Abstract
The developmental and epileptic encephalopathies (DEEs) are the most severe group of epilepsies. They usually begin in infancy or childhood with drug-resistant seizures, epileptiform EEG patterns, developmental slowing or regression, and cognitive impairment. DEEs have a high mortality and profound morbidity; comorbidities are common including autism spectrum disorders. With advances in genetic sequencing, over 400 genes have been implicated in DEEs, with a genetic cause now identified in over 50% patients. Each genetic DEE typically has a broad genotypic-phenotypic spectrum, based on the underlying pathophysiology. There is a pressing need to improve health outcomes by developing novel targeted therapies for specific genetic DEE phenotypes that not only improve seizure control, but also developmental outcomes and comorbidities. Clinical trial readiness relies firstly on a deep understanding of phenotype-genotype correlation and evolution of a condition over time, in order to select appropriate patients for clinical trials. Understanding the natural history of the disorder informs assessment of treatment efficacy in terms of both clinical outcome and biomarker utility. Natural history studies (NHS) provide a high quality, integrated, comprehensive approach to understanding a complex disease and underpin clinical trial design for novel therapies. NHS are pre-planned observational studies designed to track the course of a disease and identify demographic, genetic, environmental, and other variables, including biomarkers, that correlate with the disease’s evolution and outcomes. Due to the rarity of individual genetic DEEs, appropriately funded high-quality DEE NHS will be required, with sustainable frameworks and equitable access to affected individuals globally.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13311-021-01133-3.
Keywords: Natural history studies, Epilepsy, Developmental and epileptic encephalopathies, Clinical trials, Biomarkers, Clinical outcome assessments
What are Developmental and Epileptic encephalopathies and Why Do They Need Novel Therapies?
Definition of DEEs
A developmental and epileptic encephalopathy (DEE) is defined by a combination of two etiological processes [1]. A DEE is defined by both a ‘developmental’ encephalopathy where the developmental disorder is directly due to the underlying cause, and an ‘epileptic’ encephalopathy due to adverse effects from seizures and frequent epileptiform activity resulting in developmental slowing or regression [2].
What are the Causes of DEEs?
The aetiology of the epilepsies is heterogeneous and includes both acquired and genetic causes. DEEs are the group of epilepsies in which a genetic diagnosis is most likely to be made. In neonatal onset DEEs, the aetiology is now identified in over 60% of cases, with a range of diagnostic yields between 25 and 50% for later onset DEEs [3–9]. Over 400 DEE genes have been reported, affecting diverse cellular processes including ion transport, cell growth and differentiation, synaptic processes, transport and metabolism of small molecules, and gene transcription and translation [10].
What is the Impact of a DEE?
The impact of the epilepsy, neurodevelopmental disabilities, and other comorbidities of a DEE on the child, their family, and wider community is profound [11]. Seizures are frequent and often drug-resistant, with resultant risks of injury and death, need for emergency anti-seizure medications (ASM) and hospitalization, and negative impacts on day-to-day function. The impacts on development are immense: developmental delay resulting in intellectual disability is present in most individuals and is frequently severe [12]. Individuals with DEEs have a range of comorbidities including movement disorders, cerebral palsy, behavioural disorders, autism spectrum disorder, psychosis, sleep, gait, speech, respiratory, and gastrointestinal disorders [13, 14, 85]. The mortality of DEEs is high [15, 16]: reaching over 50% by 2 years of age for children with neonatal-onset DEE [9, 17]. These chronic complex conditions are highly challenging for families: a recent survey of caregivers of children with severe epilepsies in Denmark found that over 40% of primary caregivers had depression, anxiety, or post-traumatic stress disorder [11]. The DEEs also pose a huge economic burden on communities, with most patients being dependent on daily care and requiring support throughout life [16, 17].
What is the Treatment Gap for DEE?
In DEEs, there is a particularly urgent need for novel therapies that address all aspects of these severe disorders [18, 19]. Currently, most therapies for DEEs target individual symptoms such as seizures, and not the underlying disease mechanisms. For many individuals with DEEs, seizure control is not achieved, even when ASM are optimized for the underlying aetiology [18, 20]. However, even when seizure control is achieved, developmental impairments and other comorbidities often remain severe. Improving these is likely to require therapies which target the underlying cause of the DEE, or ‘precision medicines’ [21].
Currently, there are few aetiology-specific therapies for genetic DEEs [21, 22]. The vast majority are supplements, enzyme replacement, or special diets which target specific metabolic defects causing DEEs due to inborn errors of metabolism (IEM), with variable effectiveness. Examples include the ketogenic diet for GLUT1-deficiency encephalopathy, pyridoxine for pyridoxine-dependent epilepsies due to pathogenic variants in ALDH7A1, creatine supplements for GAMT and SLC6A8-related disorders, folinic acid for FOLR1-related disorders, galactose supplements for SLC35A2-related disorders, and tripeptidyl-peptidase I enzyme replacementff therapy for CLN2 disease [23, 24]. Beyond the metabolic DEEs, DEE-specific therapies are limited as most causes lack a ‘biochemical’ pathway to target; however, some therapies that target ion channels and glutamate receptor genes show at least partial efficacy [6, 10]. There is indeed much hope that novel genetic therapies, truly targeted to the underlying aetiology, will be transformative, and holistically address the patient’s condition [24, 25]. Options being explored include gene therapies such as gene replacement and antisense oligonucleotides, with a number of novel agents in preclinical development or in the early phases of clinical trials [26].
Individual Genetic DEEs are Rare Disorders
Epidemiological Studies of DEEs
The DEEs have an incidence of more than 1 in 2000 live births [4, 9] making them more common than many wellknown severe genetic child conditions including cystic fibrosis (1:3650) and spinal muscular atrophy (1:7000) [27, 28]. However, compared to these two diseases, which each have one genetic cause, the DEEs are highly genetically heterogeneous with over 400 monogenic causes [10].
Thus, while collectively genetic DEEs are one of the most common genetic childhood conditions, each individual genetic DEE is rare and meets criteria for being an ‘orphan’ disorder [29]. For example, SCN1A-related epilepsy has an incidence of 1 per 12,200 with the most common SCN1A-DEE phenotype, Dravet syndrome, having an incidence of 1 per 15,500; KCNQ2-related epilepsy: 1 per 17,000; and SLC2A1-related epilepsy 1 per 24,300 [4]. Globally most jurisdictions use a definition of rare or orphan disorder of being one which affects fewer than 1:2000 people [30]. The concept of rare disease is important as it implies that the cost of developing and making available a specific therapy for such conditions would not be recovered from sales in an individual country. This means special drug development pipelines and incentives are required to make orphan disease drug development an attractive option for pharmaceutical companies and researchers, for example, the Orphan Products Grants Program in the USA. Frameworks to support orphan drug development also exist in Europe and Japan, with current legislative frameworks under review in some countries including Canada and Australia [31]. High-quality global natural history studies (NHS) are acknowledged as being critical in novel therapy development and subsequent regulatory approval for rare diseases [32].
The Importance of Natural History Studies for DEEs
What is a Natural History Study?
The natural history of a disease is traditionally defined as the course a disease takes in the absence of intervention, from the disease’s onset until either its resolution or the individual’s death [32]. However, natural history studies (NHS) now commonly include patient cohorts who are receiving treatments, such as ASM. Therefore, NHS in such conditions are perhaps more accurately conceptualized as observational studies designed to track the course of a disease and to identify demographic, genetic, environmental, and other variables (e.g. treatments) that correlate with the disease’s course and outcomes. NHS are different from patient registries, which can include demographic, genotypic and certain clinical data, but do not have the same breadth and depth, nor do they track disease evolution.
