Abstract
Using an inducible transcription system which allows the regulated expression of C/EBP isoforms in tissue culture cells, we have found that the ectopic expression of C/EBPα, at a level comparable to that found in normal liver tissue, has a pronounced antimitogenic effect in mouse L cells and NIH 3T3 cells. The inhibition of cell division by C/EBPα in mouse cells cannot be reversed by simian virus 40 T antigen, by oncogenic ras, or by adenovirus E1a protein. When expressed in thymidine kinase-deficient L cells or 3T3 cells, C/EBPα is detected in a protein complex which binds to the E2F binding sites found in the promoters of the genes for E2F-1 and dihydrofolate reductase (DHFR). Bacterially expressed C/EBPα has no affinity for these E2F sites, but when recombinant C/EBPα is added to nuclear extracts from mouse fibroblasts, a new E2F binding activity appears, which contains the C/EBPα protein. Using an E2F-DP1-responsive promoter linked to a reporter gene, it can be shown that C/EBPα directly inhibits the induction of this promoter by E2F-DP1 in transient-transfection assays. Furthermore, C/EBPα can be shown to inhibit the S-phase induction of the E2F and DHFR promoters in permanent cell lines. These findings delineate a straightforward mechanism for C/EBPα-mediated cell growth arrest through repression of E2F-DP-mediated S-phase transcription.
The CCAAT enhancer binding proteins or C/EBP family of basic leucine zipper transcription factors comprises five isoforms, α to ɛ. The various isoforms show different patterns of expression in vivo, and as a group they regulate a wide variety of essential differentiation programs and cellular processes (26). C/EBPα has been implicated in the differentiation of adipocytes, hepatocytes, and lung (6, 46). C/EBPβ is required for the differentiation of myelomonocytes and may be important in the acute-phase response in liver (35). C/EBPδ may also be involved in the acute-phase response and appears to be required for the G0 growth arrest of epithelial cells in mammary tissues (34). C/EBPα has been implicated in numerous studies as an important regulator of metabolic enzymes in the cell (12, 48) and as a negative regulator of cell growth (6, 46, 49). While the mechanisms for C/EBPα-mediated control of metabolic processes have been well defined, if not as yet completely understood, the mechanism by which C/EBPα regulates cell growth has remained elusive despite attempts by several investigators to describe this process in various cell systems.
Transfection of C/EBPα into cells arrests cell growth in virtually all cell types and thereby abrogates the establishment of cell lines in which C/EBPα is stably expressed (46, 49). The use of chimeric regulatable forms of C/EBPα, by fusion to the hormone-binding domain of the estrogen receptor (46), introduces an additional potent transactivation domain into the protein (45), which may result in transcriptional activation which is different from that of native C/EBPα. 3T3-L1 cells represent a model of adipocyte differentiation in which the expression of C/EBPα is essential and sufficient for the establishment of the adipocyte phenotype as well as cell growth arrest (29). While 3T3-L1 cells provide a viable model for the study of adipocyte differentiation, the cocktail of inducers required to confer the differentiated state induces changes in these cells through several signaling pathways which undoubtedly converge on more than just one transcription factor to achieve the final effect (23, 50, 51). We have developed a system for the controlled expression of C/EBPα using an inducible expression system from a gal4-driven promoter. We have created mouse fibroblast cell lines capable of selective induction of C/EBPα via this GAL4-estrogen receptor (ER) expression system. These cells express levels of C/EBPα similar to those seen in tissues such as liver and allow the analysis of the mechanisms of C/EBPα-driven cell growth arrest when this protein is expressed at normal levels.
Cell growth is stringently regulated in a growing cell at the G1/S-phase transition of the cell cycle. Known regulators of this system have been well described, and much is known about the fashion in which cells move from G1 to S phase, including the E2F-DP-driven transcription of several genes required for DNA synthesis (17, 21). Briefly, in G1 phase, the regulatory pocket proteins, which include Rb, p107, and p130, are hypophosphorylated and, in this state, sequester the transcription factor E2F. Upon phosphorylation of retinoblastoma protein (Rb) by cyclin-dependent kinases, a process tightly regulated by both the cyclins and cyclin-dependent kinase inhibitors (CKIs), Rb loses its affinity for the transactivation domain of E2F, and this domain is released (21, 33). E2F-DP heterodimers are then able to either transactivate promoters for gene products required in S phase, such as dihydrofolate reductase (DHFR), or derepress promoters such as E2F1 (18, 25, 31, 47). Early studies of cell cycle control found that the tumor suppressor protein p53 acts to block cell cycle entry by induction of the promoter for the CK1 p21 (14). This mechanism has since been applied to other models of growth arrest at the G1/S-phase transition. In muscle cells, it was found that induction of p21 by myoD and p53 took place during differentiation and cell growth cessation (19). Also, in human HL-60 leukemia cells as well as murine erythroleukemia cells, cell growth arrest during differentiation was accompanied by elevated levels of p21 protein and mRNA (26, 30). All of these observations suggested an important role for p21 in regulating growth arrest of cells during differentiation.
C/EBPα is expressed constitutively in highly differentiated nondividing cells such as hepatocytes, adipocytes, and select cells in the lung (3). Following partial hepatectomy, C/EBPα levels in the liver begin to fall rapidly following surgery and do not return to normal levels until the liver has almost completely regenerated (20, 32). Furthermore, in preneoplastic liver nodules and in hepatoma cell lines, C/EBPα expression is low or nonexistent (16). These observations suggest that expression of C/EBPα functions in the maintenance of the differentiated and growth-arrested state of the liver. Studies in recent years have developed several potential mechanisms for C/EBPα-mediated cell growth arrest. It was first suggested that C/EBPα expression in human fibrosarcoma cell line, HT1080 cells, led to an induction of p21 promoter activity and accumulation of p21 protein, as had been observed for other systems (41). Further study indicated the potential ability of C/EBPα to stabilize p21 protein and thereby halt cell cycle progression (42). C/EBPα knockout mice were also found to express very low levels of p21 protein, while in older wild-type animals, the delayed decrease in C/EBPα expression after partial hepatectomy was concurrent with a delayed decrease in levels of p21 protein (42, 43). Works by Cram et al. (11) and by Cha et al. (7) with a hepatoma cell line describe a slightly different mechanism of p21 growth arrest mediated through the glucocorticoid receptor. Here, it was found that the p21 promoter was induced in the presence of dexamethasone and also required C/EBPα, but this effect was abolished in cells lacking a functional glucocorticoid receptor (11). While p21 clearly functions to prevent cell cycle entry into S phase, in keratinocytes it has been demonstrated that the effect is transient, while the expression of C/EBPα is maintained in highly differentiated and growth-arrested cells (3, 13). It remains unclear what the exact role of C/EBPα is in cell growth arrest and whether this effect is separate from its known activities as a transcriptional activator.
We have observed that, in mouse fibroblasts, C/EBPα has no effect on p21 promoter activity or mRNA levels or any detectable effect on p21 protein levels. Similarly, induction of C/EBPα had no effect on p27 promoter activity. Furthermore, we were unable to overcome the C/EBPα-mediated cell growth arrest with addition of simian virus 40 (SV40) T antigen or adenovirus E1A to the system. This would argue counter to p21-mediated growth arrest. On the basis of these findings, our laboratory began to study the effect of C/EBPα growth arrest on the activity of E2F-DP-responsive promoters. In this study we demonstrate that induction of C/EBPα expression in mouse fibroblasts cells leads to the appearance of C/EBPα protein in complexes bound at consensus E2F binding sites. We have also found that C/EBPα protein appears in E2F binding complexes in mouse liver and in fully differentiated 3T3-L1 cells. More significantly, we show that C/EBPα can inhibit the induction of an E2F-DP1-responsive gene in transient transfections as well as the repress the S-phase-induced transcription of DHFR and E2F-1 in permanent cell lines.
MATERIALS AND METHODS
Plasmids, cell lines, and transfections.
