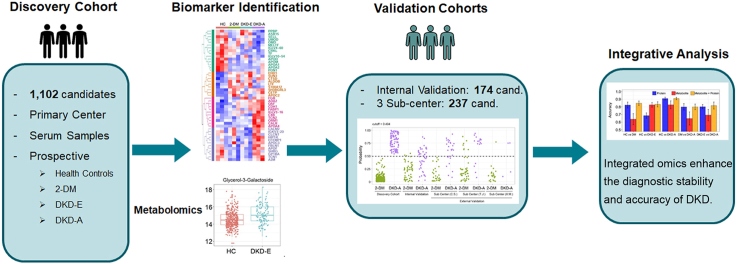
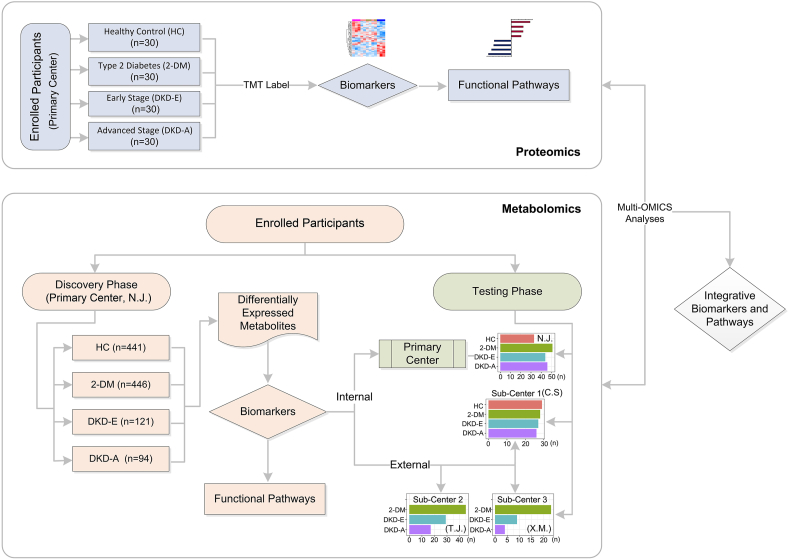
Abstract
Objective
Diabetic kidney disease (DKD) is the most common microvascular complication of type 2 diabetes mellitus (2-DM). Currently, urine and kidney biopsy specimens are the major clinical resources for DKD diagnosis. Our study proposes to evaluate the diagnostic value of blood in monitoring the onset of DKD and distinguishing its status in the clinic.
Methods
This study recruited 1,513 participants including healthy adults and patients diagnosed with 2-DM, early-stage DKD (DKD-E), and advanced-stage DKD (DKD-A) from 4 independent medical centers. One discovery and four testing cohorts were established. Sera were collected and subjected to training proteomics and large-scale metabolomics.
Results
Deep profiling of serum proteomes and metabolomes revealed several insights. First, the training proteomics revealed that the combination of α2-macroglobulin, cathepsin D, and CD324 could serve as a surrogate protein biomarker for monitoring DKD progression. Second, metabolomics demonstrated that galactose metabolism and glycerolipid metabolism are the major disturbed metabolic pathways in DKD, and serum metabolite glycerol-3-galactoside could be used as an independent marker to predict DKD. Third, integrating proteomics and metabolomics increased the diagnostic and predictive stability and accuracy for distinguishing DKD status.
Conclusions
Serum integrative omics provide stable and accurate biomarkers for early warning and diagnosis of DKD. Our study provides a rich and open-access data resource for optimizing DKD management.
Keywords: Diabetic kidney disease, Type 2 diabetes mellitus, Serum, Proteomics, Metabolomics, Machine learning
Graphical abstract
Highlight
-
•
Serum proteomics and metabolomics are novel, noninvasive approaches to detect DKD.
-
•
Integrated serum omics enhances the diagnostic stability and accuracy of DKD diagnoses.
-
•
Galactose/glycerolipid metabolism is the major disturbed metabolic pathway in DKD.
-
•
Serum metabolite glycerol-3-galactoside is an independent predictive marker of DKD.
Abbreviations
- DKD
Diabetic kidney disease
- CKD
Chronic kidney disease
- ESRD
End-stage renal disease
- UACR
Urine albumin-to-creatinine ratio
- KDIGO
Kidney disease improving global outcomes
- α2-M
α2-macroglobulin
- FABP1
Fatty acid binding protein 1
- PLTP
Phospholipid transfer protein
- SHBG
Sex hormone-binding globulin
- TIMP-1
Tissue inhibitor matrix metalloproteinase 1
- PLS-DA
Partial least squares discriminant analysis
- LDA
Linear discriminant analysis
- SVM
Support vector machine
- RF
Random forest
- Logi
Logistic regression
- ROC
Receiver operating characteristic
- AUC
Area under the curve
- DAG
Diacylglycerol
- GT
Galactosyltransferase
- Gly3P
Sn-glycerol-3-phosphate
- GK
Glycerol kinase
- TAG
Triacylglycerol
- LPA
1-Acyl-sn-glycerol-3-phosphate
- PA
1,2-Diacyl-sn-glycerol-3-phosphate
- PAP
Phosphatidate phosphatase
1. Introduction
Diabetes mellitus (DM) affects 463 million people worldwide and this number may increase to 700 million by 2045 [1]. The increasing prevalence of DM is associated with many factors such as demographic, socioeconomic, environmental, and genetic factors [2,3]. Society has made robust efforts to establish effective DM management systems. However, preventing the incidence of DM complications, such as damage to kidneys and other organs, remains an issue.
Diabetic kidney disease (DKD) is the most common microvascular complication of type 1 and type 2 DM [4]. Surpassing glomerulonephritis, DKD has become the leading cause of chronic kidney disease (CKD) and end-stage renal disease (ESRD) [5,6]. Progression of DM to DKD and ESRD is considered inevitable even when glucose is controlled [7]. Despite the progess made in this field, many aspects of DKD pathogenesis have not been addressed. For instance, in 2-DM patients, it's unclear whether the current gold-standard marker microalbuminuria or urine albumin-to-creatinine ratio (UACR) are the ideal parameters for monitoring DKD onset. It is also unknown whether pathological factors or biological processes in the circulation would help predict DKD progression from early to advanced stages, as physicians are challenged with differentiating true DKD, non-DKD, and a mixed form of DKD [8]. A meta-analysis of 48 studies revealed that non-DKD is highly prevalent in DM patients. The probability of an inaccurate DKD diagnosis reaches 49.2% if based on clinical information alone [9]. Therefore, rigorous and unbiased evidence from high-throughput analyses of multiple biological domains, including the genome, proteome, and metabolome, may be needed for precise DKD diagnosis and intervention.
Remarkable developments have been made in our capabilities to analyze omics data in large patient populations. Compared with genomics, proteomics and metabolomics have the potential to provide novel mechanistic insights into DKD progression [10,11]. Proteomics is a powerful approach for broadly profiling proteins [12], and metabolomics can broadly identify metabolites [13]. While combining these analyses could enhance DKD diagnosis, this remains untested. To this end, we integrated proteomics and metabolomics to identify biomarkers and evaluate their diagnostic values for DKD. This prospective study provides a rich, open-access resource that supports further mining of the metabolic changes in DKD patients.
2. Methods
Detailed methods are provided in the Supplemental Methods and Materials. This clinical trial was registered in the Chinese Clinical Trial Registry (ChiCTR2000028949). It was conducted in compliance with the principles of the 1975 Declaration of Helsinki and was approved by the Ethics Committees of the Affiliated Hospital of Nanjing University of Chinese Medicine (2019NL-109-02). All participants signed the consent forms to allow the extra samples to be used for academic purposes.
3. Results
3.1. Cohort characteristics, sample collection, and multiple measures
To ensure rigor, we invited the participation of four independent medical centers: one primary center (N.J.) and three subcenters (C.S., T.J., and X.M.). In each center, according to the albuminuria category classified by the Kidney Disease: Improving Global Outcomes (KDIGO) Diabetes Work Group [14], four candidate/patient groups were enrolled: healthy control (HC), 2-DM (UACR<30 mg/g), early-stage DKD (DKD-E, 30≤UACR≤300 mg/g), and advanced-stage DKD (DKD-A, UACR>300 mg/g). All recruited 2-DM patients were clinically diagnosed for ≥10 years. Participants recruited from the primary center (N.J.) were designated the discovery cohort; 1,102 total individuals were included. As a prospective study, we set up an internal testing cohort at the primary center; 174 total candidates were included. Serum samples were collected from all individuals. Meanwhile, 237 participants were recruited from three subcenters, C.S., T.J., and X.M., and designated as three independent external testing cohorts (C.S. has HC cases). Demographic and clinical characteristics of the discovery and testing cohorts were collected (Table 1 and Supplementary Table S1). The discovery cohort comprised 513 females and 589 males, 20–75 years old, with body mass index (BMI) in the 16.22–40.7 kg/m2 range. There were no significant differences in the levels of hemoglobin A1c (HbA1c), blood glucose, cholesterol, triglycerides, high-density lipoprotein, and low-density lipoprotein between patients (Table 1), but DKD-A patients had a significantly decreased estimated glomerular filtration rate (eGFR), and elevated blood urea nitrogen levels, serum creatinine, and urinary albumin versus 2-DM and DKD-E patients.
