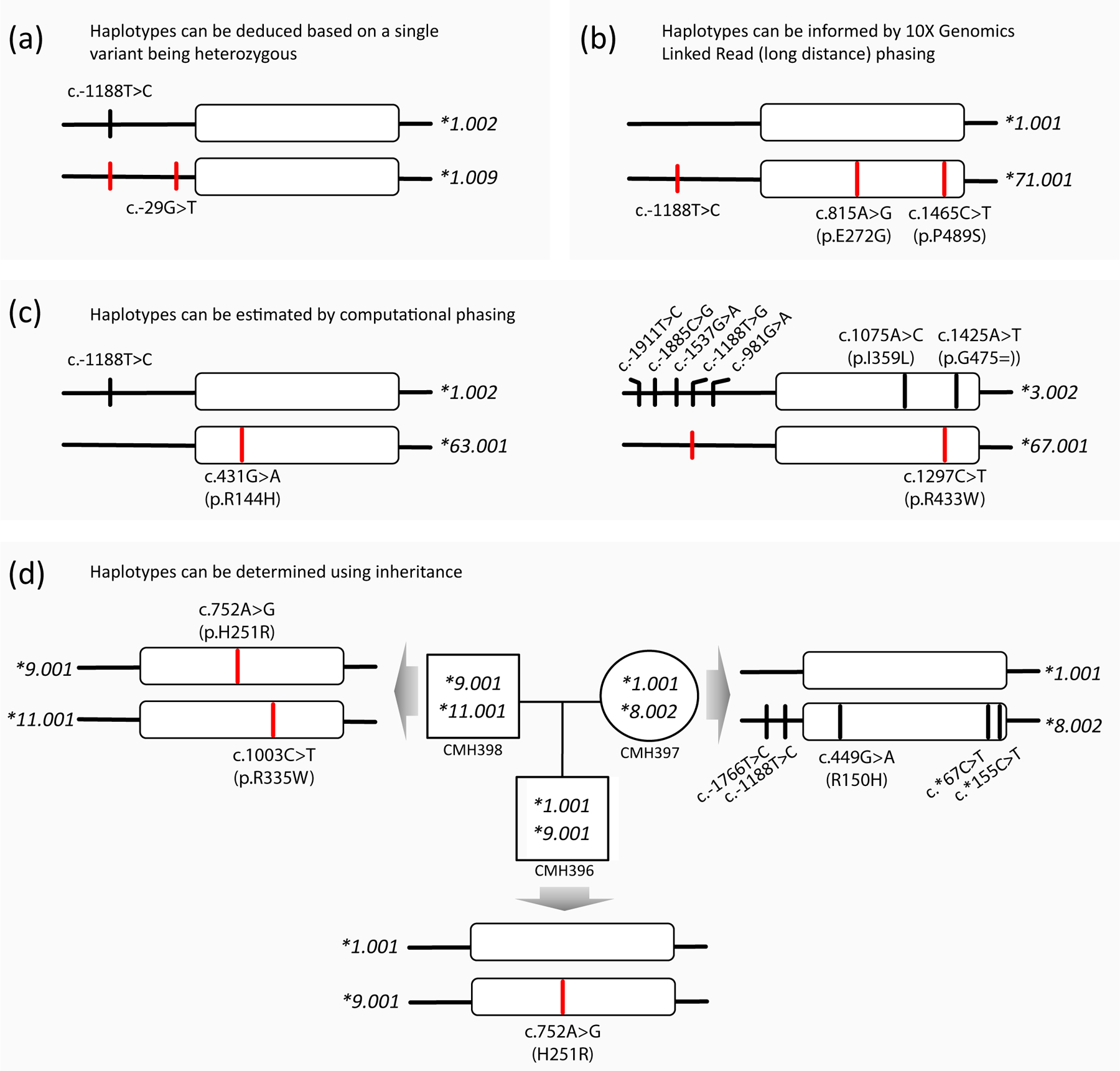
Panels (a-d) provide examples of alleles submitted to PharmVar for naming or to confirm existing allele definitions. Variants of submitted alleles are highlighted by red lines. All submissions utilized WGS data which were either confirmed by WES or targeted NGS-based sequencing panels. Panel (a) exemplifies a subject whose haplotype can be unequivocally be deduced and Panel (b) shows a subject whose three heterozygous SNVs were placed on the same chromosome using 10X Genomics Linked-Read (long-distance) phasing technology; CYP2C9*1.009 and *71 both received an evidence level of ‘Definitive’ (‘Def’). Panel (c) depicts two examples for which haplotypes were computationally inferred. Due to uncertainty regarding the phase of the variants, CYP2C9*63 and CYP2C9*67 received evidence levels of ‘Moderate’ (Mod) and ‘Limited’ (Lim), respectively. Panel (d) illustrates how haplotype can be inferred using inheritance in a family trio.