Details of the estradiol-regulatable expression system will be published elsewhere (K. D'Arigo and D. T. Kurtz, submitted for publication). Briefly, the DNA sequences encoding amino acids 1 to 147 of the yeast GAL4 gene were fused in frame to sequences corresponding to the C terminus of the human ER. This GAL4-ER fusion was then placed downstream of a constitutive promoter. Three GAL4 DNA binding sites (ACGGAGGACAGTCCTCCGA) were concatamerized and placed upstream of a minimal mouse c-fos promoter (sequences −55 to +110). The cDNA for C/EBPα (a kind gift from Stephen McKnight) was cloned downstream of this GAL4-Δfos promoter. The C/EBPα cDNA was also cloned in frame into the bacterial expression vector pET15b (Novagen) and into the mammalian expression vector pcDNA3 (InVitrogen). The gene for C/EBPβ was isolated as described previously (38) and cloned into the pcDNA3 expression vector. The cDNAs for mouse E2F1 and DP1 were cloned by reverse transcription-PCR from mouse liver RNA and cloned into pcDNA3. The promoter for mouse E2F-1, corresponding to sequences from −170 to +37, as well as a mutant E2F-1 promoter containing two disrupted E2F binding sites were a kind gift from Peggy Farnham. These promoter sequences were cloned into the chloramphenicol acetyltransferase (CAT) vector BL6 (5). The promoter for the mouse DHFR gene (−310 to +30) was cloned by PCR from mouse genomic DNA. The resulting fragment was also cloned into BL6CAT. A mutant DHFR promoter containing a disrupted E2F binding site was also a gift from Peggy Farnham. All constructs were made using standard cloning techniques (37). The plasmid 3×E2FCAT consists of three copies of the E2F-DP1 binding site from the E2F-1 promoter (see below) cloned upstream of Δfos-CAT. Plasmid 3×C/EBPCAT consists of three copies of the C/EBP binding site from the α2u globulin promoter (see below) cloned upstream of Δfos-CAT.
Tissue culture cells were transfected using the calcium phosphate method (1, 27). Mouse L (thymidine kinase [TK−] and adenine phosphoribosyltransferase deficient) cells were cotransfected with 1 μg of HSV-TK, coding for herpes simplex virus TK, 10 μg of the GAL4-ER plasmid, and 5 μg of GAL4-Δfos C/EBPα. Cells were selected in phenol red-free hypoxauthine-aminoplenin-thymidine (HAT) medium. Individual clones were tested for estradiol-induced C/EBPα expression by Western blotting and electrophoretic mobility shift analysis (EMSA). One clone, designated S6, was chosen for further study. Where indicated, S6 cells were “supertransfected” with RSV-neo (1 μg) and 10 μg of either the E2F-1-CAT, DHFR-CAT, CMV-E1a, or SV40 T antigen plasmid and selected in G418 (400 μg/ml) plus HAT in phenol red-free medium to generate the stable clones. Transient transfections of HEK293 and mouse L TK− cells were also performed using the calcium phosphate method. NIH 3T3 cells, HEK293 cells, and DU-145 cells were obtained from the American Type Culture Collection.
EMSA.
Nuclear extracts from cells or tissues were prepared as described previously with minor modifications (2). The following oligonucleotides were used for EMSA: oligonucleotides corresponding to a C/EBP binding site in the α2u-globulin promoter (TGTTTTGCGAAATGTAATG), the E2F site from the mouse E2F-1 promoter (GGATTTGGCGCGTAAAAGTG), or the mouse DHFR promoter (GCGATTTCGCGCCAAACTTC), a consensus SP1 binding site (TCGGGGCGGGGCGAGC), and an AP1 site (CTTGATGACTCAGCCGGAA). Nuclear proteins (2 to 5 μg) were incubated with a 32P-labeled oligonucleotide in an EMSA buffer containing 25 mM HEPES, 100 mM KCl, 4% Ficoll, 5 μM ZnCl2, 0.05% NP-40, 5 mM MgCl2, 1 μg of bovine serum albumin per ml, and 50 ng of poly(dI-dC) per μl at 4°C. Specific binding was inhibited using a 100-fold excess of unlabeled oligonucleotide corresponding to the labeled oligonucleotide. For antibody supershift experiments, nuclear extracts were incubated with for 20 min prior to addition of labeled probe with anti-C/EBPα antibody (Santa Cruz). For in vitro addition experiments, approximately 75 ng of hexahistidine-tagged C/EBPα protein purified from bacteria was added to nuclear extracts in EMSA buffer and incubated for 30 min at 0°C prior to any other additions. All samples were subjected to electrophoresis on 5% polyacrylamide gels at 4°C and visualized either by phosphoimaging (Molecular Dynamics) or autoradiography.
Immunoblotting.
Nuclear proteins (30 to 60 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% polyacrylamide gels and then transferred at 100 V and 250 to 350 mA to polyvinylidene difluoride (Millipore, Bedford, Mass.). Blots were probed with anti-C/EBPα antibody (Santa Cruz) and followed with a horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin secondary antibody (Amersham). The blot was developed using the ECL chemiluminescence detection system (Amersham).
[3H]thymidine incorporation.
Cells were plated in 24-well plates at a density of 5 × 104 cells/well. Cells were treated with estradiol for various times and then labeled with [3H]thymidine (1 μCi/ml) for 4 h. Incorporation of thymidine into DNA was measured by cold trichloroacetic acid precipitation. Protein concentrations were measured by the bicinchoninic acid method (Pierce), and counts were normalized for protein content.
FACS analysis.
For fluorescence-activated cell sorting (FACS), cells were analyzed for cell cycle parameters on a FACSCalibur (Becton Dickinson) flow cytometer utilizing a 488-nm argon-ion laser for excitation. Emission of the DNA cell cycle was detected through a 585-nm bandpass filter and acquired with CellQuest (Becton Dickinson) software. The data were analyzed using ModFit LT (Verity) software. Instrument performance is routinely monitored using DNA QC Particles and Calibrite beads (Becton Dickinson).
Purification of C/EBPα from bacteria.
Escherichia coli BL21(DE3) (pLysS) bacteria were transformed with pET-C/EBPα. A 2-ml culture was grown overnight and used to inoculate a 200-ml culture of 2XYT broth with ampicillin (100 μg/ml) and chloramphenicol (33 μg/ml). Five hours following amplification with a final concentration of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), cells were collected by centrifugation and lysed by sonication in ice-cold binding buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 5 mM imidazole) with a cocktail of protease inhibitors. Centrifugation at 39,000 × g was used to separate the insoluble and soluble fractions. The insoluble pellet was resuspended in 6 M guanidine HCl–50 mM phosphate (pH 8.0) and placed overnight with stirring at 4°C. The sample was then centrifuged at 100,000 × g; the supernatant was collected and run over a Biogel P10 column twice to renature the protein. Protein was stored at −80°C until used for EMSA in vitro addition experiments. Column eluant was determined to contain approximately 40% C/EBPα by densitometry analysis of silver-stained gels (data not shown).
RESULTS
Expression of C/EBPα in mouse L cells induces cell growth arrest.
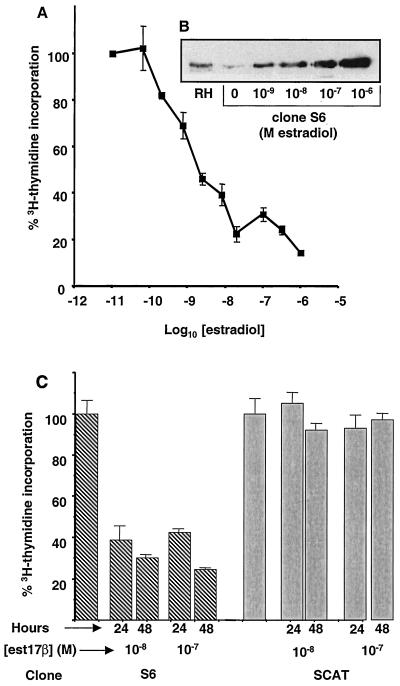
Mouse L cells were stably transfected with the estradiol-regulatable C/EBPα expression system as described in Materials and Methods. Clones surviving selection in HAT medium were tested for estradiol-inducible expression of C/EBPα by immunoblotting and EMSA. Several clones were found to display estradiol-induced expression of C/EBPα. One clone, designated S6, was further tested to determine if estradiol-dependent induction of C/EBPα led to cell cycle arrest, as seen with expression of C/EBPα in other cell lines. [3H]thymidine incorporation was analyzed at doses of estradiol ranging from 10−9 to 10−6 M and showed an estradiol dose-dependent decrease (Fig. 1A). The dose-dependent decrease in DNA synthesis correlated with an increase in the expression of C/EBPα by these cells, as shown by Western blot analysis (Fig. 1B). At 10−8 M estradiol, the level of C/EBPα protein expression in S6 cells is similar to that seen in nuclear extracts from rat hepatocytes. A marked decrease in DNA synthesis, to ∼25% of that of uninduced cells, results from a level of C/EBPα similar to that seen in hepatocytes. Thus, the cell growth suppression is not the result of gross overexpression of this transcription factor. In order to demonstrate that the growth arrest of S6 cells induced with estradiol was an effect of C/EBPα and not the GAL4-ER inducer protein, clone S6 cells were compared to clone SCAT cells. Clone SCAT cells were derived from mouse L TK− cells stably transfected with the GAL4-ER plasmid and a GAL4-driven CAT reporter gene. Cells were selected in HAT medium, and surviving clones were tested for estradiol-inducible CAT reporter gene activity. These cells express a level of the Gal4-ER protein essentially equal to that found in clone S6, as measured by Western blot analysis using an anti-ER antibody (data not shown). [3H]thymidine incorporation does not decrease following induction of SCAT cells with estradiol, while S6 cells show a marked decrease in [3H]thymidine incorporation after 24 and 48 h of treatment with estradiol (Fig. 1C).
