Summary
Gram-negative bacteria, mitochondria, and chloroplasts all possess an outer membrane populated with a host of β-barrel outer-membrane proteins (βOMPs). These βOMPs play crucial roles in maintaining viability of their hosts and therefore, it is essential to understand the biogenesis of this class of membrane proteins. In recent years, significant structural and functional advancements have been made toward elucidating this process, which is mediated by the β-barrel assembly machinery (BAM) in Gram-negative bacteria, and by the sorting and assembly machinery (SAM) in mitochondria. Structures of both BAM and SAM have now been reported, allowing a comparison and dissection of the two machineries, with other studies reporting on functional aspects of each. Together, these new insights provide compelling support for the proposed budding mechanism, where each nascent βOMP forms a hybrid-barrel intermediate with BAM/SAM in route to its biogenesis into the membrane. Here, we will review these recent studies and highlight their contributions towards understanding βOMP biogenesis in Gram-negative bacteria and in mitochondria. We will also weigh the evidence supporting each of the two leading mechanistic models for how BAM/SAM function, and offer an outlook on future studies within the field.
Keywords: Gram-negative bacteria, outer membrane, β-barrel, outer membrane protein, protein folding, envelope biogenesis, lateral gate
β-barrel outer membrane proteins
One defining feature of Gram-negative bacteria, mitochondria, and chloroplasts is that they are all enveloped by a double membrane consisting of an inner membrane (IM) and outer membrane (OM). These two membranes are home to a plethora of vitally important integral membrane proteins which play roles as gatekeepers for the cell, regulating nutrient import, virulence factor export, and maintaining the protective coat of the cell, cargo transport, membrane biogenesis and signaling transporters, enzymes, and receptors (Wimley, 2003, Koebnik et al., 2000, Fairman et al., 2011). More commonly, integral membrane proteins are observed with α-helical domains anchoring them within the membrane. In bacteria such as E. coli, however, the OM has an asymmetric composition with lipopolysaccharides in the outer leaflet and phospholipids in the inner leaflet. Here, the OM is further distinguished by the fact that it is populated by, almost exclusively, a unique class of membrane proteins called β-barrel outer membrane proteins (βOMPs) (Tamm et al., 2004, Voulhoux & Tommassen, 2004, Gentle et al., 2004, Schleiff & Soll, 2005, Gentle et al., 2005, Walther et al., 2009b). βOMPs are characterized by a transmembrane domain composed of a β-barrel fold consisting of 8 or more β-strands arranged in an anti-parallel fashion with the first and last strand coming together via a hydrogen bonding network to close the barrel (Fairman et al., 2011, Wimley, 2003). Given their surface accessibility in Gram-negative bacteria, βOMPs have gained renewed interest of late not only for vaccine development, but also as promising antibiotic targets to combat multi-drug resistance (Urfer et al., 2016, Choi & Lee, 2019, Imai et al., 2019, Luther et al., 2019, Hart et al., 2019).
The biogenesis of βOMPs into the OM is mediated by the β-barrel assembly machinery (BAM) in Gram-negative bacteria, the sorting and assembly machinery (SAM) in mitochondria, and outer envelope protein 80 (OEP80) in chloroplasts (Paschen et al., 2003, Gentle et al., 2004, Walther et al., 2009b, Webb et al., 2012a) (Figure 1). These machineries are conserved from Gram-negative bacterial ancestors to the eukaryotic organelles of endosymbiotic origin (mitochondria and chloroplasts) and have retained the core component of the folding machinery. In chloroplasts, much less is known about βOMP biogenesis and therefore, will not be discussed in detail here. We do know, however, that nascent βOMPs are imported across the OM into the intermembrane space (IMS) by the translocon of the outer membrane of chloroplasts (TOC) complex (Misra, 2012, Walther et al., 2009b, Hinnah et al., 1997, Hinnah et al., 2002, Kessler & Schnell, 2004). Then, IMS chaperones further shuttle the nascent βOMPs to OEP80 for folding/insertion into the OM. No structure of OEP80 has yet been reported, however, the structure of the POTRA domains have been reported for Toc75, the central component of the TOC complex (O’Neil et al., 2017). Toc75 is in the same family, and expected to have a similar fold, as OEP80 and the core components of BAM and SAM.
Figure 1. Paths of βOMP insertion into membranes.
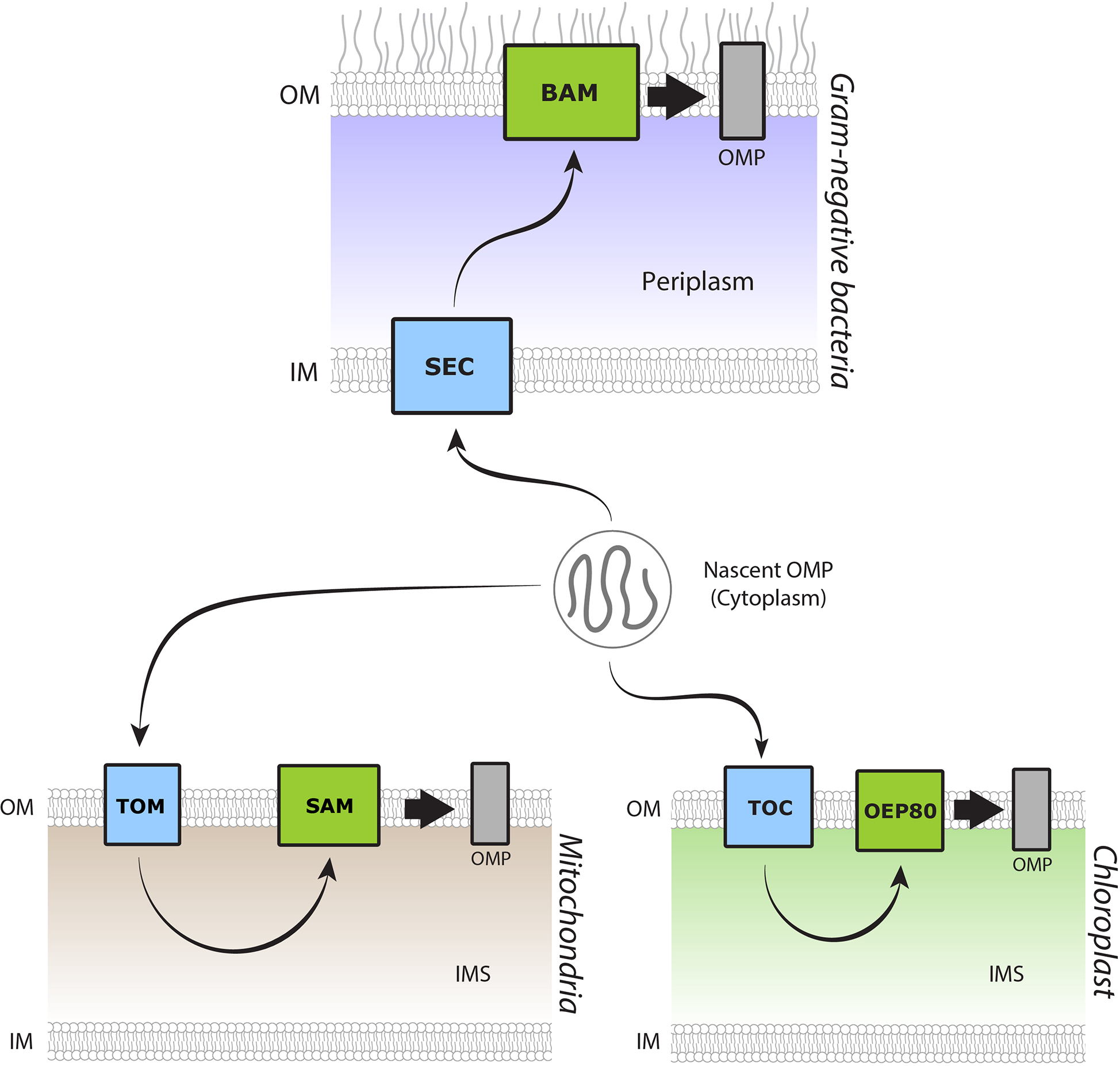
In Gram-negative bacteria, mitochondria, and chloroplasts, βOMP follow a similar general path from nascent polypeptide to insertion into the OM. βOMP are translated in the cytoplasm. They are then translocated across the IM in Gram-negative bacteria or the OM in mitochondria and chloroplasts, mediated by the SEC, TOM, and TOC complexes, respectively. After crossing the membrane, βOMPs are directed to the OM for folding and insertion, mediated by BAM, SAM, and OEP80, respectively.
