Abstract
Background
Three DNA methylation (DNAm) based algorithms, DNAm PhenoAge acceleration (AgeAccelPheno), DNAm GrimAge acceleration (AgeAccelGrim), and mortality risk score (MRscore), based on methylation in 513, 1030, and 10 CpGs, respectively, were established to predict health outcomes and mortality. We aimed to compare and validate the predictive ability of these scores and frailty in relation to mortality in a population-based cohort from Germany.
Methods
DNA methylation in whole blood was measured by the Infinium Methylation EPIC BeadChip kit (EPIC, Illumina, San Diego, CA, USA) in two random subsets of the ESTHER cohort study (n = 741 and n = 1030). AgeAccelPheno, AgeAccelGrim, and a revised MRscore to adapt EPIC, the MRscore with 8 CpGs (MRscore-8CpGs), were calculated. Frailty was assessed by a frailty index (FI).
Findings
During 17 years of follow-up, 458 deaths were observed. All DNAm algorithms and FI were positively correlated with each other. AgeAccelPheno, AgeAccelGrim, MRscore, and FI showed independent associations with all-cause mortality [hazard ratio (95% CI) per SD increase = 1·32 (1·19-1·46), 1·47 (1·32-1·64), 1·73 (1·49-2·01), and 1·31 (1·20-1·43), respectively]. Harrell's C-statistic was 0·710 for a model predicting mortality by age, sex, and leukocyte composition and increased to 0·759 in a model including MRscore-8CpGs and FI. The predictive performance was further improved (Harrell's C-statistic = 0·766) when additionally including AgeAccelPheno and AgeAccelGrim into the model.
Interpretation
The combination of a DNA methylation score based on 8 CpGs only and an easy to ascertain frailty index may strongly enhance mortality prediction beyond age and sex.
Funding
The ESTHER study was funded by grants from the Baden-Württemberg state Ministry of Science, Research and Arts (Stuttgart, Germany), the Federal Ministry of Education and Research (Berlin, Germany), the Federal Ministry of Family Affairs, Senior Citizens, Women and Youth (Berlin, Germany), and the Saarland State Ministry of Health, Social Affairs, Women and the Family (Saarbrücken, Germany). The work of Xiangwei Li was supported by a grant from Fondazione Cariplo (Bando Ricerca Malattie invecchiamento, #2017-0653).
Keywords: DNA methylation, epigenetic clock, age acceleration, frailty, mortality
Research in Context.
Evidence before this study
Recently, three DNA methylation based algorithms, DNA methylation PhenoAge acceleration, DNA methylation GrimAge acceleration, and mortality risk score were developed and shown to be strong indicators of age-related conditions and mortality. However, the predictive ability of the three DNA methylation algorithms and frailty for mortality has not been assessed in same study population, and it is therefore unclear to what extent results on their predictive performance are comparable or reflect differences in the assessed study populations.
Added value of this study
The study was conducted in two random subsets of the ESTHER cohort study (n = 741 and n = 1030) and frailty was assessed by a frailty index based on percentage of 34 selected deficits. The three DNA methylation algorithms and the frailty index were positively correlated and each of them was independently associated with mortality. The combination of the algorithms, in particular the combination of mortality risk score and frailty index may enhance the mortality prediction performance.
Implications of all the available evidence
The combination of mortality risk score by itself or in combination with a simple-to-ascertain frailty indicator could be a useful and economic measure to quantify mortality risk.
Alt-text: Unlabelled box
1. Introduction
Chronological age is a key risk factor for many chronic diseases and conditions which are leading causes of mortality, such as cardiovascular disease and cancer [1]. However, health status and mortality may vary strongly among people of the same chronological age, and complementary measures of “biological age” could be crucial to accurately describe the health status of older adults and to evaluate health-promoting interventions. Over the past decades, various approaches have been developed to estimate biological age using genetic or non-genetic indicators [2].
Frailty is a common geriatric syndrome characterized by increased vulnerability [3]. A commonly applied approach to quantify frailty is calculation of a frailty index (FI) that is defined as the presented proportion of age-related health deficits [4]. FI has been shown to be strongly associated with aging-related phenotypes [5,6] and mortality [7,8].
Biological age can also be assessed using epigenetic data. DNA methylation (DNAm)-based epigenetic clocks have been developed and shown to be indicators of age-related conditions and mortality [9,10]. Recently, using multiple methods, three promising DNAm based algorithms, including mortality risk score (MRscore) [11] and the second-generation epigenetic clocks, DNAm phenotypic age (PhenoAge) [12] and DNAm GrimAge (GrimAge) [13], were derived based on 10, 513, and 1030 CpGs, respectively. Differences between PhenoAge and GrimAge and chronological age are commonly being referred to as measures of epigenetic age acceleration and termed as AgeAccelPheno and AgeAccelGrim, respectively. AgeAccelPheno, AgeAccelGrim, and MRscore were shown to be highly correlated with all-cause mortality and cancer-specific mortality across various study populations [[14], [15], [16]]. However, the predictive performance of the three DNAm algorithms and mortality has not been evaluated in the same study population, and it is therefore unclear to what extent results on their predictive performance are comparable or reflect differences in the assessed study populations. We therefore aimed to evaluate and compare individual and joint predictive performance with respect to mortality of AgeAccelPheno, AgeAccelGrim, MRscore, and a FI, in a head-to-head comparison in a large cohort of older adults from Germany.
