Highlights
-
•
Decreased miR-376a was observed in glioma cell lines.
-
•
miR-376a directly targeted and inhibited SIRT1 in glioma.
-
•
Ectopic SIRT1 reversed miR-376a-induced the inhibition of proliferation and angiogenesis in glioma.
-
•
miR-376a inhibited YAP1/VEGF signal by binding to SIRT1.
-
•
miR-376a inhibited angiogenesis and tumour growth in vivo.
Keywords: Glioma, miR-376a, SIRT1, YAP/VEGF signalling, Angiogenesis
Abstract
Background
Glioma is the most common cancer in the central nervous system. Previous studies have revealed that the miR-376 family is crucial in tumour development; however, its detailed mechanism in glioma is not clear.
Methods
Cellular mRNA or protein levels of miR-376a, SIRT1, VEGF and YAP1 were detected via qRT–PCR or Western blotting. We analysed the proliferation, angiogenesis and migration abilities of glioma cell lines using colony formation, tube formation and Transwell assays. A luciferase assay was performed to determine whether miR-376a could recognize SIRT1 mRNA. Xenograft experiments were performed to analyse the tumorigenesis capacity of glioma cell lines in nude mice. The angiogenesis marker CD31 in xenograft tumours was detected via immunohistochemistry (IHC).
Results
miR-376a expression was lower in glioma cells than in normal astrocytes. miR-376a mimic inhibited SIRT1, YAP1, and VEGF expression and suppressed the proliferation, migration and angiogenesis abilities of the glioma cell lines LN229 and A172, whereas miR-376a inhibitor exerted the opposite functions. In a luciferase assay, miR-376a inhibited the luciferase activity of WT-SIRT1. SIRT1 overexpression upregulated YAP1 and VEGF in glioma cells and promoted proliferation, migration and angiogenesis. Xenografts with ectopic miR-376a expression exhibited lower volumes and weights and a slower growth curve. Overexpression of miR-376a inhibited YAP1/VEGF signalling and angiogenesis by inhibiting SIRT1 in xenograft tissues.
Conclusion
miR-376a directly targets and inhibits SIRT1 in glioma cells. Downregulation of SIRT1 resulted in decreased YAP1 and VEGF signalling, which led to suppression of glioma cell proliferation, migration and angiogenesis.
Introduction
Glioma is the most common primary malignant tumour in the central nervous system (representing 80% of brain tumours) and is characterized by invasive growth and early metastasis [1]. Its age-adjusted incidence is 0.59∼3.69 per 100,000 persons [2]. At present, glioma treatment primarily involves surgical resection, supplemented by radiotherapy and chemotherapy [3]. However, due to chemotherapy resistance, the mortality and recurrence rate of glioma is still very high, and the 5-year survival rate of glioma patients is only 5%. [4]. Development of a deep understanding of the molecular mechanisms driving glioma to identify effective methods for glioma diagnosis and treatment is of great significance.
MicroRNAs (miRNAs) are single-stranded RNAs approximately 22 nt in length that are involved in the regulation of many crucial biological processes in tumours. For example, cell proliferation, apoptosis, and differentiation are all regulated by miRNAs [5]. MicroRNAs inhibit the translation of target genes by binding to the 3′-untranslated region (3′-UTR) of target mRNAs. The miR-376 family is crucial in multiple cancer types. For instance, the invasion and proliferation ability of lung cancer is inhibited by miR-376a via inhibition of c-Myc expression [6]. In hepatocellular cancer, miR-376a inhibits FOXK and Snail signalling, which leads to decreased metastasis and inhibition of tumour growth [7]. A previous study revealed that expression of the miR-376 family is significantly lower in glioma patients and is crucial in cancer formation and progression [8]. In addition, it has been reported that a reduction in mature miR-376a is associated with the proliferation and invasion of glioblastoma cells [9]. However, the miR-376a-associated signalling network, especially in glioma, is not yet fully understood.
SIRT1 is a type of HDAC (histone deacetylase) that depends on NAD and regulates gene expression by altering the acetylation status of histones. In addition, SIRT1 can also deacetylate nonhistone targets, giving it a much wider spectrum of targets [10]. SIRT1 is also widely involved in the regulation of apoptosis, autophagy, senescence, proliferation, and tumour occurrence [11,12]. Several studies have shown the significance of SIRT1 in the regulation of tumours, such as melanoma, prostate cancer and colon cancer [13], [14], [15], presumably by modulating proliferation, metastasis, epithelial mesenchymal transformation (EMT) or apoptosis [12,16]. For instance, SIRT1 has been found to promote degradation of E-cadherin and subsequent EMT in melanoma; SIRT1 has also been shown to be upregulated in prostate cancer in both human patient tissues and mouse models and to promote tumorigenesis. SIRT1 can also regulate angiogenesis. Jiang and colleagues demonstrated that SIRT1 can inhibit angiogenesis in ovarian cancer [17]. Sue reported that FOXO1 signalling inhibited angiogenesis in gastric cancer by inhibiting HIF-1α-VEGF signalling and SIRT1 [18]. Another study revealed that SIRT1 is upregulated in glioma cells and associated with invasion, metastasis and cell viability [19]. However, the signals upstream and downstream of SIRT1 have not yet been described clearly in glioma. We revealed a binding sequence between miR-376a and the 3′-UTR of SIRT1 using the online database TargetScan (http://www.targetscan.org/vert_71/). Hence, a signalling axis between miR-376a/SIRT1 might exist in glioma.
YAP1 is involved in the regulation of inflammation, tumorigenesis, immunosuppression and many crucial biological processes in various cancer types [20], [21], [22]. SIRT1 was reported as an upstream signal of YAP1. For instance, activation of the c-Myc/NAMPT/SIRT1 signal was found to be crucial for YAP signal activation, and knockdown of SIRT1 resulted in decreased YAP1 expression [23]. In addition, SIRT1 is upregulated in liver cancer and promotes tumour growth by increasing YAP1 expression [24]. SIRT1 was found to deacetylate the YAP2 protein and increased the YAP2/TEAD4 association, leading to YAP2/TEAD4 transcriptional activation and cell growth promotion in hepatocellular carcinoma cells [25]. Based on these reports, SIRT1 expression can promote YAP1 expression and is positively correlated with YAP1 expression. Notably, YAP1 expression is positively correlated with VEGFA expression and secretion, which promotes angiogenesis in human renal carcinoma [26]. In glioma, miR-27v-3p can inhibit YAP1 and retard proliferation and migration while promoting apoptosis [27]. LINC00475 can promote glioma progression by regulating YAP1 expression [28]. This evidence indicates a crucial role of YAP1 in glioma development. However, whether SIRT1 exerts such functions by modulating YAP1/VEGFA signalling remains to be explored.
