Abstract
AIM
To determine if there is a difference in radiological, biochemical, or clinical severity between patients infected with Alpha-variant SARS-CoV-2 compared with those infected with pre-existing strains, and to determine if the computed tomography (CT) severity score (CTSS) for COVID-19 pneumonitis correlates with clinical severity and can prognosticate outcomes.
MATERIALS AND METHODS
Blinded CTSS scoring was applied to 137 hospital patients who had undergone both CT pulmonary angiography (CTPA) and whole-genome sequencing of SARS-CoV-2 within 14 days of CTPA between 1/12/20–5/1/21.
RESULTS
There was no evidence of a difference in imaging severity on CTPA, viral load, clinical parameters of severity, or outcomes between Alpha and preceding variants. CTSS on CTPA strongly correlates with clinical and biochemical severity at the time of CTPA, and with patient outcomes. Classifying CTSS into a binary value of “high” and “low”, with a cut-off score of 14, patients with a high score have a significantly increased risk of deterioration, as defined by subsequent admission to critical care or death (multivariate hazard ratio [HR] 2.76, p<0.001), and hospital length of stay (17.4 versus 7.9 days, p<0.0001).
CONCLUSION
There was no evidence of a difference in radiological severity of Alpha variant infection compared with pre-existing strains. High CTSS applied to CTPA is associated with increased risk of COVID-19 severity and poorer clinical outcomes and may be of use particularly in settings where CT is not performed for diagnosis of COVID-19 but rather is used following clinical deterioration.
Introduction
A novel SARS-CoV-2 Alpha (B.1.1.7) variant, was identified in September 2020 in the UK,1 and subsequently, demonstrated rapid spread, representing the dominant strain at Oxford University Hospitals NHS Trust by the end of December 2020.2 The Alpha variant (B.1.1.7 lineage) is reported to be associated with increased disease severity and mortality in the community,3, 4, 5 but there are few studies on the in-hospital population,6 , 7 and none that have evaluated imaging findings. The Alpha variant is associated with increased viral load compared to pre-existing strains in the community.8 Viral load is an important determinant of transmissibility9 and has been associated with increased clinical disease severity and mortality10 , 11; however, the relationship of viral load and computed tomography severity score (CTSS) has not been well defined.
Chest imaging has the potential to assist in stratifying COVID-19 severity, with multiple studies validating the use of visual semi-quantitative CTSS, which has been shown to correlate with clinical severity and short-term prognosis12, 13, 14, 15, 16; however, no studies address a UK population, in which the majority of the COVID-19 CT imaging comprises CT pulmonary angiography (CTPA) to evaluate for thromboembolic disease rather than high-resolution computed tomography (HRCT) or unenhanced computed tomography (CT) as part of the diagnostic work-up.17 The CTSS evaluating the lung parenchyma has not been well studied with this CTPA, in which contrast medium may confound interpretation of ground-glass opacities by increasing lung density, and which also images in arrested respiration resulting in inconsistency in the phase of the respiratory cycle. Furthermore, differential lineages affecting the UK COVID-19 patient population may limit the applicability of the findings of previous studies.
The present retrospective analysis compared radiologically determined disease severity for initial CTPA of patients infected with the Alpha variant compared with those infected with pre-existing strains. In addition, the correlation of the CTSS with viral load, symptom duration, clinical disease severity, and biochemical parameters and prognostic markers was also evaluated.
Materials and methods
Setting and patient population
Data were collected retrospectively on 137 patients at a large tertiary-referral centre in the UK who had undergone CTPA within the study time window of 1 December 2020 to 5 January 2021 and also had a positive polymerase chain reaction (PCR) for SARS-CoV-2 within 14 days of CTPA with whole-genome sequencing (WGS) of the virus.
CT protocol
The first CT pulmonary angiogram following patient presentation was performed using one of three CT systems. All examinations were performed with the standard CTPA protocol (100 Tube voltage where possible, with 70–100 ml Iodinated contrast media at a speed of 4 ml/s using pump injection, dependent upon body habitus). Images were reconstructed using sharp reconstruction kernel for parenchyma and viewed at window setting optimised for assessment of the lung parenchyma.
