Highlights
-
•
PARP1-dependent PARylation post-translationally modifies and regulates STAT5.
-
•
Catalytic PARP inhibition reduces STAT5 stability.
-
•
PARP1 loss results in reduced STAT5 signaling and activation of downstream targets.
-
•
STAT5-activated cancers are sensitive to PARP inhibition.
-
•
PARP inhibition overcomes TKI-resistance in FLT3-ITD AML.
Key words: STAT5 protein stability, Post-translational modifications, PARYlation, PARP inhibition, TKI-resistant FLT3-ITD AML
Abstract
Signal transducer and activator of transcription 5 (STAT5) signaling plays a pathogenic role in both hematologic malignancies and solid tumors. In acute myeloid leukemia (AML), internal tandem duplications of fms-like tyrosine kinase 3 (FLT3-ITD) constitutively activate the FLT3 receptor, producing aberrant STAT5 signaling, driving cell survival and proliferation. Understanding STAT5 regulation may aid development of new treatment strategies in STAT5-activated cancers including FLT3-ITD AML. Poly ADP-ribose polymerase (PARP1), upregulated in FLT3-ITD AML, is primarily known as a DNA repair factor, but also regulates a diverse range of proteins through PARylation. Analysis of STAT5 protein sequence revealed putative PARylation sites and we demonstrate a novel PARP1 interaction and direct PARylation of STAT5 in FLT3-ITD AML. Moreover, PARP1 depletion and PARylation inhibition decreased STAT5 protein expression and activity via increased degradation, suggesting that PARP1 PARylation of STAT5 at least in part potentiates aberrant signaling by stabilizing STAT5 protein in FLT3-ITD AML. Importantly for translational significance, PARPis are cytotoxic in numerous STAT5-activated cancer cells and are synergistic with tyrosine kinase inhibitors (TKI) in both TKI-sensitive and TKI-resistant FLT3-ITD AML. Therefore, PARPi may have therapeutic benefit in STAT5-activated and therapy-resistant leukemias and solid tumors.
Introduction
Aberrant signaling of signal transducer and activator of transcription 5 (STAT5) drives tumorigenesis in a variety of cancers and STAT5 constitutive activation contributes to tumor survival, growth, metastasis, and resistance to treatment [1]. In lung cancer, STAT5 acts as an oncogene and its overexpression is associated with numerous pathogenic processes including increases in nuclear BCL-xL which is anti-apoptotic and promotes metastasis [2]. In breast cancer, STAT5 acts as an oncogene through numerous mechanisms in highly aggressive triple negative breast cancer (TNBC) subtypes [1] when STAT3 is not activated [3]. However, when STAT3 is activated, STAT5 acts as a tumor suppressor in breast cancer, with loss of STAT5 promoting oncogenic STAT3 signaling [3]. Additionally, STAT5 is an essential transcription factor for normal hematopoietic development and signals for hematopoietic cell proliferation and differentiation. Thus, constitutive STAT5 activation is often a hallmark of hematological malignancies including acute myeloid leukemia (AML), for which STAT5 activation is present in AML cells of approximately 70% of patients [1].
Internal tandem duplications within the juxtamembrane domain of FMS-like tyrosine kinase 3 (FLT3-ITD) occur in the cells of 20–25% of AML patients and represent an unfavorable AML subtype that exhibits constitutive and aberrant signaling through STAT5, promoting leukemia cell growth [4], [5], [6]. The serine/threonine kinase Pim-1 is a downstream STAT5 target that not only has direct anti-apoptotic effects, but also phosphorylates and stabilizes FLT3, creating a positive feedback loop that further promotes aberrant signaling [7]. DNA damage/repair responses and acquired cytogenetic alterations are also implicated in FLT3-ITD AML disease progression [8]. We previously demonstrated that activated STAT5 induces high levels of reactive oxygen species via RAC1, leading to increased DNA double-strand breaks (DSB) [9]. We and others reported that, as a response to this genotoxic stress, FLT3-ITD upregulates activity of repair pathways, including homologous recombination (HR) via increased RAD51 [10] and a highly error-prone alternative form of non-homologous end-joining (Alt-NHEJ) via increased DNA ligase IIIα (Lig3) and poly (ADP-ribose) polymerase 1 (PARP1) [11,12]. Increased DSB induction and error-prone repair in FLT3-ITD cells results in genomic instability, promoting disease progression and drug resistance [8,9,12]. Due to the above discussed unfavorable effects of FLT3-ITD on AML prognosis, there has been a focus on development of FLT3 tyrosine kinase inhibitors (TKI) as an approach to improve treatment outcomes [13], [14], [15]. The US Food and Drug Administration has approved two TKIs, midostaurin and gilteritinib, for the treatment of FLT3-ITD AML, in combination with chemotherapy and for relapsed/refractory disease respectively [16,17]. However, TKI efficacy has been limited by the development of resistance [15,[18], [19], [20], [21]]. Resistance mechanisms include development of secondary point mutations in the FLT3 tyrosine kinase domain, most commonly in the D835 residue, reducing binding of some TKIs and upregulating STAT5 [19], [20], [21], [22], [23], and in the F691 ‘gatekeeper’ residue [[19], [20], [21],24]. Strategies are needed to prevent or overcome onset of resistance.
PARP1 is a key DNA repair factor [25,26], and, as discussed above, is overexpressed in FLT3-ITD AML [11,12], making it a potential treatment target. PARP1 functions by adding poly-ADP-ribosyl (PAR) chains to itself and other proteins to catalyze activities [27]. PARP inhibitors (PARPi) exert cytotoxic effects by inhibiting PARylation and/or by trapping PARP to DNA [28,29]. PARPi are mainly used to treat cancers with inherent HR defects such as BRCA-mutant breast, ovarian, and prostate cancer [30,31], inducing cell death by mechanisms known as synthetic lethality [31]. In FLT3-ITD AML, TKI treatment reduces expression of key HR repair proteins including RAD51, mimicking BRCA-deficiency [10,32] and TKIs have therefore been combined with PARPi to induce synthetic lethality [32]. However, this drug combination has not been examined in TKI-resistant disease and effects beyond DNA repair and synthetic lethal cytotoxicity have not yet been characterized.
PARP1 PARylation activities function in numerous processes in addition to DNA repair, including transcription, cell death, and regulation of protein stability and activity [27,[33], [34], [35], [36]]. PARP1-dependent PARylation was recently shown to regulate STAT3, another key signaling protein in cancers, by inducing STAT3 dephosphorylation and limiting its transcriptional activity [37]. In this study, we identified putative PARylation sites within STAT5 and subsequently investigated how PARP1 influences STAT5 signaling in TKI-sensitive and -resistant FLT3-ITD AML as well as other STAT5-activated cancers. We found that PARP1 binds STAT5, and PARP1 depletion abrogates STAT5 signaling in FLT3-ITD AML. Additionally, catalytic inhibition of PARylation increases STAT5 protein degradation, suggesting that PARylation aids in stabilizing STAT5. Therefore, in addition to inhibiting DNA repair, PARPi may inhibit STAT5 signaling by decreasing STAT5 protein stability and this may be relevant for developing treatment strategies in FLT3-ITD AML and other cancers with constitutively activated STAT5.