We designed this study with aim of revealing the regulatory mechanism of miR-376a and SIRT1 in glioma development and to explore new potential therapeutic targets. Our data indicated that miR-376a expression is decreased in glioma cells and that overexpression of miR-376a inhibits glioma cell proliferation, migration and angiogenesis. miR-376a directly bound SIRT1 mRNA to decrease SIRT1 expression, thereby inhibiting YAP1/VEGF signalling. These findings illustrate a new regulatory mechanism in glioma development and could provide theoretical support for future studies.
Materials and methods
Cell culture
The human umbilical vein endothelial cells (HUVECs); A172, LN229, U251, and U373 cell lines; and human astrocyte cell lines SVGP12 and NHA used in this study were obtained from Chinese Academy Sciences. A humidified incubator was used for cell culture. The incubator was supplied with 5% CO2. Cells were maintained in DMEM + 10% FBS (foetal bovine serum) + 100 U/ml P/S (penicillin +streptomycin) at 37 °C. DMEM and FBS were obtained from Gibco, USA.
Cell transfection
The miR-376a mimic and inhibitor and sh-SIRT1 were purchased from GenePharma (Shanghai, China). The SIRT1 overexpression plasmid was constructed by cloning the cDNA of SIRT1 into pcDNA3.1. We performed transfection using Lipofectamine 2000 (Invitrogen, Missouri, USA). Briefly, cells were cultured to 70% confluence. Plasmids/RNA were first incubated in Opti-MEM for 5 min, mixed with Lipofectamine 2000, incubated for 5 min with an equal amount of Opti-MEM for another 20 min, and then added to cells in serum-free medium. The serum-free medium was replaced with normal medium 6 h later, and the cells were then cultured normally. We collected cells 48 h post-transfection.
Cell counting kit-8 (CCK-8) assay
Cell proliferation was detected using a CCK-8 kit (Abcam, ab228554) according to the manufacturer's instructions. Briefly, NHC and SVGP12 cells were collected and seeded at 5 × 103 cells/well in 96-well plates. Cells were cultured for 48 h and treated with CCK-8 solution (10 μl per well) for 4 h under normal culture conditions. absorbence was measured at 450 nm using a plate reader (Tecan, Männedorf, Switzerland).
Annexin-V assay
Annexin V assays were performed using an Annexin V/FITC kit from Abcam (ab14085) following the manufacturer's instructions. Briefly, cells were collected and counted. Then, 1 × 105 cells were mixed with 500 μl Annexin V binding buffer, 5 μl Annexin V-FITC and 5 μl propidium iodide. The mixture was incubated for 5 mins at room temperature in the dark. The Annexin V-FITC signal was then analysed via flow cytometry (Ex=488 nm, Em=530 nm) using a FITC signal detector and a phycoerythrin emission signal detector to detect PI staining.
Clone formation assay
Cells were treated with trypsin for 5 min at 37 °C and then collected by centrifugation. After centrifugation for 3 min at 1000 g, the cells were resuspended. The cell concentration was determined and then adjusted to 2000 cells per mL. Cells were seeded and cultured in a 6-well plate. After 14 days, the cells were collected, washed with PBS, and stained with crystal violet (0.5%). The cells were then observed under a microscope (Olympus).
Transwell assay
First, a system with two compartments was established by placing a Transwell chamber into a 24-well plate. Cells were treated with trypsin for 5 min at 37 °C and collected via centrifugation (1000 g, 5 min). The pellet was resuspended in serum-free medium. The resuspended cells were seeded in the upper chamber, and full medium was added to the lower chamber. The system was then cultured for 24 h, and the porous membrane was collected. The membranes were then treated with methyl alcohol followed by crystal violet staining. The cell number was then analysed using a microscope (Olympus).
Tube formation assay
To perform the tube formation assay, we first pretreated plates with Matrigel Basement Membrane Matrix (Sigma–Aldrich, USA) according to the manufacturer's instructions. HUVECs were then cocultured with glioma cells with the indicated treatments or transfections for 6 h. HUVECs and glioma cells were seeded at a 1:1 ratio. After incubation for 6 h, the cells were rinsed twice with PBS and treated with 4% PFA at room temperature for 15 min. Cells were rinsed once again with PBS and observed under a microscope (Olympus).
Dual-luciferase reporter assay
We first synthesized wild-type and mutant sequences of the SIRT1 3′UTR (untranslated region) containing the miR-376a binding site. These sequences were cloned into the dual-luciferase vector system pmirGLO (Promega, USA). For the dual-luciferase reporter assay, glioma cells were transfected with the WT- or MUT-SIRT1 luciferase reporter plasmid system together with the indicated components. Cells were cultured for another 48 h and collected. A luciferase assay was then carried out using a Dual-Luciferase Reporter Assay System (Promega, USA). Experiments were performed according to the manufacturers’ instructions.
Xenograft model analysis
Ethical approval was obtained from the Animal Care and Use Committee of Hunan Provincial People's Hospital (The First Affiliated Hospital of Hunan Normal University). A total of 10 BALB/c nude mice (6 weeks old) were obtained from SJA Laboratory Animal Co., Ltd. (Hunan, China) and used in this study. The mice were randomly divided into two groups, with 5 mice in each group. The animals were bred under specific pathogen-free conditions. Glioma A172 cells stably expressing miR-376a mimic or NC mimic were collected, and 5 × 106 cells were subcutaneously inoculated into the back of the mice next to the right front limb. Every 7 days, the mice were observed, and the tumour volume was measured using a standard calliper. Tumour volume (V) was calculated as follows: V = 0.5 × L × W2, where L and W are defined as the tumour length (L) and width (W), respectively. Animals were all euthanized by the end of the experiments (5 weeks later). The tumours were isolated and weighed.
