Supplemental Digital Content is available in the text.
Keywords: endothelial cell, fibrinolysis, lipopolysaccharides, methylation, thrombosis
Abstract
Objective:
METTL3 (methyltransferase-like protein 3)-mediated N6-methyladenosine modification is the most abundant RNA modification on eukaryote mRNAs and plays a crucial role in diverse physiological and pathological processes. However, whether N6-methyladenosine modification has function in thrombosis is unknown. This study aims to determine the role of METTL3 in the endothelial cells-mediated thrombosis.
Approach and Results:
RNA-sequencing and real-time quantitative PCR revealed that the expression of PAI-1 (plasminogen activator inhibitor-1) was downregulated in METTL3 knockdown human umbilical vein endothelial cells. In vitro experiments showed that METTL3 suppressed fibrinolysis. Mechanically, RNA methylation sequencing and meRIP-quantitative real-time PCR showed that METTL3 catalyzed N6-methyladenosine modification on 3′ UTR of JUN mRNA. Western blotting analysis showed that METTL3 promoted JUN protein expression. Chromatin immunoprecipitation analysis demonstrated that JUN bound to the PAI-1 promoter in human umbilical vein endothelial cells. Furthermore, mice challenged with lipopolysaccharide resulted in higher METTL3 expression in vessels. Endothelial-specific knockdown of Mettl3 decreased expression of active PAI-1 in plasma and attenuated fibrin deposition in livers and lungs during endotoxemia.
Conclusions:
Our study reveals that METTL3-mediated N6-methyladenosine modification plays a crucial role in fibrinolysis and is an underlying target for the therapy of thrombotic disorders.
Highlights.
Endothelial METTL3 (methyltransferase-like 3) inhibits fibrinolysis by upregulating PAI-1 (plasminogen activator inhibitor-1) expression.
METTL3 promotes the expression of PAI-1 through the N6-methyladenosine-Jun proto-oncogene-YTHDF1 (YTH-domain family protein 1) pathway.
High expression of METTL3/N6-methyladenosine/Jun proto-oncogene/PAI-1 are detected in the blood vessels of endotoxemia mice.
And endothelial-specific knockdown of Mettl3 alleviated fibrin deposition by reducing the levels of active PAI-1 in lipopolysaccharide-induced endotoxemia mice.
Endothelial cells form the inner layer of the vascular endothelium and play an essential regulatory role in physiological processes, such as angiogenesis, vascular permeability, coagulation, platelet activation, and fibrinolysis.1 Normally, the endothelium regulates hemostatic balance by releasing procoagulant and anticoagulant factors including vWF (von Willebrand factor) and TFPI (tissue factor pathway inhibitor) to regulate platelet function and coagulation system.2 Besides, endothelium expresses the main fibrinolytic components tPA (tissue-type plasminogen activator), u-PA (urokinase plasminogen activator), and profibrinolytic factor PAI-1 (plasminogen activator inhibitor-1), to maintain the fibrinolytic and antifibrinolytic balance.3 Although the regulation of transcription factors4,5 and posttranscriptional mechanisms such as miRNAs6,7 are involved in the regulation of hemostatic balance have been intensely investigated, it remains obscure whether epigenetic mechanisms, particularly N6-methyladenosine (m6A) modification, are involved in this process.
m6A is one of the most abundant RNA modifications in eukaryotes.8 m6A modification is catalyzed by RNA methyltransferase complex (METTL3 [methyltransferase-like protein 3]/METTL14/WTAP),9 recognized by m6A reader proteins (YTHDF1-3 [YTH-domain family protein 1], IGF2BP1-3),10,11 and removed by m6A demethylases: (FTO and ALKBH5).12,13 METTL3 acts as the core component of the methyltransferase complex and plays a vital role in the biological process such as embryonic development,14 hematopoietic stem cell differentiation,15 spermatogenesis,16 and brain development.17 Endothelial-specific knockdown of Mettl3 in mouse embryos impairs definitive hematopoiesis.18 Knockdown of METTL3 increases endothelial cell migration and endothelial-specific Mettl3 knockdown inhibits pathological angiogenesis in vivo.19 However, the role of METTL3 in endothelial cell–mediated coagulation remains to be investigated.
In this study, we found that knockdown of METTL3 significantly decreased PAI-1 expression and promoted fibrinolysis in vitro. Furthermore, methylated RNA immunoprecipitation sequencing (MeRIP-seq), MeRIP-quantitative real-time PCR (MeRIP-qPCR), and chromatin immunoprecipitation assay revealed that METLL3 mediated the m6A modification of 3′ UTR of JUN mRNA and JUN regulated PAI-1 transcription in human umbilical vein endothelial cells (HUVECs). METTL3 promoted the translation of JUN mRNA in an m6A-YTHDF1-dependent manner. METTL3 was upregulated in vessels of lipopolysaccharide-treated mice. Endothelial-specifical METTL3 knockdown decreased plasma active PAI-1 levels and alleviated fibrin deposition in endotoxin-treated mice. Thus, METTL3 could be a potential therapeutic target for endotoxin-induced thrombosis.
Materials and Methods
Data that support the study can be obtained upon reasonable request. Additional information on materials used can be found in the Major Resources Table in the Data Supplement.
Cell Culture
HUVECs were cultured in Endothelial Cell Medium (Science Cell). HEK-293FT cells were cultured with DMEM (Gibco) supplemented with 10% FBS (Gibco). All cell lines were incubated at 37 °C and 5% CO2 in a humidified atmosphere. For lipopolysaccharide experiments, HUVECs were stimulated by 1 µg/mL lipopolysaccharide (Sigma) as previously described.20,21 After 4 hours, the cells were collected and assayed.
Fibrin Formation and Lysis Assay
Fibrin formation and lysis assay were performed as described.22–24 Cells were counted using an automated cell counter (Countstar, China), and the equal number of cells were plated in a 96-well plate. Culture medium was rapidly removed from the HUVECs. Subsequently, 2 mg/mL fibrinogen (Sigma), 5 mmol/L CaCl2 (Sangon, China), 1 nM thrombin (Sigma), 20 mmol/L HEPES (Sangon, China), and 150 mmol/L NaCl (Sangon, China) were added. For fibrinolysis assays, 1 nM tPA (Sigma) and 50 nM plasminogen (Sigma) were added after fibrin formation. Fibrin formation and lysis were detected by turbidity at 405 nm in microplate readers (TECAN, Austria).
Lentiviral Plasmid Construction and Lentiviral Infection
Overexpressed plasmids and shRNA were cloned to the pHAGE-EF1a-IRES-ZsGree-Puro or pLKO.1 vectors, respectively. To obtain lentiviral particles, HEK293T cells were transiently transfected with constructs (pLKO.1 or pHAGE-EF1a-IRES-ZsGree-puro vectors), psPAX2 and pMD2.G. HUVECs were infected with lentivirus to knockdown or overexpress target genes of interest. All shRNA sequences and protein overexpressed sequences were listed in the Data Supplement.
Western Blot Analysis
Protein sample separated on SDS-PAGE, transferred onto 0.45 μm PVDF membranes (Millipore), blocked with 3% BSA, incubated overnight at 4 °C with primary antibodies against METTL3 (ABclonal, China), PAI-1 (Abcam), YTHDF1 (Proteintech), JUN (Cell Signaling Technology), GAPDH (Proteintech), Fibrin (Merck, Germany), and detected using chemiluminescence. Densitometry analysis was performed using ImageJ software (National Institutes of Health) by measuring the band intensity for each group, and the data were normalized to GAPDH as an internal control.
RT-PCR and qPCR
Total RNA was extracted with Trizol (Thermo Fisher Scientific). Subsequently, cDNA was synthesized with a PrimeScript RT Master Mix (Takara, Japan). The mRNA expression levels were quantified by qPCR on CFX connect real-time PCR system (Bio-rad), using THUNDERBIRD SYBR qPCR Mix (Toyobo, Japan) and were normalized to the expression of GAPDH. The 2−ΔΔCt method was used to calculate the relative expression levels of mRNA. Sequences of the primers were listed in the Data Supplement.
