Abstract
Numerous therapies aimed at driving an effective anti-glioma response have been employed over the last decade; nevertheless, survival outcomes for patients remain dismal. This may be due to the expression of immune-checkpoint ligands such as PD-L1 by glioblastoma (GBM) cells which interact with their respective receptors on tumor-infiltrating effector T cells curtailing the activation of anti-GBM CD8+ T cell-mediated responses. Therefore, a combinatorial regimen to abolish immunosuppression would provide a powerful therapeutic approach against GBM. We developed a peptide ligand (CD200AR-L) that binds an uncharacterized CD200 immune-checkpoint activation receptor (CD200AR). We sought to test the hypothesis that CD200AR-L/CD200AR binding signals via he DAP10&12 pathways through in vitro studies by analyzing transcription, protein, and phosphorylation, and in vivo loss of function studies using inhibitors to select signaling molecules. We report that CD200AR-L/CD200AR binding induces an initial activation of the DAP10&12 pathways followed by a decrease in activity within 30 min, followed by reactivation via a positive feedback loop. Further in vivo studies using DAP10&12KO mice revealed that DAP10, but not DAP12, is required for tumor control. When we combined CD200AR-L with an immune-stimulatory gene therapy, in an intracranial GBM model in vivo, we observed increased median survival, and long-term survivors. These studies are the first to characterize the signaling pathway used by the CD200AR, demonstrating a novel strategy for modulating immune checkpoints for immunotherapy currently being analyzed in a phase I adult trial.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13311-021-01038-1.
Keywords: Immune checkpoints, Immunotherapy, Phase 1, GBM, CD200AR
Introduction
The unprecedented success of cancer immunotherapy has revolutionized clinical management of malignancies that previously had dismal prognoses [1]. At the forefront of immunotherapy are immune-checkpoint inhibitors [1], which have shown unparalleled success in cancer therapy [1, 2]. The 2 most clinically successful immune-checkpoint blockade strategies to date are, i.e., targeting of cytotoxic T-lymphocyte-associated protein 4 (CTLA4) and disruption of the interaction between programmed cell death 1 (PD-1) receptor and its ligand (PD-L1). However, only a small percentage of patients with certain solid tumors respond to checkpoint inhibitors including sensitive tumors such as melanoma [3, 4].
We have focused our research on the CD200 immune checkpoint to develop novel immunotherapy for glioblastoma (GBM). The CD200 protein suppresses an immune response by binding the CD200 inhibitory receptor (CD200R1) [5, 6]. Other CD200 receptors exist that were designated as CD200RLa-d by Wright in 2003 because they are coded for by CD200R-related genes in mice. These receptors will be referred to herein as CD200AR2, -3, -4, and -5 in mice, and the single CD200R-related human gene product designated is hCD200RLa [7].
We derived a peptide ligand (CD200AR-L) that elicits an immune activation response when it binds to CD200ARs on immune cells [8]. CD200AR-L was designed to be administered subcutaneously near draining lymph nodes in combination with tumor antigens. CD200AR-L activates antigen-presenting cells (APCs) resulting in CD80/86 upregulation and increased production of cytokines, leading to an enhanced anti-tumor response [9, 10] and survival benefit in mice-bearing tumors from 2 tumor cell lines [11, 12]. In addition, significantly longer survival times were seen in pet dogs with high-grade glioma when a canine-specific CD200AR-L was administered in combination with autologous tumor lysate (TL) after tumor resection compared with TL alone [13].
Although we demonstrated that targeting the CD200ARs with CD200AR-L in murine and canine high-grade glioma improved survival when used in combination with vaccination strategies, the signaling pathways induced by the CD200AR/CD200AR-L interaction that overpower the suppressive properties of the CD200 protein remain unknown. Paired receptor systems, such as the CD200 checkpoint, are cell-surface proteins that are primarily expressed on immune cells and contain conserved extracellular domains capable of eliciting either inhibitory or stimulatory signals [7, 14, 15]. The inhibitory receptors have long immunoreceptor tyrosine-based inhibitory motifs (ITIM) within the cytoplasmic region, [16] whereas activation receptors usually have short cytoplasmic tails, which associate with adaptor proteins such as DAP12, CD3, CD79, and FcR that provide immunoreceptor tyrosine-based activation motifs (ITAM) as docking sites for downstream signaling [17]. A tyrosine-based signaling motif (YINM), which couples to phosphatidyl-inositol 3-kinase (PI3K) dependent pathways, is present in the adaptor protein DNAX-activating protein of 10 kDa (DAP10), a transmembrane signaling adaptor protein predominantly expressed in CD8-T cells and monocytes [18, 19] involved in initiating cytotoxicity [18, 20] and mediating primary stimulation and costimulatory signals in NK cells.
Previously, we generated CD200AR knockout macrophages and demonstrated that CD200AR-L bound to CD200AR complexes leading to the activation of macrophages and cytokines secretion; however, the signaling pathways that result in immune activation remain unknown [21]. Here, we sought to determine whether the DAP10 and DAP12 signaling pathways play a role in the immune response initiated by CD200AR/CD200AR-L binding. We demonstrate that ligation of CD200AR-L to CD200AR upregulates transcription, and protein expression of DAP10 and DAP12 and downregulates expression of the inhibitory receptor, CD200R1. Furthermore, the effect of CD200AR-L on in vitro tumor cell proliferation and in vivo survival of mice-bearing glioma was lost when DAP10 was knocked down. Our study further demonstrates that the use of the CD200AR-L given concomitantly with gene therapy resulted in an unprecedented survival in our GBM model providing the base for the translatable development of this novel CD200 checkpoint peptide inhibitor for clinical use in glioma patients.
Materials and Methods
Cell Isolation and Stimulation
Cells were isolated from the spleens of wild-type mice, and DAP10KO and DAP12 KO mice were strained into a single cell suspension. CD11b and T cells were isolated using microbeads following the manufacturer’s protocol. Briefly, cells were magnetically labeled with CD11b or CD3 MicroBeads, (Miltenyi Biotec, Auburn, CA, USA) and loaded onto a MACS column (Miltenyi Biotec, Auburn, CA, USA) that was placed in the magnetic field of a MACS separator (Miltenyi Biotec, Auburn, California United States). Isolated cells (1 106) were seeded in 200 µL RPMI (Corning, Corning, NY) in a 48-well plate. Cells were washed 12 h later with 1× PBS and pulsed with 10 M CD200AR-L for 1 h. Unpulsed and LPS-pulsed cells were used as negative and positive controls, respectively.
Animal Studies and Cell Lines
The GL261 orthotopic transplant model was established in 6–8-week-old female C57BL/6 mice (B6, Jackson Laboratories Bar Harbor, Maine, United States); DAP10 and DAP12-deficient mice were a generous gift from Dr. Lewis Lanier (University of California) by inoculating GL261-Luc + (15,000 cells in 1 µL saline) using stereotaxis into the right striatum (coordinates 2.5-mm lateral, 0.5-mm anterior of bregma, 3-mm deep from the cortical surface of the brain) [25]. Mice received i) 65 µg tumor lysate or ii) 65 µg tumor lysate + 50 µg of CD200AR-L, iii) saline-treated group was used as a control. Mice were maintained in a pathogen-free facility according to the guidelines of the University of Minnesota Animal Care and Use Committee. Tumor burden was determined by bioluminescent imaging, using 100 mg/kg d-luciferin potassium salt (BioVision), and quantifying light emitted from tumor tissue using Living Image software (Xenogen) expressed in photons per second per square centimeter per steradian (p/s/cm2/sr). GL261 cells for tumor lysate vaccines were cultured in neural stem cell medium consisting of DMEM/F12 (1:1) with l-glutamine, sodium bicarbonate, penicillin/streptomycin (100 U/ml), B27 and N2 supplements (Life Technologies), and 0.1 mg/mL Normocin (InvivoGen, San Diego, CA). Cultures were maintained in 5% O2 and supplemented with 20 ng/mL EGF and FGF semiweekly (R&D Systems, Minneapolis, MN).
Gene Therapy
All animal experiments were performed in accordance with protocols approved by the University Committee on Use and Care of Animals (UCUCA) at the University of Michigan. Briefly, 20,000 GL261 in 1 μL DMEM were stereotactically implanted in the right striatum of syngeneic female C57BL/6 mice (6–8 weeks; Taconic, Rensselaer, NY) at the coordinates of 0.5 mm anterior and 2.0 mm lateral from the bregma and 3.5 mm ventral from the dura [22, 23]. The adenoviral vectors used in this protocol are first-generation Ad.hCMV.hsFLT3L (AdFlt3L) + Ad.hCMV.TK (Ad-TK). Gene therapy was administered 7 days post-tumor implantation, as described before [11, 24–26]. Briefly, mice received 5 × 108 plaque-forming units (pfu) of Ad-Flt3L and 1.0 × 108 pfu of Ad-TK in 1.5 μL volume in 3 locations at 3.7, 3.5, and 3.3 mm ventral from the dura, followed by ganciclovir (GCV; Biotang Inc., Albuquerque, NM) administration twice a day for 7 days [11, 24, 25].
qPCR Studies
Naïve CD11b from DAP10, DAP12 KO or wildtype were plated at a density of 1 × 106 cells per 0.2 mL per well in 48-well in RPMI medium 1 day before the experiment and then stimulated with 10 µM CD200AR-L and incubated for 1, 3, 5, 7, 15, 30, and 60 min or 6, 12, 18, and 24 h. RNA was isolated using TRIzol (Thermo Fisher, Waltham, MA) following the manufacturer’s instructions. TRIzol (0.5 mL) (Thermo Fisher, Waltham, MA) was added to 106 CD11b or T cells. Cell suspensions were incubated for 2 min at RT then lysed by adding 0.1 mL of chloroform (Fisher Scientific, Hampton, NH), mixing slowly by hand for 15 s, incubating at RT for 3 min, and cell debris was removed by centrifugation at 12,500 × g for 15 min at 4 °C. RNA in the lysate was precipitated by adding 0.25 mL of isopropanol (Fisher Scientific, Hampton, NH), incubating for 24 h, and washing with 0.5 mL of 75% of ethanol (Fisher Scientific, Hampton, NH). The concentration of the dried RNA was measured, and 1 µg of total RNA was used for the cDNA synthesis reaction.