Materials and methods
Cell culture and drugs
Parental MOLM-14 cells and MOLM-14 with D835Y and F691L mutations were generous gifts of Dr. Neil Shah, University of California at San Francisco. MDA-MB231 cells were cultured in DMEM (Corning) with 10% fetal bovine serum (FBS, Sigma) and 1% penicillin-streptomycin (P/S, Corning) and Sum159 cells were cultured in Ham's F-12 (Gibco), 5% FBS, 1% P/S and supplemented with 10 mM HEPES, 5 μg/mL insulin, and 1 μg/mL hydrocortisone. All other cell lines were cultured in RPMI-1640 (Corning) with 10% FBS and 1% P/S. For primary patient samples, mononuclear cells (MNCs) were isolated using Histopaque-1077 (Sigma-Aldrich) according to manufacturer's instructions. MNCs were incubated overnight in IMDM (Gibco) supplemented with 50 ng/ml thrombopoietin and FLT3L, 25 ng/ml stem cell factor, 10 ng/ml IL-3, IL-6, GM-CSF and G-CSF (Gemini Bio) and 20% FBS. Primary AML patient samples were obtained with informed consent on a protocol approved by the institutional review board of the University of Maryland School of Medicine (IRB # H25314). Cells were maintained at 37 °C with 5% CO2. The PARP inhibitors, veliparib (Vel, Selleck Chemicals) and talazoparib (Tal, Pfizer), were prepared as 100 mM and 5 mM stock concentrations (respectively) in DMSO and stored at −80 °C. The tyrosine kinase inhibitors quizartinib (Quiz, Enzo Life Sciences) and gilteritinib (Gilt, BioVision) were prepared at 1 mM stock concentrations in DMSO and stored at −80 °C.
Immunoblotting
Cell pellets were prepared and immunoblotted as previously described [38]. Membranes were blocked with either 5% nonfat milk or 4% bovine serum albumin (Sigma, for phosphorylated proteins) in TBST (blocking buffer). Membranes were then transferred to blocking buffer containing primary antibodies and rocked overnight at 4°C. Membranes were washed in TBST and corresponding secondary antibodies (1:10,000) in blocking buffer were applied for 1 h, rocking at room temperature. Blots were again washed in TBST and developed using a Hi/Lo Digital-ECL Western blot Detection kit and Kwik Quant Imager (KindleBio). Band densitometry was quantified using ImageJ software (NIH). For antibodies, see Supplemental Table 1.
Immunofluorescence staining
Cells were harvested, suspended in 100µL PBS with 2% FBS and cytospun onto glass slides using a Shandon Cytospin 4. Cells were then fixed and slides were processed and stained as previously described [38] using primary antibody. Cells were examined for foci and images were acquired using a Nikon fluorescent microscope Eclipse 80i (100 × /1.4 oil, Melville, NY) with a CCD (charge-coupled device) camera and the imaging software NIS Elements (BR 3.00, Nikon). 100 cells per slide were examined for foci formation and considered positive at > 20 foci. For antibodies used, see Supplemental Table 1.
Extrachromasomal alternative nonhomologous end-joining assay
Nuclear extracts of basal cells were prepared as previously described for NHEJ assay [39] for extraction of active protein. For c-NHEJ and Alt-NHEJ respectively, pimEJ5GFP and pimEJ2GFP plasmid was linearized and processed and incubated with extract as previously described [39] to allow for recombination by extract repair proteins. Relative quantity of recombined product was determined by PCR using primers spanning the repair site (c-NHEJ forward 5′-GTGCTGGTTATTGTG, reverse 5′-AACAGCTCCTCGCCC; alt-NHEJ forward 5′-CCAAGCCCGGCGTGG, reverse 5′CGTTTACGTCGCCGT) and normalized against amplification of a distant site (forward 5′-GTGCGCGGAACCCCT, reverse 5′-AGGGAATAAGGGCGA).
Immunoprecipitation
For PAR-STAT5 immunoprecipitation (IP), cell pellets were resuspended in denaturing buffer (1% SDS, 5 mM EDTA, 10 mM β-mercaptoethanol in purified water) and boiled. Supernatant was taken and diluted 1:10 in cold IP buffer (20 mM Tris 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 5 mM NEM, and protease inhibitor in purified water) then mixed on a rotator at 4°C for 10 min. Protein concentration was measured by Nanodrop and samples were diluted in IP buffer to approximately 2 µg/µL. Aliquots of diluted samples were stored at −80°C for use as input controls. Remaining lysate was rotated overnight at 4°C with pulldown antibody. A/G-coupled sepharose beads were added to lysates and rotated for 2hr at 4°C. Beads were washed 5X in IP buffer and protein was eluted using 2.5% acetic acid incubated 15 min at room temperature with periodic agitation. Samples were centrifuged and supernatant acidity was neutralized using 1 M Tris–HCl 8.0. NuPAGE LDS Sample Buffer containing 10% β-mercaptoethanol was added to eluted samples and inputs saved previously. All were boiled for 10 min prior to immunoblotting. For PARP1-STAT5 nondenaturing co-immunoprecipitation (co-IP), denaturing buffer was skipped, cell pellets were resuspended in cold IP buffer only and processed as described. For antibodies, see Supplemental Table 1.
CRISPR-Cas9 knock out
PARP1 sgRNA targeting exon 1 (5′-CGAGUCGAGUACGCCAAGAG) were designed using the BROAD GPP Web portal and purchased from Sigma. Cas9 RNP complexes were generated by incubating 200 pmol guide RNA with 200 pmol tracrRNA (Sigma) with 50 pmol SpCas9 protein (MacroLab at UC Berkeley) and then electroporated into 5 × 105 MOLM14 cells using the Invitrogen Neon electroporation system (R buffer, E2 buffer, 1050 V, 50 ms, 1 pulse). 7 days post-electroporation, cells were plated in 96 well plates at 0.5 cells/well to generate clonal cell lines. Clonal cells were harvested and analyzed by immunoblot for PARP1 protein. All experiments were conducted using 2 independent populations confirmed for PARP1 protein knockout.
Lentiviral transduction
Lentivirus production and cell infection were performed according to the pLKO.1 lentiviral vector protocol recommended by Addgene. Lentiviral PARP1 MISSION shRNA plasmids were purchased from Sigma (NM_001618 TRCN0000007928 and NM_001618 TRCN0000007929) with respective empty vector controls, SHC001 and SHC002. Briefly, the shRNA plasmids and packaging plasmids pCMB-dR8.2 and pCMV-VSVG (Addgene #8455 and #8454) were transfected into HEK293T cells with Lipofectamine®3000 (Thermo Fisher Scientific) in OPTI-MEM (Invitrogen) containing 5% FBS and 200 μM sodium pyruvate. Lentivirus was harvested after 72 h, filtered with a 0.45 μm filter, and stored at −80 °C. Lentivirus 8 μg/mL polybrene was added to target cells and 72 h after infection, knockdown cells were selected using 1–2 μg/mL puromycin.
Protein degradation
Cells were plated in quadruplicate and pretreated with 100 µM cycloheximide (CHX, Sigma) for 1 h prior to addition of veliparib to block protein translation. 30 min following CHX, 2 sets of cells were treated with 10 µM MG-132 (Cell Signaling) to inhibit the proteasome. Following another 30 min, cells were treated with and without 200 µM veliparib such that there were four treatment groups (CHX alone, CHX+Vel, CHX+MG-132, CHX+MG-132+Vel). Cells were harvested every 2 h following final treatment and samples were subsequently processed as described for immunoblotting. Protein half-life was calculated using GraphPad Prism software for nonlinear regression curve fit.
Cytotoxicity assay and determination of synergism
Cells were plated in 96-well plates with drugs at varying concentrations alone or in combination (Vel, Gilt, and Quiz alone for cytotoxicity assays, Vel and/or Gilt or Tal and/or Gilt for synergy assays). Plates were incubated for 72 h and were then terminated with CellTiter96 MTS Reagent (Promega) and incubated for an additional 2–4h for reagent metabolism. Absorbance was measured and used to determine the fraction of cells affected by treatment. Combination indices (if determining synergism) were calculated according to the Chou-Talalay method using CompuSyn software.
