Abstract
CAP-34 is a previously reported phytoprotein isolated from Clerodendrum aculeatum (syn. Volkameria aculeata), inducing systemic antiviral resistance against plant virus infection in susceptible plants. This paper compares the resistance inducing efficacy of CAP-34 and a rhizobacterial isolate P1f on tomato (systemic) and tobacco Xanthi-nc (hypersensitive), against tobacco mosaic virus (TMV). The PGPR isolate was identified as an isolate of Pseudomonas putida through molecular and biochemical characterization, and it exhibited PGPR traits such as production of auxin and siderophore. GC–MS examination of the volatiles produced by P1f included several that are implicated in antimicrobial activity, growth promotion and induced systemic resistance. Foliar treatment of tobacco plants with P1f and CAP-34 led to an induced antiviral state in hypersensitive tobacco that persisted for 5 and 3 days, post-treatment, respectively, with a percent reduction in lesion number greater than 90. A higher accumulation of hydrogen peroxide and production of peroxidase enzyme was recorded in P1f-treated leaves, in comparison to those with CAP-34 treatment. The disease incidence in tomato plants treated with CAP-34 and P1f was 30 and 60 percent, respectively, 28dpi. A significant increase was noted in growth parameters such as number of branches and flowers in CAP-34 treated plants, while a significant enhancement in plant height and dry shoot and root weight was observed in P1f-treated set, compared to the control set. ELISA values for the presence of TMV were significantly lower in the infected tomato plants in the treated sets, as compared to the control set, with CAP-34 treatment exhibiting better results as against the P1f-treated set. In the resistant plants from either set, no viral RNA or viral coat protein was detected through RT-PCR and serology. These results suggest that CAP-34 affords more pronounced protection against virus infection compared to the rhizobacterial isolate P1f.
Keywords: Induced systemic resistance, PGPR, CAP-34, Pseudomonas putida
Introduction
Crop losses due to virus infections are a matter of serious concern. Aside from breeding for resistance through classical means, induction of systemic resistance in susceptible hosts through infection by biotic agents (Fu and Dong 2013) and chemicals (Kessman et al. 1994) gained popularity as an alternative strategy to prevent virus infection. Induction of systemic resistance and growth promotion in plants by plant growth promoting rhizobacteria (PGPR) (van Loon et al. 1998; Pieterse et al. 2014; Mauch-Mani et al. 2017), or plant proteins (Prasad and Srivastava 2017), constitute other well documented methods of plant protection, advocated due to their eco-friendly tag, Thus, manipulating the resistance responses of the host to pathogen infection through various treatments has been favourably considered in the past and continues to be explored for its environment friendly and non-toxic nature (Wilkinson et al. 2019).
Antiviral resistance inducing proteins have been isolated from the leaves of Clerodendrum aculeatum and C. inerme, and roots of Boerhaavia diffusa, termed CAP-34, CIP-29 and BDP-30, respectively (Prasad et al. 1995; Verma et al. 1996; Srivastava et al. 2015b). These are highly stable basic proteins and have molecular masses in the range of 29–34 kDa. Since the first two proteins depurinate rRNA, they are also classified as ribosome-inactivating proteins (RIPs) (Olivieri et al. 1996). Peptides obtained from BDP-30 demonstrate absolute identity with Trichosanthin, an RIP from roots of Trichosanthes kirilowii (Srivastava et al. 2015b). All three proteins induce resistance in several susceptible hosts when administered as a foliar treatment prior to challenge inoculation with different viruses (Prasad and Srivastava 2017). The induced-resistant state in susceptible plants treated with such plant proteins was accompanied by the production of a virus inhibitory agent (VIA) in the host tissues, both at site and at remote-site of application (Verma and Awasthi 1980; Prasad et al. 1995; Verma et al. 1996). An in vitro incubation of leaf extracts from resistant plants with purified viruses led to inhibition of virus infectivity. Reversal in phytoprotein-mediated antiviral resistance was observed after treatment of plants with actinomycin D, implicating host transcription in VIA induction. The nature of the VIA was shown to be proteinaceous, and CT-VIA-62, a 62 kDa virus inhibitory agent, purified from Cyamopsis tetragonoloba plants treated with CIP-29, shared sequence similarity with a lectin (Verma and Dwivedi 1984; Prasad et al. 2014). Amongst the PGPR, the fluorescent Pseudomonas spp. such as P. fluorescence strain CHA0 (Maurhofer et al. 1994), P. putida strain WCS358, P aeruginosa strain 7NSK (De Meyer et al. 1999), and Bacillus subtilis strains IN937 and SE34 are very effective in controlling plant viruses by inducing resistance, and promoting plant growth, either through direct or indirect effects (Murphy et al. 2003; Maurhofer et al. 2004; Bakker et al. 2007; Beris et al. 2018).
We have evaluated the efficacy of resistance induction against tobacco mosaic virus (TMV) in the hypersensitive tobacco cv. Xanthi-nc and Solanum lycopersicum (tomato) under greenhouse conditions, following treatment with CAP-34, a basic antiviral protein from C. aculeatum, and a PGPR isolate P1f, obtained from the rhizosphere of healthy tomato plants. P1f was subsequently identified as an isolate of Pseudomonas putida. The comparative antiviral resistance induced by these two widely differing agents, a plant protein and a rhizobacterial isolate, both with the capability of eliciting similar effects in the treated plants, has been explored. Effects of the two treatments on several growth parameters of tomato plants have also been compared. Production of auxin, siderophore, and secondary metabolites/volatiles by the rhizobacterial isolate P1f, in relation to growth promotion and induced resistance, have been evaluated.
Materials and methods
Experimental hosts and maintenance of virus culture
Experimental plants were maintained in an insect-free glasshouse under natural light conditions and raised in clay pots. Nicotiana tabacum cv. Xanthi-nc, a local lesion assay host for TMV, was used for bioassay to monitor induced resistance, while Solanum lycopersicum (tomato) was employed as the experimental host to compare antiviral resistance induction by CAP-34 and the PGPR isolate P1f. Both hosts were raised from seeds and used when the tobacco plants were at a five-leaf stage, and tomato plants attained a height of 10–12 inches, supported by stakes as they grew taller. TMV was maintained on its systemic host N. tabacum cv. White Burley and the infected leaves with distinct mosaic served as a source for the TMV inoculum. The infected tissue was macerated in 10 mM phosphate buffer, pH 7.0, and the centrifuged sap was used as the inoculum after suitable dilution in distilled water.
Isolation of CAP-34 from Clerodendrum aculeatum
CAP-34 was purified from Clerodendrum aculeatum leaves through a combination of chromatographic procedures as described earlier (Verma et al. 1996). Briefly, the clarified leaf tissue homogenate was precipitated with ammonium sulfate and the precipitate was centrifuged down to be suspended in sodium acetate buffer. Following centrifugation to clarify suspended particles, the supernatant was desalted on a column of Sephadex G-25matrix. The void volume eluate was subjected to a cation-exchange chromatography step on SP Sepharose FF matrix, tested for antiviral activity and the active fraction pool was fractionated on a Superdex-75 pre-packed FPLC size-exclusion column. The matrices for chromatography were obtained from GE Healthcare. The purified protein was quantified by a modified Lowry protocol (Cadman et al.1979).
Isolation of plant growth promoting rhizobacteria
Soil was collected from the rhizosphere of healthy tomato plants growing in and around the University of Lucknow campus. Isolate P1f was obtained through soil dilution-plating on nutrient agar. Following an overnight incubation at 37 °C, single colonies were picked and inoculated on nutrient agar through the streak-plate technique. The resulting single colonies, constituting pure cultures of isolate P1f, were transferred to agar slants. Culture was maintained at 4 °C and sub-cultured periodically to retain its viability, with a loopful being transferred to a 5 mL nutrient broth tube whenever needed. CM cellulose (0.5% w/v) was added to the 24 h old broth cultures that were adjusted to a give final density of 1 × 108 cfu mL−1 using McFarland standard.