βOMP biogenesis in Gram-negative bacteria
In Gram-negative bacteria, the OM protects against harsh environments and is embedded with distinct βOMPs that fulfill many essential roles in the cell (Beveridge, 1999, Rollauer et al., 2015). The biogenesis of these βOMPs begins in the cytoplasm, where ribosomes synthesize polypeptide βOMP precursors (Tsirigotaki et al., 2017) (Figure 2). These nascent precursor βOMPs each contain an N-terminal signal peptide that routes them to the SecYEG translocon, where the signal peptide is proteolytically removed following translocation across the IM (Crane & Randall, 2017). Once in the periplasm, the nascent βOMPs interact with several chaperones including survival protein A (SurA), seventeen kilodalton protein (Skp), and FkpA to further escort the nascent βOMPs to the OM where the β-barrel assembly machinery (BAM) then mediates folding and insertion. BAM substrates can be quite diverse in their size and properties, having barrel domains ranging from 8–36 β-strands for a single monomeric βOMP, and includes the more complex multimeric βOMPs (Muhlenkamp et al., 2015, Sklar et al., 2007b, Ruiz-Perez et al., 2009).
Figure 2. Biogenesis of βOMPs in Gram-negative bacteria.
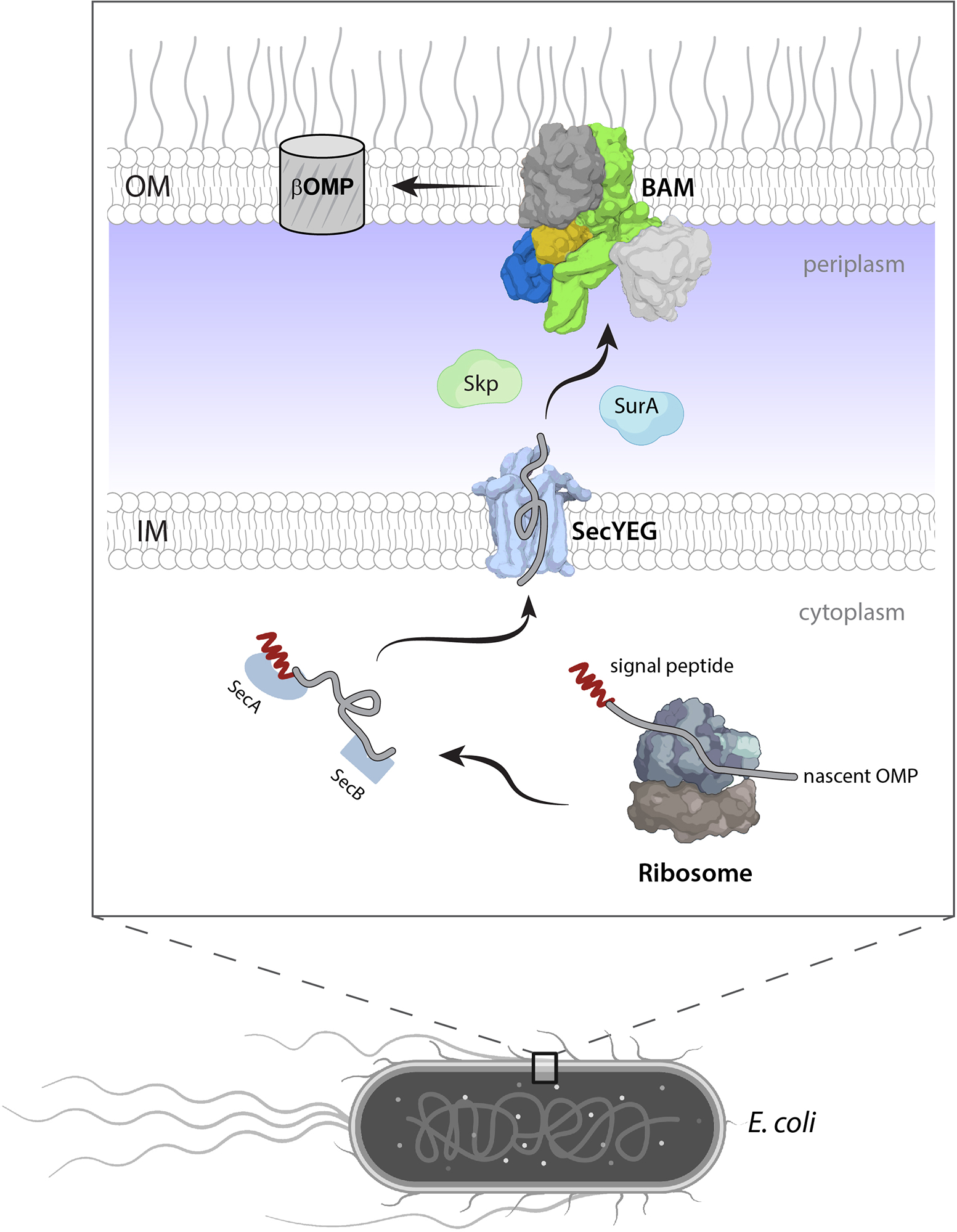
Nascent βOMPs (gray) with N-terminal signal peptide (red) are synthesized in the cytoplasm by the ribosome (gray/slate) and then routed by SecA/SecB chaperones (slate) to the SecYEG translocon (light blue) for transit into the periplasm. The periplasmic chaperones SurA (light blue) and Skp (light green) further usher nascent βOMPs to BAM, where they are folded and inserted into the OM.
In E. coli, BAM has been extensively characterized to help understand its essential role in βOMP biogenesis and consists of five components, BamA (a βOMP) and four lipoproteins termed BamB, BamC, BamD, and BamE (Kim et al., 2012, Noinaj et al., 2017, Konovalova et al., 2017) (Figure 2). However, the exact composition of BAM can vary across other bacteria (Webb et al., 2012a). BamA and BamD are essential for viability, whereas BamB, BamC, and BamE are required for efficient function of BAM, with concurrent deletion of more than one of these components also rendering cells nonviable (Wu et al., 2005, Ruiz et al., 2005, Vuong et al., 2008, Leonard-Rivera & Misra, 2012). BAM has been proposed to recognize nascent βOMPs via their terminal strand, often referred to as the β-signal, which has been shown to be species specific and trigger conformation changes in BamA (Robert et al., 2006). This β-signal contains a highly conserved phenylalanine at the C-terminus and has a consensus sequence that generally resembles HyGHyPoHyPoF (Paramasivam et al., 2012). A second targeting sequence has also been proposed in β14 that binds to BamD (Hagan et al., 2015). However, more work is needed to confirm this idea and to clarify if there may be other determinants contributing to recognition by BAM.
Structures of all the individual components of the BAM complex have now been reported for E. coli (Wu et al., 2020, Kim et al., 2012). BamA is a βOMP itself with an N-terminal periplasmic domain consisting of five polypeptide transport-associated (POTRA) domains and a 16-stranded C-terminal β-barrel domain (Noinaj et al., 2013, Albrecht et al., 2014, Ni et al., 2014). Providing the first evidence in deciphering the mechanism mediating βOMP biogenesis, the structure of BamA demonstrated that lateral gating along the barrel seam was essential for BAM function (Noinaj et al., 2014). Molecular dynamics simulations revealed that lateral gating occurred spontaneously, opening a path for nascent βOMPs to be routed from the periplasm directly into the OM. In addition, these studies highlighted that BamA is able to destabilize and thin the local membrane in proximity of the lateral seam, thereby acting as a folding catalyst to make βOMP biogenesis more energetically favorable (Moon et al., 2013, Gessmann et al., 2014, Costello et al., 2016). BamB has an eight-bladed 𝛽-propeller fold and has recently been proposed to mediate the formation of BAM clusters/precincts (Gunasinghe et al., 2018, Rassam et al., 2015). These BAM clusters have been proposed to drive OMP assembly, which is active at the cell midbody, yet becomes inactive as the precincts are pushed towards the poles during division. BamC consists of a flexible N-terminal region followed by two helix-grip domains and has been shown to be partially surface-exposed in E. coli, although the role here is not known (Webb et al., 2012b). While still a controversial notion, more recent studies have shown that in N. gonorrhoeae, BamE and even BamD (in the absence of BamE) are both also surface exposed, indicating broader studies are needed here in order to determine if this contributes to the function of BAM in βOMP biogenesis (Sikora et al., 2018). BamD contains five tetratricopeptide repeat (TPR) domains and is an essential component for viability in many bacteria (Malinverni et al., 2006, Wu et al., 2005). It interacts directly with BamA and BamC and is thought to also contribute to substrate recognition (Hagan et al., 2015, Lee et al., 2018). BamE, the smallest component, contains an α/β globular fold and has been shown to stabilize the overall complex and increase efficiency (Sklar et al., 2007a, Ryan et al., 2010).
