Abstract
The majority of the non-protein-coding RNAs are being identified with diversified functions that participate in cellular homeostasis. The circular RNAs (circRNAs) are emerging as noncoding transcripts with a key role in the initiation and development of many physiological and pathological conditions. The advancements in high-throughput RNA sequencing and bioinformatics tools help us to identify several circRNA regulatory pathways, one of which is microRNA (miRNA)-mediated regulation. Besides the direct influence over mRNA transcription, the circRNA can also control the target's expression via sponging miRNAs or the RNA-binding proteins. Studies have demonstrated the dysregulation of the circRNA-miRNA-mRNA interaction network in the pathogenesis of many diseases, including diabetes. This intricate mechanism is associated with the pathogenesis of diabetes and its complications. This review will focus on the circRNA-miRNA-mRNA interaction network that influences the gene expression in the progression of diabetes and its associated complications.
Keywords: circRNA-miRNA-mRNA, diabetes, diabetic complications
Graphical abstract
Circular RNAs (circRNAs) are closed long noncoding RNAs, and have received increasing attention in disease biology. This review highlights the circRNA-miRNA-mRNA regulatory axis in diabetes and its associated disorders. More than 20,000 novel circRNAs have been identified to date, although only a few are identified with their regulatory pathways or interaction networks.
Introduction
Diabetes mellitus (DM) is a rapidly growing epidemic and a leading metabolic and endocrine disorder caused by chronic hyperglycemia resulting from insulin resistance.1 It is estimated that 463 million people have diabetes, and this number is expected to reach 578 million by 2030 and 700 million by 2045.2 Insulin resistance and endothelial dysfunction are the two major factors that impede other vascular complications in patients with diabetes.3 Imbalanced secretion of endothelium-derived growth factors results in end-organ damage.4 Other than insulin resistance, factors like the release of free fatty acids and lipid toxicity, oxidative stress, and dyslipidemia also contribute to the impairment of endothelium.5 In diabetic patients, vascular complications such as nephropathy, retinopathy, and cardiomyopathy presents serious manifestations with poor life expectancy.6 The body is exposed to agents that produce reactive oxygen species (ROS) by transferring free unpaired electrons, causing cellular component oxidation. As a defense, the body obtains exogenous antioxidants from the diet that can neutralize these species. The imbalance between ROS and antioxidants leads to oxidative stress, creating pathological conditions like diabetes.7 Studies have also identified the play of epigenetic regulators in the pathogenesis of diabetes.
In recent times, the advent of new sequencing technologies has helped understand the transcription of human genome into RNAs, among which only 1–2% were found to code for a protein, and the rest do not.8 These noncoding RNA (ncRNA) transcripts that do not code for a protein were referred to as “junk” for a long time and later were described as a highly conserved functional molecule that regulates the gene expression in various manners.9 Based on the size of ncRNAs, they have been broadly classified as small noncoding RNAs (sncRNA) and long noncoding RNAs (lncRNAs). The transcripts such as microRNA (miRNA), small interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNAs) are below 200 nucleotides (nts) and are referred to as sncRNAs, whereas the lncRNAs above 200 nts include promoter-associated transcripts (PATs), enhancer RNAs (eRNAs), and circular RNAs (circRNAs).10 Recently, the circular lncRNAs or the circRNAs have gained focus among researchers and are extensively studied for their regulatory role in cellular signaling.11 Although circRNAs were unnoticed in the previous decades, advances in genomic sequencing, transcriptional profiling, computational tools, and structural biology have made us understand their importance in the pathogenesis of various diseases, including diabetes. Researchers have identified the functionality of circRNA to sponge its endogenous pair, the miRNA, and thereby play an important role in the progression of diabetes and its complications. For example, Zhou et al. identified a novel circRNA, circRNA 010567, that targets and increases the expression of TGF-β1 by sponging miR-141, thereby promoting myocardial fibrosis.12 In this way, this review article will summarize the findings of the circRNA-miRNA-mRNA interaction network in diabetes and its associated complications.
Regulatory functions of circRNA and miRNA
Biogenesis of circRNA
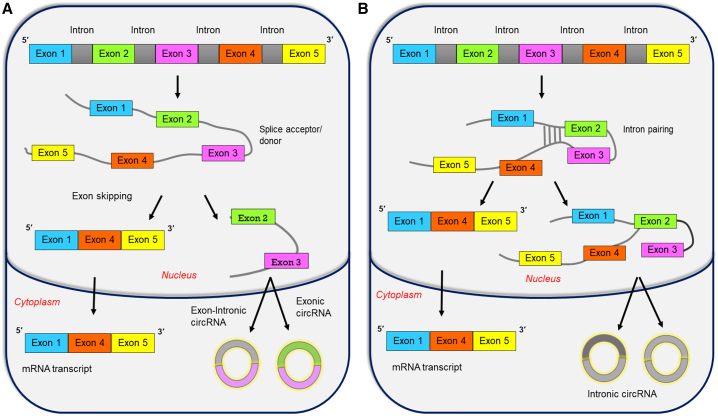
In the mid-1970s, circRNAs were discovered as viroids in RNA viruses and assumed to be an error in processing the RNA splicing mechanism. circRNAs were reported by Hsu et al. in the cytoplasm of mammalian cells using electron microscopy.13 The circRNAs are single-stranded RNA molecules that differ from their linear RNA form by forming a continuous loop covalently joined between their 5′ and 3′ ends.14 The circRNAs are formed by a back-splicing mechanism wherein the 5′ terminus of the upstream pre-mRNA is non-collinearly spliced to the downstream exons of the 3′ terminus.15
Jeck et al. (2010) have proposed two models for exon circularization: the lariat-driven circularization models and the intron pairing-driven model.16 In lariat-driven circularization (Figure 1A), two transcripts are produced due to the splicing of pre-mRNA.17 These two transcripts are mRNA that lacks skipped exons and lariat containing those skipped exons, thus creating a chance for circularization. Splicing of exon lariat results in the formation of circRNA and an intron lariat. These introns can form lasso shaped structures, but they tend to degrade due to branching enzymes.18 In intron pairing-driven circularization (Figure 1B), exons of the circRNA are complementary to introns flanking them for binding to each other. Splice sites with close proximity come close to each other, increasing the possibility of back splicing.
Figure 1.
Biogenesis of circRNA
(A) In lariat-driven circularization, the formation of exonic circRNA is through exon skipping guided by a splice acceptor/donor. (B) In intron pairing-driven circularization, the formation of intronic or exon-intronic circRNA is by the hybridization of flanking introns with close proximity and is independent of exon skipping and brings the splice site.
