Abstract
Mammalian eukaryotic initiation factor 4GI (eIF4GI) may be divided into three similarly sized regions. The central region (amino acids [aa] 613 to 1090) binds eIF3, eIF4A, and the encephalomyocarditis virus (EMCV) internal ribosomal entry site (IRES) and mediates initiation on this RNA. We identified the regions of eIF4GI that are responsible for its specific interaction with the IRES and that are required to mediate 48S complex formation on the IRES in vitro. Mutational analysis demarcated the IRES binding fragment of eIF4GI (aa 746 to 949) and indicated that it does not resemble an RNA recognition motif (RRM)-like domain. An additional amino-terminal sequence (aa 722 to 746) was required for binding eIF4A and for 48S complex formation. eIF4GI bound the EMCV IRES and β-globin mRNA with similar affinities, but association with eIF4A increased its affinity for the EMCV IRES (but not β-globin RNA) by 2 orders of magnitude. On the other hand, eIF4GI mutants with defects in binding eIF4A were defective in mediating 48S complex formation even if they bound the IRES normally. These data indicate that the eIF4G-eIF4A complex, rather than eIF4G alone, is required for specific high-affinity binding to the EMCV IRES and for internal ribosomal entry on this RNA.
The initiation phase of translation in eukaryotes is the process leading to assembly of a translation-competent 80S ribosome at the initiation codon of an mRNA. The canonical initiation process involves more than 10 initiation factors (6, 27, 38). The first stage in the initiation process is the binding of a eukaryotic initiation factor 2 (eIF2)-GTP-initiator tRNA complex, eIF1A, and eIF3 to the 40S ribosomal subunit to form a 43S complex. The second stage involves the binding of mRNA to this ribosomal complex and involves eIF3, eIF4A, eIF4B, eIF4F, and the poly(A)-binding protein (PABP). All nonorganellar cellular mRNAs have a 5′-terminal m7G cap structure that is recognized by the eIF4E (cap-binding) subunit of eIF4F. Mammalian eIF4F also contains eIF4G and eIF4A subunits. eIF4A is an RNA-dependent ATPase and RNA helicase. After binding of eIF4F to the 5′ end of an mRNA, eIF4A and eIF4B melt the RNA structure in its 5′-nontranslated region, which facilitates binding of the 43S complex to the 5′ end of the mRNA. Ribosomal binding is thought to be mediated through interactions of eIF4G and eIF4B with ribosome-bound eIF3 (22, 29). The ribosomal complex then scans to the initiating AUG codon (35). Finally, eIF5 and eIF5B mediate the displacement of factors from the 40S subunit and joining of the 60S subunit to form an active 80S ribosome (40).
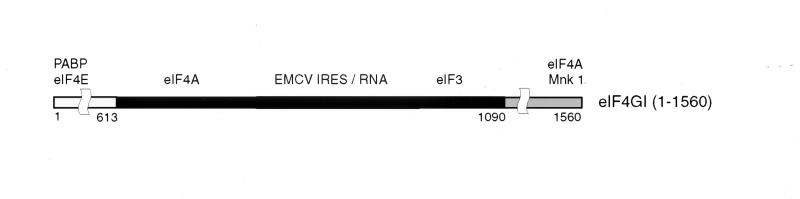
eIF4G is a large adapter protein with a modular structure that plays a key coordinating role in the early stages of initiation (19, 31). Two related eIF4G proteins (eIF4GI and eIF4GII) encoded by two different genes exist in yeast and mammals (8, 9, 15, 19). Mammalian eIF4G can be divided into three distinct functional domains. The N-terminal third (amino acids [aa] 1 to 612) contains the eIF4E and PABP binding sites (15, 24); the middle third (aa 613 to 1090) binds eIF3, eIF4A, and RNA (3, 14, 22, 30, 41); and the C-terminal third (aa 1091 to 1560) contains a second eIF4A binding site (14, 22, 30) and a binding site for the protein kinase Mnk1 (43). eIF4G therefore acts as a platform for the assembly of a multiprotein-RNA complex to recruit the ribosome to a mRNA.
Consistent with its central role in initiation, eIF4G is also an important target in the regulation of protein synthesis. The 4E binding proteins (4E-BPs) act as general inhibitors of cap-dependent translation by binding eIF4E and sequestering it from the rest of the eIF4F complex (6). Biochemical and structural studies have established that the 4E-BPs are molecular mimics of eIF4G and compete for the same binding site on the dorsal surface of eIF4E (11, 25). eIF4G is also a direct target for regulation by phosphorylation (44) and by proteolysis, both during apoptosis (26) and during infection by some picornaviruses such as poliovirus (10, 21, 22). Proteases encoded by these picornaviruses cleave eIF4G specifically, separating the eIF4E-PABP binding domain from the eIF4A and eIF3 binding sites. In contrast, other picornaviruses such as encephalomyocarditis virus (EMCV) inhibit cellular translation by dephosphorylating 4E-BP1 and thereby disrupting the eIF4E-eIF4G interaction (7). Both strategies abrogate the activity of eIF4F in initiation on capped mRNAs and thus lead to a shutoff of host cell translation.
Initiation of translation of picornavirus mRNAs occurs by a noncanonical cap-independent mechanism of internal initiation that is mediated by a ∼400-nucleotide (nt) highly structured internal ribosomal entry site (IRES) that lies immediately upstream of the initiation codon (16). The EMCV IRES epitomizes those of a large group of picornaviruses, including all members of the Aphthovirus, Cardiovirus, and Parechovirus genera. We reconstituted EMCV IRES-mediated initiation in vitro using purified translation components and found that this process is ATP dependent and utilizes the same set of canonical eIFs as does cap-mediated initiation except for eIF1, eIF1A, eIF4E, PABP, and the amino-terminal third of eIF4G, to which the last two bind (37, 39, 40). The essential region of eIF4G corresponds to the carboxy-terminal proteolytic cleavage product that is generated during poliovirus infection. It binds specifically to the J-K domain of the IRES, in close proximity to the initiation codon, and recruits eIF4A and eIF4B to the IRES (20, 41). Its interaction with the IRES is essential for 48S complex formation, and its role in IRES-mediated initiation may be analogous to that of the eIF4E-eIF4G complex in initiation on capped mRNAs, i.e., recruiting factors and promoting ribosomal attachment at a defined location on the mRNA. The requirement for eIF4G and its specific binding to the J-K domain is a general characteristic of the mechanism of initiation on all EMCV-like picornavirus IRESs, such as those of foot-and-mouth disease virus, human parechovirus 1, and Theiler's murine encephalomyelitis virus (41a; V. G. Kolupaeva, unpublished data).
In this article we define the minimum region of eIF4GI required for its specific interaction with the EMCV IRES and for support of 48S complex formation on this RNA. Our data provide evidence that the specific interaction of eIF4A with eIF4G significantly enhances its affinity for the IRES and indicate that the interaction of eIF4A and eIF4G is required to yield an active complex in IRES-mediated initiation.
MATERIALS AND METHODS
Construction of plasmids.
Plasmids pBS-globin (12), pET(His6-eIF4A) and pET(His6-4B) (39), pET28(His6-eIF4G613–1090), and pET28(His6-eIF4G613–1560) (41), pQE(His6-eIF1) and pET(His6-eIF1A) (35), and pTE1 (5) have been described previously. Truncation, insertion, and substitution mutants of eIF4GI were generated by PCR using pET28(His6-eIF4G613–1560) and Vent DNA polymerase (New England BioLabs). All PCR products were inserted between the BamHI and XhoI restriction sites of pET28b (Novagen), except for mutants eIF4GI(772–1076) and eIF4GI(800–1076), which were cloned between the EcoRI and XhoI restriction sites of pET28a. All mutations were confirmed by sequencing. p97(NAT1)[62–330] was constructed by inserting a PCR fragment corresponding to aa 62 to 330 of NAT1 (50) between the BamHI and XhoI restriction sites of pET28b. To construct pFLAG-eIF4A, cDNA corresponding to the complete eIF4A coding sequence immediately preceded by a His6 tag was generated by PCR using pET(His6-eIF4A) and was inserted into pFLAG-MAC (Sigma) between the HindIII and EcoRI restriction sites. The EMCV transcription vector pJK was constructed by inserting an EMCV nt 680 to 786 PCR fragment between the EcoRI and HindIII restriction sites of plasmid pTZ18R (Pharmacia).
RNA synthesis and purification.