Table 1.
Demographic Characteristics of the Participants for Metabolomics (Discovery Cohort).
| HC | 2-DM | DKD-E | DKD-A | |
|---|---|---|---|---|
| Age (Years) | 33.24 ± 9.08 | 54.88 ± 10.28 | 54.50 ± 11.30 | 59.62 ± 9.30 |
| Gender (F/M) | 259/182 | 166/280 | 59/62 | 29/65 |
| Body Mass Index (kg/m2) | 21.67 ± 3.58 | 25.34 ± 3.4 | 26.13 ± 3.82 | 26.15 ± 3.67 |
| Hemoglobin A1c (%) | n/a | 8.30 ± 2.00 | 9.26 ± 2.29 | 8.00 ± 2.06 |
| eGFR (ml/min/1.73m2) | n/a | 99.29 ± 14.64 | 100.18 ± 19.03 | 40.34 ± 29.97 |
| Lp-PLA2 (μg/L) | n/a | 106.55 ± 36.49 | 97.74 ± 34.76 | 125.66 ± 61.91 |
| Albumin (g/L) | 44.02 ± 3.65 | 39.03 ± 3.10 | 38.86 ± 3.56 | 31.30 ± 4.54 |
| Blood Urea Nitrogen (mg/dL) | 6.48 ± 20.41 | 6.65 ± 1.67 | 6.87 ± 2.10 | 14.91 ± 7.92 |
| Serum Creatinine (μmol/L) | 67.11 ± 13.09 | 66.78 ± 15.95 | 63.92 ± 19.22 | 244.78 ± 189.79 |
| Glucose (mmol/L) | 4.85 ± 0.41 | 7.46 ± 2.82 | 9.01 ± 3.81 | 7.13 ± 3.35 |
| Uric Acid (mg/dL) | 276.64 ± 68.30 | 301.51 ± 92.31 | 302.79 ± 98.42 | 444.39 ± 126.99 |
| Cholesterol (mmol/L) | 4.43 ± 1.12 | 4.40 ± 1.19 | 4.63 ± 1.23 | 4.83 ± 1.95 |
| Triglycerides (mmol/L) | 1.03 ± 1.34 | 2.10 ± 2.59 | 2.45 ± 2.60 | 2.11 ± 1.52 |
| High-density Lipoprotein (mmol/L) | 1.59 ± 0.3 | 1.44 ± 3.82 | 1.26 ± 0.33 | 1.30 ± 0.41 |
| Low-density Lipoprotein (mmol/L) | 2.50 ± 0.43 | 2.95 ± 0.95 | 3.24 ± 1.04 | 3.07 ± 1.49 |
| Urine Creatinine (μmol/L) | n/a | 9453.14 ± 3853.35 | 7721.95 ± 4642.14 | 4973.57 ± 2694.35 |
| Albumin to Creatinine Ratio (mg/g) | n/a | 13.47 ± 9.78 | 63.79 ± 46.99 | 2597.68 ± 1896.45 |
| Fasting C-peptide (ng/mL) | n/a | 1.55 ± 0.93 | 1.49 ± 1.04 | 1.72 ± 1.00 |
| Fasting Insulin (μU/mL) | n/a | 12.25 ± 11.98 | 17.23 ± 16.28 | 11.63 ± 7.75 |
We applied a multi-omics approach to identify biological markers in the blood of 2-DM and DKD patients (Figure 1). First, we randomly selected 30 serum samples in each group from the discovery cohort and used proteomics to identify the protein biomarkers of DKD. Second, we measured metabolites across the discovery cohort, and validated findings in the internal testing and three external testing cohorts. We then used the proteome or metabolome identified by differential expression analyses in the discovery cohort to train machine learning models, which were applied to the testing cohorts for predicting DKD status. Finally, we performed integrative analyses combining proteomics and metabolomics data. This analysis highlighted the potential biomarkers for the clinical diagnosis of DKD and the pathways involved in the onset and progression of DKD.
Figure 1.
Summary of the study design and cohort details. This clinical study recruited 1,513 participants from four independent medical centers, which includes 503 HC, 593 2-DM, 230 DKD-E, and 187 DKD-A patients in one discovery and four testing cohorts. Sera were collected and subjected to training proteomics and large-scale metabolomics. HC: healthy control; 2-DM: type 2 diabetes mellitus; DKD-E: early-stage diabetic kidney disease; DKD-A: advanced-stage diabetic kidney disease.
3.2. Proteomics deciphered DKD progression in 2-DM patients
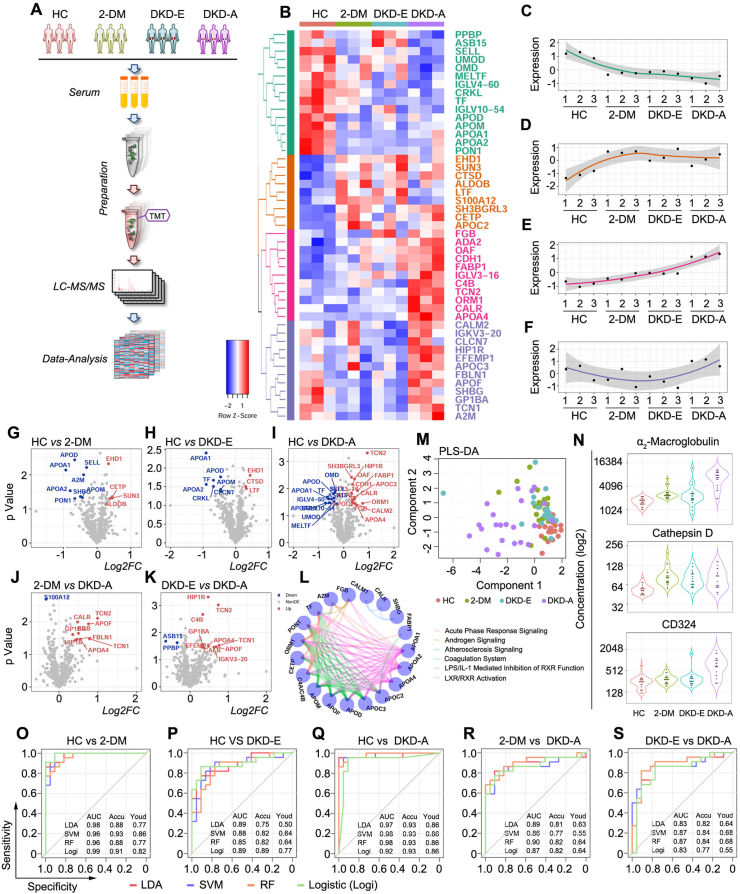
For an unbiased understanding of the underlying molecular determinants that modulate the onset and progression of DKD in circulation, we first performed a training, global-scale serum proteomic analysis. For each group, 30 serum samples were randomly selected. Sets of 10 serum samples were then mixed into 1 loading sample, resulting in 3 loading samples per group (Figure 2A). In total, 1,187 proteins were quantified across all 12 loading samples (4 groups). After pre-processing and missing value filtering, 581 proteins were used for further analysis (Supplementary Table S2). To define proteins altered in response to the change from HC to DKD, differential expression analysis was performed on all the pairwise comparisons (Supplementary Figure S1). Based on pairwise two-sample t-tests, 47 proteins were identified across four groups (Figure 2B), which were grouped into four clusters by Euclidean distance matrix hierarchical clustering based on the protein expression. Each group was graphed by average expression across the four disease subtypes. (Figure 2, C–F). We also marked differentially expressed proteins on the volcano plots of pairwise comparisons within HC, 2-DM, DKD-E, and DKD-A groups (Figure 2, G–K). Ingenuity pathway analysis (IPA) indicated that these differentially expressed proteins are involved in liver X receptor/retinoid X receptor (LXR/RXR) activation, farnesoid X receptor/retinoid X receptor (FXR/RXR) activation, and acute phase response signaling (Supplementary Figure S1). These proteins were pooled for pathway analysis, the top six pathways were selected (Figure 2L), and the involved proteins were defined as nodes. Interestingly, it was observed that among these pathways, activation of LXR/RXR signaling inhibits cell proliferation and induces apoptosis in pancreatic β-cells.
