Abstract
Sik (mouse Src-related intestinal kinase) and its orthologue BRK (human breast tumor kinase) are intracellular tyrosine kinases that are distantly related to the Src family and have a similar structure, but they lack the myristoylation signal. Here we demonstrate that Sik and BRK associate with the RNA binding protein Sam68 (Src associated during mitosis, 68 kDa). We found that Sik interacts with Sam68 through its SH3 and SH2 domains and that the proline-rich P3 region of Sam68 is required for Sik and BRK SH3 binding. In the transformed HT29 adenocarcinoma cell cell line, endogenous BRK and Sam68 colocalize in Sam68-SLM nuclear bodies (SNBs), while transfected Sik and Sam68 are localized diffusely in the nucleoplasm of nontransformed NMuMG mammary epithelial cells. Transfected Sik phosphorylates Sam68 in SNBs in HT29 cells and in the nucleoplasm of NMuMG cells. In functional studies, expression of Sik abolished the ability of Sam68 to bind RNA and act as a cellular Rev homologue. While Sam68 is a substrate for Src family kinases during mitosis, Sik/BRK is the first identified tyrosine kinase that can phosphorylate Sam68 and regulate its activity within the nucleus, where it resides during most of the cell cycle.
The Src-related intestinal kinase Sik is an intracellular tyrosine kinase that we identified in a screen for tyrosine kinases in intestinal epithelial cells (34). Although it is related to the Src family and contains SH2 and SH3 domains, it has a very short unique amino terminus and is not myristoylated (41). Sik expression is restricted to differentiating epithelial cells, and it is found in the skin and all linings of the alimentary canal. Addition of calcium to cultured primary mouse keratinocytes induces cell differentiation and rapid activation of Sik (42). Overexpression of Sik in an embryonic mouse keratinocyte cell line resulted in increased expression of the differentiation marker filaggrin during calcium-induced differentiation, suggesting that Sik is involved in a signal transduction pathway that may promote differentiation (42).
The human orthologue of Sik is called BRK (breast tumor kinase) (24, 25). Increased BRK expression has been detected in colon tumors (24), breast tumors (2, 25), and melanomas (11, 22). Neither Sik nor BRK expression has been detected in normal mammary tissue, but both proteins are expressed in normal epithelial cells that are undergoing terminal differentiation in the gastrointestinal tract (24, 41). While BRK appears to play a role in signal transduction in normal epithelial linings, its overexpression appears to be linked to the development of a variety of epithelial tumors. The seemingly paradoxical roles of Sik and BRK during differentiation and tumorigenesis are poorly understood.
To date, no substrates of Sik and BRK have been identified. Here we report that Sam68 (Src associated in mitosis; 68 kDa) is a substrate of Sik that can be phosphorylated by Sik within the nucleus. Sam68 is an RNA binding protein (47) that was first identified as a major target of Src during mitosis (14, 39). Thus far, Sam68 has been shown to be a substrate of Src family kinases (14, 29, 39, 46), ZAP70 (20), and the insulin receptor (31). Although Sam68 resides in the nucleus during most of the cell cycle, none of these tyrosine kinases colocalize with Sam68 within the nucleus. Sam68 has also been shown to be a substrate of Cdc2 during mitosis (28). Sam68 has been proposed to function as a multifunctional adapter protein for Src kinases (29, 38), and it can associate with phospholipase Cγ1, the p85 subunit of phosphatidylinositol-3-kinase (31), and the adapter proteins Grb2 (29), Nck (21), and Grap (40).
Sam68 has been shown to preferentially bind RNA with UAAA motifs (23). The RNA binding activity of Sam68 is negatively regulated by Src kinases (45), and Sam68 may function as a protein that links signaling cascades by Src kinases to RNA metabolism. The type of RNA binding motif present in Sam68 is called the hnRNP K homology (KH) domain (15, 33). Sam68 is part of a subfamily of KH domain-containing proteins, because it contains an extended KH domain embedded in a larger domain called the GSG (GRP33-Sam68-GLD1) domain (10, 19). This protein module is also referred to as the STAR (signal transduction and activation of RNA) domain (43). The GSG domain of Sam68 has been shown to be required for RNA binding (5, 23), RNA-dependent oligomerization (5), and protein localization (4). Sam68 has been observed to localize in novel nuclear bodies called Sam68-SLM nuclear bodies (SNBs) in cancer cell lines (4). Although the function of Sam68 is unknown, Sam68 has been shown to be required for cell cycle progression (3) and can function as a cellular homologue of Rev by transporting unspliced human immunodeficiency virus (HIV) RNA into the cytoplasm (27).
Here we report that both Sik and BRK colocalize with Sam68 within the nucleus. We show that Sik is active within the nucleus and that it can phosphorylate Sam68 in vivo. In addition, we demonstrate that phosphorylation of Sam68 by Sik negatively regulates its RNA binding ability and its ability to function as a Rev cellular homologue. Phosphorylation of Sam68 within the nucleus may have important physiological significance and may contribute to the posttranscriptional control of gene expression during the differentiation of epithelial linings.
MATERIALS AND METHODS
Expression constructs.
For the preparation of the mutant Sik cDNAs, we used the oligonucleotide-mediated Altered sites in vitro mutagenesis system (Promega). The Sik cDNA was cloned into the pAlter plasmid, and the oligonucleotide with the sequence 5′-CACCAGGTTTGAGAACC-3′, with a substitution of A for T resulting in substitution of the tyrosine at position 447 with phenylalanine, was used to generate the Sik Y-F construct. This type of mutation has been shown to lead to constitutive activation of the Src family of tyrosine kinases (8). Preparation of the kinase defective Sik expression construct Sik K-M was previously described (42). Wild-type Sik, Sik Y-F, and Sik K-M coding sequences were cloned into the vector pcDNA3. The GST-Sik constructs were as previously described (42).
The Fyn expression construct and Myc-Sam68 (68–347) (also called P1234) were previously described (29), as was Myc-Sam68 (5). The coding region of mouse hnRNPK was amplified using the expressed sequence tags AA544863 and AA183839 and the oligonucleotides with the sequences 5′-CAGGAATTCACTAGTCTTAGAAAA-3′ and 5′-AATGAATTCCGAACAGCCAGAAGA-3′, and it was digested with EcoRI and subcloned in frame in Myc-Bluescript (29). The DNA fragments encoding the Sam68 proline motifs P0, P1P2, P3, P4, and P5 were amplified by PCR using Myc-Sam68 as a DNA template (5). The DNA was digested with BamHI and EcoRI and subcloned in the respective sites of pGEX-KG (16). The sequences of the oligonucleotides are as follows: P0, 5′-CGT GGA TCC AAG GAC CCG TCA GGT-3′ and 5′-GCG GAA TTC TCA AGC GCC TCC TCT GGG CCC AC-3′; P1P2, 5′-CGG GGA TCC CCC GCC ACC CAG CCG CCG-3′ and 5′-GCG GAA TTC TCA CGG CTG TGG CTG ACG GGG GC-3′; P3, 5′-AAC GGA TCC CCT GAA CCC TCT CGT GGT-3′ and 5′-GCG GAA TTC TCA AGC TCC TCT AGG TGG TCC AAC-3′; P4, 5′-CGT GGA TCC CCA GTG AGA GCT CCA TCA CC-3′ and 5′-GCG GAA TTC TCA CCC AGC TGT CCG AGC TCT TG-3′. For construction of GST-Sam68 (331–443), Myc-Sam68 was digested with XhoI and the DNA fragment corresponding to amino acids 331 to 443 was subcloned in frame into Myc-BS. The resulting plasmid was digested with BamHI and KpnI, and the fragment was subcloned into the BamHI-HindIII sites of pGEX-KG. The KpnI and HindIII sites were made blunt. For construction of GST-Sam68 (354-393), Myc-Sam68 was used as a template for the following primers: forward, CCCGGATCCATTCAGAGAATACCTTTG, and reverse, ATAGAATTCTTACTCCCCTTGACTCTGGC. The DNA fragment was digested with BamHI and EcoRI and subcloned into the corresponding sites in pGEX-KG. Myc-pcDNA Sam68 was constructed by digesting Myc-BS Sam68f (5) with EcoRI and subcloning the fragment in frame in Myc-pcDNA (6). Myc-pcDNA Sam68Δ C was constructed by digesting Myc-pcDNA Sam68 with XhoI and religating. This deletes amino acids 348 to 443 of Sam68, and the translation terminates in the vector.