Immunohistochemistry (IHC)
Formalin (10%)-fixed and paraffin-embedded (FFPE) tumour tissue samples were prepared. The FFPE sample were cut into 5 μm sections. For IHC staining, sections were first treated with BSA (5%) at room temperature for 1 hour. Blocked sections were then treated with CD31 primary antibody (#77,699, Cell signalling Technology, USA) at 4 °C on a shaker overnight. Then, after washing with PBS twice, the sections were incubated at room temperature for 2 h with secondary antibodies. Sections were washed again and incubated with HRP (Dako REAL Ebision kit; DAKO, Denmark) according to the manufacturer's instructions.
qRT–PCR
Total RNA was extracted from glioma cells or xenografts using TRIzol reagent. cDNA was reverse transcribed using a PrimeScript RT reagent kit (Takara). The qRT–PCR system was prepared using SYBR Premix Ex Taq II (RR820A, Takara). Reactions were carried out in a 384-well plate under the following settings: (94 °C for 5 s, 60 °C for 34 s, and 72 °C for 30 s) for 40 cycles. The reaction was performed on an ABI 7900 system (ABI, USA). In this experiment, the internal references were U6 and GAPDH. The relative expression level was calculated using the 2−∆∆Ct method. The primers used in this study were as follows: miR-376a-5p, (forward) 5′-GCCGCGTAGATTCTCCTTCTA-3′ and (reverse) 5′- GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTACTCA-3′; SIRT1, (forward) 5′-CAGTGGCTGGAACAGTGAGA-3′ and (reverse) 5′-TCTGGCATGTCCCACTATCA-3′; YAP1, (forward) 5′-TGACCCTCGTTTTGCCATGA-3′ and (reverse) 5′-TGACCCTCGTTTTGCCATGA-3′; VEGF, (forward) 5′-GGGCAGAATCATCACGAAGT-3′ and (reverse) 5′-TGGTGATGTTGGACTCCTCA-3′; GAPDH, (forward) 5′-CCAGGTGGTCTCCTCTGA-3′ and (reverse) 5′-GCTGTAGCCAAATCGTTGT-3′; and U6, (forward) 5′-CTCGCTTCGGCAGCACA-3′ and (reverse) 5′-AACGCTTCACGAATTTGCGT-3′.
Protein extraction and western blotting
Cells or tissues were collected and lysed in RIPA buffer (Beyotime, China). Briefly, cells or homogenized tissue were mixed with RIPA buffer at a ratio of 1 mL RIPA per 107 cells. Lysis was carried out on ice. After 20 min of lysing, we centrifuged the samples at 12,000 g for 15 min at 4 °C and collected them. For western blotting, we loaded 30 μg protein into SDS–PAGE gels. The proteins were separated and transferred onto PVDF membranes. The membranes were then blocked by incubation with 5% BSA. The incubation lasted 1 hour at room temperature (RT). After washing on a shaker 3 times (5 min each time) with TBST, the membranes were incubated with primary antibody at 4 °C overnight. Membranes were washed again and incubated with secondary antibodies for 1 hour at RT in the dark. An enhanced chemiluminescence (ECL) system was used to detect the signal. Primary antibodies against VEGF (#65,373, CST), SIRT1 (#8469, CST), YAP1 (#8418, CST) and GAPDH (#5174, CST) were used.
Statistical analysis
We used Student's t-test to compare two groups and one-way ANOVA to compare multiple groups. Analysis was carried out using GraphPad Prism 7 software. All data represent the mean with the SD. P<0.05 was considered statistically significant.
Results
miR-376a suppressed the proliferation, angiogenesis and migration of glioma cells
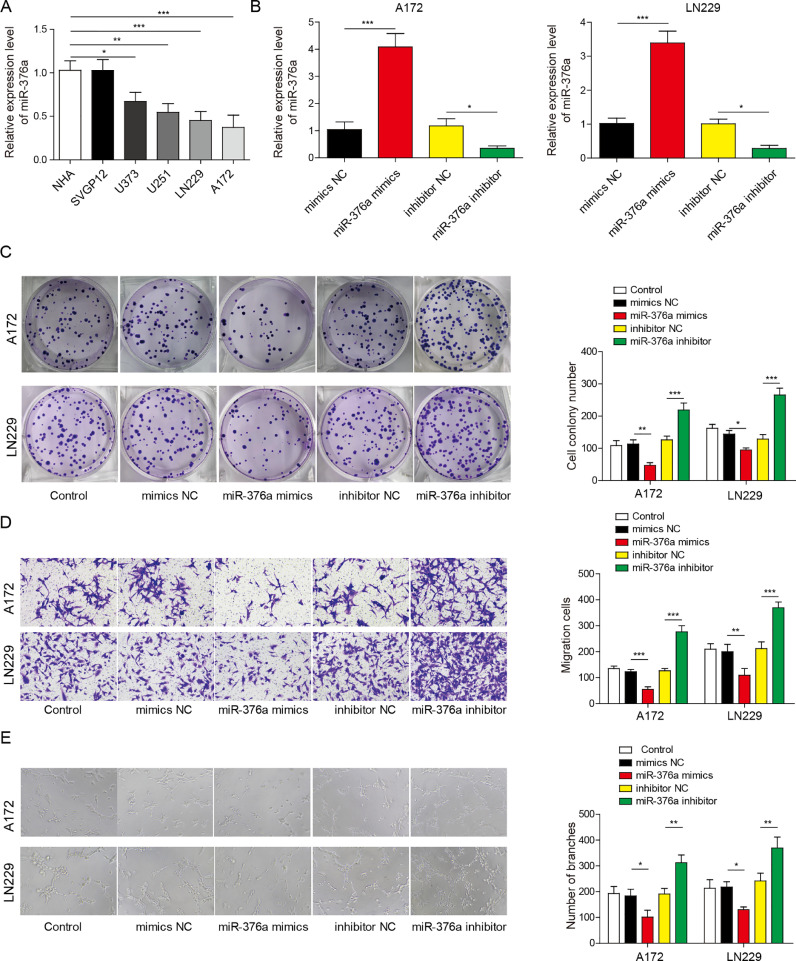
We first tried to evaluate whether the expression of miR-376a varies amongst normal human astrocytes and glioma cell lines. The immortalized normal human astrocyte cell lines SVGP12 and NHC and glioma cell lines U373, U251, LN229 and A172 were used in the experiment. qRT–PCR results indicated that the RNA level of miR-376a was significantly lower in glioma cell lines than in the SVGP12 and NHC cell lines (Fig. 1A). We thus designed a mimic and inhibitor of miR-376a to evaluate the functions of miR-376a in glioma cell lines. After transfection with miR-376a mimic, miR-376a was dramatically upregulated in both A172 and LN229 cell lines. As expected, the miR-376a inhibitor suppressed the mRNA level of miR-376a in these cell lines (Fig. 1B). Then, we analysed whether the processes associated with tumour development, such as proliferation, angiogenesis and migration, were altered by miR-376a mimic or inhibitor in A172 and LN229 cells. The results indicated that overexpression of miR-376a suppressed the proliferation, migration and angiogenesis of A172 and LN229 cells, while the miR-376a inhibitor promoted these effects (Fig. 1C–E). Taken together, these results demonstrate that miR-376a can inhibit the proliferation, migration and angiogenesis of glioma cell lines.
