Abstract
HES-1 is a Hairy-related basic helix-loop-helix protein with three evolutionarily conserved regions known to define its function as a transcription repressor. The basic region, helix-loop-helix domain, and WRPW motif have been characterized for their molecular function in DNA binding, dimer formation, and corepressor recruitment, respectively. In contrast, the function conferred by a fourth conserved region, the helix 3-helix 4 (H-3/4) domain, is not known. To better understand H-3/4 domain function, we expressed HES-1 variants under tetracycline-inducible control in PC12 cells. As expected, the induced expression of moderate levels of wild-type HES-1 in PC12 cells strongly inhibited nerve growth factor-induced differentiation. This repression was dependent on the H-3/4 domain. Unexpectedly, expression of HES-1 also arrested cell growth, an effect that could be reversed upon down regulation of HES-1. Concomitant with growth arrest, there was a strong reduction in bromodeoxyuridine incorporation and PCNA protein levels, although not in cyclin D1 expression. Expression of a HES-1 protein carrying the H-3/4 domain, but not the WRPW domain, still partially inhibited both proliferation and differentiation. Transcription assays in PC12 cells directly demonstrated that the H-3/4 domain can mediate DNA-binding-dependent transcription repression, even in the absence of corepressor recruitment by the WRPW motif. HES-1 expression strongly repressed transcription of the p21cip1 promoter, a cyclin–cyclin-dependent kinase inhibitor up regulated during NGF-induced differentiation, and the H-3/4 domain is necessary for this repression. Thus, the H-3/4 domain of HES-1 contributes to transcription repression independently of WRPW function, inhibits neurite formation, and facilitates two distinct and previously uncharacterized roles for HES-1: the inhibition of cell proliferation and the direct transcriptional repression of the NGF-induced gene, p21.
HES-1, the Hairy and Enhancer of split homologue 1 (19, 52), is a vertebrate member of a highly conserved family of Hairy-related basic helix-loop-helix (bHLH) proteins. Originally described in Drosophila melanogaster, Hairy-related proteins include Hairy (51), Deadpan (3), and the seven bHLH members of the Enhancer of split [E(Spl)] complex (14, 37). Members of this family are DNA-binding transcription repressors that antagonize the function of bHLH activators and repress neuronal development (reviewed in references 6, 21, 35, and 36). The Hairy-related proteins bind to specific DNA sites (class C sites or N-boxes) in target gene promoters by means of the conserved basic region (43, 44, 52, 56, 57, 59). The DNA-binding function of Hairy has been shown to be essential for the transcriptional repression of its downstream target, achaete, a proneural bHLH activator gene (44, 59). Transcriptional repression of target promoters is thought to occur at least partly by recruitment of a corepressor protein, Groucho, via the WRPW tetrapeptide motif conserved in the C terminus of all family members (24, 46, 61). Indeed, a fusion of the WRPW motif to the Gal-4 heterologous DNA-binding protein is sufficient by itself to repress transcription (22, 25). However, Hairy also binds to another corepressor, dCtBP (48, 65), suggesting that Hairy may have alternative repression functions in addition to the conserved Groucho recruitment mechanism. Additionally, some bHLH repressors do not share the requirement for intrinsic DNA-binding capability to repress neuronal development. A bHLH-deleted version of E(Spl) (m8) has been shown to repress neuronal development despite lacking intrinsic DNA-binding capability (24, 41, 43). Functional dissection of the E(Spl) protein in Drosophila highlighted the importance of the helix 3-helix 4 (H-3/4) domain (37) and the WRPW motif, as well as the intervening C-terminal region, for correct bristle development (24). The mechanism of repression did not appear to require the conserved basic and helix-loop-helix (HLH) regions. Moreover, while WRPW and H-3/4 deletions were generally neutral, a bHLH construct retaining just the H-3/4 region was dominant negative for bristle formation, suggesting a functional role for the H-3/4 domain.
The H-3/4 domain of Hairy (37), called the orange domain by Dawson and colleagues (13), was shown to be necessary for Hairy function in a sex determination assay in Drosophila. In the experiments of Dawson et al., the activity of the H-3/4 domain was dependent upon the presence of a DNA-binding bHLH region, but not the WRPW motif. Interestingly, a Hairy protein chimera with the H-3/4 domain replaced with the corresponding region from HES-1 retained function, whereas the corresponding E(Spl) m8 substitution was inactive. Thus, the H-3/4 domain appears to be important for function in both Hairy and E(Spl) proteins, though neither the underlying mechanism nor the basis for the apparent specificity of this region have been established.
The regulation of neuronal differentiation by HES-1 is analogous to the function of Hairy in several key respects. Like Hairy, HES-1 has been shown to be a DNA-binding transcription repressor (52, 55, 56) which recruits Groucho-related TLE family corepressors to DNA at specific sites (22, 54). Also, MASH-1, an achaete-scute homologue necessary for nervous system development (27), is transcriptionally regulated by HES-1 through a specific site (class C site) in the MASH-1 promoter (8), comparable to the transcriptional repression of achaete by Hairy (44, 59). Unlike Drosophila, where the loss of the bHLH repressor leads to additional neuronal cells, deletion of the HES-1 gene in mice results in a marked loss of neurons, apparently due to the premature differentiation of neuronal precursors (33). HES-1 has also been shown to block the transcription-activation and myogenic differentiation properties of the bHLH activator, MyoD (52). In vitro studies suggest that HES-1 interacts with the ubiquitous E2A proteins (E-proteins) E12 and E47, thereby disrupting the formation of MyoD–E-protein heterodimers. A similar inhibition of MASH-1 activity was also reported (52). The functional requirement for the H-3/4 domain in either DNA-binding-dependent or -independent repression by HES-1 has not been determined.
The linkage of bHLH repressors to cell fate specification and proliferation has been most clearly documented in the development of neuronal progenitor cells in the Drosophila peripheral nervous system (reviewed in reference 5). The differentiation of the sensory organ is promoted by bHLH activators and inhibited in the surrounding epithelial cells by the E(Spl) complex bHLH repressors, and it is dependent upon two rounds of additional cell division. The linkage between differentiation and cell cycle control is better established for the bHLH activators (10, 12, 40, 47, 66). Transcription factors such as myogenin (62) and NeuroD (38, 42) are known to coordinate the up regulation of differentiation-specific genes with exit from cell cycle. This is thought to result at least partially from the up regulation of the cyclin–cyclin-dependent kinase (CDK) inhibitor, p21cip1/WAF-1 (16, 30, 31, 40, 45, 63, 66). While it is not known if HES-1 coordinates any aspect of cell cycle control with the inhibition of differentiation, p21 could be a target for HES-1. In vivo, p21 is expressed predominantly in terminally differentiated neuronal cells, while HES-1 is expressed earlier in the neuronal precursors of the mitotically active ventricular zone (52). The p21 promoter contains multiple bHLH activator-binding sites (E-boxes) which have been shown to be functional in the up regulation of p21 (49). The Id HLH repressor protein (2), which lacks a basic region and forms non-DNA-binding heterodimers with bHLH activators (E-proteins), has also been shown to repress p21 expression (49). Similarly, HES-1 might also repress p21 transcription, either through E-protein interaction or by binding to DNA directly at specific sites.
For the analysis of H-3/4 domain function, we expressed wild-type HES-1 (WT HES-1) and several mutant forms of HES-1 in PC12 cells. PC12 cells are a rat pheochromocytoma cell line (26) that has been extensively studied in the analysis of HES-1 and neuronal differentiation (19, 55) as well as in the regulation of cell cycle by p21 (17, 50, 60, 64). We generated tetracycline-inducible stable cell lines and found that overexpression of WT HES-1-repressed nerve growth factor (NGF)-induced differentiation, as expected from previous studies (55), and that this repression was dependent upon the H-3/4 domain. Unexpectedly, we also found that overexpression of WT HES-1 also repressed proliferation. Repression of proliferation by WT HES-1 was also observed in transiently transfected neuroblastoma cells, and in colony-forming efficiency (CFE) assays in PC12 cells. Furthermore, we identified the promoter of the cyclin-CDK inhibitor, p21, as a direct target for HES-1-mediated transcriptional repression in the inducible PC12 cells, and this repression was also H-3/4 dependent. Transcription assays using the auto-regulated HES-1 promoter (56) and the p21 promoter showed that the H-3/4 domain conferred DNA-binding-dependent transcription repression function to HES-1 independently of the WRPW motif. Thus, the H-3/4 domain of HES-1 is an important component of HES-1-mediated transcription repression and the inhibition of differentiation and growth arrest.
MATERIALS AND METHODS
Expression vector construction.
The HES-1 expression plasmids used in the transient reporter assays and CFE assays were based on pCDNA3 (Invitrogen, Carlsbad, Calif.). We have previously described Flag epitope-tagged WT and basic region mutant HES-1 constructs (7). ΔR and ΔS HES-1 are deletions made by cutting the full-length construct at the most 5′ of the internal RsaI and SmaI sites, respectively and utilizing a stop codon present after the EcoRV 3′ cloning site of the pCDNA3 vector. The basic region mutation in a DNA-binding-defective mutant (B* HES-1) is from an equivalent construct previously named DN HES-1 (55), and B*ΔS HES-1 incorporates the same mutation into ΔS HES-1. The Δ3/4 HES-1 construct was made by partial RsaI and SmaI digestion to remove the H-3/4 coding region. The Gal-4 fusion proteins were generated from the full-length HES-1 pCDNA constructs. An RsaI/EarI-digested fragment was blunt-end cloned into the HindIII-digested pM vector, which contains the Gal-4 DNA-binding domain, to generate pM H3/4/C. An RsaI/SmaI-digested fragment was blunt-end cloned into the HindIII-cut pM vector to generate pM H3/4. To enable blunt-end cloning, the overhangs of the HindIII-digested pM vector were filled in with the Klenow fragment of DNA polymerase I prior to use. The construct pM C was made by first cloning the SmaI/EarI-blunted fragment of HES-1 into the EcoRV site of pCDNA3. The fragment was cut out with an EcoRI/NotI digest (sites in vector) and then blunt-end cloned into the BamHI site in pM, using Klenow to fill in the overhangs prior to ligation. The ΔbHLH expression constructs were derived from the Gal-4 fusion constructs. pM H3/4 was digested with SmaI/XbaI to release the fragment for H-3/4. A partial SmaI digest of XbaI-cut pM C was used to generate the SmaI/XbaI fragment for cloning HES C. HES 3/4/C was generated from the BamHI/XbaI fragment of pM H3/4/C. These fragments were cloned into the NotI (Klenow-filled)/XbaI sites of a pCDNA3 vector that contained the Flag epitope, followed by the nuclear localization signal derived from pVP16.
The tetracycline-inducible expression constructs were derived from pBI-EGFP (Clontech, Palo Alto, Calif.). BamHI (blunt)/XbaI-digested HES-1 fragments from the pCDNA3 constructs were cloned into the PvuII/NheI sites of the pBI vector. The ΔS and ΔR constructs utilize stop codons in the 3′ region of the pBI vector. The restriction enzymes and Klenow fragment of DNA polymerase I (for blunt-end cloning) were purchased from New England Biolabs (Beverly, Mass.) and Promega (Madison, Wis.) and were used according to the manufacturers' instructions. All of the constructs were verified, either in full or across the cloning junctions, by sequencing performed at the Cornell central sequencing facility (Cornell University, Ithaca, N.Y.). The details of the constructs and cloning procedures are available upon request.