MeRIP-Seq and MeRIP qPCR
m6A-IP and library preparation were proceeded according to previous study.25 In brief, fragmented RNA was incubated with Dynabeads Protein G (Thermo Fisher Scientific) premixed with anti-m6A antibody (Abcam) at 4 °C. The complex was washed with low-salt IP buffer and high-salt IP buffer, respectively. The m6A-enriched fragmented RNA was eluted with RLT Buffer (QIAGEN, Germany) and extracted using Dynabeads MyOne SLINE beads (Thermo Fisher Scientific). Both input and m6A IP samples were prepared for the next-generation sequencing by the Novogene (Beijing, China). The m6A-enriched motifs were identified by HOMER, and q value threshold of peak <0.05 was applied for all data sets. Sequencing data have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE158364.
For MeRIP-qPCR, cDNA was synthesized from fragmented RNA using SuperScript II Reverse Transcriptase (Thermo Fisher Scientific). GAPDH was used as a negative control as previously reported.25 Sequences of the primers for MeRIP-qPCR were listed in the Data Supplement.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation assay performed as previously described26 with some modifications. Briefly, HUVECs were fixed by formaldehyde and followed by sonication to achieve DNA shearing. Sheared chromatins incubated with Dynabeads Protein G (Thermo Fisher Scientific) coated with anti-JUN (Cell Signaling Technology) or anti-rabbit IgG (Abcam) antibody. The complex was washed with buffer (high salt wash buffer, LiCI wash buffer, and TE buffer) and eluted by elution buffer. The reverse formaldehyde crosslinking was carried by adding NaCl and DNA purified by Dynabeads MyOne SLINE beads (Thermo Fisher Scientific). Immunoprecipitated DNA was detected by PCR and agarose gel electrophoresis. Sequences of the primers for chromatin immunoprecipitation-PCR were listed in the Data Supplement.
RNA Sequencing
For RNA-seq, total RNAs from shMETTL3 and scramble HUVECs were isolated with Trizol. The library preparations were sequenced by the Novogene (Beijing, China). Adjusted P<0.05 and fold change >2 were considered as significantly differentially expressed transcripts. Gene ontology terms were considered significantly differentially expressed if showing P<0.05. The differentially expressed genes were provided in Table I in the Data Supplement. Sequencing data have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE157544.
m6A Quantification Assay
RNA was extracted using Trizol and subjected to m6A quantification using the m6A RNA Methylation Quantification Kit (Colorimetric) (Abcam) in biological triplicate. Relative m6A RNA methylation levels were calculated according to the manufacturer’s protocol.
Animals
The wild-type mice (C57BL/6J) were purchased from Experimental Animal Center, Daping Hospital of Army Medical University (Chongqing, China). Mettl3 floxed mice were kindly provided by Dr Minghan Tong (CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China). Tie2-Cre mice (Tg (Tek-Cre)1Ywa/J) were from The Jackson Laboratory. Mettl3 floxed mice and Tie2-Cre mice were mated to obtain endothelial-specific Mettl3 knockdown mice. The study was performed with male and female mice. Male and female mice were maintained under SPF conditions. Mice were under general anesthesia during experiments. We found that there was no sex difference in the results of the experiment. Data from male and female were pooled for analysis. For peripheral blood analysis, a small amount of blood was collected into EDTA tube via the retro-orbital plexus. For the endotoxemia model, mice were intraperitoneally injected with lipopolysaccharide (10 mg/kg of body weight) from Escherichia coli (O111:B4, Sigma). For ELISA sample preparation, blood was drawn from the inferior vena cava with or without an anticoagulant, followed by centrifugation. The mice were sacrificed by cervical dislocation and tissues (thoracic aorta, inferior vena cava, liver, and lung) collected and analyzed by qPCR, Western blot or immunohistochemistry, and HE-staining. The animal experiments were authorized by the Institutional Animal Care and Use Committee of Army Medical University (Chongqing, China).
Fibrin Extraction
Fibrin extraction was performed as previously described.27 In brief, tissues were homogenized in 10 volumes (mg: μl) of RIPA buffer containing PMSF. After centrifugation at 10000g for 10 minutes, the supernatant was collected for GAPDH detection. The pellet was homogenized in 3 mol/L urea and vortexed at 1500 rpm for 2 hours at 37 °C by thermomixer C (Eppendorf, Germany) and centrifuged at 14000g for 15 minutes. The pellet was suspended and vortexed at 65 °C for 30 minutes in reducing SDS buffer.
Isolation Endothelial Cells From Mouse Tissues
Endothelial cells isolated from mouse livers and lungs were performed as previously reported.28,29 Tissues were mechanically minced with scissors and digested with collagenase at 37 °C for 45 minutes. Incubated in red blood cell lysis buffer and filtered through a 70-μm strainer, the single-cell suspension was incubated with CD31-conjugated Dynabeads. Cells were washed with PBS using a magnetic rack. Cells were plated in 6-well plate at 37 °C in the incubator until confluent. Cells were resuspended and inoculated with ICAM2-coated Dynabeads. Washing and plating were performed as described.
HE Staining and IHC
Tissues were fixed by formaldehyde and embedded in paraffin. For hematoxylin-eosin staining, the sections were stained with hematoxylin and eosin and analysis of thrombi deposition. For immunohistochemistry, microvascular fibrin-rich thrombi in paraffin-embedded livers and lungs were analyzed by immunostaining using fibrin antibody (Merck, Germany) and IgG antibody (Abcam). Images were taken by microscope (OLYMPUS, Japan). The signal intensity was measured using ImageJ software (National Institutes of Health).
Immunofluorescence
Immunofluorescence was performed as previously described.30 Briefly, cells were fixed with 4% paraformaldehyde, permeabilization with 0.5% Triton X-100. Then, the cells were sequentially incubated with primary antibody for PAI-1 (Abcam) and fluorescent secondary antibody. The nuclei were counterstained with DAPI. Images were taken by fluorescent microscope (OLYMPUS, Japan). The signal intensity was measured using ImageJ software (National Institutes of Health).
ELISA
Levels of total PAI-1 and active PAI-1 in the plasma were determined using commercial ELISA kits (Total PAI-1 kit from Elabscience, China and active PAI-1 kit from Molecular Innovations, Inc).
Statistical Analysis
All statistical analyses were performed at least 3 independent biological or experimental replicates (for cell n≥3, for mice ≥6). Statistical tests were carried out using GraphPad Prism software (version 7.0). Data were tested for normality and equal variance. Data were normally distributed, parametric tests (2-tailed unpaired Student t tests) were used to determine statistical significance and presented as mean±SEM. For the data that were not normally distributed, nonparametric tests (the Mann-Whitney U test) were performed and shown as median±SD. Data are presented with a value of P<0.05 considered statistically significant.