Objectives of NHS
NHS are fundamental for enhancing patient care, for example, by informing prognostication, the counselling of affected individuals and their families about the potential future evolution of the condition, and by guiding anticipatory surveillance [32]. NHS are also required to improve future models of care by informing successful clinical trial designs. NHS achieve this primarily by (i) identifying candidate patient populations and subpopulations, (ii) providing historic control data, and, (iii) guiding choice of the most appropriate clinical outcome measures and biomarkers for clinical trials [32] (see “12” for further detail).
NHS Design
NHS vary in their design with respect to whether data are collected retrospectively or prospectively, and whether in a cross-sectional or longitudinal fashion [32, 33]. Retrospective studies compile data from existing patient records, published case, and cohort reports or from patients and parents themselves. They are relatively cheap, quick, and less labour intensive. They are very helpful in organizing information about a condition and identifying information gaps that could be addressed by prospective studies. Limitations of retrospective NHS include data gaps and omissions, inconsistencies in diagnostic criteria and assessments, and reporting biases. For example, NHS based on patient records may over-represent more severely affected patients because this patient subgroup is likely to present more frequently to a medical centre. Conversely, NHS based on published case or cohort studies may overrepresent subgroups of patients with positive response to certain trialled treatments. Another limitation of retrospective studies is that they may result in under-appreciation of the impact of recent therapeutic advances, such as fenfluramine in Dravet syndrome, on the long-term outcomes of children who have had genetic diagnoses. In addition, genetic conditions are increasingly diagnosed at a very young age, including the neonatal period; such diagnostic advances allow prompt administration of optimal therapies, and avoidance of treatments with potential negative impacts, such as sodium channel blockers in Dravet Syndrome [34]. Although innovative data-mining approaches have been developed to ‘fast-track’ retrospective NHS, caution should be taken in using this approach for conditions such as DEEs where the complexity, breadth, and severity of symptoms are likely not to be adequately captured by data mining [33].
Well-designed prospective studies can address most of the limitations of retrospective studies through the use of carefully assessed patient inclusion and exclusion criteria, approved diagnostic criteria, standardized clinical outcome assessments, and consistent assessment schedules delineating the exact data to be collected at specific time points [32]. Prospective studies do, of course, need to consider, record, and analyze the clinical features that have come before the point of enrolment for each patient, which can be provided by assessment of retrospective data.
NHS can collect data using a cross-sectional design, whereby data is collected from across a patient cohort during a specified time, or via a longitudinal design, whereby data is collected from all patients in a cohort over several time points. Cross-sectional designs are quicker and may allow inferences of the general course of a disorder by sampling a cohort of patients at various stages of the disease, gathering information on the range and severity of core features and the range of comorbidities of a disorder. However, they may not be able to fully characterize the course and the natural history of different subtypes of a condition. For these reasons, prospective longitudinal NHS are considered the gold standard, allowing for comprehensive data collection about disease onset and progression, distinction of subtypes, and phenotypic variation. However, it is acknowledged that this type of study design takes both time and intensive resourcing to ensure appropriate recruitment and informative repeated clinical data sampling, and will not be feasible, or indeed necessary, in all instances depending on how homogeneous the phenotypic spectrum of the condition is.
Many genetic DEEs are not only rare, but ‘ultra-rare’. ‘Ultra-rare’ disorders have a prevalence of less than 1 per 2,000,000 [35]. Thus, to reach patient recruitment goals, many NHS will need to be conducted as international multi-centre studies. NHS protocols and consent should ideally cover biological sample and genetic test result storage, and, where appropriate, cover possible future secondary research use of the data and collected biospecimens. This will assist in the accurate subgrouping of patients together with development and evaluation of biomarkers. A plan for dissemination of the results of NHS should also be agreed at the outset, which should be as wide as possible, including peer-reviewed journals, due to the paucity of existing natural history data for each genetic DEE.
Close liaison and co-design of NHS with patient advocacy groups has many advantages. Firstly, such engagement can optimize recruitment and patient and caregiver engagement over the length of the NHS. Secondly, co-design ensures inclusion of clinical outcome assessments (COA) [36] that measure those aspects of the disease that are most important for patients and caregivers [37]. However, NHS also require input from expert clinician-researchers with the relevant skills in phenotyping, design, and data analysis to ensure appropriate quality, interpretation, and application of the data. The potential burden of participation in NHS on families should be carefully considered, especially for families with very disabled children and for those families where there may be cultural, language, geographical, or financial barriers to participation. The psychosocial impacts on caregivers of children with DEEs are already very high, with high rates of caregiver chronic traumatic stress, anxiety, and depression [11]. Without careful consideration of how to minimize the burden of NHS on families, there is a real risk of both ethical concerns and of bias, for example over-representing more mildly affected children from metropolitan, affluent families. NHS design for DEE should thus consider innovative methods to improve access and acceptability of data collection, for example, using telehealth options, covering transport and accommodation costs when visits to tertiary centres are required, investing in appropriately trained staff such as psychologists and play therapists to support children and families, and considering mental health and wellbeing programs aimed at improving the coping and resilience of families with severely affected children [38].
How Can NHS Be Used to Enable Novel Therapeutic Drug Development and Marketing Approval?
By Identification of Patient Subgroups and Clinical Trial Candidates
NHS are required to clarify which patient population subgroups should be included in specific clinical trials. This is critical for DEEs because the phenotypic and genotypic spectrum of many genetic DEEs is complex and multifaceted. The phenotypic spectrum of specific genetic DEEs, their underlying genotypes, and pathophysiological mechanisms must be understood to appropriately develop and test the efficacy of targeted therapies.
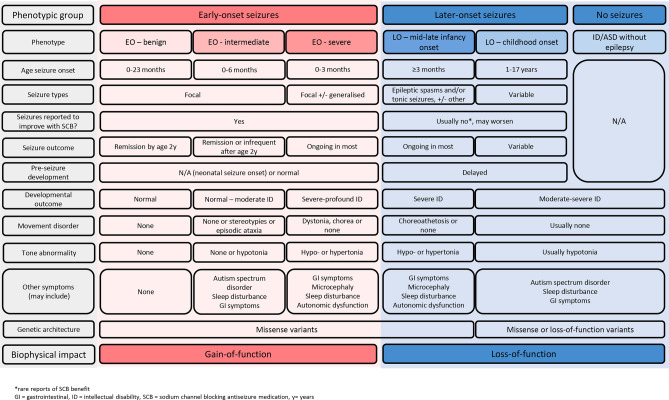
For example, as highlighted in Fig. 1, subtypes of SCN2A-related conditions are caused by different types of genetic variation which have distinct functional impacts. Understanding this complexity is required to understand rational ASM choice and novel therapeutic design. For example, patients with SCN2A-encephalopathy due to missense variants resulting in gain of Nav1.2 channel function in a single-cell model and severe increase in neuronal firing in a neuronal network model, should be separated from patients with SCN2A-encephalopathy due to variants resulting in loss of function of Nav1.2 channels. These two subtypes of SCN2A-encephalopathy differ in their clinical course, including earlier seizure onset in those with gain-of-function variants, with variable seizure control over time. Critically, they require distinct therapeutic approaches, with the potential for harm if the wrong mechanistic approach were to be employed [39–41]. For example, whereas the former patient group may be best trialled with therapies which inhibit channel function or antisense oligonucleotides which suppress expression of the mutated gene, the latter group may be best trialled with gene therapy or therapies aimed at boosting channel function [42].
Fig. 1.
Relationships between phenotypic, genotypic, and biophysical features in SCN2A-related disorders
Genotype-biophysical-phenotype correlations are often not straightforward; the same variant can be associated with different phenotypes both between and within families [43–45]. It is likely that differences in genotypic background as well as environmental factors and possibly therapeutic interventions also impact outcomes. This genotype-pathophysiology-phenotype complexity is evident in a range of DEE genes, including voltage-gated (other sodium, potassium, and calcium) and ligand-gated channelopathies (e.g. GABA, glutamatergic, cholinergic), and disorders of neurotransmitter metabolism or neurotransmission [45, 46]. The knowledge obtained from NHS of DEE subtypes and their evolution over time will inform the optimal inclusion and exclusion criteria and timing of initiation and duration of clinical trials.
