ABSTRACT
Toll-like receptors (TLRs) and interleukin-1 (IL-1) receptors regulate immune and inflammatory responses by activating the NF-κB pathway. Here, we report that B-cell‐specific loss of dynein light chain 1 (DYNLL1, LC8) or its designated transcription factor ASCIZ (ATMIN) leads to severely reduced in vivo antibody responses to TLR4-dependent but not T-cell‐dependent antigens in mice. This defect was independent of DYNLL1’s established roles in modulating BIM-dependent apoptosis and 53BP1-dependent antibody class-switch recombination. In B cells and fibroblasts, the ASCIZ-DYNLL1 axis was required for TLR4-, IL-1‐, and CD40-mediated NF-κB pathway activation but dispensable for antigen receptor and tumor necrosis factor α (TNF-α) signaling. In contrast to previous reports that overexpressed DYNLL1 directly inhibits the phosphorylation and degradation of the NF-κB inhibitor IκBα, we found here that under physiological conditions, DYNLL1 is required for signal-specific activation of the NF-κB pathway upstream of IκBα. Our data identify DYNLL1 as a signal-specific regulator of the NF-κB pathway and indicate that it may act as a universal modulator of TLR4 (and IL-1) signaling with wide-ranging roles in inflammation and immunity.
KEYWORDS: B-cell responses, BIM, DYNLL1, NF-κB, cell proliferation, immunization
INTRODUCTION
Antibody production by B cells is a defining feature of adaptive and innate immune responses (1). Antigen-specific antibody production is initiated by B-cell receptor (BCR)‐mediated activation of B lymphocytes in concert with signal-specific coactivators, including the CD40 ligand (CD40L)/CD40-mediated interaction with T cells and ligation of Toll-like receptors (TLRs) by innate immune modulators (2). A range of different TLRs are expressed at different stages of B-cell development and contribute to B-cell maturation, immunoglobulin gene (Ig) gene diversification, antibody production, and cytokine production (3, 4).
Although the different TLRs recognize a range of distinctive exogenous and endogenous signals, they—and the interleukin-1 (IL-1) receptor—all converge on MyD88 and associated signaling adapters and thereby lead to the activation of the NF-κB and mitogen-activated protein kinase (MAPK) pathways (4, 5). Activation of the NF-κB pathway by IL-1R/TLRs involves a complex signaling cascade, including the protein kinases IRAK1/4, TAK1, IKK, and ubiquitin (Ub) ligases TRAF6 and LUBAC (5). Ultimately, phosphorylation of the inhibitor of NF-κB (IκBα) by IKKβ (6) leads to its degradation and release of trapped NF-κB/Rel transcription factors into the nucleus to alter the expression of hundreds of target genes (7). In humans, mutations in TLR signaling components are associated with increased susceptibility to bacterial infections, autoimmune disorders, and B-cell lymphomas (3, 4, 8). In mice, the predominant TLR of B cells is TLR4, which plays a critical role in the innate immune response against Gram-negative bacteria by recognizing the sepsis-inducing bacterial endotoxin lipopolysaccharide (LPS) (3, 9).
DYNLL1 (also known as LC8) is a homodimeric sequence-specific chaperone that promotes the ordered (typically homo-) oligomerization of more than a hundred protein targets (10, 11). We recently reported that conditional deletion of Dynll1 or its designated transcription factor ASCIZ (also called ATMIN) during early B-cell development (via Mb1-Cre) leads to ∼10-fold reduced numbers of mature follicular B cells (B2 cells) and an >100-fold reduction of the initial pool of innate-like B-1a cells (12, 13). The B2 but not the B-1a cell developmental defects of ASCIZ- or DYNLL1-deficient mice could be suppressed by simultaneous deletion of the known DYNLL1 target BIM. In addition, DYNLL1 also contributes to the efficiency of immunoglobulin gene (Ig) class switch recombination (CSR) in mature B cells, where it modulates the oligomerization of the central DNA damage response protein 53BP1 (14).
DYNLL1 has previously been proposed to function as an inhibitor of the NF-κB pathway, based on findings that it can bind directly to the IκBα regulatory region in vitro (15) and block its phosphorylation by IKK and subsequent degradation required for NF-κB release in overexpression experiments (16). In contrast, we found here that DYNLL1 at physiological expression levels is essential for TLR4- and IL-1‐mediated activation of the NF-κB pathway in mouse B cells and fibroblasts. We show here that DYNLL1 acts upstream of IκBα by promoting the signal-specific activation of IKK and demonstrate that this is physiologically relevant for TLR4-mediated antibody responses in vivo.
RESULTS
DYNLL1 regulates B-cell activation in a signal-specific manner in vitro.
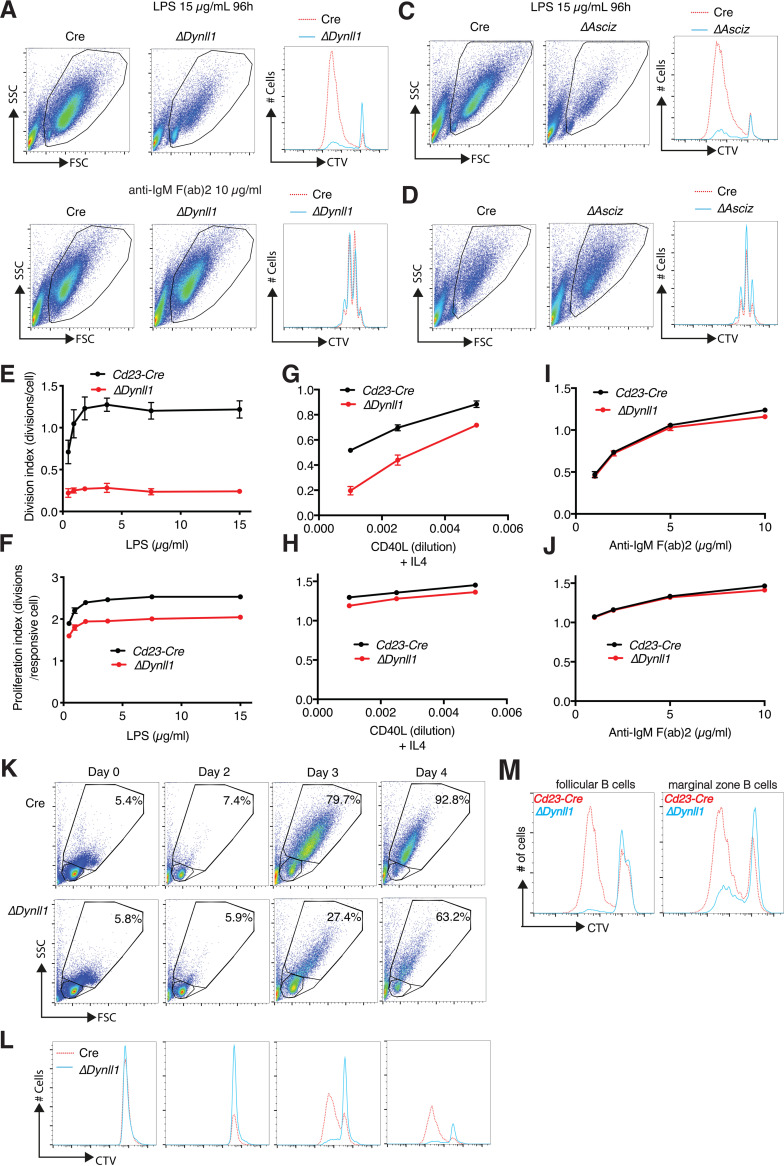
To investigate possible roles of DYNLL1 in mature B cells without confounding developmental defects, we used Cd23-Cre for its conditional deletion. This resulted in normal splenic B-cell numbers in Cd23-Cre Dynll1-deleted mice compared to those in Cd23-Cre controls (14). We then cultured purified B cells from these mice to assess their response to different activators in vitro. Stimulation with LPS led to the blasting of purified Cd23-Cre control B cells with a characteristic increase in cell size and extensive dilution of the proliferation-tracking dye CellTrace Violet (CTV) (Fig. 1A). In contrast, the emergence of larger lymphoblasts and CTV dilution was severely impaired in LPS-treated Cd23-Cre Dynll1-deleted B cells (Fig. 1A). Remarkably, in contrast to their severe activation defect in response to LPS, Dynll1-deleted B cells exhibited an entirely normal response to anti-IgM F(ab)2-mediated BCR activation (Fig. 1B). These results were independently confirmed with Cd23-Cre Asciz-deleted B cells (Fig. 1C and D), which also contain extremely low DYNLL1 levels (17).