Results
METTL3 Increases PAI-1 Expression and Suppresses Fibrinolysis In Vitro
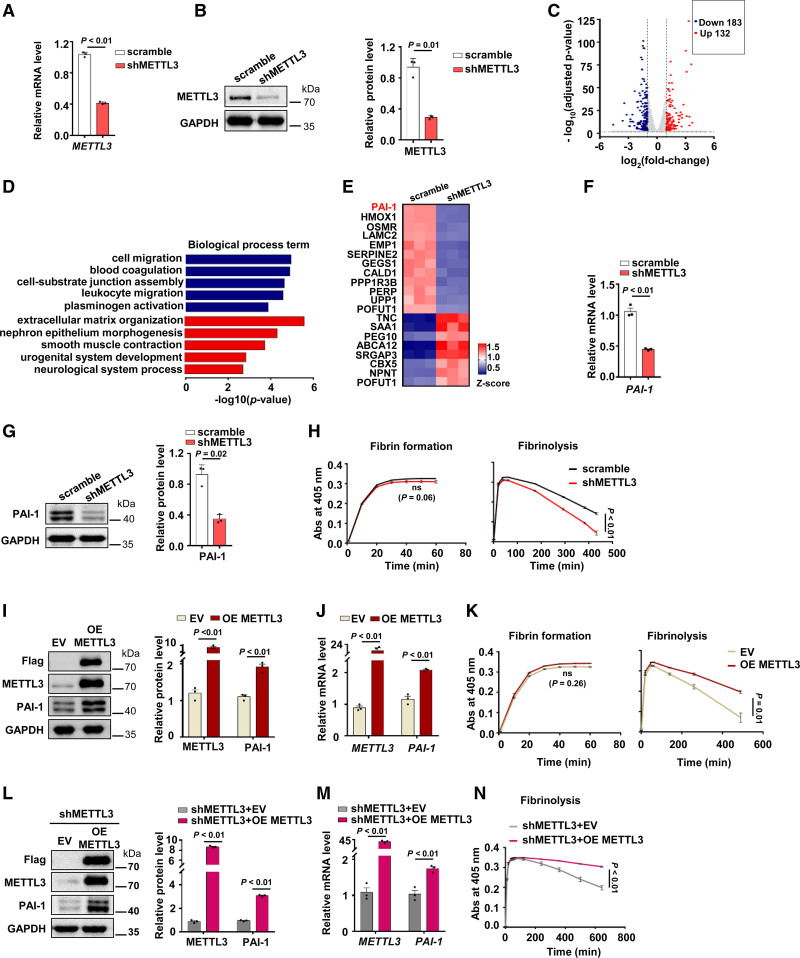
Since METTL3 plays critical role in lots of biological processes, we suspected that METTL3 could have additional functions in endothelial cells. We generated METTL3 stable knockdown HUVECs (Figure 1A and 1B) followed by RNA-seq analysis. We identified 315 differentially expressed genes (183 genes were downregulated, and 132 genes were upregulated; Figure 1C). Gene ontology enrichment analysis revealed that genes associated with cell migration and blood coagulation systems were downregulated (Figure 1D), whereas upregulated genes were enriched in matrix organization and epithelium morphogenesis (Figure 1D). Consistent with our gene ontology analysis, METTL3 promoted endothelial cell migration.19 Furthermore, blood coagulation system-related genes including ITGA2, SLC7A11, TFPI, and SERPINE 2 were reduced (Figure IA in the Data Supplement). PAI-1 was significantly decreased in METTL3 KD HUVECs (Figure 1E). Quantitative PCR (qPCR) (Figure 1F), Western blot (Figure 1G), and immunofluorescence (Figure IB in the Data Supplement) analysis showed that METTL3 knockdown impaired expression of PAI-1. PAI-1 is the principal inhibitor of tPA and u-PA and plays pivotal role in the regulation of fibrinolysis.31 Our data showed that METTL3 knockdown had little effect on tPA and u-PA expression (Figure IC in the Data Supplement) and elevated PAI-1 levels in endothelial cells impaired fibrinolytic ability (Figure ID through IF in the Data Supplement).
Figure 1.
METTL3 (methyltransferase-like protein 3) increases PAI-1 (plasminogen activator inhibitor-1) expression and suppresses fibrinolysis in vitro. A, Human umbilical vein endothelial cells (HUVECs) were infected with control (scrambled) lentivirus or METTL3 shRNA lentivirus, and the mRNA level of METTL3 was analyzed by qPCR (n=3, data are mean±SEM). B, The protein level of METTL3 in shMETTL3 HUVECs was measured by Western blot (n=3, data are mean±SEM). C, Volcano plots showed differentially expressed transcripts in shMETTL3 HUVECs vs control HUVECs, including 183 downregulated genes and 132 upregulated genes (fold change >2 and adjusted P<0.05). D, Top gene ontology (GO) biological process categories enriched in downregulated and upregulated genes (blue color represents downregulated genes and red color represents upregulated genes). E, Heat map of top 20 differentially expressed genes in shMETTL3 HUVECs. F and G, The mRNA level (F) and protein level of PAI-1 (G) were determined in control and shMETTL3 HUVECs (n=3, data are mean±SEM). H, Fibrin formation and fibrinolysis were shown in shMETTL3 HUVECs and control cells (n=3, data are mean±SEM). I and J, The expression of PAI-1 was detected by Western blot (I) and quantitative real-time PCR (qPCR) (J) in METTL3 overexpressed and control cells (empty vector: EV, n=3, data are mean±SEM). K, Fibrin formation and fibrinolysis were shown in METTL3 overexpressed and control cells (n=3, data are mean±SEM). L and M, The expression of METTL3 and PAI-1 in shMETTL3 HUVECs infected WT METTL3 (shMETTL3+OE METTL3) and control cells (shMETTL3+EV) were measured by western blot (L) and qPCR (M) (n=3, data are mean±SEM). N, Fibrinolysis was shown in shMETTL3 HUVECs infected WT METTL3 (shMETTL3+OE METTL3) and control lentivirus (shMETTL3+EV) (Abs indicates absorbance, n=3, data are mean±SEM).
To identify the role of METTL3 in fibrin formation and fibrinolysis, fibrin formation and fibrinolysis assays22,24,32 were carried out in shMETTL3 HUVECs and scramble HUVECs. As shown in Figure 1H, the final peak turbidities of clots and the time to reach the maximum turbidity were similar in shMETTL3 and scramble HUVECs, but knockdown of METTL3 enhanced fibrinolysis compared with the scramble HUVECs (Figure 1H).
In addition, we upregulated METTL3 expression via the gain-of-function analysis. Overexpression of METTL3 resulted in upregulation of PAI-1 in HUVECs (Figure 1I and 1J; Figure IG in the Data Supplement) and inhibited fibrinolysis (Figure 1K). Moreover, overexpression of METTL3 in shMETTL3 HUVECs also restored the gene expression of PAI-1(Figure 1L and 1M; Figure IH and II in the Data Supplement) and impaired fibrinolysis (Figure 1N) in METTL3 knockdown HUVECs. Therefore, these results showed that METTL3 could promote PAI-1 expression and suppress fibrinolysis in vitro.
METTL3 Regulates m6A Level of JUN mRNA and Promotes JUN Protein Expression
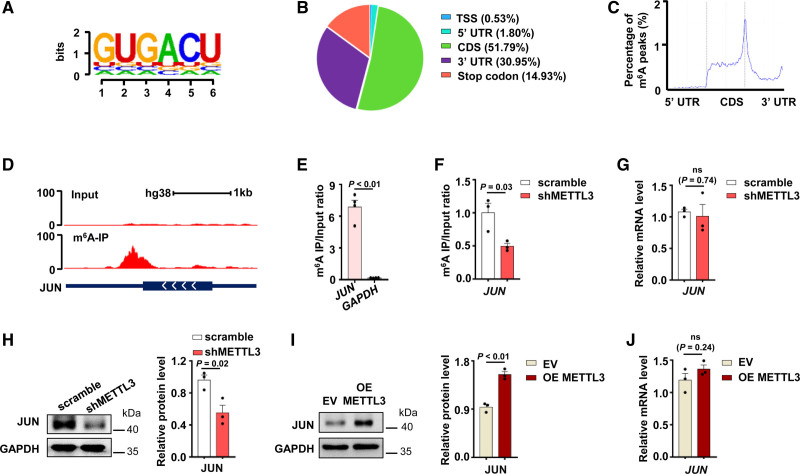
To investigate the potential mechanism of METTL3 regulating the expression of PAI-1, we carried out MeRIP-seq in HUVECs. MeRIP-seq analysis revealed that m6A peaks in HUVECs significantly enriched in the RRACH motif (Figure 2A) and predominantly localized coding sequences (CDS), 3′ untranslated regions (3′ UTRs), and stop codons (Figure 2B and 2C). mRNAs of many genes (59.82%) contain a single m6A peak (Figure IIA in the Data Supplement). Overall, the results of our m6A MeRIP-seq analysis were consistent with the published m6A features,33,34 which indicated that we successfully performed m6A MeRIP-seq and analysis in HUVECs. m6A track of individual transcripts showed that hypomethylation sites in PAI-1 mRNA (Figure IIB in the Data Supplement) but m6A peaks were enriched near the proximal 3′ UTR of JUN mRNA (Figure 2D). m6A enrichment in 3′ UTR of JUN mRNA was validated by MeRIP-qPCR assay in HUVECs (Figure 2E). Colorimetric quantification assay revealed that METTL3 silencing decreased the global m6A modification level (Figure IIC in the Data Supplement).
Figure 2.
