Abstract
Plasma proteomic profiling may aid in the discovery of novel biomarkers upstream of the development of atrial fibrillation (AF). We used data from the Atherosclerosis Risk in Communities (ARIC) study to examine the relationship between large-scale proteomics and incident AF in a cohort of older-aged adults in the US. We quantified 4877 plasma proteins in ARIC participants at visit 5 (2011-2013) using an aptamer-based proteomic profiling platform. We used Cox proportional hazards models to assess the association between protein levels and incident AF and explored relationships of selected protein biomarkers using annotated pathway analysis. Our study included 4668 AF-free participants (mean age 75 ± 5 years; 59% female; 20% black race) with proteomic measures. A total of 585 participants developed AF over a mean follow-up of 5.7 ± 1.7 years. After adjustment for clinical factors associated with AF, N-terminal pro-B-type natriuretic peptide (NT-proBNP) was associated with the risk of incident AF (hazard ratio, 1.82; 95% CI, 1.68-1.98; p-value=2.91 X 10−45 per doubling of NT-proBNP). In addition, 36 other proteins were also significantly associated with incident AF after Bonferroni correction. We further adjusted for medication use and estimated glomerular filtration rate and found 17 proteins, including Angiopoietin-2 and Transgelin, remained significantly associated with incident AF. Pathway analyses implicated the inhibition of matrix metalloproteases as the top canonical pathway in AF pathogenesis. In conclusion, using a large-scale proteomic platform we identified both novel and established proteins associated with incident AF and explored mechanistic pathways of AF development.
Keywords: atrial fibrillation, proteomics, aging, proteome
INTRODUCTION
Despite well-established risk factors and biomarkers implicated in atrial fibrillation (AF) development, there are substantial gaps in our understanding of underlying pathways for AF pathogenesis. Identification of novel biomarkers coupled with pathway analyses can advance our understanding of AF mechanisms, enhance opportunities for risk prediction, and may provide targeted preventive strategies for AF. Proteomic profiling may aid in the discovery of novel biomarkers that are upstream of the development of AF. Recently, several longitudinal cohort studies have reported associations between plasma proteomic profiling and the risk of AF. 1-5 Supplemental Table 1 lists an overview of each study along with the main results. Of these five studies, four measured N-terminal pro-B-type natriuretic peptide (NT-proBNP) in their proteomic platform, and higher NT-proBNP was significantly associated with greater incidence of AF, even after adjustment for multiple AF risk factors. However, no other similarities among study results were observed, and each study found several different proteins associated with incident AF. These prior studies may have been limited by the modest numbers of AF events and by the limited numbers of proteins included on their proteomic platforms. In this study, we used data from the Atherosclerosis Risk in Communities (ARIC) study to screen for 4877 plasma proteins and identify novel biomarkers that are associated with risk of incident AF. This community-based cohort of black and white older adults in the US has a larger number of proteins measured compared to previous studies, and nearly 600 AF events in a 6-year follow-up time, allowing us to address some limitations of previous studies.
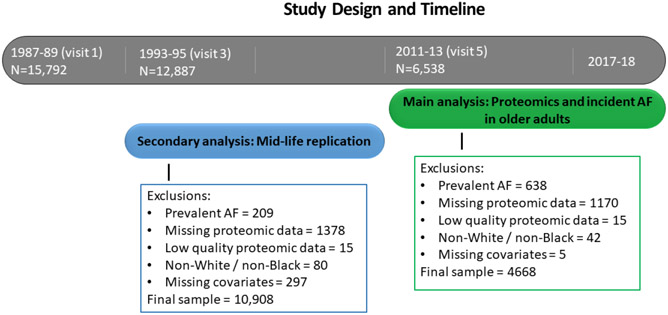
METHODS
The Atherosclerosis Risk in Communities (ARIC) study is a prospective cohort study of cardiovascular disease and atherosclerosis risk factors.6 Participants at baseline (1987-1989) included 15,792 men and women aged 45-64, recruited from 4 communities in the US (Washington County, Maryland; the northwest suburbs of Minneapolis, Minnesota; Jackson, Mississippi; and Forsyth County, North Carolina). Thus far, 7 study visits have been completed with visit 5 (baseline for our main analysis) occurring in 2011-2013. The primary analysis examined the association of ARIC visit 5 protein levels with incident AF through the end of 2017 at the Jackson field center, and through the end of 2018 at the other 3 field centers. Figure 1 contains the study design and exclusions for this study. Among the 6538 participants who attended visit 5, we excluded those with prevalent AF at visit 5 (n=638), with missing (n=1170) or low quality proteomic data (n=15), with race other than white or black and non-whites in the Minneapolis and Washington County field centers (due to low numbers; n=42), having missing covariates (n=5), resulting in a study population of 4668. We also conducted a midlife replication analysis including only those proteins significantly associated with AF risk in the visit 5 primary analyses. We examined the association of proteins measured at visit 3 (1993-1995) with incident AF through the end of 2010, which was the approximate start of visit 5. After similar exclusions, 10,908 AF-free participants with protein measures at visit 3 were included in the midlife replication analysis. This study was approved by institutional review boards at each participating center, and all study participants provided written informed consent.
Figure 1.
Study design and timeline for the main analysis and secondary analysis for the association of proteomics with incident atrial fibrillation (AF) in the ARIC study.
Incident AF was defined as in previous ARIC analyses.7 A trained abstractor obtained and recorded all International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) and ICD-10-CM hospital discharge diagnoses from each participant's hospitalizations reported in the follow-up interview. AF was defined as the presence of ICD-9-CM code 427.31 or 427.32 or ICD-10-CM code I48.xx. AF hospitalization diagnoses occurring simultaneously with heart revascularization surgery or other cardiac surgery involving heart valves or septa were not included as AF events. Deceased ARIC participants were also labeled as AF cases if their underlying cause of death was AF. AF was additionally identified by study visit ECGs, performed at visits 1-5. At each ARIC study visit, a 10-second 12-lead ECG was performed using a MAC PC cardiograph (Marquette Electronics Inc, Milwaukee, WI) and transmitted to the ARIC ECG Reading Center for coding, interpretation and storage. All ECGs automatically coded as AF were visually checked by a trained cardiologist to confirm AF diagnosis.
EDTA-plasma was obtained from blood samples that were collected during visits 3 and 5 and stored at −80 degrees C. Plasma samples were analyzed using a SOMAmer-based capture array called “SOMAscan” (Somalogic, Inc., Boulder, CO, USA). This assay was performed as described previously.8-11 Protein levels in the plasma samples were measured by the SOMAscan platform, which uses single-stranded DNA-based aptamers to capture conformational protein epitopes. Proof of principle and assay validations of specificity and intra- and inter-assay variability have been published.12-14 Briefly, protein measurements were standardized and normalized. Median signal normalization was applied to measures within plates to remove sample or assay biases due to variations in pipetting, reagent concentrations, assay timing, and other sources of systematic variability within single plate runs. Metrics of assay reproducibility have been previously reported 13 with a median coefficient of variance (quartile 1, quartile 3) of 5.0 (4.1, 6.9) and a median intraclass correlation (quartile 1, quartile 3) of 0.96 (0.92, 0.98). Four-hundred twenty-two blind duplicate plasma aliquots were included, and the median inter-assay Bland-Altman coefficient of variation was 6.3%. The median split sample reliability coefficient was 0.85 after excluding the following quality control outliers: of the 5,284 available aptamer measurements, 94 were excluded due to a Bland-Altman coefficient of variation >50% or a variance of <0.01 on the log scale; an additional 313 measurements were excluded due to non-specific binding to non-proteins. After all quality control measures were completed, 4,870 aptamer measurements were included that corresponded to 4,697 unique human proteins or protein complexes were analyzed in this study. We examined protein distributions and applied log base 2 transformation to all SOMAmer measures to correct for skewness. We winsorized outliers that were greater or less than 6 standard deviations from the sample mean on the log 2 scale.
