Abstract
Ageing leads to profound alterations in the immune system and increases susceptibility to some chronic, infectious and autoimmune diseases. In recent years, widespread application of single-cell techniques has enabled substantial progress in our understanding of the ageing immune system. These comprehensive approaches have expanded and detailed the current views of ageing and immunity. Here we review a body of recent studies that explored how the immune system ages using unbiased profiling techniques at single-cell resolution. Specifically, we discuss an emergent understanding of age-related alterations in innate and adaptive immune cell populations, antigen receptor repertoires and immune cell-supporting microenvironments of the peripheral tissues. Focusing on the results obtained in mice and humans, we describe the multidimensional data that align with established concepts of immune ageing as well as novel insights emerging from these studies. We further discuss outstanding questions in the field and highlight techniques that will advance our understanding of immune ageing in the future.
Subject terms: Inflammation, Innate immune cells, Adaptive immunity
Studying ageing at the single-cell level has provided insight into the changes that occur systemically and in tissues as we age. For example, we now have a greater appreciation of the heterogeneity and dynamic nature of immune cell ageing and of the impact of age-associated tissue remodelling on the immune system, together contributing to increased vulnerability to some diseases.
Introduction
Ageing is a multimodal process that occurs at several levels, with organs, tissues and cells undergoing substantial and simultaneous remodelling, thus limiting the power of conventional molecular biology approaches to resolve this complexity. Loss of physiological integrity, disturbed tissue homeostasis and increasing deterioration of various biological systems, including the immune system, are among the key features of ageing1. Together, these changes reshape physiological and immune landscapes as well as increase vulnerability to some diseases, frailty and multimorbidity2. Interestingly, not all immune processes are uniformly sensitive to ageing, suggesting that various immune cell populations are heterogeneously impaired in older organisms (for example, ≥65 years in humans and ≥1.5 years in mice)3. The current development of multi-omics and single-cell techniques4 (Box 1) and progress in high-throughput biology and systems immunology5 make it possible to comprehensively characterize cellular and functional heterogeneity and dynamic changes in the immune system during ageing. Novel techniques, including single-cell RNA sequencing (scRNA-seq) (Box 2), have confirmed previous findings and expanded existing paradigms in ageing and immunity built on evidence from prior targeted studies. In this Review, we summarize recent advances in our understanding of the ageing immune system and discuss how reported single-cell data and analyses align with existing concepts. We focus our analysis on accumulating single-cell results obtained from mammalian model organisms and, where possible, we discuss how these findings can be translated to immune system ageing in humans (the datasets discussed are available to browse through an Artyomov laboratory webpage).
Box 1 Multi-omics and single-cell technologies in immunology.
A comprehensive assessment of a set of molecules, such as DNAs, RNAs, proteins and metabolites, is collectively termed ‘multi-omics’. Flow cytometry is historically the first single-cell-based technique capable of simultaneously identifying 5–20 characteristics in individual cells. Typically, flow cytometry experiments use fluorescently conjugated antibodies to detect surface and intracellular epitopes and/or fluorescent probes that measure cellular characteristics such as an abundance of DNA, mitochondria, lysosomes and lipid droplets. Mass cytometry, also known as cytometry by time of flight, is a variation of flow cytometry that relies on antibodies labelled with heavy metal ion tags, which allows the measurement of up to 40 individual epitopes in individual cells. During the past decade, unbiased single-cell-based applications of omics have been extensively developed. Single-cell RNA sequencing (scRNA-seq) is a rapidly growing technology that can detect and quantitively measure 500–5,000 of the most highly expressed transcripts per cell182. Computational analysis of transcriptomes of individual cells can identify novel cell populations, activation states and differentiation programmes in immune cells. The variation of the scRNA-seq technique that identifies sequences of variable regions in the T cell receptor or B cell receptor in addition to RNA transcripts in individual cells (termed ‘single-cell T cell receptor sequencing’ and ‘single-cell B cell receptor sequencing’) has become instrumental in studying antigen receptor repertoires at single-cell resolution182. Cellular indexing of transcriptomes and epitopes by sequencing is a method in which oligonucleotide-labelled antibodies are used to integrate cellular protein and transcriptome measurements into a single-cell readout183. It exceeds the limitations of mass cytometry and, theoretically, may extend to measuring thousands of proteins and transcripts simultaneously. Single-cell assay for transposase-accessible chromatin using sequencing was recently developed, and it enables the analysis of epigenetic landscapes of individual cells184,185. Moreover, single-cell techniques that combine transcriptional information with epigenetic characteristics in individual cells have been developed186. An important limitation of these single-cell methods is that they use cell suspensions or isolated nuclei and require disaggregation of an organ, which leads to loss of spatial information relating to the anatomical localization of cells. Adding the positional information to single-cell experiments is the next frontier in the field of multi-omic immunology. Thus, spatial applications of mass cytometry (imaging mass cytometry)187 and spatial transcriptomics188 are rapidly growing fields that will help to integrate transcriptional and proteomic information in tissue sections to combine multimodal characteristics of individual cells or groups of cells with the complexity of tissue architecture.
Box 2 Single-cell RNA sequencing.
Single-cell RNA sequencing (scRNA-seq) approaches recently evolved owing to rapid progress in next-generation sequencing technologies. Single-cell isolation is the first step of scRNA-seq. Historically, fluorescence-activated cell sorting of hundreds of individual cells was one of the early strategies that rely on the availability of monoclonal antibodies that recognize proteins expressed on the cells of interest. As a more efficient variant, microfluidic technologies have recently become popular for the low-cost isolation of individual cells. One of the microfluidic technologies, Fluidigm C1, provides single-cell isolation, RNA extraction and cDNA synthesis for up to 800 cells on a single chip189. A popular alternative for single-cell isolation is microdroplet-based microfluidics190. This approach allows the dispersion of liquid droplets in an oil phase, requires lower volumes than standard microfluidic chips and can screen thousands to millions of cells. An example of a commercial microdroplet-based microfluidics system is the Chromium system from 10X Genomics, which can take a suspension of cells or nuclei as an input. Consequent steps include cell lysis, first-strand cDNA synthesis by reverse transcription of mRNA, second-strand synthesis and cDNA amplification by PCR to generate scRNA-seq libraries. Some scRNA-seq methods produce full-length coverage across transcripts (such as Smart-Seq2 (ref.191)), whereas other methods limit sequencing coverage to 3′ or 5′ regions of mRNAs (such as 10X Genomics). The most popular approaches to make scRNA-seq libraries are also limited by capturing only mRNA with poly(A) tails. Although bulk RNA-seq allows almost genome-wide detection levels of mRNA expression in pooled cell samples, scRNA-seq cannot measure the expression levels of most genes in individual cells. Indeed, the sequencing depth usually allows reliable identification of limited numbers of highly expressed transcripts per single cell (usually 1,000–5,000 individual transcripts). Because of the ongoing development of scRNA-seq techniques, different generations can differ in sequencing depth and create technical limitations to compare results from early versions versus late versions of scRNA-seq. In the latest versions, incorporation of unique molecular identifiers (random 10–16-bp sequences, also known as barcodes) in the reverse transcription step removes bias from PCR amplification steps and increases sequencing accuracy. A more detailed analysis of applications of scRNA-seq to analyse the immune system is presented in recent excellent reviews4,182.
Inflammageing
An unresolved systemic inflammation in the absence of pathogens in aged organisms is a hallmark of immune ageing, often referred to as ‘inflammageing’6,7. Inflammageing is characterized by an unbalanced production of pro-inflammatory and anti-inflammatory factors, which is profoundly predictive of death and chronic diseases8 (Fig. 1). Although the concept of inflammageing was initially built on measurements of a relatively low number of inflammatory factors, unbiased profiling studies fully corroborate this phenomenon. A recent study combined bulk RNA sequencing of 17 tissues with plasma proteomics at ten time points across the mouse lifespan to identify age-related genes clustered in consistent trajectory groups with coherent biological functions9. In accord with previous results10, this analysis identified two distinct gene clusters related to immune responses that are rapidly upregulated on transition from middle to advanced age in mice9. These transcriptional alterations likely reflect an active infiltration of immune cells into aged tissues and consequent systemic inflammation. Intriguingly, the breadth of expression changes in these clusters is greatest in adipose tissues, especially in visceral fat depots9, suggesting that fat-associated macrophages and adipocytes might be among the key regulators of inflammageing. In addition to immune cell infiltration, inflammageing might result from alterations in basic tissue architecture owing to age-associated remodelling. Broadly, there is enhanced formation of fibrotic structures and senescent cell accumulation in many tissues11–14, but some tissues are also characterized by a distinct set of immune alterations. For example, in mice, type I interferon signalling has been reported to increase with age, specifically in the choroid plexus, a tissue that produces cerebrospinal fluid. This inflammatory remodelling is associated with cognitive decline in ageing10.
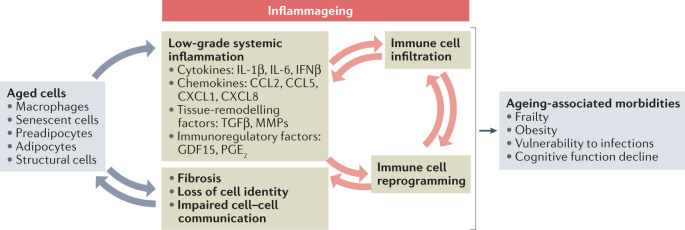
Fig. 1. Cellular and molecular components of inflammageing.
Low-grade chronic systemic inflammation, or inflammageing, comprises multiple inflammatory factors that originate from various cellular sources in aged organisms. Aged pro-inflammatory tissue macrophages produce cytokines and chemokines168, and senescent cells secrete a plethora of pro-inflammatory components and matrix metalloproteinases (MMPs) as components of the senescence-associated secretory phenotype113,114. Moreover, non-immune cells, such as adipocytes, preadipocytes and stromal cells (including fibroblasts and endothelial cells), contribute to inflammageing by secreting soluble inflammatory factors and altering the tissue microarchitecture of aged organs7. The pro-inflammatory chemokines drive excessive immune cell infiltration in tissues, where the cytokines (such as IL-6 and IL-1β) and immunoregulatory factors (such as prostaglandin E2 (PGE2)) reprogramme immune cell subsets to a more pro-inflammatory and dysfunctional state169,170. In turn, the dysfunctional immune cells, including macrophages and T cells, amplify the inflammatory and destructive processes in ageing tissues169,171,172. This crosstalk between cellular and molecular components of inflammageing ultimately results in a progressive functional decline in various organs and leads to age-associated diseases. CCL, CC-chemokine ligand; CXCL, CXC-chemokine ligand; GDF15, growth/differentiation factor 15; IFNβ, interferon-β; TGFβ, transforming growth factor-β.
Effects of ageing on myeloid cells
Ageing is associated with a gradual expansion of circulating myeloid cell populations relative to lymphoid cell populations15 (Table 1). In part, this shift is due to a decline in thymic activity required for the generation of nascent T cells16. At the same time, there is a differential effect of age on early myelopoiesis and lymphopoiesis in the bone marrow17. Changes in the bone marrow of aged mice lead to clonal skewing of haematopoietic stem cells (HSCs) towards the myeloid lineage18,19. RNA sequencing analysis of the bone marrow stromal microenvironment in mice revealed an age-driven progressive upregulation of pro-inflammatory genes (including Il6 and Il1b), which can shift the balance towards HSCs with myeloid potential20. Indeed, analysis of the young bone marrow microenvironment at single-cell resolution demonstrated that stress-induced changes in molecular pathways (such as downregulation of the Notch ligands Delta-like proteins encoded by Dll1 and Dll4) also initiate the myeloid transcriptional programme in mouse HSCs21. It is plausible that these and other similar stress-related molecular circuits underlie the accumulation of myeloid-biased HSCs in haematopoietic niches during ageing. Nevertheless, these developmental shifts do not imply the emergence of inflammageing directly. For instance, ageing of humans does not appear to be associated with an active shift of circulating monocytes to a pro-inflammatory phenotype, given that these cells show similar transcriptional profiles in young and old healthy individuals22,23. Further studies of human HSC ageing at single-cell resolution are needed to better understand how myelopoiesis is regulated in aged individuals.
Table 1.