Figure 2.
Proteomics deciphered the status of DKD. (A) Working pipeline for proteomics data collection from the participants. (B) Heatmap for biomarker detection from the proteomics data. Each row represents a protein marker, and each column represents a loading sample across the four groups, including HC, 2-DM, DKD-E, and DKD-A. Protein markers are grouped into four clusters via hierarchical clustering. (C–F) Protein expression corresponding to the four clusters shown in Figure 1B. The four clusters showed out-down, up-flat, up, and down-up patterns, respectively. (G–K) Volcano plots of the proteomics data for each pairwise comparison. The x-axis is log2 fold-change and the y-axis represents the minus log10 p-value. Overexpressed and underexpressed proteins are marked with red and blue colors, respectively. (L) Network plot illustrating the enriched pathways detected by the differentially expressed proteins. Each node represents a protein, and the proteins are connected by their involved pathways. (M) Partial least squares discriminant analysis based on the ELISA markers. (N) Violin plots illustrate the concentration of α2-macroglobulin, cathepsin D, and CD324 in each group by ELISA. (O–S) Receiver operating characteristic (ROC) curves for each pairwise prediction by four different machine learning methods. Redline for linear discriminant analysis (LDA), blue line for support vector machine (SVM), orange line for random forest (RF), and green line for logistic regression (Logi). AUC: the area under the curve; Accu: accuracy; Youd: Youden index.
To test whether these proteins could determine DKD status, eight significant proteins were validated by enzyme-linked immunosorbent assay (ELISA): adiponectin, α2-macroglobulin (α2-M), cathepsin D, CD324, fatty acid binding protein 1 (FABP1), phospholipid transfer protein (PLTP), sex hormone-binding globulin (SHBG), and tissue inhibitor matrix metalloproteinase 1 (TIMP-1). These markers were tested in 88 participants (HC = 23, 2-DM = 23, DKD-E = 20, and DKD-A = 22) from the discovery cohort across the four groups (Supplementary Table S3). Partial least squares discriminant analysis (PLS-DA) revealed that these eight proteins could differentiate the four groups (Figure 2M). Expression levels and frequency distributions of each protein were compared across all groups (Figure 2N and Supplementary Figure S2), and the panel of α2-M, cathepsin D, and CD324 showed strong differential patterns across the four groups. We then used these three markers to predict DKD stages: a 5-fold cross-validation was performed on all pairwise predictions and four machine learning algorithms were employed: linear discriminant analysis (LDA), support vector machine (SVM), random forest (RF), and logistic regression (Logi). The receiver operating characteristic (ROC) curves revealed that these three markers could predict HC versus 2-DM, DKD-E, or DKD-A, 2-DM versus DKD-A, and DKD-E versus DKD-A, as well as balance the sensitivity and specificity rates (Youden index) (Figure 2, O–S). Together, our proteomic analyses and validation data suggest that α2-M, cathepsin D, and CD324 represent a potentially valuable biomarker panel for monitoring DKD progression. However, accurately differentiating 2-DM and DKD-E remained a challenge (Supplementary Figure S3).
3.3. Pairwise comparison of differentially expressed metabolites in serum
Considering that DKD is a common metabolic disease, we hypothesize that serum metabolite profiles will reflect the DKD status more directly and accurately than current methods. To test this, we performed metabolomics analysis, which identified 349 serum metabolites. After removing exogenous metabolites identified by the untargeted database of GC–MS from Lumingbio, 207 metabolites were further analyzed. We then measured the metabolite intensities across 1,102 patients in all four groups in the discovery cohort.
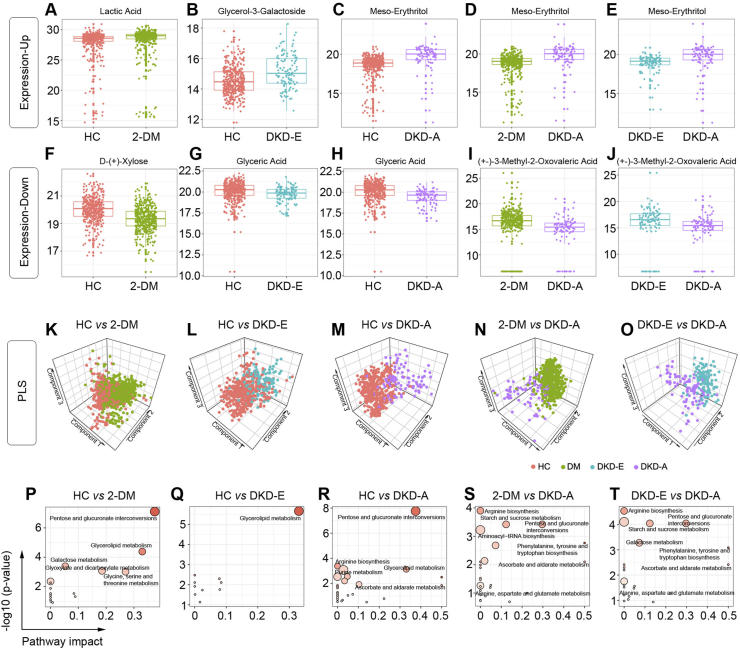
To identify differentially expressed metabolites by pairwise comparisons, a non-parametric Wilcoxon rank-sum test was performed on each metabolite (Supplementary Figures S4–S8 and Supplementary Table S4). Analysis of the top up- and down-regulated metabolites per comparison (Figure 3, A–J), identified lactic acid, glycerol-3-galactoside, meso-erythritol, D-(+)-xylose, glyceric acid, and (+-)3-methyl-2-oxovaleric acid. As a glycerolipid synthesis metabolite, glycerol-3-galactoside levels could reflect changes in glycerolipid synthesis. Also, 3-methyl-2-oxovaleric acid, a 2-oxo monocarboxylic acid, is a clinical marker for maple syrup urine disease [15]. We then applied a PLS regression analysis to these metabolites and found that they could effectively separate pairwise groups (Figure 3, K–O).
Figure 3.
Detection of differentially expressed metabolites by pairwise comparison of DKD status. (A–E) Representative box plots for top upregulated metabolites. (F–J) Representative box plots for the top downregulated metabolites. (K–O) Partial least squares discriminant analysis based on the differentially expressed metabolites. (P–T) Top significant functional pathways involved according to the differentially expressed metabolites.
We then performed pathway analyses to determine the biological significance of these metabolites in 2-DM and DKD. We generated bubble plots to illustrate the top significant pathways enriched by these biomarkers for each pairwise comparison (Figure 3, P–T). For HC versus 2-DM, metabolism changed for pentose and glucuronate interconversions, glycerolipid, and galactose; for HC versus DKD-E, glycerolipid metabolism changed; for HC versus DKD-A, glycerolipid metabolism, pentose, and glucuronate interconversions, and purine metabolism changed; for 2-DM versus DKD-A, arginine biosynthesis, starch and sucrose, and pentose and glucuronate interconversions changed; for DKD-E versus DKD-A, arginine biosynthesis, starch and sucrose metabolism, pentose and glucuronate interconversions, and galactose metabolism changed. Our analyses clearly indicate that glycerolipid metabolism, pentose and glucuronate interconversions, and galactose metabolism were the most affected pathways.