The CYP2C9*8 allele was initially defined by c.449G>T (p.R150H). After receiving submissions for this allele, its definition was revised in 2018 to include two variants in the upstream region, c.-1188T>C and c.-1766T>C and one in the 3’UTR (c.*67C>T, rs9332240). The former variants were noted in the allele’s first report (16) but omitted when it was first defined. The presence of c.-1188T>C and c.-1766T>C on the CYP2C9*8 haplotype was also described (89). Functional in vitro studies by this group suggested that c.-1766T>C impacts expression levels, and thus, c.-1766T>C was granted core SNV status. Recently, an allele was discovered which had c.449G>T but lacked c.-1766T>C; this allele would receive its own star number given the absence of the c.-1766T>C core SNV. Concerns were raised, however, whether there is indeed sufficient evidence supporting c.-1766T>C having a functional impact. The gene experts ultimately recommended to revert their initial decision and remove core SNV status from c.-1766T>C, which paved the way to designate the novel haplotype as a CYP2C9*8 suballele. This case illustrates that allele designation is not always straightforward and underscores the need to develop more concrete criteria that need to be fulfilled for non-coding SNVs to receive core SNV status.
Methods for CYP2C9 allele characterization
CYP2C9 allele characterization presents the same challenges previously discussed in the CYP2C19 PharmVar GeneFocus review (61). In this section we provide selected examples of novel alleles submitted to PharmVar and describe how they were characterized, i.e., how it was determined of which SNVs are located on each chromosome.
Figure 4a illustrates a sample that was homozygous for c.-1188T>C and heterozygous for c.-29G>T. In this scenario each haplotype can be deduced without further experimental testing (the same is true if a sample is homozygous for all SNVs); the novel allele was designated as CYP2C9*1.009 and received an evidence level of ‘Def’ indicating that the allele has been fully characterized and variants phased.
Several SNVs are often identified as heterozygous and further characterization is required to determine whether the variants are in cis (on the same chromosome) or in trans (on opposite chromosomes). WGS coupled with long-read sequencing is the most powerful and elegant approach to determine the phase of variants over long distances. As described above and shown in Figure 4b, the three SNVs found on the CYP2C9*71 allele were phased to the same chromosome using 10X Genomic Linked Read technology (10X Genomics, Pleasanton, CA); this allele also received an evidence level of ‘Def’. To characterize CYP2C9*62 (90) (not shown), a combination of methods including long-range PCR, cloning and sequencing were used to determine that the new haplotype has two upstream region SNVs (c.-1565C>T and c.-1188T>C) in addition to a novel nonsynonymous variant (c.430C>T, p.R125C). Alternatively, allele-specific PCR or single molecule real-time sequencing (e.g., Pacific Biosciences, Menlo Park, CA or Oxford Nanopore Technologies, Oxford, UK) may be utilized to demonstrate that variants are on the same allele; these approaches are however, limited by the length of the PCR fragments that can be generated.