Changes in main immune cell populations in ageing
| Immune cell population | Age-associated changes | Methods used for analysis |
|---|---|---|
| Mice | ||
| Monocytes | Increased abundance in the spleen and blood135,136 | Flow cytometry |
| Macrophages |
Decreased abundance of alveolar macrophages with reduced proliferative capacity and phagocytosis25,150,151 Increased abundance of Fn1+Ccr2+ interstitial macrophages25,27 Decreased abundance of peritoneal macrophages25,30 Increased abundance of CX3CR1+ macrophages in the liver25 and kidney152 Increased abundance of fetal-derived macrophages in mammary tissue28 |
scRNA-seq, flow cytometry, microscopy |
| DCs |
Decreased abundance of cDC1s in the spleen25 Decreased abundance of cDC2s in the peritoneum25 |
scRNA-seq, flow cytometry |
| Neutrophils | Increased abundance in the lungs and liver25,27 | scRNA-seq, flow cytometry |
| ILCs |
Decreased abundance of NK cells in the spleen, peritoneum, lungs and liver25 Increased abundance of NK cells in neurogenic niches47 Decreased abundance of ILC1s in the liver25 Decreased abundance of ILC2s in the lungs25 Increased abundance of ILC2s in the brain51 |
scRNA-seq, flow cytometry, microscopy |
| B cells |
Increased abundance of age-associated B cells in the spleen and meninges92,93,96 Increased abundance of plasma B cells in the kidney, adipose tissue and bone marrow9,26 Accumulation of Apoe+ age-associated B cells in the spleen57 Accumulation of Zbtb32+ age-associated B cells in the spleen25 Increased abundance of Zcwpw1+ B1 cells in the peritoneum25 |
scRNA-seq, flow cytometry |
| γδ T cells |
Increased abundance of Vγ6+ cells in lymph nodes82 Decreased abundance of Sox4+ cells in the spleen25 Increased abundance in the lungs and liver25 |
scRNA-seq, flow cytometry |
| CD4+ T cells |
Decreased abundance of naive cells in the spleen, peritoneum and lungs25,154 Increased abundance of exhausted-like PD1+ cells in the spleen, peritoneum, lungs and liver25,69 Increased abundance of cytotoxic cells in the spleen69 Increased abundance of activated Treg cells in the spleen25,69,154 and adipose tissue155 |
scRNA-seq, flow cytometry |
| CD8+ T cells |
Decreased abundance of naive cells in the spleen, peritoneum and lungs25,156 Increased abundance in neurogenic niches157 Increased abundance of virtual memory cells in the spleen, blood and lymph nodes156,158 Increased abundance of a CD49d+ subset in the spleen159 Accumulation of exhausted-like PD1+ cells in the spleen, peritoneum, lungs, liver, kidney, adipose tissue, meninges and blood25,160 |
scRNA-seq, flow cytometry |
| Humans | ||
| Monocytes |
Increased abundance in the blood25,60,161 Increased abundance of non-classical CD14+CD16+ cells in the blood162 |
Flow cytometry |
| DCs | Decreased abundance of pDCs in the blood162 | Flow cytometry |
| B cells |
Decreased abundance of B1 cells (CD19+CD20+CD27+CD38low/midCD43+) in the blood163 Decreased abundance of the ZBTB32+ (CD27+CD38low/mid) subset in the blood25 |
scRNA-seq, flow cytometry |
| MAIT cells | Decreased abundance in the blood25,164 | scRNA-seq, flow cytometry |
| γδ T cells |
Decreased abundance in the blood165 Decreased abundance of Vδ2+Vγ9+ cells in the blood25,166,167 Decreased abundance of SOX4+ cells in the blood25 |
scRNA-seq, flow cytometry |
| CD4+ T cells |
Decreased abundance of naive cells in the blood60,61,137,138 Increased abundance of TEM cells in the blood138 Decreased abundance of recent thymic emigrants in the blood137 Decreased abundance of an interferon-activated subset in the blood25 Increased abundance of the cytotoxic subset in the blood (supercentenarians)72 |
scRNA-seq, flow cytometry |
| CD8+ T cells |
Decreased abundance of naive cells in the blood25,61,138 Increased abundance of TEM cells in the blood25,138 Increased abundance of the GZMK+(CD28+CD57–) subset of TEM cells in the blood25 Increased abundance of the CD57+ subset in the blood149 |
scRNA-seq, flow cytometry, mass cytometry |
cDC1, conventional type 1 dendritic cell; cDC2, conventional type 2 dendritic cell; CX3CR1, CX3C chemokine receptor 1; DC, dendritic cell; GZMK, granzyme K; ILC, innate lymphoid cell; ILC1, group 1 innate lymphoid cell; ILC2, group 2 innate lymphoid cell; MAIT cells, mucosal associated invariant T cells; NK, natural killer; pDC, plasmacytoid dendritic cell; scRNA-seq, single-cell RNA sequencing; TEM cell, effector memory T cell; Treg cell, regulatory T cell.
Ageing is also associated with alterations in subpopulations of tissue-resident myeloid cells (Fig. 2). scRNA-seq analyses revealed substantial heterogeneity among tissue-resident myeloid cells, especially in response to inflammation24–27, and showed increased proportions of interstitial macrophages expressing chemokine receptor genes Ccr2 and Cx3cr1 but reduced proportions of alveolar macrophages in aged mice25 (Table 1). Moreover, alveolar macrophages in aged mice highly express genes that contribute to inflammageing, such as Spp1 (which encodes osteopontin) and Ccl3 (ref.27). In line with this, the proportions of fetal-derived tissue-resident macrophages markedly decrease in aged mouse mammary tissues. By contrast, the proportions of macrophages that arise from bone marrow-derived myeloid precursors are not changed, but these cells show increased expression of pro-inflammatory genes (such as Ccl5 and Gdf15)28. In the peritoneal cavity, scRNA-seq identified markedly reduced numbers of large peritoneal macrophages (which originate from embryonic precursors and express Icam2) in old mice versus young mice25, as previously observed by means of flow cytometry29,30. Moreover, single-cell analysis of ageing mice demonstrated increased inflammatory function (including expression of the cytokine gene Il1b) of liver-resident macrophages (also known as Kupffer cells), which likely contributes to hepatic inflammation and damage26. Thus, the tissue environment in aged mice is generally associated with a substitution of tissue-resident subsets of macrophages originating from embryonic precursors for monocyte-derived populations. Their transcriptional programmes shift towards pro-inflammatory phenotypes, contributing to inflammageing. It is crucial to further understand the fate of these macrophage lineages in various organs during human ageing, and one important resource is a recently published dataset dissecting the development of human tissue-resident macrophages at single-cell resolution31.
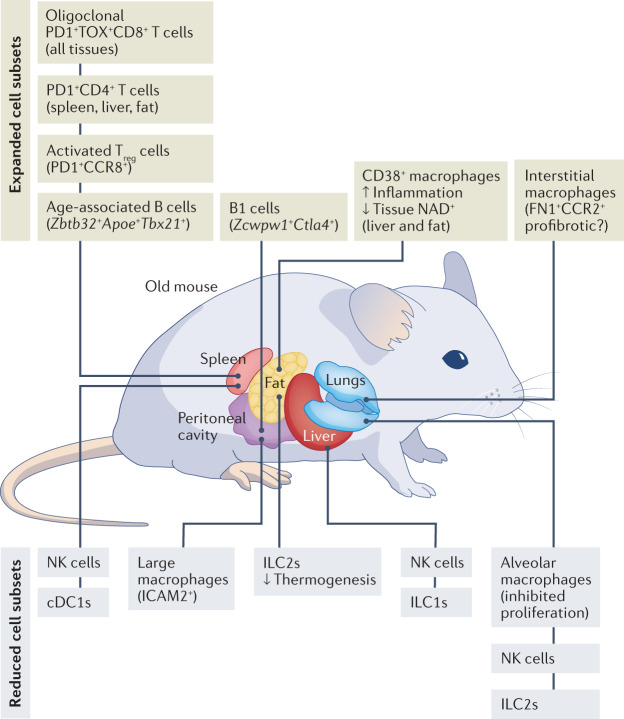
Fig. 2. Alterations of immune cell populations in ageing.
Single-cell techniques identified expanded and reduced immune cell populations with distinct phenotypes in multiple organs of old mice. Exhausted PD1+TOX+CD8+ T cells and activated PD1+CD4+ T cells accumulate across multiple tissues in ageing, whereas natural killer (NK) cells and innate lymphoid cells (ILCs) are among the cell subsets that are reduced in abundance in various organs. Functionally and phenotypically similar CD38+ macrophages expand in metabolically active organs — liver and fat — of old mice173,174. Unlike these cell subsets commonly affected by age in many tissues, alterations of other immune cell populations show organ-specific patterns in ageing. For example, activated regulatory T cells (Treg cells) increase in abundance in the spleen of old mice25,69,154. Tissue-resident alveolar macrophages decrease in abundance and fibronectin 1-positive (FN1+), CC-chemokine receptor 2-positive (CCR2+) interstitial macrophages increase in abundance in the lungs of old mice25,150. Distinct subsets of age-associated B cells accumulate in the spleen (B cells expressing Zbtb32, Apoe and Tbx21) and peritoneal cavity (B1 cells expressing Zcwpw1 and Ctla4) of aged mice. cDC1, conventional type 1 dendritic cell; ICAM1, intercellular adhesion molecule 1; ILC1s, group 1 innate lymphoid cells; ILC2s, group 2 innate lymphoid cells.
To a certain degree, pro-inflammatory shifts of myeloid cell populations in old organisms might depend on effects of ageing on physiological processes and might be ameliorated by systemic interventions, such as dietary restrictions, including caloric restriction32. Recent single-cell transcriptomics analysis of the effects of caloric restriction on multiple tissues in ageing rats showed that caloric restriction represses age-related inflammation33. Specifically, that study demonstrated that caloric restriction reverses the pro-inflammatory polarization of aged M1 macrophages, increases the proportions of anti-inflammatory macrophages with high phagocytic activity and modifies ligand–receptor-dependent communication between macrophages and stromal cells33. Further studies are needed to translate the effect of caloric restriction and similar interventions to myeloid cell populations in ageing individuals.
Neutrophils are also important myeloid cells, which although generally short-lived may also be affected by ageing. Neutrophil development and numbers seem to be unaltered systemically by ageing, yet neutrophils show a decline in phagocytic activity and killing of bacteria in elderly humans34. Furthermore, recent studies showed that age-related changes in the tissue environment could reprogramme neutrophils. Thus, in mice, ageing interferes with transepithelial migration of neutrophils in injured tissues via a process regulated by CXC-chemokine ligand 1 (CXCL1), resulting in aberrant neutrophil trafficking and subsequent remote organ damage35. CXCL1 also recruits neutrophils to the liver in aged mice, where they induce tissue senescence and inflammation via the production of reactive oxygen species36. This crosstalk between the inflammageing-associated chemokine CXCL1, tissue damage and aberrant neutrophil functions possibly contributes to the development of age-related chronic inflammation and can be further studied in the context of exacerbated pulmonary and hepatic chronic diseases in ageing individuals. Furthermore, a recent multi-omics profiling study demonstrated that age-associated neutrophil remodelling is evident in bone marrow-residing neutrophils37, suggesting a role for the aged bone marrow microenvironment in age-associated neutrophil dysregulation.
Among other cellular phenotypes linked to ageing, disturbed phagocytic activities of neutrophils, macrophages and dendritic cells (DCs) and their reduced capacity to engulf apoptotic cells via efferocytosis have been described in mice and humans38–40. In old mice, conventional type 1 dendritic cells, but not conventional type 2 dendritic cells, are decreased in abundance in the spleen25. However, it remains controversial whether aged dendritic cell subtypes demonstrate specific alterations in antigen presentation capacity41.
Effects of ageing on innate lymphocytes
Innate lymphoid cells (ILCs) include natural killer (NK) cells, group 1 ILCs (ILC1s), group 2 ILCs (ILC2s) and group 3 ILCs42. ILCs play a unique role in tissue immunity via cytokine secretion and interaction with other immune and stromal cells. However, surprisingly little is known about the role of ILCs in the homeostasis of aged tissues. Age-related changes in NK cells in older humans (≥60 years) include a shift towards a maturer CD14+CD56hi subset, coupled with diminished cytokine secretion, cytotoxicity and especially the release of perforin43–45. At least some of the age-associated alterations in NK cell functions are programmed by the aged environment in mice. Indeed, NK cell transfer experiments show that the aged environment interferes with NK cell maturation and proliferation46. In line with these observations, scRNA-seq analysis of immune cells from the spleen, lungs and liver demonstrated a decrease in NK cell proportions in old mice25. These data suggest a general ability of an aged environment to suppress NK cell homeostasis. It is possible that the decreased NK cell numbers contribute to impaired antiviral immunity in older organisms43. Intriguingly, tissue-specific proliferation and accumulation of NK cells was shown to occur in distinct aged niches, such as the dentate gyrus neurogenic niche of human and mouse aged brains. scRNA-seq analysis of NK cells in the aged dentate gyrus revealed features of increased activation and cytotoxicity47. Moreover, mouse NK cells eliminate senescent neuroblasts and mediate age-related decline in cognitive function47, providing a striking example of a deleterious effect of NK cells in old organisms.