Induction of systemic antiviral resistance in tobacco Xanthi-nc
Two basal leaves of the tobacco Xanthi-nc plants were treated with either CAP-34 (10 µg mL−1) or the PGPR isolate P1f (1 × 108 cfu mL−1) with a cotton swab. Control sets were similarly treated with either distilled water or sterile nutrient broth supplemented with 0.5% (w/v) CM cellulose. After 1, 3, 5 and 7 days of treatment, the upper untreated and the lower treated leaves were inoculated with TMV using carborundum powder (600 mesh) to facilitate mechanical transmission. Local lesions were counted 3–5 days post-inoculation (dpi) and the percentage inhibition of TMV infection was calculated for both site (treated) and remote-site (untreated) leaves as described earlier (Prasad et al. 1995).
Assay for production of the virus inhibitory agent (VIA) in the treated plants
Nicotiana tabacum cv. Xanthi-nc plants were divided into three sets, and each set received treatment with CAP-34, P1f and DW as before. After 24 h of treatment, the two treated (site) leaves and the upper untreated (remote site) leaves from each set were separately harvested, washed in DW, and homogenized in sodium acetate buffer, 0.1 M, pH 5.2. The total protein fraction (void volume eluate) was recovered from each set following chromatography of the centrifuged extracts on Sephadex G-25 matrix. These eluates were then mixed with purified TMV, incubated, and assayed for the presence of VIA as reported earlier (Prasad et al. 2014).
Peroxidase enzyme assay and in situ localization of H2O2 in the leaf tissue
N. tabacum cv. Xanthi-nc plants were given a foliar treatment with CAP-34, P1f isolate and DW (control), as described previously. Treated leaves were harvested after 24 h for assay of peroxidase enzyme. Briefly, 1 g leaf tissue was homogenized in ice-cold mortar and pestle along with 2 mL of chilled sodium phosphate buffer, 0.1 M, pH 7.0, and the homogenate was centrifuged to get a clear supernatant (enzyme extract). Reaction mixture (1 mL) contained 100 mM phosphate buffer, pH 6.5, Guaiacol 0.5% (w/v), H2O2 0.01% (v/v). Enzyme extract (50 µL) was added to initiate the reaction, and the enzymic activity was followed as an increase in absorbance at 430 nm, and expressed as µM Guaiacol oxidized min−1 mg−1 protein. Reaction mixture without H2O2 served as a blank (Hammerschmidt et al. 1982). Protein concentration in the centrifuged extract was measured, using BSA as a standard (Cadman et al. 1979).
H2O2 accumulation was detected in the leaf tissues of treated plants. Leaf discs (20 mm diameter) from each set were vacuum infiltrated for 15 min in 0.1 mg mL−1 of 3,3′-diaminobenzidine prepared in 50 mM Tris–acetate buffer, pH 5.0. Discs were incubated for 24 h at 25 °C in dark, on a shaker incubator, decolourized with ethanol, 80% (v/v), at 70 °C for 20 min, and mounted in lactic acid, phenol and water mixture (1:1:1, v/v). Nikon advanced research microscope (Model E-400) was used to photograph the discs (Hernandez et al. 2001).
16S rRNA gene sequencing and nucleotide BLAST for identification of the isolate P1f
Bacterial genome was isolated using the protocol laid down in the Bacterial DNA isolation kit (Chromous Biotech, India), and DNA was quantified using multi-mode plate reader (BioTek Instruments, USA). For the amplification of the 16S rRNA gene, the universal primer set of 27F (5′AGAGTTTGATCCTGGCTCAG3′) and 1492R (5′ACGGCTACCTTGTTACGACTT3′) were used (Frank et al. 2008). PCR was carried out using Dream Taq PCR mix (25 μL) (Thermo Fisher Scientific), comprising Taq Polymerase (1.0 unit), 2× reaction buffer and dNTPs (200 mM each), forward and reverse primers (4 µL each), RNase free water (13 μL), and 25 ng template DNA. Thermocycler (BioRad) was programmed as follows: 95 °C × 5 min (initial denaturation step), 94 °C × 45 s (denaturation), 52 °C × 45 s (annealing), 72 °C × 1 min (extension) × 30 cycles, with a final extension at 72 °C for 10 min. The PCR amplicon (1494 bp) was electrophoresed on a 1% agarose gel in Tris–borate–EDTA buffer, stained with ethidium bromide and visualised on a UV transilluminator. The amplicon was gel purified using GeneJET PCR purification kit (Thermo Fisher Scientific) and submitted for sequencing (Chromous Biotech, India). The sequence was submitted for species identification using the NCBI nucleotide BLAST.
Morphological and biochemical identification of the isolate
The isolate was cultured on nutrient agar and studied for the colony characteristics after incubating overnight at 37 °C. It was also inoculated onto King’s B medium for detection of fluorescence (King et al. 1954). KB002 HiAssorted Biochemical test kit for detection of Gram-negative bacteria was purchased from HiMedia and the reactions of the isolate to various tests were interpreted as per the Interpretation Chart supplied by the manufacturer.
Detection of siderophore, auxin, HCN production, and phosphate solubilization by the isolate P1f
P1f isolate was assessed qualitatively for siderophore production observing the protocol of Schwyn and Neilands (1987). Sterile disc dipped in the 24 h old broth culture was placed on chrome-azurol S (CAS) agar medium and incubated for up to 3 days at 37 °C. Development of orange colour around the inoculated disc indicated the production of siderophore by the bacterial isolate.
Production of Indole-3-acetic acid (IAA) was measured quantitatively (Bric et al. 1991). A loopful of the isolate was incubated for 24 h at 37 °C in 5 mL nutrient broth. The inoculum (100 µL) was transferred to King’s B broth (50 mL) containing tryptophan (500 µg mL−1) and incubated for an additional 48 h at 28 °C in a shaker incubator. Cultures were centrifuged at 10,000 rpm for 15 min and 1 mL of supernatant was transferred to a fresh tube to which 50µL of 10 mM orthophosphoric acid and 2 mL of Salkowski’s reagent (1 mL of 0.5 M FeCl3 in 50 mL of 35% HClO4) were added. Development of red colour confirmed the isolate as positive for IAA production. IAA was quantified from a standard graph plotted with 5, 10, 25, 50 and 100 µg mL−1 IAA (Gordon and Weber 1951). Phosphate solubilization was detected on the Pikovskaya agar medium (Pikovskaya 1948), while HCN production was estimated according to the protocol of Bakker and Schippers (1987).
Production of secondary metabolites by P1f and its detection by GC–MS
Secondary metabolites were separated from an overnight culture of P1f isolate using Shimadzu GC–MS -2010 Plus (Chromous Biotech, India). Briefly, the sample was mixed with chloroform, vortexed and centrifuged. The upper aqueous layer was withdrawn and the sample dried by Speed Vac. Following addition of BF3-Methanol, the sample was incubated, and ice-cold water and chloroform added. After centrifugation, the supernatant layer was collected, dried in vacuum, dissolved in chloroform and injected for GC–MS analysis. The bacterial volatile components were identified by comparison of GC retention times with those of standards and by comparison of mass spectra with spectra in the database.