METTL3 (methyltransferase-like 3) regulates N6-methyladenosine (m6A) level of JUN mRNA and promotes protein expression. A, Sequence motif identified m6A peaks in human umbilical vein endothelial cells (HUVECs). The total height of the letters depicts the information content of the position, in bits. B, Pie chart depicted the fraction of m6A peaks distribution in 5 transcript segments. C, Metagene profiles of m6A peak distribution. Each transcript was divided into 3 rescaled nonoverlapping segments: 5′ UTR, CDS, and 3′ UTR. D, The abundance of m6A in JUN mRNA was analyzed by m6A-seq. E, m6A modification of JUN mRNA in HUVECs was detected by m6A-RIP-qPCR (n=4, data are mean±SEM). F, m6A level of JUN mRNA in shMETTL3 HUVECs and control cells was measured by m6A-RIP-qPCR (n=3, data are mean±SEM). G, Quantitative real-time PCR (qPCR) analysis of JUN mRNA expression in shMETTL3 HUVECs and control cells (n=3, data are mean±SEM). H and I, JUN protein levels in shMETTL3 HUVECs (H) and METTL3 overexpressed HUVECs (I) were shown by Western blot (n=3, data are mean±SEM). J, The expression of JUN mRNA in METTL3 overexpressed HUVECs and control cells was detected by qPCR (Empty vector: EV, n=3, data are mean±SEM).
To verify m6A-modified JUN mRNA is regulated by METTL3, we performed MeRIP-qPCR in shMETTL3 and scramble HUVECs. The results showed that knockdown of METTL3 reduced the m6A modification of JUN mRNA 3′ UTR (Figure 2F). This evidence suggested that JUN mRNA is the potential target of METTL3. Then, we examined whether METTL3 regulated RNA and protein expression of JUN in HUVECs. Intriguingly, METTL3 knockdown had little effect on the abundance of JUN mRNA (Figure 2G) but reduced protein levels of JUN (Figure 2H). METTL3 overexpression increased JUN protein levels (Figure 2I) without changing JUN mRNA levels (Figure 2J). Moreover, overexpression of METTL3 in shMETTL3 HUVECs restored JUN protein levels (Figure IID in the Data Supplement). JUN regulates gene expression by interacting with specific target DNA sequences.35 We analyzed METTL3 knockdown HUVECs RNA-seq data combining PROMO36 and found that >30% of the top 50 differential expression genes in METTL3 knockdown HUVECs were regulated by JUN (Figure IIE and IIF in the Data Supplement).
METTL3 Regulates Fibrinolysis In Vitro via JUN-PAI-1 Axis
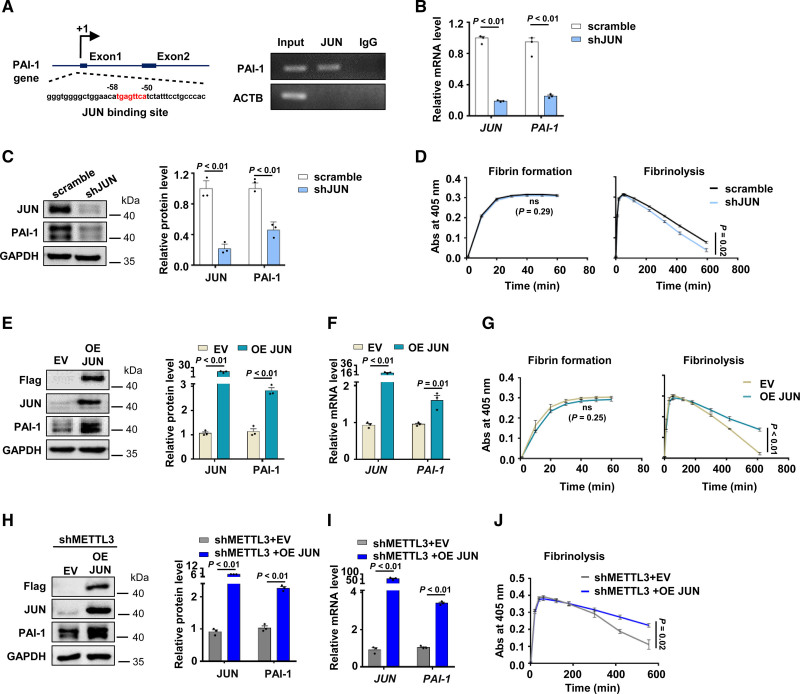
JUN-mediated PAI-1 upregulation has been reported in PMA-induced HepG2 cells.37 The PAI-1 promoter contains a consensus JUN-binding site38 (Figure 3A). Utilization of a chromatin immunoprecipitation demonstrated that JUN bound to the PAI-1 promoter area in HUVECs (Figure 3A). Thus, we hypothesized that loss of JUN downregulated gene expression of PAI-1. Indeed, knockdown of JUN significantly reduced PAI-1 expression (Figure 3B). Consistent with qPCR results, the protein expression of PAI-1 decreased upon JUN knockdown (Figure 3C, Figure IIIA in the Data Supplement). JUN depletion promoted fibrinolysis in vitro (Figure 3D). Alternatively, overexpression of JUN increased PAI-1 expression both at protein (Figure 3E, Figure IIIB in the Data Supplement) and mRNA levels (Figure 3F). The ability of fibrin degradation was decreased in JUN overexpressed HUVECs (Figure 3G).
Figure 3.
METTL3 (methyltransferase-like protein 3) regulates fibrinolysis in vitro via Jun proto-oncogene (JUN)-PAI-1 (plasminogen activator inhibitor-1) axis. A, Schematic diagram showed the JUN-binding position in PAI-1 promoter (left) and JUN binding to PAI-1 promoter was determined by ChIP-PCR (right). ACTB (β-actin) was used as a negative control to show immunoprecipitation specificity. Input: sheared chromatin was prepared before immunoprecipitation and used as a positive control for PCR. B and C, The expression of JUN and PAI-1 in shJUN HUVECs were detected by quantitative real-time PCR (qPCR; B) and Western blot (C) (n=3, data are median±SD). D, Fibrin formation and fibrinolysis were shown in shJUN HUVECs and control cells (n=3, data are mean±SEM). E and F, The expression of JUN and PAI-1 in JUN overexpressed HUVECs and control cells were analyzed by Western blot (E) and qPCR (F) (empty vector: EV, n=3, data are mean±SEM). G, Fibrin formation and fibrinolysis were shown in JUN overexpressed and control cells (n=3, data are mean±SEM). H and I, The expression of METTL3 and PAI-1 in shMETTL3 HUVECs infected with WT JUN (shMETTL3+OE JUN) and control lentivirus (shMETTL3+EV) were measured by Western blot (H) and qPCR (I) (n=3, data are mean±SEM). J, Fibrinolysis was shown in shMETTL3 HUVECs infected WT JUN (shMETTL3+OE JUN) and control lentivirus (shMETTL3+EV) (n=3, data are mean±SEM).
To further validate whether METTL3 regulates PAI-1 expression and fibrinolysis by targeting JUN, we performed the rescue experiment and found that overexpression of JUN increased protein and mRNA levels of PAI-1 in shMETTL3 HUVECs (Figure 3H and 3I; Figure IIIC and IIID in the Data Supplement). Fibrin polymerization assays showed that overexpression of JUN could inhibit fibrinolysis in METTL3-deleted HUVECs (Figure 3J). Taken together, these results revealed that METTL3 inhibited fibrinolysis by upregulating JUN-PAI-1 axis.
METTL3 Enhances the Translation of JUN Through YTHDF1
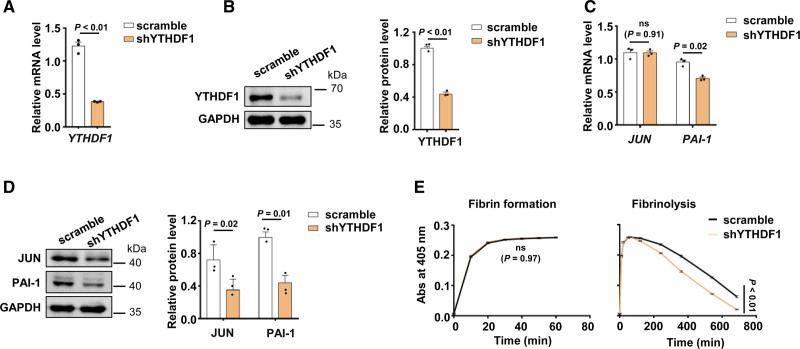
We explored the regulatory mechanism for how METTL3-mediated m6A modification regulates the translation of JUN. As previously reported, m6A modification was discriminatively bonded by specific m6A reader proteins to influence RNA fate and cell biological function.39 The YTHDF domain family proteins preferentially bind to m6A in mRNA.40 YTHDF1 enhances the translation of targeted m6A mRNA.41–43 Therefore, we used shRNA to repress YTHDF1 expression in HUVECs (Figure 4A and 4B). Knockdown of YTHDF1 decreased JUN protein expression without JUN mRNA level but reduced PAI-1 level (Figure 4C and 4D, Figure IVA in the Data Supplement). Knockdown of YTHDF1 could promote fibrinolysis (Figure 4E). In addition, knockdown of YTHDF2 had no significant effect on JUN and PAI-1 mRNA in HUVECs (Figure IVB in the Data Supplement). Taken together, our results indicated that METTL3 regulated JUN protein expression and PAI-1 expression by modulating translation in an m6A-YTHDF1-dependent manner.