FIG 1.
The ASCIZ-DYNLL1 axis is required for signal-specific B-cell activation. (A to D) Representative FACS plots and CellTrace Violet (CTV) dilution histograms of purified Cd23-Cre control, Cd23-Cre Dynll1-deleted, and Cd23-Cre Asciz-deleted splenic B cells after 4-day treatment with 15 μg/ml lipopolysaccharide (LPS) or 10 μg/ml anti-IgM F(ab)2 fragments as indicated. (E to J) Concentration-dependent responses of Cd23-Cre control and Cd23-Cre Dynll1-deleted B cells to LPS (E, F), CD40L (G, H), and BCR cross-linking (I, J) calculated from CTV FACS plots using FlowJo software. The cell division index (E, G, I) depicts the average number of divisions of all cells in the starting culture; the proliferation index (F, H, J) calculates the average number of divisions of cells that have divided at least once. Each data point represents the mean ± SEM of four independent biological replicates. (K) FACS plots of Cd23-Cre control and Cd23-Cre Dynll1-deleted splenic B cells treated for 0 to 4 days with 15 μg/ml LPS. The larger gate indicates the percentage of activated lymphoblasts, and the smaller gate represents lymphocytes. (L) CTV dilution profiles of the cultures (lymphocytes and lymphoblasts) shown above. (M) Comparison of CTV dilution profiles in FACS-purified follicular (CD23hi and CD21intermediate) and marginal zone (CD23low and CD21hi) B-cell fractions from Cd23-Cre control and Cd23-Cre Dynll1-deleted mice treated with 15 μg/ml LPS for 4 days. FSC, forward scatter; SSC, side scatter.
Severe defects of Dynll1-deleted B cells in response to LPS (Fig. 1E) but normal responses to BCR cross-linking (Fig. 1I) were consistently observed over a wide range of ligand concentrations. A mixed effect was observed in response to CD40L, with severe activation defects at low doses but progressively improved proliferation with increasing CD40L concentrations (Fig. 1G).
The normal response to BCR stimulation and the concentration-dependent improvement of the response to CD40L suggested that the in vitro proliferation defects of DYNLL1-deficient B cells more likely reflect LPS/CD40L-specific signaling defects than intrinsic cell cycle or cell viability defects. This notion was supported by modeling of the CTV dilution profiles using the FlowJo cell proliferation software package; calculation of the average division number of all cells in the culture indicated severe reductions in the cell division index of LPS- or CD40L-treated DYNLL1-deficient cells (Fig. 1E and G). However, these defects were notably attenuated when only cells that had divided at least once were taken into consideration to calculate the proliferation index of these cultures (Fig. 1F and H). In addition, time course analyses indicated that increase in B-cell volume, which precedes cell proliferation of the larger lymphoblasts after LPS stimulation (9), was considerably reduced in Cd23-Cre Dynll1-deleted cells compared to in Cd23-Cre control cells (Fig. 1K and L). Finally, similar activation defects were observed in both the follicular and marginal zone B-cell fractions (Fig. 1M). Collectively, these results indicate that DYNLL1 is particularly important for the initial signal-specific response of B cells to LPS and CD40L, but once activated, DYNLL1-deficient B cells expand in a relatively normal manner.
The ASCIZ-DYNLL1 axis is essential for TLR4-mediated antibody responses in vivo.
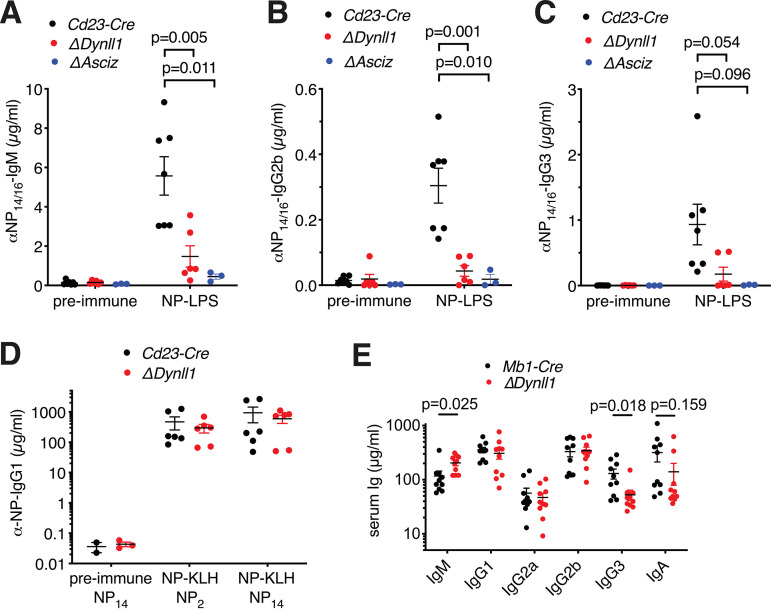
To determine whether the in vitro activation defects of ASCIZ- or DYNLL1-deficient B cells (Fig. 1) are physiologically relevant, we performed in vivo immunization experiments. In order to specifically determine the contribution of TLR4 signaling to the antibody response, we used LPS conjugated to the hapten 4-hydroxy-3-nitrophenyl-acetyl (NP) as a T-cell‐independent (TI-1) antigen to costimulate NP-specific B cells via their BCR and TLR4. In Cd23-Cre control mice, NP-LPS immunization led to >25-fold increased NP-specific IgM, IgG2b, and IgG3 antibody titers, but this response was severely attenuated in Cd23-Cre Dynll1- and Cd23-Cre Asciz-deleted mice (Fig. 2A through C). In contrast, in immunization experiments in which NP was conjugated to keyhole limpet hemocyanin as a T-cell‐dependent antigen, Cd23-Cre Dynll1-deleted mice had a largely normal antibody response (Fig. 2D), similar to what we have previously reported for Cd23-Cre Asciz-deleted mice (17). As the impaired TI-1 response includes unswitched NP-specific IgM titers (Fig. 2A), this defect is clearly independent of the recently reported role of DYNLL1 as an accessory factor in 53BP1-mediated antibody isotype switching (14). In this context, it should also be noted that conventional T-cell‐dependent antigen-specific IgG1 serum titers 4 weeks after in vivo immunization are largely maintained by selection and expansion of long-lived class-switched plasma cells (1), which would be expected to obscure the partial class-switch recombination defect of Asciz- or Dynll1-deleted B cells at much earlier time points in vitro (14).
FIG 2.
DYNLL1 is essential for TLR4-mediated T-cell-independent (TI-1) antibody responses but dispensable for T-cell-dependent antibody responses in vivo. (A to C) 4-Hydroxy-3-nitrophenyl-acetyl (NP)‐specific Ig titers before and 7 days after immunization with NP10-LPS (n = 3 to 7 mice per genotype). (D) Total (NP14) and high-affinity (NP2) antigen-specific IgG1 antibody titers (n = 6) 28 days after immunization with NP21-KLH. Preimmune sera depict separate mice. The results are from three separate immunization experiments. (E) Basal Ig titers (n = 10) of naive Mb1-Cre control and Mb1-Cre Dynll1-deleted mice at 8 weeks of age.
We also measured basal Ig titers in naive mice. Because basal antibodies are believed to be produced by the innate-like B-1a cell lineage that lacks CD23 expression, we used Mb1-Cre Dynll1-deleted mice for this purpose. For most Ig isotypes, titers were indistinguishable between groups (Fig. 2E). However, Mb1-Cre Dynll1-deleted mice contained significantly higher IgM and significantly lower IgG3 titers than Mb1-Cre control mice (Fig. 2E), with a trend toward lower IgA titers.