For toeprinting assays, EMCV RNA was transcribed in vitro from PstI-linearized pTE1 using T7 RNA polymerase. For mobility shift assays, pBS-β-globin and pJK were linearized with NcoI and HindIII, respectively, and were transcribed in vitro in the presence of [32P]UTP (3,000 Ci/mmol; ICN Radiochemicals, Irvine, Calif.) with T3 or T7 RNA polymerase as appropriate. RNA transcripts (700,000 cpm/pmol) were purified as described previously (39).
Purification of initiation factors and 40S ribosomal subunits.
40S ribosomal subunits, eIF2, eIF3, and eIF4F were purified from rabbit reticulocyte lysate (Green Hectares, Oregon, Wis.) as described previously (36, 39). Recombinant eIF1, eIF1A, eIF4A, and eIF4B were purified as described previously (35, 39). Recombinant mutant eIF4GI and p97 polypeptides were purified after expression in Escherichia coli BL21(DE3). Protein expression was induced by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) during the late log phase of growth (optical density at 600 nm, ∼0.7 to 0.8). After induction, the cells continued to grow at 37°C for an additional 3 h. Recombinant proteins were purified by chromatography using Ni2+-nitrilotriacetic acid-agarose (Qiagen) and heparin-Sepharose (Pharmacia). The concentration of proteins was measured by the Bradford assay (Bio-Rad) as specified by the manufacturer. The N-terminal deletion of the expressed eIF4GI sequence up to aa 697 very strongly increased the protein yield of eIF4GI mutants. For different eIF4GI mutants lacking aa 1 to 697 or more, the yield was in the range of 0.3 to 2 mg/liter of induced culture.
Toeprinting analysis of eIF4G-IRES and eIF4G-eIF4A-IRES complexes.
EMCV nt 315 to 1155 RNA (0.2 μg) was incubated for 5 min at 30°C with eIF4GI polypeptides (0.3 μg) in 40-μl reaction volumes that contained buffer A (2 mM dithiothreitol [DTT], 20 mM Tris-HCl [pH 7.6], 100 mM potassium acetate, 2.5 mM magnesium acetate, 0.2 mM spermidine) in the presence or absence of eIF4A (2 μg). The resulting RNA-protein complexes were analyzed by primer extension using primer 5′-GTCAATAACTCCTCTGG-3′ (complementary to EMCV nt 957 to 974) and avian myeloblastosis virus reverse transcriptase (Promega) in the presence of [α-32P]dATP (6,000 Ci/mmol; ICN Radiochemicals) essentially as described previously (39). cDNA products were analyzed by electrophoresis through 6% polyacrylamide sequencing gels and compared with appropriate dideoxynucleotide ladders. The gels were quantitated by PhosphorImager analysis to compare the relative amounts of complexes formed using different eIF4GI polypeptides. To minimize errors causing by differences in loading, all values for the stop site at C786 (caused by binding of eIF4G) in Fig. 5 were normalized relative to a stop site (N in Fig. 2) that occurs on EMCV RNA in the absence of factors, is not influenced by eIF4G, and is located closer to the primer than C786. Actual differences in loading never exceeded 70%. Theoretically, the intensity of the N stop site should be identical in all lanes if the loading was equal.
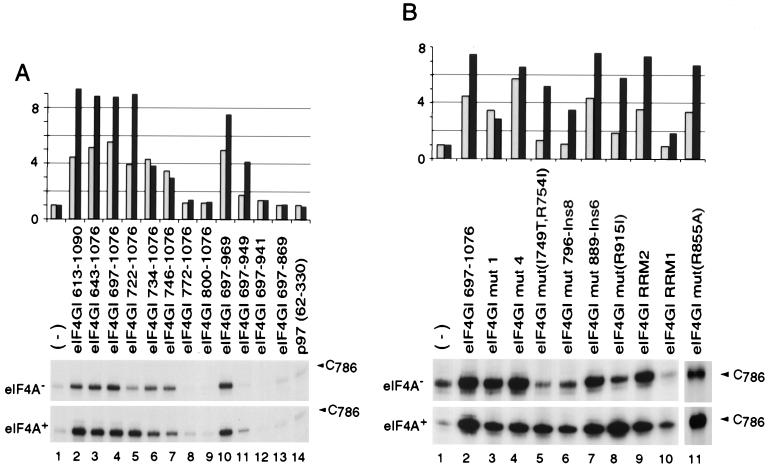
FIG. 5.
Influence of eIF4A on the interaction of eIF4GI mutants with the EMCV IRES. Toeprint analysis of ribonucleoprotein complex formation on the EMCV IRES with eIF4GI deletion mutants (A) and eIF4GI insertion-substitution mutants (B) in the presence and absence of eIF4A, as indicated, was performed. The position of the stop site due to binding of eIF4GI is indicated at C786; for greater clarity, only this part of each gel is shown. These bands were quantitated by PhosphorImager analysis and normalized as described in Materials and Methods. Values are shown schematically relative to the intensity of the C786 band in the absence of factors, which was arbitrarily assigned a value of 1; gray and black bars represent values obtained in the absence and presence of eIF4A, respectively.
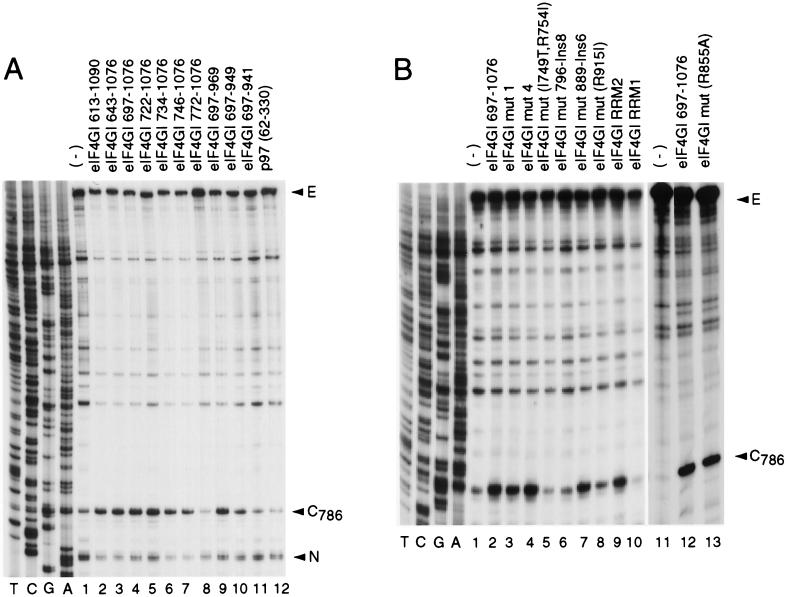
FIG. 2.
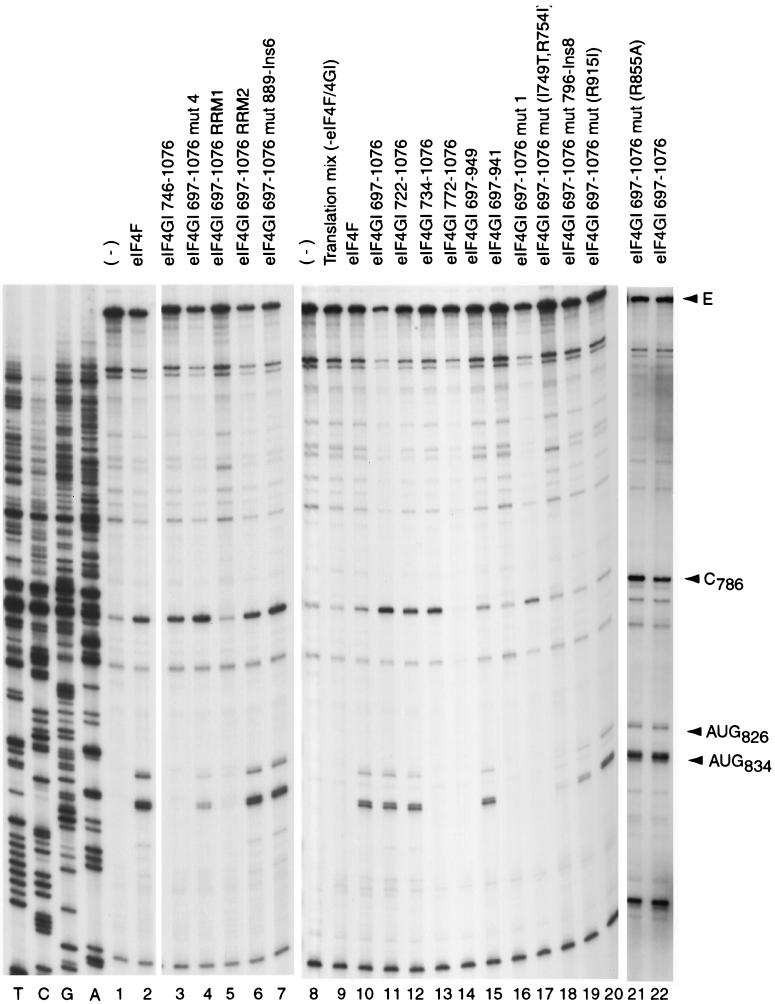
Specificity of interaction between eIF4GI mutants and the EMCV IRES. A toeprint analysis of binary-complex formation on the EMCV IRES with eIF4GI deletion mutants (A) and eIF4GI insertion-substitution mutants (B) was performed as described in Materials and Methods. The full-length cDNA extension product is marked E, the position of the stop site due to binding of eIF4G is indicated at C786, and a stop site detected on EMCV RNA irrespective of the presence or absence of eIF4GI that was used as an internal standard for quantitation is marked N. Reference lanes T, C, G, and A depict the EMCV cDNA sequence.