FIG. 1.
Cell growth arrest in cell line S6. (A) [3H]thymidine incorporation assay was performed on cells containing the GAL4-ER and GAL-C/EBPα plasmids (clone S6) treated with increasing concentrations of estradiol for 72 h. Cells were pulsed with [3H]thymidine for 4 h prior to being harvested. Samples were done in duplicate. (B) S6 cells were treated with concentrations of estradiol ranging from 10−9 to 10−6 M for 24 h. Nuclear extracts were prepared from S6 cells or rat hepatocytes (RH), and Western blotting was performed using 40 μg of nuclear protein and an anti-C/EBPα antibody (Santa Cruz). No C/EBPα protein is detectable in nuclear extracts from untransfected L cells (data not shown). (C) [3H]thymidine incorporation assay was performed on cells containing the GAL4-ER-driven C/EBPα (clone S6) or a GAL4-ER-driven CAT gene (clone SCAT). Cells were treated with 10−8 or 10−7 M estradiol (est17β) for 0, 24, or 48 h and pulsed with [3H]thymidine 4 h prior to being harvested. Samples were done in triplicate, and standard deviations are shown.
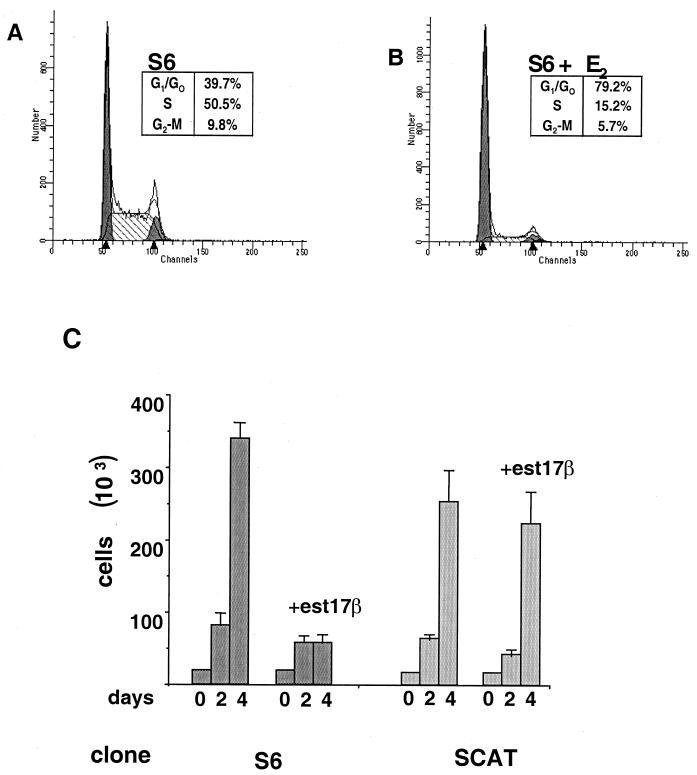
To determine the nature of the C/EBPα-induced growth arrest, cells were subjected to FACS analysis (Fig. 2A and B). As illustrated in Fig. 2A, S6 cells untreated with estradiol show a normal distribution throughout the cell cycle. S6 cells induced to express C/EBPα (Fig. 2B) show a dramatic increase in the number of cells in G0/G1, from 39.7 to 79.2%, and an equally dramatic decrease in the number of cells in S phase, from 50.5 to 15.2%. These data are indicative of a G1/G0 cell cycle arrest. Cell counts from S6 cells treated with 10−8 M estradiol showed a large decrease in cell number after 4 days compared to untreated cells, which have undergone approximately four doublings in this time, while the SCAT cells grew normally in this concentration of estradiol (Fig. 2C). To confirm that these effects were not unique to mouse L cells, NIH 3T3 cells were also stably transfected with the GAL4-ER plasmid driving C/EBPα expression, and an essentially identical pattern of cell growth arrest and [3H]thymidine incorporation was observed (data not shown).
FIG. 2.
C/EBPα-mediated growth arrest. FACS sorting of C/EBPα-expressing cells was performed. Untreated S6 cells (A) were compared to S6 cells treated with 10−7 M estradiol (E2) for 48 h (B). The percentage of cells at each phase was determined. (C) Approximately 25,000 cells of clones S6 and SCAT were plated per well of a six-well culture dish. Cells were either left untreated or treated with 10−8 M estradiol (est17β) and counted after 2 and 4 days using a hemocytometer.
C/EBPα is found in E2F binding complexes in growth- arrested S6 cells.
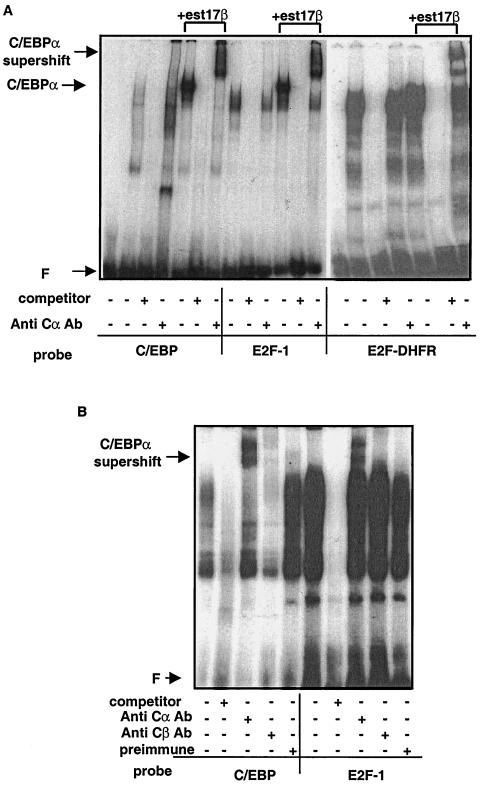
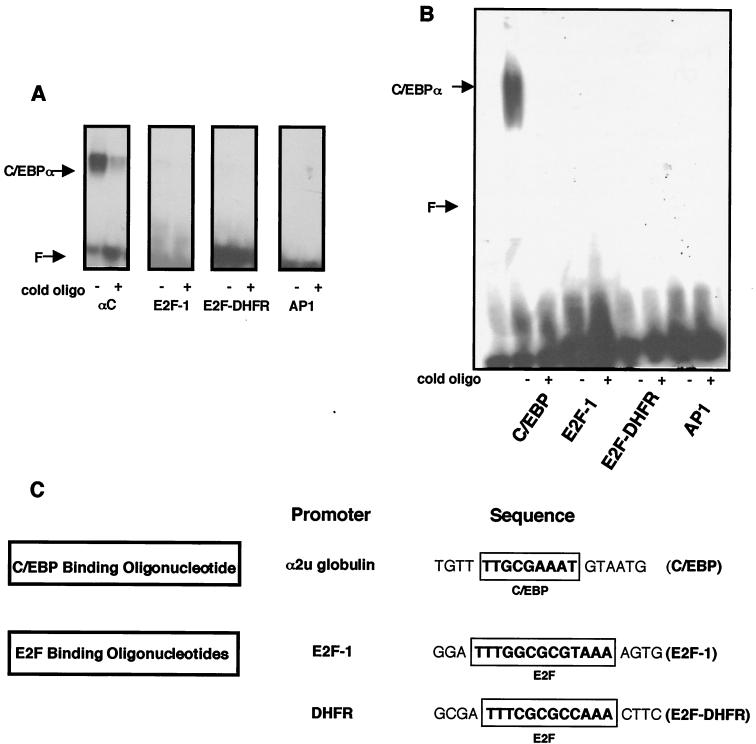
To investigate the effects of growth arrest on the pattern of E2F expression in clone S6 cells, nuclear extracts were prepared from cells following induction of C/EBPα protein and analyzed by EMSA. Extracts made from uninduced S6 cells produce a pattern of binding to a consensus C/EBP binding oligonucleotide similar to that of wild-type L TK− cells, which corresponds to the native C/EBPβ expressed in these cells. Upon induction of C/EBPα, a new shifted band appears, as expected, that supershifts with an anti-C/EBPα antibody (Fig. 3A, lanes 5 and 7). We then examined the binding of these extracts to consensus E2F binding sites from the E2F-1 and DHFR promoters, two promoters known to be regulated at the G1/S-phase transition. For both E2F oligonucleotides, a new band of binding activity appears in extracts from induced S6 cells (Fig. 3A, lanes 11 and 18). Surprisingly, when coincubated with an anti-C/EBPα antibody, the novel E2F-binding complex is supershifted, indicating that C/EBPα is present in the protein complexes which bind to a consensus E2F binding oligonucleotide (Fig. 3A, lanes 13 and 20). This novel E2F binding activity is not supershifted with nonimmune sera or with an antibody specific for C/EBPβ (data not shown).
