Abstract
The ADP-ribosylation factor 6 (ARF6) GTPase has a dual function in cells, regulating membrane traffic and organizing cortical actin. ARF6 activation is required for recycling of the endosomal membrane back to the plasma membrane (PM) and also for ruffling at the PM induced by Rac. Additionally, ARF6 at the PM induces the formation of actin-containing protrusions. To identify sequences in ARF6 that are necessary for these distinct functions, we examined the behavior of a chimeric protein of ARF1 and ARF6. The 1-6 chimera (with the amino half of ARF1 and the carboxyl half of ARF6) localized like ARF6 in HeLa cells and moved between the endosome and PM, but it did not form protrusions, an ARF6 effector function. Two residues in the amino-terminal half of ARF6, Q37 and S38, when substituted into the 1-6 chimera allowed protrusion formation, whereas removal of these residues from ARF6 resulted in an inability to form protrusions. Interestingly, expression of 1-6 in cells selectively inhibited protrusions induced by wild-type ARF6 but had no effect on ARF6-regulated membrane movement or Rac-induced ruffling. Thus, we have uncoupled two functions of ARF6, one involved in membrane trafficking, which is necessary for Rac ruffling, and another involved in protrusion formation.
The ADP-ribosylation factor (ARF) family of proteins is a subgroup of the Ras superfamily of small GTP-binding proteins. Originally identified and named for their ability to serve as cofactors in the cholera toxin-catalyzed ADP-ribosylation of the alpha subunit of Gs (25), ARFs have been shown to function in various membrane trafficking events and in the maintenance of organelle structure (8, 33). ARFs have been identified in numerous eukaryotic organisms, and ARF proteins are divided into three classes based on size, amino acid sequence (deduced from cDNA sequences), phylogenetic analysis, and gene structure (34, 44). Class I contains mammalian ARF1, -2, and -3 and yeast yArf1 and -2, class II includes mammalian ARF4 and ARF5, and class III includes mammalian ARF6, yeast yArf3, and Drosophila ARF3. Like all GTPases, ARFs exist in either an active, GTP-bound form or an inactive, GDP-bound form. Conversion between these two forms is mediated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs), which facilitate GTP exchange and hydrolysis, respectively. Although numerous ARF GEFs and GAPs have been identified in recent years, in many cases their specificity for a particular ARF and their cellular localization remain to be elucidated (34, 39).
Mammalian ARF1 and ARF6 are the least similar in amino acid sequence and the best-characterized members of the ARF family. They have been found to be expressed in all tissues and cell types examined (6, 42, 44, 46). Although both ARF1 and ARF6 have been shown to activate phospholipase D (PLD) in vitro (5, 9, 30), the localizations and functions of these ARFs in vivo are distinct (6, 35, 42). ARF1 is primarily localized to the Golgi complex, where it regulates the assembly of cytosolic coat proteins (COPI and AP adapters) and serves to regulate membrane traffic in the endoplasmic reticulum-Golgi system (29). ARF6, by contrast, localizes to a novel, membrane recycling system at the cell periphery. ARF6 is associated with a tubular endosomal compartment in its inactive GDP-bound form and with the plasma membrane (PM) in its active, GTP-bound form, and it regulates the membrane movement between these two compartments through its GTPase cycle (12, 14, 35, 37). ARF6 has also been implicated in the regulation of exocytosis of chromaffin granules (19) and recently in insulin stimulation of Glut 4 translocation (32). In HeLa cells, the endosomal recycling pathway is involved in the internalization and recycling of PM-associated proteins that are not taken up into cells by clathrin-mediated mechanisms; among the proteins that traffic through this pathway are major histocompatibility complex class I antigens (37) and Rac1 (38). In addition to this trafficking function, ARF6-GTP at the PM is associated with the formation of actin-containing protrusions (36). Furthermore, ARF6 activation is required for various processes that involve actin rearrangements such as cell spreading (41), Rac-mediated membrane ruffling (38), and Fc-mediated phagocytosis (48). Whether these actin rearrangements require the membrane trafficking function of ARF6 or the actin remodeling function of ARF6 is not clear.
We have been studying the function of ARF6 in whole cells by modulating its GTPase cycle through expression of mutant forms of ARF6 and also through the use of pharmacological reagents that induce mutant phenotypes in cells expressing wild-type ARF6. Expression of the dominant negative mutant of ARF6, T27N, in cells inhibits the ARF6-dependent movement of membrane from the endosomal compartment to the PM (37) and also inhibits cell spreading (42) and Rac-mediated ruffling (38). This phenotype is mimicked by treatment of cells expressing wild-type ARF6 with inhibitors of actin polymerization, such as cytochalasin D (CD). As with ARF1 (15, 31), treatment of cells overexpressing ARF6 with aluminum fluoride (AlF), a known activator of heterotrimeric G proteins, appears to maintain the protein in the active GTP-bound form. This results in the accumulation of ARF6-GTP at the PM, and the formation of actin-rich protrusions (36). Although this is an overexpression phenotype observed acutely with AlF treatment, these protrusions resemble those formed in untransfected HeLa cells during cell spreading, a process that requires ARF6 function (42). Studies with the ARF nucleotide binding site opener and the exchange factor for ARF6, candidate GEFs for ARF6, also report alterations of cortical actin after recruitment of these GEFs and ARF6 to the PM (16, 17).
In this study we were interested in identifying sequences in ARF6 that determine its specificity and distinguish it from other ARF proteins. Previous studies of ARF1 have identified regions in the amino-terminal half of the protein required for coat protein binding (28) and PLD activation (23, 28, 33). Interestingly, an amino acid residue (N52) in the switch I region, shown to be critical for activation of PLD (23), is conserved in all ARF proteins. To begin to understand the mechanism whereby ARF6 carries out cellular roles that are distinct from those of other ARFs, we set out to map sequences in ARF6 that confer on the molecule the ability to couple to effector functions. Following an approach that has been used by investigators studying Rab and Rho functions (7, 11, 18), we made chimeric ARFs by exchanging the amino- and carboxy-terminal halves of ARF1 and ARF6 and expressed them in HeLa cells. Here we report that the 1-6 chimera has sufficient information for targeting to ARF6 compartments and interacting with ARF6 GEFs, and yet it cannot form protrusions. Studies with this chimera have enabled us to identify residues in ARF6 near the ARF equivalent of the Ras effector loop that are required for protrusion formation but not for ARF6 regulation of membrane trafficking.
MATERIALS AND METHODS
Cells and reagents.
HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 100 IU of penicillin/ml, and 100 mg of streptomycin/ml at 37°C with 5% CO2. Brefeldin A (BFA) was obtained from Epicentre Technologies (Madison, Wis.), stored at −20°C as a stock solution of 2 mg/ml in methanol, and used at a final concentration of 2 μg/ml. Rhodamine-phalloidin was obtained from Molecular Probes, Inc. (Eugene, Oreg.) and Sigma Chemical Co. (St. Louis, Mo.). All other reagents were purchased from Sigma Chemical Co.
Antibodies.