2. Methods
2.1. Study population and data collection
Our analyses are based on data from the ESTHER study, a large population-based cohort study conducted in Germany, whose study population and design has been described in detail previously [11,17,18]. Briefly, 9940 participants aged 50-75 years were recruited by their general practitioners (GPs) during a general health screening examination between July 2000 and December 2002. The participants have been followed up every two to three years since then. At recruitment and each follow-up, standardized self-administered questionnaires were used to collect information on sociodemographic characteristics, lifestyle, and dietary factors. Self-reported smoking information at baseline was confirmed by serum cotinine measurements and was found to be highly accurate in a subgroup of 1500 study participants [19]. Results of the general health examinations were documented by the GPs on a standardized form. Blood samples were collected during the examinations and stored at -80°C for later testing. The ESTHER study population has been found to be representative of the German population of the same age (50-75) with respect to major sociodemographic variables and risk factor profiles [20].
The ESTHER study was approved by the ethics committees of the medical faculty of the University of Heidelberg and the medical board of the state of Saarland. Written informed consent was obtained from each participant.
Two independent subsets were randomly selected from the ESTHER study population for epigenome-wide DNAm measurements that were carried out in two different batches. Subsets I and II included 741 and 1030 randomly selected subjects for whom DNAm measurements were performed in August 2018 [21] and July 2019, respectively. It is worth noting that both subsets were independent of and not overlapping with a subsample of the ESTHER cohort from which the MR score had been derived in previous research [11].
2.2. Methylation assessment
DNA from whole blood samples obtained at recruitment was extracted using a salting-out procedure [22]. Genome-wide DNAm was assessed with the Infinium Methylation EPIC BeadChip kit (EPIC, Illumina, San Diego, CA, USA) [15,21]. The laboratory work was done following the manufacturer's instruction in the Genomics and Proteomics Core Facility at the German Cancer Research Center, Heidelberg, Germany (DKFZ) as previously described [23]. As reported previously [11,24], signals of probes with detection P-value >0·01, >10% missing values, and probes targeting the X and Y chromosomes were excluded.
2.3. Calculation of DNAm aging algorithms
The second-generation DNAm aging algorithms PhenoAge and GrimAge have been constructed by regressing a surrogate measure of biological age on a set of CpGs using a penalized regression analyses, such as elastic net regression [25]. The residual from the regression of DNAm algorithms on chronological age is age acceleration (Age Acc) [9]. Thus, a positive value of Age Acc indicates an accelerated aging and risk of mortality. The age acceleration of PhenoAge and GrimAge were calculated and were denoted AgeAccelPheno and AgeAccelGrim, respectively. The calculation was done using the online tool available at https://dnamage.genetics.ucla.edu/.
The original continuous MRscore (MRscore) was computed using ten CpGs from the Infinium HumanMethylation450K BeadChip Assay (450K, Illumina.Inc, San Diego, CA, USA) [11]. However, two (cg01612140 and cg 23665802) of the ten CpGs were not included in the EPIC array. We therefore adopted a new equation by regressing the remaining eight CpGs on the original MRscore in a third subset of 111 ESTHER study participants (independent of and not overlapping with the subsets MRscore was derived from) whose DNAm had been assessed by both 450K and EPIC array, and used the result to derive the original MRscore by the following equation:
The Spearman correlation coefficients of MRscore and MRscore-8CpGs in this subset are presented in Supplementary (Suppl.) Table 1.
2.4. Frailty index
The baseline FI was calculated as the percentage of deficits presented divided by 34 selected variables of deficits, as previously described by Saum et al. [7]. The included deficits included indicators of general health, diseases, symptoms, difficulties in activities of daily living, and instrumental activities of daily living [Suppl. Table 2]. Missing values (< 5% for all variables) were estimated by multiple imputation using the SAS procedure PROC MI. Age, sex, and the 34 selected deficits for construction of the FI were included in the imputation procedure. Scaled variables were dichotomized for each scale level and then the imputed values were rounded to each corresponding nearest level. Regression results for FI were based on 20 imputations and combined by the SAS procedure MIANALYZE.
2.5. Mortality ascertainment
The vital status of the subjects was followed up through record linkage with population registries until December 31, 2018. The completeness of follow-up for all-cause mortality was 99·9%. Furthermore, death certificates were obtained from local health authorities for 97·7% of participants and were utilized to define mortality from cardiovascular diseases (CVDs, ICD-10 codes I00–I99) and cancers (ICD-10 codes C00–C97 and D37–D48), respectively.
2.6. Statistical analysis
Correlations among MRscore-8CpGs, AgeAccelPheno and AgeAccelGrim, FI, and chronological age were assessed using Spearman correlation coefficients and were illustrated in a correlation matrix plot.
Associations of DNAm algorithms and FI with all-cause and cause-specific mortality were estimated using Cox proportional hazard models firstly adjusting for age, sex, batches (not for FI), and leukocyte composition [not for FI, estimated by the Houseman approach [26], model one], and additionally controlling for smoking status and alcohol consumption (Model two). Hazard ratios (HRs) and corresponding 95% confidence intervals (95% CIs) were calculated in subset I and II, separately. To assess the individual and joint predictive accuracy of age, sex, leukocyte composition, smoking, the DNAm algorithms, and FI for mortality, Harrell's C-statistics [27], a widely used approach to assess the predictive performance of the ensemble, were calculated. Harrell's C-statistics range from 0·5 to 1·0. The value of 0·5 corresponds to a non-informative prediction rule and 1·0 for a perfect association [27]. Time-dependent areas under the curve (AUCs) were additionally calculated in subset I and II. Because the DNAm profiles in the two subsets were assessed in different time periods (August 2018 for subset I and July 2019 for subset II) using different batches of DNAm assessment chips, the results from the two subsets were presented both separately and combined using fixed‐effects meta‐analysis. For all Cox models, the proportional hazards assumption was checked by scaled Schoenfeld residuals [28].