βOMP biogenesis in mitochondria
In Gram-negative bacteria, βOMPs are inserted into the membrane by a unidirectional pathway from the cytoplasm to the periplasm to the OM. In contrast, however, given the endosymbiotic nature of mitochondria and their location inside the cell, mitochondrial βOMPs are synthesized in the cytoplasm, and must first be imported across the outer mitochondrial membrane (OMM) into the IMS (Pfanner et al., 2019, Paschen et al., 2003, Hohr et al., 2015) (Figures 1 and 3). A distinction between Gram-negative bacteria and mitochondria is the number and types of βOMPs present in the OM, with mitochondria containing only a handful of βOMPs including Por1, Sam50, VDAC (1, 2, and 3), Tom40, and Mdm10 (Hohr et al., 2015). This is in contrast to Gram-negative bacteria which contain a plethora of βOMPs having a wide range of strand numbers (all even) with some containing periplasmic and/or luminal domains (Fairman et al., 2011, Koebnik et al., 2000, Schulz, 2000, Wimley, 2003). Additionally, the OMM contains the mitochondrial import machinery (MIM) complex that facilitates the insertion of α-helical membrane proteins, allowing for a diverse composition of both α-helical and β-barrel membrane proteins (Dimmer et al., 2012, Krüger et al., 2017).
Figure 3. Protein import into the mitochondria.
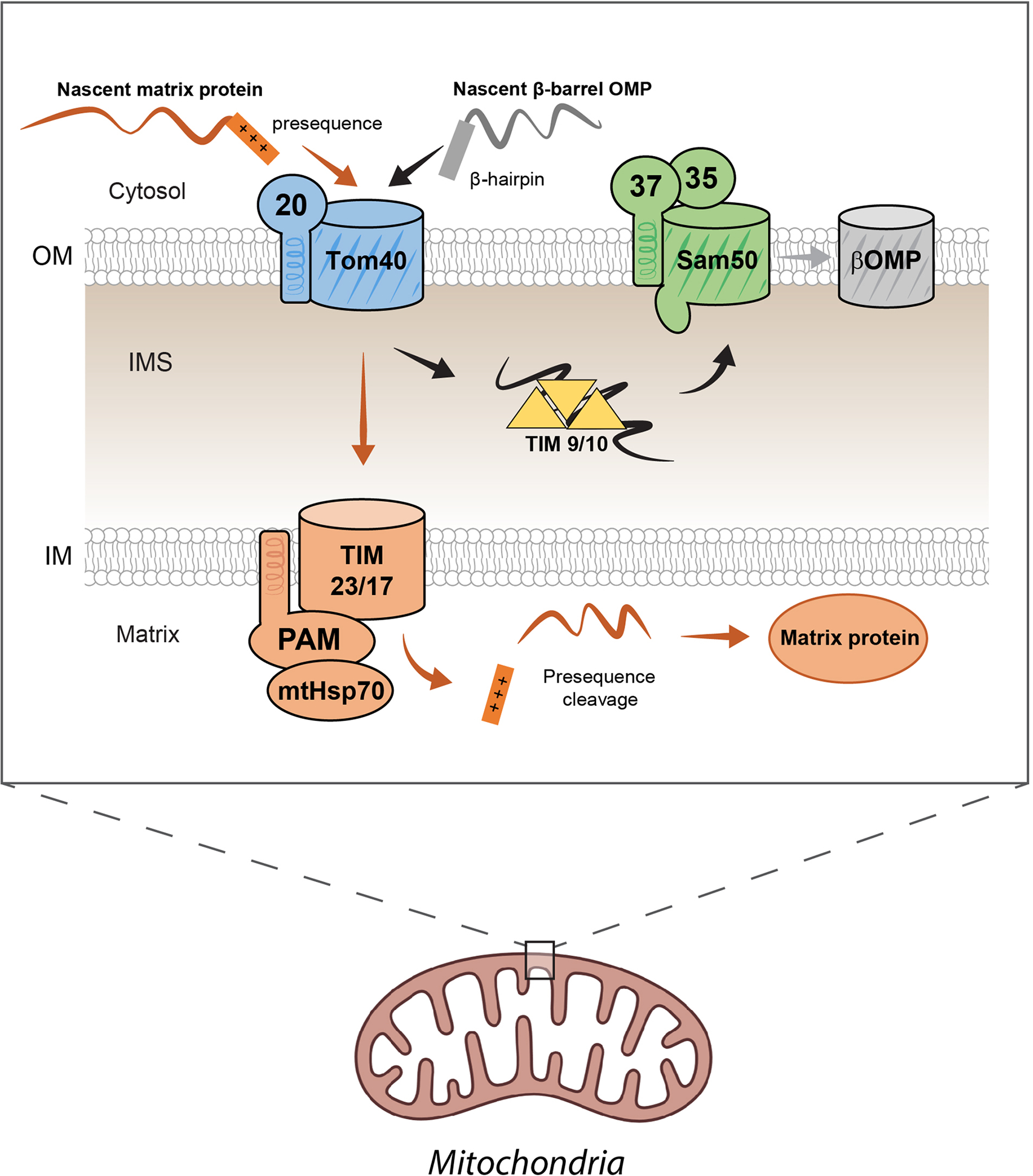
Proteins trafficked to the mitochondria are first translated in the cytoplasm. Proteins designated for import in the mitochondrial matrix contain a positively-charged presequence peptide, which is hypothesized to facilitate protein import by TOM across the outer mitochondrial membrane (OMM). After entering the intermembrane space (IMS), the polypeptide then interacts with the translocon of the inner membrane (TIM) machinery. The membrane potential across the inner mitochondrial membrane (IMM) activates the complexes’ translocation activity which is assisted by the presequence translocase associated motor (PAM) and matrix heat shock protein 70 (mtHSP70). This drives the polypeptide across the IMM towards the negatively-charged environment of the matrix. The presequence is then cleaved by the mitochondrial presequence protease and the protein is folded within the matrix. Nascent βOMPs form an N-terminal beta hairpin that facilitates their trafficking to the mitochondria. After translocation through TOM, the nascent proteins are stabilized by the TIM9/10 chaperones within the IMS and shuttled to SAM, which facilitates folding and insertion into the OMM.
Previous studies have sought to discover the distinguishing features that determine whether certain proteins over others are trafficked to the mitochondria. Mitochondrial matrix proteins all contain a positively charged presequence that facilitates their interaction with the translocase of the outer membrane (TOM) complex (Vögtle et al., 2009) (Figure 3). Recent cryo-EM structures of the TOM complex have revealed that the barrel lumen of Tom40 is largely negatively charged (Araiso et al., 2019, Tucker & Park, 2019). This has led to models where the import is facilitated by electrostatic interactions between the positively changed presequence and the negatively charged barrel lumen.
Nascent mitochondrial βOMPs contain two signals that mediate their import into the mitochondria (β-hairpin) and insertion into the OMM (β-signal) (Kutik et al., 2008, Jores et al., 2016, Hohr et al., 2018, Pfanner et al., 2019, Walther et al., 2009a). Studies have shown that these nascent βOMPs utilize the terminal β-hairpin for trafficking to the mitochondria and across the OMM (Jores et al., 2016). Studies in yeast have further demonstrated that both bacterial and chloroplast βOMPs can be targeted to the mitochondria when expressed with the C-terminal β-hairpin of mitochondrial βOMPs (Walther et al., 2009a, Kozjak-Pavlovic et al., 2011). The mode of recognition, however, that occurs between this β-hairpin and the TOM complex has not been well characterized.