FIG. 3.
E2F binding activity in S6 cells following C/EBPα induction. (A) EMSA was performed on 2.5 μg of nuclear extract protein from cell line S6. Extracts were incubated with a 32P-labeled oligonucleotide corresponding to a consensus C/EBP binding site or consensus E2F binding site. Specific binding was inhibited with an unlabeled oligonucleotide of the same sequence. Lanes 1 and 14 contain free probe; lanes 2 to 4, 8 to 10, and 15 to 17 contain nuclear extract from untreated S6 cells; and lanes 5 to 7, 11 to 13, and 18 to 20 contain nuclear extracts from S6 cells treated with 10−7 M estradiol (est17β). A rabbit anti-C/EBPα antibody (Anti Cα Ab) (Santa Cruz) was used to detect the presence of C/EBPα. (B) Nuclear extracts from mouse hepatocytes were incubated with the labeled oligonucleotides, and supershift analysis was carried out using preimmune serum, the anti-C/EBPα antibody, or an anti-C/EBPβ antibody corresponding to the N terminus of C/EBPβ (Kurtz, unpublished). F, free probe.
C/EBPα is found associated with E2F in tissues in vivo.
To determine if the appearance of C/EBPα, in these binding complexes was simply an artifact of ectopic expression of C/EBPα in these mouse fibroblasts, nuclear extracts from mouse hepatocytes were analyzed by EMSA (Fig. 3B). Lanes 1 to 5 depict the binding activity of these extracts to a consensus C/EBP binding oligonucleotide and show that binding can be supershifted with both anti-C/EBPα and anti-C/EBPβ antibodies. However, when extracts are incubated with a consensus E2F binding oligonucleotide from the E2F-1 promoter, binding activity can be supershifted in the presence of a C/EBPα antibody and does not supershift in the presence of a C/EBPβ-specific antibody (Fig. 3B, lanes 6 to 10). When this experiment was repeated using extracts from 3T3-L1 cells induced to undergo the adipocyte-like differentiation program, C/EBPα was also found associated with E2F binding complexes (data not shown). Thus, the appearance of C/EBPα in the E2F binding complex is not merely an artifact of our expression system but is also found in vivo situations where C/EBPα has been demonstrated to act as a negative regulator of cell growth. The association of C/EBPα with the E2F complex was also found in the NIH 3T3 cells that had been stably transfected with the C/EBPα expression system (data not shown).
Appearance of C/EBPα in the E2F binding complex is dose dependent in cell line S6.
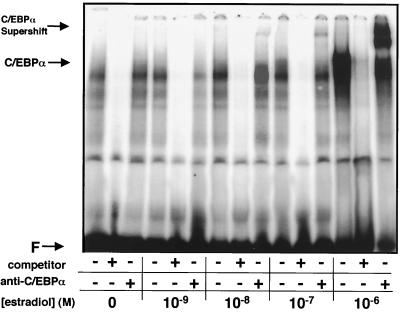
Modulating the inducing dose of estradiol can tightly regulate the level of protein expression in the Gal4-ER-inducible expression system. As shown in Fig. 1A and B, increasing doses of estradiol result in increased expression of C/EBPα protein, which correlated with a decrease in [3H]thymidine incorporation. Similarly, when these cell extracts are subjected to EMSA with an E2F-specific oligonucleotide, a band corresponding to the complex that is supershifted by the anti-C/EBPα antibody grows in intensity with increasing doses of estradiol and increased induction of C/EBPα protein expression (Fig. 4). Additionally, appearance of this binding is concurrent with an increase in the amount of C/EBPα that is seen to be supershifted when using an anti-C/EBPα antibody. This indicates a dose-dependent effect of C/EBPα in the E2F binding entities.
FIG. 4.
Dose-dependent appearance of C/EBPα in the E2F binding complex. Cell line S6 was treated with concentrations of estradiol ranging from 10−9 to 10−6 M for 24 h prior to the preparation of nuclear extracts. Extracts were incubated with a 32P-labeled oligonucleotide corresponding to the E2F binding site in the E2F-1 promoter. Specific binding was inhibited with an unlabeled oligonucleotide of the same sequence. An anti-C/EBPα antibody was used to detect the presence of C/EBPα in the binding pattern. F, free probe.
C/EBPα does not bind directly to E2F binding oligonucleotides.
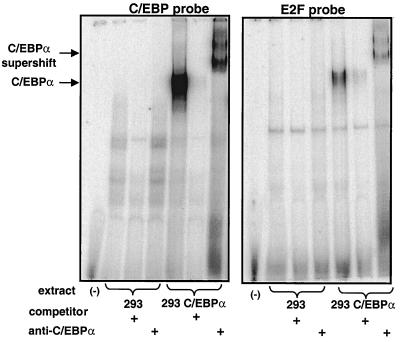
The presence of C/EBPα as a binding moiety of E2F consensus binding oligonucleotides was examined further. Neither of the E2F binding oligonucleotides derived from the E2F-1 and DHFR promoters contains a consensus C/EBP binding domain (Fig. 5C). However, we examined the possibility that C/EBPα may be binding directly to E2F consensus binding motifs. A bacterially expressed peptide fragment of C/EBPα corresponding to the C-terminal 63 amino acids (a gift from S. L. McKnight) was used in an EMSA with E2F binding oligonucleotides from the E2F-1 and DHFR promoters as well as an oligonucleotide corresponding to the TRE response element bound by AP1. The bacterially expressed fragment, corresponding to the b-zip domain of C/EBPα, bound to a consensus C/EBP binding oligonucleotide, as expected, but not to the E2F binding sites (Fig. 5A). Next, we examined the binding of a full-length C/EBPα protein expressed in bacteria to the same set of oligonucleotides. The full-length purified C/EBPα also only bound directly to a consensus C/EBP binding oligonucleotide and not to the E2F sites (Fig. 5B), indicating that the appearance of C/EBPα in E2F binding complexes found in cells (Fig. 3 and 4) may be mediated through additional protein-protein interactions and/or altered protein-DNA binding specificity. When C/EBPα was expressed in HEK293 cells by transient transfection, the expected new binding activity was found with the C/EBP binding oligonucleotide and a new band was also found with the E2F binding oligonucleotide; both of these bands supershifted upon addition of anti-C/EBPα antibody (Fig. 6). Identical results were obtained when C/EBPα was expressed in COS7 cells (data not shown). These data suggest that the binding of C/EBPα in the E2F binding complexes may involve additional proteins that are not present when the protein is purified to near homogeneity from bacteria but are present in nuclear extracts of mammalian cells when the protein is transiently expressed.
FIG. 5.
Lack of direct binding of C/EBPα to oligonucleotides with E2F consensus binding sites. (A) EMSA was used to analyze a truncated C/EBPα protein corresponding to the DNA binding and leucine zipper domains of C/EBPα (αC) purified from bacteria (gift from S. L. McKnight) and incubated with the indicated 32P-labeled oligonucleotides. Specific binding was inhibited with an unlabeled (cold) oligonucleotide (oligo) of the same sequence. (B) Full-length C/EBPα protein with a N-terminal hexahistidine tag was expressed and purified from bacteria and incubated with 32P-labeled oligonucleotides and analyzed as in panel A. (C) Comparison of oligonucleotides used in panels A and B. The E2F binding oligonucleotides derived from the DHFR and E2F-1 promoters contain only consensus E2F binding domains.
FIG. 6.