Primers for qPCR were designed using primer-BLAST and based on the following Mus musculus template sequences at the NCBI database: NM_001289726.1 for glyceraldehyde-3-phosphate dehydrogenase (mGAPDH), NM_008798.2 for programmed cell death protein 1 (mPD1), and NM_021325.3 for CD200 receptor 1 molecule (mCD200R1). The primer sets were mGAPDH forward: 5′-gtcaaggccgagaatgggaa-3′, reverse: 5′-ctcgtggttcacacccatca-3′;; mDAP10 forward: 5′-atggaccccccaggctacctc-3′, reverse: 5′-tcagcctctgccaggcatgtt-3′; DAP12 forward: 5′-cctcctgactgtgggaggat-3′, reverse: 5′-ggcgactcagtctcagcaat-3′; mCD200R1 forward: 5′-tgtgtcaagtcccagagaagtt-3′, reverse: 5′-aacttgacccagccacaaag-3′; mP38MAPK forward: 5′-cgcaaggtcactggaggaat-3′, reverse: 5′-aggttgctgggctttaggtc-3′; mERK1/2 forward: 5′-gcgattccgccatgagaatg-3′, reverse: 5′-attggaaggcttcaggtccc-3′; mSLP76 forward: 5′-cacacgcaaaccccagattc-3′, reverse: 5′-cagagcctcctcgttgttgg-3′; mPIk3 forward: 5′-agctcagtacaaccccaagc-3′, reverse: 5′-tcctgggaagtacgggtgta-3′; mcJUN forward: 5′-caagaacgtgaccgacgag-3′, reverse: 5′-gaagttgctgaggttggcgt-3′; mJAK1 forward: 5′-acaaactccggg aagagtgg-3′, reverse: 5′-gtggacaatcagtgggggag-3′; mJAK2 forward: 5′-actccgggaagagtggaaca-3′, and reverse: 5′-ccatccaccagggacacaa-3′. qPCR reactions were performed in 25-µL reaction volume in duplicate, using 5 µL cDNA diluted 1:20, using the PerfeCTa® SYBR® Green SuperMix Kit (Quantabio, Beverly, MA) and 1 µM of each primer. qPCR conditions were 35 cycles and the following cycling conditions: 95 °C, 3 min; 90 °C, 1 min; 58 °C, 1 min; 72 °C, 1 min; and 72 °C, 5 min. Four qPCR reactions were performed independently. Relative expression analysis was performed manually using the ΔΔCq method. The comparative cycle threshold (Cq) values of the samples were normalized to the housekeeping gene, mouse GAPDH, and to the positive control (LPS stimulation).
Cytokine Secretion
A total of 1 × 106 cells were grown in 0. 500 mL in RPMI-1640 in a 48-well plate for 12 h, then pulsed with 10 µM CD200AR-L and incubated for an additional 48 h. LPS (1 µM) and nonpulsed cells were used as controls. Supernatant (50 µl) were collected and analyzed for TNF-alpha levels using cytometry bead array (Biosciences). Data was analyzed using FlowJo v10.
Modeling and Sequence Analyses
All molecular modeling was carried out using Schrödinger Modeling Suite software package [27]. Multiple sequence alignment was first carried out against available protein sequences of mammalian CD200 receptors and CD200 to determine the evolutionary conservation regions. Homology modeling of the CD200AR-L-bound CD200AR complexes were carried out based on the X-ray crystallographic structural template of mouse CD200 in complex with mouse CD200R1 (PDB code: 4BFI) [14] using the mouse CD200AR2 (NP_996258.1), CD200AR3 (NP_001121604.1), and CD200AR4 (NP_997127.1) sequences obtained from the NCBI database. Because the design sequence construct of CD200AR-L was based on the mouse CD200 residue 31–44 located at the observed crystallographic structural interface, CD200AR-L was modeled based on the observed mode of CD200 binding to CD200R1 for all the modeled CD200AR complexes.
Western Analyses
3 × 106 cells were grown in 0. 200 mL in RPMI-1640 in a 48-well plate for 12 h then stimulated with 10 µM CD200AR-L for (0, 1, 3, 5, 7, 10, 15, 30, 60 min). Following stimulation, cells were lysed on ice for 10 min in 200 µl of RIPA buffer (Thermo Fisher Scientific). Protein concentrations were determined using the bicinchoninic acid colorimetric method (Pierce Biotechnology, Waltham, MA). Equal amounts of lysate (20 µg) were loaded protein concentrations were determined using the bicinchoninic acid colorimetric method (Pierce Biotechnology, Waltham, MA). Tumor lysates were diluted in reducing sample buffer (Novex), and 50 µg were loaded per lane on a 4 to 12% SDS-PAGE gel (Nu-Page) and run at 160 V (0.8 V h). Gels were then transferred to nitrocellulose at 7 V (BioRad, Hercules, CA), blocked using 5% nonfat dry milk/0.05 mM Tris-buffered saline with 0.05% Tween 20 for 1 h, incubated overnight at 4 °C with an anti-OX2 antibody, 200 μg/mL (1:1000, Santa Cruz Biotechnology, Dallas, TX) in blocking buffer, washed 3 times in TBS/Tween 20 over 15 min, and incubated with a secondary HRP-conjugated antibody for 1 h at room temperature (1:10,000, Jackson ImmunoResearch, West Grove, PA). Protein bands were visualized by ECL Plus Chemiluminescent Substrate (Pierce Biotechnology, Waltham, MA) and exposed to HyBlot CL Autoradiography film (Denville Scientific, Metuchen, NJ) for different exposure times. Blots were then washed in TBS/Tween 20 over 1 h and reincubated overnight at 4 °C with either anti-DAP10 or anti DAP12 antibodies, 200 μg/mL (1:500, Santa Cruz Biotechnology, Dallas, TX) in blocking buffer. Membranes were subsequently washed 3 times in TBS/Tween 20 and DAP10; DAP12 protein bands were visualized as indicated above. Immunoblots were analyzed quantitatively by carried out densitometry with ImageJ software (Bethesda, MD). Briefly, images were background-subtracted, and strips of the blot corresponding to each band were demarcated and analyzed for each time point/gel lane. Figures were prepared in GraphPad Prism (La Jolla, CA).
Downstream Inhibition
5 × 105 CD11b cells were pulsed with 10 μM CD200AR-L ± (10 μM) P38MAPK inhibitor (SB203580) and SLP76 or 10 nM tofacitinib to target the Jak1/Jak3, 10 μM U0126 to target MEK1/2, or 10 μM SP600125 to target JNK. Inhibition was analyzed by quantitative PCR for transcription rates or cytokine production for loss of function analysis.
Fe-IMAC Sample Preparation and LC–MS/MS Analysis
CD11b cells were pulsed with 10 μM CD200AR-L; cellular extracts were sent to Cell Signaling Technology for PTMScan Fe-IMAC analysis. Extracts were sonicated, centrifuged, reduced with DTT, and alkylated with iodoacetamide. 500 μg total protein for each sample was digested with trypsin, purified over C18 columns, and used for IMAC enrichment using Fe-NTA magnetic beads (CST #20,432) as previously described [28]. LC-MS/MS analysis was performed using an Orbitrap-Fusion Lumos Tribrid Mass Spectrometer as previously described [28, 29] with replicate injections of each sample for the IMAC analysis. Briefly, peptides were separated using a 50 cm × 100 µM PicoFrit capillary column packed with C18 reversed-phase resin and eluted with a 150-min linear gradient of acetonitrile in 0.125% formic acid delivered at 280 nl/min.
MS/MS spectra were evaluated by Cell Signaling Technology using SEQUEST and the GFY-Core platform (Harvard University, Cambridge, MA) [30–32]. Searches were performed against the most recent update of the Uniprot Mus musculus database with a mass accuracy of ± 50 ppm for precursor ions and 0.02 Da for product ions. Results were filtered to a 1% peptide-level FDR with mass accuracy ± 5 ppm on precursor ions and presence of a phosphorylated residue for IMAC enriched samples. Results were further filtered to a 1% protein level false discovery rate. All IMAC quantitative results were generated using Skyline [33] to extract the integrated peak area of the corresponding peptide assignments. Accuracy of quantitative data was ensured by manual review in Skyline or in the ion chromatogram files.