Radiological analysis (CTSS)
One radiologist with >15 years of thoracic imaging experience and one radiologist with 3.5 years of imaging experience evaluated the severity of images blinded to lineage, other radiologist scoring, and clinical information using the semi-quantitative CTSS proposed by Pan et al. 18 CTSS was determined by scoring each of the five lobes considering the extent of anatomical involvement as follows: 0, no involvement; 1, <5% involvement; 2, 5–25% involvement; 3, 26–50% involvement; 4, 51–75% involvement; and 5, >75% involvement. The resulting global CTSS was the sum of each individual lobar score and (0–25). The mean score from two CTSS raters was used for the analysis.
Clinical and biochemical data
The electronic patient records were used to establish the date of symptom onset and to determine outcome measures. Oxygen saturation and requirements were used to calculate the World Health Organization (WHO) ordinal progression score19 (Table 1 ). For the purposes of this study, severity of illness of patients admitted to hospital was derived by recording the highest point reached on this ordinal scale within 30 days of admission or from the first positive swab date if asymptomatic. The WHO score was also recorded on the day of CTPA. Patients with severe disease were defined as those patients reaching point 6 or higher during the assessment period. Mortality data were defined by death by day 30 after the first positive swab. Admission to critical care was defined as an admission to either a high-dependency unit (HDU) or intensive-care unit (ICU) within 30 days of the positive PCR swab.
Table 1.
WHO Ordinal Scale for Clinical Improvement for the assessment of COVID-19 disease severity.
| Uninfected | Uninfected; no viral RNA detected | 0 |
|---|---|---|
| Ambulatory mild disease | Asymptomatic; viral RNA detected Symptomatic; independent Symptomatic; assistance needed |
1 2 3 |
| Hospitalised: moderate disease | Hospitalised; no oxygen therapya Hospitalised; oxygen by mask or nasal prongs |
4 5 |
| Hospitalised: severe diseases | Hospitalised; oxygen by non-invasive ventilation or high flow Intubation and mechanical ventilation, pO2/FiO2 ≥150 or SpO2/FiO2 ≥200 Mechanical ventilation pO2/FiO2 <150 or (SpO2/FiO2 <200) or vasopressors Mechanical ventilation pO2/FiO2 <150 and vasopressors, dialysis, or ECMO |
6 7 8 9 |
| Dead | Dead | 10 |
Reproduced from Lancet Infect Dis 2020; 20: e192–97.19
This table gives a common outcome measure set for COVID-19 as described by the WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection.
RNA, ribonucleic acid; pO2, arterial partial pressure of oxygen; FiO2, fractional inspired oxygen; SpO2, peripheral capillary oxygen saturation; ECMO, extracorporeal membrane oxygenation.
Participants were defined as having a comorbidity if they had at least one of the following: diabetes mellitus, chronic obstructive pulmonary disease (COPD), asthma, ischaemic heart disease, hypertension. Biochemical data including C-reactive protein (CRP) and lymphocytes, both at time of CTPA and maximum level during admission was collected.
PCR and WGS
Reverse transcriptase (RT)-PCR was performed using one of five commercial assays with the majority run on the Thermo Fisher TaqPath assay. For these samples, the cycle threshold value (of test nearest CTPA) was compared using the ORF gene target. Cycle threshold is a semi-quantitative value that broadly describes the concentration of viral genetic material in a patient sample. A low cycle threshold indicates a high concentration of genetic material, associated with a high risk of infectivity, and is therefore a marker of viral load.
WGS was performed using a multiplex PCR-based approach with the ARTIC LoCost protocol and v3 primers25 using R9.4.1 flow cells (Oxford Nanopore Technologies, Oxford, UK). Consensus sequences were generated using ARTIC field bioinformatics v1.2.126 All sequences underwent quality control, requiring >50% consensus genome coverage at ≥20 depth, and agreement between Pangolin and Nextclade v2.3.24 assignments of lineage B.1.1.7.