Figure 4.
METTL3 (methyltransferase-like protein 3) enhances the translation of Jun proto-oncogene (JUN) through YTHDF1 (YTH-domain family protein 1). A and B, The decreased expression of YTHDF1 in shYTHDF1 HUVECs was confirmed by quantitative real-time PCR (qPCR; A) and Western blot (B) (n=3, data are median±SD). C and D, The expression of JUN and PAI-1 (quantitative real-time PCR) in shYTHDF1 HUVECs were measured by qPCR (C) and Western blot (D) (n=3, data are median±SD). E, Fibrin formation and fibrinolysis were shown in shYTHDF1 HUVECs and control cells (n=3, data are mean±SEM).
METTL3 Promotes Fibrin Deposition in Endotoxin-Treated Mice
Since the expression of PAI-1 is increased in lipopolysaccharide-stimulated HUVECs,44,45 we investigated whether elevated expression of PAI-1 is associated with METTL3 and JUN. We treated HUVECs with lipopolysaccharide and found that the expression of METTL3 (Figure VA and VB in the Data Supplement) and JUN (Figure VC and VD in the Data Supplement) were increased in lipopolysaccharide-stimulated HUVECs. Furthermore, we stimulated shMETTL3 HUVECs with lipopolysaccharide and found that knockdown of METTL3 prevented lipopolysaccharide-induced expression of PAI-1 in HUVECs (Figure VE and Figure VF in the Data Supplement). These results confirmed that METTL3 was upregulated and promoted PAI-1 expression in lipopolysaccharide-treated HUVECs.
Next, we explored whether METTL3 is associated with the upregulation of PAI-1 in lipopolysaccharide-treated mice. We treated wild-type (WT) mice with lipopolysaccharide and observed that fibrin (Figure VIA and VIB in the Data Supplement) and microthrombi (Figure VIC in the Data Supplement) were increased in the livers and lungs of endotoxemia mice. Consistent with the results described in previous reports,46,47 elevated plasma total PAI-1 and active PAI-1 levels were increased in lipopolysaccharide-treated mice (Figure VID in the Data Supplement). Subsequently, we assessed the expression of METTL3/m6A/JUN/ PAI-1 axis in the vasculature of lipopolysaccharide-treated mice. The relative levels of m6A modification were increased in vein and artery (Figure VIE in the Data Supplement). We found that expression of METTL3/JUN/PAI-1 in endotoxemia mice also increased in the vein (Figure VIF and VIG in the Data Supplement) and artery (Figure VIH and VII in the Data Supplement). Overall, these data indicated that elevated METTL3 is associated with increased fibrin deposition in endotoxin-treated mice.
To further elucidate the role of endothelium-resident METTL3 in fibrin deposition of endotoxin-treated mice. Mettl3- floxed mice were crossed with Tie2-Cre mice48 to obtain Mettl3fl/wt Tie2-Cre (+/−) mice (referred to as Mettl3 KD hereafter) and Mettl3fl/wt Tie2-Cre (−/−) mice (referred to as Mettl3 Con hereafter). Western blot confirmed the protein expression of METTL3 was decreased in liver and lung endothelial cells isolated from Mettl3 KD mice (Figure VIIA in the Data Supplement). Furthermore, we found that the protein expression of JUN and PAI-1 were also downregulated in liver and lung endothelial cells of Mettl3 KD mice (Figure VIIB and VIIC in the Data Supplement). This observation indicates that Mettl3 can regulate PAI-1 expression in mouse endothelial cells.
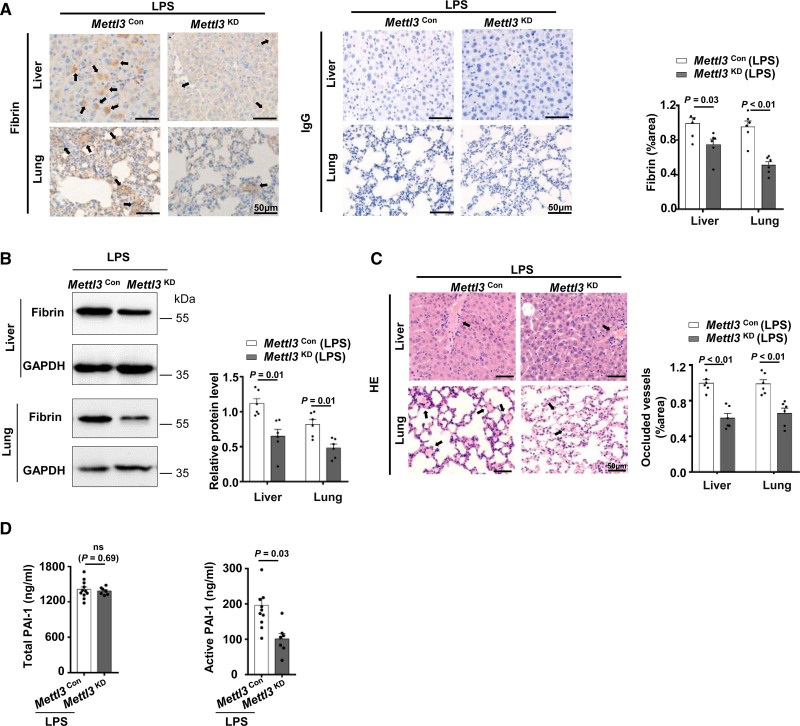
PAI-1 is the principal inhibitor of the fibrinolytic system. A prominent increase in PAI-1 results in an antifibrinolytic state in endotoxemia, further widespread fibrin deposition leads to organ dysfunction.49 We then tested whether endothelial METTL3 mediates lipopolysaccharide-induced fibrin deposition via regulating the expression of PAI-1. Immunohistochemistry and immunoblot analysis revealed that the Mettl3 KD mice alleviated fibrin deposition in livers and lungs during endotoxemia (Figure 5A and 5B). Hematoxylin and eosin staining of liver and lung sections showed that occlusion of vascular was reduced in Mettl3 KD mice compared with Mettl3 Con mice during endotoxemia (Figure 5C). The coagulation and fibrinolytic systems are highly regulated and inter-related through mechanisms that ensure balanced hemostasis.50 Therefore, we asked whether the impaired coagulation activation led to the decreased fibrin deposition in lipopolysaccharide-treated Mettl3 KD mice. However, we found the levels of thrombin antithrombin complexes in plasma of lipopolysaccharide challenged Mettl3 KD mice showed no significant change (Figure VIID in the Data Supplement). This suggested that reduced fibrin in the livers and lungs of Mettl3 KD endotoxemia mice was not due to impaired coagulation activation. Furthermore, ELISA demonstrated that plasma active PAI-1 levels, but not total PAI-1 levels, were decreased in lipopolysaccharide-treated Mettl3 KD mice (Figure 5D). PAI-1 exists in many forms in plasma, including active PAI-1, latent PAI-1, and PAI-1/tPA (u-PA) complex.51 PAI-1 active forms are mainly derived from endothelial cells and hepatocytes.52 Although platelets are a major source of PAI-1, only 3% to 5% of the PAI-1 antigen released from platelets was active.53,54 To further exclude the effect of platelets on the decreased active PAI-1 in Mettl3 KD endotoxemia mice, we used a hematology analyzer to count blood cells number in Mettl3 KD mice and observed no significant difference in platelet number between Mettl3 KD and Mettl3 Con mice (Table II in the Data Supplement). This result could explain the finding that knockdown of Mettl3 in endothelial cells impaired active PAI-1 production without affected total PAI-1 levels in lipopolysaccharide-treated Mettl3 KD mice. In summary, these findings revealed that the absence of endothelial METTL3 decreased PAI-1 expression and alleviated fibrin deposition in endotoxin-treated mice.