The circRNA contains up to five exons, and the introns bordering them are three times longer than their linear form. Research has revealed many complementary Alu repeats in the intronic region, which accelerates the splice site to locate each other and promote circularization easily.16 Due to their closed-loop structure, they lack 5′ and 3′ regions containing a poly-A tail and cap region, making them resistant to exonucleases; e.g., ribonuclease R (RNase R) mediated cleavage.19 Eunuka et al. reported that, due to this, circRNA lasts 2.5 times longer compared with their linear RNA form.20 RNase R degradation can accurately select the closed-loop nature of circRNA over their linear forms as they can degrade linear RNA and its poly-A tail. circRNAs are resistant to RNA degradation as they lack a 5′ cap and 3′ tail compared with their linear forms.21
The formation of circRNA is facilitated by reverse binding Alu elements to RNA helicases, DExH-Box Helicase 9 (DHX9) and harboring inverted repeats of long introns, which flanks the genomic structure of long exons.22 In the normal growing cells, the formation of circRNA by back-splicing event occurs due to NF90/NF110 binding to A/U-rich elements in the intronic region.23 Back splicing for circRNA is also promoted by heterogeneous nuclear ribonucleoprotein L (HNRNPL),24 muscle bind (MBL),25 and RNA-binding protein quaking I (QKI).26 It has been reported that MBL upgrades circMBL back splicing by binding to pre-mRNA and decreasing the levels of MBL, whereas QKl brings two cyclic sites closer by binding to both flanking ends of introns and combining the cyclic exons.25
Production of circRNA can also be regulated by a RNA-binding protein, fused in sarcoma (FUS),27 to the intron flanking region in back-splicing junctions, which can further be controlled by heterogeneous nuclear ribonucleoprotein (hnRNP) and serine-arginine (SR) proteins.28 RNA-editing enzymes such as adenosine deaminases acting on RNA (ADAR) can exterminate double-strand chain interaction and bind to flanking intron's double-stranded areas to inhibit circRNA formation.29 Recent studies have indicated increased circRNA synthesis by inhibiting the pre-mRNA processing mechanisms such as spliceosomes by extension of read-through of the downstream genes.30
Based on the sequence and domains of circRNA, they have been categorized as follows: (1) intronic circRNA (ciRNA), which are located in the nucleus and are joined by a 2′–5′ phosphodiester bond. This class of circRNAs features the enrichment of 3′ branch site containing an 11 C motif and a 5′ splice site containing 7 GU motif.15 (2) Exonic circRNAs (ecRNAs), which are mostly identified as an miRNA sponge, and are located in the cytoplasm. Besides being involving in gene transcription through sponging miRNAs, they also interact with RNA-binding proteins and participate in protein translation. This type of circRNA is formed by exon skipping and is joined by 3′–5′ phosphodiester bond.31 (3) Exon-intron circRNA (ElciRNA), which is also formed by exon skipping and located in the nucleus. The ends of this type of circRNA are joined by a 3′–5′ phosphodiester bond and bind with RNA to promote transcription of target genes.32
Liu et al. identified the ability of endonuclease RNase L to degrade the circRNA upon viral infection or poly (I:C) stimulation. As they lack 3′ and 5′ terminus, endoribonucleolytic cleavage was also reported to occur.33 Kim et al. identified the role of N6-Methyladenosine (m6A) methylation in the degradation of circRNAs by downregulating YTH N6-methyladenosine RNA-Binding Protein 2 (YTHDF2) through RNase P/MRp complex.34 A brief on the basic characteristics of circRNA is given in Table 1.
Table 1.
Basic characteristics of circular RNA
| RNA type | Noncoding10 |
| Splicing mechanism | back-splicing15 |
| Degradation | resistant to exonuclease cleavage19 |
| Exon and intron length | exons are in the range 1–5, while introns that flank them are three times longer compared with linear counterparts16 |
| Structure | closed loop35 |
| Types of circRNA | |
| Exonic circRNA | located in the cytoplasm and joined by 3′–5′ phosphodiester bond; exonic circRNA functions as miRNA sponge, may also function as a sponge for RBPs15 |
| Intronic circRNA | located in the nucleus and joined by 2′–5′ phosphodiester bond; intronic circRNA functions in regulating gene expression36 |
| Exon-intron circRNA | located in the nucleus and joined by 3′–5′ phosphodiester bond; exo-intron circRNA functions in regulating gene expression32 |
RBPs, RNA-binding proteins.
Functions of circRNA
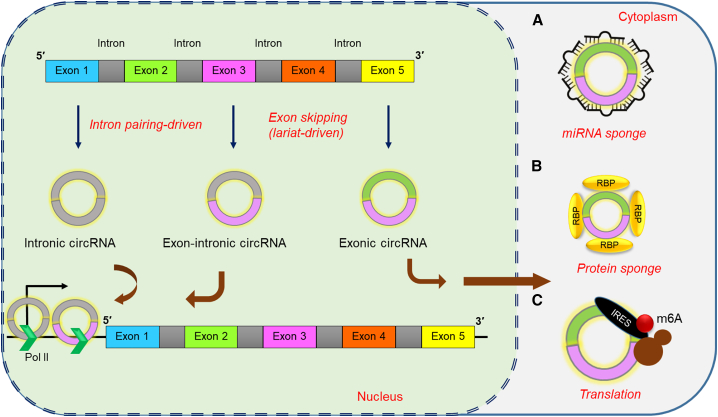
A diversified set of circRNAs have been identified for their functions like sponging, acting as decoys, or translatable elements that alter the gene or protein expression. (1) circRNAs can modulate miRNA activity by functioning as a sponge.37 A single circRNA can bind with one or many miRNAs with its circular sequence. A well-studied example of miRNA sponging is circRNA-CDR1as, which was found to harbor 63 binding sites for miR-7.38 (2) By interacting with proteins, circRNAs were found to function as decoys and alter the cellular function. Studies have demonstrated that CDK2/p21 and HIF-1α/ID1 get trapped by circFOXO3 in the cytoplasm, which is shown to block cell cycle progression and induce senescence.39 The silencing of circFOXO3 enhanced cell viability, whereas it led to apoptosis when they were ectopically expressed. Ectopic expression of circFOXO3 was reported to repress the p53 levels and increase the protein level of FOXO3. p53 levels decrease when circFOXO3 leads to MDM2-induced p53 ubiquitination, followed by degradation. circFOXO3 prevents FOXO3 ubiquitination and degradation caused by MDM2 due to its low binding affinity with FOXO3 protein.40 (3) circRNAs are a type of lncRNAs having a low potential protein-coding ability. They also contain m6A modifications or internal ribosome entry sites (IRESs) and can be translated into peptides.41 Pamudurti et al. have observed a cap-independent translation in vitro and in vivo using the ribo-circRNAs UTR. It was also reported that the circMBL isoform is likely to be regulated by FOXO. These studies provide evidences that circRNAs can also be translated.42
Dysregulated cellular functions by circular RNAs have been reported with different diseases. For example, fibroblast proliferation and migration can also be stimulated by activating alveolar macrophages, which are promoted by circZC3H443 and circHECTD1.44 Disruption of certain circRNAs, such as circHIPK245 and circHECTD1,46 can inhibit the activation of astrocytes, which may benefit stroke recovery. Some circRNAs have also been reported to regulate apoptosis.47 For example, when Itchy E3 ubiquitin protein ligase (ITCH) was increased in non-small cell lung cancer (NSCLC) cells by has_circ_0043256, it was observed to induce apoptosis, while an increase of ERBB2 in the nucleus pulposus (NP) cell by circGRB10 was reported to inhibit apoptosis.48 CDR1as have been found to have regulatory roles in specialized β cells of pancreatic islets that produce insulin. It was inferred that CDR1as might inhibit the function of miR-7 in islet cells, thus improving insulin secretion.49 The diversified functions of circRNA are depicted in Figure 2.
Figure 2.
Functions of circRNA
The intronic and exon-intronic circRNAs act on the promotor region to recruit Pol II, thereby promoting transcription of target genes. The exonic circRNA that is exported to the cytoplasm functions as an miRNA sponge and sequesters the endogenous miRNA that inhibits target mRNA (A), or binds with RNA-binding proteins and act as a protein sponge and mediates their action (B). m6A and IRES modification can promote circRNA to translate to a protein (C). RBP, RNA-binding protein.