Cell maintenance.
PC12 and tetracycline-inducible PC12 cells were maintained in Dulbecco minimum essential medium (Mediatech, Herndon, Va.) with 10% horse serum antibiotic (Gemini Bio-products, Calabasas, Calif.) and 5% fetal calf serum antibiotic (Gemini Bio-products), P-Gent antibiotic (Gemini Bio-products), and Glutamax (Life Technologies, Gaithersburg, Md.) in 10% CO2 in a humidified atmosphere at 37°C. Fungizone (Life Technologies) was added to the media during selection of the stable cell lines. PC12 tetracycline-inducible cells were maintained in the presence of 2 mg of tetracycline per ml and 100 μg of G418 per ml (Gemini Bio-products). Hygromycin B (100 μg/ml) (Life Technologies) was used to maintain selection of the cells following stable transfection with the pBI expression vector. Unless otherwise indicated, 2.5S murine NGF (Promega) was added at 100 ng/ml, human recombinant bFGF (Promega) was added at 10 ng/ml, and retinoic acid (Sigma, St. Louis, Mo.) was added to a 1 μM concentration.
Neuroblastoma cell lines were maintained in Dulbecco minimum essential medium with 15% fetal calf serum (Gemini Bio-products) plus nonessential amino acids (Life Technologies), P-Gent antibiotic (Gemini Bio-products), and Glutamax (Life Technologies) at 37°C and 10% CO2 in a humidified atmosphere.
Stable cell line generation.
The PC12 cells used to produce the stable cell lines were purchased from Clontech and are stably transfected with the tetracycline-sensitive activator protein under neomycin resistance. The PC12 cells were plated in 60-mm-diameter dishes overnight and were then transfected with 1 μg of the pBI vector (empty vector and WT, B*, and ΔS and ΔR HES-1 versions) and 0.1 μg of a Hygromycin B resistance vector using Lipofectamine Plus reagent (Life Technologies) according to the manufacturer's instructions. The cells were allowed to grow for 48 h and were then passaged to 150-mm-diameter plates in the presence of 200 μg of Hygromycin B (Life Technologies) for selection. Twenty-four colonies from each transfection were picked and plated in duplicate, either with or without 2 μg of tetracycline in the media. Clones that in the absence of tetracycline were observed to have green fluorescence under UV light (e.g., expressed enhanced green fluorescent protein [EGFP]) were identified. The corresponding uninduced clone was further analyzed for expression of the Flag epitope-tagged protein by Western analysis. Approximately 33% of the colonies selected had detectable EGFP after 48 h of induction, and all of those clones tested had detectable HES-1 or HES-1 mutant expression using anti-Flag Western analysis. Additional WT HES-1 and control cell lines were generated from a second transfection, and the cells generated were not distinguishable from the first transfection.
Growth rates were determined for the WT and control inducible PC12 cells following 3 days of maintenance in the presence or absence of 2 μg of tetracycline per ml. The cells were trypsinized and counted, and equal numbers of cells (105) were plated into 12-well dishes in duplicate. A replicate plate was made for each time point (3, 5, 7, and 9 days), at which time a plate was removed and the cells were trypsinized and recounted using a hemocytometer. The data is for the average of two independent experiments in duplicate carried out with two control and two WT HES-1 cell lines. The error is the standard deviation of the mean for the four determinations.
NGF response was determined by using cells grown at low density on collagen (rat tail type I; Sigma)-coated 100-mm-diameter dishes with or without 2 μg of tetracycline for 3 days prior to treatment with 50 ng of NGF per ml for 6 days. The percentage of cells with neurites (over two cell diameters long, with growth cone) was determined from three random fields of view (minimum, 100 cells per field) per plate. The ratio of neuritic cells in the induced state compared to the uninduced state was given as a percentage. The data is for two experiments carried out in duplicate with two cell lines for each cell type. The error is the standard deviation of the mean.
Transient transfections.
Transient transfections for the hHES p21 and Gal-4–upstream activation sequence (UAS) reporter assays were carried out in six-well tissue culture dishes in duplicate by using the Lipofectamine Plus reagent (Life Technologies) according to the manufacturer's instructions. The hHES reporter was made from the ∼1.6-kb BamHI fragment of the genomic DNA, a gift from John Feder (Mercator Genetics, Menlo Park, Calif.), fused to the luciferase gene in PGL2 basic (Promega). The PC12 cells were plated to a density of approximately 50% 24 h prior to transfection. For each transfection, 0.5 μg of the hHES reporter was transfected with 1 μg of pCDNA3 β-galactosidase (β-Gal) (internal transfection efficiency control) and 1 μg of either pCDNA3 or a pCDNA3 HES-1 construct. The p21-luciferase (p21-luc) construct was obtained from X. H. Sun (New York University, New York) and has been previously described (49). For studies of the p21 promoter, 0.25 μg of p21-luc was used with 1 μg of pCDNA3 β-Gal and either 1 μg of pCDNA3 or 0.5 mg of both pCDNA3 MASH-1 (7) and pCDNA3 E47 (a gift from Robert Benezra, Sloan-Kettering Institute, New York, N.Y.). Additionally, either 1 μg of pCDNA3 or a pCDNA3 HES-1 construct was included. For the Gal-4 assay, 1 μg each of a pM vector and the luciferase reporter with five UASΔ (22) were used per transfection. Transfections were carried out for 5 h in the presence of the lipofectamine (without serum) after which the media was replaced with fresh media (with serum), and the cells were incubated for a total of 72 h prior to assaying. The luciferase assay procedure was carried out as previously detailed (44). The data for the p21-luc and hHES-luc transfections is the average of three to five independent experiments carried out in duplicate, the data for the Gal-4–UAS experiment is for two independent experiments in duplicate. The luciferase data is corrected for β-Gal activity. The errors are the standard deviations of the means.
CFE assays.
PC12 and neuroblastoma cells plated at approximately 50% density in six-well dishes were transfected with 1 μg of either pCDNA3 or a pCDNA3 HES-1 construct. After 48 h, the cells were passaged to 100-mm-diameter dishes and selected with 400 μg of G418 per ml. Care was taken to ensure that colonies were not rinsed off or disrupted during media changes. After approximately 30 days, the plates were rinsed in phosphate-buffered saline (PBS), and the colonies were fixed and stained with 50% methanol, 10% acetic acid, and 0.05% Coomassie brilliant blue (Sigma). The colonies were counted, and the CFE was determined by dividing the number of colonies on each plate by the number obtained with the control transfection (pCDNA3 vector). The PC12 data represents the average CFE from seven (control, WT, and ΔR and ΔS HES-1), four (B* HES-1), or two (Δ3/4 HES-1) independent transfections; the error is the standard deviation of the mean. The neuroblastoma cell data is from a single experiment, representative of at least three independent transfections of the cell lines.
Western analysis.
Western analysis was performed as previously described (7). Anti-Flag monoclonal M5 antibody (Kodak, Rochester, N.Y.) was used at a 1:1,000 dilution. Anti-HES-1 N-terminal polyclonal antibody, a gift from John Feder (Mercator Genetics), has been previously described (55) and was used at a 1:1,000 dilution. Anti-p21 and anti-cyclin D1 monoclonal antibodies are from Santa Cruz (Santa Cruz, Calif.) and were used at a 1:200 dilution. Anti-PCNA was obtained from Novocastra Laboratories (Newcastle upon Tyne, United Kingdom) and was used at a 1:250 dilution. Secondary goat anti-mouse or anti-rabbit horseradish peroxidase-conjugated immunoglobulin G was purchased from Jackson Immunoresearch (West Grove, Pa.) and used at a 1:20,000 dilution. Antibodies were detected by using Pierce (Rockford, Ill.) supersignal chemiluminescent reagent on Kodak Biomax MR film. The data shown is from exposures in the linear range of detection.
Immunocytochemistry.
Anti-Flag immunocytochemistry was performed as previously described (7). The anti-Flag antibody was used at a 1:2,500 dilution, the biotin-conjugated goat anti-mouse secondary antibody (Jackson Immunoresearch) was diluted to 1:300, and the CY3-conjugated streptavidin (Jackson Immunoresearch) was diluted to 1:1,000.
Bromodeoxyuridine (BrdU) immunocytochemistry was performed on cells exposed to 50 μM BrdU for 20 h then fixed in 4% paraformaldehyde for 20 min at room temperature. Endogenous peroxidase activity was quenched with 0.01% hydrogen peroxide. The DNA was denatured in 2 M HCl then neutralized in 0.1 M Tris, pH 8.5. The cells were blocked in PBS with 5% normal goat serum and 0.1% Triton X-100 for 1 h at room temperature. Monoclonal anti-BrdU antibody (Becton Dickinson, Franklin Lakes, N.J.) was added at a 1:10 dilution in PBS with 1 mg of bovine serum albumin at 4°C overnight. Secondary goat anti-mouse horseradish peroxidase-conjugated antibody (Jackson Immunoresearch) was used at a 1:1,000 dilution, and the antibody was detected with a solution containing 2 mg of DAB, 0.02% (wt/vol) hydrogen peroxide, and 0.3% (wt/vol) NiSO4. BrdU incorporation experiments were carried out three times in duplicate, for three lines each of control, WT, and ΔR HES-1 cells. Representative fields of cells were counted, and the number of cells with BrdU staining was determined as a percentage of the total. The data is the average of all the experiments for each line, and the error given is the standard deviation of the mean.
RESULTS
Inducible expression of WT HES-1 and HES-1 deletion proteins in PC12 cells.
