Abstract
Background
Shotgun metagenomics has been used clinically for diagnosing infectious diseases. However, most technical assessments have been limited to individual sets of reference standards, experimental workflows, and laboratories.
Methods
A reference panel and performance metrics were designed and used to examine the performance of shotgun metagenomics at 17 laboratories in a coordinated collaborative study. We comprehensively assessed the reliability, key performance determinants, reproducibility, and quantitative potential.
Findings
Assay performance varied significantly across sites and microbial classes, with a read depth of 20 millions as a generally cost-efficient assay setting. Results of mapped reads by shotgun metagenomics could indicate relative and intra-site (but not absolute or inter-site) microbial abundance.
Interpretation
Assay performance was significantly impacted by the microbial type, the host context, and read depth, which emphasizes the importance of these factors when designing reference reagents and benchmarking studies. Across sites, workflows and platforms, false positive reporting and considerable site/library effects were common challenges to the assay's accuracy and quantifiability. Our study also suggested that laboratory-developed shotgun metagenomics tests for pathogen detection should aim to detect microbes at 500 CFU/mL (or copies/mL) in a clinically relevant host context (10^5 human cells/mL) within a 24h turn-around time, and with an efficient read depth of 20M.
Funding
This work was supported by National Science and Technology Major Project of China (2018ZX10102001).
Keywords: Shotgun Metagenomics for Pathogen Detection, Next-generation Sequencing, Multicenter Assessment, Diagnostic Performance
Research in Context.
Evidence before this study
Shotgun metagenomics for pathogen detection has rapidly emerged as a novel diagnostic tool for infectious diseases in various clinical settings. Its great value as an unbiased assay was also demonstrated in the early identification of the pathogen responsible for the COVID-19 pandemic. On the other hand, the agnostic features of such assays also leads to unique challenges stemming from the varied cellular and genomic characteristics of a myriad of pathogens in clinical samples. So far, assay validation in blood plasma, cerebrospinal fluid, and respiratory samples has been described. However, most evaluations have been limited to individual workflows, platforms, and laboratories. Comprehensive multicenter assessments would further our understanding of this technology.
Added value of this study
In this work, we established a pathogen reference panel that presented 30 different microorganisms of different types and assessed the assay performance in a 17-site, cross-workflow and cross-platform study. Performance of shotgun metagenomics was significantly impacted by the microbial type, the host context, and read depth. Our data support the conclusion that the relative, but not absolute, abundance of microorganisms within a sample is a key determinant of pathogen detection by metagenomics, highlighting the importance of host cell context in clinical specimens. Using the reference panel, we showed that shotgun metagenomics for pathogen detection can be a qualitative assay with indicative value for relative abundance. Precise quantitative measurement of absolute abundance will remain challenging until these deviations are better understood and more sophisticated quantitative modeling is established.
This work also indicates that caution is needed when interpreting results from shotgun metagenomics due to considerable variation in assay performance across sites and the challenge of false positive results. We also suggest the application of a precision filter to identify potential false-positive results through machine learning as a strategy for improving assay precision.
Our results also suggested that laboratory-developed shotgun metagenomic assays for pathogen detection should aim to detect microbes at 500 CFU/mL (or copies/mL) in a clinically relevant host context (10^5 human cells/mL) within a 24h turn-around time, and with a recommended cost-efficient read depth of 20M reads.
Implications of all the available evidence
Our data suggest that when given appropriate experimental and bioinformatic optimization, shotgun metagenomics for pathogen detection holds great promise as a valuable tool clinically for broad-spectrum pathogen detection. Also, this collaborative work provided a unique resource comprising nearly 600 billion reads (>5Tb) for technical evaluation in clinical and regulatory settings. We believe that our multicenter analyses could be valuable to drive further advances in shotgun metagenomics-related experimental techniques and the development of bioinformatics tools.
Alt-text: Unlabelled box
1. Introduction
Infectious diseases are a leading cause of death worldwide, attributable to a great variety of pathogens that belong to different microbial types. Rapid and precise identification of disease-causing pathogens is the key to effective clinical management but remains challenging in clinical settings [1,2]. Conventional diagnostics either rely on cultures or require a presumptive diagnosis by the clinician before testing. Recent advances in high-throughput sequencing and bioinformatics technologies have enabled rapid growth in the application of shotgun metagenomics to detect pathogens [3], [4], [5], [6], [7]. Importantly, the rapid identification of SARS-CoV-2, the causative of the COVID-19 pandemic, was highly attributable to the use of shotgun metagenomic assays [8], [9], [10], [11], [12].
Next-generation sequencing (NGS)-based assays have been widely applied in the fields of non-invasive prenatal testing and companion diagnostics for cancer treatment [13], [14], [15]. However, compared to these assays (which analyze a limited number of genetic sites within the human genome), shotgun metagenomics for pathogen detection faces unique challenges, since it involves a great variety of genomes from all organisms present in clinical samples [16], [17], [18], [19]. The cellular and genomic characteristics of these organisms require that the assay can access all genetic contents (e.g. breaking all cellular structures), and differentiate them (e.g. preventing false annotation of closely related species). So far, most assessments of shotgun metagenomics have been limited to individual sets of reference reagents, individual microbial types, or individual experimental protocols and laboratories [20,21]. A multicenter evaluation study using a common set of dedicated reference reagents and performance metrics is hence highly desirable, since it is crucial for establishing performance standards, guiding proper interpretation of results, aiding further assay development and clinical adaptations, and providing valuable information from a regulatory perspective for this newly emerging technology. Similar to the MicroArray Quality Control (MAQC) and Sequencing Quality Control (SEQC) projects, large-scale community efforts have been coordinated for assessing the performance of microarray and RNA-seq technologies across laboratories, platforms, and pipelines [22], [23], [24], [25].
In this study, we described a multicenter benchmarking study coordinated by the National Institutes for Food and Drug Control (NIFDC) of China, which included 17 independent laboratories in cross-workflow and cross-platform settings. In total, over 580 billion reads and over 5 Tb of sequencing data were generated and studied. To our knowledge, the current study represents the largest effort to date to produce and analyze comprehensive reference datasets for shotgun metagenomics for pathogen detection.
2. Methods
2.1. Preparation and validation of the reference panel
Bacterial and fungal organisms were validated by Matrix-assisted Laser Desorption/ Ionization-Time Of Flight (MALDI-TOF, Bruker, Billerica, MA), Vitek 2 (bioMérieux, Craponne, France), and a BioFire FilmArray Multiplex PCR System (bioMérieux, Craponne, France), and measured by standard plate counts as recommended by HMMD guidance (highly multiplexed microbiological/ medical countermeasure in vitro nucleic acid based diagnostic devices). Viral organisms were validated by Sanger sequencing and quantitated by droplet digital PCR (ddPCR). HeLa cells were ATCC, RRID: CVCL_0030) were validated by STR profiling (Supplementary File S1), testing for mycoplasma was not performed near the time of sample preparation. Measurement was conducted by cell counting. These microbes were then spiked into PBS solutions with 2 × 10^5/ml of HeLa cells at indicated concentrations to mimic clinical specimens from respiratory or central nervous system infections, such as CSF or BALF (Supplemental Table S1) [26], [27], [28], [29]. Pathogen reference reagents (PRHs and PRLs) were prepared by contriving microbes at high (PRH) and low (PRL) titers with HeLa cells, respectively. In the PRH group, these microorganisms were spiked at 200-350,000 CFU/ml for bacterial and 400–10,000 CFU/ml for fungal pathogens, at 660–2,000,000 copies/ml for DNA viruses and at 140-3,500,000 copies/ml for RNA viruses to represent common ranges of clinical infection.
Reference samples were sent frozen to 17 independent laboratory sites (Centers C1-C17) for metagenomic testing and bioinformatic analysis. Nucleic acid extraction was requested to be performed right after thawing to minimize cell lysis and other unexpected changes. To support quantitative assessments, all original samples in the PR panel were tested in triplicates and 10-fold diluted samples were tested in 10 replicates, except for the pathogen-free controls. All technicians were trained to follow the Standard Operating Processes (SOP) and certified to perform the assay at each site.