ILC1s are non-cytotoxic cells that produce interferon-γ (IFNγ) and function as the first line of defence against viruses and certain bacteria48. Similarly to NK cells, hepatic ILC1 numbers decrease in old mice25. Tissue-resident liver ILC1s develop locally from SCA1+MAC1+ HSCs via a mechanism driven by IFNγ49. It remains unclear which signals from the aged environment disturb ILC1 development and whether ILC1s in other organs are affected in old mice and humans. ILC2s are involved in innate immune responses against parasites and regulate tissue homeostasis42. Single-cell analyses in mice showed that ILC2s are less abundant in old lungs25 and adipose tissue50. However, the decrease in ILC2 populations is not a universal hallmark of ageing; for example, the numbers of ILC2s increase in aged mouse brains51. Ageing increases early bone marrow development of ILC2s, and the decline in tissue ILC2 populations might be due to dysfunctional migratory cues52. Functionally, ILC2s appear to be associated with healthier ageing phenotypes. In old adipose tissues, a decrease in the numbers of ILC2s is linked to a metabolic impairment and cold-induced lethality exacerbated in old mice50, whereas in the ageing brain, ILC2s function to counteract cognitive decline51. ILC2s have been found in human adipose tissue53, but it remains unclear whether they contribute to age-associated metabolic dysfunction in ageing individuals.
Effects of ageing on T cells
Age-associated CD8+ memory T cells
Essential for antiviral and antitumour immunity, CD8+ T cells are substantially affected by ageing in both mice and humans. In general, older age has been associated with a loss of naive CD8+ T cells coupled with an increase in memory subsets and an overall increase in clonally54–56. Most recently, age-associated remodelling of the mouse immune T cell compartment was comprehensively characterized by scRNA-seq and single-cell T cell receptor (TCR) sequencing of four tissues (spleen, peritoneum, lungs and liver)25. These data confirmed a decrease in the abundance of naive CD8+ T cells across the four tissues and defined a distinct age-associated CD8+ T cell subpopulation (referred to by the study authors as ‘Taa cells’) uniquely characterized by co-expression of the checkpoint receptor PD1 and the transcription factor TOX25. Although absent in young mice, these age-associated PD1+TOX+CD8+ T cells accumulate in multiple tissues in old animals, including the spleen, liver, lungs and fat. They constitute up to 60% of all CD8+ T cells in these tissues and are a striking example of systematic changes in the ageing immune system that affect multiple tissues. This observation is consistent with the organ-wide single-cell resolution profiling data produced by the Tabula Muris Senis Consortium and Calico Life Sciences26,57, although these studies did not perform a detailed analysis of CD8+ T cell ageing. The age-associated CD8+ T cells exhibit a phenotype of T cell exhaustion characterized by high expression levels of TOX, PD1, lymphocyte activation gene 3 protein (LAG3) and T cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT)25, which generally suggests a loss of classical effector functions58,59. However, upon TCR stimulation, they produce a distinct set of pro-inflammatory molecules such as granzyme K (GZMK) and CCL5 (ref.25). This observation suggests a functional role for these cells in murine ageing as a source of pro-inflammatory signals that might be particularly important in the context of old tissue remodelling or under conditions of an acute infection.
In humans, most of the studies focus on the ageing of circulating CD8+ T cells and use mass cytometry and scRNA-seq to analyse peripheral blood mononuclear cells (Table 1). For example, recent scRNA-seq studies of healthy donors reported a twofold decrease in the naive CD8+ T cell fraction of peripheral blood mononuclear cells from old individuals compared with young individuals60,61. Another study used cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) to characterize old and young peripheral blood mononuclear cells and also demonstrated an age-associated decrease in the abundance of naive CD8+ T cells and significant remodelling of the T cell memory compartment25. Specifically, in healthy lean individuals, GZMK-expressing, but not granzyme B (GZMB)-expressing, CD8+ effector memory T cells (TEM cells) proportionally increase in abundance with age relative to overall CD8+ T cells25. A recent mass cytometry study of a large cross-sectional ageing cohort of 156 healthy individuals confirmed these observations and found that GZMK-expressing CD8+ T cells gradually increase in abundance in the circulation with age along with CD8+ central memory T cells61. Of note, GZMK+CD8+ T cells express CD28 but lack CD57 and are distinct from CD45RA+ TEM cells — the age-associated expansion of which was reported in cytomegalovirus-exposed older individuals (range 21–96 years)62 but was not significant in a cohort of healthy donors (range 25–80 years)61.
Although GZMK-expressing human CD8+ TEM cells are similar to the mouse age-associated PD1+TOX+GZMK+CD8+ T cells in some aspects (such as high expression of EOMES and GZMK), in humans this cell subpopulation cannot be identified solely on the basis of expression of PD1 (refs25,61). This difference between mouse and human CD8+ T cell ageing landscapes might arise because, unlike laboratory mice, humans experience multiple immune challenges during their lifespan, which forms a broader and more phenotypically diverse pool of antigen-experienced memory T cells.
Effect of ageing on CD4+ T cells
Similarly to CD8+ T cells, the proportion of naive CD4+ T cells declines and the memory population expands with age in mice and humans25,63,64. This switch to a memory phenotype seems to be driven by a combination of reduced thymic output and cumulative effect of lifelong antigen exposure as CD4+ T cells with transgenic TCRs tend to maintain a naive phenotype even in aged mice63. Another long-known feature of age-associated CD4+ T cell remodelling is increased abundance of regulatory T cells (Treg cells) in the spleen and lymph nodes65–68. However, this increase in abundance of Treg cells was not observed in ageing lungs or liver of mice25. The accumulation of splenic Treg cells has been suggested to play a role in restricting immune responses in aged animals (such as compromising control of Leishmania major infection)66,67. A recent study used scRNA-seq to dissect the ageing of splenic CD4+ T cells in mice and reported that the age-associated increase is not a feature of all Treg cells but is instead attributed to a subpopulation of activated Treg cells69. These activated Treg cells are defined by overexpression of Foxp3, Cd81, Cd74 and genes encoding activation markers(such as Tnfrsf9)70. Activated Treg cells are approximately ten times more abundant in old mice and are superior in suppressive activities in comparison with resting Treg cells69.
The accumulation of anti-inflammatory activated Treg cells is paralleled by a marked expansion of so-called cytotoxic CD4+ T cells. These cells are characterized by the expression of Eomes and Gzmk and can produce not only IFNγ and tumour necrosis factor (TNF) but also cytotoxic GZMB and perforin upon stimulation69. Similarly to activated Treg cells, the cytotoxic CD4+ T cell subpopulation was not associated with ageing before the single-cell study but was known in other physiological contexts (reviewed by Takeuchi and Saito71). Strikingly, scRNA-seq of ageing human CD4+ T cells identified a similar subset of cytotoxic CD4+ T cells that are profoundly expanded at the extreme end of the longevity spectrum in individuals who reached the age of 110 years — supercentenarians72. The matching single-cell TCR sequencing data suggested that clonal expansion of CD4+ T cells with age is restricted primarily to the subpopulation of cytotoxic CD4+ T cells. By contrast, other T helper cell subsets remain clonally diverse, even in supercentenarians72. Interestingly, cytotoxic CD4+ T cell subpopulations are also more abundant in older patients with COVID-19 (range 32–91 years) with moderate symptoms compared with individuals with severe disease61, which might indicate that these cells can be beneficial during viral infections.
Finally, ageing is also characterized by increased expression of exhaustion markers by splenic CD4+ T cells in mice. Although the concept of T cell exhaustion is less well established for CD4+ T cells than for CD8+ T cells, an age-associated increase in the abundance of CD4+ T cells with high levels of inhibitory receptors (such as PD1 and LAG3) has been observed73,74. PD1+CD4+ T cells have a memory phenotype and are hyposensitive to TCR stimulation74, which matches the canonical features of exhausted CD8+ T cells. Exhausted-like CD4+ T cells constitute about 30% of all CD4+ T cells in the spleen of aged mice. Of note, age-associated changes in CD4+ T cell subpopulations are not universal across the tissues. scRNA-seq profiles of immune cells across four mouse tissues corroborated the data on splenic CD4+ T cell remodelling and revealed that while accumulation of PD1+ cells was observed in all tissues considered, the expansion of Treg cells was evident only in the spleen25.
Age-related cell-intrinsic deficiencies of naive CD4+ T cells have also been described in studies that used aged mice with a transgenic TCR to investigate CD4+ T cell ageing with experimentally controlled antigen exposure. These studies demonstrated that aged naive CD4+ T cells are less responsive to activation in vivo and in vitro than their young counterparts; they have decreased proliferation rates and produce less IL-2 (refs63,75,76). The defect in TCR responsiveness seems to be specific for CD4+ T cells that have spent an extended time in circulation because CD4+ T cells from young thymectomized mice exhibit this phenotype, but recent immigrants from either young or aged bone marrow do not77. Furthermore, a recent study used scRNA-seq to demonstrate that the responses of naive CD4+ T cells isolated from the spleen of old mice to in vitro stimulation are more heterogeneous and variable on the transcriptional level than those of naive CD4+ T cells from young mice, suggesting that activation programmes of aged CD4+ T cells are less tightly regulated78.
An improper activation has been suggested to drive an incomplete differentiation of aged CD4+ T cells into T helper 1 (TH1) and TH2 effector cells76. At the same time, the ability of aged CD4+ T cells to acquire a TH17-type phenotype is not compromised, creating an imbalance and a skew towards antimicrobial responses79,80. In bulk, splenic CD4+ T cells from aged mice and humans produce much more IL-17 than young controls, which might reflect an age-associated accumulation of TH17-type memory cells80,81. As TH17 cells have been implicated in the development of multiple autoimmune conditions, including inflammatory bowel diseases, one can speculate that the second peak of inflammatory bowel disease prevalence that occurs in older adults might be partially ascribed to this skewing of T cell populations. Indeed, transfer of aged CD4+ T cells to Rag1–/– recipients induced severer colitis than transfer of cells from a young donor80. However, further research is required to prove a causal relationship between these events. IL-17-producing cells are also over-represented among γδ T cells in lymph nodes of aged mice, substituting IFNγ-producing effector γδ T cells, which are more abundant in young animals82. Numbers of γδ T cells change with age in a tissue-dependent manner, as demonstrated by a recent single-cell profiling study of multiple mouse tissues. For example, γδ T cells are less frequent in the aged liver and lungs but not in the spleen25. However, under conditions of acute lung inflammation caused by a murine betacoronavirus, old mice show lower numbers of lung γδ T cells, which could be restored by a ketogenic diet83.
Altogether, the described age-associated differences in T cell populations appear to be partly responsible for altered humoral responses in old animals. It has been shown that CD4+ T cell help is required to launch a germinal centre reaction and produce high-affinity antibodies84. Eaton et al. used an adoptive transfer model to compare germinal centre responses facilitated by naive CD4+ T cells from either young mice or old mice with transgenic TCRs85. In that study, cell-intrinsic changes in naive CD4+ T cells from old donors led to fewer antigen-specific B cells in the germinal centre and reduced production of class-switched antibodies. By contrast, naive CD4+ T cells from young mice were sufficient to elicit robust responses in the aged environment. These observations highlight the importance of CD4+ T cell ageing biology for developing vaccination strategies customized to the state of the aged population.
TCR repertoire
Early studies of TCR repertoire changes during ageing — first using TCRβ chain-specific antibodies86 then by bulk TCR sequencing — suggested that TCR diversity loss is associated with the increased prevalence of memory T cells that undergo clonal expansion87,88 but can also be affected by a modest decrease in TCR diversity of naive T cells88–90. Overall, CD8+ T cells show a greater increase in clonality than CD4+ T cells, which is consistent with the more considerable reduction in naive CD8+ T cell subpopulations with ageing. scRNA-seq technology provides a unique opportunity to investigate TCR repertoire complexity via direct sequencing of paired α-chains and β-chains at single-cell resolution, which unequivocally defines TCR clones — a feature not available in bulk level analysis, which often uses only β-chain sequences. In mice, it was recently demonstrated that the accumulation of highly clonal age-associated PD1+TOX+CD8+ T cells is responsible for the clonality increase in the CD8+ T cell compartment25. The clonal expansion of these cells is ‘idiosyncratic’91, meaning that individual mice differ in the specific TCRαβ chains used by the expanded T cell clones. Therefore, there might not be a shared ‘ageing antigen’ sensed by T cells in all ageing mice. Further studies are needed to decipher single-cell clonality landscapes in older individuals and identify specific age-related antigens that drive the accumulation of distinct memory T cell subsets.