IPA Analysis
Ingenuity IPA (QIAGEN, Version 5292811, 06/02/2020) was used to run core analyses on all proteins from the Fe-IMAC analysis with at least 1 phosphopeptide that changed in abundance with treatment. Direct interactions and experimental or high confidence predicted interactions were included. For DAP10/DAP12 pathways, DAP10 and DAP12 were added to a new pathway and connected to proteins that changed with treatment in the Fe-IMAC analysis using direct or indirect interactions and experimental or high confidence predicted interactions, and direct downstream and experimental evidence only were used to map additional nodes that connect to changed proteins interacting with DAP10 and DAP12 pathways.
Results
CD200AR-L Modeling, Design, and Binding
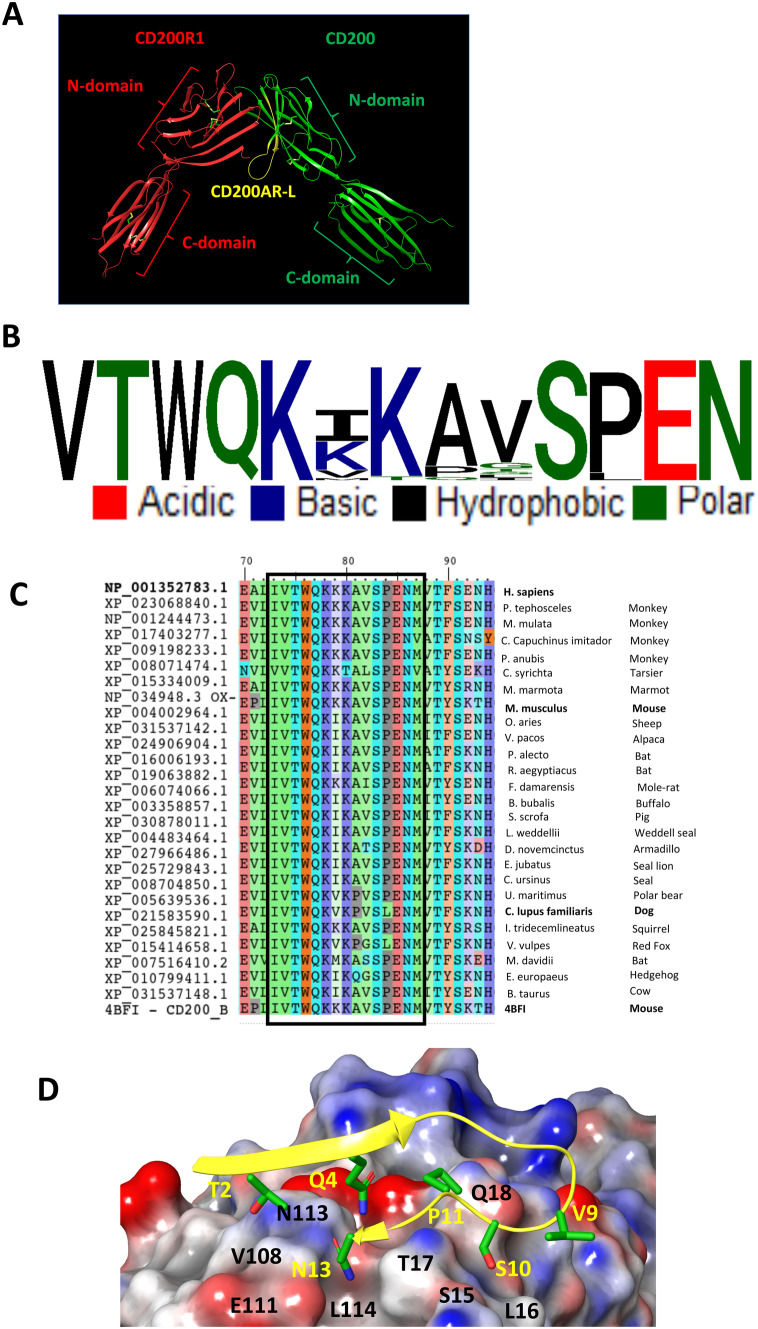
CD200AR-L originates from the region containing residues 32–44 of CD200. It accounts for 42% of the CD200 protein–protein binding site residues with CD200R1 (Fig. 1A) [14]. Multiple sequence analysis among mammalian CD200 sequences showed a highly conserved CD200AR-L sequence motif with only 3 mutable sites (Fig. 1B, C and Supplementary Fig. 1). The CD200AR-L binds to a nonconserved “LTQ” region and a highly conserved “VTPEGNF” region of the CD200R (Supplementary Figs. 2A, B). Among the murine CD200 receptors, the “LTQ” binding region is only shared between CD200R1 and CD200AR4. Homology modeling of CD200AR4 with CD200AR-L showed a highly complementary electrostatic potential surface for binding (Fig. 1C). Thus, at the molecular level, the “VTPEGNF” region is on the GFCC' of the binding site between CD200 and CD200R1 of their N-terminal, suggesting that the interaction evolved from a homotypic interaction.
Fig. 1.
CD200AR-L modeling. A Cartoon representation of the mouse CD200R1 (red) and CD200 (green) crystal structure (PDB: 4BFI) with the highlighted sequence location of CD200AR-L (yellow). Association between CD200 and CD200R1 involves the 2 N-domains which includes CD200AR-L residues 32–44 region that accounts for 42% of the CD200R1 binding site. B Sequence conservation of CD200AR-L across the 26 available mammalian CD200 sequences from NCBI. C Multiple sequence alignment of mammalian CD200 sequences from NCBI. The highly conserved CD200AR-L sequence motif (boxed) is highlighted. D Electrostatic potential surface of the binding site highlighting the electrostatic complementarity between CD200AR-L and CD200R4 residues
CD200AR-L Activates the DAP10/12 Signaling Pathways
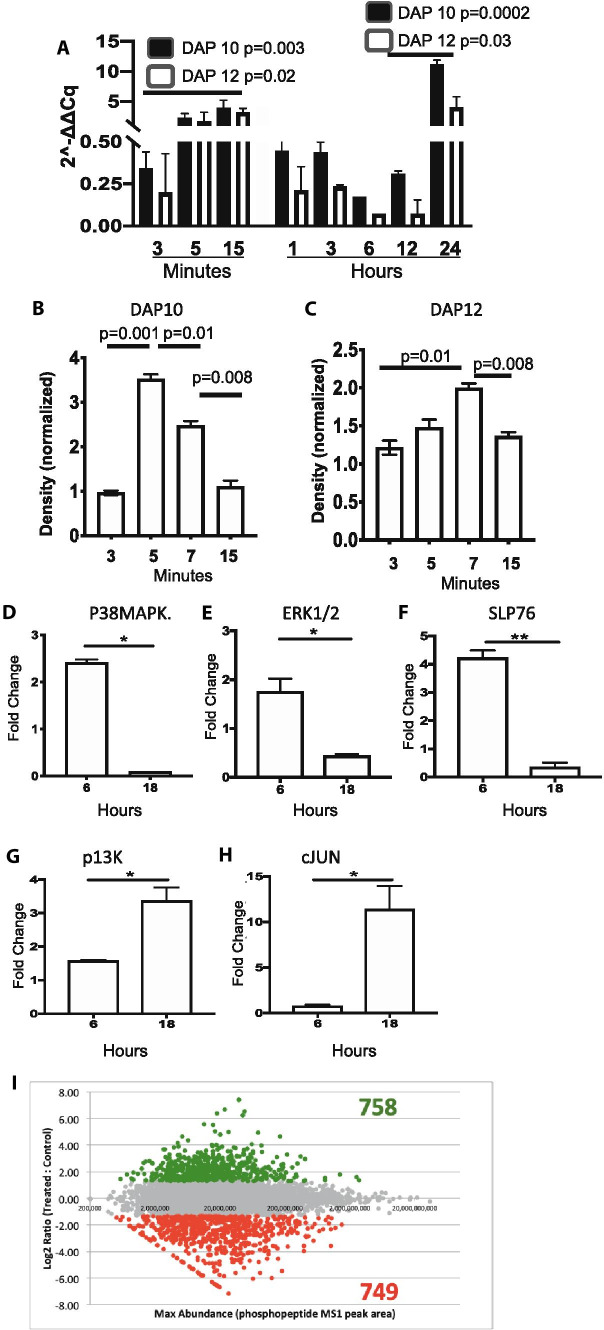
CD11b cells isolated from wild-type mice were pulsed with CD200AR-L at various time points, and subsequent transcription levels of DAP10 and DAP12 were measured (Fig. 2A). We observed a significant increase in transcription of both DAP10 (p = 0.003) and DAP12 (p = 0.02) within 15 min. In addition, we saw a significant increase of both DAP10 (p = 0.002) and DAP12 (p = 0.03) between 12 and 24 h. Transcription analysis was further validated by analyzing protein expression using Western blots density band was measured and normalized to unpulsed controls (Fig. 2B, C). These data demonstrate an increase of the DAP10 and DAP12 protein levels within minutes of CD200AR-L/CD200AR binding. Interestingly, during our Western analysis, we observed DAP 10 and DAP 12 bands at 10 and 20 kD and 12 and 25 kD, respectively; however, the 20 and 25 kD heavy bands were lost using DAP10KO tissues, indicating dimerization of the 2 proteins (Supplementary Figs. 3A, B).