All analyses were performed using SAS, version 9·4 (SAS Institute, Inc., Cary, NC). Statistical significance was defined by P-values < 0·05 in two-sided testing.
2.7. Role of funding source
All funders did not have any role in study design, data collection, data analyses, interpretation, writing of report, or decision to publish the study.
3. Results
3.1. Study population
Baseline characteristics of the two subsets are presented in Table 1. The mean (standard deviation, SD) age of the participants was 61·7 (6·6) years and 62·0 (6·7) years in subset I and subset II, respectively. The sex composition (56% women), education level (10% with ≥12 years of school education) and body mass index levels (>70% overweight or obese) were similar in both subsets (P values 0·96, 0·36, and 0·98, respectively). The levels of AgeAccelPheno, MRscore-8CpGs, and FI were likewise comparable in the two subsets (P values 0·17, 0·18, and 0·19, respectively). However, the levels of AgeAccelGrim were higher in subset II than in subset I (P = 0·01).
Table 1.
Characteristics of study population from ESTHER study
| Characteristics | Subset I (n=741) | Subset II (n=1030) | P-valuea |
|---|---|---|---|
| Age (years; mean ± SD) | 61·7±6·6 | 62·0±6·7 | 0·24 |
| Sex (N/%) | |||
| Men | 326 (44·0) | 452 (43·9) | 0·96 |
| Women | 415 (56·0) | 578 (56·1) | |
| Educational levels (N/%)b | |||
| Low (≤9 years) | 532 (74·0) | 770 (76·5) | 0·36 |
| Intermediate (10-11 years) | 112 (15·6) | 133 (13·2) | |
| High (≥12 years) | 75 (10·4) | 103 (10·2) | |
| Body mass index (N/%)c | |||
| Underweight (<18·5 kg/m2) | 4 (0·5) | 5 (0·5) | 0·98 |
| Normal weight (18·5-<25·0 kg/m2) | 191 (25·8) | 269 (26·2) | |
| Overweight (25·0-<30·0 kg/m2) | 350 (47·3) | 489 (47·7) | |
| Obesity (≥30·0 kg/m2) | 195 (26·4) | 263 (25·6) | |
| Smoking status (N/%)d | |||
| Never smoker | 353 (49·2) | 526 (52·2) | 0·38 |
| Former smoker | 235 (32·7) | 320 (31·8) | |
| Current smoker | 130 (18·1) | 162 (16·1) | |
| Alcohol consumption (grams per day) | 9·3±12·7 | 9·8±12·9 | 0·41 |
| AgeAccelPheno (mean ± SD) | -0·41±5·70 | -0·02±5·78 | 0·17 |
| AgeAccelGrim (mean ± SD) | -0·55±4·55 | 0·01±4·75 | 0·01 |
| MRscore-8CpGs (mean ± SD) | -2·44±0·45 | -2·47±0·46 | 0·18 |
| Frailty index (%; mean ± SD) | 24±14 | 23±14 | 0·19 |
Abbreviations: SD, standard deviation; AgeAccelPheno, DNA methylation phenotypic age acceleration; AgeAccelGrim, DNA methylation GrimAge acceleration; MRscore-8CpGs, revised version of continuous mortality risk score with 8 CpGs.
Chi-square test for categorical variables and analysis of variance for continuous variables.
Data missing for 46 participants.
Data missing for 5 participants.
Data missing for 45 participants.
3.2. Correlations of the DNAm algorithms, age, and FI
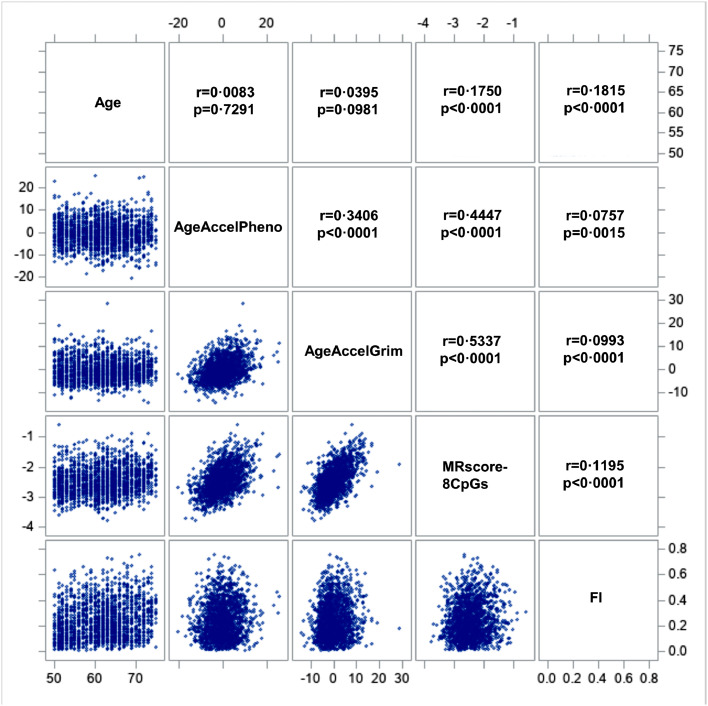
Figure 1 presents the Spearman correlation matrix of the DNAm algorithms, baseline age, and baseline FI in all participants from both subsets. AgeAccelPheno, AgeAccelGrim, and MRscore-8CpGs were significantly correlated with each other, with correlation coefficients ranging from 0·34 to 0·53. We also observed a significant correlation of age with MRscore-8CpGs (r = 0·18, P < 0·001) and FI (r = 0·18, P < 0·001). Moreover, the correlations between FI and the three DNAm algorithms were weak but statistically significant; the Spearman correlation coefficients were 0·08, 0·10, and 0·12 for AgeAccelPheno, AgeAccelGrim, and MRscore-8CpGs, respectively. Correlations were similar in subset I and subset II (Suppl. Figure 1 and Suppl. Figure 2).