Binding activity of C/EBPα expressed in HEK293 cells. HEK293 cells were transiently transfected with pcDNA3 or a pcDNA3-C/EBPα construct. Nuclear extracts from these cells were analyzed by EMSA using a 32P-labeled oligonucleotides corresponding to a C/EBP site or an E2F binding site from the E2F-1 promoter. The presence of C/EBPα in the E2F binding complex was determined by supershift analysis with an anti-C/EBPα antibody. C/EBPβ plasmid transfected into these cells results in binding to the C/EBP site but no binding to the E2F site (data not shown).
C/EBPα expressed in bacteria will combine with E2F complexes in vitro.
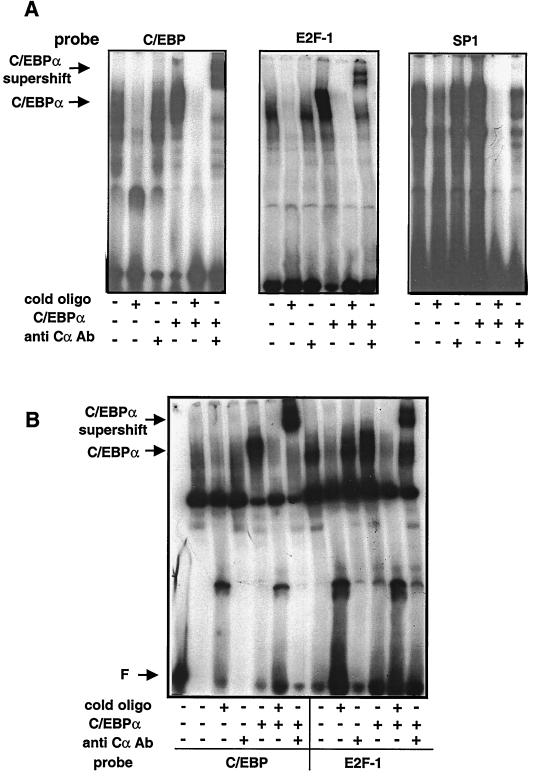
To further examine the interaction of C/EBPα with proteins binding to an E2F consensus binding sequence, the in vitro association of bacterially expressed C/EBPα with proteins from nuclear extracts was analyzed using EMSA. C/EBPα purified from bacteria was incubated with nuclear extracts from mouse L TK− cells, the parental cell line of S6 cells. The mixture was then probed with 32P-labeled oligonucleotides corresponding to binding sites for C/EBP, E2F, or SP1. In the presence of a C/EBP binding oligonucleotide, addition of C/EBPα expressed in bacteria yielded, as expected, a new band that was supershifted with the addition of anti-C/EBPα antibody (Fig. 7A, lanes 3 to 6). When an oligonucleotide containing an E2F consensus binding domain was used, a new band of binding activity was present upon addition of bacterially expressed C/EBPα and was supershifted upon addition of anti-C/EBPα antibody (Fig. 7A, lanes 10 to 12). This was observed with E2F consensus binding oligonucleotides from both the E2F-1 promoter and the DHFR promoter (data not shown). As a negative control, samples incubated with an SP1 binding oligonucleotide did not show any new binding activity upon addition of the bacterially expressed C/EBPα, nor was a supershift visible upon addition of an anti-C/EBPα antibody (Fig. 7A, lanes 16 to 18).
FIG. 7.
Bacterially expressed C/EBPα binds in the E2F complex in vitro. (A) EMSA was performed on nuclear extracts from mouse L TK− cells, the parental line of S6 cells. Where indicated, extracts were incubated with C/EBPα protein expressed in bacteria. Where indicated, the mixtures were further incubated with an anti-C/EBPα antibody (Cα Ab) and then with a probe corresponding to either a consensus C/EBP, E2F-1, or SP1 DNA binding domain. In each EMSA, the last three lanes show the addition of bacterially expressed C/EBPα protein to L TK− cell nuclear extract. Supershifts are indicated by the arrow at the left. (B) Analysis of C/EBPα binding in DU-145 cells. EMSA was performed on nuclear extracts from DU-145 cells, a human prostate cell line deficient in functional Rb protein expression. Where indicated, extracts were incubated with C/EBPα protein expressed in bacteria. Following incubation, the mixtures were further incubated with an anti-C/EBPα antibody and then with probe corresponding to either a consensus C/EBP or E2F-1 DNA binding domain. Lane 1 contains free probe only. Lanes 5 to 7 and 11 to 13 show the addition of bacterially expressed C/EBPα protein to DU-145 extracts. The arrow at left indicates supershifts in lanes 7 and 13.
Since bacterially expressed C/EBPα is not capable of binding directly to the E2F-1 or DHFR promoter-derived oligonucleotides, it appears that C/EBPα must bind to another protein(s) present in the L cell nuclear extract that allows for its appearance in complexes bound at an E2F binding site. Alternatively, C/EBPα may undergo a posttranslational modification in eukaryotic cells that enables it to bind E2F sequences. If such a posttranslational modification is responsible for the binding, it must occur at 0°C within 30 min.
Appearance of C/EBPα in E2F binding complexes does not require functional Rb protein.
Regulation of gene transcription at the G1-S transition by E2F is tightly controlled through the E2F binding protein Rb. It was first claimed that C/EBPβ can interact with Rb in U937 cells during differentiation and, by extension, that C/EBPα may also be capable of interactions with Rb, as shown in vitro glutathione S-transferase pulldown experiments, although this was never shown to occur in vivo (8, 9). These observations led us to investigate whether C/EBPα association with the E2F binding complex requires Rb. DU-145 cells are a human prostate cell line that express very low levels of a Rb protein which is truncated at amino acid 715 (36). This mutation truncates the protein in the B box required for E2F binding and renders the protein unable to bind E2F or repress cell growth (24, 39). Nuclear extracts were prepared from DU-145 cells and used in addition experiments with C/EBPα purified from bacteria. Bacterially expressed C/EBPα was incubated with DU-145 cell nuclear extracts and then analyzed by EMSA using either a C/EBP or E2F binding oligonucleotide (Fig. 7B). When an anti-C/EBPα antibody was added, a strong band was supershifted in the presence of both the C/EBP and E2F binding oligonucleotides (Fig. 7B, lanes 7 and 13). The ability of C/EBPα to bind in a protein complex in the presence of E2F binding proteins in the absence of a functional Rb protein indicates that Rb is not required for association of C/EBPα with the E2F complex.
C/EBPα represses transcriptional activation by E2F-DP1.
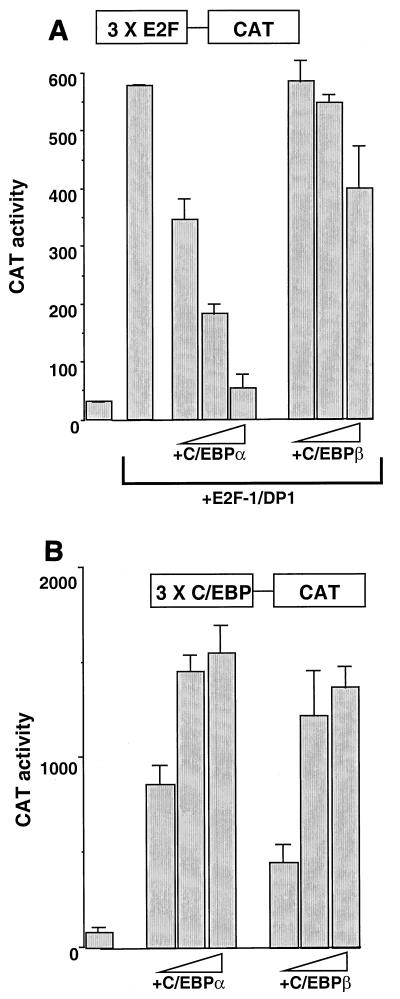
In order to determine relevance for the physical association of C/EBPα in E2F binding complexes, we examined the role that C/EBPα might play in the activation or repression of transcription of both artificial and gene-derived promoters. A concatamer of three E2F binding sites upstream of a minimal fos promoter was fused to the CAT reporter gene (3×E2F-CAT) and transiently transfected into mouse L cells. Addition of a plasmid expressing C/EBPα had no effect on the activity of this promoter (data not shown). When plasmids expressing mouse E2F1 and DP1 were included in the transfection, a large (15 to 20-fold) increase in CAT activity was seen, as expected (Fig. 8A). Titration of increasing amounts of the C/EBPα expression plasmid into the transfection resulted in a dramatic decrease in CAT activity, while addition of a plasmid expressing C/EBPβ had little if any effect. C/EBPα and -β were equally effective at inducing a C/EBP-responsive promoter (Fig. 8B). These data strongly suggest a direct effect of C/EBPα in suppressing transcription mediated by E2F-DP1.
