Abstract
Lineage tracing was originally developed by developmental biologists to identify all progeny of a single cell during morphogenesis. More recently this approach has been applied to other fields, including organ homeostasis and recovery from injury. Modern lineage tracing techniques typically rely on reporter gene expression induced by cell-specific DNA recombination. There have been important scientific advances in the last ten years that have impacted lineage tracing approaches, including intersectional genetics, optical clearing techniques and the use of sequencing-based genomic lineage tracing. The latter combines CRISPR-Cas9 based genetic scarring with single cell RNA-sequencing which in theory could allow comprehensive reconstruction of a lineage tree for an entire organism. In this review, we summarize recent advances in lineage tracing technologies and outline potential applications for kidney research.
Keywords: Cre Recombinase, lineage analysis, Crispr-Cas9
EDITOR’S NOTE
Lineage tracing has been a break-through technology, when it was first introduced to nephrology and has led to numerous fundamental insights into how kidneys develop in ontogenesis ranging to major new insights into how scars form, be it in glomeruli or the tubulointerstitium. In the present review Drs. Muto and Humphreys review the latest developments in this technology and how this will help further to understand kidney (patho)physiology.
Introduction
Lineage tracing is an experimental methodology which allows the identification of all progeny from a single cell or group of cells in a biological process. First practiced by developmental biologists in the 19th century, lineage tracing has evolved to be an essential experimental approach to dissect diverse biological processes such as tissue homeostasis, injury and repair1. A significant evolution in lineage tracing techniques occurred following the application of modern molecular biology and genetic engineering techniques around the end of the 20th century. This resulted in the establishment of genetic lineage tracing techniques which involves the heritable labeling of a cell in vivo by inducible DNA recombination. Enzymes called recombinases mediate the excision of a transcriptional stop sequence leading to heritable expression of a reporter gene (usually a fluorescent protein) from genomic DNA. Recombinase-based lineage tracing has led to much more refined understanding of stem cell biology and cell hierarchies during development, homeostasis and in pathologic states such as injury or cancer. Genetic lineage tracing has also led to many improvements in our understanding of kidney biology2
A second evolution in genetic lineage tracing technologies has occurred with the advent of CRISPR-Cas9 based gene editing3, high-throughput sequencing technology and single cell RNA-sequencing (scRNA-seq)4. Genomic lineage tracing is based on generating genomically encoded unique DNA barcodes, usually in the untranslated region of a gene, in an individual cell. These unique barcodes can then be read out by scRNA-seq. Here, we review established and emerging genetic lineage tracing techniques, first reviewing traditional recombinase-mediated approaches and next by reviewing genomic lineage tracing approaches. Finally, we discuss how these cutting-edge lineage tracing techniques might be applied to kidney research.
Established genetic lineage tracing tools
With classical genetic lineage tracing, all cells derived from a progenitor are identified and traced by expression of a reporter gene. Because this reporter gene is encoded in genomic DNA, its expression is passed on to all progeny cells and lineage relationships are recovered based on reporter gene expression. The Cre-loxP system is the most common recombinase system in genetic lineage tracing and it was identified in P1 phage5. The Cre recombinase recognize a specific DNA sequence called loxP sites. When these loxP sites are oriented the same way, Cre recombinase excises all of the intervening DNA. If the intervening DNA encodes a transcriptional STOP signal (for example, three polyadenylation signals) then any open reading frame downstream will not be transcribed. But if Cre is expressed in a cell-specific manner, the recombination of the loxP sites removes the transcriptional STOP, leading to heritable transcription of the downstream open reading frame (usually a fluorescent protein) in that cell type alone6. Temporal control of reporter gene activation can be obtained using Cre enzyme fused to a modified estrogen receptor (CreERT2) that requires tamoxifen for its activity7. There have been a number of applications of Cre-based genetic lineage tracing, including the development of the R26R-Confetti mouse by Snippert et al. for mosaic labeling to increase the resolution of lineage tracing8.
Despite the power of these genetic lineage tracing tools there are limitations as well. Often there is no appropriate Cre driver mouse line for a given cell of interest. Alternatively, Cre may be driven by a promoter that is expressed in multiple cell types, leading to challenges in accurately defining which parental cell gave rise to progeny. Another limitation of this approach is that only one reporter can be traced at a time. Finally, the dependence of lineage tracing on microscopic observations is relatively low throughput and loses three-dimensional information. More recent advances in lineage tracing technologies have addressed many of these limitations.