Ethics
The clinical information and SARS-CoV-2 PCR samples were collected as part of routine clinical care. Data were extracted and analysed using permission granted by the Institutional Review Board HRA and Health and Care Research Wales (HCRW) (IRAS 282670; REC 20/HRA/2546).
Statistical analysis
All analyses were conducted in R (version 4.0.5; https://www.r-project.org/) using statistical tests as described in figure legends. In general, for comparisons between variants, the Wilcoxon's rank sum test was used. For correlation of continuous variables, a Spearman's rank correlation was used. To test interrater reliability, the intraclass correlation coefficient (ICCs) for continuous variables was used. To explore the association of CTSS with clinical outcome a logistic regression was used controlling for age, sex, and comorbidities. For the Kaplan–Meier survival analysis, a log-rank test was used whilst Cox proportional-hazards was used to calculate hazard ratios for the composite outcome, controlling for age, sex, and comorbidities. Significance was set at p<0.05.
Results
Patient characteristics
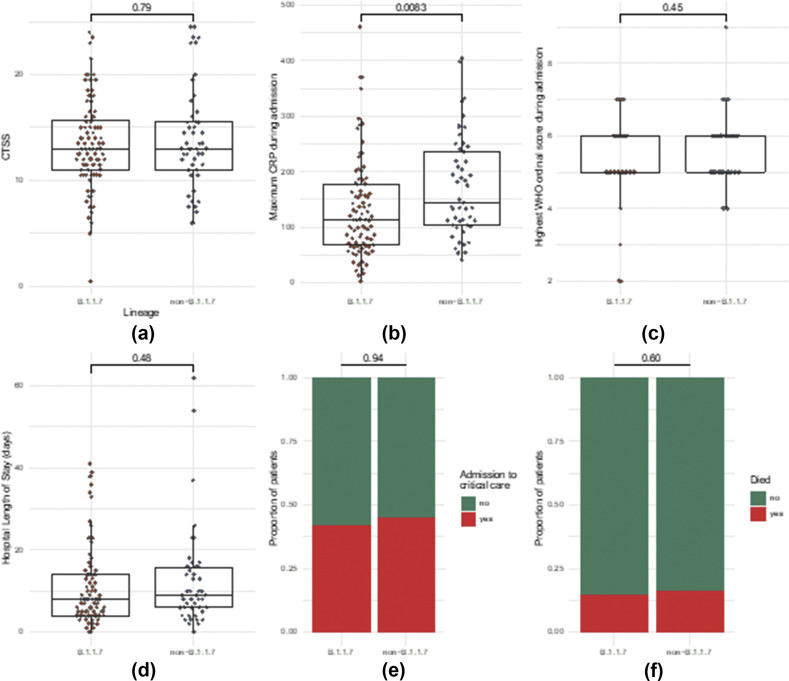
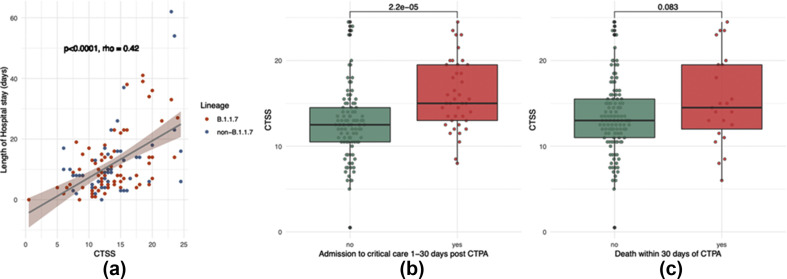
There was no evidence of difference in severity of lung involvement between variants (p=0.79; Fig 1 a). A small but statistically significance difference in maximum CRP between the Alpha and non-Alpha groups was demonstrated (134 versus 172 respectively; p=0.08); however, there was no evidence of a difference in the WHO Ordinal Score, hospital length of stay (LOS), critical care admission, or death within 30 days of positive swab (see Fig 1). Lastly, a difference in viral load as determined by cycle threshold between variants was not observed (Table 2 ).
Figure 1.
