Abstract
The nuclear body is a multiprotein complex that may have a role in the regulation of gene transcription. This structure is disrupted in a variety of human disorders including acute promyelocytic leukemia and viral infections, suggesting that alterations in the nuclear body may have an important role in the pathogenesis of these diseases. In this study, we identified a cDNA encoding a leukocyte-specific nuclear body component designated Sp110. The N-terminal portion of Sp110 was homologous to two previously characterized components of the nuclear body (Sp100 and Sp140). The C-terminal region of Sp110 was homologous to the transcription intermediary factor 1 (TIF1) family of proteins. High levels of Sp110 mRNA were detected in human peripheral blood leukocytes and spleen but not in other tissues. The levels of Sp110 mRNA and protein in the human promyelocytic leukemia cell line NB4 increased following treatment with all-trans retinoic acid (ATRA), and Sp110 localized to PML-Sp100 nuclear bodies in ATRA-treated NB4 cells. Because of the structural similarities between Sp110 and TIF1 proteins, the effect of Sp110 on gene transcription was examined. An Sp110 DNA-binding domain fusion protein activated transcription of a reporter gene in transfected mammalian cells. In addition, Sp110 produced a marked increase in ATRA-mediated expression of a reporter gene containing a retinoic acid response element. Taken together, the results of this study demonstrate that Sp110 is a member of the Sp100/Sp140 family of nuclear body components and that Sp110 may function as a nuclear hormone receptor transcriptional coactivator. The predominant expression of Sp110 in leukocytes and the enhanced expression of Sp110 in NB4 cells treated with ATRA raise the possibility that Sp110 has a role in inducing differentiation of myeloid cells.
The nuclear body (also known as nuclear domain 10, promyelocytic leukemia protein [PML] oncogenic domain, and Kr body) is a cellular structure that appears to be involved in the pathogenesis of a variety of human diseases including acute promyelocytic leukemia and acute viral infections. In addition, the nuclear body is a target of antibodies in the serum of patients with the autoimmune disease primary biliary cirrhosis (reviewed in references 17, 31, and 40). By immunohistochemical staining, nuclear bodies appear as 5 to 30 discrete, punctate regions within the nucleus. The number of nuclear bodies in the cell and the intensity of antibody staining of these structures increase in response to heat shock and viral infection, as well as exposure to interferons (IFNs) and heavy metals (3).
Although the exact role of the nuclear body in cellular biology is unknown, recent studies suggest that this structure is involved in the regulation of gene transcription. LaMorte and colleagues used an in vivo nucleic acid labeling technique to demonstrate that nascent RNA polymerase II transcripts are present near the nuclear body (23). In addition, Ishov et al. demonstrated that the nuclear body is a preferred site for transcription of viral genes (18).
A nuclear body component designated PML was identified by characterization of the t(15;17) translocation associated with acute promyelocytic leukemia (6, 9, 21, 28). In the t(15;17) translocation, the N-terminal portion of PML is fused to retinoic acid (RA) receptor α (RARα). Expression of the PML-RARα fusion protein disrupts the nuclear body, and nuclear body antigens are redistributed to numerous smaller regions in the nucleus designated “microspeckles.” Treatment of promyelocytic leukemia cells with all-trans RA (ATRA) degrades the PML-RARα fusion protein, resulting in reformation of nuclear bodies and differentiation of leukemic cells. PML has an important role in several cellular processes including regulation of cellular growth (45) and mediation of pathways of apoptosis (34, 44). Doucas et al. demonstrated that PML recruits cyclic AMP response element-binding protein (CREB)-binding protein (CBP) to the nuclear body and that PML can function as a potent nuclear hormone receptor coactivator (11).
Autoantibodies in the serum of patients with primary biliary cirrhosis were used to identify a cDNA encoding nuclear body component Sp100 (speckled, 100 kDa) (42). Two additional splice variants of Sp100 (designated Sp100b and Sp100-HMG) were subsequently reported (8, 27, 37). The Sp100 proteins interact with members of the heterochromatin protein 1 (HP1) family of nonhistone chromosomal proteins. When bound to a nonhistone promoter in transfected cells, the Sp100 proteins behave as transcriptional repressors. These observations suggest that the nuclear body in general, and the Sp100 proteins in particular, may have a role in the maintenance of chromatin architecture and in the regulation of gene transcription (27, 37).
In a previous study, we used serum from patients with primary biliary cirrhosis to identify a leukocyte-specific component of the nuclear body designated Sp140 (5). The N-terminal portion of the Sp140 sequence is homologous to the N-terminal segment in the Sp100 proteins. The middle region of Sp140 contains an amino acid sequence motif of unknown function designated a SAND domain (13). The C-terminal portion of Sp140 contains a plant homeobox domain and bromodomain and is homologous to the carboxyl portions of nuclear hormone transcription intermediary factors 1α (TIF1α), -β and -γ (25, 26, 43). When expressed in resting cells, Sp140 associated with PML-Sp100 nuclear bodies. In cells stimulated with IFN-γ, the number of PML-Sp100 nuclear bodies in each cell increased, but Sp140 associated with only a subset of these structures (4). When fused to a DNA-binding domain and transfected with a reporter plasmid into mammalian cells, Sp140 enhanced expression of a reporter gene. Taken together, these observations suggest that Sp140 may define a subset of nuclear bodies in a cell and that Sp140 may contribute to the activation of gene transcription (4).
The objective of this study was to further investigate the structure and function of the nuclear body. We observed in the National Center for Biotechnology Information database of expressed sequence tag (EST) sequences a partial cDNA that predicted an amino acid sequence that was similar to those of the N-terminal domains of Sp100 and Sp140. We used the DNA fragment to clone a full-length cDNA encoding a 110-kDa protein designated Sp110. We demonstrate that Sp110 is a leukocyte-specific component of the nuclear body and that Sp140 can recruit Sp110 to this structure. In addition, we show that Sp110 can function as an activator of gene transcription and that this protein may serve as a nuclear hormone receptor coactivator.
MATERIALS AND METHODS
Isolation and characterization of cDNA clones encoding Sp110.
A nucleotide sequence in the EST database was noted to encode a polypeptide with significant homology to the N-terminal portions of Sp100 and Sp140, and the cDNA was obtained from the IMAGE consortium (accession no. AA431918). Because the cDNA was found to be highly contaminated with unrelated cDNAs, oligonucleotides (5′-TTGAATTCATGGAAGAGGCTCTTTTTCAG-3′ and 5′-TTGAATTCCTTCTGCTAGGCCAGTTGG-3′) were prepared based on the sequence of the EST clone and PCR was used to synthesize a fragment of the cDNA. The PCR product was radiolabeled and used to screen a λGT10 cDNA library prepared from human spleen (Clontech, Palo Alto, Calif.). Six cDNA clones from among approximately one million bacteriophages hybridized with the radiolabeled probe and were isolated by plaque purification. Bacteriophage growth, DNA isolation, and subcloning into pUC19 were performed using standard procedures (35). The nucleotide sequence of the full-length cDNA was determined by the dideoxy chain termination method (36).
