Abstract
Quantifying the detection rate of the widely used quantitative RT-PCR (RT-qPCR) test for severe acute respiratory syndrome coronavirus 2 and its dependence on patient demographic characteristics and disease progression is key in designing epidemiologic strategies. Analyzing 843,917 test results of 521,696 patients, a “positive period” was defined for each patient between diagnosis of coronavirus disease 2019 and the last positive test result. The fraction of positive test results within this period was then used to estimate detection rate. Regression analyses were used to determine associations of detection with time of sampling after diagnosis, patient demographic characteristics, and viral RNA copy number based on RT-qPCR cycle threshold values of the next positive test result. The overall detection rate in tests performed within 14 days after diagnosis was 83.1%. This rate was higher at days 0 to 5 after diagnosis (89.3%). Furthermore, detection rate was strongly associated with age and sex. Finally, the detection rate with the Allplex 2019-nCoV RT-qPCR kit was associated, at the single-patient level, with viral RNA copy number (P < 10−9). These results show that the reliability of the test result is reduced in later days as well as for women and younger patients, in whom the viral loads are typically lower.
The ongoing coronavirus disease 2019 (COVID-19) pandemic has already infected tens of millions of individuals worldwide. A major tool in combating the pandemic is testing for viral carriage, which is used for both diagnostic and epidemiologic purposes. The most commonly used viral detection tests are based on quantitative RT-PCR (RT-qPCR) of viral genes. This nucleic acid test is of high specificity (ie, very low false-positive rate).1, 2, 3, 4 In contrast, the false-negative rate of these tests has often been reported as high.5, 6, 7, 8, 9 High false-negative rates may impede local and global efforts to slow down disease spread, as patients incorrectly diagnosed as noncarriers might pose an obstacle for efforts such as contact tracing.10, 11, 12 Systematically quantifying the rate of detection and its dependencies on disease progression and patient demographic characteristics is therefore critical for disease spread modeling and public health policy-making.
Various approaches have been taken to estimate the false-negative rate of COVID-19 RT-qPCR tests. Measuring the rate of false-negative results in a population of patients with highly specific pathologies (eg, chest CT imaging) has initially alerted physicians and epidemiologists of the high false-negative rate, estimated at approximately 30%.3, 4, 5, 6 , 8 , 13, 14, 15, 16 A meta-analysis of multiple such studies found that the reported rates were highly variable, with a mean false-negative rate of 11%.17 However, and as noted previously,17 , 18 these meta-analysis studies were necessarily based on a combination of variable studies of nonuniform origins and methodologies, using different kits with inherently different limits of detection and typically involving small groups of patients. Another approach compared initial RT-qPCR test results versus post hoc convalescent serologic test results, finding a false-negative rate of 14%.19 A more recent systematic approach was based on “longitudinal testing” in which the accuracy of each test is determined based on later tests of the same patient: a negative test result that is soon followed by a positive one is deemed false negative.20 , 21 Application of this approach in a hospital setting resulted in an estimation of a false-negative rate of 17.8%.18
Beyond the average false-negative rate, meta-analysis studies showed a strong association of false-negative results with time since exposure22 or time since onset of symptoms.23 These associations suggest that negative test results obtained during a “positive period” reflect waning infections and viral loads declining toward the test limit of detection. Therefore, such negative results do not necessarily indicate a false-negative result, especially as later positive results may be of high cycle threshold (CT) values and may result from fragmented RNA and not from infectious viral particles.24 Moreover, at the patient-specific level, because the viral load is associated with time since onset of symptoms, sex, and age,25, 26, 27, 28, 29, 30, 31, 32, 33 it has been proposed that detection rates might also depend on demographic characteristics; however, current studies lacked statistical power for quantifying such dependencies.18 , 34, 35, 36, 37, 38, 39
Here, using a large data set of quantitative patient-level test series performed with a single type of measurement equipment with a characterized limit of detection [Hur et al,16 and the Allplex 2019-vCoV Assay, version 2.2 (see manufacturer's instructions; Seegene, Inc., Seoul, South Korea)], with linked demographic characteristics and electronic health records (843,917 tests for 521,969 patients), a longitudinal testing–based approach was applied to quantify the detection rate of COVID-19 test results at the community level and its associations with age, sex, and time since diagnosis. Finally, we tested whether this rate is associated with viral RNA copy number at the single-patient level.
Materials and Methods
Ethical Approval
The study protocol was approved by the ethics committee of Maccabi Healthcare Services, Tel-Aviv, Israel (institutional review board number: 0066-20-MHS).
Data Collection
Anonymized clinical records of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RT-qPCR test results (test reports) were retrieved by Maccabi Healthcare Services (MHS) for the period between February 8, 2020, and September 24, 2020. Records of COVID-19 or COVID-19–related diagnoses by physicians (diagnosis reports) were retrieved for these patients. When available, CT values of the PCR test were retrieved for each test (ie, RT-qPCR measurements). Randomly generated identifiers were used to link between test results and diagnosis codes.