2.2. Comparison of bioinformatics pipelines
We used Mason (Mason – A Read Simulator for Next Generation Sequencing Data, v0.1.2) to generate simulated sequencing data for 108 microbial genomes, which including 62 bacterial, 42 viral, and 4 fungal microorganisms. A total of 100,000 single-end, 75bp reads were generated for each microbe and subjected to taxonomic identification by Centrifuge [30], Kraken [31], and CLARK [32] pipelines and a database prepared as instructed by the CLARK website and implemented with RNA viruses, which included a total of 13,879 bacteria, 6,570 viruses, and 1,429 fungi. Assessment of pipeline performance was performed at both the genus and species levels and by type of microorganism. Sensitivity was inferred by the number of reads mapped specifically to the correct taxa; and specificity was inferred by the percentage of reads mapped specifically to the incorrect taxa. Statistical comparisons were done using Wilcoxon rank tests. Comparisons among alignment algorithms of BWA [33], Bowtie2 [34], and SNAP [35] were performed using a similar strategy.
2.3. Correlation analysis of observed and theoretical abundances
Raw sequencing data from 17 sites were analyzed using the site-independent CLARK-based pipeline to obtain the observed abundance of each microorganism in each sample, except for RNA viruses. The theoretical abundance of a microorganism in a sample was proportional to the ratio of DNA of that microorganism and the size of sequencing data, calculated as below:
Where and was the copy number and genome size of microorganism i in this sample, respectively. Human cell number for each sample was constant to , and human genome size Subsequently, a linear regression model was used to estimate the correlations between the observed and theoretical abundances.
2.4. Analysis of simulated and observed coefficient of variants (CVs)
Fastq data from the 10 repeated replicates of each sample were merged, randomly re-sampled according to the original data sizes (total number of reads), and analyzed by the CLARK-based pipeline. The CVs were calculated based on the read numbers mapped to each microbe within each re-sampled replicate. This above process was repeated 10 times to obtain a total of 10 simulated CVs for each microbe. The average of these simulated CVs represented the CV derived from variations in data size. A linear regression model was used to evaluate the contribution of these CVs to the observed overall CVs. In addition, we used a linear mixed model to further evaluate whether the sequencing platform, library method, and class of microorganism affected the observed CV. The formula of the linear mixed model was defined as:
Cv_observed ∼ Cv_datasize + Library Prep + Microbial Class + Platform + (1+Cv_observed|Center) where Center was a random effect, and the read depth CV (Cv_datasize), library preparation method (Library Prep), microbial class, and sequencing platform (Platform) were fixed effects.
2.5. Analysis of read-depth requirement for pathogen detection
Fastq data from the 10 repeated replicates of each sample were merged and resampled to the desired read depth of 0.5M, 1M, 5M, 10M, 20M, 30M, and 50M total reads for each sample. The re-sampled data were analyzed by the CLARK-based pipeline in the coordinating laboratory and identification of a microorganism was defined with >4 species-specific mapped reads in a sample, a threshold at which yielded the best overall assay performance across all sites. Species-specific reads were those mapped exclusively to one microbial species, to discriminate those aligned to multiple species and not taxonomically classified in any specific species [27,36,37]. The recall performance was assessed at each indicated read depth for each site by sample or by types of microorganisms.
2.6. Statistical analysis
Besides statistical analyses, comparative analyses were conducted using Wilcoxon Rank Sum tests to evaluate the impact of various factors on the performance scores, e.g. microbial concentration, experimental factors, and bioinformatics tools. Correlations between observed abundance and expected abundance were evaluated by linear regression analyses. A linear mixed model was used to evaluate the effects of various experimental factors on the total variance. The correlation between data quality Q30 and performance scores was evaluated using Pearson's correlation analysis. All data analyses were done using R statistical software. P <0.05 was considered statistically significant, unless otherwise indicated.
2.7. Role of funding source
The funding sources did not have any role in study design, data collection, data analyses, interpretation, or writing of this manuscript.
3. Results
3.1. Study design and overview
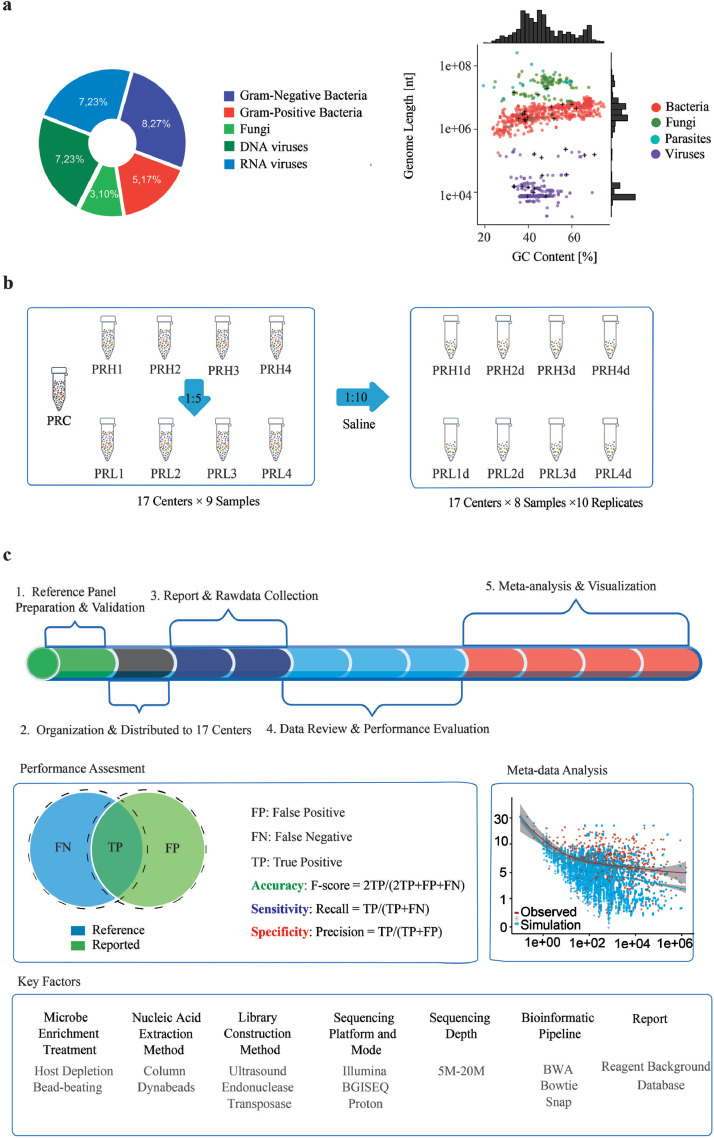
To mimic the biological context of clinical specimens from respiratory or central nervous system infections (BALF and CSF) and enable comprehensive assessments, we designed and constructed a panel of 9 pathogen reference (PR) reagents that covered 30 potentially pathogenic microorganisms of 5 different types (Gram-/+ bacteria, fungi, and DNA/RNA viruses) and included 2 × 10^5/mL human cells as the host background (Supplemental Table S1) [38,39]. These 30 species were comprised of 19 genera, with more than one species intentionally chosen from the same genus of Neisseria and Streptococcus to test the ability of the assays to discriminate closely related microbes (Supplemental Table S1). The panel also included microorganisms with a wide range of genome sizes (from 0.7 Kb to 19.05 Mb) and GC contents (from 33.2% to 70.4%, Fig. 1a, Supplemental Table S3).
Fig. 1.
Design and overview of the study. (a) Thirty microorganisms from 19 genera were chosen to represent the diversity of microbial types (left panel), genome sizes, and GC contents (right panel). (b) Pathogen reference reagents (PRHs and PRLs) were prepared by contriving microbes at high (PRH) and low (PRL) titers with HeLa cells, respectively. PRH and PRL samples were diluted 1:10 and tested in ten replicates except for the pathogen-free PRC sample. PRC details are in the Results section. (c) Overview of study design. Numbers (1-5) order the steps of analysis. F-score, Recall and Precision metrics were used to assess the performance of accuracy, sensitivity, and specificity. Key technical factors of the workflow were explored for their impact on assay performance. See also Supplemental Tables S1–S3, and S5.