Comparison of parameters between Alpha (B.1.1.7) and non-B.1.1.7 variants. (a) Difference in severity of lung involvement on CTSS. (b) Maximum CRP during admission. (c) Max WHO Ordinal Score during admission. (d) Hospital LOS. (e) Admission to critical care. (f) Death. (a–d) Statistical analysis with Wilcoxon's signed-rank test or (e–f) logistic regression controlling for age, sex and presence of a comorbidity.
Table 2.
Summary of patient characteristics.
| Characteristic | B.1.1.7 (n=88) | Non-B.1.1.7 (n=49) | p-Valuea |
|---|---|---|---|
| Sex, n (%) F M |
37 (42%) 51 (58%) |
14 (29%) 35 (71%) |
0.12 |
| Age (years), n (%) | 62 (15.3) | 66 (14.6) | 0.2 |
| Comorbidity, n (%) Hypertension Diabetes Asthma COPD Coronary artery disease |
37 (42%) 21 (24%) 19 (22%) 6 (6.8%) ]7 (8%) 11 (12%) |
25 (51%) 11 (22%) 12 (24%) 7 (14%) 3 (6.1%) 6 (12.%) |
0.3 0 0.7 0.2 >0.9 >0.9 |
| Duration of Sx from onset to CTPA (days), mean (SD) Missing data |
7.7 (3.7) 7 |
9.5 (5) 6 |
0.13 |
| Cycle threshold, mean (SD) Missing data |
17.6 (4.5) 0 |
18.9 (4.6) 7 |
0.14 |
| Admission to critical care (days), n (%) | 65 (41%) | 35 (31%) | 0.11 |
| Death, n (%) | 24 (15%) | 20 (18%) | 0.5 |
| Duration of admission (days), mean (SD) Missing data |
11 (10) 8 |
12 (12) 3 |
0.5 |
Pearson's chi-squared test, Wilcoxon's rank sum test, and Fisher's exact test. SD, standard deviation.
Evaluation of CTSS
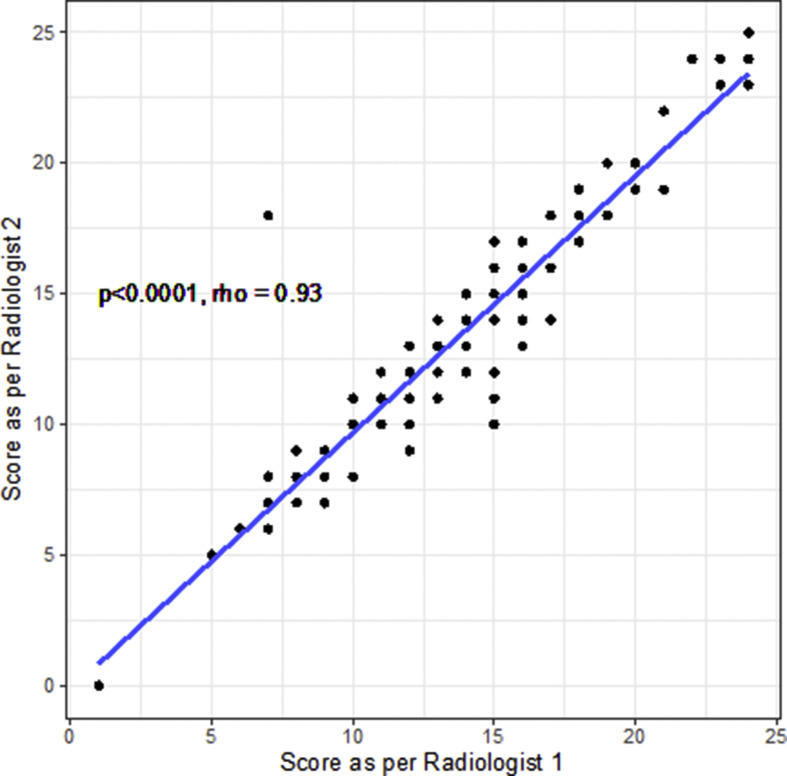
There was excellent correlation between reporters in the blinded evaluation of CT imaging using the 25 point scoring system (Spearman's rho = 0.93, p<0.0001, Fig 2 ; ICC 0.943, 0.919–0.960, p<0.0001).
