Abstract
DNA regulatory elements frequently harbor multiple recognition sites for several transcriptional activators. The response mounted from such compound response elements is often more pronounced than the simple sum of effects observed at single binding sites. The determinants of such transcriptional synergy and its control, however, are poorly understood. Through a genetic approach, we have uncovered a novel protein motif that limits the transcriptional synergy of multiple DNA-binding regulators. Disruption of these conserved synergy control motifs (SC motifs) selectively increases activity at compound, but not single, response elements. Although isolated SC motifs do not regulate transcription when tethered to DNA, their transfer to an activator lacking them is sufficient to impose limits on synergy. Mechanistic analysis of the two SC motifs found in the glucocorticoid receptor N-terminal region reveals that they function irrespective of the arrangement of the receptor binding sites or their distance from the transcription start site. Proper function, however, requires the receptor's ligand-binding domain and an engaged dimer interface. Notably, the motifs are not functional in yeast and do not alter the effect of p160 coactivators, suggesting that they require other nonconserved components to operate. Many activators across multiple classes harbor seemingly unrelated negative regulatory regions. The presence of SC motifs within them, however, suggests a common function and identifies SC motifs as critical elements of a general mechanism to modulate higher-order interactions among transcriptional regulators.
The development and physiology of higher eukaryotes rely on the accurate transcription of a large array of independent genes in response to specific temporal, spatial, and physiological cues. The information to establish such a complex program of gene expression is ultimately encoded in the regulatory DNA elements of each gene. Invariably, such cis-regulatory elements consist of clusters of recognition sites for various factors, often in multiple copies or partially overlapping each other (2). These units nucleate the cooperative assembly of multiprotein complexes or enhanceosomes (4). The final transcriptional output, however, is not the simple arithmetic addition of the independent effects of individual regulators. On the contrary, the integrated response of the gene is the result of a complex set of logical and quantitative operations that rely on the combinatorial coordination of multiple regulatory sites, factors, and signals (56–58).
A central element of this regulatory logic that serves as both an amplification and specificity mechanism is the more-than-additive, i.e., synergistic, response resulting from the recruitment of a given activator to multiple copies of a recognition site. This general form of interaction is observed even with artificial activators and thus may be an intrinsic consequence of the combinatorial design of eukaryotic transcription systems (25). The mechanisms and determinants that enable or control such synergy are poorly understood, yet they are likely to be important targets of regulation.
Steroid receptors such as the glucocorticoid receptor (GR) are useful models for studying this form of synergy, since many of their target cis-regulatory regions harbor multiple receptor binding sites (6, 21). Furthermore, different receptors display various degrees of synergy at such compound hormone response elements (HREs). Steroid receptors share a common architecture consisting of the following three major regions. (i) The first is a highly variable N-terminal domain that harbors transcriptional regulatory functions. In the case of GR, a transcriptional activation function (AF1, or enh2) (15, 20) and determinants involved in repression (36) lie within this domain. (ii) The second is a central region that harbors dual zinc fingers responsible for specific DNA recognition and DNA-induced dimerization (DNA binding domain [DBD]). Critical residues in this domain play a pivotal role in determining the regulatory outcome of interactions with other regulators (47). (iii) Finally, the third is a C-terminal ligand-binding domain (LBD) harboring a second activation function (AF2) that operates as a ligand-dependent surface for interaction with coactivators (7, 45).
Both the N- and C-terminal transcriptional activation functions of steroid receptors contribute to synergy, but it is unclear whether effects on synergy can be dissociated from effects on activation per se (31, 42, 55). The mechanism of synergy is not fully understood, but in several instances, cooperative binding to compound sites correlates with synergistic activation (49, 55). In the case of the androgen receptor (AR), interactions between the N- and C-terminal domains contribute to this effect (19, 42). For GR derivatives lacking the LBD, synergy and cooperative binding appear to depend on sequences within the N-terminal activation function (55). In certain promoter contexts, however, synergy occurs in the apparent absence of cooperative DNA binding (3, 38).
A number of observations suggest that steroid receptors harbor additional determinants that limit or control the extent of synergy. Recently, Liu et al. (27) discovered an important role for the DBD dimer interface in restraining steroid receptor synergy. Disruption of this interface, although deleterious for receptor activity at a single site, leads to a marked enhancement of activity at compound HREs (27). In addition, N-terminal determinants may also participate to restrain synergy. In contrast to GR, the closely related mineralocorticoid receptor (MR) displays weak synergy, and this difference maps to the N-terminal region. Moreover, deletion of the N-terminal regions of GR and MR produces equally strong and highly synergistic activators. This suggests that the N-terminal regions of MR (and to a lesser extent of GR) interfere with synergy (27, 40).
The characterization of many different transcription factor families indicates that in addition to DBDs and transcriptional activation regions, many regulators harbor “negative” regulatory functions. Deletion of such silencing, attenuator, or negative domains enhances transcriptional activation. Although some do function as bona fide repression domains by actively repressing transcription, the mode of action of these seemingly disparate regions remains obscure. Through our analysis of the N-terminal transcriptional regulatory region (AF1) of GR, we have now identified a novel synergy control motif (SC motif) that underlies the function of the negative regulatory regions of multiple transcription factors. These motifs operate not by affecting the intrinsic activity of an activator, but by regulating their ability to synergize at compound response elements. We define here the functional determinants of these SC motifs and explore their mechanism of action.
MATERIALS AND METHODS
Mammalian expression plasmids.
The plasmids p6RGR and p6RGRN525 (14) allow the expression of full-length rat GR and a deletion lacking amino acids 526 to 795, respectively. The original collection of multiply substituted AF1 mutants has been described previously (20). Digestion of a pBluescript derivative harboring an 860-bp BglII-PstI GR fragment with SmaI and Bsp120I, blunting, and religation produced a precise deletion of amino acids 310 to 317 that was then transferred as a 681-bp BstXI-PstI fragment into the same sites of p6RGR. Some individual amino acid changes were introduced by site-directed mutagenesis (23) and confirmed by sequencing. The K297E/I312T double mutant was transferred to the DBD dimer interface mutants p6RGR R479D and p6RGR D481R (27) as a 634-bp NcoI-ApaI fragment. The above plasmids as well as the empty (pS6R) and control β-galactosidase (p6R-βgal) (36) expression vectors are Rous sarcoma virus promoter-driven derivatives of p65. The human AR K385E, K518E, and K385E/K518E mutants were engineered into the pCMV5 derivative p5HBhAR-A (a kind gift of D. Merry, University of Pennsylvania) by PCR and confirmed by sequencing. The human ETS-1 K15E, K227E, and K15E/K227E mutants were obtained in a similar manner from pSG5hETS-1 (44).
To generate the Gal4 DBD fusion constructs, p6RGR was PCR amplified with oligonucleotides 5′cgcgaagcttggatccagcagtgtggcactgcccc3′ and 5′ctcggaattcgcggccgcttaagatctaaagcttgcctgacaataaactgggcc3′, and the product was digested with enzyme pairs BamHI and BglII or BamHI and NotI (filled in). The resulting fragments were ligated to pGal4(VP16)2 (11) that had been digested with BamHI or BamHI and XbaI (filled in). This yielded pGal4(SC)2(VP16)2 and pGal4(SC)2, respectively. In these constructs, rat GR amino acids 287 to 327 are placed between the Gal4 DBD and the tandem VP16 activation domains or fused to the Gal4 DBD alone, respectively. The corresponding derivatives with mutant SC motifs were constructed identically. Coactivator expression plasmids pSG5.HA-GRIP1 and pSG5.HA-SRC-1a have been described previously (5).
Reporter plasmids.