Fig. 2.
CD200AR-L upregulated DAP10 and DAP12 pathways. Preliminary nanostring analysis suggest that the CD200AR-L activated the DAP10 and DAP 12 pathways. To validate these results, CD11b cells isolated from wild-type mice were pulsed with the CD200AR-L, cells were harvested at various times analyzed for A transcription levels and B, C Western analysis for protein levels. In separate experiments, D-H cells were harvested at 6 and 18 h; select signaling molecules were analyzed for transcription levels. I To profile phosphorylation, cells were pulsed for 5 min and analyzed by Fe-IMAC enrichment and LC-MS/MS to derive a Sigmoidal plot of quantitative changes (log2) between CD200AR-L treatment and control. Error bars represent standard deviation, *p < 0.05, **p < 0.005, ***p < 0.0005 by t test
To further investigate involvement of the DAP10/12 pathways, CD11b cells were pulsed with CD200AR-L for 6 and 18 h and analyzed for transcription levels of specific downstream signaling molecules. The transcription levels of P38MAPK, ERK1/2, and SLP76 were significantly decreased (p = 0.02, p = 0.02 and p = 0.003, respectively) at 18 h while significant increases of PI3K (p = 0.02) and cJUN (p = 0.03) were observed (Fig. 2D–H). In a separate experiment, CD11b cells pulsed for 5 min with CD200AR-L were isolated and submitted to Cell Signaling Technology (Danvers, MA) to profile sample phosphorylation using Fe-IMAC enrichment and liquid chromatography-tandem mass spectrometry (Fig. 2I). Results from Fe-IMAC profiling were analyzed by Ingenuity IPA (QIAGEN) to find DAP10 and DAP12 interaction networks (Supplementary Figs. 4A, B). These results are consistent with a model in which immune-activating receptors recruit DAP10 and DAP12 molecules to induce a signaling cascade.
Inhibitors of Signaling Molecules Reduce CD200AR-L Activity
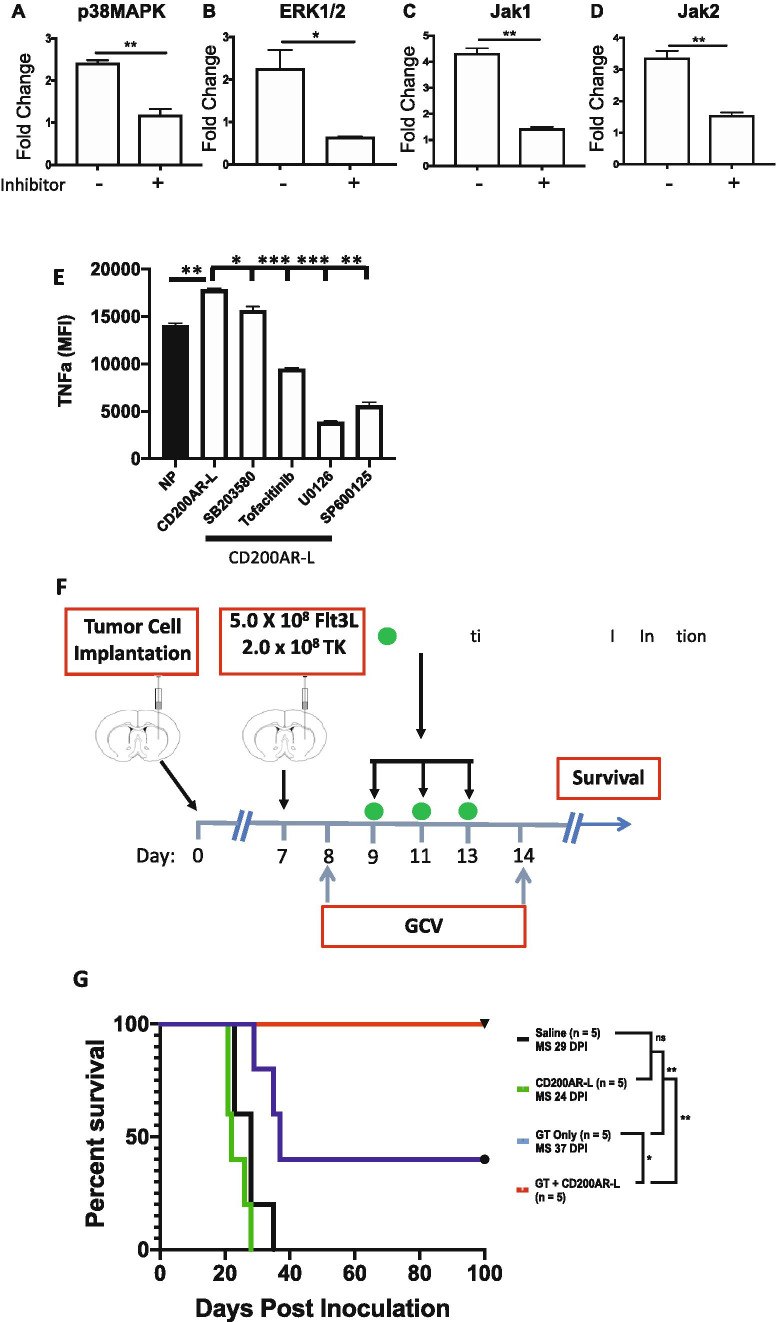
To demonstrate loss of function, CD11b cells were pulsed for 6 h with CD200AR-L with and without inhibitors of select signaling molecules. We observed significant decreases in transcription levels of P38MAPK, ERK1/2, Jak1, and Jak2 (p = 0.008, p = 0.04, p = 0.003, and p = 0.009, respectively) (Fig. 3A–D). In addition, we observed a decrease, albeit not significant, in VAV1 (data not shown). In a separate experiment, we showed that there was a significant increase in TNF-alpha secretion from CD11b cells treated with CD200AR-L compared with unpulsed cells (p = 0.006). However, we observed significant decreases in TNF-alpha production in cells treated with inhibitors of P38MAPK, SB203580 (p = 0.04); of the Jak1/Jak3 pathway, tofacitinib (p = 0.0007); of MEK1/2, U0126 (p = 0.0002); and of JNK, SP600125 (p = 0.001) compared with cells pulsed with CD200AR-L alone (Fig. 3E). These data demonstrated that CD200AR-L can signal through DAP10&12 pathways and induces immune activation on immune cells.
Fig. 3.
Signaling molecule inhibitors reduce the CD200AR-L activity. CD11b cells isolated from wild-type mice were pulsed with the CD200AR-L ± various signaling molecule inhibitor for 6 h and analyzed for transcription levels of A p38MAPK, B ERK1/2, C Jak 1, and D Jak 2. CD11b cells isolated from wild-type mice were pulsed with the CD200AR-L ± various signaling molecule inhibitor and incubated for 48 h. Supernatants were analyzed for E TNF-alpha production by bead array. CD200AR-L in combination with gene therapy (GT) results in increased efficacy. F Timeline of treatment of the combined CD200AR-L + GT survival study. 2 × 104 GL261 cells were implanted at day 0. G Kaplan–Meier GT. All mice (5/5) treated with the combination CD200AR-L + GT exhibits 100% long-term survival with no signs of residual tumor. Error bars represent standard deviation. Survival was analyzed by log-rank; Mantel–Cox test, graphs by t test; *p < 0.05, **p < 0.001, *p < 0.05, **p < 0.005, ***p < 0.0005
CD200AR-L Enhances the Therapeutic Efficacy of Immune-Stimulatory Gene Therapy
Next, to determine the therapeutic impact of blocking the CD200 immune checkpoint on the efficacy of gene-based immunotherapy, we tested the effect of CD200AR-L in combination with TK/Flt3L gene therapy in a murine glioma model [11]. CD200AR-L strongly enhanced the long-term survival of GL261-bearing mice; 100% of the mice were alive at day 60 as compared with 40% of mice (p < 0.01) from the group treated with gene therapy alone (Fig. 3F).
CD200AR-L used as monotherapy did not enhance the survival of GBM-bearing mice (median survival = 29 days for the saline-treated group versus 24 days for CD200AR-L-treated group; ( p > 0.05). Thus, our data demonstrate that the administration of CD200AR-L improves the efficacy of immune-stimulatory gene therapy, inducing a potent anti-GBM response through regulation of the CD200 immune checkpoint. Interestingly, previous murine studies reported the addition of anti-CTLA4 or anti-PD-L1 to gene therapy increased survival ~25% (30 to 55% survival) and 30% (50 to 80%, respectively) [11]. In these studies, we observed a 60% increase in survival from 40 to 100% with the addition of the CD200AR-L (Fig. 3F).