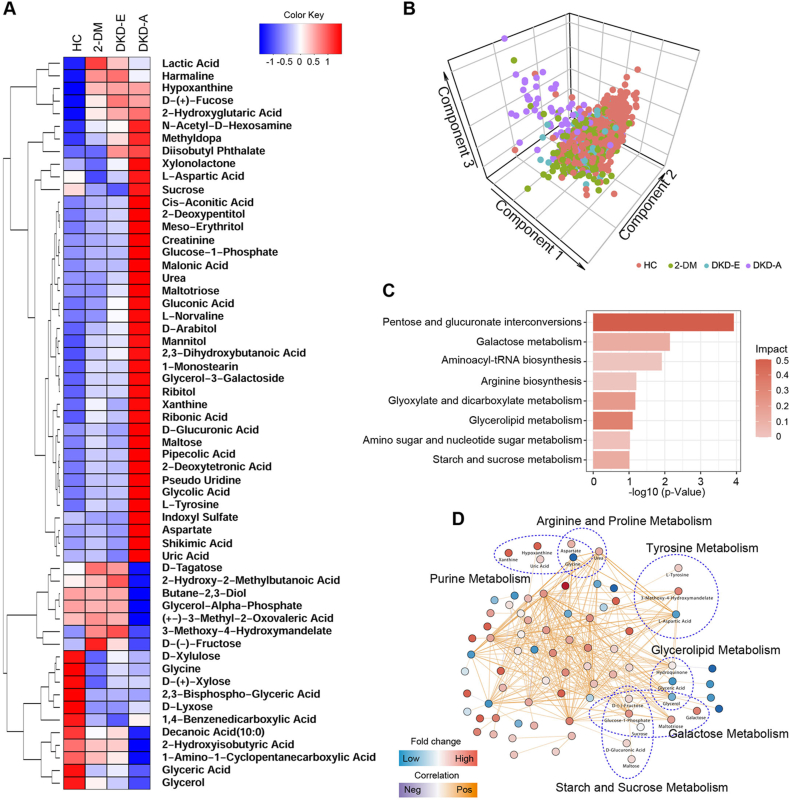
3.4. Correlation network of differential metabolites in serum
To explore the impact of metabolite alterations in response to different stages of DKD, pathway and network analyses were performed. Figure 4A shows the median expression of the 58 differentially expressed metabolites in the HC, 2-DM, DKD-E, and DKD-A groups. These biomarkers were generally split into two groups. In the first group, 40 metabolites showed comparatively lower expression in HC, 2-DM, and DKD-E but higher intensities in DKD-A patients. In the second group, 18 metabolites showed a reversed pattern, with higher expression in HC and DKD-E patients but lower expression in DKD-A patients. These metabolites were further analyzed by PLS-DA, which indicated that the four groups could be successfully separated, especially the HC and DKD-A patients (Figure 4B). The pathway analyses revealed that pentose and glucuronate interconversions, galactose metabolism, and glycerolipid metabolism were ranked at the top (Figure 4C). Consistently, our network analyses showed that many key metabolites linked to 2-DM and DKD development, including glycerol, glyceric acid, galactose, D-(−)-fucose, sucrose, and glucose-1-phosphate, were significantly altered across the four groups (Figure 4D).
Figure 4.
Metabolite biomarker identification of DKD. (A) Hierarchical clustering of differentially expressed metabolites. The median expression levels of metabolites for each group are presented in the heatmap. (B) Partial least squares discriminant analysis of the four groups. (C) Top significant pathways detected by the differentially expressed metabolites. (D) Network analysis based on the differentially expressed metabolites. Each node represents a metabolite, and the edges indicate the correlations between the metabolites. The metabolites highlighted are further grouped into metabolism pathways.
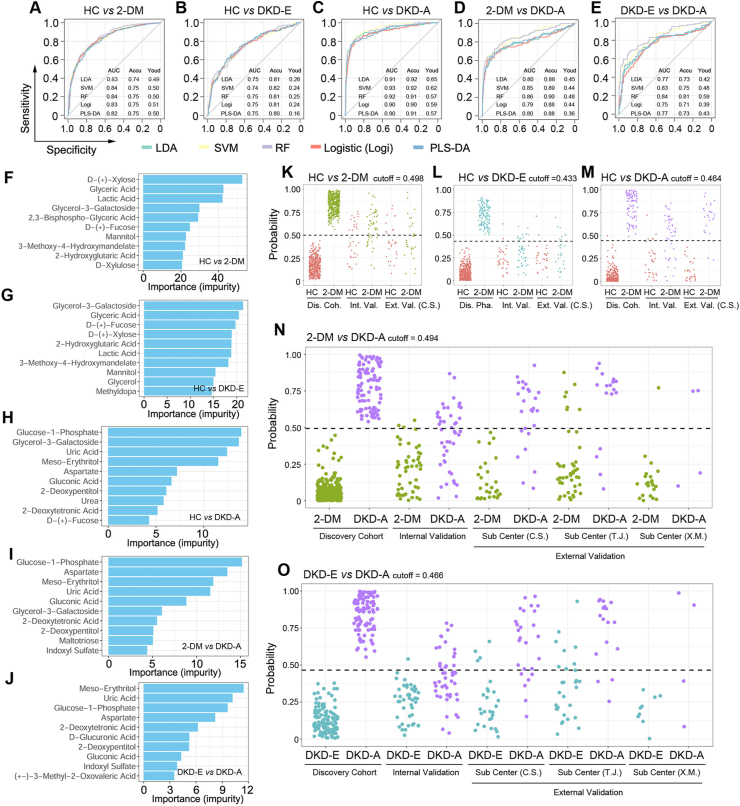
3.5. Machine learning for pairwise predictions of DKD from the expression of metabolites in serum
To verify the values of the identified metabolites in predicting DKD status, five machine learning algorithms were employed: LDA, SVM, RF, Logi, and partial least squares discriminant analysis (PLS-DA). We generated ROC curves from a 5-fold cross-validation for all pairwise predictions in the discovery cohort, and determined prediction accuracies of 75% in HC versus 2-DM, 82% in HC versus DKD-E, 93% in HC versus DKD-A, 90% in 2-DM versus DKD-A, and 84% in DKD-E versus DKD-A (Figure 5, A–E). As the five algorithms showed similar performance, RF was chosen to illustrate the results. In the RF model, the “importance” measurement is an evaluation of how the features (metabolites) can decrease the impurity in the tree splitting, where features with a higher importance score indicate a stronger contribution of the metabolites in the prediction model. We then ranked the metabolites by their importance/impurity scores for each pairwise prediction, and the top 10 are shown in Figure 5, F–J. Compared with HC, D-(+)-xylose, glyceric acid, and lactic acid were the top 3 metabolites in 2-DM patients without kidney injury (Figure 5, F–J). These metabolites directly reflected the dysregulation of glucose and lipid in 2-DM patients. On entering DKD, glycerol-3-galactoside and glucose-1-phosphate predicted kidney injury. Significantly, glycerol-3-galactoside was one of the top features in all pairwise predictions for DKD-E patients, while glucose-1-phosphate was for DKD-A patients. We also generated a binary split tree by applying all the patients in the discovery cohort to train one model (Supplementary Figures S9 and S10).
Figure 5.
Prediction of the DKD status via machine learning algorithms of the metabolomics data. (A–E) Receiver operating characteristic (ROC) curves for each pairwise prediction by five different machine learning methods. (F–J) Top prediction features selected by random forest impurity measurements. (K–O) Prediction probabilities based on the random forest algorithm. The best cutoff was trained from the discovery cohort and applied to testing cohorts for prediction. Dis. Coh. = Discovery Cohort; Int. Val. = Internal Validation; Ext. Val. = External Validation.
Besides the discovery cohort in the primary center, we measured the metabolite expression in the internal cohort and three external testing cohorts. For each pairwise prediction, classifiers trained from the discovery cohort were applied to these testing cohorts to evaluate model performance. Dot plots illustrate the probabilities of RF prediction on each patient across the discovery and testing cohorts, where the dashed-line cutoff indicates the binary split of the prediction (Figure 5, K–O). All prediction results in the testing cohorts are shown in Supplementary Table S5. Based on the highest prediction probability of the ROC in the discovery phase, the optimal cutoff values were 0.498 for HC versus 2-DM, 0.433 for HC versus DKD-E, 0.464 for HC versus DKD-A, 0.494 for 2-DM versus DKD-A, and 0.466 for DKD-E versus DKD-A. The cutoff values were then applied to predict the different stages of DKD in both internal and external testing cohorts. For HC versus 2-DM, the prediction accuracy was 64.3% in the internal testing cohort but low in the external C.S. cohort (Figure 5K and Supplementary Table S5). Similar to the prediction between HC and DKD-E (Figure 5L), the C.S. cohort showed 62.5% prediction accuracy, but the internal testing cohort showed lower accuracy. Consistent with protein markers, metabolites could not clearly distinguish 2-DM versus DKD-E (Supplementary Figure S11). All the other three binary predictions showed high performance. When comparing HCs and DKD-A, the prediction accuracy reached 73.4% (63.0% sensitivity; 87.9% specificity) for the internal testing cohort and 94.5% (88.5% sensitivity; 100% specificity) for the external C.S. cohort (Figure 5M). For 2-DM versus DKD-A, a prediction accuracy of 72.2% (47.8% sensitivity; 94.1% specificity) was achieved for the internal testing cohort and accuracies of 82.3%, 87.0%, and 88.9% were achieved for the three external testing cohorts (Figure 5N). Similar results were observed for DKD-E versus DKD-A (Figure 5O), and the prediction accuracy ranged from 66.7% to 84.6% for the four testing cohorts. The prediction results from the internal and external testing cohorts are presented in Supplementary Figure S12. Intriguingly, by employing six clinical parameters (glutamic acid decarboxylase [GAD] autoantibodies, age at diabetes onset, HbA1c, BMI, and measures of insulin resistance and insulin secretion), a recent study classified 2-DM into 5 new subtypes: severe autoimmune diabetes (SAID), severe insulin-deficient diabetes (SIDD), severe insulin-resistant diabetes (SIRD), mild obesity-related diabetes (MOD), and mild age-related diabetes (MARD) [16]. To further verify our findings, we regrouped our enrolled candidates by using this new subclassification system. As illustrated in Supplementary Figure S13, 30 differentially expressed metabolites were identified across the four 2-DM subtypes and the healthy controls. The serum metabolite distributions displayed distinguishing features within the five groups. The ROC curves also determined that the prediction accuracy could reach 91.9% in the SIDD vs the SIRD group, 61.5% in the SIDD vs the MOD group, 72.6% in the SIDD vs the MARD group, 90.3% in the SIRD vs the MOD group, 82.6% in the SIRD vs the MARD group, and 65.4% in the MOD vs the MARD group. These results indicate that select metabolites can successfully predict the 2-DM and DKD status, especially those in advanced and early stages.