Effects of ageing on B cells
Remodelling of adaptive immunity during ageing also includes altered B cell composition and function. A phenotypically and functionally separate subset of B cells has been shown to expand in the spleen and bone marrow of aged mice92,93. These cells are distinct from the conventional naive and memory B cells and have been termed ‘age-associated B cells’. Hao et al. defined age-associated B cells as CD43−CD21/CD35−CD23− B cells92, whereas Rubtsov et al. identified age-associated B cells as CD11b+CD11c+ B cells93. Interestingly, age-associated B cells express Tbx21 (which encodes T-bet), which makes them similar to the B cells that have been shown to contribute to lupus-like autoimmunity in mice94. Although the age-related factors that prime the expansion of these B cell subsets are not entirely understood, a body of evidence suggests that damage-associated molecular patterns, such as debris and chromatin from apoptotic cells, initiate age-associated B cells via the Toll-like receptor 7 (TLR7) or TLR9 axis92,93. Once age-associated B cells are generated, their survival in old mice requires IFNγ signalling95. Age-associated B cells can contribute to inflammageing by altering immune homeostasis. For example, age-associated B cells secrete IL-4 and IL-10 on activation92 and can present antigens to T cells93.
Over the past 3 years, several scRNA-seq studies reported age-associated dynamics of B cells across multiple tissues in mice9,25,26,57,96. In a study profiling both immune and non-immune cells from three tissues of young and old mice, Kimmel et al. characterized a distinct cluster of B cells that increases in abundance with age in the spleen57. This cluster is identified by the expression of the apolipoprotein gene Apoe, yet the precise identity of this B cell subset in terms of immunological nomenclature has not been specified. The aforementioned scRNA-seq profiling study of mouse immune cells in four tissues25 defined an age-associated population of conventional splenic B cells that express the transcription factor genes Tbx21 and Zbtb32 and genes encoding other markers of age-associated B cells (such as Cxcr3 (ref.97)). This Zbtb32+ B cell subset also highly expresses Apoe, suggesting a similarity with the age-associated B cell population reported by Kimmel et al.57. Single-cell analysis of B cells in the meninges revealed Apoe-expressing age-associated B cells that accumulate at the central nervous system borders in old mice96. Analysis of single-cell B cell receptor (BCR) sequencing data provided additional insights into this population. Indeed, splenic age-associated B cells have been recognized as cells displaying characteristics of antigen-experienced B cells with increased somatic hypermutation98. Consistently, the cluster of splenic Tbx21-expressing age-associated B cells demonstrates increased BCR clonality while still preserving BCR repertoire diversity25.
Another study that used a combination of scRNA-seq and single-cell BCR sequencing also reported a progressive increase in B cell clonality in ageing mice26. This increase was attributed to a cluster of cells expressing markers of plasma B cells Jchain, Xbp1 and Derl3 found in the spleen, bone marrow, kidney and fat of aged mice, albeit in tiny proportions (~0.1% in these tissues)9. Moreover, tracing the cells from this cluster across the mouse lifespan showed that, similarly to age-associated B cells, age-associated plasma B cells initially arise in the bone marrow and spleen and subsequently increase in abundance in peripheral tissues, such as adipose tissue and kidneys, later in life9. Functionally, plasma B cells can produce pro-inflammatory cytokines and enhance myelopoiesis in the ageing bone marrow99. Interestingly, a phenotypically similar population of plasma B cells increases in abundance in adipose tissue of older humans (≥70 years)9.
A focused scRNA-seq analysis of immune cells in the peritoneal cavity (a location not included in Tabula Muris Senis26) identified B1-like cells as heavily accumulating with age and characterized by a very limited BCR repertoire25. Highly enriched in VH11-2 chains, these BCRs recognize the cell membrane component phosphatidylcholine100. Similarly to splenic age-associated B cells, peritoneal B1-like cells express the transcription factor gene Zbtb32 yet represent a distinct transcriptional cluster characterized by high levels of an exclusive set of genes, including Zcwpw1 and Ctla4 (refs25,101). The Biragyn group showed an age-associated accumulation of B1-like cells, which were identified in old mice and humans by surface expression of the TNF ligand superfamily member 4-1BBL29,102,103. Functionally, B1-like cells become more activated with age, serve as potent inducers of cytotoxic CD8+ T cells and respond to commensal bacteria29,102,103. The insights from single-cell studies expand our understanding of age-associated B cell heterogeneity and point to potential antigen specificity of age-associated B cell subsets and their tissue-specific inflammatory functions in old organisms.
Role of non-immune cells in immune ageing
Immune functions of stromal cells in ageing
Immune function is not limited to cells of haematopoietic origin, and multiple types of stromal and epithelial cells are equipped to respond to pathogens and partake in immune homeostasis104,105. For instance, fibroblasts are critical structural components of most tissues and a stromal cell type extensively studied in pathological states and ageing106,107. Transcriptional analysis showed a loss of cell identity by dermal populations of old fibroblasts accompanied by upregulation of inflammatory genes in mice108. Similarly, an analysis of transcriptomic and epigenomic changes in aged fibroblasts identified an increased abundance of ‘activated fibroblasts’ (for example, fibroblasts involved in tissue repair), including pathways related to cytokine signalling and inflammation109. In line with these observations in mice, scRNA-seq analysis of human skin fibroblasts demonstrated a higher expression of genes contributing to low-grade inflammation in distinct populations of aged fibroblasts and predicted a decline in cell–cell communications between fibroblasts and other cell types in aged skin110.
Endothelial cells line vascular beds and regulate immune trafficking. Ageing appears to substantially modify the inflammatory transcriptional signatures of endothelial cells in various tissues. For instance, single-cell transcriptional profiling of endothelial cells in aged mouse brain prominently implicated altered inflammation and cytokine signalling pathways associated with neurodegenerative diseases111. Moreover, in mice, a single-cell analysis revealed transcriptional alterations in aged liver sinusoidal endothelial cells, which are essential regulators of immune defence26. Finally, communication between endothelial cells and various immune cell types (including macrophages and T cells) is markedly altered in old tissues, including liver and fat, as was revealed by a single-cell transcriptional analysis in rats33.
Senescent cell contribution to inflammageing
Ageing is characterized by a gradual accumulation of senescent cells throughout the body (Box 3). Senescence is a terminal stage of cell differentiation, often acquired by damaged or stressed cells, that leads to permanent withdrawal from the cell cycle and a switch to a distinct secretome112. This secretome, termed the ‘senescence-associated secretory phenotype’ (SASP)113–116, involves the secretion of multiple chemokines, cytokines and growth factors and contributes to low-grade systemic inflammation in ageing117. Although a core transcriptional signature of senescent cells in vitro has been described118, no robust approach to determine the transcriptional landscape of senescent cells in vivo has been developed to date. Moreover, senescent cells are profoundly variable in their expression of regulatory and SASP genes even in vitro119. Nonetheless, recent scRNA-seq analyses provided some insights into the development of senescent cells in ageing mice. The scRNA-seq atlas of ageing mouse tissues (Tabula Muris Senis) showed that the fraction of cells expressing Cdkn2a (which encodes p16INK4A, a commonly used marker of senescent cells117) increases in different tissues in old mice26. However, another single-cell analysis found no difference in Cdkn2a expression between young mice and old mice57, emphasizing current conceptual and technical limitations precluding identification of bona fide senescent cells in living organisms. A recent article reported scRNA-seq of p16INK4Ahi senescent cells with heterogeneous senescence-associated phenotypes in a reporter mouse model120. Interestingly, senescent cells include various cell types that are enriched in hepatic endothelium and renal proximal and distal tubule epithelia. Elimination of these p16INK4Ahi senescent cells decreases immune infiltration in a model of metabolic liver disease120. Further studies are needed to better understand how heterogeneous senescent cells contribute to low-grade chronic inflammation and reshape immunity in ageing.
Box 3 Cellular senescence and senescent cells.
Cellular senescence is a state of irreversible cell-cycle arrest due to cellular replicative limit, tumour suppression, inflammation, embryogenesis, wound healing or age-associated loss of regenerative capacity192. Cellular senescence has been defined as a stress response to developmental and environmental stimuli, including genotoxic agents, hypoxia, mitochondrial dysfunction, oncogene activation, epigenetic modifiers and cytokines193. Senescent cells are a product of cellular senescence and feature a combination of molecular and morphological characteristics. These cells undergo permanent cell-cycle arrest that is distinct from terminal differentiation and quiescence194. They have rewired cell-cycle regulatory pathways with a specific phosphorylation profile of cyclin-dependent kinases (CDKs) and upregulated expression of the CDK2 inhibitor p21CIP1 (also known as CDKN1A) and the CDK4/CDK6 inhibitor p16INK4A (also known as CDKN2A)193. Although increased expression of p21CIP1 and p16INK4A is not unique to senescent cells and can be found in, for example, non-senescent macrophages, p21CIP1 and p16INK4A are often used both as markers of senescent cells and as targets to develop genetic models allowing selective elimination of senescent cells132,134,195. Morphological characteristics of senescent cells include an enlarged cell size due to a diluted cytoplasmic domain196 and increased activity of lysosomal senescence-associated β-galactosidase — the most widely used histological biomarker of senescent cells197. Other features of senescent cells include DNA, protein and lipid damage, dysfunctional mitochondria and lysosomes, and an altered chromatin landscape193. Importantly, senescent cells have a specific secretory profile involving excessive secretion of pro-inflammatory cytokines and chemokines, growth modulators, angiogenic factors and matrix metalloproteinases, collectively named the ‘senescence-associated secretory phenotype’113,114. Different components of the senescence-associated secretory phenotype mediate the communication of senescent cells with immune cells, affect the functions of stromal cells in tissues and contribute to the low-grade systemic inflammatory phenotype in ageing198.
Interplay between immune and senescent cells
Recognition and elimination of senescent cells by specialized immune cells is a crucial step in maintaining tissue homeostasis and preventing maladaptive inflammation121 (Fig. 3). Components of the SASP, such as CCL2 and CCL7, attract immune cells, including macrophages, NK cells and T cells121–123. Senescent cells also recruit neutrophils36, tissue infiltration of which is one of the hallmarks of immune ageing. Macrophages can physically interact with and eliminate senescent cells in developmental and pathological processes124–128. Whether the impaired phagocytosis by aged macrophages38 is causative for senescent cell accumulation in old tissues remains to be clarified. NK cells recognize and kill senescent cells to coordinate tissue remodelling129 and limit fibrogenic responses to acute tissue damage130. Intriguingly, the cytotoxic functions of NK cells and T cells appear to control the accumulation of senescent cells in ageing because mice lacking perforin (the crucial effector of cytotoxicity) exhibit both higher senescent-cell tissue burden and chronic inflammation while they age131. Consistent with this is the observation of decreased frequencies of NK cells and replacement of tissue-resident CD8+ T cells with exhausted-like PD1+ T cells in various tissues of aged mice25. In addition to cytotoxic CD8+ T cells, at least in the context of cellular senescence in cancer, CD4+ T cells also provide immune surveillance of premalignant senescent cells and require macrophages to eliminate them128. In mice, elimination of senescent cells via genetic manipulation can prevent or delay tissue dysfunction and extend ‘healthspan’132,133 (the period of time an organism lives free of chronic diseases of ageing), suggesting that immune surveillance and elimination of senescent cells contribute to delay of age-related diseases. However, the immune cell-mediated elimination of senescent cells is context dependent and tightly regulated because a transitory accumulation of senescent cells is critical for tumour suppression and certain regenerative processes such as wound healing134.
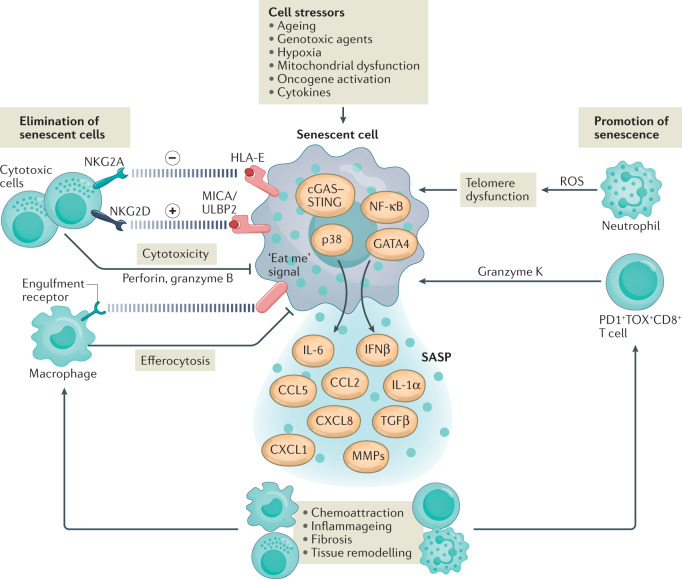
Fig. 3. Interactions between immune and senescent cells.