Statistical analysis
Unless otherwise stated, all data are presented as mean ±SEM with statistical significance calculated using two-tailed unpaired Student's t-test.
Results
TKI-resistant FLT3-ITD leukemia cells have increased STAT5 activity, DNA damage, and PARP1/Lig3-mediated error-prone DSB repair
STAT5 activity mediates aberrant signaling in FLT3-ITD AML [40] and elevated STAT5 activation has been demonstrated in mouse cells stably expressing FLT3-ITD with additional D835 mutations [23]. We inquired whether key resistance-conferring mutations occurring in human FLT3-ITD AML further altered STAT5 activity and expression of downstream targets. We compared STAT5 expression and activation in TKI-resistant MOLM-14 FLT3-ITD AML cell lines containing one of two common mutations, D835Y or F691L (Supp. 1A-B), to that in parental TKI-sensitive MOLM-14 cells and found significant increases in the level of STAT5 and activated phospho-STAT5 (pSTAT5) in TKI-resistant cells (Fig. 1A–C). The STAT5, downstream targets, Pim-1 and FLT3, were also upregulated in TKI-resistant cells (Fig. 1A, D,E).
Fig. 1.
TKI-resistance mutated FLT3-ITD leukemia cells increase STAT5 signaling and genomic instability. (A) Representative immunoblot of pSTAT5, STAT5, Pim-1, FLT3, Lig3, and PARP1 levels in TKI-resistant D835Y- and F691L-mutated MOLM-14 FLT3-ITD cells and parental MOLM-14 cells. Quantification of STAT5 (B), pSTAT5 (C), Pim-1 (D), and FLT3 (E) levels measured by immunoblot analysis compared between TKI-resistant and parental MOLM-14 cells. Graphs represent mean expression fold change ±SEM above parental MOLM-14 cells (n = 3). (F) Immunofluorescence staining for γH2AX foci was measured as an estimate of endogenous DNA DSBs in TKI-resistant and parental MOLM-14 cells. Graph represents mean foci formation ±SEM compared to parental MOLM-14 cells (n = 3). c-NHEJ (G) and Alt-NHEJ (H) repair capacity as measured by extrachromosomal assay in TKI-resistant MOLM-14 cells compared to parental MOLM-14 cells. Data represented as mean ±SEM of c-NHEJ and Alt-NHEJ capacity normalized to parental MOLM-14 cells (n = 3). Quantification of Lig3 (I) and PARP1 (J) levels measured by immunoblot analysis compared between TKI-resistant and parental MOLM-14 cells. Graphs show mean fold expression change ±SEM above parental MOLM-14 cells (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Because we previously reported that aberrant STAT5 signaling increases DNA damage and error-prone repair in FLT3-ITD AML [9,11,12], we next determined whether TKI-resistant FLT3-ITD cells demonstrated further increases in these activities compared to parental cells. We first measured levels of DSBs using immunofluorescence staining for γH2AX foci in TKI-resistant and -sensitive MOLM-14 cells. Both TKI-resistant cell lines demonstrate significantly increased foci formation, compared with parental cells (Fig. 1F, Supp. 1C).
Error-prone DSB repair can contribute significantly to the genomic instability of cells [41,42]. Our group previously reported increased activity of the highly error-prone DSB repair pathway, Alt-NHEJ but decreased activity of the less error-prone Ku-dependent classical NHEJ (c-NHEJ) pathway in FLT3-ITD AML cell lines and mouse models associated with overexpression of the Alt-NHEJ pathway constituents PARP1 and Lig3 [11,12]. Using an extrachromosomal PCR-based assay, we subsequently measured c-NHEJ and Alt-NHEJ activity in TKI-resistant and -sensitive FLT3-ITD AML and showed that c-NHEJ activity is significantly reduced (Fig. 1G) but Alt-NHEJ activity is significantly (p < 0.0001) increased in TKI-resistant cells compared with parental (Fig. 1H). Furthermore, immunoblot analysis showed that PARP1 and Lig3 expression levels were increased, compared with parental cells (Fig. 1A,I,J) while the c-NHEJ protein, Ku70 remained unchanged (Supp. 1D). These data indicate that TKI-resistant FLT3-ITD leukemia cells have higher levels of DSBs and error-prone repair and are therefore likely to be more susceptible to genomic instability and acquisition of further mutations compared to TKI-sensitive counterparts.
PARP1 binds and PARylates STAT5
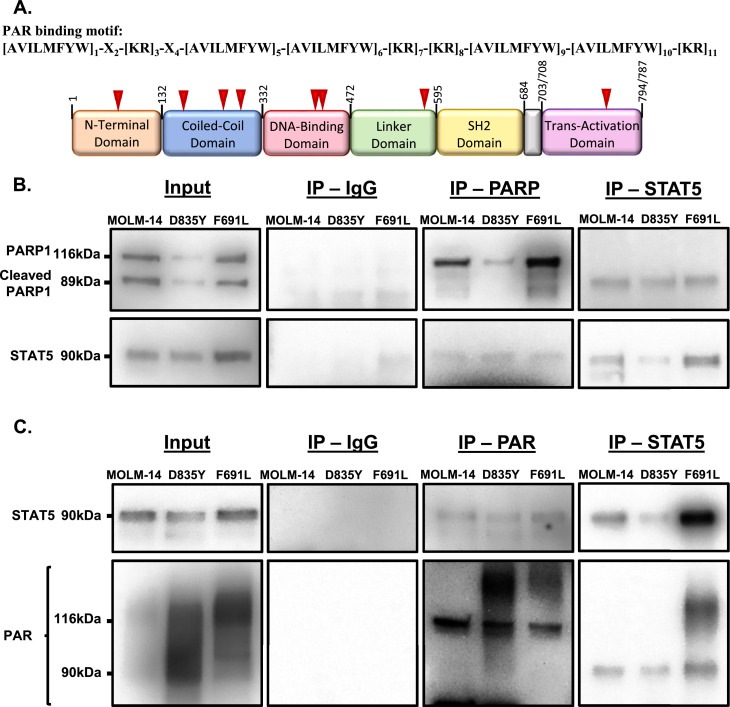
Since PARP1 is increased concordantly with STAT5 in TKI-resistant FLT3-ITD AML, we inquired whether PARP1 may contribute to AML pathogenesis by regulating STAT5 itself due to the involvement of PARylation in numerous functions outside of DNA repair [27,33]. Notably, examination of the STAT5 amino acid sequence and comparison to previously determined PAR-binding motifs [43,44] revealed multiple putative PARylation sites (Fig. 2A). Therefore, we next sought to determine whether PARP1 may interact with STAT5. Using antibodies for either PARP1 or PAR, we immunoprecipitated (IP) STAT5 in TKI-sensitive and -resistant AML cells (Fig. 2B,C) and in parallel, using antibodies to STAT5, we were able to IP both PARP1 and PAR (Fig. 2B,C). These results demonstrate a novel interaction between PARP1 PARylation and STAT5.
Fig. 2.
PARP1 and PAR immunoprecipitate with STAT5. (A) The STAT5 protein sequence was examined for potential PARylation binding motifs [43,44]. STAT5 protein is represented schematically with potential PARylation sites indicated by red arrows. (B) Co-immunoprecipitation of PARP1 and STAT5 or (C) immunoprecipitation of PAR and STAT5 from TKI-resistant and parental MOLM-14 cells. Immunoblot of input proteins shown on the left, with co-IP and IP with indicated antibody-coated beads shown on the right. IgG was used as a negative control for non-specific binding, representative images shown were taken at equal exposures to pulldown samples.