Plasmids.
A plasmid encoding Sp110 fused to the DNA-binding domain of GAL4 (amino acids 1 to 147) was prepared by ligating a cDNA encoding Sp110 into expression plasmid pBXG (pBXG-Sp110). A plasmid encoding chloramphenicol acetyltransferase (CAT) under the control of five GAL-4 binding sites, a simian virus 40 (SV40) enhancer, and an E1b TATA promoter (pG5SV-BCAT) was used as a reporter construct. Plasmids pBXG and pG5SV-BCAT were kindly provided by S. Shu and J. Bonventre (Massachusetts General Hospital, Boston, Mass.). A plasmid encoding PML was a kind gift from H. de Thé (Hopital St. Louis, Paris, France). The reporter plasmid (RARβ)3-tk-luc contains the luciferase gene under the control of a minimal herpes simplex virus thymidine kinase promoter and three copies of the RARα response element from the human RARβ promoter.
Construction of an E1-deleted, recombinant adenovirus vector containing Sp110.
The cDNA encoding Sp110 was cloned into the NotI site of pAd.RSV4 (provided by D. Dichek, Gladstone Institute for Cardiovascular Diseases, San Francisco, Calif.), which contains the Rous sarcoma virus long terminal repeat promoter and the SV40 polyadenylation signal. The plasmid containing Sp110 was cotransfected into 293 cells with pJM17 (provided by F. L. Graham, McMaster University, Hamilton, Ontario, Canada). Homologous recombination between the two plasmids resulted in an adenovirus (Ad.Sp110) that contained Sp110 sequences in place of E1 sequences. Recombinant viruses in a plaque were amplified in 293 cells, and a high-titer stock was prepared, as described previously (14). The absence of replication-competent adenovirus in the viral stock was confirmed by the failure of Ad.Sp110 to produce cytopathic changes in A549 lung carcinoma cells. In addition, PCR failed to amplify a DNA fragment corresponding to the E1 region of adenovirus using oligonucleotides and the Ad.Sp110 stock. An adenovirus vector containing the cDNA encoding Sp140 was described previously (4).
Human autoantibodies directed against nuclear body components and production of rat antiserum directed against Sp110.
Antibodies in serum from patient F111 with primary biliary cirrhosis were used to identify nuclear bodies in HEp-2 cells. The diagnosis of primary biliary cirrhosis in this patient was based on the presence of elevated liver function enzymes, high-titer antibodies directed against the mitochondrial antigen E2 pyruvate dehydrogenase complex, and characteristic histological findings in a liver biopsy specimen (22). F111 serum was previously shown to contain antibodies directed against Sp100 but lacked antibodies directed against Sp140 (4). Human, affinity-purified antibodies directed against PML were prepared as previously described (4).
To produce antibodies directed against Sp110, three male Sprague-Dawley rats were immunized with recombinant protein containing Sp110 amino acids 219 to 435 fused to glutathione S-transferase. The plasmid encoding this portion of Sp110 was prepared by ligating a BstYI/EcoRV restriction fragment of the cDNA encoding Sp110 into the BamHI/SmaI sites of pGEX (Pharmacia Biotech, Inc., Piscataway, N.J.). The plasmid was used to transform Escherichia coli, and expression of the fusion protein was induced by treatment with isopropyl-1-thio-β-d-galactopyranoside. The fusion protein was purified from E. coli proteins as described by Smith and Johnson (39). Primary immunization of three rats was performed using 50 μg of purified protein emulsified in complete Freund's adjuvant for each animal. Two subsequent booster injections consisting of 50 μg of protein were given at 2-week intervals.
Cell culture, induction of differentiation, and immunohistochemical staining.
HL60 cells, 293 cells, and HEp-2 cells were obtained from the American Type Culture Collection (Manassas, Va.). NB4 cells were a gift from M. Lanotte (Institut National de la Sante et Recherche Medicale, Paris, France). HL60 cells were maintained in RPMI medium supplemented with 10% fetal calf serum, l-glutamine (2 mM), penicillin (200 U/ml), and streptomycin (200 mg/ml). Human 293 cells were grown in low-glucose (1 g/liter) Dulbecco modified Eagle medium (DMEM) supplemented with 10% horse serum, and HEp-2 cells were maintained in high-glucose (4.5 g/liter) DMEM supplemented with 10% fetal calf serum.
The effect of IFN on the expression of Sp110 in HL60 cells was determined by incubating these cells in the presence of IFN-γ (200 U/ml). Differentiation of NB4 cells was induced by treatment for 48 h with ATRA (1 μM).
To prepare nonadherent cells for immunohistochemical staining, approximately 50,000 cells were subjected to cytospin centrifugation at 500 rpm for 5 min, followed by air drying and fixation in 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 15 min. Prior to incubation with antibodies, cells were permeabilized with 0.1% saponin in PBS and then treated with 0.6% hydrogen peroxide in 60% methanol to block endogenous peroxidase activity. Cells were stained with rat anti-Sp110 antibodies and biotinylated goat anti-rat immunoglobulin G (IgG) antiserum. Cells were subsequently incubated with horseradish peroxidase (HRP)-conjugated avidin-biotin complexes (ABC) (Vectastain Elite ABC kit; Vector Laboratories) and were exposed to 3,3′-diaminobenzimide, which resulted in brown staining.
For immunofluorescent staining, HEp-2 cells were grown in tissue culture chambers (Nunc Inc., Naperville, Ill.), fixed in 4% paraformaldehyde in PBS at room temperature for 10 min, and permeabilized by treatment with methanol for 7 min. Rat anti-Sp110 antiserum and human antibodies were incubated with substrate for 1 h at room temperature. Unbound antibodies were removed by three successive washes with PBS. Bound rat antibodies were detected using species-specific fluorescein isothiocyanate-conjugated donkey anti-rat IgG antiserum (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.). Bound human antibodies were detected using species-specific, Texas red isothiocyanate-conjugated, donkey anti-human IgG Fc antiserum (Jackson ImmunoResearch Laboratories, Inc.).
SDS-polyacrylamide gel electrophoresis and immunoblotting.