Evaluation of plant growth promotion and induced antiviral resistance in tomato plants treated with DW, CAP-34 and the isolate P1f
Isolate P1f and CAP-34 were used to induce resistance in tomato. Plants were divided into six sets, with ten replicates per treatment. CAP-34 (10 µg mL−1) and P1f (1 × 108 cfu mL−1) were administered as a foliar spray, and the spray regimen was followed for three consecutive days. The control set was similarly sprayed with DW alone. After 24 h of the final treatment, one set from each treatment was challenge inoculated with TMV on the top two compound leaves of each plant. All plants were examined regularly and evaluated for the progression of mosaic symptoms over a period of 4 weeks. Leaves were routinely harvested from the individual plants of each set that was inoculated at 7, 14 and 28 dpi for detection of TMV coat protein (CP) and RNA. The other three treated sets were left un-inoculated. Plants from both sets (inoculated and un-inoculated) were observed over a span of three months for growth promotion parameters such as plant height, number of branches, flowers and fruits per plant, fresh and dry shoot and root weight.
Detection of TMV coat protein and RNA
The three sets of treated plants (DW, CAP-34, and P1f) were examined for the presence of TMV coat protein (CP) and virus specific CP RNA at weekly intervals. The presence of the coat protein in the leaf tissue was detected initially through SDS-PAGE (Laemmli 1970) and Dot blot, and confirmed by immunoblotting (Towbin et al. 1979), while the TMV RNA was identified through RT-PCR with TMV coat protein specific primers designed using Primer BLAST on a TMV isolate Chongqing coat protein gene (GenBank ID. MT108232.1). An amplicon of 242 bp was fetched with the following primer sequences: Forward primer VP-02-F1(5′CAAGCTCGAACTGTCGCTCA-3′) and Reverse primer VP-02-R1(5′ACCGTTGCGTCGTCTACTCT-3′). DOT blot was carried out on leaf tissue samples from each treated set of plants, at 7, 14 and 28 dpi. Un-infected healthy leaf tissue samples from five plants were also included as a negative control. Immunoblot and RT-PCR were carried out using representative samples from the treated sets that did not show any TMV coat protein in the DOT blot analysis. Purified TMV was used as a positive control in the immunoblot, and RNA isolated from uninfected healthy plants served as a negative control in the RT-PCR. Preparation of the polyclonal antibody against the purified local strain of TMV was outsourced (Bangalore Genei). Dilutions of the primary antibody (1:1000) and goat anti-rabbit antibodies conjugated with alkaline phosphatase (ALP) (1:2000) were prepared in TBS-T for serological detections. Nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) served as substrate. All chemicals and the ALP conjugate were obtained from Sigma–Aldrich (USA).
Detection of viral load through ELISA
Tomato plants exhibiting mosaic symptoms were examined for the viral load through indirect ELISA (Mowat and Dawson 1987) using primary anti-TMV-antibody, and secondary goat-anti-rabbit-antibodies conjugated with ALP, and p-nitro phenyl phosphate as substrate. ELISA was carried out with samples from the treated sets which showed mosaic symptoms and that carried the TMV CP as determined by SDS–PAGE and Dot blot analysis. The leaf tissue extract from TMV resistant plants from both treatments were also considered, along with sap from uninfected healthy S. lycopersicum as the negative control. For the immunoassay, antigen (centrifuged leaf sap) was diluted to 1:100 in coating buffer, whereas primary and secondary antibodies were diluted with PBS-T to 1:2000 and 1:4000, respectively.
Statistical analysis
Data were analysed using ANOVA, Dunnett t test, treating one group as a control, and comparing all other groups against it, with SPSS software (IBM SPSS statistics version 20). All hypotheses were tested at the 95% confidence level. Growth promotion data were analyzed using multiple comparisons LSD test that tested significance at P ≤ 0.05.
Results
Induction of systemic antiviral resistance in tobacco cv. Xanthi-nc
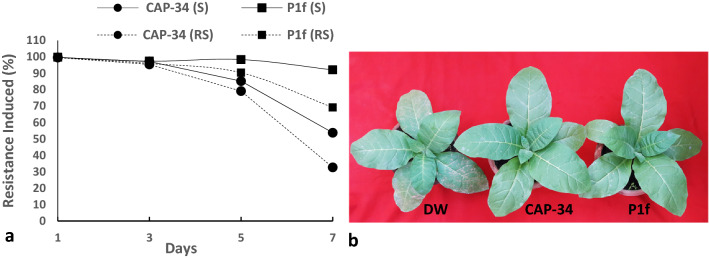
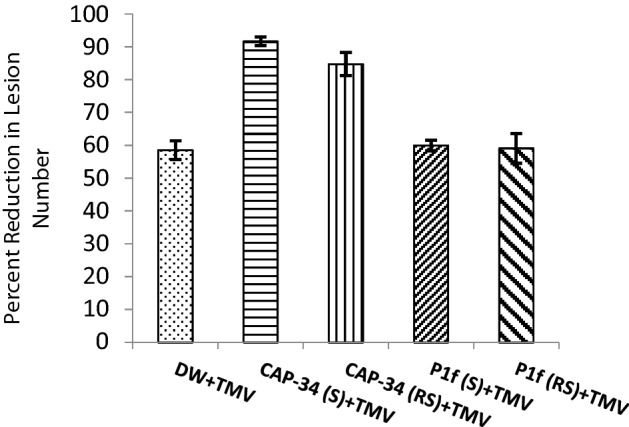
Induction of systemic antiviral resistance was evident in N. tabacum cv. Xanthi-nc plants sprayed with both the basic protein CAP-34, isolated from the leaves of Clerodendrum aculeatum (10 µg mL−1), and an inoculum preparation of a new rhizobacterial isolate P1f (1 × 108 cfu mL−1). Both treatments reduced lesion number by nearly 99%, when challenged with TMV 24 h post-treatment. The degree of resistance induced remained close to or above the 95% mark, when challenged with TMV even after 3 days of inducer application. On the fifth day, the resistance response at remote-site leaves of CAP-34 treated set dipped to 79%, while a 90% inhibition of virus was noted in the P1f-treated set. By day seven, a decline in induced resistance, at both site and remote site, was observed in the CAP-34 treated plants as well as in P1f-treated one, however, the decline was markedly greater in the set treated with CAP-34 as compared to P1f (Fig. 1a, b).
Fig. 1.
Induced antiviral resistance in Nicotiana tabacum cv. Xanthi-nc. Tobacco plants were administered foliar applications of CAP-34 (10 µg mL−1), an overnight broth culture of the rhizobacterial isolate P1f (108 cfu mL−1), and DW (control) on the two basal leaves. Challenge-inoculations with TMV were performed on the treated site leaves (S) as well as on the upper untreated remote-site leaves (RS) at days 1, 3, 5 and 7 post-treatments. Local lesions were counted 3–5 days post-inoculation, and induced resistance expressed as percent reduction in lesion number. Data represent the mean values ± SE from three replicates (a). Representative plants from the treated sets challenge inoculated with TMV 24 h post-treatment (b)
Induction of VIA
Virus inhibitory agent was detectable in the CAP-34 treated set of plants, where the total protein fraction from site as well as remote-site leaves could inhibit TMV in vitro by nearly 92% and 85%, respectively. In contrast, TMV was inhibited by approximately 58% by the protein fractions obtained from the site and remote-site leaves of plants treated with both P1f and DW (Fig. 2).
Fig. 2.