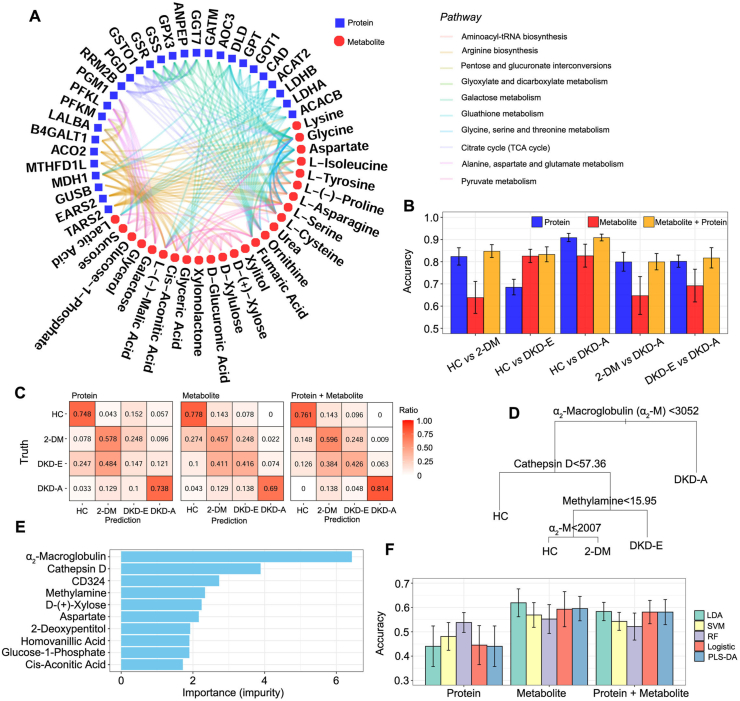
3.6. Integrating proteomics and metabolomics improves the diagnostic value for DKD
We next performed an integrated analysis to determine whether combining proteomics and metabolomics could enhance the accuracy of DKD diagnosis, compared with proteomics and metabolomics alone. We combined the top biomarkers from proteomics and metabolomics data in a pathway analysis (Figure 6A) and identified several pathways: galactose metabolism, pentose and glucuronate interconversions, citrate cycle, and pyruvate metabolism. We then combined protein and metabolite markers to predict the DKD status.
Figure 6.
Integrative analysis of proteomics and metabolomics data. (A) Significant pathways detected by the differentially expressed proteins and metabolites. Proteins and metabolites are presented in blue and red nodes, respectively. Proteins or metabolites that are involved in the same pathways are connected by the edges with the corresponding colors for each pathway. (B) Pairwise prediction accuracy based on protein features only (blue), metabolite features only (red), and integration of protein and metabolite features (orange). (C) Heatmap for the prediction accuracy. The numbers in the heatmap cells represent the ratio of true to predicted cases. (D) Prediction tree trained by the random forest model on all the discovery cohorts when integrating ELISA and metabolite features. (E) Top ELISA and metabolite features ranked by impurity measurements based on the random forest algorithm. (F) Prediction accuracy to distinguish 2-DM and DKD-E based on the five machine learning algorithms when integrating ELISA and metabolomics data.
Five machine learning models performed on protein markers only, metabolite markers only, and their combination revealed that the combinational model improved the accuracy and stability of DKD prediction. Taking the RF model results as an example, in the binary-outcome prediction (Figure 6B), the combination of proteins and metabolites led to no worse or slightly better accuracy than a single-omics marker. A contingency heatmap of the ratios of the actual diagnosis to the predicted diagnosis for all comparisons clearly showed that the combining proteins and metabolites improved the prediction accuracy over each individual marker alone (Figure 6C). The combined model especially provided a more robust separation of the 2-DM and DKD-E status. In the protein model, 48.4% of the DKD-E patients were wrongly predicted as 2-DM; however, the combined model increased the prediction accuracy of 2-DM and DKD-E to 59.6% and 42.6%, respectively. In addition, the multi-omics model successfully achieved 81.4% prediction accuracy for DKD-A patients. We also constructed a binary split tree by the RF model for the four-outcome predictions (Figure 6D), and we ranked the top markers by impurity (Figure 6E). These results suggested that α2-M, cathepsin D, and CD324 proteins, and methylamine are the top markers for DKD identification. For the prediction between 2-DM and DKD-E, the combined model showed comparable results with protein-only or metabolite-only models (Figure 6F). Collectively, the combined multi-omics model presented a robust approach for enhancing the diagnostic value of protein and metabolite markers in DKD patient blood.
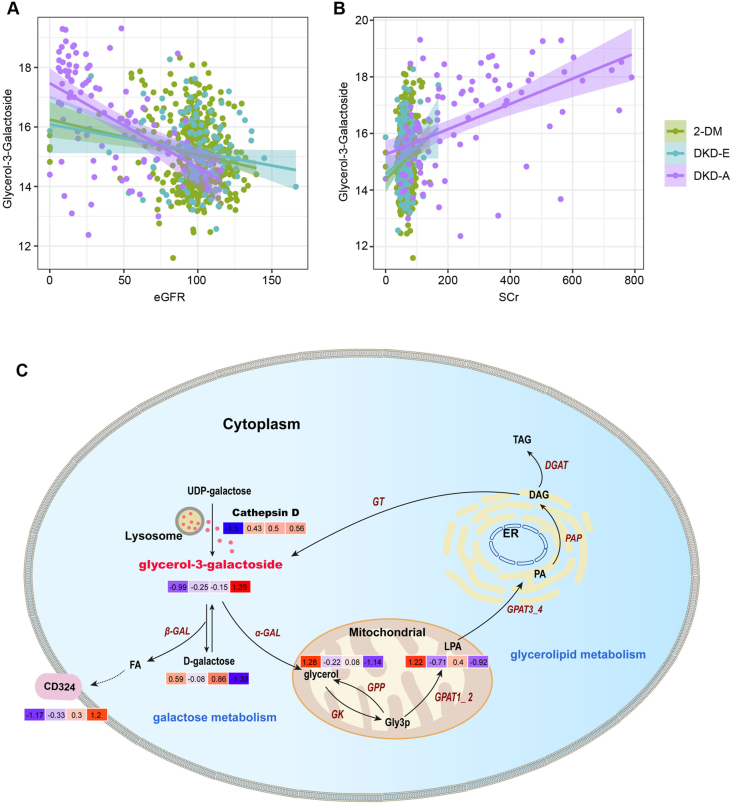
3.7. Glycerol-3-galactoside is a biomarker for monitoring DKD development
The diagnostic value of serum metabolites in DKD development are not yet known. To address this, we systemically analyzed the correlations between differentially expressed metabolites and clinical parameters of each patient. Impressively, we found that serum glycerol-3-galactoside was associated with eGFR (r = 0.437) and serum creatinine (r = 0.42) in patients (Figure 7, A–B). However, few metabolites, including glycerol-3-galactoside, exhibited associations with urinary micro- or macro-protein levels. Of particular interest, as shown in Supplementary Figure S13A, based on the new 2-DM subclassification system, glycerol-3-galactoside was also significantly induced in the SIDD, SIRD, and MARD groups, which further indicated its clinical values in 2-DM early identification. This suggested that glycerol-3-galactoside might be an independent surrogate serum biomarker for DKD development.