For immunofluorescence localization of untagged ARF6, the 1-6 chimera, and their mutants, we used rabbit polyclonal antisera raised against a C-terminal peptide of ARF6, as described previously (42). ARF6 fused to the influenza hemagglutinin (HA) epitope at its C-terminal end was localized using a mouse antibody (12CA5) against the HA epitope, purchased from BAbCo (Berkeley, Calif.). For immunofluorescence localization of Rac1, we used a mouse antibody against the nine-amino-acid epitope tag (MEYMPMEHM), which was the gift of C. J. Der (University of North Carolina, Chapel Hill). The mouse anti-Tac antibody used was the 7G7 hybridoma (40). Rhodamine- and fluorescein-labeled donkey anti-mouse and donkey anti-rabbit immunoglobulin G (IgG) were purchased from Jackson ImmunoResearch Laboratories (West Grove, Pa.).
DNA manipulations.
The cDNAs of wild-type, chimera, and mutant genes were subcloned into the modified pCDL-SRα expression vector (pXS) (43). In HeLa cells, this expression vector results in protein expression levels for ARF6 that are between 20- and 50-fold higher than endogenous protein expression (35).
The 1-6 chimera and site-specific mutations of it and of ARF6 were created by a two-step PCR procedure (2). For all PCRs, AmpliTaq DNA polymerase (Perkin-Elmer, Branchburg, N.J.) was used. PCR products and restriction fragments were purified by excising the appropriate bands from an agarose gel and recovering them with the QIAquick gel extraction kit (Qiagen, Santa Clarita, Calif.). Restriction enzymes and T4 DNA ligase were obtained from New England Biolabs (Beverly, Mass.).
The following oligonucleotides were used as primers (those in plain type are ARF1 sequences, those in boldface type are ARF6 sequences, underlined residues are mutation sites, and restriction sites are italicized): A1-5′, 5′AACAGAATTCATGGGGAACATCTTCGC3′; A6-5′, 5′AACAGAATTCATGGGGAAGGTGCTATCC3′; A6-3′, 5′CCCAGATCTTCAAGATTTGTAGTTAGAGGTTAAC3′; 1/6-A, 5′CTCCTGGCGAGCCTCATCCACACGCTCTCTGTCATTG3′; 1/6-B, 5′GCCGACCGCGACCGCATCAACGAGGCCCGTGAGGAG3′; A1QS-A, 5′GGGAATGGTGGTCACGCTTTGACCCAGCTTCAG3′; A1QS-B, 5′AAACTGAAGCTGGGTCAAAGCGTGACCACCATTC3′; A6EI-A, 5′GGGAATGGTGGTCACGATCTCGCCCAGCTTCAA3′; and A6EI-B, 5′AAGTTGAAGCTGGGCGAGATCGTGACCACCATTCCC3′.
The 1-6 chimera was constructed by fusing the N-terminal 300 bp of ARF1 to the C-terminal 237 bp of ARF6. The N-terminal portion of ARF1 was amplified using primers A1-5′ and 1/6-A, and the C-terminal portion of ARF6 was amplified using primers 1/6-B and A6-3′. The EcoRI-BglII fragment of the final PCR product was cloned into pXS.
The 1-6(T31N) chimera was constructed by fusing the N-terminal 300 bp of ARF1(T31N) to the C-terminal 237 bp of ARF6. The N-terminal portion of ARF1(T31N) was amplified using primers A1-5′ and 1/6-A, and the C-terminal portion of ARF6 was amplified using primers 1/6-B and A6-3′. The EcoRI-BglII fragment of the final PCR product was cloned into pXS.
ARF6(EI) was constructed by site-directed mutagenesis using primers A6-5′, A6EI-A, A6EI-B, and A6-3′. The EcoRI-BglII fragment of the final PCR product was cloned into pXS.
1-6(QS) was constructed by site-directed mutagenesis using primers A1-5′, A1QS-A, A1QS-B, and A6-3′. The EcoRI-BglII fragment of the final PCR product was cloned into pXS.
Sequences of all DNA constructs originating from PCR products were confirmed through the services of SeqWright (Houston, Tex.) or Veritas, Inc. (Rockville, Md.).
Transient transfection of cells.
Cells grown on coverslips were transfected in six-well dishes using the calcium phosphate procedure as previously described (4). A total of 5 μg of DNA per well was used in single-transfection experiments, such that 2.5 μg of the required plasmid was used plus 2.5 μg of pXS vector. In cotransfection experiments, 2 μg of either the ARF6-HA or Rac plasmid and 10 μg of the coexpressed plasmid were used to obtain a 1:5 ratio. Drug treatments were performed in the presence of complete medium. AlF treatment of cells was performed by supplementing the culture medium with 30 mM NaF and 50 μM AlCl3. CD was stored in dimethyl sulfoxide as a stock solution of 1 mM and used at a final concentration of 1 μM.
Immunofluorescence.
Thirty hours after transfection, the cells were treated as described above, fixed in 2% formaldehyde in phosphate-buffered saline (PBS) for 10 min, and rinsed with 10% FBS and 0.02% azide in PBS (PBS-serum). The cells were incubated with primary antibodies diluted in PBS-serum plus 0.2% saponin for 1 h, then washed (three times, 5 min each) with PBS-serum. The cells were then incubated in secondary antibodies diluted in PBS-serum plus 0.2% saponin for 1 h, washed with PBS-serum (three times, 5 min each) and once with PBS, and mounted on glass slides. A Zeiss Axioplan epifluorescence microscope and 63× Plan-Apochromat lens were used for all fluorescence microscopy, and photomicrographs were prepared using TMAX 400 film (Eastman Kodak, Rochester, N.Y.).
Internalization and recycling of anti-Tac antibodies.
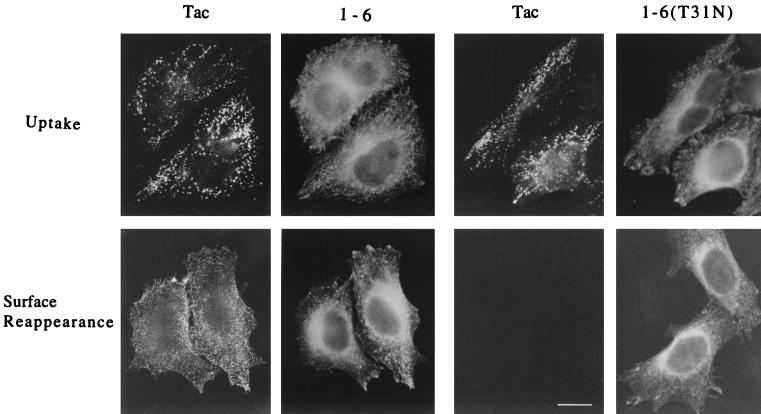
Detection of anti-Tac antibody recycling was performed as described previously (37). Cells grown on coverslips were cotransfected with Tac and either 1-6 or 1-6(T31N). Thirty hours later, cells were chilled to 4°C and incubated for 30 min in an ice-water bath with mouse anti-Tac antibodies in the absence of permeabilization to label surface Tac. Cells were rinsed with ice-cold Dulbecco's modified Eagle's medium supplemented with 10% FBS (complete medium) and prewarmed for 30 min at 37°C in the presence of CD to allow for antibody internalization. To remove anti-Tac antibodies remaining at the surface, cells were next rinsed quickly three times with a low-pH buffer (26) containing 0.5% acetic acid and 0.5 M NaCl (pH 3.0) and then three times with complete medium. Some coverslips were fixed to detect internalization, and the rest were first warmed for 30 min at 37°C to allow for antibody recycling to the surface and then fixed.