Induction of Virus Inhibitory Agent (VIA) in tobacco Xanthi-nc. Tissue homogenate (total protein fraction) from the site (S) and remote-site (RS) leaves of plants treated with CAP-34, P1f and DW, were incubated with purified TMV and assayed for reduction in virus infectivity on tobacco Xanthi-nc plants. Note that the figure depicts in vitro inactivation of TMV as against in vivo induction of resistance as depicted in Fig. 1
Assay for peroxidase enzyme and detection of H2O2 accumulation
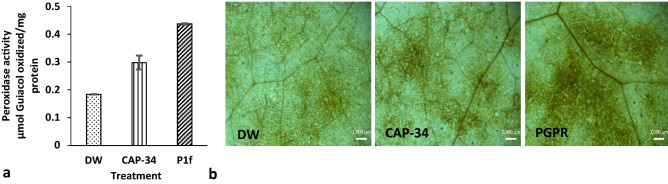
The increase in peroxidase enzyme activity in the leaves following treatment with either CAP-34 or P1f was observed. The elevation was greater for P1f-treated set as compared to the CAP-34 set, with absorbance (430 nm) values of 0.29 and 0.43, respectively (Fig. 3a). Similarly, enhanced accumulation of H2O2 was observed in the leaf discs obtained from the treated sets, with greater accumulation in the P1f-treated set as compared to the CAP-34 set (Fig. 3b).
Fig. 3.
Detection of peroxidase activity and H2O2 accumulation in leaves. Plants were treated with DW (control), CAP-34 (10 µg mL−1) and P1f isolate (1 × 108 cfu mL−1), and after 24 h of foliar application, the activity of the peroxidase enzyme was measured in the treated leaves. Data represent means of three replicates ± SEM (a). Leaf discs were stained with DAB for histo-chemical localization of H2O2 (b)
Morphological, biochemical and molecular identification of the rhizobacterial isolate P1f
The isolate P1f produced convex, punctiform, creamy white, viscid, translucent colonies with entire margins, and a prominent fluorescence when placed under UV light (Fig. 4a). The rod shaped bacteria stained Gram-negative. The identity of the bacteria was established with the HiMedia kit designed for the rapid identification of gram- negative bacteria. P1f isolate tested positive for citrate utilization, and fermentation of glucose and arabinose. However, the isolate was a non-fermenter of adonitol, lactose and sorbitol, could not utilize lysine, ornithine, did not exhibit phenylalanine deamination activity, did not produce urease and H2S, and could not reduce nitrate (data not shown). These biochemical characteristics matched those of Pseudomonas putida. The P1f isolate produced siderophore, as evident from the orange coloured zone that developed around the inoculum-saturated disc on CAS agar (Fig. 4b). The isolate produced auxin which was quantified at 20.90 µg mL−1 and 2.76 µg mL−1, with and without tryptophan, respectively (Fig. 4c). P1f was weak in solubilizing phosphate (Fig. 4d) and was deficient in an ability to produce HCN.
Fig. 4.
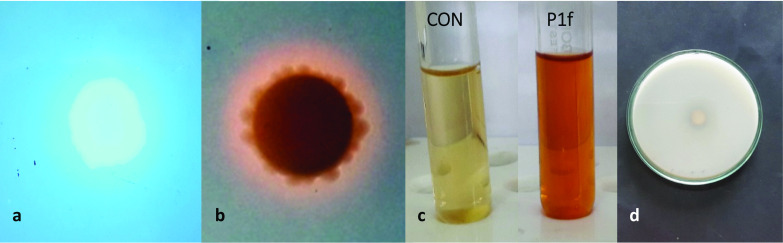
Characterization of the PGPR traits of P1f isolate. Production of fluorescence on King’s B medium (a), siderophore production on CAS-agar (b), auxin production, CON represents negative control (c), and marginal solubilization of phosphate (d)
A PCR amplicon of 1493 bp was obtained with 16S rRNA gene specific primers using genomic DNA from the bacterial isolate as the template. The amplicon was purified and the sequenced product had a size of 1359 nucleotides, exhibiting a query cover and sequence identity between 99.56 and 100% with several strains of Pseudomonas putida deposited in the GenBank database, accession no MZ569722 (Table 1).
Table 1.
Molecular identity of the PGPR isolate P1f
| Sequence ID | Description | Percent identity |
|---|---|---|
| MN318320.1 | Pseudomonas putida strain SG1 | 100% |
| MG836187.1 | Pseudomonas putida strain HRI603-1 | 100% |
| MK426804.1 | Pseudomonas putida strain uqpm04 | 100% |
| MW273908.1 | Pseudomonas putida strain QAS-2H | 100% |
| MN134487.1 | Pseudomonas putida strain PSBGB-4 | 99.78% |
| NR_043424.1 | Pseudomonas putida strain PSBGB-4 | 99.56% |
The 1359 bp sequence was subjected to nucleotide blast (NCBI). The query sequence shared a 99.56 to 100% sequence identity with several strains of Pseudomonas putida deposited with GenBank with accession no. MZ569722
Progression of mosaic symptoms in Solanum lycopersicum and detection of TMV coat protein by Dotblot, Immunoblot and RT-PCR
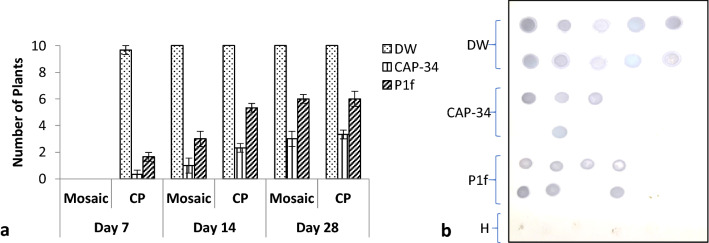
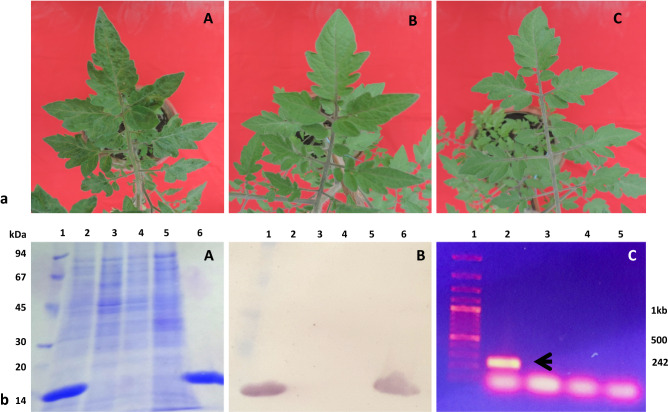
One set each of three sets of tomato plants were treated with DW, CAP-34 and P1f, and all plants in the three sets were challenge inoculated with TMV 24 h later. All treated sets were symptomless 7 days post-inoculation (dpi). At 14 dpi, all 10 plants in the DW-treated set showed prominent mosaic symptoms, while only 1 and 3 plants developed mosaic in the CAP-34 and P1f-treated sets, respectively. By the end of four weeks, the mosaic symptoms had progressed to 3 plants in the CAP-34 set and 6 plants in the P1f set, and at this point, the severity of mosaic symptoms was similar in all plants. At 7, 14, and 28 dpi, plants in each set were also screened by SDS–PAGE for the presence of the 17.5 kDa TMV coat protein. At 28 dpi, the plants with detectable TMV coat protein in the P1f set were nearly twice the number as compared to the CAP-34 treated set (Fig. 5a). In every instance of SDS–PAGE detection of TMV coat protein, a serological confirmation by Dot blot was also carried out. The result from the Dot blot analysis of one of the three experimental replicates is shown in Fig. 5b. The DW-treated plants susceptible to TMV infection (diseased-control), and CAP-34 and P1f-treated plants that were resistant to TMV infection (induced-resistant) are shown in Fig. 6a. The absence of the coat protein in induced-resistant plants at 28 dpi was reconfirmed by immunoblot and RT-PCR. No TMV coat protein specific band could be detected in the lanes carrying samples of induced-resistant plants from CAP-34 and P1f sets, and un-inoculated healthy-control plants by immunoblot and RT-PCR, while it was detectable in the samples from diseased-control set of plants, at the 17.5 kDa position in the immunoblot, and at the expected 242 bp position by RT-PCR (Fig. 6b).