PARP1 is required for STAT5 activity
PARylation of a protein can have a broad range of effects on expression and function [27,33]. We sought to determine whether PARP1-dependent PARylation influences STAT5 expression and/or activation. CRISPR-Cas9 knockout (KO) of PARP1 in parental MOLM-14 cells abolished PARP1 expression and reduced PAR presence in cells, as expected (Fig. 3A). Importantly, PARP1 loss also reduced the levels of STAT5, pSTAT5, and downstream effectors FLT3 and Pim-1 (Fig. 3A–E), suggesting that PARP1 may aid in regulating STAT5 function. Additionally, shRNA knockdown (KD) of PARP1 in TKI-resistant cells (Fig. 3F, Supp. 2) caused similar STAT5 and pSTAT5 expression loss (Fig. 3G,H, Supp. 2).
Fig. 3.
PARP1 loss and loss of PARylation reduce STAT5 expression and signaling. (A) Representative immunoblot of PAR, PARP1, pSTAT5, STAT5, Pim-1, and FLT3 in two PARP1 CRISPR-Cas9 knock out (KO) MOLM-14 cell lines and parental MOLM-14 cells. Quantification of immunoblot analysis for downstream targets STAT5 (B), pSTAT5 (C), Pim-1 (D), and FLT3 (E) in PARP1 KO MOLM-14 cells. Graphs represent average fold expression change ±SEM compared to wild type MOLM-14 cells (n = 2). (F) Quantification of immunoblot analysis for PARP1 shRNA knockdown (PARP1 KD) in TKI-resistant (red or green) and parental (blue) MOLM-14 cells. Graph represents average fold change protein expression ±SEM compared to wild type counterparts in each cell line (n = 3). Quantification of immunoblot analysis for STAT5 (G) and pSTAT5 (H) in PARP1 KD TKI-resistant (red or green) and parental (blue) MOLM-14 cells. Graphs represent average fold change in protein expression ±SEM compared to wild type counterparts of each cell line (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Loss of PARylation destabilizes STAT5 protein
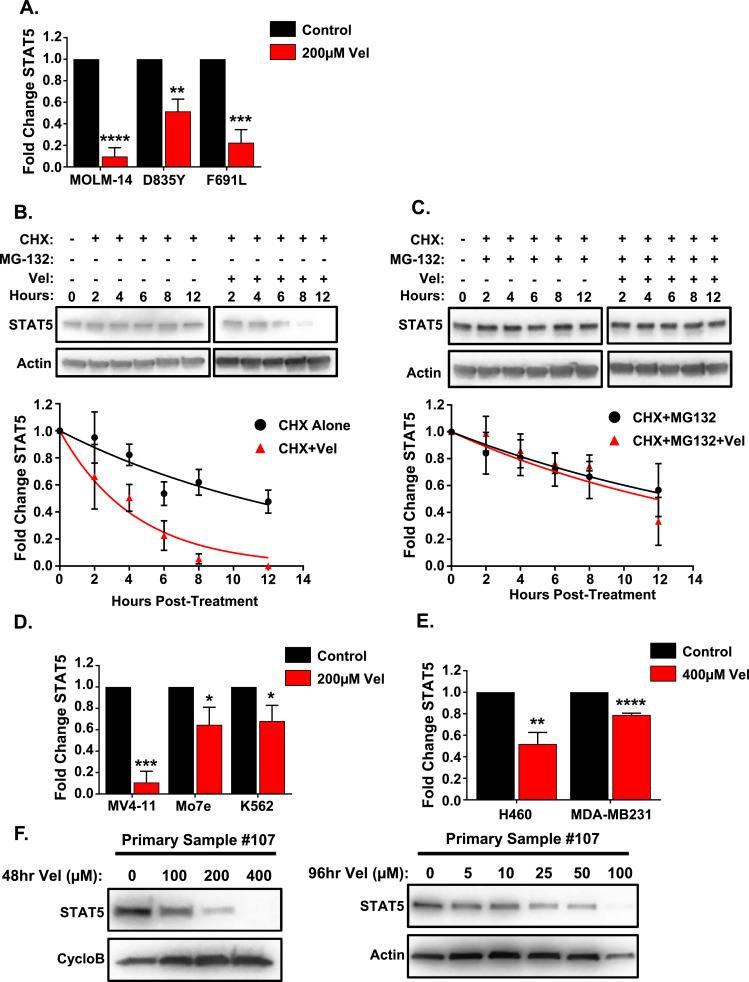
To determine PARylation effects on STAT5 in FLT3-ITD AML, we first measured STAT5 protein levels in TKI-sensitive and -resistant MOLM-14 cells treated with the PARP PARylation inhibitor, veliparib (Vel). Accordingly, Vel (200 µM, 48 hrs) leads to decreased PARylation and reduced STAT5 protein levels (Fig. 4A, Supp. 3A). Next, to determine whether this interaction affects protein stability, cells were pretreated with the protein synthesis inhibitor, cycloheximide (CHX, 100 µM), STAT5 protein half-life is approximately 10.5 h in MOLM-14 cells (9.6 and 10.4 h respectively in D835Y and F691L), but in the presence of Vel (200 µM) following CHX, protein degradation is enhanced, reducing the half-life to 3 h in MOLM-14 cells (3 and 4.1 h, respectively in D835Y and F691L) (Fig. 4B, Supp. 3B). Furthermore, pretreatment with the proteasomal inhibitor, MG-132 (10 µM), following CHX abrogates the degradation effects of CHX with Vel, increasing the STAT5 half-life to approximately 11.9 h in MOLM-14 cells (9.8 and 9.1 h, respectively in D835Y and F691L) and matching the effect of CHX and MG132 without Vel treatment (Fig. 4C, Supp. 3C). This indicates that PARylation loss increases proteasomal degradation of STAT5 in both TKI-sensitive and -resistant FLT3-ITD AML cells.
Fig. 4.