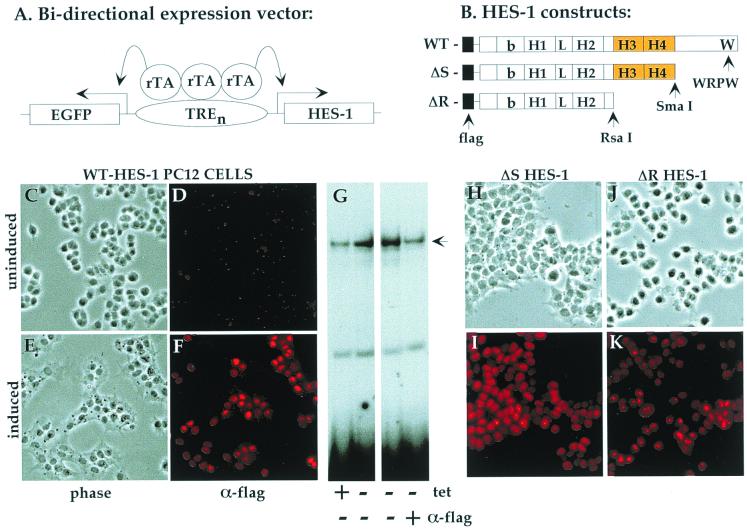
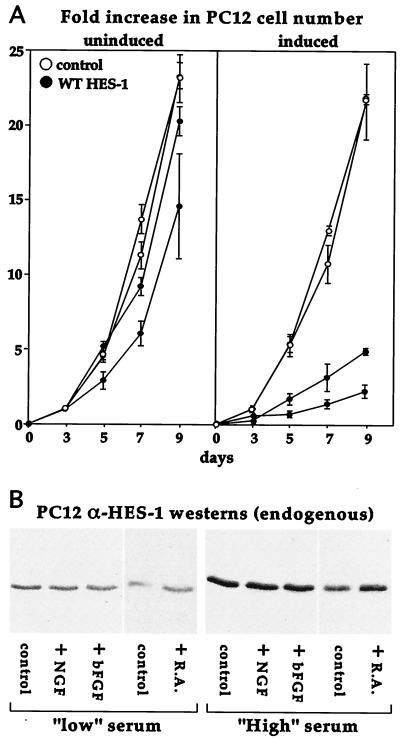
To investigate the function of the HES-1 H-3/4 domain, we generated stable PC12 cell lines that expressed Flag epitope-tagged WT HES-1 or HES-1 deletion mutants under tetracycline-inducible control (tet-off system; Clontech). We used a bidirectional expression vector (pBI-EGFP; Clontech) that coexpressed EGFP (Clontech) from the same promoter that drove expression of HES-1 (Fig. 1A). Upon withdrawal of tetracycline, the coinduction of HES-1 proteins could be monitored in the living cells by means of green fluorescence under UV light. This enabled the efficient screening of the stable clones we isolated, as only the clones that flouresced upon induction were further analyzed for exogenous HES-1 protein expression. Induction of nuclear-expressed HES-1 protein was verified by anti-Flag immunocytochemistry of the WT HES-1-inducible PC12 cells (Fig. 1F versus D). From Western blot analysis, by using anti-Flag and anti-HES-1 antibodies, we estimate that WT HES-1 levels are increased three- to fivefold over endogenous levels following 3 days of induction (see Fig. 4F and 7 for representative clones). The induction of HES-1 had a noticeable effect upon cell morphology in the absence of NGF, resulting in a noticeable flattening of the cells and an increased adhesion to the tissue culture dish (compare Fig. 1C to 1E and 2F). However, the induction of HES-1 had no apparent effect on cell viability, and the flattening was reversed upon readdition of tetracycline (data not shown). Therefore, the tetracycline-inducible PC12 cells are a useful system for examining the effects of induced expression of exogenous HES-1 protein.
FIG. 1.
HES-1-inducible expression in PC12 cells. (A) Schematic of the tetracycline-inducible bidirectional promoter used to coexpress EGFP and HES-1 variants. (B) Schematic of the WT, ΔS, and ΔR HES-1 constructs inducibly expressed in the stable PC12 cell lines. (C to F) Uninduced WT HES-1 cells, maintained in the presence of 2 μg of tetracycline per ml exhibited a normal morphology (C), and little or no Flag epitope was detected by anti-Flag immunocytochemistry (D). Withdrawal of tetracycline-induced Flag–HES-1 expression resulted in a flattened morphology in all eight cell lines examined (as shown for a representative clone in panel E) and localization of the exogenous HES-1 in the nuclei of the cells (F). A gel retardation assay (G) of WT HES-1-inducible PC12 cell nuclear extracts showed an increase in HES-1-specific DNA binding upon induction (compare lanes 2 and 1). The presence of exogenous HES-1 in the retarded complex was confirmed by the addition of anti-Flag antibodies to the binding reaction, which disrupted the binding of Flag-tagged protein (panel G, lane 4). Nuclear-localized expression of the ΔS HES-1 (H and I) and ΔR HES-1 (J and K) proteins was also determined by anti-Flag immunocytochemistry of the induced cells (I and K).
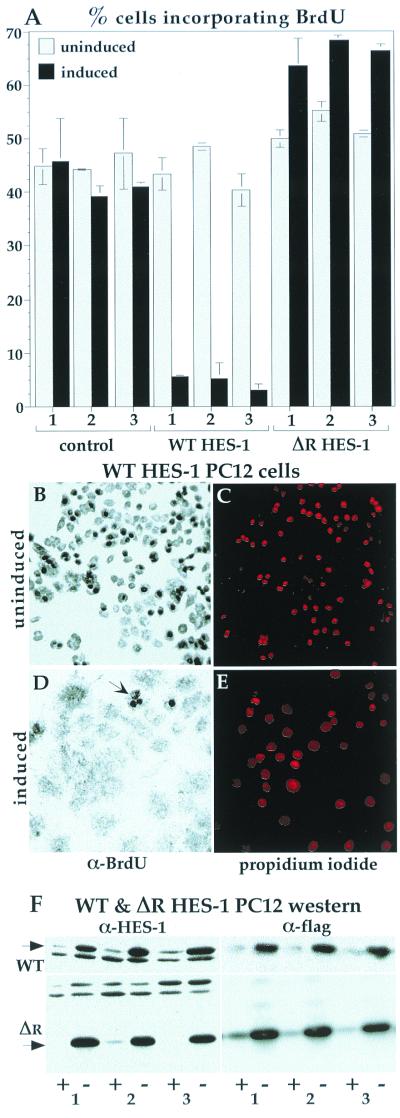
FIG. 4.
The inhibition of proliferation in HES-1-overexpressing cells was confirmed by BrdU incorporation. After 20 h of exposure to BrdU, about 40% of control cell lines had incorporated the BrdU nucleoside analogue into their DNA, as detected by anti-BrdU immunocytochemistry. This fraction of incorporation is consistent with the doubling time for this cell type of about 48 h. The level of incorporation for the three control lines was similar with or without induction of EGFP (A). The incorporation of BrdU into uninduced WT HES-1 PC12 cells was similar to control values. Upon induction of HES-1 (3 days prior to BrdU treatment), the level of BrdU incorporation dropped dramatically to about 5%, consistent with either a lower growth rate or a lower fraction of cells in S phase, and a lack of DNA synthesis (implied by the loss of PCNA). In contrast, induction of ΔR HES-1, a deletion mutant that lacks transcription repression activity (Fig. 7; discussed below) did not lower BrdU incorporation. Examples of the BrdU staining is shown for the WT HES-1 cells, both uninduced (B) and induced (D). Anti-BrdU staining is clearly seen as dark nuclei, which are mostly absent in the induced cells. The WT HES-1-induced cells that incorporated BrdU tended to have a rounded morphology (panel D, arrow), indicating either that they were in S phase or that they may have been low, or nonexpressing, subpopulations. The positions of the nuclei are shown by propidium iodide staining, which was partially obscured in the cells with high levels of BrdU incorporation due to the opacity of the DAB precipitate (C and E). (F) The induction of WT (F, upper panels) and ΔR HES-1 protein (F, lower panels) is shown by the anti-HES-1 and anti-Flag Western blots for three independent cell lines.
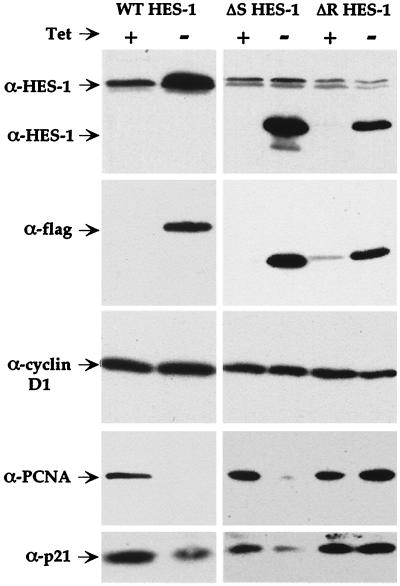
FIG. 7.
The H-3/4 domain of HES-1 mediates the repression of the cell cycle proteins PCNA and p21. Expression of induced, Flag-tagged proteins in the WT, ΔS, and ΔR HES-1 PC12 cell lines was verified by anti-HES-1 and anti-Flag Western analysis (upper two panels). Induction of both WT and ΔS HES-1 reduced the level of PCNA and p21 proteins, whereas induction of the ΔR HES-1 protein had no significant effect on either PCNA or p21 (lower 2 panels). Neither expression of WT HES-1 nor the ΔS and ΔR HES-1 mutant proteins affected the level of cyclin D1 (middle panel).
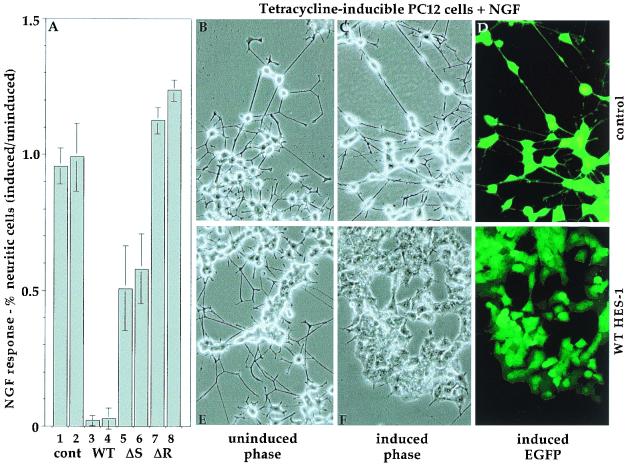
FIG. 2.
The H-3/4 domain of HES-1 partially mediates the inhibition of NGF response. The NGF response of tetracycline-inducible PC12 cells is graphed as the percentage of induced (without tetracycline) response divided by the percentage of uninduced (with tetracycline) response following 6 days of treatment with NGF (A). The NGF response for two control cell lines is unaffected by EGFP induction (columns 1 and 2). In contrast, induction of WT HES-1 greatly inhibits NGF response (columns 3 and 4). Expression of ΔS HES-1, which contains the H-3/4 domain but lacks the WRPW motif, significantly inhibits NGF response (columns 5 and 6). Expression of ΔR HES-1, which lacks both the H-3/4 and WRPW regions, does not inhibit NGF response (columns 7 and 8). Control PC12 cells, which expressed only EGFP upon induction, differentiated into a neuronal phenotype following 7 days of treatment with NGF (B to D). Induction of EGFP, seen by fluorescence microscopy in panel D, did not affect neurite outgrowth response to NGF (C). Uninduced WT HES-1 cells (E) had a NGF response similar to that of control cells (B). Upon induction of HES-1 by the removal of tetracycline, the cells no longer responded even after 7 days of NGF treatment (F and G) and have the flattened morphology of the induced cells seen in Fig. 1E. If the induced WT HES-1 cells were subsequently uninduced and then reexposed to NGF, they regained NGF-induced neurite response (data not shown).
Gel retardation analysis revealed an increase in class C (repressor specific) DNA-binding activity upon induction of HES-1 expression (Fig. 1G, compare lanes 1 and 2). From Western blot analysis, by using anti-Flag and anti-HES-1 antibodies, we estimate that WT HES-1 levels are increased three- to fivefold over endogenous levels following 3 days of induction (see Fig. 4F and 7 for representative clones). The presence of exogenous HES-1 in the DNA-binding complex was verified by disruption of DNA binding following the addition of anti-Flag antibodies to the binding reaction (Fig. 1G, lanes 3 and 4). Anti-HES-1 antibodies directed against the N-terminal region of HES-1 have been observed to similarly disrupt endogenous HES-1 DNA binding (8; our unpublished data).