Test Results
MHS aggregates all test results for all its members, regardless of whether the test itself was performed by the MHS laboratory. Test results data included, for each test, the following: random patient number, sample number, sample execution date, and test result. Test results were either “positive” (7.4%), “negative” (92%), or “borderline-positive” (0.6%, which were considered as positive in the analysis). Patients for whom two tests with different results were recorded on the same day were excluded from the analysis [274 patients (0.05%)].
Diagnosis Reports
Diagnoses are routinely recorded in the MHS database. For all patients with at least one positive test result, any symptom-based COVID-19 diagnoses recorded before their first test were retrieved. Diagnosis report data included random patient number, diagnosis date, and diagnosis code (International Classification of Diseases, Ninth Revision, and internal MHS codes).
RT-qPCR Measurements
For each test, the following data were included: sample number, PCR machine number, test well number, test date, and CT values for four channels (FAM, Cal Red 610, Quasar 670, and HEX, corresponding to the measurements of E gene, RdRp gene, N gene, and the internal control, respectively). CT values were calculated similarly for all genes using Seegene proprietary software for the Allplex 2019-nCoV assay (Seegene) after collection of oro-nasopharyngeal swab specimens.
Assigning Patient Diagnosis Date
For each patient, the earliest date of COVID-19 symptoms and/or epidemiologically based referral was considered as “date of diagnosis.” When both symptom-based diagnosis and epidemiologic-based referrals were available, they were usually recorded on the same day. For simplicity, a small number of patients for whom both a diagnosis and a referral were available but were more than 1 day apart were excluded (5.2% of diagnosed patients).
Calculating the Detection Rate
For any patient with at least one positive test result, a “positive period” was defined as the period between their date of diagnosis and their last positive test result. Negative test reports during this period were regarded as undetectable, whereas positive test reports during this period were regarded as detectable. Detection rate was calculated as: Detectable/(Detectable + UnDetectable).
Logistic Regression
Logistic regression of an undetectable result versus a detectable result was performed by using the Python statsmodels library. The probability of a detectable result was fitted to the test result (detectable, 1; undetectable, 0) for all tests within the positive period.
Linear Regression
Linear regression of CT values for each fluorescence channel was performed by using the Python statsmodels library.
Calculating Odds Ratios from Logistic Regression
Odds ratios (ORs) were calculated from the coefficients of the aforementioned logistic regression. For the binary variable sex (male, 1; female, 0), OR was defined as: OR sex = exp(C sex). For the continuous variables age and day, ORs were defined as: exp(C age × age older – C age × age younger) younger versus older, exp(C day × day early – C day × day late) early versus late, where C sex, Cday Cage are the coefficients for the sex, day, and age variables, respectively.
Differences in Detection Rate between Age Groups
Test results were divided into two groups of similar size according to patient age (<40 years and ≥40 years). Detection rate was calculated separately for each group. Statistical significance for differences in detection rate between groups was tested by using a two-sided Fisher’s exact test (SciPy in Python).
Results
Building a Large Longitudinal COVID-19 Test Data Set
Among all approximately 2 million MHS patients, 843,917 recorded tests were identified for 521,696 patients. Within this set, 51,499 patients had at least one positive result. Because quarantine discharge policy was based on test results, patients were often repeatedly tested, resulting in a series of test results for each patient. The analysis focused on 7858 patients with well-defined test series, satisfying the following conditions: i) had a defined diagnosis date with COVID-19 symptoms; ii) had at least one positive sample within 14 days after the diagnosis date; and iii) had a test series that ended with a negative result (Table 1). The majority of these test series ended with two or more negative results, in agreement with the discharge policy (68% of patients) (Supplemental Table S1).
Table 1.
Study Population Characteristics
| Characteristic | Study population (N = 7858) |
|---|---|
| Age, years | |
| Mean ± SD | 36.75 ± 20.18 |
| Distribution, no. (%) | |
| <40 years | 4391 (55.88) |
| ≥40 years | 3467 (44.12) |
| Sex, no. (%) | |
| Male | 4156 (52.89) |
| Female | 3702 (47.11) |
| Test result, no. (%) | |
| Positive | 14,559 (45.46) |
| Negative | 17,466 (54.54) |
| Median positive period length (IQR), days | 5 (1 to 16) |
| Median time to recovery (IQR), days | 18 (13 to 24) |
| Median test series length (IQR), days | 22 (16 to 30) |
IQR, interquartile range.