Peptides P0, P3, and P4 used in competition assays were synthesized by the W. M. Keck Biotechnology Resource Center, New Haven, Conn., and their sequences are as follows: P0, biotin-RLTPSRPSPLPHRPRGGGGGPRGG; P3, biotin-GVSVRGRGAAPPPPPVPRGRGVGP; P4, biotin-TRGATVTRGVPPPPTVRGAPTPR.
Cell lines.
Cell lines were obtained from the American Type Culture Collection. NMuMG cells were generally transfected using the LipofectAMINE Reagent (Gibco/BRL). HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM) with 1.0 mM sodium pyruvate and 10% bovine calf serum (HyClone, Logan, Utah) and were transfected with the vaccinia virus T7 expression system and lysed as previously described (29). COS7 cells were maintained in DMEM supplemented with 10% bovine calf serum and were transfected using the DEAE-dextran method.
Subcellular fractionation.
Cells were washed two times in 1× phosphate-buffered saline (PBS) and one time in hypotonic lysis buffer (HLB; 20 mM Tris-HCl [pH 7.5], 1 mM MnCl2, 2 mM EGTA) for 5 min on ice. Cells were then treated with 1.5 ml of HLB (with 20 μg of leupeptin/ml and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and shaken for 20 min on ice. Cells were scraped and homogenized in a Dounce homogenizer (50 to 60 strokes) and spun for 10 min at 2,300 rpm, 4°C. The supernatant from this spin was kept as cytosolic and membrane fractions. The pellet was washed in 1 ml of HLB, spun 4 min at 5,000 rpm at 4°C, and resuspended in 1 ml of Dignum buffer (20 mM HEPES [pH 7.9], 420 mM NaCl, 1.5 mM MnCl2, 0.1 mM EDTA, 25% glycerol, 0.5 mM dithiothreitol [DTT], 0.5 mM PMSF, 2 μg of leupeptin/ml, 2 μg of aprotinin/ml, 1 mM NaVO4). After shaking for 15 min at 4°C, samples were spun at 14,000 rpm for 10 min at 4°C. The supernatant was kept as the nuclear protein fraction.
Antibodies, immunoprecipitations, and immunoblotting.
Anti-Sik polyclonal antibodies N20 (sc-915) and C17 (sc-916) were purchased from Santa Cruz Biotechnology. Immunoblot analyses were performed with a combination of the two mouse Sik antibodies, N20 and C17, at a 1:5,000 dilution for increased sensitivity. BRK was detected with the Santa Cruz Biotechnology BRK antibody (C-17, sc-1188) or the Sik N20 antibody. The monoclonal antibody anti-Myc 9E10 (Santa Cruz Biotechnology) and antiphosphotyrosine antibodies 4G10 (1:10,000) and PY-20 (1:2,000) (both from Santa Cruz Biotechnology) and RC20-HRPO (1:5,000) (Transduction Laboratories) were also used. The anti-AD1 rabbit polyclonal antibody specific for Sam68 was generated using a peptide from amino acids 330 to 348 of mouse Sam68 (4). For immunoblotting, the designated primary antibody was followed by either goat anti-rabbit antibody, goat anti-mouse antibody conjugated to horseradish peroxidase (HRP) (ICN), HRP-conjugated donkey anti-rabbit antibody, or HRP-conjugated protein A (Transduction Laboratories), and chemiluminescence was used for protein detection.
Immunoprecipitations were performed as previously described (42). Anti-BRK antibodies or anti-Sam68 AD-1 and 50 μl of protein G-Sepharose (Amersham or Pharmacia Biotech) were incubated with 1 to 2 mg of cell lysate for 3 to 16 h at 4°C. As controls, lysates were incubated with Sepharose beads and rabbit serum, rabbit immunoglobulin G (IgG), or alone.
Sik-GST fusion protein in vitro binding assays.
Glutathione S-transferase–Sik (GST-Sik) and GST-Sam68 fusion proteins were prepared as described previously (29, 42). Cell lysates were precleared by incubating with GST-saturated glutathione beads for 30 min. Precipitations were performed by incubating lysates with GST, GST-SikSH2/3, Sik SH2, or Sik SH3 for 45 min at 4°C, followed by incubation with glutathione-Sepharose beads (Amersham) for 30 min. Precipitates were eluted with sample buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblotting with anti-Sam68 antibody.
Immunofluorescence.
Cells were grown on chamber slides (Falcon) and fixed in methanol at −20°C for 5 min or Carnoy's fixative for 5 min at room temperature. Cells transfected with green fluorescent protein (GFP) constructs were fixed in 4% paraformaldehyde for 5 min at room temperature and permeabilized in 50% methanol–50% acetone for 15 min at −20°C. Slides were blocked in 2% goat serum or 3% bovine serum albumin in TNT buffer (0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl, 0.05% Tween 20) for 30 to 40 min. Slides were then incubated with anti-BRK or anti-Sam68 AD-1 antibodies (1:250) overnight at 4°C, washed, incubated with biotinylated goat anti-rabbit antibody (Vector Laboratories) (1:250) for 1 h at room temperature, washed, blocked 30 min with blocking reagent (from DuPont NEN), and incubated with streptavidin-HRP (DuPont NEN) (1:100). After washing, tyramide amplification was performed using the TSA-Indirect Kit (DuPont NEN) according to manufacturer's directions. Reactions were visualized with rhodamine-avidin (Vector Laboratories) (1:500), and slides were mounted with Vectashield mounting medium (Vector Laboratories). Controls for specificity of signal were performed by preincubating BRK antibodies with the immunogenic BRK peptide (1:4) for 30 min at room temperature or by incubating sections with normal rabbit sera alone.
For double antibody labeling in HT29 and MCF-7 cells, cells were stained as above with rabbit anti-BRK antibody and visualized with rhodamine, followed by incubation with anti-Sam68 (Transduction Laboratories) (1:50) for 1 h at room temperature and anti-mouse IgG fluorescein isothiocyanate (FITC) conjugate (Sigma) (1:64), and they were analyzed by confocal microscopy.
For double antibody labeling of cells transfected with GFP-Sam68 and Sik constructs, cells were stained overnight with antiphosphotyrosine antibody conjugated to HRP (RC20-HRPO; Transduction Laboratories) at a 1:2,000 dilution at 4°C and were incubated with biotinyl tyramide and rhodamine-avidin (1:500). Next, slides were treated with 1.0% H2O2 for 15 min to inactivate HRP, followed by incubation with the second antibody, anti-Sik (C-17) at a 1:250 dilution, followed by HRP-conjugated goat anti-rabbit antibody, biotinyl tyramide, and then streptavidin Alexa 350 conjugate (Molecular Probes) (1:500) for visualization. Controls were performed by substituting rabbit serum, rabbit IgG, or blocking buffer alone for the first and second antibodies. Omission of Sik antibody in the second antibody incubation, followed by tyramide amplification, resulted in no Alexa 350 signal in these double staining experiments. Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole) (Boehringer Mannheim) for 3 min and washed before mounting.
poly(U) binding assays.
Following transfections with Sam68Δ1–67 (45) and Sik Y-F or Fyn, HeLa cells were lysed on ice in 1% Triton X-100, 25 mM Tris (pH 7.4), 150 mM NaCl, 25 mM NaF, and 100 μM sodium orthovanadate. Lysates were centrifuged to remove insoluble material, and one-fourth of the total cell lysate was added to 20 μl of agarose-poly(U) beads (Pharmacia Biotech Inc.) or agarose beads as a control for 30 min, 4°C. Beads were washed twice with lysis buffer and eluted in Laemmli sample buffer. For assessment of total protein expression, 2.5% of the cell lysate was blotted.
REV assays.