Figure 1.
Correlation matrix of DNAm algorithms and frailty. Results from meta-analyses of both subsets.
Abbreviations: AgeAccelPheno, DNA methylation phenotypic age acceleration; AgeAccelGrim, DNA methylation GrimAge acceleration; MRscore-8CpGs, revised version of continuous mortality risk score with 8 CpGs; FI, frailty index.
3.3. Associations of DNAm algorithms and FI with mortality
During 17 years of follow-up, 194 and 264 deaths were observed in subset I and II, respectively. Table 2 shows the associations of DNAm algorithms and FI with all-cause mortality and cause-specific mortality. In the meta-analysis of the two subsets, HRs (95% CIs) of all-cause mortality were 1·32 (1·19-1·46), 1·47 (1·32-1·64), 1.73 (1·49-2·01), and 1·31 (1·20-1·43) per one SD increase of AgeAccelPheno, AgeAccelGrim, MRscore-8CpGs, and FI, respectively. A one SD increase of AgeAccelPheno, AgeAccelGrim, and MRscore-8CpGs was significantly associated with 21%, 33%, and 76% increased cancer-specific mortality (P values 5·33E-07, 9·57E-07, and 4·00E-10, respectively), respectively. Furthermore, strong, statistically significant associations were observed for CVD-specific mortality, with HRs of 1·46, 1·39, 1·89, and 1·49 per SD increase in AgeAccelPheno, AgeAccelGrim, MRscore-8CpGs, and FI (P values 4·65E-07, 1·40E-05, 1·78E-15, and 1·08E-05, respectively), respectively. When including all DNAm algorithms and FI into the model (Table 3), all associations with all-cause mortality were observably attenuated but remained statistically significant, indicating that all scores were partially independently associated with all-cause mortality. Cox-proportional hazard model assumption was checked using Schoenfeld residual test (local and global test) and all variables showed P values > 0·10, which fulfilled the assumption.
Table 2.
Associations of DNAm algorithms and frailty index with mortality
| Mortality |
Subset I (n=741) HR (95% CI) |
Subset II (n=1030) HR (95% CI) |
Overall (n=1771, meta-analysis) HR (95% CI) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Model 1 | Model 2 | Cases | Model 1 | Model 2 | Cases | Model 1 | Model 2 | |
| All-cause | |||||||||
| AgeAccelPheno (per SD) | 194 | 1·39 (1·20-1·62) | 1·35 (1·16-1·57) | 264 | 1·37 (1·20-1·56) | 1·29 (1·13-1·48) | 458 | 1·38 (1·25-1·52) | 1·32 (1·19-1·46) |
| AgeAccelGrim (per SD) | 194 | 1·50 (1·31-1·73) | 1·39 (1·18-1·63) | 264 | 1·68 (1·48-1·91) | 1·56 (1·34-1·81) | 458 | 1·60 (1·46-1·75) | 1·47 (1·32-1·64) |
| MRscore-8CpGs (per SD) | 194 | 1·87 (1·55-2·26) | 1·65 (1·33-2·04) | 264 | 1·99 (1·68-2·36) | 1·82 (1·47-2·24) | 458 | 1·94 (1·71-2·20) | 1·73 (1·49-2·01) |
| FI (%; per SD) | 194 | 1·37 (1·20-1·56) | 1·35 (1·18-1·55) | 264 | 1·32 (1·18-1·48) | 1·28 (1·14-1·44) | 458 | 1·34 (1·23-1·46) | 1·31 (1·20-1·43) |
| Cancer | |||||||||
| AgeAccelPheno (per SD) | 60 | 1·38 (1·04-1·85) | 1·36 (1·02-1·81) | 78 | 1·18 (0·93-1·51) | 1·11 (0·87-1·42) | 138 | 1·26 (1·05-1·52) | 1·21 (1.00-1·46) |
| AgeAccelGrim (per SD) | 60 | 1·56 (1·23-1·99) | 1·32 (0·95-1·83) | 78 | 1·49 (1·18-1·89) | 1·34 (1·01-1·78) | 138 | 1·52 (1·29-1·80) | 1·33 (1·08-1·65) |
| MRscore-8CpGs (per SD) | 60 | 2·29 (1·65-3·19) | 1·79 (1·21-2·65) | 78 | 1·88 (1·38-2·58) | 1·72 (1·17-2·53) | 138 | 2·07 (1·65-2·59) | 1·76 (1·33-2·31) |
| FI (%; per SD) | 60 | 1·36 (1·07-1·73) | 1·29 (1.00-1·66) | 78 | 0·99 (0·79-1·25) | 0·98 (0·77-1·24) | 138 | 1·16 (0·98-1·37) | 1·11 (0·94-1·32) |
| CVD | |||||||||
| AgeAccelPheno (per SD) | 53 | 1·37 (1·03-1·83) | 1·31 (0·98-1·77) | 80 | 1·62 (1·28-2·06) | 1·57 (1·23-2·01) | 133 | 1·51 (1·26-1·82) | 1·46 (1·21-1·77) |
| AgeAccelGrim (per SD) | 53 | 1·20 (0·91-1·57) | 1·09 (0·81-1·47) | 80 | 1·76 (1·39-2·23) | 1·75 (1·31-2·33) | 133 | 1·49 (1·24-1·78) | 1·39 (1·13-1·71) |
| MRscore-8CpGs (per SD) | 53 | 1·71 (1·18-2·48) | 1·65 (1·09-2·48) | 80 | 2·09 (1·53-2·86) | 2·12 (1·45-3·12) | 133 | 1·93 (1·52-2·45) | 1·89 (1·42-2·50) |
| FI (%; per SD) | 53 | 1·49 (1·16-1·91) | 1·47 (1·14-1·90) | 80 | 1·53 (1·24-1·89) | 1·51 (1·22-1·88) | 133 | 1·51 (1·29-1·78) | 1·49 (1·27-1·76) |
Abbreviations: HR, hazard ratio; CI, confidence interval; SD, standard deviation; AgeAccelPheno, DNA methylation phenotypic age acceleration; AgeAccelGrim, DNA methylation GrimAge acceleration; MRscore-8CpGs, revised version of continuous mortality risk score with 8 CpGs; FI, frailty index; CVD, cardiovascular disease.