Figure 2.
Correlation between radiology CTSS scores, calculated with Spearman's rank correlation coefficient.
The association between CTSS score and viral load was assessed and no correlation was found between viral load (PCR cycle threshold ORF) and CTSS, when controlling for duration of time since symptom onset (p=0.73). The median symptom duration at time of CTPA was 8 days, with a range of 0–23 days. Similarly, there was no difference in CTSS for CTPAs performed in the first 7 days since symptom onset, compared to those performed after day 7 (median score 13.2 for early versus 13 for late, p=0.66).
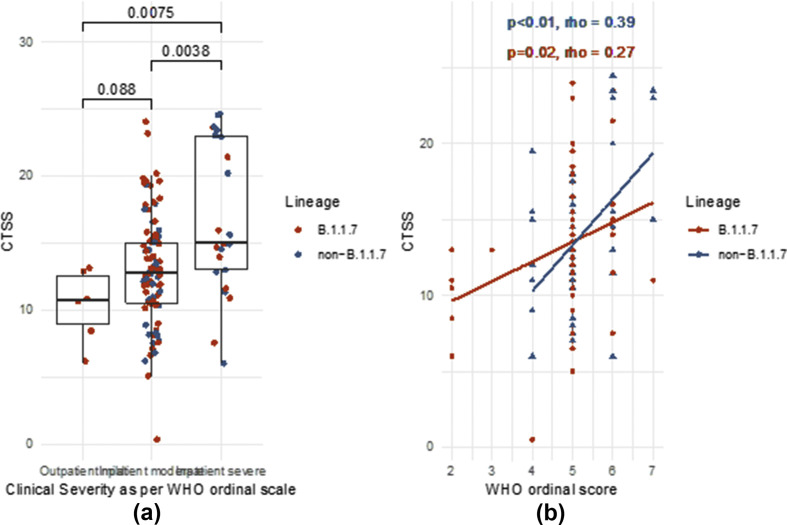
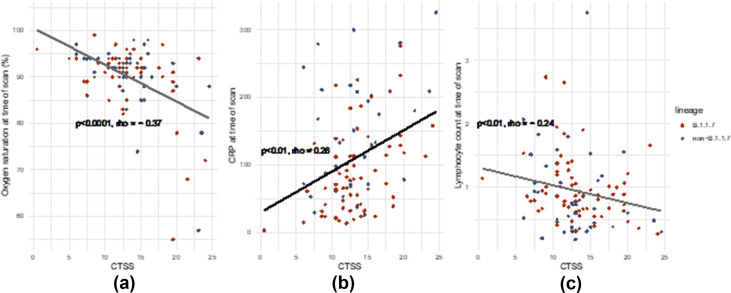
CTSS associated with clinical disease severity at the time of CTPA as demonstrated by WHO score as a categorical variable (Fig 3 a; CTSS 15 versus 12.5 for IP severe versus IP moderate, p=0.004) and weakly correlated with WHO score as a continuous measure (Fig 3B, Spearman's rho = 0.32, p<0.001) Further associations were noted with oxygen saturations (Fig 4 a, rho = –0.37, p<0.0001) and laboratory markers of severity including CRP and lymphocytes (Fig 4b and c; CRP rho = 0.26, p<0.01; lymphocytes rho = –0.24, p<0.01).
Figure 3.
Correlation of CTSS with WHO ordinal score as categorical (a) or continuous (b) variable. Comparisons are with Wilcoxon's rank sum test. Correlations are calculated with Spearman's rank correlation coefficient.
Figure 4.
Correlation of CTSS with clinical and biochemical markers of severity at time of CTPA: (a) Oxygen saturations, (b) CRP, and (c) lymphocytes. Correlations are calculated with Spearman's rank correlation.
CTSS was associated with clinical outcomes. There was also a moderate correlation between CTSS and hospital LOS (Fig 5 a, rho = 0.42, p<0.0001). Patients with subsequent admission to critical care (between 1 and 30 days post-CTPA) had a mean CTSS of 16 versus 12.6 for those who were not admitted (Fig. 5b, p<0.0001). Those who died had a mean score of 15.4 versus 13.3; however, this was not statistically significant (Fig. 5c, p=0.083).