Intersectional lineage tracing
The use of orthogonal recombinases that recognize distinct DNA sequences can lead to reporter expression only in cells that have undergone recombination from both recombinases, marking cells that lie at the intersection of both recombinase expression domains. This technique is called intersectional lineage tracing and typically combines the Cre-loxP system with another recombinase such as flippase (Flp) or Dre recombinase9. Flp was identified in yeast where it recombines DNA sequences between short flippase recognition targets (FRT), akin to the Cre-loxP system10. Flp was initially considered less useful due to its low optimum temperature (30°C) and reduced efficiency compared to Cre recombinase. However, codon optimization generated Flpo, which shows similar efficiency to Cre11. Dre recombinase, discovered in phage D6, also catalyzes site-specific recombination with target sites called rox12. Importantly, each of these recombinases do not recognize target sites of the other recombinases. Intersectional lineage tracing is thus achieved by using two orthogonal recombinases in parallel driven by unique promoters for two different genes with distinct but partially overlapping expression domains. For example, Madisen et al. generated the Ai65 reporter allele (Rosa26-CAG-FRT-STOP-FRT-loxP-STOP-loxP-tdTomato) for the dual recombinase approach13 (Fig.1a). They successfully identified a Slc32a1+ Pvalb+ GABAergic neuron subset with Cre driven by the Slc32a1 promoter and Flpo driven by the Pvalb promoter. A reporter system for intersectional lineage tracing can include more than one reporter gene as well. For example, Jensen et al. generated a dual recombinase-responsive reporter allele named RC::Fela that enabled them to trace cells that expressed Flpo alone (green fluorescent protein expression) vs. both Flpo and Cre (nuclear LacZ expression) which they used to distinguish serotonergic neuron subsets14 (Fig.1b).
Figure 1. Intersectional recombinase approach with orthogonal recombinases.
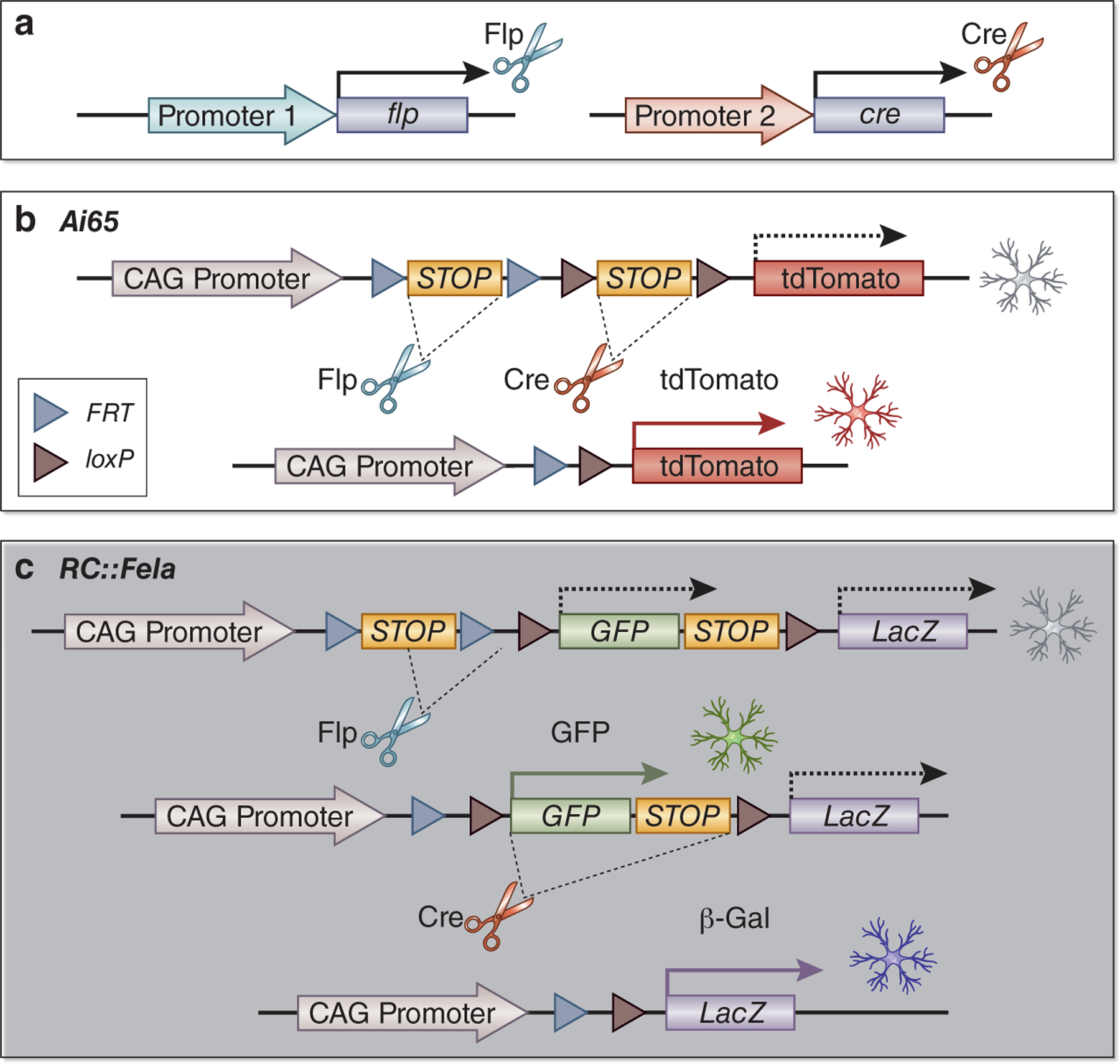
(a) Intersectional lineage tracing is achieved by using two orthogonal recombinases driven by promoters of two different genes in parallel. (b) The Ai65 reporter allele has two STOP cassettes flanked by two FRT or loxP sites. Simultaneous expressions of Flp and Cre remove two STOP cassettes to express tdTomato. (c) RC::Fela reporter allele is a dual-color reporter. Flp expression turns on only GFP expression, and concurrent expression of Flp and Cre turns on only β-galactosidase. Single Cre expression does not label a cell.
Another elegant approach to intersectional genetics is called split Cre. In this system, two separate inactive fragments of Cre enzyme are expressed under different promoters. When either fragment is expressed alone, it is catalytically inactive. However when both Cre fragments are simultaneously expressed in the same cell, the full, active-form Cre recombinase is reconstituted by either protein-protein interaction15 or protein splicing called split-interin16. Although split Cre has strong potential, it has not yet been utilized in a lineage tracing study. It could also be combined with other orthogonal recombination approaches.