Altogether, these data demonstrate that ASCIZ and DYNLL1 are specifically required for TLR4-mediated but largely dispensable for T-cell‐dependent antibody responses in vivo. The severe defect of the antibody response to NP-LPS is quantitatively comparable to the TLR4-dependent in vitro activation defects of Dynll1- or Asciz-deleted B cells. Conversely, considering that CD40L responses of DYNLL1-deficient B cells progressively improve in a concentration-dependent manner in vitro, the normal T-cell‐dependent antibody response is consistent with the notion that local CD40L concentrations are high during the direct interactions between T and B cells in in vivo (18). Finally, given that TLR4 signaling is particularly important for CSR from IgM to IgG3 (19), the specific reduction of basal IgG3 titers in naive Mb1-Cre Dynll1-deleted mice further supports an in vivo function of DYNLL1 in TLR4 signaling in B lymphoid cells.
Differential rescue of DYNLL1-related in vitro activation but not in vivo immune defects by ablation of Bim.
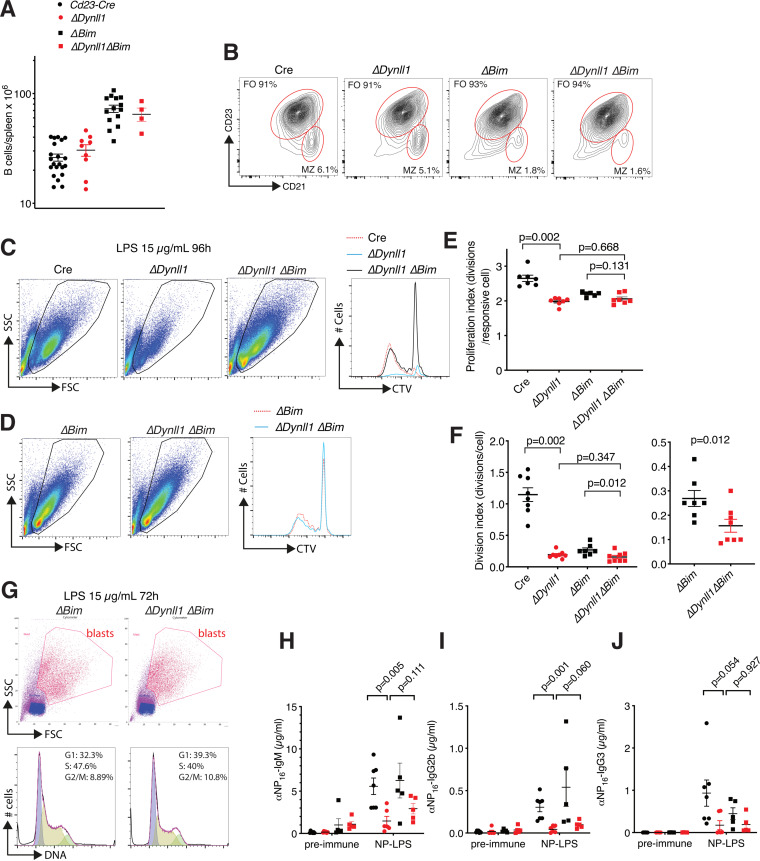
An important facet of the TLR4 response during B-cell activation is the activation of antiapoptotic mechanisms, including induction of antiapoptotic genes and posttranslational inactivation of the proapoptotic protein BIM (20). Based on the established roles of DYNLL1 in directly restraining BIM activity in vitro and in vivo (12, 21, 22), we therefore sought to assess whether and how BIM contributes to B-cell activation and antibody response defects by generating Cd23-Cre Dynll1 Bim-double-deleted mice.
Similar to what we recently reported for Cd23-Cre Bim-deleted mice (23), Cd23-Cre Dynll1 Bim-double-deleted mice contained ∼2-fold expanded mature follicular B-cell numbers compared to Cd23-Cre control and Cd23-Cre Dynll1-deleted mice (Fig. 3A and B). Interestingly, compared to Dynll1-deleted B cells, concomitant loss of Bim led to considerably increased numbers of proliferating lymphoblasts in LPS-treated cultures (Fig. 3C and D). Comparison of the proliferation index of cells in these cultures that have divided at least once indicated no differences between Bim-deleted and Dynll1/Bim-double-deleted lymphoblasts (Fig. 3E). These results indicate that BIM-mediated apoptosis is a major factor in the reduced proliferation rates of Dynll1-deficient B cells in response to LPS. However, when considering all cells in the culture, it is important to note that loss of BIM led to a marked increase in the number of surviving nonresponsive small double-deleted lymphocytes, similar to the protective effect Bim deletion by itself has on the viability of activation-resistant B cells in vitro (23). Because of the increased survival of nonresponsive cells, Bim deletion did not affect the division index of Dynll1-deleted B cells calculated by the FlowJo program (Fig. 3F). However, compared to the >5-fold reduced division index of Dynll1-deleted cells relative to Cd23-Cre control cells, the division index of Dynll1/Bim-double-deleted cells was reduced by less than 2-fold relative to Bim single mutant cells (Fig. 3D and F). This relative improvement indicates that increased BIM-mediated apoptosis in the absence of DYNLL1 is an important factor in the in vitro activation defects of Cd23 Dynll1-deleted B cells, but the residual difference (P = 0.012) (Fig. 3F) between Dynll1-deleted and Dynll1/Bim-double-deleted cells indicates that BIM is not wholly responsible for this effect. In support of additional BIM-independent mechanisms, purified LPS-treated Dynll1/Bim-double-deleted lymphoblasts contained a higher proportion of G1-phase cells compared to Bim controls (Fig. 3G), consistent with decreased mitogenic responsiveness downstream of TLR4 activation.
FIG 3.
Differential effect of Bim deletion on in vitro activation and in vivo antibody responses of Cd23-Cre Dynll1-deleted B cells and mice. (A) Total splenic B-cell numbers (B220+ IgM+) of mice of the indicated genotypes (for Cd23-Cre versus ΔDynll1, P = 0.266; for ΔBim versus ΔDynll1 ΔBim, P = 0.001). (B) Representative FACS plots and quantification of follicular and marginal zone (MZ) B-cell fractions in spleens of mice with the indicated genotypes, indicating similar cellularity changes in Cd23-Cre Bim-deleted and Cd23-Cre Dynll/Bim-double-deleted mice and in control mice. (C, D) Representative FACS plots and CTV dilution curves of Cd23-Cre control, Cd23-Cre Dynll1-deleted, Cd23-Cre Bim-deleted, and Cd23-Cre Dynll1/Bim-double-deleted B cells treated with 15 μg/ml LPS for 96 h. (E, F) FlowJo-calculated proliferation and division indices of Cd23-Cre control, Cd23-Cre Dynll1-deleted, Cd23-Cre Bim-deleted, and Cd23-Cre Dynll1/Bim-double-deleted B cells treated with 15 μg/ml LPS for 96 h. The data for Cd23-Cre Bim-deleted and Cd23-Cre Dynll1/Bim-double-deleted cells are duplicated on two different y axis for clarity. (G) FACS plots of lymphoblast purifications and their cell cycle profiles after treatment with 15 μg/ml LPS for 72 h. (H to J) NP-LPS antibody responses in Cd23-Cre Bim-deleted and Cd23-Cre Dynll1/Bim-double-deleted mice compared to the results for Cd23-Cre control and Cd23-Cre Dynll1-deleted mice from Fig. 2A to C.
To assess whether the partial rescue effect of Bim deletion on the activation of Dynll1-deficient B cells translates to improved antibody responses in vivo, we immunized Cd23-Cre Bim-deleted and Cd23-Cre Dynll1 Bim-double-deleted mice with NP-LPS. However, additional deletion of Bim only marginally—and to a much lower level than in Cd23-Cre and Bim-deleted control mice—improved the NP-specific IgM, IgG2b, and IgG3 antibody titers in Dynll1-deficient mice, and none of the small improvements were statistically significant (Fig. 3H to J). Thus, although increased BIM activity contributes to the in vitro activation defects of Dynll1-deficient B cells, their impaired antibody responses in vivo are largely mediated by other mechanisms.