Model 1, adjusted for age, sex, leukocyte composition (not for FI), and batch (not for FI).
Model 2, similar as model 1, additionally adjusted for smoking status (never smoker, former smoker, current smoker), and alcohol consumption (grams per day).
Table 3.
Associations of DNAm algorithms and frailty index with all-cause mortality after mutual control for each other
| Predictor |
Subset I (n=741) |
Subset II (n=1030) |
Overall (n=1771, meta-analysis) |
|||
|---|---|---|---|---|---|---|
| Cases | HR (95% CI)a | Cases | HR (95% CI)a | Cases | HR (95% CI)a | |
| AgeAccelPheno (per SD) | 194 | 1·18 (1·01-1·39) | 264 | 1·10 (0·95-1·27) | 458 | 1·14 (1·02-1·26) |
| AgeAccelGrim (per SD) | 194 | 1·18 (0·97-1·42) | 264 | 1·34 (1·12-1·59) | 458 | 1·26 (1·11-1·43) |
| MRscore-8CpGs (per SD) | 194 | 1·38 (1·08-1·78) | 264 | 1·43 (1·12-1·82) | 458 | 1·41 (1·18-1·67) |
| FI (%; per SD) | 194 | 1·32 (1·15-1·51) | 264 | 1·22 (1·09-1·38) | 458 | 1·26 (1·16-1·38) |
Abbreviations: FI, frailty index; HR, hazard ratio; CI, confidence interval; SD, standard deviation; AgeAccelPheno, DNA methylation phenotypic age acceleration; AgeAccelGrim, DNA methylation GrimAge acceleration; MRscore-8CpGs, revised version of continuous mortality risk score with 8 CpGs.
adjusted for age, sex, leukocyte composition (not for FI), batch (not for FI), smoking status (never smoker, former smoker, current smoker), and alcohol consumption (grams per day); results for AgeAccelPheno additionally adjusted for MRscore-8CpGs, AgeAccelGrim and frailty index; results for AgeAccelGrim additionally adjusted for MRscore-8CpGs, AgeAccelPheno and frailty index; results for MRscore-8CpGs additionally adjusted for AgeAccelPheno, AgeAccelGrim and frailty index; results for frailty index additionally adjusted for MRscore-8CpGs, AgeAccelPheno and AgeAccelGrim.
3.4. Predictive performance of FI and DNAm algorithms for all-cause mortality
Table 4 presents the performance of various combinations of markers in predicting all-cause mortality. In meta-analysis, Harrell's C-statistic (95% CI) was 0·710 (0·681-0·738) for a model including age, sex, and leukocyte composition. The predictive performance was improved after adding smoking, DNAm algorithms, or FI. C-statistics (95% CIs) were 0·736 (0·709-0·762), 0·723 (0·695-0·750), 0·751 (0·725-0·777), 0·750 (0·724-0·776), and 0·724 (0·696-0·752) for models adding smoking, AgeAccelPheno, AgeAccelGrim, MRscore-8CpGs and FI, respectively. The predictive performance was further improved by including each DNAm algorithm and FI (C-statistic was 0·735, 0·758, and 0·759 for the combination of FI with AgeAccelPheno, AgeAccelGrim, and MRscore-8CpGs, respectively) or MRscore-8CpGs (C-statistic was 0·752 and 0·759 for the combination of MRscore-8CpGs with AgeAccelPheno and AgeAccelGrim, respectively). For the model with all predictors, C-statistic reached a maximum of 0·766 (95% CI 0·741-0·791). When using these markers to predict cancer mortality (Suppl. Table 3) and CVD mortality (Suppl. Table 4), similar patterns were observed, but overall predictive performance was lower for cancer mortality and higher for CVD-specific mortality. For cancer mortality, C-statistics were 0·737 (95% CI 0·693-0·781) and 0·742 (95% CI 0·698-0·0.787) for the joint inclusion of MRscore-8CpGs and FI, and of all predictors, respectively. For CVD mortality, C-statistics were 0·820 (95% CI 0·787-0·853) for the joint inclusion of MRscore-8CpGs and FI, and 0·828 (95% CI 0·794-0·862) for the combination of all predictors (Suppl. Table 3). Consistent patterns were also demonstrated when estimating time-dependent AUCs (Suppl. Figure 3).
Table 4.