Inhibition of PARylation induces degradation of STAT5. (A) Quantification of immunoblot analysis for STAT5 expression in TKI-resistant and parental MOLM-14 cells following Vel (200 µM) treatment for 48 h. Graph represents average fold expression change ±SEM compared to untreated cells (n = 2). (B) MOLM-14 cells were pretreated with CHX (100 µM, 1hr) only or followed with Vel (200 µM) for the indicated time points and harvested for immunoblot analysis of STAT5. Representative immunoblot shown above. Graph, shown below, represents STAT5 average fold expression change ±SEM as determined by quantification of immunoblots (n = 3) at stated time points and compared to 0hr. Treatment with CHX followed by Vel (red) was compared against CHX alone (black) and nonlinear regression curve fitting was used to determine half-life of STAT5 for each treatment. (C) MOLM-14 cells were pretreated with CHX (100 µM, 1hr) and MG-132 (10 µM, 30 min) and followed with or without Vel (200 µM) for the indicated time points then harvested for immunoblot analysis of STAT5. Representative immunoblot shown above. Graph, shown below, represents STAT5 average fold expression change ±SEM as determined by quantification of immunoblots (n = 3) at stated time points and compared to 0hr. Treatment with CHX and MG-132 followed by Vel (red) was compared against CHX and MG-132 without Vel (black) and nonlinear regression curve fitting was used to determine half-life of STAT5 for each. (D) Quantification of immunoblot analysis for STAT5 in STAT5-activated leukemia cell lines (MV4–11, Mo7e, and K562) following 48 h of Vel (200 µM). Graph represents average fold expression change ±SEM compared to untreated cells (n = 2). (E) Quantification of immunoblot analysis for STAT5 in STAT5-activated lung (H460) and breast (MDA-MB231) cell lines following 48 h of Vel (400 µM). Graph represents average fold expression change ±SEM compared to untreated cells (n = 2). (F) Immunoblot of STAT5 in FLT3-ITD AML primary sample #107 treated with Vel (0–400 µM) for 48 h (left) or treated with Vel (0–100 µM) for 96 h (right). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Dysregulation of STAT5 is present in many cancers in addition to FLT3-ITD AML [1] and as such, STAT5 PARylation may affect these STAT5-activated cancers similarly. Therefore, other leukemia and solid tumor cell lines including additional MV4–11 FLT3-ITD AML cells, Mo7e acute megakaryoblastic leukemia cells, K562 chronic myeloid leukemia cells, H460 non-small cell lung cancer (NSCLC) cells, and MDA-MB231 TNBC cells, were examined and found to have elevated STAT5 and pSTAT5 (Supp. 4A). Similar to MOLM-14, Vel treatment of these cells reduced STAT5 levels however, of note, solid tumors required higher dosing which may indicate reduced reliance on PARylation for STAT5 maintenance in these cells (Fig. 4D,E, Supp. 4B). Additionally, the same Vel treatment in a FLT3-ITD AML primary sample #107 also reduced STAT5 expression and, importantly, lower doses of Vel for 96 h were able to reduce STAT5 expression in primary sample #107 (Fig. 4F). Similar STAT5 reductions in response to Vel across all these cells indicates a new potential mechanism for targeting these STAT5-activated- cancers.
STAT5-activated cancers are sensitive to PARPi
To determine the clinical implications of PARylation loss and subsequent degradation of STAT5 in response to PARPi treatment, we analyzed MOLM-14 cells as well as other cancer cells with elevated STAT5 signaling (discussed previously) for cytotoxic effects of Vel at increasing concentrations (0–400 µM). Compared to the acute monocytic leukemia cell line, THP1, which lacks substantial STAT5 protein (Supp. 4A) and demonstrates a high Vel LC50 (Supp. 5A, Supp. Table 2), TKI-resistant and -sensitive MOLM-14 cells with high STAT5 expression and activation have drastically lower LC50 values (Fig. 5A, Supp. Table 2). Additionally, our FLT3-ITD AML primary sample #107, as well as other leukemia cell lines, MV4–11, Mo7e, and K562, which also have elevated STAT5 (Supp. 4A), similarly demonstrated lower LC50 values for Vel compared to THP1 (Fig. 5B, Supp. Table 2). Furthermore, the NSCLC cell line A549, and the TNBC cell line, Sum159, demonstrate minimal STAT5 protein (Supp. 4A) and, similar to THP1, demonstrate higher Vel LC50 values (Supp. 5B,C, Supp. Table 2) compared to the NSCLC cell line, H460, and the TNBC cell line, MDA-MB231, which have substantial STAT5 protein present and subsequently lower Vel LC50 values (Fig. 5C, Supp. Table 2). Notably, response to Vel appears to be dependent on cancer subtype and basal STAT5 levels. Because STAT5 is critical in hematopoiesis, its dysregulation is highly pathogenic in many leukemias [1,45], thus these cancers are likely to rely more heavily on STAT5 signaling. As such, leukemia cells demonstrated overall increased sensitivity to Vel treatment compared to solid tumor cell lines and, interestingly, the Vel LC50 for these cells was directly correlated to the basal STAT5 expression (Fig. 5D).Furthermore, the FLT3-ITD AML primary sample #107 discussed earlier was also found to fall within this correlation curve (Fig. 5D) indicating STAT5 may be important as a biomarker for translational purposes.
Fig. 5.
PARPi induces cytotoxicity in STAT5-activated cancer. TKI-resistant and parental MOLM-14 cells (A), FLT3-ITD AML primary cells and other STAT5-activated leukemia cells (B), and STAT5-activated solid tumor cells (C), treated with Vel at increasing concentrations (0–400 µM) for 72 h, followed by MTS assay to determine cytotoxicity. Data represents percent of viable cells compared to untreated ±SEM plotted against the log of the Vel concentration (n = 4), primary sample data was taken from a single biological replicate with n = 4 technical replicates. Log curves were determined using GraphPad software and are shown with 95% confidence indicated by dotted lines. Red lines indicate time point at which 50% cytotoxicity (LC50) is achieved. (D) Vel LC50 for leukemia cell lines were plotted against STAT5 expression level determined previously. Linear regression was calculated using GraphPad prism software and association was confirmed with an R square value of 0.9646. Red indicates FLT3-ITD AML primary sample #107.
PARPi synergizes with TKI in TKI-sensitive and -resistant AML cells
While PARPi and TKI combination therapy has been reported for TKI-sensitive FLT3-ITD AML [32], this combination therapy has not been examined in TKI-resistant FLT3-ITD AML. Since we have shown that PARP1 interacts with and stabilizes STAT5 protein, PARPi may lead to destabilization of the signaling cascade that maintains TKI resistance, and resensitize cells to TKI. We therefore explored the efficacy of PARPi Vel in combination with TKIs in TKI-resistant AML using MTS assays of cells treated with the TKI gilteritinib (Gilt, 1–5 nM) and/or Vel (10–50 µM). While Gilt alone modestly reduced survival of TKI-resistant and -sensitive MOLM-14 cells as well as primary cells (Fig. 6A–D), Vel alone had a stronger effect, and the combination of Vel and Gilt further and, importantly, synergistically reduced survival of all cells regardless of TKI sensitivity status (Fig. 6A–D). This synergistic effect was further corroborated using the FDA-approved PARPi talazoparib (Tal, 5–50 nM) combined with Gilt (0.5–5 nM) in both TKI-resistant and -sensitive MOLM-14 cells (Supp. 6A-C). These data suggest that cancers without inherent or inducible defects in DNA DSB repair, such as TKI-resistant FLT3-ITD AML, may still have clinical response to treatment with PARPi due to PARP1 regulation of the transcription factor STAT5 (Fig. 7A).
Fig. 6.
PARPi synergizes with TKI in TKI-resistant and -sensitive leukemia. Parental MOLM-14 cells (A), TKI-resistant D835Y-mutated cells (B), TKI-resistant F691L-mutated cells (C) and FLT3-ITD AML primary cells (D) treated with Gilt (0.5–5 nM) or Vel (5–50 µM) or combinations of both drugs at a constant ratio. Cells were treated for 72 h followed by MTS assay to determine cytotoxicity. (Upper panels) combination indices (CI) calculated according to the Chou-Talalay method using CompuSyn software; x axis is fraction of cells affected (Fa); y axis is CI. Combinations below the black line are synergistic. (Middle and bottom panels) survival of cells treated with Gilt or Vel alone or in combination. Cell line data represented as mean ±SEM for combination compared to single treatment (n = 2). Primary sample data represented as mean ±SEM for combination compared to single treatment for a single biological replicate. Note: Data for primary sample #107 Vel treatment alone was taken from Fig. 5 cytotoxicity assays. *p < 0.05.
Fig. 7.