FIG. 8.
Direct repression of E2F-DP1-mediated transcription by C/EBPα. (A) Mouse L cells were transfected with 500 ng of plasmid 3×E2F-CAT. Where indicated, 250 ng each of E2F1 and DP1 cloned into pcDNA3 were added. pcDNA3 plasmids containing either C/EBPα or -β were added at 100, 250, or 500 ng. pcDNA3 was added to all transfections to normalize for the amount of total DNA used. An internal control RSV-luciferase plasmid was added to all transfections. After 48 h, CAT activity was measured and normalized to luciferase activity. Transfections were performed in duplicate. The data shown are representative of at least four different determinations. (B) Mouse L cells were transfected with 500 ng of plasmid 3×C/EBP-CAT. The pcDNA3-C/EBPα or -β plasmids were added at 100, 250, or 500 ng. pcDNA3 was added to all transfections to normalize for the amount of DNA used. An internal control RSV-luciferase plasmid was added to all transfections. After 48 h, CAT activity was measured and normalized to luciferase activity.
Expression of C/EBPα represses S-phase induction of the E2F-1 and DHFR promoters.
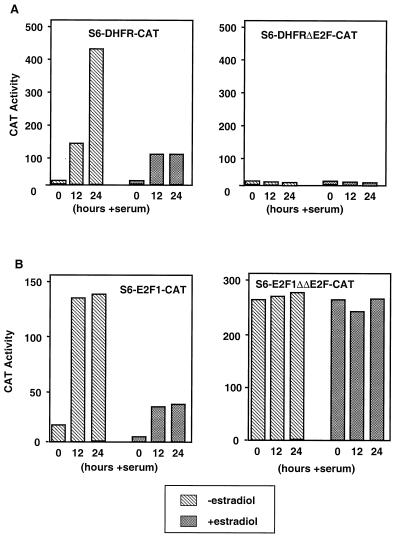
The effect of expression of C/EBPα on the transcription of promoters regulated by E2F was then examined in permanent cell lines. S6 cells were stably transfected with plasmids encoding wild-type or mutant E2F-1 and DHFR promoters linked to the CAT reporter gene. Clones positive for both estradiol-induced C/EBPα expression and serum-inducible CAT activity were identified. These clones were serum starved for 48 h and then induced to enter S phase by serum addition for 12 or 24 h. As expected, serum stimulation of these cells resulted in a large induction of CAT activity driven by the wild-type E2F-1 and DHFR promoters (Fig. 9). However, when the expression of C/EBPα was induced with estradiol, the induction of CAT was reduced to less than 25% of that in cells not treated with estradiol (Fig. 9). The mutant DHFR promoter, containing a disruption of the E2F binding site, was not serum inducible, as expected. In contrast, the mutant E2F-1 promoter, containing a disruption of both E2F binding sites, displayed a “constitutive high” phenotype, i.e., it was active in the absence of serum and this activity was not diminished by C/EBPα. These data suggest that C/EBPα, through its interaction with the E2F sites, acts to repress S-phase activation of these promoters.
FIG. 9.
Repression of S-phase-regulated promoters in S6 cells. (A) S6 cells were stably transfected with CAT reporter gene constructs containing either the −310 to +30 fragment of the murine DHFR promoter or the corresponding promoter fragment containing a mutant E2F site. (B) S6 cells were stably transfected with CAT reporter gene constructs containing either the −170 to +37 fragment of the murine E2F-1 promoter or the corresponding promoter fragment containing a two mutant E2F sites. Cells were treated with estradiol for 24 h to induce C/EBPα and then serum starved for 48 h. Serum was added back to the cells for 12 or 24 h. Cells were harvested and assayed for CAT activity, which was normalized for protein content. The result shown is one representative of several different determinations.
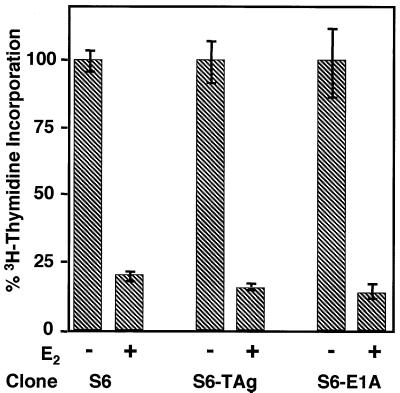
Oncogenes cannot override C/EBPα-mediated growth arrest.
Previous work in our laboratory had failed to find a link between the expression of C/EBPα in S6 cells and the induction of CKI proteins such as p21 and p27. Our data suggest the possibility that C/EBPα acts downstream of Rb as a “second-line” inhibitor of E2F-driven transcription. To test this possibility, we employed two viral proteins known to activate E2F-driven transcription through inactivation of Rb: SV40 T antigen and adenoviral E1A. S6 cells were stably transfected with constructs for either viral oncogene and selected for expression of the viral oncogene and inducible C/EBPα expression by Western blotting. Cells positive for expression of both entities were subjected to a [3H]thymidine incorporation assay. As illustrated in Fig. 10, expression of E1A or T antigen had no effect on the C/EBPα-mediated growth arrest. This was also confirmed by cell counts (data not shown). These data, coupled with the fact that Rb is not required for C/EBPα binding in the E2F complex in Rb mutant DU-145 cells (Fig. 7B), indicate to us that C/EBPα may play a role in the suppression of E2F-DP-mediated transcription at a position downstream of Rb-mediated events.
FIG. 10.
Viral oncogenes cannot override C/EBPα-mediated growth arrest. S6 cells were stably transfected with plasmids constitutively expressing either adenoviral E1A or SV40 T antigen (TAg) and reselected for their ability to express both C/EBPα and viral proteins. Cells were then assayed for [3H]thymidine incorporation 48 h following induction of C/EBPα expression. Results shown are from clones which are representative of several clones which contain E1a or T antigen.
DISCUSSION
Expression of C/EBPα in a variety of systems has been linked to growth arrest. We have used a GAL4-ER-driven expression system to examine the mechanism of growth arrest in mouse fibroblast cell lines. Using this system, we are able to express levels of C/EBPα comparable to that found in the liver in mouse L cells and NIH 3T3 cells. At these levels of C/EBPα, we observed a robust suppression of cell growth (Fig. 1). We have attempted to reverse the growth-arrested state of these cells with several known oncogenes and viral proteins known to override the antimitogenic effects of other factors. Growth arrest in S6 cells cannot be reversed by SV40 T antigen, by adenovirus E1a protein, or by oncogenic ras (data not shown). SV40 T antigen and E1a bind pocket proteins and induce rapid entry into S phase, thereby overriding the Rb-mediated control of E2F-directed S-phase transcription (15, 49). The action of p21 as a CK occurs upstream of Rb, and it would be expected that if C/EBPα-mediated control of p21 was integral to growth arrest, SV40 T antigen and E1a would override this effect.
These data, accompanied with the observation that, in mouse fibroblasts, there was no apparent change in the p21 levels induced by C/EBPα, led us to examine the E2F binding in cells before and after expression of C/EBPα. The only change in binding observed between L cells expressing C/EBPα and nonexpressing cells was the appearance of a new band migrating more slowly than the other E2F-DP complexes (Fig. 3A). This new band of binding activity could be supershifted using an anti-C/EBPα antibody. This new E2F binding activity could also be observed in NIH 3T3 cells stably transfected with the Gal4-ER-inducible C/EBPα construct. When increasing doses of estradiol were used to induce C/EBPα expression in L cells, we also observed a dose-dependent increase in appearance of this new band of binding (Fig. 4). This is in direct agreement with the dose-dependent decrease in [3H]thymidine incorporation and C/EBPα expression demonstrated in Fig. 1. This binding is also seen in nuclear extracts made from tissues as well as from terminally differentiated cells. In extracts made from mouse liver, a subset of proteins bound at an E2F binding sequence from the E2F-1 promoter are supershifted with antibodies to C/EBPα but not with antibodies to the related factor C/EBPβ (Fig. 3B). Similar results were seen in terminally differentiated 3T3-L1 cells (data not shown). While these bands represent only a small subset of the total amount of E2F expressed in liver, it is important to note that in both L cells and NIH 3T3 cells, the pattern of E2F binding is not as complex as that seen in liver and contributes to the appearance of C/EBPα as an apparently major band of E2F binding activity when expressed in L cells. We were unable to observe binding of either a pure peptide fragment or purified full-length C/EBPα to any E2F consensus binding site, although both proteins bound to a consensus C/EBP binding sequence (Fig. 5). When C/EBPα was expressed in HEK293 or COS7 cells, we observed a new band of binding using an E2F binding oligonucleotide, and as before, this band could be supershifted using an anti-C/EBPα antibody.