Reporter plasmids were constructed by inserting double-stranded oligonucleotides or PCR products harboring response elements at the indicated site(s) (in parentheses) of the basal reporter pΔODLO. This positions the elements upstream of the minimal Drosophila distal alcohol dehydrogenase promoter (−33 to +55) and the luciferase gene. The inserts for the different reporters are as follows: pΔTAT1-Luc, gtcgagcTGTACAGGATGTTCTAGCTACgtcgac; pΔTAT2-Luc, gtcgacatcagaatacagacctcAGAACATCCTGTACAgacctcAGAACATCCTGTACAacctcgtcgac; pΔTAT3-Luc, gtcgacGTAGCTAGAACATCCTGTACAGctcgacCTGTACAGGATGTTCTAGCTACgtcgagCTGTACAGGATGTTCTAGCTACcagctc; pΔTAT2-16, gtcgacgacgtcTGTACAGGATGTTCTaTGTACAGGATGTTCTgtcgag; pΔTAT2-21, gtcgacgacTGTACAGGATGTTCTactagtTGTACAGGATGTTCTaacgactgtcgac; pΔTAT2-26, gtcgacgacTGTACAGGATGTTCTactagtaacgaTGTACAGGATGTTCTaacgatgtcgac; pΔTAT1/4S, gtcgacggaggtcTGTACAGGAaTGTTCTgatgtcgac; pΔTAT3/4S, gtcgacggaggtcTGTACAGGAaTGTTCTgaggtcTGTACAGGAaTGTTCTgaggtcTGTA CAGGAaTGTTCTgatgtcgac; and pΔTAT2-SpeI,NheI, gtcgacgaggtcTGTACAGGATGTTCTgaggtcTGTACAGGATGTTCTactagtgctagcgtcgac. The SalI site of pΔODLO was used for the above inserts, and the rat TAT gene and HRE half-site sequences within them are in uppercase and underlined, respectively. pΔ(ETS-1)1DLO—aagcttcggccaagccGGAagtgagtgcctgcag (HindIII-PstI)—was also used, as was pΔENDOA-Luc, the 259-bp PCR product from mouse genomic DNA with primers cagctaagcttcctctgAGGCTTTTGCTGTT and gctagctgcagAAGTCAGGGGACTGGGAGAT (HindIII-PstI). The genomic sequences are shown in uppercase. Finally, we constructed pΔ(Gal4)1DLO (aagcttctcgagCGGAGGACTGTCCTCCGttgtcgac [HindIII-SalI] and pΔ(Gal4)2DLO (agctCGGAGGACTGTCCTCCGttctcgagaaCGGAGGACAGTCCTCCG [HindIII]).
Ligation of the 67- and 118-bp HpaII fragments from pBluescript KS(−) at the filled-in SpeI and NheI sites of pΔTAT2-SpeI,NheI generated pΔTAT2 US100 and pΔTAT2 US150, respectively. In the reporter pG5E1b-Luc, five Gal4 sites upstream of the E1b promoter drive expression of the luciferase gene.
Mammalian cell culture and transfections.
Monkey CV-1 cells were maintained in Dulbecco's modified Eagle medium (DME H16; GIBCO BRL) supplemented with 7.5% fetal bovine serum. Cells were transfected by the calcium phosphate precipitation method (Fig. 2 only) (20) or by liposome-mediated transfection as follows: 3 × 104 cells were seeded in 24-well plates (0.4 ml) 24 h prior to transfection. At a 1:1 l-α-dioleoyl phosphatidylethanolamine (DOPE)/1,2-dioleoyl-3-trimethylammonium propane (DOTAP) ratio, liposomes (16) were incubated with 0.45 μg of total plasmid DNA (15 nmol of DOTAP/μg of plasmid) for 15 min at room temperature and added to the cells (50 μl). After overnight incubation, cells were washed with phosphate-buffered saline and further incubated for 24 h in medium supplemented with 7.5% charcoal-stripped serum in the presence of agonist or vehicle (0.1% ethanol). Where indicated, Lipofectamine (2 μl) and Plus reagent (2 μl) (GIBCO BRL) were used instead of DOPE and DOTAP liposomes. Luciferase and β-galactosidase activities were determined as described previously (20). In addition to the plasmids indicated in the figure legends, all transfections included 0.25 μg (Ca-PO) or 0.05 μg (lipofection) of the p6Rβ-gal control plasmid and sufficient carrier DNA (pBluescript) to achieve a total of 4.25 μg (Ca-PO) or 0.45 μg (lipofection) of DNA. For all of the mutants described, no effect was observed in the absence of agonist, and in some cases, only the data in its presence are shown.
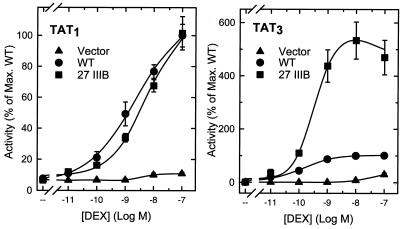
FIG. 2.
Enhanced activity mutations cause increased agonist efficacy at a compound, but not at a single HRE. CV-1 cells were transfected by the calcium phosphate method with 2 μg of reporter plasmid pΔTAT3-Luc (right) or pΔTAT1-Luc (left) and 0.2 μg of pS6R (Vector), p6RGR (WT), or p6RGR 27IIIB (27 IIIB) and treated with the indicated concentrations of dexamethasone (DEX). Data represent the averages of three independent transfections performed in duplicate and are expressed as a percentage of the maximal WT activity: 9.6 × 104 U for TAT1 and 9.4 × 105 U for TAT3.
Electrophoretic mobility shift assays.
In vitro transcription and translation reactions (SP6; Promega TNT) were programmed at a 1-mg/ml final concentration with empty pSP64T [a pSP64 derivative harboring 5′ and 3′ untranslated β-globin sequences, including a 23-bp poly(A) tract, downstream of the SP6 promoter], pSP64T-N795, or pSP64T-N795 K297E/K313E. These plasmids were constructed by inserting a 2,541-bp BamHI fragment from the corresponding p6RGR derivatives at the BglII site between the 5′ and 3′ β-globin sequences of pSP64T. Expression was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with the antibody BuGR-2 (12). For binding reactions, reticulocyte lysates (4 μl) were mixed with 3 μl of reaction mix [40 mM HEPES (pH 7.9), 140 mM KCl, 33.3% glycerol, 14.7 mM MgCl2, 10 mM dithiothreitol, 33.3 μg of poly(dI-dC) per ml, 1 μM the double-stranded blunt-ended nonspecific oligonucleotide (5′ctctcgccctcgtcgcccacgtggcgtcggc3′)] and preincubated for 20 min at 4°C. Reactions (20 min, 24°C) were initiated by the addition of 3 μl of DNA mixture containing approximately 1 nM 32P-labeled double-stranded TAT1 oligonucleotide at 3 × 106 cpm/pmol and the indicated concentrations of unlabeled TAT1 or TAT2 competitor DNA. The reactions were then resolved at room temperature on a prerun (20 min) 0.5× Tris-borate-EDTA–4% (37.5:1 acrylamide-bisacrylamide) polyacrylamide gel at 200 V. Bound and free species were quantitated without drying with a PhosphorImager (Molecular Dynamics). The TAT1 probe was prepared by annealing oligonucleotides 5′ccgggcgttgcCTGTACAGGATGTTCTAatctgag3′ and 5′gatcctcagatTAGAACATCCTGTACAGgcaacgc3′ and radiolabeling (α-[32P]dCTP) with the Klenow fragment of RNA polymerase. The TAT2 competitor, an 87-bp HindIII-XbaI fragment of pΔTAT2-Luc (5′aagcttgcatgcctgcaggtcgacatcagaatacagacctcAGAACATCCTGTACAgacctcAGAACATCCTGTACAacctcgtcgactctaga3′) was purified from a 15% preparative polyacrylamide gel. TAT2 DNA concentrations were determined by fluorometry in the presence of Hoechst 33258 with calf thymus DNA as a standard.
Yeast strains and plasmids.
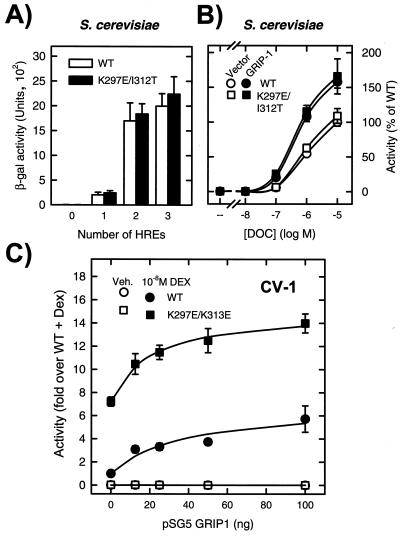
The Saccharomyces cerevisiae strain W303-1a (MATa ade2-1 trp1-1 ura3-1 leu2-3,112; his3-11,15 can1-100) was grown in minimal medium with amino acids and 2% glucose. Plasmid selection was maintained by culture in medium lacking the appropriate nutrients. The K297E/I312T mutations were transferred as 1,163-bp NcoI-SphI fragments from the p6RGR version into pRS314 G-N795 (Trp1, CEN/ARS) putting them under the control of the constitutive yeast glyceraldehyde-3-phosphate dehydrogenase promoter (24). The 2μm Ura3 reporter plasmids pUCΔSS, pUCΔS1Gs3, pUCΔS2X3S, and pΔS26X (59) consist of a minimal CYC-1 promoter linked to no HREs, a single perfectly palindromic HRE, two perfectly palindromic HREs, or three copies of the HRE from the TAT gene, respectively, and drive the expression of the Escherichia coli lacZ gene. The 2μm Leu2 GRIP1 expression plasmid pGAD424-GRIP1 has been described previously (10). Cells were cultured for 16 h in the presence or absence of the indicated amounts of deoxycorticosterone and assayed as described previously (20).