COS7 cells were transfected with a total of 3.5 μg of DNA supplemented with empty pcDNA3.1. Each transfection contained 0.125 μg of Rev response element (RRE) chloramphenicol acetyltransferase (CAT) reporter plasmid pDM128 (17), with 1.5 μg of Rev expression vector B1-SVH6Rev (7, 37), 1.5 μg of Rev mutant B1-SVH6RevM10, 1.5 μg of pcDNA-Sam68ΔC, or 1.5 μg of pcDNA-Sam68. Increasing amounts of expression vectors for Sik K-M and Sik Y-F (0.05, 0.8, and 1.6 μg) were added with pcDNA-Sam68. The β-galactosidase expression vector, pCH110 (0.125 μg; Pharmacia-Amersham Inc.) was included in all transfections for measuring the efficiency of transfection. Forty-eight hours after transfection, the cells were collected and resuspended in 150 μl of 0.25 M Tris-HCl, pH 7.8. The cell extracts were prepared by three freeze-thaw cycles, followed by a brief centrifugation to remove cell debris. CAT and β-galactosidase assays were performed as previously described (30). CAT activity was normalized to the β-galactosidase activity and did not exceed twofold.
RESULTS
BRK associates with Sam68 in human tumor cell lines.
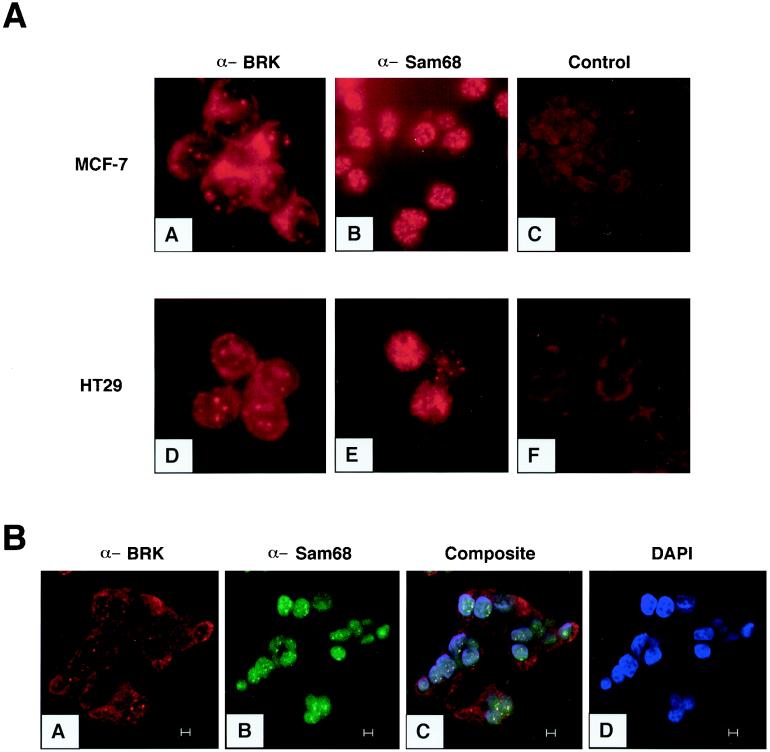
BRK is expressed in breast and colon tumors and tumor cell lines (2, 24). To better understand the role of BRK, we performed indirect immunofluorescence microscopy to visualize its cellular localization. We found that endogenous BRK localized into distinct nuclear dots in the MCF-7 and HT29 breast and colon tumor cell lines (Fig. 1A, panels A and D). The presence of BRK in nuclear dots was not due to the elevated expression of BRK in these cells because Sik overexpression in normal murine mammary gland cells was localized diffusely in the nucleus without the presence of nuclear dots (see Fig. 5 below). The RNA binding protein Sam68 was also observed in similar structures in these cells (Fig. 1A, panels B and E). When control nonimmune serum was used (Fig. 1A, panel F), or when the primary antibody was preincubated with the immunogenic peptide (Fig. 1A, panel C; BRK antibody plus BRK peptide, long exposure), no dots were observed, confirming the specificity of the signal.
FIG. 1.
BRK and Sam68 localize in nuclear structures in breast and colon tumor cell lines. (A) BRK and Sam68 are found in nuclear structures in human tumor cell lines. Immunofluorescence was used to examine the localization of endogenous BRK and Sam68 in the MCF-7 (A, B) and HT29 (C, D) tumor cell lines. Cells were fixed and stained with antibodies against BRK (A, D) or Sam68 (B, E), BRK antibody preincubated with immunogenic peptide (C), or control rabbit sera (F). (B) BRK localizes to SNBs in HT29 cells. Cells were fixed and stained with antibodies against BRK followed by staining with antibodies against Sam68 and were analyzed by confocal microscopy. BRK was visualized with rhodamine (A), and Sam68 staining was visualized with FITC (B). A composite (C) shows colocalization of BRK and Sam68 as yellow spots in the nuclei (C). Nuclei were stained with DAPI (D). Bars, 5 μm.
FIG. 5.
Wild-type Sik and Sik Y-F phosphorylate nuclear proteins that colocalize with GFP-Sam68 within the nucleus. (A) Localization of GFP-tagged Sam68, phosphotyrosine, and wild-type (WT) Sik in transfected NMuMG cells. Antiphosphotyrosine antibody was visualized with rhodamine (red), while anti-Sik antibody binding was visualized with avidin-Alexa 350 (blue). Wild-type Sik is present in the nucleus and at the membrane (panel C). (B) NMuMG cells were transfected with GFP-Sam68 and wild-type Sik (A to D), GFP-Sam68 and Sik Y-F (E to H), GFP-Sam68 and kinase-defective Sik K-M (I to L), or the GFP expression vector pEGFP-C1 and pcDNA3 (M to P). Cells were fixed 24 h after transfection, and tyrosine-phosphorylated proteins were localized using anti-phosphotyrosine antibodies (B, F, J, N). DAPI was used to stain the nuclei (D, H, L, P). In NMuMG cells, Sam68 displays diffuse, nuclear localization visible by green fluorescence (A, E, I). Cells cotransfected with GFP-Sam68 and wild-type Sik or Sik Y-F also stain strongly with the anti-phosphotyrosine antibody visualized using rhodamine (B, F), while no phosphotyrosine was detected in cells expressing kinase-defective Sik K-M (J). Panels C, G, K, and O are composites demonstrating colocalization that appears yellow. GFP alone is expressed throughout the cell (M) and is negative for anti-phosphotyrosine staining (N). (C) Increased phosphotyrosine in SNBs in HT29 cells following introduction of the activated Sik Y-F construct into HT29 cells. HT29 cells were transfected with GFP-Sam68 (A, C) and Sik Y-F, and tyrosine-phosphorylated proteins were localized using antiphosphotyrosine antibodies (B, C). Colocalization of Sam68 and the increased phosphotyrosine signal in SNBs are shown in panel C. No phosphotyrosine signal was detected in control cells transfected with kinase-defective Sik K-M (not shown). DAPI was used to stain the nuclei (D). Bars represent 5 μm.
Previously, Sam68 was reported to localize to novel nuclear structures termed SNBs that are novel and distinct from coiled bodies, gems, PML nuclear bodies, the perinucleolar compartment, and SC-35 speckles (4). SNBs contain nucleic acid that is most likely RNA, but their function remains unknown (4). We examined whether BRK colocalized with Sam68 in SNBs. HT29 cells were fixed and stained with antibodies against Sam68 (4) and BRK (BRK C-17), followed by secondary antibodies conjugated to FITC and rhodamine, respectively. The fluorescent signals were imaged using confocal microscopy. It was observed that most of the BRK nuclear dots colocalized with Sam68 SNBs (Fig. 1B). These findings demonstrate that BRK is a nuclear kinase that appears to be a component of SNBs in the HT29 colon cancer cell line.