By Providing Historic Control Data
To qualify for marketing approval under regulatory bodies such as the Federal Drug Agency (FDA), clinical trial sponsors must provide evidence that a therapy is effective compared to a valid control (FDA electronic-Code of Federal Regulations: Title 21). Control data is required to provide robust evidence that observed changes in clinical outcome assessments (COA) and/or biomarkers were specifically caused by the therapy under evaluation as opposed to other factors. For example, seizures often reduce in frequency and severity with patient age in KCNQ2, SCN2A, and PCDH19-epilepsies and, without robust control data, improvement in seizures for trials of patients with these subtypes may be misattributed to a positive impact of the novel trial therapy rather than to the natural history of the subtype itself [47, 48].
Due to the rarity of genetic conditions, under certain circumstances, the FDA will allow data from a well-conducted NHS to act as an untreated, external control group, also known as ‘historical controls’, to compare with the treatment group(s) in an investigational drug trial. Two examples of this precedent were clinical trials of enzyme replacement therapy: sebelipase alfa for patients with the severe neurometabolic condition lysosomal acid lipase deficiency and intracerebroventricular infusion of cerliponase alfa for patients with neuronal ceroid lipofuscinosis type 2 (CLN2 disease) [23, 49]. The potential use of NHS to provide historical controls for ultra-rare genetic disorders such as the genetic DEEs is a key reason to prioritize high-quality NHS for this group of diseases. This situation also means that NHS design should incorporate approved data sets from the FDA and other international regulatory bodies.
By Evaluating the Most Appropriate Clinical Outcome Measures and Biomarkers to Include in Clinical Trials
Data from NHS can also critically inform the optimal clinical outcome assessments and biomarkers that should be incorporated into targeted DEE clinical trials.
Clinical Outcome Assessments
COAs are patient, caregiver or clinician reports, specific evaluations such as psychometric assessments, or combinations of both [36]. COAs should reflect how an individual feels, functions, or survives. COA can measure the severity and evolution of a condition over time and be integrated into clinical trials [25, 50, 51]. It is ideal if clinician, patient, and caregiver feedback is incorporated into COA design to ensure that the most important and relevant aspects of the disorder are interrogated [52]. For complex conditions such as DEEs, it is therefore appropriate that clinical trials and, by extension, NHS for DEEs, choose a range of seizure and non-seizure related COAs that will appropriately and acceptably capture clinical changes at all stages of a patient’s life. Ultimately, decisions about which COAs will be ascertained in NHS will also be influenced by each assessment’s cost, logistics, and overall participant burden.
Clinical Outcome Assessment of Seizure Severity and Frequency
NHS have a critical role in delineating the nature and evolution of seizures in each genetic DEE, to inform decisions about clinical trial design. The choice of seizure-related COA for clinical trials should be tailored to genetic DEE subtypes at different patient ages.
Epilepsy clinical trials typically compare how much a novel therapy reduces ‘illness’, represented by percentage change in seizure frequency. For example, the European Medicines Agency requires the reporting of a ≥ 50% responder rate (i.e. the percentage of participants with a 50% or greater reduction in seizures) and the FDA uses a median percentage change method (i.e. the percentage by which the median seizure rate are reduced) [53]. However, reliance on seizure counts may not be sufficient for DEEs, compared with other types of epilepsy, and NHS will be useful to delineate more appropriate and reliable seizure COAs [53]. For example, it is critical to assess the range of seizure frequency, clustering, type, and evolution over time and age, to inform seizure COA design. As exemplified by SCN2A-related conditions, the above factors may vary between different subtypes of a genetic DEE (Fig. 1). For example, if it is known that it is typical in a particular subtype of DEE for seizure frequency to vary considerably, including dropping below 50% of the mean rate without any intervention, then the ≥ 50% response rate should not be chosen as it is not likely to be clinically meaningful (i.e. the inherent variability of seizures could cause patients to appear to be treatment ‘responders’ purely by chance) [54].
Long-term recording has also shown inaccuracies and inconsistencies of seizure-counting paradigms [53]. NHS must aim to distinguish which of an individual’s paroxysmal ‘episodes’ are actually seizures, and which are not, to avoid confounding COA measurement. In the DEEs, non-epileptic episodes such as inattentive staring spells in children with intellectual disability, and paroxysmal or fluctuating movement disorders, are common [55, 56]. The key role of paediatric neurologists with expertise in the characterization of seizures and differentiation from other stereotyped events, and the potential need for correlation with biomarkers such as EEG recording (as discussed below) in NHS cannot be overstated [53, 57].
Additionally, one must consider what would represent a meaningful reduction of seizures. For example, whereas for many seizure types, any reduction in seizure frequency may be clinically meaningful; for epileptic spasms, complete cessation is typically the goal given that, in children with Infantile Spasms syndrome, ongoing spasms usually herald a significant ongoing EEG abnormality that adversely impacts developmental progress. COAs may therefore need to include not only seizure counts, but measures of seizure-free periods and markers of change in the impact of seizures, such as reduction in need for emergency ASM or hospital admissions [58].
Given that many genetic DEEs are ultra-rare [35], patient cohorts in precision medicine trials are likely to be small. A major advantage of embedding NHS within clinical trial programs is the ability to study an individual over a prolonged period and develop adequate knowledge of the pre-trial seizure trajectory and expected trajectory without treatment, such that an individual patient could be their own control [32].
Decisions about the type of seizure data to be collected and analyzed in NHS of DEEs need to consider the requirement for quantitative data and datapoints that parallel those likely to be used in clinical trials so that NHS data can serve as historic or intra-patient control sets. However, they also need to facilitate exploratory and qualitative analysis to delineate seizure phenotype and evolution when this is not known, meaning that NHS may collect many datapoints which are ultimately not directly used in clinical trials. These data are nevertheless useful for understanding the disease and early recognition and diagnosis of seizure types. Data collected about seizures includes age of onset, seizure types, their frequency, impact and evolution over time, epilepsy syndrome, seizure treatments, and response. Much of this will be retrospective or baseline data and will be supplemented by prospective data collection using home seizure diaries and ongoing characterization of the epilepsy if new seizure types emerge with video and EEG studies. These data should be provided to or by a paediatric neurologist with expertise in epilepsy, and care taken to optimally characterize the epilepsy and verify the nature and frequency of reported events. This could include synchronous assessment of biomarkers such as serial periods of video-EEG monitoring to corroborate the seizure frequency reported in the seizure diaries.
Clinical Outcome Assessments of Development and Cognition
Developmental impairments are core to the definition of DEE, yet many phenotypic studies do not measure these precisely, nor at consistent time points with consistent instruments, with patients often ascertained in many centres with variable access to skilled psychometric testing. Improving developmental outcomes is a crucial goal for novel therapies; a meaningful positive impact on development will likely be essential to justify the high cost of advanced therapeutics such as gene or ASO therapy [24]. NHS offer an ideal opportunity to accurately measure developmental progress over time which will inform future clinical trial design.
Given that people with DEEs span age groups from neonates to older adults and developmental levels from normal to profoundly impaired (sometimes within a genetic DEE), a single scale will not serve all circumstances. It will be especially important to choose scales and measurements that overcome potential scale attenuation effects for lower and higher functioning individuals (floor and ceiling effects respectively). There is an opportunity for DEE NHS to guide clinical trial design by providing evidence on (i) the best assessment protocol for measuring developmental status, gains, or losses in children, adolescents, and adults with different subtypes of DEEs; (ii) the validity of developmental or neurobehavioral assessment tools for predicting later outcomes; (iii) how best to measure related aspects of neurodevelopment such as autism spectrum disorders and executive function in people with DEEs; and (iv) to quantify related morbidities, e.g. speech, gait, gastrointestinal, sleep, and mortality.