Haplotypes can also be inferred using statistical approaches such as PHASE software (91) or BEAGLE (92). PharmVar has recently updated requirements for allele definitions (81) to more readily accept haplotypes that are based on computational predictions. Alleles fulfilling the submission requirements receive an evidence level of ‘Mod’ or ‘Lim’ depending on the degree of uncertainty. Figure 4c details two examples of alleles submitted by Nizamudin et al. (93). The authors used data of 210 subjects and BEAGLE to infer haplotypes. The first example represents a subject who was heterozygous for two variants, which were phased in trans suggesting that c.431G>A (p.R144H) is the sole SNV on the novel haplotype designated as CYP2C9*63. This allele received an evidence level of ‘Mod’ because there were no other amino acid changing SNVs or other non-coding variants known to alter function. Although some uncertainty remains regarding the phase of c.-1188T>C, the function of CYP2C9*63 would be the same because c.-1188T>C does not impact function according to current knowledge. The second example provided in Figure 4c received an evidence level of ‘Lim’. As illustrated, seven SNVs were computationally inferred to represent the CYP2C9*3.002 suballele while a novel variant, c.1297C>T (p.R433W, rs776908257), was predicted to be located on the opposite chromosome forming the novel CYP2C9*67 haplotype. Due to the uncertainty of the phase of two nonsynonymous variants in this subject, the impact of the p.R433W amino acid change on enzyme activity remains to be established experimentally (in silico analyses predict p.R433W being deleterious or probably damaging (93)). Based on BEAGLE and in silico predictions, a CYP2C9*3/*67 genotype may lead to PM status while a CYP2C9*1/*3 genotype (with p.R433W being on the *3 allele) would translate into IM status. A confirmatory submission for CYP2C9*67 is needed to consolidate the definition of this allele definition.
Finally, as demonstrated in Figure 4d, haplotypes can also be delineated using inheritance information. The mother-father-child trio (data obtained from the Children’s Mercy Data Warehouse) was utilized for confirmatory submissions for CYP2C9*9.001 and *11.001, which raised their respective evidence levels from ‘Mod’ and ‘Lim’, to ‘Def’. The mother’s CYP2C9*8.002 allele was consistent with the allele’s definition which already had ‘Def’ status at the time this pedigree was analyzed.
Conclusions
This PharmVar GeneFocus on CYP2C9 provides essential information for the understanding of this highly polymorphic gene, complementing clinically relevant information provided by CPIC guidelines and other pharmacogenetic resources. We are summarizing PharmVar efforts of systematically cataloging CYP2C9 allelic variation, as well as providing examples of submissions highlighting different approaches to fully characterize novel haplotypes. In addition, we stress our collaborative efforts with the PharmGKB to make the information useful and easily accessible to the entire pharmacogenetics community.
Supplementary Material
1. CYP2C9 Genotyping Method and Data Template
2. CYP2C9 Genotype to Phenotype Transition Template
Acknowledgements
We would like to thank Ming-Ta (Mike) Lee for his time and efforts serving on the CYP2C9 expert panel.
Funding
This work was funded by the National Institutes of Health for the Pharmacogene Variation Consortium (R24 GM123930; PI, A.G.) and PharmGKB (U24 HG010615; PI, T.E.K.). We also acknowledge support: J.A.G., Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Madrid, Spain (PI18/00540, and RD16/0006/0004); M.W., Swedish Research Council (Medicine 2018-03307) and Swedish Heart-Lung Foundation (20170711 and 20200777); L.H.C. was supported in part by NIH/NCATS (UL1 TR001427); K.C.C.; NHGRI (T32 HG008958); J.D., NIH/NIMHD (U54 MD007600) and the Center for Collaborative Research in Health Disparities (RCMI-CCRHD).
Footnotes
Conflicts of Interest:
Indiana University School of Medicine Pharmacogenomics Laboratory is a fee-for-service clinical laboratory that offers clinical pharmacogenetic testing. A.L.D. is a paid employee of Acadia Pharmaceuticals; S.A.S. is a paid consultant of Sema4. All other authors declared no competing interests for this work.