Fig. 1.
miR-376a regulated the proliferation, migration and angiogenesis of glioma cells. (A) The expression level of miR-376a was detected in NHA, SVGP12, U373, U251, LN229 and A172 cells. (B) The expression level of miR-376a was detected via qRT–PCR in LN229 and A172 cells transfected with a miR-376a mimic or inhibitor. (C, D and E) The glioma cell lines A172 and LN229 were transfected with miR-376a mimic or inhibitor as indicated and subjected to a clone formation assay (C), Transwell assay to assess cell migration (D) and angiogenesis assay (E). The statistical analysis is displayed in the columns. *P< 0.05, **P< 0.01, and ***P< 0.001.
miR-376a directly recognized SIRT1 mRNA and inhibited SIRT1 expression
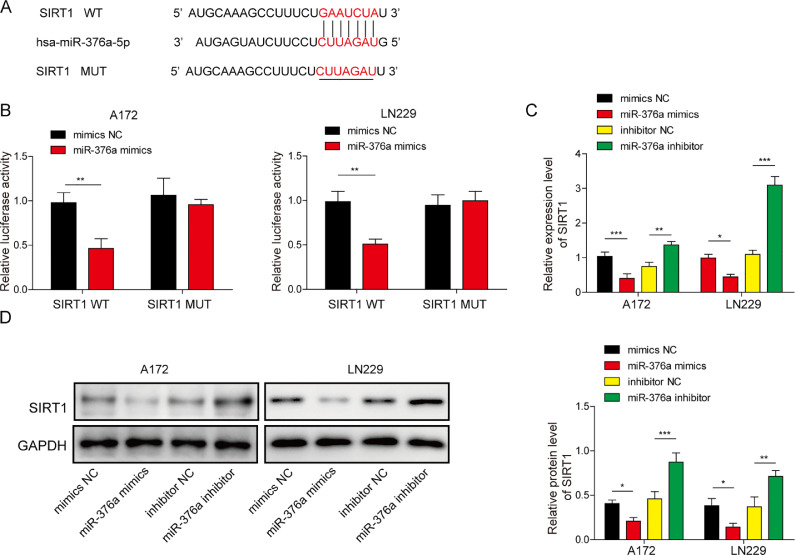
To identify the downstream factor of miR-376a in glioma, we performed bioinformatics analysis using TargetScan (http://www.targetscan.org/vert_71/), a widely used database to predict target genes of miRNAs [29], and found a spectrum of potential targets of miR-376a. amongst the spectrum of potential targets, we selected 8 potential genes that were reported to be upregulated in glioma (since miR-376a was downregulated in glioma cell lines). We detected the expression of these genes in glioma cell lines transfected with miR-376a mimic (Fig. S2). Only SIRT1 was significantly suppressed by the miR-376a mimic. Hence, SIRT1 was selected as our subject for the following study. A conserved sequence in the 3′ UTR of SIRT1 was predicted as the binding sequence of miR-376a (Fig. 2A). We designed a luciferase reporter vector system containing wild-type (WT) and mutant (MUT) binding sequences for the following experiments to evaluate whether miR-376a can recognize the 3′ UTR of SIRT1. The relative luciferase activity was significantly inhibited by the miR-376a mimic. Moreover, the miR-376a inhibitor enhanced the relative luciferase activity (Fig. 2B). The luciferase results demonstrated that miR-376a can recognize and suppress SIRT1 mRNA. To validate this finding, we also detected the mRNA and protein levels of SIRT1. As expected, both the mRNA and protein levels of SIRT1 were inhibited by the miR-376a mimic but enhanced by the miR-376a inhibitor (Fig. 2C and D). Taken together, miR-376a inhibited SIRT1 expression in the glioma cell lines LN229 and A172.
Fig. 2.
miR-376a directly regulated the expression of SIRT1 in glioma cells. (A) Schematic showing the binding sequence between miR-376a and the 3′-UTR of SIRT1. (B) The glioma cell lines A172 and LN229 were transfected with a SIRT1-WT or SIRT1-MUT dual-luciferase reporter system and subjected to a dual-luciferase reporter assay. (C) The mRNA level of SIRT1 was detected via qRT–PCR in the glioma cell lines A172 and LN229 after transfection with miR-376a mimic or inhibitor. (D) The protein level of SIRT1 was detected via WB in the glioma cell lines A172 and LN229 transfected with miR-376a mimic or inhibitor. *P< 0.05, **P< 0.01, and ***P< 0.001.
