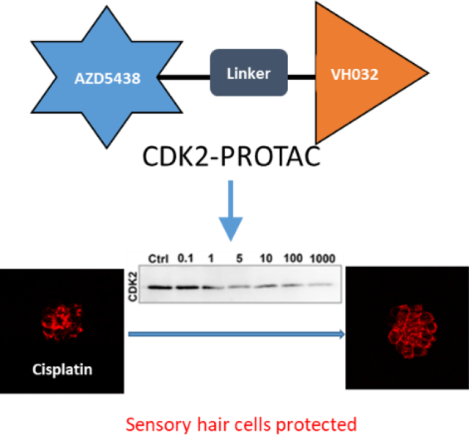
Abstract
Cyclin-dependent kinase 2 (CDK2) is a potential therapeutic target for the treatment of hearing loss and cancer. Previously, we identified AZD5438 and AT7519-7 as potent inhibitors of CDK2, however, they also targeted additional kinases, leading to unwanted toxicities. Proteolysis Targeting Chimeras (PROTACs) are a new promising class of small molecules that can effectively direct specific proteins to proteasomal degradation. Herein we report the design, synthesis, and characterization of PROTACs of AT7519-7 and AZD5438 and the identification of PROTAC-8, an AZD5438-PROTAC, that exhibits selective, partial CDK2 degradation. Furthermore, PROTAC-8 protects against cisplatin ototoxicity and kainic acid excitotoxicity in zebrafish. Molecular dynamics simulations reveal the structural requirements for CDK2 degradation. Together, PROTAC-8 is among the first-in-class PROTACs with in vivo therapeutic activities and represents a new lead compound that can be further developed for better efficacy and selectivity for CDK2 degradation against hearing loss and cancer.
Keywords: CDK2-proteolysis, PROTAC, cisplatin-induced hearing loss, noise-induced hearing loss
Graphical Abstract
Introduction:
Hearing loss is a major health problem, affecting 1.57 billion people worldwide and the number is estimated to be 2.45 billion by 2050 [1]. Hearing loss can be caused by different insults such as the use of chemotherapeutic agents, antibiotics as well as noise, and aging. The use of the chemotherapeutic agent cisplatin causes permanent high-frequency hearing loss in 40–80% of treated cancer patients [2]. Despite extensive research on the mechanisms of hearing loss, currently, there are no FDA-approved drugs available to prevent acquired or age-related hearing loss. Most of the ongoing pre-clinical or clinical trials are only focused on the use of antioxidants, vitamins, and glutathione antioxidants as putative therapeutic compounds to prevent hearing loss [3]. More recently, a phase 3 clinical trial on the usage of sodium thiosulfate (STS) to prevent cisplatin-induced hearing loss in pediatric patients with localized hepatocarcinoma has been completed but it has not yet received FDA approval [4–7]. Thus, identifying novel therapeutic interventions for acquired hearing loss is an immediate unmet need.
Towards our continuous efforts to find the plausible mechanism of cisplatin-induced ototoxicity, we have identified cyclin-dependent kinase 2 (CDK2) as a potential therapeutic target. Our previous work revealed that the CDK2 inhibitors, AZD5438, AT7519, and AT7519-7, can prevent cisplatin-induced ototoxicity in vivo, with low nanomolar CDK2 inhibitory IC50 values [8]. Furthermore, by employing a CDK2 knockout mouse model, we demonstrated that the absence of CDK2 confers resistance to cisplatin- and noise-induced hair cell loss [9]. Moreover, the inhibition or degradation of CDK2 has been the focus of many studies seeking therapies for the treatment of various types of cancers [10–15]. Studies employing AZD5438-based therapies demonstrated that hair cell loss was reduced in vivo in zebrafish lateral line neuromasts, and when delivered orally to adult FVB mice [16]. However, although AZD5438 treatment is protective, it has also been shown that systemic delivery can induce potential toxicity due to off-target effects, limiting its safety margin in humans [17–19].
Targeted protein degradation is an emerging strategy to use small molecules to knock down a specific protein by hijacking the ubiquitin-proteasome degradation system. Proteolysis targeting chimera (PROTAC) is a bifunctional molecule comprised of a ligand for the target protein and a ligand for E3 ligase recruitment, connected by a linker, representing a novel drug discovery approach [20]. After the formation of a ternary complex (targeted protein-ligand for the target protein-ligand for E3 ligase), the target protein is ubiquitinated and degraded by the proteasome [21]. An attractive feature of PROTACs is their catalytic mode of action, as one molecule can perform multiple rounds of target ubiquitination and degradation [22]. Due to this feature of PROTACs, they can function at substoichiometric receptor occupancies. Moreover, PROTACs can add an extra layer of target selectivity, thus providing highly selective degraders with reduced off-target effect [23–24]. The mounting interest in PROTAC drug discovery is also motivated by the potential to target proteins considered “undruggable” via conventional medicinal chemistry approaches [25–37]. It has been also observed that pan-CDK inhibitors have better efficacy when developed as PROTACs to degrade a specific CDK protein. Recently, the pan-CDK inhibitors SNS-032 and palbociclib were developed into highly selective CDK9 and CDK6 degraders respectively [38–39]. AT7519- and TMX-2172-PROTACs [40] have been recently identified as CDK2 degraders, but only TMX-2172 PROTAC was tested against CDK1 as well [41]. This is important since CDK1 is a key component of cell cycle regulation and proliferation, and its inhibition can lead to clinical toxicity [42].Recently Wang et al. reported the discovery of CDK2 selective PROTACs which can degrade CDK2 in acute myeloid leukemia (AML) cells in vitro but no in vivo effects were reported [43].
In this study we report the evaluation of PROTAC-8, an AZD5438-based PROTAC, that shows selective proteasomal degradation of CDK2 over CDK1, CDK5, CDK7, and CDK9, four closely related CDK proteins, in HEI-OC1 cells derived from neonatal mouse cochleae. Molecular dynamics simulations provide the structural basis for specific interactions of PROTAC-8 with CDK2 and E3 ligase. Furthermore, we show that PROTAC-8 degrades CDK2 and protects hair cells in vivo in zebrafish models for cisplatin- and excitotoxin-induced hair cell loss.
Results
From our previous study [8], we selected the two best CDK2 inhibitors, AZD5438 and AT7519-7 as our CDK2-targeting ligands for PROTAC design since they exhibit the best protection against cisplatin-induced ototoxicity in mouse cochlear explants and in vivo mouse models (Fig. 1). In the design of PROTAC, the choice of E3 ligase and the selection of target ligands and their conjugation are critical optimization variables. The most common ligases successfully used in the design of PROTAC molecules are the von Hippel–Lindau (VHL) protein complex CRL2VHL and the Cereblon (CRBN) complex CRL4CRBN [44–45]. Studies have shown that PROTACs made of the same target ligand but either VHL or CRBN ligands can exhibit different degradation selectivity and efficacy [26, 46–47]. In some systems, CRBN-based degraders showed higher degradation efficiency or selectivity than VHL-based analogs. These observations influenced us to develop VHL- and CRBN-based degraders in parallel. Accordingly, a focused library of four PROTAC degraders was synthesized by combining the CDK2 ligands with variable linkers (See Schemes 1 and 2 in supplemental material). The use of two proven E3 ligase recognition ligands, pomalidomide and VH-032, for CRBN and VHL, respectively, also ensures non-toxicity and high effectiveness of our strategy (Fig. 1) [48]. Nuclear Magnetic Resonance (NMR) spectra of 1H and HPLC analyses to corroborate identity structure and purity are presented in Supplemental Figures S1A–E.
Figure 1.
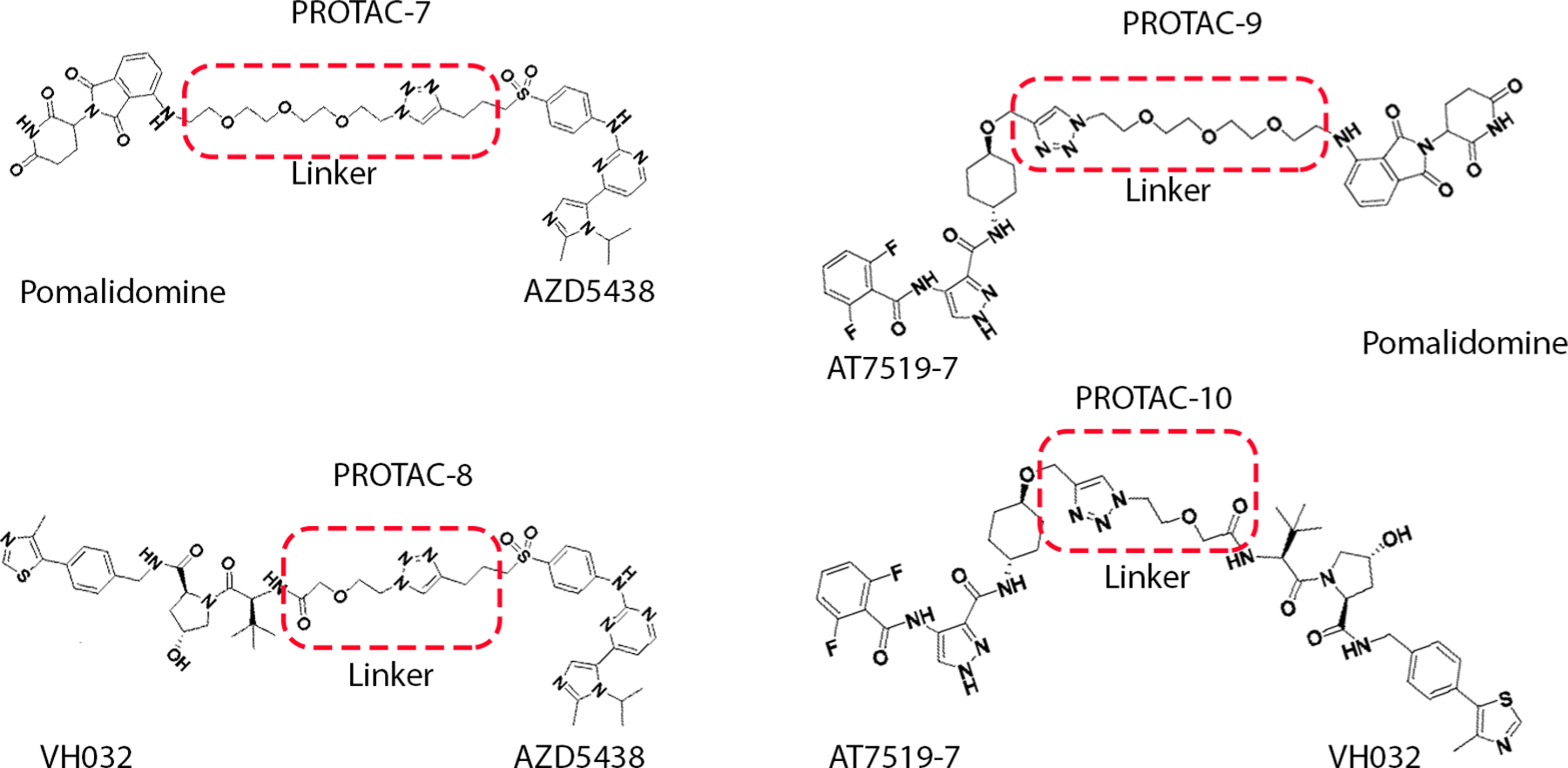
PROTACs with CDK2 inhibitors (AZD5438 and AT7519-7), linkers and E3 ligase ligands (Pomalidomide and VH-032) used in this study. Pomalidomide and VH-032, have been well studied and by themselves did not show much toxic effects while CRBN based degraders can recruit neo-substrate degradation (e.g. SALL4 for limb deformation) that can cause toxicity [60–67, 74].