Harrell's C-statistics of chronological age, DNAm algorithms, and frailty index in prediction of all-cause mortality
|
Predictors |
Overall (meta-analysis) Harrell's C statistics (95% CI) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Age+sex+LC | Smoking | AgeAccelPheno | AgeAccelGrim | MRscore-8CpGs | FI (%) | Subset I | Subset II | Overall (meta-analysis) |
| √ | 0·721 (0·661-0·780) | 0·706 (0·674-0·739) | 0·710 (0·681-0·738) | |||||
| √ | √ | 0·752 (0·698-0·806) | 0·731 (0·701-0·760) | 0·736 (0·709-0·762) | ||||
| √ | √ | 0·727 (0·668-0·786) | 0·721 (0·690-0·753) | 0·723 (0·695-0·750) | ||||
| √ | √ | 0·756 (0·700-0·812) | 0·749 (0·720-0·778) | 0·751 (0·725-0·777) | ||||
| √ | √ | 0·760 (0·707-0·812) | 0·746 (0·717-0·776) | 0·750 (0·724-0·776) | ||||
| √ a | √ | 0·741 (0·683-0·799) | 0·719 (0·687-0·751) | 0·724 (0·696-0·752) | ||||
| √ | √ | √ | 0·749 (0·692-0·806) | 0·732 (0·701-0·762) | 0·735 (0·708-0·762) | |||
| √ | √ | √ | 0·769 (0·717-0·821) | 0·756 (0·727-0·785) | 0·758 (0·734-0·784) | |||
| √ | √ | √ | 0·772 (0·722-0·822) | 0·753 (0·723-0·782) | 0·759 (0·732-0·783) | |||
| √ | √ | √ | 0·760 (0·708-0·813) | 0·749 (0·719-0·779) | 0·752 (0·726-0·778) | |||
| √ | √ | √ | 0·768 (0·715-0·822) | 0·757 (0·728-0·786) | 0·759 (0·734-0·785) | |||
| √ | √ | √ | √ | √ | 0·776 (0·724-0·828) | 0·763 (0·734-0·792) | 0·766 (0·741-0·791) | |
Abbreviations: CI, confidence interval; LC, leukocyte composition; AgeAccelPheno, DNA methylation phenotypic age acceleration; AgeAccelGrim, DNA methylation GrimAge acceleration; MRscore-8CpGs, revised version of continuous mortality risk score with 8 CpGs; FI, frailty index.
Models did not include leukocyte composition.
4. Discussion
In this study, we evaluated and compared three recently proposed aging DNAm algorithms and a FI in relation to prediction of mortality in a cohort of older adults. The three DNAm algorithms and the FI were positively correlated with each other and each of them was independently associated with all-cause and cause-specific mortality. The combination of all algorithms and the combination of MRscore-8CpGs and FI substantially enhanced the mortality prediction performance.
Whereas the first-generation epigenetic clocks were assessed solely by chronological age as the reference, PhenoAge and GrimAge were designed to better capture biological aging [9,12,13,29]. Given that AgeAccelPheno and AgeAccelGrim are reflecting differences of estimated biological age and chronological age, their lack of correlation with chronological age in our study was predictable. Moreover, AgeAccelPheno and AgeAccelGrim were observed to be weakly correlated with FI.
In a previous study from the Lothian Birth Cohort 1936, higher DNAm GrimAge was associated with lower cognitive ability and brain vascular lesions in older age [30]. Previous studies also reported that higher GrimAge and PhenoAge values were associated with an increase in physical function deficits [12,13] and were correlated with poorer fitness, such as diminished grip strength and cardio-pulmonary function [31,32] . Frailty is a consequence of a cumulative decline in many physiological systems and frail individuals are characterized by increased vulnerability to age-related disorders [33]. The observed correlations of AgeAccelPheno and AgeAccelGrim with FI in the current study may reflect declines in multiple physiological systems beyond “normal aging”. Somewhat stronger correlations with frailty were observed for MRscore-8CpGs which is additionally correlated with chronological age.
Previous studies have also assessed associations of aging DNAm algorithms with specific aging related biomarkers. AgeAccelGrim was found to be strongly associated with increased levels of plasma C-reactive protein [13], whose production is stimulated by interleukin-6 (IL-6). C-reactive protein and IL-6 are biomarkers of susceptibility to frailty, disability, morbidity, and mortality at older ages [34,35].
Our findings of the associations of AgeAccelPheno, AgeAccelGrim, and MRscore-8CpGs with all-cause and cause-specific mortality are consistent with results from other recent studies [14,36,37]. Wang et.al. conducted a study in two independent cohorts and also reported strong associations of AgeAccelGrim and MRscore-8CpGs with all-cause mortality 35. The second-generation clocks, PhenoAge and GrimAge, were designed to improve the reported weaker associations of the first-generation aging clocks with mortality [38,39] and aimed to predict mortality from aging-related diseases such as CVD and cancer [12,13]. Derived from DNAm-based surrogates for seven plasma proteins and smoking pack-years, AgeAccelGrim was found to show strong associations with all-cause and cause-specific mortality by various studies [13,40]. Another finding from our study is all algorithms are associated with all-cause mortality even when including all of them into the model. The associations suggest all scores were independently associated with all-cause mortality and all have specific pathways for all-cause mortality. In the future, more studies aimed to explore these specific pathways are needed.
Our study also highlighted the capacity of MRscore-8CpGs in the prediction of all-cause mortality beyond established DNAm aging markers and FI. A study conducted in US Normative Aging Study [14] also reported a similar conclusion that MRscore was a stronger predictor of mortality than established aging clocks. MRscore is a linear combination of 10 CpGs selected by least absolute shrinkage and LASSO regression and has been demonstrated to be a robust predictor for all-cause and CVD mortality [11,35]. Although methylation of several of the CpGs included in MRscore is strongly related to smoking. [41], strong associations with mortality persisted in our study even after adjustment for smoking. The predictive ability of the MRscore therefore goes far beyond its relationship with smoking.