Assembly and analysis of 48S ribosomal complexes on the EMCV IRES.
EMCV nt 315 to 1155 RNA (0.2 μg) was incubated for 5 min at 30°C in 40-μl reaction volumes that contained buffer A, 1 mM ATP, 0.1 mM GMP-PNP, 6 pmol of [35S]Met-tRNAiMet, 6 pmol of 40S subunits, and initiation factors eIF2 (3 μg), eIF3 (6 μg) eIF4A (2 μg), eIF4B (0.5 μg), eIF1 (0.5 μg), eIF1A (0.5 μg), and eIF4GI mutant polypeptide or eIF4F (0.5 μg) as indicated in the text. The resulting 48S initiation complexes were analyzed using the toeprinting assay described above.
Gel electrophoretic mobility shift assay.
[32P]UTP-labeled EMCV nt 680 to 786 (the J-K domain) or β-globin nt 1 to 190 RNA (2 nM concentration) as appropriate was incubated for 15 min at 30°C in 15-μl reaction volumes that contained buffer B (100 mM KCl, 20 mM Tris-HCl [pH 7.5], 4 mM DTT, 0.01% NP-40, 2 mM magnesium acetate), different amounts of eIF4GI mutant polypeptides, and eIF4A (1.5 μg) as indicated. Sample buffer (2 μl containing 15% glycerol and 0.1% bromophenol blue) was added to the reaction mixtures before they were loaded on a 6% polyacrylamide gel (acrylamide/bisacrylamide ratio, 75:1) and subjected to electrophoresis (42). The gels were quantified by PhosphorImager analysis. Binding constants were calculated by assuming 100% active protein and a 1:1 stoichiometry of RNA-protein binding and plotting 1 − unbound RNA versus eIF4G concentration, where unbound RNA is the relative amount of free RNA (obtained by quantifying the intensity of RNA bands) and eIF4G concentration is the concentration of the recombinant protein. Each Kd value obtained is the average of at least three independent experiments.
In vitro protein binding assays.
The binding between eIF4GI mutants and eIF4A was assayed essentially as described previously (32). FLAG(His6)-eIF4A (2 μg) was immobilized on 15 μl of anti-FLAG agarose beads (Sigma) by incubating for 20 min at 26°C in 60 μl of buffer C (150 mM NaCl, 10 mM Na2HPO4, 4 mM NaH2PO4 [pH 7.3], 2 mM DTT, 0.5% Triton X-100) with occasional mixing. The beads were then washed with 500 μl of buffer C. Approximately 4 μg of each eIF4GI mutant polypeptide, 20 μg of bovine serum albumin (New England BioLabs), and 20 μg of RNase A were added to the immobilized eIF4A in 60-μl reaction volumes containing buffer C and incubated for 20 min at 26°C with occasional mixing. The beads were then washed four times with 500 μl of buffer C. Bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% polyacrylamide) and either visualized by Coomassie blue staining (eIF4A) or detected by Western blotting (eIF4GI) using anti-T7-tag horseradish peroxidase-conjugated antibodies (Novagen).
To assay the binding of eIF3 to eIF4GI mutant polypeptides, 5 μg of each eIF4GI polypeptide was immobilized on 10 μl of T7-agarose beads (Novagen) by incubating for 30 min at 26°C in 40 μl of buffer D (100 mM KCl, 20 mM Tris-HCl [pH 7.5], 2 mM DTT, 0.5% Triton X-100). The beads were then washed with 500 μl of buffer D. Approximately 5 μg of eIF3, 20 μg of aprotinin (Sigma), and 20 μg of RNase A were added to the immobilized eIF4GI in 40-μl reaction volumes containing buffer D and incubated for 30 min at 26°C with occasional mixing and then for 3 h at 4°C. The beads were then washed four times with 500 μl of buffer D. Bound proteins were resolved by SDS-PAGE (12.5% polyacrylamide), and the p170 subunit of eIF3 was detected by Western blotting using a sensitive monoclonal antibody as described previously (40).
RESULTS
Construction of eIF4G mutant polypeptides.
The initiation factor eIF4G is a large protein that interacts directly with many other components of the translation initiation apparatus, including eIF4E, eIF4A, eIF3, PABP, and the Mnk1 kinase (shown schematically in Fig. 1). eIF4GI also contains two centrally located amino acid sequences (aa 855 to 862 and 757 to 762) that resemble the RNP-1 and RNP-2 motifs characteristic of RNA recognition motif (RRM) proteins (1, 8). The central domain of eIF4GI binds strongly and specifically to the J-K domain of the EMCV IRES (20, 41). Two groups of mutant eIF4GI polypeptides were constructed to identify the amino acid determinants that enable eIF4GI(613–1090) to bind to this IRES. First, a series of amino-terminal and carboxyl-terminal deletion mutants was made to localize the borders of the IRES binding domain of eIF4GI (Table 1). The amino terminus of the central domain of eIF4GI(613–1090) is close to the rhinovirus 2A protease cleavage site (21). Two amino acids (L729 and L732) that are required for eIF4A binding (14, 30) are present in eIF4GI(722–1076) but not eIF4GI(734–1076). The RNP-2 motif is present in eIF4GI(746–1076) but not eIF4GI(772–1076). Sites of C-terminal deletion were chosen to systematically remove residues that are conserved in human eIF4GI and other related proteins. eIF4GI(643–696) contains an arginine-rich region and was expressed to determine whether it could bind the EMCV IRES independently. The translation regulator p97/NAT1 (15a, 50) is related to eIF4GI, binds eIF4A (13, 15a, 30), but does not bind the EMCV IRES (T. V. Pestova, unpublished data). A p97(62–330) fragment that is homologous to eIF4GI(697–969) was also expressed and purified.
FIG. 1.
Schematic representation of eIF4GI. PABP, eIF4E, eIF4A, eIF3, EMCV IRES, and Mnk1 binding regions are shown.
TABLE 1.
Deletion mutants of eIF4GI
| eIF4GI mutant | Amino- and carboxy-terminal amino acids |
|---|---|
| eIF4GI(643–1076) | P643–P1076 |
| eIF4GI(697–1076) | P697–P1076 |
| eIF4GI(722–1076) | F722–P1076 |
| eIF4GI(734–1076) | P734–P1076 |
| eIF4GI(746–1076) | Q746–P1076 |
| eIF4GI(772–1076) | F772–P1076 |
| eIF4GI(800–1076) | F800–P1076 |
| eIF4GI(697–969) | P697–E969 |
| eIF4GI(697–949) | P697–S949 |
| eIF4GI(697–941) | P697–Q941 |
| eIF4GI(697–869) | P697–L869 |
| eIF4GI(643–696) | P643–K696 |
Amino acid substitutions and insertions were made in eIF4GI(697–1076) to identify amino acid residues that are directly involved in the interaction of eIF4G with RNA or that are responsible for the specificity of its interaction with the EMCV IRES (Table 2). Mut1 and Mut4 substitutions impair the binding of eIF4G to eIF4A (14). RRM1 and RRM2 mutants contain substitutions in putative RNP-1 and RNP-2 motifs. Other substitution mutations were made to alter residues that are conserved in eIF4GI and all related proteins; insertion mutations were made in sequences that are conserved in mammalian eIF4GI and eIF4GII and that differ from corresponding regions of 4G-like proteins that do not bind the EMCV IRES. These proteins include wheat eIF4F, wheat eIF-iso4F, and mouse p97 (38, 41).