We screened the synthesized PROTACs for CDK2 degradation in HEI-OC1 cells incubated for 24 hours with a wide range of concentrations, from 0.1 nM to 1 μM (Figure 2A). We also included the two CDK2 ligands used in the PROTAC design, AZD5438 and AT7519-7. From the initial screening, AZD5438-linked PROTAC with VH-032 as E3 ligase ligand, (PROTAC-8,) induced around 50% CDK2 degradation at 100 nM concentration (Figure 2C) while AZD5438 alone had no significant effect on the degradation. Treatment of HEI-OC1 cells with AT7519-7-linked PROTACs, i.e., PROTAC-9 and PROTAC-10, or AZD5438-PROTAC with the CRBN ligand, PROTAC-7, did not show any CDK2 degradation (Figure 2A), nor changes in the abundance of any of the closely related CDKs (SI Figure 2). We then tested whether PROTAC-8 degrades CDK1, CDK5, CDK7 or CDK9, and observed that PROTAC-8 was selective for CDK2 degradation (Figure 2B–C), with a half maximal degradation concentration (DC50) of ~100nM. PROTAC-8’s degradation effect was through the proteasomal pathway, since co-incubation of the HEI-OC1 cells with 100 nM of PROTAC-8 and 300 nM of the proteasome inhibitor, MG-132, abolished CDK2 degradation (Figure 2D).
Figure 2: Evaluation of PROTAC-7-10 treatment results in CDK2 degradation.
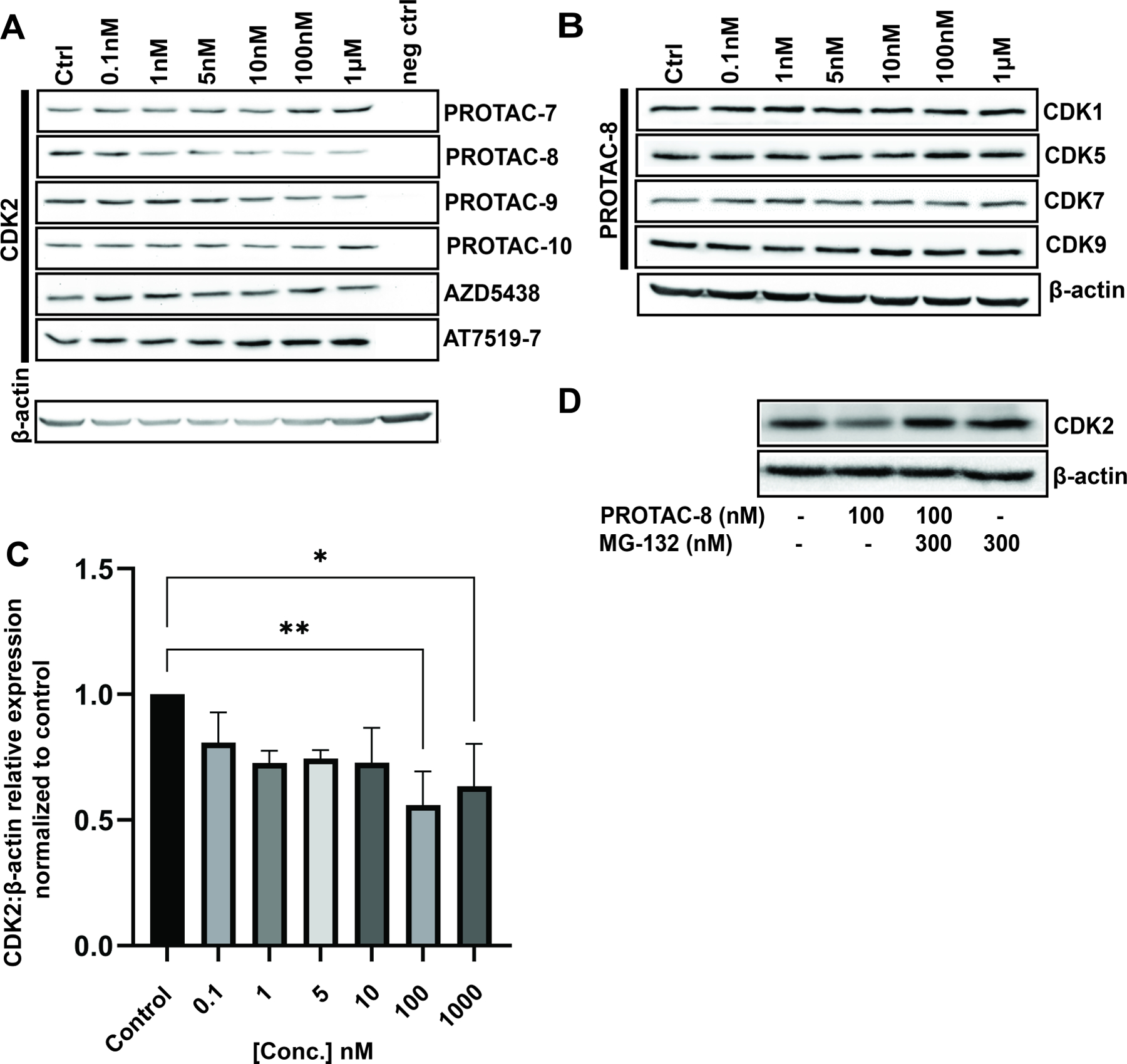
A) Representative immunoblots from HEI-OC1 cells treated with 0.1nM to 1μM of PROTACS 7–10, AZD5438, or AT7519-7 for 24 hours. neg ctrl = Kidney lysate of CDK2 knock-out mice used as a negative control for CDK2 immunoblots only; B) HEI-OC1 cell lysates were immunoassayed for CDK1, CDK5, CDK7, and CDK9. β-actin was used as loading control (similar incubation conditions were used as in 1A); C) Quantification of the corresponding CDK2 bands from three independent experiments. Kruskal-Wallis followed by Dunn’s post hoc test was used for comparison between the groups; *P < 0.05; **P < 0.01; D) Representative western blot of HEI-OC1 cell lysate after 24 hours of treatment with combinations of MG-132 and PROTAC-8. Doses higher than 1μM were not tested due to the known hook effect of PROTACs at high concentrations [20].
Next, the structure and dynamics of the ternary complex formed by the association of the E3 ligase, PROTAC-8, and CDK2 (pVHL-PROTAC-8-CDK2) were studied by five 100 ns independent molecular dynamics (MD) simulations using the AMBERff14SB force field [49] as implemented in the YASARA package (Figure 3A) [50]. During simulations, both CDK2 and pVHL retained their X-ray structure and the complexes did not dissociate in either trajectory. The relative movement of the two proteins was analyzed by calculating the bond rotation around α1, α2, α3, and α4 dihedral angles using the GROMACS analysis suite (Figure 3B) [51]. In all five MD simulations, α1, α2, and α3 showed high flexibility by interchanging between trans, gauche(+) and gauche(−) conformations and they had similar distribution (SI Figure 3A). The flexibility of the linker present in the PROTAC-8 molecule provided an independent movement of pVHL about CDK2. In contrast, in the case of the α4 dihedral angle, the rotation about the bond that connects the linker to VH-032, was rigid and stayed in gauche(−) conformation in four simulations (SI Figure 3B). This rigidity most likely contributes to the stable binding between the E3 ligase pVHL and its ligand, VH-032, present in PROTAC-8. The independent movement of the two proteins docked to PROTAC-8 is further supported by the dynamic cross-correlation matrix (DCCM) analysis [50, 52] of the MD simulations (SI Figure 4). In MD simulations 1–4, no correlated movement was observed between pVHL and CDK2 (SI Figure 4A), only intradomain correlated movements were observed as indicated by the red lines between Cα atom pairs within each protein. On the contrary, in MD simulation 5 beyond the intradomain correlated movements, the two proteins moved in an anti-correlated manner as indicated by the blue lines between Cα atom pairs between the two proteins (SI Figure 4B). This anti-correlated movement coincided with gauche(+) conformation of α4 angle and indicates that such conformation is most likely not advantageous for CDK2 ubiquitination. The independent, noncorrelated movement of the protein provides the opportunity for pVHL to reach various ubiquitination sites on CDK2. Such MD simulation analysis also provides a framework for future design of optimized PROTACs specific for CDK2.