Covariates for this analysis include AF risk factors from the CHARGE-AF score,15 namely age, sex, race, cigarette smoking status, height, weight, systolic and diastolic blood pressure, anti-hypertensive medication use, diabetes, prevalent myocardial infarction, and prevalent heart failure. We additionally included several medications and estimated glomerular filtration rate (eGFR) as covariates, reasoning that, in addition to being associated with the risk of AF, these variables could also affect protein levels. Covariates measured at visit 5 were used in the main analysis and those measured at visit 3 were used in the midlife replication analysis.
Baseline characteristics were described as mean (SD) for continuous covariates and counts (%) for dichotomous covariates. Our primary analysis used Cox proportional hazards regression models to relate each log base-2 protein level to incident AF (censored at the last follow-up time, death, or the end of 2017 / 2018). We used a series of models to examine the associations and to compare results with results from previous studies. A minimally adjusted model 0 accounted for age, sex, and race/center and provided comparisons with previous cohorts’ results. Model 1 adjusted for the variables in CHARGE-AF score.15 Model 2 additionally adjusted for the confounders of eGFR, anticoagulant use, beta blocker use, and antiarrhythmic (Class I and III) medication use. Bonferroni correction was used to correct for multiple tests; we considered P<0.05 / 4877 = 1.025 x 10−5 to be statistically significant.
We performed additional analyses on the 40 proteins that were statistically significant in either model 1 or model 2. We explored interactions by age, sex and race using a multiplicative term in model 2. We additionally adjusted for NT-proBNP to determine the association of protein levels with incident AF, independent of the level of NT-proBNP. We assessed the proportional hazards assumption in the top 40 proteins with scaled Schoenfeld residuals using both graphical and numerical tests and found no evidence of modeling violations.
In the midlife replication analysis, we used ARIC visit 3 as baseline (1993-95) and examined the association of the 40 proteins with the risk of incident AF through the end of 2010, which was approximately the start of visit 5. We applied the same exclusion criteria as for the visit 5 analysis and used covariates measured at visit 3. For all of these analyses using the top 40 proteins, Bonferroni correction was used to account for multiple tests; we considered P<0.05 / 40 = 1.25 x 10−3 to be statistically significant. We performed statistical analyses using SAS v 9.4 (SAS Inc, Cary, NC).
We performed network pathway analysis to 1) further explore biological mechanisms connected to the proteins associated with incident AF and 2) to identify factors upstream to AF. We analyzed data using of Ingenuity Pathway Analysis (IPA; QIAGEN Inc).14 We included estimates and p-values from model 2 and restricted the IPA to the proteins associated with incident AF at a false discovery rate (FDR) corrected threshold of P < 0.05, resulting in 60 associated proteins. Of these, 56 were successfully mapped to genes in the Ingenuity Pathways Knowledge Base; in some cases duplicated SOMAmers mapped to a single gene (e.g., SVEP1a and SVEP1b) and in other cases, more than one gene product corresponded to a single gene ID (e.g., NT-proBNP and natriuretic peptide B). In the case of duplicates, the maximum expression value of the two was used in the analysis. Further details regarding IPA can be found elsewhere.14 In brief, we used IPA Core Analysis to estimate the degree to which specific canonical pathways, protein networks, and upstream regulators were implicated based on the set of proteins found to be associated with AF risk. For all of the IPA analyses, only statistically significant canonical pathways, physiological systems, upstream regulators, and causal networks are reported, and only a subset are provided in our results.
RESULTS
A total of 4,688 participants with protein levels measured at visit 5 were included in the main analysis (mean age = 75 ± 5 years; 59% female; 20% black race). A total of 585 (13%) participants developed incident AF during a mean (SD) follow-up time of 5.7 (2) years. Descriptive characteristics are provided in Table 1 based on incident AF status. Those who developed AF were older, more likely to be male and white, and had a worse cardiovascular profde compared to those who did not develop AF.
Table 1.
Baseline Clinical Characteristics of Participants by Incident Atrial Fibrillation Status, ARIC, 2011-2013
| Variable | No incident atrial fibrillation through 2018 (n=4083) |
Incident atrial fibrillation through 2018 (n=585) |
|---|---|---|
| Age (years) | 75.2 (5.1) | 77.0 (5.4) |
| Women | 2434 (60%) | 304 (52%) |
| Black | 846 (21%) | 73 (12%) |
| Height (cm) | 165.4 (9.3) | 166.7 (9.9) |
| Weight (kg) | 78.2 (17.1) | 81.0 (18.0) |
| Current smoker | 229 (6%) | 36 (6%) |
| Systolic blood pressure (mmHg) | 130.3 (17.8) | 130.0 (19.0) |
| Diastolic blood pressure (mmHg) | 66.5 (10.5) | 64.0 (10.9) |
| Antihypertensive medication use | 2939 (72%) | 484 (83%) |
| Diabetes mellitus | 1257 (31%) | 204 (35%) |
| Myocardial infarction | 269 (7%) | 67 (11%) |
| Heart failure | 131 (3%) | 60 (10%) |
| Estimated glomerular filtration rate (mL/min per m2) | 65.5 (17.7) | 60.8 (17.9) |
| Anticoagulation medication use | 90 (2%) | 37 (6%) |
| Beta blocker medication use | 1213 (30%) | 285 (49%) |
| Antiarrhythmic use, class I and III | 11 (0.3%) | 13 (2%) |
Values correspond to mean (standard deviation) or N (%)
After adjustment for age, sex, and race/center, 126 protein were significantly associated (p<1.025 x 10−5) with incident AF as listed in Supplementary Table 2. After adjustment for variables included in the CHARGE-AF risk score (model 1), and further adjustment for eGFR and medication use (model 2) 37 and 17 proteins, respectively, remained significantly associated with incident AF. These proteins are listed in Table 2 and ordered by the p-value (from smallest to largest) of Model 2 with p-values <1.025 x 10−5 considered significant. After multivariable adjustment, NT-proBNP had the most significant association; for each doubling of its protein level, the risk of AF was 1.75 times higher (95% CI = 1.60-1.91). Transgelin had the strongest effect size in regards to the risk of incident AF; for every doubling of the protein level, the risk of AF was 2 times higher. Several proteins were inversely associated with incident AF including Protein delta homolog 1 (DLK1) and ATS 13 (ADAMTS13). Protein SET had the strongest inverse effect size; for every doubling of Protein SET the risk of AF decreased by approximately 55% (HR=0.45, 95% CI = 0.28-0.71). Two of the top proteins, SVEP1 and DLK1, are listed twice due to distinct aptamers binding to the same protein. The top 100 proteins associated with incident AF after adjustment for model 2, along with the FDR p-values are presented in Supplementary Table 3.