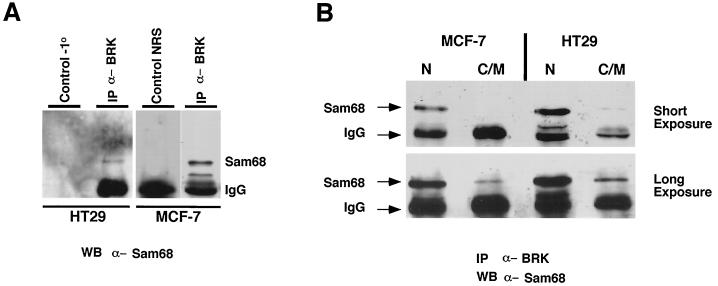
The colocalization of BRK and Sam68 in SNBs suggested that these proteins might associate. To examine whether endogenous BRK associates with Sam68 in HT29 and MCF-7 cells, we performed coimmunoprecipitation experiments. Anti-BRK immunoprecipitates from HT29 and MCF-7 cells contained a band corresponding to Sam68 that was not detected when normal rabbit serum was used as a control or when the primary antibody was omitted (Fig. 2A). A significant increase in the amount of Sam68 that coprecipitated with BRK was detected when nuclear protein fractions were used for the immunoprecipitations (Fig. 2B). These findings suggest that BRK is a nuclear tyrosine kinase that associates with Sam68 in SNBs in cancer cell lines.
FIG. 2.
BRK and Sam68 associate within the nuclei of HT29 and MCF-7 cells. (A) Sam68 coimmunoprecipitates with BRK from lysates of the HT29 colon and MCF-7 breast carcinoma cell lines. Cells were lysed, and 1 mg of total-cell lysate was incubated with anti-BRK antibodies, normal rabbit serum (NRS), or Sepharose beads alone as a control for nonspecific binding to beads. The immunoprecipitate was resolved by SDS-PAGE followed by immunoblotting with anti-Sam68 AD1 polyclonal antibodies. (B) Nuclear (N) and cytosolic and membrane (C/M) fractions from MCF-7 and HT29 cells were immunoprecipitated with anti-BRK antibodies, followed by immunoblotting with anti-Sam68 antibodies. A short exposure (10 s) shows interaction in the nuclear fraction only. A longer exposure (45 s) shows much weaker interaction in the cytosolic and membrane fraction.
Sik phosphorylates Sam68 in vivo.
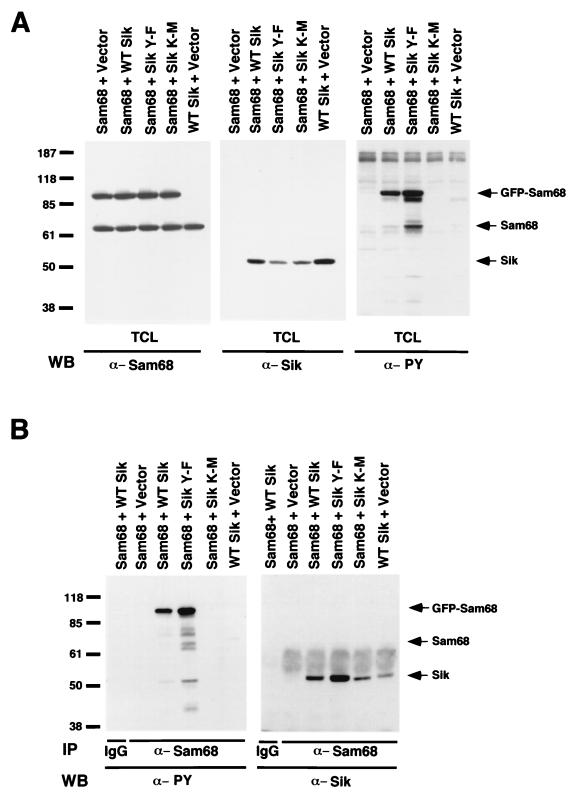
Colocalization of BRK with Sam68 suggested that Sam68 may be a substrate for the Sik and BRK tyrosine kinase within the nucleus. To determine if Sam68 is a substrate of Sik, the normal murine mammary gland cell line NMuMG (18) was transiently transfected with wild-type, putative activated (Y-F), and kinase-defective (K-M) Sik. The putative activated form of Sik contains a Tyr-to-Phe substitution of the potential regulatory tyrosine at position 447 of Sik, while kinase-defective Sik contains a substitution of a conserved Lys at position 219 within the kinase catalytic domain with Met (42). Previously, we showed that Sik K-M has no kinase activity and acts as a dominant negative protein, but no enzymatic regulatory role has been demonstrated for the carboxy-terminal tyrosine of Sik (42). The different Sik expression constructs were cotransfected with a GFP-Sam68 fusion construct (4), and tyrosine phosphorylation of Sam68 was examined by immunoblotting with anti-phosphotyrosine antibodies. The NMuMG cell line does not express endogenous Sik (24) (Fig. 3A, middle panel, Sam68 + Vector). Tyrosine-phosphorylated GFP-Sam68 was detected in total-cell lysates from NMuMG cells cotransfected with wild-type Sik or Sik Y-F and the GFP-Sam68 expression construct, but not in cells cotransfected with vector alone or the kinase-defective Sik K-M construct (Fig. 3A, right panel). Higher levels of phosphorylated GFP-Sam68 and endogenous Sam68 were detected in cells expressing Sik Y-F than cells expressing wild-type Sik (Fig. 3A, right panel). The elevated tyrosine phosphorylation of Sam68 in Sik Y-F-transfected cells was not a result of elevated expression of Sik Y-F or GFP-Sam68 (Fig. 3A, left and middle panels). The association between Sik and Sam68 was further investigated in NMuMG cells transfected with GFP-Sam68 and Sik. Anti-Sam68 immunoprecipitates contained a coimmunoprecipitated phosphotyrosine protein with a molecular mass of 50 kDa that migrates in the expected position for autophosphorylated Sik (Fig. 3B, left panel). Moreover, Sik coimmunoprecipitated with both GFP-Sam68 and endogenous Sam68, because Sik could be detected in Sam68 immunoprecipitates from cells transfected with only wild-type Sik (Fig. 3B, right panel). Greater levels of Sik Y-F coimmunoprecipitated with Sam68 than wild-type Sik or Sik K-M. This may be explained by the increased ability of Sik Y-F to phosphorylate and then bind phosphorylated Sam68 through its SH2 domain (see Fig. 6A). No band corresponding to Sik was present in Sam68 precipitates from cells transfected with GFP-Sam68 and pcDNA3 (Vector) or in immunoprecipitations with IgG.
FIG. 3.
Sik phosphorylates Sam68 in vivo. (A) Sam68 is tyrosine phosphorylated in NMuMG cells expressing wild-type Sik and Sik Y-F. NMuMG cells were cotransfected with GFP-Sam68 and Vector alone, wild-type (WT) Sik, Sik Y-F, or kinase-defective Sik K-M. Total-cell lysates were divided equally and immunoblotted with antibodies against Sam68, Sik, and phosphotyrosine. Tyrosine-phosphorylated Sam68 was detected only in lysates containing wild-type Sik or Sik Y-F. Although lower levels of Sik Y-F were expressed than wild-type Sik, the highest levels of tyrosine-phosphorylated Sam68 were detected in cells cotransfected with Sik Y-F, suggesting that tyrosine 447 of Sik negatively regulates its activity. (B) Sik associates with Sam68. Immunoprecipitations were performed with Sam68 antibody or IgG as a control and with lysates from the transfected cells in panel A. Immunoblotting was performed with antiphosphotyrosine or anti-Sik antibodies. Sik was observed to coprecipitate with Sam68 from lysates of cells transfected with Sik expression constructs. Sik antibody binding was detected using HRP-conjugated protein A. Tyrosine-phosphorylated Sik could also be detected in the Sam68 immunoprecipitates (left panel).
FIG. 6.