Figure 3: Structure of pVHL - PROTAC-8 - CDK2 ternary complex.
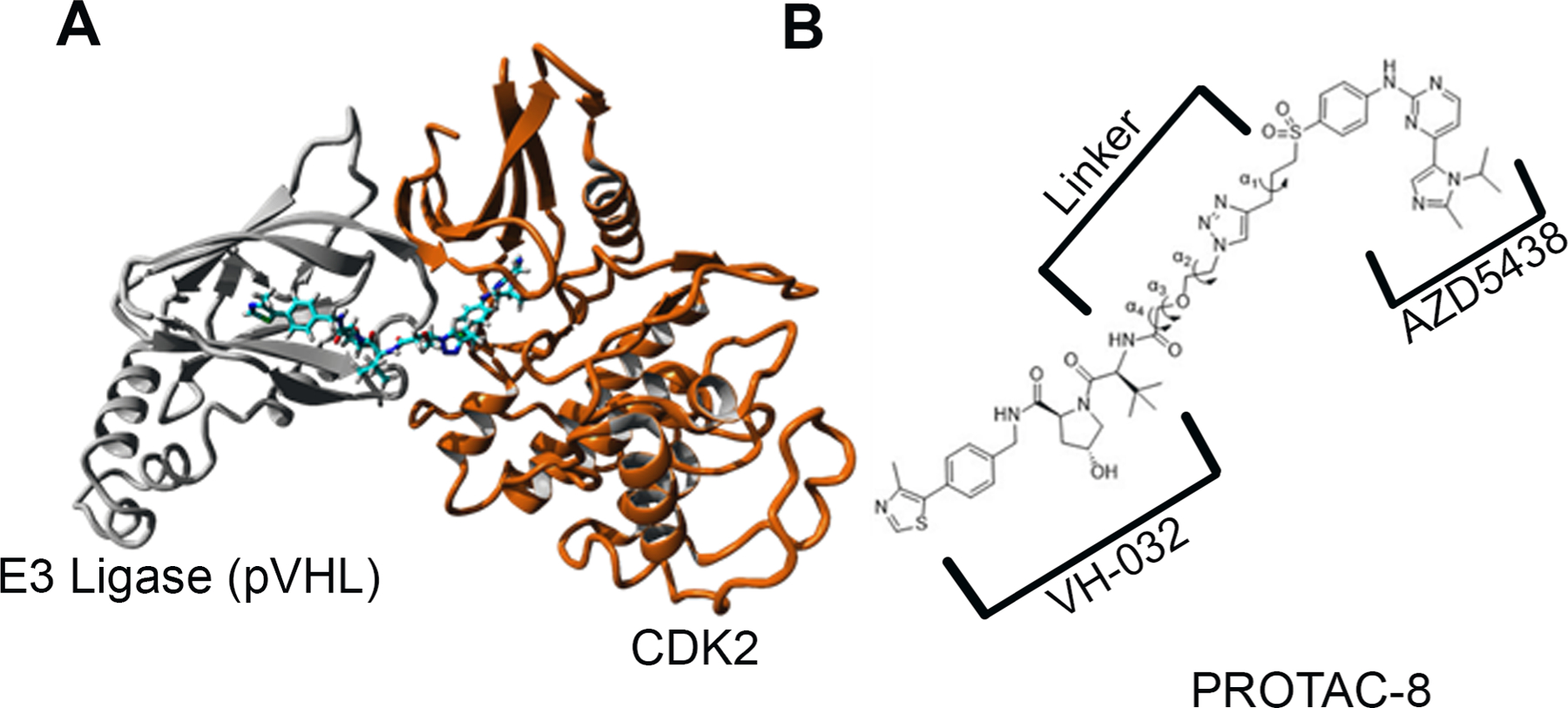
A) Ribbon representation of the energy-minimized structure of the pVHL - PROTAC-8 – CDK2 ternary complex. pVHL (grey) connected through PROTAC-8 to CDK2 (orange); B) Definition of α1, α2, α3, and α4 dihedral angles used for determination of the relative movement of the two proteins in the ternary complex
Finally, we decided to test whether PROTAC-8-mediated CDK2 degradation occurs in vivo and whether it can prevent hair cell loss. For this purpose, 5 days post-fertilization (dpf) zebrafish were treated with PROTAC-8 at different concentrations for 24 hours and then, processed for degradation or protection studies (Figure 4). Experiments with AZD5438 were run in parallel and used as controls for hair cell protection without CDK2 degradation. Results from these studies showed that fish exposed to 1μM of PROTAC-8 showed a significant decrease in CDK2 abundance (Figure 4A–B), with a DC50 of ~100nM. Moreover, PROTAC-8 treatment protected neuromast hair cells from cisplatin- and kainic acid-induced ototoxicity (Figure 4C–E). For cisplatin, PROTAC-8 showed significant protection at doses doses ranging from 1nM to 1μM (Figure 4C, 4E) while for kainic acid the protection was significant at doses ranging from 5nM to 100nM (Figure 4D–E). Overall, both AZD5438 and PROTAC-8 were able to protect the zebrafish lateral line neuromasts from cisplatin and kainic acid damage. While inhibition of CDK2 by AZD5438 might contribute to the protective effect observed with PROTAC-8, the specific degradation of CDK2 by PROTAC-8 provides a significant advantage over AZD5438 and promises a better future for CDK2-PROTAC optimization against cisplatin- and noise-induced hearing loss.
Figure 4. In vivo evaluation of PROTAC-8.
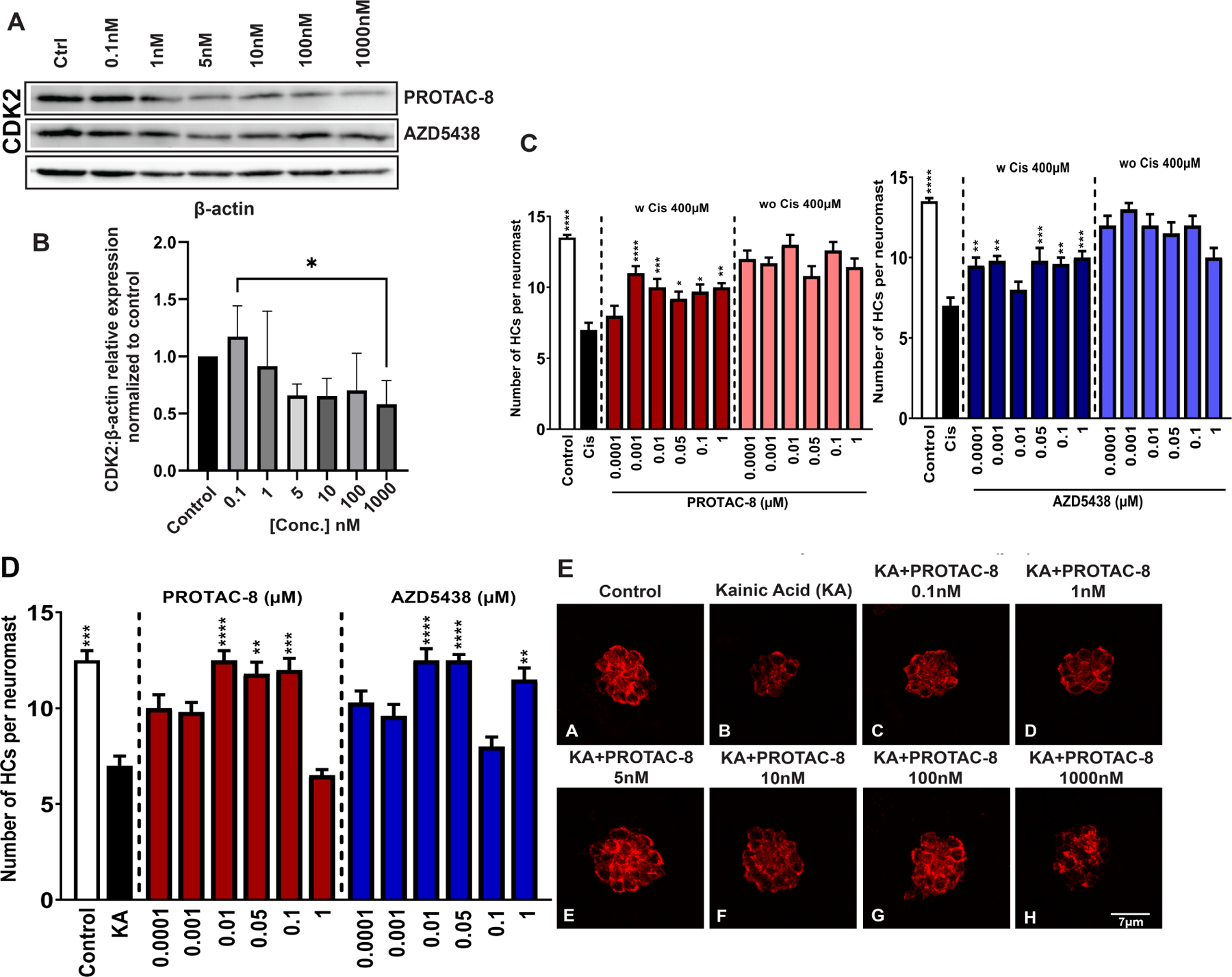
A) Representative immunoblots from the treatment of zebrafish with PROTAC-8 from 0.1nM to 1μM for 24 hours. Although CDK2 abundance seems to be reduced for some of AZD5438 concentrations (i.e., 5 nM and 10 nM), the normalization and quantification data did not show any significant differences compared to control samples.; B) Quantification of the corresponding CDK2 bands from three independent experiments. Kruskal-Wallis followed by Dunn’s post hoc test was used for comparison between the groups where *P < 0.05; **P < 0.01; C) The number of hair cells per neuromast was quantified after treatment with PROTAC-8 (left) or AZD5438 (right) in the presence or absence of 400μM of Cisplatin. Results are expressed as mean ± SEM. Statistical analysis: 1-way ANOVA with correction for Dunnett’s multiple comparisons test. **P < 0.01; ***P < 0.001; ****P < 0.0001 versus cisplatin treatment; Black bar: media-treated control; white bar: cisplatin-treated control; D) The number of hair cells per neuromast was quantified after pre-treatment with PROTAC-8 or AZD5438 followed by kainic acid (KA) 300μM for 50min. Results are expressed as mean ± SEM. Statistical analysis: 1-way ANOVA with correction for Dunnett’s multiple comparisons test. **P < 0.01; ***P < 0.001; ****P < 0.0001 versus KA alone; E) Representative images of 5 dpf zebrafish pretreated with PROTAC-8 (0.1nM-1μM) followed by incubation with KA.