To detect antibody internalization, cells were processed for immunofluorescence as described above, using rabbit anti-ARF6 antiserum, fluorescently labeled donkey anti-rabbit IgG (to detect anti-ARF6 antibodies), and donkey anti-mouse IgG (to detect anti-Tac antibodies).
To detect Tac antibody recycled to the surface, cells were incubated with fluorescently labeled donkey anti-mouse IgG in the absence of saponin. To label ARF6, cells were incubated with anti-ARF6 antibodies and the appropriate secondary antibodies in the presence of saponin.
RESULTS
The 1-6 chimera localizes and traffics like wild-type ARF6 yet cannot form protrusions.
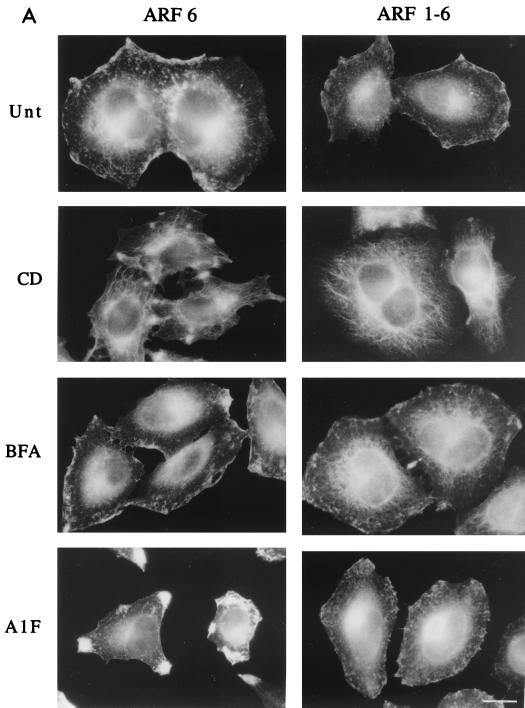
We constructed a 1-6 chimera of ARF consisting of the amino half of ARF1 (residues 1 to 100) and the carboxyl half of ARF6 (residues 97 to 175). HeLa cells transiently transfected with a plasmid encoding either ARF6 or the 1-6 chimera were left untreated or were treated with either CD, BFA, or AlF and then fixed and processed for indirect immunofluorescence localization of ARF6 or the 1-6 chimera using a polyclonal antibody raised to the C-terminal end of ARF6 (42). In untreated cells, the 1-6 chimera localized to the PM and associated with a tubular endosomal compartment, similar to the localization of ARF6 (Fig. 1A). CD treatment shifted the distribution of the 1-6 chimera and ARF6 from the PM to the tubular endosomal compartment, and BFA treatment did not affect the distribution of either the chimera or ARF6 (Fig. 1A). In cells expressing ARF6, AlF treatment resulted in the formation of protrusions at the PM (Fig. 1A), a manifestation of the actin rearrangement effector function for ARF6 (38, 42). In contrast, the 1-6 chimera did not form protrusive structures at the PM upon treatment with AlF (Fig. 1A). The failure to make protrusions was not due to differences in expression levels, as all constructs were expressed in a vector using the same promoter and similar elevated levels of ARF expression were observed (Fig. 1B). Furthermore, looking at individual cells under the microscope, we found that individual cells expressing very high levels of the 1-6 chimera still did not make protrusions. Thus, the carboxyl half of ARF6 contains sufficient information for localization both at the PM and on the endosomal compartment. Furthermore, the information for protrusion formation is in the amino-terminal half of ARF6, but this information is absent from the same region in ARF1.
FIG. 1.
The ARF1-6 chimera localizes like ARF6 yet cannot form protrusions. (A) HeLa cells were transfected with plasmids encoding either ARF6 or the ARF1-6 chimera and were untreated (Unt) or incubated in the presence of either 1 μM CD for 30 min (CD), 2 μg of BFA/ml for 10 min (BFA), or 30 mM NaF and 50 μM AlCl3 for 30 min (AlF). Cells were fixed and immunolabeled with polyclonal anti-ARF6 antibodies that recognize a C-terminal peptide of the protein. Bar, 15 μm. (B) Expression levels of ARF6 in untransfected and transfected HeLa cells. HeLa cells were not transfected (Endog.) or were transfected with 2.5 μg of plasmid encoding ARF6, 1-6, 1-6(T31N), or 1-6(QS). Cell extracts were loaded onto sodium dodecyl sulfate–13% polyacrylamide gel electrophoresis, transferred to nitrocellulose, blotted with rabbit anti-ARF6 antiserum, and visualized by enhanced chemiluminescence as described previously (42). For untransfected HeLa cells, 10 μg of protein was loaded per lane, whereas for transfected HeLa lysates, 2 μg of protein was loaded per lane.
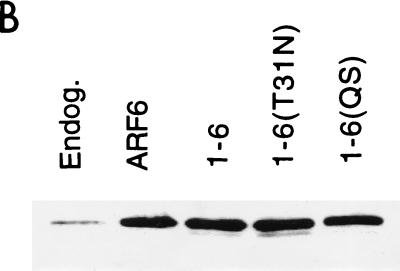
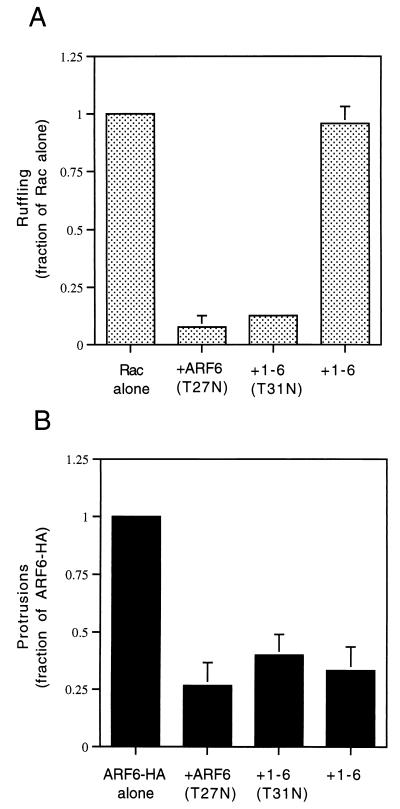
One possible explanation for the inability of the 1-6 chimera to form protrusions is that it is unable to interact effectively with the GEF that activates ARF6. Although the ARF6-specific GEF is not known in HeLa cells, GTP-binding-defective mutants of GTPases can be generated that result in an inhibitory phenotype in cells by sequestering the GEFs (3). Such an ARF6 mutant, ARF6(T27N), when overexpressed in HeLa cells accumulates on the tubular endosomal compartment and inhibits recycling of membrane to the PM (37), Rac-induced membrane ruffling (38), and cell spreading (42). To test whether the 1-6 chimera can interact with the ARF6 GEF, we mutated the threonine at position 31 to an asparagine and asked whether the resulting mutant chimera, 1-6(T31N), can exert inhibitory effects similar to those generated by expression of ARF6(T27N). We first tested whether 1-6(T31N) could inhibit Rac-mediated ruffling. HeLa cells were either transfected with plasmids encoding epitope-tagged Rac1 alone or cotransfected with 1-6(T31N) or ARF6(T27N). Following treatment with AlF, cells were fixed and immunolabeled with monoclonal anti-EE antibodies to detect Rac1 and polyclonal anti-ARF6 antibodies to detect 1-6(T31N) or ARF6(T27N). Cells expressing Rac1 alone exhibited extensive PM ruffles along the edges of the cells (Fig. 2A), as previously observed (38). Coexpression of 1-6(T31N) with Rac1 inhibited ruffling similarly to coexpression of ARF6(T27N) with Rac1 (Fig. 2A) (38). The inhibition of ruffling was quantitated by scoring the fraction of ruffling cells in the total transfected pool for each treatment (see the legend to Fig. 3). Both ARF6(T27N) and 1-6(T31N) were effective at inhibiting Rac ruffling to approximately 8 and 12%, respectively, of that of cells expressing Rac1 alone (Fig. 3A). Similar to ARF6(T27N), the 1-6(T31N) chimera was localized to the endosomal compartment and was also observed to inhibit recycling of membrane to the PM (see Fig. 6) and cell spreading (data not shown).