Table 2.
Protein Biomarkers Associated with Incident Atrial Fibrillation in Late-life, ARIC, 2011-2018
| Model 1 | Model 2 | ||||
|---|---|---|---|---|---|
| Protein Name | Gene Name | HR (95% CI) |
p value | HR (95% CI) |
p value |
| N-terminal pro-BNP | NPPB | 1.82 (1.68-1.98) | 2.91E-45 ‡ | 1.75 (1.60-1.91) | 4.59E-35 ‡ |
| Sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 | SVEP1 | 2.01 (1.71-2.36) | 2.39E-17 ‡ | 1.89 (1.61-2.23) | 2.47E-14 ‡ |
| Sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 | SVEP1 | 1.92 (1.65-2.24) | 2.90E-17 ‡ | 1.84 (1.57-2.16) | 3.31E-14 ‡ |
| Natriuretic peptides B | NPPB | 1.52 (1.36-1.70) | 3.10E-13 ‡ | 1.46 (1.30-1.65) | 4.58E-10 ‡ |
| Transgelin | TAGLN | 1.88 (1.54-2.29) | 3.21E-10 ‡ | 2.01 (1.56-2.59) | 6.41E-08 ‡ |
| Angiopoietin-2 | ANGPT2 | 1.86 (1.53-2.25) | 2.88E-10 ‡ | 1.74 (1.42-2.14) | 1.62E-07 ‡ |
| Protein delta homolog 1 | DLK1 | 0.72 (0.63-0.84) | 1.73E-05 | 0.68 (0.58-0.79) | 7.22E-07 ‡ |
| Slit homolog 2 protein | SLIT2 | 1.44 (1.25-1.65) | 2.90E-07 ‡ | 1.41 (1.23-1.62) | 7.66E-07 ‡ |
| CMRF35-like molecule 2 | CD300E | 1.51 (1.27-1.80) | 2.28E-06 ‡ | 1.52 (1.28-1.80) | 1.68E-06 ‡ |
| Protein delta homolog 1 | DLK1 | 0.73 (0.63-0.85) | 3.33E-05 | 0.68 (0.55-0.80) | 1.81E-06 ‡ |
| Antileukoproteinase | SLPI | 1.97 (1.54-2.51) | 6.66E-08 ‡ | 1.92 (1.46-2.52) | 2.43E-06 ‡ |
| Bone sialoprotein 2 | IBSP | 1.37 (1.22-1.54) | 1.05E-07 ‡ | 1.33 (1.18-1.50) | 2.59E-06 ‡ |
| Microfibril-associated glycoprotein 4 | MFAP4 | 1.54 (1.31-1.80) | 1.22E-07 ‡ | 1.47 (1.25-1.72) | 3.13E-06 ‡ |
| Shadow of prion protein | SPRN | 1.53 (1.26-1.84) | 1.14E-05 | 1.57 (1.30-1.90) | 3.50E-06 ‡ |
| R-spondin-4 | RSPO4 | 1.65 (1.35-2.02) | 1.45E-06 ‡ | 1.63 (1.33-2.01) | 3.67E-06 ‡ |
| Chordin-like protein 1 | CHRDL1 | 1.86 (1.47-2.37) | 3.17E-07 ‡ | 1.79 (1.39-2.31) | 7.64E-06 ‡ |
| Spondin-1 | SPON1 | 1.93 (1.49-2.49) | 6.24E-07 ‡ | 1.81 (1.39-2.34) | 7.70E-06 ‡ |
| Endothelial cell-specific molecule 1 | ESM1 | 1.76 (1.40-2.20) | 7.77E-07 ‡ | 1.66 (1.32-2.07) | 1.08E-05 |
| R-spondin-1 | RSPO1 | 1.65 (1.36-1.99) | 2.64E-07 ‡ | 1.57 (1.28-1.92) | 1.26E-05 |
| Macrophage-capping protein | CAPG | 1.53 (1.31-1.77) | 3.28E-08 ‡ | 1.44 (1.22-1.70) | 1.59E-05 |
| Scavenger receptor class F member 1 | SCARF1 | 1.81 (1.42-2.32) | 2.51E-06 ‡ | 1.78 (1.37-2.31) | 1.77E-05 |
| Atrial natriuretic factor | NPPA | 1.72 (1.42-2.09) | 3.03E-08 ‡ | 1.54 (1.26-1.88) | 2.30E-05 |
| Insulin-like growth factor-binding protein 2 | IGFBP2 | 1.40 (1.21-1.62) | 7.27E-06 ‡ | 1.35 (1.16-1.57) | 9.30E-05 |
| Growth/differentiation factor 11/8 | GDF11 MSTN |
0.55 (0.42-0.72) | 9.54E-06 ‡ | 0.59 (0.45-0.77) | 9.78E-05 |
| Triggering receptor expressed on myeloid cells 1 | TREM1 | 1.56 (1.30-1.87) | 1.14E-06 ‡ | 1.50 (1.22-1.84) | 1.00E-04 |
| A disintegrin and metalloproteinase with thrombospondin motifs 13 | ADAMTS13 | 0.55 (0.43-0.71) | 2.56E-06 ‡ | 0.60 (0.46-0.71) | 1.15E-04 |
| Metalloproteinase inhibitor 4 | TIMP4 | 1.52 (1.26-1.82) | 7.03E-06 ‡ | 1.43 (1.19-1.73) | 1.58E-04 |
| Ribonuclease pancreatic | RNASE1 | 1.29 (1.17-1.44) | 1.49E-06 ‡ | 1.38 (1.17-1.64) | 1.60 E-04 |
| EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | 2.13 (1.57-2.90) | 1.24E-06 ‡ | 1.94 (1.37-2.75) | 1.70E-04 |
| Regenerating islet-derived protein 3-alpha | REG3A | 1.30 (1.16-1.46) | 4.81E-06 ‡ | 1.26 (1.12-1.43) | 2.01E-04 |
| Lysosomal Pro-X carboxypeptidase | PRCP | 0.56 (0.43-0.72) | 9.07E-06 ‡ | 0.60 (0.46-0.79) | 2.13E-04 |
| Cartilage intermediate layer protein 2 | CILP2 | 0.64 (0.53-0.78) | 6.27E-06 ‡ | 0.69 (0.57-0.84) | 2.15E-04 |
| Sodium/potassium-transporting ATPase subunit beta-1 | ATP1B1 | 0.62 (0.50-0.76) | 9.74E-06 ‡ | 0.66 (0.53-0.82) | 2.50E-04 |
| Hepatitis A virus cellular receptor 2 | HAVCR2 | 1.60 (1.31-1.95) | 3.57E-06 ‡ | 1.51 (1.21-1.88) | 3.05E-04 |
| Endostatin | COL18A1 | 1.93 (1.47-2.55) | 3.14E-06 ‡ | 1.90 (1.33-2.72) | 4.07E-04 |
| Protein SET | SET | 0.36 (0.23-0.55) | 4.36E-06 ‡ | 0.45 (0.28-0.71) | 5.88E-04 |
| Gamma-aminobutyric acid receptor-associated protein-like 1 | GABARAPL1 | 1.80 (1.41-2.31) | 2.60E-06 ‡ | 1.65 (1.23-2.21) | 8.44E-04 |
| Gamma-aminobutyric acid receptor-associated protein | GABARAP | 1.95 (1.46-2.60) | 6.88E-06 ‡ | 1.73 (1.22-2.47) | 2.30E-03 |
| Coagulation Factor X | F10 | 0.51 (0.40-0.64) | 3.09E-08 ‡ | 0.69 (0.47-0.99) | 4.50E-02 |
| Coagulation factor Xa | F10 | 0.52 (0.41-0.66) | 8.99E-08 ‡ | 0.71 (0.50-1.01) | 5.95E-02 |
Model 1: adjusted for age, sex, race/center, current cigarette smoking, height, weight, systolic and diastolic blood pressure, the use of hypertension medications, diabetes, prevalent myocardial infarction and prevalent heart failure. Model 2: adjusted for Model 1 + estimated glomerular filtration rate, antiarrhythmic medication use, beta blocker medication use, and anticoagulation use
Hazard ratio (HR) expressed as the risk of incident AF per doubling of the protein value
Significance level of P<0.05/4877 = 1.025 x 10−5. These 40 proteins are ordered by smallest to largest p-value for Model 2.