DAP10, But Not DAP12 Pathway Is Required for CD200AR-L Ability to Control Tumor Growth
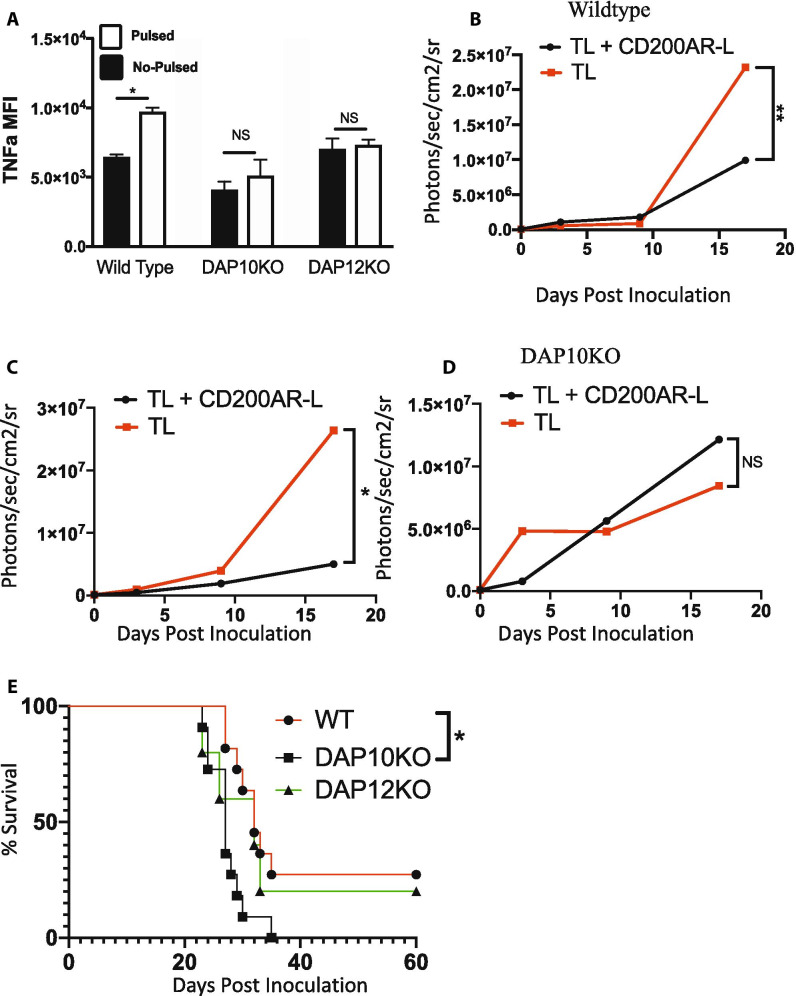
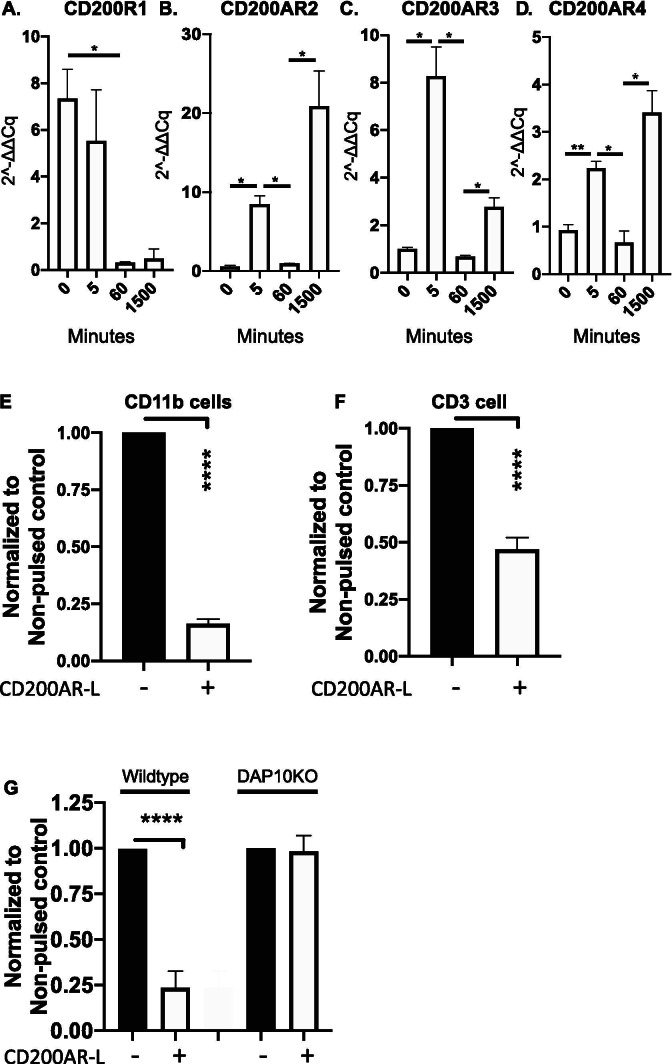
To assess whether both DAP10 and DAP12 are required for CD200AR-L to elicit an anti-glioma response, CD11b cells were isolated from DAP10KO and DAP12KO mice and stimulated with CD200AR-L. CD200AR-L significantly increased TNF-alpha production in wild-type C57B/6 CD11b cells (p = 0.03); however, there were no changes in TNF-alpha production in DAP10KO and DAP12KO CD11b cells (Fig. 4A) demonstrating the CD200AR-L is DAP10&12 dependent for TNF alpha.
Fig. 4.
Knocking out the DAP10 and DAP12 pathways inhibits ability the of CD200AR-L to activate CD11b cells. A CD11b cells isolated from wildtype, DAP 10, and DAP12 mice were pulsed with the CD200AR-L and incubated for 48 h. Supernatants were analyzed for TNF-alpha production by bead array. B Wildtype, C DAP12KO, and D DAP10K0 tumor-bearing mice were vaccinated with tumor lysate or CD200AR-L + tumor lysate and monitored for tumor growth and E survival. Error bars represent standard deviation, *p < 0.05, **p < 0.005, ***p < 0.0005 by t test or 2-way ANOVA
We next sought to determine the roles of DAP10 and DAP12 in vivo. Wildtype, DAP10KO, and DAP12KO mice with GL261 tumors were vaccinated with tumor lysates (TL) with and without CD200AR-L. As previously reported, tumor growth was significantly reduced in wild-type mice vaccinated with TL + CD200AR-L (p < 0.0001) (Fig. 4B) as well as in DAP12KO mice (p = 0.03) (Fig. 4C). In contrast, the anti-glioma response was lost in the DAP10KO mice (Fig. 4D), and survival was significantly decreased compared with wild-type mice receiving TL + CD200AR-L (p = 0.023) (Fig. 4E). We conclude that activation of DAP10 signaling pathways allowed DAP12KO mice to respond to CD200AR-L treatment. These experiments demonstrate that the DAP10 pathway, not DAP12, is required to control tumor growth through the ligation of the activation receptor CD200AR.
CD200AR-L Modulates CD200 Checkpoint Receptor Expression
Next, we investigated whether DAP10 plays a role in the regulation of the CD200 checkpoint receptors. CD11b cells were pulsed with the CD200AR-L at various time points and transcription levels of the inhibitory CD200AR and activation receptors CD200AR2, CD200AR3, and CD200AR4 were measured. We observed a downregulation of the inhibitory CD200R1 within 60 min (p = 0.03), which remained low throughout the experiment (Fig. 5A). In contrast, transcription levels of the activation receptors CD200AR2, CD200AR3, and CD200AR4 were upregulated within 5 min post-CD200AR-L pulsing (p = 0.01, p = 0.02, and p = 0.03, respectively), then downregulated by 30 min (p = 0.02, p = 0.02, and p = 0.007, respectively), followed by upregulation via the positive feedback loop (p = 0.04, p = 0.02, and p = 0.03 respectively) (Fig. 5B–D).
Fig. 5.
CD200AR-L modulates CD200 checkpoint receptor expression. CD11b cells isolated from wild-type mice were pulsed with the CD200AR-L; cells were harvested at various times analyzed for transcription levels for the A inhibitory receptor, CD200R1, and the B activation receptors, CD200AR2, C CD200AR3, and D CD200AR4 expression levels. CD11b cells isolated from wild-type mice were pulsed with the CD200AR-L; cells were E analyzed by flow cytometry for the inhibitory CD200R1 expression. F In a separate experiment, CD11b cells isolated from wildtype mice were pulsed with the CD200AR-L + OVA, washed, and added to T-cells. CD3+CD200R1+ cells were analyzed by flow cytometry. G Wildtype and DAP10KO mice were vaccinated with CD200AR-L for 3 consecutive days over the inguinal lymph nodes. Mice were sacrificed on day 4; lymphocytes isolated from inguinal lymph nodes were analyzed for CD11b+CD200R1+ cells were analyzed by flow cytometry. MFIs were normalized to no pulsed controls. Error bars represent standard deviation, *p < 0.05, **p < 0.005, ***p < 0.0005 by t test
To determine the effect of CD200AR-L on CD200R1 protein levels, CD11b cells pulsed with CD200AR-L were analyzed by flow cytometry, and a dramatic decrease in CD200R1 expression (p < 0.0001) was observed (Fig. 5E). To determine the effect on CD200R1 expression on T cells, CD11b cells were stimulated with ovalbumin ± CD200AR-L and washed, then T cells were added back to the CD11b cells significantly downregulating the inhibitory CD200R1 on T cells (p < 0.0001) (Fig. 5F).