TABLE 2.
eIF4GI(697–1076) mutants
| eIF4GI(697–1076) mutant | Substitution or insertion |
|---|---|
| eIF4GI mut1 | L729A, L732A, F737A |
| eIF4GI mut4 | R935A, F938A |
| eIF4GI RRM1 | L857A, I860A |
| eIF4GI RRM2 | V758A, L761A |
| eIF4GI mut(I749T, R754I) | I749T, R754I |
| eIF4GI mut796-Ins8 | V796-EGEQGEAG-T797 |
| eIF4GI mut(R855A) | R855A |
| eIF4GI mut889-Ins6 | D889-KKACPD-E890 |
| eIF4GI mut(R915I) | R915I |
Binding of eIF4GI mutant polypeptides to the EMCV IRES.
The specific interaction of the EMCV IRES with eIF4G results in the formation of a stable complex that can be detected by primer extension inhibition (toeprinting). Bound eIF4G yields a toeprint at C786 near the base of the J-K domain of the IRES (41). Toeprinting was used to assay the interaction of the eIF4GI mutant polypeptides described above with the EMCV IRES (Fig. 2). The central domain (aa 613 to 1090) of eIF4GI and all derivatives of it deleted from its N terminus (D613) to Q746 bound stably to the IRES (Fig. 2A, lanes 1 to 7). An additional deletion to F772 in eIF4GI(772–1076) abrogated this interaction (lane 8). From these data, we conclude that the N-terminal border of the domain of eIF4GI that binds to the IRES lies between residues 746 and 772. A C-terminal deletion mutant, eIF4GI(697–969), bound stably to the IRES, eIF4GI(697–949) bound weakly, and eIF4GI(697–941) did not bind at all (lanes 9 to 11). A p97(62–330) fragment that corresponds to eIF4GI(697–969) did not bind to the EMCV IRES (lane 12).
Variants of eIF4GI(697–1076) containing mut1 or mut4 substitutions (14) bound to the EMCV IRES as strongly as the corresponding wild-type polypeptide did (Fig. 2B, lanes 2 to 4). The mut889-Ins6, R855A, and RRM2 mutant eIF4GI(697–1076) polypeptides also retained wild-type activity in this assay (lanes 7, 9, and 13). The IRES-binding activity of mut(I749T, R754I), mut796-Ins8, and R915I mutant eIF4GI(697–1076) polypeptides was strongly reduced compared to that of the wild-type polypeptide (lanes 5, 6, and 8), and binding of the eIF4GI(697–1076) RRM1 mutant to the IRES was not detectable (lane 10). The lack of effect of the RRM2 mutation on the IRES-binding activity of eIF4GI suggests that the central domain of eIF4G does not resemble an RRM domain.
Binding of eIF4GI mutant polypeptides to β-globin mRNA.
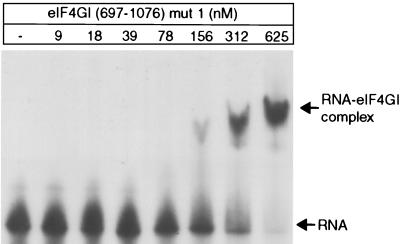
The borders of the IRES binding domain of eIF4GI were determined by deletion analysis, and several mutations were identified that impaired this specific interaction, as described above. To distinguish between determinants of the general RNA binding and specific IRES binding activities of eIF4GI, the interaction of these eIF4GI polypeptides with an uncapped 190-nt long 5′-terminal fragment of β-globin mRNA was also analyzed. We assume that this interaction is representative of the general RNA binding properties of eIF4G. The formation of binary complexes between a low concentration of [32P]-labeled RNA (∼2 nM) with increasing concentrations of eIF4GI polypeptides was analyzed using a quantitative mobility shift assay. The results of a typical mobility shift assay done using mut1 eIF4GI(697–1076) are shown in Fig. 3. The protein concentration at half-saturation is equal to the equilibrium dissociation constant (Kd) for the reaction, assuming that one molecule of eIF4GI polypeptide binds one molecule of RNA, that all protein was active, that all of the sample was recovered, and that there was no cooperativity in binding. Binding data are summarized in Table 3. Deletion of 54 N-terminal amino acid residues from eIF4GI(643–1076), yielding eIF4GI(697–1076), caused a fivefold reduction in binding to β-globin RNA. These 54 residues contain a region (aa 643 to 675) that comprises mostly Arg, Gly, and Pro residues (68%). Short arginine-rich motifs are found in some sequence-specific RNA binding proteins (49), and repeated RGG boxes have been identified as a domain that binds RNA (1). Although eIF4GI(643–675) does not correspond to canonical forms of either motif, its influence on the general RNA binding activity of eIF4GI prompted us to assay the RNA binding activity of a polypeptide, eIF4GI(643–696), which contains this sequence. This fragment bound β-globin RNA relatively strongly (Kd ≈ 200 nM). This result indicates that residues 643 to 696 contribute to the general RNA binding activity of eIF4GI and may even correspond to a separate RNA binding domain. Additional N-terminal deletions from P697 to F800 did not result in any additional loss of binding affinity of eIF4GI for β-globin mRNA.
FIG. 3.
Interaction of eIF4GI(697–1076) mut1 with β-globin RNA as assayed by an electrophoretic mobility shift assay. The positions of free RNA and of the RNA-eIF4GI complex are indicated.
TABLE 3.
Binding affinities of β-globin RNA to eIF4GI mutants
| eIF4GI mutant | Kd (nM) |
|---|---|
| eIF4GI(643–1076) | 100 |
| eIF4GI(697–1076) | 500 |
| eIF4GI(722–1076) | 300 |
| eIF4GI(734–1076) | 550 |
| eIF4GI(746–1076) | 500 |
| eIF4GI(772–1076) | 500 |
| eIF4GI(800–1076) | 600 |
| eIF4GI(697–969) | 1,000 |
| eIF4GI(697–949) | 2,800 |
| eIF4GI(697–941) | 2,600 |
| eIF4GI(697–869) | 6,500 |
| eIF4GI(643–696) | 200 |
| eIF4GI mut1 | 400 |
| eIF4GI mut4 | 300 |
| eIF4GI RRM1 | 800 |
| eIF4GI RRM2 | 550 |
| eIF4GI mut(I749T, R754I) | 1,500 |
| eIF4GI mut796-Ins8 | 700 |
| eIF4GI mut(R855A) | 800 |
| eIF4GI mut889-Ins6 | 1,100 |
| eIF4GI mut(R915I) | 1,000 |
| p97(NAT1) 62–330 | >4,000 |
C-terminal truncations decreased the general RNA binding properties of eIF4GI(697–1076). Mutant eIF4GI(697–969) bound β-globin RNA about half as strongly as eIF4GI(697–1076) (Table 3). The low general RNA binding activity of eIF4GI(697–949) may account for the weaker specific EMCV IRES binding activity of this mutant compared to eIF4GI(697–1076) (Fig. 2A, lanes 4 and 10).
The two amino acid substitutions I749T and R754I in mut(I749T, R754I) eIF4GI(697–1076) reduced the binding to β-globin RNA about threefold. The 6-aa mut889-Ins6 insertion and the single R915I substitution both reduced the binding of eIF4GI(697–1076) about twofold. Surprisingly, mutations in both RNP-1 and RNP-2 motifs had no significant effect on the general RNA binding properties of eIF4GI. This result casts further doubt on the existence of a central RRM domain in eIF4GI.
Although many eIF4GI mutants behaved similarly in the general (β-globin) RNA binding assay and in the specific EMCV IRES toeprinting assay, it is important to note that the RRM1 substitution mutation and all N-terminal deletion mutations starting from Q746 had a significantly greater effect on the EMCV IRES binding activity of eIF4GI than on its general RNA binding activity. Specific binding of eIF4GI to the IRES was effectively abrogated as a result of deletion from P697 to F772, whereas eIF4GI polypeptides with deletions to P697, F772, and even F800 all bound β-globin mRNA with the same affinity. These mutations therefore altered regions of eIF4GI required for specific recognition of the EMCV IRES.
Activity of eIF4GI mutants in promoting 48S complex formation on the EMCV IRES.