Figure 5.
METTL3 (methyltransferase-like protein 3) promotes fibrin deposition in endotoxin-treated mice. A and B, Fibrin in livers and lungs of LPS-treated Mettl3 Con (n=6, 3 males and 3 females) and Mettl3 KD mice (n=6, 3 males and 3 females) was detected by immunohistochemistry (A) and Western blot (B). Data are mean±SEM and the black arrow indicates fibrin. C, Thrombus in livers and lungs of LPS-treated Mettl3 Con and Mettl3 KD mice (n=6, 3 males and 3 females) was confirmed by HE-staining. Data are mean±SEM, and the black arrow indicates thrombus. D, The levels of total and active PAI-1 in Mettl3 Con (n=11, 6 males and 5 females) and Mettl3 KD mice (n=7, 4 males and 3 females) with stimulation of LPS were detected by ELISA, data are mean±SEM.
Discussion
METTL3-mediated mRNA m6A modification plays pivotal roles in several biological processes including embryonic development14 and embryonic neural stem cell self-renew.55 METTL3 also binds chromatin to regulate heterochromatin in mouse embryonic stem cells.56 METTL3-mediated m6A modification determines cell fate during embryogenesis.57 Endothelial-specific METTL3-mediated m6A regulates hematopoietic stem cell development through Notch signaling18 and angiogenesis via Wnt signaling.19 In this study, we reported that the knockdown of METTL3 in endothelial cells promoted fibrinolysis by decreasing the expression of PAI-1. Furthermore, MeRIP-seq showed that hypomethylation sites in PAI-1 mRNA and m6A peaks significantly enriched in JUN mRNA. KIAA1429, recruiter and guider of methyltransferase complex, regulated JUN mRNA stability in an m6A-independent manner.58 Here, we revealed that knockdown of METTL3 decreased the m6A level of JUN mRNA and reduced JUN protein level but had little effect on JUN mRNA. Our study suggested that METTL3 regulated JUN translation in an m6A-dependent manner. In further investigations, we confirmed that METTL3-m6A-JUN regulated PAI-1 transcription in HUVECs.
mRNA m6A modification is recognized by many m6A reader proteins. YT521-B homology (YTH) domain-containing proteins are the extensively studied readers including YTHDF1-3 and YTHDC2.59 YTHDF1 increases the translation efficiency of targeted m6A mRNA by interacting with translation initiation machinery and ribosome.41 YTHDF2 recruits CCR4-NOT deadenylation machinery to its targets resulting in a decrease in RNA stability and RNA degradation.60,61 YTHDF3 encourages protein synthesis interacting with YTHDF1 and affects RNA decay via YTHDF2.62 YTHDC2 promotes the translation efficiency and also decreases mRNA enrichment.63 Besides, other reader proteins also can recognize m6A sites, including the IGF2BP1-3 (insulin-like growth factor 2 mRNA-binding protein family),11 HNRNP (heterogeneous nuclear ribonucleoprotein) protein family,64 PPRC2A (proline-rich coiled-coil 2A),65 and eIF3 (eukaryotic initiation factor3).66 Knockdown of METTL3 had little effect on JUN mRNA abundance but reduced JUN protein level; therefore, we focused on YTHDF1 that regulates translation. However, whether other reader proteins binding to m6A modified transcripts in HUVECs needs further investigation.
High PAI-1 concentrations are indeed associated with various thrombotic disorders.67 Sepsis, defined as organ dysfunction resulting from a dysregulated host response to infection, is accompanied by coagulation dysfunction. Upregulated PAI-1 expression leads to disseminated intravascular coagulation, circulatory hypoperfusion, and organ dysfunction in septic patients. And PAI-1 is a significant predictor of disease severity and all-cause mortality in sepsis.68 Here, we found that METTL3-m6A-JUN-PAI-1 increased in the blood vessel of lipopolysaccharide-treated WT mice. Furthermore, we used Tie2-Cre mice to obtain endothelial knockdown of Mettl3 mice. Tie2-Cre mouse models exhibit various extent of Cre activity in the hematopoietic lineage.69 For example, >85% circulating blood cells show Cre activity in Tg (Tek-cre)12Flv mice.70 Using Tg (Tek-cre)12Flv allele mice to knockdown Mettl3 may lead to a decrease in the number of monocytes due to METTL3 affects hematopoietic cell differentiation.15 Monocytes are the primary hematopoietic cells that express TF in endotoxemia mice.71 Therefore, TF levels from monocytes maybe decreased, which lead to less fibrin deposition in endothelial Mettl3 knock down mice generated by Tg (Tek-cre)12Flv allele. However, we used the Tg (Tek-cre)1Ywa/J allele to generate Mettl3 KD mice in our study. And we found that no significant changes in the circulating blood cell count in Mettl3 KD and Mettl3 Con mice generated by Tg (Tek-cre)1Ywa/J allele. Consistent with previous reports that with the Tg (Tek-cre)1Ywa/J allele, only a little circulating cell show Cre activity in the adult mouse.48,69
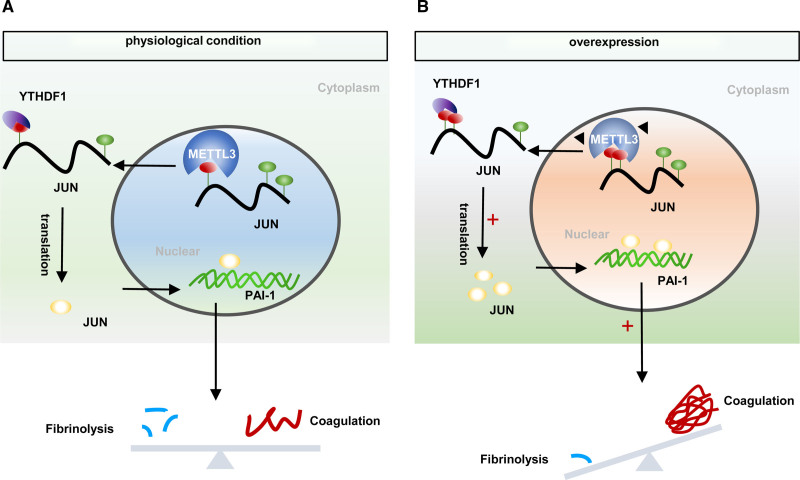
In the lipopolysaccharide-induced septic mouse model, PAI-1 deficiency could ameliorate fibrin deposition.72 Our study revealed that knockdown of endothelial METTL3 decreased plasma active PAI-1 levels and alleviated lipopolysaccharide-induced fibrin deposition. Fibrinolytic system is regulated by various cofactors, receptors, and inhibitors.50 We found that plasma thrombin antithrombin complex levels remain unchanged in endotoxemia Mettl3 KD mice. However, whether other molecules might regulate fibrin deposition in lipopolysaccharide-treated Mettl3 KD mice require further investigation. In summary, our results showed knockdown of METTL3 in HUVECs decreased PAI-1 expression and promoted fibrinolysis in vitro. Moreover, METTL3 regulated JUN/PAI-1 expression in an m6A-YTHDF1-dependent manner (Figure 6). Endothelial cell-specific knockdown of Mettl3 alleviated fibrin deposition in lipopolysaccharide-induced endotoxemia mice by reducing the levels of active PAI-1, which might provide novel insight into the treatment of thrombotic disease.
Figure 6.
Model of METTL3 (methyltransferase-like protein 3) promotes fibrin deposition by increasing the expression of PAI-1 (plasminogen activator inhibitor-1). A, In nuclear, the N6-methyladenosine (m6A) methyltransferase METTL3 mediates m6A methylation at JUN mRNA. After exporting JUN to cytoplasm, YTHDF1 (YTH-domain family protein 1) recognizes m6A-modified mRNA enhanced translation. Further, the transcription factor JUN binds to PAI-1 DNA promoter and initiates its transcription. B, Overexpression of METTL3 increases the methylated modification and translation of JUN mRNA, further facilitating the transcriptional effect of JUN on PAI-1. The increased expression of PAI-1 strengthens its inhibitory effect on fibrinolysis, which ultimately results in fibrin deposition and breaks the hemostatic balance.