Senescent cells differentiate from various cell types in response to stress signals in ageing tissues. Senescent cells differ in their transcriptional signatures and show heterogeneity in the senescence-associated secretory phenotype (SASP)119. Innate immune signalling pathways (involving p38 mitogen-activated protein kinase and cGAS–STING pathways) and transcriptional factors (such as NK-κB and GATA4) induce cytokines and tissue remodelling factors that constitute the SASP175–179. Among the SASP components, growth factors (such as transforming growth factor-β (TGFβ)) and matrix metalloproteinases (MMPs) increase tissue fibrosis, whereas chemokines and cytokines create a pro-inflammatory environment in ageing tissues. Chemokines of the SASP attract immune cells that can interact with, identify and eliminate senescent cells. Cytotoxic natural killer (NK) cells and CD8+ T cells recognize and kill senescent cells via a perforin-dependent mechanism129–131. This elimination of senescent cells is orchestrated by specific activating (MICA/ULBP2–NKG2D) and inhibitory (HLA-E–NKG2A) ligand–receptor interactions between senescent cells and cytotoxic cells180,181. Moreover, tissue macrophages can find and eliminate damaged and dying senescent cells by efferocytosis. By contrast, chemoattraction of neutrophils by the SASP components CXC-chemokine ligand 1 (CXCL1) and CXCL8 expand senescent cell load in ageing tissues. Neutrophils drive telomere dysfunction and induce senescence in bystander non-immune cells by generating reactive oxygen species (ROS)36. Accumulation of exhausted-like PD1+TOX+CD8+ T cells associated with ageing in old tissues can enhance expression of the SASP via pro-inflammatory granzyme K (GZMK)25. CCL, CC-chemokine ligand; IFNβ, interferon-β.
Immune ageing in mice versus humans
Despite tremendous progress in single-cell techniques and their application to mouse immune ageing in various tissues, current single-cell data in humans are generally limited to immune cell populations from the blood (Table 1). Nevertheless, the comparison of available datasets shows a certain degree of similarity in how distinct immune cell subsets change with ageing in mice and humans. For example, monocytes increase in abundance whereas plasmacytoid dendritic cells decrease in abundance in both species when they age25,135,136. Similarly, naive CD4+ T cells and CD8+ T cells and populations of SOX4-expressing γδ T cells are less abundant in old mice and humans25,137,138. Furthermore, cytotoxic CD4+ T cells and GZMK+CD8+ TEM cells universally accumulate during ageing in both species25,69,72. These findings suggest that mice and humans share unidentified age-related factors that similarly affect these immune cell populations. One of the candidates for this role might be a pro-inflammatory component of the SASP, a shared characteristic of old mice and humans. Importantly, unlike in laboratory mice, which usually live in specific-pathogen-free conditions, the immune system in humans is constantly reshaped by a history of infections, and a role for some chronic viral infections (such as cytomegalovirus infection) in immune ageing in humans is well documented139. This and other infections could explain the more complex pattern of age-associated alterations among CD8+ T cell subsets in older humans compared with mice. Moreover, this might be why there is no direct phenotypic similarity between age-associated B cells revealed by scRNA-seq in these species25. Furthermore, our understanding of how tissue-resident immune cells (including macrophages, T cells and ILCs) age in humans is minimal and will significantly benefit from future single-cell studies.
Expanding the ageing paradigm
Single-cell technologies have already significantly expanded the current paradigm of immune ageing. For example, they have made it possible to better understand the heterogeneity of human CD8+ TEM cells and distinguish between GZMK+ and GZMB+ TEM cell populations, revealing the GZMK+ cells as the crucial age-associated CD8+ TEM cell subset in humans25. Single-cell analyses also defined cytotoxic CD4+ T cells as a key feature of immune ageing conserved between mice and humans69,72. In addition, combined single-cell sequencing of RNA and antigen receptors expanded our capability to study TCR and BCR repertoires to an unprecedented level. One of the recent discoveries stemming from this expanded toolbox is that the age-associated loss of TCR repertoire diversity in mouse CD8+ T cells appears to be driven primarily by the replacement of polyclonal naive T cells and TEM cells by oligoclonal PD1+TOX+CD8+ T cells25. Finally, unbiased single-cell profiling of immune cells across multiple tissues led to the demonstration that a dietary intervention with a beneficial effect on healthspan not only reshapes age-disturbed immune landscapes but also specifically reprogrammes distinct pro-inflammatory macrophage subsets in rodents33. These results suggest an intriguing possibility that some immune cell populations might be a future target for anti-ageing therapies.
The next-generation profiling techniques will allow further exploration of the many remaining questions in the field of immune ageing. For instance, the interaction between immune cells and the ageing environment is an important avenue for investigation, including the need to understand which processes are driven by immune cells and which reflect adaptation of immune cells to the aged environment. Spatial transcriptomics and imaging cytometry can reveal spatial relationships between immune cells and non-immune cells and promise to solve this puzzle in an unbiased manner. Also, tissue-resident immune cell populations are not sufficiently studied in ageing humans. Thus, spatial techniques will be an essential foundation for human immune ageing research. This approach is especially important for a better understanding of immune responses to pathogens in older individuals, in which the severity of some (but not all) viral and bacterial infections — such as pneumococcal disease, influenza virus and COVID-19 — is profoundly increased compared with that in younger adults140–144. Single-cell analyses across individuals of different ages can discriminate between the effects of ageing and the effects of the infectious disease itself61, which will help to distinguish between age-associated changes and the immune mechanisms of diseases such as COVID-19 (ref.145).
Another frontier in single-cell immunology is multiplexing of orthogonal approaches to characterize the ageing immune system. For instance, a simultaneous characterization of hundreds and thousands of genes and proteins in individual immune cells (for example, using CITE-seq) will deepen the immune profiling and facilitate prediction and validation of immune cell functions in ageing. The same approach is valid for a combination of transcriptomics and epigenomics of ageing immune cells. Parallel application of scRNA-seq and single-cellassay for transposase-accessible chromatin using sequencing (scATAC-seq) has already demonstrated encouraging potential to decipher new regulatory programmes in age-associated immune cell populations of CD8+ T cells in mice and humans25 (Table 2). Therefore, combined scRNA-seq and scATAC-seq in individual cells can identify crucial functional states and the underlying epigenetic landscapes that adapt the immune system to the aged environment.
Table 2.
Single-cell transcriptomic and epigenetic datasets of immune ageing
| Dataseta | Species and organs | Details | Ref. |
|---|---|---|---|
| Profiling of single cells across the lifespan in mice (Tabula Muris Senis) | Mouse immune and non-immune cells from the bladder, bone marrow, brain, fat, heart and aorta, kidney, large intestine, limb muscle, diaphragm, liver, lung, mammary gland, pancreas, skin, spleen, thymus, tongue and trachea | scRNA-seq of total cells from male and female C57BL/6JN mice from six age groups: 1 month (the equivalent of early human childhood) to 30 months (the equivalent of a human centenarian). This dataset contains scTCR-seq and scBCR-seq data | 26 |
| Profiling of immune cells in young and old mice | Sorted mouse immune cells from the spleen, liver, lung and peritoneal cavity | scRNA-seq of CD45+ cells from young (3–4 months) and aged (17–18 months) male C57BL/6J mice. This dataset contains scTCR-seq and scBCR-seq data. Because of selective enrichment for CD45+ cells, this dataset captures rare immune cell populations in tissues | 25 |
| Mouse ageing cell atlas in three organs | Mouse immune and non-immune cells from the spleen, lung and kidney | scRNA-seq of total cells from young (7 months) and aged (22–23 months) male C57BL/6J mice. This dataset contains scTCR-seq and scBCR-seq data | 57 |
| Mouse ageing lung atlas | Mouse single-cell suspensions of whole lungs | scRNA-seq of total cells from young (3 months) and aged (24 months) male C57BL/6N mice | 27 |
| Immune cells in young and old mice | Mouse cells from the meninges, bone marrow and blood | scRNA-seq of cells from young (3 months) and aged (25 months) female C57BL/6J mice. This dataset contains scBCR-seq data | 96 |
| Analysis of immune cells in young and aged mice | Mouse immune cells from the spleen and dentate gyrus | scRNA-seq of CD45+ cells from young (3 months) and old (18 months) C57BL/6 mice | 47 |
| Ageing-associated alterations in mammary cells in mice | Epithelial and stromal cells from mouse mammary tissues | scRNA-seq of cells from young (3–4 months) and aged (13–14 months) virgin female C57BL/6J mice | 28 |
| Ageing-associated alterations in adipose tissue in mice | Tissue-resident immune cells from young and old gonadal adipose tissue | scRNA-seq of CD45+ cells from young (2–3 months) and aged (18–24 months) male C57BL/6J mice | 50 |
| Effect of ageing on CD4+ T cells in mice | Mouse CD4+ T cells from the spleen | scRNA-seq of CD4+ T cells from young (2–3 months) and aged (22–24 months) C57BL/6J mice | 9 |
| Single-cell atlas of ageing in rats | Rat immune and non-immune cells from the liver, fat, kidney, aorta, skin and bone marrow | scRNA-seq of total cells from young (5 months) and old (27 months) male and female Rattus norvegicus. One group of old rats was calorie restricted (70% of calories) starting at 18 months | 33 |
| Cells from wounds of old mice | Mouse cells from entire wounds | scRNA-seq of cells from skin wounds from aged (24 months) C57BL/6JN mice with fast-healing and slow-healing trajectories | 109 |
| Cell composition in old neurogenic niches | Mouse cells from subventricular zone neurogenic niche | scRNA-seq of cells from subventricular zone from young (3 months) and aged (28–29 months) male C57BL/6JN mice | 157 |
| PBMCs of healthy young and older individuals | Human PBMCs from healthy non-obese individuals | scRNA-seq and CITE-seq (39 antibodies) of PBMCs from 11 young (25–29 years) and 10 older (62–70 years) male donors. This dataset contains scTCR-seq data | 25 |
| PBMCs of young and older healthy adults and young and older adults with COVID-19 | Human PBMCs | scRNA-seq and scATAC-seq of PBMCs from young (20–45 years) and older (60–80 years) male and female donors — healthy or with COVID-19. This dataset contains scTCR-seq and scBCR-seq data | 60 |
| PBMCs of supercentenarians and control adult individuals | Human PBMCs from older individuals (controls) and supercentenarians | scRNA-seq of PBMCs from five controls (50–80 years) and seven supercentenarians (110 years). This dataset contains scTCR-seq data | 72 |
| Epigenetic landscapes of aged immune cells in mice and humans | Mouse splenocytes and human PBMCs | scATAC-seq of mouse CD45+ cells from the spleen of young (3–4 months) and aged (17–18 months) male C57BL/6J mice and of human PBMCs from three young (25–29 years) and three older (62–70 years) healthy male donors | 25 |
CITE-seq, cellular indexing of transcriptomes and epitopes by sequencing; PBMCs, peripheral blood mononuclear cells; scATAC-seq, single-cell assay for transposase-accessible chromatin using sequencing; scBCR-seq, single-cell B cell receptor sequencing; scRNA-seq, single-cell RNA sequencing; scTCR-seq, single-cell T cell receptor sequencing. aDatasets can be explored online through an Artyomov laboratory webpage.
Cellular senescence has been associated with various age-related phenotypes and diseases, but further studies are needed to determine the causative roles of senescent cells in this context112. Overcoming the current conceptual and technical limitations of identifying bona fide senescent cells at the single-cell level, especially in vivo, is a significant challenge limiting our ability to capture the interactions between senescent and immune cells and better understand the role of the SASP in inflammageing. Possible approaches might rely on combined measurements of mRNAs and proteins alongside the specific functional markers of macromolecular damage in individual cells. Furthermore, despite the well-documented beneficial effect of targeted elimination of senescent cells133,146,147, it remains to be understood how this affects the ageing immune system.
Finally, a challenging caveat in studying immune ageing is the profound diversity of ageing phenotypes in mice and humans. Because physiological manifestations of ageing depend on a combination of genotype, medical history and environment, it is crucial to understand how these factors impact the immune system during ageing. Furthermore, immune ageing can be studied in the context of various risk factors that affect healthspan. For example, genetic factors driving the variability of functional decline in ageing are recognized in multiple genetic association studies148. Future single-cell research can deepen our understanding of how these and other factors affect distinct populations of immune cells at different ages. In mouse models of immune ageing, this should prompt researchers to strictly control effects of ageing that are derivatives from associated physiological processes (for example, decreased activity and accumulation of adipose tissue). Similarly, single-cell analysis of the immune system from phenotypically and genetically diverse individuals of different ages will help to reveal important mechanisms of immune system adaptation to the aged environment. Adding the temporal dimension to single-cell studies (both cross-sectional and longitudinal) across the lifespan is a major future direction to deeply characterize individual trajectories of immune ageing in mice and humans. For example, following up time-resolved multidimensional changes in healthy immune ageing149 or different stages of the immune response to an infection in young hosts versus older hosts is a crucial future goal.