FIG. 2.
The GTP-binding-defective mutant of 1-6 acts like a dominant negative ARF6 mutant, inhibiting ARF6 protrusions and Rac ruffling. (A) HeLa cells were transfected with wild-type Rac1 (Rac) or with Rac and either 1-6(T31N) or ARF6(T27N) (1:5 ratio) and then incubated in the presence of AlF for 30 min. (B) HeLa cells were transfected with either wild-type HA-tagged ARF6 (ARF6-HA) or with ARF6-HA and either 1-6(T31N) or ARF6(T27N) (1:5 ratio) and then incubated in the presence of AlF for 30 min. The overexpressed proteins were then localized by immunofluorescence. Bar, 15 μm.
FIG. 3.
Quantitation of Rac ruffling and ARF6 protrusion formation. (A) HeLa cells were transfected with either plasmids encoding Rac1 alone or those encoding Rac1 and either ARF6(T27N), 1-6(T31N), or 1-6 (1:5 ratio). (B) HeLa cells were transfected with either ARF6-HA alone or with ARF6-HA and either ARF6(T27N), 1-6(T31N), or 1-6 (1:5 ratio). Cells were incubated for 30 min in the presence of AlF and fixed, and the overexpressed proteins were labeled by immunofluorescence. For each condition, over 500 transfected cells were counted, and the fraction of Rac- or ARF6-transfected cells that were ruffling or forming protrusions, respectively, was noted. For cells overexpressing only Rac1 or ARF6, the fraction of ruffling or protruding cells was normalized to 1.0, and the other conditions were then expressed as a fraction of 1.0. Data shown are the means and standard errors of three independent experiments.
FIG. 6.
The 1-6 chimera does not block internalization or recycling of surface Tac into the tubular compartment. HeLa cells expressing Tac and either the 1-6 chimera or 1-6(T31N) were incubated with anti-Tac antibodies (7G7) at 4°C to bind to surface Tac. Excess antibodies were washed off, and the cells were then incubated at 37°C for 30 min in the presence of CD to allow internalization of the Tac antibodies into the endosomal compartment. Cells were then rinsed to remove remaining surface anti-Tac antibodies and either fixed immediately and assessed for Tac antibody internalized by immunofluorescence (Uptake) or warmed to 37°C for 30 min in the absence of CD before fixation. Tac antibody that reappeared on the cell surface was detected by incubation with fluorescently labeled secondary antibodies in the absence of detergent permeabilization (Surface Reappearance). The 1-6 chimera or 1-6(T31N) was subsequently localized in these cells after permeabilization. Bar, 15 μm.
An additional test for the effectiveness of a GTP-binding-defective mutant is to assess whether the mutant is able to inhibit directly a function of the wild-type protein. We therefore analyzed the ability of 1-6(T31N) and ARF6(T27N) to inhibit protrusions formed by cells overexpressing wild-type ARF6. HeLa cells were either transfected with plasmids encoding influenza HA epitope-tagged ARF6 (ARF6-HA) alone or cotransfected with the 1-6(T31N) or ARF6(T27N) mutant. Following AlF treatment, cells were fixed and immunolabeled with both monoclonal anti-HA antibodies to detect overexpressed, wild-type ARF6 and with polyclonal anti-ARF6 antibodies to additionally detect 1-6(T31N) or ARF6(T27N). Cells expressing ARF6-HA alone formed protrusions upon addition of AlF, whereas coexpression with either 1-6(T31N) or ARF6(T27N) inhibited protrusion formation (Fig. 2B). The inhibition of ARF6-induced protrusions was quantitated by scoring the fraction of protruding cells in the total transfected pool (see the legend to Fig. 3). ARF6(T27N) and 1-6(T31N) inhibited protrusions to approximately 27 and 40%, respectively, of that of cells expressing ARF6-HA alone (Fig. 3B).
The experiments described above demonstrate that the 1-6(T31N) chimera displays all the inhibitory effects of ARF6(T27N) and behaves like an effective dominant negative ARF6 mutant. By contrast, expression of the dominant negative ARF1 mutant, T31N, which disassembles the Golgi complex and blocks secretion (10), had no appreciable effect on any of these ARF6 functions (data not shown). These observations suggest that 1-6, like ARF6, cycles between the endosome and the PM and is capable of effective interaction with the ARF6 GEF. Therefore, the inability of 1-6 to induce protrusions (an ARF6 effector function at the PM) is likely due to the absence of an ARF6 effector domain in the 1-6 chimera.
The effector domain in ARF6 includes residues Q37 and S38.