These observations suggested that, in order for C/EBPα to be found in the E2F-DP binding complexes, this association must be mediated either through a unique DNA-protein contact or a novel protein-protein interaction or, conceivably, a posttranslational modification that occurs only in eukaryotic cells. A precedent exists for the alteration of a consensus binding sequence for another member of the C/EBP family of transcription factors. When C/EBPβ forms a heterodimer with a Rel homology domain, the binding specificity of the heterodimer is no longer a consensus C/EBP binding site (40). It is also possible that C/EBPα may be involved in direct protein-protein interactions with some E2F and/or DP isoform.
Mixing experiments were performed in which bacterially expressed full-length C/EBPα was added to nuclear extracts from growing L cells. When a mixture of bacterially expressed C/EBPα and L cell nuclear extracts was resolved by EMSA using an E2F binding oligonucleotide, we again observed a new band of binding that corresponded to C/EBPα by supershift analysis (Fig. 7). These results suggest that if the association of C/EBPα with the E2F-DP complex is the result of posttranslational modification, this modification must occur at 0°C in a relatively short period of time in vitro. The same mixing experiment was performed using nuclear extracts from cells deficient in a functional Rb protein: when nuclear extracts made from DU-145 cells were used in the mixing experiments, we saw no alteration in the appearance of C/EBPα in E2F-DP binding complexes. While this observation does not rule out an association of C/EBPα with another pocket protein such as p107 or p130, it does indicate that Rb is not a key player in the association of C/EBPα with the E2F-DP complex. We observed evidence for the physical association of C/EBPα with E2F binding complexes not only in our tissue culture model, but also in mouse liver extracts and in fully differentiated 3T3-L1 cells. Currently in our laboratory we are assessing the domains of C/EBPα required for this interaction and what proteins are observed in this complex.
C/EBPα cannot activate transcription from a promoter containing a concatamer of E2F sites. When a 3×E2F-CAT construct was cotransfected into L cells with plasmids encoding C/EBPs, no induction was seen, indicating that, in the cellular context, C/EBPα is not functioning as a simple transcriptional activator. However, this artificial promoter is inducible by E2F-DP1, and significantly, C/EBPα is able to suppress this transcriptional activation.
Furthermore, C/EBPα was able to repress the cell cycle-mediated activation of both the E2F-1 and DHFR promoters in permanent cell lines, and this suppression occurred only in promoters containing functional E2F binding sites. These observations lend a mechanistic significance to the physical association of C/EBPα with E2F binding complexes and demonstrate a straightforward mechanism for C/EBPα-mediated control of cell growth, namely, growth arrest through transcriptional repression. Consistent with this model is our observation that the induction of C/EBPα in S6 cells leads to a rapid (within 12 h) downregulation of the mRNAs for several cell cycle-regulated genes, including cyclin D1 and E2F-1 (data not shown).
While this paper was in preparation, a report by Timchenko et al. pointed to a role for C/EBPα in disrupting the association of E2F4 with the pocket protein p107 (44) and reported that the induction of C/EBPα in HT1080 cells resulted in a loss of E2F binding. No mention was made in this latest report concerning the stabilization of p21 protein by C/EBPα. The results presented herein are not in agreement with these observations. We find that the induction of C/EBPα in mouse fibroblasts leads to the gain of a new E2F binding activity which contains C/EBPα and almost certainly is not mediated by p107. Growth arrest in our cells is not rescued by addition of SV40 T antigen or E1a, which bind not only to Rb but also the related pocket proteins p107 and p130 (52), and would argue against C/EBPα acting by disrupting E2F associations with the pocket protein family as a mechanism of growth arrest. Indeed, in our cell lines containing inducible C/EBPα and expressing T antigen or E1A, C/EBPα is still found in the E2F binding complex (data not shown).
It is still not clear if there is any specificity in the activation of transcription by discrete E2F or DP isoforms at individual S-phase-inducible promoters or whether this specificity is dependent on cell type. It therefore remains possible that the disruption by C/EBPα of p107-E2F4 complexes may occur in mouse liver; however, how this disruption would suppress E2F-mediated transcription is not clear.
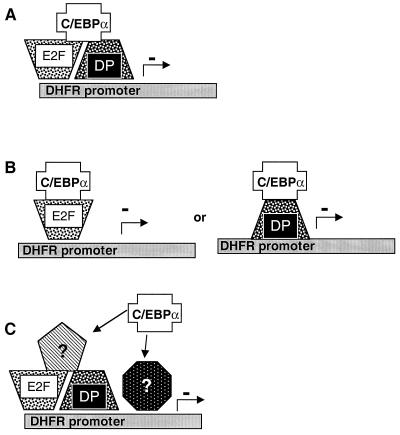
Figure 11 illustrates several possible models for C/EBPα-mediated suppression of cell growth. Most simply, C/EBPα may act as a “surrogate” Rb and prevent E2F-DP-mediated transcriptional activation even when these proteins are bound at E2F sites (Fig. 11A). Alternatively, C/EBPα may be able to form a dimer with some E2F or DP isoform. Both E2F and DP proteins contain a leucine zipper domain and may interact in some way with the leucine zipper domains of C/EBPα, with repression of transcription resulting from an improper transactivation domain for the context of the promoter (Fig. 11B). Another possibility is that C/EBPα acts in concert with another as yet unknown protein, completely unrelated to E2F or DP, which binds E2F sites directly and with increased avidity over E2F-DP pairs, resulting in no transactivation.
FIG. 11.
Models of C/EBPα action on an S-phase-regulated promoter. At least three potential models can explain the role of C/EBPα in cell growth arrest. (A) C/EBPα may act in place of Rb as a negative regulator of the transactivation potential of the E2F-DP heterodimer. (B) Alternatively, C/EBPα may bind either E2F or DP, resulting in a heterodimer incapable of activating E2F-driven promoters. (C) Potentially, C/EBPα acts through an as yet uncharacterized protein to effect its results on E2F-driven transcription, either by competing with E2F-DP heterodimers for DNA binding or by interfering with transactivation.
Our results indicate that expression of C/EBPα in cells at physiologically relevant levels leads to cell growth arrest. While expression of C/EBPα may correlate with other events in the cell during the establishment of this cell cycle block, these events may be secondary to the physical association of C/EBPα with S-phase promoters and its activity as a transcriptional repressor.
ACKNOWLEDGMENTS
This work was supported by NIH grant DK46446, DOE grant DE-FC02-98CH10902, and an MUSC University Research Committee grant to D.T.K.
The expert technical assistance of Susan Brady and Stephanie Cook is gratefully acknowledged. We also wish to acknowledge Candace Enockson, MT(ASCP), operator of the Analytical Flow Cytometry Shared Facility at the Medical University of South Carolina, as well as the MUSC Biotechnology Resource Laboratory for DNA sequencing and the MUSC BioMolecular Computing Resource for DNA sequence analysis.