Among this reference panel of 9 PR samples, one served as control (pathogen reference control or PRC) and had no contrived microbes. The other 8 samples can be grouped into two sets (PRH1-PRH4 and PRL1-PRL4) or four pairs (e.g. PRH1 and PRL1). Each pair of PR samples comprised the same contrived microorganisms at two different titers. The one in the PRH group had microbes contrived at a 5-fold higher titer compared to their PRL counterpart (Fig. 1b). For instance, PR1H and PR1L both contained the same microorganisms (Escherichia coli K1, Streptococcus pneumoniae, Cryptococcus neoformans, Echovirus 11, Herpes simplex virus 1, Human betaherpesvirus 5, Human herpesvirus 6B), but each microorganism in PR1H was 5-fold greater in titer than in PRL1. Every reference reagent in the PR panel was verified by polymerase chain reaction (PCR)-based methods and distributed to 17 independent laboratory sites (centers C1-C17) for blinded metagenomic testing and bioinformatic analysese (Fig. 1c, Supplemental Methods). These laboratories employed various experimental procedures, bioinformatic pipelines, and sequencing platforms to set up independent metagenomic assays, as detailed in Supplemental Table S2. By breaking down the assay workflow into technical steps, our study included a total of 4 sample preprocessing methods, 2 nucleic acid extraction techniques, 3 library preparation approaches, 6 sequencing platforms, and 4 bioinformatic pipelines for alignment.
To support quantitative assessments, the PR panel was tested undiluted and in 10 replicates at 1:10 dilution, except for the pathogen-free PRC sample. There was a total of 2,641 libraries sequenced (Fig. 1c, Supplemental Table S2, Supplemental Table S4), generating 587 billion reads and 5.51 TB of data. The trial was performed single-blinded. Results along with raw sequencing data were submitted for further analyses. Given the unique, agnostic nature of this assay, we assessed the results using performance metrics including the measures of Recall, Precision, and F-score to indicate the assay sensitivity, specificity, and overall accuracy (Fig. 1c).
3.2. Multicenter assessment of shotgun metagenomics
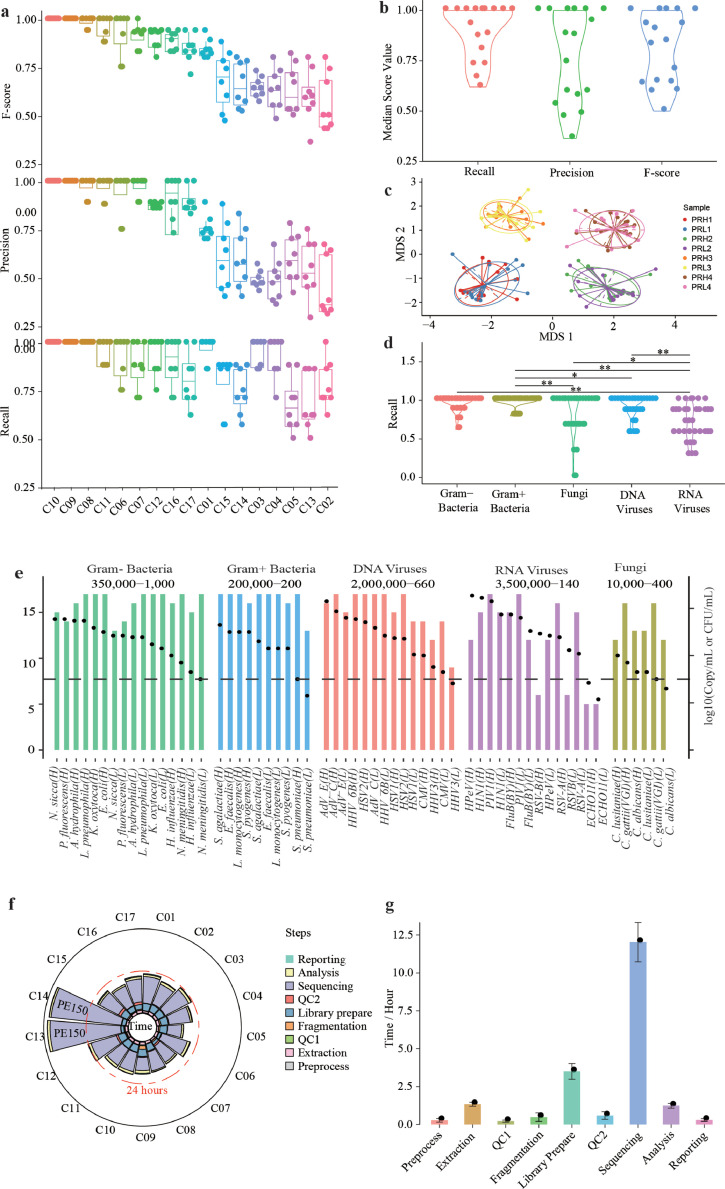
F-scores varied considerably across the 17 laboratory sites, with a range from 0.5-1.0 and an average of 0.81. Although only 2of the 17 sites achieved an overall F-score of 1.0, 59% (10 sites) achieved an F-score of >0.83 (Fig. 2a, B). When analyzed by samples Nearly 40% of sites achieved F-scores of over 0.9, nearly 70% were over 0.75, and only 4% were lower than 0.5 (Supplemental Fig. S1a). Visualization of the similarity in detected microbes demonstrated that results were clearly grouped by the reference sample, despite variances across sites (Fig. 2c).
Fig. 2.
Assessment of assay performance across sites. (a) Summary of assay performance as measured by recall, precision, and F-score by site (n = 136). (b) Summary of assay performance by measure (n = 17). (c) Visualization of the similarity in microbial compositions across sites and reference reagents (n = 136). A nMDS plot of a Bray-Curtis dissimilarity matrix was constructed from the species composition of each reference reagent at each site (n = 136). (d) Summary of assay performance by microbial type, (n = 136), *, P < 0.05, **, P < 0.01 by Wilcoxon rank sum test. (e) Detection of each microorganism in the reference panel. Bacteria and fungi were measured in CFU/ml, and viruses were measured in copies/ml. Columns indicate the number of detection sites, and black dots indicate titers of microorganisms (n = 136). Ranges of abundances of each microbial type are displayed at the top. (f) Assay turn-around times by site (n = 17). (g) Assay turn-around times by step (n = 17). See also Supplemental Figs. S1, S2.
Recall and Precision contributed differentially to the site-to-site variation in F-score (Fig. 2a, Supplemental Fig. S1b). While Recall levels remained relatively consistent (average=0.88, range: 0.75-1.0), Precision varied significantly across sites (average=0.77, range: 0.45-1.0). Similar observations were made at the sample level (Supplemental Fig. S1a). To further dissect the cross-site variation in diagnostic performance, we analyzed the true positive (TP), false positive (FP), and false negative (FN) results at each site. FP results were the most variable across sites, ranging from 0 to 35 counts per site (Supplemental Fig. S1c), while TP and FN results appeared to be relatively consistent, ranging from 21-30 to 0-9 counts, respectively. These results suggest that the overall assay performance across workflows and sites was differentiated more by their ability to reduce FP results, rather than their ability toto reduce FN results. Intriguingly, despite measuring two different aspects of assay performance, we observed a significant positive, instead of negative correlation between Recall and Precision (P=0.013, Wilcoxon rank sum test, Supplemental Fig. S1d).