A limitation of intersectional lineage tracing is the small number of orthogonal recombinase drivers available. Notably, Han et al recently developed over 70 orthogonal drivers with homologous recombination using CRISPR-Cas9 to trace diverse cell types17. Increasing efficiency of genetic engineering by CRISPR-Cas9 technology thus enhances the evolution of genetic lineage tracing strategies. These approaches also require considerable mouse breeding, since a minimum of three alleles is required (two cell-specific recombinase lines and one reporter line).
-. Optical clearing techniques for 3D lineage tracing
The application of optical clearing techniques to lineage tracing has allowed investigators to analyze cell hierarchies in three dimensions. Optical clearing is a methodology to improve the depth of optical microscopy to enable observation in the three-dimensional (3D) structures. Established genetic lineage tracing has been traditionally performed on tissue sections. These two-dimensional approaches did not accurately reflect 3D positional information like vasculature or kidney tubules. Optical clearing techniques have been shown to allow 3D exploration of cellular movement or cell lineages in a tissue or organ, or even a whole organism.
Optical clearing is achieved by removing pigment, lipids and calcium phosphate to match the refractive indices (RI) of all the structures in a sample. This results in transparency of the tissue or organ by minimizing scattered light. There are three major categories of optical clearing techniques: solvent-, aqueous- and hydrogel-based approaches. Solvent-based methods aim to dehydrate a sample and then match RI with solvents. The disadvantages of this approach are use of toxic or corrosive chemical and sample shrinkage18. Aqueous-based methods depend on hydrophilic reagents usually with high levels of biocompatibility, biosafety and preservation of fluorescent proteins. For example, Davis et al. and Rios et al. performed 3D lineage tracing of mammary stem cells in mice with aqueous-based optical clearing strategies19,20. The third approach, hydrogel-based techniques rely on cross-linking of the biomolecules like proteins and RNA to a hydrogel which efficiently remove lipids with minimal loss of structures and molecules. This approach including CLARITY is compatible with RNA detection21 and it may be applicable to sequencing-based genomic lineage tracing with 3D spatial information in the future.
Genomic lineage tracing
The development of high-throughput sequencing technologies has led to a new paradigm in lineage tracing: sequencing-based genomic lineage tracing.