Identifying Undetectable and Detectable Test Results
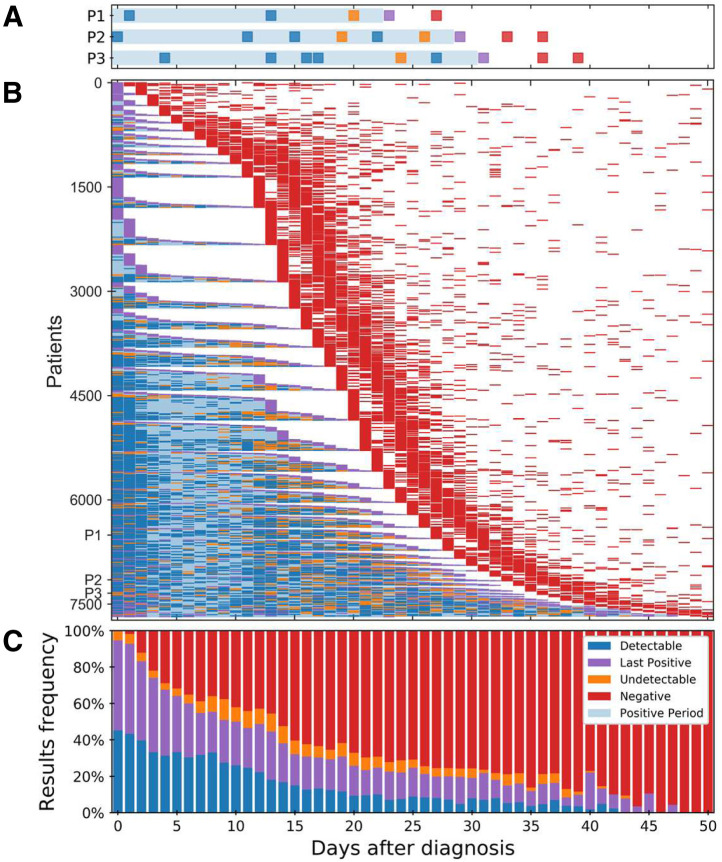
Undetectable and detectable test results were defined based on their context within a patient series of test results. For each patient, the series of test results after diagnosis were considered (Figure 1, A and B). The median day since diagnosis until the first negative result, indicating recovery, was 18 (interquartile range, 13-24 days) (Table 1). For few patients, a negative result came only after >50 days (2.5% of patients). For each patient, the period from diagnosis date to the last positive sample was then considered as a period in which the patient was carrying viral RNA fragments, even if not yet at detectable loads (ie, the positive period). Taking an epidemiologic stand, only negative results outside of the positive period were regarded as negative, whereas negative test results within this patient-specific positive time period were regarded as undetectable results. Similarly, positive test results within this period were regarded as detectable (Figure 1A). To avoid bias for detectable results, the last positive result, used for defining the end of the positive period, was not counted toward detectable results. The rate of negative results increased over time, and at 20 days after diagnosis, the number of negative test results first surpassed the number of positive or undetectable results (Figure 1, B and C). Relative to time of the first negative test result, undetected test results were extremely rare during the two preceding days but were otherwise relatively equally distributed, indicating that undetectable test results were not restricted to the short time period of the end of carriage (Supplemental Figure S1). Focusing on days 0 to 14 after diagnosis, representing the typical duration of the disease, 1047 test results defined as undetectable and 5143 test results defined as detectable were identified, indicating an overall detection rate of 83.1%.
Figure 1.
Longitudinal quantitative RT-PCR severe acute respiratory syndrome coronavirus 2 test results for patients diagnosed with coronavirus disease 2019. A: Test results for three representative patients (P1, P2, and P3). The day of diagnosis and the last positive test result (purple) demarcate the “positive period” (light blue shading). Negative test results within this individually determined period were regarded as “undetectable” (orange). Similarly, positive test results within the positive period were regarded as detectable (blue). All test series end with a sequence of one or more negative results (red). B: Longitudinal severe acute respiratory syndrome coronavirus 2 test results for the study population (Table 1). [For clarity, 191 patients for whom the first negative sample (red) was obtained >50 days after the day of diagnosis were omitted.] Patients are sorted according to the dates, relative to diagnosis, of their first negative result, then by the relative date of the last positive result. C: Frequency of test results per day relative to diagnosis.
Age, Sex, and Time after Diagnosis Are Associated with Detection Rate
To identify personalized features associated with detection rate, multivariate logistic regression for the odds of an undetectable versus detectable result was performed (as discussed in Logistic Regression and Calculating Odds Ratios from Logistic Regression). Patient age, sex, and number of days since diagnosis were all associated with an undetectable result (Supplemental Table S2). Patient age was strongly anticorrelated with an undetectable result, with an OR of 1.36 (95% CI, 1.34 to 1.39) for patients 10 years younger. The number of days from diagnosis was positively correlated with an undetectable result, with an OR of 1.98 (95% CI, 1.78 to 2.20) for samples taken at day 14 compared with samples taken at day 0. Lastly, patient sex was also strongly associated with undetectable results, with a female to male OR of 1.73 (95% CI, 1.58 to 1.9).
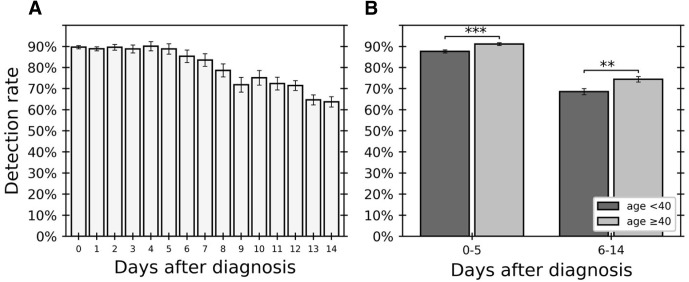
Following the observed association between time after diagnosis and detectable result, the detection rate during disease progression was characterized. Calculating detection rate per day after diagnosis (as discussed in Calculating the Detection Rate), that detection rate during the first few days was found to be fairly constant and high (89.3%, days 0 to 5) and to then gradually decrease over days 6 to 14 (Figure 2 A).