All proteins listed are novel associations with incident AF except for NT-proBNP and ADAMTS13 which have been previously published.
We examined interactions by age, sex, and race in the 40 proteins listed in Table 2 and we did not find any statistically significant interactions. We additionally adjusted for NT-proBNP to determine the association of protein levels with incident AF independent of NT-proBNP, and results for the main 40 proteins are listed in Supplementary Table 4. Eight of the proteins remained significantly associated with incident AF and include CMRF35-like molecule 2 (CD300E), Growth/differentiation factor 11/8 (GDF11 MSTN), DLK1 (2 aptamers), Antileukoproteinase (SLPI), Cartilage intermediate layer protein 2 (CILP2), Scavenger receptor class F member 1 (SCARF1), and Gamma-aminobutyric acid receptor-associated protein-like 1 (GABARAPL1).
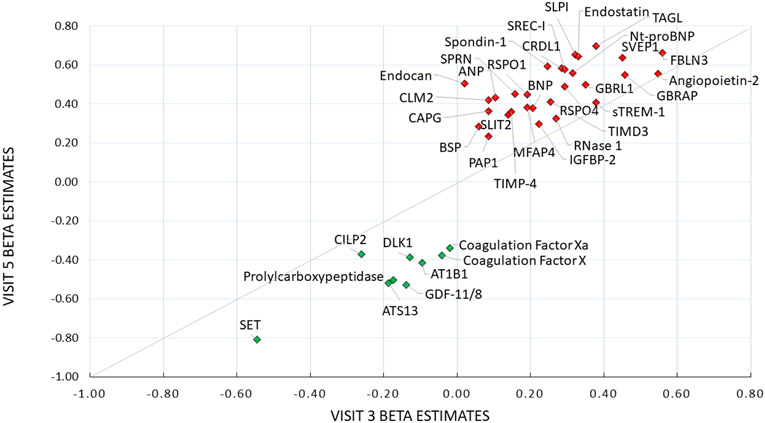
We ran a secondary analysis as an internal validation with 10,908 AF-free participants with protein measures at visit 3 and followed them until the end of 2010. At this visit, participants were younger with fewer comorbidities and on fewer medications (mean age = 60 ± 6 years; 55% female; 21% black race). A total of 1397 (13%) participants developed incident AF during a mean (SD) follow-up time of 13.9 (4) years. Of the top 40 proteins from the main analysis, 21 were significantly associated with incident AF (Table 3) in mid-life replication model 1, and 17 remained significant after adjustment for factors in model 2. NT-proBNP, SVEP1, Natriuretic peptides B, Transgelin, and Angiopoietin-2 were the proteins most strongly associated with incident AF in both mid-life and later-life. Figure 2 depicts the beta estimates from model 2 for the top 40 proteins measured at mid-life (visit 3) plotted against those measured in later life (visit 5) for the association with incident AF. Several proteins maintained relatively consistent effect sizes at both visits, including CILP2, IGFBP-2, and Angiopoietin-2, among others.
Table 3.
Replication Analysis of Associations of the Top 40 Late-Life Protein Biomarkers Measured in Mid-life with Incident Atrial Fibrillation, ARIC, 1993-2010
| Model 1 | Model 2 | ||||
|---|---|---|---|---|---|
| Protein Target Name | Gene Name | HR (95% CI) |
p value |
HR (95% CI) |
p value |
| N-terminal pro-BNP | NPPB | 1.40 (1.32-1.47) | 3.29E-35 ‡ | 1.37 (1.30-1.45) | 2.92E-31 ‡ |
| Angiopoietin-2 | ANGPT2 | 1.77 (1.55-2.02) | 3.54E-17 ‡ | 1.73 (1.51-1.98) | 6.42E-16 ‡ |
| Sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 | SVEP1 | 1.57 (1.36-1.80) | 3.11E-10 ‡ | 1.57 (1.36-1.81) | 4.20E-10 ‡ |
| Sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 | SVEP1 | 1.52 (1.33-1.74) | 6.99E-10 ‡ | 1.52 (1.33-1.74) | 9.91E-10 ‡ |
| Triggering receptor expressed on myeloid cells 1 | TREM1 | 1.49 (1.30-1.71) | 1.14E-08 ‡ | 1.46 (1.26-1.68) | 1.73E-07 ‡ |
| Insulin-like growth factor-binding protein 2 | IGFBP2 | 1.26 (1.16-1.37) | 7.11E-08 ‡ | 1.25 (1.15-1.36) | 1.91E-07 ‡ |
| Ribonuclease pancreatic | RNASE1 | 1.32 (1.21-1.45) | 2.49E-09 ‡ | 1.31 (1.18-1.45) | 3.80E-07 ‡ |
| EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | 1.83 (1.47-2.27) | 3.86E-08 ‡ | 1.75 (1.40-2.18) | 6.58E-07 ‡ |
| Transgelin | TAGLN | 1.50 (1.30-1.74) | 6.20E-08 ‡ | 1.46 (1.25-1.70) | 2.33E-06 ‡ |
| Natriuretic peptides B | NPPB | 1.25 (1.13-1.38) | 7.82E-06 ‡ | 1.23 (1.12-1.36) | 2.52E-05 ‡ |
| Protein SET | SET | 0.56 (0.42-0.73) | 2.12E-05 ‡ | 0.58 (0.44-0.75) | 7.90E-05 ‡ |
| Gamma-aminobutyric acid receptor-associated protein | GABARAP | 1.69 (1.36-2.11) | 2.37E-06 ‡ | 1.58 (1.25-2.00) | 1.17E-04 ‡ |
| Gamma-aminobutyric acid receptor-associated protein-like 1 | GABARAPL1 | 1.48 (1.24-1.76) | 1.14E-05 ‡ | 1.42 (1.18-1.70) | 2.49E-04 ‡ |
| Hepatitis A virus cellular receptor 2 | HAVCR2 | 1.34 (1.16-1.54) | 5.11E-05 ‡ | 1.29 (1.12-1.49) | 4.43E-04 ‡ |
| Microfibril-associated glycoprotein 4 | MFAP4 | 1.21 (1.08-1.34) | 5.41E-04 ‡ | 1.21 (1.09-1.35) | 4.56E-04 ‡ |
| Cartilage intermediate layer protein 2 | CILP2 | 0.77 (0.67-0.89) | 3.87E-04 ‡ | 0.77 (0.67-0.89) | 4.98E-04 ‡ |
| Endostatin | COL18A1 | 1.45 (1.21-1.74) | 7.41E-05 ‡ | 1.39 (1.15-1.68) | 7.77E-04 ‡ |
| Antileukoproteinase | SLPI | 1.46 (1.21-1.76) | 8.97E-05 ‡ | 1.38 (1.13-1.68) | 1.32E-03 |