Currently most clinical trials in early-onset epilepsies use standardized intelligence or developmental quotient scales, such as the Wechsler Scales of Intelligence or the Stanford Binet Intelligence Scales (measuring intelligence quotient: IQ), or, especially for younger children, the Bayley Scales of Infant and Toddler Development (BSID) (measuring developmental quotient: DQ) [59, 60]. Many traditional methods of generating IQ or DQ scores in lower functioning individuals are inaccurate and inadequate. Different statistical approaches can be more discerning: for example, using raw rather than composite scores and measuring the true deviation in performance from standardization sample norms [61, 62]. A recent large study of the BSID in Angelman syndrome used raw scores to derive domain-specific age-equivalent and growth-equivalent scores (change over time). Although limited by cross-sectional rather than longitudinal design, this study showed that the BSID was sensitive to changes in development with increasing age. In addition, a floor effect did not appear to be a significant limiting factor. Despite these positive findings, caution should be demonstrated in assuming it may be similarly sensitive in other DEEs, particularly those associated with more profound developmental impairments [63].
Adaptive skills are defined as the ‘effectiveness with which the individual copes with the natural and social demands of [their] environment’. For example, a commonly used adaptive behaviour scale is the Vineland Adaptive Behavior Scale (VABS) which uses semi-structured interviews or parent/caregiver questionnaires to assess adaptive function and has been successfully integrated into clinical trial design for some early-onset epilepsies [58]. However, while it has been demonstrated to help overcome the floor effect in individuals with moderate to severe intellectual disability, concerns have recently been raised over floor effects in some domains in individuals with SCN2A-related disorders, and further work will be required to determine the optimal and appropriate uses of this scale [62].
Scales included in NHS need to consider more than the range of ages and developmental levels of participants. Assessing development in an NHS for a rare DEE presents major logistical issues: due to small patient numbers, NHS are likely to be multi-site and include patients from many countries who live in settings ranging from large to small cities to rural and remote locations. This has practical implications; for example, with preference for scales that can be administered via telehealth, especially during a pandemic, and are standardized in different languages. Inclusions of subscales that are less reliant on verbal IQ are also important for patients with relative deficits in expressive language, or where standardized forms in a range of languages are not available. Standardized scales also have cost, time, patient burden, and personnel implications that need to be pragmatically considered in NHS design to ensure viability.
Robust developmental data from NHS may have important implications for clinical trials. Meaningful change in development will take considerably longer to become apparent than change in seizure frequency, meaning that longer duration trials are needed in order to measure developmental benefit. The existence of ‘historic control’ developmental data may, in some instances, obviate the need for a placebo control group, mitigating potential ethical implications of trial participants being subject to a prolonged duration of non-active treatment.
Disease-Specific Severity Measures
Composite measures of disease severity have been developed for some DEEs, including CDKL5 deficiency disorder and tuberous sclerosis [64–67]. Such measures, which may include seizures, developmental, or other features, have potential utility in clinical trials, not only for measuring change over time, but also for ensuring, in conditions with variable severity, that severity at baseline is similar in treatment and control groups. Prior to use in clinical trials, both content validity and predictive validity of developed instruments are ideal. Natural history studies have the capacity to identify clinical features which are common, important, and associated with disease severity. They may serve as a potential basis for the development of severity scales and subsequent validity testing. It is important to note, however, that such scales may be disease-specific, with limited utility for broader use across the DEEs, given the variable presence and severity of symptoms in different genetic epilepsies.
Clinical Outcome Assessment of Other Comorbidities, Global Impression of Change and Quality of Life
In addition to seizures and development, NHS must also collect data on other DEE morbidities and the impact of the condition on the individual, family, and society, as illustrated by the marked impact of these comorbidities on child and caregiver quality of life [11, 52, 68].COAs exist for common DEE comorbidities such as movement disorders, sleep, gastrointestinal symptoms, autism spectrum disorders, psychiatric symptoms, and behavioural disorders [69, 70]. However, few have been tested in DEEs to determine whether they are meaningful, appropriate, validated, and easy to apply [50, 51, 71]. Specific caregiver measures are also required such as measures of quality of life, and global impression of improvements; such measures, the Caregiver Global Impression of Change (CGIC) and the Quality of Life in Childhood Epilepsy Questionnaire, are being increasingly incorporated into NHS and clinical trial design [58, 72]. Research, in collaboration with families, also needs to focus on the development of measures that most effectively capture the wider impact of the DEE on the daily life of the child and family. Economic measures such as hospital and resource use should also be incorporated into NHS, as these ‘real-world’ impacts need to be evaluated to provide evidence of economic benefit in precision medicine trials for genetic DEEs.
Biomarkers for DEEs
A biomarker is a characteristic that is measured as an indicator of normal biological or pathological processes, or responses to an exposure or intervention, including therapeutic interventions [73]. Biomarkers can be diagnostic (facilitating early detection of disease), prognostic (providing information on likely health outcomes and assisting in stratification of phenotypic severity), or pharmacological (providing information on therapeutic responses of patient subgroups). They need to have high degrees of sensitivity, specificity, precision, and reproducibility; reflect true disease pathophysiology rather than epiphenomena; and be able to be easily and cost-effectively measured. The FDA have highlighted that NHS provide an opportunity to collect specimens and data that could be used to identify, develop, and robustly validate biomarkers to act as surrogate endpoints in clinical trials.
There are currently many challenges in designing and validating robust biomarkers for epilepsy in general and DEEs in particular. Just as with COAs, NHS offer an opportunity to explore and validate the most appropriate biomarkers for genetic DEEs and their subtypes, to improve the quality, reliability, and outcome assessment of clinical trials.
Electrophysiological Biomarkers
The major biomarker used in epilepsy, EEG, has a critical role in DEE diagnosis and management in clinical practice. Given interictal epileptiform EEG abnormalities adversely impact development, and improvement in clinical state may be paralleled by an improvement in the interictal EEG, EEG could be an important trial outcome measure. Unfortunately, there are few scales to measure EEG abnormalities, and thus, NHS provide an opportunity to develop and test novel EEG scales. Inter-rater reliability is a major consideration; this has been poor in a number of previous studies of EEG in infantile spasms but improved when scales were simplified, and criteria clearly defined [74]. Considerable work on EEG has been done in studies of tuberous sclerosis, culminating in the EPISTOP study where EEG was successfully used as a biomarker, although the authors noted the burden of frequent EEG studies contributed to participant drop out [60]. Understanding the evolution of EEG for a genetic DEE with age through NHS will inform the design of regimes (frequency and duration) of EEG recordings for clinical trials. Development of an interictal EEG scale that takes into account both the background rhythms and epileptiform activity, and which could be used across all DEEs, would be ideal. However, the utility and applicability of a single scale may vary between different genetic DEEs, given variation in EEG abnormalities and their relationship to disease severity. For example, in many young individuals with Dravet syndrome, the interictal EEG background is often normal even in a child having frequent or severe seizures [75]; therefore, a scale that incorporates a measure of the degree of background abnormality may not be a useful measure of disease improvement with novel therapies in the Dravet population.
Alternative proposed epilepsy biomarkers include ‘quantitative’ EEG and transcranial magnetic stimulation, but there is limited evidence of their utility as biomarkers in DEEs to date [76, 77]. Novel, less resource intensive, methods to accurately capture and record epileptic activity (such as wearable seizure detection devices) are also being developed and hold promise as biomarkers for clinical trials [78]. However, they still need to be objectively assessed and validated in comparison with video-telemetry and home seizure diaries considering such issues as sensitivity, false alarms, latency, adherence, and impact on patient and caregiver quality of life. Incorporation of such assessments into NHS may prove useful [78].
