Abstract
Background
Although active surveillance SARS-CoV-2 variants of concern (VOCs) is required for proper outbreak control measures, many lower income countries find it challenging to detect VOCs by carrying genomic sequencing alone, due to limited resources.
Methods
VOCs can also be identified by the unique mutations in the spike protein by real-time PCR that detect these single nucleotide polymorphisms (SNPs). We used a multiplex, real-time PCR assay for detection of these SNPs for identification of the prevalence of different SARS-CoV-2 VOCs in 16/26 districts in Sri Lanka.
Results
Of the 664/934 that were subjected to the multiplex qRT-PCR, 638 (96.1 %) detected L452R and K417 in the channels and were identified as the delta variant. 25 samples (3.9 %) detected N501Y, with K417 were considered as the alpha variant. Of 10/16 districts in Sri Lanka, the delta variant was the only VOC detected.
Conclusions
This multiplex real-time qRT-PCR which identifies certain SNPs specific to the VOCs appears to be a fast, cheaper and less technically demanding method to generate data regarding the spread of different SARS-CoV-2 variants, and is a suitable method for lower income countries, to supplement the data generated by genomic sequencing.
Keywords: SARS-CoV-2, Variants of concern, Single nucleotide polymorphisms, Mutations, Multiplex PCR, Sequencing
1. Introduction
Emergence of highly transmissible SARS-CoV-2 variants of concern (VOCs) such as the delta variant or VOCs that evade vaccine induced or natural immunity, has given rise to massive outbreaks in many countries. To make policy decisions regarding outbreak control, imposing and lifting movement restrictions and to plan health care resources, it is crucial for countries to carry out active surveillance to monitor the prevalence of these VOCs. The WHO has recommended that countries should sequence at least 1% of their SARS-CoV-2 positive samples, in order detect emerging VOCs and significant mutations that may occur in the virus (WHO, 2021a). Although many higher income countries have been able to sequence more than 1 % of their positive samples, lower-middle income countries have only been able to sequence less than 0.2 % of their positive samples (GISAID, 2021), due to limitations in infrastructure and funding. This has significantly hampered surveillance of SARS-CoV-2 VOCs in different countries, especially in remote areas.
The SARS-CoV-2 VOCs can also be identified by the unique mutations in the spike protein that are associated with each variant and these mutations can be identified by assays that detect these single nucleotide polymorphisms (SNPs) (WHO, 2021b). Here we describe the use of a multiplex real time PCR (Babiker et al., 2021)that detects the SNPs associated with the four VOCs and how it was utilized to carry out surveillance for the VOCs in Sri Lanka and to take policy decisions regarding outbreak control measures (Table 1 ).
Table 1.
List of primers and probes for the SNP assay for variants.
| Primer Name | Sequence |
|---|---|
| SpikeSNP Ext | TGAAGTCAGACAAATCGCTCC |
| SpikeSNP R1 | TGGTGCATGTAGAAGTTCAAAAG |
| Probe Name | Sequence |
|---|---|
| 417 K Pr2 | CAAACTGGA+A + A + G+ATTGCTG |
| N501Y Pr1.S2 | CCCAC+T + T + ATGGTGTTGG |
| E484 K Probe | ACCTTGTAATGGTGT+T + A + AAGGTTTT |
| 452R_unmod | ATAATTACCGGTATAGATTGTTTAGGAAGT |
2. Materials and methods
Sri Lanka has 25 districts and samples from positive individuals were obtained from 19/25 districts from 1st to 10th September based on the prevalence of SARS-CoV-2 in each district. The multiplex real-time RT-PCR was carried out in 934 samples that gave a positive result by SARS-CoV-2 qRT-PCR in different laboratories in Sri Lanka. 664 samples gave a positive result and were included in further analysis.
2.1. Multiplex real time PCR
Viral RNA in nasopharyngeal samples was extracted using Maelstrom 9600 automated extraction instrument (TANBead, Taiwan) with their nucleic acid extraction kit (TANBead, Taiwan). Details of the primers and duel labelled probes used are previously described and used to detect the presence of the following spike protein mutations: N501Y, E484 K and L452R with K417 as reference: N501Y, E484 K, K417 and L452R (Babiker et al. (2021)). The multiplex real-time PCR was performed using GoTaq® Probe 1-Step RT-qPCR System (Promega, Cat: A6120) in CFX96 Touch Real-Time PCR Detection System (BIORAD, USA). The reactions consisted of 25 μl volumes and contained the following reagents: GoScript™ RT Mix for 1-Step RT-qPCR (1X), GoTaq® Probe qPCR Master Mix with dUTP (1X), 400 nM of each primer, 200 nM of each probe, 10 μl of RNA, and PCR grade water. Following Reverse transcription for 20 min at 50 °C and Reverse transcriptase inactivation and GoTaq® DNA Polymerase activation at 95 °C for 15 min, the reaction was carried out for 45 cycles of 15 s at 94 °C and 1 min at 60 °C.