Article Information
Acknowledgments
We thank Dr Minghan Tong (State Key Laboratory of Molecular Biology, Shanghai Key Laboratory of Molecular Andrology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, China) for providing Mettl3 floxed mice.
Sources of Funding
This work was supported by the National Natural Science Foundation of China (Grant numbers 81971815 to A. Wen) and The Talent Project of Army Medical University (Grant numbers D-3149 to H. Wu).
Disclosures
None.
Supplemental Materials
Data Supplement Major Resources
Data Supplement Figures I–VII
Data Supplement Tables I and II
Supplementary Material
Nonstandard Abbreviations and Acronyms
- eIF3
- eukaryotic initiation factor3
- HNRNP
- heterogeneous nuclear ribonucleoprotein
- HUVEC
- human umbilical vein endothelial cell
- IGF2BP1-3
- insulin-like growth factor 2 mRNA-binding protein family
- JUN
- jun proto-oncogene
- m6A
- N6-methyladenosine
- METTL3
- methyltransferase-like protein 3
- PAI-1
- plasminogen activator inhibitor-1
- PPRC2A
- proline-rich coiled-coil 2A
- qPCR
- quantitative real-time PCR
- TFPI
- tissue factor pathway inhibitor
- tPA
- tissue-type plasminogen activator
- u-PA
- urokinase plasminogen activator
- vWF
- von Willebrand factor
- WT
- wild-type
- YTHDF1
- YTH-domain family protein 1
The Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.121.316414.
For Sources of Funding and Disclosures, see page 2887 and 2888.
Contributor Information
Qin Bai, Email: QinBai123@outlook.com.
Yao Lu, Email: 381127011@qq.com.
Yanhua Chen, Email: 409747351@qq.com.
Han Zhang, Email: zhanghan5650@126.com.
Weiwei Zhang, Email: 363349457@qq.com.
Huang Wu, Email: wuhuang2019@126.com.
References
- 1.Favero G, Paganelli C, Buffoli B, Rodella LF, Rezzani R. Endothelium and its alterations in cardiovascular diseases: life style intervention. Biomed Res Int. 2014;2014:801896. doi: 10.1155/2014/801896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madoiwa S. Recent advances in disseminated intravascular coagulation: endothelial cells and fibrinolysis in sepsis-induced DIC. J Intensive Care. 2015;3:8. doi: 10.1186/s40560-015-0075-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collen D, Lijnen HR. The tissue-type plasminogen activator story. Arterioscler Thromb Vasc Biol. 2009;29:1151–1155. doi: 10.1161/ATVBAHA.108.179655 [DOI] [PubMed] [Google Scholar]
- 4.Lin Z, Kumar A, SenBanerjee S, Staniszewski K, Parmar K, Vaughan DE, Gimbrone MA, Jr, Balasubramanian V, García-Cardeña G, Jain MK. Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circ Res. 2005;96:e48–e57. doi: 10.1161/01.RES.0000159707.05637.a1 [DOI] [PubMed] [Google Scholar]
- 5.Zhou G, Hamik A, Nayak L, Tian H, Shi H, Lu Y, Sharma N, Liao X, Hale A, Boerboom L, et al. Endothelial Kruppel-like factor 4 protects against atherothrombosis in mice. J Clin Invest. 2012;122:4727–4731. doi: 10.1172/JCI66056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiang Y, Cheng J, Wang D, Hu X, Xie Y, Stitham J, Atteya G, Du J, Tang WH, Lee SH, et al. Hyperglycemia repression of miR-24 coordinately upregulates endothelial cell expression and secretion of von Willebrand factor. Blood. 2015;125:3377–3387. doi: 10.1182/blood-2015-01-620278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali HO, Arroyo AB, González-Conejero R, Stavik B, Iversen N, Sandset PM, Martínez C, Skretting G. The role of microRNA-27a/b and microRNA-494 in estrogen-mediated downregulation of tissue factor pathway inhibitor α. J Thromb Haemost. 2016;14:1226–1237. doi: 10.1111/jth.13321 [DOI] [PubMed] [Google Scholar]
- 8.Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014;11:669–672. doi: 10.4161/rna.28829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- 15.Lee H, Bao S, Qian Y, Geula S, Leslie J, Zhang C, Hanna JH, Ding L. Stage-specific requirement for Mettl3-dependent m6A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol. 2019;21:700–709. doi: 10.1038/s41556-019-0318-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q, Zhang KJ, Zhang X, Zhou Y, Zhang T, et al. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017;27:1216–1230. doi: 10.1038/cr.2017.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang CX, Cui GS, Liu X, Xu K, Wang M, Zhang XX, Jiang LY, Li A, Yang Y, Lai WY, et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16:e2004880. doi: 10.1371/journal.pbio.2004880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv J, Zhang Y, Gao S, Zhang C, Chen Y, Li W, Yang YG, Zhou Q, Liu F. Endothelial-specific m6A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res. 2018;28:249–252. doi: 10.1038/cr.2017.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao MD, Jiang Q, Ma Y, Liu C, Zhu CY, Sun YN, Shan K, Ge HM, Zhang QY, Zhang HY, et al. Role of METTL3-Dependent N6-Methyladenosine mRNA modification in the promotion of angiogenesis. Mol Ther. 2020;28:2191–2202. doi: 10.1016/j.ymthe.2020.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugin J, Schürer-Maly CC, Leturcq D, Moriarty A, Ulevitch RJ, Tobias PS. Lipopolysaccharide activation of human endothelial and epithelial cells is mediated by lipopolysaccharide-binding protein and soluble CD14. Proc Natl Acad Sci USA. 1993;90:2744–2748. doi: 10.1073/pnas.90.7.2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Read MA, Cordle SR, Veach RA, Carlisle CD, Hawiger J. Cell-free pool of CD14 mediates activation of transcription factor NF-kappa B by lipopolysaccharide in human endothelial cells. Proc Natl Acad Sci USA. 1993;90:9887–9891. doi: 10.1073/pnas.90.21.9887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell RA, Overmyer KA, Selzman CH, Sheridan BC, Wolberg AS. Contributions of extravascular and intravascular cells to fibrin network formation, structure, and stability. Blood. 2009;114:4886–4896. doi: 10.1182/blood-2009-06-228940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puy C, Ngo ATP, Pang J, Keshari RS, Hagen MW, Hinds MT, Gailani D, Gruber A, Lupu F, McCarty OJT. Endothelial PAI-1 (Plasminogen Activator Inhibitor-1) Blocks the Intrinsic Pathway of Coagulation, Inducing the Clearance and Degradation of FXIa (Activated Factor XI). Arterioscler Thromb Vasc Biol. 2019;39:1390–1401. doi: 10.1161/ATVBAHA.119.312619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wileman SM, Booth NA, Moore N, Redmill B, Forrester JV, Knott RM. Regulation of plasminogen activation by TGF-beta in cultured human retinal endothelial cells. Br J Ophthalmol. 2000;84:417–422. doi: 10.1136/bjo.84.4.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng Y, Wang S, Gao S, Soares F, Ahmed M, Guo H, Wang M, Hua JT, Guan J, Moran MF, et al. Refined RIP-seq protocol for epitranscriptome analysis with low input materials. PLoS Biol. 2018;16:e2006092. doi: 10.1371/journal.pbio.2006092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xing YH, Yao RW, Zhang Y, Guo CJ, Jiang S, Xu G, Dong R, Yang L, Chen LL. SLERT Regulates DDX21 rings associated with Pol I transcription. Cell. 2017;169:664–678.e16. doi: 10.1016/j.cell.2017.04.011 [DOI] [PubMed] [Google Scholar]
- 27.Tabrizi P, Wang L, Seeds N, McComb JG, Yamada S, Griffin JH, Carmeliet P, Weiss MH, Zlokovic BV. Tissue plasminogen activator (tPA) deficiency exacerbates cerebrovascular fibrin deposition and brain injury in a murine stroke model: studies in tPA-deficient mice and wild-type mice on a matched genetic background. Arterioscler Thromb Vasc Biol. 1999;19:2801–2806. doi: 10.1161/01.atv.19.11.2801 [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Niu N, Xu S, Jin ZG. A simple protocol for isolating mouse lung endothelial cells. Sci Rep. 2019;9:1458. doi: 10.1038/s41598-018-37130-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vila Ellis L, Cain MP, Hutchison V, Flodby P, Crandall ED, Borok Z, Zhou B, Ostrin EJ, Wythe JD, Chen J. Epithelial vegfa specifies a distinct endothelial population in the mouse lung. Dev Cell. 2020;52:617–630.e6. doi: 10.1016/j.devcel.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H, Yin QF, Luo Z, Yao RW, Zheng CC, Zhang J, Xiang JF, Yang L, Chen LL. Unusual processing generates SPA LncRNAs that sequester multiple RNA binding proteins. Mol Cell. 2016;64:534–548. doi: 10.1016/j.molcel.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 31.Longstaff C, Kolev K. Basic mechanisms and regulation of fibrinolysis. J Thromb Haemost. 2015;13Suppl 1S98–105. doi: 10.1111/jth.12935 [DOI] [PubMed] [Google Scholar]