Biogenesis and functions of miRNA
miRNAs are one of the classes of sncRNAs, typically 20–23 nt in length. They have been reported to alter mRNA and protein expressions by regulating their transcriptional and post-transcriptional levels.50 Among all the identified miRNAs to date, about half of them are processed more from introns compared with exons of protein-coding genes; i.e., they are intragenic,51 while regulation of other miRNAs is controlled by their promoters and is independently transcribed of host gene; i.e., they are intergenic.52 miRNAs can be transcribed into clusters as well.53
The majority of mature miRNAs are transcribed from pri-miRNA processing with the help of a microprocessor complex consisting of Drosha (ribonuclease III enzymes) and an RNA-binding protein, DiGeorge Syndrome Critical Region 8 (DGCR8) in the nucleus.54 DGCR8 recognizes the m6A GGAC region in the pri-miRNA motif, which is then cleaved by Drosha, leading to 3′ overhang. The pre-miRNA, which is generated from 3′ overhang, is exported to the cytoplasm through exportin 5 (XPO5)/RanGTP complex.55 The miRNA duplex is then loaded onto the argonaute (AGO) in an ATP-dependent manner, generating the guide and passenger strands based on the degree of complementarity.56 The guide strand and the miRNA response element form the miRNA-induced silencing complex (miRISC), which induces AGO2 endonuclease activity to cleave the target mRNA site.57,58
Some miRNAs prefer Drosha/DGCR8, an independent pathway to produce pre-miRNAs from the introns of mRNA during splicing and debranching (e.g., mitrons).59,60 This type of processed miRNAs contains 3′ overhang produced by terminal uridylyltransferase (TUTase), which undergoes monouridylation for efficient dicer processing. These pre-miRNAs are then exported to the cytoplasm through exportin 1. The shorter miRNAs or endogenous short hairpin RNAs (shRNAs) are exported via exportin 5 to the cytoplasm after being recognized and cleaved by Drosha/DGCR8 complexes. These shorter transcripts are then loaded by AGO2 in the cytoplasm and turn into mature miRNA by poly(A)-specific ribonuclease (PARN)-mediated 3′–5′ trimming of 5p strand.61
circRNA-miRNA-mRNA interaction in diabetes and associated complications
miRNAs are recorded to play a significant role in the pathogenesis of diabetes by controlling the upregulation or downregulation of the genes involved in signaling cascade.62 For example, miR375 is involved in the development of pancreatic β cells. Studies have found the altered expression of this particular miRNA in T2DM patients alters glucose homeostasis by decreasing insulin secretion.63,64 Characterizing miRNAs based on their altered expression in diabetes and its associated complications has emerged from experimental studies with the aid of in silico databases. Several hyperglycemia-induced miRNAs, such as miR21, 192, 216, 217, and 377, are linked with the pathogenesis of diabetes and its complications. Therapeutic interventions of miRNA can inhibit the function of miRNA or repair its diminished activity. Experimentally, gene knockouts and antisense oligonucleotides have been widely used to inhibit the activity of miRNA, whereas replacing the function of miRNA can be achieved by introducing an miRNA mimic. Although the development of the delivery system for miRNAs to reach specific cells is complicated, an increasing number of patents have been applied for recently for miRNA-based therapeutics, especially in cancer. In this connection, BalkrishenBhat et al. have filed a patent targeting miRNAs for metabolic disorders through Regulus Therapeutics (US 20,180,171,334 A1). Many modifications for the delivery of miRNA into the muscle to reach a specific target are being carried out. For example, a 2′-OMe-modified anti-miR with phosphorothioate linkage for miR-103/107 to improve glucose homeostasis has been reported by Trajkovski et al.65 The miRNA-based therapeutics for clinical use in diabetic patients are still in the pipeline.
Besides regulating their targets, miRNAs were found to be controlled by several intrinsic factors, and circRNAs are one of them. circRNAs partly inhibit the miRNA activity by interacting with them, suggesting their possible role in gene and protein expression.66
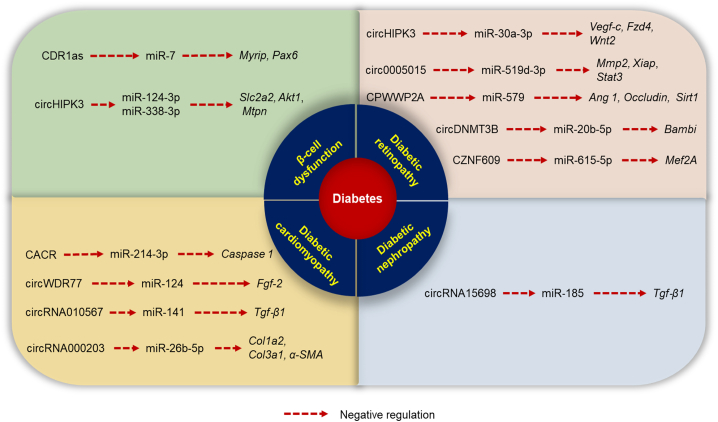
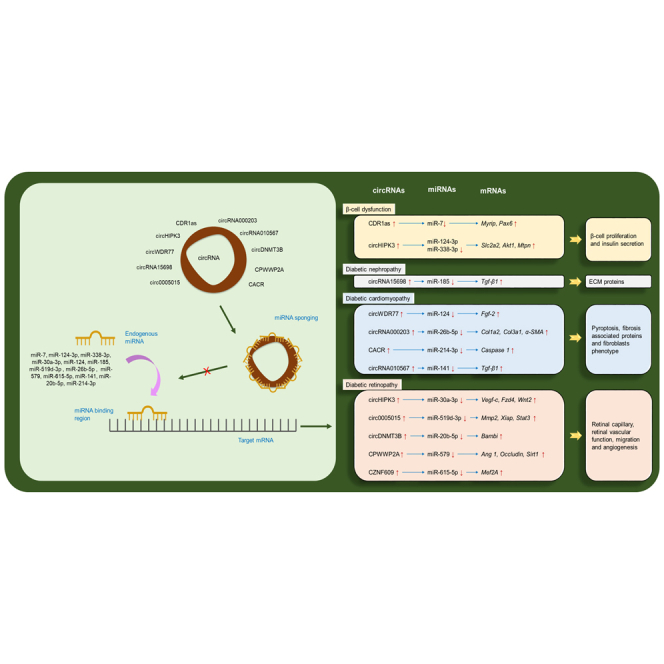
circRNAs play a role in gene expression as they can partly inhibit miRNA activity. They bind to miRNA like a sponge and act as competing endogenous RNA, regulating the function of the target miRNA, thus indirectly targeting mRNA levels.67 The next section describes in detail the role of this interactive network in regulating diabetes and its associated complications (Figure 3). Table 2 represents the overview of this interaction network.
Figure 3.
circRNA-miRNA-mRNA interaction in diabetes and its complications
The circRNAs negatively regulate the miRNA, which inhibits the expression of mRNA.
Table 2.