Figure 2.
Detection rate changes along time after day of diagnosis and differs between age groups. Detection rate per day after diagnosis was calculated for 6190 tests. A: Daily detection rate for days between day of diagnosis (day 0) to 14 days after diagnosis. B: Difference in detection rate between two age groups (<40 years and ≥40 years [dark and light gray, respectively]) calculated separately for early and late days after diagnosis. Fisher’s exact test (as discussed in Differences in Detection Rate between Age Groups) was used. Error bars indicate SD. ∗∗P < 0.01, ∗∗∗P < 0.001.
The earlier days after diagnosis (days 0 to 5), in which the detection rate was relatively high, and in which a precise diagnosis is most critical for epidemiologic needs and contact tracing were further studied. Multivariate logistic regression analysis of test results for these days alone identified an association of undetectable results during these days with sex and age. The risk of undetectable result was again associated with age (OR of 1.56 for patients 10 years younger; 95% CI, 1.51 to 1.61), and sex (female to male OR of 2.02; 95% CI, 1.70 to 2.40) (Supplemental Table S3). Dividing the patients into two age groups of similar size (those aged <40 years and ≥40 years) (Table 1), the detection rate during this initial period was found to be significantly lower for the younger age group (P = 0.0004, Fisher’s exact test; OR, 0.72; 95% CI, 0.59 to 0.88) (Figure 2B). Similarly, the detection rate during the later period (days 6 to 14) also significantly increased with age (P = 0.003, Fisher’s exact test).
Age, Sex, and Time after Diagnosis Are Associated with Viral RNA Copy Number Similarly to Detection Rate
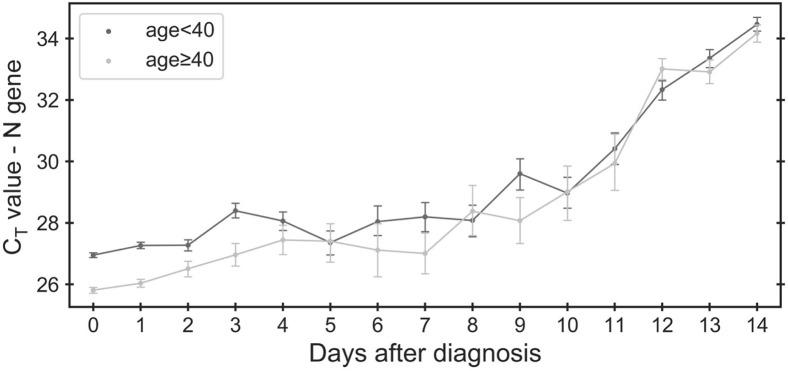
Based on previous reports of viral load differences between male subjects and female subjects, among age groups and along disease progression,4 , 25, 26, 27, 28 , 30 , 40, 41, 42, 43, 44, 45, 46 we hypothesized that differences in detection rate across demographic factors and disease progression may stem from changes in viral load, which would be reflected in the measured CT values. To test this hypothesis, associations of CT values of the three viral genes (N, E, and RdRp) and the internal control gene were studied with patient age and sex and with number of days after diagnosis (Figure 3 and Supplemental Figure S2). Indeed, a linear regression model revealed positive correlation of the CT of viral genes with the number of days after diagnosis, and negative correlation with age and sex (male; as discussed in Linear Regression) (Supplemental Table S4). An opposite association was found with the internal control gene, in agreement with within-tube competition for reagents between the multiplexed reactions (Supplemental Figure S2C).47 The viral RNA copy number association with demographic characteristics and time therefore mirrored the associations of the detection rate with these same parameters.
Figure 3.
Differential change in cycle threshold (CT) value of the N gene along time after day of diagnosis for different age groups. In the first 4 days after diagnosis, CT values of the N gene are lower for older patients (age ≥40 years, light gray) than for younger patients (age <40 years, dark gray). Error bars indicate SEM.