In addition to WT HES-1, we also generated cell lines expressing the deletion mutants ΔS HES-1 and ΔR HES-1 (Fig. 1B). ΔS HES-1 is a deletion of HES-1 from a SmaI restriction site 3′ to the H-3/4 domain that removes the known repression motif WRPW. ΔR HES-1 is a truncation of HES-1 from an RsaI site that deletes the C-terminal region from the start of the H-3/4 domain, producing essentially a bHLH-only protein construct. Therefore, the contribution of both the WRPW-containing region and the H-3/4 domain to HES-1 function could be analyzed in the inducible PC12 cell system. Expression and nuclear localization of the ΔS HES-1 and ΔR HES-1 proteins were verified by anti-Flag immunocytochemistry of induced cells (Fig. 1H to K) and by Western analysis (see Fig. 7).
The H-3/4 domain contributes to HES-1-mediated inhibition of NGF response in PC12 cells.
We have previously shown that transient and low-level constitutive expression of WT HES-1 inhibits the NGF-induced differentiation of PC12 cells (55). Accordingly, WT HES-1 expression in the inducible cell lines also inhibited the NGF-dependent neurite response. Uninduced WT HES-1 cells respond to NGF by extending neurites (Fig. 2E) indistinguishable from those in control cells (Fig. 2A, bars 1 and 2, and B). Upon the withdrawal of tetracycline and induction of WT HES-1 (as indicated by EGFP expression) (Fig. 2G), the cells no longer become neuritic in the presence of NGF (Fig. 2A, lanes 3 and 4, and 2F and G). In contrast, tetracycline induction of EGFP in control cells had no effect on NGF response (Fig. 2A, lanes 1 and 2, B, and C).
To test the requirement of the WRPW-containing and H-3/4 regions for HES-1-mediated inhibition of NGF response, we compared the response of WT, ΔS and ΔR HES-1, and control tetracycline-inducible PC12 cells to 50 ng of NGF per ml for 6 days, both in the presence and absence of 2 μg of tetracycline per ml. The fold change in NGF response (neurite outgrowth) upon tetracycline induction is shown for two lines of each cell type (Fig. 2A). Induction of EGFP alone has essentially no effect on the rate of neurite outgrowth in the two lines of control cells, whereas neurite outgrowth is almost completely inhibited by induction of WT HES-1 (compare Fig. 2A lanes 1 and 2 to lanes 3 and 4). Induction of ΔR HES-1, which lacks both the WRPW-containing and H-3/4 regions, does not inhibit NGF-induced neurite formation and, indeed, may slightly potentiate neurite outgrowth (Fig. 2A, lanes 7 and 8). Induction of the ΔS HES-1 protein, which contains the H-3/4 domain but not the WRPW motif, partially inhibits neurite formation (Fig. 2A, lanes 5 and 6). Thus, the H-3/4 domain of HES-1 confers partial repression of neurite outgrowth even in the absence of the WRPW motif, defining the H-3/4 domain as an effector of HES-1 function.
HES-1 induction inhibits proliferation in PC12 cells.
We unexpectedly found that the induction of moderate levels of WT HES-1 in the inducible cells strongly inhibited proliferation. We quantified this in a proliferation assay: Fig. 3A shows the rate of proliferation for two WT HES-1 clones and two control clones (empty vector, EGFP only), with and without induction. The uninduced WT HES-1 cells had a rate of proliferation similar to uninduced control cells. Induction of EGFP in control cells had no significant effect upon proliferation. In marked contrast, the induction of HES-1 greatly lowered the rate of proliferation of the cells.
FIG. 3.
Induction of WT HES-1 inhibits PC12 cell proliferation. (A) Two lines of control and two lines of WT HES-1 PC12 cells were maintained either with or without tetracycline for 3 days, then equal numbers of cells were passaged in triplicate to measure proliferation. The cells were maintained up to 9 additional days (with or without tetracycline), and a set of plates for each cell type was counted at the 3-, 5-, 7-, and 9-day time points. The experiment was repeated, and the data for both experiments is shown in the graph as the average fold-increase in cell number with the error as the standard deviation (A). The growth of uninduced WT HES-1 cells was slightly lower than control lines, perhaps due to leakage of the exogenous HES-1 (A, left panel). In contrast, the growth of the WT HES-1 cells was markedly lower upon induction of HES-1 (A, right panel). Induction of EGFP did not greatly affect the growth of control cells. The rate of proliferation of the WT HES-1 cells may be overestimated in the induced panel, since prolonged induction could select for the lower-inducing, faster-growing cells. (B) Western analysis of endogenous HES-1 in parental PC12 cells showed that HES-1 was induced by serum in the media (B, compare the panels of low-serum-level-maintained cells to the panels of normal (high)-serum-level-maintained cells). Addition of the differentiation agents NGF or basic FGF did not appear to affect HES-1 levels in either serum condition. In contrast, retinoic acid (R.A.), which halts PC12 cell proliferation but does not differentiate the cells, did raise HES-1 protein levels slightly.
HES-1 mRNA is transiently induced by a number of growth factors, including NGF, fibroblast growth factor (FGF), and, to a lesser extent, epidermal growth factor (19). To establish the relationship between these factors and endogenous HES-1 in the regulation of proliferation, we examined the level of HES-1 following exposure to growth factors in standard and low-serum conditions. We noted that HES-1 levels respond to the level of serum in the culture media (Fig. 3B). The level of HES-1 protein increased when the cells were grown in a normal (high)-serum environment (10% horse serum, 5% fetal bovine serum) (Fig. 3B, right panel) compared to a low serum environment (1/10 normal level) (Fig. 3B, left panel). In contrast, the addition of the differentiating agents NGF (100 ng/ml) or basic FGF (10 ng/ml) to the media for 3 days had no apparent effect on HES-1 expression in either serum condition. However, the addition of retinoic acid, which halts proliferation without inducing differentiation in PC12 cells (53), resulted in a modest increase in HES-1 levels.
To further investigate the growth-suppressing properties of HES-1, we examined the extent of BrdU uptake by inducible WT HES-1 and control PC12 cells. The cells were grown for 3 days, either with or without tetracycline, were supplemented with BrdU for 20 h, and were then fixed. BrdU incorporation was detected by using an anti-BrdU antibody and visualized by DAB staining of the fixed cells. Three control lines had equivalent BrdU uptake (expressed as the percentage of cells that were BrdU positive) (Fig. 4A), whether induced or uninduced. The uninduced WT HES-1 cell lines had a degree of BrdU uptake similar to that of control cells, but BrdU incorporation was greatly reduced upon induction of HES-1 (Fig. 4A). The loss of BrdU incorporation is clearly seen by comparison of the photographs of the WT HES-1 cells, with and without induction (Fig. 4B versus D). The positions of the nuclei are indicated by propidium iodide staining (Fig. 4C and E). The nuclear staining reveals that the nuclei also increased in area following HES-1 induction, a change in morphology that reflected the induced flattening of the cell bodies. The level of exogenous HES-1 protein induced for the three cell lines can be seen in the anti-HES-1 and anti-Flag Western blots (Fig. 4F, top panels). These experiments clearly demonstrate that the overexpression of HES-1 results in a dramatic inhibition of DNA synthesis, consistent with the concomitant loss of PCNA expression (shown in Fig. 7) and a halt in cell cycle at G1.
The H-3/4 domain mediates HES-1-induced growth inhibition.
In contrast to WT HES-1, PC12 cells that expressed the deletion-mutant ΔR HES-1 were not inhibited in proliferation. ΔR HES-1 is deleted C-terminal to the HLH region, and the removal of the WRPW motif in ΔR HES-1 was expected to abolish transcription repression function, as well as any H-3/4 domain function. Indeed, analysis of three independent lines of tetracycline-inducible ΔR HES-1 expressing cells (Fig. 4F) showed a modest increase in BrdU uptake upon induction (Fig. 4A). In addition, induction of ΔR HES-1 resulted in a more rounded cell morphology (data not shown) and a decreased adhesion to the tissue culture plastic surface. This is a phenotype apparently opposite to that resulting from WT HES-1 induction. Analysis of growth rates was complicated by the tendency of the ΔR HES-1-induced cells to detach from the dish and grow as clumps (data not shown). The BrdU incorporation data, however, clearly indicated that the inhibition of proliferation was dependent upon the region of HES-1 C terminal to the HLH domain, which contains the H-3/4 domain and also the WRPW motif.
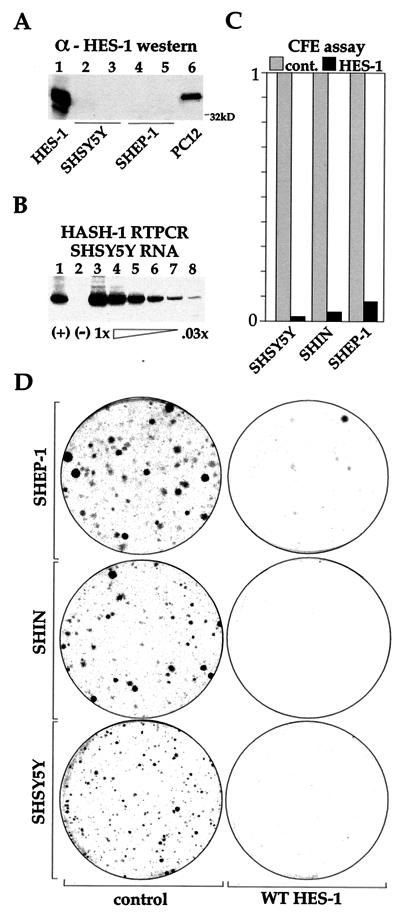
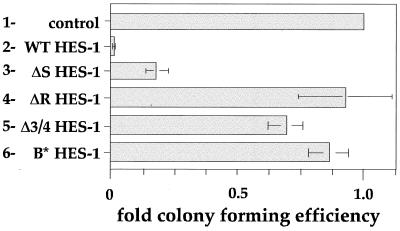
Initial experiments to establish the role of the H-3/4 domain in regulating proliferation rates in ΔS HES-1- and ΔR HES-1-expressing cells were complicated by the very different adhesion properties of the cell lines, which created plating and counting inconsistencies (data not shown). Therefore, we performed CFE assays instead, which allowed for a more direct comparison of the growth-inhibiting properties of the various HES-1 constructs. Because individual transfected colonies contained only a few hundred cells, the local growth environment would be similar in all transfections, and proliferation effects could be readily identified. PC12 cells were transfected with the HES-1 vector containing the neomycin resistance gene and allowed to grow in G418 selection media until colonies were clearly visible (see Materials and Methods) (for examples of CFE assay plates, see Fig. 6). The numbers of colonies formed by populations either expressing HES-1 or HES-1 mutant proteins were compared to the number of colonies in the empty vector control plates (normalized as 1.0). The CFE of WT HES-1 (Fig. 5, bar 2) was substantially lower than the CFE for the control (empty vector) (Fig. 5, bar 1), supporting the finding of lower growth rates and reduced BrdU incorporation in WT HES-1-overexpressing cells (Fig. 3 and 4). This result could explain the documented difficulty in generating stable clones with high levels of HES-1 expression (34, 55). Furthermore, we were unable to detect any WT HES-1 by Western analysis (of the Flag epitope) from the small number of clones that were present in duplicate transfections (data not shown). In contrast to WT expression, expression of the ΔR HES-1 mutant did not affect the ability to generate clones in PC12 cells (Fig. 5, bar 4). This result is consistent with the normal or slightly raised incorporation of BrdU seen in the ΔR HES-1 stable cell lines (Fig. 4). However, the ΔS construct, which contains the H-3/4 domain, had a significantly reduced CFE (Fig. 5, bar 3). Expression of Δ3/4 HES-1, which has the H-3/4 domain internally deleted (see Fig. 8A for structure) did not significantly reduce the CFE, a surprising result considering that the protein retained the WRPW motif. Together, the CFE data for the ΔS, ΔR, and Δ3/4 HES-1 constructs strongly suggest that the H-3/4 domain mediates the growth arrest function of HES-1. The ability to bind to DNA would also appear to be important to HES-1 function in the inhibition of proliferation, since expression of B* HES-1 only slightly lowered the CFE (Fig. 5, bar 6). Thus, growth inhibition is dependent upon the DNA-binding function and the presence of the H-3/4 domain in the proteins being expressed.