As described previously, eIF4A and eIF4GI(613–1090) have the same activity as eIF4F holo-factor in promoting 48S complex formation on the EMCV IRES (41). To investigate the correlation between the ability of eIF4GI to bind specifically to the EMCV IRES and to promote formation of 48S complexes, the activity of the eIF4GI mutants described above in this process was investigated, using toeprinting to assay the formation of 48S complexes at the EMCV initiation codon AUG834 in a fully reconstituted system. Toeprinting involves cDNA synthesis by reverse transcriptase on a template RNA to which a ribosomal complex is bound. cDNA synthesis is arrested by the bound complex, yielding toeprints at its leading edge. Eukaryotic 48S complexes inhibit primer extension on the EMCV IRES at positions nt 15 to 17 3′ to the A of the initiation codon (39, 41).
N-terminal deletions made in eIF4GI up to F722 did not affect its activity in 48S complex formation (Fig. 4, lanes 10 to 12). Deletion of another 12 aa to P734 abrogated the activity of eIF4GI in this assay (lane 13). However, this deletion mutant eIF4GI(734–1076) was still able to bind specifically to the EMCV IRES (Fig. 2A, lane 6; Fig. 4, lane 13). Thus, aa 722 to 734 are involved in an interaction other than IRES recognition that is important for the function of eIF4GI in 48S complex formation. This interaction is likely to involve eIF4A, since eIF4GI mut1 has substitutions L729A and L732A in this region and is defective in binding eIF4A (14).
FIG. 4.
Primer extension analysis of 48S initiation complexes assembled on EMCV RNA using translation mix (eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, initiator tRNA, and 40S subunits) (lanes 3 to 7 and 9 to 21) with eIF4F (lanes 2 and 10) or eIF4GI mutants (lanes 3 to 7 and 11 to 21) as indicated. The full-length cDNA extension product is marked E, the position of the stop site due to binding of eIF4GI is indicated at C786, and cDNA products labelled AUG826 and AUG834 terminated at stop sites 15 to 17 nt downstream of the stated initiation codon. Reference lanes T, C, G, and A depict the EMCV cDNA sequence.
C-terminal deletions made in eIF4GI up to S949 did not affect its activity in 48S complex formation (Fig. 4, lanes 10 and 15). Deletion of another 8 aa to Q941 in eIF4GI completely abrogated its activity in this assay (lane 16). This effect may be accounted for by the observation that this additional deletion abrogated the specific binding of eIF4GI to the IRES (Fig. 2A, lane 11). We conclude that the borders of the minimum active core of eIF4GI that is required to promote 48S complex formation on the EMCV IRES lie between residues 722 and 949.
The eIF4GI(697–1076) RRM2, mut(R855A), and mut889-Ins6 mutants bound to the EMCV IRES (Fig. 2B, lanes 2, 7, 9, and 13) and promoted 48S complex formation (Fig. 4, lanes 2, 6, 7, 11, and 21) as well as the equivalent wild-type polypeptide did. The eIF4GI(697–1076) mut1 mutant also bound as well to the EMCV IRES as wild-type eIF4GI(697–1076) did (Fig. 2B, lanes 2 and 3) but was absolutely inactive in promoting 48S complex formation (Fig. 4, lane 17). This eIF4GI mutant is unable to bind eIF4A (14), and this result therefore indicates that binding of eIF4GI to the IRES is not sufficient for 48S complex formation on this mRNA in the absence of a stable interaction between eIF4GI and eIF4A. The eIF4GI(697–1076) mut4 mutant bound stably to the EMCV IRES (Fig. 2B, lane 4) but promoted 48S complex formation on it much more weakly than the equivalent wild-type eIF4GI did (Fig. 4, lanes 2, 4, and 11). This result is consistent with the reported defect of this mutant in binding eIF4A (14). However, its activity was sufficiently greater than that of the mut1 eIF4GI mutant to be detected in our assay. The eIF4GI(697–1076) mut(R915I) mutant bound weakly to the EMCV IRES (Fig. 2B, lane 8) but had near-wild-type activity in promoting 48S complex formation (Fig. 4, lanes 11 and 20). The eIF4GI(697–1076) mut(I749T, R754I) and mut796-Ins8 mutants bound to the EMCV IRES significantly less strongly than the equivalent wild-type polypeptide did (Fig. 2B, lanes 2, 5, and 6) but were still able to promote 48S complex formation on this RNA, albeit less efficiently than the wild-type polypeptide did (Fig. 4, lanes 11, 18, and 19). Binding of the eIF4GI(697–1076) RRM1 mutant to the EMCV IRES was undetectable by toeprinting (Fig. 2B, lane 10), but this polypeptide nevertheless promoted very low levels of 48S complex formation (Fig. 4, lane 5).
The activities of eIF4GI mut1, mut4, and eIF4GI(734–1076) mutant polypeptides led us to conclude that the ability of eIF4GI to bind specifically to the EMCV IRES is not sufficient for its activity in promoting 48S complex formation on this RNA and that an interaction with eIF4A is also required. In addition, the activity of a number of other eIF4GI mutants [in particular mut(R915I), RRM1, and mut796-Ins8] in promoting 48S complex formation was greater than would be expected on the basis of their ability to bind to the EMCV IRES. This conclusion suggests that other components of the translation apparatus may enhance the IRES binding activity of eIF4G.
eIF4A and eIF4GI bind synergistically to the EMCV IRES.
To quantitate the interaction of eIF4GI(697–1076) and mutant derivatives thereof with the IRES, binding constants for these polypeptides were determined using RNA transcripts corresponding to the EMCV J-K domain (nt 680 to 786) in a mobility shift assay essentially as described above for the interaction of eIF4GI polypeptides with β-globin RNA. This 107-nt fragment of the EMCV IRES binds to eIF4GI with the same specificity as the intact IRES does (Kolupaeva, unpublished). Binding data are summarized in Table 4.
TABLE 4.
Binding affinities of EMCV nt 680 to 786 RNA to eIF4GI mutants in the absence and in the presence of eIF4A
| eIF4GI mutant |
Kd (nM)
|
|
|---|---|---|
| −eIF4A | +eIF4A | |
| eIF4GI(643–1076) | 170 | 5 |
| eIF4GI(697–1076) | 400 | 6 |
| eIF4GI(722–1076) | 600 | 80 |
| eIF4GI(734–1076) | 700 | 650 |
| eIF4GI(746–1076) | 1,400 | 1,300 |
| eIF4GI(772–1076) | >6,000 | >6,000 |
| eIF4GI(697–969) | 2,300 | 40 |
| eIF4GI(697–949) | 2,800 | 400 |
| eIF4GI(697–941) | 3,000 | 3,000 |
| eIF4GI(697–869) | >8,000 | >8,000 |
| eIF4GI mut1 | 200 | 200 |
| eIF4GI mut4 | 120 | 30 |
| eIF4GI RRM1 | 1,800 | 400 |
| eIF4GI RRM2 | 480 | 10 |
| eIF4GI mut(I749T, R754I) | 1,000 | 200 |
| eIF4GI mut796-Ins8 | 1,200 | 400 |
| eIF4GI mut(R855A) | 500 | 8 |
| eIF4GI mut889-Ins6 | 800 | 5 |
| eIF4GI mut(R915I) | 1,700 | 300 |
| p97(NAT1) 62–330 | >4,000 | >4,000 |
Surprisingly, binding constants for the interaction of eIF4GI polypeptides with the EMCV J-K domain were of the same order of magnitude as for their interaction with β-globin RNA (Table 3). The values obtained correlated well with the binding data obtained using the toeprinting assay on the intact IRES (Fig. 2): mutants with lower specificity for the EMCV IRES (toeprinting assay) showed lower binding constants. It is therefore not clear how EMCV RNA can compete with cellular mRNAs for eIF4F. Since eIF4G is bound to eIF4A in the eIF4F complex and this interaction is important for the ability of eIF4G to promote 48S complex formation on the EMCV IRES, the influence of eIF4A on the binding constants for binding of eIF4GI polypeptides to EMCV J-K and β-globin RNA transcripts was assayed using the same mobility shift assay. Data for the J-K domain are summarized in Table 4.
Inclusion of eIF4A in binding reaction mixtures decreased the binding constants to the EMCV J-K domain for eIF4GI(697–1076) and for some mutant derivatives thereof by up to 2 orders of magnitude. These derivatives included deletion mutants eIF4GI(722–1076), eIF4GI(697–969), and eIF4GI(697–949), substitution mutants RRM2 and mut(R855A), and insertion mutant mut889-Ins6. eIF4A alone did not have a detectable binding affinity for this RNA (data not shown). No enhancement of binding by inclusion of eIF4A was detected for eIF4GI(734–1076), eIF4GI(746–1076), eIF4GI(772–1076), and eIF4GI(697–941) and substitution mutant mut1 (Table 4). A modest (two- to fivefold) increase in binding by inclusion of eIF4A was observed for RRM1, mut4, mut(I749T, R754I), and mut(R915I) substitution mutants and for the mut796-Ins8 insertion mutant.