-. Lineage tracing with cell barcoding and high-throughput sequencing
In contrast to traditional genetic lineage tracing where cells are tracked by expression of a reporter gene, in genomic lineage tracing cells are labeled with unique DNA barcodes that can be traced by high-throughput sequencing22. Furthermore, transcribable cell barcode systems have been devised. In such systems, rather than a fluorescent protein, cells express a unique RNA barcode encoded by their DNA. This mRNA barcode – located for example in the 3’ untranslated region of a gene – can be captured by scRNA-seq permitting simultaneous lineage assignment with transcriptomic information in the same cell. Biddy et al. reported an elegant approach named CellTagging, lineage tracing with combinatorial barcode labeling in vitro followed by single-cell transcriptomic analysis23. This involved generation of a lentiviral library driving expression of green fluorescent protein with unique barcodes in the 3’ untranslated region. They transduced fibroblasts with this library and could successfully reconstruct lineage trees during reprogramming of these fibroblasts into induced endoderm progenitors. A limitation of this approach is that it requires lentiviral transduction so has limited applicability in vivo. For in vivo genomic lineage tracing, Pei et al. developed a mouse called Polylox with an artificial DNA recombination locus consisting of multiple loxP sites in alternating orientations (Fig. 2). Tamoxifen induced Cre-mediated partial excision and inversion of these loxP sites could then be read out by single molecule real time sequencing of fluorescence-activated cell sorted populations of cells. The concept is that Cre exhibits incomplete and random enzymatic activity when confronted with multiple loxP sites, generating several hundred thousand unique cell barcodes in vivo. This study provided proof of principle for genomic lineage tracing in vivo.24 Furthermore, a newer version entitled PolyloxExpress has been developed which allows simultaneous lineage analysis with single cell transcriptomics.25 The joint profiling of fate and transcriptome in vivo is an exciting new development for the field.
Figure 2. Temporal and tissue-specific generation of barcodes in vivo with Polylox system.
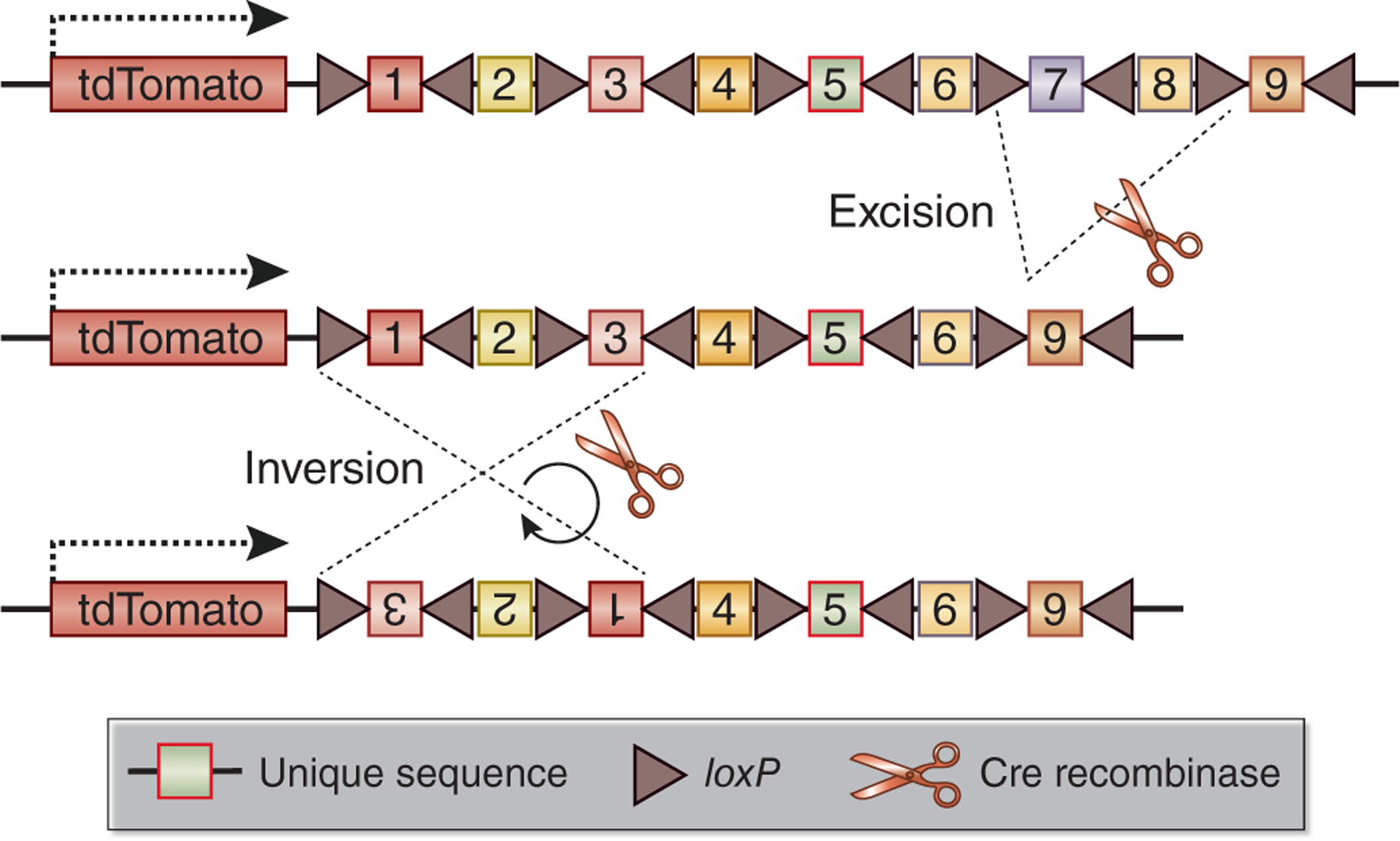
The Polylox system is characterized by nine tandem, unique DNA sequences flanked by loxP sites. In a PolyloxExpress allele, the barcode sequence is transcribed as the 3’ untranslated region of a tdTomato transcript. The Polylox cassettes are diversified by unique combinations of Cre-mediated excisions and inversions during Cre induction.