RESULTS
GR N-terminal mutants with enhanced activity.
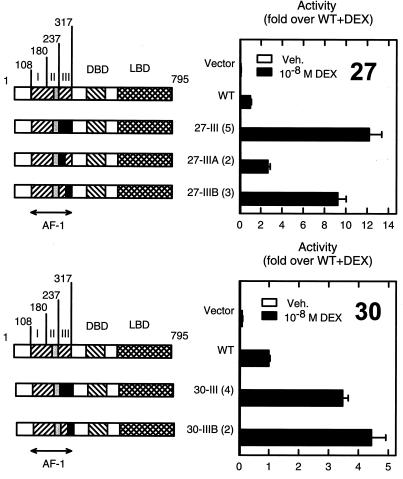
One of us previously generated a library of mutants designed to contain multiple random substitutions (12 on average) throughout the AF1 region (amino acids 108 to 317) of rat GR and through a genetic screen identified critical residues within a small central subregion (amino acids 219 to 234) essential for transcriptional activation (20). Unexpectedly, during the characterization of mutants with substitutions outside of the critical activation region, we identified two with enhanced activity (Fig. 1). Mutants AF1-27III and AF1-30III are 12- and 3-fold more active than the wild-type (WT) receptor at a compound HRE (three copies of a tyrosine aminotransferase gene HRE, TAT3). These mutants harbor eight and four substitutions, respectively, C terminal to the previously defined activation function. Notably, for both mutants, the effect is recapitulated by three (K297E/F303S/I312T; AF1-27IIIB) or two (Q295H/K313E; AF1-30IIIB) substitutions present within the last 24 residues of AF1 (Fig. 1).
FIG. 1.
AF1 mutants that enhance activity map C terminal to the activation function. CV-1 cells were transfected with the reporter plasmid pΔTAT3-Luc (0.2 μg) and vectors for the expression of full-length GR (0.02 μg), either WT or containing segments (in black) from mutants 27 (top) or 30 (bottom) in an otherwise WT background. Cells were treated with vehicle (Veh.) or 10 nM dexamethasone (DEX). Left, diagram of the constructs used. The number of substitutions in each construct is shown in parentheses. The critical region for transcriptional activation is boxed in gray. The boundary between regions IIIA and IIIB lies at amino acid 289. Right, activity relative to that of the WT. Data are the averages of three to four independent transfections performed in triplicate and are normalized to the WT activity in the presence of dexamethasone (2.0 × 105 U). In this and other figures, the measure of dispersion used is the standard error of the mean.
The enhanced activity is due to an increase in synergy at compound HREs.
At a compound HRE, both the intrinsic receptor activity and the ability of adjacently bound receptors to synergize contribute to the final response. To explore whether the mutations affect either property, we generated agonist dose response curves for either single (TAT1) or compound (TAT3) HREs. At the compound HRE (Fig. 2, right), the AF1-27IIIB mutant displayed enhanced activity compared to the WT throughout the dose response. Half-maximal stimulation, however, occurred at the same agonist concentration. Thus, it is likely that the mutations render GR a more effective activator without altering ligand binding. If this were due to a higher intrinsic activation potential, the activity at a single HRE should also be enhanced. On the contrary, the mutant was indistinguishable from the WT at the TAT1 reporter (Fig. 2, left). These surprising results indicate that rather than increasing the expression or intrinsic activity of the receptor, the mutations enhance the ability of individual receptors to engage in synergy. Thus, the ratio of activities at TAT3 and TAT1, an index of synergy, is 9.7 for the WT receptor, whereas for the mutant, it reaches 61.5. This effect was preserved across a wide concentration range of expression plasmid, suggesting that it is unlikely to be due to a relief of squelching (data not shown).
A short motif defines a synergy control domain.
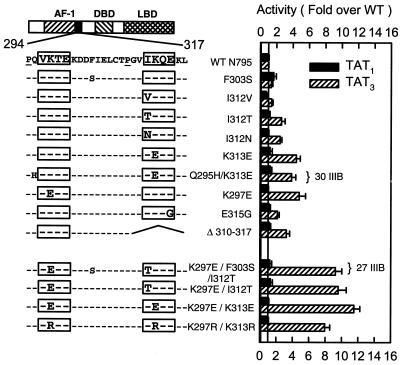
To define the determinants responsible for the enhanced synergy phenotype, we characterized the individual substitutions present in mutants AF1-27IIIB and AF1-30IIIB as well as additional mutations in this region. The analysis at single or compound HREs (Fig. 3) coupled with sequence comparison across species revealed that the substitutions that enhance synergy lie within two copies of a conserved motif. This short SC motif has the form (I/V)K(T/Q)E and is preceded within three residues by a Pro residue (Fig. 3). For the second motif, replacement of Ile 312 at the first position with Val, which is found in other species and in the first motif, has no consequences. Thr or Asn substitutions, however, cause a two- to threefold enhancement in activity. At the second position, replacing Lys with Glu in either the first or second motif yields a more pronounced four- to fivefold effect. Interestingly, Lys features other than charge are important, since Arg substitutions at this position are equally disruptive (see below). A Glu315Gly substitution at the fourth position of the second motif causes a twofold increase, whereas deletion of the entire motif enhances activity threefold. Other substitutions outside of the motifs (Q295H and F303S) are without effect. The two motifs cooperate functionally, since double mutants lead to a much more pronounced 8- to 12-fold effect. This is why the original AF1-27III mutant (K297E/I312T) has a more dramatic effect than AF1-30III (K313E). None of the alterations influenced receptor activity at a single site (Fig. 3) or in the absence of hormone (not shown). Taken together, these results indicate that many different substitutions are effective, implying that the normal role of the motifs is to restrain synergy and that the mutations disable their function.
FIG. 3.
SC mutants identify two copies of a short amino acid motif. CV-1 cells were transfected with 0.2 μg of reporter plasmid pΔTAT1-Luc (black bars) or pΔTAT3-Luc (hatched bars) and 0.02 μg of expression plasmids for WT full-length GR or the indicated mutants. Cells were treated with 10 nM dexamethasone and assayed as described in Materials and Methods. At left is a diagram of the location and sequence of the relevant region with the motifs boxed. The mutations tested are shown below. Data represent averages of three to four independent transfections performed in triplicate and are expressed as a percentage of the WT activity: 7.6 × 104 U for TAT1 and 2.7 × 105 U for TAT3.
SC motifs are functional in other factors and operate at natural response elements.
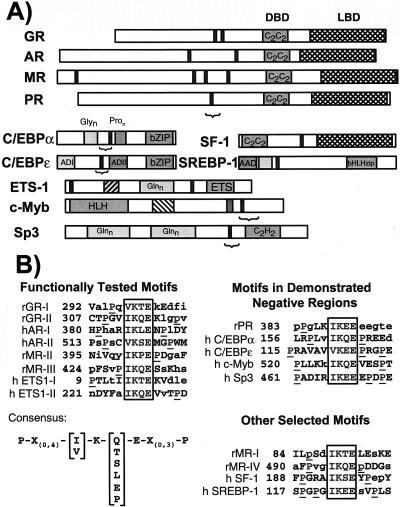
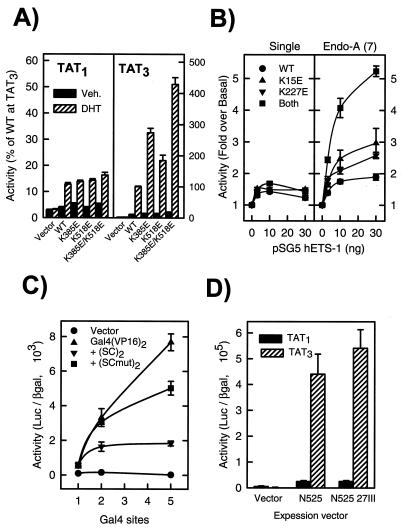
By examining other steroid receptors, we identified several SC motifs throughout their otherwise very divergent N-terminal regions: four in MR, two in AR, a single one in the progesterone receptor (PR), and none in the estrogen receptors α and β (see Fig. 5A). In the case of AR, disruption of either motif led to a two- to threefold enhancement at TAT3 and together yielded a fivefold increase in activity (Fig. 4A). As for GR, the mutations had no significant effect at a single HRE. Similarly, when analyzed in the context of an MR construct lacking motifs 1 and 4, Lys-to-Glu substitutions in motifs 2 and 3 enhanced receptor activity at compound HREs (not shown). Interestingly, the motif in PR lies within and is likely to be responsible for the function of a recently proposed negative modulation domain (307 to 427 in human PR). Deletion of this region enhances activity, whereas its duplication renders PR a very poor activator at compound HREs (18).