After entering the IMS, nascent βOMPs interact with the Tim9/Tim10 chaperone complex, shuttling them to SAM, which mediates their biogenesis into the OMM (Weinhäupl et al., 2018) (Figure 3). All mitochondrial βOMPs share a conserved β-signal found within the terminal strand, which contains a consensus “PoXGXXHyXHy” motif, where Po is any polar residue, X is any residue, and Hy is any hydrophobic residue (Kutik et al., 2008). Previous studies have found that this β-signal alone is sufficient for interaction with SAM, interacting directly with the lateral seam of Sam50 (Kutik et al., 2008, Hohr et al., 2015, Hohr et al., 2018).
SAM is comprised of three components termed Sam50, Sam35, and Sam37 (1:1:1 ratio), with Sam50 and Sam37 being essential for βOMP biogenesis (Waizenegger et al., 2004, Milenkovic et al., 2004, Kutik et al., 2008, Wiedemann et al., 2003, Paschen et al., 2005, Stroud et al., 2011) (Figure 3). Sam50 is the mitochondrial ortholog of BamA (consisting of a 16-stranded C-terminal β-barrel domain and a single POTRA domain) and is directly responsible for the insertion of mitochondrial βOMPs into the OMM (Kutik et al., 2008, Wiedemann et al., 2003). However, in contrast to the lipoproteins in BAM, Sam35 and Sam37 completely reside on the cytosolic side of the OMM and are not homologous to any of the BAM accessory proteins. Instead they have a GST-like folds with Sam37 containing an N-terminal α-helical domain that anchors it to the OMM, opposite to the side of substrate recognition and insertion (Waizenegger et al., 2004, Milenkovic et al., 2004). Given that SAM receives nascent mitochondrial βOMPs from the IMS, it is unlikely that Sam35 and Sam37 play a direct role in substrate recognition, but certainly do contribute to other aspects of βOMP biogenesis by SAM including substrate release (Chan & Lithgow, 2008, Kutik et al., 2008).
Structures of BAM
The first structures of BAM from E. coli were reported in 2016 (Gu et al., 2016, Han et al., 2016, Bakelar et al., 2016). These structures revealed two main differences based primarily on the conformational state of the barrel domain of BamA: outward-open (PDB IDs 5D0Q and 5EKQ) or inward-open (PDB IDs 5D0O and 5AYW) (Wu et al., 2020) (Figure 4A, B, and C). The inward-open state is characterized by canonical pairing of the β-strands at the BamA barrel seam, with POTRA5 positioned away from the center of the barrel. The outward-open state is characterized by a BamA barrel seam that is open toward the extracellular side, with positioning of POTRA5 directly beneath the BamA barrel, and accompanied by a 45° counterclockwise rotation of the periplasmic ring relative to that in the inward-open state. These inward-open structures were solved with BamB bound, while the outward-open structures were solved in the absence of BamB, suggesting that conformational switching from the outward-open to the inward-open state may be mediated by BamB association. However, the cryo-EM structure of BAM (with BamB) was reported soon after in the outward-open conformation, debunking the notion that BamB may be regulating conformational states of BAM, and leaving the field with a set of conformational states with unknown function (Iadanza et al., 2016). While all structures to date of BAM have been monomeric, studies have indicated that within the cell, BAM may instead function as localized precincts, which might be formed through BamB, which contains WD40-like motifs commonly associated with scaffolding proteins (Noinaj et al., 2011, Gunasinghe et al., 2018, Jansen et al., 2012, Rassam et al., 2015).
Figure 4. The structure of the β-barrel assembly machinery (BAM).
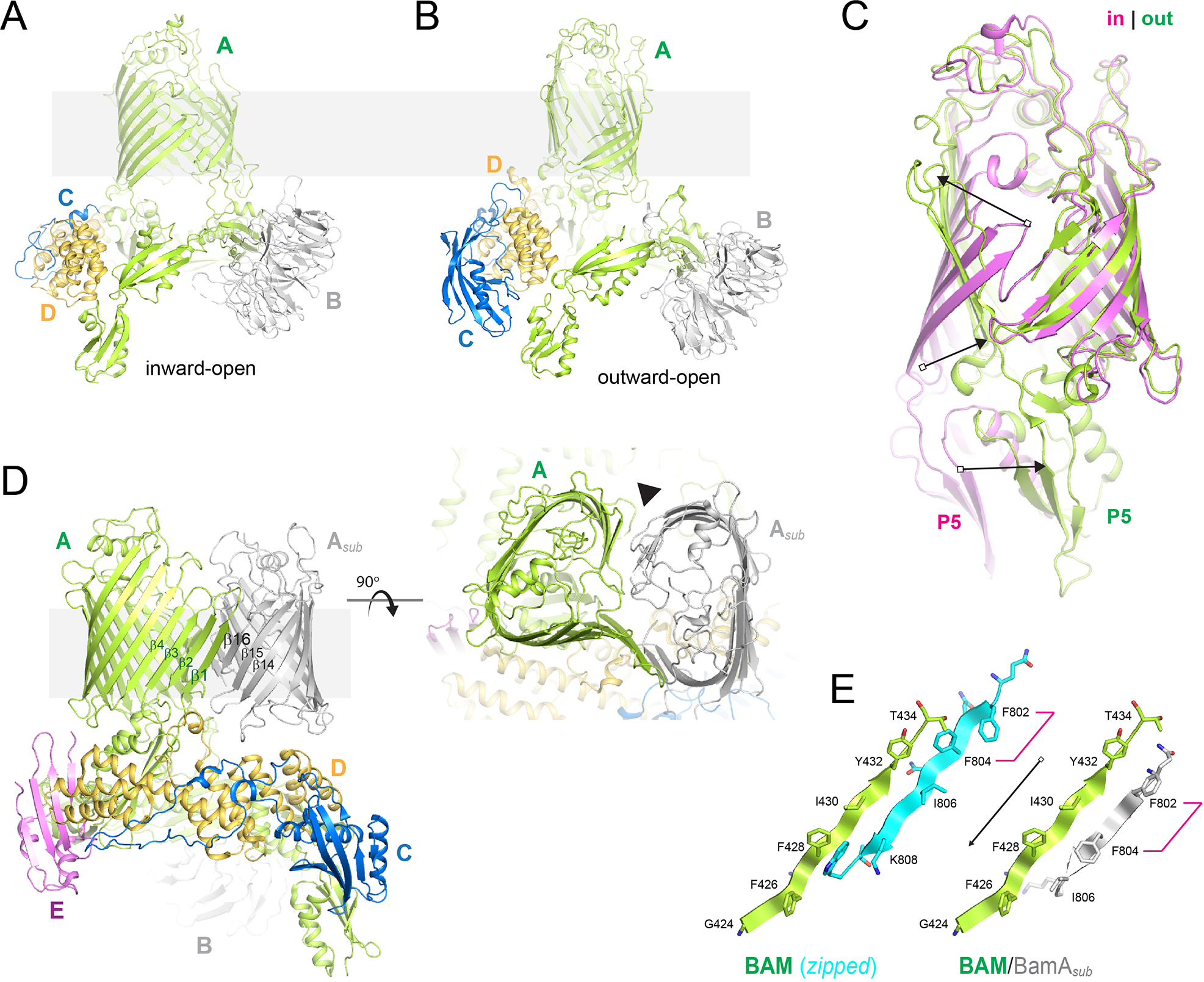
A. Shown is BAM in the inward-open (PDB ID 5D0O) and outward-open states (PDB ID 5LJO; panel B), with BamA in green, BamB in gray, BamC in blue, and BamD in gold. C. A zoomed view of the barrel domain of BamA comparing the conformational changes observed in the inward-open (magenta) and outward-open states (green); POTRA5 is indicated by ‘P5’, while the arrows indicate the conformational changes. D. The structure of the hybrid-barrel BAM/BamAsub complex (PDB ID 6V05) showing orthogonal views; BamA from BAM is in green, while the BamAsub in in gray. Strands at the interface are labeled, while the outer edges of the barrels mediating the curling of the BamAsub is indicated by the black solid triangle. E. A structural alignment at the BamA barrel seam for the BamAsub (right; green and gray), compared to BamA-zipped (left; green and cyan), which shows a four residue shift producing a C-terminal extension on the substrate that may play a role in the final step of βOMP biogenesis by BAM.