HEp-2 cells were lysed in cold PBS containing phenylmethylsulfonyl fluoride (1 mM), leupeptin (2 μM), and pepstatin A (1 μM). Cellular extracts were fractionated in an SDS–8% polyacrylamide gel and transferred to nitrocellulose membranes. Membranes were incubated in blocking solution (PBS containing 5% nonfat dry milk) and then with rat antiserum diluted 1:1,000 in blocking solution. Bound rat antibodies were detected using HRP-conjugated goat anti-rat antiserum (Amersham) and chemiluminescence.
RNA blot hybridization.
The level of Sp110 mRNA in human tissues was determined by hybridizing membranes containing 2.5 μg of poly(A)+-selected RNA from human tissues (multiple-tissue Northern blots; Clontech Laboratories) with a 32P-radiolabeled 1.4-kb XbaI restriction fragment of the Sp110 cDNA. Membranes were washed under stringent conditions and exposed to autoradiography for 1 h. To confirm the presence of poly(A)+-selected RNA in each lane, the membranes were hybridized with a 32P-radiolabeled β-actin cDNA. The membranes were washed under stringent conditions and exposed to autoradiography for 30 min.
RNA was extracted from human cell lines HL60 and NB4 using the guanidinium isothiocyanate-cesium chloride method (35). RNA was fractionated in formaldehyde-agarose gels (5 μg/lane), and equal loading of RNA was confirmed by staining 28S and 18S rRNA with ethidium bromide. RNA was transferred to nylon membranes, and membranes were hybridized with the radiolabeled XbaI restriction fragment of the Sp110 cDNA or the EcoRI/BamHI restriction fragment of the cDNA encoding human Sp100 (5). Membranes were washed and subjected to autoradiography.
Mammalian cell transfection and reporter assays.
COS cells were transfected using the SuperFect transfection system (Qiagen, Inc., Valencia, Calif.). Forty-eight hours after transfection, cells were washed twice with PBS, and cell extracts were prepared and assayed for CAT or luciferase activity, as described previously (2, 33).
A plasmid encoding growth hormone was included in each transfection for normalization of transfection efficiencies. The concentration of growth hormone in tissue culture medium 48 h after transfection was determined using a commercial radioimmunoassay kit (Nichols Institute, San Juan Capistrano, Calif.).
RESULTS
Isolation and characterization of a cDNA encoding Sp110.
We observed in the EST database a nucleotide sequence encoding a polypeptide fragment with significant amino acid sequence homology with the N-terminal regions of nuclear body components Sp100 and Sp140. Nucleotide sequences derived from the EST clone were used to screen a λgt10 cDNA library prepared from human spleen. Six cDNA clones encoding portions of Sp110 were isolated, sequenced, and assembled to prepare a full-length Sp110 cDNA (GenBank accession no. AF280094).
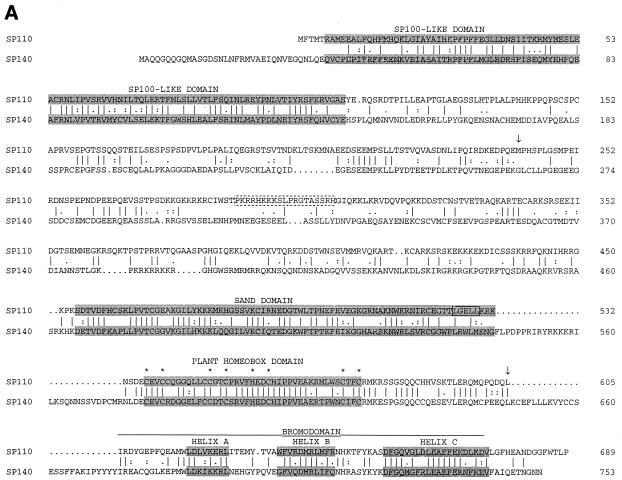
The cDNA encoding Sp110 was 2,336 bp in length with an open reading frame from nucleotides 78 to 2146 encoding a protein containing 689 amino acids (Fig. 1A). The start codon was preceded by an in-frame stop codon, suggesting that this was a full-length cDNA. Amino acids 241 to 605 of Sp110 were almost identical to amino acids 1 to 365 of a previously reported polypeptide designated nuclear phosphoprotein 72 (20). In this region, the amino acid sequence of Sp110 differed from that of nuclear phosphoprotein 72 at amino acid 523.
FIG. 1.
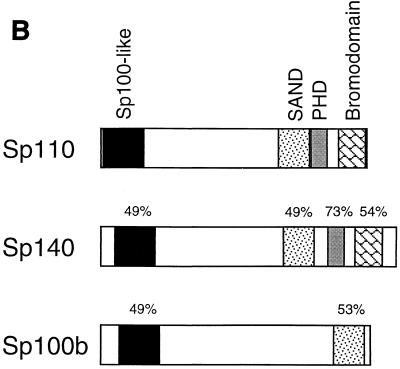
(A) The predicted amino acid sequence of Sp110 and comparison with Sp140. Sp110 has a modular structure that includes the “Sp100-like domain,” SAND domain, plant homeobox domain, and bromodomain (shaded). Dashed box, presumed nuclear localization sequence in Sp110, between amino acids 288 and 306; asterisks, conserved cysteine and histidine residues in the plant homeobox domain; solid box, LXXLL-type nuclear hormone receptor interaction domain in Sp110. The amino acid sequence of the IFN-inducible protein nuclear phosphoprotein 72 is contained within the sequence of Sp110 beginning at methionine 241 and extending to leucine 605 (arrows). (B) Amino acid sequence homology between Sp110 and Sp140 and between Sp110 and Sp100b. Regions of homology in the Sp100-like region, SAND domain, plant homeobox domain, and bromodomain are indicated.
Several features of the predicted amino acid sequence of Sp110 were of particular interest (Fig. 1B). The N-terminal portion of Sp110, between amino acids 6 and 159, was 49% identical to the N-terminal portions of both Sp100 (42) and Sp140 (5). A second region of homology between Sp110 and both Sp100b and Sp140 was present between amino acids 452 and 532. In this region, Sp110 was 53% identical to Sp100b (8) and 49% identical to Sp140. This portion of Sp100b and Sp140 was previously designated a SAND domain (13). Sp110 amino acids 537 to 577 spanned a plant homeobox domain (1), and amino acids 606 to 674 contained the A, B, and C helices of a bromodomain (19). The plant homeobox domain and bromodomain of Sp110 were 73 and 54% identical, respectively, to the corresponding regions in Sp140. In addition, these portions of Sp110 were 56 and 46% identical, respectively, to the corresponding regions in murine TIF1α (not shown). A putative nuclear localization sequence was present between amino acids 288 and 306 (38), and an LXXLL-type nuclear hormone receptor interaction motif was present between amino acids 525 and 529 (25).