FIG. 5.
SC motifs in various transcription factors. (A) Solid vertical bars indicate SC motifs, whereas a bracket indicates regions of demonstrated negative function. (B) Sequence alignment of functional SC motifs. For each entry, core motif residues are boxed, flanking Pro residues are underlined, and residues absolutely conserved in different species or that match the consensus (lower left) are in uppercase. Motifs found in negative regulatory regions are at right, and other selected conserved motifs are shown below. r, rat; h, human.
FIG. 4.
Effect of SC mutations in various activators. CV-1 cells were transfected with the indicated plasmids and assayed as described in Materials and Methods. (A) AR. Cells were transfected with 0.2 μg of pΔTAT1-Luc (black bars) or pΔTAT3-Luc (hatched bars) and 0.2 μg of either p5HBhAR expression plasmid or the indicated mutants and treated with vehicle (Veh.) or 100 nM dihydrotestosterone (DHT). Data from four to eight independent transfections performed in triplicate are expressed as a percentage of the activity of the WT at TAT3 in the presence of DHT (4.5 × 104 U). (B) ETS-1. Cells were transfected (Lipofectamine) with the indicated amounts of pSG5 ETS-1 or the indicated mutants and 0.1 μg of pΔ(ETS-1)1DLO (left) or pΔENDOA-Luc (right). Data are expressed as fold induction over reporter alone (1.6 × 103 and 5.3 × 103 U, respectively) and represent three independent transfections performed in quadruplicate. (C) Gal4-VP16 fusions. Cells were transfected with 0.1 ng of the indicated Gal4 DBD fusion proteins and 0.1 μg of reporter plasmids harboring either one, [pΔ(GAL4)1DLO], two [pΔ(GAL4)2DLO], or five (pG5E1b) Gal4 sites. Data are the average of four replicates from a representative experiment. Similar results were obtained in three other transfections using other amounts of the activators. (D) GR N525 derivatives. Cells were transfected with 0.2 μg of reporter plasmid pΔTAT1-Luc or pΔTAT3-Luc and 0.1 μg of expression plasmid p6RGR N525 derivatives. Data represent the average of three independent transfections performed in quadruplicate.
Initial database searching revealed that transcription factors from other families harbor conserved SC motifs. We identified two of them in the ETS-1 proto-oncogene: one near the extreme N terminus and another upstream of the DBD, but outside of the “autoinhibitory” domain (46) (Fig. 5A). To our knowledge, no function has been assigned to these regions. To examine their role, we disrupted the motifs individually and in combination and assayed the mutants at a single high-affinity binding site (33) or at a natural compound response element from the Endo-A cytokeratin gene that harbors seven tandem ETS-1 binding sites (44). As shown in Fig. 4B, the mutations did not alter the activity of ETS-1 significantly at a single site. In contrast, either mutation increased the activity at the Endo-A enhancer and, when combined, yielded a fourfold enhancement. These results indicate that SC motifs limit synergy in multiple classes of regulators and that they function at natural compound elements.
Based on the eight SC motifs we have defined functionally and by comparing their across-species conservation, we obtained a more refined definition for SC motifs (Fig. 5B). The core of the motif is composed of an Ile or Val residue at the first position followed by invariant Lys and Glu residues at positions 2 and 4. At the third position, a small subset of amino acids can be accommodated. In addition, a Pro residue is usually present within the four or five residues preceding or following the core of the motif. In fact, in most cases both upstream and downstream, Pro residues are present. The additional motifs in MR and PR (Fig. 5B) match the consensus and hence are likely to be functional.
A search for the occurrence of SC motifs in the SwissProt database revealed that it appears at a frequency of 3.1% of protein sequences (SwissProt release 38). This value is marginally higher than the predicted random occurrence (2.7%). Notably ∼20% of them are transcriptional regulatory factors. This fraction reaches 63% (38 of 60 matches) if the search is restricted to human proteins with motifs having both upstream and downstream prolines. Furthermore, in many cases, the motifs, but not the adjacent sequences, are conserved in different species, suggesting a selective pressure for their maintenance. For example, several orphan intracellular receptors (ERRα, -β and -γ, OR-1, TR-2, RORβ, and SF-1), as well as other factors, like SREBP-1 and multiple members of the SOX family of developmental regulators (SOX 1, 2, 3, 9, 10, and 11), harbor conserved SC motifs (Fig. 5A). Notably, for several factors across multiple activator classes, like PR, Sp3, C/EBPɛ, and c-Myb, the motifs reside in demonstrated negative regulatory regions (1, 9, 18, 54). Since these negative regions were defined at compound response elements and share several properties (addressed in Discussion) with the motifs we have described, it is likely that rather than being conventional repression domains, they represent examples of synergy control. Taken together, these results suggest that SC motifs are critical elements of a general mechanism to control the synergy of regulators at compound response elements. By using GR as a paradigm, we have begun to define the features of both the regulator and the response element required for appropriate control of synergy and to dissect the underlying mechanisms.
SC motifs limit synergy autonomously without altering intrinsic DNA binding.
Our analysis of multiple activators indicates that SC motifs function in different contexts and are required for appropriate synergy control. For a sufficiency test, we incorporated a small region of GR encompassing both SC motifs into the synthetic activator Gal4(VP16)2. As shown in Fig. 4C and observed previously (11), Gal4(VP16)2 displays a high degree of synergy. Introduction of the WT SC motifs, however, is sufficient to selectively reduce activity at compound but not single Gal4 sites. This effect requires functional SC motifs, since disabling mutations in them restore synergy (Fig. 4C). Notably, when fused to the Gal4 DBD alone, both the WT and mutant motifs failed to activate or repress transcription directly (not shown). These fusions, however, retained DNA binding, since they were able to displace Gal4(VP16)2 (not shown). Furthermore, in contexts in which GR functions as a repressor of other factors, like AP-1 or NF-κB, disruption of the SC motifs has no consequences (20; data not shown). Thus, SC motifs are transcription regulatory sequences that operate unlike conventional repression regions; they are silent on their own, but limit the degree of synergy mediated by an independent activation function.
Two such functions (AF1 and AF2) have been identified in GR. Removal of the LBD and its associated AF2 function by truncation at position 525 generates a ligand-independent activator that relies on AF1 for function. Although this derivative displays substantial synergy (TAT3/TAT1 ratio of ∼20), it is insensitive to disruption of the SC motifs (Fig. 4D). On the other hand, although disruption of AF1 by three clustered substitutions reduces full-length receptor activity (20), inactivation of the SC motifs in this context still enhances synergy (not shown). Hence, it appears that the SC motifs limit synergy emanating from activation functions other than AF1. Together with the data presented above, this implies that the effect of SC motifs may be restricted to a subset of activation functions that include VP16, one or more activation functions in ETS-1, and, likely, the AF2 (but not AF1) of steroid receptors.
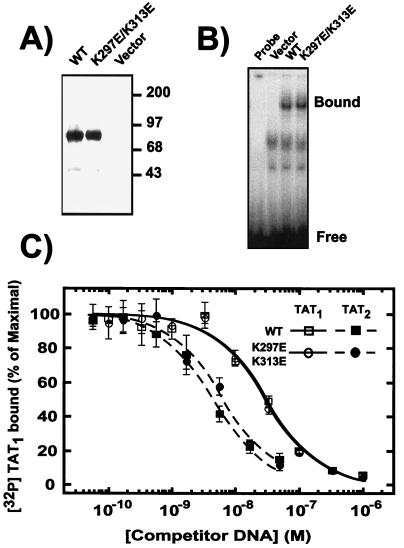
One of the proposed mechanisms for synergy is cooperative binding to compound sites (49, 55). In fact, GR displays a substantially higher apparent binding affinity for a pair versus a single binding site, especially when the sites are closely positions on the same side of the DNA helix (43). We therefore examined the effect of SC motif mutations on the ability of GR to bind to single or compound response elements. As shown in Fig. 6, the apparent affinity of the receptor to a pair of sites spaced by 21 bp is substantially higher than the affinity for a single copy. Notably, disruption of the SC motifs did not significantly alter this behavior, even though it enhanced activity at this specific compound element (see below). These results suggest that the mutations do not alter the intrinsic DNA binding properties of the receptor, and therefore SC motif function is likely to involve events subsequent to DNA recognition.