- 32.Valladolid C, Martinez-Vargas M, Sekhar N, Lam F, Brown C, Palzkill T, Tischer A, Auton M, Vijayan KV, Rumbaut RE, et al. Modulating the rate of fibrin formation and clot structure attenuates microvascular thrombosis in systemic inflammation. Blood Adv. 2020;4:1340–1349. doi: 10.1182/bloodadvances.2020001500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 34.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohmann D, Bos TJ, Admon A, Nishimura T, Vogt PK, Tjian R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987;238:1386–1392. doi: 10.1126/science.2825349 [DOI] [PubMed] [Google Scholar]
- 36.Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333 [DOI] [PubMed] [Google Scholar]
- 37.Arts J, Grimbergen J, Bosma PJ, Rahmsdorf HJ, Kooistra T. Role of c-Jun and proximal phorbol 12-myristate-13-acetate-(PMA)-responsive elements in the regulation of basal and PMA-stimulated plasminogen-activator inhibitor-1 gene expression in HepG2. Eur J Biochem. 1996;241:393–402. doi: 10.1111/j.1432-1033.1996.00393.x [DOI] [PubMed] [Google Scholar]
- 38.Olman MA, Hagood JS, Simmons WL, Fuller GM, Vinson C, White KE. Fibrin fragment induction of plasminogen activator inhibitor transcription is mediated by activator protein-1 through a highly conserved element. Blood. 1999;94:2029–2038. [PubMed] [Google Scholar]
- 39.Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, Li J, Hao P, Zhang Y, Zhang F, et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249–253. doi: 10.1038/s41586-018-0666-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schleef RR, Loskutoff DJ, Podor TJ. Immunoelectron microscopic localization of type 1 plasminogen activator inhibitor on the surface of activated endothelial cells. J Cell Biol. 1991;113:1413–1423. doi: 10.1083/jcb.113.6.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louise CB, Obrig TG. Human renal microvascular endothelial cells as a potential target in the development of the hemolytic uremic syndrome as related to fibrinolysis factor expression, in vitro. Microvasc Res. 1994;47:377–387. doi: 10.1006/mvre.1994.1030 [DOI] [PubMed] [Google Scholar]
- 46.Numaguchi Y, Ishii M, Kubota R, Morita Y, Yamamoto K, Matsushita T, Okumura K, Murohara T. Ablation of angiotensin IV receptor attenuates hypofibrinolysis via PAI-1 downregulation and reduces occlusive arterial thrombosis. Arterioscler Thromb Vasc Biol. 2009;29:2102–2108. doi: 10.1161/ATVBAHA.109.195057 [DOI] [PubMed] [Google Scholar]
- 47.Yang X, Cheng X, Tang Y, Qiu X, Wang Z, Fu G, Wu J, Kang H, Wang J, Wang H, et al. The role of type 1 interferons in coagulation induced by gram-negative bacteria. Blood. 2020;135:1087–1100. doi: 10.1182/blood.2019002282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106 [DOI] [PubMed] [Google Scholar]
- 49.Gando S, Levi M, Toh CH. Disseminated intravascular coagulation. Nat Rev Dis Primers. 2016;2:16037. doi: 10.1038/nrdp.2016.37 [DOI] [PubMed] [Google Scholar]
- 50.Chapin JC, Hajjar KA. Fibrinolysis and the control of blood coagulation. Blood Rev. 2015;29:17–24. doi: 10.1016/j.blre.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iba T, Thachil J. Clinical significance of measuring plasminogen activator inhibitor-1 in sepsis. J Intensive Care. 2017;5:56. doi: 10.1186/s40560-017-0250-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yarmolinsky J, Bordin Barbieri N, Weinmann T, Ziegelmann PK, Duncan BB, Inês Schmidt M. Plasminogen activator inhibitor-1 and type 2 diabetes: a systematic review and meta-analysis of observational studies. Sci Rep. 2016;6:17714. doi: 10.1038/srep17714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Declerck PJ, Alessi MC, Verstreken M, Kruithof EK, Juhan-Vague I, Collen D. Measurement of plasminogen activator inhibitor 1 in biologic fluids with a murine monoclonal antibody-based enzyme-linked immunosorbent assay. Blood. 1988;71:220–225. [PubMed] [Google Scholar]
- 54.Booth NA, Simpson AJ, Croll A, Bennett B, MacGregor IR. Plasminogen activator inhibitor (PAI-1) in plasma and platelets. Br J Haematol. 1988;70:327–333. doi: 10.1111/j.1365-2141.1988.tb02490.x [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G, et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21:195–206. doi: 10.1038/s41593-017-0057-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu W, Li J, He C, Wen J, Ma H, Rong B, Diao J, Wang L, Wang J, Wu F, et al. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature. 2021;591:317–321. doi: 10.1038/s41586-021-03210-1 [DOI] [PubMed] [Google Scholar]
- 57.Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–276. doi: 10.1038/nature23883 [DOI] [PubMed] [Google Scholar]
- 58.Miao R, Dai CC, Mei L, Xu J, Sun SW, Xing YL, Wu LS, Wang MH, Wei JF. KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer. J Cell Physiol. 2020;235:7420–7432. doi: 10.1002/jcp.29645 [DOI] [PubMed] [Google Scholar]
- 59.Liao S, Sun H, Xu C. YTH Domain: A Family of N6-methyladenosine (m6A) Readers. Genomics Proteomics Bioinformatics. 2018;16:99–107. doi: 10.1016/j.gpb.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, Chen Y, Xiao Y, Gao Y, Zhang Q, et al. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41. doi: 10.1038/s41422-018-0113-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5’ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thögersen AM, Jansson JH, Boman K, Nilsson TK, Weinehall L, Huhtasaari F, Hallmans G. High plasminogen activator inhibitor and tissue plasminogen activator levels in plasma precede a first acute myocardial infarction in both men and women: evidence for the fibrinolytic system as an independent primary risk factor. Circulation. 1998;98:2241–2247. doi: 10.1161/01.cir.98.21.2241 [DOI] [PubMed] [Google Scholar]
- 68.Tipoe TL, Wu WKK, Chung L, Gong M, Dong M, Liu T, Roever L, Ho J, Wong MCS, Chan MTV, et al. Plasminogen Activator Inhibitor 1 for Predicting Sepsis Severity and Mortality Outcomes: A Systematic Review and Meta-Analysis. Front Immunol. 2018;9:1218. doi: 10.3389/fimmu.2018.01218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Payne S, De Val S, Neal A. Endothelial-specific cre mouse models. Arterioscler Thromb Vasc Biol. 2018;38:2550–2561. doi: 10.1161/ATVBAHA.118.309669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang Y, Harrington A, Yang X, Friesel RE, Liaw L. The contribution of the Tie2+ lineage to primitive and definitive hematopoietic cells. Genesis. 2010;48:563–567. doi: 10.1002/dvg.20654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pawlinski R, Pedersen B, Schabbauer G, Tencati M, Holscher T, Boisvert W, Andrade-Gordon P, Frank RD, Mackman N. Role of tissue factor and protease-activated receptors in a mouse model of endotoxemia. Blood. 2004;103:1342–1347. doi: 10.1182/blood-2003-09-3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poole LG, Massey VL, Siow DL, Torres-Gonzáles E, Warner NL, Luyendyk JP, Ritzenthaler JD, Roman J, Arteel GE. Plasminogen activator inhibitor-1 is critical in alcohol-enhanced acute lung injury in mice. Am J Respir Cell Mol Biol. 2017;57:315–323. doi: 10.1165/rcmb.2016-0184OC [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.