Recent cryo-EM studies of E. coli BAM in complex with a substrate BamA molecule (PDB ID 6V05) have revealed the first structural evidence supporting a direct interaction between the barrel domain of BamA with substrate βOMPs during biogenesis (Tomasek et al., 2020) (Figure 4D and E). Here, the C-terminal strand (β16) of the substrate pairs with the first strand (β1) of BamA of BAM, forming a continuous β-sheet consisting of an elongated asymmetric hybrid-barrel. Interestingly, the registry observed here is shifted by four residues compared to that observed for the fully closed form of a single BamA, indicating that BamA can somehow decipher substrate from self. These studies also postulate that features found in both the initial strands and last strands of substrates help stabilize the nascent βOMPs during biogenesis. Additionally, an observed overhang of the terminal strand of the substrate may trigger strand-exchange to pair with its first strand, finalizing the folding process and promoting release from BAM.
Structures of SAM
Another exciting report from 2020 was the elucidation of the first structures of SAM from Myceliophthora thermophila (Diederichs et al., 2020). Here, Sam35 mediates the primary interaction with Sam50 (buried surface area, 1944 Å2) and also forms an extensive interaction with Sam37 (buried surface area, 1892 Å2). Sam37, however, has a non-essential minimal interaction with Sam50 (buried surface area, 927 Å2) (Figure 5A). These structures were determined both in nanodiscs and in detergent, with the structure in nanodiscs having a monomeric form, and the detergent structures having a mix of monomers and inverted dimers (Figure 5A, B, and C). Monomeric forms were observed with the barrel in a mostly closed form with few interactions along the first and last stands. The dimers form an extended β-sheet along the barrel domain interface that extended between the dimer subunits and exhibit quasi-two-fold symmetry, with the barrel domain of Sam50 in a laterally open state. This is mostly attributed to conformational shifts along strands β1-β4, with the remainder of the barrel domain remaining relatively static. Whether or not these conformations observed in the dimeric SAM structures truly represent physiological states remains to be determined.
Figure 5. The structure of the sorting and assembly machinery (SAM).
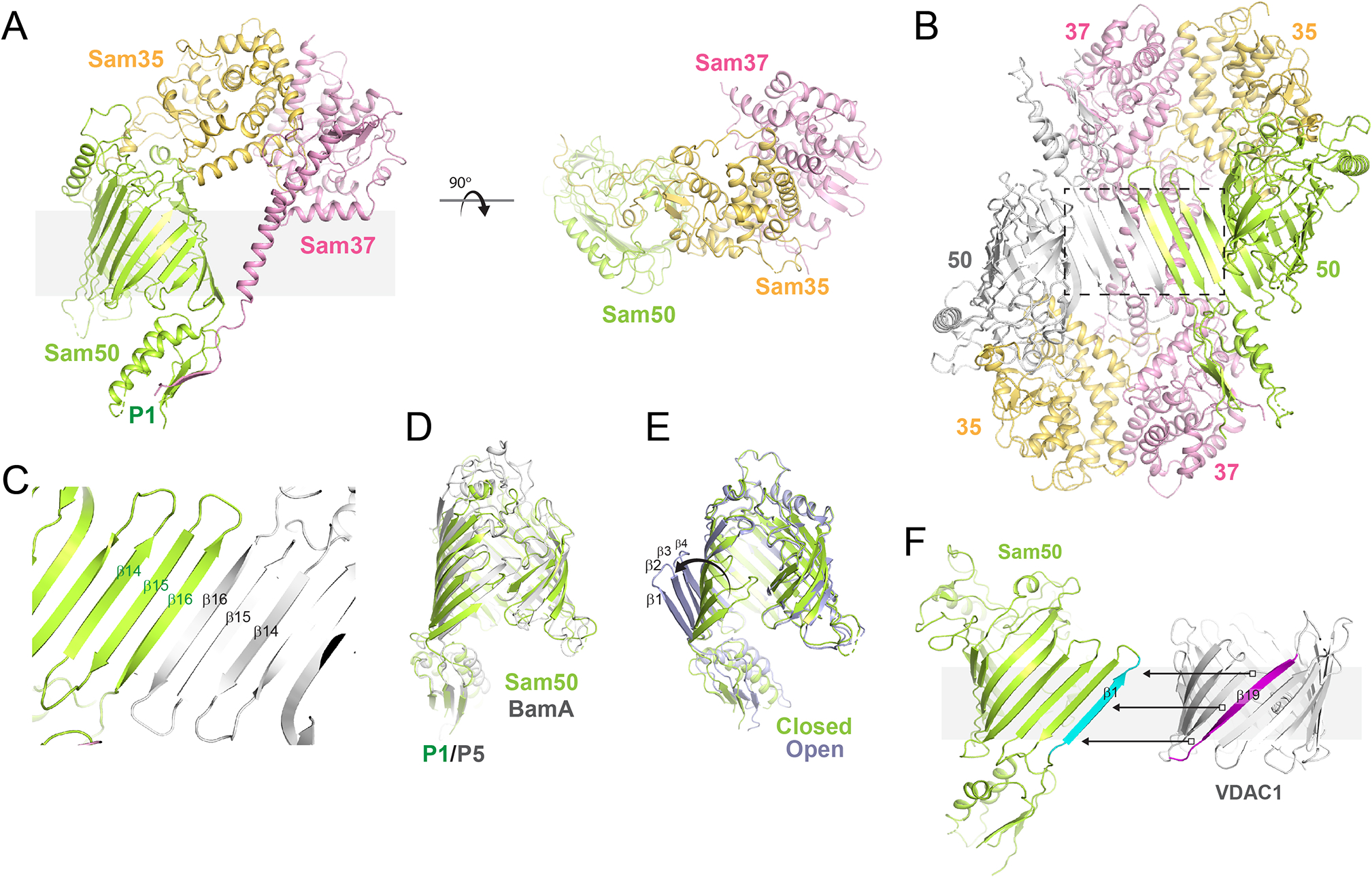
A. Shown are orthogonal views of SAM (PDB ID 6WUL) with Sam50 in green, Sam35 in gold, and Sam37 in magenta. B. Several dimer forms were reported of SAM and shown here is the form which may mimic β-strand templating (PDB ID 6WUM), similar to what was observed for BAM in complex with BamAsub (PDB ID 6V05), albeit with the dimer form having pseudo two-fold symmetry (inverted). C. Zoomed view of the templating interface, a view turned 180° along y-axis from the region within the dashed box in panel B. D. Orthogonal view of the structural alignment of BamA (gray) and Sam50 (green). E. A structural comparison of the open (gray) and closed (green) states of Sam50, depicting the movement of β1-β4. F. A model showing how templating may occur between Sam50 (β1 shown in cyan) and VDAC1, which contains 19 β-strands, with the nineteenth strand (magenta) being required to ensure proper topology during biogenesis.
Structural comparison of BAM and SAM
The structures of BAM and SAM share little outside of the conserved core components BamA and Sam50 (Paschen et al., 2003, Jiang et al., 2012, Ulrich & Rapaport, 2015, Gentle et al., 2005). Yet, even with these differences, a commonality in BAM and SAM is observed, as some bacterial OMPs are able to utilize the mitochondrial SAM/TOM architecture for insertion into the OM and vice versa (Jiang et al., 2012). A structural alignment of monomeric Sam50 with BamA reveals a mostly closed state for the barrel domain which best compares to the inward-open state of BamA found within the BAM structures (RMSD of ~2.2 Å), however, with fewer hydrogen bonds, if any, between the first and last strands of the Sam50 barrel (Figure 5D). It may be that without the presence of IMS accessory proteins, that Sam50 has evolved an intermediate state where the POTRA domain does not need to move at all and bypasses the requirement of a significantly sheared β-barrel domain observed in the BamA outward-open state. This may be possible, in part, because the catalog of substrates that SAM needs to assemble is much less than that of BAM. To date, all substrates of SAM, except for Sam50 itself, have only 19 β-strands (Tom40, VDAC (1, 2, and 3), and Mdm10) (Araiso et al., 2019, Tucker & Park, 2019, Ujwal et al., 2008, Imai et al., 2011), whereas BAM needs to insert a larger pool of higher diversity substrates with monomeric βOMPs ranging from 8 to 36 strands. It is worth also noting that without the nineteenth strand, the SAM substrates could possibly be inserted inverted, somewhat mimicking what was observed in the dimer structures (Figure 5C and F). Further, despite the interface between the dimers being inverted, they still closely resemble the interface observed in the recent BAM/BamA cryo-EM structure, possibly indirectly demonstrating a shared mechanism for βOMP biogenesis with BAM, as is expected. Additionally, the open state of Sam50 may represent a state analogous to the BamA outward-open state, but with some notable differences. For example, the closed state of Sam50 is nearly identical to its open state, except for the first four strands of the β-barrel, which are curled out ~35–40° to open the Sam50 barrel creating an ~20 Å opening (Figure 5E). Additionally, there is only ~3–4 Å shift in the position of the POTRA domain between the two states. Despite the observed differences discussed here, BAM and SAM, as well as OEP80, are all thought to share a common conserved mechanism for the biogenesis of βOMPs into the membrane.