Proposed mechanism for PARPi use in STAT5-activated cancers. (A) TKI-resistance mutated FLT3-ITD receptor signals strongly through activation of STAT5. STAT5 promotes a state of genomic instability in these cells, leading to leukemia cell survival and future mutations, while also signaling to stabilize FLT3 receptor via activation of Pim-1 kinase [7]. In this proposed model, PARP1 will feed back to PARylate and stabilize STAT5, making these cells susceptible to PARP1 inhibition. Use of an inhibitor of PARylation will subsequently destabilize the STAT5 signaling loop and we theorize this may cause preferential loss of mutated FLT3 receptor, thus allowing for functional TKI use and further synergism with PARPi ultimately leading to cell death. (B) Schematic representation of the broad effects of PARPi in STAT5-regulated cancers. PARP1 PARylates STAT5, leading to stabilization and thus allowing for activation in cancer and downstream oncogenic effects. Use of PARPi in these cells reduces PARylation of STAT5, causing proteasomal degradation of the protein, which decreases pathogenesis, resistance, and induces cell death.
Discussion
Constitutive activation of STAT5 is an important mechanism for aberrant and pathogenic signaling in both hematologic malignancies and solid tumors [1]. In addition to its canonical activation by tyrosine phosphorylation, other post-translational modifications, including acetylation and sumoylation, have been demonstrated to affect STAT5 signaling [46] however, little is known about how STAT5 protein stability is maintained. Here, we show for the first time that PARP1 PARylates STAT5 and is at least in part required for maintaining STAT5 protein function in FLT3-ITD AML and other STAT5-activated cancers. Importantly, we demonstrate that loss of STAT5 PARylation through PARP1 KO, KD, or PARylation inhibition decreases STAT5 expression due to increased proteasomal degradation. This not only decreases STAT5 signaling to downstream targets, but induces cytotoxicity, suggesting that PARP1 could be a therapeutic target in STAT5-activated cancers (Fig. 7B). Indeed, although PARPi doses used as short duration treatments in our studies are relatively high, we also demonstrate efficacy in FLT3-ITD AML primary cells, and we show that lower doses over longer time periods are able to reduce STAT5 in these samples indicating translational capability for this treatment.
PARP1 is involved in many processes beyond DNA repair through attaching PAR residues to target proteins [27,[33], [34], [35], [36]]. While PARP1 has been implicated in the transcriptional regulation of proteins, few studies have examined the role of PARP1 in protein stability. One such study demonstrated that PARP1 aids in maintaining the stability of the regularly spliced tumor suppressor, p53 [35], and PARP1 was also recently shown to be involved in regulating stability and function of HIPK2, a nuclear kinase that functions in DNA damage response [36]. Additionally, PARP1 was found to PARylate the STAT family protein STAT3, and to decrease STAT3-dependent PD-L1 transcription by reducing STAT3 phosphorylation [37]. Our studies build on the effects of PARP1 on STAT proteins by demonstrating that PARP1 regulates STAT5 protein stability, thereby influencing downstream signaling. This provides further proof that PARP1 plays an important role in the cellular homeostasis of proteins and suggests that the stability of other STAT proteins, as well as other transcription factors may be regulated by this mechanism. Furthermore, we demonstrate that c-NHEJ repair is reduced in TKI-resistant cells, yet despite c-NHEJ loss being associated with resistance to PARPi [47], these cells were highly sensitive, further indicating that the inhibition of non-repair related pathways is critical for the application of PARPi.
Based on the classical synthetic lethal approach for using PARPi [31], other groups have successfully used PARPis with TKIs to induce synergistic cytotoxicity in FLT3-ITD AML due to TKI-induced downregulation of DNA repair proteins mimicking a BRCA-deficient phenotype [10,32]. However, we now show that PARP1 plays a role in regulating the primary FLT3-ITD signaling protein, STAT5, which suggests that this novel PARP1 activity should be considered when assessing mechanistic PARPi effects in FLT3-ITD AML and possibly in all STAT5-activated cancers. Furthermore, PARPi effects on STAT5 protein stability is perhaps of greater importance in TKI-resistant AML subtypes. With TKI-resistance, TKIs will minimally downregulate DSB repair proteins through FLT3 receptor signaling and therefore synthetic lethality mechanisms may not be operative with added PARPi treatment. However, we demonstrate that in TKI-resistant FLT3-ITD AML, PARPis decrease STAT5 protein and activity, reducing downstream Pim-1 and FLT3. These changes potentially resensitize cells to TKIs by preferentially destabilizing resistant FLT3 receptor and thus result in synergistic cytotoxicity, however, more work is needed to fully elucidate this mechanism (Fig. 7A). Also, while PARP1 may primarily affect STAT5 protein stability, it is plausible that PARP1 also directly regulates Pim-1 and FLT3 via PARylation. Future investigation into the mechanistic role of PARylation on these proteins is critical in fully understanding PARP1 functions in FLT3-ITD AML.
STAT5 can act as either a tumor suppressor or an oncogene driving disease progression in AML and many other cancers, with its dysregulation most frequently due to constitutive activation through mechanisms including overexpression, increased receptor signaling, and/or loss of negative regulators [1]. Significantly, many of these STAT5-activated cancers, including AML, NSCLC, and TNBC, are among those with the highest mortality rates and have the highest unmet needs therapeutically. Relatedly, although not examined in this work, STAT5 is directly associated with poor prognosis in colorectal cancer [1], and thus we suspect that PARP1 inhibition might be important in treating advanced colorectal cancer, as well as many other STAT5-activated cancers. Our work suggests that examination of STAT5 levels may be an important biomarker for determining sensitivity to PARPi in broad range of cancers without repair deficiencies and that PARP1 PARylation of a larger variety of proteins must be examined for these novel post-translational effects as these may be key for expanding the clinical PARPi use.
Disclosures
This article was prepared while T. Kingsbury was employed at University of Maryland School of Medicine. The opinions expressed in this article are the author's own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
CRediT authorship contribution statement
Anna J. Dellomo: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization, Project administration. Rachel Abbotts: Methodology, Investigation, Writing – review & editing, Visualization. Christian L. Eberly: Methodology, Investigation. Mariusz Karbowski: . Maria R. Baer: . Tami J. Kingsbury: Conceptualization, Methodology, Writing – review & editing, Supervision. Feyruz V. Rassool: Conceptualization, Writing – original draft, Supervision, Project administration, Funding acquisition.
CRediT authorship contribution statement
Anna J. Dellomo: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization, Project administration. Rachel Abbotts: Methodology, Investigation, Writing – review & editing, Visualization. Christian L. Eberly: Methodology, Investigation. Mariusz Karbowski: . Maria R. Baer: . Tami J. Kingsbury: Conceptualization, Methodology, Writing – review & editing, Supervision. Feyruz V. Rassool: Conceptualization, Writing – original draft, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Our studies have been supported by funding from NCIR21 (1R21 CA208937–01A1) (F. V. R., A. J. D.), Adelson Medical Research Foundation (2002469473) (F. V. R.), Van Andel Institute – Stand Up to Cancer (F. V. R.), Maryland Cigarette Restitution funds (CRF) (F. V. R.), UMGCCC P30 CA134274 (F. V. R. and M. R. B.) and the Leukemia Lymphoma Society (LLS) award P-TRP-5885–15 R (F. V. R. and M. R. B.). This research was also supported in part by the Graduate Program in life Sciences at University of Maryland, Baltimore (A. J. D.).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101283.