-. Retrospective lineage tree reconstitution with genetic scarring using CRISPR
CRISPR-Cas9 technology has been leveraged to achieve genomic lineage tracing in vivo. In this system, a cell barcode has several single guide RNA (sgRNA) target sites. Random edits on a barcode induced by Cas9 are accumulated in each cell during a biological process in which barcodes are passed on to the descendants. By sequencing these uniquely scarred barcodes, one can infer which cells are derived from a common ancestor based on a shared scar, enabling lineage tree reconstitution throughout an entire organism (Fig.3a).
Figure 3. Genomic lineage tracing with CRISPR-Cas9 based genetic scarring and the CARLIN system.
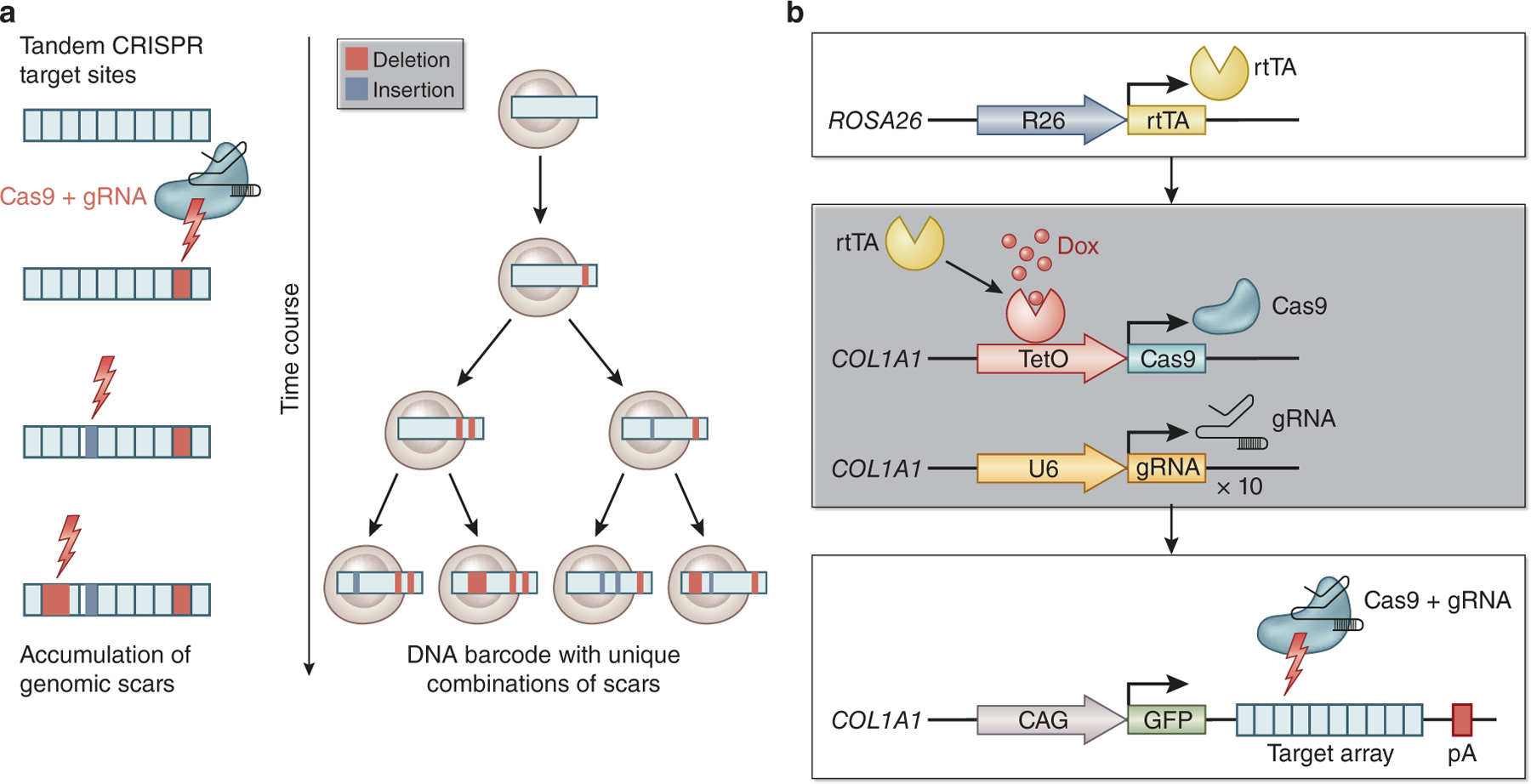
(a) Genomic lineage tracing with CRISPR-Cas9 based genetic scarring system. Intrinsic DNA barcodes are scarred by Cas9 with deletion or insertion to render them unique (left). As a result, lineage trees are recovered based on shared scars (right). (b) CRISPR array repair lineage tracing (CARLIN). CARLIN adopted “Tet-ON” system with the reverse tetracycline-controlled transactivator (rtTA) for induction of scars (top). Cas9 expression is induced by doxycycline (Dox) treatment. Ten gRNAs are also constitutively expressed (middle). Cas9 protein and gRNA generate genetic scars in each target sequence on target array (CARLIN array). CARLIN array is expressed as a 3’ UTR of a GFP transcript, enabling capture in scRNA-seq (bottom).
For example, McKenna et al. designed a genomic barcode with several Cas9 target sites. They infused Cas9 proteins and sgRNAs into the transgenic zebrafish fertilized eggs that harbor the genomic barcodes26, and following genome sequencing in adult fish identified thousands of scarred barcodes. This method was named GESTALT; “genome editing of synthetic target arrays for lineage tracing”. Their results suggested that most organs in adult fish are derived from progeny from a small number of progenitor cells. Raj