References
- (1).Daly AK, Rettie AE, Fowler DM & Miners JO Pharmacogenomics of CYP2C9: Functional and Clinical Considerations. J Pers Med 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Miners JO & Birkett DJ Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol 45, 525–38 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Scott J & Poffenbarger PL Pharmacogenetics of tolbutamide metabolism in humans. Diabetes 28, 41–51 (1979). [PubMed] [Google Scholar]
- (4).Miners JO, Wing LM & Birkett DJ Normal metabolism of debrisoquine and theophylline in a slow tolbutamide metaboliser. Aust N Z J Med 15, 348–9 (1985). [DOI] [PubMed] [Google Scholar]
- (5).Yasumori T, Kawano S, Nagata K, Shimada M, Yamazoe Y & Kato R Nucleotide sequence of a human liver cytochrome P-450 related to the rat male specific form. J Biochem 102, 1075–82 (1987). [DOI] [PubMed] [Google Scholar]
- (6).Meehan RR et al. Human cytochrome P-450 PB-1: a multigene family involved in mephenytoin and steroid oxidations that maps to chromosome 10. Am J Hum Genet 42, 26–37 (1988). [PMC free article] [PubMed] [Google Scholar]
- (7).Veronese ME, Mackenzie PI, Doecke CJ, McManus ME, Miners JO & Birkett DJ Tolbutamide and phenytoin hydroxylations by cDNA-expressed human liver cytochrome P4502C9. Biochem Biophys Res Commun 175, 1112–8 (1991). [DOI] [PubMed] [Google Scholar]
- (8).Romkes M, Faletto MB, Blaisdell JA, Raucy JL & Goldstein JA Cloning and expression of complementary DNAs for multiple members of the human cytochrome P450IIC subfamily. Biochemistry 30, 3247–55 (1991). [DOI] [PubMed] [Google Scholar]
- (9).Rettie AE et al. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem Res Toxicol 5, 54–9 (1992). [DOI] [PubMed] [Google Scholar]
- (10).Leemann T, Transon C & Dayer P Cytochrome P450TB (CYP2C): a major monooxygenase catalyzing diclofenac 4’-hydroxylation in human liver. Life Sci 52, 29–34 (1993). [DOI] [PubMed] [Google Scholar]
- (11).Stubbins MJ, Harries LW, Smith G, Tarbit MH & Wolf CR Genetic analysis of the human cytochrome P450 CYP2C9 locus. Pharmacogenetics 6, 429–39 (1996). [DOI] [PubMed] [Google Scholar]
- (12).Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF & Korzekwa KR Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics 4, 39–42 (1994). [DOI] [PubMed] [Google Scholar]
- (13).Sullivan-Klose TH et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics 6, 341–9 (1996). [DOI] [PubMed] [Google Scholar]
- (14).Dickmann LJ et al. Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol Pharmacol 60, 382–7 (2001). [DOI] [PubMed] [Google Scholar]
- (15).Kidd RS, Curry TB, Gallagher S, Edeki T, Blaisdell J & Goldstein JA Identification of a null allele of CYP2C9 in an African-American exhibiting toxicity to phenytoin. Pharmacogenetics 11, 803–8 (2001). [DOI] [PubMed] [Google Scholar]
- (16).Blaisdell J et al. Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics 14, 527–37 (2004). [DOI] [PubMed] [Google Scholar]
- (17).Johnson JA et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin Pharmacol Ther 102, 397–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Theken KN et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clin Pharmacol Ther 108, 191–200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Karnes JH et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and HLA-B Genotypes and Phenytoin Dosing: 2020 Update. Clin Pharmacol Ther 109, 302–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).PharmGKB CYP2C9 gene page <https://www.pharmgkb.org/gene/PA126>.
- (21).Ingelman-Sundberg M, Oscarson M, Daly AK, Garte S & Nebert DW Human cytochrome P-450 (CYP) genes: a web page for the nomenclature of alleles. Cancer Epidemiol Biomarkers Prev 10, 1307–8 (2001). [PubMed] [Google Scholar]
- (22).Gaedigk A et al. The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharmacol Ther 103, 399–401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).PharmVar Standards <https://www.pharmvar.org/genes>.
- (24).PharmGKB CYP2C9 Drug Label Annotations <https://www.pharmgkb.org/gene/PA126/labelAnnotation>.
- (25).FDA Pharmacogenomic Biomarkers in Drug Labeling <https://www.fda.gov/drugs/science-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling>.
- (26).FDA Table of Pharmacogenetics Associations <https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations>.
- (27).CPIC Gene/drug pairs <https://cpicpgx.org/genes-drugs/>.