One primary aim of developing DNAm biomarkers is finding an accurate, simple, and feasible method to predict mortality or lifespan. In that respect, MRscore, requiring methylation quantification at a much lower number of CpGs, by itself or in combination with some easy-to-determine frailty measure, such as FI, has potential capacity to be a practical and economic indicator for mortality risk stratification. However, potential additional costs including the assessment of leukocyte composition need to be kept in mind. Such mortality risk prediction might serve several purposes. One obvious purpose could be identification of elderly people at higher risks of death and diseases who are in particular need of targeted intervention or earlier medical care. Another important application could be use of DNAm algorithms such as MRscore as early indicators of effectiveness of specific interventions which could be most valuable both in intervention research as well as in routine medical practice. However, as with other risk markers, individual risk assessment should be done with due caution, given that risk assessment remains far from perfect.
There are several limitations of this study that need to be kept in mind. First, the cross-sectional approach precludes drawing conclusions on temporal and potentially causal directions of the correlations between methylation algorithms and frailty. Although the temporal direction of associations with the mortality outcomes is self-evident, it remains to be established to what extent the DNAm algorithms are mere indicators of mortality risks or DNA methylation changes might also be causally related to increased mortality. Second, the original MRscore had been derived from an epigenome-wide screening for mortality-related DNAm using the Infinium HumanMethylation450K BeadChip Assay and included ten CpGs. Because two of the ten CpGs are missing in the EPIC microarray data, only a proxy of the original MRscore, predicted by eight CpGs could be used in our analyses. One of the two missing CpGs (cg06126421) was one of the top hits related to smoking in previous EWAS from various study populations [42,43] and was found to be highly predictive of lung cancer risk (odds ratio per 1 SD lower value: 2·11) [44]. Although MRscore and MRscore-8CpGs were very highly correlated, their predictive performance for mortality might slightly differ. Third, as MRscore was initially derived from the ESTHER cohort (albeit from a different subset of the cohort), the capacity of MRscore in predicting mortality might be lower in different cohorts, and the comparison of predictive performance of the three methylation scores might be slightly biased in favor of the MRscore in our study. More independent cohorts are therefore essential to validate the findings in our study. However, our results are consistent with findings of comparative analyses of various scores from other independent cohorts including two conducted in United States [14,36] and one in Germany [11]. Fourth, the current analysis was based on a population based cohort study that enrolled participants 50-75 years of age in Germany. The findings need to be validated in further independent studies, including studies from other countries, studies in different age groups and studies in population with different risk factor profiles.
In conclusion, all DNAm algorithms and the FI were positively correlated with each other, and all of them, were independently associated with all-cause and cause-specific mortality. The MRscore-8CpGs by itself or in combination with a simple-to-ascertain FI could be a particularly useful and economic measure to quantify mortality risk. Further research should aim for evaluating its use in both observational and interventional aging research and clinical practice.
Contributors
Conception and design: X. Li, Y. Zhang, H. Brenner
Development of methodology: X. Li, Y. Zhang, X. Gào, H. Brenner
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): B. Holleczek, B. Schöttker, H. Brenner
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): X. Li, Y. Zhang, X. Gào, B. Schöttker
Writing, review, and/or revision of the manuscript: X. Li, Y Zhang, X Gào, B. Holleczek, B. Schöttker, H. Brenner
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): X. Li, Y Zhang, X Gào
Study supervision: B. Holleczek, H. Brenner
All authors read and approved the final version of the manuscript.
Data sharing statement
Data available on request from the authors.
Declaration of Competing Interest
All authors confirmed the full access to all the data in the study and accepted responsibility to submit for publication. No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank the study participants and their general practitioners as well as laboratory and administrative staff of the ESTHER study team. The authors gratefully acknowledge contributions of DKFZ Genomics and Proteomics Core Facility in the processing of DNA samples and performing the laboratory work.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103686.
Appendix. Supplementary materials
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Moskalev A. The challenges of estimating biological age. Elife. 2020;9 doi: 10.7554/eLife.54969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue QL. The frailty syndrome: definition and natural history. Clin Geriatr Med. 2011;27(1):1–15. doi: 10.1016/j.cger.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitnitski AB, Mogilner AJ, Rockwood K. Accumulation of deficits as a proxy measure of aging. ScientificWorldJournal. 2001;1:323–336. doi: 10.1100/tsw.2001.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.At J, Bryce R, Prina M, Acosta D, Ferri CP, Guerra M, et al. Frailty and the prediction of dependence and mortality in low- and middle-income countries: a 10/66 population-based cohort study. BMC Med. 2015;13:138. doi: 10.1186/s12916-015-0378-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S, Jazwinski SM. Quantitative measures of healthy aging and biological age. Healthy Aging Res. 2015;4 doi: 10.12715/har.2015.4.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saum KU, Dieffenbach AK, Muller H, Holleczek B, Hauer K, Brenner H. Frailty prevalence and 10-year survival in community-dwelling older adults: results from the ESTHER cohort study. Eur J Epidemiol. 2014;29(3):171–179. doi: 10.1007/s10654-014-9891-6. [DOI] [PubMed] [Google Scholar]