circRNA-miRNA-mRNA interaction network
| Name of the circular RNA | Target miRNA | Target mRNA | Study model | Diabetic complication | Outcome | Reference |
|---|---|---|---|---|---|---|
| CDR1as ↑ | miR-7↓ | Myrip and Pax6 ↑ | MIN6 cells (rat islet cells) | diabetes | affected insulin secretion | 49 |
| circHIPK3 ↑ | miR-124-3p and miR-338-3p↓ | Slc2a2, Akt1, and Mtpn ↑ | human islets, MIN6B1 cells (rat islet cells) | diabetes | regulated essential β cell activities, altered expression in diabetes models | 68 |
| circRNA15698 ↑ | miR-185 ↓ | Tgf-β1 ↑ | diabetic mice C57BL/KsJ-db/db | diabetic nephropathy | promoted ECM-related protein synthesis | 69 |
| circWDR77 ↑ | miR-124 ↓ | Fgf-2 ↑ | human VSMCs and HEK 293T cells | diabetic cardiomyopathy | suppressed VSMC proliferation and migration | 70 |
| circRNA000203 ↑ | miR-26b-5p ↓ | Col1a2, Col3a1, and α-SMA ↑ | diabetic db/db mice, C57BL/6 mice | diabetic cardiomyopathy | the enhanced fibrotic phenotype in cardiac fibroblasts | 71 |
| CACR ↑ | miR-214-3p ↓ | Caspase 1 ↑ | human cardiomyocyte AC16 cells | diabetic cardiomyopathy | alleviated pyroptosis in cardiomyocytes | 72 |
| circRNA010567 ↑ | miR-141 ↓ | Tgf-β1 ↑ | diabetic db/db mice, C57BL/6 mice | diabetic cardiomyopathy | suppressed fibrosis-associated protein resection in CFs | 12 |
| circHIPK3 ↑ | miR-30a-3p ↓ | Vegf-c, Fzd4, and Wnt2 ↑ | C57BL/6 mice | DR | altered retinal endothelial cell function and microvascular dysfunction | 73 |
| circ0005015 ↑ | miR-519d-3p ↓ | Mmp2, Xiap and Stat3 ↑ | HRVECs | DR | facilitated retinal endothelial angiogenic function | 74 |
| circDNMT3B ↓ | miR-20b-5p ↑ | Bambi ↓ | Sprague-Dawley rats, HRMECs | DR | reduced retinal acellular capillary number and alleviated visual damage | 75 |
| CPWWP2A ↑ | miR-579 ↓ | Ang 1, Occludin, and Sirt1 ↑ | C57BL/6 mice, HRVECs and human retinal pericytes | DR | affected retinal vascular dysfunction | 76 |
| CZNF609 ↑ | miR-615-5p ↓ | Mef2A ↑ | Human umbilical vein endothelial cells (HUVECs), C57BL/6 mice | DR | increased the migration and tube formation ability of HUVECs, and rescued HUVECs from oxidative stress or hypoxia-induced apoptosis | 77 |
CDR1as-miR7 interaction in regulating insulin homeostasis
Insulin homeostasis is essential to bridge the balance between glucose and insulin. As per the World Health Organization (WHO), more than 420 million adults live with diabetes, and most of them do not have insulin homeostasis.78 Pax6 is one of the genes responsible for insulin production in β cells of pancreatic islets.79 In this connection, CDR1as (ciRS-7) has been reported as the target of Pax6 mRNA and one of the most promising miRNA targets among several human circRNAs consisting of 71 binding sites or 26 clusters corresponding to miR-7.80 Hansen et al. demonstrated the regulatory function of CDR1as (ciRS-7) as an miR-7 sponge whose overexpression induced midbrain developmental defects in zebrafish. This phenotype also reported the alteration in the expression of miR-7 in the central nervous system.81,82 Dysregulated expression of miR-7 has been proved with the loss of insulin secretion and insulin exocytosis, resulting in diabetes.83,84 Xu et al. reported the increased expression of insulin corresponding with overexpression of CDR1as, as demonstrated in min-6 and mouse islet cells. Further, it was also demonstrated that forskolin (labdane diterpene) induced CDR1as overexpression and phorbol myristate acetate (PMA) induced miR-7 downregulation, whereas only glucose was observed to regulate neither CDR1as nor miR-7.49 This study has reported that CDR1as overexpression with increased mRNA expressions of insulin and Pax6 (known for increasing insulin secretion by binding to promoters of insulin gene). This study has also shown the decreased mRNA expression of Myrip, which is involved in insulin exocytosis, forming CDR1as-miR7-Pax6/Myrip interaction network in regulating insulin homeostasis.49
circHIPK3-miR-124-3p and miR-338-3p interaction in β cell function
The main function of β-cells is to secrete insulin, thereby regulate glucose homeostasis in the body. At the time of digestion and where there is an increased availability of glucose, insulin starts to secrete from β-cells of pancreatic islets. The mRNAs, such as Akt, Mtpn, and Slc2a2, were reported to have a strong association with insulin secretion. circHIPK3, which targets the above-mentioned mRNAs, is one of the abundantly expressed circRNAs in the pancreatic islets that originates from the Hipk3 gene and is reported to reduce expression in the patients’ islets.85 Stoll et al. confirmed that circHIPK3 controls insulin secretion and has activity on glucose conversion in β-cells of db/db mice. The expression of mRNAs such as Akt, Mtpn, and Slc2a2 was found to be downregulated upon inhibiting circHIPK3. Further, the authors also confirmed a drop in the luciferase activity of the construct containing the 3′ UTR of Mtpn upon silencing this particular circRNA. Using computational tools, miR-124-3p and miR-338-3p were found to target circHIPK3 involved in β-cell function by controlling insulin secretion and β-cell proliferation, respectively.68
circRNAWDR77-miR124 interaction in atherosclerosis
High blood glucose levels in patients with diabetes are more susceptible to proliferation and migration of vascular smooth muscle cells (VSMCs), ultimately ending up in atherosclerosis.86 In 2017, Chen et al. proved the involvement of circRNA in regulating atherosclerosis. This study identified the differentially expressed circRNAs through microarray analysis and reported circWDR77 to be largely expressed in diabetic patients. Through bioinformatic analysis, the interaction between circRNA WDR77 and miR-124 was predicted and confirmed with qPCR. The expression of miR-124 was observed to be decreased in VSMCs treated with high glucose. Further, the mRNA Fgf2 was also predicted to be a target of miR-124, which was then confirmed using a luciferase reporter assay. This study also proved the loop with circRNA and Fgf2 mRNA by silencing circRNA WDR77, which inhibited the expression of Fgf2.70
circRNA15698-miR185 interaction in diabetic nephropathy
Diabetic nephropathy (DN) is characterized by the proliferation of mesangial cells (MCs) and accumulation of extracellular matrix (ECM).87When exposed to hyperglycemic conditions, the expression of inflammatory cytokines in MCs is reported to progressively increase, which contributes to the progression of chronic kidney diseases like fibrosis.70 Hu et al. (2019) demonstrated the upregulation of circRNA15698 expression in total RNA extracted from the kidney cortex tissue of a db/db mice model. Further, upon knockdown of this particular circRNA, a reduction in ECM accumulation along with a decreased expression of fibrosis-related proteins in MCs were observed. The interaction of miR-185 with this circRNA was analyzed through a bioinformatics approach and validation using a luciferase reporter assay. This study also confirmed a reduced expression of miR-185 in mice models. The relation between circRNA and miRNA was examined upon knockdown of circRNA, which significantly increased miRNA level, suggesting a negative role. Tgf-β1 was chosen as a target mRNA in this interactive network through bioinformatics studies and validated with luciferase reporter assay. Together, the results of this study have confirmed the role of the circRNA15698-miR185-Tgf-β1 interaction network in the regulation of DN.69
circRNA000203-miR-26b-5p interaction in diabetic cardiomyopathy
One of the risks of diabetes, diabetic cardiomyopathy (DCM) is attributed to insulin resistance, hyperglycemia, increased fatty acids, and myocardial fibrosis.88 Tang et al. (2017) demonstrated the upregulation of circRNA000203 and its parental genes in the myocardium of diabetic mice (db/db) compared with non-diabetic mice (db/m). The study also demonstrated that circRNA000203 increases the expression of Col1a2, Col3a1, and α-SMA in mouse cardiac fibroblasts induced by Ang-II. This particular circRNA was screened to bind with miR-26b-5p through bioinformatics tools and was analyzed to suppress the effects of Col1a2 and CTGF in cardiac fibroblasts. This study presented that circRNA000203 suppressed the interaction between miR-26b-5p and Col1a2 and Ctgf. This leads to impaired miR-26b-5p inhibition effects on the fibrosis-associated genes Col1a2, Col3a1, α-SMA, and Ctgf, in cardiac fibroblasts. The results of this study together highlighted the role of circRNA000203-miR-26b-5p-Col1a2/Col3a1/α-SMA interaction network in the regulation of DCM.71