Figure 7.
Glycerol-3-galactoside synthesis is an independent metabolic event amid DKD progression. (A–-B) The correlations between glycerol-3-galactoside and eGFR or serum creatinine (Scr) levels. (C) Schematic diagram depicting the process of glycerol-3-galactoside synthesis and its roles in the onset of DKD. Each color box (left to right) indicates the normalized expression value of the corrsponding metabolite in the HC, 2-DM, DKD-E, and DKD-A groups, respectively.
Our integrative omics analysis indicated enhanced the stability of diagnostic values if proteomes and metabolites were combined. While α2-M is a well-characterized glycemic control marker [17], the role of glycerol-3-galactoside in DKD pathogenesis is largely unknown. Glycerol-3-galactoside is synthesized from UDP-galactose and diacylglycerol (DAG) by a microsomal galactosyltransferase (GT), and the lysosomal enzyme cathepsin D helps produce UDP-galactose (Figure 7C). Glycerol-3-galactoside is then hydrolyzed by α- or β-galactosidase (GAL) to galactose, glycerol, or fatty acid (FA). FA can induce CD324 expression to amplify DKD progression. In the mitochondria, glycerol is converted to sn-glycerol-3-phosphate (Gly3P) by glycerol kinase (GK) or potentially through a Gly3P phosphatase (GPP). Gly3P is an essential precursor for lipid synthesis and the accumulation of triacylglycerol (TAG) in response to nutrient starvation [18]. Gly3P is then converted to TAG for neutral lipid storage, or through other modifications to phospholipids, including 1-acyl-sn-glycerol-3-phosphate (LPA) and 1, 2-diacyl-sn-glycerol-3-phosphate (PA). PA is a prominent activator of several signaling pathways and can become DAG through phosphatidate phosphatase (PAP) [19]. DAG is a vital second messenger that regulates numerous physiological activities in DKD. In addition, DAG can be acylated by DGAT to form TAG. DAG is also a substrate for the synthesis of glycerol-3-galactoside [20]. Collectively, our data implicate that changes in serum protein α2-M/cathepsin D/CD324 and metabolite glycerol-3-galactoside may be effective biomarkers for determining the onset and progression of DKD.
4. Discussion
Early detection, diagnosis, and treatment of DKD are challenging [21]. Currently, microalbuminuria alone determines DKD onset, and kidney biopsy differentiates true DKD, non-DKD, and mixed-form DKD [9,[22], [23], [24]]. Therefore, identification of non-invasive serum biomarkers could enhance DKD management.
Facilitated by recent advances in high-throughput technologies, proteomics and metabolomics have been rapidly translating into clinical use to demonstrate how to leverage biomarkers to improve accuracy, enhance diagnostics, and reduce errors [25,26]. Functionally, proteomics provides a powerful tool to rapidly identify and quantify proteins present in cells, tissues, biofluids, or other biological samples [27,28]. In comparison, metabolomics is mainly employed to determine the small molecule fingerprints of cellular processes [29]. As the end products of cellular regulatory processes, the levels of metabolites are considered to be the ultimate response of biological systems to pathophysiological changes in various metabolic disorders [30]. Metabolites can closely reflect the disease phenotype to address a critical clinical need because they represent the downstream expression of genome, transcriptome, and proteome [31]. Indeed, the proteome and metabolome are not disjointed. Protein levels influence the metabolic profile, and the concentrations of the metabolites, in turn, affect protein expression [32]. Therefore, integrative omics is expected to provide us with unprecedented insight into biological entities such as circulating biomarkers of 2-DM and DKD [33].
Here, we integrated the proteomics and metabolomics of serum from 1,513 HC, 2-DM, DKD-E, and DKD-A patients from four independent medical centers. Our training proteomic analysis identified serum α2-M, cathepsin D, and CD324 proteins as a robust biomarker panel for monitoring DKD progression. Our full-scale metabolomics analysis identified pentose and glucuronate interconversions as well as galactose and glycerolipid metabolism as the key disturbed pathways in DKD. We also identified glycerol-3-galactoside as an independent biomarker for predicting DKD. Impressively, by integrating serum proteomics and metabolomics, we improved the value of these biomarkers for predicting DKD onset and progression.
Proteomics has clinical value for treating various diseases [34], but early DKD detection using preteomics has been challenging [35]. One proteomic study identified 273 urinary peptides differentially expressed between healthy controls and CKD patients, which when combined as the classifier CKD273 [36], was validated as a biomarker for CKD stratification, CKD early endpoint determination, and progression of DM patients with normoalbuminuria to microalbuminuria [[37], [38], [39], [40], [41]]. However, few trials have been proposed to evaluate the predictive value of blood for the course of human DKD.
Our training proteomics data revealed the difficulties in selecting a circulating surrogate DKD biomarker, especially for advanced DKD. Nevertheless, among 47 serum proteins, a combined panel of α2-M, cathepsin D, and CD324 was strongly associated with DKD diagnosis, indicating the role of multi-protein markers in monitoring DKD progression, especially advanced stages (Figure 2).
What are the mechanisms of these serum proteins in DKD? α2-M is a large plasma protein mainly produced by the liver. Due to its large size, the release of increased α2-M in urine is prevented and it is retained in circulation in diseased bodies. Increased or decreased α2-M has no apparent adverse effects on the physiological functions, [42] but α2-M can inactivate various proteinases and bind many cytokines or growth factors, such as platelet-derived growth factor, insulin, and transforming growth factor-β. Cathepsin D is an aspartic endoprotease ubiquitously distributed in lysosomes [43,44] that can regulate extracellular matrix homeostasis, glomerular permeability, endothelial function, and inflammation in kidney diseases [45]. CD324 (AKA E-cadherin) is a cell–cell adhesion protein increased during microalbuminuria progression after diabetes [46]. Soluble CD324 fragments in the urine of patients with early-stage DKD is considered to be a sign of kidney deterioration.
Surprisingly, this combinational biomarker shows modest performance in distinguishing 2-DM and DKD-E patients (Supplementary Figure S3). Whether glycemia-induced kidney damage is reflected in blood protein levels remains unclear from our training proteomic analyses. Therefore, the sensitivity and specificity of serum proteins indirectly predicting kidney damage may be a concern, especially for identifying early-stage DKD.
Compared with serum proteins, serum metabolites can increase the accuracy of determining the kidney's status in DKD [47]. Across the four groups, we identified 58 serum metabolites, most of which fell into 3 broad groups (pentose and glucuronate interconversions, galactose metabolism, and glycerolipid metabolism), indicating broad metabolic disturbances during DKD progression (Figure 3, Figure 4). Most of these serum metabolites were upregulated or downregulated in DKD-A patients compared with healthy adults. These changes might be caused by nephron absolute loss because increases in blood urea and creatinine levels were observed. Under these conditions, the damaged kidney has little ability to maintain metabolite distributions. Other diseased organs, due to diabetes or lower glomeruli filtration rate, would also be expected to disrupt the metabolic microenvironment in circulation.
Our original hypothesis was that we would identify protein or metabolite biomarkers for monitoring the onset of early stage DKD. Interestingly, lactic acid, glycerol-3-galactoside, meso-erythritol, D-(+)-xylose, and (+-)-3-methoxy-4-hydroxymandelate were altered in 2-DM and DKD-E patients, but not in DKD-A patients. Fructose, galactose, and glucose can be metabolized to generate energy [48]. However, long-term increased serum galactose and d-fructose, together with high lactic acid, can cause blood pH changes, which might play etiologic roles in diabetes and DKD. In contrast, in 2-DM and DKD-E patients, glycerol and glyceric acid were dramatically reduced in serum, versus healthy adults. However, changes in glucose or lipids have little correlation with clinical parameters. In addition, when comparing healthy adults with DKD-E patients, only the glycerolipid metabolism pathway was significantly altered. The difficulty in distinguishing 2-DM and DKD-E can be attributed to two possible causes. One is that the specificity of the expressed proteins/metabolites reflecting kidney damage was relatively low, as their levels in serum were not determined by kidney dysfunction only. The second is that we speculated that at early DKD stages, serum proteins/metabolites were completely disorganized due to renal damage causing too much noise to identify specific expression patterns.