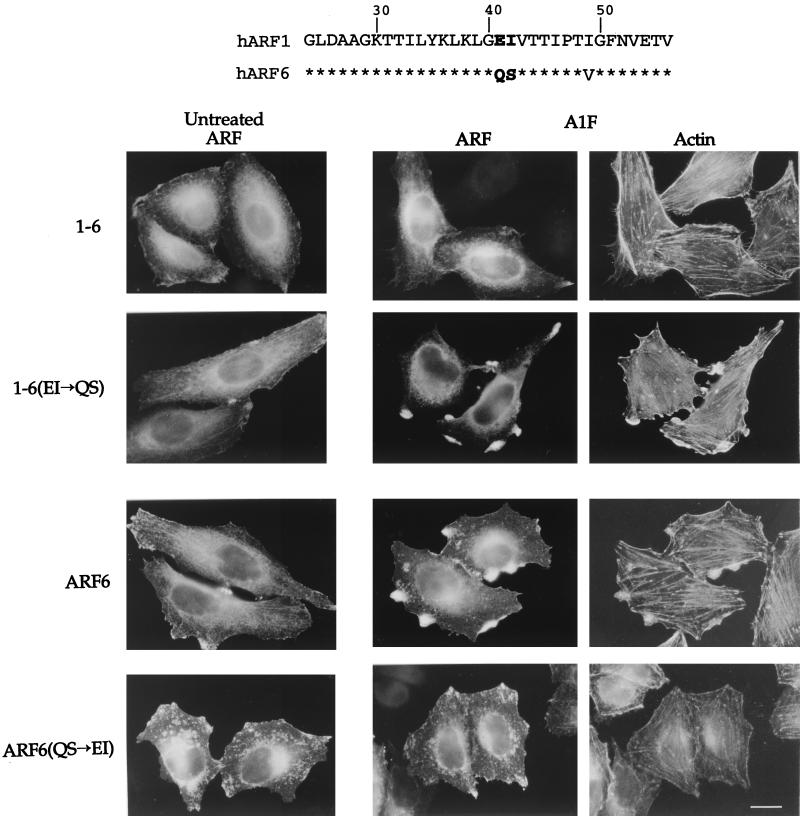
The failure of the 1-6 chimera to form protrusions suggests that residues in the amino-terminal half of ARF6 are required for protrusion formation. Interestingly, we have observed that the Saccharomyces cerevisiae protein yArf3 localizes in HeLa cells in a manner similar to ARF6, but like the 1-6 chimera it does not form protrusions in response to AlF (O. Al-Awar et al., unpublished data) (Fig. 1). These observations suggest that both yArf3 and the 1-6 chimera lack information in their amino-terminal halves that is required for this ARF6 effector function. We therefore searched for a unique sequence in the amino-terminal half of ARF6 that is not present in either ARF1 or yArf3. Residues Q37 and S38 in ARF6 fit this criterion (Fig. 4, top).
FIG. 4.
ARF6 effector domain includes residues Q37 and S38. (Top) Amino acid sequence comparison between human ARF1 and ARF6 from amino acid 24 in ARF1 (20 in ARF6) to amino acid 56 in ARF1 (52 in ARF6). Asterisks indicate identity. Note the conservation in the switch I region encompassing residues 45 to 54. (Bottom) HeLa cells were transfected with plasmids encoding either the 1-6 chimera, 1-6(EI→QS), ARF6, or ARF6(QS→EI). Cells were then left untreated or incubated for 30 min with AlF. The expressed proteins were labeled with ARF6-specific antiserum followed by rhodamine-conjugated phalloidin to visualize F-actin. Bar, 15 μm.
To assess whether residues Q37 and S38 are critical for the formation of protrusions, we replaced the equivalent amino acids in the 1-6 chimera, E41 and I42, with QS. The resulting mutant, 1-6(QS), was transiently transfected into HeLa cells. 1-6(QS) localized similarly to ARF6 and to 1-6 in cells. In untreated cells, 1-6(QS), like 1-6 and ARF6, was present at the PM and on the tubular juxtanuclear compartment (Fig. 4). CD treatment shifted both 1-6 and 1-6(QS) to the tubular compartment, as observed in Fig. 1 (and data not shown). In contrast to cells expressing 1-6, cells expressing 1-6(QS) did form protrusions enriched in F-actin upon the addition of AlF, similar to protrusions seen in cells expressing wild-type ARF6 (Fig. 4). We next mutated residues Q37 and S38 in the full-length ARF6 to EI, their ARF1 equivalents. Cells expressing the resulting mutant, ARF6(EI), did not form protrusions upon AlF treatment (Fig. 4), although ARF(EI) did at times accumulate at sites along the PM. The gain of function by 1-6(QS) and the loss of function by ARF6(EI) demonstrated that residues Q37 and S38 are critical for the ARF6 effector function of protrusion formation.
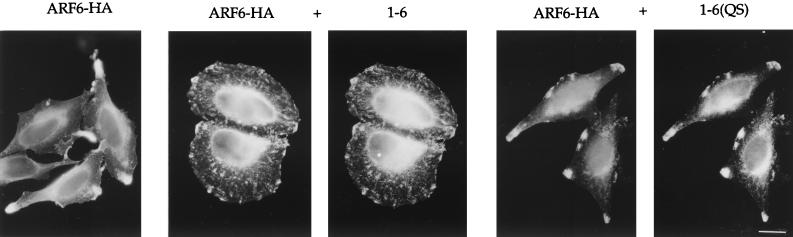
The 1-6 chimera antagonizes selected ARF6 functions.
The observation that the 1-6 chimera localized like ARF6 in the cells yet could not form protrusions upon AlF addition led us to investigate whether the expression of this effector function-negative chimera would interfere with the ability of wild-type ARF6 to induce protrusions. To test this possibility, we examined whether coexpression of 1-6 with ARF6-HA would inhibit protrusions formed in the presence of AlF. As shown in Fig. 5, cells expressing ARF6-HA alone formed protrusions after addition of AlF, whereas cells coexpressing 1-6 with ARF6-HA did not. Inhibition of protrusion formation was also observed in cell coexpressing ARF6(EI) and ARF6-HA (data not shown). As expected, cells coexpressing ARF6-HA and 1-6(QS) formed protrusions. We quantitated the inhibitory effect of 1-6, and remarkably, the extent of inhibition observed with 1-6 was comparable to that observed with the GTP-binding-defective mutants, 1-6(T31N) and ARF6(T27N) (Fig. 3B). However, the mechanism by which the 1-6 chimera inhibits protrusion formation is distinct from that observed with these mutants (see below).
FIG. 5.
The 1-6 chimera inhibits protrusion formation. HeLa cells were transfected with plasmids encoding either ARF6-HA alone (ARF6-HA), or with ARF6-HA and either 1-6 or 1-6(QS) (1:5 ratio). Cells were treated with AlF for 30 min, fixed, and processed for immunofluorescence. Bar, 15 μm.
The ability of the 1-6 chimera to inhibit protrusion formation led us to ask whether it also inhibited two other ARF6 functions: membrane trafficking (37) and Rac-mediated ruffling (38). To assess the effect of the 1-6 chimera on the recycling of endosomal membrane back to the PM, we followed the internalization and recycling of Tac, the interleukin-2 receptor α subunit, a membrane marker for the ARF6 endosomal compartment (37). We have previously demonstrated that expression of ARF6(T27N) blocks the ARF6-regulated recycling of Tac from the endosomal compartment back out to the PM (37); we predicted that 1-6(T31N) would act similarly. Cells coexpressing Tac and either 1-6 or 1-6(T31N) were treated as described in Materials and Methods. Cells expressing the 1-6 chimera internalized and accumulated anti-Tac antibodies into the ARF6-labeled endosomal compartment during the internalization period, similar to Tac internalization in cells expressing 1-6(T31N) (Fig. 6, Uptake). Tac was recycled back to the PM after removal of CD, as detected by surface reappearance of anti-Tac antibodies, in cells expressing 1-6 (Fig. 6, Surface Reappearance) in a pattern and time course similar to that previously observed for cells expressing ARF6 (37). By contrast, 1-6(T31N) expression inhibited the recycling step (Fig. 6, Surface Reappearance). Thus, expression of the 1-6 chimera does not perturb the functioning of the ARF6-regulated membrane recycling pathway.