| R-spondin-4 | RSPO4 | 1.37 (1.14-1.64) | 6.87E-04 ‡ | 1.34 (1.12-1.61) | 1.44E-03 |
| Scavenger receptor class F member 1 | SCARF1 | 1.37 (1.13-1.66) | 1.10E-03 ‡ | 1.34 (1.11-1.63) | 2.91E-03 |
| Chordin-like protein 1 | CHRDL1 | 1.38 (1.13-1.68) | 1.52E-03 | 1.33 (1.09-1.63) | 5.16E-03 |
| R-spondin-1 | RSPO1 | 1.23 (1.08-1.40) | 1.64E-03 | 1.21 (1.06-1.38) | 5.36E-03 |
| Spondin-1 | SPON1 | 1.29 (1.07-1.56) | 6.51E-03 | 1.28 (1.06-1.55) | 9.41E-03 |
| Protein delta homolog 1 | DLK1 | 0.91 (0.83-1.00) | 4.96E-02 | 0.88 (0.80-0.97) | 1.10E-02 |
| Protein delta homolog 1 | DLK1 | 0.91 (0.82-1.00) | 5.22E-02 | 0.88 (0.79-0.97) | 1.24E-02 |
| A disintegrin and metalloproteinase with thrombospondin motifs 13 | ADAMTS13 | 0.81 (0.70-0.93) | 3.70E-03 | 0.83 (0.72-0.96) | 1.39E-02 |
| Metalloproteinase inhibitor 4 | TIMP4 | 1.19 (1.04-1.35) | 1.06E-02 | 1.16 (1.02-1.33) | 2.29E-02 |
| Slit homolog 2 protein | SLIT2 | 1.15 (1.00-1.31) | 4.96E-02 | 1.15 (1.01-1.32) | 3.96E-02 |
| Shadow of prion protein | SPRN | 1.15 (0.99-1.35) | 6.63E-02 | 1.17 (1.00-1.36) | 4.56E-02 |
| Lysosomal Pro-X carboxypeptidase | PRCP | 0.81 (0.69-0.96) | 1.42E-02 | 0.84 (0.71-1.00) | 4.84E-02 |
| Regenerating islet-derived protein 3-alpha | REG3A | 1.12 (1.02-1.22) | 1.19E-02 | 1.09 (1.00-1.19) | 5.64E-02 |
| Growth/differentiation factor 11/8 | GDF11 MSTN |
0.88 (0.74-1.04) | 1.30E-01 | 0.87 (0.73-1.03) | 1.05E-01 |
| Macrophage-capping protein | CAPG | 1.12 (1.01-1.25) | 2.72E-02 | 1.09 (0.98-1.21) | 1.06E-01 |
| Sodium/potassium-transporting ATPase subunit beta-1 | ATP1B1 | 0.90 (0.79-1.02) | 8.61E-02 | 0.91 (0.80-1.03) | 1.26E-01 |
| Bone sialoprotein 2 | IBSP | 1.05 (0.95-1.15) | 2.78E-01 | 1.06 (0.97-1.17) | 1.98E-01 |
| CMRF35-like molecule 2 | CD300E | 1.08 (0.93-1.24) | 3.06E-01 | 1.09 (0.93-1.26) | 2.39E-01 |
| Atrial natriuretic factor | NPPA | 1.11 (0.92-1.34) | 2.82E-01 | 1.11 (0.91-1.34) | 2.99E-01 |
| Coagulation Factor X | F10 | 0.87 (0.71-1.06) | 1.71E-01 | 0.96 (0.77-1.20) | 7.30E-01 |
| Endothelial cell-specific molecule 1 | ESM1 | 1.03 (0.89-1.18) | 7.15E-01 | 1.02 (0.89-1.17) | 7.52E-01 |
| Coagulation factor Xa | F10 | 0.88 (0.72-1.08) | 2.08E-01 | 0.98 (0.78-1.22) | 8.24E-01 |
Model 1: adjusted for age, sex, race/center, current cigarette smoking, height, weight, systolic and diastolic blood pressure, the use of hypertension medications, diabetes, prevalent myocardial infarction and prevalent heart failure. Model 2: adjusted for Model 1 + estimated glomerular filtration rate, antiarrhythmic medication use, beta blocker medication use, and anticoagulation use
Hazard ratio (HR) expressed as the risk of incident AF per doubling of the protein value
Significance level of P<0.05/40 = 1.25 x 10−3. These 40 proteins are ordered by smallest to largest p-value for Model 2.
Figure 2.
Beta estimates for Associations of the Top 40 Protein Biomarkers Measured in Mid-life (visit 3) and Late-Life (visit 5) with Incident Atrial Fibrillation, ARIC, 1993-2018. Green diamonds represent inverse associations with the risk of AF and red diamonds indicate an increased risk of incident AF. The fully-adjusted model adjusted for age, sex, race/center, current cigarette smoking, height, weight, systolic and diastolic blood pressure, the use of hypertension medications, diabetes, prevalent myocardial infarction, prevalent heart failure, estimated glomerular filtration rate, antiarrhythmic medication use, beta blocker medication use, and anticoagulation use
Proteins associated with AF in late-life model 2 with an FDR P value <0.05 (listed in Supplemental Table 3) were brought into the IPA environment. Of those, 56 proteins were mapped into 9 main networks, with the top network centered around MMP-2, which was an upregulated protein in our analysis. IPA identified canonical pathways, which are well-characterized metabolic and cell-signaling pathways, using known associations of our uploaded proteins. The 10 canonical pathways that were most significantly associated with our proteins are listed in Supplemental Table 5. The top canonical pathway was the inhibition of MMPs, followed by axonal guidance signaling, and factors promoting cardiogenesis. The identified downstream diseases and functions were found to be centered around common themes of inflammatory response suppression, cardiac dysfunction, kidney failure, and cell movement of cancer cells.