Discussion
Our previous studies [8–9] implicated CDK2 as a target for the prevention of acquired hearing loss. However, CDK2 inhibitors can have off-target effects due to high similarities among the different CDKs’ active sites [41]. Furthermore, CDK2 may have kinase independent, noncanonical activities underlying hearing loss and neutrophil migration that cannot be targeted by the use of small molecule kinase inhibitors [9, 53]. We thus decided to employ a targeted protein degradation strategy to identify compounds that will specifically mark CDK2 for proteasomal degradation. For this purpose, we generated a focused library of PROTACs. PROTACs are bifunctional molecules containing a ligand domain for the protein of interest linked to a domain that corresponds to a ligand for E3 ligase [20]. In the design of PROTACs, properties of the linker, such as length, composition, and site of attachment, are known to be important but often their impact on activity varies in a target- and context-dependent fashion [46, 54–56]. Moreover, small-molecule binders for both the protein of interest and the E3 ligase are required. There are about six hundred different E3 ubiquitin ligases in the human genome, but only several were successfully used in the design of PROTACs [44–45]. The most common ligases successfully used in the design of PROTAC are VHL and CRBN [44–45]. Given this information, we initially synthesized four PROTAC molecules, two containing the VHL’s recognition ligand VH-032, and two containing the CRBN’s recognition ligand pomalidomide [48] linked to AZD5438 or AT7519-7. Of these four PROTACs, only PROTAC-8 (AZD5438-VHL), significantly decreased CDK2 abundance without any effect on the related CDKs, CDK1, CDK5, CDK7, and CDK9. Surprisingly, PROTAC-7, a AZD5438-PROTAC containing the CRBN ligand, did not show any effect on CDK2 abundance, suggesting that while AZD5438 contributes to CDK2 specificity, it is the E3 ligand, VH-032, the one that properly positions CDK2 for E3 ligase-mediated ubiquitination and degradation. This was confirmed by the requirement of MG132 for PROTAC-8’s proteasomal degradation of CDK2. MD simulations and computational analysis of the structure and dynamics of the ternary complex formed by the CDK2, PROTAC-8, and E3 ligase confirmed that, while the linker present in PROTAC-8 provides the necessary rigidity for the E3 ligase to bind to its ligand, it also allows the independent movement of the E3 ligase and CDK2. This independent movement provides the opportunity for the E3 ligase to ubiquitinate CDK2, marking it for proteasomal degradation. Based on this MD simulation analysis, we will be able to optimize the design and synthesis of more efficacious PROTAC-8 derivatives for CDK2 degradation.
The CDK2 specificity of AZD5438 is most likely the contributing factor for the specificity of PROTAC-8 to CDK2 [42]. Although the degradation in HEI-OC1 cells was only around 50%, the selectivity of PROTAC-8 encouraged us to go further in its characterization. Thus, we tested PROTAC-8 in vivo in zebrafish models for ototoxicity. We found that similar to AZD5438, PROTAC-8 was able to protect neuromast hair cells from cisplatin- and kainic acid-induced ototoxicity [57–58]. Moreover, while AZD5438 did not show any reduction in CDK2 abundance at the different doses tested in our experiments, treatment with 1μM of PROTAC-8 resulted in the significant degradation of CDK2. Although we cannot completely rule out the possibility that PROTAC-8 is protecting the hair cells through CDK2 inhibition via AZD5438, the fact that we also observed degradation of CDK2, strongly points to a dual function for PROTAC-8 that includes both inhibition and degradation of CDK2.
In summary, to our knowledge, this is the first time a PROTAC strategy has been used as a treatment to prevent acquired hearing loss. Although PROTAC has been the focus of many studies associated with cancer therapies [25–37], nothing has been done in the hearing field. Moreover, very few PROTACS have demonstrated in vivo physiological activities. In the present work, we showed that when CDK2 is specifically degraded, hair cells became resistant to ototoxic insults both in vitro and in vivo. Future studies employing MD and zebrafish testing will help in the development of better PROTAC-8 derivatives with higher CDK2 degradation properties at lower concentrations.
Conclusions
PROTACs are a new promising class of small molecules that can effectively degrade the specific protein targets; however, few have been demonstrated to have in vivo physiological effects. Here, we designed and synthesized a library of PROTACs based on known CDK2 inhibitors. Of this library, PROTAC-8 acts as a specific CDK2 degrader with no effect on other closely related CDKs. In vivo zebrafish models for cisplatin- and kainic acid-induced ototoxicity, demonstrated PROTAC-8 protects hair cells from their deleterious effects, underscoring PROTAC-8’s therapeutic potential. MD simulations reveal structural basis for PROTAC-8 as a CDK2-specific degrader and provide a framework for the development of novel and more potent PROTAC-8 derivatives that aim to prevent and treat hearing loss and various cancers.
Experimental Section
Overall strategy
We selected the well-known PEG and alkyl type linkers because of their versatility and flexibility, and we incorporated a triazole into the linker to take advantage of a streamlined synthetic approach utilizing click chemistry in order to access many linker lengths along with various combinations of CDK2 ligand and E3 ligand [59]. We used crystal structures of the known CDK2 ligand (AT7519 (Selleckchem Cat: S1524), pdb code: 2VU3 and AZD5438 (Selleckchem, Cat: S2621), pdb code: 6GUH) bound in the CDK2 active site in order to determine where the linker should be attached, and what parts of the bound molecule were solvent exposed and could potentially serve as an anchor point for the linker. In PROTAC-7 and PROTAC-8, we did not directly attach the triazole to the methyl sulfone of AZD5438 because of the potential for a steric clash between the triazole and the protein surface. Instead, we extended the alkyl chain by two additional carbons to allow for some breathing room away from the CDK2 protein surface.. This short alkyl chain would also allow for greater flexibility for the linker after clearing the protein surface. Similar principles were applied to PROTAC-9 and PROTAC-10, although synthetic feasibility and commercial material availability were also in consideration for this case. For linker length, we used a mix of long and short linkers (9–17 atoms) according to multiple publications on the optimization of PROTAC linkers [60–67].
Chemistry
All reactions were carried out in flame-dried flasks with magnetic stirring. Unless otherwise noted, all experiments were performed under a nitrogen atmosphere. All reagents were purchased from Sigma Aldrich, Acros Organics, Fisher Scientific, or Alfa Aesar. Solvents were treated with 4 Å molecular sieves and distilled before use. Purifications of reaction products were carried out by column chromatography using Chem Lab silica gel (230–400 mesh). 1H NMR spectra were recorded with tetramethylsilane (TMS) as internal standard at ambient temperature unless otherwise indicated Bruker 500 MHz for 1H NMR (SI Figures 1). Chemical shifts are reported in parts per million (ppm) and coupling constants are reported as Hertz (Hz). Splitting patterns are designated as singlet (s), broad singlet (bs), doublet (d), triplet (t). Splitting patterns that could not be interpreted or easily visualized are designated as multiple (m). Final compounds used in the study were more than 95% pure.
2-(2,6-dioxopiperidin-3-yl)-4-((2-(2-(2-(2-(4-(3-((4-((4-(1-isopropyl-2-methyl-1H-imidazol-5-yl)pyrimidin-2-yl)amino)phenyl)sulfonyl)propyl)-1H-1,2,3-triazol-1-yl)ethoxy)ethoxy)ethoxy)ethyl)amino)isoindoline-1,3-dione (PROTAC-7):
To a mixture of 4-(1-isopropyl-2-methyl-1H-imidazol-5-yl)-N-(4-(pent-4-yn-1-ylsulfonyl)phenyl)pyrimidin-2-amine (8 mg) and 4-((2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)amino)-2-(2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione (9 mg) in DMSO (450 μL) and water (300 μL) was added copper sulfate (3.4 mg), followed by a solution of sodium (R)-5-((S)-1,2-dihydroxyethyl)-4-hydroxy-2-oxo-2,5-dihydrofuran-3-olate (8.5 mg) in water (150 μL). The mixture was stirred at rt for 20 h. The mixture was filtered rinsing with a small amount of DMSO (1 mL) and then purified by prep HPLC to give the product as a yellow solid (3.6 mg, 21% yield). 1H NMR (500 MHz, Methanol-d4) δ 8.45 (d, J = 5.2 Hz, 1H), 8.42 (bs, 1H), 7.96 − 7.94 (m, 2H), 7.79 − 7.76 (m, 3H), 7.50 (dd, J = 8.5, 7.1 Hz, 1H), 7.43 (s, 1H), 7.13 (d, J = 5.2 Hz, 1H), 7.02 (dd, J = 14.6, 7.8 Hz, 2H), 5.75 (p, J = 7.1 Hz, 1H), 5.02 (dt, J = 12.8, 6.3 Hz, 1H), 4.48 − 4.46 (t, J = 5.2 Hz, 2H), 3.82 − 3.80 (t, J = 5.2 Hz, 2H), 3.68 (t, J = 5.2 Hz, 2H), 3.62 − 3.59 (m, 2H), 3.58 − 3.56 (m, 2H), 3.55 (s, 4H), 3.45 (t, J = 5.2 Hz, 2H), 3.23 − 3.19 (m, 2H), 2.88 − 2.81 (m, 1H), 2.78–2.74 (m, 2H), 2.71 − 2.68 (m, 1H), 2.58 (s, 3H), 2.11 − 1.97 (m, 4H), 1.54 (d, J = 7.1 Hz, 6H).HRMS (ESI) m/z calcd for C43H51N11O9S (M+H)+ 898.3670, found 898.3677.