Viral RNA Copy Number and Detection Rate Are Associated at the Individual Patient Level
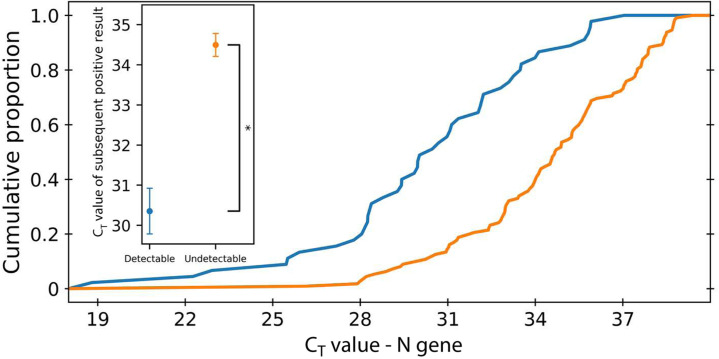
Finally, an association of detection rate with viral RNA copy number at the individual patient level was tested. Because CT values are, by definition, not available for undetectable results, the CT values of the next positive result were used as a proxy (provided that such a sample existed within the 14 days after diagnosis). Comparing the distribution of CT values of positive test results after undetectable results versus the CT values of positive test results after detectable results, it was found that undetectable results were indeed associated with reduced viral RNA copy number for all three viral genes (Mann-Whitney U test; P -values of 1.65 × 10−9, 1.03 × 10−9, and 8.99 × 10−10 for N, E, and RdRp genes, respectively) (Figure 4 and Supplemental Figure S3). Importantly, these CT values, despite being higher, were on average well below the limit of detection and within the observed CT distribution of each gene (Figure 4, inset, and Supplemental Figure S4).
Figure 4.
Difference between cycle threshold (CT) values of the N gene of positive results after undetectable and detectable test. Positive test results that followed an undetectable test result have a higher Ct value than positive test results after undetectable test results. Error bars indicate SEM. ∗P < 10−10.
Discussion
The current analysis of a large data set of electronic health records of patients with COVID-19 showed that although, on average, the detection rate is about 83%, this rate varies strongly with age, sex, and time after diagnosis. At the first few days after diagnosis, the detection rate is only 90% on average and even higher for men and older patients. Combining these data with CT values of RT-qPCR tests provides evidence that detection rates possibly stem from low viral loads at the single-patient level.
The study has several limitations. First, all positive test results are treated as true positives. Although errors may occur, the rate of false-positive results is very low1, 2, 3, 4 and is not expected to significantly affect results. Future studies can further improve the reliability of confirmation of positive cases by combining PCR test results with serology test results. Second, negative results at the end of test series are treated as “true negative” and not an undetectable result, whereas it is possible that if further tests were performed, additional positive tests might have been detected. Again, this is not expected to significantly affect results as most series in this study end with two consecutive negative results. Moreover, this bias will mostly affect the calculated detection rate at later days after diagnosis. Third, because viral loads after infection may first increase and only later decrease,4 , 26 , 41 , 46 it is possible that detection rates follow a similar pattern: first increasing and only later decreasing. Analyzing the cohort, only the later phase of the decreasing detection rate could be identified. However, it is possible that with different cohorts or inclusion criteria, the earlier phase of decreasing viral load and increasing detection rate can also be observed. Fourth, despite all samples with CT values being treated in the same central facility, which increases the uniformity of the data set, multiple other factors such as the type of swab and swab transport conditions (eg, time, temperature), as well as operator-dependent factors regarding sample collection and handling, can affect test sensitivity. These factors are not recorded and are therefore not included in our analysis. Fifth, because the beginning of the positive period is based on the date of diagnosis, it does not directly represent the onset of disease. However, it is the best available proxy for it. Finally, this study was limited to unvaccinated symptomatic patients; therefore, these results may not represent the detection rate for asymptomatic or vaccinated patients for whom viral load may be lower.48 , 49
Despite these limitations, these results provide important epidemiologic input as to the patient-specific sensitivity of tests, with important implications for epidemiologic policy-making, contact tracing, and disease prevention and control. The results underscore that the risk of an undetectable infection at the very early days after diagnosis might be lower than previously thought, reinforcing the usefulness of these tests for epidemiologic decisions.
Footnotes
Supported by the Israel Science Foundation (grant 3633/19 to R.Ki. and G.C.) within the KillCorona–Curbing Coronavirus Research Program. The funding source was not involved in the writing of the manuscript.
M.L.-T., I.Y., and H.U. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2021.10.010.
Author Contributions
M.L.-T., I.Y., T.P., G.C., and R.Ki. designed the study; I.Y., L.S., E.H., and R.Ka. collected data; M.L.-T., I.Y., H.U., and R.Ki. analyzed data; M.L.-T., I.Y., H.U., J.K., L.S., A.B.-T., T.P., S.G., G.C., and R.Ki. interpreted data; M.L.-T., I.Y., H.U., and R.Ki. wrote the manuscript, with comments from all authors.
Supplemental Data
Longitudinal quantitative RT-PCR severe acute respiratory syndrome coronavirus 2 test results for patients diagnosed with coronavirus disease 2019. A: Longitudinal severe acute respiratory syndrome coronavirus 2 test results for the study population (Table 1). [For clarity, 191 patients for whom the first negative sample (red) was obtained more than 50 days after the day of diagnosis were omitted.] The day of diagnosis and the last positive test result (purple) demarcate the “positive period” (light blue shading). Negative test results within this individually determined period were regarded as “undetectable” (orange). Similarly, positive test results within the positive period were regarded as detectable (blue). All test series end with a sequence of one or more negative results (red). Patients are sorted by the dates, relative to their first negative result, then by the relative date of the last positive result. B: Frequency of test results per day relative to first negative result. P1, P2, and P3 indicate three representative patients.