FIG. 6.
WT HES-1 inhibits proliferation in neuroblastoma cell lines. Three neuroblastoma cell lines (SHSY5Y, SHEP1, and SHIN) were tested for CFE following transfection with WT HES-1. These cell lines do not have significant expression of HES-1 protein, as shown by Western analysis of two of the lines in panel A. Transiently expressed HES-1 (lane 1) and endogenous PC12 cell (lane 6) lysates were run as positive controls for the detection of HES-1 protein. Lanes 3 and 5 are from lysates of NGF-treated cells. As might be anticipated by the lack of HES-1, expression of the MASH-1 gene can be detected in these cells, shown by reverse transcription-PCR of the SHSY5Y line in panel B. The (+) lane (lane 1) is a cDNA-positive control, the (−) lane (lane 2) is a reverse-transcriptase-negative control, and lanes 3 through 8 are serial dilutions of the input reverse transcription reaction for PCR amplification. MASH-1 mRNA has also been detected by reverse transcription-PCR in the SHEP and SHIN cell lines, as well as in LAI 5S, LAI 55N, and BEI YC neuroblastoma cell lines (data not shown). The greatly reduced colony formation of the HES-1-transfected cell lines is shown in panel C, and representative plates with Coomassie blue-stained cell colonies are shown in panel D. The near absence of colonies expressing WT HES shows that the inhibition of cell proliferation is a general property of WT HES-1 overexpression.
FIG. 5.
The H-3/4 domain mediates the inhibition of proliferation by HES-1. To determine if the growth inhibition phenotype resulting from WT HES-1 overexpression was a function of the H-3/4 domain, we performed CFE assays in PC12 cells using expression vectors encoding WT, ΔR, ΔS, Δ3/4, and B∗ HES-1 proteins. The empty expression vector (pCDNA3) was used as the control. The cells were grown in the presence of 200 μg of G418 per ml to select cells containing the expression construct, which has a neomycin resistance gene. The number of colonies present after 1 month of growth was determined and normalized to the control value (1.0). Six separate transfections with the same amount of vector were performed for all except Δ3/4 HES-1, which was transfected twice.
FIG. 8.
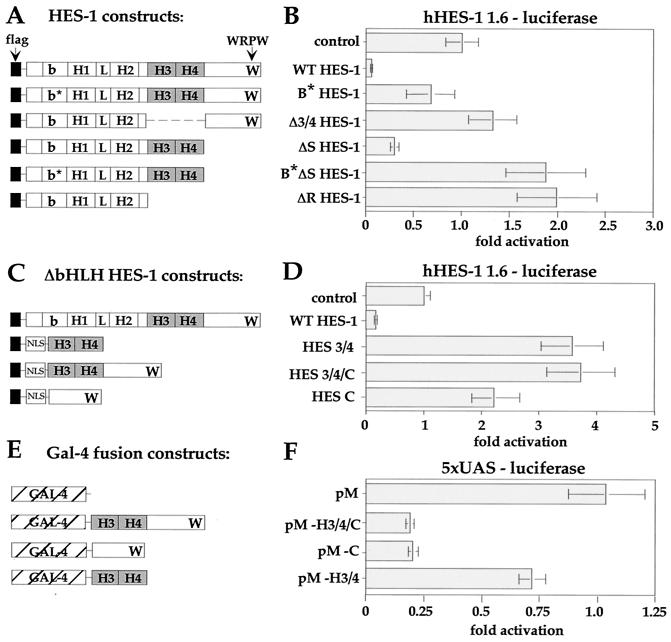
Transcriptional analysis of HES-1. The hHES-1 promoter was incorporated upstream of a luciferase reporter gene (hHES-luc) in order to analyze the regulation of the hHES-1 gene by HES-1 in PC12 cells. The luciferase activity of the reporter was corrected for the level of β-Gal activity from a cotransfected internal control plasmid and expressed as fold activity, with the control value normalized as 1.0. (A and B) The function of the H-3/4 domain in HES-1-mediated repression. WT HES-1 strongly repressed transcription from the hHES-1 promoter compared to the control transfected with empty expression vector. In comparison, loss of DNA binding due to mutation of the basic region (B∗ HES-1) resulted in essentially no transcription repression. Furthermore, removal of the H-3/4 domain in Δ 3/4 HES-1 also resulted in a loss of repression activity of the protein, despite the presence of the WRPW motif in the DNA-binding-competent protein. Indeed, HES-1 with the H-3/4 domain (ΔS HES-1), but not the rest of the C terminus (including WRPW), was functional as a modest repressor. The repression activity of this protein was also impaired by the inability to bind to DNA (ΔB∗S HES-1), and instead functioned as a weak activator, or derepressor, of the promoter. Similarly, ΔR HES-1, which lacks both the H-3/4 and the WRPW structures, was also a modest functional activator. (C and D) The role of DNA binding in HES-1-mediated repression. Comparison of the transcription activity of ΔbHLH HES-1 constructs to WT HES-1. Expression of the H-3/4 domain alone (HES 3/4), as well as the WRPW-containing constructs (HES 3/4/C and HES C) with a nuclear-localization signal increased the activity of the promoter. (E and F) Repression function of H-3/4 and WRPW heterologous fusion proteins. The ΔbHLH HES-1 constructs were fused to a heterologous Gal-4 DNA-binding protein (E) and tested for their ability to repress a Gal-4-binding-site-containing reporter construct (with five UAS and a simian virus 40 minimal promoter) (F). The WRPW-containing domains of HES-1 (pM-h3/4/C and pM-C) acted as transcription repressors when fused to Gal-4, consistent with the recruitment to DNA of the corepressor, TLE.
HES-1 inhibits proliferation in neuroblastoma.
To determine if HES-1 also inhibits the growth of other neuronal cell types, we performed CFE assays on three neuroblastoma cell lines, SHEP1, SHSY5Y, and SHIN. Neuroblastoma are neuronal tumor cell lines and, at least for these three types, do not express detectable levels of HES-1 protein (shown for SHEP and SHSY5Y in Fig. 6A). Accordingly, they express mRNA of the HES-1-regulated gene, MASH-1 (Fig. 6B). The expression of exogenous WT HES-1 in these cell lines results in the formation of essentially no stable clones (Fig. 6C and D). Furthermore, we were unable to generate NIH 3T3 cells constitutively expressing HES-1 (data not shown), suggesting that growth inhibition is a general property of HES-1 overexpression.
The H-3/4 domain mediates HES-1 repression of p21 protein expression in PC12 cells.
To determine whether HES-1 represses the expression of cell cycle proteins and, if so, whether the WRPW motif and/or the H-3/4 domain mediate this repression, we compared the effect of expression of WT HES-1, ΔS HES-1, and ΔR HES-1 on the expression of three cell cycle proteins: p21CIP1, PCNA, and cyclin D1. Expression of the WT HES-1, ΔS HES-1, and ΔR HES-1 proteins in induced cells was verified by anti-HES-1 and anti-Flag Western analysis of cell lysates (Fig. 7, top two panels). The induction of WT HES-1 resulted in a significant loss of the S-phase marker PCNA (Fig. 7, second panel from bottom), consistent with the lack of BrdU incorporation and growth arrest in G1. The level of the cyclin-CDK inhibitor, p21, was also considerably reduced by HES-1 induction (Fig. 7, bottom panel). In contrast, the level of the G1 progression cyclin, D1, remained constant (Fig. 7, middle panel).
As observed for WT HES-1 overexpression, the expression of neither ΔS nor ΔR HES-1 affected cyclin D1 protein levels (Fig. 7, middle panel). Expression of the ΔS HES-1 construct significantly reduced the level of PCNA and p21 protein in the cells, similar to the result of WT HES-1 expression (Fig. 7, lower two panels). In contrast, induced expression of the ΔR HES-1 protein, which lacks the H-3/4 domain, did not significantly affect the level of either p21 or PCNA protein (Fig. 7, lower two panels). Together, these data indicate that the repression of PCNA and p21 expression by HES-1 is at least partially dependent upon the H-3/4 domain.
The H-3/4 domain of HES-1 contributes to HES-1 function in transcription repression.