Fig. 5.
Progression of mosaic and detection of TMV coat protein in Solanum lycopersicum (tomato). Plants in each set received a foliar treatment with CAP-34 (10 µg mL−1), P1f (1 × 108 cfu mL−1) and DW (control) for three consecutive days, and were challenged with TMV 24 h after the final treatment. On days 7, 14, and 28 post-inoculation, plants exhibiting mosaic were recorded and each plant was also screened for the TMV coat protein (CP). Ten replicates were used per treatment and the data represent mean ± SEM for each set (a). DotBlot detection of TMV coat protein in the three sets of plants (28 dpi), constituting one experimental replicate set. Five plants (H) were included as a healthy-control (b)
Fig. 6.
Induction of resistance against TMV in tomato and detection of TMV coat protein/RNA in induced-resistant tomato plants. Plants in each set received a foliar treatment with CAP-34 (10 µg mL−1), P1f (1 × 108 cfu mL−1) and DW for three consecutive days, and were challenged with TMV 24 h after the final treatment. At 28 dpi, representative plants from the DW-treated control set exhibiting mosaic symptoms (A) and the symptomless induced-resistant plants treated with CAP-34 (B) and P1f (C) are shown (Fig. 6a). Plants that were exhibiting resistance to TMV infection at 28dpi were evaluated for the presence or absence of the TMV coat protein by SDS–PAGE (A), Immunoblotting (B), and RT-PCR (C). TMV coat protein (17.5 kDa) was detectable in lanes loaded with DW-treated extract (disease-control) and purified TMV (A, B, lanes 2 and 6, respectively). Leaf extracts from induced-resistant CAP-34, P1f, and un-inoculated plants (healthy-control) were devoid of the TMV CP (A and B, lanes 3, 4, and 5, respectively). RT-PCR result is shown in C. Lanes 2, 3, 4, and 5 were loaded with the disease-control, induced-resistant CAP-34, P1f, and un-inoculated plants (healthy-control) samples, respectively. A 242 bp product, specific to the TMV coat protein was fetched following RT-PCR using TMV-CP specific primer pair, and detectable only in the disease-control set, showing systemic mosaic (C, lane2). A, B lane 1, protein markers; C lane 1, DNA ladder (Fig. 6b)
Estimation of comparative viral load in the plants exhibiting detectable mosaic by ELISA
A comparative estimation of the viral load in the tomato plants treated with DW, CAP-34 and P1f was made through indirect ELISA, by measuring absorbance at 405 nm, using plants showing mosaic at 28 dpi, and including an un-inoculated healthy-control. Using leaf extract at a 1:100 dilution, the absorbance values recorded were 2.55, 0.92, 1.26, and 0.04 for the disease-control, CAP-34, P1f-treated plants, and healthy-control, respectively. Both CAP-34 and P1f-treated plants showed considerably reduced TMV protein as compared to DW-treated, disease-control. The resistant plants gave a negligible absorbance, comparable to the healthy-control (Fig. 7).
Fig. 7.
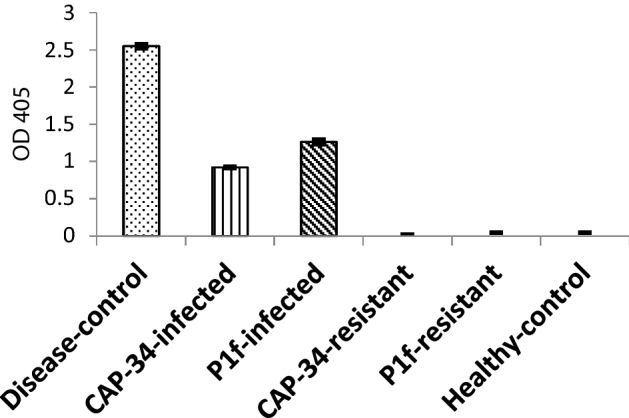
TMV accumulation detected in tomato plants by indirect ELISA at 405 nm. All plant tissues were extracted at a 1:100 dilution. The mean ELISA values ± SEM from three replicates of the experiment are plotted. CAP-34-infected and P1f-infected represent plants that were treated with CAP-34 and P1f, but developed mosaic upon challenge inoculation with TMV, while CAP-34-resistant and P1f-resistant represent samples from treated plants that showed resistance to TMV infection. The ELISA values were found significant at the 0.05 level compared to the DW-treated (disease-control) set
CAP-34 and P1f induced growth promotion in Solanum lycopersicum
Plant growth promoting effects on tomato were observed on the TMV inoculated plants, and also in the remaining three sets of plants that were treated with both CAP-34 and P1f, but left un-inoculated. Both treatments increased the fruit yield to similar levels. A statistically significant enhancement in the number of branches and flowers was observed in the set of plants treated with CAP-34, whereas a substantive increase in plant height and dry shoot and root weights was noted in the P1f-treated set. The number of fruits, and fresh weights of the fruit, shoot and root were equally enhanced by the two treatments. The set of plants that were challenged with TMV post-treatment, exhibited all values higher than the diseased-control plants, however, these values were somewhat less than those of the un-inoculated plants (Table 2).
Table 2.
Comparison of CAP-34 and P1f isolate treatments on the plant growth parameters of Solanum lycopersicum (tomato)
| Treatment* | Height (cm) | Branching (number) |
Flower (number) | Fruit (number) |
Fruit weight (g) |
Shoot fresh weight (g) | Shoot dry weight (g) | Root fresh weight (g) | Root dry weight (g) |
|---|---|---|---|---|---|---|---|---|---|
| DW | 82.10 ± 1.53a | 6.90 ± 0.37a | 8.80 ± 0.44a | 7.50 ± 0.37a | 133.00 ± 4.22a | 124.00 ± 3.22a | 42.97 ± 1.32a | 22.20 ± 1.19a | 10.11 ± 0.45a |
| DW/TMV | 56.40 ± 0.65b | 6.30 ± 0.633a | 5.90 ± 0.62b | 5.00 ± 0.51b | 84.16 ± 2.28b | 84.47 ± 3.45b | 29.51 ± 2.03b | 16.24 ± 0.77b | 6.47 ± 0.33b |
| CAP-34 | 116.80 ± 2.48c | 9.40 ± 0.60b | 11.50 ± 0.77c | 9.50 ± 0.65ac | 169.60 ± 3.42c | 151.20 ± 4.2c | 50.61 ± 2.46ac | 32.90 ± 1.07c | 12.66 ± 0.60c |
| CAP-34/TMV | 84..40 ± 2.00a | 8.10 ± 0.78c | 9.20 ± 0.53a | 7.80 ± 0.38a | 140.65 ± 6.96d | 135.74 ± 2.37d | 37.43 ± 2.18d | 25.97 ± 1.16d | 12.19 ± 0.71c |
| P1f | 123.40 ± 1.92d | 8.60 ± 0.61c | 10.70 ± 0.68d | 8.20 ± 0.44a | 168.40 ± 6.133c | 155.90 ± 3.33c | 53.52 ± 1.71de | 32.10 ± 1.69c | 14.88 ± 0.66d |
| P1 f /TMV | 90.20 ± 2.46e | 7.30 ± 0.59a | 10.2 ± 0.68a | 8.40 ± 0.42a | 141.04 ± 2.09d | 138.01 ± 2.59d | 41.76 ± 1.41ad | 29.70 ± 0.92c | 12.66 ± 0.71c |
*Six sets of plants, with ten plants in each set, were treated with DW, CAP-34 (10 µg mL−1), and P1f isolate (1 × 108 cfu mL−1). DW-treated plants served as a control. Three sets of treated plants were inoculated with TMV 24 h later, while three remaining sets of treated plants were left un-inoculated. Growth parameters were measured at the end of 3 months for each plant
Values represent mean of ten replicates ± SE. The values in the same column having different letters (a–e) are significantly different (P ≤ 0.05) in multiple comparisons LSD test
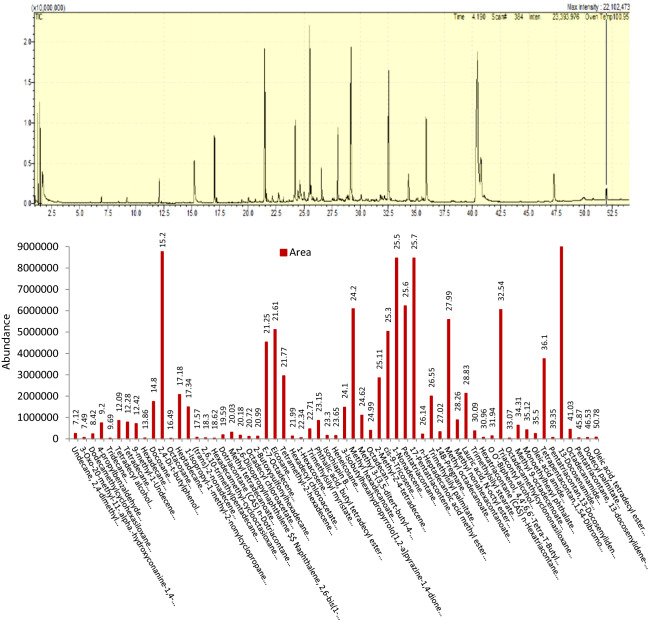
Study of the volatiles produced by P1f
Nearly 61 metabolites were produced by the overnight culture of the isolate P1f as determined through GC–MS analysis (Fig. 8). A few were produced in relatively larger amounts by the bacterial isolate P1f, and these included several alkenes, alkanes, esters, alcohols, ketones, amides and aromatic compounds viz., 2,4-Di-t-butylphenol, E-7-Octadecene, Eicosane, Methyl hexadecanoate, Cis-9-tricosene, 1-Nonadecene, Pentatricontane, 17 Pentatriacontene, Methyl octadecanoate, Troicontyl alcohol and 13-Docasenamide.