Differential change in cycle threshold (Ct) value along time after day of diagnosis for different age groups. In the first 4 days after diagnosis, Ct values of the E gene (A), RdRp gene (B) and N gene (main text, Figure 3) are lower for older patients (age ≥40 years, light gray) than for younger patients (age <40 years, dark gray). The opposite trend appears for the internal control gene (C), in agreement with within-tube competition for reagents between the multiplexed reactions. Error bars indicate SEM.
Difference between cycle threshold (Ct) values of positive results after undetectable and detectable test results. Positive test results that followed an undetectable test result had a higher Ct value for the E gene (A), RdRp gene (B), and N gene (main text, Figure 4), whereas no significant difference was observed for the internal control gene (C). Error bars indicate SEM. ∗P value <10−9.
Distribution of cycle threshold (Ct) values. The frequency of Ct values for positive test results. A:E gene. B:RdRp gene. C:N gene.
References
- 1.Bisoffi Z., Pomari E., Deiana M., Piubelli C., Ronzoni N., Beltrame A., Bertoli G., Riccardi N., Perandin F., Formenti F., Gobbi F., Buonfrate D., Silva R. Sensitivity, specificity and predictive values of molecular and serological tests for COVID-19: a longitudinal study in emergency room. Diagnostics (Basel) 2020;10:669. doi: 10.3390/diagnostics10090669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfefferle S., Reucher S., Nörz D., Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25:2000152. doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu S.L., Mertens A.N., Crider Y.S., Nguyen A., Pokpongkiat N.N., Djajadi S., Seth A., Hsiang M.S., Colford J.M., Jr., Reingold A., Arnold B.F., Hubbard A., Benjamin-Chung J. Substantial underestimation of SARS-CoV-2 infection in the United States. Nat Commun. 2020;11:4507. doi: 10.1038/s41467-020-18272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 5.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296:E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Paolo M., Iacovelli A., Olmati F., Menichini I., Oliva A., Carnevalini M., Graziani E., Mastroianni C.M., Palange P. False-negative RT-PCR in SARS-CoV-2 disease: experience from an Italian COVID-19 unit. ERJ Open Res. 2020;6:00324-2020. doi: 10.1183/23120541.00324-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y., Yang M., Yuan J., Wang F., Wang Z., Li J., Zhang M., Xing L., Wei J., Peng L., Wong G., Zheng H., Wu W., Shen C., Liao M., Feng K., Li J., Yang Q., Zhao J., Liu L., Liu Y. Laboratory diagnosis and monitoring the viral shedding of SARS-CoV-2 infection. Innovation (N Y) 2020;1:100061. doi: 10.1016/j.xinn.2020.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J.-F., Yan K., Ye H.-H., Lin J., Zheng J.-J., Cai T. SARS-CoV-2 turned positive in a discharged patient with COVID-19 arouses concern regarding the present standards for discharge. Int J Infect Dis. 2020;97:212–214. doi: 10.1016/j.ijid.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection—challenges and implications. N Engl J Med. 2020;383:e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 11.Çubukçu H.C., Coşkun A. False negative results and tolerance limits of SARS-CoV-2 laboratory tests. Pathog Glob Health. 2021;115:137–138. doi: 10.1080/20477724.2021.1881370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watson J., Whiting P.F., Brush J.E. Interpreting a covid-19 test result. BMJ. 2020;369:m1808. doi: 10.1136/bmj.m1808. [DOI] [PubMed] [Google Scholar]
- 13.Kohli A., Joshi A., Shah A., Jain R.D., Gorlawar A., Dhapare A., Desai J., Shetty A., Shah C., Ostwal P., Talraja A. Does CT help in reducing RT-PCR false negative rate for COVID-19? Indian J Radiol Imaging. 2021;31(Suppl 1):S80–S86. doi: 10.4103/ijri.IJRI_739_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., Liu X., Xia C., Liu J., Zhou X.-H. More accurate estimates of the accuracy of RT-PCR and chest CT tests for COVID-19. Res Square. 2021 doi: 10.21203/rs.3.rs-182065/v1. [Preprint] doi: [DOI] [Google Scholar]
- 15.Reich N., Lowe C.F., Puddicombe D., Matic N., Greiner J., Simons J., Leung V., Chu T., Naik H., Myles N., Burns L., Romney M.G., Ritchie G., Champagne S., Dooley K., Sekirov I., Stefanovic A. Diagnostic accuracy of RT-PCR for detection of SARS-CoV-2 compared to a “composite reference standard” in hospitalized patients. medRxiv. 2021 doi: 10.1101/2021.02.18.21252016. [Preprint] doi: [DOI] [Google Scholar]