These data show that inclusion of eIF4A in binding reaction mixtures increased the affinity of eIF4GI for the EMCV J-K domain to an extent that would make the EMCV IRES competent to compete with cellular capped mRNAs for eIF4F. The extent to which eIF4GI mutant polypeptides responded to inclusion of eIF4A in binding reaction mixtures correlated directly with their activity in promoting 48S complex formation on the EMCV IRES. eIF4GI mutants, such as substitution mutant mut1 and deletion mutant eIF4GI(734–1076), whose binding to the EMCV IRES did not respond at all to inclusion of eIF4A in binding reactions were unable to promote 48S complex formation on this IRES.
The enhancement of the binding of eIF4GI to the EMCV J-K domain did not depend on the ATPase activity of eIF4A. Essentially the same level of stimulation was obtained in the presence or absence of ATP and when wild-type eIF4A was replaced by the negative trans-dominant R362Q eIF4A mutant (reference 34 and data not shown).
Mobility shift analysis indicated quantitatively that eIF4A enhanced the binding of eIF4GI to the IRES but gave no indication of the site of the interaction of eIF4GI on this RNA. Toeprinting analysis was used to confirm that inclusion of eIF4A in binding reaction mixtures enhanced the toeprint at C786 caused by specific binding of eIF4GI. Toeprinting assays were done exactly as described above for analysis of binary eIF4GI-IRES complexes, except that in parallel reactions, eIF4A was included together with EMCV RNA and derivatives of eIF4GI(697–1076). Although toeprinting is not appropriate for the determination of binding constants because it has low sensitivity and involves reverse transcription (which has the potential to displace bound protein, thus falsely increasing the Kd of formation of the RNA-protein complex), toeprinting is a reliable assay for the localization of specific protein binding sites on an mRNA. The results of toeprinting analyses (Fig. 5) and mobility shift analyses were qualitatively similar. The intensity of the C786 toeprint was not enhanced by inclusion of eIF4A in reaction mixtures that contained the mut1 eIF4G(697–1076) substitution mutant or the eIF4GI(734–1076), eIF4GI(746–1076), eIF4GI(772–1076), eIF4GI(800–1076), eIF4GI(697–941), or eIF4GI(697–869) deletion mutants (Fig. 5A, lanes 6 to 9, 12, and 13; Fig. 5B, lane 3). The prominence of this toeprint was strongly increased by inclusion of eIF4A in reaction mixtures with eIF4GI(613–1090), eIF4GI(643–1076), eIF4GI(697–1076), eIF4GI(722–1076), eIF4GI(697–969), and eIF4GI(697–949) deletion mutants (Fig. 5A, lanes 2 to 5, 10, and 11), and mut(I749T, R754I), mut796-Ins8, mut(R855A), mut889-Ins6, mut(R915I), and RRM2 insertion or substitution mutants (Fig. 5B, lanes 5 to 9 and 11). The strong binding of mut4 eIF4GI(697–1076) to the EMCV IRES was very weakly enhanced by eIF4A (Fig. 5B, lane 4). The poor binding of the RRM1 eIF4GI(697–1076) substitution mutant to the IRES was also only weakly enhanced by eIF4A (lane 10). The weak enhancement by eIF4A of the binding of this eIF4GI mutant to the IRES could be due to disruption of functional interactions between eIF4GI and eIF4A or to the weak initial interaction of this mutant with the IRES.
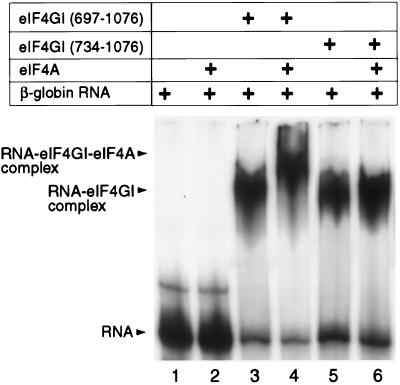
Inclusion of eIF4A with derivatives of eIF4GI(697–1076) did not alter their binding constants of interaction with β-globin RNA in mobility shift assays (data not shown). The enhancement by eIF4A of the binding of eIF4GI to the EMCV IRES is therefore specific for this RNA. Nevertheless, mobility shift analysis done using β-globin RNA in the presence of eIF4A and derivatives of eIF4GI was useful because it enabled us to assay the interaction of these two polypeptides. The addition of eIF4A to a reaction mixture that contained eIF4GI(697–1076) resulted in a specific supershift of β-globin RNA (Fig. 6, lanes 3 and 4). No supershift was detected when eIF4A was included in a similar assay mixture containing eIF4GI(734–1076) (lanes 5 and 6). No binding of eIF4A alone to β-globin RNA was detected using this assay (lanes 1 and 2). We conclude that residues 697 to 734 contain determinants of the interaction of eIF4GI with eIF4A.
FIG. 6.
Interaction between eIF4A and eIF4G determined by the electrophoretic mobility shift assay, showing the specific supershift of the β-globin mRNA–eIF4GI complex in the presence of eIF4A. The positions of free RNA, the RNA-eIF4GI complex, and the RNA-eIF4GI-eIF4A complex are indicated.
Protein-protein interactions between eIF4A and eIF4GI.
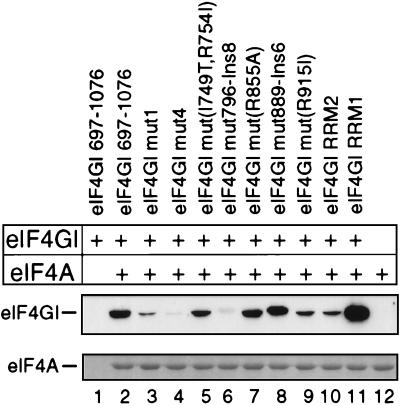
Mammalian eIF4G contains two separate binding sites for eIF4A, located in the central and C-terminal thirds of the protein (14, 22, 30). Yeast eIF4G contains a single eIF4A binding site, located at a position that corresponds to the central eIF4A binding site in the mammalian factor (4, 32). The effect of mutations in eIF4GI(697–1076) on its binding to eIF4A was assayed. The interaction of mut1 and mut4 substitution mutants with eIF4A was dramatically reduced (Fig. 7, lanes 3 and 4), consistent with previous reports (14). A similar phenotype was observed for the mut796-Ins8 eIF4GI(697–1076) mutant (lane 6). Although mut1 eIF4GI(697–1076) bound to eIF4A slightly more strongly than did either of these other two mutants, its binding to the EMCV IRES was not enhanced by eIF4A and it was unable to promote 48S complex formation on this IRES, whereas the mut4 eIF4GI(697–1076) mutant and, to a greater extent, the mut796-Ins8 eIF4GI(697–1076) mutant retained low level activity in both assays. The binary eIF4A-mut1 eIF4GI(697–1076) complex therefore does not have an active conformation sufficient to promote 48S complex formation on the EMCV IRES.
FIG. 7.
Interaction of insertion and substitution mutant eIF4GI(697–1076) polypeptides with immobilized eIF4A in a direct binding assay, as described in Materials and Methods. eIF4A was visualized by Coomassie blue staining, and eIF4GI polypeptides were detected by Western blotting with anti-T7 tag antibodies.
The ability of the mut(I749T, R754I) and RRM1 eIF4GI(697–1076) mutant polypeptides to bind eIF4A was not impaired, and the ability of mut(R915I) and RRM2 eIF4GI(697–1076) mutant polypeptides to bind eIF4A was reduced but not abolished (Fig. 7, lanes 5, 9, 10, and 11). Although these mutants all had a low affinity for the IRES, their ability to bind eIF4A was sufficient for it to enhance their binding to the IRES and to enable them to promote very low levels of 48S complex formation on it. These mutations therefore primarily affect the specific interaction of eIF4GI with the EMCV IRES rather than its binding with eIF4A. The interactions of the wild-type, mut(R855A), and mut889-Ins6 eIF4GI(697–1076) polypeptides with eIF4A were similar (lanes 2, 7, and 8). The interaction of these polypeptides was strongly enhanced by eIF4A (Table 4), and they were all equally active in promoting 48S complex formation on the IRES (Fig. 4, lanes 6, 7, and 21).