DYNLL1 is required for and upregulated in response to LPS-induced NF-κB pathway activation in B cells.
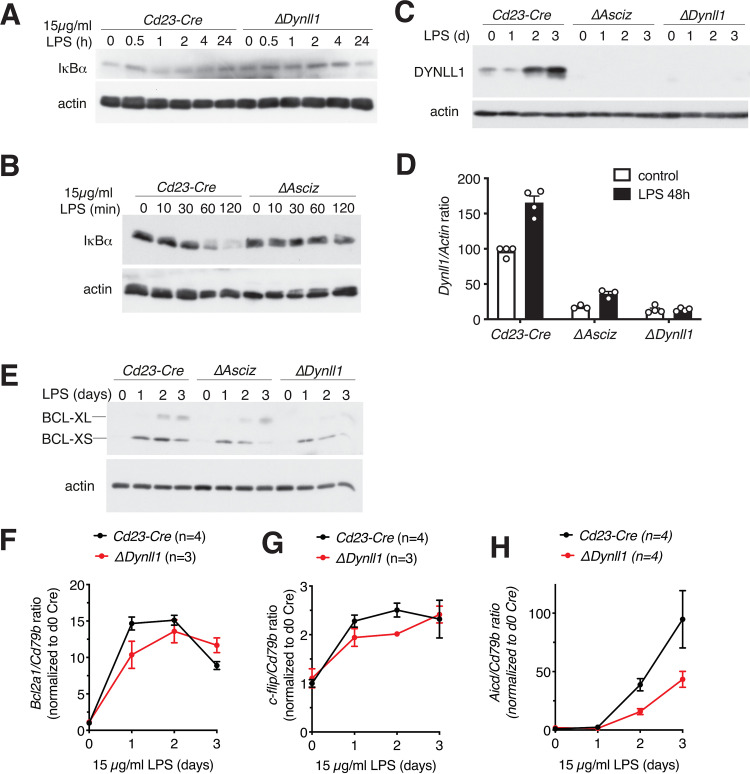
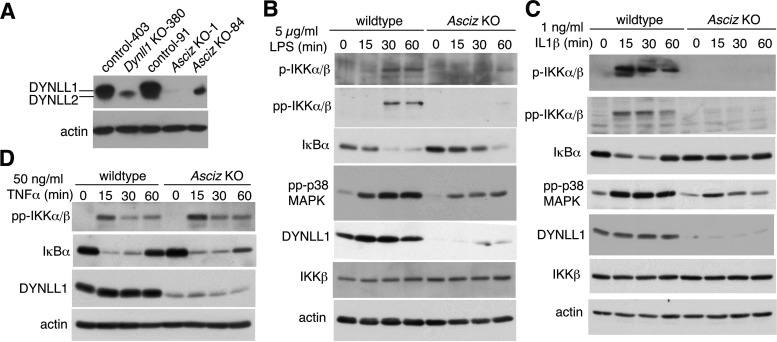
Because LPS and TLR4 are prototypical activators of the canonical NF-κB pathway, our findings that DYNLL1 was essential for TLR4-mediated B-cell activation and antibody responses seemed contradictory to its previously proposed function as an inhibitor of IκBα degradation and NF-κB activation (16). To resolve this conundrum, we performed immunoblot analyses to assess IκBα protein levels in LPS-treated B cells. As expected, IκBα protein amounts were transiently reduced after 1- to 2-h LPS treatment in Cd23-Cre control B cells, but in contrast to the current model, IκBα levels remained largely stable in Cd23-Cre Dynll1-deleted B cells (Fig. 4A). These results were independently confirmed using Cd23-Cre Asciz-deleted B cells (Fig. 4B), which also lacked detectable DYNLL1 expression (Fig. 4C). Interestingly, we noticed that DYNLL1 itself was significantly upregulated—at both the protein and mRNA levels—during the LPS-induced lymphoblast differentiation (Fig. 4C and D).
FIG 4.
The ASCIZ-DYNLL1 axis is required for LPS-induced NF-κB pathway activation in B cells. (A to C) Western blot analysis of Cd23-Cre control, Cd23-Cre Dynll1-deleted, and Cd23-Cre Asciz-deleted splenic B cells treated for the indicated times with 15 μg/ml LPS. (D) Quantitative RT-PCR analysis of Dynll1 mRNA expression, normalized to Actin mRNA, after treatment of Cd23-Cre control, Cd23-Cre Dynll1-deleted, and Cd23-Cre Asciz-deleted splenic B cell with 15 μg/ml LPS for 48 h (for Cd23-Cre, n = 4, P = 0.00004; for ΔAsciz, n = 3, P = 0.0071; for ΔDynll1, n = 4, P > 0.999). (E) Western blot analysis of the NF-κB target BCL-X-S/L in Cd23-Cre control, Cd23-Cre Dynll1-deleted, and Cd23-Cre Asciz-deleted splenic B cells treated for 0 to 3 days with 15 μg/ml LPS. (F to H) qPCR quantitation of NF-κB targets Bcl2a1, c-flip, and Aicd in Cd23-Cre control and Cd23-Cre Dynll1-deleted splenic B cells for 0 to 3 days with 15 μg/ml LPS.
We also monitored the response of selected NF-κB target genes (BCL-XL/S, Bcl2a1, c-flip, and Aicd genes) over a 3-day LPS treatment time course. While the targets exhibited different induction kinetics, in all cases their response was modestly delayed in Cd23-Cre Dynll1-deleted B cells compared to the response of Cd23-Cre controls (Fig. 4E to H). Collectively, these results indicate that in contrast to ectopic overexpression experiments in cell lines (16), under physiological conditions, including moderate upregulation during the LPS response, DYNLL1 may promote LPS-induced activation of the NF-κB pathway in primary B cells.
DYNLL1 is required for signal-specific activation of the NF-κB pathway upstream of IκBα.
To expand our findings beyond B cells, we turned to fibroblasts. Because germ line loss of Dynll1 leads to early embryonic lethality in mice, we used limb buds, which are highly enriched in fibroblasts, from mesenchyme-specific conditional Prx1-Cre Dynll1-deleted and Prx1-Cre control embryonic day 12.5 (E12.5) embryos to prepare murine embryonic fibroblast (MEF) cell lines (24). However, during the immortalization process, the closely related DYNLL2 protein (>95% amino acid identity to DYNLL1), which runs just below DYNLL1 on Western blots (25) and is normally expressed at >100-fold lower levels (24), was noticeably upregulated in Dynll1−/− MEFs. ASCIZ is an extremely specific transcription factor for the Dynll1 gene with general congruence between DYNLL1-null and ASCIZ-null loss of function phenotypes from Drosophila to mammals (12, 14, 17, 24, 26–29). In order to avoid interference from the upregulated DYNLL2 in Dynll1−/− MEFs, we therefore chose an Asciz−/− MEF clone that contained the lowest levels of DYNLL1 and DYNLL2 for these experiments (Fig. 5A) (note that only Dynll1 and not Dynll2 is directly regulated by ASCIZ [30]). Again, IκBα degradation in response to LPS treatment was notably attenuated in the Asciz−/− cells compared to the response in isogenic wild-type control MEFs (Fig. 5B), demonstrating that the importance of the ASCIZ-DYNLL1 axis for TLR4 signaling extends beyond the B-cell lineage.
FIG 5.
The ASCIZ-DYNLL1 axis is required for LPS- and IL-1–induced NF-κB pathway activation upstream of IκBα in MEFs. (A) Western blot analysis of DYNLL1 and DYNLL2 levels in isogenic control, Dynll1−/−, and Asciz−/− MEFs with a DYNLL1/2-cross-reactive antibody. Note that DYNLL2 has faster mobility than DYNLL1 and higher affinity for this antibody (25). MEF cell line numbers correspond to the originating embryo. (B to D) Western blots of isogenic wild-type control and DYNLL1-deficient Asciz−/− MEFs (clone 1). The treatment conditions and antibodies used are indicated within the figure panels. IL1, interleukin-1; TNF-α, tumor necrosis factor α.