CACR-miR-214-3p interaction in diabetic cardiomyopathy
The expression of caspase-1-associated circRNA (CACR) has been reported to be increased in high-glucose-induced cardiomyocytes. The authors of this study induced diabetic conditions by exposing the cells to high glucose condition and confirmed the activation of pyroptosis with increased mRNA and protein expression of Nlrp3, Caspase-1, and IL-1β. As per the previous literature reports, miR-214-3p was found to be one of the targets of Caspase-1 and was confirmed to be reduced under high glucose conditions. Through a computational approach, CACR was predicted to bind with miR-214-3p. Hence the expression of CACR was inhibited using an Antisense Oligonucleotide (ASO), confirmed using western blotting. Further, silencing CACR was found to alleviate high-glucose-induced pyroptosis by increasing miR-214-3p and subsequently inhibiting its endogenous target, Caspase-1. The expressions of Nlrp3, and IL-1β were also low on silencing CACR but did not have many changes on silencing miR-214-3p, unlike Caspase-1. So, this study in detail demonstrated that CACR regulated pyroptosis through miR-214-3p/Caspase-1 pathway in high-glucose-induced cardiomyocytes, suggesting its plausible role.72
circRNA 010567-miR-141 interaction in myocardial fibrosis
A study conducted by Zhou et al. in 2017 has identified the circRNA010567 as one of the differentially expressed circRNAs in db/db mouse myocardium using microarray analysis. This study reported the expression of circRNA010567 to be significantly high among the other upregulated circRNAs. RNAi-mediated knockdown of this specific circRNA partly affected the expression of miR-141 in mouse cardiac fibroblast cells. A decreased expression of miR-141 was observed in the db/db mice myocardium, suggesting a negative regulation. Further, this study had found a higher affinity of Tgf-β1 mRNA with miR-141 using bioinformatics, which was then confirmed with dual-luciferase reporter assays. Further, miR-141 was observed to increase the expression of TGF-β1 significantly. Diabetic mice myocardial fibrosis is thus mediated by the circRNA010567-miR-141-Tgf-β1 axis and thus was proposed as a novel target.12
circHIPK3-miR-30a-3p interaction in retinal dysfunction
Diabetic retinopathy (DR) or retinal dysfunction is characterized by vascular permeability, capillary occlusion at the early stage (non-proliferative DR), and neovascularization at the later stage (proliferative DR).89 Although most of the cases have been reported with aberrant angiogenesis, the less appreciated contact of retinal dysfunction in diabetic patients is the impaired angiogenesis.90 In this context, Shan et al. have deciphered the increased expression of circHIPK3 in mouse retinal endothelial cells and human retinal vascular endothelial cells (HRVECs). This increase in circHIPK3 expression was observed to sponge the endogenous miR-30, which was confirmed using a luciferase reporter and fluorescence in situ hybridization (FISH) techniques. Angiogenic markers such as Vegf-c, Fzd4, and Wnt2 were the potential targets and seemed to be reduced on silencing circHIPK3 with increased miR-30a-3p expression. This study has demonstrated the role of circHIPK3 in DR by inhibiting miR-30a, thereby increasing the target angiogenic genes’ expression and promoting angiogenesis.73
circRNA0005015-miR-519d-3p interaction in DR
DR is characterized by increased vascular permeability and the growth of new blood vessels in the retina and posterior surface of the vitreous,91 which results in neurodegeneration and dysfunction of the microvascular retina.92 Retinal vasculature is characterized by the migration and proliferation of endothelial cells, which are largely affected by hyperglycemia.93 circRNA expression profiling carried out by Zhang and his research group in 2017 have identified 365 circRNAs to be upregulated in diabetic human retinas of among 529 differentially expressed ones. They have also identified an upregulated expression of circ0005015 from Has2 gene locus in the plasma, vitreous, and preretinal fibrovascular samples among DR patients. Through bioinformatics tools, the regulatory expression of circ0005015 with miR-519d-3p was identified and was confirmed using a luciferase reporter assay. This study also demonstrated the decreased expression of Mmp2, Stat3, and Xiap genes in HRVECs upon overexpression of miR-519d-3p, and the ability of tube formation was decreased. This study has suggested the existence of the circRNA0005015-miR-519d-3p-Mmp2/Stat3/Xiap interaction network in DR.74
circRNA DNMT3B - miR-20b-5p interaction in DR
Recently, the role of circRNA DNMT3B has been demonstrated by Zhu et al. (2019) in high-glucose-induced human retinal microvascular endothelial cells (HRMECs). As per previous literature reports, the expression of miR-20b-5p was examined in the retina of diabetic rats. The study showed an increased expression of miR-20b-5p in the retina of diabetic rats compared with the control ones. Further, this was validated in vitro in high-glucose-induced HRMECs. Using the bioinformatics approach and as per the previous reports, the research team further investigated the effect of Bambi in high-glucose conditions.
Also, silencing Bambi increased proliferation, migration, and tube formation of HRMECs and counteracted the effects caused by inhibiting miR-20b-5p. The expression of circRNA DNMT3B was decreased and was in line with fibrovascular membranes of DR patients. Using the bioinformatics tools, this particular circRNA was found to sponge miR-20b-5p. This was validated in vitro and overexpression of circRNA DNMT3B attenuated high-glucose-induced effects of miR-20b-5p and Bambi. Hence, the results of this study showed that circRNA DNMT3B regulated high-glucose-induced HRMEC function by targeting miR-20b-5p and Bambi, suggesting its role in DR.75
CPWWP2A-miR-579 interaction in DR
Dysregulated functions of retinal microvasculature that comprise endothelial cells and pericytes have been reported to be a major cause of vascular leakage in patients with DR.94 An elevated expression of cPWWP2A in pericytes was reported by Liu et al. in 2018, suggesting their role in DR. This study reported 89% similarity of circRNA0000254 with the human genome in db/db mice by performing microarray analysis, which was then named CPWWP2A as its host gene is PWWP2A. The exosome carrying this particular circRNA from endothelial cells to pericytes has been assessed. With bioinformatics aid, the interaction of cPWWP2A with miR-579 was confirmed using a luciferase reporter assay. Co-localization of circRNA with miRNA has been reported by the RNA-FISH hybridization technique. The expressions of target genes, such as Angiopoietin1 (Ang1), Occludin, and Sirt1, were found to be low upon overexpression of miR-579 and silencing CPWWP2A. This study witnessed the interaction network between cPWWP2A-miR-579-Ang1/Occludin/Sirt1 in the regulation of vascular dysfunction in DR.76
cZNF609- miR-615-5p interaction in DR
Vascular dysfunction, impaired angiogenesis, and vessel loss are the major hallmarks of ischemic progressions and are also persistent in DR.95 The C57BL/6 mice were exposed to a hypoxic environment with streptozotocin injection to make them retinopathic, and the regulation of cZNF609 was analyzed by Liu and his research group in 2017. The study revealed the cytoplasmic localization of cZNF609 and also explored the pro-angiogenic role of this particular circRNA on silencing cZNF609, which decreased retinal vessel loss and suppressed angiogenesis. Further, the inhibition and sequestration of miR-615-5p by cZNF609 to improve the Mef2A levels has been studied. This study has provided insight on cZNF609-miR-615-5p-Mef2A in the pathogenesis of vascular dysfunction.77
Database for detecting circRNA and miRNA interaction with mRNA
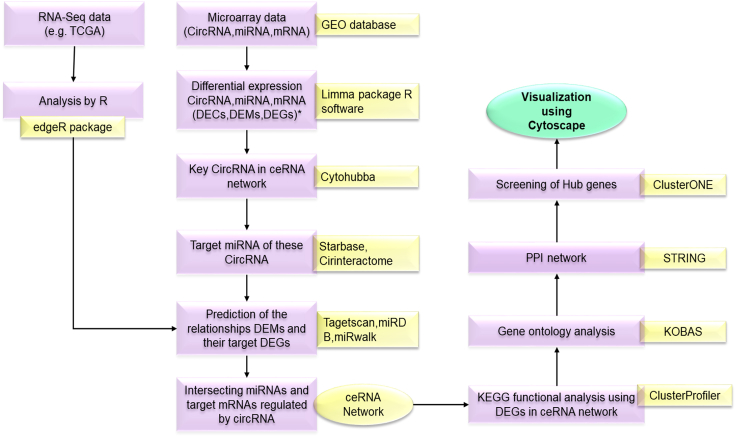
To study the various aspects of circRNA and miRNA, multiple algorithms have been developed to detect both and their target interactions from the retrieved RNA sequences. These algorithms have helped develop various databases in which particular circRNAs or miRNAs can be explored with respect to their functions. Some of these are freely accessible for curating circRNAs or miRNAs from different species and providing more information about their role in various diseases.96 Tables 3 and 4 represent the available circRNA and miRNA databases to get information on their target interaction networks. A step-wise construction of this interaction network with the aid of bioinformatics tools is shown in Figure 4.