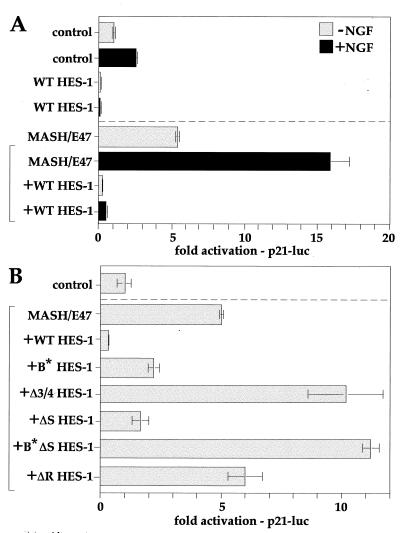
HES-1 has been previously shown to be autoregulatory (56), and, therefore, the HES-1 promoter represents a good model for assaying the transcription repression function of HES-1 proteins. The activity of various HES-1 constructs was assessed by using a HES-1 responsive reporter, incorporating 1.6-kb of human HES-1 genomic DNA (20) upstream of the start codon, fused to a luciferase reporter gene (hHES-luc). In addition to the WT HES-1, ΔS HES-1, ΔR HES-1, Δ3/4 HES-1, and B* HES-1 constructs described above, we also generated a DNA-binding-defective version of ΔS HES-1 (B*ΔS HES-1) (Fig. 8A). The HES-1 constructs each had a Flag epitope fused at the N terminus, and expression of the constructs was confirmed by Western blots of parallel transfections (data not shown). Equivalent amounts of the pCDNA3 expression vectors were transfected into PC12 cells, and the activity of the hHES-luc promoter was determined. Wild-type HES-1 strongly repressed the hHES promoter (Fig. 8B). The DNA-binding-defective (B*) form of HES-1 only weakly repressed the promoter, confirming the importance of the HES-1 DNA binding for repression of this promoter (Fig. 8B). DNA binding is thought to be necessary to recruit the TLE-related family of corepressors to the promoter, and this is facilitated by the C-terminal WRPW motif of HES-1. Expression of ΔR HES-1, which lacks the WRPW motif and the H-3/4 domain, resulted in mild activation of the promoter. This may result from a competitive inhibition for the DNA-binding sites by ΔR HES-1 protein, which would prevent endogenous HES-1 from occupying the DNA and repressing transcription. In contrast, transfection of ΔS HES-1 resulted in repression of the hHES promoter, albeit less effectively than the wild type (∼3-fold compared to ∼14-fold repression). The ΔS HES-1 data demonstrates that the H-3/4 domain functions in transcription repression, because ΔS HES-1 differs from ΔR HES-1 only by inclusion of this region. Comparison of the activity between these two mutants showed an approximate sixfold difference in activity due to the H-3/4 domain. The requirement for the H-3/4 domain was supported by the use of an internal-deletion HES-1 mutant (Δ3/4 HES-1). Despite the presence of a WRPW motif (and despite expression at levels comparable to WT HES-1 [data not shown]), the absence of the H-3/4 domain renders this protein inactive as a repressor. Like the ΔR construct, the Δ3/4 HES-1 mutant is a weak functional activator (or derepressor) of the hHES-1 promoter, although the mechanisms may not be the same in each case.
Further analysis of the function of the H-3/4 domain on the hHES-1 promoter was performed with constructs deleted of the bHLH region, allowing us to test the requirement for intrinsic DNA binding in H-3/4 and WRPW-mediated repression. Three constructs were generated (Fig. 8C), one with the complete C-terminal region beyond the end of helix II (HES 3/4/C), another encoding just the H-3/4 domain (HES 3/4), and the third containing the C-terminal region beyond helix IV (HES C). These constructs correspond to bHLH deletions of the WT, ΔS, and Δ3/4 HES-1 constructs, respectively. To ensure nuclear localization of the basic region-truncated proteins, a nuclear localization signal from VP16 was fused at the N-terminal end after the Flag epitope (see Materials and Methods). Expression of all the ΔbHLH mutants resulted in transcriptional activation, rather than repression, of the hHES-1 reporter (Fig. 8D). This activation suggests that both of the repression domains (WRPW and H-3/4) were able to titrate repressor function away from the DNA. Thus, bHLH region function (i.e., DNA binding) is necessary for HES-1 repressor activity on this promoter.
Unlike the WRPW motif, which is a portable repression domain (22), fusion of the H-3/4 region to a heterologous Gal-4 DNA-binding protein (pM H-3/4) (Fig. 8E) conferred only a slight repression to a promoter containing multimerized Gal-4-binding sites (with five UAS) (Fig. 8F) upstream of a minimal simian virus 40 promoter. A Gal-4 fusion of the entire protein from helix 3 to WRPW (pM H3/4/C) strongly repressed the promoter and a similar repression activity to the same construct deleted of the H-3/4 region (pM C) (Fig. 8F). This suggests that either (i) recruitment of cofactors is limiting in the case of H-3/4-mediated interactions and not for WRPW-mediated recruitment of TLE or (ii) that the repression conferred by the H-3/4 domain is markedly more constrained than the WRPW motif by structural or contextual requirements.
HES-1 repression of transcription of the p21 promoter in PC12 cells.
The reduced expression of p21 in the stable cell lines following induction of wild-type and ΔS HES-1, but not ΔR HES-1 (Fig. 7), suggests that p21 may be a direct target for transcriptional repression by HES-1. Indeed, the promoter of p21 contains a consensus class C site, as well as several nonconsensus sites to which HES-1 can bind specifically (data not shown). In addition, HES-1 is known to inhibit the bHLH activator from binding to E-boxes, enhancer elements that are also necessary for up regulation of p21 transcription (49). Thus, p21 is a plausible target for HES-1-mediated transcriptional regulation, whether by class C site or E-box-directed mechanisms. To examine the mechanism of p21 regulation by HES-1, we used a p21 promoter construct with 2.4 kb of DNA upstream of the start site, fused to a luciferase reporter gene (49). The constitutive expression of p21 was increased by NGF treatment, and in both instances the transcription activity was strongly repressed by HES-1 (Fig. 9A). The addition of MASH-1 and E47 bHLH activators increased both the basal and the NGF-induced activity several-fold, presumably by binding to the E-box enhancers present in the p21 promoter. Again, in both instances HES-1 expression inhibited the activation to below basal levels.
FIG. 9.
HES-1 represses p21 transcription. (A) The endogenous activity of the p21 promoter in PC12 cells (upper half of panel A) was modestly increased by NGF treatment (dark shading), consistent with the induction of p21 by NGF in PC12 cells. Both the endogenous and NGF-induced promoter activities were strongly repressed by HES-1 to below basal levels. Activation of the E-box-enhanced p21 promoter by coexpression of MASH-1 and E47 (lower half of panel A, bracketed) was also further increased by NGF treatment (dark shading). Again, both the MASH-1–E47 and the MASH-1–E47–NGF-activated promoters were strongly repressed to below basal levels by the expression of HES-1. (B) MASH-1–E47-enhanced activation of the p21 promoter (lower section of panel B, bracketed) was repressed by HES-1 in an H-3/4 domain-dependent manner. WT HES-1 strongly, and ΔS HES-1 modestly, repressed the activated p21 promoter, whereas Δ 3/4 HES-1 modestly activated the promoter. ΔR HES-1 conferred no repression activity, and B∗ΔS HES-1 was a modest activator.
The ability of HES-1 mutants to repress the MASH-1–E47-activated p21 promoter was examined (Fig. 9B). Again, WT HES-1 strongly repressed the E47–MASH-1-activated promoter. ΔS HES-1, but not ΔR HES-1, was also able to repress the promoter, to around threefold below its activated state. In contrast, the DNA-binding-defective form of ΔS HES-1 (B*ΔS HES-1) slightly activated the promoter, consistent with the titration of repressor activity away from DNA. The requirement for the H-3/4 domain was further demonstrated by the Δ3/4 construct, which was nonfunctional as a transcription repressor and instead activated (derepressed) the promoter roughly twofold above the MASH-1–E47-enhanced level. Interestingly, the DNA-binding-defective mutant (B*) also repressed the p21 promoter, though considerably more weakly than WT HES-1. However, both the ΔS and B* HES-1 constructs showed significant losses of repression activity compared to their normal DNA-binding equivalents, demonstrating that a functional basic region is necessary for full repression activity.
Together, the above results show that HES-1 represses p21 transcription in PC12 cells. Moreover, the transcription repression data from both the hHES-1 and the p21 promoters highlights the importance of the H-3/4 domain in DNA-binding-dependent transcription repression by HES-1.
DISCUSSION
In this study, we have demonstrated a functional role for the H-3/4 domain in the class C site (N-box)-dependent transcription repression by HES-1. In addition, we have shown that HES-1 expression inhibits PC12 cell proliferation as well as differentiation and that the H-3/4 domain is important for both of these inhibitory activities. The repression of p21 transcription is unlikely to mediate the inhibition of proliferation by HES-1, since p21 is a cyclin-CDK inhibitor that negatively regulates proliferation. Instead, p21 repression by HES-1 may contribute to the repression of differentiation by HES-1, as discussed below.
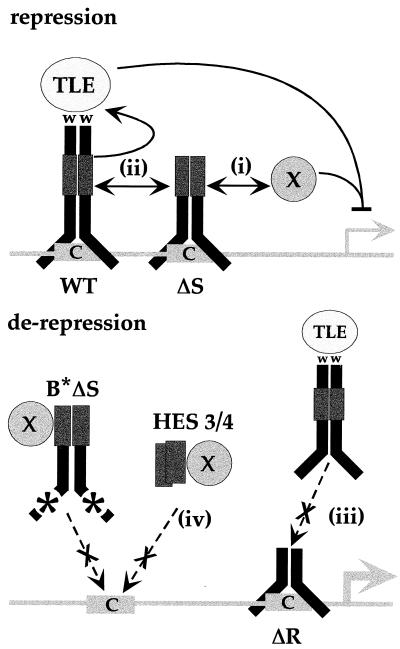
In Fig. 10, we present a model to illustrate the potential mechanisms by which the H-3/4 domain contributes to the DNA-binding-dependent regulation of transcription by HES-1. This model contains elements that serve to explain the observed ability of HES-1 or HES-1 mutants to repress, or in some cases to derepress, transcription. We propose that the H-3/4 domain, a putative protein interaction motif (37), is necessary for either (Fig. 10, i) the direct recruitment of an unknown corepressor and/or (Fig. 10, ii) the stabilization or regulation of WRPW-mediated repression function through intra- or intermolecular interaction. While we have suggested that the H-3/4 domain interacts with an unknown corepressor, repression could also result from the interaction of the H-3/4 domain with bHLH activators bound to adjacent sites. This is analogous to a model proposed by Dawson and colleagues (13) in which the corepressor would be the bHLH activator protein, Scute, bound to an E-box. The ability of some HES-1 mutant proteins to derepress the hHES-1 promoter could result from the occupancy of DNA sites (Fig. 10, iii) by proteins deficient in transcription repression activity (i.e., ΔR HES-1 and Δ3/4 HES-1). Derepression also resulted from the expression of HES-1 mutants that lack an intrinsic DNA-binding function, either due to deletion of the bHLH region (all Δ bHLH proteins) or due to a basic region mutation (B* and B*ΔS HES-1) that disrupts DNA binding. This derepression is likely to result from the titration of corepressors such as X (or TLE) away from DNA (Fig. 10, iv).
FIG. 10.
Model of DNA binding, H-3/4-dependent transcription repression by HES-1. The H-3/4 domain may be required to recruit unknown corepressors (X) to DNA (i) to interact with other promoter-bound proteins to either stabilize repression (ii) or inhibit activators (not shown), or to serve as a heterodimer with endogenous HES-1, providing an active conformation for TLE function (ii). Presumably, Δ 3/4 HES-1, lacking the H-3/4 domain, acts instead to inhibit these functions (data not shown). ΔR HES-1 as a homodimer, or as an inactive heterodimer with endogenous HES-1, could derepress the promoter by binding to DNA without forming repressor complexes (iii). This would also result in the competitive inhibition of any endogenous HES-1 homodimers for the DNA sites. As a ΔbHLH protein, the H-3/4 domain could rerepress the promoter by titrating unknown corepressors (shown as X) away from DNA or by forming DNA-binding-deficient heterodimers with endogenous HES-1 to the same effect (iv).