Expression of Sp110 in human tissues and cell lines.
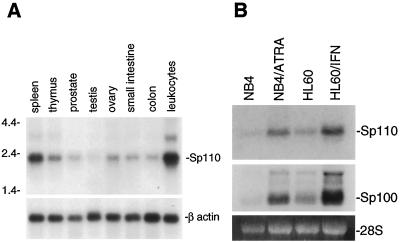
The expression of the gene encoding Sp110 in human tissues was examined by RNA blot hybridization. High levels of Sp110 mRNA were detected in human peripheral blood leukocytes and spleen (Fig. 2A). In contrast, lower levels of Sp110 mRNA were observed in thymus, prostate, testis, ovary, small intestine, and colon. In addition, low levels of Sp110 mRNA were observed in human heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas (data not shown). The tissue distribution of Sp110 mRNA was similar to that observed for Sp140 (4); both Sp110 and Sp140 are predominantly expressed in human leukocytes.
FIG. 2.
(A) Identification of Sp110 mRNA in human tissues by RNA blot hybridization. A membrane containing 2.5 μg of poly(A)+ RNA per lane from human spleen, thymus, prostate, testis, ovary, small intestine, colon, and peripheral blood leukocytes was hybridized with a 32P-radiolabeled XbaI restriction fragment of the Sp110 cDNA. After being washed under stringent conditions, the membrane was exposed to autoradiography. High levels of mRNA encoding Sp110 were detected in human spleen and peripheral blood leukocytes. To confirm the presence of RNA in each lane, the membrane was subsequently hybridized with a radiolabeled human β-actin cDNA probe. (B) Expression of Sp110 mRNA in myeloid precursor cell lines. Low levels of Sp110 mRNA were detected in NB4 cells (lane NB4) and in HL60 cells (lane HL60). Treatment of NB4 cells with ATRA (1 μM) for 48 h induced expression of Sp110 (lane NB4/ATRA). Treatment of HL60 cells with IFN-γ (200 U/ml) for 48 h also markedly increased Sp110 gene expression (lane HL60/IFN). Similar changes in Sp100 mRNA were observed in ATRA-treated NB4 cells and IFN-γ-treated HL60 cells. Ethidium bromide staining of 28S RNA confirmed equal loading of RNA samples.
To investigate the expression of Sp110 in cells of the monocyte/granulocyte lineage, RNA was prepared from the myeloid precursor cell lines HL60 and NB4 (7, 24). Low levels of Sp110 mRNA were detected in NB4 cells (Fig. 2B, lane NB4) and HL60 cells (Fig. 2B, lane HL60). To examine the effect of cellular differentiation on Sp110 mRNA, NB4 cells were treated for 48 h with ATRA (1 μM). The level of Sp110 mRNA was increased in ATRA-treated NB4 cells (Fig. 2B, lane NB4/ATRA). These results demonstrated that differentiation of NB4 cells was associated with increased expression of Sp110. To examine the effect of IFN-γ treatment on Sp110 mRNA levels, HL60 cells were treated with IFN-γ (200 U/ml) for 48 h. A marked increase in Sp110 mRNA was observed (Fig. 2B, lane HL60/IFN). These results demonstrated that, as with Sp100, PML, and Sp140, IFN-γ treatment enhances expression of Sp110.
Cellular location of Sp110.
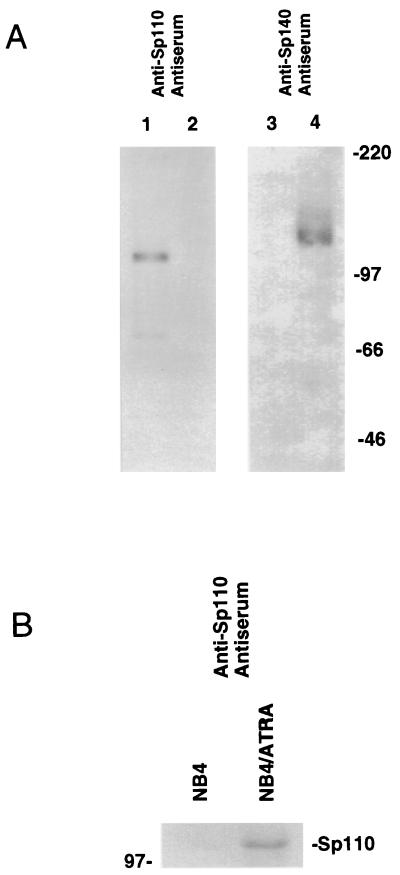
The structural similarities between Sp110 and nuclear body components Sp100 and Sp140 suggested that Sp110 may also be a component of the nuclear body. To facilitate studies of the cellular location of Sp110, antiserum directed against a recombinant fragment of Sp110 (amino acids 219 to 324) was generated in rats and an adenovirus vector encoding Sp110 (Ad.Sp110) was prepared. The rat anti-Sp110 antiserum reacted with Sp110 in extracts prepared from Ad.Sp110-infected HEp-2 cells (Fig. 3A, lane 1) but not with Sp140 in extracts prepared from Ad.Sp140-infected HEp-2 cells (Fig. 3A, lane 2) or with Sp100, which is normally expressed in HEp-2 cells. In contrast, rat anti-Sp140 antiserum, previously prepared against Sp140 amino acids 131 to 391 (5), reacted with Sp140 (Fig. 3A, lane 4) but not with Sp110 (Fig. 3A, lane 3) or Sp100. These results demonstrated that the rat anti-Sp110 antiserum was specific for Sp110. Immunoblotting was used to confirm that the ATRA-mediated increase in Sp110 mRNA in NB4 cells was accompanied by a corresponding increase in the level of Sp110 protein (Fig. 3B).
FIG. 3.
(A) Immunoblotting of adenovirus-infected HEp-2 cells using anti-Sp110 and anti-Sp140 antisera. Immunoblots were prepared from extracts of HEp-2 cells infected with Ad.Sp110 (lanes 1 and 3) or Ad.Sp140 (lanes 2 and 4). Anti-Sp110 antiserum reacted with Sp110 in Ad.Sp110-infected HEp-2 cells (lane 1) but not with Sp140 in Ad.Sp140-infected HEp-2 cells (lane 2) or Sp100 (normally expressed in HEp-2 cells). Anti-Sp140 antiserum reacted with Sp140 in Ad.Sp140-infected HEp-2 cells (lane 4) but not with Sp110 in Ad.Sp110-infected HEp-2 cells (lane 3). (B) Immunoblot of ATRA-treated and control NB4 cells. Immunoblots were prepared from extracts of NB4 cells treated for 48 h with ATRA (1 μM; lane NB4/ATRA) or control NB4 cells (lane NB4). ATRA treatment increased the level of immunoreactive Sp110.