Overexpression of SIRT1 antagonized the effects of miR-376a in glioma cells
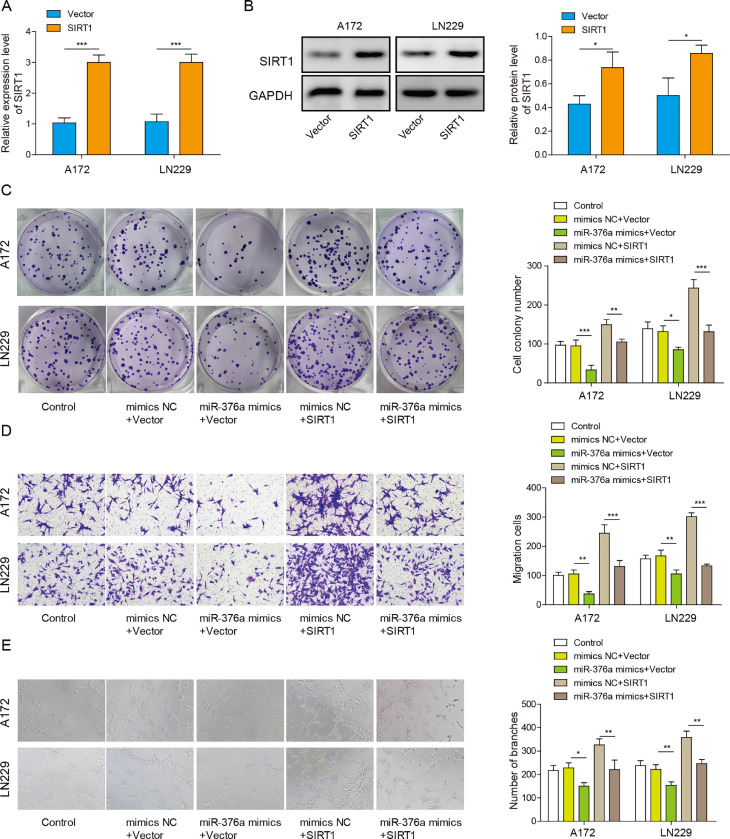
To evaluate the function of SIRT1 in glioma cells, we overexpressed SIRT1 in the glioma cell lines A172 and LN229 (Fig. 3A and B). Then, the clone formation, migration and angiogenesis abilities of these glioma cells were detected. miR-376a mimic inhibited the clonal formation ability of the glioma cells. Meanwhile, SIRT1 overexpression promoted clone formation (Fig. 3C). Furthermore, the clone formation ability of cells transfected with both SIRT1 and miR-376a was similar to that of the control group. Overexpression of SIRT1 abolished the effect of the miR-376a mimic, indicating that SIRT1 overexpression antagonized the function of the miR-376a mimic. (Fig. 3C). Similar trends were observed in migration and angiogenesis. While the miR-376a mimic suppressed migration and angiogenesis, overexpression of SIRT1 remarkably promoted migration and angiogenesis (Fig. 3D and E). In addition, overexpression of SIRT1 abolished the inhibitory effect of the miR-376a mimic on glioma cell migration and angiogenesis (Fig. 3D and E). These results demonstrate that miR-376a inhibits glioma proliferation, migration and angiogenesis by targeting SIRT1.
Fig. 3.
Overexpression of SIRT1 antagonized the effects of miR-376a in glioma cells. (A) The mRNA level of SIRT1 was analysed via qRT–PCR in the glioma cell lines A172 and LN229 transfected with SIRT1 overexpression vector. (B) The protein level of SIRT1 was analysed via Western blotting in LN229 and A172 cells transfected with SIRT1 overexpression vector. (C-E) The glioma cell lines A172 and LN229 were transfected with miR-376a mimic or a SIRT1 overexpression vector as indicated. Clone formation assays (C), Transwell assays (D) and angiogenesis assays (E) were then performed in these cells. *P< 0.05, **P< 0.01, and ***P< 0.001.
To explore whether SIRT1 and miR-376a exert the same functions in normal human astrocytes, the following experiments were performed. The miR-376a inhibitor suppressed the expression level of miR-376a (Fig. S1A) while upregulating the mRNA and protein levels of SIRT1 (Fig. S1B and C). Knockdown of SIRT1 using shSIRT1 successfully suppressed both the mRNA and protein levels of SIRT1 in NHA and SVGP12 cells (Fig. S1D and E). These effects were similar in the glioma cell lines LNN229 and A172. Furthermore, the miR-376a inhibitor promoted proliferation and inhibited apoptosis of NHA and SVGP12 cells (Fig. S1F and G), and SIRT1 silencing abolished these functions of the miR-376a inhibitor. These data indicate that miR-376a can regulate cell proliferation and apoptosis in normal astrocytes through SIRT1.
miR-376a inhibited the YAP1/VEGF signalling axis by suppressing SIRT1 in glioma cells
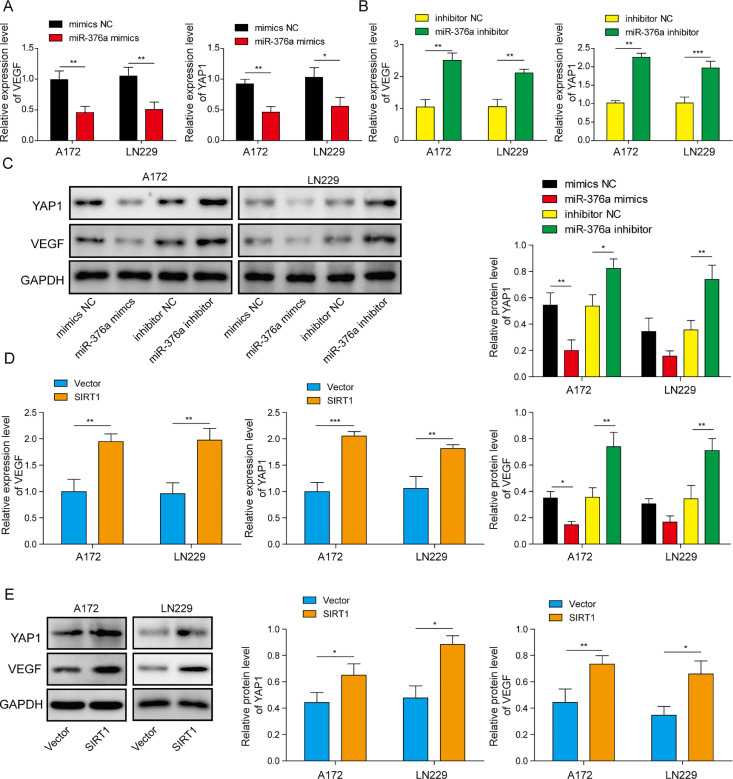
YAP1/VEGF signalling participates in tumour development in multiple cancer types, including glioma [26,30]. In addition, SIRT1 was reported to be an upstream regulator of the YAP1/VEGF pathway [31]. To determine whether YAP1/VEGF signalling is regulated by SIRT1 and miR-376a in glioma cell lines, the following experiments were performed. We first transfected the glioma cell lines A172 and LN229 with miR-376a mimic or inhibitor and detected the mRNA levels of YAP1 and VEGF in these cells. Both YAP1 and VEGF mRNA expression was suppressed by the miR-376a mimic. Moreover, the miR-376a inhibitor enhanced these mRNA levels (Fig. 4A and B). A similar trend was observed in protein expression. The miR-376a mimic dramatically suppressed the protein expression of YAP1 and VEGF, while the miR-376a inhibitor exerted the opposite effects, upregulating YAP1/VEGF expression (Fig. 4C). We also validated whether SIRT1 is an upstream regulator of YAP1/VEGF. SIRT1 overexpression in glioma cell lines resulted in enhanced mRNA and protein levels of YAP1 and VEGF (Fig. 4D and E). Given that SIRT1 is a target of miR-376a in glioma cells, we concluded that miR-376a can inhibit YAP1/VEGF expression by inhibiting SIRT1 in the glioma cell lines A172 and LN229.