We also monitored activation of the IKK complex, which acts upstream of IκBα. Remarkably, while TAK1-mediated priming (p-IKK) and autoactivating diphosphorylation (pp-IKK) of IKK was evident after 30 to 60 min of LPS treatment in control cells, it was barely detectable in the DYNLL1-deficient Asciz−/− MEFs (Fig. 5B). In addition to IKK, TAK1 also activates the p38 MAPK pathway in response to LPS treatment in a bifurcation from the NF-κB pathway (5, 6). Although activating p38 diphosphorylation (pp-p38) was still detectable in Asciz−/− cells, it was notably attenuated compared to diphosphorylation in control MEFs (Fig. 5B). These results imply that DYNLL1 acts at the level of or upstream of TAK1 by either promoting TAK1 activation or its interaction with IKK and p38.
Comparable results were also obtained for treatment with IL-1β, whose receptor activates the NF-κB pathway similar to TLR4 via MyD88 as a shared signaling adaptor (5). Similar to the LPS response, IKK activation and IκBα degradation were severely impaired, and p38 activation was noticeably diminished in IL-1‐treated Asciz−/− MEFs compared to p38 activation in control cells (Fig. 5C). Interestingly, in contrast to the impaired response to LPS and IL-1β, IKK activation and IκBα degradation was unaffected in Asciz−/− MEFs in response to treatment with tumor necrosis factor-α (TNF-α) (Fig. 5D), the receptor for which does not interact with MyD88 and uses different signaling components to activate TAK1 and IKK (31). Taken together, these results indicate that, similar to its role in B-cell activation, the ASCIZ-DYNLL1 axis regulates the NF-κB pathway in MEFs in a signal-specific manner by promoting the activation of the TAK1-IKK cascade upstream of IκBα.
DISCUSSION
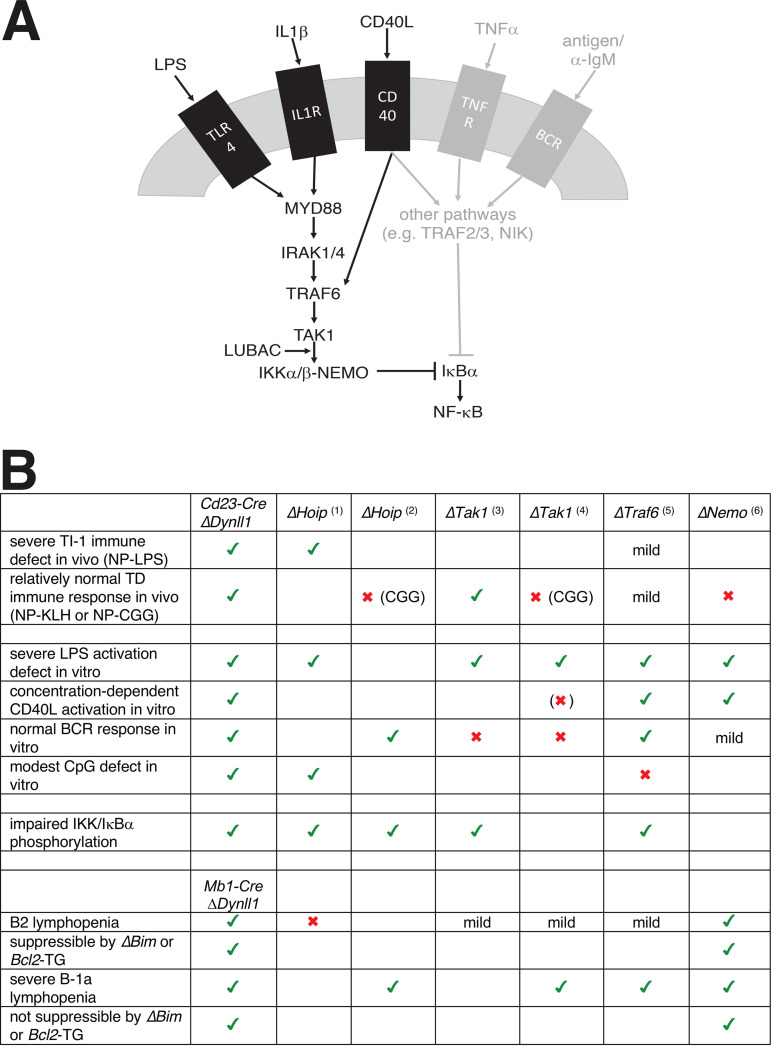
Collectively, our data from B cells and MEFs show that the ASCIZ-DYNLL1 axis is required for the signal-specific activation of the NF-κB pathway in response to LPS, IL-1, and low-dose CD40L but not to TNF-α and BCR ligation. A common and distinguishing feature of the affected pathways is that they signal at least in part through the Ub ligase TRAF6 (Fig. 6A). In contrast, the unaffected ligand TNF and the BCR pathway utilize the related Ub ligase TRAF2 or the TAK1-independent noncanonical NF-κB pathway, respectively (or both TRAF6 and TRAF2 in case of the partially impaired CD40L response), to promote IκBα phosphorylation and degradation (31). While the precise mechanism of DYNLL1’s action in this context remains to be determined, we speculate that it may contribute to the assembly of TRAF6‐TAK1‐IKK activation complexes. As DYNLL1 typically promotes the oligomerization of its targets by facilitating the alignment of their oligomerization domains, including often coiled-coil domains, into an ordered register (10, 11), it is interesting to note that coil-coiled mediated oligomerization has recently been shown to regulate TRAF6 catalytic processivity (32).
FIG 6.
Role of the ASCIZ-DYNLL1 axis in NF-κB signaling. (A) Schematic diagram of ASCIZ/DYNLL1-dependent signaling in B cells and MEFs. Pathways that require DYNLL1 for IκBα degradation are shown in black, and DYNLL1-independent pathways are shown in gray. (B) Comparison of B-cell phenotypes of Dynll1-deficient mice to published Traf6, LUBAC (Hoip1), Tak1, and Nemo conditional KO mice. Green ticks indicate similar phenotypes; red crosses indicate different phenotypes. Phenotypes for other mice are compiled from Mb1-Cre ΔHoip (Sasaki et al. [37]), Mb1-Cre ΔHoip (Sasaki et al. [38]), Cd19-Cre ΔTak1 (Schuman et al. [34]), Mb1-Cre ΔTak1 (Shinohara et al. [35]), Cd19-Cre ΔTraf6 (Kobayashi et al. [33]), and Mb1-Cre and Cd21-Cre ΔNemo (Derudder et al. [36]).
Consistent with the biochemical support for a role of DYNLL1 upstream of IκBα, the B-cell defects we have reported here are overall remarkably similar to the phenotypes of B lymphoid-specific conditional deletions of Traf6 (33), Tak1 (34, 35), the IKK subunit Nemo (36), and the LUBAC catalytic subunit Hoip (37, 38). These similarities (Fig. 6B) include severely impaired LPS-induced B-cell activation in vitro (Traf6, Tak1, Nemo, and Hoip); impaired IKK activation or IκBα degradation in response to LPS (Traf6, Hoip, and Tak1); concentration-dependent CD40L-induced activation defects (Traf6, Tak1, and Nemo); and impaired in vivo antibody responses to NP-LPS (Traf6 and Hoip). The main differences between our findings and the other models relate to the in vivo response to T-cell‐dependent antigens, which were also largely normal in NP‐keyhole limpet hemocyanin (KLH)‐immunized Tak1-deleted mice but impaired in Traf6-deleted (albeit mildly), Hoip-deleted, or Nemo-deleted mice (as well as NP-CGG‐immunized Tak1-deleted mice), and in vitro responses to BCR ligation, which were impaired in the absence of Tak1 or Nemo but also normal in Hoip- or Traf6-deleted B cells. It should be noted that the Traf6, Tak1, Nemo, and Hoip deletions were driven by Mb1-Cre or Cd19-Cre, which become active during early B lymphoid stages (39) and therefore also lead to developmental defects that may indirectly contribute to some of the differences compared to our postdevelopmental Cd23-Cre‐driven Asciz and Dynll1 deletions (14, 17). Indeed, when considering the previously reported developmental defects of Mb1-Cre Dynll1- or Asciz-deleted mice (12, 13), the similarities between the mouse models extend even further. Traf6-, Tak1-, and Nemo-deleted mice also contain mildly to severely reduced B-2 cell numbers, which in the case of Nemo and Asciz/Dynll1 deletion could be suppressed by transgenic BclII overexpression or Bim deletion, respectively; and Traf6-, Tak1-, Nemo-, and Hoip-deficient mice contain extremely reduced B-1a cell numbers, which similar to Mb1-Cre Dynll1- or Asciz-deleted mice cannot be rescued by the BclII transgene or Bim deletion.