The Sik SH2 and SH3 domains bind Sam68. (A) The ability of Sam68 to bind the SH2 and SH3 domains of Sik was tested. NMuMG cells were transfected with GFP-Sam68 and the expression vector pcDNA3 (Vector) or the wild-type Sik, activated Sik Y-F, or kinase-defective Sik K-M expression constructs. Cell lysates were divided equally and incubated with GST, GST-Sik SH2+SH3, GST-Sik SH2, and GST-SH3 covalently coupled to beads. Bound proteins as well as an aliquot of total-cell lysate from GFP-Sam68-transfected cells were separated by SDS-PAGE and immunoblotted with Sam68 AD1 polyclonal antibody. Positions of GFP-Sam68 and the endogenous Sam68 protein are indicated with arrows. GFP-Sam68 and endogenous Sam68 protein bound to the GST-Sik, SH2+SH3, and GST-Sik SH3 fusion proteins in all of the cell lysates. GFP-Sam68 binding to the GST-Sik SH2 domain was detected only in cells transfected with wild-type Sik or Sik Y-F, suggesting that phosphorylation by Sik is required for Sik SH2 binding. (B) Sam68 proline motif 3 (P3) associates with the SH3 domain of Sik. HeLa cell lysates were incubated with GST, GST-Sam68 P0, GST-Sam68 P1P2, GST-Sam68 P3, or GST Sam68 P4 covalently coupled to beads. The beads were washed, and the bound BRK was observed by immunoblotting. An aliquot of the HeLa cell lysate was used to represent total-cell lysate. BRK binding was only detected with the GST-Sam68 P3 fusion protein. (C) Beads containing covalently coupled GST-Sik SH3 protein were preincubated for 15 min at room temperature with the indicated concentration of Sam68 proline-rich peptide. Subsequently, HeLa cell lysates were added to each tube for 30 min at 4°C. The beads were washed extensively, and the bound Sam68 was quantitated by immunoblotting. Binding of Sam68 with the GST-Sik SH3 fusion protein was efficiently competed with the P3 peptide.
The ability of the different Sik constructs to phosphorylate Sam68 in HeLa cells transfected with wild-type Sik, Sik Y-F, and Sik K-M and Myc-tagged Sam68 was further examined using the vaccinia virus T7 expression system. Transfected HeLa cells were lysed, and the proteins were analyzed by immunoblotting with anti-Sik, anti-Myc, and anti-phosphotyrosine antibodies. Several proteins, including one comigrating with Sam68, appeared heavily phosphorylated by the Sik Y-F construct (Fig. 4A, right panel). These data provide additional evidence that Sik is negatively regulated by phosphorylation of the carboxy-terminal tyrosine at position 447 and that substitution of Sik Y447F activates the kinase. The anti-Sik and anti-Myc immunoblots show equivalent expression of the proteins (Fig. 4A, left and middle panels).
FIG. 4.
Sik Y-F has increased kinase activity and specifically phosphorylates Sam68. (A) The carboxy-terminal tyrosine of Sik functions as a negative regulatory site. HeLa cells were cotransfected with Myc-tagged Sam68 and wild-type (WT) Sik, Sik Y-F, or Sik K-M (kinase defective). Cells were lysed, the proteins were separated by SDS-PAGE, and immunoblotting with anti-Sik, anti-Myc, and antiphosphotyrosine antibodies was performed. Increased phosphorylation of Myc-Sam68 was visible in total-cell lysates from Sik Y-F-transfected cells (right panel). (B) HeLa cells were transfected with Sik Y-F or were cotransfected with Sik Y-F and Myc-Sam68, Myc-hnRNPK, or truncated Myc-Sam68 (68-347). The cells were lysed and immunoprecipitated with IgG (control) or anti-Myc antibodies. The bound proteins were analyzed by immunoblotting with antiphosphotyrosine antibodies (left panel). The same membrane was subsequently immunoblotted with anti-Myc antibodies (middle panel). Total-cell extracts (TCL) were immunoblotted with anti-Sik antibodies (right panel). The band at ∼55 kDa in lanes 1 to 16 represents the heavy chain of the immunoprecipitating antibodies. (C) Sik phosphorylates the carboxy terminus of Sam68. In vitro kinase assays were performed using full-length GST-Sik, [γ-32P]ATP, and 2 μg of the following substrates: GST-Grb2-N-SH3 (negative control), GST alone, GST-Sam68 (331–443), or GST-Sam68 (354–393). The proteins were separated by SDS-PAGE and stained with Coomassie blue (left). The gel was dried, and the phosphorylated proteins were visualized by autoradiography (right).
The ability of Sik Y-F to phosphorylate other nuclear KH domain proteins, such as hnRNPK, was examined. A truncated form of Sam68, Sam68 (68–347), which contains amino acids 68 to 347 and lacks part of the amino terminus and the tyrosine-rich carboxy terminus, was also investigated as a substrate for Sik. HeLa cells were transfected with Sik Y-F alone or were cotransfected with Sik Y-F and Myc-Sam68, Myc-hnRNPK, or Myc-Sam68 (68–347). The cells were lysed and immunoprecipitated with control IgG or anti-Myc antibodies. The bound proteins were analyzed by immunoblotting with antiphosphotyrosine, anti-Myc, and anti-Sik antibodies (Fig. 4B). A phosphotyrosine-containing protein with a molecular mass of 68 kDa (lane 6) was observed in anti-Myc immunoprecipitates from extracts transfected with wild-type Myc-tagged Sam68, but not with Myc-hnRNPK or Myc-Sam68 (68–347) (Fig. 4B, left panel). The membrane was reimmunoblotted with anti-Myc antibodies to confirm the equivalent expression of Myc-Sam68, Myc-hnRNPK, or Myc-Sam68 (68–347) (Fig. 4B, middle panel). Total-cell extracts were also verified for the equivalent expression of Sik Y-F by immunoblotting with anti-Sik antibodies (Fig. 4B, right panel). These data suggest that the C terminus of Sam68 is the target for the Sik tyrosine kinase.
To confirm that the carboxy terminus of Sam68 is directly phosphorylated by Sik, we incubated full-length GST-Sik with GST-Sam68 (331-443) and GST-Sam68 (354-393), two Sam68 carboxy terminus fusion proteins containing amino acids 331 to 443 and 354 to 393, respectively, in the presence of [γ-32P]ATP. These two carboxy-terminal fragments of Sam68 were efficiently phosphorylated by Sik, whereas a control GST protein containing the amino-terminal SH3 domain of Grb2 and GST alone was not phosphorylated (Fig. 4C, right panel). The amounts of proteins used were determined to be equivalent as visualized by Coomassie blue staining (left panel). These data suggest that Sik directly and specifically phosphorylates the C terminus of Sam68.
Colocalization of Sik, Sam68, and phosphotyrosine in the nuclei of transfected cells.
The localization of Sik and Sam68 was investigated in the NMuMG cell line. NMuMG cells were transfected with both wild-type Sik and GFP-Sam68 and were analyzed by confocal microscopy. The pattern of wild-type Sik expression was visualized by avidin-Alexa 350 (blue) (Fig. 5A, panel C) and was detected in the nucleus and at the membrane. Expression of the Sik Y-F and Sik K-M expression constructs was also detected in the nuclei and at the membranes of transfected cells (data not shown). GFP-Sam68, wild-type Sik, and phosphotyrosine colocalized in the nucleoplasm of the cells (Fig. 5A, panel D). Nuclear bodies were more commonly seen in cancer cell lines, but were not generally observed in the nontransformed NMuMG cell line (Fig. 5A and B). In experiments with control IgG, no specific fluorescent signal was detected.
The localization and tyrosine phosphorylation of Sam68 in the presence of wild-type Sik, Sik Y-F, and Sik K-M were examined (Fig. 5B). Phosphotyrosine was readily detected only in the nuclei of cells transfected with active Sik and Sam68, suggesting that Sik is active within the nuclei and phosphorylates Sam68. Phosphotyrosine was detected in the nuclei of cells transfected with wild-type Sik and Sik Y-F (Fig. 5B, panels B and F), but not in kinase-defective Sik K-M-transfected cells (Fig. 5B, panel J). The majority of Sik-phosphorylated protein colocalized with Sam68 (Fig. 5B, panels C and G). We also observed that the intensity of the antiphosphotyrosine staining was greatest in cells transfected with Sik Y-F and Sam68. Cotransfection of the GFP expression vector pEGFP-C1 and the empty Sik expression vector pcDNA3 resulted in diffuse GFP fluorescence throughout the cell (Fig. 5B, panel M) and no detectable antiphosphotyrosine staining (Fig. 5B, panel N). These data provide further support that Sam68 is a substrate for Sik in vivo.