- 16.Hur K.-H., Park K., Lim Y., Jeong Y.S., Sung H., Kim M.-N. Evaluation of four commercial kits for SARS-CoV-2 real-time reverse-transcription polymerase chain reaction approved by emergency-use-authorization in Korea. Front Med (Lausanne) 2020;7:521. doi: 10.3389/fmed.2020.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H., Hong H., Yoon S.H. Diagnostic performance of CT and reverse transcriptase polymerase chain reaction for coronavirus disease 2019: a meta-analysis. Radiology. 2020;296:E145–E155. doi: 10.1148/radiol.2020201343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams T.C., Wastnedge E., McAllister G., Bhatia R., Cuschieri K., Kefala K., Hamilton F., Johannessen I., Laurenson I.F., Shepherd J., Stewart A., Waters D., Wise H., Templeton K.E. Sensitivity of RT-PCR testing of upper respiratory tract samples for SARS-CoV-2 in hospitalised patients: a retrospective cohort study. Wellcome Open Res. 2020;5:254. doi: 10.12688/wellcomeopenres.16342.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holborow A., Asad H., Porter L., Tidswell P., Johnston C., Blyth I., Bone A., Healy B. The clinical sensitivity of a single SARS-CoV-2 upper respiratory tract RT-PCR test for diagnosing COVID-19 using convalescent antibody as a comparator. Clin Med (Lond) 2020;20:e209–e211. doi: 10.7861/clinmed.2020-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson E.E., Rosenthal J., Wren J., Seetao E., Olson N.H. Repeated testing necessary: assessing negative predictive value of SARS-CoV-2 qPCR in a population of young adults. bioRxiv. 2021 doi: 10.1101/2021.03.10.21253292. [Preprint] doi: [DOI] [Google Scholar]
- 21.Kanji J.N., Zelyas N., MacDonald C., Pabbaraju K., Khan M.N., Prasad A., Hu J., Diggle M., Berenger B.M., Tipples G. False negative rate of COVID-19 PCR testing: a discordant testing analysis. Virol J. 2021;18:13. doi: 10.1186/s12985-021-01489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kucirka L.M., Lauer S.A., Laeyendecker O., Boon D., Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 2020;173:262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wikramaratna P.S., Paton R.S., Ghafari M., Lourenço J. Estimating the false-negative test probability of SARS-CoV-2 by RT-PCR. Euro Surveill. 2020;25:2000568. doi: 10.2807/1560-7917.ES.2020.25.50.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefferson T., Spencer E.A., Brassey J., Heneghan C. Viral cultures for coronavirus disease 2019 infectivity assessment: a systematic review. Clin Infect Dis. 2021;73:e3884–e3899. doi: 10.1093/cid/ciaa1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H., Lau D.P.-L., Choi C.Y.-C., Chen L.-L., Chan W.-M., Chan K.-H., Ip J.D., Ng A.C.-K., Poon R.W.-S., Luo C.-T., Cheng V.C.-C., Chan J.F.-W., Hung I.F.-N., Chen Z., Chen H., Yuen K.-Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 27.Yonker L.M., Neilan A.M., Bartsch Y., Patel A.B., Regan J., Arya P., Gootkind E., Park G., Hardcastle M., St. John A., Appleman L., Chiu M.L., Fialkowski A., De la Flor D., Lima R., Bordt E.A., Yockey L.J., D’Avino P., Fischinger S., Shui J.E., Lerou P.H., Bonventre J.V., Yu X.G., Ryan E.T., Bassett I.V., Irimia D., Edlow A.G., Alter G., Li J.Z., Fasano A. Pediatric severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): clinical presentation, infectivity, and immune responses. J Pediatr. 2020;227:45–52.e5. doi: 10.1016/j.jpeds.2020.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones T.C., Biele G., Mühlemann B., Veith T., Schneider J., Beheim-Schwarzbach J., Bleicker T., Tesch J., Schmidt M.L., Sander L.E., Kurth F., Menzel P., Schwarzer R., Zuchowski M., Hofmann J., Krumbholz A., Stein A., Edelmann A., Corman V.M., Drosten C. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021;373:eabi5273. doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lennon N.J., Mina M., Rehm H.L., Hung D.T., Smole S., Woolley A.E., Lander E., Gabriel S. LB-11. Comparison of viral loads in individuals with or without symptoms at time of COVID-19 testing among 32,480 residents and staff of nursing homes and assisted living facilities in Massachusetts. Open Forum Infect Dis. 2020;7:S848–S849. [Google Scholar]
- 30.Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P., Ahern S., Carty P.G., O’Brien K.K., O’Murchu E., O’Neill M., Smith S.M., Ryan M., Harrington P. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi F., Wu T., Zhu X., Ge Y., Zeng X., Chi Y., Du X., Zhu L., Zhu F., Zhu B., Cui L., Wu B. Association of viral load with serum biomarkers among COVID-19 cases. Virology. 2020;546:122–126. doi: 10.1016/j.virol.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alidjinou E.A., Poissy J., Ouafi M., Caplan M., Benhalima I., Goutay J., Tinez C., Faure K., Chopin M.-C., Yelnik C., Lambert M., Hober D., Preau S., Nseir S., Engelmann I. on behalf of the Lille Covid Research Network Licorne: Spatial and temporal virus load dynamics of SARS-CoV-2: a single-center cohort study. Diagnostics (Basel) 2021;11:427. doi: 10.3390/diagnostics11030427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahallawi W.H., Alsamiri A.D., Dabbour A.F., Alsaeedi H., Al-Zalabani A.H. Association of viral load in SARS-CoV-2 patients with age and gender. Front Med (Lausanne) 2021;8:608215. doi: 10.3389/fmed.2021.608215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao A.T., Tong Y.X., Gao C., Zhu L., Zhang Y.J., Zhang S. Dynamic profile of RT-PCR findings from 301 COVID-19 patients in Wuhan, China: a descriptive study. J Clin Virol. 