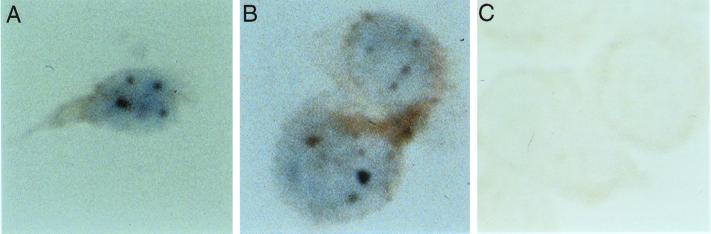
To investigate the cellular location of Sp110, rat anti-Sp110 antibodies and immunohistochemistry were used to stain NB4 cells before and after treatment with ATRA. Anti-Sp110 antiserum stained nuclear body-like structures in NB4 cells that were treated for 48 h with ATRA (Fig. 4A and B). We previously demonstrated that a mouse monoclonal antibody directed against PML and rat anti-Sp140 antibodies produced the same pattern of staining in ATRA-treated NB4 cells (5). As expected, based on mRNA blot (Fig. 2B) and immunoblot results (Fig. 3B), Sp110 was not detected in the nuclei of untreated NB4 cells (Fig. 4C).
FIG. 4.
Immunohistochemical localization of Sp110 in NB4 cells. Control NB4 cells and NB4 cells treated for 48 h with ATRA (1 μM) were subjected to cytospin centrifugation, fixed, and stained with rat anti-Sp110 antiserum. Staining was observed within dot-like regions in the nuclei of ATRA-treated NB4 cells (A and B). Anti-Sp110 antiserum did not react with untreated NB4 cells (C).
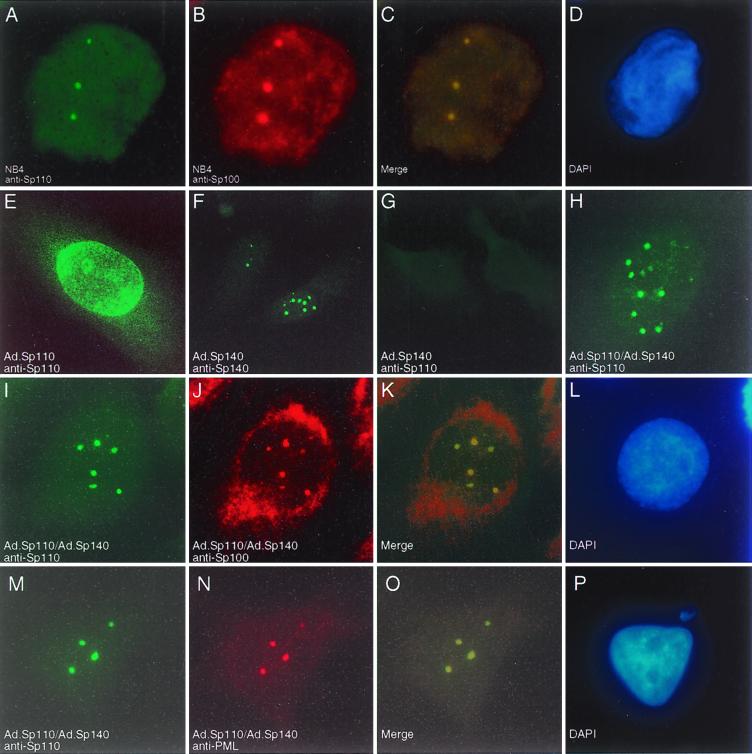
To determine the location of Sp110 with respect to the PML-Sp100 nuclear body, NB4 cells were treated with ATRA and stained with rat anti-Sp110 antiserum and human serum containing antibodies directed against Sp100 (Fig. 5A to D). Sp110 colocalized with Sp100 in nuclear bodies. To further investigate the cellular location of Sp110, adenovirus-mediated gene transfer was used to express Sp110 in human cell lines that normally do not have this protein. We previously used a similar approach to examine the location of Sp140 with respect to Sp100 and PML (4). At a multiplicity of infection (MOI) of 25 viruses per cell, approximately 25% of HEp-2 cells expressed levels of Sp110 that were detectable by indirect immunofluorescence. Surprisingly, Sp110 did not localize to nuclear bodies in these cells but instead appeared to produce a granular nuclear staining pattern with prominent staining near the nuclear membrane (Fig. 5E). Cytoplasmic staining was also observed in a few cells (not shown). To reconcile the different results obtained using the leukocyte cell line NB4 and Ad.Sp110-infected HEp-2 cells, we considered the possibility that the leukocyte-specific nuclear body component Sp140 recruits Sp110 to the nuclear body. HEp-2 cells were infected with both Ad.Sp140, at a MOI of 50, and Ad.Sp110, at a MOI of 25. At a MOI of 50, essentially all of the HEp-2 cells expressed detectable Sp140 within nuclear bodies (Fig. 5F). In cells infected with Ad.Sp140 alone, anti-Sp110 antiserum did not stain nuclear bodies, confirming that anti-Sp110 antiserum did not cross-react with Sp140 (Fig. 5G). In cells infected with both Sp110 and Sp140, Sp110 localized to nuclear bodies (Fig. 5H) and colocalized with Sp100-containing nuclear bodies (Fig. 5I to L). In addition, in cells infected with both Sp110 and Sp140, Sp110 colocalized with PML-containing nuclear bodies (Fig. 5M to P). These results demonstrated that Sp140 enhances the localization of Sp110 to the PML-Sp100 nuclear body.
FIG. 5.
Immunofluorescence microscopy of ATRA-treated NB4 cells and adenovirus-infected HEp-2 cells. NB4 cells were treated for 48 h with ATRA (1 μM) and were stained with rat anti-Sp110 antiserum (A) and human serum containing anti-Sp100 antibodies (B). Sp110 colocalized with Sp100 in nuclear bodies in ATRA-treated NB4 cells (merging of green and red fluorescence and DAPI [4′,6′-diamidino-2-phenylindole] staining are shown in panels C and D, respectively). To further investigate the cellular location of Sp110, HEp-2 cells, which normally do not express either Sp110 or Sp140, were infected with Ad.Sp110 and stained with anti-Sp110 antiserum. Sp110 was observed in a granular pattern within the nucleus and appeared to associate with the nuclear membrane (E). In contrast, HEp-2 cells infected with Ad.Sp140 and stained with anti-Sp140 antiserum revealed a typical nuclear body staining pattern (F). No fluorescence was seen in cells infected with Ad.Sp140 and stained with anti-Sp110 antiserum (G), confirming the specificity of this antiserum for Sp110. In cells infected with both Ad.Sp110 and Ad.Sp140, Sp110 localized to nuclear bodies (H, I, and M). Staining of cells infected with both Ad.Sp110 and Ad.Sp140 with anti-Sp110 antiserum (I) and anti-Sp100 antibodies (J) revealed colocalization of the two proteins (K). In addition, staining of infected cells with anti-Sp110 (M) and anti-PML (N) antibodies demonstrated colocalization of the two proteins (O). (L and P), DAPI staining.