REFERENCES
- 1.Addison W R, Kurtz D T. Nucleotide sequences required for the regulation of a rat alpha2u-globulin gene by glucocorticoids. Mol Cell Biol. 1986;6:2334–2346. doi: 10.1128/mcb.6.7.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addison W R, Kurtz D T. Identification of nuclear proteins that bind to the glucocorticoid regulatory region of a rat alpha 2u-globulin gene. J Biol Chem. 1989;264:21891–21895. [PubMed] [Google Scholar]
- 3.Birkenmeier E H, Gwynn B, Howard S, Jerry J, Gordon J I, Landschulz W H, McKnight S L. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev. 1989;3:1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- 4.Boshart M, Kluppel M, Schmidt A, Schutz G, Luckow B. Reporter constructs with low background activity utilizing the CAT gene. Gene. 1992;110:129–130. doi: 10.1016/0378-1119(92)90456-y. [DOI] [PubMed] [Google Scholar]
- 5.Brookstein R, Shew J-Y, Chen P-L, Scully P, Lee W-H. Suppression of tumorgenicity of human prostate carcinoma cells by replacing a mutated Rb gene. Science. 1989;247:712–715. doi: 10.1126/science.2300823. [DOI] [PubMed] [Google Scholar]
- 6.Cao Z, Umek R M, McKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 7.Cha H H, Cram E J, Wang E C, Huang A J, Kasler H G, Firestone G L. Glucocorticoids stimulate p21 gene expression by targeting multiple transcriptional elements within a steroid responsive region of the p21waf1/cip1 promoter in rat hepatoma cells. J Biol Chem. 1998;273:1998–2007. doi: 10.1074/jbc.273.4.1998. [DOI] [PubMed] [Google Scholar]
- 8.Chen P L, Riley D J, Chen-Kiang S, Lee W H. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P L, Riley D J, Chen Y, Lee W H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 10.Chow K N, Dean D C. Domains A and B in the Rb pocket interact to form a transcriptional repressor motif. Mol Cell Biol. 1996;16:4862–4868. doi: 10.1128/mcb.16.9.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cram E J, Ramos R A, Wang E C, Cha H H, Nishio Y, Firestone G L. Role of the CCAAT/enhancer binding protein-α transcription factor in the glucocorticoid stimulation of p21waf1/cip1 gene promoter activity in growth-arrested rat hepatoma cells. J Biol Chem. 1998;273:2008–2014. doi: 10.1074/jbc.273.4.2008. [DOI] [PubMed] [Google Scholar]
- 12.Darlington G J, Wang N, Hanson R W. C/EBP alpha: a critical regulator of genes governing integrative metabolic processes. Curr Opin Genet Dev. 1995;5:565–570. doi: 10.1016/0959-437x(95)80024-7. [DOI] [PubMed] [Google Scholar]
- 13.Di Cunto F, Topley G, Calautti E, Hsiao J, Ong L, Seth P K, Dotto G P. Inhibitory function of p21Cip1/WAF1 in differentiation of primary mouse keratinocytes independent of cell cycle control. Science. 1998;280:1069–1072. doi: 10.1126/science.280.5366.1069. [DOI] [PubMed] [Google Scholar]
- 14.El Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1 a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 15.Endo T, Goto S. Retinoblastoma gene product Rb accumulates during myogenic differentiation and is deinduced by the expression of SV40 large T antigen. J Biochem. 1992;112:427–430. doi: 10.1093/oxfordjournals.jbchem.a123916. [DOI] [PubMed] [Google Scholar]
- 16.Friedman A D, Landschulz W H, McKnight S L. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- 17.Fry C J, Farnham P J. Context-dependent transcriptional regulation. J Biol Chem. 1999;274:29583–29586. doi: 10.1074/jbc.274.42.29583. [DOI] [PubMed] [Google Scholar]
- 18.Fry C J, Pearson A, Malinowski E, Bartley S M, Greenblatt J, Farnham P J. Activation of the murine dihydrofolate reductase promoter by E2F1: a requirement for CBP recruitment. J Biol Chem. 1999;274:15883–15891. doi: 10.1074/jbc.274.22.15883. [DOI] [PubMed] [Google Scholar]
- 19.Guo K, Wang J, Andres V, Smith R C, Walsh K. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol Cell Biol. 1993;15:3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haber B A, Mohn K L, Diamond R H, Taub R. Induction patterns of 70 genes during nine days after hepatectomy define the temporal course of liver regeneration. J Clin Investig. 1993;91:1319–1326. doi: 10.1172/JCI116332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 23.Hemati N, Ross S E, Erickson R L, Groblewski G E, MacDougald O A. Signaling pathways through which insulin regulates CCAAT/enhancer binding protein alpha (C/EBPα) phosphorylation and gene expression in 3T3-L1 adipocytes: correlation with GLUT4 gene expression. J Biol Chem. 1997;272:25913–25919. doi: 10.1074/jbc.272.41.25913. [DOI] [PubMed] [Google Scholar]
- 24.Herwig S, Strauss M. The retinoblastoma protein: a master regulator of cell cycle, differentiation, and apoptosis. Eur J Biochem. 1997;246:581–601. doi: 10.1111/j.1432-1033.1997.t01-2-00581.x. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao K-M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 26.Jiang H, Lin J, Su Z Z, Collart F R, Huberman E, Fisher P B. Induction of differentiation in human promyelocytic HL-60 leukemia cells activates p21, WAF1/CIP1, expression in the absence of p53. Oncogene. 1994;9:3397–3406. [PubMed] [Google Scholar]
- 27.Kurtz D T. Hormonal inducibility of rat α2u globulin genes in transfected mouse cells. Nature. 1981;291:629–632. doi: 10.1038/291629a0. [DOI] [PubMed] [Google Scholar]
- 28.Lekstrom-Himes J, Xanthopoulos K G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 29.Lin F T, Lane M D. CCAAT/enhancer-binding protein alpha is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc Natl Acad Sci USA. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacLeod K F, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 31.Means A L, Slansky J E, McMahon S L, Knuth M W, Farnham P J. The HIP binding site is required for growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1992;16:1054–1063. doi: 10.1128/mcb.12.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mischoulon D, Rana B, Bucher N L, Farmer S R. Growth-dependent inhibition of CCAAT enhancer-binding protein (C/EBPα) gene expression during hepatocyte proliferation in the regenerating liver and in culture. Mol Cell Biol. 1992;12:2553–2560. doi: 10.1128/mcb.12.6.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 34.O'Rourke J P, Newbound G C, Hutt J A, DeWille J. CCAAT/enhancer-binding protein delta regulates mammary epithelial cell G(0) growth arrest and apoptosis. J Biol Chem. 1999;274:16582–16589. doi: 10.1074/jbc.274.23.16582. [DOI] [PubMed] [Google Scholar]
- 35.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273:29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- 36.Rubin S J, Hallahan D E, Ashman C R, Brachman D G, Beckett M A, Virudachalam S, Yandell D W, Weichselbaum R R. Two prostate carcinoma cell lines demonstrate abnormalities in tumor suppressor genes. J Surg Oncol. 1991;46:31–36. doi: 10.1002/jso.2930460108. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schwartz D A, Kurtz D T. Sequence requirements for secondary glucocorticoid inducibility of rat alpha 2u globulin genes. Mol Cell Endocrinol. 1996;120:153–159. doi: 10.1016/0303-7207(96)03833-6. [DOI] [PubMed] [Google Scholar]
- 39.Sellers W R, Kaelin W G., Jr Role of the retinoblastoma protein in the pathogenesis of human cancer. J Clin Oncol. 1997;15:3301–3312. doi: 10.1200/JCO.1997.15.11.3301. [DOI] [PubMed] [Google Scholar]
- 40.Stein B, Cogswell P C, Baldwin A S., Jr Functional and physical associations between NF-kappa B and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timchenko N A, Wilde M, Nakanishi M, Smith J R, Darlington G J. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 42.Timchenko N A, Harris T E, Wilde M, Bilyeu T A, Burgess-Beusse B L, Finegold M J, Darlington G J. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol Cell Biol. 1997;17:7353–7361. doi: 10.1128/mcb.17.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Timchenko N A, Wilde M, Kosai K I, Heydari A, Bilyeu T A, Finegold M J, Mohamedali K, Richardson A, Darlington G J. Regenerating livers of old rats contain high levels of C/EBPalpha that correlate with altered expression of cell cycle associated proteins. Nucleic Acids Res. 1998;26:3293–3299. doi: 10.1093/nar/26.13.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timchenko N A, Wilde M, Darlington G J. C/EBPalpha regulates formation of S-phase-specific E2F-p107 complexes in livers of newborn mice. Mol Cell Biol. 1999;19:2936–2945. doi: 10.1128/mcb.19.4.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 46.Umek R M, Friedman A D, McKnight S L. CCAAT-enhancer binding proteins: a component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- 47.van Ginkel P R, Hsiao K-M, Schjerven H, Farnham P J. E2F-mediated growth regulation requires transcription factor cooperation. J Biol Chem. 1997;272:18367–18374. doi: 10.1074/jbc.272.29.18367. [DOI] [PubMed] [Google Scholar]
- 48.Wang N D, Finegold M J, Bradley A, Ou C N, Abdelsayed S V, Wilde M D, Taylor L R, Wilson D R, Darlington G J. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 49.Watkins P J, Condreay J P, Huber B E, Jacobs S J, Adams D J. Impaired proliferation and tumorigenicity induced by CCAAT/enhancer-binding protein. Cancer Res. 1996;56:1063–1067. [PubMed] [Google Scholar]
- 50.Wu Z, Rosen E D, Brun R, Hauser S, Adelmant G, Troy A E, McKeon C, Darlington G J, Spiegelman B M. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 51.Yeh W C, Cao Z, Classon M, McKnight S L. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- 52.Zalvide J, DiCaprio J A. Role of pRb-related proteins in simian virus 40 large T-antigen-mediated transformation. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]