Since BRK and Sam68 were observed to colocalize in SNBs (Fig. 1), the presence of phosphotyrosine in these SNBs following transfection of the Sik expression constructs was examined. HT29 cells were transfected with GFP-Sam68 and wild-type Sik, Sik Y-F, or kinase-defective Sik K-M. The transfected cells were fixed, stained with antiphosphotyrosine antibodies, and analyzed by confocal microscopy. Tyrosine-phosphorylated protein colocalized with Sam68 in SNBs in these transformed cells transfected with wild-type Sik and Sik Y-F, but not with Sik K-M. The Sik Y-F transfection is shown in Fig. 5C. These data demonstrate that Sik most likely targets Sam68 in SNBs in cancer cell lines.
Sik-Sam68 interaction is mediated by both the SH3 and SH2 domains.
To determine which part of Sik interacts with Sam68, different domains of Sik were expressed in bacteria as GST-fusion proteins and used in GST pull down assays. Lysates from NMuMG cells transfected with wild-type Sik, Sik Y-F, or Sik K-M and GFP-Sam68 expression constructs were incubated with GST alone, GST-Sik SH2+SH3, GST-Sik SH2, and GST-Sik SH3 bound to beads. Bound proteins were detected by immunoblotting with anti-Sam68 AD1 antibody (Fig. 6A). GFP-Sam68 and the endogenous Sam68 were observed to associate with the Sik SH3 and Sik SH2+SH3 domain fusion proteins (Fig. 6A). Association of GFP-Sam68 with the Sik SH2 domain was observed only when it was coexpressed with wild-type Sik or Sik Y-F, but not kinase-defective Sik K-M (Fig. 6A). We also observed association of endogenous phosphorylated Sam68 protein following longer exposures of the immunoblots. These data demonstrate that Sik associates with Sam68 through both its SH3 and SH2 domains.
Sam68 contains at least five proline motifs and interacts with the SH3 domains of several proteins, including Src, Fyn, and PLCγ-1 (14, 29, 39, 46). To determine which proline motif within Sam68 mediates interaction with the Sik SH3 domain, we used a GST pull down approach, with lysates from HeLa cells that express human BRK and GST fusion proteins representing the five proline (P0, P1P2, P3, and P4) motifs in Sam68. We found that GST-P3 was the main polypeptide that interacted with BRK (Fig. 6B). To further confirm the specific interaction of Sam68 P3 with the Sik SH3 domain, we tested the ability of peptides representing the proline-rich sequences P0, P3, and P4 to compete with binding of Sam68 with the GST-Sik SH3 domain. The Sam68 P3 peptide competed with the binding between Sam68 and the Sik SH3 domain. No significant competition was observed with the P0 and P4 peptides. Thus, the Sik SH3 domain appears to interact with one major proline motif in Sam68, P3, that is neither a type I nor a type II proline motif (13).
Sik inhibits the RNA binding ability of Sam68.
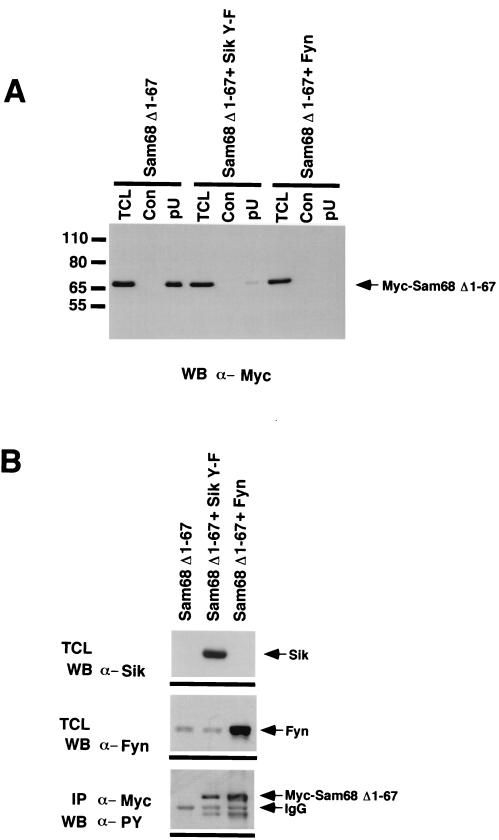
Tyrosine phosphorylation of Sam68Δ1–67 negatively regulates its ability to bind to homopolymeric RNA (45). To determine whether Sik can regulate the ability of Sam68 to bind RNA, HeLa cells were transfected with Myc-Sam68Δ1–67 or cotransfected with Myc-Sam68Δ1–67 and Sik Y-F or Fyn. The expression of Fyn served as a positive control, as we have shown previously that Fyn can negatively regulate Sam68Δ1–67 homopolymeric RNA binding (45). HeLa cell lysates were divided equally and incubated with either poly(U) immobilized to agarose or agarose alone. Sam68 bound poly(U) homopolymeric RNA, when expressed alone (Fig. 7A). However, little or no RNA binding was detected when Sam68 was coexpressed with Sik Y-F or Fyn (Fig. 7A). The reduction of bound Myc-Sam68 was not due to poor expression of Myc-Sam68 (Fig. 7A). Total-cell lysates were immunoblotted with anti-Sik, anti-Fyn, and anti-phosphotyrosine antibodies, and we found that Sik Y-F and Fyn were both overexpressed and Myc-Sam68 was tyrosine phosphorylated (Fig. 7B). These data suggest that Sik is able to negatively regulate the ability of Sam68 to bind RNA.
FIG. 7.
Sik negatively regulates the ability of Sam68 to bind RNA. (A) HeLa cell lysates from cells either transfected with Myc-Sam68Δ1–67 alone or cotransfected with Myc-Sam68Δ1–67 and Sik Y-F or Fyn were divided equally and precipitated with agarose (Con) or poly(U)-agarose (pU), followed by anti-Myc immunoblotting. An aliquot of total-cell lysate (TCL) was also included to monitor expression of Myc-Sam68Δ1–67. The ability of Myc-Sam68Δ1–67 to bind poly(U) agarose was inhibited in cells transfected with either Sik Y-F or Fyn. (B) Sik and Fyn are efficiently expressed and phosphorylate Myc-Sam68Δ1–67 in transfected cells. Cell lysates from transfected cells were immunoblotted with anti-Sik and anti-Fyn antibodies to demonstrate that these kinases were efficiently expressed. Immunoprecipitation of Myc-Sam68Δ1–67 followed by immunoblotting with antiphosphotyrosine confirmed that Myc-Sam68Δ1–67 was tyrosine phosphorylated by both Sik and Fyn.
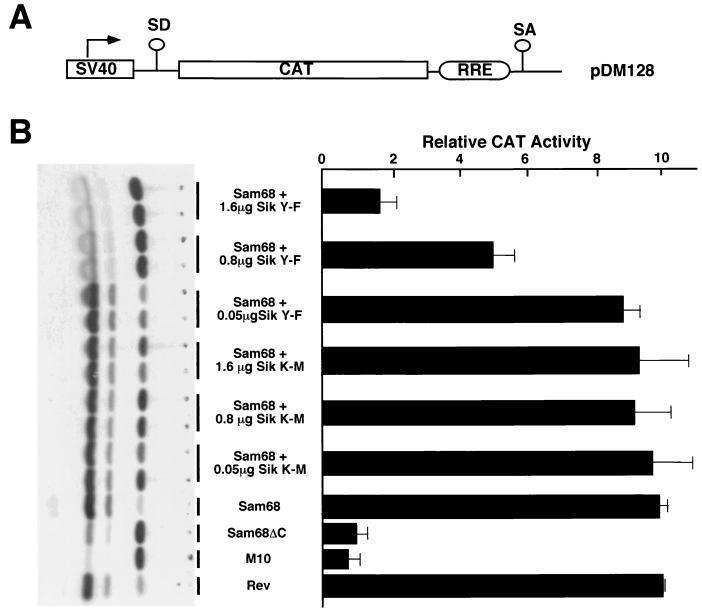
Sam68 has been shown to function as a cellular homologue of Rev in transporting HIV RNA (27). We examined whether Sik could regulate this nuclear function of Sam68 by determining if Sik could modulate RRE-directed reporter gene expression (Fig. 8). COS7 cells were transfected with an RRE-CAT reporter plasmid in the presence of Sam68 and increasing amounts of kinase-active Sik Y-F or kinase-inactive Sik K-M. The transfection of Sam68 or Rev with the RRE-CAT reporter resulted in a 10-fold increase in CAT activation in comparison to Sam68ΔC and RevM10, two proteins shown to be inactive in Rev function. These data are consistent with previously published data (27). The cotransfection of Sam68 with Sik Y-F, but not Sik K-M, decreased CAT activity in a dose-dependent manner. These findings indicate that Sik can regulate a nuclear function of Sam68 and that its kinase activity is required.