et al. developed mutable barcodes that are transcribed as RNA barcodes27. With this lineage tracing system called scGESTALT, they identified over 100 cell types from ~60,000 single-cell transcriptomes with unique barcodes from zebrafish juvenile brains, successfully reconstructing a lineage tree with hundreds of branches. Similar genomic scarring approaches have been applied in mammals. For example, Bowling et al. generated a CRISPR array repair lineage tracing (CARLIN) mouse line that carries ten copies of guide RNAs (gRNAs), their respective target sequences (CARLIN array) and a tetracycline-inducible (Tet-ON) Cas9 gene (Fig. 3b). When combined with a reverse tetracycline-controlled transactivator (rtTA) expressing mouse line, tetracycline treatment induces Cas9 expression, scarring the CARLIN array in the 3’ untranslated region (3’ UTR) of GFP at any time point in the biological process. The CARLIN mouse system thus enables genomic lineage tracing with simultaneous transcriptomic analysis at a single-cell level in vivo28. They demonstrated heterogeneity in HSC expansion after myeloablation using this mouse model as a proof-of-principle.
As very new technologies, important limitations still need to be overcome for these cell barcoding approaches. Most notably, cell barcoding only provides hierarchical lineage trees, but not the topological and morphological information for the cell lineage. Other limitations include inefficiency and artifacts in barcode capture. For example, detection of transcribed cell barcodes using single-cell RNA sequencing (scRNA-seq) will generate errors such as barcode dropouts, artifacts induced in PCR amplification or sequencing29. Another concern is a bias of cell sampling due to inadequate tissue dissociation. This problem is relevant to solid organs including the kidney. Advance in computational data correction and the application of spatial genomics technology may circumvent these limitations in genomic lineage tracing30,31.