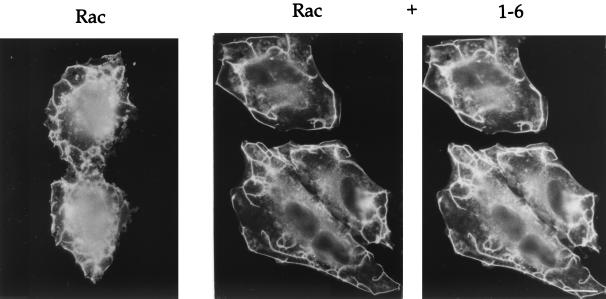
Since both the trafficking of Rac and the ability of Rac to form PM ruffles is dependent upon ARF6 in HeLa cells (38), we next asked whether the 1-6 chimera would interfere with the ability of wild-type Rac to induce PM ruffles in response to AlF treatment. Cells were transfected with either Rac1 alone or with the 1-6 chimera and Rac1. Following treatment with AlF, cells were fixed and immunolabeled to detect 1-6 and Rac1. PM ruffling was readily observed around the edges of cells expressing Rac1 either alone or with 1-6 (Fig. 7). Similarly, coexpression of ARF6(EI) with Rac also did not inhibit ruffling (data not shown). We next quantitated the ruffling response in cells coexpressing Rac1 and 1-6 and found that the fraction of cells showing PM ruffling was nearly identical to that observed in cells expressing Rac1 alone (Fig. 3A). Thus, expression of the 1-6 chimera selectively inhibits protrusion formation but does not interfere with the ARF6-regulated membrane-trafficking events, namely the recycling of membrane back to the PM. By contrast, 1-6(T31N) behaves like the ARF6 dominant negative mutant, ARF6(T27N), inhibiting all known ARF6 activities including both membrane trafficking and formation of actin-containing protrusions at the PM (Fig. 3 and 8).
FIG. 7.
The 1-6 chimera does not inhibit Rac-induced ruffling. HeLa cells were transfected with plasmids encoding either Rac1 alone or Rac and the 1-6 chimera (1:5 ratio). Cells were incubated in the presence of AlF for 30 min and fixed, and the expressed proteins were localized by immunofluorescence. Bar, 15 μm.
FIG. 8.
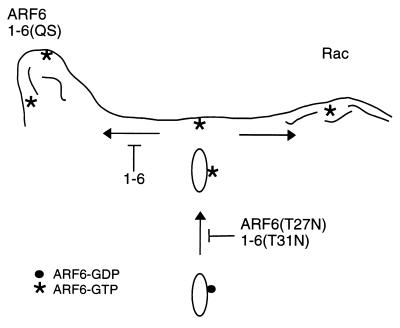
Working model for ARF6 action on trafficking and cortical actin structures. ARF6-GTP (asterisks) at the PM is involved in the generation of actin filament-containing protrusions (left side) or in the presence of Rac, actin-containing PM ruffles (right side). 1-6(QS) can generate protrusions whereas 1-6 cannot. Both ARF6(T27N) and 1-6(T31N) inhibit activation of ARF6 and therefore the recycling of endosomal membrane back to the PM. Expression of 1-6 inhibits ARF6-mediated protrusions but not Rac-mediated ruffling or membrane recycling back to the PM.
DISCUSSION
All of the human ARF family members are expressed, to various extents, in all cell types. Although the different ARFs share sequence similarity and common biochemical activities, in cells they likely have distinct functions. Their specificity in vivo may be mediated through targeting to distinct membrane compartments and through coupling to specific effectors. In this study, we analyzed the localization and activities of 1-6, a chimeric ARF molecule containing the amino-terminal half of ARF1 and carboxyl-terminal half of ARF6. 1-6 contained sufficient information from the carboxyl-terminal half to target it like ARF6 i.e., to the PM and endosomal compartment and not to the Golgi complex where ARF1 localizes. This molecule behaved similarly to ARF6 in its ability to cycle between the endosomal compartment and the PM, and it appeared to interact with an ARF6-specific GEF. Yet unlike ARF6, the 1-6 chimera could not induce protrusion formation upon AlF treatment, an ARF6 effector function at the PM. Thus, we concluded that the sequences in ARF6 that are necessary for induction of protrusion formation are present in the amino-terminal half of the protein, and that the amino-terminal half of ARF1 could not substitute for ARF6 in this regard.
The overall similarity between ARF1 and ARF6 in their amino-terminal halves led us to attempt to identify unique sequences in ARF6 necessary for this effector function. The identification of sequences critical for specific effector functions and the subsequent generation of various effector mutants has aided in the understanding of GTPases with multiple effector functions, in particular for Rho proteins (18, 24, 27, 45). We searched for a sequence in ARF6 that is missing in the amino-terminal half of the 1-6 chimera which would be necessary for formation of protrusions. We found that two residues in ARF6, Q37 and S38, were critical for this ARF6 effector function. Substitution of residues QS for residues EI in the analogous position in the 1-6 chimera was sufficient to result in a gain of function, allowing the chimera to form protrusions in response to AlF. We have recently identified the target of AlF in our cells as the heterotrimeric G protein alpha subunit, Gαq. Coexpression of constitutively active Gq(Q209L) with ARF6 induces protrusions in the absence of AlF (H. Radhakrishna and J. G. Donaldson, unpublished data). We have observed that coexpression of Gq(Q209L) with 1-6, like AlF treatment of cells expressing 1-6 alone, also did not result in protrusion formation, whereas coexpression of Gq(Q209L) with 1-6(QS) did form protrusions (O. Awar, unpublished observations). Taken together, these data indicate that residues Q37 and S38 in ARF6 represent a site of interaction with effector molecules which is necessary for protrusion formation. A recent study suggests that ARF6 may influence cortical actin through activation of phosphatidylinositol 4-phosphate 5-kinase α (22). It will be interesting to examine whether residues Q37 and S38 are required for this activity.