Transcriptional activation by Sp110.
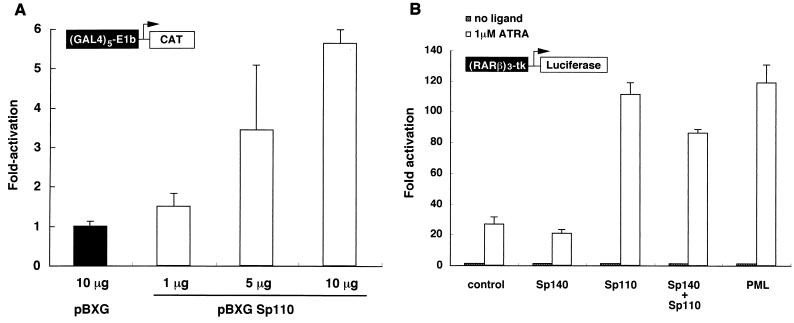
The amino acid sequence motifs in Sp110, including the SAND domain, plant homeobox domain, and bromodomain, raised the possibility that Sp110 may have a role in the regulation of gene transcription. To examine the potential effect of Sp110 on gene transcription, a eukaryotic expression plasmid encoding Sp110 fused to the DNA-binding domain of GAL4 (pBXG-Sp110) was cotransfected with a CAT reporter plasmid containing five GAL4 binding sites and an SV40 enhancer region (pG5SV-BCAT) into COS cells. There was a dose-dependent increase in CAT activity in cells transfected with increasing amounts of pBXG-Sp110 (Fig. 6A). These results were similar to those observed using pBXG-Sp140 (4) but were in contrast to those obtained with pBXG-Sp100. The GAL4-Sp100 fusion protein was previously shown to inhibit CAT activity when cotransfected with the reporter plasmid (4, 27, 37). These results demonstrated that Sp110 is capable of modulating gene transcription and can act in these cells as a transcriptional activator.
FIG. 6.
(A) Sp110 acts as a transcriptional activator when tethered to DNA. Plasmids encoding the GAL4 DNA-binding domain fused to Sp110 (pBXG-Sp110) or the GAL4 DNA-binding domain alone were transfected into COS cells together with a reporter plasmid directing expression of CAT under the control of a GAL4 DNA-binding domain response element. A plasmid encoding growth hormone was included as a control for efficiency of transfection, and transfections were performed in triplicate. The total amount of plasmid DNA was the same in each transfection. Results are means ± standard errors of the means (SEM). CAT activity in pBXG-Sp110-transfected cells was expressed as the fold increase compared with the activity in pBXG-transfected cells. Production of growth hormone in cells transfected with pBXG did not differ from that in cells transfected with pBXG-Sp110. Transfection of COS cells with 1, 5, and 10 μg of pBXG-Sp110 increased CAT activity in a DNA dose-dependent manner. (B) Sp110 may function as a nuclear hormone receptor transcriptional coactivator. Plasmids encoding Sp110, Sp140, Sp110 and Sp140, or PML or vector alone was transfected into COS cells together with a reporter plasmid containing the luciferase gene driven by three copies of the RARα response element derived from the RARβ promoter region. Results are fold increases in luciferase activity in ATRA-treated versus untreated transfected cells. Transfections were performed in triplicate, and results are presented as means ± SEM. The results are representative of three separate experiments. Expression of Sp110 enhanced ATRA-induced responsiveness compared with vector alone. The extent of enhanced ATRA responsiveness by Sp110 was similar to that induced by PML. In contrast, Sp140 did not increase ATRA-induced expression of the reporter gene and expression of both Sp140 and Sp110 did not enhance luciferase activity to a greater extent than did expression of Sp110 alone.
Because of the structural similarities between Sp110 and TIFα, the possibility that Sp110, like TIFα, functions as a RAR transcriptional coactivator was considered. When cotransfected into COS cells with a reporter gene containing three copies of the RARα response element, Sp110 significantly enhanced ATRA-induced expression of the reporter gene (Fig. 6B). Similar results were observed in studies using HeLa cells instead of COS cells (data not shown). The extent of reporter gene activation by Sp110 was similar to that induced by overexpression of nuclear body component PML. In contrast, Sp140 did not enhance ATRA-induced expression of the reporter gene and the combination of Sp110 and Sp140 did not produce an effect greater than that of Sp110 alone (Fig. 6B). These results demonstrated that Sp110 can function as a nuclear hormone receptor coactivator.
DISCUSSION
In this study, we identified a novel nuclear body component designated Sp110. As with other nuclear body components, expression of Sp110 was enhanced by IFN treatment. In addition, expression of Sp110 was increased in the acute promyelocytic cell line NB4 following treatment with ATRA. In studies using a reporter gene driven by a RAR response element, Sp110 was shown to enhance ATRA-mediated signal transduction.
Structural features of Sp110.
Sp110 has significant homology with nuclear body components Sp100 and Sp140. Review of the National Center for Biotechnology Information Unigene database (http://www.ncbi.nlm.nih.gov/UniGene/index.html) revealed that the genes encoding Sp100, Sp110, and Sp140 are closely linked on 33 centimorgans of human chromosome 2 between markers D2S2158 and D2S125. The proximity of these three genes suggests that Sp100, Sp110, and Sp140 may have arisen via local gene duplication events.
Sp110 has a modular structure that is common in proteins that are components of larger complexes. The Sp100-like domain in the N-terminal portion of Sp110 has a potential α-helical motif. This portion of Sp100 was previously reported to be capable of mediating homodimerization (37). It is possible that Sp110 interacts with either Sp100 or Sp140 (or with itself to form a homodimer) in this portion of the protein.