In conclusion, adding space, time, depth and diversity to single-cell research of immune ageing is the next goal of the field.
Acknowledgements
The authors thank L. Arthur for editing the manuscript. The work was supported by a grant from the Aging Biology Foundation (to M.N.A.).
Glossary
- Multi-omics
An integrative analysis approach using datasets that broadly cover data generated from the genome, epigenome, transcriptome, proteome and metabolome.
- Inflammageing
Chronic low-grade systemic inflammation occurring in the absence of a pathogen or exogenous inflammatory agent that develops with advanced age owing to endogenous pro-inflammatory processes.
- Senescent cell
A terminally differentiated cell characterized by irreversible cell cycle arrest and a specific secretory profile that develops in response to various stress triggers and accumulates in aged organisms.
- Caloric restriction
A dietary intervention that reduces average daily caloric intake below typical or habitual levels, while containing essential nutrients and not leading to malnutrition.
- M1 macrophages
An historical, overly simplistic, classification of pro-inflammatory macrophages activated in vitro with interferon-γ (IFNγ) and lipopolysaccharide that only partially reflects the heterogeneity of highly specialized, dynamic and heterogeneous macrophage subsets in vivo, the activation of which is continuously shaped by multiple matrix and cellular signals in the tissue microenvironment.
- T cell exhaustion
A specific state of T cell dysfunction driven by a persistent antigen stimulation and chiefly defined by reduced cytotoxicity and inability to produce pro-inflammatory cytokines (such as interferon-γ (IFNγ) and tumour necrosis factor (TNF)) and proliferate on T cell receptor stimulation.
- Cellular indexing of transcriptomes and epitopes by sequencing
(CITE-seq). A method that uses oligonucleotide-labelled antibodies to integrate cellular protein and transcriptome measurements into an integrated single-cell readout.
- Damage-associated molecular patterns
Endogenous molecules released from damaged or dying cells, such as mitochondria and chromatin components, which activate the innate immune system by interacting with pattern recognition receptors such as Toll-like receptors.
- Spatial transcriptomics
Methods designed to map mRNA expression profiles in individual cells or groups of cells to their locations in histological tissue sections.
- Imaging cytometry
Methods designed to combine flow cytometry or mass cytometry features with morphological analysis of thousands of cellular events in histological tissue sections.
- Single-cell assay for transposase-accessible chromatin using sequencing
(scATAC-seq). A method for mapping the accessible genome of individual cells that allows epigenetic profiling with single-cell resolution.
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Immunology thanks G. Pawelec, S. Shen-Orr and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Calico Life Sciences: https://www.calicolabs.com/
Immune ageing datasets: https://artyomovlab.wustl.edu/immune-aging/explore.html
Tabula Muris: https://tabula-muris.ds.czbiohub.org/
Tabula Muris Senis: https://tabula-muris-senis.ds.czbiohub.org/
These authors contributed equally: Denis A. Mogilenko, Irina Shchukina.
References
- 1.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Partridge L, Deelen J, Slagboom PE. Facing up to the global challenges of ageing. Nature. 2018;561:45–56. doi: 10.1038/s41586-018-0457-8. [DOI] [PubMed] [Google Scholar]
- 3.Nikolich-Žugich J. The twilight of immunity: emerging concepts in aging of the immune system. Nat. Immunol. 2018;19:10–19. doi: 10.1038/s41590-017-0006-x. [DOI] [PubMed] [Google Scholar]
- 4.Gomes T, Teichmann SA, Talavera-López C. Immunology driven by large-scale single-cell sequencing. Trends Immunol. 2019;40:1011–1021. doi: 10.1016/j.it.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Davis MM, Tato CM, Furman D. Systems immunology: just getting started. Nat. Immunol. 2017;18:725–732. doi: 10.1038/ni.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franceschi C, et al. Inflamm-aging: an evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2006;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 7.Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A. Inflammaging: a new immune–metabolic viewpoint for age-related diseases. Nat. Rev. Endo. 2018;14:576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- 8.Morrisette-Thomas V, et al. Inflamm-aging does not simply reflect increases in pro-inflammatory markers. Mech. Ageing Dev. 2014;139:49–57. doi: 10.1016/j.mad.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaum N, et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nature. 2020;583:596–602. doi: 10.1038/s41586-020-2499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baruch K, et al. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science. 2014;346:89–93. doi: 10.1126/science.1252945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato Y, Yanagita M. Immunology of the ageing kidney. Nat. Rev. Neph. 2019;15:625–640. doi: 10.1038/s41581-019-0185-9. [DOI] [PubMed] [Google Scholar]
- 12.Karin O, Agrawal A, Porat Z, Krizhanovsky V, Alon U. Senescent cell turnover slows with age providing an explanation for the Gompertz law. Nat. Commun. 2019;10:5495. doi: 10.1038/s41467-019-13192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson HL, Smithey MJ, Surh CD, Nikolich-Žugich J. Functional and homeostatic impact of age-related changes in lymph node stroma. Front. Immunol. 2017 doi: 10.3389/fimmu.2017.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yousefzadeh MJ, et al. Tissue specificity of senescent cell accumulation during physiologic and accelerated aging of mice. Aging Cell. 2020;19:e13094. doi: 10.1111/acel.13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiger H, de Haan G, Florian MC. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 2013;13:376–389. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- 16.Elyahu Y, Monsonego A. Thymus involution sets the clock of the aging T-cell landscape: Implications for declined immunity and tissue repair. Ageing Res. Rev. 2021;65:101231. doi: 10.1016/j.arr.2020.101231. [DOI] [PubMed] [Google Scholar]
- 17.Dorshkind K, Höfer T, Montecino-Rodriguez E, Pioli PD, Rodewald H-R. Do haematopoietic stem cells age? Nat. Rev. Immunol. 2020;20:196–202. doi: 10.1038/s41577-019-0236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi DJ, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl Acad. Sci. USA. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beerman I, et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl Acad. Sci. USA. 2010;107:5465–5470. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helbling PM, et al. Global transcriptomic profiling of the bone marrow stromal microenvironment during postnatal development, aging, and inflammation. Cell Rep. 2019;29:3313–3330.e4. doi: 10.1016/j.celrep.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Tikhonova AN, et al. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569:222–228. doi: 10.1038/s41586-019-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shchukina I, et al. Enhanced epigenetic profiling of classical human monocytes reveals a specific signature of healthy aging in the DNA methylome. Nat. Aging. 2021;1:124–141. doi: 10.1038/s43587-020-00002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds LM, et al. Transcriptomic profiles of aging in purified human immune cells. BMC Genomics. 2015;16:333. doi: 10.1186/s12864-015-1522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassler K, Schulte-Schrepping J, Warnat-Herresthal S, Aschenbrenner AC, Schultze JL. The myeloid cell compartment — cell by cell. Ann. Rev. Immunol. 2019;37:269–293. doi: 10.1146/annurev-immunol-042718-041728. [DOI] [PubMed] [Google Scholar]
- 25.Mogilenko DA, et al. Comprehensive profiling of an aging immune system reveals clonal GZMK+CD8+ T cells as conserved hallmark of inflammaging. Immunity. 2021;54:99–115.e12. doi: 10.1016/j.immuni.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Almanzar N, et al. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 2020;583:590–595. doi: 10.1038/s41586-020-2496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angelidis I, et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 2019;10:1–17. doi: 10.1038/s41467-019-08831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li CM, et al. Aging-associated alterations in mammary epithelia and stroma revealed by single-cell RNA sequencing. Cell Rep. 2020;33:108566. doi: 10.1016/j.celrep.2020.108566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodogai M, et al. Commensal bacteria contribute to insulin resistance in aging by activating innate B1a cells. Sci. Transl. Med. 2018;10:eaat4271. doi: 10.1126/scitranslmed.aat4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linehan E, et al. Aging impairs peritoneal but not bone marrow-derived macrophage phagocytosis. Aging Cell. 2014;13:699–708. doi: 10.1111/acel.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bian Z, et al. Deciphering human macrophage development at single-cell resolution. Nature. 2020;582:571–576. doi: 10.1038/s41586-020-2316-7. [DOI] [PubMed] [Google Scholar]
- 32.Fontana L, Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell. 2015;161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma S, et al. Caloric restriction reprograms the single-cell transcriptional landscape of Rattus norvegicus aging. Cell. 2020;180:984–1001.e22. doi: 10.1016/j.cell.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Butcher S, Chahel H, Lord JM. Ageing and the neutrophil: no appetite for killing? Immunology. 2000;100:411–416. doi: 10.1046/j.1365-2567.2000.00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barkaway A, et al. Age-related changes in the local milieu of inflamed tissues cause aberrant neutrophil trafficking and subsequent remote organ damage. Immunity. 2021;54:1494–1510.e7. doi: 10.1016/j.immuni.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagnado A, et al. Neutrophils induce paracrine telomere dysfunction and senescence in ROS-dependent manner. EMBO J. 2021;40:e106048. doi: 10.15252/embj.2020106048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu RJ, et al. Multi-omic profiling of primary mouse neutrophils predicts a pattern of sex- and age-related functional regulation. Nat. Aging. 2021;1:715–733. doi: 10.1038/s43587-021-00086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aprahamian T, Takemura Y, Goukassian D, Walsh K. Ageing is associated with diminished apoptotic cell clearance in vivo. Clin. Exp. Immunol. 2008;152:448–455. doi: 10.1111/j.1365-2249.2008.03658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chougnet CA, et al. Loss of phagocytic and antigen cross-presenting capacity in aging dendritic cells is associated with mitochondrial dysfunction. J. Immunol. 2015;195:2624–2632. doi: 10.4049/jimmunol.1501006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butcher SK, et al. Senescence in innate immune responses: reduced neutrophil phagocytic capacity and CD16 expression in elderly humans. J. Leukoc. Biol. 2001;70:881–886. doi: 10.1189/jlb.70.6.881. [DOI] [PubMed] [Google Scholar]
- 41.Wong C, Goldstein DR. Impact of aging on antigen presentation cell function of dendritic cells. Curr. Opin. Immunol. 2013;25:535–541. doi: 10.1016/j.coi.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vivier E, et al. Innate lymphoid cells: 10 years on. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 43.Hazeldine J, Lord JM. The impact of ageing on natural killer cell function and potential consequences for health in older adults. Ageing Res. Rev. 2013;12:1069–1078. doi: 10.1016/j.arr.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manser AR, Uhrberg M. Age-related changes in natural killer cell repertoires: impact on NK cell function and immune surveillance. Cancer Immunol. Immunother. 2016;65:417–426. doi: 10.1007/s00262-015-1750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hazeldine J, Hampson P, Lord JM. Reduced release and binding of perforin at the immunological synapse underlies the age-related decline in natural killer cell cytotoxicity. Aging Cell. 2012;11:751–759. doi: 10.1111/j.1474-9726.2012.00839.x. [DOI] [PubMed] [Google Scholar]
- 46.Chiu B-C, Martin BE, Stolberg VR, Chensue SW. The host environment Is responsible for aging-related functional NK cell deficiency. J. Immunol. 2013;191:4688–4698. doi: 10.4049/jimmunol.1301625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin W-N, et al. Neuroblast senescence in the aged brain augments natural killer cell cytotoxicity leading to impaired neurogenesis and cognition. Nat. Neurosci. 2021;24:61–73. doi: 10.1038/s41593-020-00745-w. [DOI] [PubMed] [Google Scholar]
- 48.Bal SM, Golebski K, Spits H. Plasticity of innate lymphoid cell subsets. Nat. Rev. Immunol. 2020;20:552–565. doi: 10.1038/s41577-020-0282-9. [DOI] [PubMed] [Google Scholar]
- 49.Bai L, et al. Liver type 1 innate lymphoid cells develop locally via an interferon-γ–dependent loop. Science. 2021;371:eaba4177. doi: 10.1126/science.aba4177. [DOI] [PubMed] [Google Scholar]
- 50.Goldberg EL, et al. IL-33 causes thermogenic failure in aging by expanding dysfunctional adipose ILC2. Cell Metab. 2021 doi: 10.1016/j.cmet.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fung ITH, et al. Activation of group 2 innate lymphoid cells alleviates aging-associated cognitive decline. J. Exp. Med. 2020;217:e20190915. doi: 10.1084/jem.20190915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’Souza SS, et al. Compartmentalized effects of aging on group 2 innate lymphoid cell development and function. Aging Cell. 2019;18:e13019. doi: 10.1111/acel.13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brestoff JR, et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. 2015;519:242–246. doi: 10.1038/nature14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mittelbrunn M, Kroemer G. Hallmarks of T cell aging. Nat. Immunol. 2021;22:687–698. doi: 10.1038/s41590-021-00927-z. [DOI] [PubMed] [Google Scholar]