2.2. Sensitivity and specificity of detecting the alpla and delta variant with the multiplex real time PCR
In order to determine the sensitivity and specificity of detection of the alpha and delta variants, we carried out the real-time qPCR assay in 20 samples which were previously identified as the alpha variant and 20 samples which were previously identified by us as the delta variant. RNA was re-extracted from the stored nasopharyngeal swabs of individuals who were found to be infected with the alpha variant during the May 2021 (Jeewandara et al., 2021a) and the delta variant in July 2021 (Jeewandara et al., 2021b). The cycle threshold of these 40 samples were <25. These 20 samples were then tested with the multiplex real time PCR assay to determine accuracy and sensitivity.
3. Results
Only 664/934 of the positive samples, sent from different laboratories from Sri Lanka were amplified. Although these samples gave a positive result with the conventional real-time qRT-PCR assays for detection of SARS-CoV-2, only samples with Ct values of <30 were amplified by the multiplex SNP detection qRT-PCR. At Ct values of ≤30, this multiplex real-time PCR assay showed a sensitivity of 100 %
Sri Lanka has been experiencing a massive outbreak since mid-April 2021, which began with the introduction of the alpha variant (B.1.1.7) (Jeewandara et al., 2021a) and later the case numbers further increased with introduction of the delta variant (B.1.617.2), into the community from May. From mid-April 2021 to 31st August 2021, Sri Lanka reported 338,242 cases, of which 59.5 % were reported in the Western Province (Epidemiology unit MoH, Sri Lanka, 2021) represented by the three districts: Colombo, Kalutara and Gampaha. 176 samples were amplified from the Western Province and all of them were identified as delta by sequencing of the SARS-CoV-2 viruses. We randomly selected 20 samples identified as alpha during May 2021 by sequencing and all 20 of these samples had genes targeting N501Y and K417 were amplified whereas there was no amplification of other genes. We also tested 20 samples that were assigned the delta lineage by sequencing and the only genes targeting L452R was amplified. Therefore, this assay seems to have 100 % specific in identifying the presence of these two SNPs in the virus.
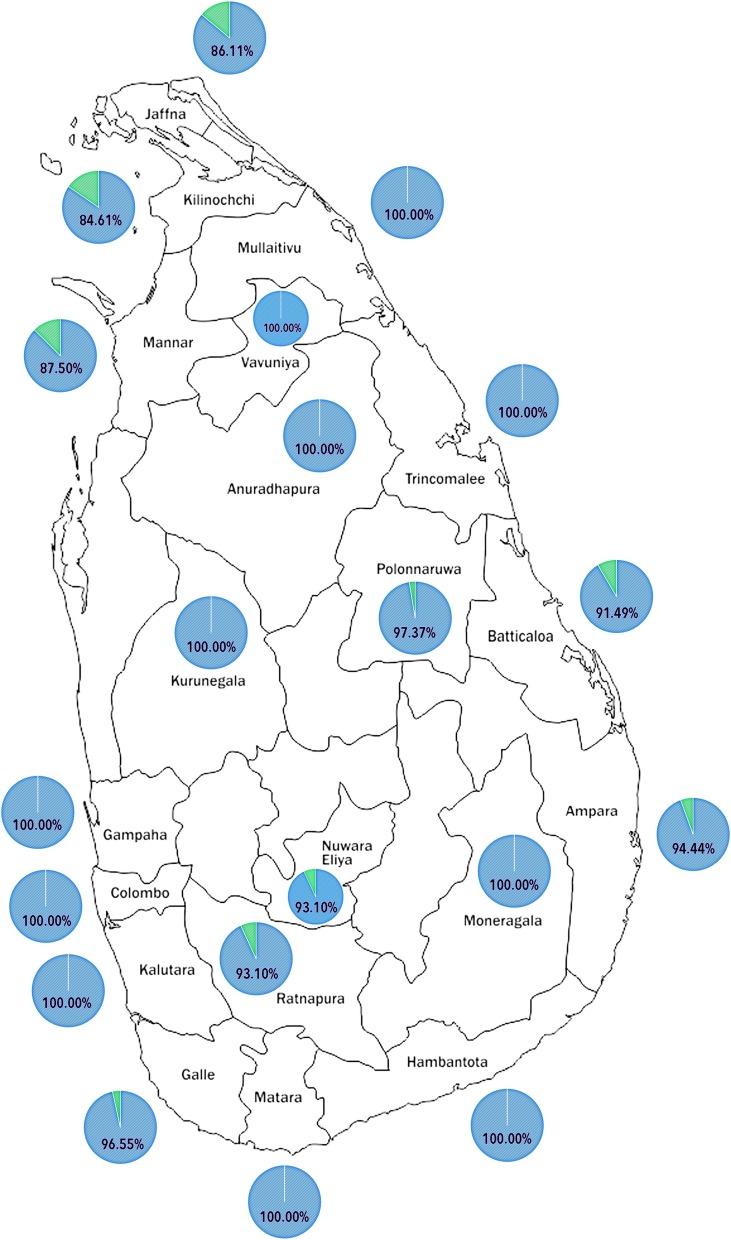
638 (96.1 %) samples showed amplification for the genes targeting L452R and K417 and therefore, were considered to be the delta variant. 25 (3.9 %) showed amplification of genes for N501Y with K417 and therefore, were considered to belong to the alpha variant. The distribution of the different VOCs in the 16 districts in Sri Lanka is shown in Fig. 1 . Although case numbers were lower, only the delta variant could be identified from several other districts such as Kurunegala, Anuradhapura, Polonnaruwa in the North Western and North Central region and Matara, Hambantota districts in the Southern Province. The alpha variant was the only SARS-CoV-2 variant that was seen distributed across Sri Lanka by mid-May (Jeewandara et al., 2021a). We detected the delta variant in the community, by random sampling by the 3rd week of May 2021 (Hadfield et al., 2018) and it appears to have completely displaced the alpha variant, within a period of 12 weeks.