Biological Biomarkers
For genetic DEEs, there is the potential for biomarkers which reflect the levels of gene or protein expression, as shown in other neurological conditions where gene or ASO therapies are utilized. For example, biomarkers included in clinical trials for spinal muscular atrophy include not only electrophysiological measures (e.g. compound motor action potentials, electrical impedance myography), but also genetic and biomolecular markers (e.g. survival motor neuron (SMN2) mRNA and protein expression) which were ascertained and validated during NHS [79]. The development and validation of biomarkers have been a major focus of translational research in paediatric cancers, which are also a target for ‘precision medicine’ approaches [80–83]. Thus, in order to ensure collection of appropriate biospecimens and their analysis, it is important that NHS design is carefully discussed with a multidisciplinary team of molecular pathologists, physiologists and functional scientists.
A major limitation of the measurement of such biomarkers is the need for repeated invasive specimen collection. Nevertheless, it is vital that potential biologic biomarkers be explored, as they can provide an important and objective adjunct to clinical observations in the clinical trial setting. For example, they could determine whether lack of effect of a novel treatment was in fact due to lack of ‘target engagement’ (e.g. an ASO designed to downregulate expression did not in fact reduce protein levels) rather than true lack of effect of a treatment with the proposed mechanism of action.
Conclusions
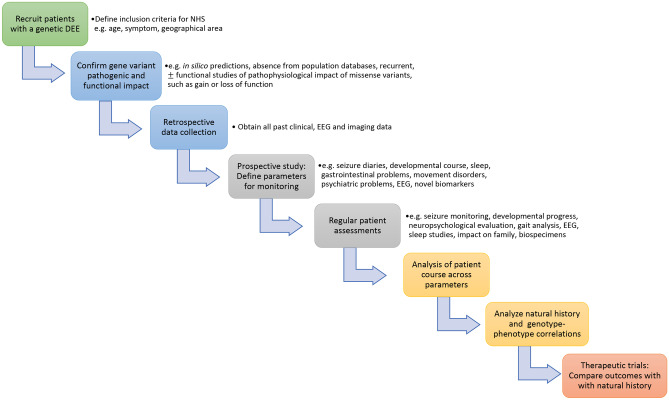
There are global calls to prioritize investment in NHS in view of their central role in assessing therapeutic developments for rare disorders such as DEEs [33, 84]. Existing patient registries, retrospective case-based studies, and novel ‘big data’ mining approaches will help with optimal study design [33]. However, longitudinal NHS, incorporating high-quality genotype–phenotype data collection, will be required to adequately evaluate COAs and biomarkers for DEEs in order to optimize clinical trial design and provide historical control data. The recommended elements that should be included in NHS protocols for DEEs, adapted from FDA guidelines, are summarized in Table 1, and a proposed conceptual framework for NHS study design provided in Fig. 2.
Table 1.
Elements that should be included in NHS designs for DEEs (adapted from Rare Diseases: Natural History Studies for Drug Development Guidance for Industry; draft document March 2019) (https://www.fda.gov/media/122425/download)
| Element | Features | Consideration for genetic DEEs |
|---|---|---|
| Disease definition and diagnostic criteria for study entry | ||
| Demographic details | ||
| Disease related information | Clinical signs and symptoms | Input of expert (pediatric) neurologist, ideally epileptologist, to phenotyping |
| Age at diagnosis and onset and offset of symptoms | Particular attention to accuracy in detection of onset, type and changes in seizure patterns | |
| Genotypic, biophysical and phenotypic features important in identifying disease subpopulations or types | Include accurate genotyping and ideally evaluation of functional impact (e.g. in vitro studies) of causal variant to ensure accurate subtyping | |
| Clinically meaningful disease effects and outcomes including focus on those important to patient and families | Clinical outcome assessments of seizures: may include assessment of seizure diaries, use of antiseizure medication, need for hospitalization, time to and length of seizure-free periods | |
| Clinical outcome assessments of development tailored to age and developmental level of individual: likely to require both measures of intelligence/developmental quotient, relevant subscales, and measures of adaptive functioning | ||
| Clinical outcome assessments of relevant comorbidities for individual DEE: including sleep, behaviour, autism spectrum disorder, psychosis, movement disorders | ||
| Overall assessments of clinical severity | ||
| ‘Real-world’ impacts of DEEs: e.g. number of hospitalizations, cost of medication, investigations, therapies, education, transport, parental psychosocial impacts | ||
| Measurement of biomarkers | What biomarkers of seizures will be measured: may include EEG, telemetry, wearable devices | |
| Collection of biospecimens | Which biological biomarkers specific to a genetic DEE will be collected and measured: e.g. protein/ gene levels | |
| Methods for data collection | Assessments schedules, inter- and intra-rater reliability scoring for clinical outcome assessments and quality control for biomarker measurement | Tailor to each genetic DEE |
| Analysis plan | Multidisciplinary approach | Input of experts according to field of evaluation e.g. neurologists, speech pathologists, psychiatrists, physiologists regarding biomarkers |
| Treatment guidelines or algorithms, tailored to the region | Changes in standard of care over time | Mechanisms to evaluate changes over time in multicenter studies |
Fig. 2.
Conceptual framework for a natural history study
Given that well designed, prospective, longitudinal NHS are lengthy and resource intensive, and that the number of individual genetic DEEs is large, and ever increasing, it is critical that adequate funding and support for these studies is provided. Emerging partnerships between clinicians, patient and family groups, not-for-profit organizations, industry, and regulatory bodies to support equitable opportunities for NHS for rare diseases should be applauded and expanded. Consideration of novel means of accurately and acceptably capturing essential clinical outcome and biomarker data across international multi-centre studies will be important [32]. Development of cross-disease platforms that are flexible enough to capture NHS for a range of genetic DEE should be prioritized, as well as information technology solutions to allow high-quality national, and indeed global, data entry and curation. Such collaborations are required to minimize the risk of multiple small studies ‘reinventing the wheel' and being too short, underpowered, and under-resourced to fulfil their primary objectives. Critically, this will reduce duplication of efforts, inefficiencies, and the risk that ultra-rare DEEs are not studied due to the lack of a critical mass of expert and family interest. Expertly designed and executed DEE NHS will be a critical step in the development, assessment and implementation of novel therapies required to revolutionize patient outcomes.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge the other principal investigators of the Australasian Genetic Epilepsy Natural History Study: Steven Petrou, Claire Wakefield, Suncica Lah, Michael Hildebrand, and Kim Dalziel.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Funding