The C-EBPα transcription factor has previously been shown to regulate p21 at the translational rather than transcriptional level (58). While it is possible that HES-1 may also repress p21 at the translational level, the direct transcriptional repression of the p21 promoter by HES-1 (Fig. 9) indicates that this previously characterized transcriptional repressor protein does repress p21 at the transcriptional level.
DNA-binding-dependent, H-3/4-domain-mediated mechanisms of transcription repression.
Our results for the DNA-binding and non-DNA-binding HES-1 variants complement each other and together indicate that HES-1-mediated transcription repression is primarily dependent upon binding to cognate class C DNA sites. First, the loss of DNA-binding ability in B* HES-1 and B*ΔS HES-1 results in a substantially impaired ability to repress transcription in comparison to the corresponding DNA-binding-competent protein (WT and ΔS HES-1, respectively) (Fig. 8B). Second, the ΔR and Δ 3/4 HES-1 proteins have the ability to bind to DNA, but since they lack the H-3/4 and/or WRPW regions, they do not facilitate repression (Fig. 8B). The transcription derepression resulting from ΔR HES-1 expression (Fig. 8B) is consistent with the hypothesis that the ΔR HES-1 protein is a competitive inhibitor of DNA binding by endogenous HES-1 (Fig. 10, iii). The ΔR HES-1 protein is essentially the same as a bHLH-only form of HES-1 that we have previously shown binds to HES-1-specific DNA sequences in vitro (55) as well as in nuclear extracts from mammalian cells (data not shown). PC12 cells contain endogenous HES-1 (19, 55), which can inhibit the hHES-1 reporter and lower its basal activity. Exogenous ΔR HES-1 could therefore compete for the same class C sites as the endogenous HES-1 protein. Since ΔR HES-1 lacks the ability to recruit corepressors, occupancy of the class C sites would effectively derepress the system. Furthermore, the lack of repression by Δ3/4 HES-1 (Fig. 8B), which has a functional WRPW motif and is expressed equivalently to the WT and B* HES-1 proteins in transient transfections (data not shown), suggests that the recruitment of factors to DNA via the H-3/4 domain may be critical to WRPW-mediated repression in this particular assay. A similar loss of function was reported for Δ H-3/4 deletions in both E(Spl) (24) and in Hairy (13), as determined by genetic assays in Drosophila. Together, the data from the DNA-binding and nonbinding HES-1 variants indicate that direct binding of HES-1 to class C sites is necessary for transcription repression activity.
Previous studies have shown that the WRPW domain is sufficient to mediate transcriptional repression when fused to the heterologous DNA-binding protein, Gal-4 and thus can repress in the absence of an H-3/4 domain (22; see above). By contrast, when the H-3/4 domain was fused to Gal-4 (pM-H3/4), it was not sufficient to significantly mediate transcription repression (Fig. 8F). However, expression of a free H-3/4 domain (HES 3/4) can derepress the hHES promoter effectively (Fig. 8D). This may reflect the ability of the H-3/4 domain to interact with a corepressor protein. If so, the lack of repression observed for the Gal-4 H-3/4 domain fusion protein (pM H3/4) may result instead from structural limitations imposed by the fusion to the Gal-4 protein. For example, the amphipathic helices of the H-3/4 domain may dimerize when normally fused to bHLH domain, but not when fused to Gal-4. Nevertheless, in the context of the native protein, H-3/4 can mediate repression independently of the WRPW motif.
The indirect binding of HES-1 to DNA via interactions with non-bHLH corepressor proteins may also contribute to the transcription repression activity of HES-1. Such a mechanism has been specifically proposed to explain the repression by the HES-related E(Spl) proteins of the Drosophila scute SMC enhancer (11). Here, E(Spl) proteins have been proposed to interact with a putative corepressor protein (called Xα) bound to an essential NF-κB-like alpha site conserved in the enhancer. Thus, HES-1 and related bHLH repressor proteins may repress transcription by multiple mechanisms, with the relative contribution of each mechanism being dependent upon the overall context of the promoter binding sites and the specific types and relative concentrations of activator and repressor proteins present in a given cell.
The repression of a p21-dependent cell cycle exit by HES-1 may inhibit differentiation.
The ability of HES-1 to repress NGF-induced neurite outgrowth in PC12 cells is consistent with the role of HES-1 as a repressor of neuronal differentiation. This repression is at least partly dependent upon the H-3/4 domain, suggesting that transcriptional repression of differentiation-specific genes is mediated in part through this domain. In principle, the ability of HES-1 to block proliferation through repression of as-yet-undefined target genes may, in itself, be sufficient to inhibit differentiation by precluding a cell cycle exit program essential for differentiation. In particular, previous studies have suggested that such a differentiation-specific cell cycle arrest program may be mediated by p21: several studies have indicated that the differentiation of certain cell types may require the up regulation of p21 expression to induce exit from cell cycle (28, 39, 45). NGF signaling in PC12 cells induces p21 expression as part of a peripheral-neuron-like differentiation process (64), and overexpression of p21 alone is sufficient to arrest cell cycle in PC12 cells (18). In addition to cell cycle exit, p21 overexpression results in differentiation-specific cell cycle changes and potentiates differentiation in response to growth factor signaling (17). Furthermore, the growth arrest mediated by p21 is a necessary precondition for the differentiation of NGF-treated PC12 cells grown in serum (50, 60). Therefore, expression of p21 may be critical for establishing specific cell cycle exit conditions that are necessary for differentiation. Thus, the prevention of a p21-dependent exit from cell cycle, and any differentiation-specific cell cycle changes associated with p21, may be part of the pathway by which HES-1 inhibits differentiation.
In vivo, the repression of p21 may contribute to the repression of neural differentiation by HES-1. The phenotype of HES-1 knockout mice (33) is an extensive reduction in neural tissue proposed to result from the premature differentiation of the neuronal precursors and the concomitant exit from cell cycle. Such a premature exit from the cell cycle could result from the inappropriate up regulation of p21. At present, it is not known if the premature induction of p21, which is sufficient to induce growth arrest in cultured cells (31), is also sufficient to arrest growth in vivo. If so, an important function of HES-1 in vivo would be the inhibition of differentiation through the transcriptional repression of p21.
Induction of growth arrest by HES-1.
We have found that a moderate overexpression of WT HES-1 strongly inhibits proliferation in PC12 cells. We previously reported our inability to obtain stable HES-1-expressing PC12 cell lines when using an expression vector with a strong promoter (cytomegalovirus) to constitutively express HES-1 (55). HES-1-expressing cell lines were obtained only by transfection with a vector containing a low-activity (uninduced) mouse mammary tumor virus promoter to drive expression, and even these lines were slow growing and difficult to maintain. Subsequently, Issack and Ziff (34) noted an inability to maintain cells transfected with a HES-1 expression vector in culture. By generating tetracycline-inducible stable cell lines, we have now shown that the expression of moderate levels of exogenous HES-1 results in a marked inhibition of proliferation (Fig. 3B and 4A). The induced cells do not undergo increased cell death, and proliferation is restored if the induction is halted. Thus, the effect of HES-1 on proliferation is likely to result from the regulation of cell cycle effector genes and not from either nonspecific, toxic effects or from the induction of apoptosis.
The strong correlation of growth arrest with transcription repression (compare Fig. 5 and 7B) and the importance of the H-3/4 domain for both phenomena is consistent with the established role of HES-1 as a transcription repressor and suggests that the repression of cell cycle control genes is part of the function of endogenous HES-1 in PC12 cells. HES-1 may also repress proliferation in vivo, although it is difficult to extrapolate an antiproliferative phenotype in a tumor-cell-based overexpression system to the in vivo function of HES-1. An alternative possibility is that growth arrest is caused by HES-1 expression squelching, or sequestering a limiting factor specifically needed for proliferation but not for transcription repression, and that endogenous HES-1 normally induces proliferation in PC12 cells. However, a direct interpretation of our data suggests that induced HES-1 acts in PC12 cells to repress cell cycle progression genes, thereby controlling cell cycle as a function of differentiation.
While we have identified the cell cycle inhibitor p21 as a target for HES-1 regulation, the role of p21 regulation in HES-1-mediated growth arrest is unclear. Typically, induction of p21 (to a level equimolar to cyclin D1 concentration [31, 32]), rather than repression of p21, is associated with a halt in proliferation. However, the repression of p21 by HES-1 may interfere with proliferation if p21 is required at low levels to promote cell cycle, perhaps as an assembly factor for cyclin-CDK (9, 23; reviewed in reference 1). It is also possible that the loss of p21 results in an effective increase in (active) cyclin D1, which has been shown to halt proliferation in epithelial cells by extending S phase (29). Clearly though, the halt in proliferation resulting from HES-1 expression is independent of p21 up regulation, since HES-1 represses p21 expression. Further work in p21-deficient cells, which are not growth inhibited (9, 15), will be needed to determine whether HES-1-mediated growth arrest is completely independent of p21. Alternatively, the repression of PCNA—an essential DNA replication factor that is also down regulated upon HES-1 induction—could account for the halt in PC12 cell proliferation. However, loss of PCNA expression may occur indirectly because the PCNA proximal promoter lacks consensus class C sites (unpublished data), and we have not determined if down regulation occurs due to direct repression by HES-1.
The ability of HES-1 to inhibit proliferation in PC12 and neuroblastoma tumor cell lines suggests that the misregulation of HES-1 and HES-1-regulated genes may play a role in the development of neuronal tumors. During development, HES-1 functions to negatively regulate a cascade of bHLH activators that control the commitment and differentiation of neuronal precursors (reviewed in reference 36). For example, HES-1 directly represses the transcription of MASH-1, a neuronal commitment gene (8). The direct repression of MASH-1 by HES-1 is consistent with the correlated up regulation of MASH-1 and down regulation of HES-1 observed in highly metastatic small-cell lung cancer (SCLC) tumor cells (8). The neuroendocrine phenotype associated with SCLC is believed to be dependent upon MASH-1 expression (4). Given the strong repression of proliferation in neuronal tumor lines by HES-1, it will be interesting and important to determine whether the loss of HES-1 expression contributes to the metastatic proliferation of SCLC cells.
In conclusion, the data presented here provide novel and direct evidence that the H-3/4 domain is required for transcription repression by HES-1, in addition to the previously identified repression motif, WRPW. We also have identified a novel target gene for HES-1, the cyclin-CDK inhibitor, p21cip1, which may partially mediate HES-1 repression of differentiation. Moreover, we have shown that HES-1 expression inhibits PC12 cell proliferation as well as differentiation and that the H-3/4 domain is important for both of these inhibitory activities. The importance of the H-3/4 domain in direct transcriptional repression suggests that the downstream targets of HES-1 are essential for both neurite formation and proliferation. The discovery and analysis of additional HES-1-regulated genes will provide additional insights to the mechanism by which HES-1 mediates the regulation of differentiation and the cell cycle.
ACKNOWLEDGMENTS
We thank John Feder (Mercator Genetics) for the kind gifts of the anti-HES-1 antibodies and the hHES-1 genomic DNA and Anders Ström (Karolinska Institute, Sweden) for cloning the hHES-1 luciferase reporter construct.