(2S,4R)-4-hydroxy-1-((S)-2-(2-(2-(4-(3-((4-((4-(1-isopropyl-2-methyl-1H-imidazol-5-yl)pyrimidin-2-yl)amino)phenyl)sulfonyl)propyl)-1H-1,2,3-triazol-1-yl)ethoxy)acetamido)-3,3-dimethylbutanoyl)-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (PROTAC-8):
To a mixture of 4-(1-isopropyl-2-methyl-1H-imidazol-5-yl)-N-(4-(pent-4-yn-1-ylsulfonyl)phenyl)pyrimidin-2-amine (8 mg) and (2S,4R)-1-((S)-2-(2-(2-azidoethoxy)acetamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (10 mg) in DMSO (450 μL) and water (300 μL) was added copper sulfate (3.2 mg), followed by a solution of sodium (R)-5-((S)-1,2-dihydroxyethyl)-4-hydroxy-2-oxo-2,5-dihydrofuran-3-olate (8 mg) in water (150 μL). The mixture was filtered, rinsing with a small amount of DMSO, and purified by prep HPLC. White solid (5.4 mg, 31% yield). 1H NMR (500 MHz, Methanol-d4) δ 8.84 (s, 1H), 8.45 (d, J = 5.2 Hz, 1H), 7.96 (d, J = 8.8 Hz, 2H), 7.82 (s, 1H), 7.78 (d, J = 8.9 Hz, 2H), 7.47 − 7.41 (m, 3H), 7.39 (d, J = 8.4 Hz, 2H), 7.13 (d, J = 5.2 Hz, 1H), 5.82 − 5.67 (m, 1H), 4.67 − 4.56 (m, 4H), 4.50 (d, J = 15.4 Hz, 1H), 4.37 (d, J = 15.5 Hz, 1H), 4.07 − 3.97 (m, 2H), 3.97 − 3.90 (m, 2H), 3.87 − 3.75 (m, 2H), 3.23 − 3.15 (m, 2H), 2.79 (t, J = 7.5 Hz, 2H), 2.59 (d, J = 3.6 Hz, 3H), 2.44 (s, 3H), 2.28 − 2.19 (m, 1H), 2.12 − 1.97 (m, 3H), 1.56 − 1.52 (m, 6H), 0.98 (s, 9H). HRMS (ESI) m/z calcd for C48H60N12O7S2 (M+H)+ 981.4228, found 981.4227.
4-(2,6-difluorobenzamido)-N-((1r,4r)-4-((1-(2-(2-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)ethyl)-1H-1,2,3-triazol-4-yl)methoxy)cyclohexyl)-1H-pyrazole-3-carboxamide (PROTAC-9):
To a mixture of 4-(2,6-difluorobenzamido)-N-((1r,4r)-4-(prop-2-yn-1-yloxy)cyclohexyl)-1H-pyrazole-3-carboxamide (8 mg) and 4-((2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)amino)-2-(2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione (10.4 mg) in DMSO (450 μL) and water (300 μL) was added copper sulfate (3.6 mg), followed by a solution of sodium (R)-5-((S)-1,2-dihydroxyethyl)-4-hydroxy-2-oxo-2,5-dihydrofuran-3-olate (8.9 mg) in water (150 μL). The mixture was stirred at rt for 20 h. Water (5 mL) was added, and the mixture was extracted with EtOAc (3 × 5 mL). The organics were dried over Na2SO4, filtered, and concentrated. Flash chromatography (CH2Cl2:MeOH (0 to 10%)) gave the title compound as a yellow solid (5 mg, 29%). 1H NMR (400 MHz, Methanol-d4) δ 8.33 (s, 1H), 7.99 (s, 1H), 7.61 − 7.46 (m, 2H), 7.16 − 7.10 (m, 2H), 7.05 (dd, J = 13.3, 7.8 Hz, 2H), 5.03 (dd, J = 12.4, 5.5 Hz, 1H), 4.60 (s, 2H), 4.57 − 4.51 (m, 2H), 3.88 − 3.83 (m, 2H), 3.71 (t, J = 5.2 Hz, 2H), 3.66 − 3.60 (m, 4H), 3.59 (s, 4H), 3.48 (t, J = 5.2 Hz, 2H), 3.43 (dd, J = 9.7, 5.4 Hz, 1H), 2.92 − 2.78 (m, 1H), 2.78 − 2.62 (m, 2H), 2.11 − 2.07 (m, 2H), 1.96 (s, 2H), 1.48 − 1.27 (m, 6H). HRMS (ESI) m/z calcd for C41H46F2N10O10 (M+H)+ 877.3445, found 877.3444.
4-(2,6-difluorobenzamido)-N-((1S,4r)-4-((1-(2-(2-(((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-2-oxoethoxy)ethyl)-1H-1,2,3-triazol-4-yl)methoxy)cyclohexyl)-1H-pyrazole-3-carboxamide (PROTAC-10):
To a mixture of 4-(2,6-difluorobenzamido)-N-((1r,4r)-4-(prop-2-yn-1-yloxy)cyclohexyl)-1H-pyrazole-3-carboxamide (8 mg) and (2S,4R)-1-((S)-2-(2-(2-azidoethoxy)acetamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (10 mg) in DMSO (450 μL) and water (300 μL) was added copper sulfate (3.2 mg), followed by a solution of sodium (R)-5-((S)-1,2-dihydroxyethyl)-4-hydroxy-2-oxo-2,5-dihydrofuran-3-olate (8 mg) in water (150 μL). The mixture was diluted with EtOAc (10 mL) and water (10 mL) and the layers were separated. The aqueous layer was extracted with EtOAc (3 × 10 mL). The combined organics were washed with brine (10 mL), dried over Na2SO4, filtered, and concentrated. Flash chromatography in CH2Cl2/MeOH (1–12%) provided the desired product as colorless oil (4.1 mg, 24% yield). 1H NMR (500 MHz, Methanol-d4) δ 8.87 (s, 1H), 8.33 (s, 1H), 8.05 (s, 1H), 7.57 (tt, J = 8.4, 6.3 Hz, 1H), 7.48 − 7.40 (m, 4H), 7.13 (t, J = 8.4 Hz, 2H), 4.70 − 4.61 (m, 5H), 4.61 − 4.47 (m, 3H), 4.37 (d, J = 15.4 Hz, 1H), 4.07 − 3.99 (m, 2H), 3.99 − 3.94 (m, 2H), 3.88 − 3.77 (m, 3H), 3.47 − 3.40 (m, 1H), 2.47 (s, 3H), 2.26 − 2.18 (m, 1H), 2.14 − 2.01 (m, 4H), 1.96 (d, J = 12.8 Hz, 1H), 1.44 − 1.35 (m, 4H), 1.01 (s, 9H). HRMS (ESI) m/z calcd for C46H55F2N11O8S (M+H)+ 960.4002, found 960.4002.
HEI-OC1 experiments
HEI-OC1 cells were maintained in DMEM with 10% of Fetal bovine serum at 33°C in a 10% CO2 atmosphere. Cells were plated in 6-well plates (60,000 cells per well) 24 hours before the initiation of the experiment and then incubated for an additional 24 hours with 0.1 nM to 1 μM of PROTACS 7–10, AZD5438 or AT519-7. Cells were harvested in RIPA buffer (NaCl 150 mM, Tris-HCl 50 mM, Nonidet-40 1%, Sodium deoxycholate 0.5%, SDS 0.1%, pH 7.4) containing protease (Sigma-Aldrich, MO P8340) and phosphatase inhibitors (ThermoFisher, CA, A32957) for immunoblot analysis.
In the case of the proteasome inhibitory studies, HEI-OC1 cells were incubated with PROTAC-8 100 nM with or without MG-132 (300 nM, Calbiochem, MO, 474790), for 24 hours. Lysates were prepared after a 24-hour incubation and processed for Western blot analysis.
Animals
Four days post-fertilization (dpf) zebrafish (Danio rerio) larvae were obtained by pair mating of adult TuAB fish maintained at Creighton University by standard methods approved by the Institutional Animal Care and Use Committee. Experimental fish were maintained at 28.5°C in E3 media (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2 and 0.33 mM MgSO4, pH 7.2) until the day of the experiment.
In vivo experiments
Cisplatin studies:
Four days post-fertilization (dpf) fish were pre-incubated with different concentrations of PROTAC-8 or AZD5438 (0.1 nM to 1 μM) for 24hrs in E3 media. The next day animals were co-incubated with PROTAC-8 or AZD5438 and cisplatin (Millipore Sigma, 479306) 400 μM for 6 hours [57]. At the end of the incubation, fish were transferred to fresh E3 media, led to recover for 1 hour, and fixed overnight with 4% paraformaldehyde. Animals were immunostained for the hair cell marker, otoferlin (HCS-1, DSHB), and process for fluorescence microscopy. Otic, middle, and opercular neuromasts were identified, and hair cells were manually counted using a Zeiss AxioSkop 2 fluorescence microscope with a 40x oil objective.
Kainic acid studies:
Fish was pre-incubated with PROTAC-8 or AZD5438 as described above. The next day fish were transferred to a fresh solution of kainic acid 300 μM for 50 min, led recover in fresh E3 media for 2 hours, and fixed with 4% paraformaldehyde. Animals were processed for immunofluorescence analysis as described for the cisplatin treatments.
After the 24 hours incubation with PROTAC-8 and before the incubation with the ototoxin, some fish were collected for immunoblot analysis of CDK2 abundance.
Stock solutions for PROTAC-8, AZD5438, and cisplatin were prepared in DMSO. Control animals were exposed to 0.1% DMSO in E3 media. Images were taken employing a Zeiss LSM 710 confocal microscope with a 63x oil objective.