We identified galactose and glycerolipid metabolism as the most disturbed pathways in DKD, indicating the critical role of glycerol-3-galactoside, the final product of these pathways. Glycerol-3-galactoside is not well explored in kidney diseases. This is the first report of glycerol-3-galactoside in non-uremic concentrations associated with DKD onset. In serum, glycerol-3-galactoside levels were tightly associated with eGFR and serum creatinine levels, but independent of albuminuria and microalbuminuria levels (Figure 7). Our training proteomic analyses indicated that several proteases were released by lysosomes (Figure 2). As mentioned, glycerol-3-galactoside is synthesized from UDP-galactose and DAG by microsomal GT, with lysosomal enzymes involved in the upstream process (Figure 7). Glycerol-3-galactoside can then be hydrolyzed into galactose, FA, and glycerol, which can induce kidney damage. The excretion rates of glycerol-3-galactoside in urine and blood from HCs and 2-DM and DKD patients are unknown. We speculate that glycerol-3-galactoside can be toxic, contributing to insulin resistance and rendering patients susceptible to more severe diabetes and its complications, including DKD.
Although untargeted metabolomics revealed abundant associations in DKD development, it's unclear if they're causal [49]. Pathological roles of glycerol-3-galactoside in diabetic kidneys and other diabetes-targeted organs should help illuminate new diagnoses and treatments. In addition, metabolomics demonstrated that 2-DM versus DKD-A and DKD-E versus DKD-A were well-predicted in internal or external testing cohorts by the trained model in the discovery cohort. Compared with the machine learning proteomics results, metabolome-based prediction showed improved monitoring performance for DKD onset in 2-DM patients. Nevertheless, the boundary of 2-DM and DKD-E remains unsatisfactory for diagnostic purposes.
A novel aspect of this study is that we integrated serum proteomics and metabolomics to enhance the prediction of DKD, such as for 2-DM vs DKD-E. Integrative omics stabilized the diagnostic values for 2-DM and all stages of DKD, and machine learning further increased the accuracy. Our integrative approach is a new attempt to develop a signature for personalized DKD care, but clinical translation is expensive [50].
Our study has some limitations: 1) Because kidney damage may be observed in 2-DM patients with normal microalbuminuria, and the microalbuminuria levels in most of our DKD-E patients ranged from 50 to 100 mg/g, we may have missed proteome- or metabolome-based values. 2) Although we applied five machine learning methods, further deep mining or algorithms may be needed. 3) Our analyses excluded some classes of lipids and amino acids in serum. 4) Proteome- or metabolome-based landscapes in the serum of non-diabetic CKD were not profiled. Thus, we could not fully determine whether metabolite or protein changes in DKD patients are due to absolute nephron loss or diabetes itself. Future studies should compare urine and serum for DKD.
5. Conclusion
Our study illustrated that serum integrative omics provides a high confidence interval for identifying biomarkers for the diagnosis of DKD or biochemical insights to monitor the status of DKD. Although it is too early to conclude that these markers will replace the invasive approach for DKD differentiation in the clinic, our findings indeed shed light on the development of valuable diagnostic tools for effective clinical implementation and designing of novel therapeutic targets to achieve earlier prevention, earlier diagnosis, and earlier treatment of DKD. We believe that this promising serum-based integrative omics approach will have widespread applications in the diagnosis of DKD in the clinic after further rigorous, prospective longitudinal studies.
Data availability
Raw mass spectrometry data were deposited in MassIVE with the data set identifier MSV000087487.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authorship contribution statement
ZD and LS conceived the project. LS, AX, WM, XT, ZL, ZK, YY, XL, and QY participated in the human sample collection. AX performed the clinical trial registration. LS, GY, XT, PYC, HT, YY, ZR, TXJ, LS, and ZD participated in the data collection and analyses. ZD and LS wrote and revised the manuscript. BC, HG, KD, HT, YY, TJ, ZY, WY, FH, LS, and ZD edited the manuscript. All authors approved the final version for publication. ZD and LS supervised the entire project.
Acknowledgments
This investigator-initiated clinical study was supported in part by the University of Pittsburgh Center for Research Computing through the resources provided. We are grateful to Dr. Justin Radolf at the University of Connecticut, School of Medicine, for reviewing the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101367.
Contributor Information
Xiaofei An, Email: anxiaofei2000@163.com.
Silvia Liu, Email: shl96@pitt.edu.
Dong Zhou, Email: dzhou@uchc.edu.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Saeedi P., Petersohn I., Salpea P., Malanda B., Karuranga S., Unwin N., et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the international diabetes federation diabetes atlas. Diabetes Research and Clinical Practice. 2019;157:107843. doi: 10.1016/j.diabres.2019.107843. 9(th) edition. [DOI] [PubMed] [Google Scholar]
- 2.Fu H., Liu S., Bastacky S.I., Wang X., Tian X.J., Zhou D. Diabetic kidney diseases revisited: a new perspective for a new era. Molecular Metabolism. 2019;30:250–263. doi: 10.1016/j.molmet.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reidy K., Kang H.M., Hostetter T., Susztak K. Molecular mechanisms of diabetic kidney disease. Journal of Clinical Investigation. 2014;124(6):2333–2340. doi: 10.1172/JCI72271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkins R.C., Zimmet P., Committee I-IWKDS Diabetes: diabetic kidney disease: act now or pay later. Nature Reviews Nephrology. 2010;6(3):134–136. doi: 10.1038/nrneph.2010.10. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y., Lee K., Ni Z., He J.C. Diabetic kidney disease: challenges, advances, and opportunities. Kidney Disease. 2020;6(4):215–225. doi: 10.1159/000506634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuttle K.R., Bakris G.L., Bilous R.W., Chiang J.L., de Boer I.H., Goldstein-Fuchs J., et al. Diabetic kidney disease: a report from an ADA Consensus Conference. Diabetes Care. 2014;37(10):2864–2883. doi: 10.2337/dc14-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alicic R.Z., Rooney M.T., Tuttle K.R. Diabetic kidney disease: challenges, progress, and possibilities. Clinical Journal of the American Society of Nephrology. 2017;12(12):2032–2045. doi: 10.2215/CJN.11491116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anders H.J., Huber T.B., Isermann B., Schiffer M. CKD in diabetes: diabetic kidney disease versus nondiabetic kidney disease. Nature Reviews Nephrology. 2018;14(6):361–377. doi: 10.1038/s41581-018-0001-y. [DOI] [PubMed] [Google Scholar]
- 9.Fiorentino M., Bolignano D., Tesar V., Pisano A., Biesen W.V., Tripepi G., et al. Renal biopsy in patients with diabetes: a pooled meta-analysis of 48 studies. Nephrology Dialysis Transplantation. 2017;32(1):97–110. doi: 10.1093/ndt/gfw070. [DOI] [PubMed] [Google Scholar]
- 10.Eddy S., Mariani L.H., Kretzler M. Integrated multi-omics approaches to improve classification of chronic kidney disease. Nature Reviews Nephrology. 2020;16(11):657–668. doi: 10.1038/s41581-020-0286-5. [DOI] [PubMed] [Google Scholar]
- 11.Komorowsky C.V., Brosius F.C., 3rd, Pennathur S., Kretzler M. Perspectives on systems biology applications in diabetic kidney disease. Journal of Cardiovascular Translational Research. 2012;5(4):491–508. doi: 10.1007/s12265-012-9382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubin R.F., Rhee E.P. Proteomics and metabolomics in kidney disease, including insights into etiology, treatment, and prevention. Clinical Journal of the American Society of Nephrology. 2020;15(3):404–411. doi: 10.2215/CJN.07420619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clish C.B. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harbor Molecular Case Studies. 2015;1(1):a000588. doi: 10.1101/mcs.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kidney Disease: Improving Global Outcomes Diabetes Work G. KDIGO 2020 clinical practice guideline for diabetes management in chronic kidney disease. Kidney International. 2020;98(4S):S1–S115. doi: 10.1016/j.kint.2020.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Wendel U., Even G., Langenbeck U., Schadewaldt P., Hummel W. Determination of (S)- and (R)-2-oxo-3-methylvaleric acid in plasma of patients with maple syrup urine disease. Clinica Chimica Acta. 1992;208(1–2):85–91. doi: 10.1016/0009-8981(92)90024-k. [DOI] [PubMed] [Google Scholar]