-. Lineage tree reconstruction with inherent genomic scarring
These emerging genetic lineage tracing tools depend on genetic engineering, which cannot be applied to human tissue. However spontaneous somatic mutations accumulate during aging, and can be used as inherent genomic barcodes to reconstruct lineage relationships and this has been done the human brain32 and in colon cancer33. Since somatic mutagenesis is infrequent, reconstruction of high-resolution lineage trees is not compatible with this approach. Mitochondrial DNA has a higher somatic mutation rate compared with nuclear DNA and also has the advantage of a large number of copies per cell. Ludwig et al. have shown that single-cell data can be used for retrospective lineage tracing in human tissue by using somatic mutations on mitochondrial DNA as natural cell barcodes34.
Application of emerging lineage tracing tools to kidney
A number of questions in kidney research may be solved using these emerging genetic and genomic lineage tracing approaches. For example, conflicting data and opinions about the cell types responsible for renal fibrosis exist35. Intersectional lineage tracing approaches may be well suited to dissect mesenchymal cell heterogeneity, which is an emerging area in fibrosis research. Kaverina et al. adopted a dual lineage tracing strategy to show transdifferentiation of glomerular parietal epithelial cells (eGFP+ cells) to the podocytes (tdTomato+ eGFP+ cells) after podocyte depletion36. Also, optical clearing followed by 3D imaging will enhance the accuracy in lineage tracing, as the kidney is composed of complicated three-dimensional structures. Puelles et al. applied tissue clearing technology to a mouse model of rapidly progressive glomerulonephritis, and they succeeded in capturing crescent formation by parietal epithelial cells37. New orthogonal driver lines for kidney research are required for intersectional recombinase strategies it must be noted.
Although genomic lineage tracing has not been demonstrated in kidney thus far, pseudo-temporal trajectories generated from scRNA-seq data have suggested lineage relationships in human kidney organoids38. There is an immediate need to apply genomic lineage tracing strategies to deconvolve lineage hierarchies in kidney organoid differentiation. Improved methods for gene transfer and genetic engineering are needed as well. For example, we have developed a method by which synthetic adeno-associated viral vectors can be used to efficiently transduce genes to renal cell types such as mouse pericytes, fibroblasts, and mesangial cells39. This technique has a potential to be applied to introduction of cell barcodes into the renal mesenchymal cell types in vivo.
Conclusion
Exciting advances in gene editing, mouse genetics, next generation sequencing and single cell biology are now being applied to lineage tracing approaches (Table 1). Although these strategies have not yet been applied to kidney, we suggest that these new approaches have a great potential to answer many critical questions in this field.
Table 1 |.
Genetic and genomic lineage tracing tools
| Class | Method | Model | Labelling | Analysis | Reference |
|---|---|---|---|---|---|
| Genetic | Single-color | One cell-specific recombinase line and one reporter line (mouse) | Constitutive or tamoxifen induction | Microscope | 1,2 |
| Intersectional | Two cell-specific recombinase lines and one reporter line (mouse) | Constitutive or tamoxifen induction | Microscope | 9,13,14,17 | |
| 3D lineage tracing | Single-color or intersectional lineage tracing mouse model | Constitutive or tamoxifen induction | Optic clearing and Microscope | 19,20,21 | |
| Genomic | CellTagging | Culture cells | Lentiviral tag transduction | scRNA-seq | 23 |
| Polylox | Cell-specific, inducible Cre line and Rosa26-Polylox knock-in mouse line | Tamoxifen induction | DNA-seq | 24 | |
| PolyloxExpress | Cell-specific, inducible Cre line and Rosa26-PolyloxExpress knock-in mouse line | Tamoxifen induction | scRNA-seq | 25 | |
| GESTALT | GESTALT barcode transgenic zebrafish | Cas9 protein and sgRNAs injection | DNA-seq | 26 | |
| scGESTALT | scGESTALT barcode transgenic zebrafish line and inducible Cas9 zebrafish line | Cas9 protein and sgRNAs injection /Heat shock | scRNA-seq | 27 | |
| CARLIN | CARLIN mouse line and rtTA transgenic mouse line (Tet-ON) | Doxycycline | scRNA-seq | 28 |
Acknowledgements
We apologize to those authors whose works we could not include due to space constraints. BDH is supported by grants DK103740 and UC2DK126024 from NIDDK and from grants from the Chan Zuckerberg Initiative and the Alport Syndrome Foundation. YM is supported by a research fellowship from the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowships for Research Abroad. The figures were generated with BioRender.com.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
The authors declare no competing interests.