Among different microbial types, RNA viruses appeared to be the most challenging type to detect, with an average Recall of only 0.71 across all sites, significantly lower than that of other pathogens. Both Gram-positive and Gram-negative bacteria had the highest Recall among all microbial types (0.96 and 0.94), followed by DNA viruses and fungi at 0.89 and 0.80, respectively (Fig. 2d). Similar Recall patterns were observed between PRH and PRL panels (Supplemental Fig. S2a). Most microorganisms at titers above 200 CFU/mL or copies/mL could be detected by >50% of the sites, despite some RNA viruses that were missed at even above 100,000 copies/mL (Fig. 2e). Among all the microorganisms in our panel, fungi, and RNA viruses (including Echovirus 11, Human respiratory syncytical virus B, Human parecho virus, Candida albicans\, and Candida lusitaniae) were the most prevalent causes of FN results (Supplemental Fig. S2b). In line with these findings, the ability to detect fungi and RNA viruses varied widely across sites, whereas the ability to detect Gram-positive and Gram-negative bacteria was relatively consistent (Fig. 2e). These results show the importance of using a reference panel specifically designed for shotgun metagenomics for pathogen detection to cover all microbial types, as many reference reagents for microbiome studies only include bacteria [40].
Fourteen sites had a technical turnaround time (TAT) between 20 and 24 h, ranging from 15.4 to 40.0 h (Fig. 2f, Supplemental Table S5). The sequencing reaction took up the largest portion of the workflow, followed by library construction, nucleic acid extraction, and data analysis, with each constituting 66.6%, 14.3%, 5.9%, and 5.4% of the accumulated TATs, respectively (Fig. 2g, Supplemental Table S5).
3.3. Assay sensitivity depends on microbe: host abundance ratio
In clinical specimens, pathogens almost always exist amid a variable abundance of host cells. Conventional molecular diagnostics, such as PCR-based assays, often work by detecting specific pathogens with limited interference from human or other microorganisms. Unlike these targeted assays, shotgun metagenomic assays involve unbiased analyses of all nucleic acid molecules within a sample. Thus, we posited that both the absolute pathogen abundance and the relative microbe:host abundance ratio may affect assay performance and should be built into the design of the reference reagents.
Since all samples in our reference panel included the same titer of human cells (2 × 10^5/mL), PRL therefore represented a 5-fold higher abundance than PRH in both absolute abundance and relative microbe:host abundance. On the other hand, 1:10 dilution of any sample represented a 10-fold decrease in absolute abundance, and the relative microbe:host abundance remained unchanged compared to its undiluted counterpart (Fig. 1b).
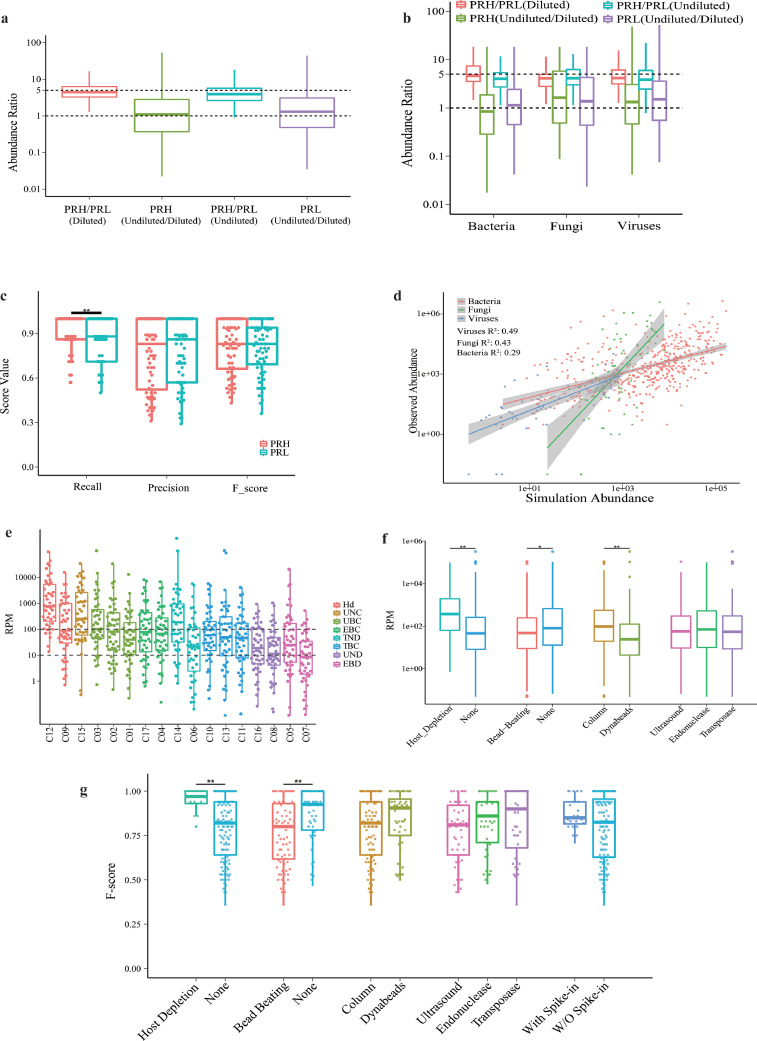
When we compared the observed abundances (as indicated by the number of mapped reads) between PRHs and their PRL counterparts, undiluted and their diluted samples, we saw a 5-fold difference in median observed abundance between PRH and PRL, and 10-fold sample dilution did not result in lower observed abundances (Fig. 3a). Consistent observations were made when bacteria, viruses, and fungi were analyzed separately (Fig. 3b). In agreement with these findings, a lowered relative abundance in PRL resulted in a lower Recall performance (Fig. 3c), while solely reducing the absolute abundance through sample dilution did not significantly affect the performance (Supplemental Fig. S3). These data show that the relative microbe:host abundance ratio, but not absolute microbial abundance, is a key determinant of assay sensitivity by shotgun metagenomics. Therefore, the limit of detection of this assay should be assessed and defined with the relative abundance ratio, rather than the absolute microbial abundance, as used for most conventional assays such as PCR-based diagnostics.
Fig. 3.
Microbial abundance and pathogen detection by metagenomics. (a,b) Observed abundance ratios between PRH and their PRL counterparts, and undiluted samples and their diluted counterparts, analyzed together (a) or by microbial type (b). (c) Correlations between relative abundance and performance (n = 136). (d) Correlations between observed and expected abundances in three microbial types, using the reads per million (RPM) of human papilloma virus within each sample as an internal control for normalization. (e,f) RPM of microbial detection varied across sites grouped by workflow, Hd: Host-deplete, UNC: Ultrasound, None, Column; UBC: Ultrasound, Bead−beating, Column; EBC: Endonuclease, Bead−beating, Column; TND: Transposase, None, Dynabeads; TBC: Transposase, Bead−beating, Column; UND: Ultrasound, None, Dynabeads; EBD: Endonuclease, Bead−beating, Dynabeads. (e) or key technical variables (f). (n = 136), *, P < 0.05, **, P < 0.01 by Wilcoxon rank sum test. See also Supplemental Figs. S3–S5. (g) Correlations between F-score and major workflow-dependent technical variables, (n = 270). **, P < 0.01 by Wilcoxon rank sum test.
3.4. Intra- and cross-site comparison of microbial abundance
We set out to assess the assay's potential in inferring the expected abundance from the number of reads. We defined the expected pathogen abundance in a sample as (pathogen genome size x pathogen titer) / (human genome size x human cell titer) x the total number of clean reads, and the observed abundance as the actual number of reads. We reasoned that for the assay to allow relative quantification of pathogens, there should be a linear correlation between the observed and expected abundances. Linear regression analysese showed significant correlations between the expected and observed abundances, either when all the pathogens were analyzed as a whole or separately according to the types of microbes (P < 0.001, Wilcoxon rank sum test, Fig. 3d). A similar correlation was observed when the abundance of human papilloma virus contained in HeLa cells was used as an internal control for normalization (Supplemental Fig. S4). It was not unexpected that the observed abundance was generally lower than the theoretical expectation (Supplemental Fig. S5), which might reflect the loss of microbial nucleic acids during the experimental processes such as cell wall breaking. The significant correlation between the observed and expected abundances, along with the recovery of the microbe:host ratio, suggested the assay's ability to measure intra-site relative abundance.