Direct binding of eIF3 to eIF4GI is not required for 48S complex formation on the EMCV IRES.
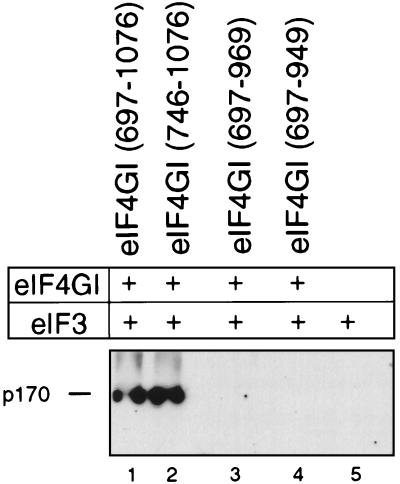
The middle third of eIF4G binds directly to eIF3 (14, 22, 30), and this interaction may be important for ribosomal recruitment to mRNAs. In the course of the studies reported here, a series of N- and C-terminal deletion mutations in eIF4GI was made that may affect its interaction with eIF3. For this reason, a binding assay was used to investigate the ability of these mutant polypeptides to bind eIF3. N-terminal deletions made in eIF4GI(697–1076) up to Q746 did not abrogate its ability to bind eIF3 (Fig. 8, lanes 1 and 2). However, a C-terminal deletion to E969 abrogated the interaction of eIF4GI with eIF3 (lanes 3 and 4). These results were obtained using a sensitive monoclonal antibody against the p170 subunit of eIF3. The eIF4GI deletion mutants eIF4GI(697–969) and eIF4GI(697–949), which did not bind eIF3 in this assay, were both active in promoting 48S complex formation on the EMCV IRES (Fig. 4, lane 15, and data not shown). However, the immobilization of eIF4G may affect its ability to bind eIF3. For example, if the interaction of eIF4G and eIF3 involves multiple contacts, some of them might be hidden as a result of the immobilization of eIF4G. Hiding of some contacts may not abolish the binding of immobilized full-length eIF4G with eIF3 but could prevent the interaction with eIF3 of some immobilized eIF4G deletion mutants. For this reason, we cannot exclude the possibility that some of those eIF4G mutants, which in immobilized form lost the ability to bind eIF3, may retain eIF3 binding activity in solution.
FIG. 8.
Interaction of eIF3 with immobilized eIF4GI deletion mutant polypeptides in a direct-binding assay, as described in Materials and Methods. The eIF3 p170 subunit is indicated on the left and was visualized by Western blotting with a specific monoclonal antibody.
DISCUSSION
eIF4G is an adapter protein with a modular structure that plays a key coordinating role in the early stages of initiation by acting as a platform for the assembly of a multiprotein complex to recruit the ribosome to an mRNA. In the translation of capped mRNAs, eIF4G plays this role as a subunit of the heterotrimeric factor eIF4F and the specificity of its interaction with mRNAs is initially determined by binding of the eIF4E subunit of eIF4F to the mRNA 5′-terminal cap. The core sequence of eIF4G that is necessary and sufficient for cap-dependent translation has recently been defined and shown to include the N-terminal eIF4E binding site (30). eIF4G plays an analogous role in the initiation of translation by internal ribosomal entry, as exemplified by initiation on the EMCV IRES (39, 41). In this instance, specific binding to the IRES is a property of eIF4G itself (20, 41) and is necessary for internal ribosomal entry (41). We have now defined the core sequence of eIF4G that is required for specific binding to the EMCV IRES, for interaction with eIF4A, and for mediation of binding of a 43S preinitiation complex to the IRES (Table 5). These results identify the eIF4G-eIF4A complex (rather than eIF4G alone) as the moiety responsible for specific high-affinity binding to the IRES and indicate that the interactions between eIF4G and eIF4A as well as between eIF4G and the IRES are essential for subsequent recruitment of the 43S ribosomal complex to the EMCV initiation codon.
TABLE 5.
Activities of eIF4GI mutant polypeptides
| eIF4GI mutant | Binary-complex formation | Binding to eIF4A | Ternary-complex formation | 48S complex formation | Binding to eIF3 |
|---|---|---|---|---|---|
| eIF4GI(613–1090) | +++ | +++ | +++ | ||
| eIF4GI(643–1076) | +++ | +++ | |||
| eIF4GI(697–1076) | +++ | +++ | +++ | +++ | +++ |
| eIF4GI(722–1076) | +++ | +++ | +++ | ||
| eIF4GI(734–1076) | +++ | − | − | ||
| eIF4GI(746–1076) | + | − | +++ | ||
| eIF4GI(772–1076) | − | − | − | ||
| eIF4GI(697–969) | ++ | +++ | − | ||
| eIF4GI(697–949) | + | ++ | ++ | − | |
| eIF4GI(697–941) | − | − | − | ||
| eIF4GI mut1 | +++ | −/+ | − | − | |
| eIF4GI mut4 | +++ | −/+ | + | + | |
| eIF4GI RRM1 | −/+ | +++ | ± | −/+ | |
| eIF4GI RRM2 | +++ | + | +++ | +++ | |
| eIF4GI mut(I749T, R754I) | ± | ++ | + | ± | |
| eIF4GI mut796-Ins8 | ± | −/+ | + | + | |
| eIF4GI mut(R855A) | +++ | +++ | +++ | +++ | |
| eIF4GI mut889-Ins6 | +++ | +++ | +++ | +++ | |
| eIF4GI mut(R915I) | ± | + | ++ | ++ |
A core sequence of about 300 aa whose amino- and carboxyl-terminal borders lie between aa 746–772 and aa 941–949, respectively, binds specifically to the IRES. It has been suggested that this region might correspond to an RRM-like domain (2, 8), but the lack of effect of mutations in its putative RNP-2 motif on IRES binding suggests that this is unlikely. In addition, mutations in putative RNP-1 and RNP-2 motifs have no effect on the general RNA binding activity of eIF4G. Specific binding of eIF4G to the IRES was also unaffected by groups of mutations (L729A L732A F737A and R935A F938A) which impair the interaction of eIF4G with eIF4A (14), by the substitution R855A, and by insertion of 8 aa after D899. However, the substitutions I749T R754I, L857A I860A (in eIF4GI RRM1), and R915I and the insertion of 6 aa after V796 affected IRES binding considerably more than they affected binding to β-globin mRNA. These observations indicate a specific requirement for residues in eIF4GI for tight binding to the IRES independent of the ability to interact with eIF4A and to bind cooperatively. Surprisingly, derivatives of eIF4G(697–1076) that bound specifically to the EMCV IRES did so with an affinity (Kd = 120 to 800 nM) that did not differ significantly from their affinity for uncapped globin mRNA; this is clearly not sufficient to account for the ability of the IRES to compete successfully with other mRNAs for eIF4F.
Significantly, inclusion of eIF4A in binding reaction mixtures increased the affinity of eIF4G for the IRES by up to 2 orders of magnitude without affecting the affinity of its binding to globin RNA. The interaction of the IRES with the eIF4G-eIF4A complex rather than eIF4G alone is sufficient for EMCV IRES-containing mRNAs to be competitive with other mRNAs. EMCV has therefore developed a novel alternative to the cap-eIF4E interaction as a mechanism for recruiting 43S complexes to a specific location on an mRNA, by exploiting the affinity of the IRES for the eIF4G-eIF4A complex. We consider that eIF4A may provide an additional site of contact with the IRES and/or alter the structure of eIF4G so that it binds the IRES with higher affinity. The ATP binding and hydrolysis activities of eIF4A are not important for this interaction, since the IRES-eIF4G-eIF4A complex assembled with equal specificity and affinity in the absence and presence of ATP and on replacement of wild-type eIF4A by the trans-dominant R362Q mutant, which has defects in ATP binding, RNA binding, and RNA helicase activities (33). Whatever the mechanism by which eIF4A enhances the IRES binding affinity of eIF4G, it is clear that the IRES has evolved to bind the eIF4G-eIF4A complex rather than eIF4G alone. Mutations in eIF4G that impair its interaction with eIF4A render it unable to mediate 48S complex formation, even if the ability of these mutants to bind to the IRES is unaffected.