Models for βOMP biogenesis by BAM and SAM
Despite the wealth of structural information for BAM reported in 2016, and more recently for SAM in 2020, direct structural evidence reporting the molecular details of βOMP assembly has remained elusive for the most part. Reported biochemical and structural data narrowed the field down to two overarching models describing how βOMPs may be assembled into the OM (Noinaj et al., 2017, Wu et al., 2020, Konovalova et al., 2017). Variations have been proposed, but the most distinguishing feature among models is whether or not β-strand templating at the BamA/Sam50 β-barrel seam plays a significant role in the assembly of βOMP substrates. In the model sometimes referred to as the budding model, the βOMP substrate utilizes the exposed β-strand backbone at the barrel seam of BamA/Sam50 as a template onto which it attaches its own β-strands, beginning with its C-terminal strand (β-signal) and systematically adding β-strands until the entire protein is assembled and buds away (Kim et al., 2012, Gruss et al., 2013). In the non-templating model, also referred to as the assisted model, the nascent βOMPs utilize BAM/SAM for localization and possibly stabilization to the membrane, but then spontaneously inserts into a primed portion of the membrane produced by a thinned hydrophobic region flanking the BamA/Sam50 lateral gate. While evidence exists in support of both models, recent structural and functional reports have provided new and exciting data directly supporting the budding model, tipping the scales in favor of this previously controversial hypothesis.
Critics of the budding model point out that just because disulfide crosslinking at the BamA seam is lethal is not conclusive evidence that lateral gating exists or that templating must occur. Rather, alternative explanations are that it slows down the folding of βOMPs and/or causes assembly defects to a sufficient degree to result in an overaccumulation of unfolded βOMPs in the periplasm. Another explanation is that crosslinking BamA prevents conformational changes in BAM which are essential for its role as a catalyst on the membrane. Evidence supporting the assisted model includes the fact that some βOMPs readily assemble into liposomes spontaneously, and that locking the lateral gate of BamA closed does not prevent its ability to accelerate the folding of small OMPs (Burgess et al., 2008, Iadanza et al., 2016, Kleinschmidt, 2015, Doerner & Sousa, 2017). However, several of these studies suffer from the challenging task of finding proper experimental conditions such as: (i) having suitable controls to account for the presence of a membrane destabilization factor itself, which is another separate role of the barrel domain of BamA, and (ii) reporting the percentage of crosslinking efficiency in BamA mutants. Other in vitro studies have shown that OmpA can fold spontaneously into diverse membrane environments and that the thickness of the membrane can have a significant effect on the rate of folding (Burgess et al., 2008). In vivo crosslinking studies with LptD and EspP have also been presented as supporting evidence of the assisted model, where folding of both were proposed to have been mostly folded within the periplasm in route to BAM, rather than directly by BAM itself (Lee et al., 2016, Pavlova et al., 2013, Ieva et al., 2011). These observations together seemingly support the notion that BAM was simply needed as a membrane disruptase rather than serving as a conductor orchestrating the entire biogenesis process.
In order for BamA/Sam50-mediated β-strand templating to occur, as has been proposed in the budding model, the interaction between the β1 and β16 strands of the barrel domain would need to first be disrupted, thus creating a separation generically referred to as the lateral gate. While normally this would be energetically unfavorable, the structures of BamA/Sam50 have demonstrated that each is tailored to be stable despite few to no hydrogen bonds at the barrel seam. Further, crosslinking studies initially performed on SAM and more recently for BAM, have demonstrated direct crosslinking of the barrel domains of Sam50 and BamA and their respective substrates, VDAC1 and EspP (Ieva et al., 2011, Pavlova et al., 2013, Doyle & Bernstein, 2019, Hohr et al., 2018). In both cases, the crosslinks were consistent with trapping the substrates in intermediate states which supported a hybrid-barrel intermediate. Further, dimers of Sam50 directly demonstrated β-templating between subunits, albeit unconventionally given the inverted topologies (Diederichs et al., 2020). The most striking evidence published to date in favor of the budding model, however, is the recent structure of a late-stage assembly intermediate of BAM in complex with a substrate BamA molecule consisting of 14 strands bound at the lateral gate; the first direct structural evidence of a hybrid-barrel being formed during βOMP biogenesis (Tomasek et al., 2020). This structure alone, though, does not allow differentiation between whether the substrate is inserted as a partial or full barrel, or if the substrate is inserted systematically strand by strand; more studies are needed here to make this distinction.
Summary and future outlook
βOMPs serve essential roles in Gram-negative bacteria, mitochondria, and chloroplasts, however, the process for the biogenesis of these βOMPs has remained elusive. Now, more than two decades since the first component of BAM was initially discovered, we are finally at the cusp of being able to describe the molecular mechanism for how these folding machineries function. Recent in vivo and in vitro reports have provided a wealth of evidence demonstrating a hybrid-barrel intermediate, directly supporting the budding model. But due to accumulating evidence for both the budding and the assisted models, the notion that perhaps these folding machineries are able to apply each mechanism in a βOMP-dependent manner cannot be disregarded. Other questions about the mechanism that remain unanswered include: how are substrates initially recognized, are substrates inserted partially folded or strand by strand, and what is the role of the accessory proteins? These questions will likely be answered through the elucidation of a larger number of insertion intermediates, as well as, new biochemical studies that pinpoint the molecular features of BAM that are essential for folding βOMP substrates. While certainly an exciting time for the field, more studies are needed to solidify exactly how BAM and SAM function and to address the lingering questions outlined above. With the innovations in experimental design within these systems and the advancements in structural biology methodologies, there is reason to be optimistic that we may soon be able to describe step-by-step exactly how these fascinating machineries operate.
Acknowledgements
We would like to acknowledge funding supporting this work through grants GM127884 (N.N.), GM127896 (N.N.), and GM132024 (T32 Molecular Biophysics Training Program to E.B.) from the National Institute of General Medical Sciences (NIGMS).
Footnotes
Competing interests
Authors declare no competing interests.