As microbial abundance was inferred by the fraction of mapped reads, we wondered if the numbers of mapped reads could be relevant across sites. Numbers of mapped reads per million (RPM) varied significantly across sites, with differences of up to two orders of magnitudes. Such a difference in RPM was not just a result of applying different techniques, as substantial variation was still observed when sites using similar technical workflows were grouped and compared (Fig. 3e). By analyzing each key technical component in the experimental procedures, our data revealed that host depletion and column-based extraction methods were associated with higher RPMs than other technical variables, whereas library preparation by ultrasound, endonuclease, or transposase did not show significant effects on RPM (Fig. 3f). Adaptation of a bead-beating step was associated with a lower RPM, in agreement with its negative correlation with F-score (Fig. 3g). Subgroup analyses further showed potential associations between host depletion and improved Recall among all microbial types, also between bead-beating pretreament and lowered Recall for RNA viruses (Supplemental Fig. S6)
These results suggest that pathogen abundance can be inferred by RPM within each site, but without a way to normalize the “site effect”, cross-site comparisons provided limited information when conducting cross-center evaluation.
3.5. Library effect impacts assay variation
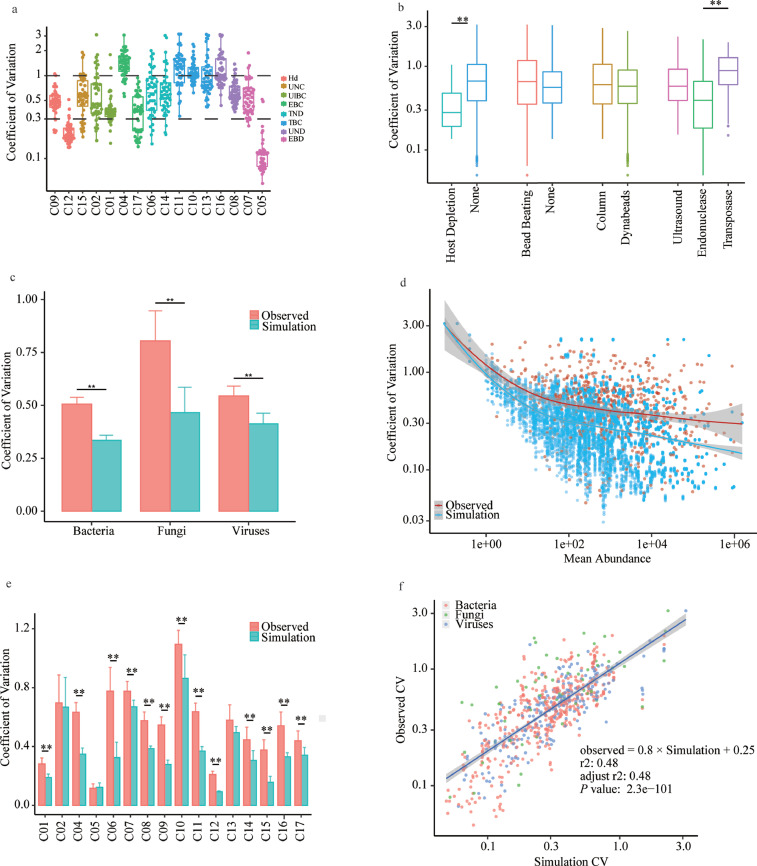
To understand the assay's reproducibility, we took advantage of our large replicated dataset to measure the coefficient of variations (CV) of mapped reads at each site. The average CV was 0.65 and ranged between 0.12 and 1.10; 75% of the sites had CVs below 0.5. This variation remained at a comparable level among sites with similar technical workflows (Fig. 4a). A host depletion step appeared to be associated with a lower CV of 0.28, which might be due to its higher RPM. While no differences were observed in other processes based on cell wall breaking and various nucleic acid extraction methods, endonuclease- and transpose-based library preparation demonstrated the lowest and highest CVs of 0.4 and 1.0 (Fig. 4b), respectively. This finding implies that such preparation was an important step that introduced variances, possibly because transpose-based protocols were more sensitive to changes in the DNA input in a library preparation reaction [41]. We found significantly higher CVs for fungal detection versus bacterial or viral detection (0.80, 0.51, and 0.54, respectively) (P < 0.001, Wilcoxon rank sum test, Fig. 4c).
Fig. 4.
Assessment of assay reproducibility of pathogen metagenomics. (a–c) Coefficients of variance (CV) of microbial mapped reads across sites sorted by workflows (a), or grouped by technical variables (b), or microbial type (c). (d,e) Comparison between the overall CV (observed) and the sampling CV (simulated), with data from all sites analyzed together (d) or individually (e). (f) Linear regression with sampling simulated CV as the independent variable. **, P < 0.01 by Wilcoxon rank sum test. See also Supplemental Table S6.
To examine how much these fluctuations stemmed from read depth-dependent sampling noise, we performed random re-sampling from the pooled reads to represent such a variance and calculated the CVs of these simulated and experimental datasets. As shown in Fig. 4d, the overall CVs were significantly greater than the simulated CVs regardless of pathogen types. This difference in CV was consistent when each laboratory site or microbial type was assessed individually (Fig. 4c, e), suggesting that besides read depth-dependent sampling, other experimental variables also contributed considerably to the observed fluctuations in metagenomic results.
We then attempted to determine how much each of the read depth-dependent variance and other experimental variables contributed to the total variance. We identified a significant linear correlation with an adjusted R2 of 0.48 and a slope of 0.8 (P < 0.001, Wilcoxon rank sum test, Fig. 4f), indicating that both read depth-dependent and experimental variances contributed significantly to overall fluctuation. Among the potential experimental variances, a linear mixed model identified fungal pathogens and transposase-based library construction as significant contributors (Supplemental Table S6), which was consistent with our previous observations.
Our data suggest that these variations should be considered when designing studies to evaluate such an assay, and also imply that precise and quantitative measurement of pathogen abundance by metagenomics will remain challenging until these variations are better understood and controlled.
3.6. Read-depth dependency of assay recall
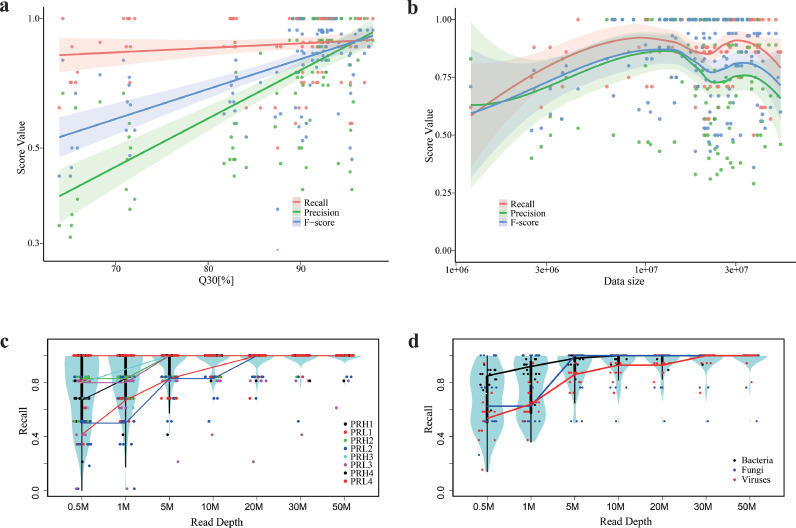
Next, we sought to understand how workflow-dependent technical variables may lead to varied site performance. Among the experimental steps, sample pre-treatment had a greater impact on assay performance compared to nucleic acid extraction, library preparation, or use of internal controls. Preprocessing the samples with host cell depletion was significantly associated with improved F-scores (P < 0.001, Wilcoxon rank sum test). Unexpectedly, a bead-beating step designed for breaking cell walls did not always result in greater performance but was associated with overall reduced F-scores (P < 0.001, Wilcoxon rank sum test). Also, higher Recall scores in all microbial types with host depletion, and lower Recall scores in RNA viruses with bead-beating (Supplemental Fig. S6). Different technical methods for nucleic acid purification and library preparation, different sequencing platforms, and the use of spike-in internal controls were not correlated with overall assay performance (Fig. 3g, Supplemental Fig. S7). Using the Q30 score as a quality indicator, F-score and Precision (but not Recall) were positively correlated with higher sequencing data quality (P < 0.05, Wilcoxon rank sum test, Fig. 5a).