Next, we investigated if CD200AR-L downregulated CD200R1 in vivo. In these experiments, wildtype and DAP10KO mice were vaccinated in the hind leg for drainage into the inguinal lymph nodes for 3 consecutive days. Lymphocytes isolated from the inguinal lymph nodes were analyzed by flow cytometry for CD11b+CD200AR-L+ expression (Fig. 5E, Supplementary Fig. 5). We observed a significant downregulation of the inhibitory CD200R1 (p < 0.0001) in wild-type mice but not in DAP10KO mice, demonstrating that CD200AR-L downregulated the inhibitory receptor in vivo through the DAP10 pathway.
Discussion
In this study, we have uncovered a key signaling pathway used by the CD200 immune checkpoint. Specifically, we establish that this novel peptide ligand (CD200AR-L) activates both DAP10 and DAP12 pathways, but only the DAP10 pathway is required for tumor control. These results represent a paradigm shift for immunotherapy. There have been rigorous studies by several groups providing evidence that targeting the CD200 checkpoint enhances the effects of immunotherapy [34–38]. In the most advanced of these studies, a monoclonal antibody against the CD200 protein (samalizumab) was evaluated in a clinical trial initiated in 2008 for relapsed or refractory B-cell chronic lymphocytic leukemia (B-CLL) and multiple myeloma (NCT00648739) [39]. However, there was a negligible (10%) reduction in bulk disease in 36% of the patients in the study. This poor efficacy may have been caused by CD200 protein that is shed by tumors, including glioma, that promoted an immunosuppressive microenvironment prior to treatment with the monoclonal antibody [6, 40–42].
The use of monoclonal antibodies to block suppressive proteins elucidated by tumors resulted in extended survival in patients with certain tumors; however, this therapy can be associated with severe toxicities and adverse events. We sought to develop a peptide ligand (CD200AR-L) to be delivered in combination with antigen-source tumor lysates (TL) to reduce the suppressive effects of the inhibitory CD200 protein and stimulate an anti-glioma response. Subsequent studies demonstrated that survival was significantly extended when an additional injection of CD200AR-L was given 24 h prior to the CD200AR-L + TL vaccine [9]. The data suggest that the enhanced survival is due to the ability of the CD200AR-L to heighten cytokine and chemokine production [9] and upregulate CD80/86 in antigen-presenting cells and induction of monocyte differentiation to immature dendritic cells [8, 10]. Herein, we report a positive feedback loop 24 h after CD200AR-L/CD200AR ligation (Fig. 2A) that reactivates APCs and upregulates CD200-activating receptors whereas downregulating expression of the inhibitory receptor, CD200R1 (Fig. 5A–C). Therefore, vaccinating with CD200ARL + TL following CD200AR-L/CD200AR ligation allows for a potent antigen-specific anti-tumor response [8].
Numerous studies have shown that the extent of T cell infiltration into the tumor microenvironment (TME) positively correlates with a good prognosis in cancer patients [43–45]. However, there are many ways to limit the body’s ability to stimulate an anti-tumor response and to inhibit induced CD8+ T cells from extravasating into the TME. We reported that CD200 inhibited the ability to mount an anti-tumor response through 3 separate mechanisms: 1) vaccines derived from tumors contain the inhibitory CD200 protein, and thus, vaccination introduces the protein and reduces the ability to mount an effective anti-tumor response; 2) CD200 solubilized from tumors induces immunosuppression in the TME and draining lymph nodes; and 3) CD200 expression is upregulated on the tumor vasculature cells that may inhibit activated-T cells from extravasating into the TME [9, 10]. Thus, downregulation of the inhibitory CD200R1 by CD200AR-L protects the immune cells from all potential suppressive mechanisms of the CD200 protein.
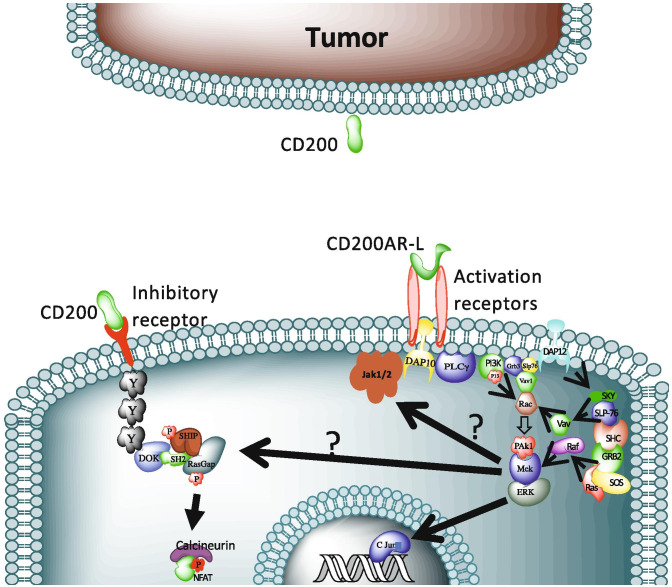
In this study, we report that the activity of CD200AR-L is mediated through the activation of the DAP10 and DAP12 pathways, which is demonstrated by upregulation of the 2 pathways upon CD200AR-L ligation of the activation receptors and loss of APC activation in DAP10 and DAP12 knockout cells. However, in addition to the protein bands for DAP10 and DAP12 in the expected locations of 10 and 12 kD, respectively, we found another band at 25 kD (Supplementary Fig. 3A) that suggests that the 2 proteins form a dimer. This supposition was confirmed in experiments using DAP10KO cells that showed a strong DAP12 band at 12 kD, but no bands at 10 kD or 25 kD compared with wild-type cells (Supplementary Fig. 3B). Although the DAP10 and DAP12 pathways have also been reported to work independently [46], these studies confirm the findings of Rabinovich et al. that suggested the 2 pathways work together by forming dimers, possibly by the creation of cysteine bonds [47]. We suggest that this is also true for the anti-glioma response, as CD200AR-L activation is shown by decreased TNF-alpha production, and the ability of CD200AR-L to control tumor growth was only lost in DAP10KO mice (Fig. 4A). This may be explained by the presence of an intact DAP10 pathway in the DAP12KO mice. These data demonstrate developed an experimental model that CD200AR-L activates both DAP10 and DAP12 pathways for the secretion of cytokines like TNF alpha (Fig. 6), but the effect of CD200AR-L on tumor control is only dependent on the DAP10 pathway.
Fig. 6.
Model of the CD200AR-L signaling pathway. CD200AR-L binds to the CD200AR complex activating the DAP10 pathway and associated signaling molecules (PLCy, PI3K, Grb3, Slp76, Vav1, Rac, Pak1, Mek, ERK, and C Jun) DAP12 pathway and associated signaling molecules (SKY, SLP76, SHC, GRB2, and Ras) and Jak1/2 molecules
Powerful immunosuppressive mechanisms occurring within the GBM TME are mediated by the engagement of immune-checkpoint blockade receptors like PD-1, CTLA-4, LAG-3, and TIM-3, and others. Recent clinical trials have demonstrated remarkable results with the use of inhibitors of immune checkpoints, such as anti-CTLA-4 and anti-PD-1 antibodies, in patients with late-stage melanoma and squamous cell lung cancer. Albeit their efficacy in the clinic, when used to treat GBM as monotherapies were not significant [48]. Our data, combining the administration of CD200AR-L with the TK/Flt3L gene therapy in GBM-bearing mice, showed an impressive 100% long-term survivorship [4]. It has been demonstrated that CD200 signaling has negative effects on dendritic cell function and antigen presentation [5, 49]. Thus, the CD200AR-L was administered to coincide with the peak of the TK/Flt3L-induced dendritic cell migration and antigen presentation. At this point, inhibition of the CD200 pathway is more critical and is possibly why CD200AR-L administration shows such high efficacy when used in combination with TK/Flt3L-mediated gene therapy. To summarize, our data indicate that blocking CD200 immune checkpoint enhances the efficacy of immune-mediated gene therapy in an intracranial mouse GBM model. Additionally, our data also highlight the importance of developing multipronged strategies, because immune-checkpoint blockade as single treatment modality did not elicit therapeutic benefit. Previous data showing maximal survival benefit when PD-L1, CTLA-4, and IDO are blocked simultaneously support this assertion [50].
In conclusion, we are the first to develop a peptide ligand that targets an immune checkpoint to actively stimulate antigen-presenting cells, priming them to take up tumor antigens, whereas downregulating the inhibitory CD200 receptor to protect against an immune suppression by CD200 protein secreted by tumors. We report here that CD200AR-L binds to the CD200AR complex activating the DAP10 and DAP12 pathways to overcome the suppressive properties of the CD200 protein. We previously reported that using tumor lysates in combination with a canine CD200AR-L elicits improved median survival in a spontaneous high-grade glioma clinical trial [9, 10, 13]. In this study, we report that CD200AR-L used in combination with immune-stimulatory gene therapy, which elicits reprograming of the glioma immune microenvironment, leads to 100% long-term survival in an aggressive intracranial glioma mouse model (Fig. 3F, G). This is a new paradigm for cancer treatment; using the CD200AR-L peptide designed to overpower the suppressive CD200 protein whereas simultaneously activating the immune system for an antigen-specific response should be effective in many solid tumors [9].