- 16.Ahlqvist E., Prasad R.B., Groop L. Subtypes of type 2 diabetes determined from clinical parameters. Diabetes. 2020;69(10):2086–2093. doi: 10.2337/dbi20-0001. [DOI] [PubMed] [Google Scholar]
- 17.Aitken J.P., Ortiz C., Morales-Bozo I., Rojas-Alcayaga G., Baeza M., Beltran C., et al. alpha-2-macroglobulin in saliva is associated with glycemic control in patients with type 2 diabetes mellitus. Disease Markers. 2015;2015:128653. doi: 10.1155/2015/128653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Driver T., Trivedi D.K., McIntosh O.A., Dean A.P., Goodacre R., Pittman J.K. Two glycerol-3-phosphate dehydrogenases from chlamydomonas have distinct roles in lipid metabolism. Plant Physiology. 2017;174(4):2083–2097. doi: 10.1104/pp.17.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pascual F., Carman G.M. Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochimica et Biophysica Acta. 2013;1831(3):514–522. doi: 10.1016/j.bbalip.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prentki M., Madiraju S.R. Glycerolipid metabolism and signaling in health and disease. Endocrine Reviews. 2008;29(6):647–676. doi: 10.1210/er.2008-0007. [DOI] [PubMed] [Google Scholar]
- 21.Thomas M.C., Brownlee M., Susztak K., Sharma K., Jandeleit-Dahm K.A., Zoungas S., et al. Diabetic kidney disease. Nature Review Disease Primers. 2015;1:15018. doi: 10.1038/nrdp.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haller H., Ito S., Izzo J.L., Jr., Januszewicz A., Katayama S., Menne J., et al. Olmesartan for the delay or prevention of microalbuminuria in type 2 diabetes. New England Journal of Medicine. 2011;364(10):907–917. doi: 10.1056/NEJMoa1007994. [DOI] [PubMed] [Google Scholar]
- 23.Espinel E., Agraz I., Ibernon M., Ramos N., Fort J., Seron D. Renal biopsy in type 2 diabetic patients. Journal of Clinical Medicine. 2015;4(5):998–1009. doi: 10.3390/jcm4050998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caramori M.L. Should all patients with diabetes have a kidney biopsy? Nephrology Dialysis Transplantation. 2017;32(1):3–5. doi: 10.1093/ndt/gfw389. [DOI] [PubMed] [Google Scholar]
- 25.Mann M., Kumar C., Zeng W.F., Strauss M.T. Artificial intelligence for proteomics and biomarker discovery. Cells and Systems. 2021;12(8):759–770. doi: 10.1016/j.cels.2021.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Zhang A., Sun H., Yan G., Wang P., Wang X. Metabolomics for biomarker discovery: moving to the clinic. BioMed Research International. 2015;2015:354671. doi: 10.1155/2015/354671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angel T.E., Aryal U.K., Hengel S.M., Baker E.S., Kelly R.T., Robinson E.W., et al. Mass spectrometry-based proteomics: existing capabilities and future directions. Chemical Society Reviews. 2012;41(10):3912–3928. doi: 10.1039/c2cs15331a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 29.Kell D.B., Oliver S.G. The metabolome 18 years on: a concept comes of age. Metabolomics. 2016;12(9):148. doi: 10.1007/s11306-016-1108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant Molecular Biology. 2002;48(1–2):155–171. [PubMed] [Google Scholar]
- 31.Rinschen M.M., Ivanisevic J., Giera M., Siuzdak G. Identification of bioactive metabolites using activity metabolomics. Nature Reviews Molecular Cell Biology. 2019;20(6):353–367. doi: 10.1038/s41580-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanchet L., Smolinska A., Attali A., Stoop M.P., Ampt K.A., van Aken H., et al. Fusion of metabolomics and proteomics data for biomarkers discovery: case study on the experimental autoimmune encephalomyelitis. BMC Bioinformatics. 2011;12:254. doi: 10.1186/1471-2105-12-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z.Z., Gerszten R.E. Metabolomics and proteomics in type 2 diabetes. Circulation Research. 2020;126(11):1613–1627. doi: 10.1161/CIRCRESAHA.120.315898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortiz A. Proteomics for clinical assessment of kidney disease. Proteomics - Clinical Applications. 2019;13(2) doi: 10.1002/prca.201900004. [DOI] [PubMed] [Google Scholar]
- 35.Tofte N., Persson F., Rossing P. Omics research in diabetic kidney disease: new biomarker dimensions and new understandings? Journal of Nephrology. 2020;33(5):931–948. doi: 10.1007/s40620-020-00759-4. [DOI] [PubMed] [Google Scholar]
- 36.Good D.M., Zurbig P., Argiles A., Bauer H.W., Behrens G., Coon J.J., et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Molecular & Cellular Proteomics. 2010;9(11):2424–2437. doi: 10.1074/mcp.M110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pontillo C., Mischak H. Urinary peptide-based classifier CKD273: towards clinical application in chronic kidney disease. Clinical Kidney Journal. 2017;10(2):192–201. doi: 10.1093/ckj/sfx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molin L., Seraglia R., Lapolla A., Ragazzi E., Gonzalez J., Vlahou A., et al. A comparison between MALDI-MS and CE-MS data for biomarker assessment in chronic kidney diseases. Journal of Proteomics. 2012;75(18):5888–5897. doi: 10.1016/j.jprot.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 39.Pontillo C., Jacobs L., Staessen J.A., Schanstra J.P., Rossing P., Heerspink H.J.L., et al. A urinary proteome-based classifier for the early detection of decline in glomerular filtration. Nephrology Dialysis Transplantation. 2017;32(9):1510–1516. doi: 10.1093/ndt/gfw239. [DOI] [PubMed] [Google Scholar]
- 40.Zurbig P., Mischak H., Menne J., Haller H. CKD273 enables efficient prediction of diabetic nephropathy in nonalbuminuric patients. Diabetes Care. 2019;42(1):e4–e5. doi: 10.2337/dc18-1322. [DOI] [PubMed] [Google Scholar]
- 41.Tofte N., Lindhardt M., Adamova K., Bakker S.J.L., Beige J., Beulens J.W.J., et al. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): a prospective observational study and embedded randomised placebo-controlled trial. Lancet Diabetes Endocrinology. 2020;8(4):301–312. doi: 10.1016/S2213-8587(20)30026-7. [DOI] [PubMed] [Google Scholar]
- 42.Yoshino S., Fujimoto K., Takada T., Kawamura S., Ogawa J., Kamata Y., et al. Molecular form and concentration of serum alpha2-macroglobulin in diabetes. Scientific Reports. 2019;9(1):12927. doi: 10.1038/s41598-019-49144-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wildenthal K. Inhibition by insulin of cardiac cathepsin D activity. Nature. 1973;243(5404):226–227. doi: 10.1038/243226a0. [DOI] [PubMed] [Google Scholar]
- 44.Hilfiker-Kleiner D., Kaminski K., Podewski E., Bonda T., Schaefer A., Sliwa K., et al. A cathepsin D-cleaved 16 kDa form of prolactin mediates postpartum cardiomyopathy. Cell. 2007;128(3):589–600. doi: 10.1016/j.cell.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 45.Cocchiaro P., De Pasquale V., Della Morte R., Tafuri S., Avallone L., Pizard A., et al. The multifaceted role of the lysosomal protease cathepsins in kidney disease. Frontiers Cell Development Biology. 2017;5:114. doi: 10.3389/fcell.2017.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koziolek M., Mueller G.A., Dihazi G.H., Jung K., Altubar C., Wallbach M., et al. Urine E-cadherin: a marker for early detection of kidney injury in diabetic patients. Journal of Clinical Medicine. 2020;9(3):639. doi: 10.3390/jcm9030639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niewczas M.A., Sirich T.L., Mathew A.V., Skupien J., Mohney R.P., Warram J.H., et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney International. 2014;85(5):1214–1224. doi: 10.1038/ki.2013.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beckerman P., Susztak K. Sweet debate: fructose versus glucose in diabetic kidney disease. Journal of the American Society of Nephrology. 2014;25(11):2386–2388. doi: 10.1681/ASN.2014050433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poesen R., Evenepoel P., Meijers B. The metabolomics grail: promising although not yet holy. Kidney International. 2015;87(4):864. doi: 10.1038/ki.2014.396. [DOI] [PubMed] [Google Scholar]
- 50.Critselis E., Vlahou A., Stel V.S., Morton R.L. Cost-effectiveness of screening type 2 diabetes patients for chronic kidney disease progression with the CKD273 urinary peptide classifier as compared to urinary albumin excretion. Nephrology Dialysis Transplantation. 2018;33(3):441–449. doi: 10.1093/ndt/gfx068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry data were deposited in MassIVE with the data set identifier MSV000087487.