- (28).Yasar U et al. Pharmacokinetics of losartan and its metabolite E-3174 in relation to the CYP2C9 genotype. Clin Pharmacol Ther 71, 89–98 (2002). [DOI] [PubMed] [Google Scholar]
- (29).Limdi NA et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther 83, 312–21 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pirmohamed M et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med 369, 2294–303 (2013). [DOI] [PubMed] [Google Scholar]
- (31).Gage BF et al. Effect of Genotype-Guided Warfarin Dosing on Clinical Events and Anticoagulation Control Among Patients Undergoing Hip or Knee Arthroplasty: The GIFT Randomized Clinical Trial. JAMA 318, 1115–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kimmel SE et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med 369, 2283–93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Drozda K et al. Poor warfarin dose prediction with pharmacogenetic algorithms that exclude genotypes important for African Americans. Pharmacogenet Genomics 25, 73–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Pratt VM et al. Recommendations for Clinical CYP2C9 Genotyping Allele Selection: A Joint Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn 21, 746–55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bousman CA, Zierhut H & Muller DJ Navigating the Labyrinth of Pharmacogenetic Testing: A Guide to Test Selection. Clin Pharmacol Ther 106, 309–12 (2019). [DOI] [PubMed] [Google Scholar]
- (36).PharmGKB Gene-specific Information Tables for CYP2C9 <https://www.pharmgkb.org/page/cyp2c9RefMaterials>.
- (37).Macias Y, Gomez Tabales J, Garcia-Martin E & Agundez JAG An update on the pharmacogenomics of NSAID metabolism and the risk of gastrointestinal bleeding. Expert Opin Drug Metab Toxicol 16, 319–32 (2020). [DOI] [PubMed] [Google Scholar]
- (38).Selmaj K et al. Siponimod for patients with relapsing-remitting multiple sclerosis (BOLD): an adaptive, dose-ranging, randomised, phase 2 study. Lancet Neurol 12, 756–67 (2013). [DOI] [PubMed] [Google Scholar]
- (39).FDA Siponimod Drug label <https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/209884s000lbl.pdf>.
- (40).Drug Interactions Flockhart Table™ <https://drug-interactions.medicine.iu.edu/Main-Table.aspx>.
- (41).O’Reilly RA, Trager WF, Rettie AE & Goulart DA Interaction of amiodarone with racemic warfarin and its separated enantiomorphs in humans. Clin Pharmacol Ther 42, 290–4 (1987). [DOI] [PubMed] [Google Scholar]
- (42).McDonald MG, Au NT, Wittkowsky AK & Rettie AE Warfarin-amiodarone drug-drug interactions: determination of [I](u)/K(I,u) for amiodarone and its plasma metabolites. Clin Pharmacol Ther 91, 709–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Bertilsson G et al. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci U S A 95, 12208–13 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT & Kliewer SA The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102, 1016–23 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Merwick A, Hannon N, Kelly PJ & O’Rourke K Warfarin-flucloxacillin interaction presenting as cardioembolic ischemic stroke. Eur J Clin Pharmacol 66, 643–4 (2010). [DOI] [PubMed] [Google Scholar]
- (46).Pottegard A, Henriksen DP, Madsen KG, Hellfritzsch M, Damkier P & Stage TB Change in International Normalized Ratio Among Patients Treated With Dicloxacillin and Vitamin K Antagonists. JAMA 314, 296–7 (2015). [DOI] [PubMed] [Google Scholar]
- (47).Sahi J, Shord SS, Lindley C, Ferguson S & LeCluyse EL Regulation of cytochrome P450 2C9 expression in primary cultures of human hepatocytes. J Biochem Mol Toxicol 23, 43–58 (2009). [DOI] [PubMed] [Google Scholar]
- (48).Honkakoski P, Sueyoshi T & Negishi M Drug-activated nuclear receptors CAR and PXR. Ann Med 35, 172–82 (2003). [DOI] [PubMed] [Google Scholar]
- (49).Gomez-Tabales J, Garcia-Martin E, Agundez JAG & Gutierrez-Merino C Modulation of CYP2C9 activity and hydrogen peroxide production by cytochrome b5. Sci Rep 10, 15571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zanger UM & Schwab M Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138, 103–41 (2013). [DOI] [PubMed] [Google Scholar]
- (51).Yang X et al. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res 20, 1020–36 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Zhang H et al. Content and activity of human liver microsomal protein and prediction of individual hepatic clearance in vivo. Sci Rep 5, 17671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Dorado P et al. Losartan hydroxylation phenotype in an Ecuadorian population: influence of CYP2C9 genetic polymorphism, habits and gender. Pharmacogenomics 13, 1711–7 (2012). [DOI] [PubMed] [Google Scholar]