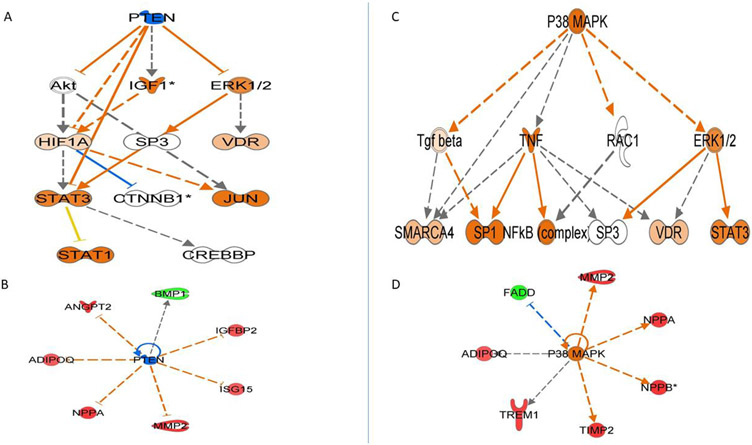
Upstream regulator analysis identified molecules upstream of the proteins that potentially explain the up-regulated and down-regulated proteins observed in our dataset. Our top 20 identified upstream regulators are listed in Supplemental Table 6. Figure 3 depicts the mechanistic networks that are associated with the top 2 identified upstream regulators and links the upstream regulator to our observed proteins via the intermediary molecules depicted in the figure. PTEN (phosphatase and tensin homolog) was predicted to be significantly inhibited based on the observed protein expressions in our data. P38 MAPK was predicted to be our strongest activated upstream regulator.
Figure 3.
The top upstream regulators identified using IPA, based on experimentally observed relationships between regulators and genes or gene products. Panel A depicts the hierarchical associations between PTEN and its expected downstream regulators to produce the associations we observe in the proteins in Panel B. Panel C depicts the hierarchical associations we would expect between P38 MAPK and intermediate regulators to produce the effects on the proteins observed in Panel D. Orange nodes are when leading the activation of the downstream node, blue nodes are predicted to be inhibited, and white nodes represent IPA predicted molecules with non-consistent activation patterns. Red nodes are upregulated proteins, green nodes are downregulated proteins. Edges connecting the nodes are colored orange when leading to activation, blue when leading to inhibition, and yellow if the findings underlying the relationship are inconsistent with the state of the downstream node.
DISCUSSION
In this community-based prospective population study of older adults, we tested 4,877 plasma proteins and observed that 37 proteins were associated with risk of incident AF over a nearly 6 year follow-up period at a Bonferonni corrected significance level and after adjustment for known AF risk factors. After additional adjustment for eGFR and medication use, 17 proteins remained significantly associated with an increased risk of AF. In a midlife replication sample that used proteins measured at an early ARIC visit, nearly half of the top proteins from the main analysis also demonstrated a robust association with non-overlapping incident AF events. Several proteins maintained relatively consistent effect sizes at both visits, including CILP2, IGFBP-2, and Angiopoietin-2, among others. For all analyses, NT-proBNP showed the strongest association with incident AF. Using a less stringent FDR-corrected threshold, we performed network pathway analysis on the top 56 unique proteins mapped to genes and determined that the inhibition of matrix metalloproteases was the primary canonical pathway represented by our results. We identified several potential upstream regulators that may provide insight into biological mechanisms involved in AF pathogenesis.
Natriuretic peptides (both NT-proBNP and mid-regional atrial natriuretic peptide) are markers of cardiac overload. Multiple prospective population-based cohort studies and previous proteomic analyses have reported that higher baseline NT-proBNP concentrations predict increased incident AF.1,3-5,15-18 We also corroborated several other proteins that have been associated with incident AF from other proteomic analyses including ATS13 (ADAMTS13) and Angiopoietin-2.3 Additionally, previously reported BMP-1,3 MMP-2, and IGFBP-74 associations with AF met our less-stringent FDR p value cutoff and were included in IPA. Angiopoietins are endothelial growth factors that regulate angiogenesis and vascular function and increased levels of angiopoietin-2 have been observed in several types of prevalent cardiovascular disease. Similarly, the BMP signaling pathway plays an important role in the development of myocardial remodeling.19 ADAMTS13 is a von Willebrand factor protease that has been positively associated with incident MI, stroke, AF, the risk of stroke in patients with AF, indicating a role as a potential marker of a prothrombotic environment.20,21 The peptic hormone insulin-like growth factor 1 (IGF-1) and several of its binding proteins are associated positively with cardiovascular disease incidence,22 and have additionally been linked to AF.4
Our study reports several novel associations between circulating protein levels and incident AF, which we confirmed in the mid-life replication analysis. Transgelin, a 22- kD protein of the calponin family, is exclusively and abundantly expressed in the cytoskeleton of visceral and vascular smooth muscle cells. SVEP1 is a cell-adhesion molecule that acts as a ligand for integrin α9β1 and is believed to facilitate cellular adhesion in the context of pro-inflammatory signaling.23 The identification of a disease-associated missense variant in SVEP1 has been hypothesized to play a role in the development of atherosclerosis and coronary heart disease,24 but the contribution of SVEP1 in AF remains to be clarified. Additional prospective studies, using immunoassays, should verify whether Transgelin and SVEP1 are associated with AF incidence and whether the associations are causal.
Pathway analysis indicated our top canonical pathway was the suppression of MMPs and that pathway included higher detected levels of TIMP-2, TIMP-4 and MMP-2. Atrial fibrosis is considered a key element of the AF substrate, with extracellular matrix (ECM) remodeling playing a major role in this process.25 The MMPs are a family of twenty zinc-dependent enzymes that together with their specific endogenous inhibitors (TIMPs), regulate the degradation of collagen and other ECM molecules. Several case-control studies have observed relationships between MMPs and AF, with the most significant associations related to MMP-9,26,27 and mixed results between MMP-2 and incident AF.28 Observational studies of TIMP levels and AF have mainly shown no association, although higher TIMP-4 levels were found to be associated with prevalent AF in a few studies.28-30 We found increased levels of both TIMP-2 and MMP-2 were associated with greater incident AF, and appear to be activated by several different regulators in our network analysis.
IPA identified potential relationships upstream of our target molecules along with the predicted activated / inhibited state of genes and gene products. The top upstream molecule was PTEN, which is involved in aging and tumor suppression and was predicted to be significantly inhibited based on the observed protein expressions in our data. PTEN negatively regulates intracellular levels of phosphatidylinositol-3,4,5-trisphosphate in cells and functions as a tumor suppressor by negatively regulating the AKT/PKB signaling pathway. P38 MAPK was predicted by the IPA to be activated and plays a role in apoptosis and cell differentiation. This protein kinase is also involved in a variety of binding steps, including magnesium ion binding, phosphatase binding, and transcription factor binding among other functions.
The main strengths of this study are the plethora of proteomic data in a community-based prospective sample, the quality of risk factor variables measured, and the number of AF events during follow-up. The ARIC study also includes black individuals, which have not been included in proteomic - AF analyses to date. We found no evidence of race interaction, indicating that the observed associations did not differ between blacks and whites. We were able to perform an internal mid-life replication analysis which strengthened our findings in older adults, however, replication in an external cohort would further strengthen the reproducibility and particularly establish the generalizability of these findings. Our study has several additional limitations. Incident AF was identified mainly from hospitalization discharges, and we could be missing asymptomatic AF or AF managed exclusively in an outpatient setting. However, we and others have previously shown that the validity of AF ascertainment using hospitalizations is acceptable, and that incidence rates of AF in the ARIC study are consistent with other population-based studies. The possibility of protein degradation during long-term storage cannot be excluded; however, a validation study in ARIC did not support widespread protein degradation across visits.13 Proteins in this study were measured from plasma and protein origin cannot be confirmed; therefore our IPA pathway results should be viewed within this limitation and considered speculative. Although our proteomic platform is the largest to date in cardiovascular research, we are only able to detect proteins included on this platform. Finally, SOMAscan measurements were semi-quantitative and need replication in other prospective studies.