Fig. 4.
miR-376a regulated the YAP1/VEGF signalling axis by inhibiting SIRT1 in glioma cells. (A) The mRNA levels of YAP1 and VEGF were detected by qRT–PCR in the glioma cell lines A172 and LN229 transfected with miR-376a mimic. (B) The mRNA levels of YAP1 and VEGF were detected by qRT–PCR in the glioma cell lines A172 and LN229 transfected with miR-376a inhibitor. (C) The protein levels of YAP1 and VEGF were detected by Western blotting in the glioma cell lines A172 and LN229 transfected with miR-376a mimic or inhibitor. The mRNA (D) and protein (E) levels of YAP1 and VEGF were detected by qRT–PCR (D) and Western blotting (E) in the glioma cell lines A172 and LN229 transfected with SIRT1 overexpression vector. *P< 0.05, **P< 0.01, and ***P< 0.001.
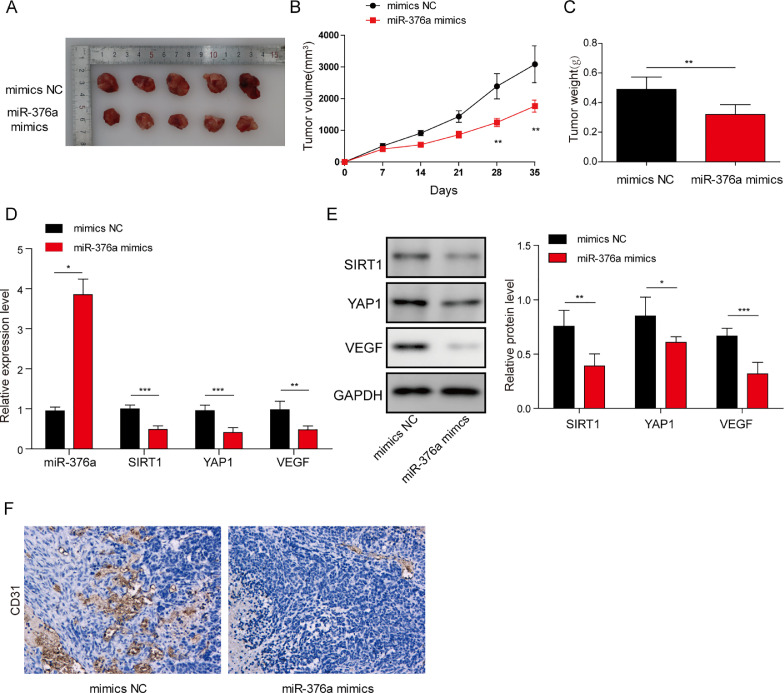
miR-376a inhibited tumorigenesis and angiogenesis in nude mice by regulating the SIRT1/YAP1/VEGF signalling axis
Next, we tried to identify the functions of miR-376a and SIRT1 in a mouse model. The glioma cell line LN229 was transfected with miR-376a mimic or control (NC) and subjected to tumorigenesis analysis in nude mice. The tumour volume, weight and growth curve were all inhibited in the miR-376a mimic group compared to the NC group (Fig. 5A–C). We then analysed the expression of miR-376a, SIRT1, YAP1 and VEGF in these tumour tissues. In tumour tissues from the miR-376a mimic group, suppression of YAP1, SIRT1 and VEGF was observed, together with enhanced miR-376a expression (Fig. 5D and E). Because VEGF usually promotes tumour development by promoting angiogenesis [32], we also detected the angiogenesis marker CD31 in these tumour tissues. As expected, the miR-376a group showed suppressed CD31 expression, which indicated hampered angiogenesis (Fig. 5F). Taken together, miR-376a inhibited tumorigenesis and angiogenesis in the glioma cell line LN229 by inhibiting SIRT1 and suppressing YAP1/VEGF signalling.
Fig. 5.
miR-376a inhibited tumorigenesis and angiogenesis in nude mice by regulating the SIRT1/YAP1/VEGF signalling axis. (A) Representative images of tumours from nude mice after subcutaneous injection of A172 cells stably expressing miR-376a mimic or mimic NC (5 mice per group). (B) Quantifications of tumour volume. (C) Quantification of tumour weight. (D) The expression levels of miR-376a, SIRT1, YAP1 and VEGF in tumour tissues were detected by qRT–PCR (n = 5). (E) The protein levels of SIRT1, YAP1 and VEGF in tumour tissues were detected by Western blotting (n = 5). (F) Immunohistochemistry analysis of CD31 levels in tumour tissues from mice in different groups. *P< 0.05, **P< 0.01, and ***P< 0.001.
Discussion
Glioma is the most common nervous system cancer. However, the prognosis after traditional treatment options, such as surgical resection, chemotherapy or radiotherapy, is still poor [33]. Therefore, a detailed understanding of the molecular mechanism underlying glioma development is necessary for development of new therapeutic methods. In this study, we revealed that miR-376a inhibits tumour development, growth and angiogenesis in the glioma cell lines A172 and LN229. miR-376a exerted these functions by inhibiting the expression of its target SIRT1, which led to suppression of YAP1/VEGF signalling. These findings illustrate a portion of the complex mechanism of glioma development and can contribute to future studies. miRNAs are involved in the regulation of a variety of key cellular processes related to tumorigenesis and development, including cell migration, invasion, EMT, apoptosis, and proliferation [34]. miRNAs can recognize and bind their target mRNAs to mediate their degradation through the RNA-induced silencing complex (RISC), thereby suppressing the expression of their target genes [35]. In glioma, miRNAs have also been reported to be critical regulators. For example, miR-451 inhibited the proliferation and invasion of glioma by inhibiting the expression of cab39, which led to a decrease in mTOR signalling [30]. miR-124 inhibited glioma proliferation and metastasis by inhibiting ROCK1 signalling [36]. miR-376 has been described as a tumour suppressor in lung cancer and hepatocellular cancer by inhibiting proliferation, invasion and metastasis [6,7]; however, the molecular mechanism is not yet fully understood in glioma. In our results, miR-376a was found to be a critical regulator of glioma development. Overexpression of miR-376a inhibited cell proliferation, migration and angiogenesis in glioma. These findings were consistent with those of previous reports. For instance, decreased mature miR-376a is usually associated with increased glioma invasion[9]. Restoration of miR-376a in lung cancer leads to decreased proliferation and invasion[6]. All this evidence demonstrates that miR-376a is a key regulator in glioma.