FIG. 6.
DNA binding by GR is not affected by SC mutations. (A) In vitro transcription-translation reactions (3 μl) programmed with empty vector or vectors for the expression of either WT or K297E/K313E full-length GR were analyzed for receptor expression by immunoblotting with the monoclonal antibody BuGR-2. (B) Electrophoretic mobility shift assays were performed as described in Materials and Methods in the absence (Probe) or presence of reticulocyte lysates harboring no receptor (Vector), full-length WT GR, or the K297E/K313E mutant. The positions of the free TAT1 probe and GR-DNA complex are shown to the left. (C) Electrophoretic mobility shift assays were carried out with lysates containing WT receptor (squares) or the K297E/K313E mutant (circles) as in panel B in the presence of the indicated concentrations of unlabeled DNA harboring one (TAT1 [open symbols]) or two (TAT2 [solid symbols]) receptor binding sites (21-bp spacing). No competition was observed with a nonspecific oligonucleotide. Data represent the average of three to four independent experiments and are expressed as a percentage of the maximal binding observed. This value did not exceed 5% of the total amount of labeled probe and was comparable to the binding observed in the absence of cold specific competitor.
Synergy control is functional at various compound HRE configurations.
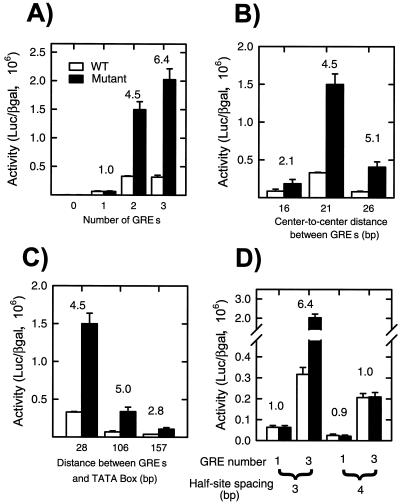
For both natural and synthetic enhancers, the extent and direction of functional interactions between regulators can depend strongly on specific spatial relationships between their binding sites (13, 22, 35). We thus explored the consequences of varying a number of binding site parameters on the activity of GR bearing either WT or mutant (K297E/I312T) SC motifs. As seen in Fig. 7A, the phenotype of the mutant requires more than one binding site and becomes more prominent as the number of sites is increased. This confirms that GR's potential to synergize is restricted and that the mutations relieve such constraints. We observed similar effects when using a perfectly palindromic site or an HRE derived from the phosphofructokinase gene instead of the TAT sequence (not shown).
FIG. 7.
Comparison of the effects of SC mutations on GR activity at reporters with various numbers, spacings, and locations of GR binding sites. GRE, GR element. CV-1 cells were transfected with 0.02 μg of the expression plasmid p6RGR (WT) or p6RGR K297E/I312T (Mutant) and 0.2 μg of the following reporter plasmids: pΔODLO, pΔTAT1-Luc, pΔTAT2-Luc, and pΔTAT3-Luc (A); pΔTAT2-16, pΔTAT2-21, and pΔTAT2-26 (B); pΔTAT2-Luc, pΔTAT2-US100, and pΔTAT2-US150 (C); and pΔTAT1-Luc, pΔTAT3-Luc, pΔTAT1/4S, and pΔTAT3/4S (D). Cells were treated with vehicle or 10 nM dexamethasone and assayed as described in Materials and Methods. Data represent averages of 3 to 11 independent transfections performed in triplicate. The ratio of mutant over WT activity is indicated above each set of bars.
The spacing between two adjacent HREs alters their phasing along the DNA and influences receptor binding and activity (43). In keeping with previous results, when two HREs are presented on the same face of the DNA (center-to-center distance of 21 bp), the receptor activity was highest (Fig. 7B). When the sites are brought closer or separated by a half-turn of B-DNA (16- and 26-bp spacing, respectively), and thus put out of register, receptor activity was reduced. In all cases, however, disruption of the SC motifs led to enhanced activity (Fig. 7B). Like for other regulators, the distance between HREs and the transcription start site influences steroid receptor activity (32, 41). As a pair of HREs is separated from the TATA box, receptor activity is reduced. Nevertheless, the mutant receptor invariably displayed enhanced activity (Fig. 7C). Taken together, these results indicate that within the range examined, the effect of the SC motifs depends neither on the spacing between binding sites nor on the distance to the TATA box.
Individual steroid receptor binding sites are composed of two half-sites separated by 3 bp. DNA recognition involves the cooperative binding of receptor monomers to each half-site via a DNA-induced dimerization interface in the DBD. Increasing the spacing to 4 bp severely compromises WT and mutant receptor activities at a single site, presumably by disfavoring the dimer interface (Fig. 7D). When this site is present in multiple copies, however, receptor activity is surprisingly high and approaches that of the receptor at the compound HRE with the appropriate 3-bp spacing (Fig. 7D). This is consistent with the observation that disruption of the DBD dimer interface via receptor mutations strongly compromises activity at a single site but leads to an enhanced receptor synergy at compound HREs (27). Interestingly, in the context of a 4-bp spacing, mutation of the SC motifs has no enhancing effect. These results suggest that at certain compound HREs that disfavor dimerization, a relief from SC may compensate for the loss in DNA binding. A natural example of this may be the mouse mammary tumor virus compound HRE. This element harbors multiple receptor binding sites, but in each case, one of the half-sites diverges substantially from the consensus. GR activity at this element is highly synergistic, and SC mutations are ineffectual in this context (not shown). Taken together, these results suggest that in steroid receptors, both the SC motifs and the DBD dimer interface play critical roles in restraining synergy.
Functional relationship between the synergy control domain and the DBD dimer interface.
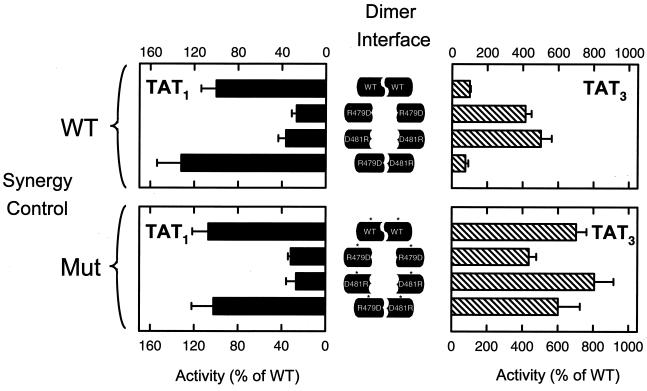
The results presented above indicate that disruption of the DBD dimer interface, like mutations in the SC motifs, leads to increased activity at compound HREs. In contrast to the dimer mutants, however, disruption of the SC motifs does not affect activity at a single site. To explore the interaction of both types of mutations, we examined the effect of DBD dimer interface mutants in the context of WT or mutant SC motifs. Both crystallographic (28) and genetic (27) data revealed that complementary salt bridges between Arg 479 of each monomer with Asp 481 of the opposite partner are integral parts of the dimer interface. As seen in Fig. 8, disruption of this interface by replacement of Arg 479 with Asp or Asp 481 with Arg leads to a dramatic loss of activity at a single site. The response of these mutants at a compound HRE, however, is enhanced compared to that of the WT receptor (Fig. 8, top) (27). Although each substitution disfavors homodimerization, the reciprocal nature of the salt bridges suggests that the two mutant receptors could reestablish a dimer interface through heterodimerization. In fact, when both mutant receptors are coexpressed, receptor activity is restored at a single site and synergy is reduced to WT levels at the compound HRE (Fig. 8, top) (27). These results show that, like for the SC motifs, disruption of the dimer interface leads to a loss of synergy control. The same series of experiments, but in the context of mutant SC motifs (Fig. 8, bottom), indicated that at a single site, the SC motifs have no effect on their own and they do not affect the behavior of the dimer mutants (Fig. 8, bottom left). At the compound HRE, and similar to the dimer mutants, disruption of the SC motifs causes an enhancement of activity. Combination of both types of mutations, however, does not lead to any additional enhancement (Fig. 8, bottom right).
FIG. 8.
Functional interaction between the DBD dimer interface and SC motifs. CV-1 cells were transfected with 0.2 μg of reporter plasmid pΔTAT1-Luc (left) or pΔTAT3-Luc (right) and 0.02 μg of expression plasmids for WT full-length GR or the indicated mutants (Mut). Coexpression was performed with 0.01 μg of each mutant expression plasmid. The top and bottom panels represent the effect of dimer mutants in the context of WT and the K297E/I312T mutant, respectively. Cells were treated with 10 nM dexamethasone and assayed as described in Materials and Methods. Data represent averages of three to seven independent transfections performed in triplicate and are expressed as a percentage of the corresponding WT activity: 4.7 × 104 U for TAT1 and 2.6 × 105 U for TAT3.