- (54).Hatta FH et al. Differences in CYP2C9 Genotype and Enzyme Activity Between Swedes and Koreans of Relevance for Personalized Medicine: Role of Ethnicity, Genotype, Smoking, Age, and Sex. OMICS 19, 346–53 (2015). [DOI] [PubMed] [Google Scholar]
- (55).Sandberg M, Johansson I, Christensen M, Rane A & Eliasson E The impact of CYP2C9 genetics and oral contraceptives on cytochrome P450 2C9 phenotype. Drug Metab Dispos 32, 484–9 (2004). [DOI] [PubMed] [Google Scholar]
- (56).Cherala G, Pearson J, Maslen C & Edelman A An ethinyl estradiol-levonorgestrel containing oral contraceptive does not alter cytochrome P4502C9 in vivo activity. Drug Metab Dispos 42, 323–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Mwinyi J, Cavaco I, Yurdakok B, Mkrtchian S & Ingelman-Sundberg M The ligands of estrogen receptor alpha regulate cytochrome P4502C9 (CYP2C9) expression. J Pharmacol Exp Ther 338, 302–9 (2011). [DOI] [PubMed] [Google Scholar]
- (58).Koukouritaki SB et al. Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther 308, 965–74 (2004). [DOI] [PubMed] [Google Scholar]
- (59).Zane NR, Chen Y, Wang MZ & Thakker DR Cytochrome P450 and flavin-containing monooxygenase families: age-dependent differences in expression and functional activity. Pediatr Res 83, 527–35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Magliocco G, Rodieux F, Desmeules J, Samer CF & Daali Y Toward precision medicine in pediatric population using cytochrome P450 phenotyping approaches and physiologically based pharmacokinetic modeling. Pediatr Res 87, 441–9 (2020). [DOI] [PubMed] [Google Scholar]
- (61).Botton MR et al. PharmVar GeneFocus: CYP2C19. Clin Pharmacol Ther 109, 352–66 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Whirl-Carrillo M et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92, 414–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Sangkuhl K et al. Pharmacogenomics Clinical Annotation Tool (PharmCAT). Clin Pharmacol Ther 107, 203–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Caudle KE et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab 15, 209–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM & Caudle KE The Clinical Pharmacogenetics Implementation Consortium: 10 Years Later. Clin Pharmacol Ther 107, 171–5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Caudle KE et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med 19, 215–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Caudle KE et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci 13, 116–24 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Nofziger C et al. PharmVar GeneFocus: CYP2D6. Clin Pharmacol Ther 107, 154–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Cavallari LH et al. Multi-site investigation of strategies for the clinical implementation of CYP2D6 genotyping to guide drug prescribing. Genet Med 21, 2255–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Caraballo PJ, Bielinski SJ, St Sauver JL & Weinshilboum RM Electronic Medical Record-Integrated Pharmacogenomics and Related Clinical Decision Support Concepts. Clin Pharmacol Ther 102, 254–64 (2017). [DOI] [PubMed] [Google Scholar]
- (71).Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE & Hoffman JM Integrating pharmacogenomics into electronic health records with clinical decision support. Am J Health Syst Pharm 73, 1967–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Gray KA, Yates B, Seal RL, Wright MW & Bruford EA Genenames.org: the HGNC resources in 2015. Nucleic Acids Res 43, D1079–85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Pratt VM et al. Characterization of 137 Genomic DNA Reference Materials for 28 Pharmacogenetic Genes: A GeT-RM Collaborative Project. J Mol Diagn 18, 109–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Moyer AM, Rohrer Vitek CR, Giri J & Caraballo PJ Challenges in Ordering and Interpreting Pharmacogenomic Tests in Clinical Practice. Am J Med 130, 1342–4 (2017). [DOI] [PubMed] [Google Scholar]
- (75).Gaedigk A, Whirl-Carrillo M, Pratt VM, Miller NA & Klein TE PharmVar and the Landscape of Pharmacogenetic Resources. Clin Pharmacol Ther 107, 43–6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).GTR: Genetic Testing Registry. Central location for voluntary submission of genetic test information by providers <https://www.ncbi.nlm.nih.gov/gtr/>.