Table 3.
Database for circRNA prediction
| Online database | Remarks | Reference |
|---|---|---|
| circBase | annotation of circRNAs from the eukaryotic cell | 113 |
| starBase | contains information about different types of RNAs. This tool can be used for the detection of miRNA-circRNA interaction | 97 |
| circlncRNAnet | this tool aims to test in silico hypotheses of ncRNA-based functions by keeping a record of RNA sequencing data | 98 |
| circ2Traits | this online tool gives data about the positions of circRNAs in the genome and associated diseases | 99 |
| DeepBase | this tool provides a platform based on next-generation sequencing for annotation and discovery of ncRNAS | 31 |
| CircInteractome | this tool coordinates circRNA with RBPs | 100 |
| CirCpedia | this database gives information about various human and mouse samples through reverse splicing | 101 |
| circRNADb | this online tool gives information about more than 30,000 exons with circRNA in the human genome | 102 |
| TSCD | this tool helps in the characterization of tissue-specific circRNAs | 103 |
Table 4.
Database for miRNA prediction
| Online database | Remarks | Reference |
|---|---|---|
| Target scan | searches for conserved 8mer and 7mer sites matching the seed region of each miRNA for predicting their biological targets | 104 |
| miRDB | online target prediction database; analyzes thousands of miRNAs and their targets through SVM learning machine by miRtarget | 105 |
| miRanda | identifies target genes and their level of downregulation at mRNA level | 106 |
| RNAhybrid | online prediction tool with unique features such as speed up of seed match, seed region G:U base pairing | 107 |
| MirGator | online portal based on deep sequencing and mRNA target prediction | 108 |
| miRecords | provides validates data regarding miRNA mRNA targets among seven animal species | 109 |
| miRTarBase | contains experimentally validated miRNA target interactions data | 110 |
SVM, support vector machine.
Figure 4.
Construction of circRNA-miRNA-mRNA interaction network
The step-wise construction of circRNA-miRNA-mRNA interaction network using bioinformatics tools. ∗Differentially expressed circRNAs (DECs), differentially expressed miRNAs (DEMs), and differentially expressed genes (DEGs).
Discussion and future prospectus
In this review, we have analyzed the literature on the circRNA-miRNA-mRNA interaction network in diabetes and its associated complications. Recently, research in the field of noncoding RNAs has received increasing attention in disease biology. Of note, more studies focusing on the mechanism of miRNA sponge are being elucidated. The inhibitory effects of miRNAs have been revealed in many studies and are reported to be strongly associated with various diseases, including diabetes. Hence, exogenous tools like synthetic miRNA mimics and agomiRs that bind to the 3′ UTR to modify the expression of native genes have started to gain interest in the research community. In parallel, a few researchers also focus on the upstreams of miRNA to inhibit its expression. circRNA is one of them that mainly act as an miRNA or protein sponge. The use of circRNAs in therapeutic avenues has been reviewed by Holdt et al.111 The circRNAs can be synthesized or modified chemically as miRNA mimics using RNA ligases or ribozymes and introducing photolabile linkers. This could prevent the linearization and further inactivation of circRNAs. Further, chemical modifications to improve stability and binding affinity or coating circRNAs with proteins make the system easier to recognize. The delivery of any naked RNA and its half-life into any living system is, however, tricky. Research in the field of encapsulating RNAs into nanovesicles is ongoing.
The physiological functions of most of the circRNAs are yet to be identified, and may reveal some of their abilities to act as protein counterparts. Important identified circRNAs are exonic circRNAs that function as miRNA sponges to counteract and alleviate the miRNA-induced changes. The role of intronic circRNAs is less explored. Advances in RNA technologies would help us foresee many developments in circRNA research in the near future.
Conclusion
The focus of research has been expanded in identifying several epigenetic tools involved in the pathology and progression of various diseases. This review has summarized the circRNA-miRNA-mRNA interaction network in diabetes and its associated disorders. More than 20,000 novel circRNAs have been identified to date,112 although only a few are identified with their regulatory pathways or interaction networks. The importance of identifying the interaction network between them is that it would shed light on the holistic and mechanistic picture of the regulation of various genes and their upstream elements in response to different pathological conditions. In the future, extensive research in this field would allow better diagnostics and provide more knowledge on the interactive mechanisms.
Acknowledgments
One of the authors, RJ acknowledges the Council of Scientific and Industrial Research (CSIR), New Delhi, India, for the award of Senior Research Fellowship (SRF) [File no.: 09/1045(11089)/2020-EMR-I]. Funding: the authors gratefully acknowledge the Indian Council of Medical Research (grant no. 2020-9621), Government of India, for financial assistance. This project is also supported by the grant (project code: UIC202007) from BNU-HKBU United International College.
Author contributions
The study was conceptualized and designed by SS, RJ, and KMR. Collection and assessment of data by SS. Writing original manuscript by SS and RJ. Further reviewing and necessary editing were provided by RJ, KG, BX, and KMR. Supervision for this study, and final approval to this manuscript, was provided by BX and KMR. All data were generated in-house, and no paper mill was used. All authors agree to be accountable for all aspects of work, ensuring integrity and accuracy.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Baojun Xu, Email: baojunxu@uic.edu.cn.
Kunka Mohanram Ramkumar, Email: ramkumak@srmist.edu.in.