CDKs immunoblot assays
For the calculation of CDKs abundance, immunoblots were performed in HEI-OC1 cells and zebrafish treated with PROTACS as described before [57]. Briefly, 20–30 μg of protein were used per lane. Membranes were blocked for 1 hour at room temperature with 3% milk followed by overnight incubation with the primary antibody in 3% milk. After several washes and the secondary incubation for 1 hour, membranes were developed employing an iBright FL1000 (ThermoFisher, CA). Immunoblots were stripped and probed for β-actin as the loading control. Specific bands were quantified using ImageJ (NIH).
To be noted: Since AZD5438 was developed as a specific inhibitor for CDK1, CDK2 and CDK9 [18], we characterized PROTAC-8’s degradative effect against these three CDKs. Additionally, CDK5 was included because of its closer phylogenetical origin, while CDK7 served as a control for off-target effect since it is distantly related to CDK2
Primary antibodies: rabbit anti-CDK2 dilution 1:500 (CST, MA, #2546), rabbit anti-CDK5 dilution 1:1,000 (CST, MA, #2506), rabbit anti-CDK9 dilution 1:1,000 (CST, MA, #2316), rabbit anti-CDK1 dilution 1:1,000 (ABclonal, MA, #A11420), mouse anti-β-actin dilution 1:2,000 (Sigma-Aldrich, MO, A5441).
Confocal imaging
For the screening of the effect of PROTAC-8, neuromast hair cell counts were performed manually, employing a Zeiss AxioSkop 2 fluorescence microscope with a 40x oil objective.
Confocal imaging was performed using a Zeiss LSM 710 confocal laser scanning image system with a 63x oil objective. Images were captured at room temperature with automatically set sectioning. The acquired images were processed with ZEN black edition software. Z-stack images are presented as flat Z-projections.
Molecular dynamics (MD) simulations
Using the X-ray structures of the CDK2-AZD5438 complex (PDB i.d. 6GUE) and the pVHL-VH-032 complex (PDB i.d. 5NW0) PROTAC-8 was built on the 6GUE structure in YASARA [50] and the apo-pVHL was docked on the complex. The structure of the pVHL - PROTAC-8 - CDK2 ternary complex was then energy-minimized using the AMBERff14SB [49] and the GAFF [70] parameter sets. After energy minimization, the pVHL - PROTAC-8 - CDK2 ternary complex was solvated with 154 mM aqueous NaCl solution in an 11.3 nm × 11.3 nm × 11.3 nm cubic box and the energy of the system was minimized again. MD simulations (100 ns) were performed in an NPT ensemble at 1 atm pressure and 310 K. Integration time was 2 fs, bonds for pVHL - PROTAC-8 - CDK2 ternary complex and water were constrained with the LINCS [71] and SETTLE [72] algorithm, respectively. The non-bonded cut-off was 0.8 nm and the long-range electrostatic was treated with the PME method [73].
Statistics
For HEI-OC1 cells and fish experiments, one-way ANOVA followed by Dunnett’s multiple comparisons test was performed using GraphPad Prism version 8.2.0 software. P values less than 0.05 were considered significant. Results are expressed as mean ± SEM. For the experiments performed with fish, 6–8 fish were used per experiment. Western blots were performed 2 to 3 times employing 3 different biological replicates. Zebrafish experiments were performed 3 times.
Supplementary Material
Highlights:
PROTAC-based hearing loss therapy.
In vivo CDK2 degradation.
Prevention of hair cell damage
Acknowledgments.
We thank the Zuo lab members for their critical comments on the study. The study is supported in part by NIH-R01DC015444, NIH-R01DC015010, USAMRMC-RH170030, ONR-N00014-18-1-2507, DoD-RH190050, LB692/Creighton, 1P20GM139762-01, ALSAC, P30CA21765, and the Bellucci Foundation.
Abbreviations used
- AML
acute myeloid leukemia
- CDK
cyclin-dependent kinase
- CDK1
cyclin-dependent kinase 1
- CDK2
cyclin-dependent kinase 2
- CDK5
cyclin-dependent kinase 5
- CDK7
cyclin-dependent kinase 7
- CDK9
cyclin-dependent kinase 9
- CRBN
Cereblon
- DCCM
dynamic cross-correlation matrix
- FDA
food and drug administration
- FVB
friend leukemia virus B
- HEI-OC1
House Ear Institute-organ of Corti 1
- IC50
half maximal inhibitory concentration
- MD
molecular dynamics
- PROTAC
proteolysis targeting chimeras
- STS
sodium thiosulfate
- VHL
von Hippel–Lindau
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting information
Additional figures illustrating NMR spectra, HPLC profiles and western blots for the four PROTACs as well as dynamic cross-correlation matrix analysis.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].GBD Collaborators, Hearing loss prevalence and years lived with disability, 1990–2019: findings from the Global Burden of Disease Study 2019, Lancet 397 (2021) 996–1009. doi: 10.1016/S0140-6736(21)00516-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Knight KR, Chen L, Freyer D, Aplenc R, Bancroft M, Bliss B, Dang H, Gillmeister B, Hendershot E, Kraemer DF, Lindenfeld L, Meza J, , Neuwelt EA, Pollock BH, Sung L, Group-Wide, Prospective Study of Ototoxicity Assessment in Children Receiving Cisplatin Chemotherapy (ACCL05C1): A Report From the Children’s Oncology Group, J. Clin. Oncol 35 (2017) 440–445. doi: 10.1200/JCO.2016.69.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hazlitt RA, Min J, Zuo J, Progress in the Development of Preventative Drugs for Cisplatin-Induced Hearing Loss, J. Med. Chem 61 (2018) 5512–5524. doi: 10.1021/acs.jmedchem.7b01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brock PR, Maibach R, Childs M, Rajput K, Roebuck D, Sullivan MJ, Laithier V, Ronghe M, Dall’Igna P, Hiyama E, Brichard B, Skeen J, Mateos ME, Capra M, Rangaswami AA, Ansari M, Rechnitzer C, Veal GJ, Covezzoli A, Brugieres L, Perilongo G, Czauderna P, Morland B, A Neuwelt E, Sodium Thiosulfate for Protection from Cisplatin-Induced Hearing Loss, N. Engl. J. Med 378 (2018) 2376–2385. doi: 10.1056/NEJMoa1801109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Freyer DR, Chen L, Krailo MD, Knight K, Villaluna D, Bliss B, Pollock BH, Ramdas J, Lange B, Van Hoff D, VanSoelen ML, Wiernikowski J, Neuwelt EA, Sung L, Effects of sodium thiosulfate versus observation on development of cisplatin-induced hearing loss in children with cancer (ACCL0431): a multicentre, randomised, controlled, open-label, phase 3 trial, Lancet Oncol. 18 (2018) 63–74. doi: 10.1016/S1470-2045(16)30625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kechai NE, Agnely F, Mamelle E, Nguyen Y, Ferrary E, Bochot A, Recent advances in local drug delivery to the inner ear, Int. J. Pharm 494 (2015) 83–101. doi: 10.1016/j.ijpharm.2015.08.015. [DOI] [PubMed] [Google Scholar]
- [7].Muller U, Barr-Gillespie PG, New treatment options for hearing loss, Nat. Rev. Drug Discov 14 (2015) 346–365. doi: 10.1038/nrd4533. [DOI] [PubMed] [Google Scholar]
- [8].Hazlitt RA, Teitz T, Bonga JD, Fang J, Diao S, Iconaru L, Yang L, Goktug AN, Currier DG, Chen T, Rankovic Z, Min J, Zuo J, Development of Second-Generation CDK2 Inhibitors for the Prevention of Cisplatin-Induced Hearing Loss, J. Med. Chem 61 (2018) 7700–7709. doi: 10.1021/acs.jmedchem.8b00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Teitz T, Fang J, Goktug AN, Bonga JD, Diao S, Hazlitt RA, Iconaru L, Morfouace M, Currier D, Zhou Y, Umans RA, Taylor MR, Cheng C, Min J, Freeman B, Peng J, Roussel MF, Kriwacki R, Guy RK, Chen T, Zuo J, CDK2 inhibitors as candidate therapeutics for cisplatin- and noise-induced hearing loss, J. Exp. Med 215 (2018) 1187–1203. doi: 10.1084/jem.20172246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang J, Yang T, Xu G, Liu H, Ren C, Xie W, Wang M, Cyclin-Dependent Kinase 2 Promotes Tumor Proliferation and Induces Radio Resistance in Glioblastoma, Transl. Oncol 9 (2016) 548–556. doi: 10.1016/j.tranon.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yin X, Yu J, Zhou Y, Wang C, Jiao Z, Qian Z, Sun H, Chen B, Identification of CDK2 as a novel target in treatment of prostate cancer, Future Oncol. 14 (2018) 709–718. doi: 10.2217/fon-2017-0561 [DOI] [PubMed] [Google Scholar]
- [12].Faber AC, Chiles TC, Inhibition of cyclin-dependent kinase-2 induces apoptosis in human diffuse large B-cell lymphomas, Cell Cycle 6 (2007) 2982–2989. doi: 10.4161/cc.6.23.4994 [DOI] [PubMed] [Google Scholar]
- [13].Li Y, Yang XH, Fang SJ, Qin CF, Sun RL, Liu ZY, Jiang BY, Wu X, Li G, HOXA7 stimulates human hepatocellular carcinoma proliferation through cyclin E1/CDK2, Oncol. Rep 33 (2015) 990–996. doi: 10.3892/or.2014.3668 [DOI] [PubMed] [Google Scholar]