Finally, the seemingly contradictory observations that DYNLL1 at physiological expression levels promotes but at excessively high overexpression levels inhibits NF-κB signaling (16) may highlight the intriguing dual activity of ASCIZ in the regulation of the Dynll1 gene. While ASCIZ is essential for the activation of Dynll1 expression, it also functions as a feedback sensor of DYNLL1 protein levels because DYNLL1 can bind directly to >10 different sites in the transcription activation domain of ASCIZ and thereby repress its activity (30, 40). It has remained an enigma why DYNLL1 requires such a designated bimodal transcription factor. The opposing effects of very low versus very high DYNLL1 protein levels on NF-κB signaling suggest that one important function of this feedback mechanism may be to ensure that the positive role of DYNLL1 in the upstream activation of the NF-κB pathway is not counteracted by an inadvertent inhibitory effect on the interaction of activated IKK with its target IκBα.
An important question for future work will be to determine the precise molecular mechanism of how DYNLL1 promotes the activation of IKK and which protein it binds to in this context. Finally, based on the similar findings for the important role of the ASCIZ-DYNLL1 axis in the regulation of TLR4-mediated NF-κB pathway activation in B cells and MEFs, we would predict that this role is universally conserved across different cell types. Given the importance of TLR4 signaling in inflammation and immunity, including sepsis, it will therefore be interesting to assess its functions in other cell types and disease processes.
MATERIALS and METHODS
Mice.
Animal experiments were performed according to the Australian Code for the Care and Use of Animals for Scientific Purposes, 8th Edition (2013), and approved by the St. Vincent’s Hospital Melbourne Animal Ethics Committee (approval 002/17). Cd23-Cre (41), Mb1-Cre (39), Dynll1-floxed (17), Asciz-floxed (13), and Bim-floxed (42) mice had been generated or backcrossed for at least 10 generations on a C57BL6 background and were housed in specific pathogen-free microisolators. Cd23-CreTg/+ Dynll1fl/+ mice were intercrossed with Dynll1fl/+ mice to generate Cd23-CreTg/+ Dynll1fl/fl deleted mice and isogenic Cd23-CreTg/+ Dynll1+/+ controls. Similar heterozygous intercrosses were used to generate Cd23-Cre Asciz-deleted and Mb1-Cre Dynll1-deleted mice. For BIM experiments, Cd23-CreTg/+ Dynll1fl/+ Bimfl/fl mice were intercrossed with Dynll1fl/+ Bimfl/fl mice. Mice were used at ≥8 weeks of age for in vitro B-cell analyses and basal Ig titer assays and at 3 to 6 months of age for in vivo immunization experiments; they were not selected on gender.
Immunization.
The mice were intraperitoneally (i.p.) injected with 10 μg NP10-LPS or with 100 μg NP21-KLH in alum. Serum was collected before, 1 week after (NP-LPS), and 4 weeks after (NP-KLH) immunization. NP-specific Ig titers were determined using Nunc Maxisorb plates coated with 50 μl per well of 5 μg/ml NP2‐bovine serum albumin (BSA) or NP14/NP16-BSA, and horseradish peroxidase (HRP)‐labeled goat anti-mouse IgM, IgG1, IgG2, or IgG3 antibodies (Southern Biotech, 5300-05). For quantification, reference wells were coated with goat anti-mouse Ig capture antibody and purified mouse IgM, IgG1, IgG2, or IgG3 (Southern Biotech, 5300-01). Enzyme-linked immunosorbent assay (ELISA) plates were developed using 2,2'-azinobis(3-ethylbenzothiazolinesulfonic acid) (ABTS) and read using a Polarstar at 410 nm.
B-cell isolation and in vitro activation.
B cells were isolated from single-cell splenocyte suspensions using B-cell isolation kits (Miltenyi Biotec, 130090862) and magnetically activated cell sorting (MACS) separation LS columns (Miltenyi Biotec, 130042401) following the manufacturer’s instructions. Ten million isolated B cells were incubated with CellTrace Violet (CTV; Thermo Fisher Scientific, C34557) at 1:1,000 dilution at 37°C in the dark for 20 min and centrifuged at 300 × g for 10 min, and the cell pellet was washed once with 10 ml of MACS buffer. CTV-stained B cells were cultured in the B-cell culture media (RPMI 1640 [Sigma-Aldrich, R8758] supplemented with 5% (vol/vol) fetal bovine serum (FBS; Assay Matrix, ASFBS-F, batch A18C016], 50 μM β-mercaptoethanol, 100 U/ml penicillin G, and 100 μg/ml streptomycin sulfate) at a seeding density of 1 million cells/ml. The cells were stimulated with lipopolysaccharide (LPS; Invivogen, tlrl-3pelps), recombinant CD40L plus 1/100 conditioned mouse IL-4 supernatant (kind gift from Andreas Strasser, Walter and Eliza Hall Institute, Melbourne, Australia), or anti-IgM F(ab)2 fragment (eBioscience 16-5092-85) at the concentrations and treatment times indicated in the figure legends and panels.
Flow cytometry.
The following reagents were used for cell staining: B220-FITC (Biolegend, 103206), CD19-APC eFluor780 (eBioscience, 47-0193-82), CD21-PE (Biolegend, 123410), CD23-biotin (BD, 553137), Brilliant Violet 605 Streptavidin (Biolegend, 405229), and propidium iodide (Sigma, P4864-10ML). Cells were applied to flow cytometry using a BD LSRFortessa with FACSDiva software (BD) and analyzed using FlowJo 10.3 software (Tree Star). Follicular and marginal zone B cells (Fig. 1M) and activated lymphoblasts (Fig. 3G) were isolated using a BD Influx cell sorter. For follicular and marginal zone B-cell proportions, splenic single cell suspensions were first gated on live B220+ CD19+ lymphocytes, followed by gating on CD21/35 and CD23 (follicular B cells: CD21/35lo CD23hi; marginal zone B cells: CD21/35hi CD23lo). Cell division and proliferation indices of CTV-stained cells were determined using the cell proliferation module, and cell cycle stages were determined using the cell cycle module of FlowJo 10.3 software (Tree Star).
MEF experiments.
Dynll1−/− MEFs and isogenic controls were prepared from E12.5 Prx1-Cre Dynll1fl/fl and Prx1-Cre Dynll1+/+ embryonic limb buds (24) and immortalized randomly using the same standard 3T3 protocol as for the preparation of isogenic wild-type and Asciz−/− MEFs from E12.5 embryos (43). MEFs were seeded at 1.5 × 105 cells per well in 6-well plates in 3 ml Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich, R8758) with 10% fetal calf serum (FCS) or at 106 MEFs per 10-cm dish in 10 ml DMEM-FCS and grown for 2 days at 37°C with ambient O2 and 5% CO2. LPS, IL-1β (Propo Tech 200-01B-10UG), or TNF-α (In Vitro Technologies, RDS410TRNC010) was added with fresh medium at the doses and times indicated in the figure legends and panels.
Western blots and quantitative PCR.