SIRT1 is class-III histone deacetylase. Previous studies have shown that the function of SIRT1 can vary in different cells. It can be either a tumour suppressor or a tumour promoter, determined by cell type and subcellular localization [37]. SIRT1 has been reported to be responsible for proliferation, metastasis, invasion and other aspects of glioma [[38], [39]]. The regulation of SIRT1 also involves a complex network. For instance, it can be regulated by circRNA, miR-326, miR-133 and many other factors [38], [39], [40]. In this study, SIRT1 was found to be a direct target of miR-376a in glioma. Overexpression of SIRT1 promoted glioma proliferation, migration and angiogenesis and partially abolished the effects of miR-376a in glioma cells.
SIRT1 can directly promote the expression of a spectrum of genes by modulating the acetylation status of histones [12]. In our study, overexpression of SIRT1 resulted in upregulation of YAP1 and VEGFA, which promoted glioma cell proliferation, migration and angiogenesis. The results of this study suggest that overexpression of SIRT1 results in upregulation of YAP1 and VEGF, which promotes glioma cell proliferation, migration and angiogenesis.
Conclusions
In this study, we demonstrated that miR-376a is suppressed in glioma and first revealed that SIRT1 is a direct target of miR-376a in glioma. In addition, we found that miR-376a inhibits glioma cell proliferation, migration, and angiogenesis by suppressing YAP1/VEGFA signalling through targeting of SIRT1. These findings shed new light on our understanding of the molecular regulatory network in glioma and could provide potential therapeutic targets in the future.
Funding
None.
Ethical approval
Ethical approve was obtained from the Animal Care and Use Committee of Hunan Provincial People's Hospital (The first affiliated hospital of Hunan normal university).
Consent for publication
Not Applicable. This article does not contain any studies with human participants performed by any of the authors.
Availability of data and material
All data generated or analysed during this study are included in this article. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
CRediT authorship contribution statement
Yong-Wen Deng: Conceptualization, Funding acquisition, Writing – original draft. Yu-Gao Shu: Data curation, Resources, Methodology, Formal analysis. Sheng-Li Sun: Investigation, Software, Visualization, Project administration, Supervision, Validation, Writing – review & editing.
CRediT authorship contribution statement
Yong-Wen Deng: Conceptualization, Funding acquisition, Writing – original draft. Yu-Gao Shu: Data curation, Resources, Methodology, Formal analysis. Sheng-Li Sun: Investigation, Software, Visualization, Project administration, Supervision, Validation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to give our sincere gratitude to the reviewers for their constructive comments.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101270.
Appendix. Supplementary materials
References
- 1.Molinaro A.M., Taylor J.W., Wiencke J.K., Wrensch M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019;15:405–417. doi: 10.1038/s41582-019-0220-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom Q.T., Bauchet L., Davis F.G., Deltour I., Fisher J.L., Langer C.E., Pekmezci M., Schwartzbaum J.A., Turner M.C., Walsh K.M., Wrensch M.R., Barnholtz-Sloan J.S. The epidemiology of glioma in adults: a "state of the science" review. Neuro. Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gusyatiner O., Hegi M.E. Glioma epigenetics: from subclassification to novel treatment options. Semin. Cancer Biol. 2018;51:50–58. doi: 10.1016/j.semcancer.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Chen R., Smith-Cohn M., Cohen A.L., Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14:284–297. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hrovatin K., Kunej T. Classification of miRNA-related sequence variations. Epigenomics. 2018;10:463–481. doi: 10.2217/epi-2017-0126. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Cong W., Wu G., Ju X., Li Z., Duan X., Wang X., Gao H. miR-376a suppresses the proliferation and invasion of non-small-cell lung cancer by targeting c-Myc. Cell Biol. Int. 2018;42:25–33. doi: 10.1002/cbin.10828. [DOI] [PubMed] [Google Scholar]
- 7.Meng F., Liu J., Lu T., Zang L., Wang J., He Q., Zhou A. SNHG1 knockdown upregulates miR-376a and downregulates FOXK1/Snail axis to prevent tumor growth and metastasis in HCC. Mol. Ther. Oncolytics. 2021;21:264–277. doi: 10.1016/j.omto.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Q., Wang C., Hou Z., Wang G., Lv J., Wang H., Yang J., Zhang Z., Zhang H. Serum microRNA-376 family as diagnostic and prognostic markers in human gliomas. Cancer Biomark. 2017;19:137–144. doi: 10.3233/CBM-160146. [DOI] [PubMed] [Google Scholar]
- 9.Choudhury Y., Tay F.C., Lam D.H., Sandanaraj E., Tang C., Ang B.T., Wang S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig. 2012;122:4059–4076. doi: 10.1172/JCI62925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang H.C., Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C., Zhou M., Ge Y., Wang X. SIRT1 and aging related signaling pathways. Mech. Ageing Dev. 2020;187 doi: 10.1016/j.mad.2020.111215. [DOI] [PubMed] [Google Scholar]
- 12.Alves-Fernandes D.K., Jasiulionis M.G. The role of SIRT1 on DNA damage response and epigenetic alterations in cancer. Int. J. Mol. Sci. 2019;20:3153. doi: 10.3390/ijms20133153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun T., Jiao L., Wang Y., Yu Y., Ming L. SIRT1 induces epithelial-mesenchymal transition by promoting autophagic degradation of E-cadherin in melanoma cells. Cell Death Dis. 2018;9:136. doi: 10.1038/s41419-017-0167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huffman D.M., Grizzle W.E., Bamman M.M., Kim J.S., Eltoum I.A., Elgavish A., Nagy T.R. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 15.Stünkel W., Peh B.K., Tan Y.C., Nayagam V.M., Wang X., Salto-Tellez M., Ni B., Entzeroth M., Wood J. Function of the SIRT1 protein deacetylase in cancer. Biotechnol. J. 2007;2:1360–1368. doi: 10.1002/biot.200700087. [DOI] [PubMed] [Google Scholar]