References
- Albrecht R, Schutz M, Oberhettinger P, Faulstich M, Bermejo I, Rudel T, Diederichs K, and Zeth K (2014) Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallographica Section D 70: 1779–1789. [DOI] [PubMed] [Google Scholar]
- Araiso Y, Tsutsumi A, Qiu J, Imai K, Shiota T, Song J, Lindau C, Wenz LS, Sakaue H, Yunoki K, Kawano S, Suzuki J, Wischnewski M, Schütze C, Ariyama H, Ando T, Becker T, Lithgow T, Wiedemann N, Pfanner N, Kikkawa M, and Endo T (2019) Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature 575: 395–401. [DOI] [PubMed] [Google Scholar]
- Bakelar J, Buchanan SK, and Noinaj N (2016) The structure of the beta-barrel assembly machinery complex. Science 351: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ (1999) Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol 181: 4725–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess NK, Dao TP, Stanley AM, and Fleming KG (2008) Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. The Journal of biological chemistry 283: 26748–26758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, and Lithgow T (2008) The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol Biol Cell 19: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi U, and Lee CR (2019) Antimicrobial Agents That Inhibit the Outer Membrane Assembly Machines of Gram-Negative Bacteria. Journal of microbiology and biotechnology 29: 1–10. [DOI] [PubMed] [Google Scholar]
- Costello SM, Plummer AM, Fleming PJ, and Fleming KG (2016) Dynamic periplasmic chaperone reservoir facilitates biogenesis of outer membrane proteins. Proceedings of the National Academy of Sciences of the United States of America 113: E4794–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane JM, and Randall LL (2017) The Sec System: Protein Export in. EcoSal Plus 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs KA, Ni X, Rollauer SE, Botos I, Tan X, King MS, Kunji ERS, Jiang J, and Buchanan SK (2020) Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis. Nature communications 11: 3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer KS, Papić D, Schumann B, Sperl D, Krumpe K, Walther DM, and Rapaport D (2012) A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins. J Cell Sci 125: 3464–3473. [DOI] [PubMed] [Google Scholar]
- Doerner PA, and Sousa MC (2017) Extreme Dynamics in the BamA beta-Barrel Seam. Biochemistry 56: 3142–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MT, and Bernstein HD (2019) Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA beta-barrel. Nature communications 10: 3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman JW, Noinaj N, and Buchanan SK (2011) The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol 21: 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, and Lithgow T (2004) The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol 164: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle IE, Burri L, and Lithgow T (2005) Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol 58: 1216–1225. [DOI] [PubMed] [Google Scholar]
- Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, Zaccai NR, and Fleming KG (2014) Outer membrane beta-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proceedings of the National Academy of Sciences of the United States of America 111: 5878–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss F, Zahringer F, Jakob RP, Burmann BM, Hiller S, and Maier T (2013) The structural basis of autotransporter translocation by TamA. Nat Struct Mol Biol 20: 1318–1320. [DOI] [PubMed] [Google Scholar]
- Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, and Dong C (2016) Structural basis of outer membrane protein insertion by the BAM complex. Nature 531: 64–69. [DOI] [PubMed] [Google Scholar]
- Gunasinghe SD, Shiota T, Stubenrauch CJ, Schulze KE, Webb CT, Fulcher AJ, Dunstan RA, Hay ID, Naderer T, Whelan DR, Bell TDM, Elgass KD, Strugnell RA, and Lithgow T (2018) The WD40 Protein BamB Mediates Coupling of BAM Complexes into Assembly Precincts in the Bacterial Outer Membrane. Cell reports 23: 2782–2794. [DOI] [PubMed] [Google Scholar]
- Hagan CL, Wzorek JS, and Kahne D (2015) Inhibition of the beta-barrel assembly machine by a peptide that binds BamD. Proceedings of the National Academy of Sciences of the United States of America 112: 2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, Zhao Y, and Huang Y (2016) Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol 23: 192–196. [DOI] [PubMed] [Google Scholar]
- Hart EM, Mitchell AM, Konovalova A, Grabowicz M, Sheng J, Han X, Rodriguez-Rivera FP, Schwaid AG, Malinverni JC, Balibar CJ, Bodea S, Si Q, Wang H, Homsher MF, Painter RE, Ogawa AK, Sutterlin H, Roemer T, Black TA, Rothman DM, Walker SS, and Silhavy TJ (2019) A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proceedings of the National Academy of Sciences of the United States of America 116: 21748–21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah SC, Hill K, Wagner R, Schlicher T, and Soll J (1997) Reconstitution of a chloroplast protein import channel. The EMBO journal 16: 7351–7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah SC, Wagner R, Sveshnikova N, Harrer R, and Soll J (2002) The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophys J 83: 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohr AI, Straub SP, Warscheid B, Becker T, and Wiedemann N (2015) Assembly of beta-barrel proteins in the mitochondrial outer membrane. Biochim Biophys Acta 1853: 74–88. [DOI] [PubMed] [Google Scholar]
- Hohr AIC, Lindau C, Wirth C, Qiu J, Stroud DA, Kutik S, Guiard B, Hunte C, Becker T, Pfanner N, and Wiedemann N (2018) Membrane protein insertion through a mitochondrial beta-barrel gate. Science 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, and Ranson NA (2016) Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nature communications 7: 12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R, Tian P, Peterson JH, and Bernstein HD (2011) Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. Proceedings of the National Academy of Sciences of the United States of America 108: E383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Fujita N, Gromiha MM, and Horton P (2011) Eukaryote-wide sequence analysis of mitochondrial beta-barrel outer membrane proteins. BMC Genomics 12: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Meyer KJ, Iinishi A, Favre-Godal Q, Green R, Manuse S, Caboni M, Mori M, Niles S, Ghiglieri M, Honrao C, Ma X, Guo JJ, Makriyannis A, Linares-Otoya L, Bohringer N, Wuisan ZG, Kaur H, Wu R, Mateus A, Typas A, Savitski MM, Espinoza JL, O’Rourke A, Nelson KE, Hiller S, Noinaj N, Schaberle TF, D’Onofrio A, and Lewis K (2019) A new antibiotic selectively kills Gram-negative pathogens. Nature 576: 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen KB, Baker SL, and Sousa MC (2012) Crystal structure of BamB from Pseudomonas aeruginosa and functional evaluation of its conserved structural features. PloS one 7: e49749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JH, Tong J, Tan KS, and Gabriel K (2012) From evolution to pathogenesis: the link between beta-barrel assembly machineries in the outer membrane of mitochondria and gram-negative bacteria. Int J Mol Sci 13: 8038–8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jores T, Klinger A, Groß LE, Kawano S, Flinner N, Duchardt-Ferner E, Wöhnert J, Kalbacher H, Endo T, Schleiff E, and Rapaport D (2016) Characterization of the targeting signal in mitochondrial β-barrel proteins. Nat Commun 7: 12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F, and Schnell DJ (2004) Chloroplast protein import: solve the GTPase riddle for entry. Trends Cell Biol 14: 334–338. [DOI] [PubMed] [Google Scholar]
- Kim KH, Aulakh S, and Paetzel M (2012) The bacterial outer membrane beta-barrel assembly machinery. Protein Sci 21: 751–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt JH (2015) Folding of beta-barrel membrane proteins in lipid bilayers - Unassisted and assisted folding and insertion. Biochimica et biophysica acta 1848: 1927–1943. [DOI] [PubMed] [Google Scholar]
- Koebnik R, Locher KP, and Van Gelder P (2000) Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol 37: 239–253. [DOI] [PubMed] [Google Scholar]
- Konovalova A, Kahne DE, and Silhavy TJ (2017) Outer Membrane Biogenesis. Annual review of microbiology 71: 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozjak-Pavlovic V, Ott C, Götz M, and Rudel T (2011) Neisserial Omp85 protein is selectively recognized and assembled into functional complexes in the outer membrane of human mitochondria. J Biol Chem 286: 27019–27026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger V, Becker T, Becker L, Montilla-Martinez M, Ellenrieder L, Vögtle FN, Meyer HE, Ryan MT, Wiedemann N, Warscheid B, Pfanner N, Wagner R, and Meisinger C (2017) Identification of new channels by systematic analysis of the mitochondrial outer membrane. J Cell Biol 216: 3485–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Kruger V, Prinz C, Meisinger C, Guiard B, Wagner R, Pfanner N, and Wiedemann N (2008) Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell 132: 1011–1024. [DOI] [PubMed] [Google Scholar]
- Lee J, Sutterlin HA, Wzorek JS, Mandler MD, Hagan CL, Grabowicz M, Tomasek D, May MD, Hart EM, Silhavy TJ, and Kahne D (2018) Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion. Proceedings of the National Academy of Sciences of the United States of America 115: 2359–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, Gronenberg LS, Sutterlin HA, Davis RM, Ruiz N, Silhavy TJ, and Kahne DE (2016) Characterization of a stalled complex on the beta-barrel assembly machine. Proceedings of the National Academy of Sciences of the United States of America 113: 8717–8722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard-Rivera M, and Misra R (2012) Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of beta-barrel outer membrane proteins, including that of BamA itself. Journal of bacteriology 194: 4662–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther A, Urfer M, Zahn M, Muller M, Wang SY, Mondal M, Vitale A, Hartmann JB, Sharpe T, Monte FL, Kocherla H, Cline E, Pessi G, Rath P, Modaresi SM, Chiquet P, Stiegeler S, Verbree C, Remus T, Schmitt M, Kolopp C, Westwood MA, Desjonqueres N, Brabet E, Hell S, LePoupon K, Vermeulen A, Jaisson R, Rithie V, Upert G, Lederer A, Zbinden P, Wach A, Moehle K, Zerbe K, Locher HH, Bernardini F, Dale GE, Eberl L, Wollscheid B, Hiller S, Robinson JA, and Obrecht D (2019) Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 576: 452–458. [DOI] [PubMed] [Google Scholar]
- Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, and Silhavy TJ (2006) YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Molecular microbiology 61: 151–164. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Kozjak V, Wiedemann N, Lohaus C, Meyer HE, Guiard B, Pfanner N, and Meisinger C (2004) Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J Biol Chem 279: 22781–22785. [DOI] [PubMed] [Google Scholar]
- Misra R (2012) Assembly of the beta-Barrel Outer Membrane Proteins in Gram-Negative Bacteria, Mitochondria, and Chloroplasts. ISRN Mol Biol 2012: 708203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon CP, Zaccai NR, Fleming PJ, Gessmann D, and Fleming KG (2013) Membrane protein thermodynamic stability may serve as the energy sink for sorting in the periplasm. Proceedings of the National Academy of Sciences of the United States of America 110: 4285–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenkamp M, Oberhettinger P, Leo JC, Linke D, and Schutz MS (2015) Yersinia adhesin A (YadA)--beauty & beast. International journal of medical microbiology : IJMM 305: 252–258. [DOI] [PubMed] [Google Scholar]
- Ni D, Wang Y, Yang X, Zhou H, Hou X, Cao B, Lu Z, Zhao X, Yang K, and Huang Y (2014) Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. The FASEB Journal 28: 2677–2685. [DOI] [PubMed] [Google Scholar]
- Noinaj N, Fairman JW, and Buchanan SK (2011) The Crystal Structure of BamB Suggests Interactions with BamA and Its Role within the BAM Complex. J Mol Biol 407: 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N, Gumbart JC, and Buchanan SK (2017) The beta-barrel assembly machinery in motion. Nature reviews. Microbiology 15: 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N, Kuszak Adam J., Balusek C, Gumbart James C., and Buchanan Susan K. (2014) Lateral Opening and Exit Pore Formation Are Required for BamA Function. Structure 22: 1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, and Buchanan SK (2013) Structural insight into the biogenesis of beta-barrel membrane proteins. Nature 501: 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil PK, Richardson LGL, Paila YD, Piszczek G, Chakravarthy S, Noinaj N, and Schnell D (2017) The POTRA domains of Toc75 exhibit chaperone-like function to facilitate import into chloroplasts. Proceedings of the National Academy of Sciences of the United States of America 114: E4868–E4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramasivam N, Habeck M, and Linke D (2012) Is the C-terminal insertional signal in Gram-negative bacterial outer membrane proteins species-specific or not? BMC Genomics 13: 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen SA, Neupert W, and Rapaport D (2005) Biogenesis of beta-barrel membrane proteins of mitochondria. Trends in biochemical sciences 30: 575–582. [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, and Neupert W (2003) Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature 426: 862–866. [DOI] [PubMed] [Google Scholar]
- Pavlova O, Peterson JH, Ieva R, and Bernstein HD (2013) Mechanistic link between beta barrel assembly and the initiation of autotransporter secretion. Proceedings of the National Academy of Sciences of the United States of America 110: E938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Warscheid B, and Wiedemann N (2019) Mitochondrial proteins: from biogenesis to functional networks. Nat Rev Mol Cell Biol 20: 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassam P, Copeland NA, Birkholz O, Toth C, Chavent M, Duncan AL, Cross SJ, Housden NG, Kaminska R, Seger U, Quinn DM, Garrod TJ, Sansom MS, Piehler J, Baumann CG, and Kleanthous C (2015) Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature 523: 333–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, and Tommassen J (2006) Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol 4: e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollauer SE, Sooreshjani MA, Noinaj N, and Buchanan SK (2015) Outer membrane protein biogenesis in Gram-negative bacteria. Philos Trans R Soc Lond B Biol Sci 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, and Nataro JP (2009) Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. Journal of bacteriology 191: 6571–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Falcone B, Kahne D, and Silhavy TJ (2005) Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121: 307–317. [DOI] [PubMed] [Google Scholar]
- Ryan KR, Taylor JA, and Bowers LM (2010) The BAM complex subunit BamE (SmpA) is required for membrane integrity, stalk growth and normal levels of outer membrane {beta}-barrel proteins in Caulobacter crescentus. Microbiology 156: 742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, and Soll J (2005) Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep 6: 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz GE (2000) beta-Barrel membrane proteins. Current opinion in structural biology 10: 443–447. [DOI] [PubMed] [Google Scholar]
- Sikora AE, Wierzbicki IH, Zielke RA, Ryner RF, Korotkov KV, Buchanan SK, and Noinaj N (2018) Structural and functional insights into the role of BamD and BamE within the beta-barrel assembly machinery in Neisseria gonorrhoeae. The Journal of biological chemistry 293: 1106–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, and Silhavy TJ (2007a) Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proceedings of the National Academy of Sciences 104: 6400–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Kahne D, and Silhavy TJ (2007b) Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes & Development 21: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud DA, Becker T, Qiu J, Stojanovski D, Pfannschmidt S, Wirth C, Hunte C, Guiard B, Meisinger C, Pfanner N, and Wiedemann N (2011) Biogenesis of mitochondrial β-barrel proteins: the POTRA domain is involved in precursor release from the SAM complex. Mol Biol Cell 22: 2823–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm LK, Hong H, and Liang B (2004) Folding and assembly of beta-barrel membrane proteins. Biochimica et biophysica acta 1666: 250–263. [DOI] [PubMed] [Google Scholar]
- Tomasek D, Rawson S, Lee J, Wzorek JS, Harrison SC, Li Z, and Kahne D (2020) Structure of a nascent membrane protein as it folds on the BAM complex. Nature 583: 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirigotaki A, De Geyter J, Šoštaric N, Economou A, and Karamanou S (2017) Protein export through the bacterial Sec pathway. Nat Rev Microbiol 15: 21–36. [DOI] [PubMed] [Google Scholar]
- Tucker K, and Park E (2019) Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat Struct Mol Biol 26: 1158–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, Ping P, and Abramson J (2008) The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proceedings of the National Academy of Sciences of the United States of America 105: 17742–17747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich T, and Rapaport D (2015) Biogenesis of beta-barrel proteins in evolutionary context. International journal of medical microbiology : IJMM 305: 259–264. [DOI] [PubMed] [Google Scholar]
- Urfer M, Bogdanovic J, Lo Monte F, Moehle K, Zerbe K, Omasits U, Ahrens CH, Pessi G, Eberl L, and Robinson JA (2016) A Peptidomimetic Antibiotic Targets Outer Membrane Proteins and Disrupts Selectively the Outer Membrane in Escherichia coli. The Journal of biological chemistry 291: 1921–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, and Meisinger C (2009) Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139: 428–439. [DOI] [PubMed] [Google Scholar]
- Voulhoux R, and Tommassen J (2004) Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res Microbiol 155: 129–135. [DOI] [PubMed] [Google Scholar]
- Vuong P, Bennion D, Mantei J, Frost D, and Misra R (2008) Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. Journal of bacteriology 190: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger T, Habib SJ, Lech M, Mokranjac D, Paschen SA, Hell K, Neupert W, and Rapaport D (2004) Tob38, a novel essential component in the biogenesis of beta-barrel proteins of mitochondria. EMBO Rep 5: 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Papic D, Bos MP, Tommassen J, and Rapaport D (2009a) Signals in bacterial beta-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc Natl Acad Sci U S A 106: 2531–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Rapaport D, and Tommassen J (2009b) Biogenesis of beta-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cellular and molecular life sciences : CMLS 66: 2789–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CT, Heinz E, and Lithgow T (2012a) Evolution of the beta-barrel assembly machinery. Trends Microbiol 20: 612–620. [DOI] [PubMed] [Google Scholar]
- Webb CT, Selkrig J, Perry AJ, Noinaj N, Buchanan SK, and Lithgow T (2012b) Dynamic association of BAM complex modules includes surface exposure of the lipoprotein BamC. J Mol Biol 422: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhäupl K, Lindau C, Hessel A, Wang Y, Schütze C, Jores T, Melchionda L, Schönfisch B, Kalbacher H, Bersch B, Rapaport D, Brennich M, Lindorff-Larsen K, Wiedemann N, and Schanda P (2018) Structural Basis of Membrane Protein Chaperoning through the Mitochondrial Intermembrane Space. Cell 175: 1365–1379.e1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Kozjak V, Chacinska A, Schönfisch B, Rospert S, Ryan MT, Pfanner N, and Meisinger C (2003) Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424: 565–571. [DOI] [PubMed] [Google Scholar]
- Wimley WC (2003) The versatile beta-barrel membrane protein. Curr Opin Struct Biol 13: 404–411. [DOI] [PubMed] [Google Scholar]
- Wu R, Stephenson R, Gichaba A, and Noinaj N (2020) The big BAM theory: An open and closed case? Biochimica et biophysica acta. Biomembranes 1862: 183062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, and Kahne D (2005) Identification of a Multicomponent Complex Required for Outer Membrane Biogenesis in Escherichia coli. Cell 121: 235–245. [DOI] [PubMed] [Google Scholar]