Fig. 1.
The prevalence of alpha variant (green) compared to delta variant (blue) in each district of Sri Lanka. The different SARS-CoV-2 variants of concern were identified by the multiplex real-time PCR assay for detection of SNPs.
4. Discussion
Sri Lanka has been experiencing a massive outbreak of COVID-19 from mid-April 2021 onwards, which was further exacerbated with the rapid spread of delta variant in the country. The delta variant was first detected in the community in the Colombo district (in the Western Province) on a sample obtained on 22nd of May 2021, when Sri Lanka was under a strict lockdown. Although the lockdown ended by 21st of June, to curtail the spread of the delta variant to other provinces, interprovincial travel restrictions were imposed. However, due to the rapid rise of cases and deaths, and with the health care systems being overwhelmed, the authorities imposed another lockdown on the 20th of August (2 months later). Although the genomic sequencing carried out by us showed delta was the predominant variant in the Western Province, due to limitations in resources, it was not feasible to carry out genomic sequencing for the whole country, especially when very high case numbers were reported from all districts. However, in order to make policy decisions regarding the expected rise in the number of cases in district, to decide when to lift the lockdown in each area and whether to further impose interprovincial travel restrictions, it is crucial that data regarding the prevalence of different variants in these locations are available.
In this study, we identified the delta variant in all districts of Sri Lanka, based on the presence of the L425R in the virus. However, the SNPs resulting in L452R and K417 mutations is not unique to delta alone and is detected in several variants of interest such as Kappa (B.1.617.1), Epsilon (B.1.427/9) and C.36 (Covariants -EH, 2021). Therefore, this multiplex real-time PCR approach would not be useful to identify emergence of new SARS-CoV-2 variants or to differentiate between the variants which have L425R, N501Y or E484 K, which again is seen in different SARS-CoV-2 variants (Covariants -EH, 2021). However, in situations where there is a dominant variant with L452R or N501Y, confirmed by whole genomic sequencing, this would be an easy to use, low-cost assay to rapidly identify the prevalence of VOCs such as delta. Furthermore, it would also help to monitor mutations of concern such as the E484 K that could arise in the delta variant itself. In fact, we have recently detected the E484 K mutation in the delta variant itself (Jeewandara et al., 2021b) and the use of this multi-plex RT-q PCR would help us to further monitor the spread of this variants, in a larger sample cohort.