Fig. 8.
GC–MS analysis of volatiles and secondary metabolites produced by the Pseudomonas putida isolate P1f
Discussion
Antiviral resistance inducing protein CAP-34, isolated from the leaves of Clerodendrum aculeatum (syn. Volkameria aculeata), and a new rhizobacterial isolate, P1f, identified as Pseudomonas putida based on 16S rRNA gene sequence and metabolic capability, were evaluated for their antiviral resistance inducing ability in Nicotiana tabacum (tobacco) cultivar, Xanthi-nc and Solanum lycopersicum (tomato), against TMV. Interest in this comparison arose from the fact that the two differed widely in their nature as inducers, but led to the development of antiviral resistance, albeit to different extents. Since manipulation of either of the two modes for disease control would require an understanding of the mechanism that led to observed effects, we considered it prudent to carry out this comparative study. Although P1f was identified as an isolate of Pseudomonas putida, a definitive identification of Pseudomonas species, especially within the same group, has always presented a problem, and many sequences deposited in the database have been erroneously identified (Morimoto et al. 2020).
Rhizobacteria-mediated resistance in susceptible hosts against plant viruses, especially CMV, has been extensively demonstrated (Raupach et al. 1996; Murphy et al. 2000; Zehnder et al.2000; 2001). 10 µg mL−1 (CAP-34) and 1 × 108 cfu mL−1 (P1f) were the lowest concentrations resulting in maximum resistance induction. Hence, all experiments were conducted using these concentrations. In the present study, the most effective form of treatment for induction of resistance against TMV on tobacco was in the form of a foliar spray and not a soil drench (data not shown), hence this became the method of choice for induction of resistance against TMV in tomato and tobacco. The duration of induced-resistant state in the treated tobacco plants was greater with the rhizobacterial isolate, as compared to that with CAP-34. Previous results have shown that CAP-34 induced resistance typically lasts for a few days, before dying out (Prasad et al. 2014). Hence, for effective virus control, the treatment needed to be repeated to prolong the antiviral state in the plants (Srivastava et al. 2009). PGPR induced resistance responses are known to persist (van Loon et al. 1998). For comparative evaluation, tomato plants were challenge inoculated with TMV 24 h after treatment, when both treatments (CAP-34 and P1f) yielded a similar resistance response on hypersensitive tobacco.
The number of plants that succumbed to mosaic in the CAP-34 treated set was approximately 50% of the number that were infected within the set treated with P1f isolate, along with a corresponding difference in ELISA values. The visual intensity of mosaic was comparable in all symptomatic plants at 28 dpi. The complete absence of the coat protein or viral RNA in the resistant plants within the CAP-34- and P1f-treated sets, indicated an interference with the infection process of TMV. The slow increase in the number of symptomatic plants or a delay in symptom development in some plants within the treated sets could be due to an impediment in the movement of the virus, or virus replication itself.
The protection afforded against virus infection in plants treated by PGPR in a few instances could be due to enhanced growth of plants, which helped them respond better to the infection stress, or a consequence of ISR (Murphy et al. 2003; Beneduzi et al. 2012). Growth promotion studies have been carried out earlier using CAP-34, but a comparative study with rhizobacteria was never undertaken. P. putida isolate P1f-treated tomato exhibited better height and dry shoot weights. In comparison, however, the branching and flowering in CAP-34 treated sets was noticeably higher than the P1f. CAP-34 has been previously studied for its resistance induction against PRSV and TMV in papaya and the susceptible Xanthi cultivar of tobacco, respectively, and effective virus inhibition in both cases was shown to be due to systemic resistance induction (Srivastava et al. 2009, 2015b). Rhizobacteria protect susceptible plants from multiple pathogen infections. Pseudomonas species, especially strains of P. fluorescens, P. aeruginosa, and P. putida have been reported to protect plants against viral, bacterial and fungal pathogens (Ahn et al. 2007; Verhagen et al. 2010). The potential of PGPR to curb plant RNA virus infections became evident recently (Maksimov et al. 2019), while the bacterial determinants of ISR produced by fluorescent pseudomonads have been known since before (Bakker et al. 2007). Pseudobactin, a fluorescent siderophore, could promote plant growth and increase yield (Kloepper et al. 1980). It has since been reported to be a determinant of ISR, being produced by P. fluorescens WCS374r and P. putida WCS358 (Ran et al. 2005; Meziane et al. 2005). Another Fe-regulated elicitor of ISR produced by P. putida BTP1 has been characterized as N-alkylated benzylamine derivative (Ongena et al. 2005). The siderophore produced by the P. putida isolate P1f has not been characterized, but it could contribute to resistance induction, especially since the cell-free extract itself could induce resistance in tobacco, though not to the same extent as the bacterial cells in an overnight culture (data not shown).
An analysis of the volatile organic compounds (VOCs)/secondary metabolite) profile of P1f was carried out since bacterial volatile components are known to serve as agents for triggering growth promotion in plants and inducing systemic resistance in susceptible hosts (Ryu et al. 2003, 2004; Gouda et al. 2018; Netzker et al. 2020). B. subtilis GB03 that induced ISR released volatile compounds that were identified as 3-hydroxy-2-butanone, 2,3-butanediol, tetramethyl pyrazine, and the hydrocarbons decane, undecane, decanal, dodecane, decanol, 2-undecanone, 2-tridecanone, and 2-tridecanol. GC–MS identified VOCs produced by P. putida P1f, several of which reportedly induce ISR, alter bacterial gene expression or show antimicrobial activity (Netzker et al. 2020). Amongst these, of special interest is the toxic lipophilic phenol, 2,4-di-t-butylphenol that is also known to control quorum sensing mediated biofilm formation by bacteria (Viszwapriya et al. 2016) and hence could have a potential application in spheres where biofilm formation poses a risk.