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file10 (PDF 7200 KB) Supplemental Figure 1. Multiple sequence alignment of CD200 across mammals. Multiple sequence alignment of mammalian CD200 sequences from NCBI. The highly conserved CD200AR-L sequence motif (boxed) is highlighted. .Supplemental Figure 2. Multiple sequence alignment of CD200R across mammals. (A) Multiple sequence alignment of mammalian CD200 sequences from NCBI. The binding site regions for CD200 (solid line boxes) and CD200AR-L (dash line boxes) highlight the non-conserved “LTQ” region and a highly conserved “VTPEGNF” region of the CD200R. The blue and orange bars correspond to the N- and C- domain sequence regions. CD200 binding site boxed in solid line. CD200AR-L boxed in dashed line. (B) Multiple sequence alignment of CD200R1 and CD200AR2-4 highlighting the “LTQ” binding region for CD200AR-L is only shared between CD200R1 and CD200AR4. The blue and orange bars correspond to the N- and C- domain sequence regions. Supplemental Figure 3. CD200AR-L induces DAP10/DAP12 dimers. CD11b cells isolated from spleen of either wild type C57BL/6 or DAP10KO mice, were pulsed with 10 M of CD200AR-L at various time points (1, 3, 5, 7, 15, 30 and 60 minutes). Tissue was then lysed with RIPA buffer and analyzed by western blot for the presence of DAP10 and DAP12. (A) Besides a 10KD (DAP10) and a 12KD (DAP12) bands, a ~25KD band is consistently detected in the wild type CD11b lysate with either anti-DAP10 or anti-DAP12 antibody. (B) DAP10 and DAP12 bands, but not the 25KD dimer band, were detected in the DAP10KO lysate by anti-DAP10 or \ anti-DAP12 antibodies, respectively. Supplemental Figure 4. CD200AR-L activates the DAP10 and DAP12 pathways. Wildtype CD11b cells were pulsed for 5 minutes with the CD200AR-L. (A) IPA analysis was used to construct interaction maps proteins between sites that changed in the Fe-IMAC mass spectrometry analysis DAP10 and DAP12 pathways. (B) heat map of CD11b cells were pulsed for 5 minutes with the CD200AR-L. Cells were subjected to Fe-IMAC enrichment and LC-MS/MS and resulting quantitative data was input into IPA for pathway analysis of activation of DAP10/12 signaling as well as IPA canonical ERK/MAPK, PI3K/AKT and PLC pathways. Red indicates sites with decreased phosphorylation relative to control, green indicates increased, and yellow indicates different sites on one protein that both increased and decreased. Direct and indirect IPA interactions with experimental or high confidence scores were considered in constructing the pathway. Supplemental Figure 5. Flow cytometry gating. lymphocytes isolated from wildtype or DAP10KO mice were pulsed with the CD200AR-L and analyzed for (A) CD45 population, then (B) CD11b population. CD11b cell population from (C&D) wildtype and (D) DAP10KO mice were analyzed for CD200R1 expression. MFIs were normalized to no pulsed controls. Error bars represent standard deviation, *P<0.05, **P<0.005, ***P<0.0005 by t-test.
Funding
This work was financially supported by the Randy Shaver Cancer Research and Community Fund, Hyundai Hope on Wheels, Musella Foundation, Children’s Cancer Research Foundation, Humor to Fight the Tumor, and Shepherd Trust for financial support. This work was partially funded through NIH/NCI (U54CA210190-5) and NIH/NINDS (R21-NS091555; R37-NS094804 and R01-NS074387).
Footnotes
The original online version of this article was revised to update a co-author’s name. Maria Castro will be updated to appear as Maria G. Castro.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Elisabet Ampudia-Mesias and Francisco Puerta-Martinez contributed equally to the work.
Change history
4/30/2021
A Correction to this paper has been published: 10.1007/s13311-021-01062-1
References
- 1.Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A. 2010;107(9):4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dine J, Gordon R, Shames Y, Kasler MK, Barton-Burke M. Immune checkpoint Inhibitors: an innovation in immunotherapy for the treatment and management of patients with cancer. Asia Pac J Oncol Nurs. 2017;4(2):127–135. doi: 10.4103/apjon.apjon_4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with iambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369(2):134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorczynski R, Chen Z, Kai Y, Lee L, Wong S, Marsden PA. CD200 is a ligand for all members of the CD200R family of immunoregulatory molecules. J Immunol. 2004;172(12):7744–7749. doi: 10.4049/jimmunol.172.12.7744. [DOI] [PubMed] [Google Scholar]
- 6.Gorczynski RM. CD200 and its receptors as targets for immunoregulation. Curr Opin Investig Drugs. 2005;6(5):483–488. [PubMed] [Google Scholar]
- 7.Wright GJ, Cherwinski H, Foster-Cuevas M, Brooke G, Puklavec MJ, Bigler M, et al. Characterization of the CD200 receptor family in mice and humans and their interactions with CD200. J Immunol. 2003;171(6):3034–3046. doi: 10.4049/jimmunol.171.6.3034. [DOI] [PubMed] [Google Scholar]
- 8. Z. Xiong et al., Tumor-derived vaccines containing CD200 inhibit immune activation: implications for immunotherapy. Immunotherapy 2016;8:1059-1071 (2016). http://biorxiv.org/cgi/content/short/726778v1. [DOI] [PMC free article] [PubMed]
- 9.Moertel CL, Xia J, LaRue R, Waldron NN, Andersen BM, Prins RM, et al. CD200 in CNS tumor-induced immunosuppression: the role for CD200 pathway blockade in targeted immunotherapy. J Immunother Cancer. 2014;2(1):46. doi: 10.1186/s40425-014-0046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong Z, Ampudia-Mesias E, Shaver R, Horbindki C, Moertel C, Olin M. Tumor-derived vaccines containing CD200 inhibit immune activation: implications for immunotherapy. Immunotherapy. 2016;8(9):1059–1071. doi: 10.2217/imt-2016-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamran N, Kadiyala P, Saxena M, Candolfi M, Li Y, Moreno-Ayala MA, et al. Immunosuppressive myeloid cells' blockade in the glioma microenvironment enhances the efficacy of immune-stimulatory gene therapy. Mol Ther. 2017;25(1):232–248. doi: 10.1016/j.ymthe.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiesner SM, Decker SA, Larson JD, Ericson K, Forster C, Gallardo JL, et al. De novo induction of genetically engineered brain tumors in mice using plasmid DNA. Cancer Res. 2009;69(2):431–439. doi: 10.1158/0008-5472.CAN-08-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olin MR, Ampudia-Mesias E, Pennell CA, Sarver A, Chen CC, Moertel CL, et al. Treatment combining CD200 immune checkpoint inhibitor and tumor-lysate vaccination after surgery for pet dogs with high-grade glioma. Cancers (Basel). 2019;11(2). [DOI] [PMC free article] [PubMed]
- 14.Hatherley D, Lea SM, Johnson S, Barclay AN. Structures of CD200/CD200 receptor family and implications for topology, regulation, and evolution. Structure. 2013;21(5):820–832. doi: 10.1016/j.str.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Borne P, Rygiel TP, Hoogendoorn A, Westerlaken GH, Boon L, Quax PH, et al. The CD200-CD200 receptor inhibitory axis controls arteriogenesis and local T lymphocyte influx. PLoS One. 2014;9(6):e98820. doi: 10.1371/journal.pone.0098820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daeron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- 17.Isakov N. Immunoreceptor tyrosine-based activation motif (ITAM), a unique module linking antigen and Fc receptors to their signaling cascades. J Leukoc Biol. 1997;61(1):6–16. doi: 10.1002/jlb.61.1.6. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Song Y, Bakker AB, Bauer S, Spies T, Lanier LL, et al. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285(5428):730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 19.Chang C, Dietrich J, Harpur AG, Lindquist JA, Haude A, Loke YW, et al. Cutting edge: KAP10, a novel transmembrane adapter protein genetically linked to DAP12 but with unique signaling properties. J Immunol. 1999;163(9):4651–4654. [PubMed] [Google Scholar]
- 20.Billadeau DD, Upshaw JL, Schoon RA, Dick CJ, Leibson PJ. NKG2D-DAP10 triggers human NK cell-mediated killing via a Syk-independent regulatory pathway. Nat Immunol. 2003;4(6):557–564. doi: 10.1038/ni929. [DOI] [PubMed] [Google Scholar]
- 21.Xiong Z, Ampudia-Mesias E, Pluhar GE, Rathe S, Largaespada D, Sham Y, et al. CD200 immune checkpoint reversal at the site of tumor vaccine inoculation: A novel approach to glioblastoma immunotherapy. Clinical Cancer Research. 2019;26(1):232–241. doi: 10.1158/1078-0432.CCR-19-2234. [DOI] [PubMed] [Google Scholar]
- 22.Candolfi M, Yagiz K, Foulad D, Alzadeh GE, Tesarfreund M, Muhammad AK, et al. Release of HMGB1 in response to proapoptotic glioma killing strategies: efficacy and neurotoxicity. Clin Cancer Res. 2009;15(13):4401–4414. doi: 10.1158/1078-0432.CCR-09-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Candolfi M. Temozolomide does not impair gene therapy-mediated antitumor immunity in syngeneic brain tumor models. Clinical cancer research. 2014;20:1555–1565. doi: 10.1158/1078-0432.CCR-13-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali S, Curtin JF, Zirger JM, Xiong W, King GD, Barcia C, et al. Inflammatory and anti-glioma effects of an adenovirus expressing human soluble Fms-like tyrosine kinase 3 ligand (hsFlt3L): treatment with hsFlt3L inhibits intracranial glioma progression. Mol Ther. 2004;10(6):1071–1084. doi: 10.1016/j.ymthe.2004.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtin JF, Liu N, Candolfi M, Xiong W, Assi H, Yagiz K, et al. HMGB1 mediates endogenous TLR2 activation and brain tumor regression. PLoS Med. 2009;6(1):e10. doi: 10.1371/journal.pmed.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Sanderson N, Wawrowsky K, Puntel M, Castro M, Lowenstein P. Kupfer-type immunological synapse characteristics do not predict anti-brain tumor cytolytic T-cell function in vivo. Proc Natl Acad Sci U S A. 2010;107(10):4716–4721. doi: 10.1073/pnas.0911587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrodinger L, New York, NY. Schrodinger Modeling Suite Package. 2018.