In conclusion, we conducted proteomic profiling in a community-based population to assess the relationship between proteomics and incident AF in a cohort of older-aged black and white adults. The current results reinforced previous findings but offered novel observations into the biological changes that may precede AF onset and provided insight into mechanistic pathways of AF development. If replicated, these proteins may prove to be novel biomarkers for AF, allow for the development of AF risk scores, or serve as possible pharmacologic targets in AF treatment or prevention.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the staff and participants of the ARIC study for their important contributions.
FUNDING
The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700005I, HHSN268201700004I).
This work was also supported by grants from the National Heart Lung and Blood Institute [R01HL126637-01A1 (LYC), R01HL141288 (LYC), K24 HL148521 (AA), and the American Heart Association [16EIA26410001 (AA)].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Disclosures: None
REFERENCES
- 1.Lind L, Sundstrom J, Stenemo M, Hagstrom E, Arnlov J. Discovery of new biomarkers for atrial fibrillation using a custom-made proteomics chip. Heart 2017;103:377–382. [DOI] [PubMed] [Google Scholar]
- 2.Willeit K, Pechlaner R, Willeit P, Skroblin P, Paulweber B, Schernthaner C, Toell T, Egger G, Weger S, Oberhollenzer M, Kedenko L, Iglseder B, Bonora E, Schett G, Mayr M, Willeit J, Kiechl S. Association Between Vascular Cell Adhesion Molecule 1 and Atrial Fibrillation. JAMA Cardiol 2017;2:516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko D, Benson MD, Ngo D, Yang Q, Larson MG, Wang TJ, Trinquart L, McManus DD, Lubitz SA, Ellinor PT, Vasan RS, Gerszten RE, Benjamin EJ, Lin H. Proteomics Profiling and Risk of New-Onset Atrial Fibrillation: Framingham Heart Study. J Am Heart Assoc 2019;8:e010976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molvin J, Jujic A, Melander O, Pareek M, Rastam L, Lindblad U, Daka B, Leosdottir M, Nilsson P, Olsen M, Magnusson M. Exploration of pathophysiological pathways for incident atrial fibrillation using a multiplex proteomic chip. Open Heart 2020;7:e001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staerk L, Preis SR, Lin H, Lubitz SA, Ellinor PT, Levy D, Benjamin EJ, Trinquart L. Protein Biomarkers and Risk of Atrial Fibrillation: The FHS. Circulation Arrhythmia and Electrophysiology 2020;13:e007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol 1989;129:687–702. [PubMed] [Google Scholar]
- 7.Alonso A, Agarwal SK, Soliman EZ, Ambrose M, Chamberlain AM, Prineas RJ, Folsom AR. Incidence of atrial fibrillation in whites and African-Americans: the Atherosclerosis Risk in Communities (ARIC) study. Am Heart J 2009;158:111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, Flather D, Forbes A, Foreman T, Fowler C, Gawande B, Goss M, Gunn M, Gupta S, Halladay D, Heil J, Heilig J, Hicke B, Husar G, Janjic N, Jarvis T, Jennings S, Katilius E, Keeney TR, Kim N, Koch TH, Kraemer S, Kroiss L, Le N, Levine D, Lindsey W, Lollo B, Mayfield W, Mehan M, Mehler R, Nelson SK, Nelson M, Nieuwlandt D, Nikrad M, Ochsner U, Ostroff RM, Otis M, Parker T, Pietrasiewicz S, Resnicow DI, Rohloff J, Sanders G, Sattin S, Schneider D, Singer B, Stanton M, Sterkel A, Stewart A, Stratford S, Vaught JD, Vrkljan M, Walker JJ, Watrobka M, Waugh S, Weiss A, Wilcox SK, Wolfson A, Wolk SK, Zhang C, Zichi D. Aptamer-based multiplexed proteomic technology for biomarker discovery. PloS one 2010;5:e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gold L, Walker JJ, Wilcox SK, Williams S. Advances in human proteomics at high scale with the SOMAscan proteomics platform. New biotechnology 2012;29:543–549. [DOI] [PubMed] [Google Scholar]
- 10.Kim CH, Tworoger SS, Stampfer MJ, Dillon ST, Gu X, Sawyer SJ, Chan AT, Libermann TA, Eliassen AH. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci Rep 2018,8:8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Candia J, Cheung F, Kotliarov Y, Fantoni G, Sellers B, Griesman T, Huang J, Stuccio S, Zingone A, Ryan BM, Tsang JS, Biancotto A. Assessment of Variability in the SOMAscan Assay. Sci Rep 2017;7:14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Z, Xiao Z, Kalantar-Zadeh K, Moradi H, Shafi T, Waikar SS, Quarles LD, Yu Z, Tin A, Coresh J, Kovesdy CP. Validation of a Novel Modified Aptamer-Based Array Proteomic Platform in Patients with End-Stage Renal Disease. Diagnostics (Basel) 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tin A, Yu B, Ma J, Masushita K, Daya N, Hoogeveen RC, Ballantyne CM, Couper D, Rebholz CM, Grams ME, Alonso A, Mosley T, Heiss G, Ganz P, Selvin E, Boerwinkle E, Coresh J. Reproducibility and Variability of Protein Analytes Measured Using a Multiplexed Modified Aptamer Assay. J Appl Lab Med 2019;4:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alonso A, Krijthe BP, Aspelund T, Stepas KA, Pencina MJ, Moser CB, Sinner MF, Sotoodehnia N, Fontes JD, Janssens AC, Kronmal RA, Magnani JW, Witteman JC, Chamberlain AM, Lubitz SA, Schnabel RB, Agarwal SK, McManus DD, Ellinor PT, Larson MG, Burke GL, Launer LJ, Hofman A, Levy D, Gottdiener JS, Kaab S, Couper D, Harris TB, Soliman EZ, Stricker BH, Gudnason V, Heckbert SR, Benjamin EJ. Simple risk model predicts incidence of atrial fibrillation in a racially and geographically diverse population: the CHARGE-AF consortium. J Am Heart Assoc 2013;2:e000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patton KK, Ellinor PT, Heckbert SR, Christenson RH, DeFilippi C, Gottdiener JS, Kronmal RA. N-terminal pro-B-type natriuretic peptide is a major predictor of the development of atrial fibrillation: the Cardiovascular Health Study. Circulation 2009;120:1768–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patton KK, Heckbert SR, Alonso A, Bahrami H, Lima JA, Burke G, Kronmal RA. N-terminal pro-B-type natriuretic peptide as a predictor of incident atrial fibrillation in the Multi-Ethnic Study of Atherosclerosis: the effects of age, sex and ethnicity. Heart 2013;99:1832–1836. [DOI] [PubMed] [Google Scholar]