Figure 5.
CTSS score varies by clinical outcome: (a) LOS in hospital, (b) admission to critical care, and (c) death. Correlations are calculated with Spearman's rank correlation. Comparisons performed using Wilcoxon's rank sum test.
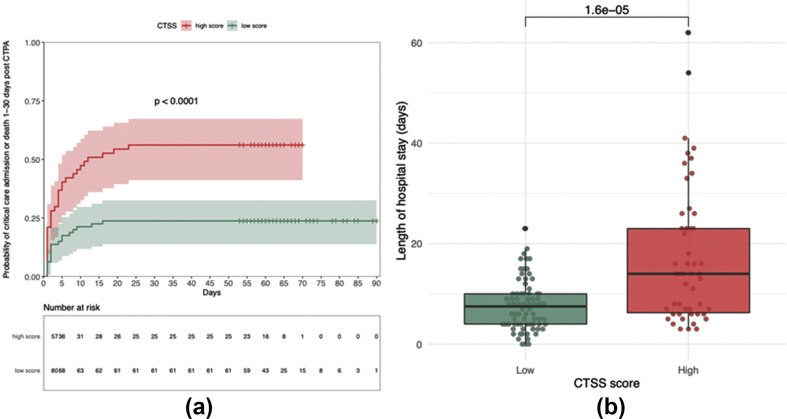
To explore the association with clinical outcome further, a logistic regression model was performed controlling for age, gender, comorbidities, and variant. A strongly significant association was found between CTSS score and both admission to critical care and death with each point increase in CTSS giving an odds ratio for critical care admission (1–30 days after CTPA) of 1.21 (1.1–1.34, p<0.0001) and death (1–30 days after CTPA) of 1.15 (1.034–1.3, p<0.01). CTSS was then classified into a binary outcome of “high” and “low” and used a receiver operating characteristic (ROC) curve to define an optimal cut-off for CTSS on CTPA. Using a composite outcome of admission to critical care or death, a cut-off of 13.5 was obtained (sensitivity = 0.69, specificity = 0.65, AUC = 0.72). Taking a threshold score of ≥14 to be “high”, Kaplan–Meier survival analysis was performed and demonstrated significant divergence in the composite outcome of critical care admission or death (log rank p<0.0001; Fig 6 a). Using Cox proportional hazards analysis, controlling for age, sex, and comorbidities demonstrated a significant hazard ratio (HR) for the composite outcome (HR=2.76 [1.53–4.96], p<0.001). Finally, using this cut-off of 14, hospital LOS was also prolonged significantly at a mean stay of 17.4 days for those with a high score versus 7.9 days for those with a low score (Fig 6b).
Figure 6.
(a) Kaplan–Meier survival curves for those with a high versus low CTSS for admission to critical care or and death. (b) Hospital LOS for those with a high versus low CTSS. Statistics with (a) log-rank or (b) Wilcoxon's rank sum test.
Discussion
The present retrospective study compares the severity of lung disease, short-term prognosis, and viral load between patients hospitalised with the Alpha variant and pre-existing variants of SARS-CoV-2 in a UK tertiary-referral centre. There was no evidence of a difference between variants in disease severity, either radiologically on clinical outcomes or on viral load. The cause for apparent low but significant higher CRP in patients infected with non-Alpha variants is unclear. CTSS performed during CTPA can effectively stratify patients into those that are at increased risk of deterioration (leading to critical care admission or death) with a cut-off score of 14 giving a significant HR (2.76, p<0.001) for this outcome.
The present findings are consistent with published work6 that describes no difference in severity or death between variants in a hospitalised cohort, despite larger community-based studies detecting a difference in mortality outcome.5 The present study is believed to be the first to compare lineage of hospital presenters undergoing CTPA and report no evidence of a difference in radiological severity on CTPA between the Alpha variant and non-Alpha variant infection; however, the study was not able to replicate the finding of a difference in viral load.6
Furthermore, the present study did not find an association between PCR cycle threshold and CT determined severity of COVID-19. Cycle threshold is used as a marker of viral load, with lower cycle threshold suggesting higher viral load. Only one study to date has correlated cycle threshold with CT findings, and demonstrated that high CTSS were associated with lower viral load,20 a finding that was not replicated in the present study.