B cells were placed on ice at the indicated time points and collected by centrifugation at 375 × g, washed in ice-cold phosphate-buffered saline (PBS), and lysed in cold modified radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 20 mM Tris [pH 7.4] 1 mM EDTA, 1 mM EGTA, 10 mM NaF, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× protein inhibitor cocktail [Sigma]). MEFs were scraped directly into RIPA buffer on ice. Lysed cells were sonicated and centrifuged for 10 min at 4°C. Protein (∼25 μg per lane) was electrophoresed using Laemmli buffered SDS polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes for detection using ECL (GE Healthcare) or Super-ECL reagents (Pierce) and X-ray film (Fuji). Films were scanned in trans-illumination mode using a dual lens Epson V700 photoscanner. The following antibodies were used: actin‐horseradish peroxidase (Santa Cruz Biotechnology, sc-1616, 1:10,000 dilution), BCL-X-L/S (BD Biosciences, 61021, 1:1,000), DYNLL1/2 (Abcam, ab51603, 1:5,000), IκBα (Cell Signaling Technology, 4814, 1:1,000), IKKβ (Cell Signaling Technology, 8943, 1:1,000), phospho-IKKα (S176)/IKKβ (S177) (Cell Signaling Technology, 2078, 1:1,000), di-phospho-IKKα/β (Ser176/180) (Cell Signaling Technology, 2697, 1:1,000), and phospho-p38 MAPK (Thr180/Tyr182) (Cell Signaling Technology, 4511, 1:1,000).
For quantitative PCR (qPCR) analyses, 3 × 106 MACS-purified B cells were cultured with LPS (15 μg/ml) for 0 to 3 days. After treatment, the cells were washed once with ice-cold PBS, and cell pellets were snap-frozen on dry ice and stored at −80°C before RNA extraction. Total RNA was extracted using an ISOLATE II RNA micro kit (Bioline BIO-52075) following the manufacturer’s protocol. 300 ng purified RNA was subjected to reverse transcription with oligo(dT) and random primers using the SuperScript III first-strand synthesis system (Invitrogen, 18080-051). qPCR reagents were mixed as follows: 4 μl cDNA (6 ng), 0.5 μl primer mix (300 nM each), 5 μl SYBR Select Master mix (Applied Biosystems, 4472908), and 0.5 μl RNase/DNase free water. qPCR conditions were set as follows: 1 cycle (10 min at 95°C), 45 cycles (15 s at 95°C), and 60 s at 60°C. After each PCR run, a disassociation curve program was added to validate the primer specificity. The standard curve method of comparative quantification was employed to quantify the results (44). The results were normalized to Cd79b as an internal control and expressed as percentages of maximum expression. The following primers were used (37, 45): Cd79b_forward (5′-TGTTGGAATCTGCAAATGGA-3′), Cd79b_reverse (5′-TAGGCTTTGGGTGATCCTTG-3′); Aicd_forward (5′-AGAAAGTCACGCTGGAGACC-3′), Aicd_reverse (5′-CTCCTCTTCACCACGTAGCA-3′), Bcl2a1a_forward (5′-TCCACAAGAGCAGATTGCCCTG3′), Bcl2a1a_reverse (5′-GCCAGCCAGATTTGGGTTCAAAC-3′), c-flip_forward (5′-GCTCTACAGAGTGAGGCGGTTT-3′), and c-flip_reverse (5′-CACCAATCTCCATCAGCAGGAC-3′).
Statistical analysis.
Data from independent experimental replicates were analyzed using GraphPad Prism software. The numbers of independent samples are indicated in the figures. Error bars indicate the mean ± the standard error of the mean (SEM). The P values were calculated using the two-tailed unpaired Student’s t test.
ACKNOWLEDGMENTS
We thank Nora Tenis, Emily Derrick, Lingli Li, and Kristy O’Donnell for help with mouse genotyping, ELISAs, and antigen preparation; the St. Vincent’s Institute fluorescence-activated cell sorter (FACS) facility for help with flow cytometry and cell sorting; the St. Vincent’s Hospital Bioresources Centre for animal care; and Carl Walkley and Tom Brodnicki for manuscript comments.
This work was supported by Worldwide Cancer Research grant 16-0156 (J.H.), a research fellowship 1060675 from the National Health and Medical Research Council (NHMRC) of Australia (D.T.), NHMRC Independent Research Institutes Infrastructure Support, and Victorian State Government Operational Infrastructure Support grants.
Author contribution: R.L., A.K., and J.H. carried out experiments. R.L., A.K., D.T., and J.H. analyzed experimental data. D.T. provided essential reagents and advice. R.L. and J.H. designed experiments. J.H. wrote the paper.
We declare no competing financial interests.
REFERENCES
- 1.LeBien TW, Tedder TF. 2008. B lymphocytes: how they develop and function. Blood 112:1570–1580. 10.1182/blood-2008-02-078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hobeika E, Nielsen PJ, Medgyesi D. 2015. Signaling mechanisms regulating B-lymphocyte activation and tolerance. J Mol Med 93:143–158. 10.1007/s00109-015-1252-8. [DOI] [PubMed] [Google Scholar]
- 3.Buchta CM, Bishop GA. 2014. Toll-like receptors and B cells: functions and mechanisms. Immunol Res 59:12–22. 10.1007/s12026-014-8523-2. [DOI] [PubMed] [Google Scholar]
- 4.Isaza-Correa JM, Liang Z, van den Berg A, Diepstra A, Visser L. 2014. Toll-like receptors in the pathogenesis of human B cell malignancies. J Hematol Oncol 7:57. 10.1186/s13045-014-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D, Cohen P. 2013. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci USA 110:15247–15252. 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Clark K, Lawrence T, Peggie MW, Cohen P. 2014. An unexpected twist to the activation of IKKbeta: TAK1 primes IKKbeta for activation by autophosphorylation. Biochem J 461:531–537. 10.1042/BJ20140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Lenardo MJ, Baltimore D. 2017. 30 years of NF-kappaB: a blossoming of relevance to human pathobiology. Cell 168:37–57. 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, Shaffer AL, Romesser P, Wright G, Powell J, Rosenwald A, Muller-Hermelink HK, Ott G, Gascoyne RD, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Staudt LM. 2011. Oncogenically active MYD88 mutations in human lymphoma. Nature 470:115–119. 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerondakis S, Grumont RJ, Banerjee A. 2007. Regulating B-cell activation and survival in response to TLR signals. Immunol Cell Biol 85:471–475. 10.1038/sj.icb.7100097. [DOI] [PubMed] [Google Scholar]
- 10.Barbar E. 2008. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry 47:503–508. 10.1021/bi701995m. [DOI] [PubMed] [Google Scholar]
- 11.Rapali P, Szenes A, Radnai L, Bakos A, Pal G, Nyitray L. 2011. DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J 278:2980–2996. 10.1111/j.1742-4658.2011.08254.x. [DOI] [PubMed] [Google Scholar]
- 12.King A, Li L, Wong DM, Liu R, Bamford R, Strasser A, Tarlinton DM, Heierhorst J. 2017. Dynein light chain regulates adaptive and innate B cell development by distinctive genetic mechanisms. PLoS Genet 13:e1007010. 10.1371/journal.pgen.1007010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurado S, Gleeson K, O’Donnell K, Izon DJ, Walkley CR, Strasser A, Tarlinton DM, Heierhorst J. 2012. The zinc-finger protein ASCIZ regulates B cell development via DYNLL1 and Bim. J Exp Med 209:1629–1639. 10.1084/jem.20120785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker JR, Cuella-Martin R, Barazas M, Liu R, Oliveira C, Oliver AW, Bilham K, Holt AB, Blackford AN, Heierhorst J, Jonkers J, Rottenberg S, Chapman JR. 2018. The ASCIZ-DYNLL1 axis promotes 53BP1-dependent non-homologous end joining and PARP inhibitor sensitivity. Nat Commun 9:5406. 10.1038/s41467-018-07855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crepieux P, Kwon H, Leclerc N, Spencer W, Richard S, Lin R, Hiscott J. 1997. I kappaB alpha physically interacts with a cytoskeleton-associated protein through its signal response domain. Mol Cell Biol 17:7375–7385. 10.1128/MCB.17.12.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung Y, Kim H, Min SH, Rhee SG, Jeong W. 2008. Dynein light chain LC8 negatively regulates NF-kappaB through the redox-dependent interaction with IkappaBalpha. J Biol Chem 283:23863–23871. 