- 18.Sinner MF, Stepas KA, Moser CB, Krijthe BP, Aspelund T, Sotoodehnia N, Fontes JD, Janssens AC, Kronmal RA, Magnani JW, Witteman JC, Chamberlain AM, Lubitz SA, Schnabel RB, Vasan RS, Wang TJ, Agarwal SK, McManus DD, Franco OH, Yin X, Larson MG, Burke GL, Launer LJ, Hofman A, Levy D, Gottdiener JS, Kaab S, Couper D, Harris TB, Astor BC, Ballantyne CM, Hoogeveen RC, Arai AE, Soliman EZ, Ellinor PT, Stricker BH, Gudnason V, Heckbert SR, Pencina MJ, Benjamin EJ, Alonso A. B-type natriuretic peptide and C-reactive protein in the prediction of atrial fibrillation risk: the CHARGE-AF Consortium of community-based cohort studies. Europace : European pacing, arrhythmias, and cardiac electrophysiology : journal of the working groups on cardiac pacing, arrhythmias, and cardiac cellular electrophysiology of the European Society of Cardiology 2014;16:1426–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrell NW, Bloch DB, ten Dijke P, Goumans MJ, Hata A, Smith J, Yu PB, Bloch KD. Targeting BMP signalling in cardiovascular disease and anaemia. Nat Rev Cardiol 2016;13:106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uemura T, Kaikita K, Yamabe H, Soejima K, Matsukawa M, Fuchigami S, Tanaka Y, Morihisa K, Enomoto K, Sumida H, Sugiyama S, Ogawa H. Changes in plasma von Willebrand factor and ADAMTS13 levels associated with left atrial remodeling in atrial fibrillation. Thromb Res 2009;124:28–32. [DOI] [PubMed] [Google Scholar]
- 21.Hijazi Z, Wallentin L, Lindback J, Alexander JH, Connolly SJ, Eikelboom JW, Ezekowitz MD, Granger CB, Lopes RD, Pol T, Yusuf S, Oldgren J, Siegbahn A. Screening of Multiple Biomarkers Associated With Ischemic Stroke in Atrial Fibrillation. J Am Heart Assoc 2020;9:e018984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourron O, Le Bouc Y, Berard L, Kotti S, Brunel N, Ritz B, Leclercq F, Tabone X, Drouet E, Mulak G, Danchin N, Simon T. Impact of age-adjusted insulin-like growth factor 1 on major cardiovascular events after acute myocardial infarction: results from the fast-MI registry. J Clin Endocrinol Metab 2015;100:1879–1886. [DOI] [PubMed] [Google Scholar]
- 23.Nakada TA, Russell JA, Boyd JH, Thair SA, Walley KR. Identification of a nonsynonymous polymorphism in the SVEP1 gene associated with altered clinical outcomes in septic shock. Crit Care Med 2015;43:101–108. [DOI] [PubMed] [Google Scholar]
- 24.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators, Stitziel NO, Stirrups KE, Masca NG, Erdmann J, Ferrario PG, Konig IR, Weeke PE, Webb TR, Auer PL, Schick UM, Lu Y, Zhang H, Dube MP, Goel A, Farrall M, Peloso GM, Won HH, Do R, van Iperen E, Kanoni S, Kruppa J, Mahajan A, Scott RA, Willenberg C, Braund PS, van Capelleveen JC, Doney AS, Donnelly LA, Asselta R, Merlini PA, Duga S, Marziliano N, Denny JC, Shaffer CM, El-Mokhtari NE, Franke A, Gottesman O, Heilmann S, Hengstenberg C, Hoffman P, Holmen OL, Hveem K, Jansson JH, Jockel KH, Kessler T, Kriebel J, Laugwitz KL, Marouli E, Martinelli N, McCarthy MI, Van Zuydam NR, Meisinger C, Esko T, Mihailov E, Escher SA, Alver M, Moebus S, Morris AD, Muller-Nurasyid M, Nikpay M, Olivieri O, Lemieux Perreault LP, AlQarawi A, Robertson NR, Akinsanya KO, Reilly DF, Vogt TF, Yin W, Asselbergs FW, Kooperberg C, Jackson RD, Stahl E, Strauch K, Varga TV, Waldenberger M, Zeng L, Kraja AT, Liu C, Ehret GB, Newton-Cheh C, Chasman DI, Chowdhury R, Ferrario M, Ford I, Jukema JW, Kee F, Kuulasmaa K, Nordestgaard BG, Perola M, Saleheen D, Sattar N, Surendran P, Tregouet D, Young R, Howson JM, Butterworth AS, Danesh J, Ardissino D, Bottinger EP, Erbel R, Franks PW, Girelli D, Hall AS, Hovingh GK, Kastrati A, Lieb W, Meitinger T, Kraus WE, Shah SH, McPherson R, Orho-Melander M, Melander O, Metspalu A, Palmer CN, Peters A, Rader D, Reilly MP, Loos RJ, Reiner AP, Roden DM, Tardif JC, Thompson JR, Wareham NJ, Watkins H, Willer CJ, Kathiresan S, Deloukas P, Samani NJ, Schunkert H. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med 2016;374:1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nattel S, Burstein B, Dobrev D. Atrial remodeling and atrial fibrillation: mechanisms and implications. Circulation Arrhythmia and Electrophysiology 2008;1:62–73. [DOI] [PubMed] [Google Scholar]
- 26.Huxley RR, Lopez FL, MacLehose RF, Eckfeldt JH, Couper D, Leiendecker-Foster C, Hoogeveen RC, Chen LY, Soliman EZ, Agarwal SK, Alonso A. Novel association between plasma matrix metalloproteinase-9 and risk of incident atrial fibrillation in a case-cohort study: the Atherosclerosis Risk in Communities study. PloS one 2013;8:e59052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano Y, Niida S, Dote K, Takenaka S, Hirao H, Miura F, Ishida M, Shingu T, Sueda T, Yoshizumi M, Chayama K. Matrix metalloproteinase-9 contributes to human atrial remodeling during atrial fibrillation. J Am Coll Cardiol 2004;43:818–825. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Xu B, Wu N, Xiang Y, Wu L, Zhang M, Wang J, Chen X, Li Y, Zhong L. Association of MMPs and TIMPs With the Occurrence of Atrial Fibrillation: A Systematic Review and Meta-analysis. Can J Cardiol 2016;32:803–813. [DOI] [PubMed] [Google Scholar]
- 29.Wakula P, Neumann B, Kienemund J, Thon-Gutschi E, Stojakovic T, Manninger M, Scherr D, Scharnagl H, Kapl M, Pieske B, Heinzel FR. CHA2DS2-VASc score and blood biomarkers to identify patients with atrial high-rate episodes and paroxysmal atrial fibrillation. Europace : European pacing, arrhythmias, and cardiac electrophysiology: journal of the working groups on cardiac pacing, arrhythmias, and cardiac cellular electrophysiology of the European Society of Cardiology 2017;19:544–551. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Zhang HT, Yang XL. Effect of matrix metalloproteinase and their inhibitors on atrial myocardial structural remodeling. J Cardiovasc Med (Hagerstown) 2013;14:265–269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.