No difference was demonstrated in CTSS between early and late-phase disease as determined by date from symptom onset. Although many studies have demonstrated increased CTSS with later phase disease,13 , 21 , 22 other studies have demonstrated severity of COVID-19 to be variable at different temporal windows of the epidemic curve.23 The relationship between CTSS and duration of symptoms is therefore complex.
The finding that CTSS evaluated on CTPA can provide insight into likelihood of subsequent deterioration or prolonged hospital stay is of particular value in settings where CT does not perform part of the diagnostic pathway for COVID-19, such as the UK. Here, CT is performed due to clinical deterioration and evaluation for pulmonary embolism, for which there is very limited validation of the CTSS. The present study demonstrates that these CTPA can be used to predict likely deterioration.
CTPA may lead to more challenging interpretation of ground-glass opacities due to COVID-19, as there may be ground-glass opacities due to expiration and contrast medium. The present study shows a higher mean score than previous studies, which may reflect this difference. Although there are numerous different proposed CTSS, the 25 point scale originally used for SARS24 first used by Pan et al. 18 for COVID-19 has been most widely used, and is likely to be more acceptable in clinical practice than the 40 point scale proposed by Feng et al. 14 Demonstration that CTSS on CTPA may help risk stratify and is associated with increased risk of HDU/ITU admission and LOS and may help further define its role.
The present study is limited by its retrospective design, small sample size, and lack of an external validation cohort. Furthermore, CTSS is a semi-quantitative scoring system, which is not necessarily an accurate surrogate for percentage lung involvement, as scoring of all lobes are equally weighted despite not comprising equal proportion of ventilatory capacity. A strength of this study is full genome sequencing to establish B.1.1.7 lineage, rather than the S-gene target failure (SGTF) status as proxy. It is also the first to perform blinded comparison of severity on imaging between the groups.
Future studies may assess if radiological patterns, including features such as ground-glass or crazy-paving are similar between emerging variants as well as if the longer-term sequelae, for example, of fibrosis, is similar between the two groups. Validation of the CTSS cut-off described on a separate cohort would also substantially build on these current findings. Software analysis of percentage lung involvement may also allow more quantitative evaluation.
This study is only the second such study to compare in-hospital disease severity in the Alpha variant to wild-type viruses, and it is the first work to correlate lineage with imaging findings. CTSS on CTPA was also correlated with clinical and biochemical markers of severity and short-term prognosis in a UK population, whereby demonstrating higher scores are associated critical care admission and death.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
The authors thank Avneet Gill, Oxford University Hospitals NHS Trust, Oxford, UK.