This study was supported by National Health and Medical Research Council of Australia (Centre for Research Excellence in Developmental and Epileptic Encephalopathies Diagnosis, Australian Medical Research Future Fund, and the Australian Epilepsy Research Fund).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58(4):512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheffer IE, Liao J. Deciphering the concepts behind “Epileptic encephalopathy” and “Developmental and epileptic encephalopathy”. Eur J Paediatr Neurol. 2020;24:11–14. doi: 10.1016/j.ejpn.2019.12.023. [DOI] [PubMed] [Google Scholar]
- 3.Papuc SM, Abela L, Steindl K, Begemann A, Simmons TL, Schmitt B, et al. The role of recessive inheritance in early-onset epileptic encephalopathies: a combined whole-exome sequencing and copy number study. Eur J Hum Genet. 2019;27(3):408–421. doi: 10.1038/s41431-018-0299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symonds JD, Zuberi SM, Stewart K, McLellan A, O‘Regan M, MacLeod S, Jollands A, Joss S, Kirkpatrick M, Brunklaus A, Pilz DT, Shetty J, Dorris L, Abu-Arafeh I, Andrew J, Brink P, Callaghan M, Cruden J, Diver LA, Findlay C, Gardiner S, Grattan R, Lang B, MacDonnell J, McKnight J, Morrison CA, Nairn L, Slean MM, Stephen E, Webb A, Vincent A, Wilson M. Incidence and phenotypes of childhood-onset genetic epilepsies: a prospective population-based national cohort. Brain. August 2019;142(8):2303–2318. 10.1093/brain/awz195. [DOI] [PMC free article] [PubMed]
- 5.Møller RS, Hammer TB, Rubboli G, Lemke JR, Johannesen KM. From next-generation sequencing to targeted treatment of non-acquired epilepsies. Expert Rev Mol Diagn. 2019;19(3):217–228. doi: 10.1080/14737159.2019.1573144. [DOI] [PubMed] [Google Scholar]
- 6.Bayat A, Bayat M, Rubboli G, Møller RS. Epilepsy Syndromes in the First Year of Life and Usefulness of Genetic Testing for Precision Therapy. Genes (Basel). 2021;12(7):1051. doi: 10.3390/genes12071051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer EE, Sachdev R, Macintosh R, Melo US, Mundlos S, Righetti S, et al. Diagnostic Yield of Whole Genome Sequencing After Nondiagnostic Exome Sequencing or Gene Panel in Developmental and Epileptic Encephalopathies. Neurology. 2021;96(13):e1770–e1782. doi: 10.1212/WNL.0000000000011655. [DOI] [PubMed] [Google Scholar]
- 8.Shellhaas RA, Wusthoff CJ, Tsuchida TN, Glass HC, Chu CJ, Massey SL, et al. Profile of neonatal epilepsies: Characteristics of a prospective US cohort. Neurology. 2017;89(9):893–899. doi: 10.1212/WNL.0000000000004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howell KB, Eggers S, Dalziel K, Riseley J, Mandelstam S, Myers CT, McMahon JM, Schneider A, Carvill GL, Mefford HC. Victorian Severe Epilepsy of Infancy Study Group, Scheffer IE, Harvey AS. A population-based cost-effectiveness study of early genetic testing in severe epilepsies of infancy. Epilepsia. 2018 Jun; 59(6):1177-87. 10.1111/epi.14087. Epub 2018 May 11. PMID: 29750358; PMCID: PMC5990455. [DOI] [PMC free article] [PubMed]
- 10.Symonds JD, McTague A. Epilepsy and developmental disorders: Next generation sequencing in the clinic. Eur J Paediatr Neurol. 2020;24:15–23. doi: 10.1016/j.ejpn.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Jakobsen AV, Moller RS, Nikanorova M, Elklit A. The impact of severe pediatric epilepsy on experienced stress and psychopathology in parents. Epilepsy Behav. 2020;113:107538. [DOI] [PubMed]
- 12.Nickels KC, Wirrell EC. Cognitive and Social Outcomes of Epileptic Encephalopathies. Semin Pediatr Neurol. 2017;24(4):264–275. doi: 10.1016/j.spen.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Nevin S, Wakefield CE, Schilstra C, McGill BC, Bye A, Palmer EE. The information needs of parents of children with severe early-onset epilepsy (SEE): A systematic review. 2020;112:107382 [DOI] [PubMed]
- 14.Rodda JM, Scheffer IE, McMahon JM, Berkovic SF, Graham HK. Progressive gait deterioration in adolescents with Dravet syndrome. Arch Neurol. 2012;69(7):873–878. doi: 10.1001/archneurol.2011.3275. [DOI] [PubMed] [Google Scholar]
- 15.Cooper MS, Mcintosh A, Crompton DE, McMahon JM, Schneider A, Farrell K, et al. Mortality in Dravet syndrome. Epilepsy Res. 2016 Oct 26;128:43–47. [DOI] [PubMed]
- 16.Harini C, Nagarajan E, Bergin AM, Pearl P, Loddenkemper T, Takeoka M, et al. Mortality in infantile spasms: A hospital-based study. Epilepsia. 2020;61(4):702–713. doi: 10.1111/epi.16468. [DOI] [PubMed] [Google Scholar]
- 17.Burgess R, Wang S, McTague A, Boysen KE, Yang X, Zeng Q, et al. Genetic Landscape of Epilepsy of Infancy with Migrating Focal Seizures. Ann Neurol. 2019;86(6):821–831. doi: 10.1002/ana.25619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15(3):304–316. doi: 10.1016/S1474-4422(15)00250-1. [DOI] [PubMed] [Google Scholar]
- 19.Riikonen R. Infantile Spasms: Outcome in Clinical Studies. Pediatr Neurol. 2020;108:54–64. doi: 10.1016/j.pediatrneurol.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Stamberger H, Nikanorova M, Willemsen MH, Accorsi P, Angriman M, Baier H, et al. STXBP1 encephalopathy: A neurodevelopmental disorder including epilepsy. Neurology. 2016;86(10):954–962. doi: 10.1212/WNL.0000000000002457. [DOI] [PubMed] [Google Scholar]
- 21.Consortium E A roadmap for precision medicine in the epilepsies. Lancet Neurol. 2015;14(12):1219–1228. doi: 10.1016/S1474-4422(15)00199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moller RS, Hammer TB, Rubboli G, Lemke JR, Johannesen KM. From next-generation sequencing to targeted treatment of non-acquired epilepsies. Expert Rev Mol Diagn. 2019;19(3):217–228. doi: 10.1080/14737159.2019.1573144. [DOI] [PubMed] [Google Scholar]
- 23.Schulz A, Ajayi T, Specchio N, de Los Reyes E, Gissen P, Ballon D, et al. Study of Intraventricular Cerliponase Alfa for CLN2 Disease. N Engl J Med. 2018;378(20):1898–1907. doi: 10.1056/NEJMoa1712649. [DOI] [PubMed] [Google Scholar]
- 24.Striano P, Minassian BA. From Genetic Testing to Precision Medicine in Epilepsy. Neurotherapeutics. 2020;17(2):609–615. doi: 10.1007/s13311-020-00835-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steriade C, French J, Devinsky O. Epilepsy: key experimental therapeutics in early clinical development. Expert Opin Investig Drugs. 2020;29(4):373–383. doi: 10.1080/13543784.2020.1743678. [DOI] [PubMed] [Google Scholar]
- 26.Steriade C, French J, Devinsky O. Epilepsy: key experimental therapeutics in early clinical development. Expert Opin Investig Drugs. 2020 Apr;29(4):373–83. [DOI] [PubMed]
- 27.Susannah Ahern GS, Mark Tacey, Michael Esler, John Oldroyd, Joanna Dean SBobotACFD, Registry. The Australian Cystic Fibrosis Data Registry Annual Report, 2015. Monash University, Department of Epidemiology and Preventive Medicine; 2017.