Reference
- 1.Kretzschmar K & Watt FM Lineage Tracing. Cell 148, 33–45 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Humphreys BD & DiRocco DP Lineage-tracing methods and the kidney. Kidney International 86, 481–488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adli M The CRISPR tool kit for genome editing and beyond. Nat Commun 9, 1911 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H & Humphreys BD The promise of single-cell RNA sequencing for kidney disease investigation. Kidney International 92, 1334–1342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauer B Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol 7, 2087–2096 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewandoski M Conditional control of gene expression in the mouse. Nat Rev Genet 2, 743–755 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Seibler J et al. Rapid generation of inducible mouse mutants. Nucleic Acids Res 31, e12 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snippert HJ et al. Intestinal Crypt Homeostasis Results from Neutral Competition between Symmetrically Dividing Lgr5 Stem Cells. Cell 143, 134–144 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Liu K, Jin H & Zhou B Genetic lineage tracing with multiple DNA recombinases: A user’s guide for conducting more precise cell fate mapping studies. Journal of Biological Chemistry 295, 6413–6424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senecoff JF, Rossmeissl PJ & Cox MM DNA recognition by the FLP recombinase of the yeast 2 μ plasmid. Journal of Molecular Biology 201, 405–421 (1988). [DOI] [PubMed] [Google Scholar]
- 11.Raymond CS & Soriano P High-Efficiency FLP and ΦC31 Site-Specific Recombination in Mammalian Cells. PLoS ONE 2, e162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauer B DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Research 32, 6086–6095 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madisen L et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen P et al. Redefining the serotonergic system by genetic lineage. Nat Neurosci 11, 417–419 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirrlinger J et al. Split-Cre Complementation Indicates Coincident Activity of Different Genes In Vivo. PLoS ONE 4, e4286 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P et al. Intersectional Cre Driver Lines Generated Using Split-Intein Mediated Split-Cre Reconstitution. Sci Rep 2, 497 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X et al. A suite of new Dre recombinase drivers markedly expands the ability to perform intersectional genetic targeting. Cell Stem Cell S1934590921000072 (2021) doi: 10.1016/j.stem.2021.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Ariel P A beginner’s guide to tissue clearing. Int J Biochem Cell Biol 84, 35–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis FM et al. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat Commun 7, 13053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rios AC et al. Intraclonal Plasticity in Mammary Tumors Revealed through Large-Scale Single-Cell Resolution 3D Imaging. Cancer Cell 35, 618–632.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Ueda HR et al. Tissue clearing and its applications in neuroscience. Nat Rev Neurosci 21, 61–79 (2020). [DOI] [PubMed] [Google Scholar]
- 22.VanHorn S & Morris SA Next-Generation Lineage Tracing and Fate Mapping to Interrogate Development. Dev Cell 56, 7–21 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Biddy BA et al. Single-cell mapping of lineage and identity in direct reprogramming. Nature 564, 219–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei W et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pei W et al. Resolving Fates and Single-Cell Transcriptomes of Hematopoietic Stem Cell Clones by PolyloxExpress Barcoding. Cell Stem Cell 27, 383–395.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 26.McKenna A et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raj B et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol 36, 442–450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowling S et al. An Engineered CRISPR-Cas9 Mouse Line for Simultaneous Readout of Lineage Histories and Gene Expression Profiles in Single Cells. Cell 181, 1410–1422.e27 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner DE & Klein AM Lineage tracing meets single-cell omics: opportunities and challenges. Nat Rev Genet 21, 410–427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frieda KL et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Askary A et al. In situ readout of DNA barcodes and single base edits facilitated by in vitro transcription. Nat Biotechnol 38, 66–75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodato MA et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung ML et al. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res 27, 1287–1299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig LS et al. Lineage Tracing in Humans Enabled by Mitochondrial Mutations and Single-Cell Genomics. Cell 176, 1325–1339.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphreys BD Mechanisms of Renal Fibrosis. Annu Rev Physiol 80, 309–326 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Eng DG et al. Detection of renin lineage cell transdifferentiation to podocytes in the kidney glomerulus with dual lineage tracing. Kidney Int 93, 1240–1246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puelles VG et al. Novel 3D analysis using optical tissue clearing documents the evolution of murine rapidly progressive glomerulonephritis. Kidney Int 96, 505–516 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Wu H et al. Comparative Analysis and Refinement of Human PSC-Derived Kidney Organoid Differentiation with Single-Cell Transcriptomics. Cell Stem Cell 23, 869–881.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikeda Y, Sun Z, Ru X, Vandenberghe LH & Humphreys BD Efficient Gene Transfer to Kidney Mesenchymal Cells Using a Synthetic Adeno-Associated Viral Vector. J Am Soc Nephrol 29, 2287–2297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