Contributor Information
Modernising Medical Microbiology Group:
References
- 1.Wise J. COVID-19: new coronavirus variant is identified in UK. BMJ. 2020;371:m4857. doi: 10.1136/bmj.m4857. [DOI] [PubMed] [Google Scholar]
- 2.Lumley S.F., Rodger G., Constantinides B., et al. An observational cohort study on the incidence of SARS-CoV-2 infection and B.1.1.7 variant infection in healthcare workers by antibody and vaccination status. MedRxiv. 2021:2021. doi: 10.1101/2021.03.09.21253218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies N.G., Jarvis C.I., van Zandvoort K., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593(7858):270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grint D.J., Wing K., Williamson E., et al. Case fatality risk of the SARS-CoV-2 variant of concern. Eurosurveillance. 2021;26(11):1–6. doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Challen R., Brooks-Pollock E., Read J.M., et al. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:1–10. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frampton D., Rampling T., Cross A., et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK : a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis. 2021;3099(21):1–11. doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martina Patone A., Thomas K., Hatch R., et al. Analysis of severe outcomes associated with the SARS-CoV-2 variant of concern 202012/01 in england using ICNARC case mix programme and QResearch databases. MedRxiv. 2021:2021. doi: 10.1101/2021.03.11.21253364. [DOI] [Google Scholar]
- 8.Kidd M., Richter A., Best A., et al. S-Variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral load in samples tested by TaqPath polymerase chain reaction. J Infect Dis. 2021 May 28;223(10):1666–1670. doi: 10.1093/infdis/jiab082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawasuji H., Takegoshi Y., Kaneda M., et al. Transmissibility of COVID-19 depends on the viral load around onset in adult and symptomatic patients 2020. PLoS One. 2020;15(12) doi: 10.1371/journal.pone.0243597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fajnzylber J., Regan J., Coxen K., et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020 Oct 30;11(1):5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y., Yan L.M., Wan L., et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20(6):656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieveld A.W.E., Azijli K., Teunissen B.P., et al. Chest CT in COVID-19 at the ED: validation of the COVID-19 Reporting and Data System (CO-RADS) and CT severity score: a prospective, multicenter, observational study. Chest. 2021;159(3):1126–1135. doi: 10.1016/j.chest.2020.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francone M., Iafrate F., Masci G.M., et al. Chest CT score in COVID-19 patients: correlation with disease severity and short-term prognosis. Eur Radiol. 2020;30(12):6808–6817. doi: 10.1007/s00330-020-07033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Z., Yu Q., Yao S., et al. Early prediction of disease progression in COVID-19 pneumonia patients with chest CT and clinical characteristics. Nat Commun. 2020 Oct 2;11(1):4968. doi: 10.1038/s41467-020-18786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou S., Chen C., Hu Y., et al. Chest CT imaging features and severity scores as biomarkers for prognostic prediction in patients with COVID-19. Ann Transl Med. 2020;8(21):1449. doi: 10.21037/atm-20-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang R., Li X., Liu H., et al. Chest CT Severity Score: an imaging tool for assessing severe COVID-19. Radiol Cardiothorac Imaging. 2020;2(2) doi: 10.1148/ryct.2020200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Royal College of Radiologists The role of CT in patients suspected with COVID-19 infection. https://www.rcr.ac.uk/college/coronavirus-covid-19-what-rcr-doing/clinical-information/role-ct-chest/role-ct-patients
- 18.Pan F., Ye T., Sun P., et al. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295(3):715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall J.C., Murthy S., Diaz J., et al. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis. 2020;20(8):e192–e197. doi: 10.1016/S1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karahasan Yagci A., Sarinoglu R.C., Bilgin H., et al. Relationship of the cycle threshold values of SARS-CoV-2 polymerase chain reaction and total severity score of computerized tomography in patients with COVID 19. Int J Infect Dis. 2020;101:160–166. doi: 10.1016/j.ijid.2020.09.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding X., Xu J., Zhou J., et al. Chest CT findings of COVID-19 pneumonia by duration of symptoms. Eur J Radiol. 2020;127:109009. doi: 10.1016/j.ejrad.2020.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawoud M.M., Dawoud T.M., Ali N.Y.A., et al. Chest CT in COVID-19 pneumonia: a correlation of lung abnormalities with duration and severity of symptoms. Egypt J Radiol Nucl Med. 2020;51(1):246. doi: 10.1186/s43055-020-00359-z. [DOI] [Google Scholar]
- 23.Gumus T., Cengiz D., Kartal F., et al. Changes in computed tomography findings of COVID-19 pneumonia: less extensive lung involvement with decreasing disease prevalence. J Med Virol. 2021;93(4):2056–2064. doi: 10.1002/jmv.26573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang Y.C., Yu C.J., Chang S.C., et al. Pulmonary sequelae in convalescent patients after severe acute respiratory syndrome: evaluation with thin-section CT. Radiology. 2005;236(3):1067–1075. doi: 10.1148/radiol.2363040958. [DOI] [PubMed] [Google Scholar]
- 25.Quick J. nCoV-2019 sequencing protocol v3 (LoCost) https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye
- 26.Loman N., Rowe W., Rambaut A. Artic network. https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html