Appendix. Supplementary materials
References
- 1.Halim C.E., Deng S., Ong M.S., Yap C.T. Involvement of STAT5 in Oncogenesis. Biomedicines. 2020;8(9) doi: 10.3390/biomedicines8090316. Epub 2020/08/28PubMed PMID: 32872372; PubMed Central PMCID: PMC7555335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sánchez-Ceja S.G., Reyes-Maldonado E., Vázquez-Manríquez M.E., López-Luna J.J., Belmont A., Gutiérrez-Castellanos S. Differential expression of STAT5 and Bcl-xL, and high expression of Neu and STAT3 in non-small-cell lung carcinoma. Lung Cancer. 2006;54(2):163–168. doi: 10.1016/j.lungcan.2006.07.012. Epub 2006/09/07PubMed PMID: 16959370. [DOI] [PubMed] [Google Scholar]
- 3.Walker S.R., Nelson E.A., Zou L., Chaudhury M., Signoretti S., Richardson A., et al. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol. Cancer Res. 2009;7(6):966–976. doi: 10.1158/1541-7786.MCR-08-0238. Epub 2009/06/02PubMed PMID: 19491198. [DOI] [PubMed] [Google Scholar]
- 4.Gilliland D.G., Griffin J.D. Role of FLT3 in leukemia. Curr. Opin. Hematol. 2002;9(4):274–281. doi: 10.1097/00062752-200207000-00003. PubMed PMID: 12042700. [DOI] [PubMed] [Google Scholar]
- 5.Gilliland D.G., Griffin J.D. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100(5):1532–1542. doi: 10.1182/blood-2002-02-0492. PubMed PMID: 12176867. [DOI] [PubMed] [Google Scholar]
- 6.Choudhary C., Brandts C., Schwable J., Tickenbrock L., Sargin B., Ueker A., et al. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110(1):370–374. doi: 10.1182/blood-2006-05-024018. Epub 2007/03/13PubMed PMID: 17356133. [DOI] [PubMed] [Google Scholar]
- 7.Natarajan K., Xie Y., Burcu M., Linn D.E., Qiu Y., Baer M.R. Pim-1 kinase phosphorylates and stabilizes 130 kDa FLT3 and promotes aberrant STAT5 signaling in acute myeloid leukemia with FLT3 internal tandem duplication. PLoS ONE. 2013;8(9):e74653. doi: 10.1371/journal.pone.0074653. Epub 2013/09/05PubMed PMID: 24040307; PubMed Central PMCID: PMC3764066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gourdin T.S., Zou Y., Ning Y., Emadi A., Duong V.H., Tidwell M.L., et al. High frequency of rare structural chromosome abnormalities at relapse of cytogenetically normal acute myeloid leukemia with FLT3 internal tandem duplication. Cancer Genet. 2014;207(10–12):467–473. doi: 10.1016/j.cancergen.2014.09.001. Epub 2014/09/16PubMed PMID: 25441683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sallmyr A., Fan J., Datta K., Kim K.T., Grosu D., Shapiro P., et al. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 2008;111(6):3173–3182. doi: 10.1182/blood-2007-05-092510. Epub 2008/01/11PubMed PMID: 18192505. [DOI] [PubMed] [Google Scholar]
- 10.Seedhouse C.H., Hunter H.M., Lloyd-Lewis B., Massip A.M., Pallis M., Carter G.I., et al. DNA repair contributes to the drug-resistant phenotype of primary acute myeloid leukaemia cells with FLT3 internal tandem duplications and is reversed by the FLT3 inhibitor PKC412. Leukemia. 2006;20(12):2130–2136. doi: 10.1038/sj.leu.2404439. Epub 2006/10/26PubMed PMID: 17066094. [DOI] [PubMed] [Google Scholar]
- 11.Muvarak N., Kelley S., Robert C., Baer M.R., Perrotti D., Gambacorti-Passerini C., et al. c-MYC generates repair errors via increased transcription of alternative-NHEJ factors, LIG3 and PARP1, in Tyrosine Kinase Activated Leukemias. Mol. Cancer Res.. 2015;13(4):699–712. doi: 10.1158/1541-7786.MCR-14-0422. Epub 2015/03/31PubMed PMID: 25828893; PubMed Central PMCID: PMC4398615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan J., Li L., Small D., Rassool F. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood. 2010;116(24):5298–5305. doi: 10.1182/blood-2010-03-272591. Epub 2010/08/31PubMed PMID: 20807885; PubMed Central PMCID: PMC3012544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanada M., Matsuo K., Suzuki T., Kiyoi H., Naoe T. Prognostic significance of FLT3 internal tandem duplication and tyrosine kinase domain mutations for acute myeloid leukemia: a meta-analysis. Leukemia. 2005;19(8):1345–1349. doi: 10.1038/sj.leu.2403838. PubMed PMID: 15959528. [DOI] [PubMed] [Google Scholar]
- 14.Smith C.C., Wang Q., Chin C.S., Salerno S., Damon L.E., Levis M.J., et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485(7397):260–263. doi: 10.1038/nature11016. Epub 2012/04/15PubMed PMID: 22504184; PubMed Central PMCID: PMC3390926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daver N., Schlenk R.F., Russell N.H., Levis M.J. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33(2):299–312. doi: 10.1038/s41375-018-0357-9. Epub 2019/01/16PubMed PMID: 30651634; PubMed Central PMCID: PMC6365380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone R.M., Mandrekar S.J., Sanford B.L., Laumann K., Geyer S., Bloomfield C.D., et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N. Engl. J. Med. 2017;377(5):454–464. doi: 10.1056/NEJMoa1614359. Epub 2017/06/23PubMed PMID: 28644114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perl A.E., Altman J.K., Cortes J., Smith C., Litzow M., Baer M.R., et al. Selective inhibition of FLT3 by gilteritinib in relapsed or refractory acute myeloid leukaemia: a multicentre, first-in-human, open-label, phase 1-2 study. Lancet Oncol. 2017;18(8):1061–1075. doi: 10.1016/S1470-2045(17)30416-3. Epub 2017/06/20PubMed PMID: 28645776PubMed Central PMCID: PMC5572576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piloto O., Wright M., Brown P., Kim K.T., Levis M., Small D. Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood. 2007;109(4):1643–1652. doi: 10.1182/blood-2006-05-023804. Epub 2006/10/17PubMed PMID: 17047150; PubMed Central PMCID: PMC1794049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daver N., Cortes J., Ravandi F., Patel K.P., Burger J.A., Konopleva M., et al. Secondary mutations as mediators of resistance to targeted therapy in leukemia. Blood. 2015;125(21):3236–3245. doi: 10.1182/blood-2014-10-605808. Epub 2015/03/20PubMed PMID: 25795921; PubMed Central PMCID: PMC4440880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith C.C., Lasater E.A., Zhu X., Lin K.C., Stewart W.K., Damon L.E., et al. Activity of ponatinib against clinically-relevant AC220-resistant kinase domain mutants of FLT3-ITD. Blood. 2013;121(16):3165–3171. doi: 10.1182/blood-2012-07-442871. Epub 2013/02/21PubMed PMID: 23430109; PubMed Central PMCID: PMC3630831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith C.C., Zhang C., Lin K.C., Lasater E.A., Zhang Y., Massi E., et al. Characterizing and Overriding the Structural Mechanism of the Quizartinib-Resistant FLT3 "Gatekeeper" F691L Mutation with PLX3397. Cancer Discov. 2015;5(6):668–679. doi: 10.1158/2159-8290.CD-15-0060. Epub 2015/04/06PubMed PMID: 25847190; PubMed Central PMCID: PMC4522415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Bubnoff N., Engh R.A., Aberg E., Sänger J., Peschel C., Duyster J. FMS-like tyrosine kinase 3-internal tandem duplication tyrosine kinase inhibitors display a nonoverlapping profile of resistance mutations in vitro. Cancer Res. 2009;69(7):3032–3041. doi: 10.1158/0008-5472.CAN-08-2923. Epub 2009/03/24PubMed PMID: 19318574. [DOI] [PubMed] [Google Scholar]