References
- 1.Shepherd P.R., Kahn B.B. Glucose transporters and insulin action--implications for insulin resistance and diabetes mellitus. N. Engl. J. Med. 1999;341:248–257. doi: 10.1056/NEJM199907223410406. [DOI] [PubMed] [Google Scholar]
- 2.Cheng L.J., Chen J.H., Lin M.Y., Chen L.C., Lao C.H., Luh H., Hwang S.J. A competing risk analysis of sequential complication development in Asian type 2 diabetes mellitus patients. Sci. Rep. 2015;5:15687. doi: 10.1038/srep15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadi H.A., Suwaidi J.A. Endothelial dysfunction in diabetes mellitus. Vasc. Health Risk Manag. 2007;3:853–876. [PMC free article] [PubMed] [Google Scholar]
- 4.Tan K.C., Chow W.S., Ai V.H., Lam K.S. Effects of angiotensin II receptor antagonist on endothelial vasomotor function and urinary albumin excretion in type 2 diabetic patients with microalbuminuria. Diabetes Metab. Res. Rev. 2002;18:71–76. doi: 10.1002/dmrr.255. [DOI] [PubMed] [Google Scholar]
- 5.Tangvarasittichai S. Oxidative stress, insulin resistance, dyslipidemia and type 2 diabetes mellitus. World J. Diabetes. 2015;6:456–480. doi: 10.4239/wjd.v6.i3.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chawla A., Chawla R., Jaggi S. Microvasular and macrovascular complications in diabetes mellitus: distinct or continuum? Indian J. Endocrinol. Metab. 2016;20:546–551. doi: 10.4103/2230-8210.183480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asmat U., Abad K., Ismail K. Diabetes mellitus and oxidative stress–A concise review. Saudi Pharm. J. 2016;24:547–553. doi: 10.1016/j.jsps.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang P., Wu W., Chen Q., Chen M. Non-coding RNAs and their integrated networks. J. Integr. Bioinform. 2019;16 doi: 10.1515/jib-2019-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao C., Yu B. Role of long noncoding RNAs and circular RNAs in nerve regeneration. Front Mol. Neurosci. 2019;12:165. doi: 10.3389/fnmol.2019.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou B., Yu J.W. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem. Biophys. Res. Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 13.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 14.Holdt L.M., Kohlmaier A., Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell. Mol. Life Sci. 2018;75:1071–1098. doi: 10.1007/s00018-017-2688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santer L., Bar C., Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. 2019;27:1350–1363. doi: 10.1016/j.ymthe.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogozin I.B., Carmel L., Csuros M., Koonin E.V. Origin and evolution of spliceosomal introns. Biol. Direct. 2012;7:11. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucl. Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Z., Wang K., Wu F., Wang W., Zhang K., Hu H., Liu Y., Jiang T. circRNA disease: a manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis. 2018;9:475. doi: 10.1038/s41419-018-0503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aktas T., Avsar Ilik I., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 23.Li X., Liu C.X., Xue W., Zhang Y., Jiang S., Yin Q.F., Wei J., Yao R.W., Yang L., Chen L.L. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell. 2017;67:214–227.e217. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 24.Fei T., Chen Y., Xiao T., Li W., Cato L., Zhang P., Cotter M.B., Bowden M., Lis R.T., Zhao S.G., et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. U S A. 2017;114:E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D., Rosa A., De Santis R., Scarfo R., Peruzzi G., et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C., et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell. Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Liang D., Tatomer D.C., Luo Z., Wu H., Yang L., Chen L.L., Cherry S., Wilusz J.E. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell. 2017;68:940–954.e943. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Liu H., Li W., Yu J., Li J., Shen Z., Ye G., Qi X., Li G. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY) 2017;9:1585–1594. doi: 10.18632/aging.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 33.Liu C.X., Li X., Nan F., Jiang S., Gao X., Guo S.K., Xue W., Cui Y., Dong K., Ding H., et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880.e821. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 34.Park O.H., Ha H., Lee Y., Boo S.H., Kwon D.H., Song H.K., Kim Y.K. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol. Cell. 2019;74:494–507.e498. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 35.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 37.Ren S., Lin P., Wang J., Yu H., Lv T., Sun L., Du G. Circular RNAs: promising molecular biomarkers of human aging-related diseases via functioning as an miRNA sponge. Mol. Ther. Methods Clin. Dev. 2020;18:215–229. doi: 10.1016/j.omtm.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu C.Y., Kuo H.C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019;26:29. doi: 10.1186/s12929-019-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucl. Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abe N., Matsumoto K., Nishihara M., Nakano Y., Shibata A., Maruyama H., Shuto S., Matsuda A., Yoshida M., Ito Y., et al. Rolling circle translation of circular RNA in living human cells. Sci. Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21.e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X., Wang J., Zhou Z., Jiang R., Huang J., Chen L., Cao Z., Chu H., Han B., Cheng Y., et al. Silica-induced initiation of circular ZC3H4 RNA/ZC3H4 pathway promotes the pulmonary macrophage activation. FASEB J. 2018;32:3264–3277. doi: 10.1096/fj.201701118R. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Z., Jiang R., Yang X., Guo H., Fang S., Zhang Y., Cheng Y., Wang J., Yao H., Chao J. circRNA mediates silica-induced macrophage activation via HECTD1/ZC3H12A-dependent ubiquitination. Theranostics. 2018;8:575–592. doi: 10.7150/thno.21648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang R., Zhang Y., Han B., Bai Y., Zhou R., Gan G., Chao J., Hu G., Yao H. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13:1722–1741. doi: 10.1080/15548627.2017.1356975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han B., Zhang Y., Zhang Y., Bai Y., Chen X., Huang R., Wu F., Leng S., Chao J., Zhang J.H., et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14:1164–1184. doi: 10.1080/15548627.2018.1458173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo W., Zhang B., Mu K., Feng S.Q., Dong Z.Y., Ning G.Z., Li H.R., Liu S., Zhao L., Li Y., et al. Circular RNA GRB10 as a competitive endogenous RNA regulating nucleus pulposus cells death in degenerative intervertebral disk. Cell Death Dis. 2018;9:319. doi: 10.1038/s41419-017-0232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian F., Yu C.T., Ye W.D., Wang Q. Cinnamaldehyde induces cell apoptosis mediated by a novel circular RNA hsa_circ_0043256 in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017;493:1260–1266. doi: 10.1016/j.bbrc.2017.09.136. [DOI] [PubMed] [Google Scholar]
- 49.Xu H., Guo S., Li W., Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng J., Xing W., Xie L. Regulatory roles of microRNAs in diabetes. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17101729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isik M., Korswagen H.C., Berezikov E. Expression patterns of intronic microRNAs in Caenorhabditis elegans. Silence. 2010;1:5. doi: 10.1186/1758-907X-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Rie D., Abugessaisa I., Alam T., Arner E., Arner P., Ashoor H., Astrom G., Babina M., Bertin N., Burroughs A.M., et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017;35:872–878. doi: 10.1038/nbt.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanzer A., Stadler P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 54.Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 56.Yoda M., Kawamata T., Paroo Z., Ye X., Iwasaki S., Liu Q., Tomari Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galagali H., Kim J.K. miRISC composition determines target fates in time and space. Dev. Cell. 2018;47:142–143. doi: 10.1016/j.devcel.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 59.Babiarz J.E., Ruby J.G., Wang Y., Bartel D.P., Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruby J.G., Jan C.H., Bartel D.P. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 63.Lovis P., Gattesco S., Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol. Chem. 2008;389:305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- 64.Zhao H., Guan J., Lee H.-M., Sui Y., He C., Siu J., Tse P., Tong P., Lai F., Chan J. Up-regulated pancreatic tissue microRNA-375 associates with human type 2 diabetes through β-cell deficit and islet amyloid deposition. Pancreas. 2010;39:843–846. doi: 10.1097/MPA.0b013e3181d12613. [DOI] [PubMed] [Google Scholar]
- 65.Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M., Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 66.Verduci L., Strano S., Yarden Y., Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol. Oncol. 2019;13:669–680. doi: 10.1002/1878-0261.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitra A., Pfeifer K., Park K.S. Circular RNAs and competing endogenous RNA (ceRNA) networks. Transl Cancer Res. 2018;7:S624–S628. doi: 10.21037/tcr.2018.05.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stoll L., Sobel J., Rodriguez-Trejo A., Guay C., Lee K., Veno M.T., Kjems J., Laybutt D.R., Regazzi R. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol. Metab. 2018;9:69–83. doi: 10.1016/j.molmet.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu W., Han Q., Zhao L., Wang L. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-beta1. J. Cell Physiol. 2019;234:1469–1476. doi: 10.1002/jcp.26959. [DOI] [PubMed] [Google Scholar]