- 55.Lerner A, Yamada T, Miller RA. Pgp-1hi T lymphocytes accumulate with age in mice and respond poorly to concanavalin A. Eur. J. Immunol. 1989;19:977–982. doi: 10.1002/eji.1830190604. [DOI] [PubMed] [Google Scholar]
- 56.Ku CC, Kappler J, Marrack P. The growth of the very large CD8+T cell clones in older mice is controlled by cytokines. J. Immunol. 2001;166:2186–2193. doi: 10.4049/jimmunol.166.4.2186. [DOI] [PubMed] [Google Scholar]
- 57.Kimmel JC, et al. Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging. Genome Res. 2019;29:2088–2103. doi: 10.1101/gr.253880.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T cell exhaustion during chronic viral infection and cancer. Ann. Rev. Immunol. 2019;37:457–495. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 59.Blank CU, et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 2019;19:665–674. doi: 10.1038/s41577-019-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng Y, et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell. 2020;11:740–770. doi: 10.1007/s13238-020-00762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arthur L, et al. Cellular and plasma proteomic determinants of COVID-19 and non-COVID-19 pulmonary diseases relative to healthy aging. Nat. Aging. 2021;1:535–549. doi: 10.1038/s43587-021-00067-x. [DOI] [PubMed] [Google Scholar]
- 62.Wertheimer AM, et al. Aging and cytomegalovirus infection differentially and jointly affect distinct circulating T cell subsets in humans. J. Immunol. 2014;192:2143–2155. doi: 10.4049/jimmunol.1301721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linton PJ, Haynes L, Klinman NR, Swain SL. Antigen-independent changes in naive CD4 T cells with aging. J. Exp. Med. 1996;184:1891–1900. doi: 10.1084/jem.184.5.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ernst DN, et al. Differences in the expression profiles of CD45RB, Pgp-1, and 3G11 membrane antigens and in the patterns of lymphokine secretion by splenic CD4+ T cells from young and aged mice. J. Immunol. 1990;145:1295–1302. doi: 10.4049/jimmunol.145.5.1295. [DOI] [PubMed] [Google Scholar]
- 65.Han G-M, Zhao B, Jeyaseelan S, Feng J-M. Age-associated parallel increase of Foxp3+CD4+ regulatory and CD44+CD4+ memory T cells in SJL/J mice. Cell. Immunol. 2009;258:188–196. doi: 10.1016/j.cellimm.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lages CS, et al. Functional regulatory T cells accumulate in aged hosts and promote chronic infectious disease reactivation. J. Immunol. 2008;181:1835–1848. doi: 10.4049/jimmunol.181.3.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishioka T, Shimizu J, Iida R, Yamazaki S, Sakaguchi S. CD4+CD25+Foxp3+ T cells and CD4+CD25-Foxp3+ T cells in aged mice. J. Immunol. 2006;176:6586–6593. doi: 10.4049/jimmunol.176.11.6586. [DOI] [PubMed] [Google Scholar]
- 68.Sharma S, Dominguez AL, Lustgarten J. High accumulation of T regulatory cells prevents the activation of immune responses in aged animals. J. Immunol. 2006;177:8348–8355. doi: 10.4049/jimmunol.177.12.8348. [DOI] [PubMed] [Google Scholar]
- 69.Elyahu Y, et al. Aging promotes reorganization of the CD4 T cell landscape toward extreme regulatory and effector phenotypes. Sci. Adv. 2019;5:eaaw8330. doi: 10.1126/sciadv.aaw8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo X, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 71.Takeuchi A, Saito T. CD4 CTL, a cytotoxic subset of CD4+ T cells, their differentiation and function. Front. Immunol. 2017;8:194. doi: 10.3389/fimmu.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hashimoto K, et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc. Natl Acad. Sci. USA. 2019;116:24242–24251. doi: 10.1073/pnas.1907883116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Channappanavar R, Twardy BS, Krishna P, Suvas S. Advancing age leads to predominance of inhibitory receptor expressing CD4 T cells. Mech. Ageing Dev. 2009;130:709–712. doi: 10.1016/j.mad.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 74.Shimada Y, Hayashi M, Nagasaka Y, Ohno-Iwashita Y, Inomata M. Age-associated up-regulation of a negative co-stimulatory receptor PD-1 in mouse CD4+ T cells. Exp. Gerontol. 2009;44:517–522. doi: 10.1016/j.exger.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 75.Haynes L, Eaton SM, Burns EM, Rincon M, Swain SL. Inflammatory cytokines overcome age-related defects in CD4 T cell responses in vivo. J. Immunol. 2004;172:5194–5199. doi: 10.4049/jimmunol.172.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haynes L, Linton PJ, Eaton SM, Tonkonogy SL, Swain SL. Interleukin 2, but not other common gamma chain-binding cytokines, can reverse the defect in generation of CD4 effector T cells from naive T cells of aged mice. J. Exp. Med. 1999;190:1013–1024. doi: 10.1084/jem.190.7.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsukamoto H, et al. Age-associated increase in lifespan of naïve CD4 T cells contributes to T-cell homeostasis but facilitates development of functional defects. Proc. Natl Acad. Sci. USA. 2009;106:18333–18338. doi: 10.1073/pnas.0910139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martinez-Jimenez CP, et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science. 2017;355:1433–1436. doi: 10.1126/science.aah4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maue AC, et al. Proinflammatory adjuvants enhance the cognate helper activity of aged CD4 T cells. J. Immunol. 2009;182:6129–6135. doi: 10.4049/jimmunol.0804226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ouyang X, et al. Potentiation of Th17 cytokines in aging process contributes to the development of colitis. Cell Immunol. 2011;266:208–217. doi: 10.1016/j.cellimm.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang M-C, Liao J-J, Bonasera S, Longo DL, Goetzl EJ. Nuclear factor-kappaB-dependent reversal of aging-induced alterations in T cell cytokines. FASEB J. 2008;22:2142–2150. doi: 10.1096/fj.07-103721. [DOI] [PubMed] [Google Scholar]
- 82.Chen H-C, et al. IL-7-dependent compositional changes within the γδ T cell pool in lymph nodes during ageing lead to an unbalanced anti-tumour response. EMBO Rep. 2019;20:e47379. doi: 10.15252/embr.201847379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ryu S, et al. Ketogenic diet restrains aging-induced exacerbation of coronavirus infection in mice. eLife. 2021;10:e66522. doi: 10.7554/eLife.66522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Foy TM, Aruffo A, Bajorath J, Buhlmann JE, Noelle RJ. Immune regulation by CD40 and its ligand GP39. Annu. Rev. Immunol. 1996;14:591–617. doi: 10.1146/annurev.immunol.14.1.591. [DOI] [PubMed] [Google Scholar]
- 85.Eaton SM, Burns EM, Kusser K, Randall TD, Haynes L. Age-related defects in CD4 T cell cognate helper function lead to reductions in humoral responses. J. Exp. Med. 2004;200:1613–1622. doi: 10.1084/jem.20041395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Callahan JE, Kappler JW, Marrack P. Unexpected expansions of CD8-bearing cells in old mice. J. Immunol. 1993;151:6657–6669. doi: 10.4049/jimmunol.151.12.6657. [DOI] [PubMed] [Google Scholar]
- 87.Britanova OV, et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J. Immunol. 2014;192:2689–2698. doi: 10.4049/jimmunol.1302064. [DOI] [PubMed] [Google Scholar]
- 88.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc. Natl Acad. Sci. USA. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Egorov ES, et al. The changing landscape of naive T cell receptor repertoire with human aging. Front. Immunol. 2018;9:1618. doi: 10.3389/fimmu.2018.01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Greef PC, et al. The naive T-cell receptor repertoire has an extremely broad distribution of clone sizes. eLife. 2020;9:e49900. doi: 10.7554/eLife.49900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miller RA. The aging immune system: primer and prospectus. Science. 1996;273:70–74. doi: 10.1126/science.273.5271.70. [DOI] [PubMed] [Google Scholar]
- 92.Hao Y, O’Neill P, Naradikian MS, Scholz JL, Cancro MP. A B-cell subset uniquely responsive to innate stimuli accumulates in aged mice. Blood. 2011;118:1294–1304. doi: 10.1182/blood-2011-01-330530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rubtsov AV, et al. Toll-like receptor 7 (TLR7)–driven accumulation of a novel CD11c+B-cell population is important for the development of autoimmunity. Blood. 2011;118:1305–1315. doi: 10.1182/blood-2011-01-331462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rubtsova K, et al. B cells expressing the transcription factor T-bet drive lupus-like autoimmunity. J. Clin. Invest. 2017;127:1392–1404. doi: 10.1172/JCI91250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cancro MP. Age-associated B cells. Annu. Rev. Immunol. 2020;38:315–340. doi: 10.1146/annurev-immunol-092419-031130. [DOI] [PubMed] [Google Scholar]
- 96.Brioschi S, et al. Heterogeneity of meningeal B cells reveals a lymphopoietic niche at the CNS borders. Science. 2021;373:eabf9277. doi: 10.1126/science.abf9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rubtsova K, Rubtsov AV, Cancro MP, Marrack P. Age-associated B cells: a T-bet–dependent effector with roles in protective and pathogenic Immunity. J. Immunol. 2015;195:1933–1937. doi: 10.4049/jimmunol.1501209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Russell Knode LM, et al. Age-associated B cells express a diverse repertoire of VH and Vκ genes with somatic hypermutation. J. Immunol. 2017;198:1921–1927. doi: 10.4049/jimmunol.1601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pioli PD, Casero D, Montecino-Rodriguez E, Morrison SL, Dorshkind K. Plasma cells are obligate effectors of enhanced myelopoiesis in aging bone marrow. Immunity. 2019;51:351–366.e6. doi: 10.1016/j.immuni.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Singh SP, et al. Cell lines generated from a chronic lymphocytic leukemia mouse model exhibit constitutive Btk and Akt signaling. Oncotarget. 2017;8:71981–71995. doi: 10.18632/oncotarget.18234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang Y, et al. CTLA-4 expression by B-1a B cells is essential for immune tolerance. Nat. Commun. 2021;12:525. doi: 10.1038/s41467-020-20874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee-Chang C, et al. Aging converts innate B1a cells into potent CD8+ T cell inducers. J. Immunol. 2016;196:3385–3397. doi: 10.4049/jimmunol.1502034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee-Chang C, et al. Accumulation of 4-1BBL+B cells in the elderly induces the generation of granzyme-B+ CD8+ T cells with potential antitumor activity. Blood. 2014;124:1450–1459. doi: 10.1182/blood-2014-03-563940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Buckley CD, Barone F, Nayar S, Bénézech C, Caamaño J. Stromal cells in chronic inflammation and tertiary lymphoid organ formation. Annu. Rev. Immunol. 2015;33:715–745. doi: 10.1146/annurev-immunol-032713-120252. [DOI] [PubMed] [Google Scholar]
- 105.Krausgruber T, et al. Structural cells are key regulators of organ-specific immune responses. Nature. 2020;583:296–302. doi: 10.1038/s41586-020-2424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tigges J, et al. The hallmarks of fibroblast ageing. Mech. Ageing Dev. 2014;138:26–44. doi: 10.1016/j.mad.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 107.Lynch MD, Watt FM. Fibroblast heterogeneity: implications for human disease. J. Clin. Invest. 2018;128:26–35. doi: 10.1172/JCI93555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Salzer MC, et al. Identity noise and adipogenic traits characterize dermal fibroblast aging. Cell. 2018;175:1575–1590.e22. doi: 10.1016/j.cell.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 109.Mahmoudi S, et al. Heterogeneity in old fibroblasts is linked to variability in reprogramming and wound healing. Nature. 2019;574:553–558. doi: 10.1038/s41586-019-1658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Solé-Boldo L, et al. Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Comm. Biol. 2020;3:1–12. doi: 10.1038/s42003-020-0922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao L, et al. Pharmacologically reversible zonation-dependent endothelial cell transcriptomic changes with neurodegenerative disease associations in the aged brain. Nat. Commun. 2020;11:4413. doi: 10.1038/s41467-020-18249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Childs BG, et al. Senescent cells: an emerging target for diseases of ageing. Nat. Rev. Drug. Disc. 2017;16:718–735. doi: 10.1038/nrd.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Coppé J-P, Desprez P-Y, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. Mech. Dis. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Faget DV, Ren Q, Stewart SA. Unmasking senescence: context-dependent effects of SASP in cancer. Nat. Rev. Cancer. 2019;19:439–453. doi: 10.1038/s41568-019-0156-2. [DOI] [PubMed] [Google Scholar]