- [14].Alexander A, Karakas C, Chen X, Carey JP, Yi M, Bondy M, Thompson P, Cheung KL, Ellis IO, Gong Y, Krishnamurthy S, Alvarez RH, Ueno NT, Hunt KK, Keyomarsi K, Cyclin E overexpression as a biomarker for combination treatment strategies in inflammatory breast cancer, Oncotarget 8 (2017) 14897–14911. doi: 10.18632/oncotarget.14689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yang L, Fang D, Chen H, Lu Y, Dong Z, Ding HF, Jing Q, Su SB, Huang S, Cyclin-dependent kinase 2 is an ideal target for ovary tumors with elevated cyclin E1 expression, Oncotarget 6 (2015) 20801–20812. doi: 10.18632/oncotarget.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ingersoll MA, Malloy EA, Caster LE, Holland EM, Xu Z, Zallocchi M, Currier D, Liu H, He DZZ, Min J, Chen T, Zuo J, Teitz T, BRAF inhibition protects against hearing loss in mice, Sci. Adv 6 (2020). doi: 10.1126/sciadv.abd0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Boss DS, Schwartz GK, Middleton MR, Amakye DD, Swaisland H, Midgley RS, Ranson M, Danson S, Calvert H, Plummer R, Morris C, Carvajal RD, Chirieac LR, Schellens JHM, Shapiro GI, Safety, tolerability, pharmacokinetics and pharmacodynamics of the oral cyclin-dependent kinase inhibitor AZD5438 when administered at intermittent and continuous dosing schedules in patients with advanced solid tumours, Ann. Oncol 21 (2010) 884–894. doi: 10.1093/annonc/mdp377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Camidge DR, Pemberton M, Growcott J, Amakye D, Wilson D, Swaisland H, Forder C, Wilkinson R, Byth K, Hughes A, A phase I pharmacodynamic study of the effects of the cyclin-dependent kinase-inhibitor AZD5438 on cell cycle markers within the buccal mucosa, plucked scalp hairs and peripheral blood mononucleocytes of healthy male volunteers, Cancer Chemother. Pharmacol 60 (2007) 479–488. doi: 10.1007/s00280-006-0387-2. [DOI] [PubMed] [Google Scholar]
- [19].Camidge DR, Smethurst D, Growcott J, Barrass NC, Foster JR, Febbraro S, Swaisland H, Hughes A, A first-in-man phase I tolerability and pharmacokinetic study of the cyclin-dependent kinase-inhibitor AZD5438 in healthy male volunteers, Cancer Chemother. Pharmacol 60 (2007) 391–398. doi: 10.1007/s00280-006-0371-x. [DOI] [PubMed] [Google Scholar]
- [20].Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ, Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation, Proc. Natl. Acad. Sci. U. S. A. 98 (2001) 8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nowak RP, DeAngelo SL, Buckley D, He Z, Donovan KA, An J, Safaee N, Jedrychowski MP, Ponthier CM, Ishoey M, Zhang T, Mancias JD, Gray NS, Bradner JE, Fischer ES, Plasticity in binding confers selectivity in ligand-induced protein degradation, Nat. Chem. Biol 14 (2018) 706–714. doi: 10.1038/s41589-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bondeson DP, MaresSmith AIE, Ko E, Campos S, Miah AH, Mulholland KE, Routly N, Buckley DL, Gustafson JL, Zinn N, Grandi P, Shimamura S, Bergamini G, Faelth-Savitski M, Bantscheff M, Cox C, Gordon DA, Willard RR, Flanagan JJ, Casillas LN, Votta BJ, den Besten W, Famm K, Kruidenier L, Carter PS, Harling JD, Churcher I, Crews CM, Catalytic in vivo protein knockdown by small-molecule PROTACs, Nat. Chem. Biol 11 (2015) 611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gadd MS, Testa A, Lucas X, Chan KH, Chen W, Lamont DJ, Zengerle M, Ciulli A, Structural basis of PROTAC cooperative recognition for selective protein degradation, Nat. Chem. Biol 13 (2017) 514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lai AC, Toure M, Hellerschmied D, Salami J, Jaime-Figueroa S, Ko E, Hines J, Crews CM, Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL, Angew. Chem. Int. Ed. Engl 55 (2016) 807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang X, Dixit VM, Drugging the undruggables: exploring the ubiquitin system for drug development, Cell Res. 26 (2016) 484–498. doi: 10.1038/cr.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE, DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation, Science 348 (2015) 1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lebraud H, Wright DJ, Johnson CN, Heightman TD, Protein Degradation by In-Cell Self-Assembly of Proteolysis Targeting Chimeras, ACS Cent. Sci 2 (2016) 927–934. doi: 10.1021/acscentsci.6b00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wurz RP, Dellamaggiore K, Dou H, Javier N, Lo MC, McCarter JD, Mohl D, Sastri C, Lipford JR, Cee VJ, A “Click Chemistry Platform” for the Rapid Synthesis of Bispecific Molecules for Inducing Protein Degradation, J. Med. Chem 61 (2018) 453–461. doi: 10.1021/acs.jmedchem.6b01781. [DOI] [PubMed] [Google Scholar]
- [29].Toure M, Crews CM, Small-Molecule PROTACS: New Approaches to Protein Degradation, Angew. Chem. Int. Ed. Engl 55 (2016) 1966–1973. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- [30].Schiedel M, Herp D, Hammelmann HS, Swyter S, Lehotzky A, Robaa D, Olah J, Ovadi J, Sippl W, Jung M, Chemically Induced Degradation of Sirtuin 2 (Sirt2) by a Proteolysis Targeting Chimera (PROTAC) Based on Sirtuin Rearranging Ligands (SirReals), J. Med. Chem 61 (2018) 482–491. doi: 10.1021/acs.jmedchem.6b01872. [DOI] [PubMed] [Google Scholar]
- [31].Khan S, Zhang X, Lv D, Zhang Q, He Y, Zhang P, X Liu, Thummuri D, Yuan Y, Wiegand JS, Pei J, Zhang W, Sharma A, McCurdy CR, Kuruvilla VM, Baran N, Ferrando AA, Kim YM, Rogojina A, Houghton PJ, Huang G, Hromas R, Konopleva M, Zheng G, Zhou D, A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity, Nat. Med 25 (2019) 1938–1947. doi: 10.1038/s41591-019-0668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gechijian LN, Buckley DL, Lawlor MA, Reyes JM, Paulk J, Ott CJ, Winter GE, Erb MA, Scott TG, Xu M, Seo HS, Dhe-Paganon S, Kwiatkowski NP, Perry JA, Qi J, Gray NS, Bradner JE, Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands, Nat. Chem. Biol 14 (2018) 405–412. doi: 10.1038/s41589-018-0010-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Remillard D, Buckley DL, Paulk J, Brien GL, Sonnett M, Seo HS, Dastjerdi S, Wuhr M, Dhe-Paganon S, Armstrong SA, Bradner JE, Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands, Angew. Chem. Int. Ed. Engl 56 (2017) 5738–5743. doi: 10.1002/anie.201611281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zoppi V, Hughes SJ, Maniaci C, Testa A, Gmaschitz T, Wieshofer C, Koegl M, Riching KM, Daniels DL, Spallarossa A, Ciulli A, Iterative Design and Optimization of Initially Inactive Proteolysis Targeting Chimeras (PROTACs) Identify VZ185 as a Potent, Fast, and Selective von Hippel-Lindau (VHL) Based Dual Degrader Probe of BRD9 and BRD7, J. Med. Chem 62 (2019) 699–726. doi: 10.1021/acs.jmedchem.8b01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, McEachern D, Chen J, Yang CY, Liu Z, Wang M, Liu L, Jiang H, Wen B, Kumar P, Meagher JL, Sun D, Stuckey JA, Wang S, A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo, Cancer Cell 36 (2019) 498–511 e17. doi: 10.1016/j.ccell.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhou H, Bai L, Xu R, Zhao Y, Chen J, McEachern D, Chinnaswamy K, Wen B, Dai L, Kumar P, Yang CY, Liu Z, Wang M, Liu L, Meagher JL, Yi H, Sun D, Stuckey JA, Wang S, Structure-Based Discovery of SD-36 as a Potent, Selective, and Efficacious PROTAC Degrader of STAT3 Protein, J. Med. Chem 62 (2019) 11280–11300. doi: 10.1021/acs.jmedchem.9b01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Robb CM, Contreras JI, Kour S, Taylor MA, Abid M, Sonawane YA, Zahid M, Murry DJ, Natarajan A, Rana S, Chemically induced degradation of CDK9 by a proteolysis targeting chimera (PROTAC), Chem. Commun. (Camb) 53 (2017) 7577–7580. doi: 10.1039/c7cc03879h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jiang B, Wang ES, Donovan KA, Liang Y, Fischer ES, Zhang T, Gray NS, Development of Dual and Selective Degraders of Cyclin-Dependent Kinases 4 and 6, Angew. Chem. Int. Ed. Engl 58 (2019) 6321–6326. doi: 10.1002/anie.201901336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, Zhang T, Kwiatkowski N, Boukhali M, Green JL, Haas W, Nomanbhoy T, Fischer ES, Young RA, Bradner JE, Winter GE, Gray NS, Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation, Nat. Chem. Biol 14 (2018) 163–170. doi: 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Teng M, Jiang J, He Z, Kwiatkowski NP, Donovan KA, Mills CE, Victor C, Hatcher JM, Fischer ES, Sorger PK, Zhang T, Gray NS, Development of CDK2 and CDK5 Dual Degrader TMX-2172, Angew. Chem. Int. Ed. Engl 59 (2020) 13865–13870. doi: 10.1002/anie.202004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhou F, Chen L, Cao C, Ju Y, Luo X, Zhou P, Zhao L, Du W, Cheng J, Xie Y, Chen Y, Development of selective mono or dual PROTAC degrader probe of CDK isoforms, Eur. J. Med. Chem 187 (2020) 111952. doi: 10.1016/j.ejmech.2019.111952 [DOI] [PubMed] [Google Scholar]
- [42].Wood DJ, Korolchuk S, Tatum NJ, Wang LZ, Endicott JA, Noble ME, Martin MP, Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition, Cell Chem. Biol 26 (2019) 121–130 e5. doi: 10.1016/j.chembiol.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang L, Shao X, Zhong T, Wu Y, Xu A, Sun X, Gao H, Liu Y, Lan T, Tong Y, Tao X, Du W, Wang W, Chen Y, Li T, Meng X, Deng H, Yang B, He Q, Ying M, Rao Y, Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy, Nat. Chem. Biol (2021). doi: 10.1038/s41589-021-00742-5. [DOI] [PubMed] [Google Scholar]