Thus, it appears that appropriate synergy control requires both the SC motifs and a WT dimer interface. Moreover, since the SC motif and dimer interface effects are not additive, it is likely that they involve common or converging mechanisms.
Synergy control requires additional factor(s), but does not alter receptor sensitivity to p160 coactivators.
Reconstitution of a regulatory pathway in a heterologous system is a powerful approach to the identification and characterization of its components. We therefore examined whether the function of the SC motifs can be recapitulated in S. cerevisiae. In contrast to their behavior in mammalian cells, disruption of the SC motifs has no detectable phenotype in yeast (Fig. 9A). Given that the SC motifs in GR appear to target synergy afforded by the LBD, we examined whether sensitivity to the SC motifs in yeast could be restored by coexpression of the AF2 coactivator GRIP-1. As shown in Fig. 9B, however, this is not the case, suggesting that additional components may be required for SC motif function. Consistent with this idea, the activity of both the WT and mutant receptors was enhanced in mammalian cells by coexpression of GRIP-1 (Fig. 9C) and SRC-1a (not shown), suggesting that the motifs do not alter the sensitivity of the receptor to these p160 coactivators. Taken together, these results suggest that the effect of SC motifs is not an intrinsic property of the receptor, but rather, the expression of this regulatory feature requires an appropriate cellular environment.
FIG. 9.
SC motifs are silent in yeast and do not alter the effect of p160 coactivators. (A) Yeast cells harboring Trp1-based expression plasmids for either full-length WT GR or the K297E/K313E mutant were transformed with Ura3-based reporter plasmid harboring no HREs (pUCΔSS), a single palindromic HRE (pUCΔS1Gs3), two palindromic HREs (pUCΔS2X3S), or three TAT HREs (pΔs26X). (B) Yeast cells harboring the above receptor plasmids and the pΔs26X reporter were transformed with the GRIP1 expression vector pGAD424-GRIP1 or a control plasmid. Cells were grown in the presence of 10 μM (A) or the indicated amounts (B) of deoxycorticosterone (DOC) and assayed as described in Materials and Methods. The data represent averages of four to six independent transformants. (C) CV-1 cells were transfected by lipofection with 0.2 μg of pΔTAT3-Luc, 0.02 μg of p6RGR WT or K297E/K313E, and 0.1 μg of a mixture of pSG5 vector and the indicated amounts of pSG5.HA-GRIP1. Cells were treated with vehicle (Veh.) or 10 nM dexamethasone (DEX) and assayed as described in Materials and Methods. Data represent averages of three independent transfections performed in triplicate.
DISCUSSION
Synergy as a target for regulation.
We have uncovered a novel functional region defined by one or more copies of a short amino acid motif that restrains the ability of regulators to engage in synergy. Importantly, these SC motifs influence neither the intrinsic transcriptional activation nor the DNA binding properties of the activator. SC motifs appear to be devoid of intrinsic activation or repression properties and map to regions other than activation functions. This reveals that features involved in controlling synergy are distinct from transcriptional activation per se. Like activation functions, however, these motifs cooperate functionally and can be transplanted to heterologous proteins. These features, coupled with their ability to operate in many compound response element configurations, make SC motifs well suited for the general control of transcription factor synergy.
SC motifs as a common feature of many negative regions.
SC motifs occur frequently within documented negative regulatory regions of numerous transcription factors and may account for a number of functional differences among members of individual families. In the cases we have explored and describe below, several common features point toward the importance of SC motifs and suggest that a common synergy control mechanism participates in the function of these seemingly disparate regions.
(i) Steroid receptors.
To our knowledge, SC motifs are the first example of a regulatory function that can be assigned to a common primary structure feature within the otherwise highly divergent N-terminal regions of steroid receptors and may account for some of their distinct properties. For example, the presence of four SC motifs in MR versus only two in GR correlates with the considerably weaker synergy of MR (27, 40). For genes in which receptor synergy plays a major role, this difference may contribute to receptor selectivity. Similarly, the single motif found in PR lies within and is likely to be responsible for the function of a recently proposed negative modulation domain (307 to 427 in the human PR). Deletion of this region enhances activity, whereas its duplication renders PR a very poor activator at compound HREs (18).
(ii) Sp1 family.
Sp1 and Sp3 are related factors that recognize similar DNA sequences and harbor glutamine-rich activation domains. Curiously, although Sp3 displays activity at a single site, it fails to activate at compound response elements, suggesting a compromised ability to engage in synergy. Dennig et al. (9) demonstrated that a negative regulatory region unique to Sp3 restricts its activation domains. A single SC motif resides in this region, and deletions or mutations within the SC motif relieve inhibition and render Sp3 a potent activator at compound Sp1 sites (9).
(iii) Myb family.
Several parallels can also be established in the case of the Myb family. The two SC motifs in c-Myb map to the C-terminal region and lie within a negative regulatory domain absent in the viral oncogenic forms and in certain tumor-derived cell lines (48, 52). Deletion of this region leads to enhanced transactivation at compound response elements and is sufficient for oncogenic transformation (17). As in the case of GR, these effects are not observed in yeast. Similar negative effects of the C-terminal region have been observed for the related A-Myb but not B-Myb proteins (34). Strikingly, the SC motifs in A- and c-Myb are evolutionarily conserved across multiple vertebrates, but are absent in B-Myb.
(iv) C/EBP family.
Similarly, within the C/EBP family, an attenuator domain in C/EBPα has been mapped to a region between two N-terminal transcription activation functions (37). This region decreases transactivation in multiple compound promoter contexts and, consistent with our results, does so without affecting DNA binding. Notably, this region includes a highly conserved SC motif. In the case of C/EBPɛ, an SC motif is located in a similar functional region. Importantly, mutations within this region that enhance activity (1) map to the SC motif.
The predictive value of SC motifs is exemplified in our analysis of ETS-1, since a negative function for the regions harboring the SC motifs was, to our knowledge, previously unrecognized. It is unlikely, however, that all matches to the consensus constitute functional SC motifs. In fact, so far, we have not detected a functional role for the SC motif in SF-1 (not shown).
For the cases described above, the effects of these seemingly unrelated negative regions have been examined at both natural and artificial compound response elements, attesting to the generality and relevance of this mechanism. Although effects at a single site have not been examined in all cases, the striking similarities strongly suggest that the SC motifs contribute to the function of these regions, likely by limiting synergy. We are expanding our analysis to some of these factors and to the more complex and prevalent functional interactions involving synergy between different classes of regulators.
Activator functions involved in synergy control.
Our analysis indicates that SC motifs can influence synergy afforded by the activation functions of multiple factors from diverse families. SC motif influence is not totally promiscuous, however, since the N-terminal AF1 function of GR appears to be insensitive. This is not simply due to its moderately acidic character, since other “acidic” activators such as VP16 are regulated by SC motifs, and likely reflects either a different mechanism of synergy or an intrinsic property of AF1.
The SC motifs identified here can limit synergy when embedded in synthetic activators. Within their natural context, however, additional features of the specific activator can influence synergy control. In the case of steroid receptors, the DBD dimer interface plays an important role, since specific receptor mutations or noncanonical spacing of half-sites that disfavor this dimer interface lead to enhanced synergy. Furthermore, simultaneous disruption of the SC motifs and the DBD dimer interface produces an effect no greater than that resulting from altering either one alone. Hence, it is likely that a common SC mechanism requires both the SC motifs and an engaged DBD dimer interface for function. These properties would therefore constrain the effect of SC motifs to multiple, properly dimerized receptor pairs bound to compound HREs. A relationship between SC motifs and DBDs is also present in the case of c-Myb, since N-terminal deletions adjacent to the DBD have phenotypes similar to deletion of the C-terminal negative region. Furthermore, the DBD and regions adjacent to one of the SC motifs can interact (8). Whether dimerization per se is an obligate requirement for SC function, however, awaits further examination in other activators.
An SC motif.