Due to limitations in resources, many lower income and lower-middle income countries are unable to carryout genomic sequencing in order to make these vital decisions. The real-time PCR to identify the different SARS-CoV-2 variants based on their SNPs in the spike protein, is a fast, cheaper and less technically demanding method to generate data regarding the spread of different SARS-CoV-2 variants. For instance, based on the prices of these assays in Sri Lanka (based on custom taxes etc…), the cost of Illumina sequencing is US$200 per sample, for Oxford Nanopore sequencing technology the cost is US$20/sample, while the cost per sample of this in-house muiltiplex real-time PCR is US$ 8 per sample.
In Sri Lanka, despite the interprovincial travel restrictions imposed from May 2021, and despite being under a lockdown until 21st of June, which was gradually relaxed afterwards, the delta variant appeared to have completely replaced the alpha variant within 12 weeks. This is not surprising given the higher transmissibility of delta variant (Callaway, 2021). Delta variant is also shown to associate with higher hospitalization rates and more severe disease, especially in unvaccinated populations (Sheikh et al., 2021). In many resource-poor countries such as Sri Lanka, there is a vast difference in the health care resources available in metropolitan areas and rural areas. Therefore, when planning the allocation of health care resources, to make decisions regarding relaxing lockdowns, vaccination and to implement other infection control measures, it is crucial to have a bird’s eye view of the prevalence of the delta variant and other VOCs in different parts of the country. The availability of these data has enabled policy makers to make scientific decisions regarding COVID-19 control in Sri Lanka.
Author statement
Conceptualization: CJ, GNM, JW
Data curation: DG, VW, PKBM, WR, RR, VD, IP, AS, PB, GN, VRF, AK, DAE, KDS, RW
Formal analysis: LG, TPJ, OD
Project administration: CJ
Funding acquisition: CJ, GNM, GSO, JW
Investigation: LG, MH, TPJ, OD
Validation: JW
Writing original draft: GNM
Writing: reviewing and editing: GSO, JW
Declaration of Competing Interest
None of the authors have any conflicts of interest.
Acknowledgements
We are grateful to the NIH, USA (grant number 5U01AI151788-02) and the WHO for supporting this study.
Data availability
No data was used for the research described in the article.
Data will be made available on request.
All data is within the manuscript and figure
References
- Babiker A., Immergluck K., Stampfer S.D., Rao A., Bassit L., Su M., et al. Single-amplicon, multiplex real-time RT-PCR with tiled probes to detect SARS-CoV-2 spike mutations associated with variants of concern. J. Clin. Microbiol. 2021 doi: 10.1128/JCM.01446-21. JCM0144621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Delta coronavirus variant: scientists brace for impact. Nature. 2021;595:17–18. doi: 10.1038/d41586-021-01696-3. [DOI] [PubMed] [Google Scholar]
- Covariants -EH . 2021. Covariants. CoVariants: 2020-2021 Emma Hodcroft. [Google Scholar]
- Epidemiology unit MoH, Sri Lanka . Ministry of Health; Sri Lanka: 2021. COVID-19 Epidemiological Summary-august Epidemiology Unit. [Google Scholar]
- GISAID . 2021. SARS-CoV-2 Sequence Entries With Complete Collection Date Information Shared Via GISAID. [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeewandara C., Jayathilaka D., Ranasinghe D., Hsu N.S., Ariyaratne D., Jayadas T.T., et al. Genomic and epidemiological analysis of SARS-CoV-2 viruses in Sri Lanka. Front. Microbiol. 2021:12. doi: 10.3389/fmicb.2021.722838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeewandara C.J.D., Ranasinghe D., Hsu N.S., Ariyaratne D., Jayadas T.T., Panambara Arachchige D.M., Lindsey B.B., Gomes L., Parker M.D., Wijewickrama A., Karunaratne M., Ogg G.S., de Silva T.I., Malavige G.N. 2021. Genomic Epidemiology of Novel Coronavirus in Sri Lanka. Allergy Immunology and Cell Biology Unit (AICBU) at University of Sri Jayewardenepura. [Google Scholar]
- Sheikh A., McMenamin J., Taylor B., Robertson C. Public Health S, the EIIC. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397:2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . WHO Headquarters (HQ); 2021. Guidance for Surveillance of SARS-CoV-2 Variants: Interim Guidance. 9 August 2021. [Google Scholar]
- WHO . World Health Organization. Regional Office for Europe; 2021. Methods for the Detection and Identification of SARS-CoV-2 Variants; p. 6. March 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.
Data will be made available on request.
All data is within the manuscript and figure