- 16.Maiese K. Moving to the rhythm with clock (Circadian) genes, autophagy, mTOR, and SIRT1 in degenerative disease and cancer. Curr. Neurovasc. Res. 2017;14:299–304. doi: 10.2174/1567202614666170718092010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang W., Jiang P., Yang R., Liu D.F. Functional role of SIRT1-induced HMGB1 expression and acetylation in migration, invasion and angiogenesis of ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:4431–4439. doi: 10.26355/eurrev_201807_15494. [DOI] [PubMed] [Google Scholar]
- 18.Kim S.Y., Ko Y.S., Park J., Choi Y., Park J.W., Kim Y., Pyo J.S., Yoo Y.B., Lee J.S., Lee B.L. Forkhead transcription factor FOXO1 inhibits angiogenesis in gastric cancer in relation to SIRT1. Cancer Res. Treat. 2016;48:345–354. doi: 10.4143/crt.2014.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Chen X., Cui Y., Wei Q., Chen S., Wang X. Effects of SIRT1 silencing on viability, invasion and metastasis of human glioma cell lines. Oncol. Lett. 2019;17:3701–3708. doi: 10.3892/ol.2019.10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raj N., Bam R. Reciprocal crosstalk between YAP1/hippo pathway and the p53 family proteins: mechanisms and outcomes in cancer. Front. Cell Dev. Biol. 2019;7:159. doi: 10.3389/fcell.2019.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shibata M., Ham K., Hoque M.O. A time for YAP1: tumorigenesis, immunosuppression and targeted therapy. Int. J. Cancer. 2018;143:2133–2144. doi: 10.1002/ijc.31561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zanconato F., Cordenonsi M., Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H., Liu N., Zhao Y., Zhu X., Wang C., Liu Q., Gao C., Zhao X., Li J. Oncogenic USP22 supports gastric cancer growth and metastasis by activating c-Myc/NAMPT/SIRT1-dependent FOXO1 and YAP signaling. Aging (Albany NY) 2019;11:9643–9660. doi: 10.18632/aging.102410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Cui R., Zhang X., Qiao Y., Liu X., Chang Y., Yu Y., Sun F., Wang J. SIRT1 increases YAP- and MKK3-dependent p38 phosphorylation in mouse liver and human hepatocellular carcinoma. Oncotarget. 2016;7:11284–11298. doi: 10.18632/oncotarget.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao B., Hu F., Cheng J., Wang P., Xu M., Yuan F., Meng S., Wang Y., Yuan Z., Bi W. SIRT1 regulates YAP2-mediated cell proliferation and chemoresistance in hepatocellular carcinoma. Oncogene. 2014;33:1468–1474. doi: 10.1038/onc.2013.88. [DOI] [PubMed] [Google Scholar]
- 26.Xu S., Zhang H., Chong Y., Guan B., Guo P. YAP promotes VEGFA expression and tumor angiogenesis though Gli2 in human renal cell carcinoma. Arch. Med. Res. 2019;50:225–233. doi: 10.1016/j.arcmed.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Miao W., Li N., Gu B., Yi G., Su Z., Cheng H. MiR-27b-3p suppresses glioma development via targeting YAP1. Biochem. Cell. Biol. 2020;98:466–473. doi: 10.1139/bcb-2019-0300. [DOI] [PubMed] [Google Scholar]
- 28.Yu M., Yi B., Zhou W., Gong W., Li G., Yu S. Linc00475 promotes the progression of glioma by regulating the miR-141-3p/YAP1 axis. J. Cell. Mol. Med. 2021;25:463–472. doi: 10.1111/jcmm.16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nan Y., Guo H., Guo L., Wang L., Ren B., Yu K., Huang Q., Zhong Y. MiRNA-451 inhibits glioma cell proliferation and invasion through the mTOR/HIF-1α/VEGF signaling pathway by targeting CAB39. Hum. Gene Ther. Clin. Dev. 2018;29:156–166. doi: 10.1089/humc.2018.133. [DOI] [PubMed] [Google Scholar]
- 31.Zheng X.W., Shan C.S., Xu Q.Q., Wang Y., Shi Y.H., Wang Y., Zheng G.Q. Buyang huanwu decoction targets SIRT1/VEGF pathway to promote angiogenesis after cerebral ischemia/reperfusion injury. Front. Neurosci. 2018;12:911. doi: 10.3389/fnins.2018.00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan Z., Chen K., Wu W., Zhou Y., Zhu J., Wu G., Cao L., Zhang X., Guan H., Yang Y., Zhang W., Li J. Overexpression of HOXC10 promotes angiogenesis in human glioma via interaction with PRMT5 and upregulation of VEGFA expression. Theranostics. 2018;8:5143–5158. doi: 10.7150/thno.27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu S., Tang L., Li X., Fan F., Liu Z. Immunotherapy for glioma: current management and future application. Cancer Lett. 2020;476:1–12. doi: 10.1016/j.canlet.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Lu T.X., Rothenberg M.E. MicroRNA. J. Allergy Clin. Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michlewski G., Cáceres J.F. Post-transcriptional control of miRNA biogenesis. RNA. 2019;25:1–16. doi: 10.1261/rna.068692.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao C., Shen J., Meng Z.X., He X.F. Sevoflurane inhibits glioma cells proliferation and metastasis through miRNA-124-3p/ROCK1 axis. Pathol. Oncol. Res. 2020;26:947–954. doi: 10.1007/s12253-019-00597-1. [DOI] [PubMed] [Google Scholar]
- 37.Ong A.L.C., Ramasamy T.S. Role of Sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Res. Rev. 2018;43:64–80. doi: 10.1016/j.arr.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Li C., Liu Z., Yang K., Chen X., Zeng Y., Liu J., Li Z., Liu Y. miR-133b inhibits glioma cell proliferation and invasion by targeting SIRT1. Oncotarget. 2016;7:36247–36254. doi: 10.18632/oncotarget.9198. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Wang B., Li B., Si T. Knockdown of circ0082374 inhibits cell viability, migration, invasion and glycolysis in glioma cells by miR-326/SIRT1. Brain Res. 2020;1748 doi: 10.1016/j.brainres.2020.147108. [DOI] [PubMed] [Google Scholar]
- 40.Fang D.Z., Wang W.J., Li F.Y., Liu J., Hui X.B., Liu D., Wang X.D. Circ_0005075 stimulates the proliferation and metastasis of glioma via downregulating SIRT1. Eur. Rev. Med. Pharmacol. Sci. 2020;24:258–266. doi: 10.26355/eurrev_202001_19918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this article. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.