- 8.Zaslavsky O, Walker RL, Crane PK, Gray SL, Larson EB. Glucose Levels and Risk of Frailty. J Gerontol A Biol Sci Med Sci. 2016;71(9):1223–1229. doi: 10.1093/gerona/glw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Wilson R, Heiss J, Breitling LP, Saum KU, Schottker B, et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat Commun. 2017;8:14617. doi: 10.1038/ncomms14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10(4):573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019;11(2):303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao X, Colicino E, Shen J, Just AC, Nwanaji-Enwerem JC, Wang C, et al. Comparative validation of an epigenetic mortality risk score with three aging biomarkers for predicting mortality risks among older adult males. Int J Epidemiol. 2019;48(6):1958–1971. doi: 10.1093/ije/dyz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao X, Zhang Y, Boakye D, Li X, Chang-Claude J, Hoffmeister M, et al. Whole blood DNA methylation aging markers predict colorectal cancer survival: a prospective cohort study. Clin Epigenetics. 2020;12(1):184. doi: 10.1186/s13148-020-00977-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Ploner A, Wang Y, Magnusson PK, Reynolds C, Finkel D, et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. Elife. 2020;9 doi: 10.7554/eLife.51507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schottker B, Muhlack DC, Hoppe LK, Holleczek B, Brenner H. Updated analysis on polypharmacy and mortality from the ESTHER study. Eur J Clin Pharmacol. 2018;74(7):981–982. doi: 10.1007/s00228-018-2445-1. [DOI] [PubMed] [Google Scholar]
- 18.Holleczek B, Schottker B, Brenner H. Helicobacter pylori infection, chronic atrophic gastritis and risk of stomach and esophagus cancer: Results from the prospective population-based ESTHER cohort study. Int J Cancer. 2020;146(10):2773–2783. doi: 10.1002/ijc.32610. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Florath I, Saum KU, Brenner H. Self-reported smoking, serum cotinine, and blood DNA methylation. Environ Res. 2016;146:395–403. doi: 10.1016/j.envres.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 20.Low M, Stegmaier C, Ziegler H, Rothenbacher D, Brenner H, study E. [Epidemiological investigations of the chances of preventing, recognizing early and optimally treating chronic diseases in an elderly population (ESTHER study)] Dtsch Med Wochenschr. 2004;129(49):2643–2647. doi: 10.1055/s-2004-836089. [DOI] [PubMed] [Google Scholar]
- 21.Gao X, Zhang Y, Burwinkel B, Xuan Y, Holleczek B, Brenner H, et al. The associations of DNA methylation alterations in oxidative stress-related genes with cancer incidence and mortality outcomes: a population-based cohort study. Clin Epigenetics. 2019;11(1):14. doi: 10.1186/s13148-018-0604-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florath I, Butterbach K, Muller H, Bewerunge-Hudler M, Brenner H. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet. 2014;23(5):1186–1201. doi: 10.1093/hmg/ddt531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehne B, Drong AW, Loh M, Zhang W, Scott WR, Tan ST, et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015;16:37. doi: 10.1186/s13059-015-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 26.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrell FE, Jr., Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247(18):2543–2546. [PubMed] [Google Scholar]
- 28.Grambsch PM, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81(3):11. [Google Scholar]
- 29.Bocklandt S, Lin W, Sehl ME, Sanchez FJ, Sinsheimer JS, Horvath S, et al. Epigenetic predictor of age. PLoS One. 2011;6(6):e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillary RF, Stevenson AJ, Cox SR, McCartney DL, Harris SE, Seeboth A, et al. An epigenetic predictor of death captures multi-modal measures of brain health. Mol Psychiatry. 2019 doi: 10.1038/s41380-019-0616-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gale CR, Marioni RE, Harris SE, Starr JM, Deary IJ. DNA methylation and the epigenetic clock in relation to physical frailty in older people: the Lothian Birth Cohort 1936. Clin Epigenetics. 2018;10(1):101. doi: 10.1186/s13148-018-0538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breitling LP, Saum KU, Perna L, Schottker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics. 2016;8:21. doi: 10.1186/s13148-016-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clegg A, Young J, Iliffe S, Rikkert MO, Rockwood K. Frailty in elderly people. Lancet. 2013;381(9868):752–762. doi: 10.1016/S0140-6736(12)62167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin L, Park S, Lakatta EG. RAGE signaling in inflammation and arterial aging. Front Biosci (Landmark Ed) 2009;14:1403–1413. doi: 10.2741/3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Saum KU, Schottker B, Holleczek B, Brenner H. Methylomic survival predictors, frailty, and mortality. Aging (Albany NY) 2018;10(3):339–357. doi: 10.18632/aging.101392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, Ni W, Yao Y, Just A, Heiss J, Wei Y, et al. DNA methylation-based biomarkers of age acceleration and all-cause death, myocardial infarction, stroke, and cancer in two cohorts: The NAS, and KORA F4. EBioMedicine. 2021;63 doi: 10.1016/j.ebiom.2020.103151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCrory C, Fiorito G, Hernandez B, Polidoro S, O'Halloran AM, Hever A, et al. GrimAge outperforms other epigenetic clocks in the prediction of age-related clinical phenotypes and all-cause mortality. J Gerontol A Biol Sci Med Sci. 2020 doi: 10.1093/gerona/glaa286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics. 2016;8:64. doi: 10.1186/s13148-016-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15(1):149–154. doi: 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fahy GM, Brooke RT, Watson JP, Good Z, Vasanawala SS, Maecker H, et al. Reversal of epigenetic aging and immunosenescent trends in humans. Aging Cell. 2019;18(6):e13028. doi: 10.1111/acel.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, et al. Epigenetic Signatures of Cigarette Smoking. Circ Cardiovasc Genet. 2016;9(5):436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teschendorff AE, Yang Z, Wong A, Pipinikas CP, Jiao Y, Jones A, et al. Correlation of Smoking-Associated DNA Methylation Changes in Buccal Cells With DNA Methylation Changes in Epithelial Cancer. JAMA Oncol. 2015;1(4):476–485. doi: 10.1001/jamaoncol.2015.1053. [DOI] [PubMed] [Google Scholar]
- 43.Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, et al. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22(5):843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Elgizouli M, Schottker B, Holleczek B, Nieters A, Brenner H. Smoking-associated DNA methylation markers predict lung cancer incidence. Clin Epigenetics. 2016;8:127. doi: 10.1186/s13148-016-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.