10.1074/jbc.M803072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong DM, Li L, Jurado S, King A, Bamford R, Wall M, Walia MK, Kelly GL, Walkley CR, Tarlinton DM, Strasser A, Heierhorst J. 2016. The transcription factor ASCIZ and its target DYNLL1 are essential for the development and expansion of MYC-driven B cell lymphoma. Cell Rep 14:1488–1499. 10.1016/j.celrep.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Dustin ML. 2014. What counts in the immunological synapse? Mol Cell 54:255–262. 10.1016/j.molcel.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Z, Zan H, Pone EJ, Mai T, Casali P. 2012. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol 12:517–531. 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee A, Grumont R, Gugasyan R, White C, Strasser A, Gerondakis S. 2008. NF-kappaB1 and c-Rel cooperate to promote the survival of TLR4-activated B cells by neutralizing Bim via distinct mechanisms. Blood 112:5063–5073. 10.1182/blood-2007-10-120832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A. 1999. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 3:287–296. 10.1016/S1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- 22.Singh PK, Roukounakis A, Frank DO, Kirschnek S, Das KK, Neumann S, Madl J, Romer W, Zorzin C, Borner C, Haimovici A, Garcia-Saez A, Weber A, Hacker G. 2017. Dynein light chain 1 induces assembly of large Bim complexes on mitochondria that stabilize Mcl-1 and regulate apoptosis. Genes Dev 31:1754–1769. 10.1101/gad.302497.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, King A, Bouillet P, Tarlinton DM, Strasser A, Heierhorst J. 2018. Proapoptotic BIM impacts B lymphoid homeostasis by limiting the survival of mature B cells in a cell-autonomous manner Front Immunol 9:592. 10.3389/fimmu.2018.00592. 10.3389/fimmu.2018.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King A, Hoch NC, McGregor NE, Sims NA, Smyth IM, Heierhorst J. 2019. Dynll1 is essential for development and promotes endochondral bone formation by regulating intraflagellar dynein function in primary cilia. Hum Mol Genet 28:2573–2588. 10.1093/hmg/ddz083. [DOI] [PubMed] [Google Scholar]
- 25.Singh PK, Roukounakis A, Weber A, Das KK, Sohm B, Villunger A, Garcia-Saez AJ, Hacker G. 2020. Dynein light chain binding determines complex formation and posttranslational stability of the Bcl-2 family members Bmf and Bim. Cell Death Differ 27:434–450. 10.1038/s41418-019-0365-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaytseva O, Tenis N, Mitchell N, Kanno S, Yasui A, Heierhorst J, Quinn LM. 2014. The novel zinc finger protein dASCIZ regulates mitosis in Drosophila via an essential role in Dynein Light-Chain Expression. Genetics 196:443–453. 10.1534/genetics.113.159541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goggolidou P, Stevens JL, Agueci F, Keynton J, Wheway G, Grimes DT, Patel SH, Hilton H, Morthorst SK, DiPaolo A, Williams DJ, Sanderson J, Khoronenkova SV, Powles-Glover N, Ermakov A, Esapa CT, Romero R, Dianov GL, Briscoe J, Johnson CA, Pedersen LB, Norris DP. 2014. ATMIN is a transcriptional regulator of both lung morphogenesis and ciliogenesis. Development 141:3966–3977. 10.1242/dev.107755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He YJ, Meghani K, Caron MC, Yang C, Ronato DA, Bian J, Sharma A, Moore J, Niraj J, Detappe A, Doench JG, Legube G, Root DE, D’Andrea AD, Drane P, De S, Konstantinopoulos PA, Masson JY, Chowdhury D. 2018. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature 563:522–526. 10.1038/s41586-018-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zalmas LP, Lu WT, Kanu N. 2019. An emerging regulatory network of NHEJ via DYNLL1-mediated 53BP1 redistribution. Ann Transl Med 7:S93. 10.21037/atm.2019.04.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jurado S, Conlan LA, Baker EK, Ng JL, Tenis N, Hoch NC, Gleeson K, Smeets M, Izon D, Heierhorst J. 2012. ATM substrate Chk2-interacting Zn2+ finger (ASCIZ) Is a bi-functional transcriptional activator and feedback sensor in the regulation of dynein light chain (DYNLL1) expression. J Biol Chem 287:3156–3164. 10.1074/jbc.M111.306019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayden MS, Ghosh S. 2012. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 26:203–234. 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu L, Xu J, Xie X, Zhou Y, Tao P, Li H, Han X, Wang C, Liu J, Xu P, Neculai D, Xia Z. 2017. Oligomerization-primed coiled-coil domain interaction with Ubc13 confers processivity to TRAF6 ubiquitin ligase activity. Nat Commun 8:814. 10.1038/s41467-017-01290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi T, Kim TS, Jacob A, Walsh MC, Kadono Y, Fuentes-Panana E, Yoshioka T, Yoshimura A, Yamamoto M, Kaisho T, Akira S, Monroe JG, Choi Y. 2009. TRAF6 is required for generation of the B-1a B cell compartment as well as T cell-dependent and -independent humoral immune responses. PLoS One 4:e4736. 10.1371/journal.pone.0004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuman J, Chen Y, Podd A, Yu M, Liu HH, Wen R, Chen ZJ, Wang D. 2009. A critical role of TAK1 in B-cell receptor-mediated nuclear factor kappaB activation. Blood 113:4566–4574. 10.1182/blood-2008-08-176057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shinohara H, Kurosaki T. 2016. Negative role of TAK1 in marginal zone B-cell development incidental to NF-kappaB noncanonical pathway activation. Immunol Cell Biol 94:821–829. 10.1038/icb.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derudder E, Herzog S, Labi V, Yasuda T, Kochert K, Janz M, Villunger A, Schmidt-Supprian M, Rajewsky K. 2016. Canonical NF-kappaB signaling is uniquely required for the long-term persistence of functional mature B cells. Proc Natl Acad Sci USA 113:5065–5070. 10.1073/pnas.1604529113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki Y, Iwai K. 2018. Crucial role of linear ubiquitin chain assembly complex-mediated inhibition of programmed cell death in TLR4-mediated B cell responses and B1b cell development. J Immunol 200:3438–3449. 10.4049/jimmunol.1701526. [DOI] [PubMed] [Google Scholar]
- 38.Sasaki Y, Sano S, Nakahara M, Murata S, Kometani K, Aiba Y, Sakamoto S, Watanabe Y, Tanaka K, Kurosaki T, Iwai K. 2013. Defective immune responses in mice lacking LUBAC-mediated linear ubiquitination in B cells. EMBO J 32:2463–2476. 10.1038/emboj.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, Reth M. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA 103:13789–13794. 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark S, Myers JB, King A, Fiala R, Novacek J, Pearce G, Heierhorst J, Reichow SL, Barbar EJ. 2018. Multivalency regulates activity in an intrinsically disordered transcription factor. Elife 7:e36258. 10.7554/eLife.36258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M. 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28:751–762. 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Herold MJ, Stuchbery R, Merino D, Willson T, Strasser A, Hildeman D, Bouillet P. 2014. Impact of conditional deletion of the pro-apoptotic BCL-2 family member BIM in mice. Cell Death Dis 5:e1446. 10.1038/cddis.2014.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu R, King A, Hoch NC, Chang C, Kelly GL, Deans AJ, Heierhorst J. 2017. ASCIZ/ATMIN is dispensable for ATM signaling in response to replication stress. DNA Repair 57:29–34. 10.1016/j.dnarep.2017.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29:e45. 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pone EJ, Zhang J, Mai T, White CA, Li G, Sakakura JK, Patel PJ, Al-Qahtani A, Zan H, Xu Z, Casali P. 2012. BCR-signalling synergizes with TLR-signalling for induction of AID and immunoglobulin class-switching through the non-canonical NF-κB pathway. Nat Commun 3:767. 10.1038/ncomms1769. [DOI] [PMC free article] [PubMed] [Google Scholar]