The crystal structures of both the GDP- and GTP-bound forms of ARF1 have been solved (1, 20, 21). A comparison of these structures reveals that, like Ras, upon GTP binding ARF1 undergoes a conformational change that involves significant shifts in the positioning of the switch I and switch II regions. In ARF1, switch I encompasses residues 45 to 54 and switch II encompasses residues 70 to 80 (20). Assuming similarity in structure, for ARF6 the switch I and switch II regions would encompass residues 41 to 50 and 66 to 76, respectively. It is noteworthy that there is high conservation in the amino acid sequence among all ARFs in the switch I and switch II regions (see Fig. 4 for residues in the switch I region in ARF1 and ARF6). Intriguingly, residues Q37 and S38, which result in a gain of function when substituted into the 1-6 chimera, are positioned a few residues prior to the predicted switch I region. Significantly, all the ARF proteins in class I and class II, in all organisms, contain residues EI at this site. Only mammalian ARF6 and its class III homologues in Drosophila (accession no. P40946), Caenorhabditis elegans (accession no. CAB55153), Schizosaccharomyces pombe (accession no. CAB51340), and Xenopus (accession no. P51645) contain residues QS at this site. The S. cerevisiae yArf3 protein (accession no. P40994), a class III member that localizes like ARF6 when expressed in mammalian cells, does not contain these residues and, consistent with the data in this paper, cannot induce protrusion formation (O. Al-Awar et al., unpublished data). These observations further suggest that residues QS are critical for ARF6-specific functions involved in actin reorganization at the PM, and that conserved ARF sequences in the switch I and switch II regions may be used for regulation of membrane trafficking, a function common to all ARFs. Future investigations will focus on testing whether antibodies or peptides specific to the QS region of ARF6 interfere with its functioning and on identifying target molecules that interact with ARF6 in this region.
Studies in a variety of systems have highlighted a dual function for ARF6 as a regulator of membrane traffic and as a modulator of actin dynamics at the PM (34). Separating these two effector functions for ARF6 has proven difficult since the GTP-binding-defective mutant of ARF6, ARF6(T27N), inhibits both trafficking (12, 37) and cortical actin functions (13, 36). The phenotype of the expressed 1-6 chimera appears to separate these two activities of ARF6, because the 1-6 chimera selectively inhibits protrusion formation while not affecting the membrane trafficking function of ARF6 (Fig. 8). One explanation for these observations is that in cells, the 1-6 chimera is targeted correctly to the ARF6 compartment, interacts with the ARF6 GEFs and GAPs, and carries out the ARF6 trafficking functions via shared effector domains between ARF1 and ARF6 in their amino halves. However, the 1-6 chimera cannot induce protrusion formation, and it inhibits the ability of wild-type ARF6 to form protrusions due to the absence of residues QS near the switch I region. Similar observations have been made for Rac effector domain mutants. An effector domain mutant of Rac that activates p21-activated kinase but does not induce ruffling also functions as a dominant negative for Rac ruffling (41).
The failure of the 1-6 chimera to affect Rac ruffling in HeLa cells partially resolves the issue of the role of ARF6 in Rac-mediated ruffling. We previously demonstrated that Rac colocalizes with ARF6 on the endosome and at the PM, that ARF6 regulates the trafficking of Rac to the PM, and that ARF6 activity was required for Rac-mediated PM ruffling (38). At that time, we could not determine whether the ARF6 requirement for Rac ruffling was due to ARF6 regulation of trafficking or due to an ARF6-dependent effect on cortical actin at the PM, since we were inhibiting both functions with ARF6(T27N). The lack of inhibition of Rac ruffling by the 1-6 chimera suggests that it may be the trafficking function of ARF6, and not the specific actin remodeling function, that is necessary for the formation of Rac ruffles. This trafficking function may extend beyond that of the trafficking of Rac itself to include trafficking or recruitment of components required for Rac ruffling. This was recently suggested for ARF6 and Rac functions in macrophages (47).
Like 1-6, ARF6(EI) does not form protrusions, blocks protrusions induced by ARF6, and does not block Rac ruffling. Selective inhibition of certain ARF6 functions by the 1-6 chimera and ARF6(EI) suggests that these molecules are acting as dominant-negative ARF6 mutants by a mechanism different from that of ARF6(T27N). Although 1-6 and ARF6(EI) lack residues that allow protrusion formation, they might act as inhibitors by sequestering factors that wild-type ARF6 requires for protrusions. These factors are presumably not required for Rac ruffling. Identification of these limiting factors and also the molecules that specifically interact with the QS residues in ARF6 should provide us with insight into how ARF6 alters actin dynamics at the PM.
ACKNOWLEDGMENTS
We thank C. Der for reagents and F. Brown, E. Korn, and P. Randazzo for discussions and critical reading of the manuscript.
N. N. Powell was supported by the Biomedical Research Training Program for Underrepresented Minorities, NHLBI, NIH.
REFERENCES
- 1.Amor J C, Harrison D H, Kahn R A, Ringe D. Structure of the human ADP ribosylation factor 1 complexed with GDP. Nature. 1994;372:704–708. doi: 10.1038/372704a0. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1995. [Google Scholar]
- 3.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino J S, Suzuki C K, Lippincott-Schwartz J, Weissman A M, Klausner R D. Pre-Golgi degradation of newly synthesized T cell antigen receptor chains: intrinsic sensitivity and the role of subunit assembly. J Cell Biol. 1989;10:73–83. doi: 10.1083/jcb.109.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown H A, Gutowski S, Moomaw C R, Slaughter C, Sternweis P C. ADP ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 6.Cavenagh M M, Whitney J A, Carroll K, Zhang C-J, Boman A L, Rosenwald A G, Mellman I, Kahn R A. Intracellular distribution of Arf proteins in mammalian cells. J Biol Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- 7.Chavrier P, Gorvel J P, Stelzer E, Simons K, Gruenberg J, Zerial M. Hypervariable C-terminal domain of rab proteins acts as a targeting signal. Nature. 1991;353:769–772. doi: 10.1038/353769a0. [DOI] [PubMed] [Google Scholar]
- 8.Chavrier P, Goud B. The role of ARF and Rab GTPases in membrane transport. Curr Opin Cell Biol. 1999;11:466–475. doi: 10.1016/S0955-0674(99)80067-2. [DOI] [PubMed] [Google Scholar]
- 9.Cockroft S, Thomas G M H, Fensome A, Geny B, Cunningham E, Gout I, Hiles I, Totty N F, Truong O, Hsuan J J. Phospholipase D: a downstream effector of ARF in granulocytes. Science. 1994;263:523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- 10.Dascher C, Balch W E. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J Biol Chem. 1994;269:1437–1448. [PubMed] [Google Scholar]