- 70.Chen C., Gong W., Li C., Xiong F., Wang S., Huang J., Wang Y., Chen Z., Chen Q., Liu P., et al. Sphingosine kinase 1 mediates AGEs-induced fibronectin upregulation in diabetic nephropathy. Oncotarget. 2017;8:78660–78676. doi: 10.18632/oncotarget.20205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang C.M., Zhang M., Huang L., Hu Z.Q., Zhu J.N., Xiao Z., Zhang Z., Lin Q.X., Zheng X.L., Yang M., et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang F., Li A., Qin Y., Che H., Wang Y., Lv J., Li Y., Li H., Yue E., Ding X., et al. A novel circular RNA mediates pyroptosis of diabetic cardiomyopathy by functioning as a competing endogenous RNA. Mol. Ther. Nucleic Acids. 2019;17:636–643. doi: 10.1016/j.omtn.2019.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shan K., Liu C., Liu B.H., Chen X., Dong R., Liu X., Zhang Y.Y., Liu B., Zhang S.J., Wang J.J., et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 74.Zhang S.J., Chen X., Li C.P., Li X.M., Liu C., Liu B.H., Shan K., Jiang Q., Zhao C., Yan B. Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest Ophthalmol. Vis. Sci. 2017;58:6500–6509. doi: 10.1167/iovs.17-22698. [DOI] [PubMed] [Google Scholar]
- 75.Zhu K., Hu X., Chen H., Li F., Yin N., Liu A.L., Shan K., Qin Y.W., Huang X., Chang Q., et al. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. EBioMedicine. 2019;49:341–353. doi: 10.1016/j.ebiom.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu C., Ge H.M., Liu B.H., Dong R., Shan K., Chen X., Yao M.D., Li X.M., Yao J., Zhou R.M., et al. Targeting pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction. Proc. Natl. Acad. Sci. U S A. 2019;116:7455–7464. doi: 10.1073/pnas.1814874116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu C., Yao M.D., Li C.P., Shan K., Yang H., Wang J.J., Liu B., Li X.M., Yao J., Jiang Q., et al. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7:2863–2877. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ojha A., Ojha U., Mohammed R., Chandrashekar A., Ojha H. Current perspective on the role of insulin and glucagon in the pathogenesis and treatment of type 2 diabetes mellitus. Clin. Pharmacol. 2019;11:57–65. doi: 10.2147/CPAA.S202614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Swisa A., Avrahami D., Eden N., Zhang J., Feleke E., Dahan T., Cohen-Tayar Y., Stolovich-Rain M., Kaestner K.H., Glaser B., et al. PAX6 maintains beta cell identity by repressing genes of alternative islet cell types. J. Clin. Invest. 2017;127:230–243. doi: 10.1172/JCI88015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mouradian M.M. MicroRNAs in Parkinson's disease. Neurobiol. Dis. 2012;46:279–284. doi: 10.1016/j.nbd.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 82.Pollock A., Bian S., Zhang C., Chen Z., Sun T. Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell. Rep. 2014;7:1184–1196. doi: 10.1016/j.celrep.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bravo-Egana V., Rosero S., Molano R.D., Pileggi A., Ricordi C., Dominguez-Bendala J., Pastori R.L. Quantitative differential expression analysis reveals miR-7 as major islet microRNA. Biochem. Biophys. Res. Commun. 2008;366:922–926. doi: 10.1016/j.bbrc.2007.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Latreille M., Hausser J., Stutzer I., Zhang Q., Hastoy B., Gargani S., Kerr-Conte J., Pattou F., Zavolan M., Esguerra J.L., et al. MicroRNA-7a regulates pancreatic beta cell function. J. Clin. Invest. 2014;124:2722–2735. doi: 10.1172/JCI73066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Locke J.M., da Silva Xavier G., Dawe H.R., Rutter G.A., Harries L.W. Increased expression of miR-187 in human islets from individuals with type 2 diabetes is associated with reduced glucose-stimulated insulin secretion. Diabetologia. 2014;57:122–128. doi: 10.1007/s00125-013-3089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mukohda M., Lu K.T., Guo D.F., Wu J., Keen H.L., Liu X., Ketsawatsomkron P., Stump M., Rahmouni K., Quelle F.W., et al. Hypertension-causing mutation in peroxisome proliferator-activated receptor gamma impairs nuclear export of nuclear factor-kappaB p65 in vascular smooth muscle. Hypertension. 2017;70:174–182. doi: 10.1161/HYPERTENSIONAHA.117.09276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Su X., Li T., Liu Z., Huang Q., Liao K., Ren R., Lu L., Qi X., Wang M., Chen J., et al. Licochalcone A activates Keap1-Nrf2 signaling to suppress arthritis via phosphorylation of p62 at serine 349. Free Radic. Biol. Med. 2018;115:471–483. doi: 10.1016/j.freeradbiomed.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 88.Murarka S., Movahed M.R. Diabetic cardiomyopathy. J. Card Fail. 2010;16:971–979. doi: 10.1016/j.cardfail.2010.07.249. [DOI] [PubMed] [Google Scholar]
- 89.Wang W., Lo A.C.Y. Diabetic retinopathy: pathophysiology and treatments. Int. J. Mol. Sci. 2018;19:1816. doi: 10.3390/ijms19061816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stitt A.W., McGoldrick C., Rice-McCaldin A., McCance D.R., Glenn J.V., Hsu D.K., Liu F.T., Thorpe S.R., Gardiner T.A. Impaired retinal angiogenesis in diabetes: role of advanced glycation end products and galectin-3. Diabetes. 2005;54:785–794. doi: 10.2337/diabetes.54.3.785. [DOI] [PubMed] [Google Scholar]
- 91.Fong D.S., Aiello L., Gardner T.W., King G.L., Blankenship G., Cavallerano J.D., Ferris F.L., 3rd, Klein R., American Diabetes A. Diabetic retinopathy. Diabetes Care. 2003;26:S99–S102. doi: 10.2337/diacare.26.2007.s99. [DOI] [PubMed] [Google Scholar]
- 92.Stitt A.W., Curtis T.M., Chen M., Medina R.J., McKay G.J., Jenkins A., Gardiner T.A., Lyons T.J., Hammes H.P., Simo R., et al. The progress in understanding and treatment of diabetic retinopathy. Prog. Retin. Eye Res. 2016;51:156–186. doi: 10.1016/j.preteyeres.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 93.Shin E.S., Sorenson C.M., Sheibani N. Diabetes and retinal vascular dysfunction. J. Ophthalmic Vis. Res. 2014;9:362–373. doi: 10.4103/2008-322X.143378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang H. Pericyte-endothelial interactions in the retinal microvasculature. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21197413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carmeliet P. Angiogenesis in health and disease. Nat. Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 96.Xu S., Zhou L., Ponnusamy M., Zhang L., Dong Y., Zhang Y., Wang Q., Liu J., Wang K. A comprehensive review of circRNA: from purification and identification to disease marker potential. PeerJ. 2018;6:e5503. doi: 10.7717/peerj.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aldred K.J., McPherson S.A., Turnbough C.L., Jr., Kerns R.J., Osheroff N. Topoisomerase IV-quinolone interactions are mediated through a water-metal ion bridge: mechanistic basis of quinolone resistance. Nucl. Acids Res. 2013;41:4628–4639. doi: 10.1093/nar/gkt124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu S.M., Liu H., Huang P.J., Chang I.Y., Lee C.C., Yang C.Y., Tsai W.S., Tan B.C. circlncRNAnet: an integrated web-based resource for mapping functional networks of long or circular forms of noncoding RNAs. Gigascience. 2018;7:1–10. doi: 10.1093/gigascience/gix118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hancock J.M. Circles within circles: commentary on Ghosal et al. (2013) "Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits". Front Genet. 2014;5:459. doi: 10.3389/fgene.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang X.O., Dong R., Zhang Y., Zhang J.L., Luo Z., Zhang J., Chen L.L., Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen X., Han P., Zhou T., Guo X., Song X., Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luo Y., Wang C., Yong P., Ye P., Liu Z., Fu Z., Lu F., Xiang W., Tan W., Xiao J. Decreased expression of the long non-coding RNA SLC7A11-AS1 predicts poor prognosis and promotes tumor growth in gastric cancer. Oncotarget. 2017;8:112530–112549. doi: 10.18632/oncotarget.22486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang X., El Naqa I.M. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 106.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kruger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucl. Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cho S., Jang I., Jun Y., Yoon S., Ko M., Kwon Y., Choi I., Chang H., Ryu D., Lee B., et al. MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucl. Acids Res. 2013;41:D252–D257. doi: 10.1093/nar/gks1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T. miRecords: an integrated resource for microRNA-target interactions. Nucl. Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., Huang W.C., Sun T.H., Tu S.J., Lee W.H., et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucl. Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holdt L., Kohlmaier A., Teupser D. Circular RNAs as therapeutic agents and targets. Front. Physiol. 2018;9:1262. doi: 10.3389/fphys.2018.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tang M., Kui L., Lu G., Chen W. Disease-associated circular RNAs: from biology to computational identification. Biomed. Res. Int. 2020;2020:6798590. doi: 10.1155/2020/6798590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Glazar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]