2020;127:104346. doi: 10.1016/j.jcv.2020.104346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arevalo-Rodriguez I., Buitrago-Garcia D., Simancas-Racines D., Zambrano-Achig P., Del Campo R., Ciapponi A., Sued O., Martinez-García L., Rutjes A.W., Low N., Bossuyt P.M., Perez-Molina J.A., Zamora J. False-negative results of initial RT-PCR assays for COVID-19: a systematic review. PLoS One. 2020;15:e0242958. doi: 10.1371/journal.pone.0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J.-J., Cao Y.-Y., Dong X., Wang B.-C., Liao M.-Y., Lin J., Yan Y.-Q., Akdis C.A., Gao Y.-D. Distinct characteristics of COVID-19 patients with initial rRT-PCR-positive and rRT-PCR-negative results for SARS-CoV-2. Allergy. 2020;75:1809–1812. doi: 10.1111/all.14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020;92:903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao A.T., Tong Y.X., Zhang S. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. 2020;92:1755–1756. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y., Chen S., Yang Z., Guan W., Liu D., Lin Z., Zhang Y., Xu Z., Liu X., Li Y. SARS-CoV-2 viral load in clinical samples from critically ill patients. Am J Respir Crit Care Med. 2020;201:1435–1438. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W.-D., Chang S.-Y., Wang J.-T., Tsai M.-J., Hung C.-C., Hsu C.-L., Chang S.-C. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. 2020;81:318–356. doi: 10.1016/j.jinf.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z., Doan K., Bastien N., Li Y., Van Caeseele P.G., Poliquin G. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cevik M., Kuppalli K., Kindrachuk J., Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. 2020;371:m3862. doi: 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- 47.Hoorfar J., Malorny B., Abdulmawjood A., Cook N., Wagner M., Fach P. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J Clin Microbiol. 2004;42:1863–1868. doi: 10.1128/JCM.42.5.1863-1868.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petter E., Mor O., Zuckerman N., Oz-Levi D., Younger A., Aran D., Erlich Y. Initial real world evidence for lower viral load of individuals who have been vaccinated by BNT162b2. medRxiv. 2021 doi: 10.1101/2021.02.08.21251329. [Preprint] doi: [DOI] [Google Scholar]
- 49.Levine-Tiefenbrun M., Yelin I., Katz R., Herzel E., Golan Z., Schreiber L., Wolf T., Nadler V., Ben-Tov A., Kuint J., Gazit S., Patalon T., Chodick G., Kishony R. Initial report of decreased SARS-CoV-2 viral load after inoculation with the BNT162b2 vaccine. Nat Med. 2021;27:790–792. doi: 10.1038/s41591-021-01316-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Longitudinal quantitative RT-PCR severe acute respiratory syndrome coronavirus 2 test results for patients diagnosed with coronavirus disease 2019. A: Longitudinal severe acute respiratory syndrome coronavirus 2 test results for the study population (Table 1). [For clarity, 191 patients for whom the first negative sample (red) was obtained more than 50 days after the day of diagnosis were omitted.] The day of diagnosis and the last positive test result (purple) demarcate the “positive period” (light blue shading). Negative test results within this individually determined period were regarded as “undetectable” (orange). Similarly, positive test results within the positive period were regarded as detectable (blue). All test series end with a sequence of one or more negative results (red). Patients are sorted by the dates, relative to their first negative result, then by the relative date of the last positive result. B: Frequency of test results per day relative to first negative result. P1, P2, and P3 indicate three representative patients.
Differential change in cycle threshold (Ct) value along time after day of diagnosis for different age groups. In the first 4 days after diagnosis, Ct values of the E gene (A), RdRp gene (B) and N gene (main text, Figure 3) are lower for older patients (age ≥40 years, light gray) than for younger patients (age <40 years, dark gray). The opposite trend appears for the internal control gene (C), in agreement with within-tube competition for reagents between the multiplexed reactions. Error bars indicate SEM.
Difference between cycle threshold (Ct) values of positive results after undetectable and detectable test results. Positive test results that followed an undetectable test result had a higher Ct value for the E gene (A), RdRp gene (B), and N gene (main text, Figure 4), whereas no significant difference was observed for the internal control gene (C). Error bars indicate SEM. ∗P value <10−9.
Distribution of cycle threshold (Ct) values. The frequency of Ct values for positive test results. A:E gene. B:RdRp gene. C:N gene.