FIG. 8.
Sik negatively regulates the ability of Sam68 to function as a cellular Rev homologue. (A) A schematic diagram of the Rev responsive element reporter CAT construct is shown. The splice acceptor and donor sites are indicated as SA and SD. CAT indicates the chloramphenicol acetyltransferase cDNA, and RRE is the HIV Rev responsive element. (B) COS7 cells were transfected with the RRE-CAT reporter plasmid in the presence of the indicated expression vectors and pCH110. CAT activity was normalized to β-galactosidase activity. Each bar represents CAT activity from at least eight samples from at least three separate experiments and the standard deviation indicated. The autoradiogram shown on the left represents a typical experiment: the two rows of dots on the left represent monoacetylated [14C]chloramphenicol and the row on the right represents unacetylated [14C]chloramphenicol.
DISCUSSION
The tyrosine kinases represent a large family of diverse proteins that play important roles in the regulation of growth and differentiation. Thus far, only a small number of nuclear tyrosine kinases have been identified, including Abl, Rak, Fes, Fer, and the dual-specificity kinase Wee1 (reviewed in references 26 and 44). In these studies, we showed that Sik and BRK are present in the nucleus. The mechanisms by which Sik and BRK localize to the nucleus are unknown, as they lack a clear nuclear localization signal (41). It is possible that Sik and BRK are transported to the nucleus through association with other proteins containing nuclear localization signals. Like Abl, Sik and BRK are not nuclear specific. In addition to being present in the nucleus, BRK was detected in the cytoplasm of HT29 cells (Fig. 1). Sik protein and kinase activity were also detected at the membrane of transfected NMuMG cells (Fig. 5A), consistent with the earlier observation that Sik can associate with a GAP-associated protein (37).
We demonstrate that the RNA binding protein Sam68 is a substrate for Sik within the nucleus and that Sik can inhibit the ability of Sam68 to bind RNA. RNA binding proteins may regulate gene expression by a number of mechanisms (reviewed in reference 32). They may alter RNA structure to regulate interaction with trans-acting factors or provide localization or targeting signals. Although its cellular function is unknown, Sam68 has been shown to be able to functionally substitute for the HIV-1 Rev protein, which plays an essential role in the nuclear export of unspliced and partially spliced viral transcripts and export of the HIV genome (27). We show that Sik kinase activity can negatively regulate the ability of Sam68 to function as a cellular homologue of Rev. These data strengthen the possibility that signaling cascades can regulate the RNA function of GSG domain-containing proteins or STAR proteins. The ability of Sam68 to act in RNA transport suggests a role in posttranscriptional regulation of gene expression, which may be negatively regulated by Sik within the nucleus. Sam68 may also serve as an adaptor for Sik, bringing it into proximity of other, as of yet unidentified, substrates.
Sik and its human orthologue BRK have only 80% amino acid sequence identity (24). Nevertheless, the genes have been mapped to regions of the mouse and human genomes that share conservation of synteny, and we have found that the mouse and human proteins are expressed in similar patterns in differentiated epithelial tissues (24). Here we show that the mouse and human proteins both localize to the nucleus and that they both associate with Sam68. These data provide further evidence that the functions of Sik and BRK in the two species are conserved.
NMuMG cells were isolated from the mammary glands of Namru mice and have epithelial growth characteristics, and they do not form malignant lesions when introduced into nude mice (18). Sik localization is diffuse within the nuclei of immortalized NMuMG cells, while BRK appears in SNBs in the HT29 colon adenocarcinoma cell line (Fig. 5). These data complement earlier studies by Chen et al. (4), who found that SNBs were predominant in transformed cells. SNBs are novel unique dynamic structures that disassemble when transcription is inhibited with actinomycin D (4). When GFP-Sam68 and wild-type Sik are introduced into HT29 cells, they localize to the SNBs, which become tyrosine phosphorylated (Fig. 5C). Tyrosine phosphorylation of Sam68 by Sik does not appear to alter its localization because Sam68 coexpressed with active Sik Y-F is retained in SNBs (Fig. 5).
We have shown that Sik can bind Sam68 through both its SH3 and SH2 domains. It was previously shown that Sam68 also binds SH3 and SH2 domains of Src kinases (14, 29, 39, 46). The binding affinities of specific SH2 domains are influenced by sequence context. For example, Src family members prefer the sequence pYEEI, while the SH2 domains of p85 and PLC-γ select the general motif pY-hydrophobic-X-hydrophobic (35, 36). Using a technique employing degenerate phosphopeptide libraries to predict the specificity of individual SH2 domains, it was determined that the Sik SH2 domain may bind to phosphorylated proteins with p-YEEY, YEDY, YDEY, and YDDY motifs (Z. Songyang and L. C. Cantley, personal communication). Interestingly, Sam68 contains the sequence YEDY in its carboxy terminus, and this is a putative binding site for the Sik SH2 domain. This sequence may also be the target of Sik, as we show here that Sik can phosphorylate the carboxy terminus of Sam68.
Sam68 is the first substrate identified for the Sik kinase. Related KH domain-containing proteins have been shown to play important roles in development, and these include human FMR1 (fragile X mental retardation syndrome) (9); mouse Qk1 (quaking), required for myelination (12); Caenorhabditis elegans GLD-1, required for germ cell differentiation (19); and Drosophila Who/How, required for muscle differentiation (1). The tissue- and differentiation-specific expression and activation of Sik (41, 42) suggest that Sik is involved in regulating epithelial cell differentiation. Sik is unique in that it is the only known tyrosine kinase that can phosphorylate Sam68 within the nucleus, where it can modulate its RNA binding ability and perhaps the pattern of gene expression associated with epithelial cell differentiation. Since Sam68 has also been shown to be involved in cell cycle regulation (3), overexpression of Sik and BRK may contribute to the development of epithelial cancers by altering the ability of Sam68 to regulate cell growth.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant DK44525, Department of the Army grant DAMD17-96-1-6175 (A.L.T.), and Medical Research Council of Canada grant MT 13377 S.R. is a Scholar of the MRC, and T.C. is supported by a Doctoral Research Award from the MRC. J.J.D. is supported by the Signal Transduction and Cellular Endocrinology NIH training grant DK07739.
We thank Michael Serfas and Shahab Uddin for helpful discussions and critical reading of the manuscript.
J.J.D. and S.R. contributed equally to this work.