- (77).Botton MR et al. Structural variation at the CYP2C locus: Characterization of deletion and duplication alleles. Hum Mutat 40, e37–e51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Huddart R et al. Standardized Biogeographic Grouping System for Annotating Populations in Pharmacogenetic Research. Clin Pharmacol Ther 105, 1256–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Zhang F & Finkelstein J Inconsistency in race and ethnic classification in pharmacogenetics studies and its potential clinical implications. Pharmgenomics Pers Med 12, 107–23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).CPIC. SOP for Assigning Allele Function.
- (81).PharmVar Allele Designation and Evidence Level Criteria document <www.pharmvar.org/documents/Allele_Designation_and_Evidence_Level_Criteria_V2.1.pdf>.
- (82).PharmVar CYP2C9 Gene Expert Panel <https://www.pharmvar.org/expert-panels>.
- (83).NCBI Reference Sequences database <https://www.ncbi.nlm.nih.gov/refseq/>.
- (84).Gaedigk A, Casey S, Whirl-Carrillo M, Miller NA & Klein TE PharmVar: A Global Resource and Repository for Pharmacogene Variation. Clin Pharmacol Ther accepted, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Gaedigk A et al. The Evolution of PharmVar. Clin Pharmacol Ther 105, 29–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).PharmGKB Publication Tips <https://www.pharmgkb.org/page/curationTips>
- (87).Genetic Testing Reference Materials Coordination Program (GeT-RM). <https://www.cdc.gov/labquality/get-rm/index.html>.
- (88).Pratt VM et al. Characterization of Reference Materials for CYP2C9, CYP2C19, VKORC1, CYP2C Cluster Variant, GGCX, and Other Pharmacogenetic Alleles with an Association for Molecular Pathology (AMP) Pharmacogenetics Working Group Tier 2 Status - A GeT-RM Collaborative Project. J Mol Diagn, May 18, doi: 10.1016/j.jmoldx.2021.04.012. Online ahead of print. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Cavallari LH et al. CYP2C9 promoter region single-nucleotide polymorphisms linked to the R150H polymorphism are functional suggesting their role in CYP2C9*8-mediated effects. Pharmacogenet Genomics 23, 228–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Chen H et al. An identification and functional evaluation of a novel CYP2C9 variant CYP2C9*62. Chem Biol Interact 327, 109168 (2020). [DOI] [PubMed] [Google Scholar]
- (91).Stephens M & Donnelly P A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73, 1162–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Browning BL & Browning SR A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84, 210–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Nizamuddin S, Dubey S, Singh S, Sharma S, Machha P & Thangaraj K CYP2C9 Variations and Their Pharmacogenetic Implications Among Diverse South Asian Populations. Pharmgenomics Pers Med 14, 135–47 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).PharmVar CYP2C9 gene page < https://www.pharmvar.org/gene/CYP2C9 >.
- (95).Pharm P450 Nomenclature site – Archive; previoulsy at cypalleles.ki/se <https://www.pharmvar.org/htdocs/archive/index_original.htm; previoulsy at cypalleles.ki/.
- (96).CPIC Guidelines <https://cpicpgx.org/guidelines/>.
- (97).Human Genome Variation Society. Nomenclature for the description of sequence variations <http://www.hgvs.org/content/guidelines>.
- (98).Locus Reference Genomic (LRG) Records <https://www.lrg-sequence.org/>.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. CYP2C9 Genotyping Method and Data Template
2. CYP2C9 Genotype to Phenotype Transition Template