- 11.Diekmann D, Nobes C D, Burbelo P D, Abo A, Hall A. Rac GTPase interacts with GAPs and target proteins through multiple effector sites. EMBO J. 1995;14:5297–5305. doi: 10.1002/j.1460-2075.1995.tb00214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Souza-Schorey C, Li G, Colombo M I, Stahl P D. A regulatory role for ARF6 in receptor-mediated endocytosis. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- 13.D'Souza-Schorey C, Boshans R L, McDonough M, Stahl P D, Van Aelst L. A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 1997;16:5445–5454. doi: 10.1093/emboj/16.17.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Souza-Schorey C, van Donselaar E, Hsu V W, Yang C, Stahl P D, Peters P J. ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J Cell Biol. 1998;140:603–616. doi: 10.1083/jcb.140.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finazzi D, Cassel D, Donaldson J G, Klausner R D. Aluminum fluoride acts on the reversibility of ARF-dependent coat protein binding to Golgi membranes. J Biol Chem. 1994;269:13325–13330. [PubMed] [Google Scholar]
- 16.Franco M, Peters P J, Boretto J, van Donselaar E, Neri A, D'Souza-Schorey C, Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank S R, Hatfield J C, Casanova J E. Remodeling of the actin cytoskeleton is coordinately regulated by protein kinase C and the ADP-ribosylation factor nucleotide exchange factor ARNO. Mol Biol Cell. 1998;9:3133–3146. doi: 10.1091/mbc.9.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujisawa K, Madaule P, Ishizaki T, Watanabe G, Bito H, Saito Y, Hall A, Narumiya S. Different regions of Rho determine Rho-selective binding of different classes of Rho target molecules. J Biol Chem. 1998;273:18943–18949. doi: 10.1074/jbc.273.30.18943. [DOI] [PubMed] [Google Scholar]
- 19.Galas M-C, Helms J B, Vitale N, Thierse D, Aunis D, Bader M-F. Regulated exocytosis in chromaffin cells: a potential role for a secretory granule-associated ARF6 protein. J Biol Chem. 1997;272:2788–2793. doi: 10.1074/jbc.272.5.2788. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 21.Greasley S, Jhoti H, Fensome A C, Cockcroft S, Thomas G M, Bax B. Crystallization and preliminary X-ray diffraction studies on ADP-ribosylation factor 1. J Mol Biol. 1994;244:651–653. doi: 10.1006/jmbi.1994.1759. [DOI] [PubMed] [Google Scholar]
- 22.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris A J, Frohman M A, Kanaho Y. Phosphatidylinositol 4-phosphate 5-kinase α is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 23.Jones D H, Bax B, Fensome A, Cockcroft S. ADP ribosylation factor 1 mutants identify a phospholipase D effector region and reveal that phospholipase D participates in lysosomal secretion but is not sufficient for recruitment of coatomer I. Biochem J. 1999;341:185–192. [PMC free article] [PubMed] [Google Scholar]
- 24.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. Rac regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 25.Kahn R A, Gilman A G. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP-binding protein. J Biol Chem. 1986;261:7906–7911. [PubMed] [Google Scholar]
- 26.Klausner R D, Van Renswoude J, Ashwell G, Kempf C, Schechter A N, Dean A, Bridges K R. Receptor-mediated endocytosis of transferrin in K562 cells. J Biol Chem. 1983;258:4715–4724. [PubMed] [Google Scholar]
- 27.Lamarche N, Tapon N, Stowers L, Burbelo P D, Aspenstrom P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and junk/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 28.Liang J O, Sung T C, Morris A J, Frohman M A, Kornfeld S. Different domains of mammalian ADP-ribosylation factor 1 mediate interaction with selected target proteins. J Biol Chem. 1997;272:33001–33008. doi: 10.1074/jbc.272.52.33001. [DOI] [PubMed] [Google Scholar]
- 29.Lippincott-Schwartz J, Cole N B, Donaldson J G. Building a secretory apparatus: role of ARF1/COPI in Golgi biogenesis and maintenance. Histochem Cell Biol. 1998;109:449–462. doi: 10.1007/s004180050247. [DOI] [PubMed] [Google Scholar]
- 30.Massenburg D, Han J-S, Liyanage M, Patton W A, Rhee S G, Moss J, Vaughan M. Activation of rat brain phospholipase D by ADP-ribosylation factors 1, 5, and 6: separation of ADP-ribosylation factor-dependent and oleate-dependent enzymes. Proc Natl Acad Sci USA. 1994;91:11718–11722. doi: 10.1073/pnas.91.24.11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melançon P, Glick B S, Malhotra V, Weidman P J, Serafini T, Gleason M L, Orci L, Rothman J E. Involvement of the GTP-binding “G” proteins in transport through the Golgi stack. Cell. 1987;58:329–336. doi: 10.1016/0092-8674(87)90591-5. [DOI] [PubMed] [Google Scholar]
- 32.Millar C A, Powell K A, Hickson G R X, Bader M-F, Gould G W. Evidence for a role for ADP-ribosylation factor 6 in insulin-stimulated glucose transporter-4 (GLUT4) trafficking in 3T3-L1 adipocytes. J Biol Chem. 1999;274:17619–17625. doi: 10.1074/jbc.274.25.17619. [DOI] [PubMed] [Google Scholar]
- 33.Moss J, Vaughan M. Structure and function of ARF proteins: activators of cholera toxin and critical components of intracellular vesicular transport processes. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- 34.Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 35.Peters P J, Hsu V W, Ooi C E, Finazzi D, Teal S B, Oorschot V, Donaldson J G, Klausner R D. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radhakrishna H, Klausner R D, Donaldson J G. Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J Cell Biol. 1996;134:935–947. doi: 10.1083/jcb.134.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radhakrishna H, Donaldson J G. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radhakrishna H, Al-Awar O, Khatchikian Z, Donaldson J G. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J Cell Sci. 1999;112:855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- 39.Roth M G. Snapshots of ARF1: implications for mechanisms of activation and inactivation. Cell. 1999;97:149–152. doi: 10.1016/s0092-8674(00)80723-0. [DOI] [PubMed] [Google Scholar]
- 40.Rubin L A, Kurman C C, Biddison W E, Goldman N D, Nelson D L. A monoclonal antibody 7G7/B6 binds to an epitope of the human interleukin-2 (IL-2) receptor that is distinct from that recognized by IL-2 or anti-Tac. Hybridoma. 1985;4:91–102. doi: 10.1089/hyb.1985.4.91. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz M A, Meredith J E, Kiosses W B. An activated Rac mutant functions as a dominant negative for membrane ruffling. Oncogene. 1998;17:625–629. doi: 10.1038/sj.onc.1201977. [DOI] [PubMed] [Google Scholar]
- 42.Song J, Khachikian Z, Radhakrishna H, Donaldson J G. Localization of endogenous ARF6 to sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J Cell Sci. 1998;111:2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- 43.Takebe Y, Seiki M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuchiya M, Price S R, Tsai S-C, Moss J, Vaughan M. Molecular identification of ADP-ribosylation factor mRNAs and their expression in mammalian cells. J Biol Chem. 1991;266:2772–2777. [PubMed] [Google Scholar]
- 45.Westwick J K, Lambert Q T, Clark G J, Symons M, van Aelst L, Pestell R G, Der C J. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol Cell Biol. 1997;17:1324–1335. doi: 10.1128/mcb.17.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang C Z, Heimberg H, D'Souza-Schorey C, Mueckler M M, Stahl P D. Subcellular distribution and differential expression of endogenous ADP-ribosylation factor 6 in mammalian cells. J Biol Chem. 1998;273:4006–4011. doi: 10.1074/jbc.273.7.4006. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Q, Calafat J, Janssen H, Greenberg S. ARF6 is required for growth factor- and Rac-mediated membrane ruffling in macrophages at a stage distal to Rac membrane targeting. Mol Cell Biol. 1999;19:8158–8168. doi: 10.1128/mcb.19.12.8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Q, Cox D, Tseng C-C, Donaldson J G, Greenberg S. A requirement for ARF6 in Fcγ receptor-mediated phagocytosis in macrophages. J Biol Chem. 1998;273:19977–19981. doi: 10.1074/jbc.273.32.19977. [DOI] [PubMed] [Google Scholar]