The middle portion of Sp110 contains the SAND domain. Other proteins that have this motif include Sp100b and Sp140, AIRE-1 (encoded by a gene disrupted in an autosomal recessive autoimmune disease involving endocrine glands [12, 32]), nuclear phosphoprotein 72, and DEAF-1 (a transcription factor in Drosophila melanogaster [15]). The SAND domain contains conserved hydrophobic residues and a potential globular fold and is predicted to have eight β strands (13). The function of the SAND domain remains to be determined. Because the amino acid sequence of Sp110 contains that of nuclear phosphoprotein 72, it seems likely that the previously reported sequence (20) represents the product of a partial cDNA. The cDNA encoding nuclear phosphoprotein 72 lacks the nucleotides encoding the N-terminal 240 amino acids of Sp110. In addition, a frameshift mutation at the 3′ end of the coding sequence may have resulted in a premature termination codon resulting in loss of the bromodomain of Sp110.
Sp110 has a plant homeobox domain, which is a cysteine-rich region that spans 50 to 80 amino acids and contains the motif Cys4-His-Cys3 (1). Although the precise function of the plant homeobox domain is unknown, many of the more than 40 proteins that contain this motif are involved in chromatin-mediated control of gene transcription.
The bromodomain is an α-helical motif that, like the plant homeobox domain, is present in many proteins involved in the regulation of gene transcription (19). Dhalluin et al. reported the solution structure of the bromodomain of the histone acetyltransferase (HAT) coactivator p300/CBP-associated factor (10). The bromodomain was shown to specifically interact with acetylated lysine residues, and the authors suggested that the bromodomain is functionally linked to the HAT activity of transcriptional coactivators.
Although the original description of the bromodomain reported a conserved motif spanning approximately 60 amino acids and containing two α helices (16), Le Douarin and colleagues suggested that this domain may span 110 amino acids and contain two additional α helices (25). The four predicted α helices were designated Z, A, B, and C. Le Douarin et al. noted that the A, B, and C helices are conserved among all bromodomains and suggested that the more-variable sequences of the Z helix may confer a specific protein-protein interaction. Sp110 contains an unusual bromodomain in that, although it has A, B, and C helices, it completely lacks the Z helix. The functional significance of this “partial” bromodomain in Sp110 remains to be determined.
Sp110 is a potential bridge between the nuclear body and the nuclear membrane.
Rat antiserum directed against Sp110 stained nuclear bodies in ATRA-treated NB4 cells. In contrast, Sp110 was detected throughout the nucleus and prominently near the nuclear membrane in HEp-2 cells infected with Ad.Sp110. Infection of HEp-2 cells with both Ad.Sp110 and Ad.Sp140 resulted in localization of Sp110 to the PML-Sp100 nuclear body and suggested that Sp140 may recruit Sp110 to the nuclear body. Because Sp110 appears to be able to interact with both the nuclear membrane and the nuclear body, Sp110 may serve to bridge these two nuclear compartments. Nuclear body components SUMO-1 and Sp100 also appear to interact with proteins in the nuclear membrane. SUMO-1 covalently modifies the nuclear membrane protein RanGAP1, as well as nuclear body components Sp100 and PML (29, 30, 41). Sp100 interacts with HP1 and appears to recruit this protein to the nuclear body. HP1, in turn, interacts with the lamin B receptor, which is an integral component of the nuclear envelope (46). Sp110 joins SUMO-1 and Sp100 as a third link between the nuclear matrix-associated nuclear body components and the nuclear envelope.
Sp110 and the regulation of gene transcription.
Previous investigators demonstrated that Sp100 inhibits gene transcription when bound to the promoter region of a reporter gene in transfected mammalian cells. In contrast, we have demonstrated that the leukocyte-specific nuclear body components Sp140 (4) and Sp110 activate transcription of a reporter gene. These results suggest that the presence of Sp110 and Sp140 within nuclear bodies may change the function of these structures from inhibitors to activators of gene transcription.
Recent studies suggest that the nuclear body may have an important role in nuclear hormone receptor signal transduction. CBP, which is a coactivator for the glucocorticoid and retinoid X nuclear hormone receptors, was shown to localize to the nuclear body (11). In addition, PML, through its association with CBP, enhanced nuclear hormone receptor transcriptional activity (11). In this study, expression of Sp110 in mammalian cells enhanced the expression of a reporter gene under the control of a RARα response element in an ATRA-dependent manner. These results suggest that Sp110, like PML, can function as a coactivator of signal transduction through the RAR.
Despite the structural similarities between Sp140 and Sp110, Sp140 did not enhance ATRA-induced expression of the reporter gene. The functional differences between Sp110 and Sp140 may be a result of the presence of an LXXLL nuclear hormone receptor interaction domain adjacent to the plant homeobox domain and bromodomain in Sp110. In contrast, the LXXLL domain is not present in Sp140. A comparable situation exists in the TIF1 family of proteins: TIF1α (which contains an LXXLL motif adjacent to the plant homeobox domain and bromodomain) enhances RA-induced signal transduction; TIF1β (which lacks this motif) does not enhance RA-induced signaling (25).
Coexpression of Sp140 and Sp110 did not further increase RA responsiveness above that produced by expression of Sp110 alone. Because Sp110 appears to require the presence of Sp140 to localize to the nuclear body and because Sp140 is not present in COS cells, these results raise the possibility that localization of Sp110 to the nuclear body may not be critical for the activity of Sp110 in RA-mediated signal transduction.
Sp110 and acute promyelocytic leukemia.
In acute promyelocytic leukemia, a translocation between chromosomes 15 and 17 results in the fusion of nuclear body protein PML to RARα. Expression of the fusion protein disrupts the nuclear body and inhibits normal myeloid maturation. The fusion protein may disrupt the signal transduction pathways of either RARα or PML or both. These hypotheses do not explain the apparent specificity of the t(15;17) translocation for acute promyelocytic leukemia. Despite the fact that both PML and RARα are expressed in a wide variety of tissues and cell lines, acute promyelocytic leukemia is the only known malignancy associated with the t(15;17) translocation. One possible explanation for the specificity of the t(15;17) translocation for acute promyelocytic leukemia is that the fusion protein disrupts the normal function of leukocyte-specific proteins such as Sp110 and Sp140. The observations that expression of Sp110 is induced by ATRA and that Sp110 enhances signal transduction mediated by RARα raise the possibility that Sp110 has an important role in the effectiveness of ATRA therapy in patients with acute promyelocytic leukemia.
In summary, we have identified a novel member of the Sp100/Sp140 family of proteins. Sp110 is an ATRA- and IFN-inducible, leukocyte-specific nuclear body component. Sp110, like Sp140, activates gene transcription when tethered to the promoter region of a reporter gene. In addition, Sp110 enhances signal transduction mediated by the RAR. These studies suggest that leukocyte-specific nuclear body components can modulate gene expression and may participate in the differentiation of myeloid cells.