Rapid defence responses of the host involving production of callose and phenolic compounds along with stronger and faster expression of defense-related genes underlies the phenomenon of ISR. This preparedness, termed defence priming (Verhagen et al. 2004; Martinez-Madina et al. 2016; Mauch-mani et al. 2017), involves local and systemic transcriptional reprogramming (Schenk et al. 2014). Enhanced peroxidase enzyme activity and H2O2 accumulation was notably greater for P1f-treated tobacco plants as compared to CAP-34, and may have participated in the inhibition of virus infection in tobacco, and an increase in the duration of resistance in P1f-treated plants.
Induced systemic resistance by CIP-29, CAP-34 and BDP-30 is host mediated, known to be abolished by simultaneous application of actinomycin D. A virus inhibitory agent (VIA) was always found associated with this form of resistance that could inactivate plant viruses in vitro (Verma and Awasthi 1980; Verma and Dwivedi 1984; Prasad et al. 1995). Two virus inhibitory proteins, CT-VIA-62 and CT-VIA-32 were purified from Cyamopsis tetragonoloba plants subsequent to treatment with CIP-29. CT-VIA-62 shared sequence homology with a lectin that possessed a mannose-binding domain (Prasad et al. 2014). CP-VIA-34, isolated from induced-resistant papaya plants, affords yet another example of a VIA that inactivated TMV, but was devoid of protease, DNase and RNase activities in vitro (Srivastava et al. 2015a). As the VIA is synthesized in every instance of resistance induction by CAP-34, CIP-29 or BDP-30, the leaf extract from the PGPR induced-resistant tobacco plants was evaluated for VIA production. The absence of virus inhibitory activity in the total protein fraction obtained from the P1f-treated set were perhaps indicative of an alternative mechanism, different from that of the VIA.
Acknowledgements
AKG gratefully acknowledges financial support as CSIR NET-JRF no. 19/06/2016(i)EU-V. Rajesh K Tewari, Associate Professor, is acknowledged for help.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Footnotes
ACCESSION NO.: GenBank MZ569722.
References
- Ahn IP, Lee SW, Suh SC. Rhizobacteria-induced priming in Arabidopsis is dependent on ethylene, jasmonic acid, and NPR1. Mol Plant Microbe Interact. 2007;20:759–768. doi: 10.1094/MPMI-20-7-0759. [DOI] [PubMed] [Google Scholar]
- Bakker AW, Schippers B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp-mediated plant growth-stimulation. Soil Biol Biochem. 1987;19:451–457. [Google Scholar]
- Bakker PAHM, Pieterse CMJ, van Loon LC. Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology. 2007;97:239–243. doi: 10.1094/PHYTO-97-2-0239. [DOI] [PubMed] [Google Scholar]
- Beneduzi A, Ambrosini A, Passaglia MP. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Gen Mol Biol. 2012;35(4):1044–1051. doi: 10.1590/s1415-47572012000600020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beris D, Theologidis I, Skandalis N, Vassilakos N. Bacillus amyloliquefaciens strain MBI600 induces salicylic acid dependent resistance in tomato plants against Tomato spotted wilt virus and Potato virus Y. Sci Rep. 2018;8:10320. doi: 10.1038/s41598-018-28677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bric JM, Bostock RM, Silversone SE. Rapid in situ assay for indole acetic acid production by bacteria immobilization on a nitrocellulose membrane. App Environ Microbiol. 1991;57:535–538. doi: 10.1128/aem.57.2.535-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman E, Bostwick JR, Eichberg J. Determination of protein by modified Lowry procedure in the presence of some commonly used detergents. Anal Biochem. 1979;96:21–23. doi: 10.1016/0003-2697(79)90548-7. [DOI] [PubMed] [Google Scholar]
- De Meyer G, Capieau K, Audenaert K, Buchala A, Métraux JP, Höfte M. Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol Plant Microbe Interact. 1999;12:450–458. doi: 10.1094/MPMI.1999.12.5.450. [DOI] [PubMed] [Google Scholar]
- Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Dong X. Systemic acquired resistance: TURNING local infection into global defence. Annu Rev Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- Gordon SA, Weber RP. Colorimetric estimation of indole acetic acid. Plant Physiol. 1951;26:192–195. doi: 10.1104/pp.26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouda S, Kerry RG, Das G, Paramithiotis S, Shin H-S, Patra JK. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol Res. 2018;206:131–140. doi: 10.1016/j.micres.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt R, Nuckles E, Kuc J. Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiol Plant Pathol. 1982;20:73–82. [Google Scholar]
- Hernandez JA, Ferrer MA, Jimenez A, BarceloAR SF. Antioxidant systems and O2¯/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol. 2001;127:817–831. doi: 10.1104/pp.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessmann H, Staub T, Hofmann C, Maetzke T, Herzog J, Ward E, Uknes S, Ryals J. Induction of systemic acquired disease resistance in plants by chemicals. Annu Rev Phytopathol. 1994;32:439–459. doi: 10.1146/annurev.py.32.090194.002255. [DOI] [PubMed] [Google Scholar]
- King ED, Ward MK, Raney DE. Two simple media for the demonstration ofpyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- Kloepper JW, Leong J, Teintze M, Schroth MN. Pseudomonas siderophores: a mechanism explaining disease-suppressive soils. Curr Microbiol. 1980;4:317–320. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maksimov IV, Sorokan AV, Burkhanova GF, Vesselova SV, Yu AV, Yu SM, Avalbaev AM, Dhaware PD, Mehetre GT, Singh BP, Khairullin RM. Mechanisms of plant tolerance to RNA viruses induced by plant growth-promoting microorganisms. Plants (basel) 2019;8:575. doi: 10.3390/plants8120575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Medina A, Flors V, Heil M, Mauch-Mani B. Recognizing plant defense priming. Trends Plant Sci. 2016 doi: 10.1016/j.tplants.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Baccelli I, Luna E, Flors V. Defense priming: an adaptive part of induced resistance. Annu Rev Plant Biol. 2017;68:485–512. doi: 10.1146/annurev-arplant-042916-041132. [DOI] [PubMed] [Google Scholar]
- Maurhofer M, Hase C, Meuwly P, Metraux J-P, Defago G. Induction of systemic resistance of tobacco to tobacco necrosis virus by the root colonizing Pseudomonas fluorescens strain CHA0: influence of the gacA gene and of pyoverdine production. Phytopathology. 1994;84:139–146. [Google Scholar]
- Maurhofer M, Baehler E, Notz R, Martinez V, Keel C. Cross talk between 2,4-Diacetylphloroglucinol—producing biocontrol Pseudomonads on wheat roots. Appl Environ Microbiol. 2004;70:1990–1998. doi: 10.1128/AEM.70.4.1990-1998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meziane H, Van der Sluis I, Van Loon LC, Höfte M, Bakker PAHM. Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol Plant Pathol. 2005;6:177–185. doi: 10.1111/j.1364-3703.2005.00276.x. [DOI] [PubMed] [Google Scholar]