Fig. 5.
Associations between assay performance and major technical variables. (a) Correlations between performance metrics and sequencing quality as inferred by Q30 score. (b) Correlations between site performance metrics and read depth. (c,d) Performance of pathogen detection as measured by Recall as a function of read depth. Data were grouped and analyzed by each reference reagent (c) or microbial type (d). See also Fig. S6–S8.
We further explored the impact of read depth on assay performance. Although initially, the diagnostic performance improved as the read depth increased, further increase in dataset size beyond 10 million did not consistently result in higher scores (Fig. 5b). This observation supports the interpretation that the contribution of read depth to assay performance plateaus after a certain read depth. Leveraging our data, which constitute the deepest sequencing of any sample set yet reported, we next set out to determine the optimal read depth by assessing how well the pathogens in our panel could be detected as a function of read depth. To allow raw data analyses, we chose a CLARK-based pipeline for subsequent site-independent bioinformatics analyses by study coordinating lab because it demonstrated good performance in both simulated and experimental sequencing datasets [30], [31], [32] (Supplemental Fig. S8, and more details in Methods). As shown in Fig. 5c, some pathogens could be detected with only 0.5 million total reads. For instance, site C12 achieved a full Recall of 1.0 at a read depth of 0.5 million in 6 of the 8 PR samples. Nonetheless, when considering the data from all sites, a read depth of 20 million reads enabled detection of most microorganisms in our panel; above that point, benefits from deeper sequencing decreased significantly (Fig. 5c).
We performed sub-analyses by different microbial types of bacteria, fungi, and viruses. The performance of fungal detection plateaued at 5 million reads, while the performance of bacterial and viral detection plateaued at 10 and 20 million reads, respectively (Fig. 5d). These observations were also in line with the interpretation that the sensitivity of shotgun metagenomics decreases as the size of the microbial genome decreases (virus<bacterium<fungus), as smaller genomes result in fewer numbers of nucleic acid fragments that can be sequenced.
These findings suggest read depth as a critical variable that impacts assay Recall when both developing and assessing performance of metagenomic tests. Our results also indicate that although a metagenomic assay requires as few as 0.5 million reads per sample for pathogen detection under optimal conditions, in general, a read depth of 20 million was appropriate under most assay settings.
3.7. Assay precision is challenged by background microbes
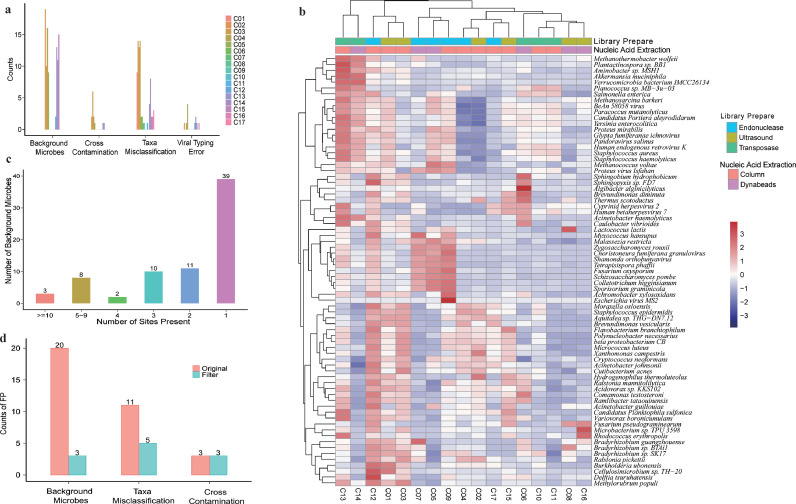
To better understand the causes of FP results, which substantially impacted assay performance, we further categorized the causes of FP results into four groups: cross-contamination, background microorganisms, species misclassification, and viral typing error (Fig. 6a). Among these, background microbes and misclassification of species were the leading causes of FP results (49% and 39%, respectively). These two causes also varied the most among sites (Supplemental Fig. S9).
Fig. 6.
Reducing false-positive results in pathogen metagenomics. (a) Four main causes of false-positive results (n = 17). (b) Patterns of background microbes clustered dependent on methods of library preparation and nucleic acid extraction. (c) Summary of the prevalence of background microbes (n = 17). (d) Improved assay specificity after applying filters to remove false-positive results.
We then sought to evaluate how much taxonomic misclassification could be attributed to the alignment algorithms. To ensure comprehensive microbial coverage, we included 100,000 reads each derived from a total of 108 species comprising 62 bacteria, 42 viruses, and 4 fungi into our simulated dataset and compared the alignment methods employed by the sites in this study (bwa, bowtie, and SNAP) [33], [34], [35] by measuring the percentages of simulated reads that were correctly or incorrectly classified. We found no significant differences at both the genus or species levels, or by microbial type (Supplemental Fig. S10), implying that the alignment method was not a critical performance-differentiating factor.
To gain more insights into FP results caused by background microorganisms, we compared the background patterns from all sites (Fig. 6b). Nine prevalent microorganisms (Staphylococcus haemolyticus, Yersinia enterocolitica, Paracoccus mutanolyticus, Cutibacterium acnes, Malassezia restricta, Human endogenous retrovirus K, Moraxella osloensis, BAV virus and Proteus virus Isfahan) were presented in >5 sites, while others were more site-specific (Fig. 6c). Level of background microbes as represented by their read counts were independent of the reference reagents, but significantly affected by all wet-lab procedures (including sample pretreatment. nucleic acid extraction and library preparation) (Supplemental Table S7). Patterns of these background microbes clustered partially but clearly depended on the methods of library construction (Fig. 6b). These findings imply that microbial backgrounds can be derived from both common and workflow-specific sources, and that addressing such issues to improve assay precision would require site-dependent approaches. Surprisingly, we found that higher levels of background microbes did not associate with lower site performance, as measured by either precision or F-scores (Supplemental Fig. S11), suggesting that to improve assay performance, more efficient data filtering methods would be necessary apart from lowering the background levels.
We explored whether genome coverage and regional sequencing depth (besides read counts) could be informative in discriminating TP from FN. We defined genome coverage as the fraction of genome covered by metagenomic sequencing, and the regional sequencing depth as the total sequencing length divided by the covered genomic fraction. TP results were associated with significantly lower regional sequencing depth and higher genome coverage (Supplemental Fig. S12), which was consistent with the fact that these microorganisms exist in the samples as full and uniform genomes. Similar observations were also made for FN results that were missed originally but discovered by our site-independent bioinformatics pipeline. All FP detections showed significantly lower levels of genome coverage. A significant increase in the level of regional depth was also found in background microbes, implying that they presented as genomic fragments instead of whole microbial cells or genomes in the samples.
Taking all these factors into consideration, we built a precision filter that identified potential FP results through machine learning and applied it to the data derived from site C14, the site with the highest level of FP results. Integrating RPM, RPM ratio (sample:control), and genome coverage, our method reduced FP results from 34 to 11 counts (Fig. 6d). Importantly, applying such a filter did not compromise Recall, suggesting a potential strategy for improving the precision of shotgun metagenomics for pathogen detection.
4. Discussion
In this 17-site study, nine reference samples that mimicked the context of clinical specimens respiratory or central nervous system infections were profiled with various workflows. The data presented here provide one of the deepest assessments of shotgun metagenomics to date.
Currently, shotgun metagenomics is mostly applied for acute and severe infections that cannot be diagnosed by conventional approaches, under clinical scenarios that are highly sensitive to testing TAT [5,7,[42], [43], [44], [45], [46], [47]]. Our study showed that read length of SE75 was sufficient to achieve high or even full F-scores, and longer read length such as PE150 did not always result in high performance (Supplemental Fig. S13). Considering sequencing as the major time-consuming step in the assay (Fig. 2f, g), we recommend a read length of no longer than 75 bp when developing metagenomic assays, which could serve as an effective strategy to reduce TAT. Other factors that affect real-world TAT (e.g. such as sample logistics, testing volume, skillfulness of the technicians) should also be optimized. Lowering the cost of assays may facilitate wider application of shotgun metagenomics [48]. Given the wide range of pathogen abundance in clinical specimens, false positive results could also stem from index miss assignment. Such an issue may vary depending on sequencing technologies and should be considered when establishing an assay [49].