- [44].Bulatov E, Ciulli A, Targeting Cullin-RING E3 ubiquitin ligases for drug discovery: structure, assembly and small-molecule modulation, Biochem. J 467 (2015) 365–86. doi: 10.1042/BJ20141450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lucas X, Ciulli A, Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies, Curr. Opin. Struct. Biol 44 (2017) 101–110. doi: 10.1016/j.sbi.2016.12.015. [DOI] [PubMed] [Google Scholar]
- [46].Chan KH, Zengerle M, Testa A, Ciulli A, Impact of Target Warhead and Linkage Vector on Inducing Protein Degradation: Comparison of Bromodomain and Extra-Terminal (BET) Degraders Derived from Triazolodiazepine (JQ1) and Tetrahydroquinoline (I-BET726) BET Inhibitor Scaffolds, J. Med. Chem 61 (2018) 504–513. doi: 10.1021/acs.jmedchem.6b01912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Goracci L, Desantis J, Valeri A, Castellani B, Eleuteri M, Cruciani G, Understanding the Metabolism of Proteolysis Targeting Chimeras (PROTACs): The Next Step toward Pharmaceutical Applications, J. Med. Chem 63 (2020) 11615–11638. doi: 10.1021/acs.jmedchem.0c00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Steinebach C, Kehm H, Lindner S, Vu LP, Kopff S, Lopez Marmol A, Weiler C, Wagner KG, Reichenzeller M, Kronke J, Gutschow M, PROTAC-mediated crosstalk between E3 ligases, Chem. Commun. (Camb) 55 (2019) 1821–1824. doi: 10.1039/c8cc09541h. [DOI] [PubMed] [Google Scholar]
- [49].Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C, ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB, J. Chem. Theory. Comput 11 (2015) 3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Krieger E, Vriend G, New ways to boost molecular dynamics simulations, J. Comput. Chem 36 (2015) 996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Abraham MJ, Murtola T, Schulz R, Pálla S, Smith JC, Hess B, Lindahl E, GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX 1 (2015) 19–25. [Google Scholar]
- [52].Hunenberger PH, Mark AE, van Gunsteren WF, Fluctuation and cross-correlation analysis of protein motions observed in nanosecond molecular dynamics simulations, J. Mol. Biol 252 (1995) 492–503. doi: 10.1006/jmbi.1995.0514. [DOI] [PubMed] [Google Scholar]
- [53].Hsu AY, Wang D, Liu S, Lu J, Syahirah R, Bennin DA, Huttenlocher A, Umulis DM, Wan J, Deng Q, Phenotypical microRNA screen reveals a noncanonical role of CDK2 in regulating neutrophil migration, Proc. Natl. Acad. Sci. U. S. A 116 (2019) 18561–18570. doi: 10.1073/pnas.1905221116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cyrus K, Wehenkel M, Choi EY, Lee H, Swanson H, Kim KB, Jostling for position: optimizing linker location in the design of estrogen receptor-targeting PROTACs, ChemMedChem 5 (2010) 979–85. doi: 10.1002/cmdc.201000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Maniaci C, Hughes SJ, Testa A, Chen W, Lamont DJ, Rocha S, Alessi DR, Romeo R, Ciulli A, Homo-PROTACs: bivalent small-molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation, Nat. Commun 8 (2017) 830. doi: 10.1038/s41467-017-00954-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zorba A, Nguyen C, Xu Y, Starr J, Borzilleri K, Smith J, Zhu H, Farley KA, Ding W, Schiemer J, Feng X, Chang JS, Uccello DP, Young JA, Garcia-Irrizary CN, Czabaniuk L, Schuff B, Oliver R, Montgomery J, Hayward MM, Coe J, Chen J, Niosi M, Luthra S, Shah JC, El-Kattan A, Qiu X, West GM, Noe MC, Shanmugasundaram V, Gilbert AM, Brown MF, Calabrese MF, Delineating the role of cooperativity in the design of potent PROTACs for BTK, Proc. Natl. Acad. Sci. U. S. A 115 (2018) E7285–E7292. doi: 10.1073/pnas.1803662115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zallocchi M, Hati S, Xu Z, Hausman W, Liu H, He DZ, Zuo J, Characterization of quinoxaline derivatives for protection against iatrogenically induced hearing loss, JCI Insight 6 (2021). doi: 10.1172/jci.insight.141561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sheth S, Mukherjea D, Rybak LP, Ramkumar V, Mechanisms of Cisplatin-Induced Ototoxicity and Otoprotection, Front. Cell Neurosci 11 (2017) 338. doi: 10.3389/fncel.2017.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Xia LW, Ba MY, Liu W, Cheng W, Hu CP, Zhao Q, Yao YF, Sun MR, Duan YT, Triazol: a privileged scaffold fo proteolysis targeting chimeras, Future Med. Chem 11 (2019) 2019–2073. doi: 10.4155/fmc-2019-0159. [DOI] [PubMed] [Google Scholar]
- [60].Donoghue C, Cubillos-Rojas M, Gutierrez-Prat N, Sanchez-Zarzalejo C, Verdaguer X, Riera A, Nebreda AR, Optimal linker length for small molecule PROTACs that selectively target p38α and p38β for degradation, Eur. J. Med. Chem 201 (2020) 112451. doi: 10.1016/j.ejmech.2020.112451 [DOI] [PubMed] [Google Scholar]
- [61].Zagidullin A, Milyukov V, Rizvanov A, Bulatov E, Novel approaches for the rational design of PROTAC linkers, Explor. Target. Anti-tumor Ther 1 (2020) 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Shaheer M, Singh R, Sobhia ME, Protein degradation: a novel computational approach to design protein degrader probes for main protease of SARS-CoV-2, Truct Dyn. 30 (2021) 1–13. doi: 10.1080/07391102.2021.1953601 [DOI] [PubMed] [Google Scholar]
- [63].Fischer PD, Papadopoulos E, Dempersmier JM, Wang ZF, Nowak RP, Donovan KA, Kalabathula J, Gorgulla C, P M Junghanns P, Kabha E, Dimitrakakis N, Petrov OI, Mitsiades C, Ducho C, Gelev V, Fischer ES, Wagner G, Arthanari H, A biphenyl inhibitor of eIF4E targeting an internal binding site enables the design of cell-permeable PROTAC-degraders, Eur J Med Chem. 219 (2021) 113435. doi: 10.1016/j.ejmech.2021.113435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zorba A, Nguyen C, Xu Y, Starr J, Borzilleri K, Smith J, Zhu H, Farley KA, Ding W, Schiemer J, Feng X, Chang JS, Uccello DP, Young JA, Garcia-Irrizary CN, Czabaniuk L, Schuff B, Oliver R, Montgomery J, Hayward MM, Coe J, Chen J, Niosi M, Luthra S, Shah JC, El-Kattan A, Qiu X, West GM, Noe MC, Shanmugasundaram V, Gilbert AM, Brown MF, Calabrese MF, Delineating the role of cooperativity in the design of potent PROTACs for BTK, Proc Natl Acad Sci U S A. 115(31) (2018) E7285–E7292. doi: 10.1073/pnas.1803662115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jaime-Figueroa S, Buhimschi AD, Toure M, Hines J, Crews CM, Design, synthesis and biological evaluation of Proteolysis Targeting Chimeras (PROTACs) as a BTK degraders with improved pharmacokinetic properties, Bioorg. Med. Chem. Lett 30(3 (2020)126877. doi: 10.1016/j.bmcl.2019.126877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Liang J, Xie H, Yang R, Wang N, Zheng Z, Zhou C, Wang Y, Wang Z, Liu H, Shan L, Ke Y, Designed, synthesized and biological evaluation of proteolysis targeting chimeras (PROTACs) as AR degraders for prostate cancer treatment, Bioorg. Med. Chem 4 (2021)116331. doi: 10.1016/j.bmc.2021.116331 [DOI] [PubMed] [Google Scholar]
- [67].Li Y, Zhang S, Zhang J, Hu Z, Xiao Y, Huang J, Dong C, Huang S, Zhou H, Exploring the PROTAC degron candidates: OBHSA with different side chains as novel selective estrogen receptor degraders (SERDs), Eur. J. Med. Chem 15 (2019) 172:48–61. doi: 10.1016/j.ejmech.2019.03.058 [DOI] [PubMed] [Google Scholar]
- [68].Pan C, Lei Z, Wang S, Wang X, Wei D, Cai X, Luoreng Z, Wang L, Ma Y, Genome-wide identification of cyclindependent kinase (CDK) genes affecting adipocyte differentiation in cattle, BMC Genomics 22 (2021) 532. 10.1186/s12864-021-07653-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, Morgan DO, Tsai L, Wolgemuth DJ, Cyclin-dependent kinases: a family portrait, Nat. Cell Biol 11(11) (2009) 1275–1276. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA, Development and testing of a general amber force field, J. Comput. Chem 25 (2004) 1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- [71].Hess B, Bekker H, Berendsen HJC, Fraaije JGEM, LINCS: A linear constraint solver for molecular simulations, J. Comput. Chem 18 (1997) 1463–1472. [Google Scholar]
- [72].Miyamoto S, Kollman PA, Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models, J. Comput. Chem 13 (1992) 952–962. [Google Scholar]
- [73].Essman U, Perera L, Berkowitz ML, A smooth particle mesh Ewald method, J. Chem. Phys 103 (1995) 8577–8593. [Google Scholar]
- [74].Belair DG, Lu G, Waller LE, Gustin JA, Collins ND, Kolaja KL, Thalidomide Inhibits Human iPSC Mesendoderm Differentiation by Modulating CRBN-dependent Degradation of SALL4, Sci. Rep 10(1) (2020) 2864. doi: 10.1038/s41598-020-59542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