- 115.Kuilman T, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 116.Acosta JC, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Di Micco R, Krizhanovsky V, Baker D, d’Adda di Fagagna F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021;22:75–95. doi: 10.1038/s41580-020-00314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hernandez-Segura A, et al. Unmasking transcriptional heterogeneity in senescent cells. Curr. Biol. 2017;27:2652–2660.e4. doi: 10.1016/j.cub.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wiley CD, et al. Analysis of individual cells identifies cell-to-cell variability following induction of cellular senescence. Aging Cell. 2017;16:1043–1050. doi: 10.1111/acel.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Omori S, et al. Generation of a p16 reporter mouse and its use to characterize and target p16high cells in vivo. Cell Metab. 2020;32:814–828.e6. doi: 10.1016/j.cmet.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 121.Kale A, Sharma A, Stolzing A, Desprez P-Y, Campisi J. Role of immune cells in the removal of deleterious senescent cells. Immun. Ageing. 2020;17:16. doi: 10.1186/s12979-020-00187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Freund A, Orjalo AV, Desprez P-Y, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol. Med. 2010;16:238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Antonangeli F, Zingoni A, Soriani A, Santoni A. Senescent cells: living or dying is a matter of NK cells. J. Leukoc. Biol. 2019;105:1275–1283. doi: 10.1002/JLB.MR0718-299R. [DOI] [PubMed] [Google Scholar]
- 124.Storer M, et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell. 2013;155:1119–1130. doi: 10.1016/j.cell.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 125.Muñoz-Espín D, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013;155:1104–1118. doi: 10.1016/j.cell.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 126.Yun MH, Davaapil H, Brockes JP. Recurrent turnover of senescent cells during regeneration of a complex structure. eLife. 2015;4:e05505. doi: 10.7554/eLife.05505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kang T-W, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 129.Brighton PJ, et al. Clearance of senescent decidual cells by uterine natural killer cells in cycling human endometrium. eLife. 2017;6:e31274. doi: 10.7554/eLife.31274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ovadya Y, et al. Impaired immune surveillance accelerates accumulation of senescent cells and aging. Nat. Commun. 2018;9:5435. doi: 10.1038/s41467-018-07825-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Baker DJ, et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell. 2014;31:722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Puchta A, et al. TNF drives monocyte dysfunction with age and results in impaired anti-pneumococcal immunity. PLoS Pathog. 2016;12:e1005368. doi: 10.1371/journal.ppat.1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Loukov D, Naidoo A, Puchta A, Marin JLA, Bowdish DME. Tumor necrosis factor drives increased splenic monopoiesis in old mice. J. Leuk. Biol. 2016;100:121–129. doi: 10.1189/jlb.3MA0915-433RR. [DOI] [PubMed] [Google Scholar]
- 137.Tong Q-Y, et al. Human thymic involution and aging in humanized mice. Front. Immunol. 2020;11:1399. doi: 10.3389/fimmu.2020.01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li M, et al. Age related human T cell subset evolution and senescence. Immun. Ageing. 2019;16:24. doi: 10.1186/s12979-019-0165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jergović M, Contreras NA, Nikolich-Žugich J. Impact of CMV upon immune aging: facts and fiction. Med. Microbiol. Immunol. 2019;208:263–269. doi: 10.1007/s00430-019-00605-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gordon A, Reingold A. The burden of influenza: a complex problem. Curr. Epidemiol. Rep. 2018;5:1–9. doi: 10.1007/s40471-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Johnson RW, et al. The impact of herpes zoster and post-herpetic neuralgia on quality-of-life. BMC Med. 2010;8:37. doi: 10.1186/1741-7015-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wroe PC, et al. Aging population and future burden of pneumococcal pneumonia in the United States. J. Infect. Dis. 2012;205:1589–1592. doi: 10.1093/infdis/jis240. [DOI] [PubMed] [Google Scholar]
- 143.Salje H, et al. Estimating the burden of SARS-CoV-2 in France. Science. 2020;369:208–211. doi: 10.1126/science.abc3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chowers MY, et al. Clinical characteristics of the West Nile fever outbreak, Israel, 2000. Emerg. Infect. Dis. 2001;7:675–678. doi: 10.3201/eid0704.017414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Montgomery RR, Steen H. Using ‘big data’ to disentangle aging and COVID-19. Nat. Aging. 2021;1:496–497. doi: 10.1038/s43587-021-00078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xu M, et al. Senolytics improve physical function and increase lifespan in old age. Nat. Med. 2018;24:1246–1256. doi: 10.1038/s41591-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Amor C, et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature. 2020;583:127–132. doi: 10.1038/s41586-020-2403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Melzer D, Pilling LC, Ferrucci L. The genetics of human ageing. Nat. Rev. Genet. 2020;21:88–101. doi: 10.1038/s41576-019-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Alpert A, et al. A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat. Med. 2019;25:487–495. doi: 10.1038/s41591-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McQuattie-Pimentel AC, et al. The lung microenvironment shapes a dysfunctional response of alveolar macrophages in aging. J. Clin. Invest. 2021;131:e140299. doi: 10.1172/JCI140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wong CK, et al. Aging impairs alveolar macrophage phagocytosis and increases influenza-induced mortality in mice. J. Immunol. 2017;199:1060–1068. doi: 10.4049/jimmunol.1700397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ide S, et al. Yolk-sac-derived macrophages progressively expand in the mouse kidney with age. eLife. 2020;9:e51756. doi: 10.7554/eLife.51756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wong CP, Magnusson KR, Ho E. Aging is associated with altered dendritic cells subset distribution and impaired proinflammatory cytokine production. Exp. Gerontol. 2010;45:163–169. doi: 10.1016/j.exger.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 154.Pieren DKJ, Smits NAM, van de Garde MDB, Guichelaar T. Response kinetics reveal novel features of ageing in murine T cells. Sci. Rep. 2019;9:5587. doi: 10.1038/s41598-019-42120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bapat SP, et al. Depletion of fat-resident Treg cells prevents age-associated insulin resistance. Nature. 2015;528:137–141. doi: 10.1038/nature16151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Quinn KM, et al. Age-related decline in primary CD8+ T cell responses is associated with the development of senescence in virtual memory CD8+ T cells. Cell Rep. 2018;23:3512–3524. doi: 10.1016/j.celrep.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 157.Dulken BW, et al. Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature. 2019;571:205–210. doi: 10.1038/s41586-019-1362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chiu B-C, Martin BE, Stolberg VR, Chensue SW. Cutting edge: central memory CD8 T cells in aged mice are virtual memory cells. J. Immunol. 2013;191:5793–5796. doi: 10.4049/jimmunol.1302509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Clambey ET, White J, Kappler JW, Marrack P. Identification of two major types of age-associated CD8 clonal expansions with highly divergent properties. Proc. Natl Acad. Sci. USA. 2008;105:12997–13002. doi: 10.1073/pnas.0805465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee K-A, et al. Characterization of age-associated exhausted CD8+ T cells defined by increased expression of Tim-3 and PD-1. Aging Cell. 2016;15:291–300. doi: 10.1111/acel.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Seidler S, Zimmermann HW, Bartneck M, Trautwein C, Tacke F. Age-dependent alterations of monocyte subsets and monocyte-related chemokine pathways in healthy adults. BMC Immunol. 2010;11:30. doi: 10.1186/1471-2172-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stervbo U, et al. Effects of aging on human leukocytes (part I): immunophenotyping of innate immune cells. Age. 2015;37:92. doi: 10.1007/s11357-015-9828-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rodriguez-Zhurbenko N, Quach TD, Hopkins TJ, Rothstein TL, Hernandez AM. Human B-1 cells and B-1 cell antibodies change with advancing age. Front. Immunol. 2019;10:483. doi: 10.3389/fimmu.2019.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van der Geest KSM, et al. Impact of aging on the frequency, phenotype, and function of CD161-expressing T cells. Front. Immunol. 2018 doi: 10.3389/fimmu.2018.00752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Colonna-Romano G, et al. Impairment of gamma/delta T lymphocytes in elderly: implications for immunosenescence. Exp. Gerontol. 2004;39:1439–1446. doi: 10.1016/j.exger.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 166.Tan CTY, et al. Vδ2+ and α/β T cells show divergent trajectories during human aging. Oncotarget. 2016;7:44906–44918. doi: 10.18632/oncotarget.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Michishita Y, et al. Age-associated alteration of γδ T-cell repertoire and different profiles of activation-induced death of Vδ1 and Vδ2 T cells. Int. J. Hematol. 2011;94:230–240. doi: 10.1007/s12185-011-0907-7. [DOI] [PubMed] [Google Scholar]
- 168.Oishi Y, Manabe I. Macrophages in age-related chronic inflammatory diseases. NPJ Aging Mech. Dis. 2016;2:1–8. doi: 10.1038/npjamd.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Minhas PS, et al. Restoring metabolism of myeloid cells reverses cognitive decline in ageing. Nature. 2021;590:122–128. doi: 10.1038/s41586-020-03160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ferrucci L, Fabbri E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardio. 2018;15:505–522. doi: 10.1038/s41569-018-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Desdín-Micó G, et al. T cells with dysfunctional mitochondria induce multimorbidity and premature senescence. Science. 2020;368:1371–1376. doi: 10.1126/science.aax0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shenoy AT, Mizgerd JP. Seedy CD8+ T RM cells in aging lungs drive susceptibility to pneumonia and sequelae. Cell. Mol. Immunol. 2021;18:787–789. doi: 10.1038/s41423-020-00629-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Covarrubias AJ, et al. Senescent cells promote tissue NAD+ decline during ageing via the activation of CD38+ macrophages. Nat. Metab. 2020;2:1265–1283. doi: 10.1038/s42255-020-00305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chini CCS, et al. CD38 ecto-enzyme in immune cells is induced during aging and regulates NAD+ and NMN levels. Nat. Metab. 2020;2:1284–1304. doi: 10.1038/s42255-020-00298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Freund A, Patil CK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 2011;30:1536–1548. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Acosta JC, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 177.Kang C, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349:aaa5612. doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Glück S, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017;19:1061–1070. doi: 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dou Z, et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 2017;550:402–406. doi: 10.1038/nature24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sagiv A, et al. NKG2D ligands mediate immunosurveillance of senescent cells. Aging. 2016;8:328–344. doi: 10.18632/aging.100897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pereira BI, et al. Senescent cells evade immune clearance via HLA-E-mediated NK and CD8+ T cell inhibition. Nat. Commun. 2019;10:1–13. doi: 10.1038/s41467-019-10335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018;18:35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 183.Stoeckius M, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cusanovich DA, et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lareau CA, et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotech. 2019;37:916–924. doi: 10.1038/s41587-019-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cao J, et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361:1380–1385. doi: 10.1126/science.aau0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 188.Ståhl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 189.Gong H, Do D, Ramakrishnan R. Single-cell mRNA-seq using the Fluidigm C1 system and integrated fluidics circuits. Methods Mol. Biol. 2018;1783:193–207. doi: 10.1007/978-1-4939-7834-2_10. [DOI] [PubMed] [Google Scholar]
- 190.Utada AS, et al. Monodisperse double emulsions generated from a microcapillary device. Science. 2005;308:537–541. doi: 10.1126/science.1109164. [DOI] [PubMed] [Google Scholar]
- 191.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 192.Sharpless NE, Sherr CJ. Forging a signature of in vivo senescence. Nat. Rev. Cancer. 2015;15:397–408. doi: 10.1038/nrc3960. [DOI] [PubMed] [Google Scholar]
- 193.Gorgoulis V, et al. Cellular senescence: defining a path forward. Cell. 2019;179:813–827. doi: 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 194.He S, Sharpless NE. Senescence in health and disease. Cell. 2017;169:1000–1011. doi: 10.1016/j.cell.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Grosse L, et al. Defined p16high senescent cell types are indispensable for mouse healthspan. Cell Metab. 2020;32:87–99.e6. doi: 10.1016/j.cmet.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 196.Neurohr GE, et al. Excessive cell growth causes cytoplasm dilution and contributes to senescence. Cell. 2019;176:1083–1097.e18. doi: 10.1016/j.cell.2019.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lee BY, et al. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 198.Furman D, et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019;25:1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]