Using our reference reagents which mimicked the clinical specimens from respiratory or central nervous system infections [27,39,50,51], a read depth of 20M generally sufficient and cost-efficient for pathogen detection [52]. However, read depth used should be carefully considered when developing the assay for other sample types, or atypical specimens such as those with higher abundance of host cells.
Abundance of human cells was a critical factor affecting pathogen detection by metagenomics. Our data supported a mathematical model in which a sample comprising 10^5/mL each of human cells and bacterial cells would only yield 0.1% of total reads mapped to the bacteria, assuming a human genome of 3Gb and a bacterial genome of 3Mb [16,39]. Therefore, sensitivity of shotgun metagenomic assays is affected by the host nucleic acids, and therefore varied across samples [16]. Host depletion is highly valuable in improving assay sensitivity via increasing the relative microbial abundance. A variety of approaches have been reported for host cell depletion [53], [54], [55], [56]. However, thorough validation is needed before applying these methods to ensure that pathogens are not unintentionally removed along with the host cells. Indeed, in a previous study, differential lysis could significantly reduce human cells but at the same time compromise the detection of viral and certain bacterial pathogens [54]. Additionally, HPV-18 (known to be contained in HeLa cells) was detected by our metagenomic assays, illustrating that integration of microbial sequences in the host genome can also be a source of false-positive results and should be carefully analyzed.
By including various types of pathogens and human cells, our reference panel represents the common context of clinical specimens, such as cerebrospinal fluid and bronchoalveolar lavage fluid, where infiltration of immune cells is often found with occurring lying infection. Although sharing common characteristics, our reference samples may not precisely represent plasma specimens where human cell-free nucleic acids are believed to be more prevalent [3]. For instance, it remains to be determined whether assay variation would be influenced by the cell wall-breaking step, and how each library preparation method fits in the context of cell-free nucleic acids. We spiked in multiple microorganisms in each reference sample to achieve more comprehensive assessments. Although this might differ from scenarios of mono-microbial infections, we did not expect much interference among microbes based on the fact that microbial DNA normally accounted for only a very small portion (<1%) of our reference samples as well as in clinical specimens.
There are limitations of our study. As a complexed assay, the performance of a shotgun metagenomic assay depends on affected by many technical variables. By benchmarking workflows used across 17 laboratories, we aimed to evaluate their differences, to provide insight into which technical processes of the assay might need improvements, and to pave the way towards increased quality and developing common best practices for clinical metagenomics. As the workflows assessed in our trial were developed independently, further studies designed with single-variable, parallel experiments of a fixed workflow and laboratory would be needed to validate the impact of each individual technical process.
Another limitation of our study is that there was only a 5-fold decrease in the microbial abundance from the PRH to the PRL samples. Although significantly lowered F-scores were observed in PRL, adding lower-titered samples would be valuable in representing the substantial variation in pathogen concentration. Unlike other targeted assays, shotgun metagenomics is also unique in its potential for unbiased detection of novel pathogenic microbes, as was shown in the discovery of COVID-19 [8], [9], [10], [11], [12]. This unbiased detection also heavily depends on bioinformatics analyses to discriminate between novel and previously identified pathogens [8,57,58], as well as closely related ones, for instance, between SARS-CoV-2 and SARS-CoV. Evaluating such an unusual aspect of assay performance would require new designs of the reference reagents that represent potential novel species.
Data in this study included sequencing results generated from different platforms and workflows using the same set of reference samples. This information is a unique resource that could be valuable for the development and optimization of bioinformatics pipelines for rapid pathogen detection. Current bioinformatics pipelines mostly rely on the number of mapped reads for pathogen identification [59], [60], [61]. With our dataset, more sophisticated identification algorithms could be explored by integrating more variables, such as genome coverage and phylogenetic relationships, to improve specificity. These data also provide a general overview of the current performance of shotgun metagenomics, which could aid in establishing regulatory or technical references.
Contributors
YC Wang and CT Zhang conceived, designed and supervised the experiments; DL Liu, T Xu, HW Zhou, QW Yang, X Mo and YL Wu wrote the manuscript; DW Shi, JW Ai, JJ Zhang, Y TAO, DH Wen, YG Tong, LL Ren, W Zhang, SM Xie, WJ Chen, WL Xing, JY Zhao, YL Wu, XF Meng, C Ouyang, Z Jiang, ZK Liang, HQ Tan, Y Fang, N Qin, YL Guan, and W Gai performed the experiments. DL Liu, T Xu, HW Zhou, QW Yang, X Mo, YL Wu and YC Wang verified the underlying data. All of the authors have read and approved the final manuscript. SH Xu, WJ Wu and WH Zhang helped with design of experiments, supervising a specific platform, and proofreading the manuscript.
Data sharing statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Following complete publication, the data generated in this study are available to researchers through GISAID accession ID CNP0001292.
Declaration of Competing Interest
SM Xie, WJ Chen, JY Zhao, YL Wu, XF Meng, C Ouyang, Z Jiang, ZK Liang, HQ Tan, Y Fang, N Qin, YL Guan, and W Gai were employed by Vision Medicals Center for Infectious Diseases, BGI PathoGenesis Pharmaceutical Technology, Dalian GenTalker Clinical Laboratory, Guangzhou Sagene Biotech Co., Ltd., Guangzhou Kingmed Diagnostics, Hangzhou MatriDx Biotechnology Co., Ltd, Genskey Medical Technology, Co., Ltd., Guangzhou Darui Biotechnology, Co., Ltd., Hangzhou IngeniGen XunMinKang Biotechnology Co., Ltd., Dinfectome Inc, Realbio Genomics Institute, Hugobiotech Co., Ltd., WillingMed Technology (Beijing) Co., Ltd., respectively, outside the submitted work. WL Xing was employed by School of Medicine Tsinghua University and CapitalBio Technology Co., Ltd.
Acknowledgments
This work was supported by National Science and Technology Major Project of China (2018ZX10102001).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103649.
Contributor Information
Sihong Xu, Email: xushong@nifdc.org.cn.
Wenjuan Wu, Email: wwj3289@easthospital.cn.
Wenhong Zhang, Email: zhangwenhong@fudan.edu.cn.
Chuntao Zhang, Email: zhangchuntao@nifdc.org.cn.
Youchun Wang, Email: wangyc@nifdc.org.cn.