Even though the affinity of the eIF4G-eIF4A complex for the IRES is unaffected by ATP, assembly of 48S complexes on the EMCV IRES is absolutely ATP dependent (39, 41). Although the mechanism of 48S complex formation on the IRES is not yet known, several explanations for this ATP requirement can be proposed. The simplest possibility is that binding of the 43S complex to a defined location on the IRES may require local unwinding of mRNA, possibly to create an unstructured region around the initiation codon. Initiation on the EMCV IRES has previously been found to occur by direct ribosomal attachment to this area without prior scanning (18). In this model, specific binding of eIF4G to the J-K domain directs the helicase activity of eIF4A to a defined region of the IRES. A second, more speculative hypothesis can also be suggested. The fate of different translation components, in particular of eIF4F, during and after the binding of 43S complexes to mRNAs is not known for either the cap-dependent or IRES-mediated modes of initiation. If eIF4F should be displaced from its initial binding site to allow binding of the 43S complex to mRNA, the ATPase activity of eIF4A could play a role in this process by inducing conformational changes. An impaired interaction between eIF4A and eIF4G may impair either of these possible functions of eIF4A.
We anticipate that there are multiple points of interaction between eIF4G and eIF4A and suggest that the correct pattern of interactions between them must be established for initiation on the EMCV IRES to occur. For example, although mut1 eIF4G(697–1076) bound eIF4A more strongly than the equivalent mut4 and mut796-Ins8 polypeptides did, it was absolutely inactive in mediating 48S complex formation on the IRES whereas the mut4 and mut796-Ins8 polypeptides had residual activity in this assay. This observation underscores the requirement for correct assembly of the eIF4G-eIF4A complex for participation in 48S complex formation on the EMCV IRES.
The central domain of eIF4G binds eIF3 (22, 30), and this association has been considered likely to be of fundamental importance in initiation as a bridging interaction between the 43S complex and mRNA. In experiments reported here, aa 969 to 1076 of eIF4GI was found to contain essential determinants of the interaction with eIF3, and eIF4GI polypeptides truncated at their carboxy terminus to E969 were found to be unable to bind eIF3. However, such polypeptides are nevertheless active in mediating 48S complex formation on the EMCV IRES. Direct interaction between eIF3 and eIF4GI is therefore not necessary for 48S complex formation on the EMCV IRES. This conclusion does not rule out the possibility that the 43S ribosomal preinitiation complex is recruited to this mRNA through an intermediate interaction, for example involving eIF4B. This factor binds directly to eIF3 and to the 40S subunit and interacts functionally with eIF4A and eIF4F (17, 23, 28, 29, 33, 45). Alternatively, it is possible that other components of the 43S complex interact directly with the EMCV IRES. We have previously described that eIF3 and 40S subunits are able to bind specifically to noncontiguous regions of hepatitis C virus, classical swine fever virus, and bovine viral diarrhea virus IRESs (36, 37, 48). Although these IRESs are unrelated to the EMCV IRES, we cannot exclude the possibility that specific interactions between the EMCV IRES and components of the 43S complex have so far escaped our attention. We have previously noted that deletion of the I domain of the EMCV IRES abrogates its activity without impairing the interaction of eIF4G-eIF4F with the J-K domain (20), possibly suggesting a role for this domain in potential interactions of the EMCV IRES with the 43S complex.
ACKNOWLEDGMENTS
We thank N. Sonenberg and T. Innerarity for plasmids, D. Etchison for a monoclonal antibody, and N. Sonenberg for very helpful discussions.
This work was supported by grant MCB9726958 from the NSF.
REFERENCES
- 1.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 2.De Gregorio E, Preiss T, Hentze M W. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Gregorio E, Preiss T, Hentze M W. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999;18:4865–4874. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez D, Altmann M, Benz J, Baumann U, Trachsel H. Interaction of translation initiation factor eIF4G with eIF4A in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:26720–26726. doi: 10.1074/jbc.274.38.26720. [DOI] [PubMed] [Google Scholar]
- 5.Evstafieva A G, Ugarova T Y, Chernov B K, Shatsky I N. A complex RNA sequence determines the internal initiation of encephalomyocarditis virus RNA translation. Nucleic Acids Res. 1991;19:665–671. doi: 10.1093/nar/19.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gingras A-C, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 7.Gingras A C, Svitkin Y, Belsham G J, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyer C, Altmann M, Lee H S, Blanc A, Deshmukh M, Woolford Jr J L, Trachsel H, Sonenberg N. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellen C U T, Witherell G W, Schmidt M, Shin S H, Pestova T V, Gil A, Wimmer E. A cytoplasmic 57kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henis-Korenblit S, Strumpf N L, Goldstaub D, Kimchi A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol. 2000;20:496–506. doi: 10.1128/mcb.20.2.496-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Imataka H, Olsen H S, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 17.Jaramillo M, Dever T E, Merrick W C, Sonenberg N. RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol Cell Biol. 1991;11:5992–5997. doi: 10.1128/mcb.11.12.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminski A, Howell M T, Jackson R J. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keiper B D, Gan W, Rhoads R E. Protein synthesis initiation factor 4G. Int J Biochem Cell Biol. 1999;31:37–41. doi: 10.1016/s1357-2725(98)00130-7. [DOI] [PubMed] [Google Scholar]
- 20.Kolupaeva V G, Pestova T V, Hellen C U T, Shatsky I N. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 21.Lamphear B J, Yan R, Yang F, Waters D, Liebig H-D, Klump H, Kuechler E, Skern T, Rhoads R E. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human Coxsackievirus and rhinovirus. J Biol Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- 22.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 23.Lawson T G, Lee K A, Maimone M M, Abramson R D, Dever T E, Merrick W C, Thach R E. Dissociation of double-stranded polynucleotide helical structures by eukaryotic initiation factors, as revealed by a novel assay. Biochemistry. 1989;28:4729–4734. doi: 10.1021/bi00437a033. [DOI] [PubMed] [Google Scholar]
- 24.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcotrigiano J, Gingras A-C, Sonenberg N, Burley S K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 26.Marissen W E, Lloyd R E. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrick W C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Méthot N, Song M S, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol. 1996;16:5328–5334. doi: 10.1128/mcb.16.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Méthot N, Pickett G, Keene J D, Sonenberg N. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA recognition motif. RNA. 1996;2:38–50. [PMC free article] [PubMed] [Google Scholar]
- 30.Morino S, Imataka H, Svitkin Y V, Pestova T V, Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol. 2000;20:468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morley S J, Curtis P S, Pain V M. eIF4G: translation's mystery factor begins to yield its secrets. RNA. 1997;3:1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 32.Neff C L, Sachs A B. Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol Cell Biol. 1999;19:5557–5564. doi: 10.1128/mcb.19.8.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pause A, Méthot N, Svitkin Y, Merrick W C, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pestova T V, Borukhov S I, Hellen C U T. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 36.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U T. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal initiation of translation of hepatitis C virus and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestova T V, Hellen C U T. Internal initiation of translation of bovine viral diarrhea virus RNA. Virology. 1999;258:249–256. doi: 10.1006/viro.1999.9741. [DOI] [PubMed] [Google Scholar]
- 38.Pestova T V, Hellen C U T. The structure and function of initiation factors in eukaryotic protein synthesis. Cell Mol Life Sci. 2000;57:651–674. doi: 10.1007/PL00000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pestova T V, Hellen C U T, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestova T V, Lomakin I B, Lee J H, Choi S K, Dever T E, Hellen C U T. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 41.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poporechnaya, V. I. Agol, and C. U. T. Hellen. A cell-cycle dependent protein serves as a template-specific translation initiation factor. Genes Dev., in press. [PMC free article] [PubMed]
- 42.Predki P F, Nayak L M, Gottlieb M B, Regan L. Dissecting RNA-protein interactions: RNA-RNA recognition by Rop. Cell. 1995;80:41–50. doi: 10.1016/0092-8674(95)90449-2. [DOI] [PubMed] [Google Scholar]
- 43.Pyronnet S, Imataka H, Gingras A C, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raught B, Gingras A-C, Gygi S P, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 2000;19:434–444. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray B K, Lawson T G, Kramer J C, Cladaras M H, Grifo J A, Abramson R D, Merrick W C, Thach R E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 46.Rogers G W, Jr, Richter N J, Merrick W C. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 47.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sizova D V, Kolupaeva V G, Pestova T V, Shatsky I N, Hellen C U T. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan R, Frankel A D. Structural variety of arginine-rich RNA-binding peptides. Proc Natl Acad Sci USA. 1995;92:5282–5286. doi: 10.1073/pnas.92.12.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamanaka S, Poksay K S, Arnold K S, Innerarity T L. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]