We have identified eight examples of functional SC motifs in different activators. Sequence comparison across different species coupled with mutational analysis revealed that a branched aliphatic residue at the first position followed by invariant Lys and Glu residues at positions 2 and 4 are critical features for SC motif function. Several features suggest that these motifs are likely to be solvent exposed and accessible to interactions. First, the SC motif is highly charged. Second, limited proteolytic cleavage indicates that Glu 295, within the second motif of human GR, is a preferred V8 cleavage site (B. Darimont, personal communication). Third, the motif is preceded and/or followed by a Pro residue, and the sequence in the vicinity of the upstream Pro often varies in size by a few residues in different species. This suggests that the motif may lie near the end of secondary structure elements or within a loop that can tolerate insertions. Fourth, a search of the PDB structure database revealed that, in seven of the nine intracellular proteins of known three-dimensional structure that harbor a match to the motif, it lies within an extended loop or a β-sheet strand with the conserved charged residues of the motif exposed to solvent. The propensity of these sequences to adopt such an exposed configuration is consistent with the view that these motifs could operate as protein-protein interaction surfaces.
A model for synergy control.
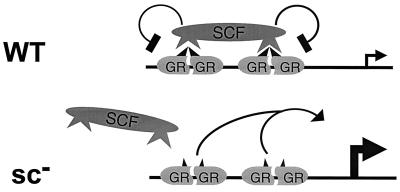
Our characterization indicates that SC motifs do not alter intrinsic DNA binding and that they fail to function in S. cerevisiae. In addition, they are functional in a wide variety of contexts and affect multiple activation domains. These observations and similar ones from other negative regulatory regions harboring SC motifs suggest that SC is unlikely to be due to a direct intramolecular interaction. Rather, they suggest that additional cellular factors are involved. One possibility is that a synergy control factor (SCF) is recruited to compound HREs by recognizing SC motifs when presented in the appropriate context of compound response elements (Fig. 10). The selective effect of such a factor at compound sites could be the result of its multivalent binding to adjacently bound regulators. In this model, mutations in the SC motifs would prevent interaction with SCF and thus lead to constitutive high levels of synergy (Fig. 10). Consistent with this idea, overexpression of the C-terminal Myb domain (50) and C/EBPɛ negative region (1) enhances the activity of the respective proteins at compound sites, possibly by titrating SCF or other components. Successful competition, however, has not been achieved with Sp3 (9) or GR (not shown). We are currently searching for SC motif interacting factors.
FIG. 10.
Model for the mechanism of action of SC motifs. Selective recruitment of SCF to compound response elements through interactions with the SC motifs limits transcriptional activity. See text for details.
The mechanisms involved in synergy, especially those involving steps subsequent to DNA binding, are not well understood, but effects beyond DNA binding imply the alteration of the transcription complex (53). An SCF could thus interfere with or favor the disassembly of an active transcription complex. SC motif function, however, does not appear to involve an alteration in the sensitivity to certain coactivators. Both GRIP1 and SRC-1 enhance the activity of the WT and mutant GR. Similarly, disruption of the SC motif in Sp3 does not alter the interaction of the glutamine-rich regions with TAF 110 (9).
The recruitment of cofactors to sequence-specific regulators via short amino acid motifs has already been observed in the case of the interaction of the p160 class of coactivators with the LBD of steroid receptors (7) or CREB and CREB-binding protein (39). Furthermore, the function of SC motifs could also be regulated through specific posttranslational modifications. The critical Lys at position 2 could be the target of acetylation or ubiquitination, especially since Arg is not functional at this position. These regulatory modifications have been recently detected in a number of transcription factors (26, 30, 60). Similarly, phosphorylation of Ser 532 immediately downstream of the second SC motif of human c-Myb attenuates the function of the negative regulatory domain and enhances activity at compound response elements (29, 51). To date, however, we have not detected these modifications within the SC motifs of GR.
According to this general model, synergy control through SC motifs may permit the selective deployment or utilization of functional synergy surfaces of transcription factors at the appropriate promoter context or developmental stage. Synergy could thus be controlled through the regulation of the activity of SC motifs or through the expression and/or function of SCF. For example, as part of a developmental program, selective loss of SCF only when a specific tissue reaches a terminally differentiated state would permit the characteristic high-level expression of the appropriate set of regulated genes. A similar effect could be achieved if a given factor harboring SC motifs is developmentally replaced by a close relative lacking them.
The presence of multiple binding sites for many transcription factors at natural regulatory DNA elements coupled with the combinatorial nature of transcriptional control makes the potential number of interactions enormous. The SC motifs described here may constitute an example of cellular regulatory devices that, by following simple rules, could limit functional interactions to a more tractable number. Clearly, defining the rules that govern such higher-order interactions will be essential to understand and manipulate the regulatory logic of complex transcriptional systems.
ACKNOWLEDGMENTS
We thank Keith R. Yamamoto and the members of his laboratory for their support; Beatrice Darimont for unpublished results; Michael Stallcup, A. Seth, Dianne Merry, and Alex Lange for kindly providing expression and reporter plasmids; and Marc Diamond, Carol Gross, Holly Ingraham, Inez Rogatsky, and Keith R. Yamamoto for insightful discussions and comments on the manuscript.
J. A. Iñiguez-Lluhí is a Special Fellow of the Leukemia Society of America, and D. Pearce acknowledges NIH support through grant DK-R29-51151.
REFERENCES
- 1.Angerer N D, Du Y, Nalbant D, Williams S C. A short conserved motif is required for repressor domain function in the myeloid-specific transcription factor CCAAT/enhancer-binding protein epsilon. J Biol Chem. 1999;274:4147–4154. doi: 10.1074/jbc.274.7.4147. [DOI] [PubMed] [Google Scholar]
- 2.Arnone M I, Davidson E H. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- 3.Bailly A, Rauch C, Cato A C, Milgrom E. In two genes, synergism of steroid hormone action is not mediated by cooperative binding of receptors to adjacent sites. Mol Cell Endocrinol. 1991;82:313–323. doi: 10.1016/0303-7207(91)90045-t. [DOI] [PubMed] [Google Scholar]
- 4.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen D, Ma H, Hong H, Koh S S, Huang S M, Schurter B T, Aswad D W, Stallcup M R. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 6.Danesch U, Gloss B, Schmid W, Schutz G, Schule R, Renkawitz R. Glucocorticoid induction of the rat tryptophan oxygenase gene is mediated by two widely separated glucocorticoid-responsive elements. EMBO J. 1987;6:625–630. doi: 10.1002/j.1460-2075.1987.tb04800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dash A B, Orrico F C, Ness S A. The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev. 1996;10:1858–1869. doi: 10.1101/gad.10.15.1858. [DOI] [PubMed] [Google Scholar]
- 9.Dennig J, Beato M, Suske G. An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J. 1996;15:5659–5667. [PMC free article] [PubMed] [Google Scholar]
- 10.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 11.Emami K H, Carey M F. A synergistic increase in potency of a multimerized VP16 transcriptional activation domain. EMBO J. 1992;11:5005–5012. doi: 10.1002/j.1460-2075.1992.tb05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gametchu B, Harrison R W. Characterization of a monoclonal antibody to the rat liver glucocorticoid receptor. Endocrinology. 1984;114:274–279. doi: 10.1210/endo-114-1-274. [DOI] [PubMed] [Google Scholar]
- 13.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCRalpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 14.Godowski P J, Picard D, Yamamoto K R. Signal transduction and transcriptional regulation by glucocorticoid receptor-LexA fusion proteins. Science. 1988;241:812–816. doi: 10.1126/science.3043662. [DOI] [PubMed] [Google Scholar]
- 15.Hittelman A B, Burakov D, Iñiguez-Lluhí J A, Freedman L P, Garabedian M J. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong K, Zheng W, Baker A, Papahadjopolous D. Stabilization of cationic liposome-plasmid DNA complex by polyamines and poly(ethylene glycol)-phospholipid conjugates for efficient in vivo gene delivery. FEBS Lett. 1997;400:233–237. doi: 10.1016/s0014-5793(96)01397-x. [DOI] [PubMed] [Google Scholar]