- 28.Vill K, Kolbel H, Schwartz O, Blaschek A, Olgemoller B, Harms E, et al. One Year of Newborn Screening for SMA - Results of a German Pilot Project. J Neuromuscul Dis. 2019;6(4):503–515. doi: 10.3233/JND-190428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter T, Nestler-Parr S, Babela R, Khan ZM, Tesoro T, Molsen E, et al. Rare Disease Terminology and Definitions-A Systematic Global Review: Report of the ISPOR Rare Disease Special Interest Group. Value Health. 2015;18(6):906–914. doi: 10.1016/j.jval.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 30.In: Field MJ, Boat TF, editors. Rare Diseases and Orphan Products: Accelerating Research and Development. The National Academies Collection: Reports funded by National Institutes of Health. Washington (DC)2010. [PubMed]
- 31.Miller KL, Mueller C, Liu G, Miller Needleman KI, Maynard J. FDA orphan products clinical trial grants: assessment of outcomes and impact on rare disease product development. Orphanet J Rare Dis. 2020;15(1):234. doi: 10.1186/s13023-020-01514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gavin P. The importance of natural histories for rare diseases. Expert Opinions Orphan Drugs. 2015;3(8):855–857. [Google Scholar]
- 33.Garbade SF, Zielonka M, Komatsuzaki S, Kolker S, Hoffmann GF, Hinderhofer K, et al. Quantitative retrospective natural history modeling for orphan drug development. Journal of inherited metabolic disease. 2021;44(1):99–109. doi: 10.1002/jimd.12304. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan J, Scheffer IE, Lagae L, Nabbout R, Pringsheim M, Talwar D, et al. Fenfluramine HCl (Fintepla® ) provides long‐term clinically meaningful reduction in seizure frequency: Analysis of an ongoing open‐label extension study. Epilepsia. 2020 Nov;61(11):2396‐2404. 10.1111/epi.16722. Epub 2020 Oct 19. PMID: 33078386; PMCID:PMC7756901. [DOI] [PMC free article] [PubMed]
- 35.Hennekam RC. Care for patients with ultra-rare disorders. Eur J Med Genet. 2011;54(3):220–224. doi: 10.1016/j.ejmg.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Walton MK, Powers JH, 3rd, Hobart J, Patrick D, Marquis P, Vamvakas S, et al. Clinical Outcome Assessments: Conceptual Foundation-Report of the ISPOR Clinical Outcomes Assessment - Emerging Good Practices for Outcomes Research Task Force. Value Health. 2015;18(6):741–752. doi: 10.1016/j.jval.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morel T, Cano SJ. Measuring what matters to rare disease patients - reflections on the work by the IRDiRC taskforce on patient-centered outcome measures. Orphanet J Rare Dis. 2017;12(1):171. doi: 10.1186/s13023-017-0718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKeon G, Palmer EE, Macintosh R, Nevin SM, Wheatley L, Rosenbaum S. Feasibility of a mental health informed physical activity intervention for the carers of children with developmental and epileptic encephalopathy. Epilepsy Behav. 2021;121(Pt A):108022. [DOI] [PubMed]
- 39.Lauxmann S, Verbeek NE, Liu Y, Zaichuk M, Muller S, Lemke JR, et al. Relationship of electrophysiological dysfunction and clinical severity in SCN2A-related epilepsies. Hum Mutat. 2018;39(12):1942–1956. doi: 10.1002/humu.23619. [DOI] [PubMed] [Google Scholar]
- 40.Berecki G, Howell KB, Deerasooriya YH, Cilio MR, Oliva MK, Kaplan D, et al. Dynamic action potential clamp predicts functional separation in mild familial and severe de novo forms of SCN2A epilepsy. Proc Natl Acad Sci USA. 2018;115(24):E5516–E5525. doi: 10.1073/pnas.1800077115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ben-Shalom R, Keeshen CM, Berrios KN, An JY, Sanders SJ, Bender KJ. Opposing Effects on NaV1.2 Function Underlie Differences Between SCN2A Variants Observed in Individuals With Autism Spectrum Disorder or Infantile Seizures. Biol Psychiatry. 2017;82(3):224–32. [DOI] [PMC free article] [PubMed]
- 42.Wolff M, Johannesen KM, Hedrich UBS, Masnada S, Rubboli G, Gardella E, et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017;140(5):1316–1336. doi: 10.1093/brain/awx054. [DOI] [PubMed] [Google Scholar]
- 43.Scheffer IE, Berkovic SF. Generalized epilepsy with febrile seizures plus. A genetic disorder with heterogeneous clinical phenotypes. Brain. 1997;120 (Pt 3):479–90. [DOI] [PubMed]
- 44.Moller RS, Heron SE, Larsen LH, Lim CX, Ricos MG, Bayly MA, et al. Mutations in KCNT1 cause a spectrum of focal epilepsies. Epilepsia. 2015;56(9):e114–e120. doi: 10.1111/epi.13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer EE. Potassium Channel Mutations in Epilepsy. The Oxford Handbook of Neuronal Ion Channels. Edited by Arin Bhattacharjee. Subject: Neuroscience, Molecular and Cellular Systems. Online Publication Date: Aug 2020. 10.1093/oxfordhb/9780190669164.013.13.
- 46.Mastrangelo M. Epilepsy in inherited neurotransmitter disorders: Spotlights on pathophysiology and clinical management. Metab Brain Dis. 2021;36(1):29–43. doi: 10.1007/s11011-020-00635-x. [DOI] [PubMed] [Google Scholar]
- 47.Berkovic SF, Heron SE, Giordano L, Marini C, Guerrini R, Kaplan RE, et al. Benign familial neonatal-infantile seizures: characterization of a new sodium channelopathy. Ann Neurol. 2004;55(4):550–557. doi: 10.1002/ana.20029. [DOI] [PubMed] [Google Scholar]
- 48.Kolc KL, Sadleir LG, Depienne C, Marini C, Scheffer IE, Moller RS, et al. A standardized patient-centered characterization of the phenotypic spectrum of PCDH19 girls clustering epilepsy. Transl Psychiatry. 2020;10(1):127. doi: 10.1038/s41398-020-0803-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frampton JE. Sebelipase Alfa: A Review in Lysosomal Acid Lipase Deficiency. Am J Cardiovasc Drugs. 2016;16(6):461–468. doi: 10.1007/s40256-016-0203-2. [DOI] [PubMed] [Google Scholar]
- 50.Myers KA, Davey MJ, Ching M, Ellis C, Grinton BE, Roten A, et al. Randomized Controlled Trial of Melatonin for Sleep Disturbance in Dravet Syndrome: The DREAMS Study. J Clin Sleep Med. 2018;14(10):1697–1704. doi: 10.5664/jcsm.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moavero R, Benvenuto A, Emberti Gialloreti L, Siracusano M, Kotulska K, Weschke B, et al. Early Clinical Predictors of Autism Spectrum Disorder in Infants with Tuberous Sclerosis Complex: Results from the EPISTOP Study. J Clin Med. 2019;8(6). [DOI] [PMC free article] [PubMed]
- 52.Nabbout R, Auvin S, Chiron C, Thiele E, Cross H, Scheffer IE, et al. Perception of impact of Dravet syndrome on children and caregivers in multiple countries: looking beyond seizures. Dev Med Child Neurol. 2019;61(10):1229–1236. doi: 10.1111/dmcn.14186. [DOI] [PubMed] [Google Scholar]
- 53.Karoly P, Goldenholz DM, Cook M. Are the days of counting seizures numbered? Curr Opin Neurol. 2018;31(2):162–168. doi: 10.1097/WCO.0000000000000533. [DOI] [PubMed] [Google Scholar]
- 54.Goldenholz DM, Moss R, Scott J, Auh S, Theodore WH. Confusing placebo effect with natural history in epilepsy: A big data approach. Ann Neurol. 2015;78(3):329–336. doi: 10.1002/ana.24470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hughes R, Poon WY, Harvey AS. Limited role for routine EEG in the assessment of staring in children with autism spectrum disorder. Arch Dis Child. 2015;100(1):30–33. doi: 10.1136/archdischild-2014-306400. [DOI] [PubMed] [Google Scholar]