- Morimito Y, Tohya M, Aibibula Z, Baba T, Hiroyuki D, Kirikae T. Re-identification of strains deposited as Pseudomonas aeruginosa, Pseudomonas fluorescens and Pseudomonas putida in GenBank based on whole genome sequences. Int J Syst Evol Microbiol. 2020;70:5958–5963. doi: 10.1099/ijsem.0.004468. [DOI] [PubMed] [Google Scholar]
- Mowat WP, Dawson S. Detection of plant viruses by ELISA using crude sap extracts unfractionated antisera. J Virol Methods. 1987;15:233–247. doi: 10.1016/0166-0934(87)90101-7. [DOI] [PubMed] [Google Scholar]
- Murphy JF, Zehnder GW, Schuster DJ, Sikora EJ, Polston JE, Kloepper JW. Plant growth-promoting rhizobacterial mediated protection in tomato against Tomato mottle virus. Plant Dis. 2000;84:779–784. doi: 10.1094/PDIS.2000.84.7.779. [DOI] [PubMed] [Google Scholar]
- Murphy JF, Reddy MS, Ryu C-M, Kloepper JW, Li R. Rhizobacteria mediated growth promotion of tomato leads to protection against cucumber mosaic virus. Phytopathology. 2003;93:1301–1307. doi: 10.1094/PHYTO.2003.93.10.1301. [DOI] [PubMed] [Google Scholar]
- Netzker T, Shepherdson EMF, Zambri MP, Elliot MA. Bacterial volatile compounds: functions in communication, cooperation, and competition. Annu Rev Microbiol. 2020;74:409–430. doi: 10.1146/annurev-micro-011320-015542. [DOI] [PubMed] [Google Scholar]
- Olivieri F, Prasad V, Valbonesi P, Srivastava S, Ghosal- Chowdhury P, Barbieri L, Bolognesi A, Stirpe F. A systemic antiviral resistance inducing protein isolated from Clerodendrum inerme Gaertn. is a polynucleotide: adenosine glycosidase (ribosome-inactivating protein) FEBS Lett. 1996;396:132–134. doi: 10.1016/0014-5793(96)01089-7. [DOI] [PubMed] [Google Scholar]
- Ongena M, Jourdan E, Schäfer M, Kech C, Budzikiewicz H, Luxen A, Thonart P. Isolation of an N-alkylated benzylamine derivative from Pseudomonas putida BTP1 as elicitor of induced systemic resistance in bean. Mol Plant-Microbe Interact. 2005;18:562–569. doi: 10.1094/MPMI-18-0562. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PA. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- Pikovskaya RI. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Mikrobiologiya. 1948;17:362–370. [Google Scholar]
- Prasad V, Srivastava S, Verma HN. Two basic proteins isolated from Clerodendrum inerme Gaertn. are inducers of systemic antiviral resistance in susceptible plants. Plant Sci. 1995;110:73–82. [Google Scholar]
- Prasad V, Srivastava S (2017) Phytoproteins and induced antiviral defence in susceptible plants: The Indian context. In: Mandal B, Rao GP, Baranwal V, Jain R (eds.) A century of plant virology in India, pp 689–728. 10.1007/978-981-10-5672-7_28
- PrasadV MSK, Srivastava S, Srivastava A. A virus inhibitory protein isolated from Cyamopsis tetragonoloba upon induction of systemic antiviral resistance shares partial amino acid sequence homology with a lectin. Plant Cell Rep. 2014;33:1467–1478. doi: 10.1007/s00299-014-1630-7. [DOI] [PubMed] [Google Scholar]
- Ran LX, Van Loon LC, Bakker PAHM. No role for bacterially produced salicylic acid in rhizobacterial induction of systemic resistance in Arabidopsis. Phytopathology. 2005;95:1349–1355. doi: 10.1094/PHYTO-95-1349. [DOI] [PubMed] [Google Scholar]
- Raupach GS, Liu L, Murphy JF, Tuzun S, Kloepper JW. Induced systemic resistance of cucumber and tomato against cucumber mosaic cucumovirus using plant growth-promoting rhizobacteria (PGPR) Plant Dis. 1996;80:891–894. [Google Scholar]
- Ryu C-M, Farag MA, Hu C-H, Reddy MS, Wei HX, PareKloepper PWJW. Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C-M, Farag MA, Hu C-H, Reddy MS, Kloepper JW, Pare PW. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004;134:1017–1026. doi: 10.1104/pp.103.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk ST, Hernandez-Reyes C, Samans B, Stein E, Neumann C, et al. N-acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell. 2014;26:2708–2723. doi: 10.1105/tpc.114.126763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for detection and determination of siderophore. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Trivedi S, Krishna SK, Verma HN, Prasad V. Suppression of papaya ringspot virus infection in Carica papaya with CAP-34, a systemic antiviral resistance inducing protein from Clerodendrum aculeatum. Eur J Plant Pathol. 2009;123:241–246. [Google Scholar]
- Srivastava A, Srivastava S, Prasad V. Antiviral resistance induced in papaya by CAP-34, a resistance inducing protein from Clerodendrum aculeatum, is associated with a proteinaceous virus inhibitory activity. J Plant Pathol. 2015;97:45–54. [Google Scholar]
- Srivastava S, Verma HN, Srivastava A, Prasad V. BDP-30, a systemic resistance inducer from Boerhaavia diffusa L., suppresses TMV infection, and displays homology with ribosome inactivating proteins. J Biosci. 2015;40(1):1–12. doi: 10.1007/s12038-014-9494-0. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of protein from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon LC, Bakker PAHM, Pieterse MJ. Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol. 1998;36:453–483. doi: 10.1146/annurev.phyto.36.1.453. [DOI] [PubMed] [Google Scholar]
- Verhagen BWM, Glazebrook J, Zhu T, Chang H-S, van Loon LC, Pieterse CMJ. The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol Plant-Microbe Interact. 2004;17:895–908. doi: 10.1094/MPMI.2004.17.8.895. [DOI] [PubMed] [Google Scholar]
- Verhagen BWM, Trotel-Aziz P, Couderchet M, Hofte M, Aziz A. Pseudomonas spp.-induced systemic resistance to Botrytis cinerea is associated with induction and priming of defence responses in grapevine. J Exp Bot. 2010;61:249–260. doi: 10.1093/jxb/erp295. [DOI] [PubMed] [Google Scholar]
- Verma HN, Awasthi LP. Occurrence of a highly antiviral agent in plants treated with Boerhaavia diffusa inhibitor. Can J Bot. 1980;58:2141–2144. [Google Scholar]
- Verma HN, Dwivedi SD. Properties of a virus inhibiting agent isolated from plants following treatment with Bougainvillea spectabilis leaf extract. Physiol Plant Pathol. 1984;25:93–101. [Google Scholar]
- Verma HN, Srivastava S, VarshaKumar D. Induction of systemic resistance in plants against viruses by a basic protein from Clerodendrum aculeatum leaves. Phytopathology. 1996;86:485–492. [Google Scholar]
- Viszwapriya D, Prithika U, Deebika S, Balamurugan K, Pandian SK. In vitro and in vivo antibiofilm potential of 2, 4-di-tert-butylphenol from seaweed surface associated bacterium Bacillus subtilis against group A Streptococcus. Microbiol Res. 2016;191:19–31. doi: 10.1016/j.micres.2016.05.010. [DOI] [PubMed] [Google Scholar]
- Wilkinson SW, Mageroy MH, Sanchez AL, Smith LM, Furci L, Anne Cotton TE, Krokene P, Ton J. Surviving in a hostile world: plant strategies to resist pests and diseases. Annu Rev Phytopath. 2019;57:505–529. doi: 10.1146/annurev-phyto-082718-095959. [DOI] [PubMed] [Google Scholar]
- Zehnder GW, Yao C, Murphy JF, Sikora EJ, Kloepper JW. Induction of resistance in tomato against cucumber mosaic cucumovirus by plant growth-promoting rhizobacteria. Bio Control. 2000;45:127–137. [Google Scholar]
- Zehnder GW, Murphy JF, Sikora EJ, Kloepper JW. Application of rhizobacteria for induced resistance. Eur J Plant Pathol. 2001;107:39–50. [Google Scholar]