REFERENCES
- 1.Baehrecke E H. who encodes a KH RNA binding protein that functions in muscle development. Development. 1997;124:1323–1332. doi: 10.1242/dev.124.7.1323. [DOI] [PubMed] [Google Scholar]
- 2.Barker K T, Jackson L E, Crompton M R. BRK tyrosine kinase expression in a high proportion of human breast carcinomas. Oncogene. 1997;15:799–805. doi: 10.1038/sj.onc.1201241. [DOI] [PubMed] [Google Scholar]
- 3.Barlat I, Maurier F, Duchesne M, Guitard E, Tocque B, Schweighoffer F. A role for Sam68 in cell cycle progression antagonized by a spliced variant within the KH domain. J Biol Chem. 1997;272:3129–3132. doi: 10.1074/jbc.272.6.3129. [DOI] [PubMed] [Google Scholar]
- 4.Chen T, Boisvert F M, Bazett-Jones D P, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T, Damaj B B, Herrera C, Lasko P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen T, Richard S. Structure-function analysis of Qk1: a lethal point mutation in mouse quaking prevents homodimerization. Mol Cell Biol. 1998;18:4863–4871. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochrane A W, Chen C H, Kramer R, Tomchak L, Rosen C A. Purification of biologically active human immunodeficiency virus rev protein from Escherichia coli. Virology. 1989;173:335–337. doi: 10.1016/0042-6822(89)90252-3. [DOI] [PubMed] [Google Scholar]
- 8.Cooper J A, Howell B. The when and how of Src regulation. Cell. 1993;73:1051–1054. doi: 10.1016/0092-8674(93)90634-3. [DOI] [PubMed] [Google Scholar]
- 9.De Boulle K, Verkerk A J, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra B A, Willems P J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 10.Di Fruscio M, Chen T, Bonyadi S, Lasko P, Richard S. The identification of two Drosophila K homology domain proteins. Kep1 and SAM are members of the Sam68 family of GSG domain proteins. J Biol Chem. 1998;273:30122–30130. doi: 10.1074/jbc.273.46.30122. [DOI] [PubMed] [Google Scholar]
- 11.Easty D J, Mitchell P J, Patel K, Florenes V A, Spritz R A, Bennett D C. Loss of expression of receptor tyrosine kinase family genes PTK7 and SEK in metastatic melanoma. Int J Cancer. 1997;71:1061–1065. doi: 10.1002/(sici)1097-0215(19970611)71:6<1061::aid-ijc24>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 12.Ebersole T A, Chen Q, Justice M J, Artzt K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- 13.Feng S, Kasahara C, Rickles R J, Schreiber S L. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc Natl Acad Sci USA. 1995;92:12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fumagalli S, Totty N F, Hsuan J J, Courtneidge S A. A target for Src in mitosis. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- 15.Gibson T J, Thompson J D, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993;324:361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- 16.Guan K L, Dixon J E. Eucaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 17.Hope T J, Huang X J, McDonald D, Parslow T G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hynes N E, Jaggi R, Kozma S C, Ball R, Muellener D, Wetherall N T, Davis B W, Groner B. New acceptor cell for transfected genomic DNA: oncogene transfer into a mouse mammary epithelial cell line. Mol Cell Biol. 1985;5:268–272. doi: 10.1128/mcb.5.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones A R, Schedl T. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 1995;9:1491–1504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- 20.Lang V, Mege D, Semichon M, Gary-Gouy H, Bismuth G. A dual participation of ZAP-70 and scr protein tyrosine kinases is required for TCR-induced tyrosine phosphorylation of Sam68 in Jurkat T cells. Eur J Immunol. 1997;27:3360–3367. doi: 10.1002/eji.1830271235. [DOI] [PubMed] [Google Scholar]
- 21.Lawe D C, Hahn C, Wong A J. The Nck SH2/SH3 adaptor protein is present in the nucleus and associates with the nuclear protein SAM68. Oncogene. 1997;14:223–231. doi: 10.1038/sj.onc.1200821. [DOI] [PubMed] [Google Scholar]
- 22.Lee S T, Strunk K M, Spritz R A. A survey of protein tyrosine kinase mRNAs expressed in normal human melanocytes. Oncogene. 1993;8:3403–3410. [PubMed] [Google Scholar]
- 23.Lin Q, Taylor S J, Shalloway D. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J Biol Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 24.Llor X, Serfas M S, Bie W, Vasioukhin V, Polonskaia M, Derry J, Abbott C M, Tyner A L. BRK/Sik expression in the gastrointestinal tract and in colon tumors. Clin Cancer Res. 1999;5:1767–1777. [PubMed] [Google Scholar]
- 25.Mitchell P J, Barker K T, Martindale J E, Kamalati T, Lowe P N, Page M J, Gusterson B A, Crompton M R. Cloning and characterisation of cDNAs encoding a novel non-receptor tyrosine kinase, brk, expressed in human breast tumours. Oncogene. 1994;9:2383–2390. [PubMed] [Google Scholar]
- 26.Pendergast A M. Nuclear tyrosine kinases: from Abl to WEE1. Curr Opin Cell Biol. 1996;8:174–181. doi: 10.1016/s0955-0674(96)80063-9. [DOI] [PubMed] [Google Scholar]
- 27.Reddy T R, Xu W, Mau J K, Goodwin C D, Suhasini M, Tang H, Frimpong K, Rose D W, Wong-Staal F. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat Med. 1999;5:635–642. doi: 10.1038/9479. [DOI] [PubMed] [Google Scholar]
- 28.Resnick R J, Taylor S J, Lin Q, Shalloway D. Phosphorylation of the Src substrate Sam68 by Cdc2 during mitosis. Oncogene. 1997;15:1247–1253. doi: 10.1038/sj.onc.1201289. [DOI] [PubMed] [Google Scholar]
- 29.Richard S, Yu D, Blumer K J, Hausladen D, Olszowy M W, Connelly P A, Shaw A S. Association of p62, a multifunctional SH2- and SH3-domain-binding protein, with src family tyrosine kinases, Grb2, and phospholipase C gamma-1. Mol Cell Biol. 1995;15:186–1897. doi: 10.1128/mcb.15.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard S, Zingg H H. Analysis of cis-acting elements of the oxytocin gene by DNA-mediated gene transfer. In: Conn P M, editor. Methods in neuroscience. Orlando, Fla: Academic Press Inc; 1992. pp. 324–343. [Google Scholar]
- 31.Sanchez-Margalet V, Najib S. p68 Sam is a substrate of the insulin receptor and associates with the SH2 domains of p85 PI3K. FEBS Lett. 1999;455:307–310. doi: 10.1016/s0014-5793(99)00887-x. [DOI] [PubMed] [Google Scholar]
- 32.Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- 33.Siomi H, Matunis M J, Michael W M, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siyanova E Y, Serfas M S, Mazo I A, Tyner A L. Tyrosine kinase gene expression in the mouse small intestine. Oncogene. 1994;9:2053–2057. [PubMed] [Google Scholar]
- 35.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 36.Songyang Z, Shoelson S E, McGlade J, Olivier P, Pawson T, Bustello X R, Barbacid M, Sabe H, Hanafusa H, Yi T, Ren R, Baltimore D, Ratnofsky S, Feldman R A, Cantley L C. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swenarchuk L, Harakidas P, Cochrane A. Regulated expression of HIV-1 Rev function in mammalian cell lines. Can J Microbiol. 1999;45:480–490. [PubMed] [Google Scholar]
- 38.Taylor S J, Anafi M, Pawson T, Shalloway D. Functional interaction between c-Src and its mitotic target, Sam 68. J Biol Chem. 1995;270:10120–10124. doi: 10.1074/jbc.270.17.10120. [DOI] [PubMed] [Google Scholar]
- 39.Taylor S J, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 40.Trub T, Frantz J D, Miyazaki M, Band H, Shoelson S E. The role of a lymphoid-restricted, Grb2-like SH3-SH2-SH3 protein in T cell receptor signaling. J Biol Chem. 1997;272:894–902. doi: 10.1074/jbc.272.2.894. [DOI] [PubMed] [Google Scholar]
- 41.Vasioukhin V, Serfas M S, Siyanova E Y, Polonskaia M, Costigan V J, Liu B, Thomason A, Tyner A L. A novel intracellular epithelial cell tyrosine kinase is expressed in the skin and gastrointestinal tract. Oncogene. 1995;10:349–357. [PubMed] [Google Scholar]
- 42.Vasioukhin V, Tyner A L. A role for the epithelial-cell-specific tyrosine kinase Sik during keratinocyte differentiation. Proc Natl Acad Sci USA. 1997;94:14477–14482. doi: 10.1073/pnas.94.26.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- 44.Wang J Y. Nuclear protein tyrosine kinases. Trends Biochem Sci. 1994;19:373–376. doi: 10.1016/0968-0004(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 45.Wang L L, Richard S, Shaw A S. P62 association with RNA is regulated by tyrosine phosphorylation. J Biol Chem. 1995;270:2010–2013. doi: 10.1074/jbc.270.5.2010. [DOI] [PubMed] [Google Scholar]
- 46.Weng Z, Thomas S M, Rickles R J, Taylor J A, Brauer A W, Seidel-Dugan C, Michael W M, Dreyfuss G, Brugge J S. Identification of Src, Fyn, and Lyn SH3-binding proteins: implications for a function of SH3 domains. Mol Cell Biol. 1994;14:4509–4521. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong G, Muller O, Clark R, Conroy L, Moran M F, Polakis P, McCormick F. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell. 1992;69:551–558. doi: 10.1016/0092-8674(92)90455-l. [DOI] [PubMed] [Google Scholar]