Appendix. Supplementary materials
References
- 1.Barlam T.F., Cosgrove S.E., Abbo L.M., et al. Implementing an antibiotic stewardship program: guidelines by the infectious diseases society of america and the society for healthcare epidemiology of America. Clin Infect Dis. 2016;62(10):e51–e77. doi: 10.1093/cid/ciw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liesenfeld O., Lehman L., Hunfeld K.P., Kost G. Molecular diagnosis of sepsis: New aspects and recent developments. Eur J Microbiol Immunol (BP) 2014;4(1):1–25. doi: 10.1556/EuJMI.4.2014.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blauwkamp T.A., Thair S., Rosen M.J., et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 4.Ye S.H., Siddle K.J., Park D.J., Sabeti P.C. Benchmarking metagenomics tools for taxonomic classification. Cell. 2019;178(4):779–794. doi: 10.1016/j.cell.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson M.R., Naccache S.N., Samayoa E., et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370(25):2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simner P.J., Miller H.B., Breitwieser F.P., et al. Development and optimization of metagenomic next-generation sequencing methods for cerebrospinal fluid diagnostics. J Clin Microbiol. 2018;56(9) doi: 10.1128/JCM.00472-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong G.L., MacCannell D.R., Taylor J., et al. Pathogen genomics in public health. N Engl J Med. 2019;381(26):2569–2580. doi: 10.1056/NEJMsr1813907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulut C., Kato Y. Epidemiology of COVID-19. Turk J Med Sci. 2020;50(Si-1):563–570. doi: 10.3906/sag-2004-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic
- 11.Zhang H., Ai J.W., Yang W., et al. Metatranscriptomic characterization of COVID-19 identified a host transcriptional classifier associated with immune signaling. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. 271-80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber S., Spiegl B., Perakis S.O., et al. Technical evaluation of commercial mutation analysis platforms and reference materials for liquid biopsy profiling. Cancers. 2020;12(6) doi: 10.3390/cancers12061588. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salk J.J., Schmitt M.W., Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018;19(5):269–285. doi: 10.1038/nrg.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng D.T., Mitchell T.N., Zehir A., et al. Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westermann A.J., Gorski S.A. Vogel. dual RNA-seq of pathogen and host. Nat Rev Microbiol. 2012;10(9):618–630. doi: 10.1038/nrmicro2852. [DOI] [PubMed] [Google Scholar]
- 17.Consortium TIHiRN The integrative human microbiome project. Nature. 2019;569(7758):641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fettweis J.M., Serrano M.G., Brooks J.P., et al. The vaginal microbiome and preterm birth. Nat Med. 2019;25(6):1012–1021. doi: 10.1038/s41591-019-0450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd-Price J., Arze C., Ananthakrishnan A.N., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Junier T., Huber M., Schmutz S., et al. Viral metagenomics in the clinical realm: lessons learned from a swiss-wide ring trial. Genes. 2019;10(9) doi: 10.3390/genes10090655. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Vries J.J.C., Brown J.R., Fischer N., et al. Benchmark of thirteen bioinformatic pipelines for metagenomic virus diagnostics using datasets from clinical samples. J Clin Virol. 2021;141 doi: 10.1016/j.jcv.2021.104908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi L., Reid L.H., Jones W.D., et al. The microarray quality control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium SMI. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the sequencing quality control consortium. Nat Biotechnol. 2014;32(9):903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi L., Campbell G., Jones W.D., et al. The microarray quality control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28(8):827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu L. RNA-Seq reproducibility assessment of the sequencing quality control project. Cancer Inform. 2020;19 doi: 10.1177/1176935120922498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z.Y., Dang D., Wu H. Next-generation sequencing of cerebrospinal fluid for the diagnosis of unexplained central nervous system infections. Pediatr Neurol. 2021;115:10–20. doi: 10.1016/j.pediatrneurol.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Miller S., Naccache S.N., Samayoa E., et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genom Res. 2019;29(5):831–842. doi: 10.1101/gr.238170.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing X.W., Zhang J.T., Ma Y.B., et al. Metagenomic next-generation sequencing for diagnosis of infectious encephalitis and meningitis: a large, prospective case series of 213 patients. Front Cell Infect Microbiol. 2020;10:88. doi: 10.3389/fcimb.2020.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domagała-Kulawik J., Skirecki T., Maskey-Warzechowska M., Grubek-Jaworska H., Chazan R. Bronchoalveolar lavage total cell count in interstitial lung diseases–does it matter? Inflammation. 2012;35(3):803–809. doi: 10.1007/s10753-011-9378-5. [DOI] [PubMed] [Google Scholar]
- 30.Kim D., Song L., Breitwieser F.P., Salzberg S.L. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genom Res. 2016;26(12):1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genom Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ounit R., Wanamaker S., Close T.J., Lonardi S. CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genom. 2015;16(1):236. doi: 10.1186/s12864-015-1419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korf I. Gene finding in novel Genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlaberg R., Chiu C.Y., Miller S., Procop G.W., Weinstock G. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141(6):776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 37.Zhang F., Chen J., Huang H., et al. Application of metagenomic next-generation sequencing in the diagnosis and treatment guidance of pneumocystis Jirovecii pneumonia in renal transplant recipients. Eur J Clin Microbiol Infect Dis. 2021;40(9):1933–1942. doi: 10.1007/s10096-021-04254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahlbrecht J., Hillebrand L.K., Schwenkenbecher P., et al. Cerebrospinal fluid features in adults with enteroviral nervous system infection. Int J Infect Dis. 2018;68:94–101. doi: 10.1016/j.ijid.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 39.Wilson M.R., Sample H.A., Zorn K.C., et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380(24):2327–2340. doi: 10.1056/NEJMoa1803396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amos G.C.A., Logan A., Anwar S., et al. Developing standards for the microbiome field. Microbiome. 2020;8(1):98. doi: 10.1186/s40168-020-00856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Picelli S., Björklund A.K., Reinius B., Sagasser S., Winberg G., Sandberg R. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24(12):2033–2040. doi: 10.1101/gr.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M., Yang F., Lu Y., Huang W. Identification of Enterococcus faecalis in a patient with urinary-tract infection based on metagenomic next-generation sequencing: a case report. BMC Infect Dis. 2020;20(1):467. doi: 10.1186/s12879-020-05179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann B., Tappe D., Höper D., et al. A variegated squirrel bornavirus associated with fatal human encephalitis. N Engl J Med. 2015;373(2):154–162. doi: 10.1056/NEJMoa1415627. [DOI] [PubMed] [Google Scholar]
- 44.Gu L., Liu W., Ru M., et al. The application of metagenomic next-generation sequencing in diagnosing chlamydia psittaci pneumonia: a report of five cases. BMC Pulm Med. 2020;20(1):65. doi: 10.1186/s12890-020-1098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang M., Weng X., Chen L., et al. Fulminant central nervous system varicella-zoster virus infection unexpectedly diagnosed by metagenomic next-generation sequencing in an HIV-infected patient: a case report. BMC Infect Dis. 2020;20(1):159. doi: 10.1186/s12879-020-4872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Z., Zhang C., Li W., et al. Metagenomic next-generation sequencing contribution in identifying prosthetic joint infection due to Parvimonas micra: a case report. J Bone Jt Infect. 2019;4(1):50–55. doi: 10.7150/jbji.30615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L., Liu W., Zhang Q., et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9(1):313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiu C.Y., Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Illumina https://www.illumina.com/content/dam/illumina-marketing/documents/products/whitepapers/index-hopping-white-paper-770-2017-004.pdf?linkId=36607862
- 50.Langelier C., Kalantar K.L., Moazed F., et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci U S A. 2018;115(52):E12353–E12e62. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodhead M., Blasi F., Ewig S., et al. Guidelines for the management of adult lower respiratory tract infections–full version. Clin Microbiol Infect. 2011;17(Suppl 6):E1–59. doi: 10.1111/j.1469-0691.2011.03672.x. Suppl 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oyola S.O., Gu Y., Manske M., et al. Efficient depletion of host DNA contamination in malaria clinical sequencing. J Clin Microbiol. 2013;51(3):745–751. doi: 10.1128/JCM.02507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu W., Crawford E.D., O'Donovan B.D., et al. Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17:41. doi: 10.1186/s13059-016-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson M.T., Pope C.E., Marsh R.L., et al. Human and extracellular DNA depletion for metagenomic analysis of complex clinical infection samples yields optimized viable microbiome profiles. Cell Rep. 2019;26(8) doi: 10.1016/j.celrep.2019.01.091. 2227-40.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marotz C.A., Sanders J.G., Zuniga C., Zaramela L.S., Knight R., Zengler K. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome. 2018;6(1):42. doi: 10.1186/s40168-018-0426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matranga C.B., Andersen K.G., Winnicki S., et al. Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol. 2014;15(11):519. doi: 10.1186/s13059-014-0519-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ai J.W., Zhang Y., Zhang H.C., Xu T., Zhang WH. Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg Microbes Infect. 2020;9(1):597–600. doi: 10.1080/22221751.2020.1738905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren L.L., Wang Y.M., Wu Z.Q., et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020;133(9):1015–1024. doi: 10.1097/CM9.0000000000000722. (Engl) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Breitwieser F.P., Baker D.N., Salzberg SL. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018;19(1):198. doi: 10.1186/s13059-018-1568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang D.D., Froula J., Egan R., Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corvelo A., Clarke W.E., Robine N., Zody M.C. taxMaps: comprehensive and highly accurate taxonomic classification of short-read data in reasonable time. Genome Res. 2018;28(5):751–758. doi: 10.1101/gr.225276.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.