- 28.Stokes MP, Farnsworth CL, Gu H, Jia X, Worsfold CR, Yang V, et al. Complementary PTM profiling of drug response in human gastric carcinoma by immunoaffinity and IMAC methods with total proteome analysis. Proteomes. 2015;3(3):160–183. doi: 10.3390/proteomes3030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Possemato A, Paulo J, Mulhern D, Guo A, Gygi S, Beausoleil S. Multiplexed phosphoproteomic profiling using titanium dioxide and immunoaffinity enrichments reveals complementary phosphorylation events. J Proteome Res. 2017;16(4):1506–1514. doi: 10.1021/acs.jproteome.6b00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 31.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104(5):1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kretz-Rommel A, Qin F, Dakappagari N, Ravey EP, McWhirter J, Oltean D, et al. CD200 expression on tumor cells suppresses antitumor immunity: new approaches to cancer immunotherapy. J Immunol. 2007;178(9):5595–5605. doi: 10.4049/jimmunol.178.9.5595. [DOI] [PubMed] [Google Scholar]
- 35.Gorczynski RM, Clark DA, Erin N, Khatri I. Role of CD200 expression in regulation of metastasis of EMT6 tumor cells in mice. Breast Cancer Res Treat. 2011;130(1):49–60. doi: 10.1007/s10549-010-1259-3. [DOI] [PubMed] [Google Scholar]
- 36.Gorczynski RM, Chen Z, Khatri I, Podnos A, Yu K. Cure of metastatic growth of EMT6 tumor cells in mice following manipulation of CD200:CD200R signaling. Breast Cancer Res Treat. 2013;142(2):271–282. doi: 10.1007/s10549-013-2735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Copland DA, Calder CJ, Raveney BJ, Nicholson LB, Phillips J, Cherwinski H, et al. Monoclonal antibody-mediated CD200 receptor signaling suppresses macrophage activation and tissue damage in experimental autoimmune uveoretinitis. Am J Pathol. 2007;171(2):580–588. doi: 10.2353/ajpath.2007.070272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rygiel TP, Karnam G, Goverse G, van der Marel AP, Greuter MJ, van Schaarenburg RA, et al. CD200-CD200R signaling suppresses anti-tumor responses independently of CD200 expression on the tumor. Oncogene. 2012;31(24):2979–2988. doi: 10.1038/onc.2011.477. [DOI] [PubMed] [Google Scholar]
- 39.Adler I. Alexion Reports Results from First Clinical Trial of Novel Anti-Cancer Antibody Samalizumab at ASH Annual Meeting 2010.
- 40.Gorczynski R, Chen Z, Khatri I, Yu K. sCD200 present in mice receiving cardiac and skin allografts causes immunosuppression in vitro and induces Tregs. Transplantation. 2013;95(3):442–447. doi: 10.1097/TP.0b013e3182754c30. [DOI] [PubMed] [Google Scholar]
- 41.Twito T, Chen Z, Khatri I, Wong K, Spaner D, Gorczynski R. Ectodomain shedding of CD200 from the B-CLL cell surface is regulated by ADAM28 expression. Leuk Res. 2013;37(7):816–821. doi: 10.1016/j.leukres.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Wong KK, Brenneman F, Chesney A, Spaner DE, Gorczynski RM. Soluble CD200 is critical to engraft chronic lymphocytic leukemia cells in immunocompromised mice. Cancer Res. 2012;72(19):4931–4943. doi: 10.1158/0008-5472.CAN-12-1390. [DOI] [PubMed] [Google Scholar]
- 43.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29(15):1949–1955. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 44.Li J, Wang J, Chen R, Bai Y, Lu X. The prognostic value of tumor-infiltrating T lymphocytes in ovarian cancer. Oncotarget. 2017;8(9):15621–15631. doi: 10.18632/oncotarget.14919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geng F, Bao X, Dong L, Guo QQ, Guo J, Xie Y, et al. Doxorubicin pretreatment enhances FAPalpha/survivin co-targeting DNA vaccine anti-tumor activity primarily through decreasing peripheral MDSCs in the 4T1 murine breast cancer model. Oncoimmunology. 2020;9(1):1747350. doi: 10.1080/2162402X.2020.1747350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voehringer D, Rosen DB, Lanier LL, Locksley RM. CD200 receptor family members represent novel DAP12-associated activating receptors on basophils and mast cells. J Biol Chem. 2004;279(52):54117–54123. doi: 10.1074/jbc.M406997200. [DOI] [PubMed] [Google Scholar]
- 47.Rabinovich B, Li J, Wolfson M, Lawrence W, Beers C, Chalupny J, et al. NKG2D splice variants: a reexamination of adaptor molecule associations. Immunogenetics. 2006;58(2–3):81–88. doi: 10.1007/s00251-005-0078-x. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, Chu T, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25(3):462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmannova D, Kolackova M, Kondelkova K, Kunes P, Krejsek J, Andrys C. CD200/CD200R paired potent inhibitory molecules regulating immune and inflammatory responses; part I: CD200/CD200R structure, activation, and function. Acta Medica (Hradec Kralove). 2012;55(1):12–17. doi: 10.14712/18059694.2015.68. [DOI] [PubMed] [Google Scholar]
- 50. Wainwright DA, Chang AL, Dey M, Balyasnikova IV, Kim C, Tobias AL, et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4 and PD-L1 in mice with brain tumors. Clin Cancer Res. 2014. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file10 (PDF 7200 KB) Supplemental Figure 1. Multiple sequence alignment of CD200 across mammals. Multiple sequence alignment of mammalian CD200 sequences from NCBI. The highly conserved CD200AR-L sequence motif (boxed) is highlighted. .Supplemental Figure 2. Multiple sequence alignment of CD200R across mammals. (A) Multiple sequence alignment of mammalian CD200 sequences from NCBI. The binding site regions for CD200 (solid line boxes) and CD200AR-L (dash line boxes) highlight the non-conserved “LTQ” region and a highly conserved “VTPEGNF” region of the CD200R. The blue and orange bars correspond to the N- and C- domain sequence regions. CD200 binding site boxed in solid line. CD200AR-L boxed in dashed line. (B) Multiple sequence alignment of CD200R1 and CD200AR2-4 highlighting the “LTQ” binding region for CD200AR-L is only shared between CD200R1 and CD200AR4. The blue and orange bars correspond to the N- and C- domain sequence regions. Supplemental Figure 3. CD200AR-L induces DAP10/DAP12 dimers. CD11b cells isolated from spleen of either wild type C57BL/6 or DAP10KO mice, were pulsed with 10 M of CD200AR-L at various time points (1, 3, 5, 7, 15, 30 and 60 minutes). Tissue was then lysed with RIPA buffer and analyzed by western blot for the presence of DAP10 and DAP12. (A) Besides a 10KD (DAP10) and a 12KD (DAP12) bands, a ~25KD band is consistently detected in the wild type CD11b lysate with either anti-DAP10 or anti-DAP12 antibody. (B) DAP10 and DAP12 bands, but not the 25KD dimer band, were detected in the DAP10KO lysate by anti-DAP10 or \ anti-DAP12 antibodies, respectively. Supplemental Figure 4. CD200AR-L activates the DAP10 and DAP12 pathways. Wildtype CD11b cells were pulsed for 5 minutes with the CD200AR-L. (A) IPA analysis was used to construct interaction maps proteins between sites that changed in the Fe-IMAC mass spectrometry analysis DAP10 and DAP12 pathways. (B) heat map of CD11b cells were pulsed for 5 minutes with the CD200AR-L. Cells were subjected to Fe-IMAC enrichment and LC-MS/MS and resulting quantitative data was input into IPA for pathway analysis of activation of DAP10/12 signaling as well as IPA canonical ERK/MAPK, PI3K/AKT and PLC pathways. Red indicates sites with decreased phosphorylation relative to control, green indicates increased, and yellow indicates different sites on one protein that both increased and decreased. Direct and indirect IPA interactions with experimental or high confidence scores were considered in constructing the pathway. Supplemental Figure 5. Flow cytometry gating. lymphocytes isolated from wildtype or DAP10KO mice were pulsed with the CD200AR-L and analyzed for (A) CD45 population, then (B) CD11b population. CD11b cell population from (C&D) wildtype and (D) DAP10KO mice were analyzed for CD200R1 expression. MFIs were normalized to no pulsed controls. Error bars represent standard deviation, *P<0.05, **P<0.005, ***P<0.0005 by t-test.