ACKNOWLEDGMENTS
This work was supported by grants from the Arthritis Foundation (D.B.B.), Massachusetts Biomedical Research Corporation, and the Phillippe Foundation (J.-D.C.) and by National Institutes of Health grants AR-01866 and DK-051179 (D.B.B.) and HL-55377 (K.D.B.). K. D. Bloch is an Established Investigator of the American Heart Association.
We thank K. J. Bloch, L. Diller, and A. Rosenzweig for advice, S. Schlutsmeyer, and A. Brown for technical assistance, and J. Bonventre and S. Shu for gifts of plasmids and adenovirus.
REFERENCES
- 1.Aasland R, Gibson T J, Stewart A F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 2.Alam J, Cook J L. Reporter genes: application to the study of mammalian gene transcription. Anal Biochem. 1990;188:245–254. doi: 10.1016/0003-2697(90)90601-5. [DOI] [PubMed] [Google Scholar]
- 3.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloch D B, Chiche J-D, Orth D, Rosenzweig A, Bloch K D. Structural and functional hetereogeneity of nuclear bodies. Mol Cell Biol. 1999;19:4423–4430. doi: 10.1128/mcb.19.6.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloch D B, de la Monte S M, Guigaouri P, Filippov A, Bloch K D. Identification and characterization of a leukocyte-specific component of the nuclear body. J Biol Chem. 1996;46:29198–29204. doi: 10.1074/jbc.271.46.29198. [DOI] [PubMed] [Google Scholar]
- 6.Borrow J, Goddard A, Sheer D, Solomon E. Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science. 1990;249:1577–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- 7.Collins S J, Ruscetti F W, Gallagher R E, Gallo R C. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc Natl Acad Sci USA. 1978;75:2458–2462. doi: 10.1073/pnas.75.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dent L D, Yewdell J, Puvion-Dutilleul F, Koken M H M, de The H, Staudt L M. LySp100-associated nuclear domains (LANDS): description of a new class of subnuclear structures and their relationship to PML nuclear bodies. Blood. 1996;88:1423–1436. [PubMed] [Google Scholar]
- 9.de The H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor α gene to a novel transcribed locus. Nature (London) 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 10.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M-M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 11.Doucas V, Tini M, Egan D A, Evans R M. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc Natl Acad Sci USA. 1999;96:2627–2632. doi: 10.1073/pnas.96.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finnish-German APECED Consortium. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- 13.Gibson T J, Ramu C, Gemund C, Aasland C. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem Sci. 1998;23:242–244. doi: 10.1016/s0968-0004(98)01231-6. [DOI] [PubMed] [Google Scholar]
- 14.Graham F L, Preyec L. Manipulation of adenovirus vectors. In: Murray E, editor. Methods in molecular biology: gene transfer and expression protocols. Clifton, N.J: Humana Press; 1991. pp. 109–128. [DOI] [PubMed] [Google Scholar]
- 15.Gross C T, McGinnis W. DEAF-1, a novel protein that binds an essential region in a Deformed response element. EMBO J. 1996;15:1961–1970. [PMC free article] [PubMed] [Google Scholar]
- 16.Haynes S R, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodges M, Tissot C, Howe K, Girmwade D, Freemont P S. Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am J Hum Genet. 1998;63:297–304. doi: 10.1086/301991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcription environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeanmougin F, Wurtz J-M, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 20.Kadereit S, Gewert D R, Galabru J, Hovanessian A G, Meurs E F. Molecular cloning of two new interferon-induced, highly-related nuclear phosphoproteins. J Biol Chem. 1993;268:24432–24441. [PubMed] [Google Scholar]
- 21.Kakizuka A, Miller W H, Umesono K, Warrell R P, Frankel S R, Murty V V V S, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan M M. Primary biliary cirrhosis. N Engl J Med. 1996;335:1570–1580. doi: 10.1056/NEJM199611213352107. [DOI] [PubMed] [Google Scholar]
- 23.LaMorte V J, Dyck J A, Ochs R L, Evans R E. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanotte M, Matrin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3) Blood. 1991;77:1080–1086. [PubMed] [Google Scholar]
- 25.Le Douarin B, Nielson A L, Garnier J-M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 26.Le Douarin B, Zechel C, Garnier J-M, Lutz Y, Tora L, Pierrat B, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehming N, Saux A L, Schuller J, Ptashne M. Chromatin components as part of a putative transcriptional repressing complex. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longo L, Pandolfi P P, Biondi A, Rombaldi A, Mencarelli A, Lo Coco F, Direrio D, Pegoraro L, Avanzi G, Tabilio A, Zangrilli D, Alcalay M, Donti E, Grignani F, Pelicci P G. Rearrangements and aberrant expression of the retinoic acid receptor α gene in acute promyelocytic leukemias. J Exp Med. 1990;172:1571–1575. doi: 10.1084/jem.172.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 30.Matunis M J, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melnick A, Licht J D. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 32.Nagamine K, Peterson P, Scott H S, Kudoh J, Minoshima S, Heino M, Krohr K J E, Lalioti M D, Mullis P E, Antonarakis S E, Kawasaki K, Asakawa S, Ito F, Shimizu N. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–397. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 33.Neumann J R, Mornecy C A, Russian K O. A novel rapid assay for chloramphenicol acetyltransferase gene expression. BioTechniques. 1987;5:444–447. [Google Scholar]
- 34.Quignon F, De Bels F, Koken M, Feunteun J, Ameisen J-C, de The H. PML induces a novel caspase-independent death process. Nat Genet. 1998;10:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sanger F. Determination of nucleotide sequence in DNA. Science. 1981;214:1205–1210. doi: 10.1126/science.7302589. [DOI] [PubMed] [Google Scholar]
- 37.Seeler J-S, Marchia A, Sitterlin D, Transy C, Dejean A. Interaction of Sp100 and HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silver P A. How proteins enter the nucleus. Cell. 1991;64:489–497. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- 39.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione-S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 40.Sternsdorf T, Grotzinger T, Jensen K, Will H. Nuclear dots: actors on many stages. Immunobiology. 1997;198:307–331. doi: 10.1016/S0171-2985(97)80051-4. [DOI] [PubMed] [Google Scholar]
- 41.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szostecki C, Guldner H H, Netter H J, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 43.Venturini L, You J, Stadler M, Galien R, Lallemand V, Koken M H M, Mattei M G, Ganser A, Chambon P, Losson R, de The H. TIF1γ, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 1999;18:1209–1217. doi: 10.1038/sj.onc.1202655. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z-G, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi P P. Pml is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z G, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi P P. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 46.Ye Q, Worman H J. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]