- 17.Hu Y L, Ramsay R G, Kanei-Ishii C, Ishii S, Gonda T J. Transformation by carboxyl-deleted Myb reflects increased transactivating capacity and disruption of a negative regulatory domain. Oncogene. 1991;6:1549–1553. [PubMed] [Google Scholar]
- 18.Huse B, Brenz Verca S, Mattey P, Rusconi S. Definition of a negative modulation domain in the human progesterone receptor. Mol Endocrinol. 1998;12:1334–1342. doi: 10.1210/mend.12.9.0164. [DOI] [PubMed] [Google Scholar]
- 19.Ikonen T, Palvimo J J, Janne O A. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 20.Iñiguez-Lluhí J A, Lou D Y, Yamamoto K R. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N-terminus. J Biol Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- 21.Kelly E J, Sandgren E P, Brinster R L, Palmiter R D. A pair of adjacent glucocorticoid response elements regulate expression of two mouse metallothionein genes Proc. Natl Acad Sci USA. 1997;94:10045–10050. doi: 10.1073/pnas.94.19.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 23.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 24.Lefstin J A, Thomas J R, Yamamoto K R. Influence of a steroid receptor DNA-binding domain on transcriptional regulatory functions. Genes Dev. 1994;8:2842–2856. doi: 10.1101/gad.8.23.2842. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y S, Carey M, Ptashne M, Green M R. How different eukaryotic transcriptional activators can cooperate promiscuously. Nature. 1990;345:359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- 26.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W, Wang J, Yu G, Pearce D. Steroid receptor transcriptional synergy is potentiated by disruption of the DNA-binding domain dimer interface. Mol Endocrinol. 1996;10:1399–1406. doi: 10.1210/mend.10.11.8923466. [DOI] [PubMed] [Google Scholar]
- 28.Luisi B F, Xu W X, Otwinowski Z, Freedman L P, Yamamoto K R, Sigler P B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 29.Miglarese M R, Richardson A F, Aziz N, Bender T P. Differential regulation of c-Myb-induced transcription activation by a phosphorylation site in the negative regulatory domain. J Biol Chem. 1996;271:22697–22705. doi: 10.1074/jbc.271.37.22697. [DOI] [PubMed] [Google Scholar]
- 30.Mitsui A, Sharp P A. Ubiquitination of RNA polymerase II large subunit signaled by phosphorylation of carboxyl-terminal domain. Proc Natl Acad Sci USA. 1999;96:6054–6059. doi: 10.1073/pnas.96.11.6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller M, Baniahmad C, Kaltschmidt C, Renkawitz R. Multiple domains of the glucocorticoid receptor involved in synergism with the CACCC box factor(s) Mol Endocrinol. 1991;5:1498–1503. doi: 10.1210/mend-5-10-1498. [DOI] [PubMed] [Google Scholar]
- 32.Nordeen S K, Ogden C A, Taraseviciene L, Lieberman B A. Extreme position dependence of a canonical hormone response element. Mol Endocrinol. 1998;12:891–898. doi: 10.1210/mend.12.6.0118. [DOI] [PubMed] [Google Scholar]
- 33.Nye J, Petersen J, Gunther C, Jonsen M, Graves B. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 34.Oh I-H, Reddy E P. The C-terminal domain of B-Myb acts as a positive regulator of transcription and modulates its biological functions. Mol Cell Biol. 1998;18:499–511. doi: 10.1128/mcb.18.1.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearce D, Matsui W, Miner J N, Yamamoto K R. Glucocorticoid receptor transcriptional activity determined by spacing of receptor and nonreceptor DNA sites. J Biol Chem. 1998;273:30081–30085. doi: 10.1074/jbc.273.46.30081. [DOI] [PubMed] [Google Scholar]
- 36.Pearce D, Yamamoto K R. Mineralocorticoid and glucocorticoid receptor activities are distinguished by nonreceptor factors at a composite element. Science. 1993;259:1161–1165. doi: 10.1126/science.8382376. [DOI] [PubMed] [Google Scholar]
- 37.Pei D, Shih C. An “attenuator domain” is sandwiched by two distinct transactivation domains in the transcription factor C/EBP. Mol Cell Biol. 1991;11:1480–1487. doi: 10.1128/mcb.11.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perlmann T, Eriksson P, Wrange Ö. Quantitative analysis of the glucocorticoid receptor-DNA interaction at the mouse mammary tumor virus glucocorticoid response element. J Biol Chem. 1990;265:17222–17229. [PubMed] [Google Scholar]
- 39.Radhakrishnan I, Pérez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 40.Rupprecht R, Arriza J L, Spengler D, Reul J M, Evans R M, Holsboer F, Damm K. Transactivation and synergistic properties of the mineralocorticoid receptor: relationship to the glucocorticoid receptor. Mol Endocrinol. 1993;7:597–603. doi: 10.1210/mend.7.4.8388999. [DOI] [PubMed] [Google Scholar]
- 41.Sathya G, Li W, Klinge C M, Anolik J H, Hilf R, Bambara R A. Effects of multiple estrogen responsive elements, their spacing, and location on estrogen response of reporter genes. Mol Endocrinol. 1997;11:1994–2003. doi: 10.1210/mend.11.13.0039. [DOI] [PubMed] [Google Scholar]
- 42.Scheller A, Hughes E, Golden K L, Robins D M. Multiple receptor domains interact to permit, or restrict, androgen-specific gene activation. J Biol Chem. 1998;273:24216–24222. doi: 10.1074/jbc.273.37.24216. [DOI] [PubMed] [Google Scholar]
- 43.Schmid W, Strahle U, Schutz G, Schmitt J, Stunnenberg H. Glucocorticoid receptor binds cooperatively to adjacent recognition sites. EMBO J. 1989;8:2257–2263. doi: 10.1002/j.1460-2075.1989.tb08350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seth A, Robinson L, Panayiotakis A, Thompson D M, Hodge D R, Zhang X K, Watson D K, Ozato K, Papas T S. The EndoA enhancer contains multiple ETS binding site repeats and is regulated by ETS proteins. Oncogene. 1994;9:469–477. [PubMed] [Google Scholar]
- 45.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 46.Skalicky J J, Donaldson L W, Petersen J M, Graves B J, McIntosh L P. Structural coupling of the inhibitory regions flanking the ETS domain of murine Ets-1. Protein Sci. 1996;5:296–309. doi: 10.1002/pro.5560050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starr D B, Matsui W, Thomas J R, Yamamoto K R. Intracellular receptors use a common mechanism to interpret signalling information at response elements. Genes Dev. 1996;10:1271–1283. doi: 10.1101/gad.10.10.1271. [DOI] [PubMed] [Google Scholar]
- 48.Tomita A, Watanabe T, Kosugi H, Ohashi H, Uchida T, Kinoshita T, Mizutani S, Hotta T, Murate T, Seto M, Saito H. Truncated c-Myb expression in the human leukemia cell line TK-6. Leukemia. 1998;12:1422–1429. doi: 10.1038/sj.leu.2401113. [DOI] [PubMed] [Google Scholar]
- 49.Tsai S Y, Tsai M J, O'Malley B W. Cooperative binding of steroid hormone receptors contributes to transcriptional synergism at target enhancer elements. Cell. 1989;57:443–448. doi: 10.1016/0092-8674(89)90919-7. [DOI] [PubMed] [Google Scholar]
- 50.Vorbrueggen G, Kalkbrenner F, Guehmann S, Moelling K. The carboxyterminus of human c-myb protein stimulates activated transcription in trans. Nucleic Acids Res. 1994;22:2466–2475. doi: 10.1093/nar/22.13.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vorbrueggen G, Lovric J, Moelling K. Functional analysis of phosphorylation at serine 532 of human c-Myb by MAP kinase. Biol Chem. 1996;377:721–730. doi: 10.1515/bchm3.1996.377.11.721. [DOI] [PubMed] [Google Scholar]
- 52.Wang D-M, Dubendorff J W, Woo C H, Lipsick J S. Functional analysis of carboxy-terminal deletion mutants of c-Myb. J Virol. 1999;73:5875–5886. doi: 10.1128/jvi.73.7.5875-5886.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J, Ellwood K, Lehman A, Carey M F, She Z S. A mathematical model for synergistic eukaryotic gene activation. J Mol Biol. 1999;286:315–325. doi: 10.1006/jmbi.1998.2489. [DOI] [PubMed] [Google Scholar]
- 54.Williamson E A, Xu H N, Gombart A F, Verbeek W, Chumakov A M, Friedman A D, Koeffler H P. Identification of transcriptional activation and repression domains in human CCAAT/enhancer-binding protein epsilon. J Biol Chem. 1998;273:14796–14804. doi: 10.1074/jbc.273.24.14796. [DOI] [PubMed] [Google Scholar]
- 55.Wright A P, Gustafsson J A. Mechanism of synergistic transcriptional transactivation by the human glucocorticoid receptor. Proc Natl Acad Sci USA. 1991;88:8283–8287. doi: 10.1073/pnas.88.19.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto K R, Darimont B D, Wagner R L, Iñiguez-Lluhí J A. Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harbor Symp Quant Biol. 1998;63:587–598. doi: 10.1101/sqb.1998.63.587. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto K R, Pearce D, Thomas J, Miner J N. Combinatorial regulation at a mammalian composite response element. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. p. 1169. [Google Scholar]
- 58.Yanofsky C. Transcriptional regulation: elegance in design and discovery. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 3–26. [Google Scholar]
- 59.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 60.Zhang W, Bieker J J. Acetylation and modulation of erythroid Krüppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]