P.C. was supported by the Graduate Program in Cell Biology and Genetics at the W. G. S. M. S. C. U. and by a National Institutes of Health (NIH) predoctoral training grant (NS07384-05). S.S. was supported by the Pew Scholars in Biomedical Research Program. K.N. was supported by a grant from the NIH (NS28652). J.A.W. was funded by grants from the NIH (EY06454 and NS31728). M.C. was funded by grants from the NIH (NS28652), Sloan Foundation, and the Pew Scholars in Biomedical Research Program.
REFERENCES
- 1.Ball K L. p21: structure and functions associated with cyclin-CDK binding. Vol. 3. New York, N.Y: Plenum Press; 1997. [DOI] [PubMed] [Google Scholar]
- 2.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 3.Bier E, Vaessin H, Younger-Shepherd S, Jan L Y, Jan Y N. deadpan, an essential pan-neural gene in Drosophila, encodes a helix-loop-helix protein similar to the hairy gene product. Genes Dev. 1992;6:2137–2151. doi: 10.1101/gad.6.11.2137. [DOI] [PubMed] [Google Scholar]
- 4.Borges M, Linnoila R I, van de Velde H J, Chen H, Nelkin B D, Mabry M, Baylin S B, Ball D W. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature. 1997;386:852–855. doi: 10.1038/386852a0. [DOI] [PubMed] [Google Scholar]
- 5.Campos-Ortega J A. Early neurogenesis in Drosophila melanogaster. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 1091–1129. [Google Scholar]
- 6.Campuzano S, Modolell J. Patterning of the Drosophila nervous system: the achaete-scute gene complex. Trends Genet. 1992;8:202–208. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]
- 7.Castella P, Wagner J A, Caudy M. Regulation of hippocampal neuronal differentiation by the basic helix-loop-helix transcription factors HES-1 and MASH-1. J Neurosci Res. 1999;56:229–240. doi: 10.1002/(SICI)1097-4547(19990501)56:3<229::AID-JNR2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Thiagalingam A, Chopra H, Borges M W, Feder J N, Nelkin B D, Baylin S B, Ball D W. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc Natl Acad Sci USA. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng M, Olivier P, Diehl J A, Fero M, Roussel M F, Roberts J M, Sherr C J. The p21(Cip1) and p27(Kip1) CDK “inhibitors” are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crescenzi M, Fleming T P, Lassar A B, Weintraub H, Aaronson S A. MyoD induces growth arrest independent of differentiation in normal and transformed cells. Proc Natl Acad Sci USA. 1990;87:8442–8446. doi: 10.1073/pnas.87.21.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Culi J, Modolell J. Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 1998;12:2036–2047. doi: 10.1101/gad.12.13.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis R L, Weintraub H, Lassar A B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 13.Dawson S R, Turner D L, Weintraub H, Parkhurst S M. Specificity for the Hairy/Enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol Cell Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delidakis C, Artavanis-Tsakonas S. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc Natl Acad Sci USA. 1992;89:8731–8735. doi: 10.1073/pnas.89.18.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 16.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 17.Erhardt J A, Pittman R N. Ectopic p21(WAF1) expression induces differentiation-specific cell cycle changes in PC12 cells characteristic of nerve growth factor treatment. J Biol Chem. 1998;273:23517–23523. doi: 10.1074/jbc.273.36.23517. [DOI] [PubMed] [Google Scholar]
- 18.Erhardt J A, Pittman R N. p21WAF1 induces permanent growth arrest and enhances differentiation, but does not alter apoptosis in PC12 cells. Oncogene. 1998;16:443–451. doi: 10.1038/sj.onc.1201577. [DOI] [PubMed] [Google Scholar]
- 19.Feder J, Sheng M, Jan L Y, Jan Y N. A rat gene with sequence homology to the Drosophila gene hairy is rapidly induced by growth factors known to influence neuronal differentiation. Mol Cell Biol. 1993;13:105–113. doi: 10.1128/mcb.13.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feder J N, Li L, Jan L Y, Jan Y N. Genomic cloning and chromosomal localization of HRY, the human homolog of the Drosophila segmentation gene, hairy. Genomics. 1994;20:56–61. doi: 10.1006/geno.1994.1126. [DOI] [PubMed] [Google Scholar]
- 21.Fisher A L, Caudy M. The function of Hairy-related bHLH proteins in cell fate decisions. BioEssays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Fisher A L, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fotedar R, Fitzgerald P, Rousselle T, Cannella D, Doree M, Messier H, Fotedar A. p21 contains independent binding sites for cyclin and cdk2: both sites are required to inhibit cdk2 kinase activity. Oncogene. 1996;12:2155–2164. [PubMed] [Google Scholar]
- 24.Giebel B, Campos-Ortega J A. Functional dissection of the Drosophila Enhancer of split protein, a suppressor of neurogenesis. Proc Natl Acad Sci USA. 1997;94:6250–6254. doi: 10.1073/pnas.94.12.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grbavec D, Stifani S. Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem Biophys Res Commun. 1996;223:701–705. doi: 10.1006/bbrc.1996.0959. [DOI] [PubMed] [Google Scholar]
- 26.Greene L A, Tischler S A. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Cell. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guillemot F, Lo L C, Johnson J E, Auerbach A, Anderson D J, Joyner A L. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 28.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 29.Han E K, Sgambato A, Jiang W, Zhang Y J, Santella R M, Doki Y, Cacace A M, Schieren I, Weinstein I B. Stable overexpression of cyclin D1 in a human mammary epithelial cell line prolongs the S-phase and inhibits growth. Oncogene. 1995;10:953–961. [PubMed] [Google Scholar]
- 30.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interaction protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 31.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hengst L, Gopfert U, Lashuel H A, Reed S I. Complete inhibition of Cdk/cyclin by one molecule of p21(Cip1) Genes Dev. 1998;12:3882–3888. doi: 10.1101/gad.12.24.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishibashi M, Ang S L, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 34.Issack P S, Ziff E B. Altered expression of helix-loop-helix transcriptional regulators and cyclin D1 in Wnt-1-transformed PC12 cells. Cell Growth Differ. 1998;9:837–845. [PubMed] [Google Scholar]
- 35.Jan Y N, Jan L Y. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- 36.Kageyama R, Nakanishi S. Helix-loop-helix factors in growth and differentiation of the vertebrate nervous system. Curr Opin Genet Dev. 1997;7:659–665. doi: 10.1016/s0959-437x(97)80014-7. [DOI] [PubMed] [Google Scholar]
- 37.Knust E, Schrons H, Grawe F, Campos-Ortega J A. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics. 1992;132:505–518. doi: 10.1093/genetics/132.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee E J, Hollenberg M S, Snider L, Turner L D, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 39.Macleod K F, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 40.Mutoh H, Fung B P, Naya F J, Tsai M J, Nishitani J, Leiter A B. The basic helix-loop-helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc Natl Acad Sci USA. 1997;94:3560–3564. doi: 10.1073/pnas.94.8.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakao K, Campos-Ortega J A. Persistent expression of genes of the enhancer of split complex suppresses neural development in Drosophila. Neuron. 1996;16:275–286. doi: 10.1016/s0896-6273(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 42.Naya F J, Stellrecht C M, Tsai M J. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 43.Oellers N, Dehio M, Knust E. bHLH proteins encoded by the Enhancer of split complex of Drosophila negatively interfere with transcriptional activation mediated by proneural genes. Mol Gen Genet. 1994;244:465–473. doi: 10.1007/BF00583897. [DOI] [PubMed] [Google Scholar]
- 44.Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- 45.Parker S B, Eichele G, Zhang P, Rawls A, Sands A T, Bradley A, Olson E N, Harper J W, Elledge S J. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 46.Paroush Z, Finley R L, Jr, Kidd T, Wainwright S M, Ingham P W, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 47.Peverali F A, Ramqvist T, Saffrich R, Pepperkok R, Barone M V, Philipson L. Regulation of G1 progression by E2A and Id helix-loop-helix proteins. EMBO J. 1994;13:4291–4301. doi: 10.1002/j.1460-2075.1994.tb06749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poortinga G, Watanabe M, Parkhurst S M. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prabhu S, Ignatova A, Park S T, Sun X H. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qu Z, Wolfraim L A, Svaren J, Ehrengruber M U, Davidson N, Milbrandt J. The transcriptional corepressor NAB2 inhibits NGF-induced differentiation of PC12 cells. J Cell Biol. 1998;142:1075–1082. doi: 10.1083/jcb.142.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rushlow C A, Hogan A, Pinchin S M, Howe K M, Lardelli M, Ish-Horowicz D. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 1989;8:3095–3103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 53.Scheibe R J, Wagner J A. Retinoic acid regulates both expression of the nerve growth factor receptor and sensitivity to nerve growth factor. J Biol Chem. 1992;267:17611–17616. [PubMed] [Google Scholar]
- 54.Stifani S, Blaumueller C M, Redhead N J, Hill R E, Artavanis-Tsakonas S. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- 55.Ström A, Castella P, Rockwood J, Wagner J, Caudy M. Mediation of NGF signaling by post-translational inhibition of HES-1, a basic helix-loop-helix repressor of neuronal differentiation. Genes Dev. 1997;11:3168–3181. doi: 10.1101/gad.11.23.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takebayashi K, Sasai Y, Sasai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- 57.Tietze K, Oellers N, Knust E. Enhancer of splitD, a dominant mutation of Drosophila, and its use in the study of functional domains of a helix-loop-helix protein. Proc Natl Acad Sci USA. 1992;89:6152–6156. doi: 10.1073/pnas.89.13.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Timchenko N, Harris T, Wilde M, Bilyeu T, Burgess-Beusse B, Finegold M, Darlington G. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol Cell Biol. 1997;17:7353–7361. doi: 10.1128/mcb.17.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Doren M, Bailey A M, Esnayra J, Ede K, Posakony J W. Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes Dev. 1994;8:2729–2742. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- 60.van Grunsven L A, Billon N, Savatier P, Thomas A, Urdiales J L, Rudkin B B. Effect of nerve growth factor on the expression of cell cycle regulatory proteins in PC12 cells: dissection of the neurotrophic response from the anti-mitogenic response. Oncogene. 1996;12:1347–1356. [PubMed] [Google Scholar]
- 61.Wainwright S M, Ish-Horowicz D. Point mutations in the Drosophila hairy gene demonstrate in vivo requirements for basic, helix-loop-helix, and WRPW domains. Mol Cell Biol. 1992;12:2475–2483. doi: 10.1128/mcb.12.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wright W E, Sassoon D A, Lin V K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 63.Xiong Y, Zhang H, Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- 64.Yan G Z, Ziff E B. Nerve growth factor induces transcription of the p21 WAF1/CIP1 and cyclin D1 genes in PC12 cells by activating the Sp1 transcription factor. J Neurosci. 1997;17:6122–6132. doi: 10.1523/JNEUROSCI.17-16-06122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H, Levine M. Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc Natl Acad Sci USA. 1999;96:535–540. doi: 10.1073/pnas.96.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang P, Wong C, Liu D, Finegold M, Harper J W, Elledge S J. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 1999;13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]