- 56.Gburek-Augustat J, Beck-Woedl S, Tzschach A, Bauer P, Schoening M, Riess A. Epilepsy is not a mandatory feature of STXBP1 associated ataxia-tremor-retardation syndrome. Eur J Paediatr Neurol. 2016;20(4):661–665. doi: 10.1016/j.ejpn.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 57.Mullen SA, Carney PW, Roten A, Ching M, Lightfoot PA, Churilov L, et al. Precision therapy for epilepsy due to KCNT1 mutations: A randomized trial of oral quinidine. Neurology. 2018;90(1):e67–e72. doi: 10.1212/WNL.0000000000004769. [DOI] [PubMed] [Google Scholar]
- 58.Devinsky O, Cross JH, Wright S. Trial of Cannabidiol for Drug-Resistant Seizures in the Dravet Syndrome. N Engl J Med. 2017;377(7):699–700. doi: 10.1056/NEJMc1708349. [DOI] [PubMed] [Google Scholar]
- 59.Lux AL, Edwards SW, Hancock E, Johnson AL, Kennedy CR, Newton RW, et al. The United Kingdom Infantile Spasms Study (UKISS) comparing hormone treatment with vigabatrin on developmental and epilepsy outcomes to age 14 months: a multicentre randomised trial. Lancet Neurol. 2005;4(11):712–717. doi: 10.1016/S1474-4422(05)70199-X. [DOI] [PubMed] [Google Scholar]
- 60.Kotulska K, Kwiatkowski DJ, Curatolo P, Weschke B, Riney K, Jansen F, et al. Prevention of Epilepsy in Infants with Tuberous Sclerosis Complex in the EPISTOP Trial. Ann Neurol. 2021;89(2):304–314. doi: 10.1002/ana.25956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sansone SM, Schneider A, Bickel E, Berry-Kravis E, Prescott C, Hessl D. Improving IQ measurement in intellectual disabilities using true deviation from population norms. J Neurodev Disord. 2014;6(1):16. doi: 10.1186/1866-1955-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berg AT, Palac H, Wilkening G, Zelko F, Schust Meyer L. SCN2A-Developmental and Epileptic Encephalopathies: Challenges to trial-readiness for non-seizure outcomes. Epilepsia. 2021 Jan;62(1):258–68. [DOI] [PubMed]
- 63.Sadhwani A, Wheeler A, Gwaltney A, Peters SU, Barbieri-Welge RL, Horowitz LT, et al. Developmental Skills of Individuals with Angelman Syndrome Assessed Using the Bayley-III. J Autism Dev Disord. 2021; 10.1007/s10803-020-04861-1. [DOI] [PMC free article] [PubMed]
- 64.Saldaris J, Weisenberg J, Pestana-Knight E, Marsh ED, Suter B, Rajaraman R, et al. Content Validation of Clinician-Reported Items for a Severity Measure for CDKL5 Deficiency Disorder. J Child Neurol. 2021;36(11):998–1006. doi: 10.1177/08830738211019576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Demarest S, Pestana-Knight EM, Olson HE, Downs J, Marsh ED, Kaufmann WE, et al. Severity Assessment in CDKL5 Deficiency Disorder. Pediatr Neurol. 2019;97:38–42. doi: 10.1016/j.pediatrneurol.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Demarest ST, Olson HE, Moss A, Pestana-Knight E, Zhang X, Parikh S, et al. CDKL5 deficiency disorder: Relationship between genotype, epilepsy, cortical visual impairment, and development. Epilepsia. 2019;60(8):1733–1742. doi: 10.1111/epi.16285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Humphrey A, Ploubidis GB, Yates JR, Steinberg T, Bolton PF. The Early Childhood Epilepsy Severity Scale (E-Chess) Epilepsy Res. 2008;79(2–3):139–145. doi: 10.1016/j.eplepsyres.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 68.Licheni SH, McMahon JM, Schneider AL, Davey MJ, Scheffer IE. Sleep problems in Dravet syndrome: a modifiable comorbidity. Dev Med Child Neurol. 2018;60(2):192–198. doi: 10.1111/dmcn.13601. [DOI] [PubMed] [Google Scholar]
- 69.Hanratty J, Livingstone N, Robalino S, Terwee CB, Glod M, Oono IP, et al. Systematic Review of the Measurement Properties of Tools Used to Measure Behaviour Problems in Young Children with Autism. PLoS One. 2015;10(12):e0144649. [DOI] [PMC free article] [PubMed]
- 70.Randall M, Egberts KJ, Samtani A, Scholten RJ, Hooft L, Livingstone N, et al. Diagnostic tests for autism spectrum disorder (ASD) in preschool children. Cochrane Database Syst Rev. 2018;7:CD009044. [DOI] [PMC free article] [PubMed]
- 71.Bruni O, Ottaviano S, Guidetti V, Romoli M, Innocenzi M, Cortesi F, et al. The Sleep Disturbance Scale for Children (SDSC). Construction and validation of an instrument to evaluate sleep disturbances in childhood and adolescence. J Sleep Res. 1996;5(4):251–61. [DOI] [PubMed]
- 72.Goodwin SW, Lambrinos AI, Ferro MA, Sabaz M, Speechley KN. Development and assessment of a shortened Quality of Life in Childhood Epilepsy Questionnaire (QOLCE-55) Epilepsia. 2015;56(6):864–872. doi: 10.1111/epi.13000. [DOI] [PubMed] [Google Scholar]
- 73.Pitkanen A, Ekolle Ndode-Ekane X, Lapinlampi N, Puhakka N. Epilepsy biomarkers - Toward etiology and pathology specificity. Neurobiol Dis. 2019;123:42–58. doi: 10.1016/j.nbd.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mytinger JR, Hussain SA, Islam MP, Millichap JJ, Patel AD, Ryan NR, et al. Improving the inter-rater agreement of hypsarrhythmia using a simplified EEG grading scale for children with infantile spasms. Epilepsy Res. 2015;116:93–98. doi: 10.1016/j.eplepsyres.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 75.Lee HF, Chi CS, Tsai CR, Chen CH, Wang CC. Electroencephalographic features of patients with SCN1A-positive Dravet syndrome. Brain Dev. 2015;37(6):599–611. doi: 10.1016/j.braindev.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 76.Stern WM, Sander JW, Rothwell JC, Sisodiya SM. Impaired intracortical inhibition demonstrated in vivo in people with Dravet syndrome. Neurology. 2017;88(17):1659–1665. doi: 10.1212/WNL.0000000000003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sueri C, Gasparini S, Balestrini S, Labate A, Gambardella A, Russo E, et al. Diagnostic Biomarkers of Epilepsy. Curr Pharm Biotechnol. 2018;19(6):440–450. doi: 10.2174/1389201019666180713095251. [DOI] [PubMed] [Google Scholar]
- 78.Van Ness PC. Are Seizure Detection Devices Ready for Prime Time? Epilepsy Curr. 2019;19(1):36–37. doi: 10.1177/1535759719827430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kariyawasam DST, D’Silva A, Lin C, Ryan MM, Farrar MA. Biomarkers and the Development of a Personalized Medicine Approach in Spinal Muscular Atrophy. Front Neurol. 2019;10:898. doi: 10.3389/fneur.2019.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blattner-Johnson M, Jones DTW, Pfaff E. Precision medicine in pediatric solid cancers. Semin Cancer Biol. 2021;S1044-579X(21)00178-4. [DOI] [PubMed]
- 81.Ben-Hamo R, Jacob Berger A, Gavert N, Miller M, Pines G, Oren R, et al. Predicting and affecting response to cancer therapy based on pathway-level biomarkers. Nat Commun. 2020;11(1):3296. doi: 10.1038/s41467-020-17090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wong M, Mayoh C, Lau LMS, Khuong-Quang DA, Pinese M, Kumar A, et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nature medicine. 2020;26(11):1742–1753. doi: 10.1038/s41591-020-1072-4. [DOI] [PubMed] [Google Scholar]
- 83.Tsoli M, Wadham C, Pinese M, Failes T, Joshi S, Mould E, et al. Integration of genomics, high throughput drug screening, and personalized xenograft models as a novel precision medicine paradigm for high risk pediatric cancer. Cancer Biol Ther. 2018;19(12):1078–1087. doi: 10.1080/15384047.2018.1491498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Austin CP, Cutillo CM, Lau LPL, Jonker AH, Rath A, Julkowska D, et al. Future of Rare Diseases Research 2017–2027: An IRDiRC Perspective. Clin Transl Sci. 2018;11(1):21–27. doi: 10.1111/cts.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Licheni SH, Mcmahon JM, Schneider AL, Davey MJ, Scheffer IE. Sleep problems in Dravet syndrome: a modifiable comorbidity. Dev Med Child Neurol. 2018;60(2):192–198. doi: 10.1111/dmcn.13601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.