- 23.Bagrintseva K., Geisenhof S., Kern R., Eichenlaub S., Reindl C., Ellwart J.W., et al. FLT3-ITD-TKD dual mutants associated with AML confer resistance to FLT3 PTK inhibitors and cytotoxic agents by overexpression of Bcl-x(L) Blood. 2005;105(9):3679–3685. doi: 10.1182/blood-2004-06-2459. Epub 20041230PubMed PMID: 15626738. [DOI] [PubMed] [Google Scholar]
- 24.Lee L.Y., Hernandez D., Rajkhowa T., Smith S.C., Raman J.R., Nguyen B., et al. Preclinical studies of gilteritinib, a next-generation FLT3 inhibitor. Blood. 2017;129(2):257–260. doi: 10.1182/blood-2016-10-745133. Epub 2016/12/01PubMed PMID: 27908881; PubMed Central PMCID: PMC5234222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Vos M., Schreiber V., Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem. Pharmacol. 2012;84(2):137–146. doi: 10.1016/j.bcp.2012.03.018. Epub 2012/03/31PubMed PMID: 22469522. [DOI] [PubMed] [Google Scholar]
- 26.Audebert M., Salles B., Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279(53):55117–55126. doi: 10.1074/jbc.M404524200. Epub 2004/10/21PubMed PMID: 15498778. [DOI] [PubMed] [Google Scholar]
- 27.Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012;13(7):411–424. doi: 10.1038/nrm3376. Epub 2012/06/20PubMed PMID: 22713970. [DOI] [PubMed] [Google Scholar]
- 28.Murai J., Huang S.Y., Renaud A., Zhang Y., Ji J., Takeda S., et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 2014;13(2):433–443. doi: 10.1158/1535-7163.MCT-13-0803. Epub 2013/12/19PubMed PMID: 24356813; PubMed Central PMCID: PMC3946062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murai J., Huang S.Y., Das B.B., Renaud A., Zhang Y., Doroshow J.H., et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72(21):5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. PubMed PMID: 23118055; PubMed Central PMCID: PMC3528345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtin N. PARP inhibitors for anticancer therapy. Biochem. Soc. Trans. 2014;42(1):82–88. doi: 10.1042/BST20130187. PubMed PMID: 24450632. [DOI] [PubMed] [Google Scholar]
- 31.Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. PubMed PMID: 15829967. [DOI] [PubMed] [Google Scholar]
- 32.Maifrede S., Nieborowska-Skorska M., Sullivan-Reed K., Dasgupta Y., Podszywalow-Bartnicka P., Le B.V., et al. Tyrosine kinase inhibitor-induced defects in DNA repair sensitize FLT3(ITD)-positive leukemia cells to PARP1 inhibitors. Blood. 2018;132(1):67–77. doi: 10.1182/blood-2018-02-834895. Epub 2018/05/21PubMed PMID: 29784639; PubMed Central PMCID: PMC6034642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morales J., Li L., Fattah F.J., Dong Y., Bey E.A., Patel M., et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit. Rev. Eukaryot. Gene Expr. 2014;24(1):15–28. doi: 10.1615/critreveukaryotgeneexpr.2013006875. PubMed PMID: 24579667; PubMed Central PMCID: PMC4806654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiber V., Dantzer F., Ame J.C., de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7(7):517–528. doi: 10.1038/nrm1963. PubMed PMID: 16829982. [DOI] [PubMed] [Google Scholar]
- 35.Wesierska-Gadek J., Wang Z.Q., Schmid G. Reduced stability of regularly spliced but not alternatively spliced p53 protein in PARP-deficient mouse fibroblasts. Cancer Res. 1999;59(1):28–34. PubMed PMID: 9892179. [PubMed] [Google Scholar]
- 36.Choi J.R., Shin K.S., Choi C.Y., Kang S.J. PARP1 regulates the protein stability and proapoptotic function of HIPK2. Cell Death Dis. 2016;7(10):e2438. doi: 10.1038/cddis.2016.345. Epub 2016/10/27PubMed PMID: 27787517; PubMed Central PMCID: PMC5134000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding L., Chen X., Xu X., Qian Y., Liang G., Yao F., et al. PARP1 suppresses the transcription of PD-L1 by Poly(ADP-Ribosyl)ating STAT3. Cancer Immunol. Res. 2019;7(1):136–149. doi: 10.1158/2326-6066.CIR-18-0071. Epub 2018/11/06PubMed PMID: 30401677. [DOI] [PubMed] [Google Scholar]
- 38.McLaughlin L.J., Stojanovic L., Kogan A.A., Rutherford J.L., Choi E.Y., Yen R.C., et al. Pharmacologic induction of innate immune signaling directly drives homologous recombination deficiency. Proc. Natl. Acad. Sci. U. S. A. 2020;117(30):17785–17795. doi: 10.1073/pnas.2003499117. Epub 2020/07/10PubMed PMID: 32651270; PubMed Central PMCID: PMC7395437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abbotts R., Topper M.J., Biondi C., Fontaine D., Goswami R., Stojanovic L., et al. DNA methyltransferase inhibitors induce a BRCAness phenotype that sensitizes NSCLC to PARP inhibitor and ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 2019 doi: 10.1073/pnas.1903765116. Epub 2019/10/07PubMed PMID: 31591209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grafone T., Palmisano M., Nicci C., Storti S. An overview on the role of FLT3-tyrosine kinase receptor in acute myeloid leukemia: biology and treatment. Oncol. Rev. 2012;6(1):e8. doi: 10.4081/oncol.2012.e8. Epub 2012/04/17PubMed PMID: 25992210; PubMed Central PMCID: PMC4419636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dellomo A.J., Baer M.R., Rassool F.V. Partnering with PARP inhibitors in acute myeloid leukemia with FLT3-ITD. Cancer Lett. 2019;454:171–178. doi: 10.1016/j.canlet.2019.03.048. Epub 2019/04/04PubMed PMID: 30953707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rassool F.V., Tomkinson A.E. Targeting abnormal DNA double strand break repair in cancer. Cell Mol. Life Sci. 2010;67(21):3699–3710. doi: 10.1007/s00018-010-0493-5. Epub 2010/08/10PubMed PMID: 20697770; PubMed Central PMCID: PMC3014093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pleschke J.M., Kleczkowska H.E., Strohm M., Althaus F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275(52):40974–40980. doi: 10.1074/jbc.M006520200. PubMed PMID: 11016934. [DOI] [PubMed] [Google Scholar]
- 44.Gagné J.P., Isabelle M., Lo K.S., Bourassa S., Hendzel M.J., Dawson V.L., et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36(22):6959–6976. doi: 10.1093/nar/gkn771. Epub 2008/11/03PubMed PMID: 18981049; PubMed Central PMCID: PMC2602769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brachet-Botineau M., Polomski M., Neubauer H.A., Juen L., Hédou D., Viaud-Massuard M.C., et al. Pharmacological Inhibition of Oncogenic STAT3 and STAT5 Signaling in Hematopoietic Cancers. Cancers (Basel). 2020;12(1) doi: 10.3390/cancers12010240. Epub 2020/01/18PubMed PMID: 31963765; PubMed Central PMCID: PMC7016966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krämer O.H., Moriggl R. Acetylation and sumoylation control STAT5 activation antagonistically. JAKSTAT. 2012;1(3):203–207. doi: 10.4161/jkst.21232. PubMed PMID: 24058773; PubMed Central PMCID: PMC3670247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel A.G., Sarkaria J.N., Kaufmann S.H. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108(8):3406–3411. doi: 10.1073/pnas.1013715108. Epub 20110207PubMed PMID: 21300883; PubMed Central PMCID: PMC3044391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.